Abstract
Osteosarcoma (OS) is the most common primary malignant bone tumor in pediatric and adolescent patients. The calcyclin-binding protein/Siah-1-interacting protein (CacyBP/SIP) performs an essential function in cell proliferation and apoptosis. The present study investigated the effect of CacyBP/SIP in OS cell proliferation and apoptosis. CacyBP/SIP mRNA expression levels were evaluated in four OS cell lines by quantitative PCR. CacyBP/SIP expression was downregulated in Saos-2 cells using a lentivirus transfection system and the transfection efficiency was analyzed. The effects of CacyBP/SIP downregulation on Saos-2 cell proliferation and colony-formation ability were evaluated by MTT and colony-formation assays. The effect of CacyBP/SIP knockdown on Saos-2 cell cycle and apoptosis was analyzed by flow cytometry cell sorting. The Cancer Genome Atlas (TCGA) data was analyzed for validation. Human OS cell lines Saos-2, MG-63, HOS and U20S expressed CacyBP/SIP mRNA. CacyBP/SIP knockdown significantly inhibited cell proliferation and colony-formation ability. G1/S phase arrest was induced by CacyBP/SIP downregulation, which also resulted in the downregulation of CDK and cyclins and the upregulation of p21. In addition, CacyBP/SIP downregulation induced Saos-2 cell apoptosis mediated by Bax and Bcl-2. High expression of CacyBP/SIP was significantly associated with poor prognosis in TCGA sarcoma database. Thus, CacyBP/SIP performs important functions in the proliferation and apoptosis of human OS cells.
Keywords: osteosarcoma, proliferation, cell cycle, apoptosis
Introduction
Osteosarcoma (OS) is the most common primary malignant bone tumor in children and adolescents. The incidence of OS is ~4-5 per year per million people (1). Treatment for OS has made notable progress with the development of surgery and the combination of neoadjuvant chemotherapy. However, the problems of tumor metastasis, recurrence and multi-chemoresistance have yet to be solved (2). In recent years, targeted therapy has been developed as a treatment for human malignant tumors (3). These therapeutic drugs always specifically target several molecules, such as growth factor receptors or intracellular signaling proteins that are involved with tumor proliferation, migration and/or invasion (4,5).
Calcyclin-binding protein/Siah-1-interacting protein (CacyBP/SIP) was initially identified in Ehrlich ascites tumor cells as a target of calcyclin (6,7). Later, CacyBP/SIP was defined as a binding partner for Siah-1(8). Several studies revealed that CacyBP/SIP is wildly expressed in a range of human tissues (9), where it influences diverse cellular processes, including proliferation (10), differentiation (11), multidrug resistance (12) and tumorigenicity (13). Other studies have indicated that CacyBP/SIP plays a role as an oncogene with increased expression in human glioma and colorectal cancer (10,14). Nevertheless, CacyBP/SIP is expressed at low levels in renal cell carcinoma and gastric cancer (15,16). Notably, CacyBP/SIP has also been reported to serve a role in promoting apoptosis in acute lymphocytic leukemia where it appears to function as a tumor suppressor (13). These data indicate that the effect of CacyBP/SIP on tumorigenic progression may differ depending on the tumor type. It has been demonstrated that CacyBP/SIP, as a novel phosphatase, targets ERK1/2 and MAPK p38(17). It has also been shown that CacyBP/SIP is expressed in the nuclei of osteogenic sarcoma cells (9). However, the function of CacyBP/SIP in OS cells remains to be elucidated.
The present study evaluated the expression of CacyBP/SIP in OS cell lines and investigated the roles of CacyBP/SIP in OS cell proliferation and apoptosis. It was demonstrated that downregulation of CacyBP/SIP inhibited cellular proliferation, and induced G1/S phase cell arrest and cell apoptosis. All these data suggest that CacyBP/SIP may play a key role in OS progression and that CacyBP/SIP might be a target for OS treatment.
Materials and methods
Cell lines and regents
Saos-2, MG-63, HOS, U20S OS cells and 293T cells were obtained from the American Type Culture Collection and cultured in DMEM medium (Corning, Inc.). All media contained 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.), penicillin G (100 U/ml) and streptomycin (100 μg/ml; Sigma-Aldrich; Merck KGaA). All cell cultures were maintained as a monolayer at 37˚C in a humidified atmosphere containing 5% CO2.
Construction of recombinant lentivirus and gene silencing
The short hairpin (sh)RNA (5'-TTACCTGACCCAGGTTG AA-3') for human CacyBP/SIP gene was inserted into the lentivirus expression plasmid pGCSIL-GFP (Shanghai GeneChem Co., Ltd.) and non-silencing shRNA (5'-TTCTC CGAACGTGTCACGT-3') was used as a negative control. For virus packaging, 0.5 µg CacyBP/SIP-shRNA vector or control (Ctrl) vector together with 0.5 µg pHelper 1.0 and 0.5 µg pHelper 2.0 (Shanghai GeneChem Co., Ltd.), were added to 293T cells with Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Saos-2 cells were transduced with either the CacyBP/SIP-shRNA lentivirus or Ctrl lentivirus at a multiplicity of infection of 10 for 72 h. The transduced cells expressing GFP protein were observed by using fluorescence microscopy to determine the transduction efficiency. CacyBP/SIP expression was confirmed by reverse transcription-quantitative PCR (RT-qPCR) and western blot analysis after lentivirus transduction.
RNA extraction and RT-qPCR
After 3 days of transduction, total RNA was isolated from Saos-2, MG-63, HOS and U20S OS cells (2x105 cells/well) using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Total isolated RNA (2 µg) was used to synthesize cDNA using reverse transcription system composed by M-MLV reverse transcriptase (Promega Corporation), M-MLV reverse transcriptase 5X reaction buffer (Promega Corporation), dNTPs (Promega Corporation) and random primers (Invitrogen; Thermo Fisher Scientific, Inc.) for 60 min at 37˚C according to the manufacturer's protocol. CacyBP/SIP mRNA expression was evaluated by RT-qPCR on Roche LightCycler 480 platform with SYBR Master Mixture (Takara Bio, Inc.). The following thermocycling conditions were used for qPCR: Initial denaturation at 95˚C for 30 sec; followed by 40 cycles of denaturation at 95˚C for 5 sec and extension at 60˚C for 30 sec. The absorbance value was obtained at the extension stage. GAPDH was used as the internal reference control. The following PCR primers were used: CacyBP/SIP forward, 5'-CTCCCATTACAACGGGCTATAC-3' and reverse, 5'-GAACTGCCTTCCACAGAGATG-3'; GAPDH forward, 5'-TGACTTCAACAGCGACACCCA-3' and reverse, 5'-CACCCTGTTGCTGTAGCCAAA-3'. Data were analyzed using the 2-∆∆Cq method (18). Results were presented as CT values, which were defined as the threshold PCR cycle number at which an amplified product was first detected. The average CT was calculated for both CacyBP/SIP and GAPDH, and ΔCT was determined as the ratio of the mean of the triplicate CT values for CacyBP/SIP to the mean of the triplicate CT values for GAPDH. Each experiment was performed in triplicate and repeated three times.
Western blotting
Cells were washed with ice cold PBS and then lysed in 2X lysis buffer (100 mM Tris-HCl, pH 6.8; 2% mercaptoethanol; 20% glycerinum; 4% SDS) on ice for 15 min. The lysates were clarified by centrifugation at 12,000 x g for 15 min at 4˚C and the supernatants were employed for further analysis. The total protein concentration was estimated using a bicinchoninic acid protein assay kit (Beyotime Institute of Biotechnology). Protein samples (20 µg) were loaded and electrophoresed in an SDS-PAGE (10% gel) at 120 mA for 1 h and subsequently transferred to PVDF membranes (EMD Millipore) at 300 mA for 120 min. After being blocked with TBS with Tween-20 (TBST) containing 5% (w/v) non-fat dried skim milk powder for 24 h at 4˚C, membranes were incubated with a CacyBP/SIP antibody (cat. no. 3354; 1:1,000) or a GAPDH antibody (cat. no. 3683; 1:1,000; both from Cell Signaling Technology, Inc.) overnight at 4˚C. p21 antibody (ab188224; 1:1,000), cyclin-dependent kinase (CDK) 2 antibody (ab32147; 1:1,000), CDK4 antibody (ab199728; 1:2,000), Cyclin D1 antibody (ab226977; 1:2,000), Cyclin E1 antibody (ab33911; 1:2,000), Bax antibody (ab32503; 1:2,000) and Bcl-2 antibody (ab692; 1:500) were purchased from Abcam. After washing with TBST, the membranes were incubated with anti-rabbit horseradish peroxidase-conjugated secondary antibody (cat. no. 7074; 1:2,000; Cell Signaling Technology, Inc.) at room temperature for 2 h. The membranes were analyzed and visualized by Pierce ECL Western Blotting Substrate (cat. no. 32109; Pierce; Thermo Fisher Scientific, Inc.). Each experiment was repeated three times. The quantification of proteins was performed in ImageJ software (version 1.7.9; National Institutes of Health).
Cell growth assay
Cell growth was measured using a Celigo Imaging Cytometer (Nexcelom Bioscience LLC). Briefly, 3 days after Saos-2 cells were transfected with the negative Ctrl lentivirus or CacyBP/SIP-shRNA lentivirus, cells in logarithmic phase were digested, resuspended, counted and inoculated in 96-well plates for 5 days (2,000 cells/well). Cell growth rate was defined as cell count on day n / cell count on day 1, where n=2, 3, 4 and 5. Plates were analyzed with using the Celigo image cytometer (4.1.3.0; Nexcelom Bioscience LLC).
MTT assay
Following a previous report (19), Saos-2 cells from different groups (shCacyBP/SIP, shCtrl) were seeded in 96-well plates at a density of 3,000 cells/well after 72 h of lentivirus transduction. On days 1, 3 and 5, MTT was added into each well at a final concentration of 5 mg/ml for 4 h. Acidic isopropanol [10% SDS, 5% (v/v) isopropanol and 0.01 mol/l HCl] was subsequently added to stop the reaction. Absorbance was measured with an ELISA reader (Bio-Rad Laboratories, Inc.) at a wavelength of 595 nm. Viability of cells was calculated from absorbance. Each experiment was performed in triplicate and repeated three times.
Colony formation assay
In order to assay monolayer colony formation, stably transduced Saos-2 cells from shCacyBP/SIP and shCtrl groups after a 72-h transfection were seeded into 6-well plates at a density of 600 cells/well. After culture for 16 days, cells were fixed with 4% paraformaldehyde (1 ml/well) for 30 min at room temperature. The cells were washed with PBS and then stained with 500 µl Giemsa staining solution at room temperature for 15 min. Colonies (>50 cells/colony) were photographed by using a fluorescence microscope under x100 magnification (IX71; Olympus Corporation) and counted by using the ImageJ software (version 1.7.9; National Institutes of Health). Each experiment was performed in triplicate and repeated three times.
Cell cycle analysis
A total of 3 days after transfection, the cells were seeded in 60-mm-diameter plates for 72 h and were collected and fixed and permeabilized in 70% ethanol on ice for 1 h. After washing with PBS twice, the cells were incubated with staining buffer (including 10 µg/ml RNase, propidium iodide) at room temperature for 10 min and were subsequently subject to cell cycle analysis by flow cytometry (Guava easyCyte HT; EMD Millipore). The data were analyzed using the ModFit LT™ 3.3 software (Verity Software House, Inc.).
Cell apoptosis detection
For the apoptosis assay, after 3 days of CacyBP/SIP lentiviral transfection, the cells were seeded in a 6-well plate for 72 h. Subsequently, the cells were harvested and washed with binding buffer, resuspended in staining buffer and then incubated with Annexin V-APC (cat. no. 88-8007; eBioscience; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions and detected by a flow cytometer (Guava easyCyte HT; EMD Millipore). The data were analyzed by CytoSoft 5.3.1 software (EMD Millipore).
The Cancer Genome Atlas (TCGA) database
The expression profiles of CacyBP/SIP as well as clinical information of sarcoma (SARC) samples were obtained from TCGA database (https://cancergenome.nih.gov/), including level 2 RNA-seq data from 262 SARC patients (20,21). The K-means clustering analysis with K=2 was performed using R 3.6.1 (https://www.r-project.org/) to categorize the patients into two groups based on the expression levels of CacyBP/SIP. The K-means clustering is an unsupervised clustering, mainly used to group unlabeled data, where K represents the number of packets. Packet represents the grouping condition of unsupervised clustering. In the present study, K=2 means clustering into two groups after unsupervised clustering. The K-means algorithm iteratively assigns each data point to K packets, so that the data points are aggregated based on feature similarity (22).
Statistical analysis
All statistical analyses were performed using SPSS 13.0 software (SPSS, Inc.). The differences between groups were compared using Student's t-test and data are presented as the mean ± standard deviation of three independent experiments. The log-rank test and Kaplan-Meier curve were employed to evaluate the association between CacyBP/SIP expression and prognosis. P<0.05 was considered to indicate a statistically significant difference.
Results
Lentivirus-mediated shRNA effectively decreases CacyBP/SIP expression in Saos-2 cells
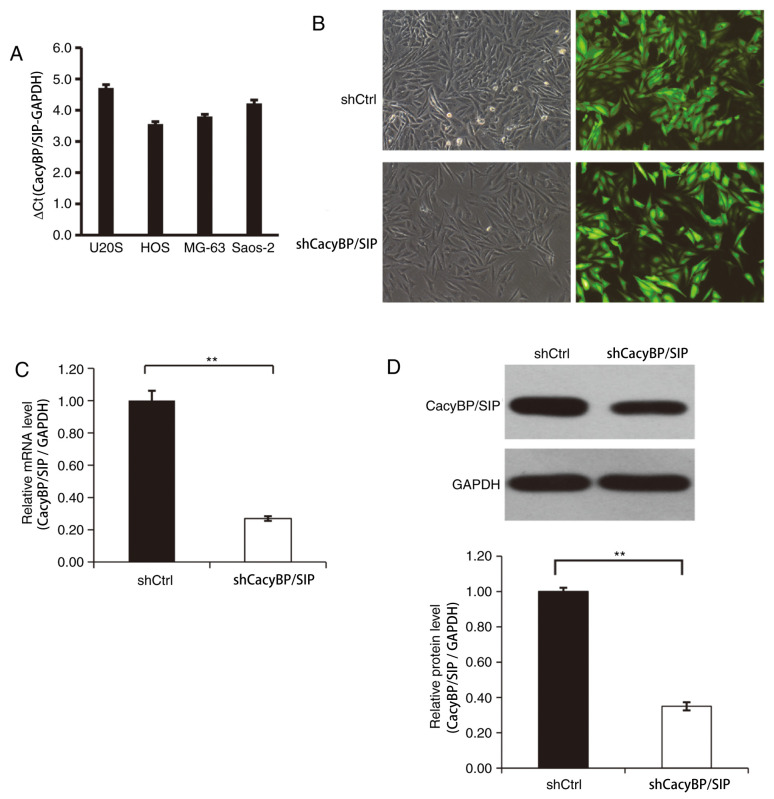
The levels of CacyBP/SIP expression in a variety of OS cell lines was first evaluated and CacyBP/SIP mRNA expression was detected in all four OS cell lines including Saos-2, MG-63, HOS and U20S (Fig. 1A). After 3 days of CacyBP/SIP lentiviral transfection, the transfection efficiency was found to be ~80% for the CacyBP/SIP-shRNA lentivirus and the Ctrl shRNA lentivirus (Fig. 1B). The fluorescence level of GFP protein was observed to determine the transfection efficiency. CacyBP/SIP mRNA level was confirmed by RT-qPCR. The result indicted that CacyBP/SIP-shRNA lentivirus transfection significantly reduced CacyBP/SIP expression compared the Ctrl lentivirus-transfected cells (Fig. 1C). Decreased CacyBP/SIP protein level was detected in CacyBP/SIP-shRNA lentivirus-transfected Saos-2 cells and the knockdown efficiency was quantitatively evaluated (Fig. 1D).
Figure 1.
Lentivirus-mediated shRNA effectively decreases CacyBP/SIP expression in Saos-2 cells. (A) The levels of CacyBP/SIP expression in a variety of osteosarcoma cell lines. After 3 days of CacyBP/SIP lentiviral transfection, the transfection efficiency was (B) observed with fluorescence and (C) determined with reverse transcription-quantitative PCR. Magnification, x100. (D) Decreased CacyBP/SIP protein level was detected in CacyBP/SIP-shRNA lentivirus-transfected Saos-2 cells. **P<0.01. sh, short hairpin RNA; CacyBP/SIP, calcyclin-binding protein/Siah-1-interacting protein; Ctrl, control.
Knockdown of CacyBP/SIP inhibits Saos-2 cell proliferation and colony formation in vitro
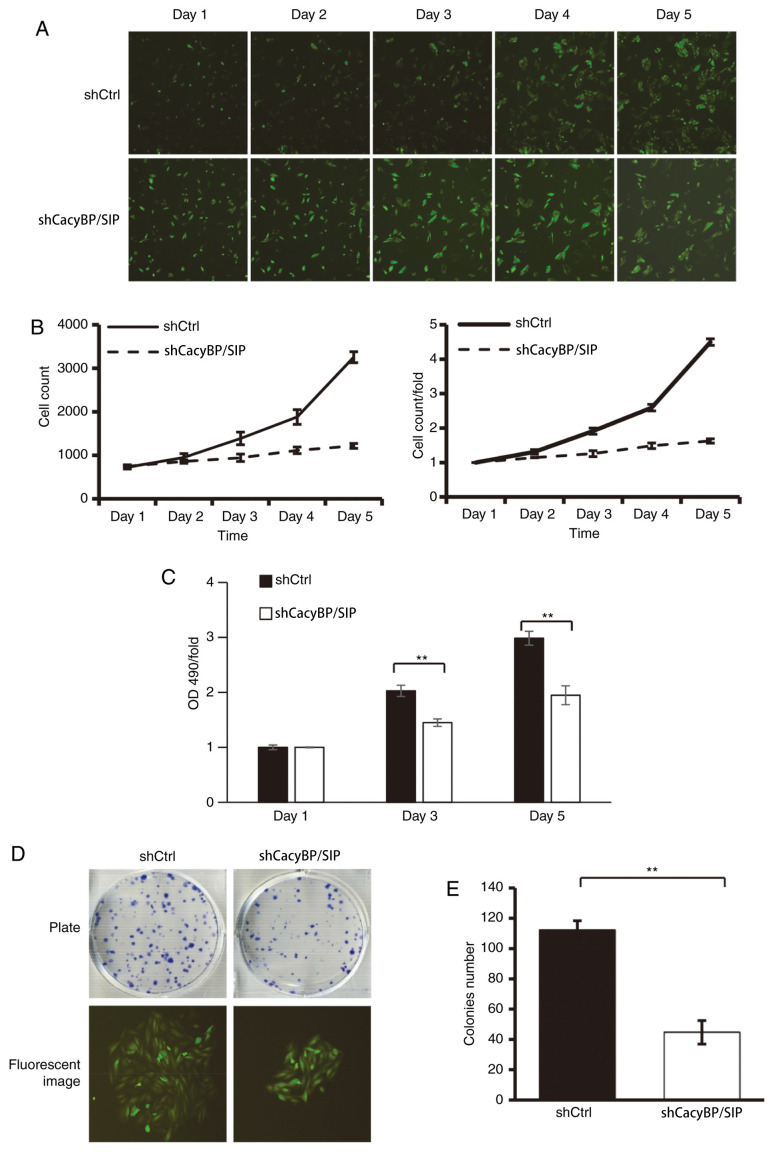
In order to explore the function of CacyBP/SIP on cell growth, Saos-2 cells expressing either CacyBP/SIP-shRNA lentivirus or Ctrl lentivirus were seeded in 96-well plates and cell growth was monitored with a Celigo Imaging Cytometer daily for 5 days. The results demonstrated that following the downregulation of CacyBP/SIP expression, the total number of cells remained almost unchanged from day 2 to day 5. The cell growth rate was stalled compared with that in the shCtrl group (Fig. 2A and B). There was a consistent result from the MTT assay (Fig. 2C).
Figure 2.
Knockdown of CacyBP/SIP inhibits Saos-2 cell proliferation and colony formation in vitro. Saos-2 cells expressing either CacyBP/SIP-shRNA lentivirus or Ctrl lentivirus were seeded in 96-well and 6-well plates and cell growth was monitored by a Celigo Imaging Cytometer daily for 5 days. The (A) total number of cells and (B) cell growth rate were determined. Magnification, x40. (C) MTT assay and (D) colony formation assay was performed to evaluate cell proliferation. Magnification, x100. (E) Statistical analysis of colony numbers between two groups. **P<0.01. CacyBP/SIP, calcyclin-binding protein/Siah-1-interacting protein; sh, short hairpin RNA; Ctrl, control.
The colony formation was assayed to determine CacyBP/SIP knockdown in Saos-2 and U20S cells tumorigenesis in vitro. The results demonstrated that CacyBP/SIP knockdown in Saos-2 cells caused a substantial reduction in colony formation compared with the Ctrl cells (Fig. 2D). Similarly, CacyBP/SIP knockdown in U20S cells also caused a substantial reduction in colony formation (Fig. S1). The number of colonies in the CacyBP/SIP-shRNA lentivirus-transduced cells was significantly decreased compared with the Ctrl group, with shCtrl 112±6 vs. shCacyBP/SIP 45±8 in Saos-2 cells (P<0.01; Fig. 2E).
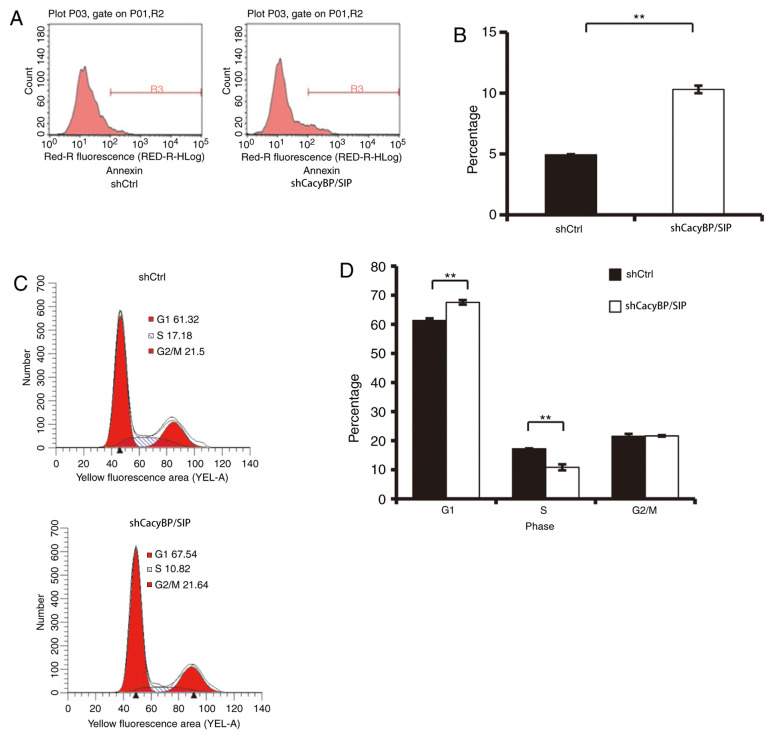
Knockdown of CacyBP/SIP induces Saos-2 cell apoptosis and cell cycle arrest
Alterations in cell proliferation is frequently caused by cell apoptosis (23). To further explore this mechanism, cell apoptosis was examined by Annexin V-APC. As illustrated in Fig. 3A and B, 6 days after cell seeding, the percentage of apoptotic Saos-2 cells was significantly increased in the shCacyBP/SIP group compared with that in the shCtrl group (shCtrl, 4.93±0.06%; shCacyBP/SIP 10.3±0.31%; P<0.01). This result indicated that CacyBP/SIP may be associated with the apoptosis of Saos-2 cells.
Figure 3.
Knockdown of CacyBP/SIP induces Saos-2 cell apoptosis and cell cycle arrest. (A and B) The percentage of apoptotic cells was determined with Annexin V-APC. (C and D) Flow cytometry was performed to evaluate G1/S arrest in Saos-2 cells following knockdown of CacyBP/SIP. **P<0.01. CacyBP/SIP, calcyclin-binding protein/Siah-1-interacting protein; sh, short hairpin RNA; Ctrl, control; OD, optical density.
To further explore the mechanism by which CacyBP/SIP promoted Saos-2 cell growth, the effects of silencing CacyBP/SIP expression on the cell cycle were investigated by flow cytometry. A total of 67.54±0.76% of shCacyBP/SIP cells were in the G1 phase, compared with 61.32±0.69% of Ctrl cells (Fig. 3C and D; P<0.01). In addition, S phase distribution of Ctrl cells and shCacyBP/SIP cells were 17.18±0.16 and 10.82±1.01%, respectively (Fig. 3C and D; P<0.01). The results indicated that downregulation of CacyBP/SIP induced G1/S phase cell cycle arrest.
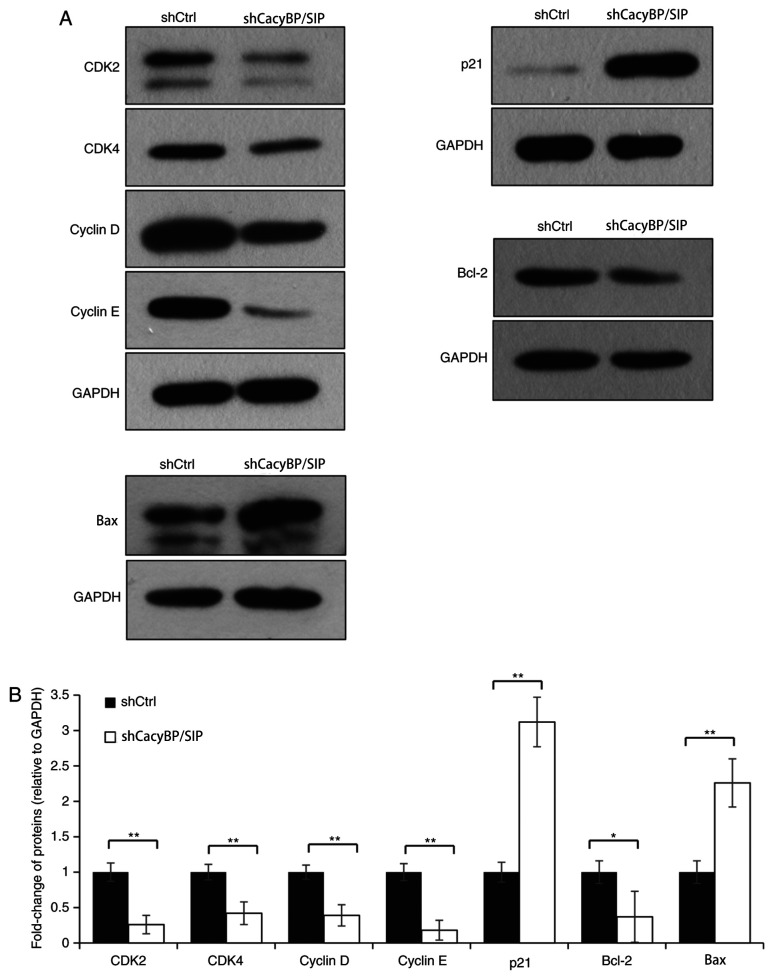
Effects of CacyBP/SIP knockdown on CDK/cyclin/P21 and Bcl-2/Bax axis
Next, the levels of key proteins involved in the intracellular mechanisms of the cell cycle and apoptosis were evaluated. The data demonstrated that knockdown of CacyBP/SIP protein was associated with an increase in p21 levels and a decrease in CDK2, CDK4, cyclin D and cyclin E levels (P<0.01; Fig. 4), indicating that CacyBP/SIP arrested the process of cell cycle G1 to S phase transition. Knockdown of CacyBP/SIP resulted in an increase of Bax (P<0.01; Fig. 4), and decrease of Bcl-2 (P<0.05; Fig. 4), which are two key regulators in the apoptosis process. The results demonstrated that CDK/cyclin/P21 axis and Bcl-2/Bax axis were activated by CacyBP/SIP.
Figure 4.
Analysis of CDK/cyclin/P21 and Bcl-2/Bax axis. (A and B) Western blot analysis of levels of key proteins involved in the intracellular mechanisms of the cell cycle and apoptosis. *P<0.05 and **P<0.01. CDK, cyclin-dependent kinases; sh, short hairpin RNA; Ctrl, control.
Prognosis analysis based on TCGA data
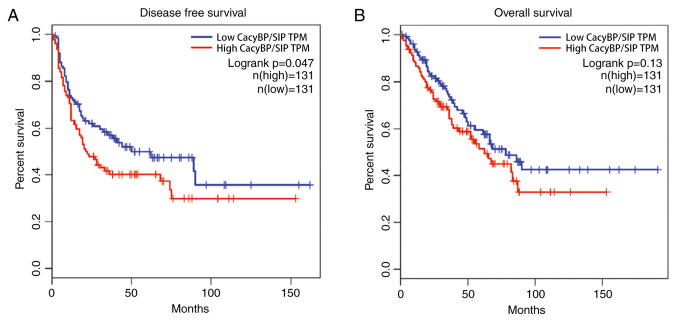
Patients with high CacyBP/SIP mRNA levels demonstrated worse prognosis compared with patients with low CacyBP/SIP mRNA levels, from both the disease free survival (Fig. 5A) and overall survival (Fig. 5B) data. The results demonstrated that CacyBP/SIP had a tumor-driving role in TCGA samples.
Figure 5.
Prognosis analysis based on TCGA data. TCGA database analysis demonstrated that patients with high CacyBP/SIP mRNA levels demonstrated worse prognosis compared with patients with low CacyBP/SIP mRNA levels. (A) Disease free survival analysis. (B) Overall survival analysis. TCGA, The Cancer Genome Atlas; CacyBP/SIP, calcyclin-binding protein/Siah-1-interacting protein; sh, short hairpin RNA; Ctrl, control.
Discussion
CacyBP/SIP is an important protein in tumorigenesis and is widely detected in a variety of human tissues, where it regulates cell proliferation, differentiation, chemotherapy resistance and tumorigenicity (8). However, the exact role of CacyBP/SIP in cancer remains unclear. CacyBP/SIP has been reported to be an oncogene in a number of malignant tumors. In human glioma, colorectal cancer and pancreatic cancer patients, increased expression of CacyBP/SIP was significantly associated with tumor cell proliferation and metastasis (10,15,24). In addition, decreased expression of CacyBP/SIP was found in renal cell carcinoma, gastric cancer and chronic lymphocytic leukemia cells where it appeared to be a tumor suppressor (13,15,16,25,26). Therefore, CacyBP/SIP cannot be regarded only as an oncogene or tumor suppressor. Its function appears to vary according to cell lines and the organs involved, or the different modes of tumor progression.
In the present study, OS cell lines Saos-2, MG-63, HOS and U20S expressed CacyBP/SIP. Lentivirus-mediated CacyBP/SIP knockdown markedly inhibited cell proliferation in both Saos-2 and U20S cells and induced Sao-2 cell apoptosis in vitro. The total cell number in the shCacyBP/SIP group remained unchanged, whilst the total cell number of shCtrl group were significantly increased during these tested days. In addition, the cell growth rate in the CacyBP/SIP-shRNA lentivirus-transduced group were significantly decreased compared with the Ctrl lentivirus-transduced group from days 2 to 5. Colony formation assay demonstrated that CacyBP/SIP knockdown in Saos-2 cells caused a substantial reduction in colony formation numbers. In addition, the data demonstrated that knockdown of CacyBP/SIP inhibited the cell cycle by inducing G1/S phase arrest. p21, also known as CDK inhibitor 1, is capable of binding and inhibiting numerous cyclin/CDK complexes, such as cyclin D/CKD4(27), cyclin E/CDK2(28) and cyclin A/CDK2(29). The results of the present study demonstrated that knockdown of CacyBP/SIP protein was associated with an increase in p21 levels and decrease in CDK2, CDK4, cyclin D and cyclin E levels. In accordance with this result, several studies have identified that overexpression of p21 can induce cell cycle arrest at G1, G2 and S phases (13,30). It has been previously reported that p21 serves as a tumor suppressor (31). The present study indicated that CacyBP/SIP was associated with the CDK/cyclin/P21 axis. All of the data from the present study suggested that knockdown of CacyBP/SIP may inhibit OS cell proliferation by regulating specific cell factors involved in cell cycle and apoptosis processes. Furthermore, CacyBP/SIP knockdown induced cell apoptosis. Taken together, the present study demonstrated that CacyBP/SIP serves an important role in OS cell proliferation and apoptosis, suggesting an oncogenic role for CacyBP/SIP in OS. Further research is required to explore the complex molecular mechanisms of CacyBP/SIP in OS cells.
In conclusion, the results of the present study suggested that the downregulation of CacyBP/SIP by shRNA could markedly suppress proliferation and tumorigenesis by inducing apoptosis in OS cells. To the best of the authors' knowledge, this is the first study to investigate the role of CacyBP/SIP in OS cells. Further investigation is required to elucidate the mechanisms of CacyBP/SIP in OS cells. The present study revealed the function of CacyBP/SIP in OS cells and indicated that CacyBP/SIP might be a potential molecular target in the treatment of OS.
Supplementary Material
Acknowledgements
Not applicable.
Funding
This work was supported by grants from the Hebei Province Scientific Research Foundation for the Returned Overseas Chinese Scholars (grant no. CY201715) and a grant from the Key R&D Plan Project of Hebei Science and Technology Department (grant no. 18277798D).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
MZ, RZZ and GCZ designed all the experiments, analyzed the data and revised the paper; MZ, RZZ, DWQ and HYC performed experiments, drafted and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 2.Bernthal NM, Federman N, Eilber FR, Nelson SD, Eckardt JJ, Eilber FC, Tap WD. Long-term results (>25 years) of a randomized, prospective clinical trial evaluating chemotherapy in patients with high-grade, operable osteosarcoma. Cancer. 2012;118:5888–5893. doi: 10.1002/cncr.27651. [DOI] [PubMed] [Google Scholar]
- 3.Grignani G, Palmerini E, Dileo P, Asaftei SD, D'Ambrosio L, Pignochino Y, Mercuri M, Picci P, Fagioli F, Casali PG, et al. A phase II trial of sorafenib in relapsed and unresectable high-grade osteosarcoma after failure of standard multimodal therapy: An Italian Sarcoma Group study. Ann Oncol. 2012;23:508–516. doi: 10.1093/annonc/mdr151. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, Yang J, Zhao N, Wang C, Kamar S, Zhou Y, He Z, Yang J, Sun B, Shi X, et al. Progress in the chemotherapeutic treatment of osteosarcoma. Oncol Lett. 2018;16:6228–6237. doi: 10.3892/ol.2018.9434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grignani G, Palmerini E, Ferraresi V, D'Ambrosio L, Bertulli R, Asaftei SD, Tamburini A, Pignochino Y, Sangiolo D, Marchesi E, et al. Italian Sarcoma Group: Sorafenib and everolimus for patients with unresectable high-grade osteosarcoma progressing after standard treatment: A non-randomised phase 2 clinical trial. Lancet Oncol. 2015;16:98–107. doi: 10.1016/S1470-2045(14)71136-2. [DOI] [PubMed] [Google Scholar]
- 6.Filipek A, Kuźnicki J. Molecular cloning and expression of a mouse brain cDNA encoding a novel protein target of calcyclin. J Neurochem. 1998;70:1793–1798. doi: 10.1046/j.1471-4159.1998.70051793.x. [DOI] [PubMed] [Google Scholar]
- 7.Filipek A, Wojda U. p30, a novel protein target of mouse calcyclin (S100A6) Biochem J. 1996;320:585–587. doi: 10.1042/bj3200585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Topolska-Woś AM, Chazin WJ, Filipek A. CacyBP/SIP - Structure and variety of functions. Biochim Biophys Acta. 2016;1860:79–85. doi: 10.1016/j.bbagen.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 9.Zhai H, Shi Y, Jin H, Li Y, Lu Y, Chen X, Wang J, Ding L, Wang X, Fan D. Expression of calcyclin-binding protein/Siah-1 interacting protein in normal and malignant human tissues: An immunohistochemical survey. J Histochem Cytochem. 2008;56:765–772. doi: 10.1369/jhc.2008.950519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi H, Gao Y, Tang Y, Wu Y, Gong H, Du J, Zheng B, Hu J, Shi Q, Yu R. CacyBP/SIP protein is important for the proliferation of human glioma cells. IUBMB Life. 2014;66:286–291. doi: 10.1002/iub.1263. [DOI] [PubMed] [Google Scholar]
- 11.Rosińska S, Leśniak W, Filipek A. Distinct effect of CacyBP/SIP on the ERK1/2-CREB-BDNF pathway in undifferentiated and differentiated neuroblastoma NB2a cells. Neurochem Int. 2016;97:65–72. doi: 10.1016/j.neuint.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Chen X, Zheng P, Xue Z, Li J, Wang W, Chen X, Xie F, Yu Z, Ouyang X. CacyBP/SIP enhances multidrug resistance of pancreatic cancer cells by regulation of P-gp and Bcl-2. Apoptosis. 2013;18:861–869. doi: 10.1007/s10495-013-0831-9. [DOI] [PubMed] [Google Scholar]
- 13.Fu C, Wan Y, Shi H, Gong Y, Wu Q, Yao Y, Niu M, Li Z, Xu K. Expression and regulation of CacyBP/SIP in chronic lymphocytic leukemia cell balances of cell proliferation with apoptosis. J Cancer Res Clin Oncol. 2016;142:741–748. doi: 10.1007/s00432-015-2077-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhai H, Shi Y, Chen X, Wang J, Lu Y, Zhang F, Liu Z, Lei T, Fan D. CacyBP/SIP promotes the proliferation of colon cancer cells. PLoS One. 2017;12(e0169959) doi: 10.1371/journal.pone.0169959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ning X, Sun S, Hong L, Liang J, Liu L, Han S, Liu Z, Shi Y, Li Y, Gong W, et al. Calcyclin-binding protein inhibits proliferation, tumorigenicity, and invasion of gastric cancer. Mol Cancer Res. 2007;5:1254–1262. doi: 10.1158/1541-7786.MCR-06-0426. [DOI] [PubMed] [Google Scholar]
- 16.Sun S, Ning X, Liu J, Liu L, Chen Y, Han S, Zhang Y, Liang J, Wu K, Fan D. Overexpressed CacyBP/SIP leads to the suppression of growth in renal cell carcinoma. Biochem Biophys Res Commun. 2007;356:864–871. doi: 10.1016/j.bbrc.2007.03.080. [DOI] [PubMed] [Google Scholar]
- 17.Topolska-Woś AM, Rosińska S, Filipek A. MAP kinase p38 is a novel target of CacyBP/SIP phosphatase. Amino Acids. 2017;49:1069–1076. doi: 10.1007/s00726-017-2404-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Chai R, Yu X, Tu S, Zheng B. Depletion of UBA protein 2-like protein inhibits growth and induces apoptosis of human colorectal carcinoma cells. Tumour Biol. 2016;37:13225–13235. doi: 10.1007/s13277-016-5159-y. [DOI] [PubMed] [Google Scholar]
- 20.Henderson T, Chen M, Darrow MA, Li CS, Chiu CL, Monjazeb AM, Murphy WJ, Canter RJ. Alterations in cancer stem-cell marker CD44 expression predict oncologic outcome in soft-tissue sarcomas. J Surg Res. 2018;223:207–214. doi: 10.1016/j.jss.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. doi: 10.1016/j.cell.2017.10.014. Comprehensive and Integrated Genomic Characterization of Adult Soft Tissue Sarcomas. Cell 171: 950-965.e928, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartigan JA, Wong MA. A K-means clustering algorithm. Appl Stat. 2013;28:100–108. [Google Scholar]
- 23.Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411:342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- 24.Vats P, Ray K, Majumadar D, Amitabh Joseph DA, Bayen S, Akunov A, Sarbaev A, Singh SB. Changes in cardiovascular functions, lipid profile, and body composition at high altitude in two different ethnic groups. High Alt Med Biol. 2013;14:45–52. doi: 10.1089/ham.2012.1071. [DOI] [PubMed] [Google Scholar]
- 25.Zhai HH, Meng J, Wang JB, Liu ZX, Li YF, Feng SS. CacyBP/SIP nuclear translocation induced by gastrin promotes gastric cancer cell proliferation. World J Gastroenterol. 2014;20:10062–10070. doi: 10.3748/wjg.v20.i29.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ning X, Sun S, Zhang K, Liang J, Chuai Y, Li Y, Wang X. S100A6 protein negatively regulates CacyBP/SIP-mediated inhibition of gastric cancer cell proliferation and tumorigenesis. PLoS One. 2012;7(e30185) doi: 10.1371/journal.pone.0030185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Wang S, Qian W, Ji D, Wang Q, Zhang Z, Wang S, Ji B, Fu Z, Sun Y. uc.338 targets p21 and cyclin D1 via PI3K/AKT pathway activation to promote cell proliferation in colorectal cancer. Oncol Rep. 2018;40:1119–1128. doi: 10.3892/or.2018.6480. [DOI] [PubMed] [Google Scholar]
- 28.Merli M, Benassi MS, Gamberi G, Ragazzini P, Sollazzo MR, Molendini L, Magagnoli G, Ferrari C, Maltarello MC, Picci P. Expression of G1 phase regulators in MG-63 osteosarcoma cell line. Int J Oncol. 1999;14:1117–1121. doi: 10.3892/ijo.14.6.1117. [DOI] [PubMed] [Google Scholar]
- 29.Adams PD, Sellers WR, Sharma SK, Wu AD, Nalin CM, Kaelin WG Jr. Identification of a cyclin-cdk2 recognition motif present in substrates and p21-like cyclin-dependent kinase inhibitors. Mol Cell Biol. 1996;16:6623–6633. doi: 10.1128/mcb.16.12.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song Z, Lin J, Sun Z, Ni J, Sha Y. RNAi-mediated downregulation of CDKL1 inhibits growth and colony-formation ability, promotes apoptosis of human melanoma cells. J Dermatol Sci. 2015;79:57–63. doi: 10.1016/j.jdermsci.2015.03.020. [DOI] [PubMed] [Google Scholar]
- 31.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.