Abstract
Autoimmune diseases are a group of heterogeneous disorders characterized by damage to various organs caused by abnormal innate and adaptive immune responses. The pathogenesis of autoimmune diseases is extremely complicated and has not yet been fully elucidated. Long noncoding RNAs (lncRNAs), which are defined as transcripts containing more than 200 nucleotides with no protein-coding capacity, are emerging as important regulators of gene expression via epigenetic modification, transcriptional regulation and posttranscriptional regulation. Accumulating evidence has demonstrated that lncRNAs play a key role in the regulation of immunological functions and autoimmunity. In this review, we discuss various molecular mechanisms by which lncRNAs regulate gene expression and recent findings regarding the involvement of lncRNAs in many human autoimmune diseases, including rheumatoid arthritis (RA), systemic lupus erythematosus (SLE), idiopathic inflammatory myopathy (IIM), systemic sclerosis (SSc) and Sjögren’s syndrome (pSS).
Keywords: Long noncoding RNAs, Epigenetic regulation, Autoimmune diseases
Highlights
-
•
lncRNAs are observed to be differentially expressed in various autoimmune diseases.
-
•
lncRNAs are involved in abnormal immune regulation and inflammatory responses in autoimmune diseases, which provides new insight into disease pathogenesis.
-
•
LncRNAs may have the potential of biomarkers for diagnosis and prognosis of autoimmune diseases.
1. Introduction
Autoimmune diseases are a group of heterogeneous disorders that are caused by inappropriate immune responses to ‘self’ antigens; these immune responses attack normal molecules, cells and tissues of the human body, thereby causing damage to various organs and systems. Autoimmune diseases comprise a wide spectrum of complicated diseases, mainly including rheumatoid arthritis (RA), ankylosing spondylitis (AS), systemic lupus erythematosus (SLE), idiopathic inflammatory myopathy (IIM), systemic sclerosis (SSc) and Sjögren’s syndrome (pSS), and the clinical manifestations of these diseases vary from mild skin rashes to severe multiple organ dysfunction. As the pathological process of autoimmune diseases is complicated and not yet fully elucidated, the early diagnosis and efficient treatment of most autoimmune diseases have been a challenge for clinicians. Hence, further understanding of the underlying molecular mechanisms and defining the crucial regulators in autoimmune diseases are imperative (see Table 1).
Table 1.
Summary of lncRNAs involved in autoimmune diseases.
| Autoimmune disease | lncRNA | Species | Tissue/cell | Expression level | Function | Ref |
|---|---|---|---|---|---|---|
| RA | Hotair, LUST, anti-NOS2A, MEG9, SNHG4, TUG1, NEAT1 | Human | Serum exosome | Up-regulated | – | [51] |
| Malat1, SNHG1, mascRNA, PR antisense transcripts, PRINS, HOXA3as |
Human | Serum exosome | Down-regulated | [51] | ||
| Hotair, LUST, H19 antisense, anti-NOS2A, MEG9, SNHG4, HAR1B, TUG1, NEAT1, and GAS5 |
Human | PBMC | Up-regulated | [51] | ||
| DHFR upstream transcripts, Jpx, KRASP1, mascRNA, PR antisense transcripts, PRINS, and HOXA3as |
Human | PBMC | Down-regulated | [51] | ||
| ENST00000483588 | Human | FLSs | Up-regulated | Positively correlated with CRP and the Simplified Disease Activity Index score(SDAI). | [53] | |
| lnc-AL928768.3, lnc-AC091493.1 | Human | Synovium tissue | Up-regulated | Positively associated with CRP and disease activity score in 28 joints(DAS28) | [54] | |
| U75927, XR_008357, MRAK046251, | AA rat | Synovium tissue | Up-regulated | [59] | ||
|
DQ266363, XR_006457, MRAK003448 |
AA rat | Synovium tissue | Down-regulated | [59] | ||
| HOTAIR | Human | PBMC, Serum exosome |
Up-regulated | Regulates macrophage migration. | [51] | |
| Human | Osteoclast, FLS |
Down-regulated | Regulates MMP-2 and MMP-13 level. | [51] | ||
| Human, rat | LPS-treated chondrocytes | Down-regulated | Regulates proliferation and inflammation of chondrocytes. | [63] | ||
| HIX003209 | Human | PBMC, macrophage | Up-regulated | Increase inflammation by sponging miR-6089 | [64] | |
| NTT | Human | PBMC, monocyte | Up-regulated | Promotes monocyte differentiation by regulating nearby gene PBOV1. | [65] | |
| Linc-P21 | Human | Whole blood | Down-regulated | Regulates NF-kB activity. | [67] | |
| LOC100652951, LOC100506036 |
Human | T cell | Up-regulated | Regulates inflammation response. | [68] | |
| GAS5, THRIL, RMRP | Human | T cell | Up-regulated | – | [69] | |
| H19 | Human | Synovial tissue, FLS, macrophage | Up-regulated | Induced by serum starvation and under control of PI3K and ERK pathway. | [74] | |
| LERFS | Human | Synovial tissue, FLS | Down-regulated | Regulates migration, invasion and proliferation by interacting with hnRNPQ. | [41] | |
| AFAS1 | Human | FLS | Up-regulated | Regulates migration, invasion by suppressing miR-27a. | [76] | |
| GAPLINC | Human | FLS | Up-regulated | Regulates migration, invasion by suppressing miR-382–5p and miR-575. | [77] | |
| PICSAR | Human | FLS | Up-regulated | Regulates migration, invasion and proliferation by sponging miR-4701–5p. | [78] | |
| FER1L4 | Human | Synovial tissue, FLS | Downregulated | Regulates inflammation via targeting NLRC5. | [79] | |
| MALAT1 | Human | Quercetin-treated FLS | Up-regulated | Responsible for the quercetin-induced apoptosis. | [80] | |
| UCA1 | Human | FLS | Down-regulated | Induces apoptosis by Wnt6 | [81] | |
| DILC | Human | FLS, plasma | Down-regulated | Regulates apoptosis and IL-6 expression. | [82] | |
| Lnc-IL7R | Human | FLS | – | Promotes growth of RA FLS through interaction with EZH2 | [83] | |
| LINC00152 | Human | FLS | Up-regulated | Regulates proliferation and apoptosis via Wnt/beta-catenin signaling pathway | [84] | |
| GAS5 | Human | Tanshinone IIA -treated FLS, plasma | Down-regulated | Responsible for apoptosis and IL-18 expression. | [85,86] | |
| ITSN1-2 | Human | FLS | Up-regulated | Regulates apoptosis, proliferation and inflammation by NOD2/RIP2 pathway. | [87] | |
| PVT1 | Rat | FLS | Up-regulated | Regulates inflammation response and apoptosis. | [90] | |
| MEG3 | Rat | FLS | Down-regulated | Modulates inflammation by targeting NLRC5 | [88] | |
| MEG3 | Human | FLS, chonroncyte | Down-regulated | Inhibit proliferation and inflammation by miR-141 and AKT/mTOR pathway | [84] | |
| C5T1lncRNA | Human | Various tissues, PBMC | Up-regulated | Influences the transcript levels of C5. | [71] | |
| NR024118 | Mouse | FLS | Up-regulated | Regulates inflammation response. | [91] | |
| SLE | uc001ykl.1 | Human | T cell | Down-regulated | Correlates with ESR and CRP. | [95] |
| ENST00000448942 | Human | T cell | Down-regulated | Correlates with ESR and anti-Sm antibodies. | [95] | |
| linc0597, lnc0640, and lnc5150 | Human | Plasma | Up-regulated | Biomarkers of SLE | [97] | |
| GAS5 and lnc7074 | Human | Plasma | Down-regulated | Biomarkers of SLE | [97] | |
| ENST00000604411.1, ENST00000501122.2 lnc-HSFY2-3:3 |
Human | DC | Up-regulated | Positively correlated with the SLEDAI score. | [96] | |
| lnc-HSFY2-3:3, lnc-SERPINB9-1:2 | Human | DC | Down-regulated | – | [96] | |
| NEAT1 | Human | PBMCs, monocytes | Up-regulated | Correlates with SLEDAI. Regulates expression of IL-6, CCL2, and CXCL10. |
[98] | |
| GAS5 | Human, mouse | CD4+ T cells, B cells, plasma | Down-regulated | Correlates with ESR and SLEDAI. | [100,[104], [105], [106]] | |
| MALAT 1 | Human | Monocytes | Up-regulated | Regulate IL21 expression. | [109] | |
| Linc0949 | Human | PBMC | Down-regulated | Correlates with C3 level and SLEDAI and incidence of LN. | [107] | |
| Lnc5150, lnc3643,lnc7514 | Human | PBMC | Down-regulated | Associated with laboratory features in SLE | [108] | |
| lncRNA CYP2C9 | Human | Interacts with transcription factor PU.1 (SPI1), MSR1 and CCR1 | [110] | |||
| Lnc-DC | Human | plasma | Down-regulated | Significantly higher expressed in LN compared with SLE without nephritis. Showed weakly negative correlation with C3 level. | [106] | |
| Linc0597 | Human | plasma | Up-regulated | Negative correlates with C3 level | [106] | |
| Human | PBMC | Down-regulated | – | [107] | ||
| TUG1 | Human | PBMC | Down-regulated | Correlates with ESR, SLEDAI, disease duration and LN. | [111] | |
| TUG1 | Pyrrolidine dithiocarbamate treated mouse | Kidney | Upregulated | Involved in the protection of NF-kappaB inhibition on kidney injury. | [112] | |
| RP11-2B6.2 | Human | Kidney biopsies from LN patients | Up-regulated | Regulates IFN-I pathway through epigenetic inhibition of SOCS1. | [113] | |
| IMMs | lncRNA 7 S L | Human | Serum | – | Autoantibody against lncRNA 7 S L is related to PM/DM. | [115] |
| ENST00000541196.1, uc011ihb.2, linc-DGCR6-1, | Human | Muscle | Down-regulated | – | [116] | |
| ENST00000551761.1, ENST00000583156.1 | Human | Muscle | Down-regulated | – | [116] | |
| H19, lncMyoD, MALAT1 |
Human | Muscle | Up-regulated | – | [117] | |
| SSc | TSIX | Human | Serum, skin fibroblast | Up-regulated | Increases type I collagen mRNA stability | [119] |
| CTBP1-AS2, AGAP2-AS1 | Human | Skin | Up-regulated | – | [120]. | |
| OTUD6B-AS1 | Human | Skin | Down-regulated | – | [120]. | |
| ncRNA00201 | Human | PBMC | Down-regulated | Regulates genes involved in vasculopathy, fibrosis and autoimmunity in SSc. | [121] | |
| NRIR | Human | PBMC | Up-regulated | Regulates IFN response. | [122] | |
| pSS | IFNGAS | Human | CD4+ T cell | Up-regulated | Correlated with SSA, ESR and IgG. | [124] |
| ENST00000455309.1, n336161, NR_002712, ENST00000546086.1, Lnc-UTS2D-1:1, n340599, and TCONS_l2_00014794 | Human | labial salivary glands | Up-regulated | Correlates with beta2 microglobulin, disease course, ESR, rheumatoid factor (RF), IgA, IgM, visual analogue scale (VAS) of parotid swelling and VAS of dry eyes | [125] | |
| LINC00657, CTD-2020K17.1 |
Human | PBMC | Up-regulated | Involved in pathogenesis of pSS. | [114] | |
| LINC00511 | Human | PBMC | Down-regulated | Involved in pathogenesis of pSS. | [114] | |
| PVT1 | Human | CD4+ T cell | Up-regulated | Controls proliferation and effector functions of CD4 T cells | [127] | |
| AS | NDRG1-AS6, CSNK1D-AS8 | Human | Hip joint ligament tissue | Up-regulated | – | [128] |
| CD46-AS9, SMYD5-AS2, NR_045553 | Human | Hip joint ligament tissue | Down-regulated | – | ||
| H19 | Human | PBMC | Up-regulated | Increases expression of IL-17 A and IL-23 via interaction with miR22–5p and miR675–5p | [129] |
The pathogenesis process of autoimmune diseases involves abnormal regulation of the immune system and cellular activity. For decades, our understanding of autoimmunity has been limited to the world of proteins; however, with the advance of the noncoding RNA (ncRNA) research field, it is increasingly apparent that this knowledge is currently incomplete. Emerging evidence indicates that long noncoding RNAs (lncRNAs) play important roles in epigenetic modification, transcriptional regulation and posttranscriptional regulation and that these molecules participate in various human diseases, especially cancers and inflammatory disorders [[1], [2], [3]].
Here, we will review and summarize the lncRNAs involved in the pathogenesis of autoimmune diseases and the underlying molecular mechanisms. In addition, we will emphasize the clinical relevance of lncRNAs in the diagnosis and prognosis of autoimmune diseases and the possibility that some lncRNAs may be promising targets for novel treatment strategies.
2. Long noncoding RNAs (lncRNAs)
With the completion of the human genome project and the development of high-throughput genomic sequencing technologies, ncRNA has attracted increasing attention. Previous studies have surprisingly revealed that although the majority of the human genome is transcribed, less than 2% of the human genome encodes proteins, and the remainder encodes ncRNAs [4]. For a long time, ncRNAs, which account for the majority of various transcripts, have been considered to be the ‘noise’ produced in the process of transcription and have been overlooked. Recently, observations made by human genome-wide association studies (GWASs) found that more than 90% of disease-related SNPs are associated with noncoding elements of the genome, indicating that mutations in ncRNAs may explain some disease phenotypes [5]. Among the various kinds of ncRNAs, lncRNAs have recently become a major focus of research about disorders including autoimmune diseases.
lncRNAs, which are defined as transcripts at least 200 nucleotides in length without protein-coding potential, were first described as a new kind of transcript during the large-scale sequencing of full-length murine cDNA libraries in 2002 [6]. Most lncRNAs are transcribed by RNA polymerase (Pol) Pol II/Pol I, and some are transcribed by RNA Pol III [7,8]. The number of lncRNAs in humans has been estimated to be more than 100,000 [9]. Compared to messenger RNAs (mRNAs), lncRNAs tend to be expressed at relatively low levels [10,11], causing difficulty in detection and analysis. As shown by recent studies, most lncRNAs exhibit weak primary sequence conservation. Instead, many lncRNAs are observed to form well-conserved RNA secondary structures [12], which participate in coordinating RNA–RNA, RNA–protein and RNA–DNA interactions.
lncRNAs are exquisitely regulated and specifically expressed in many different organs, tissues, cell types and subcellular compartments [13]. Moreover, the expression of lncRNAs varies between different developmental stages [14] or disease states [2], suggesting that lncRNAs are critical regulators in cellular processes and disease progression. In fact, dozens of lncRNAs have been proven to be involved in the etiology of many human diseases [2,15,16].
2.1. Classification of lncRNAs
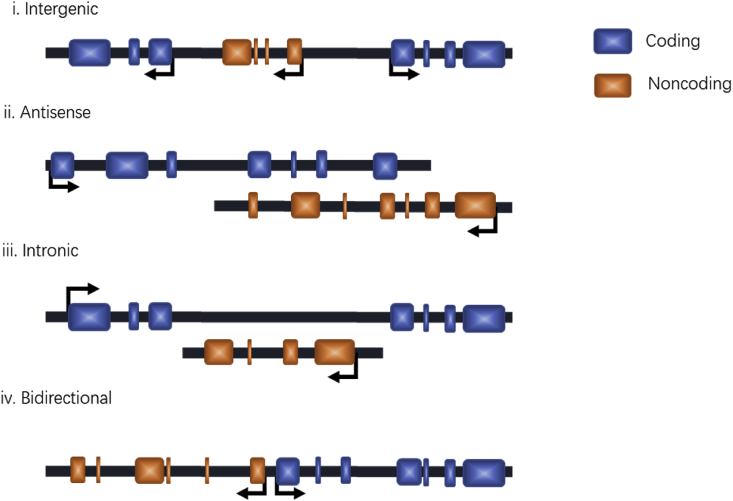
lncRNAs are often categorized by their position relative to nearby protein-coding genes, i.e., intergenic, antisense, intronic, or bidirectional (Fig. 1). Intergenic lncRNAs (also termed large intervening noncoding RNAs or lincRNAs) are lncRNAs located between protein-coding genes with separate transcriptional units. Antisense lncRNAs are lncRNAs that are transcribed in the opposite direction of nearby protein-coding genes and span at least one exon. Intronic lncRNAs are lncRNAs that originate from intronic regions without overlapping any exons. Finally, bidirectional lncRNAs are transcripts that initiate in a divergent fashion from the promoter of nearby protein-coding genes [[17], [18], [19]].
Fig. 1.
Classification of by their genomic location relative to nearby protein-coding genes. (i) Intergenic lncRNAs: lncRNAs located between protein-coding genes; (ii)Antisense lncRNAs: lncRNAs transcribed in the opposite direction of nearby protein-coding genes, and span over at least one exon; (iii)Intronic lncRNAs: lncRNAs that originate from intronic regions of protein-coding genes; (iv) Bidirectional lncRNAs: lncRNAs that initiate transcription in a divergent fashion from the promoter of nearby protein-coding gene.
lncRNA transcripts can directly interact with proteins, DNA, and other RNAs to regulate their target molecules. Nevertheless, not all lncRNAs exert their regulatory function depending on the lncRNA molecule itself, and the processes of lncRNA transcription and/or splicing also exhibit functional roles [20,21]. According to the targets regulated by lncRNAs, we can classify lncRNAs into 2 different groups: (i) lncRNAs acting in cis, which influence the expression and/or chromatin state of nearby genes, and (ii) lncRNAs acting in trans, which leave the site of transcription and execute regulatory functions throughout the cell [22]. Recent studies have demonstrated that some lncRNAs are also capable of encoding peptides such as myoregulin [23] and SPAR [24], which execute biological functions. This type of lncRNA is not discussed further in this review.
2.2. Molecular mechanisms of lncRNAs
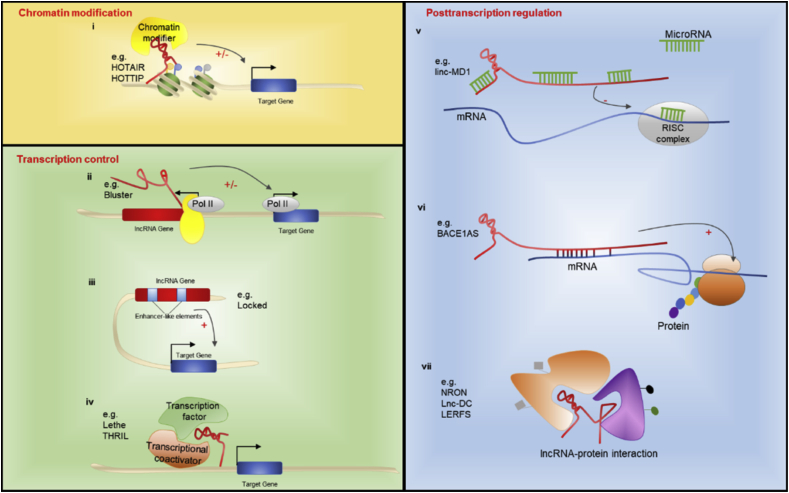
The molecular functions of lncRNAs are diverse and complicated, and several mechanisms of lncRNA regulation have been proposed to date. It is well accepted that lncRNAs can serve as signals, decoys, guides, and scaffolds to perform various functions in a wide array of biological processes at the epigenetic, transcriptional and posttranscriptional levels [25,26] (Fig. 2).
Fig. 2.
Mechanisms of lncRNA functions. (i) lncRNAs mediate recruitment of chromatin modifying complexes to specific genome loci; (ii) The process of splicing and transcription of lncRNAs regulate nearby protein-coding gene; (iii) Enhancer-like elements existing in lncRNA loci regulate gene expression; (iv) lncRNAs regulate transcription by interaction with transcription factors and transcriptional coactivators; (v) lncRNAs act as ceRNA to modulate expression of genes targeted by microRNAs; (vi) lncRNAs bind to mRNA and alter its stability and translation activity; (vii) lncRNAs regulate posttranslational modification, subcellular localization, activation and degradation of protein by forming lncRNA-protein complexes. Pol II: RNA polymerase-II; RISC complex: RNA-induced silencing complex.
2.2.1. Chromatin modification
Chromatin modification plays an important role in the regulation of gene expression. One of the first lncRNAs reported to regulate epigenetic modification was.
HOTAIR (HOX antisense intergenic RNA); this lncRNA acts as a scaffold that recruits chromatin modifying complexes PRC2 (polycomb repressive complex 2) to the HOXD locus, coordinates H3K27 methylation and H3K4me2 demethylation of target genes, and contributes to maintaining a repressive chromatin state [27], which has been implicated in the pathogenesis of several kinds of cancers, including breast cancer, lung cancer, and colon cancer [28]. On the other hand, HOTTIP, which is transcribed from a locus upstream of HOXA genes, recruits the MLL-1/WDR5 complex to the 5’ region of HOXA genes, mediates H3K4me3 modification and activates transcription of the HOXA locus [29].
2.2.2. Transcription control
The initiation of transcription involves the coordination of various processes and factors, including the recruitment of RNA polymerase-II (Pol II), localization and function of transcription factors, integration of different kinds of coregulators at the promoter regions of specific genes, and function of transcriptional enhancers. lncRNAs have been described to modulate transcription processes in both lncRNA transcript sequence-dependent and transcription- or splicing-dependent manners.
As demonstrated before, the splicing and transcription of the lncRNA Blustr regulates the expression of its nearby protein-coding gene Sfmbt2 by recruiting polymerases and chromatin modifiers in a mechanism that is independent of the sequence of Blustr itself [30]. In addition, emerging evidence has shown that DNA elements within the lncRNA promoter or gene locus may function as important cis-acting enhancer elements [30]. The promoter of the lncRNA downstream of the Cdkn1b (Lockd) locus was found to harbor many enhancer-like elements, through which the transcription of its neighboring gene Cdkn1b is positively regulated [31]. Another mechanism of lncRNA function is that lncRNAs interact with transcription factors or transcriptional coactivators, thereby influencing their localization and ability to bind to the promoter region of target genes. For example, the lncRNA Lethe, derived from pseudogenes, associates with the NF-kB p65 (RelA) subunit to inhibit its binding to the promoters of TNF-α, IL-6 and IL-8 [32]. The lncRNA THRIL modulates TNFα expression by interacting with hnRNP L, which is critical for the transcriptional activation of TNF-α gene [33].
2.2.3. Posttranscription regulation
In addition to being critical regulators of gene expression, lncRNAs are also important players in posttranscriptional regulatory networks. One of the most recognized mechanisms is that lncRNAs work as competitive endogenous RNAs (ceRNAs), or ‘microRNA sponges’, to reduce the levels of microRNAs and therefore regulate the expression of their target genes [34]. For instance, by binding to miR-133 and miR-135, the lncRNA linc-MD1 exquisitely modulates the expression of MAML1 and MEF2C, which control muscle differentiation [35]. Likewise, lncRNAs can also bind to mRNAs and control mRNA stability and translation activity. Previous research shows that the antisense RNA BACE1AS interacts with the BACE1 transcript, increases the stability and abundance of the BACE1 mRNA, and thus increases the expression level of the BACE1 protein [36]. In addition, lncRNAs also associate with proteins and influence posttranslational modifications such as phosphorylation [37,38] and ubiquitination [39], consequently influencing the localization, activity and degradation of the corresponding proteins. For instance, the lncRNA NRON regulates the dephosphorylation and nuclear import of activated T-cell nuclear factor (NFAT) by forming a NRON-NEAT complex [40]. Another classic example of this mechanism is the regulation of STAT3 phosphorylation by lnc-DC [37]. A recent study reported a novel lncRNA, LERFS, that regulates the expression and activity of RhoA, Rac1 and CDC42, probably by binding to hnRNP Q [41]; this finding reinforces the notion that lncRNAs may be the critical ‘reprogrammer’ at the posttranscriptional level.
3. Important roles of lncRNAs in autoimmune diseases
lncRNAs are widely accepted as emerging players in gene regulation and disease pathogenesis and have been intensively studied in the context of cancer [42,43], inflammation [44], and innate and adaptive immunity [18]. Herein, we provide a brief overview of the lncRNAs involved in the pathogenesis of common autoimmune diseases and discuss their potential as targets of new strategies for the diagnosis and treatment of autoimmune diseases.
3.1. lncRNAs in RA
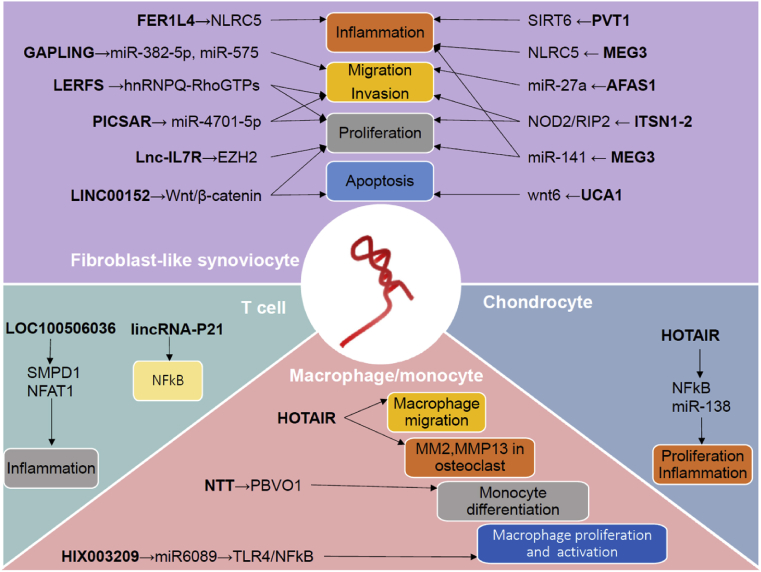
RA is a chronic systematic autoimmune disease that is characterized by multiple symmetrical joint inflammation and progressive bone and cartilage destruction [45]. As one of the most common rheumatic diseases, RA has been a heavy burden to human health because it causes joint destruction and physical disability; however, despite intensive research in the field, the etiology and pathogenesis of RA is not yet fully understood [46]. Dysregulated immune responses [47], release of proinflammatory cytokines (such as IL-6, TNF-α and IL-1β) [48], and abnormal activation of fibroblast-like synoviocytes (FLSs) [49] are thought to contribute to RA pathogenesis (Fig. 3).
Fig. 3.
Important lncRNAs involved in the pathogenesis of RA.
Recently, the involvement of lncRNAs in the pathogenesis of RA has attracted increasing attention. High-throughput analyses by microarray or sequencing revealed that the expression profiles of lncRNAs are altered in the PBMCs [[50], [51], [52]], serum exosomes [51], osteoclasts [51], FLSs [51,53], synovial tissues [54], plasma [55,56], and synovium of a rat model of RA [57]. Some of these differentially expressed lncRNAs are related to disease activity. In CD14 (+) monocytes from RA patients, a large number of lncRNAs exhibited significantly altered expression after either IL-6 or TNF-α inhibition [58]. Aside from human lncRNAs, lncRNA expression in the synovium of rats with adjuvant-induced arthritis (AA) differs from that in the synovium of control rats [59]. In addition, treatment with astragalosides (AST), a traditional Chinese medicine used in the treatment of RA, greatly changes the expression profile of lncRNAs in an AA rat model [60].
HOTAIR is a lncRNA that is widely studied in the cancer field as an important regulator of epigenetic modification; HOTAIR functions by recruiting chromatin modifying complexes [61,62]. The expression of HOTAIR is upregulated in the PBMCs and serum exosomes of patients with RA compared to those of healthy controls, this upregulated expression promotes the migration of macrophages and enhances the recruitment of macrophages to target tissues [51]. On the other hand, HOTAIR expression is decreased in differentiated osteoclasts and RA FLSs, and overexpression of HOTAIR decreases the levels of MMP-2 and MMP-13, which are important players in the process of matrix destruction and joint damage [51]. Further research showed that HOTAIR expression is significantly reduced in chondrocytes after LPS treatment. Overexpression of HOTAIR inhibited the LPS-induced reduction in the cell proliferation rate and IL-17 and IL-23 production by modulating the miR-138 and NF-κB pathways and regulating IL-1β and TNF-α. In addition, similar results have been observed in an RA rat model [63]. These results suggest that HOTAIR may be a promising biomarker and therapeutic target for RA.
Another example of a functional lncRNA is HIX003209, which acts as a ceRNA and exacerbates inflammation by sponging miR-6089 through the TLR4/NF-kappaB pathway in macrophages [64]. The lncRNA NTT, a lncRNA regulated by the monocyte key transcription factor C/EBPβ, is elevated in rheumatoid arthritis and promotes monocyte differentiation by regulating the nearby gene PBOV1 [65].
lincRNA-p21, which functions as a key repressor of the TP53 target by interacting with ING1b and MDM2, is associated with tumor development and progression and may predict treatment response [66]. In RA, lincRNA-p21 was found to be expressed at a lower level in whole blood samples, while the expression of phosphorylated p65 (RelA), a marker of NF-κB activation, was higher than that in the control. Methotrexate (MTX) treatment in RA patients increases the expression levels of lincRNA-p21 and decreases the levels of p65. An in vitro study using either activated primary T cells or Jurkat cells indicated that lincRNA-p21 was induced via the DNA-dependent protein kinase catalytic subunit (DNA PKcs). Furthermore, MTX modulated NF-κB activity through the induction of lincRNA-p21 [67], suggesting a role of lincRNA-p21 in regulating NF-κB activity.
In T cells from patients with RA, compared with their expression in healthy controls, the lncRNAs LOC100652951, LOC100506036 [68], THRIL, and RMRP [69] are upregulated. RA patients treated with biological agents such as abatacept and tocilizumab show lower expression levels of LOC100652951. The expression of LOC100506036 increases in activated Jurkat cells, and silencing LOC100506036 reduces the expression of IFN-γ. In addition, the knockdown of LOC100506036 leads to decreased expression of NEAT1, which is a critical regulator of various cytokines [68]. The results above indicate that LOC100506036 could contribute to the inflammatory responses in RA.
The TRAF1-C5 region has been identified as an RA susceptibility gene [70]. The expression level of the lncRNA C5T1, a lncRNA transcribed from the 3’ untranslated region (UTR) of C5, predominantly in the nucleus, shows a positive correlation with C5 mRNA in various tissues and in PBMCs, indicating that C5T1 influences the transcription levels of C5 [71].
The lncRNA H19 is abundantly expressed in embryonal tissue and a number of different tumors [72,73]. In RA synovial tissues, H19 expression is significantly increased relative to that in osteoarthritis (OA) or normal/joint trauma control tissues and is located in the lining layer, diffuse infiltrates, and stroma regions [74]. Synovial macrophages and fibroblasts cultured in vitro also showed elevated expression of H19. Moreover, the expression of H19 is induced in RA FLSs by starvation, which is independent of stimulation with TNF-α, IL-1β or platelet-derived growth factor-BB (PDGF-BB). The expression of H19 is regulated by the PI3K and ERK-1/2 pathways, which are observed to play pivotal roles in the activation of RA FLSs [74]. In contrast, in a Chinese Han population, no association of single nucleotide polymorphisms (SNPs) within H19 with genetic susceptibility to RA was found [75]. Therefore, the roles of H19 in RA pathogenesis still need to be further studied.
Abnormal activation and tumor-like transformation of RA-FLSs have been regarded as key steps in joint destruction. AK309896, termed as LERFS (lowlyexpressed in rheumatoid fibroblast–like synoviocytes), is a novel 1729-nucleotide lncRNA transcribed from chromosome 9q13 and predominantly located in the cytoplasm; LERFS was found to be significantly downregulated in RA FLSs by a microarray screen. PDGFBB treatment induced significant downregulation of LERFS; on the other hand, MTX treatment enhanced the expression of LERFS. In vitro experiments in cultured FLSs proved that LERFS negatively regulates the migration, invasion, and proliferation of FLSs through interaction with heterogeneous nuclear ribonucleoprotein Q (hnRNPQ), which modulates the mRNA stability or translation of RhoA, Rac1, and CDC42 by binding to target mRNAs. In vivo experiments using a nude mouse migration model and a SCID mouse co-implantation invasion model further supported the regulation of RA FLS migration and invasion by LERFS [41].
ZFAS1 is another lncRNA that is involved in the abnormal activation of RA FLSs. Increased expression of ZFAS1 promotes RA FLS migration and invasion by suppressing miR-27a [76]. Similarly, lncRNA GAPLINC [77] and PICSAR [78] participate in the regulation of proliferation, migration and invasion by sponging microRNA molecules. In addition, FER1L4 regulates inflammation in RA FLSs by potentially targeting NLRC5 [79]. Several lncRNAs, including MALAT1 [80], UCA1 [81], DILC [82], LncIL7R [83] and LINC00152 [84], have been reported to contribute to apoptosis and proliferation in RA-FLSs. lncRNA GAS5 [85,86] and ITSN1-2 [87] are implicated in apoptosis and inflammation in RA FLSs. MEG3, a lncRNA that participates in cell proliferation in cancer tissues, also modulates inflammation by targeting NLRC5 [88], miR-141 and the AKT/mTOR pathway [89]. In addition, in an RA rat model, PVT1 regulates both inflammation and apoptosis in RA-FLSs through the demethylation of sirt6 [90].
In a murine anti-collagen monoclonal antibody-induced model of RA, shikonin, a major active ingredient isolated from zicao, suppressed the secretion and expression of IL-6, IL-8, and MMPs by enhancing the expression of the lncRNA NR024118 [91], implying a suppressive role of this lncRNA in inflammation; however, the role of the human homologue of lncRNA NR024118 remains to be established.
Accumulating findings provide novel and strong evidence that lncRNAs may be important regulators and potential therapeutic targets in RA. Nevertheless, the role of lncRNAs in RA is not yet fully understood and requires further investigation.
3.2. lncRNAs in SLE
SLE is a common, chronic autoimmune disease with a high incidence rate in women of childbearing age. SLE is characterized by the production of autoantibodies and deposition of immune complexes in various tissues and organs, causing damage to almost all organs and systems of the human body, most commonly the skin, kidney, lung, nervous system and circulatory system. The etiology of SLE, which involves genetic susceptibility, environmental factors, loss of immune tolerance to autoantigens and perturbations of both innate and adaptive immune systems, is complicated and not yet fully elucidated [92]. Accumulating evidence indicates that lncRNAs might contribute to the pathogenesis of SLE. Comprehensive analyses using microarray or RNA sequencing revealed greatly altered lncRNA expression profiles in the whole blood [93], PBMCs [94], T cells [95], monocyte-derived dendritic cells (moDCs) [96] and plasma [97] of SLE patients compared to those of healthy controls, and the expression of some of these differentially expressed lncRNAs might correlate with the disease activity of SLE patients.
Nuclear enriched abundant transcript 1 (NEAT1), a constitutively and widely expressed lncRNA that is involved in viral infection and innate immunity, is more highly expressed in peripheral blood mononuclear cells (PBMCs) and monocytes in SLE patients compared to those in healthy controls. The expression level of NEAT1 in SLE patients positively correlates with the SLE Disease Activity Index (SLEDAI) score. Further in vitro studies proved that NEAT1 regulates LPS-induced expression of IL-6, CCL2, and CXCL10, which play critical roles in the pathogenesis of SLE by recruiting inflammatory cells through the regulation of the MAPK signaling pathways [98]. A recent study demonstrated that the lncRNA NEAT1-BAFF axis contributes to the activation of B cells and disease progression in SLE [99].
GWASs have identified chromosomal region 1q25 as an SLE susceptibility locus, and this locus harbors the gene that encodes the lncRNA growth arrest-specific transcript 5 (GAS5) [100]. GAS5 is a widely expressed lncRNA with many functions in tumorigenesis [101], glucocorticoid responses [102] and growth arrest in human T cells [103]. In BXSB mice, which spontaneously develop lupus nephritis, 6 SNPs have been found to be correlated with susceptibility to nephritis [104]. Further research shows that the expression levels of GAS5 are lower in the CD4+ T cells and B cells from patients with SLE compared to those from healthy controls; however, this differential expression is not seen in whole blood leukocytes, indicating that GAS5 might be differentially controlled in immune cells and function as a regulator in the immune response [105]. Moreover, the expression of GAS5 is decreased in the plasma of SLE patients compared with that in healthy controls. Furthermore, the expression levels of GAS5 in plasma are negatively correlated with erythrocyte sedimentation rates (ESR) and SLEDAI-2K scores in SLE patients [106], suggesting an important role of GAS5 in the pathogenesis of SLE. However, the exact role of GAS5 in immune cell function and abnormal autoimmunity in SLE needs further clarification.
Linc0949 is a large intergenic noncoding RNA (lincRNA) associated with disease activity and organ damage in SLE. The expression of linc0949 is significantly decreased in the PBMCs of SLE patients compared to those of controls, reduced in SLE patients with cumulative organ damage, and significantly increased after treatment. The expression level of linc0949 is correlated with the SLEDAI score, levels of C3 and incidence of lupus nephritis [107], indicating that linc0949 could be a potential biomarker for diagnosis and for assessing disease activity, organ damage and therapeutic responses in SLE. Further research found that the expression of linc0949 is reduced in PBMCs obtained from healthy donors after treatment with Pam3CSK4, a TLR2 ligand, which is unlike the results observed in PBMCs from SLE patients [107], suggesting an abnormal pattern of lin0949 regulation in SLE. In addition, the aberrant expression of several other lncRNAs, including Lnc5150, Lnc3643 and lnc7514, was found to be associated with laboratory results and disease activity in SLE [108].
In addition to the previous examples, researchers have found several other candidates that are possibly involved in SLE. MALAT-1 expression is abnormally increased in monocytes from SLE patients compared with those from healthy controls. Additionally, silencing MALAT-1 significantly reduced the expression of IL-21 by regulating the SIRT1 pathway in monocytes of SLE patients compared with those of healthy controls [109]. Bioinformatics analysis revealed that lncRNA CYP2C91 is implicated in SLE pathogenesis by interacting with transcription factor PU.1 (SPI1), MSR1 and CCR1 [110]. lncRNA TUG1 expression is markedly lower in PBMCs of SLE patients compared with that in healthy controls and even lower in SLE patients with LN. The level of TUG1 is correlated with SLEDAI, ESR, disease duration and 24-h urinary protein [111]. In a mouse SLE model, TUG1 was observed to be involved in the protective effect of NF-κB inhibition on kidney injury [112]. Elevated lncRNA RP11-2B6.2 was observed in kidney biopsies from LN patients and positively correlated with disease activity and IFN scores. The knockdown of the lncRNA RP11-2B6.2 in renal cells inhibits the expression of IFN stimulated genes (ISGs) and the phosphorylation of JAK1, TYK2, and STAT1 in the IFN-I pathway via the epigenetic regulation of SOCS1 [113]. The precise role of these lncRNAs and their underlying biological mechanisms in SLE still require further investigation.
3.3. lncRNAs in IMM
IIMs are a group of heterogeneous disorders that cause damage to multiple organs because of chronic inflammation of skeletal muscle. IIMs mainly comprise four subgroups: dermatomyositis (DM), polymyositis (PM), immune-mediated necrotic myopathy and inclusion body myositis (IBM) [114]. In a study conducted in PM/DM patients, autoantibodies against lncRNA 7 S L, the RNA component of signal recognition particle (SRP), were found to be related to ethnic background, clinical manifestations, and seasonal disease onset [115], suggesting that lncRNA 7 S L is related to the pathogenesis of PM/DM. Transcriptional profiling using microarray analysis identified more than 1000 differentially expressed lncRNAs in DM patients. Bioinformatics prediction suggested that linc-DGCR6-1 may regulate the type 1 interferon-inducible gene USP18 [116]. Fifty-five and 46 lncRNAs are differentially expressed in muscle biopsies obtained from IBM and anti-Jo-1-associated myositis (Jo-1) patients, respectively. H19, lncMyoD and MALAT1 are upregulated in both IBM and Jo-1 myositis compared with healthy controls [117]. Taken together, lncRNA expression profiles are aberrantly regulated in IMMs, and further investigation of lncRNA function remains to be done.
3.4. lncRNAs in SSc
SSc is a complex autoimmune disease characterized by changes in the microvasculature and fibrosis of the skin and internal organs. The excessive production of various extracellular matrix proteins, mainly type I collagen, by skin dermal fibroblasts is a critical step in the pathological process in SSc [118]. The lncRNA TSIX is increased in the serum and skin fibroblasts of patients with SSc compared with SLE patients and healthy controls, which may be regulated by the TGF-β signaling pathway. Further studies revealed that TSIX siRNA significantly reduced type I collagen mRNA stability [119], implying a regulatory role of lncRNAs in the tissue fibrosis of SSc. RNA sequencing of skin biopsy samples found that 257 antisense lncRNAs are differentially expressed in early SSc patients compared to healthy individuals [120]. Recently, a comprehensive analysis of lncRNA expression in PBMCs from 20 SSc patients and 20 healthy donors revealed that the lncRNA ncRNA00201 is significantly downregulated in SSc. NcRNA00201 has been reported to regulate tumor proliferation and target the hnRNPC gene, which encodes an SSc-associated autoantigen. Further bioinformatic analysis of ncRNA00201-targeted microRNAs and genes indicated that ncRNA00201 regulates genes involved in vasculopathy, fibrosis and autoimmunity in SSc [121], providing new insight into disease pathogenesis. TLR activation and IFN-stimulated genes (ISGs) upregulation in PBMCs have been identified as possible contributors to the pathogenesis of SSc. The lncRNA named the Negative Regulator of the IFN Response (NRIR) was found to be significantly upregulated in SSc. In NRIR-silenced monocytes, the expression of CXCL10 and CXCL11, which are two IFN-related chemokines associated with SSc pathogenesis, was reduced, showing that the dysregulation of NRIR in SSc monocytes may be involved in the IFN response pathway in SSc patients [122]. To date, however, knowledge of the lncRNA function and molecular mechanism is still lacking and needs to be further studied.
3.5. lncRNAs in pSS
pSS is an autoimmune disease that occurs predominantly in middle-aged women and is characterized by chronic inflammation in salivary and lacrimal glands; this inflammation is caused by lymphocyte and plasma cell infiltration [123]. The lncRNA IFNGAS (also known as TMEVPG1), which is encoded by a gene located near the Ifn gene, contributes to IFNγ expression. IFNGAS expression is increased in CD4+ T cells from pSS patients compared with those from healthy controls. In addition, the expression level of IFNGAS is correlated with the levels of SSA, ESR and IgG [124]. Examination of the expression profile of lncRNAs in labial salivary glands (LSGs) by microarray identified 1243 differentially expressed lncRNAs in pSS patients. Eight of these differentially expressed lncRNAs, including ENST00000420219.1, ENST00000455309.1, n336161, NR_002712, ENST00000546086.1, Lnc-UTS2D-1:1, n340599, and TCONS_l2_00014794, were confirmed to be upregulated and found to correlate with beta2 microglobulin, disease course, ESR, rheumatoid factor (RF), IgA, IgM, visual analogue scale (VAS) of parotid swelling and VAS of dry eyes [125]. The transcriptome analysis of PBMCs from pSS patients and healthy controls identified 199 differentially expressed lncRNAs. Through a complex network analysis of lncRNA–miRNA–gene functional interactions, three lncRNAs, namely, LINC00657, LINC00511 and CTD-2020K17.1, were identified to be involved in the pathogenesis of pSS [126]. Recently, a study demonstrated that the lncRNA PVT1 is upregulated in the CD4+ T cells of SS patients. PVT1 could maintain the expression of Myc, thus controlling the proliferation and function of CD4+ T cells by regulating the reprogramming of glycolysis, providing a novel mechanistic function of lncRNA PVT1 in the pathogenesis of SS [127].
3.6. lncRNAs in AS
AS is a heritable chronic inflammatory autoimmune disease that leads to the fusion of vertebrae and sacroiliac joints and is characterized by back pain, arthralgia and disability. A total of 661 lncRNAs were found to be differentially expressed in hip joint ligament tissue samples obtained from AS patients compared with those from healthy controls [128]. Recently, lncRNA H19 was found to be upregulated in the PBMCs of AS compared with those of healthy controls. Moreover, H19 modulates the expression of IL-17 A and IL-23 via interaction with miR22–5p and miR675–5p [129], suggesting an important role of lncRNAs in controlling inflammation in AS.
4. Conclusion and future perspectives
As an emerging topic in the field of epigenetic regulation research, lncRNAs have been demonstrated to play critical roles in many physiological and pathological processes via various exquisite mechanisms. With great advances in tumor biology research, lncRNAs are widely regarded as promising biomarkers and therapeutic targets in various types of tumors. However, compared to cancer, lncRNA research in autoimmune diseases is a nascent and developing field. In this review, we summarize the most current knowledge on the function of lncRNAs in the pathogenesis of autoimmune diseases and the underlying molecular mechanisms. A wide spectrum of lncRNAs is observed to be differentially expressed in various autoimmune diseases, including RA, SLE, IMMs, SSc and pSS, and some of these molecules were found to be correlated to clinical presentations and disease activity, showing the potential of these lncRNAs as biomarkers for diagnosis, treatment and prognosis. However, only a few of these lncRNAs were verified to execute confirmed functions in the process of diseases, and the precise molecular basis of these functions remains largely unknown. Much more intensive research is needed in the future to explore the regulatory pattern responsible for the altered lncRNA expression profiles between different developmental stages, different disease states, different cell types and tissues and the precise functional roles of these lncRNAs. To date, the vast majority of lncRNAs remain poorly defined, and the challenge going forward will be to annotate lncRNA sequences, to identify functional structures and domains, and to elucidate the lncRNA-centered mechanisms and their interaction partners. With advances in molecular biological techniques, the unknown mechanisms by which lncRNA regulate autoimmune diseases will be identified, and disease-related functional lncRNAs could be translated into clinical practice.
Declaration of competing interest
The authors declare that they have no competing interests.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81871275, 81671591, U1401222); the Fundamental Research Funds for the Central Universities of China (17ykjc07).
References
- 1.Martin L., Chang H.Y. Uncovering the role of genomic "dark matter" in human disease. J. Clin. Invest. 2012;122(5):1589–1595. doi: 10.1172/JCI60020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maass P.G., Luft F.C., Bahring S. Long non-coding RNA in health and disease. J. Mol. Med. (Berl.) 2014;92(4):337–346. doi: 10.1007/s00109-014-1131-8. [DOI] [PubMed] [Google Scholar]
- 3.Kazemzadeh M., Safaralizadeh R., Orang A.V. LncRNAs: emerging players in gene regulation and disease pathogenesis. J. Genet. 2015;94(4):771–784. doi: 10.1007/s12041-015-0561-6. [DOI] [PubMed] [Google Scholar]
- 4.Djebali S. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mt M. Systematic localization of common disease-associated variation in regulatory DNA. Science (New York, N.Y.) 2012;337(6099):1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okazaki Y. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420(6915):563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 7.Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat. Struct. Mol. Biol. 2007;14(2):103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- 8.Bierhoff H. Noncoding transcripts in sense and antisense orientation regulate the epigenetic state of ribosomal RNA genes. Cold Spring Harbor Symp. Quant. Biol. 2010;75:357–364. doi: 10.1101/sqb.2010.75.060. [DOI] [PubMed] [Google Scholar]
- 9.Zhao Y. NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 2016;44(D1):D203–D208. doi: 10.1093/nar/gkv1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quinn J.J., Chang H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016;17(1):47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 11.Derrien T. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nitsche A., Stadler P.F. Evolutionary clues in lncRNAs. Wiley Interdiscip Rev RNA. 2017;8(1) doi: 10.1002/wrna.1376. [DOI] [PubMed] [Google Scholar]
- 13.Mercer T.R. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. U. S. A. 2008;105(2):716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dinger M.E. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18(9):1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi X. Cancer Lett; 2013. Long Non-coding RNAs: A New Frontier in the Study of Human Diseases. [DOI] [PubMed] [Google Scholar]
- 16.Marques-Rocha J.L. Noncoding RNAs, cytokines, and inflammation-related diseases. Faseb. J. 2015;29(9):3595–3611. doi: 10.1096/fj.14-260323. [DOI] [PubMed] [Google Scholar]
- 17.Smith M.A., Mattick J.S. Structural and functional annotation of long noncoding RNAs. Methods Mol. Biol. 2017;1526:65–85. doi: 10.1007/978-1-4939-6613-4_4. [DOI] [PubMed] [Google Scholar]
- 18.Fitzgerald K.A., Caffrey D.R. Long noncoding RNAs in innate and adaptive immunity. Curr. Opin. Immunol. 2014;26C:140–146. doi: 10.1016/j.coi.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L.L. Linking long noncoding RNA localization and function. Trends Biochem. Sci. 2016;41(9):761–772. doi: 10.1016/j.tibs.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 20.Engreitz J.M. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539(7629):452–455. doi: 10.1038/nature20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson K.M. Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature. 2016;539(7629):433–436. doi: 10.1038/nature20128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kopp F., Mendell J.T. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson D.M. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160(4):595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsumoto A. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature. 2017;541(7636):228–232. doi: 10.1038/nature21034. [DOI] [PubMed] [Google Scholar]
- 25.Wang K.C., Chang H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rinn J.L., Chang H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gupta R.A. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan C., Ning Y., Pan Y. Emerging roles of HOTAIR in human cancer. J. Cell. Biochem. 2020 doi: 10.1002/jcb.29591. In press. [DOI] [PubMed] [Google Scholar]
- 29.Wang K.C. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472(7341):120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Engreitz J.M. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539(7629):452–455. doi: 10.1038/nature20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paralkar V.R. Unlinking an lncRNA from its associated cis element. Mol. Cell. 2016;62(1):104–110. doi: 10.1016/j.molcel.2016.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rapicavoli N.A. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife. 2013;2 doi: 10.7554/eLife.00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z. The long noncoding RNA THRIL regulates TNF expression through its interaction with hnRNPL. Proc. Natl. Acad. Sci. Unit. States Am. 2014;111(3):1002–1007. doi: 10.1073/pnas.1313768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomson D.W., Dinger M.E. Endogenous microRNA sponges: evidence and controversy. Nat. Rev. Genet. 2016;17(5):272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 35.Cesana M. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Faghihi M.A. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 2008;14(7):723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang P. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344(6181):310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 38.Liu B. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Canc. Cell. 2015;27(3):370–381. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 39.Taniue K. Long noncoding RNA UPAT promotes colon tumorigenesis by inhibiting degradation of UHRF1. Proc. Natl. Acad. Sci. U. S. A. 2016;113(5):1273–1278. doi: 10.1073/pnas.1500992113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma S. Dephosphorylation of the nuclear factor of activated T cells (NFAT) transcription factor is regulated by an RNA-protein scaffold complex. Proc. Natl. Acad. Sci. U. S. A. 2011;108(28):11381–11386. doi: 10.1073/pnas.1019711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zou Y. Long noncoding RNA LERFS negatively regulates rheumatoid synovial aggression and proliferation. J. Clin. Invest. 2018;128(10):4510–4524. doi: 10.1172/JCI97965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silva A., Bullock M., Calin G. The clinical relevance of long non-coding RNAs in cancer. Cancers. 2015;7(4):2169–2182. doi: 10.3390/cancers7040884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang G., Lu X., Yuan L. LncRNA: a link between RNA and cancer. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 2014;1839(11):1097–1109. doi: 10.1016/j.bbagrm.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 44.Chew C.L. Noncoding RNAs: master regulators of inflammatory signaling. Trends Mol. Med. 2018;24(1):66–84. doi: 10.1016/j.molmed.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 45.Cooles F.A., Isaacs J.D. Pathophysiology of rheumatoid arthritis. Curr. Opin. Rheumatol. 2011;23(3):233–240. doi: 10.1097/BOR.0b013e32834518a3. [DOI] [PubMed] [Google Scholar]
- 46.McInnes I.B., Schett G. The pathogenesis of rheumatoid arthritis. N. Engl. J. Med. 2011;365(23):2205–2219. doi: 10.1056/NEJMra1004965. [DOI] [PubMed] [Google Scholar]
- 47.McGonagle D., Watad A., Savic S. Mechanistic immunological based classification of rheumatoid arthritis. Autoimmun. Rev. 2018;17(11):1115–1123. doi: 10.1016/j.autrev.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 48.Brzustewicz E., Bryl E. The role of cytokines in the pathogenesis of rheumatoid arthritis – practical and potential application of cytokines as biomarkers and targets of personalized therapy. Cytokine. 2015;76(2):527–536. doi: 10.1016/j.cyto.2015.08.260. [DOI] [PubMed] [Google Scholar]
- 49.Lefevre S. Role of synovial fibroblasts in rheumatoid arthritis. Curr. Pharmaceut. Des. 2015;21(2):130–141. doi: 10.2174/1381612820666140825122036. [DOI] [PubMed] [Google Scholar]
- 50.Yuan M. Long noncoding RNA profiling revealed differentially expressed lncRNAs associated with disease activity in PBMCs from patients with rheumatoid arthritis. PloS One. 2017;12(11) doi: 10.1371/journal.pone.0186795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song J. PBMC and exosome-derived Hotair is a critical regulator and potent marker for rheumatoid arthritis. Clin. Exp. Med. 2015;15(1):121–126. doi: 10.1007/s10238-013-0271-4. [DOI] [PubMed] [Google Scholar]
- 52.Luo Q. Comprehensive analysis of long non-coding RNA and mRNA expression profiles in rheumatoid arthritis. Exp Ther Med. 2017;14(6):5965–5973. doi: 10.3892/etm.2017.5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y. Long noncoding RNA expression profile in fibroblast-like synoviocytes from patients with rheumatoid arthritis. Arthritis Res. Ther. 2016;18(1):227. doi: 10.1186/s13075-016-1129-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun L. Inflammopharmacology; 2019. Analysis of lncRNA Expression Profiles by Sequencing Reveals that lnc-AL928768.3 and lnc-AC091493.1 Are Novel Biomarkers for Disease Risk and Activity of Rheumatoid Arthritis. [DOI] [PubMed] [Google Scholar]
- 55.Qin W. Plasma long non-coding RNA expression profiles in patients with rheumatoid arthritis. Clin. Lab. 2019;65(8) doi: 10.7754/Clin.Lab.2019.190144. [DOI] [PubMed] [Google Scholar]
- 56.Gong X. Circulating lnc-ITSN1-2 expression presents a high value in diagnosis of rheumatoid arthritis and correlates with disease activity. Int. J. Clin. Exp. Pathol. 2017;10(10):10451–10458. [PMC free article] [PubMed] [Google Scholar]
- 57.Jiang H. LncRNAs expression in adjuvant-induced arthritis rats reveals the potential role of LncRNAs contributing to rheumatoid arthritis pathogenesis. Gene. 2016;593(1):131–142. doi: 10.1016/j.gene.2016.08.012. [DOI] [PubMed] [Google Scholar]
- 58.Mueller N. Interleukin-6 and Tumour Necrosis Factor-alpha differentially regulate lincRNA transcripts in cells of the innate immune system in vivo in human subjects with rheumatoid arthritis. Cytokine. 2014;68(1):65–68. doi: 10.1016/j.cyto.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 59.Jiang H. LncRNAs expression in adjuvant-induced arthritis rats reveals the potential role of LncRNAs contributing to rheumatoid arthritis pathogenesis. Gene. 2016;593(1):131–142. doi: 10.1016/j.gene.2016.08.012. [DOI] [PubMed] [Google Scholar]
- 60.H J. Effect of astragalosides on long non-coding RNA expression profiles in rats with adjuvant-induced arthritis. Int. J. Mol. Med. 2019;44(4):1344–1356. doi: 10.3892/ijmm.2019.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bhan A., Mandal S.S. LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer. Biochim. Biophys. Acta Rev. Canc. 2015;1856(1):151–164. doi: 10.1016/j.bbcan.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rajagopal T. HOTAIR LncRNA: a novel oncogenic propellant in human cancer. Clin. Chim. Acta. 2020;503:1–18. doi: 10.1016/j.cca.2019.12.028. [DOI] [PubMed] [Google Scholar]
- 63.Hj Z. LncRNA HOTAIR alleviates rheumatoid arthritis by targeting miR-138 and inactivating NF-κB pathway. Int. Immunopharm. 2017;50:283–290. doi: 10.1016/j.intimp.2017.06.021. [DOI] [PubMed] [Google Scholar]
- 64.Yan S. Long non-coding RNA HIX003209 promotes inflammation by sponging miR-6089 via TLR4/NF-kappaB signaling pathway in rheumatoid arthritis. Front. Immunol. 2019;10:2218. doi: 10.3389/fimmu.2019.02218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang C.A. lncRNA NTT/PBOV1 Axis promotes monocyte differentiation and is elevated in rheumatoid arthritis. Int. J. Mol. Sci. 2018;19(9) doi: 10.3390/ijms19092806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen S. LincRNa-p21: function and mechanism in cancer. Med. Oncol. 2017;34(985) doi: 10.1007/s12032-017-0959-5. [DOI] [PubMed] [Google Scholar]
- 67.Spurlock C.F. Methotrexate inhibits NF-κB activity via long intergenic (noncoding) RNA-p21 induction. Arthritis & Rheumatology. 2014;66(11):2947–2957. doi: 10.1002/art.38805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu M. Increased expression of long noncoding RNAs LOC100652951 and LOC100506036 in T cells from patients with rheumatoid arthritis facilitates the inflammatory responses. Immunol. Res. 2016;64(2):576–583. doi: 10.1007/s12026-015-8756-8. [DOI] [PubMed] [Google Scholar]
- 69.Moharamoghli M. The expression of GAS5, THRIL, and RMRP lncRNAs is increased in T cells of patients with rheumatoid arthritis. Clin. Rheumatol. 2019;38(11):3073–3080. doi: 10.1007/s10067-019-04694-z. [DOI] [PubMed] [Google Scholar]
- 70.Kurreeman F.A.S. A candidate gene approach identifies the TRAF1/C5 region as a risk factor for rheumatoid arthritis. PLoS Med. 2007;4(e2789):1515–1524. doi: 10.1371/journal.pmed.0040278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Messemaker T.C. A novel long non-coding RNA in the rheumatoid arthritis risk locus TRAF1-C5 influences C5 mRNA levels. Gene Immun. 2016;17(2):85–92. doi: 10.1038/gene.2015.54. [DOI] [PubMed] [Google Scholar]
- 72.Ayesh S. Possible physiological role of H19 RNA. Mol. Carcinog. 2002;35(2):63–74. doi: 10.1002/mc.10075. [DOI] [PubMed] [Google Scholar]
- 73.Matouk I.J. The oncofetal H19 RNA connection: hypoxia, p53 and cancer. Biochim. Biophys. Acta. 2010;1803(4):443–451. doi: 10.1016/j.bbamcr.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 74.Stuhlmuller B. Detection of oncofetal h19 RNA in rheumatoid arthritis synovial tissue. Am. J. Pathol. 2003;163(3):901–911. doi: 10.1016/S0002-9440(10)63450-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Af H. No association of single nucleotide polymorphisms within H19 and HOX transcript antisense RNA (HOTAIR) with genetic susceptibility to systemic lupus erythematosus, rheumatoid arthritis, and primary Sjögren’s syndrome in a Chinese Han population. Clin. Rheumatol. 2017;36(11):2447–2453. doi: 10.1007/s10067-017-3833-0. [DOI] [PubMed] [Google Scholar]
- 76.Ye Y., Gao X., Yang N. LncRNA ZFAS1 promotes cell migration and invasion of fibroblast-like synoviocytes by suppression of miR-27a in rheumatoid arthritis. Hum. Cell. 2018;31(1):14–21. doi: 10.1007/s13577-017-0179-5. [DOI] [PubMed] [Google Scholar]
- 77.Mo B.Y. Long non-coding RNA GAPLINC promotes tumor-like biologic behaviors of fibroblast-like synoviocytes as MicroRNA sponging in rheumatoid arthritis patients. Front. Immunol. 2018;9:702. doi: 10.3389/fimmu.2018.00702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bi X. LncRNA PICSAR promotes cell proliferation, migration and invasion of fibroblast-like synoviocytes by sponging miRNA-4701-5p in rheumatoid arthritis. EBioMedicine. 2019;50:408–420. doi: 10.1016/j.ebiom.2019.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.H Y. Clinical and experimental rheumatology; 2019. Long Noncoding RNA FER1L4 Regulates Rheumatoid Arthritis via Targeting NLRC5. [PubMed] [Google Scholar]
- 80.Pan F. Quercetin promotes the apoptosis of fibroblast-like synoviocytes in rheumatoid arthritis by upregulating lncRNA MALAT1. Int. J. Mol. Med. 2016;38(5):1507–1514. doi: 10.3892/ijmm.2016.2755. [DOI] [PubMed] [Google Scholar]
- 81.Yan Z.F. UCA1 impacts progress of rheumatoid arthritis by inducing the apoptosis of fibroblast-like synoviocyte. Eur. Rev. Med. Pharmacol. Sci. 2018;22(4):914–920. doi: 10.26355/eurrev_201802_14370. [DOI] [PubMed] [Google Scholar]
- 82.Wang G. LncRNA DILC participates in rheumatoid arthritis by inducing apoptosis of fibroblast-like synoviocytes and down-regulating IL-6. Biosci. Rep. 2019;39(5) doi: 10.1042/BSR20182374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Z Y. Lnc-IL7R promotes the growth of fibroblast-like synoviocytes through interaction with enhancer of zeste homolog 2 in rheumatoid arthritis. Mol. Med. Rep. 2017;15(3):1412–1418. doi: 10.3892/mmr.2017.6150. [DOI] [PubMed] [Google Scholar]
- 84.Wang W. FOXM1/LINC00152 feedback loop regulates proliferation and apoptosis in rheumatoid arthritis fibroblast-like synoviocytes via Wnt/beta-catenin signaling pathway. Biosci. Rep. 2020;40(1) doi: 10.1042/BSR20191900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li G. Tanshinone IIA promotes the apoptosis of fibroblast-like synoviocytes in rheumatoid arthritis by up-regulating lncRNA GAS5. Biosci. Rep. 2018;38(5) doi: 10.1042/BSR20180626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ma C., Wang W., Li P. LncRNA GAS5 overexpression downregulates IL-18 and induces the apoptosis of fibroblast-like synoviocytes. Clin. Rheumatol. 2019;38(11):3275–3280. doi: 10.1007/s10067-019-04691-2. [DOI] [PubMed] [Google Scholar]
- 87.Yue T. Downregulation of lncRNA ITSN1-2 correlates with decreased disease risk and activity of rheumatoid arthritis (RA), and reduces RA fibroblast-like synoviocytes proliferation and inflammation via inhibiting NOD2/RIP2 signaling pathway. Am J Transl Res. 2019;11(8):4650–4666. [PMC free article] [PubMed] [Google Scholar]
- 88.Yr L. Long noncoding RNA MEG3 regulates rheumatoid arthritis by targeting NLRC5. J. Cell. Physiol. 2019;234(8):14270–14284. doi: 10.1002/jcp.28126. [DOI] [PubMed] [Google Scholar]
- 89.Li G. LncRNA MEG3 inhibits rheumatoid arthritis through miR-141 and inactivation of AKT/mTOR signalling pathway. J. Cell Mol. Med. 2019;23(10):7116–7120. doi: 10.1111/jcmm.14591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang C.W. Long non-coding RNA PVT1 knockdown suppresses fibroblast-like synoviocyte inflammation and induces apoptosis in rheumatoid arthritis through demethylation of sirt6. J. Biol. Eng. 2019;13:60. doi: 10.1186/s13036-019-0184-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang K.Y., Chen D.L. Shikonin inhibits inflammatory response in rheumatoid arthritis synovial fibroblasts via lncRNA-NR024118. Evid Based Complement Alternat Med. 2015;2015 doi: 10.1155/2015/631737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rahman A., Isenberg D.A. Systemic lupus erythematosus. N. Engl. J. Med. 2008;358(9):929–939. doi: 10.1056/NEJMra071297. [DOI] [PubMed] [Google Scholar]
- 93.Zhang Q. Integrated analysis of lncRNA, miRNA and mRNA expression profiling in patients with systemic lupus erythematosus. Arch. Med. Sci. 2019;15(4):872–879. doi: 10.5114/aoms.2018.79145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ye H. Full high-throughput sequencing analysis of differences in expression profiles of long noncoding RNAs and their mechanisms of action in systemic lupus erythematosus. Arthritis Res. Ther. 2019;21(1):70. doi: 10.1186/s13075-019-1853-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lj L. Comprehensive long non-coding RNA expression profiling reveals their potential roles in systemic lupus erythematosus. Cell. Immunol. 2017;319:17–27. doi: 10.1016/j.cellimm.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 96.Wang Y. Long noncoding RNA expression profile and association with SLEDAI score in monocyte-derived dendritic cells from patients with systematic lupus erythematosus. Arthritis Res. Ther. 2018;20(1):138. doi: 10.1186/s13075-018-1640-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wu G.C. Differential plasma expression profiles of long non-coding RNAs reveal potential biomarkers for systemic lupus erythematosus. Biomolecules. 2019;9(6) doi: 10.3390/biom9060206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang F. Identification of the long noncoding RNA NEAT1 as a novel inflammatory regulator acting through MAPK pathway in human lupus. J. Autoimmun. 2016;75:96–104. doi: 10.1016/j.jaut.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 99.Dong G. Granulocytic myeloid-derived suppressor cells contribute to IFN-I signaling activation of B cells and disease progression through the lncRNA NEAT1-BAFF axis in systemic lupus erythematosus. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2020;1866(1) doi: 10.1016/j.bbadis.2019.165554. [DOI] [PubMed] [Google Scholar]
- 100.Suarez-Gestal M. Replication of recently identified systemic lupus erythematosus genetic associations: a case-control study. Arthritis Res. Ther. 2009;11(3):R69. doi: 10.1186/ar2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ghaforui-Fard S., Taheri M. Growth arrest specific transcript 5 in tumorigenesis process: an update on the expression pattern and genomic variants. Biomed. Pharmacother. 2019;112 doi: 10.1016/j.biopha.2019.108723. [DOI] [PubMed] [Google Scholar]
- 102.Kino T. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010;3(107):ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Williams G.T., Mourtada-Maarabouni M., Farzaneh F. A critical role for non-coding RNA GAS5 in growth arrest and rapamycin inhibition in human T-lymphocytes. Biochem. Soc. Trans. 2011;39(2):482–486. doi: 10.1042/BST0390482. [DOI] [PubMed] [Google Scholar]
- 104.Haywood M.E.K. Overlapping BXSB congenic intervals, in combination with microarray gene expression, reveal novel lupus candidate genes. Gene Immun. 2006;7(3):250–263. doi: 10.1038/sj.gene.6364294. [DOI] [PubMed] [Google Scholar]
- 105.Mayama T., Marr A.K., Kino T. Differential expression of glucocorticoid receptor noncoding RNA repressor Gas5 in autoimmune and inflammatory diseases. Horm. Metab. Res. 2016;48(8):550–557. doi: 10.1055/s-0042-106898. [DOI] [PubMed] [Google Scholar]
- 106.Wu G.C. Identification of long non-coding RNAs GAS5, linc0597 and lnc-DC in plasma as novel biomarkers for systemic lupus erythematosus. Oncotarget. 2017;8(14):23650–23663. doi: 10.18632/oncotarget.15569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wu Y. Association of large intergenic noncoding RNA expression with disease activity and organ damage in systemic lupus erythematosus. Arthritis Res. Ther. 2015;17:131. doi: 10.1186/s13075-015-0632-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang J.B. Expression of several long noncoding RNAs in peripheral blood mononuclear cells of patients with systemic lupus erythematosus. Adv. Med. Sci. 2019;64(2):430–436. doi: 10.1016/j.advms.2019.08.002. [DOI] [PubMed] [Google Scholar]
- 109.Yang H. Long noncoding RNA MALAT-1 is a novel inflammatory regulator in human systemic lupus erythematosus. Oncotarget. 2017;8(44):77400–77406. doi: 10.18632/oncotarget.20490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Suravajhala P. Potential role of lncRNA cyp2c91-protein interactions on diseases of the immune system. Front. Genet. 2015;6:255. doi: 10.3389/fgene.2015.00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cao H.Y. Clinical significance of reduced expression of lncRNA TUG1 in the peripheral blood of systemic lupus erythematosus patients. Int. J. Rheum. Dis. 2020;23(3):428–434. doi: 10.1111/1756-185X.13786. [DOI] [PubMed] [Google Scholar]
- 112.Cao H.Y. The protection of NF-kappaB inhibition on kidney injury of systemic lupus erythematosus mice may be correlated with lncRNA TUG1. Kaohsiung J. Med. Sci. 2020 doi: 10.1002/kjm2.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liao Z. Identification of renal long non-coding RNA RP11-2B6.2 as a positive regulator of type I interferon signaling pathway in lupus nephritis. Front. Immunol. 2019;10:975. doi: 10.3389/fimmu.2019.00975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lundberg I.E., de Visser M., Werth V.P. Classification of myositis. Nat. Rev. Rheumatol. 2018;14(5):269–278. doi: 10.1038/nrrheum.2018.41. [DOI] [PubMed] [Google Scholar]
- 115.Satoh T. Novel autoantibodies against 7SL RNA in patients with polymyositis/dermatomyositis. J. Rheumatol. 2005;32(9):1727–1733. [PubMed] [Google Scholar]
- 116.Peng Q. Transcriptomic profiling of long non-coding RNAs in dermatomyositis by microarray analysis. Sci. Rep. 2016;6(1) doi: 10.1038/srep32818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pd H. Transcriptional profiling identifies differential expression of long non-coding RNAs in Jo-1 associated and inclusion body myositis. Sci. Rep. 2017;7(1):8024. doi: 10.1038/s41598-017-08603-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Denton C.P., Khanna D. Systemic sclerosis. Lancet. 2017;390(10103):1685–1699. doi: 10.1016/S0140-6736(17)30933-9. [DOI] [PubMed] [Google Scholar]
- 119.Wang Z. Long non-coding RNA TSIX is upregulated in scleroderma dermal fibroblasts and controls collagen mRNA stabilization. Exp. Dermatol. 2016;25(2):131–136. doi: 10.1111/exd.12900. [DOI] [PubMed] [Google Scholar]
- 120.Messemaker T.C. Antisense long non-coding RNAs are deregulated in skin tissue of patients with systemic sclerosis. J. Invest. Dermatol. 2018;138(4):826–835. doi: 10.1016/j.jid.2017.09.053. [DOI] [PubMed] [Google Scholar]
- 121.Dolcino M. In systemic sclerosis, a unique long non coding RNA regulates genes and pathways involved in the three main features of the disease (vasculopathy, fibrosis and autoimmunity) and in carcinogenesis. J. Clin. Med. 2019;8(3) doi: 10.3390/jcm8030320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mariotti B. The long non-coding RNA NRIR drives IFN-response in monocytes: implication for systemic sclerosis. Front. Immunol. 2019;10:100. doi: 10.3389/fimmu.2019.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Brito-Zeron P. Sjogren syndrome. Nat Rev Dis Primers. 2016;2:16047. doi: 10.1038/nrdp.2016.47. [DOI] [PubMed] [Google Scholar]
- 124.Wang J. Upregulation of long noncoding RNA TMEVPG1 enhances T helper type 1 cell response in patients with Sjogren syndrome. Immunol. Res. 2016;64(2):489–496. doi: 10.1007/s12026-015-8715-4. [DOI] [PubMed] [Google Scholar]
- 125.Shi H. Long non-coding RNA expression profile in minor salivary gland of primary Sjogren’s syndrome. Arthritis Res. Ther. 2016;18(1):109. doi: 10.1186/s13075-016-1005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dolcino M. Long non-coding RNAs modulate sjogren’s syndrome associated gene expression and are involved in the pathogenesis of the disease. J. Clin. Med. 2019;8(9) doi: 10.3390/jcm8091349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.J F. LncRNA PVT1 links Myc to glycolytic metabolism upon CD4 T cell activation and Sjögren’s syndrome-like autoimmune response. J. Autoimmun. 2019 doi: 10.1016/j.jaut.2019.102358. [DOI] [PubMed] [Google Scholar]
- 128.Zhang C. Differentially expressed mRNAs, lncRNAs, and miRNAs with associated co-expression and ceRNA networks in ankylosing spondylitis. Oncotarget. 2017;8(69):113543–113557. doi: 10.18632/oncotarget.22708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Z X. H19 increases IL-17a/IL-23 releases via regulating VDR by interacting with miR675-5p/miR22-5p in ankylosing spondylitis. Molecular therapy. Nucleic acids. 2019;19:393–404. doi: 10.1016/j.omtn.2019.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]