Abstract
Investigation of the fungal communities in animal models of Inflammatory Bowel Diseases (IBD) showed a controversial role of Saccharomyces cerevisiae and Candida spp. In health and disease. These conflicting observations could be ascribed to immunogenic differences among co-specific strains. To assess the relevance of intra-strains differences on yeast immunogenicity and impact on the microbiota, we screened S. cerevisiae and Candida spp. Strains isolated from fecal samples of IBD patients. We compared the cytokine profiles, obtained upon stimulation of Peripheral Blood Mononuclear Cells (PBMCs) and Dendritic Cells with different yeast strains, and evaluated the relationship between strain’s cell wall sugar amount and immune response. Moreover, the gut microbiota composition was explored in relation to fungal isolation from fecal samples by metabarcoding analysis. The comparison of cytokine profiles showed strain dependent rather than species-dependent differences in immune responses. Differences in immunogenicity correlated with the cell wall composition of S. cerevisiae intestinal strains. Stimulation of human healthy PBMCs with different strains showed a pro-inflammatory IL-6 response counterbalanced by IL-10 production. Interestingly, Crohn’s (CD) patients responded differently to “self” and “non-self” strains, eliciting pure Th1 or Th17 cytokine patterns. The differences observed in vitro were recapitulated in vivo, where different strains contributed in dramatically different ways to local epithelial activity and to the inflammation of wild type and Interleukin-deficient mice. Furthermore, we observed that the gut microbiota profiles significantly differentiated according to the presence of Saccharomyces or Candida spp. or the absence of fungal isolates in fecal samples. Our results show the importance to deepen metagenomics and immunophenotyping analyses to the strain level, to elucidate the role of fungal and bacterial communities in health and disease.
Keywords: Mycobiome, Microbiome, S. cerevisiae, Candida spp., Inflammatory bowel disease
Highlights
-
•
Previous studies indicated an involvement of gut mycobiome in IBD pathogenesis.
-
•
We screened for immunomodulatory properties S. cerevisiae and Candida strains from IBD patients.
-
•
The fungal immunomodulation depends on strain-rather than species-specific traits.
-
•
Differences in immunogenicity correlate with the cell wall composition of gut strains.
-
•
CD patients responded differently to “self” and “non-self” strains.
1. Introduction
Several studies have investigated the contribution of bacterial communities in the etiology of Inflammatory Bowel Disease (IBD), revealing an alteration of the microbiota in occurrence of these conditions [[1], [2], [3], [4], [5], [6], [7]]. Yet, despite the large quantity of metagenomics and clinical information, studies on gut microbiota have not led to discrimination of the cause-effect relationships between alterations of microbiota and IBD. The most advanced studies suggest that commensal fungi have a crucial role in IBD pathogenesis and chronicity [[8], [13]]. It is worth to consider that first clues on the potential involvement of fungi in IBD came from the observation of an abnormal response to Saccharomyces cerevisiae in Crohn’s disease (CD) patients. Main and coworkers [9] described for the first time in 1988 the presence of antibodies against S. cerevisiae in CD patients’ blood, but not in Ulcerative Colitis (UC) patients, suggesting the diagnostic role of these antibodies to discriminate the two IBDs. Anti-Saccharomyces cerevisiae antibody (ASCA) recognizes cell wall peptidomannans of this yeast [10], although, later on, also Candida albicans was proven a potent immunogen for ASCA [11,12]. The relevance of fungal communities on human health has been further confirmed in recent studies [[14], [15], [16]], which highlighted the role of gut fungi in shaping both innate and adaptive immunity [[17], [18], [19], [20]]. Studies in mouse models showed that gut inflammation promotes fungal proliferation [21], and that Dextran Sulphate Sodium (DSS)-induced gut inflammation is associated with an increase in Candida species [22].
Furthermore, recent studies reported clear differences between adult and pediatric IBD patients’ gut fungal communities [8] and more heterogeneous fungal communities in CD patients compared to healthy subjects [[23], [24], [25], [26]], suggesting a role of altered mycobiota in inflammatory diseases and “leaky gut” syndrome. Sokol and collaborators [23] observed a clear fungal dysbiosis in CD patients with an enrichment in Candida spp. and a reduction of S. cerevisiae in disease versus remission, thus proposing a beneficial effect of S. cerevisiae colonization on host health. Liguori and co-workers [32], assessing the microbiota and mycobiota composition in CD patients’ gut mucosa, observed that S. cerevisiae was enriched in CD patients’ non-inflamed gut mucosa. On the other hand, a recent study reported that S. cerevisiae is able to exacerbate DSS-induced colitis, and affects gut barrier permeability by inducing overproduction of uric acid as a result of host purine metabolism [33]. All these studies have highlighted the relevance of S. cerevisiae in gut inflammation, but the association of different species with flare or remission is controversial [[34], [35], [36]]. The wide genetic and phenotypic variability observed for S. cerevisiae [[38], [39], [40], [41]] could explain the inconsistencies in the results of different studies.
Aiming to dissect the yeast-host relationship, we performed a screening for immunomodulatory properties on different S. cerevisiae strains isolated from fecal samples of IBD and healthy subjects, previously characterized for genotypic and phenotypic traits [41]. We then compared immune responses to fecal S. cerevisiae strains to those elicited by strains isolated from different sources [41] or by fecal Candida spp. Strains, and correlated them with cell wall sugar amount. The immune responses to selected strains were also tested in vivo. Finally, through metabarcoding analysis, we also evaluated the inter-kingdom relationships between fungal and bacterial gut communities in fecal samples of CD patients.
2. Methods
2.1. Enrolment of patients and healthy subjects
A total of 93 pediatric subjects, encompassing 34 CD (age average: 15.1 years; range: 10.5–18.9 years), 27 UC patients (age average 12.4 years; range: 3.25–18.9 years), and 32 healthy children (HC, age average: 12.8 years; range: 4.5–19 years), were enrolled at the Meyer Children’s Hospital (Florence, Italy). Clinical data of IBD patients including localization, disease activity, inflammation indexes and ongoing treatment are reported in Table 1. The inflammatory status was assessed routinely in all IBD patients through clinical parameters, such as blood erythrocyte sedimentation rate (ESR), C Reactive Protein (CRP), fecal calprotectin [42] and endoscopy. For CD patients, disease activity was scored using the Pediatric Crohn’s Disease Activity Index (PCDAI). For UC patients, the Pediatric Ulcerative Colitis Activity Index (PUCAI) was used. Positivity for Anti-Saccharomyces cerevisiae Antibodies, measured as IgA and IgG levels, was also assessed (Table 1). All enrolled patients and healthy children were Italian and had a Mediterranean diet (generally eating bread and dairy products). Patients did not take yeast-based probiotics over the three months prior the stool sampling.
Table 1.
Clinical data of IBD patients investigated in this study.
| CD | UC | |
|---|---|---|
| Number of patients | 34 | 27 |
| Gender: M/F ratio | 20/14 | 13/14 |
| Age average (range) | 15.1 (10.5–18.9) | 12.4 (3.25–18.9) |
| LOCALIZATION OF DISEASE (number of patients; %) | ||
| Ileum | 14 (41.1%) | 0 (0%) |
| Colon | 0 (0%) | 19 (70.3%) |
| Ileum-colon | 20 (58%) | 0 (0%) |
| Sigma-rectum | 0 (0%) | 8 (29.6%) |
| DISEASE ACTIVITY (number of patients; %) | ||
| PCDAI <10 | 20 (58.8%) | |
| PCDAI >10 | 14 (41.1%) | |
| PUCAI <10 | 23 (85.2%) | |
| PUCAI>10 | 4 (14.8%) | |
| Anti S. cerevisiae antibody (number of patients; %) | ||
| ASCA+ | 27 (79.4%) | 3 (11.1%) |
| ASCA- | 7 (20.6%) | 24 (88.9%) |
| INFLAMMATION INDICES (number of patients; %) | ||
| Calprotectin >100 | 18 (53%) | 17 (62.9%) |
| Calprotectin <100 | 16 (47%) | 10 (37%) |
| ESR>31 | 12 (35.3%) | 6 (22.2%) |
| ESR<31 | 19 (55.9%) | 19 (0%) |
| ESR (NA) | 3 (8.8%) | 2 (7.4%) |
| CRP >0.5 mg/L | 16 (47%) | 6 (22.2%) |
| CRP <0.5 mg/L | 15 (44.2%) | 19 (70.3%) |
| CRP (NA) | 3 (8.8%) | 2 (7.4%) |
| THERAPY AT SAMPLING (number of patients; %) | ||
| Enteral Nutrition + azathioprine | 10 (29.4%) | |
| Enteral Nutrition + 6-MP | 2 (5.9%) | |
| Methotrexate | 2 (5.9%) | |
| Infliximab | 16 (47%) | |
| Talidomide | 4 (11.7%) | |
| 5-ASA (mesalazine) | 3 (11.1%) | |
| 5-ASA + steroids | 2 (7.4%) | |
| 5-ASA + azathioprine | 11 (40.7%) | |
| 5-ASA+ 6 MP | 2 (7.4%) | |
| 5-ASA + methotrexate | 1 (3.7%) | |
| 5-ASA + Infliximab | 8 (29.6%) | |
All parents and caregivers of the enrolled patients were made aware of the nature of the experiment conducted in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki), and gave written informed consent for stool and blood sample collection in accordance with the sampling protocol approved by the Ethical Committees of the Azienda Ospedaliera Universitaria (AOU) Careggi and AOU Meyer Children’s Hospital, Florence, Italy (Ref. n. 87/10).
2.2. Isolation and identification of yeast species from fecal samples
Feces were collected from all pediatric subjects. A 1 ml feces aliquot was plated on Yeast Extract-Peptone-Dextrose (YPD) agar medium supplemented with chloramphenicol (1 mg/ml) and incubated at 28 °C. After 2–3 days, fungal colonies were observed. Yeast genomic DNA was extracted as previously described by Sebastiani and collaborators [43]. Fungal isolates were identified by Sanger sequencing of the ITS1-5.8S-ITS2 regions (ribosomal Internal Transcribed Spacer, ITS), using ITS1 (5′-GTTTCCGTAGGTGAACTTGC-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC -3′) primers, compared to the sequenced deposited in the NCBI database using megaBLAST algorithm. Long-term storage of the isolates was carried out in 20% glycerol at −80 °C.
Each isolate was named with a three characters-code: the first letter (Y) identified the isolate as a yeast, the second character (a letter) corresponded to the ID of the patient from whose feces the strain was isolated, the third character (a number) was specific of the isolate, in an arbitrarily numbering of the isolates found in the given fecal sample.
For host-fungal interaction studies, isolates were cultured overnight at 37 °C in Sabouraud dextrose agar (SDA, Oxoid, Hampshire, UK). Then, yeast cells were washed twice with sterile Phosphate Buffered Saline (PBS, EuroClone, Wetherby, UK), counted and suspended in Roswell Park Memorial Institute (RPMI) 1640 medium at the desired concentration.
2.3. Human peripheral blood mononuclear cells and dendritic cells preparation, fungal challenge and cytokine assays
Peripheral Blood Mononuclear Cells (PBMCs) were isolated from fresh blood obtained from 5 pediatric CD patients and 6 healthy donors, as controls (from Meyer Children Hospital and the Transfusion Unit of the Careggi Hospital, Florence, Italy, respectively), by Ficoll-Hypaque density gradient centrifugation (Biochrom AG). Monocytes were isolated from low density PBMCs by magnetic enrichment with anti-CD14 beads (Milteny Biotec). Cells were cultured in the presence of GM-CSF (800 U/ml) and recombinant Interleukin [IL-4] (1000 U/ml) for 6 days to allow Dendritic Cells (DC) differentiation as previously described[44].
All stimulations were carried out by challenging PBMCs with live fungi at 106 cell/ml concentration. After 24 h or 7 days of incubation, supernatants were collected and stored at −20 °C until assayed by means of cytokine detection. Human Milliplex® assay for Tumor Necrosis Factor alpha [TNF-α], Interferon [IFN]-γ IL-1β, IL-6, IL-10, IL-23, IL-12p70 and IL-17A production was performed according to the manufacturer’s instructions using Luminex technology.
2.4. Cell wall extraction and quantification of sugars by HPAEC-PAD
The sugar composition of S. cerevisiae and C. albicans cell walls was analyzed as previously described [45] applying the following modifications. Briefly, about 200 mg of cell cultures at the stationary phase were harvested and washed with deionized water. Cells were suspended in 1 ml of 10 mM Tris-HCl (pH 8.5) and subsequently disrupted by three rounds of vortex at maximum speed (30 s) and chilling on ice (1 min) using glass beads (0.45–0.55 mm). Cell pellets were subjected to extraction with 100 μl of 72% H2SO4 (w/w) for 1 h at room temperature. The resulting slurry was diluted with MilliQ water, to a final volume of 1 ml and heated for 4 h at 100 °C. The hydrolysate was then diluted to 9 ml with MilliQ water, neutralized with saturated Ba(OH)2 and left overnight at 4 °C to allow the precipitation of sulphate ions. After centrifugation at 3800g for 5 min, the supernatant was subjected to monosaccharide analysis with High-Performance Anion-Exchange Chromatography coupled with Pulsed Amperometric Detector (HPAEC-PAD). All samples were filtered through a 0.2 mm Spartan 13 filter (Schleicher & Schuell Microscience, Dassel, Germany) prior to analysis on a Dionex HPAEC equipped with a CarboPac PA10 (4 × 50 mm) guard column and a CarboPac PA10 (4 × 250 mm) analytical column. Separation was performed as previously described[45]. Sugars were quantified with a pulsed amperometric detector (PAD) with gold electrode. Glucosamine, galactose, glucose and mannose (for chitin, glucan and mannan content determination, respectively) were identified by comparison with reference compounds and quantified according to calibration curves obtained for each sugar.
2.5. Mice infection and cytokine detection
C57BL/6 female mice (6–8 weeks old) were purchased from Charles River (Calco, Milan) and maintained under specific pathogen-free conditions at the Animal Facility of the University of Perugia (Perugia, Italy). Experiments were performed according to the Italian Approved Animal Welfare Assurance A 245/2011-B. Homozygous Il-17a−/−, Ifng−/− and Il-10−/− mice on a C57BL/6 background, were bred under specific pathogen-free conditions.
2.5.1. Fungal infection and histology
For gastrointestinal infection, 108 yeast cells were injected intragastrically. Mice were monitored for fungal growth [colony forming units (log10 CFU), per organ, ± standard error of the mean (SEM) obtained by serially diluting homogenates on Sabouraud dextrose agar plates incubated at 35 °C for 48 h] and histology.
For histology, paraffin-embedded tissue sections (3–4 μm) were stained with periodic acid-Schiff (PAS) reagent. Sections of frozen tissues, cut at 4 μm, were fixed for 60 s in methanol. Photographs were taken using the Olympus BX51 microscope at 10 or 40 (insets) × objective.
2.5.2. ELISA assay
Cytokine content was assessed by enzyme-linked immunosorbent assays (R&D Systems) on stomach homogenates or supernatants of cultured stomachs.
The double-tailed Student’s t-test was used to compare the significance of differences between groups. Comparison with a p-value < 0.05 were considered significant. The data reported are representative of four independent experiments, with similar results. The in vivo groups consisted of six mice/group.
2.6. Statistical analysis
To evaluate variables that might influence the presence of yeast in fecal samples, such as clinical parameters, sex, age and location of the inflammation and treatment, we performed logistic regression using automated model selection. To observe correlations between yeast isolates from IBD, clinical features and ASCA, we used the Wald test. Chi Square statistics were applied to associate significant correlations with the variables mentioned above. Yates correction was applied in the case of expected frequencies less than 5. G test was applied to evaluate significant correlation between fungal isolates and mucosal indices.
Spearman’s correlations among sugar cell wall and cytokine release upon fungal stimulation were performed using the R software through the stats R package (version 3.1.2). P-values were corrected for multiple test comparison by using the false discovery rate correction.
2.7. Bacterial DNA extraction from fecal samples and pyrosequencing
Fecal samples were preserved in RNAlater (Qiagen) at 4 °C for the first 48 h, and kept at −80 °C until DNA extraction. Bacterial genomic DNA extraction and quality control were carried out following our previous protocol [46]. For each sample, we amplified the 16S rRNA gene using the special fusion primer set specific for V5–V6 hypervariable regions and corresponding to primers 784F and 1061R described by Andersson et al. [47], and using the FastStart High Fidelity PCR system (Roche Life Science, Milano, Italy). The 454 pyrosequencing was carried out on the GS FLX + system using the Titanium chemistry following the manufacturer recommendations.
2.8. Metabarcoding data analysis
Raw 454 files were demultiplexed using Roche’s.sff file software. Reads were pre-processed using the MICCA pipeline (version 1.5, http://compmetagen.github.io/micca/)[48]. Pyrosequencing resulted in a total of 212,614 16S rDNA reads with a mean of 13,288 sequences per sample. Average sequence lengths were 273 nt (±SD 4) and 256 nt (±SD 1) for the first and second run, respectively. Raw 454 files were made available at the European Nucleotide Archive (http://www.ebi.ac.uk/ena/data/view) under the accession study PRJEB22036 and PRJEB22343. Forward and reverse primer trimming and quality filtering were performed using micca-preproc truncating reads shorter than 250 nt (quality threshold = 18). Denovo sequence clustering, chimera filtering and taxonomy assignment were performed by micca-otu-denovo (parameters -s 0.97 -c). Operational Taxonomic Units (OTUs) were assigned by clustering the sequences with a threshold of 97% pairwise identity, and their representative sequences were classified using the RDP software version 2.7 [49]. Template-guided multiple sequence alignment was performed using PyNAST57 (version 0.1)[50] against the multiple alignment of the Greengenes 13_8 16S rRNA gene database [51] filtered at 97% similarity. Rarefaction was performed in order to reduce the sampling heterogeneity. A total of 8500 sequences per sample was obtained.
For diversity analysis, OTU table was summarized at three different phylogenetic levels: Phylum, Family, and Genus. Alpha diversity was evaluated using the genus-level OTUs table, measuring richness (S), as the number of observed OTUs (using specnumber function in the Vegan R package), and Shannon–Wiener index (H) (using diversity function in the Vegan R package). Considering the presence of S. cerevisiae (Sc group), other yeasts, such as Candida (group Other Y), or absence of yeast (group Y-) in the feces, pairwise Wilcoxon rank sum test (using pairwise. wilcox.test function in the Stats R package) was used to assess significant differences between samples.
Beta diversity of gut microbiota was evaluated at genus-level using beta dispersion (calculated on the Bray-Curtis dissimilarity matrix) as a measure of within-group heterogeneity (using function betadisper in the Vegan R package). To test if bacterial community in the feces of different groups (i.e. groups of samples Sc, Other Y, and Y-) had a different heterogeneity, a permutational test with 999 permutations, was used (using permutest function in the Vegan R package). To compare the relative abundances of OTUs among the three groups of subjects, the two-sided unpaired Wilcoxon test was performed, removing taxa not having a relative abundance of at least 0.1% in at least 20% of the samples, and using the function mt () in the phyloseq library. The p-values were adjusted for multiple comparison controlling the family-wise Type I error rate (minP procedure). Ternary plots were drawn using R package (ggtern) to depict the distribution of bacterial communities among the three different groups of fecal samples: (i) Sc group, in which S. cerevisiae was isolated; (ii) Other Y group, in which other yeasts, such as Candida, were isolated; (iii) Y- group, in which no yeasts were isolated.
Based on relative abundances, the metagenomic biomarker discovery and related statistical significance were assessed using the linear discriminant analysis (LDA) effect size (LEfSe) method[52]. LEfSe uses the Kruskal–Wallis rank-sum test to identify features with significantly different abundances between assigned taxa compared to the groups, and LDA to estimate the size effect of each feature. An alpha significance level of 0.05, either for the factorial Kruskal-Wallis test among classes or for the pairwise Wilcoxon test between subclasses, was used. A size-effect threshold of 2.0 on the logarithmic LDA score was used for discriminative microbial biomarkers.
3. Results
3.1. The mycobiota of CD patients is enriched for S. cerevisiae strains
We used selective media with bacterial growth inhibitors to characterize the cultivable gut mycobiota present in feces from 34 pediatric CD, 27 pediatric UC, and 32 healthy children (HC). Sequencing of the rDNA ITS1-5.8S-ITS4 region allowed us to classify a total of 112 yeast isolates as belonging to 20 different species (Table 2). The CD group showed the highest number of isolates (N = 78), belonging to 12 different species, compared to UC and HC (N = 12 and N = 22 isolates, respectively; Table 1). S. cerevisiae, C. albicans and C. parapsilosis composed the largest part of the CD cultivable mycobiota (Table 2). Considering the most abundant fungal species, we isolated 27 S. cerevisiae strains, 22 C. albicans strains and 16 C. parapsilosis strains in 26.4% (9 out of 34), 14.7% (5 out of 34) and 17.6% (6 out of 34) of CD patients, respectively. We observed a significantly higher fungal richness in CD compared to UC and HC (p < 0.0001 Kruskal-Wallis test FDR correction; figure S1). By stratifying CD patients according to clinical markers useful for monitoring mucosal disease activity (e.g. fecal calprotectin dosage, Table 1), we observed that 52.9% (18 out of 34) of patients showed high mucosal inflammation indices. We found fungal isolates in 61% (11 out of 18) of CD patients with mucosal inflammation, 45% of which (5 out of 11) were S. cerevisiae. Yeasts were isolated in 44.4% (12/27) of UC patients, and in only 7.4% (2 out of 27) of UC patients with no mucosal inflammation we were able to identify S. cerevisiae strains. To evaluate variables that might influence the presence of yeasts in fecal samples, we evaluated potential correlations among presence/absence of yeasts and clinical parameters, sex, age or location of the inflammation and treatment. We observed that, in CD patients, the presence of yeasts significantly correlated only with mucosal inflammation indices (G test, G adj. 6.280; p = 0.012), in agreement with previous observations [26].
Table 2.
Yeast species isolated and identified by sequencing of the ITS1-5.8S-ITS2 region. For each yeast species, the number of isolates and the number of subjects from which the corresponding yeast species was isolated are reported. For each donor group, the total number of isolated strains for each species, as well as the overall number of isolates, are indicated. CD= Crohn’s Disease patients, UC=Ulcerative Colitis patients, HC= Healthy Children.
| N° of yeast isolates |
N° of subjects with yeast isolates |
|||||||
|---|---|---|---|---|---|---|---|---|
| Phylum | Yeast Species | CD | UC | HC | Total | CD | UC | HC |
| Ascomycetes | Saccharomyces cerevisiae | 27 | 2 | 2 | 31 | 9 | 2 | 2 |
| S. delbrueckii | 2 | 0 | 0 | 2 | 1 | 0 | 0 | |
| Candida albicans | 22 | 7 | 10 | 39 | 5 | 6 | 10 | |
| C. ernobii | 0 | 1 | 0 | 1 | 0 | 1 | 0 | |
| C. glabrata | 0 | 1 | 0 | 1 | 0 | 1 | 0 | |
| C. pararugosa | 1 | 0 | 0 | 1 | 1 | 0 | 0 | |
| C. parapsilosis | 16 | 1 | 1 | 18 | 6 | 1 | 1 | |
| C. tropicalis | 0 | 0 | 1 | 1 | 0 | 0 | 1 | |
| C.zeylanoides | 0 | 0 | 1 | 1 | 0 | 0 | 1 | |
| Clavispora lusitaniae | 2 | 0 | 2 | 4 | 2 | 0 | 2 | |
| Issatchenkia orientalis | 1 | 0 | 0 | 1 | 1 | 0 | 0 | |
| Pichia caribbica | 0 | 0 | 1 | 1 | 0 | 0 | 1 | |
| P. kluyveri | 0 | 0 | 1 | 1 | 0 | 0 | 1 | |
| P. membranifaciens | 1 | 0 | 0 | 1 | 1 | 0 | 0 | |
| Yarrowia lipolytica | 0 | 0 | 1 | 1 | 0 | 0 | 1 | |
| Basidiomycetes | Cryptococcus adeliensis | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| C. liquefaciens | 1 | 0 | 0 | 1 | 1 | 0 | 0 | |
| C. saitoi | 0 | 0 | 1 | 1 | 0 | 0 | 1 | |
| Rhodotorula mucilaginosa | 3 | 0 | 1 | 4 | 3 | 0 | 1 | |
| Trichosporon faecale | 1 | 0 | 0 | 1 | 1 | 0 | 0 | |
| Total isolates | 78 | 12 | 22 | 112 | ||||
| Total species | 12 | 5 | 11 | 20 | ||||
3.2. Immunophenotypic screening of S. cerevisiae and Candida spp.
To investigate the host immune reactivity to different yeast strains isolated from gut samples, we screened a large set of S. cerevisiae and Candida spp. Strains, isolated from IBD and HC feces. Aiming at this, we measured the cytokine profiles produced in vitro by PBMCs from healthy donors and CD patients (Fig. 1, Fig. 2 and figures S2-S3-S4), and by human healthy DCs (Fig. 3 and figures S5-S6) when challenged with fungal cells. In particular, we compared the immunomodulatory properties of a set of 23 S. cerevisiae strains isolated from human gut of IBD and HC and strains from other sources (Table S1).
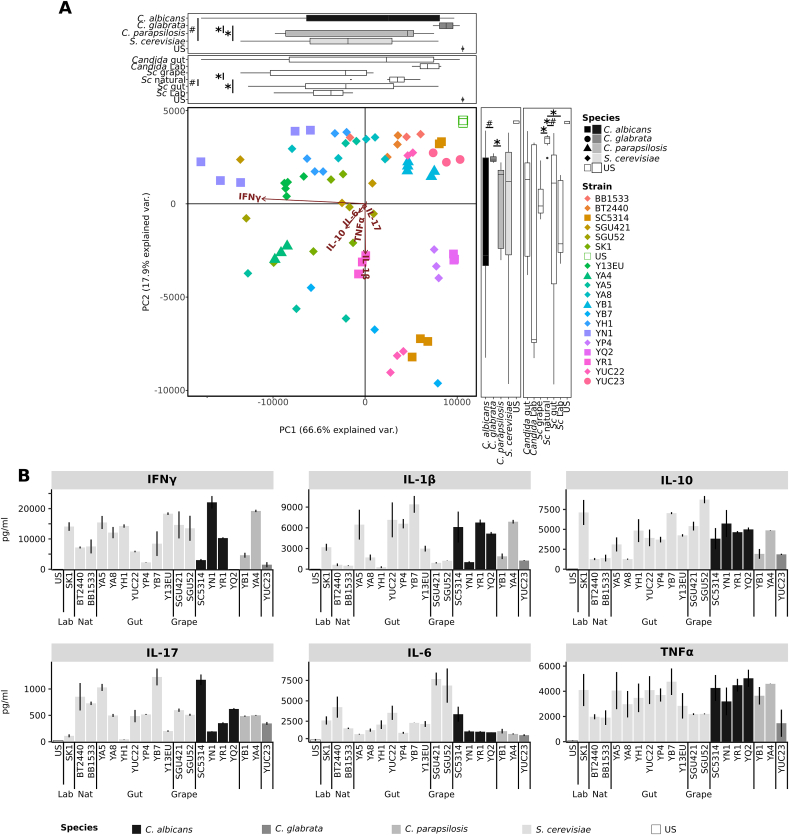
Fig. 1.
Innate and adaptive immune response of PBMCs from healthy donors to different yeasts strains. Selected S. cerevisiae (light grey) and Candida spp. (grey scales) strains were tested for the ability to induce inflammatory (IL-6, IL-1β, TNF-α, IFN-γ, IL-17A) and inflammatory responses (IL-10), and specifically cytokines deriving by Th1 (IFN-γ), Th17 (IL-17) and Treg (IL-10) priming. Healthy PBMCs from 5 donors were stimulated with live yeast cells for 24 h or 5 days, and cytokine levels were measured. (A) Principal component Analysis of cytokines profiles of yeast strains according to the different species and sources. Boxplots recapitulate the coordinates (horizontal boxplots show PC1, vertical box plots show PC2) of strains grouped according to their species or isolation source (Lab = laboratory strains, Gut, Grape, and Nat = natural environment, such as soil, tree). * and # indicate p-values< 0.05, fdr corrected, calculated with Wilcoxon and Levene tests respectively. (B) Cytokines produced by PBMCs from healthy donors in response to the challenge with the selected yeast isolates. Bars indicate the average; error bars indicate the standard error.
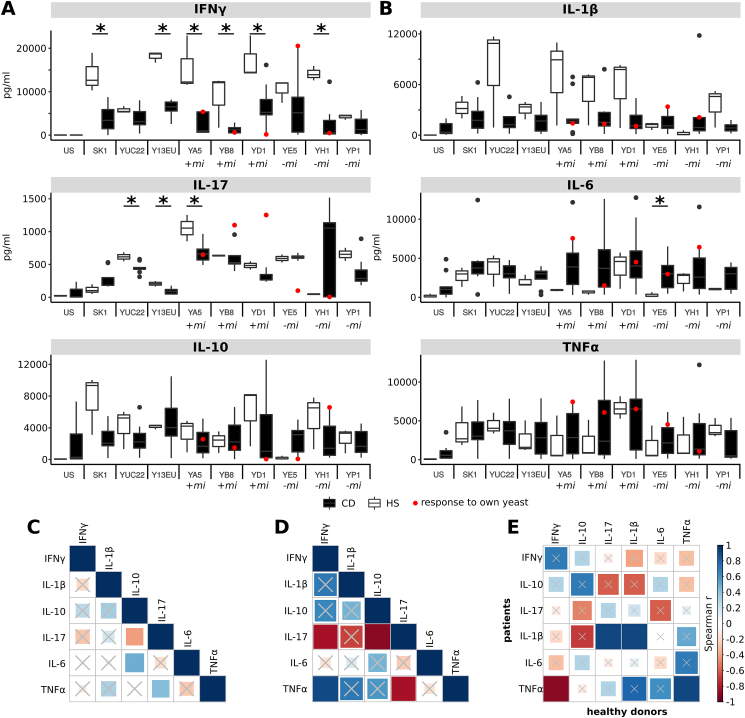
Fig. 2.
Stimulation of cytokine production upon challenge of PBMCs from healthy donors to different yeasts strains. (A–B) Individual cytokine patterns for S. cerevisiae strains isolated from IBD patients (n = 7) and healthy donors (n = 6). PBMC T-polarizing (A) and pro-inflammatory (B) cytokine release from HS (white boxplots) and CD patients (black boxplots), upon stimulation with 9 selected S. cerevisiae strains (indicated in horizontal axis). In each scatter-plot, red points represent CD patient PBMC cytokine release after stimulation with their own isolate. +mi and -mi indicate the yeast was isolated when the corresponding patients had mucosal inflammation or not mucosal inflammation, respectively. * indicate significant differences between the cytokine levels expressed by healthy subjects’ or CD′ PBMCs (Wilcoxon p < 0.05). (C–D) Statistics of Spearman correlations among levels of cytokines produced by (C) PBMCs from healthy donors and (D) patients in response to S. cerevisiae isolates. (E) Correlations among cytokine levels by CD patients and healthy donors. The color in each square indicates the Spearman correlation r among the variables reported in the two coordinates, as indicated by colored scale bar. Crossed squares lack of statistical significance (p > 0.05). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
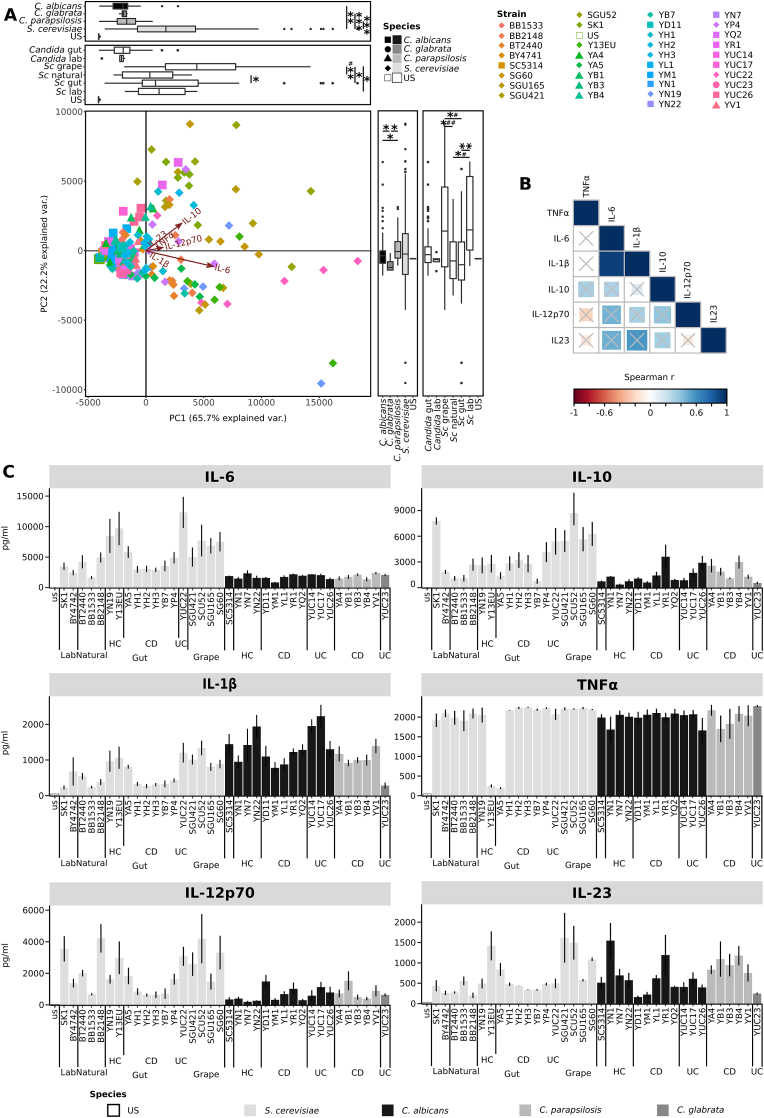
Fig. 3.
Immune response of DCs from healthy donors to different yeasts strains. (A) Principal component Analysis of cytokines profiles of yeast strains according to the different species and sources. Boxplots recapitulate the coordinates (horizontal boxplots show PC1, vertical box plots show PC2) of strains grouped according to their species or isolation source. * and # indicate p-values< 0.05, fdr corrected, calculated with Wilcoxon and Levene tests respectively. (B) Strain-specific cytokine profiles of DCs, challenged with S. cerevisiae (light grey) and Candida spp. (black and dark grey) strains. DCs from healthy donors (n = 6) were stimulated with fungal cells for 24 h and cultures supernatants used for IL-6, IL-10,IL-1β,TNF-α, IL12p70 and IL-23 measurement. For S. cerevisiae strains, different sources of isolation are indicated: laboratory, natural environment (such as soil, tree), gut, and grape. Gut isolates are indicated as isolated from CD, UC patients or healthy children (HC). Bars indicate the average, error bars indicate the standard error.
A set of S. cerevisiae and Candida spp. Strains was preliminarily tested for the ability to induce inflammatory (IL-1β, IL-6, TNF-α, IFN-γ, IL-17A) and anti-inflammatory (IL-10) cytokines upon challenge of PBMCs isolated from healthy donors (Fig. 1). By comparing the general profile of cytokine production by healthy PBMCs in response to the tested strains through PCA (Fig. 1A), we observed that the vast majority of the samples variance was explained in the first component (66.6%), which was driven by IFN-γ levels. The cytokines profiles induced by S. cerevisiae strains were significantly different from those induced by C. glabrata strains (Wilcoxon p < 0.05) and the responses induced by C. albicans isolates were more variable than those induced by S. cerevisiae (Levene p < 0.05) (Fig. 1A). S. cerevisiae strains isolated from human gut induced a more variable response than co-specific strains isolated from natural sources (Levene p < 0.05, Fig. 1A). In many cases, the pro-inflammatory responses (IFN-γ, IL-1β, IL-6, and IL-17) were counterbalanced by anti-inflammatory IL-10 induction to a different extent (Fig. 1B). As observed for S. cerevisiae, cytokines production differed among strains upon stimulation with different Candida spp. Strains, except for a general low level of IL-6 and a tendency of high level of TNF-α (Fig. 1B and Table S3). Generalizing at species level, S. cerevisiae strains induced higher amounts of IL-6 compared to C. glabrata and C. parapsilosis, and of IFN-γ compared to C. parapsilosis strains (Wilcoxon p < 0.05, figure S2). Regarding stimulation with different Candida spp., other differences at the species level included higher IFN-γ and IL-17 in response to C. parapsilosis compared to C. glabrata, higher IL-6 in response to C. albicans compared to C. glabrata and C. parapsilosis, and higher TNF-α in response to C. albicans compared to C. glabrata (Wilcoxon p < 0.05, figure S2). Interestingly, the differences in IL-1β, IL-6, IL-17, and TNF-α levels produced by PBMCs were higher among co-specific strains than among strains of different species (Wilcoxon p < 0.05, Fig. 1B and figure S3).
Considering that the inflammatory status of CD patients may induce different immune responses to fungi compared to healthy subjects, we evaluated the cytokine profiles elicited by fungal isolates in a set of CD patients’ PBMCs and compared them with that of healthy donors (Fig. 2). In general, we observed a tendency for strain-specific pattern in immune response, and a trend of higher level of the pro-inflammatory cytokines IFN-γ IL-17 and IL-6 secreted by healthy PBMCs than patients’ ones (Fig. 2A; Wilcoxon p-value <0.05). To note, the response of PBMCs from 5 CD patients to S. cerevisiae strains isolated from the corresponding fecal samples (“self”) showed different cytokine profiles compared to the response to strains isolated from other patients (“non-self”; Fig. 2, red dots). For example, PBMCs of the CD patient E, in clinical remission and without mucosal inflammation (Fig. 2; Table S3), expressed more IFN-γ, IL-1β and TNF-α, and less IL-17A and IL-10 in response to the “self” strains (YE5) compared to other CD patients in response to the same strain (Fig. 2A and B, red dot). In addition, PBMCs from the CD patient H, in clinical remission but with active mucosal inflammation (Table S3), released lower IL-17A, IFN-γ, IL-6 and IL-1β and higher IL-10 upon challenge with the “self” (YH1) compared to other patients’ PBMCs in response to the same isolate (Fig. 2A and B, red dot). YB8 and YD1 strains, isolated from CD patients’ B and D in active disease and mucosal inflammation, induced the production of high levels of IL-17A and low levels of IFN-γ and IL-10 by PBMCs of donors B and D, respectively (Fig. 2A, red dots). The cytokine pattern observed in response to S. cerevisiae strains differed, and in some cases showed an opposite trend, when we compared healthy subjects and patients. Spearman correlation analysis between the different cytokine responses showed that, unlike for healthy donors, the inflammatory and clinical status of the patient differently affect the immune response against yeast strains (Fig. 2C and D). IL-10 produced by healthy PBMCs upon challenge with S. cerevisiae strains positively correlated with IL-6 and negatively with IL-17, while the latter positively correlated with TNF-α. On the other hand, IL-17 secreted by CD patients PBMCs (Fig. 2D) negatively correlated with IFN-γ, TNF-α and IL-10, and TNF-α correlated positively with IFN-γ. When comparing cytokine profiles induced by healthy and CD PBMCs, positive correlations were found among levels of IL-1β and TNF-α produced by healthy PBMCs and the same cytokines secreted by CD patients PBMCs, respectively (Fig. 2E). A negative correlation was found between TNF-α produced by patients’ PBMCs and IFN-γ expressed by healthy subjects’ PBMCs, and a positive correlation between IL-1β expressed by patients’ PBMCs and IL-17 by healthy PBMCs (Fig. 2E).
We also assessed the response of DCs to different strains and species. TNF-α, IL-6, IL-1β, IL-10, IL-12p70 and IL-23 cytokines levels were measured from the supernatant of DC challenged with a larger set of different fungal strains (Fig. 3 and figs. S5-S6). By comparing the general profile of cytokine production through PCA (Fig. 3A), we observed that the vast majority of samples variance was explained in the first component (65.7%). According to this component, the cytokine profiles induced by S. cerevisiae strains were significantly different from those induced by all Candida spp. Strains (Wilcoxon p < 0.01 S. cerevisiae strains vs C. albicans strains; p < 0.001 S. cerevisiae vs C. glabrata and S. cerevisiae and C. parapsilosis, Table S2). Considering the different sources of the strains, the cytokine profiles of S. cerevisiae strains resulted significantly different from strains collected in natural environments (such as soil, tree) (Wilcoxon p < 0.05) and grape (Wilcoxon p < 0.001, Table S1) (Fig. 3A). At species level, we observed higher levels of IL-6, IL-12p70, and anti-inflammatory IL-10 upon stimulation with S. cerevisiae compared to the three tested Candida species (figure S4, Wilcoxon signed-rank test fdr corrected p < 0.05 complete results in Table S2). These findings were in accordance with the previous observations on tolerogenic response (high IL-10 levels) upon stimulation of DCs with S. cerevisiae compared to C. albicans, as observed by Sokol and co-workers [23]. Contrarily, IL-1β levels were higher upon stimulation with Candida spp. compared with S. cerevisiae strains (Fig. 4, Wilcoxon Student’s t signed-rank test fdr corrected, Table S2). Both when considering the cytokine profiles and individual cytokines, greater differences were observed at the strain-level than at the species level (figure S5). As observed for cytokine profiles (Fig. 3A), the production of individual cytokines varied according to the isolation source of the tested strain (Fig. 3C and Table S2). The levels of IL-6 and IL-1β were lower in response to S. cerevisiae strains isolated from CD patients than from HC donors or from grapes (Fig. 3C). In general, Spearman correlation analysis showed a positive correlation between IL-6 and IL-1β (Fig. 3B). IL-10 was lower in DCs challenged with S. cerevisiae strains from CD patients than with lab, grape, or UC strains, while S. cerevisiae grape strains induced more IL-10 than HC and strains from natural environment (Fig. 3C, Table S2). This suggests that S. cerevisiae strains from CD patients are in general less immunogenic than the other strains. The levels of IL-12p70 were higher in DCs challenged with S. cerevisiae strains isolated from CD patients compared to strains isolated from any other source, while CD Candida spp. Strains induced a higher expression of IL-12p70 compared to C. albicans strains isolated from HC donors (Fig. 3C, Table S2). IL-23 was higher in DC challenged with S. cerevisiae grape isolates and lower in DCs challenged with laboratory and natural compared to CD co-specific strains (Fig. 3C, Table S2). On the other hand, Candida spp. Strains isolated from CD patients induced a lower expression of IL-23 in contrast with Candida spp. Strains isolated from HC donors (Fig. 3C, Table S2). Similar amounts of TNF-α were quantified from DCs cells challenged with different S. cerevisiae or Candida spp. Isolates, except for two S. cerevisiae gut isolates (Y13EU and YA5), which showed a markedly low induction of this cytokine (Fig. 3C).
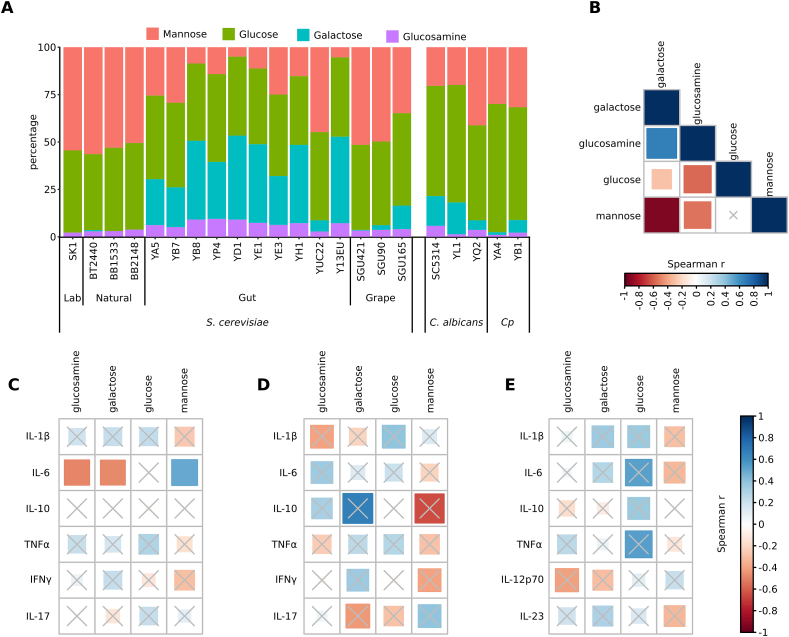
Fig. 4.
Cell wall sugar components of S. cerevisiae and Candida spp. Strains and correlation with cytokine induction. (A) Percentage of mannan (measured as mannose moieties), glucan (glucose), galactose and chitin (glucosamine) amounts in S. cerevisiae and of Candida spp strains. S. cerevisiae strains isolated from human gut in comparison with strains from other sources (laboratory, natural environment, wine, and wasp’s gut). Candida spp. Strains isolated from CD patients in comparison with SC5314 reference strain. Ca= C. albicans and Cp = C. parapsilosis. (B) Statistics of Spearman correlations among the amount of cell wall components of S. cerevisiae gut isolates. (C–E) Statistics of Spearman correlations among the amount of cell wall components of S. cerevisiae gut isolates and cytokine production by (C) PBMCs from healthy donors, (D) PBMCs from DC patients, and (E) DCs from healthy donors upon challenge with yeast strains. The color in each square indicates the Spearman correlation r among the variables reported in the two coordinates, as indicated by colored scale bar. Crossed squares lack of statistical significance (p > 0.05). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Cell wall composition of S. cerevisiae and Candida spp. Strains
The sugar moieties of fungal cell wall are the principal antigens sensed by the host immune system during host-fungal interaction. Investigations on the importance of fungal cell wall in promoting immune response indicated that a multilayered cell wall controls the exposure of pathogen-associated molecular patterns to immune cells [53]. Given the different cytokine profiles among the screened strains, we compared the cell wall sugar composition of a selection of 10 IBD S. cerevisiae strains, with a set of 7 strains isolated from other sources (laboratory, natural environment, and grapes), and 4 Candida spp. Stains isolated from fecal samples of CD patients and the SC5314 C. albicans reference strain (Fig. 4A and Table S1).
HPAEC-PAD analysis (see Methods) revealed that S. cerevisiae gut isolates showed inverse mannose/galactose ratio, with a significantly lower percentage of mannose (measured as mannan moieties) compared to all the other strains (Fig. 4A; Kruskal-Wallis test, FDR correction p < 0.05). Gut strains showed a higher amount of glucosamine (chitin moieties), and a significantly higher galactose content compared to strains isolated from other sources (Kruskal-Wallis test, FDR correction p < 0.05, Fig. 4A). When compared with gut Candida spp. Strains, we observed a higher amount of galactose and glucosamine, and lower amount of glucose (glucan moieties) in S. cerevisiae gut strains (Fig. 4; Mann-Whitney test p = 0.048 for galactose, p = 0.01 for glucosamine, and p = 0.0016 for glucose). Spearman correlation analysis (Fig. 4B) confirmed that in S. cerevisiae strains, the amount of galactose positively correlated with glucosamine (Spearman r = 0.7, fdr<0.05), and that mannose and glucose negatively correlated with galactose and glucosamine (Spearman r = 0.7 and r = 0.4, fdr<0.05). Thus, the cell wall composition of S. cerevisiae strains isolated from human intestinal tract appears to be peculiar for this set of strains compared to co-specific strains isolated from other sources and to other yeast species.
We then investigated potential correlations among the yeast cell wall sugar components and the levels of cytokines produced by PBMCs (either of healthy donors’ or CD patients’), and by healthy DCs upon strain’s challenge (Spearman correlation; Fig. 4CE). Interestingly, we found correlations with IL-6, produced by healthy PBMCs, which negatively correlated with the amount of glucosamine and galactose and positively with mannose (Spearman r = −0.47, r = −0.48 and r = 0.40, fdr<0.05; Fig. 4C). Despite below the statistical significance threshold, it is worthy to note that in CD patients IL-17 responses were anti-correlated with galactose and correlated with mannose levels.
3.4. In vivo challenge of S. cerevisiae strains
To corroborate the in vitro results, we assessed the susceptibility of C57BL/6 mice to gastrointestinal colonization and/or infection with different gut S. cerevisiae strains (Fig. 5) found to elicit distinct cytokine patterns in human healthy DCs and PBMCs. At 3 and 7 days after intra-gastric inoculation, we evaluated the fungal growth, the patterns of cytokine production in the colon, and the local inflammatory pathology. We found that the tested S. cerevisiae strains differed in their ability to grow and colonize the colon at 3 days after inoculation (figure S6).
Fig. 5.
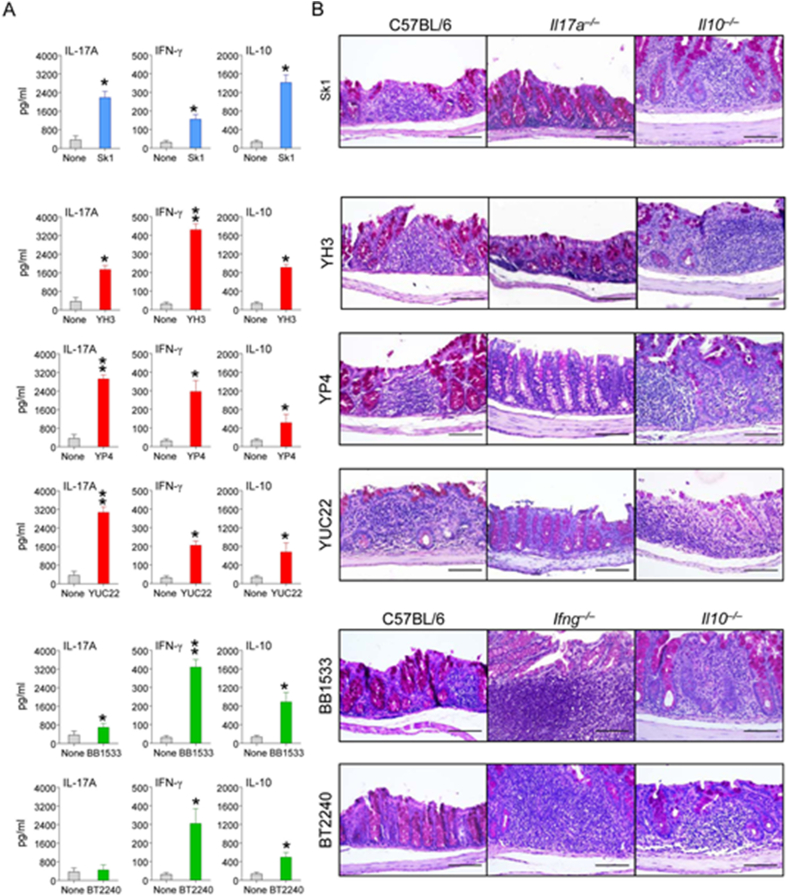
S. cerevisiae infection in mice. (A) Cytokine release (pg/ml) upon challenge with selected S. cerevisiae strains at 3 days post-inoculation with SK1 reference strain (blue), YH3, YP4, YUC22 CD gut strains (red) and BB1533, BT2240 natural strains (green). Student t-Test *p < 0.05, **p < 0.01, naïve versus inoculated C57BL/6 mice (6 mice/group). (B) Local epithelial activity and inflammation in the gut of C57BL/6 mice, in Il-17a−/−, Ifnγ −/−, and Il-10−/− mice induced by the 6 different S. cerevisiae strains inoculation. Inflammatory cell recruitment and positive mucin staining in mice 3 days after inoculation. Scale bars, 200 mm. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
To correlate the fungal growth with local cytokine production in the colon, we measured the levels of cytokines known to afford colonization resistance (IL-17A and IFN-γ) or immune tolerance (IL-10) in mice. The cytokine profiles were measured in wild type C57BL/6 mice after inoculation with a set of S. cerevisiae strains chosen to recapitulate the diversity in immune responses observed in patients and HC, such as gut strains (YH3, YUC22 and YP4), natural strains (BT2240, BB1533) or the laboratory strain SK1. The relative levels of each cytokine differed significantly among isolates (Fig. 5A). IL-17A was the cytokine most highly produced in response to the YUC22 and YP4 strains, IFN-γ in response to BB1533 or BT2240 natural strains, and IL-10 with the SK1 reference strain (Fig. 5A). These results corroborated the hypothesis that fungal immune-reactivity is strain-dependent. To define the functional role, if any, of these cytokines in yeast colonization and/or infection, we comparatively evaluated parameters of inflammatory pathology in the colon of wild-type C57BL/6 or cytokine-deficient mice inoculated with the different strains. The histology analysis on tissues of wild type mice infected with different S. cerevisiae strains revealed that colonization was associated with inflammatory influx of the mucosa and positive mucin staining. Interestingly, inflammation and hypertrophy of the colon mucosa was drastically reduced in Il-17a−/−mice inoculated with the IL-17A-producing strains (YUC22 and YP4), and similarly in Ifnγ−/− mice inoculated with the IFN-γ-producing isolates (YH3, BB1533 and BT2240) (Fig. 5B). As expected, increased inflammation was observed in Il-10−/− mice, as opposed to wild type mice, inoculated with the IL-10-inducing SK1. Thus, S. cerevisiae strains contributed differently to local epithelial activity and inflammation in the colon, corroborating the hypothesis of a strain-specific rather than a species-specific immune reactivity.
3.5. Fungal interaction with gut bacterial communities
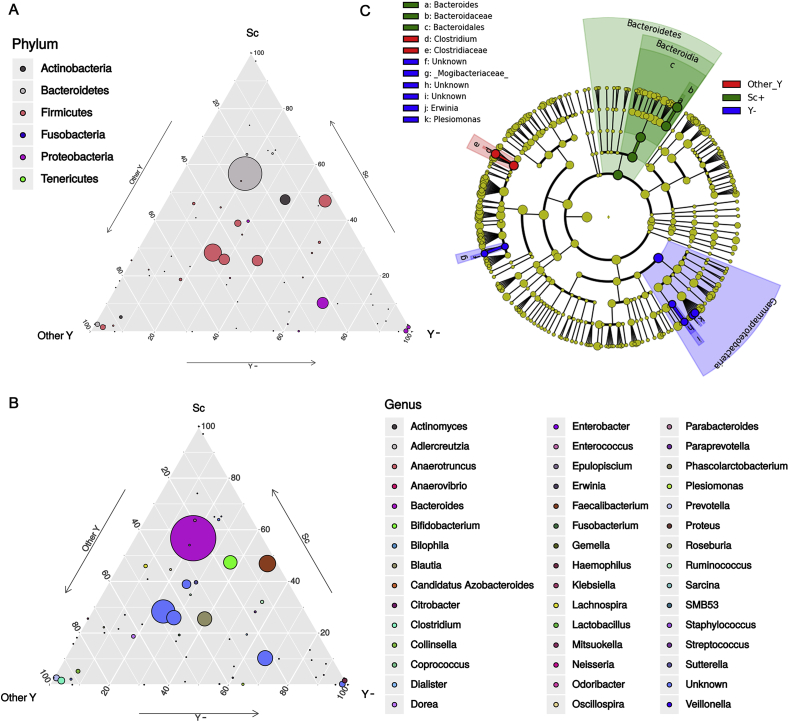
In order to evaluate potential inter-microbial kingdom relationships in the gut environment, we explored the gut microbiota composition in relation with fungal isolation from fecal samples. We performed metabarcoding analysis on a selection of 19 fecal samples from CD patients (Table S4) from which we isolated S. cerevisiae (n = 7 fecal samples, Sc + group) or other yeasts (Candida spp.; n = 5 samples, other Y group) (figure S8). We compared microbial profiles with those of CD fecal samples (n = 7) from which we did not isolate any yeasts (Y- group; figure S8). Alpha- and beta-diversities were computed in order to estimate within-sample richness and between-sample dissimilarity, respectively. We found higher bacterial richness (alpha diversity) and diversity (beta diversity) in samples from which neither Candida spp. nor S. cerevisiae strains were isolated (Y- group) compared to the other fecal samples (figure S8A-C), although differences were not statistically significant. The taxa distribution in the three groups of samples (Other Y, Sc and Y-) showed variations in gut microbiota composition associated with presence/absence of fungal isolates (figure S8). Ternary plot (Fig. 6A and B) showed generalist or sample-specific bacterial taxa among the three sample groups (circles in the middle of the triangle, and circles in the vertices, respectively). At the phylum level, we observed that the Bacteroidetes phylum was generally present in all sample groups, but one OTU (grey circle in Fig. 6A) was more abundant in fecal samples in which S. cerevisiae was isolated. Two OTUs identified as Bacteroidetes and Firmicutes had specific distributions for samples in which other yeasts were isolated (grey and pink circles in the vertex of the triangle indicated as Other Y, Fig. 6A). Three OTUs belonging to Proteobacteria were specific for samples in which no yeast were isolated (fuchsia circles near or in the vertex of the triangles indicated in Y-). At the genus level, we observed that the abundance of Faecalibacterium, a well-known bacterium with immunoregulatory properties, discriminated samples in which S. cerevisiae was isolated; while Clostridium, Collinsella and Prevotella were associated with samples from which we isolated other yeasts. Finally, in samples from which we did not isolate any yeast, we observed typically high levels of Haemophilus and Plesiomonas, the latter a genus very close to a known species named as Aeromonas shigelloides, which was found to be responsible for diarrhea and gastroenteritis in humans [54].
Fig. 6.
Gut microbiota composition and relationships with fungal isolates. (A–B) Ternary plots reveal OTUs relative abundance (dot size) at (A) phylum and (B) genus level among the three sample groups (Sc: presence of S. cerevisiae, Other Y: presence of Candida spp., and Y-: absence of fungal isolates). Generalist taxa are represented as circles in the middle of the triangle, sample-specific bacterial taxa are represented as circles in the summit or along the edges of the triangle. (C) Cladogram representing the differences in bacterial taxa in relation with presence/absence of cultivable fungi, by LEfSe analysis. Significant ranking among groups are obtained by alpha value = 0.05 for the factorial Kruskal-Wallis test among classes. The threshold for the logarithmic LDA score was 2.0.
By linear discriminant analysis effect size (LEfSe) confirmed that, when S. cerevisiae was isolated from fecal samples (Sc group), the Bacteroidetes phylum and in particular Bacteroides genus were significantly enriched (Fig. 6C). Gammaproteobacteria, including Erwinia and Plesiomonas, and Mogibacteriaceae were associated with the absence of fungal isolates in fecal samples (Y- group). When we isolated Candida spp. (Other Y group), we found an enrichment of the Clostridiaceae, including Clostridium genus (Fig. 6C). Interestingly, an enrichment of the Porphyromonadaceaeand of the Faecalibacterium genus was observed in samples with S. cerevisiae isolation compared to samples positive for other yeasts, while in the presence of other yeasts an enrichment of Clostridiaceae (Clostridium), Plesiomonas, and Erwinia was confirmed (figure S9).
Overall, these results highlight that the presence of S. cerevisiae is associated with a favorable gut environment for beneficial bacterial genera, such as Faecalibacterium. On the other hand, the absence of yeasts or the presence of other yeast species is associated with potential pathogenic bacteria.
4. Discussion
Our analysis of cultivable fungi shows that S. cerevisiae and Candida spp. are relevant components of the gut mycobiota, in agreement with previous studies [8,55]. We found that S. cerevisiae is enriched in gastrointestinal tract of pediatric CD patients compared to UC and HC, and this result suggests that a CD-specific gut environment may favor expansion of fungi, through the onset of peculiar features that are likely to affect the yeasts fitness and the interaction with the host. Hence, similarly to diagnostic ASCA marker, it is possible to infer a differentiation of the role of the fungi in CD versus UC pathogenesis. Our recent study [41] provided evidence of genetic and phenotypic differences between strains isolated from gut and non-gut environments (e.g. natural sources, fermentation, grapes). Here, we show how genetic differences among strains may reflect the strain-specific differences in eliciting host immune reactivity. We observed a strain-specific pattern of pro- and anti-inflammatory cytokines (T-polarizing cytokines and T-regulatory IL-10, respectively) upon stimulation of human PBMCs and DCs. The majority of S. cerevisiae strain elicited IL-10 production to counterbalance IL-6 induction in PBMCs. This anti-inflammatory effect of S. cerevisiae was supported by Spearman correlation and was in agreement with previous results obtained in mice DCs showing an immunoregulatory response of S. cerevisiae compared with C. albicans [23]. The levels of cytokines produced in response to yeast strains varied significantly among healthy subjects and CD patients, possibly because of an effect of the inflammatory and clinical status of the patient on the immune response to yeasts. In DCs, the levels of IL-6, IL-1β, and IL-10 were lower in response to S. cerevisiae strains isolated from CD patients than from HC donors or from grapes. This is in agreement with the finding that S. cerevisiae strains from CD patients induce higher level of IL-12p70 and IFN-γ, suggesting that these strains are able to train the immune system properly tuning innate and adaptive immunity [56]. We also observed negative correlations between induction of IL-17 and IFN-γ secreted by healthy PBMCs, and IL-1β and TNF-α produced by CD patient PBMCs. Contrarily, the production of IL-1β and TNF-α was positively correlated between healthy donors’ and CD patients’ PBMCs. Interestingly, some CD patients showed a particular immune response to proprietary strain, suggesting that recognition of a yeast strain previously encountered, contributes in shaping a patient-specific immune response. This result could be affected by several variables, such as disease status, therapy, etc. that have not been controlled in the present study.
Sugar components of yeast cell wall are the main antigens of the host-immune recognition, and differences in cell wall components may subtend differences in cytokine profiles. We confirmed our previous finding [41] of the peculiar composition of the gut strains cell wall, characterized by high levels of galactose and glucosamine (chitin moieties) and low levels of glucose (beta-glucan) and mannose (mannans). The different cell wall composition in human gut S. cerevisiae isolates is partially associated with strain-specific differences in the cytokine patterns induced in healthy donors, with the amount of galactose and glucosamine in S. cerevisiae strains being negatively correlated with the levels of IL-6 expressed in challenged healthy PBMCs. We also noticed a trend indicating that IL-17 production in patients were anti-correlated with galactose and correlated with mannose levels. This finding is also in agreement with the result indicating that S. cerevisiae strains from CD patients induce higher level of IL12p70 and IFN-γ.
The results of the in vitro screening were corroborated by in vivo experiments in which S. cerevisiae strains differently contributed to local epithelial activity and inflammation on wild type C57BL/6 mice and in Interleukin (Il)-17A-, Ifnγ- and Il-10- deficient mice.
In agreement with our previous study [41], our results underline the importance to investigate the interplay between fungal cell wall and gut immune function. These considerations are particularly relevant to understand the role of these strains in health and disease, since the molecular mechanisms used by yeasts to colonize the host as a harmless commensal appear as a continuum with the strategies used for evading immune surveillance and can thus potentially turn a friend into a foe.
It is noteworthy that in this study strains from CD patients were less prone to colonize the mouse gut than strains from natural sources. However, despite S. cerevisiae is rare in the human gut, present in less than 2% of healthy subjects [57], yet we report S. cerevisiae in 26.4% of the patients, a ten-fold increase compared to HC. We acknowledge that our results are limited to single time points and further studies should be aimed at assessing the persistence over time of specific yeast species or strains. Based on our results, we could hypothesize that some CD S. cerevisiae strains may be passenger, introduced with the diet, and could be capable of colonizing the host in the presence of a leaky gut like the one of CD patients. Archetypical opportunistic fungal pathogens, such as the yeast C. albicans, commonly occur as commensals on mucosal surfaces of humans, but invade the host when epithelial barriers are disturbed or the host immune system is impaired. Pathogenicity possibly evolved from commensalism, by acquisition of traits suited to colonize and then invade the host.
In the gut environment, yeast survival is difficult and competition is intense if we consider coexistence with bacteria. Therefore, we explored the equilibrium between fungal and bacterial communities and the mutual influence between gut microbiota and presence/absence of commensal fungi in the context of IBD. Our results show that beneficial bacterial genera, such as Bacteroides and Faecalibacterium, are associated with the presence of S. cerevisiae. On the contrary, the gut microbiota of CD patients lacking of fungi is enriched in potential pathogenic bacteria, especially Proteobacteria including the Enterobacteriaceae family, or, in some cases, the presence of Candida spp. Is associated to an enrichment in potentially pathogenic bacteria involved in colitis. Furthermore, our observation of drastic changes in the gut microbiota in the presence of S. cerevisiae strains suggests that in certain cases the yeast strains could interfere with the gut microbiota and mycobiota, by priming the immune system against potential pathogens possibly by inducing a trained immunity [58].
Altogether, these results encourage for in-depth, strain-level extensive studies on human gut mycobiota and integration of metagenomic data with culturomics and immunology to further establish the relevance of fungi in host physiology and host-microbe interaction, as well as the interaction with microbial communities. Understanding the impact of different fungal strains on the host’s immune system provides useful insights on how the modulation of intestinal mycobiome could influence the microbiome in health and disease, the results on the variability in the response to “self” versus “not-self” strains suggest the importance of performing these analyses on PBMCs from the patients. This finding may be particularly relevant when evaluating the possibility to use S. cerevisiae strains in probiotic interventions or when planning protocols for fecal transplantation.
Consent for publication
Informed consent was obtained from the enrolled subjects or tutors.
Availability of supporting data
Raw sequences are available in the European Nucleotide Archive (ENA) with accession number PRJEB22036 and PRJEB22343 (http://www.ebi.ac.uk/ena/data/view/PRJEB22036 and http://www.ebi.ac.uk/ena/data/view/PRJEB22343).
Funding acquisition
Duccio Cavalieri, Paolo Lionetti.
Credit authors statement
Conceptualization: Duccio Cavalieri, Carlotta De Filippo, Paolo Lionetti, Monica Di Paola. Methodology: Duccio Cavalieri, Carlotta De Filippo, Paolo Lionetti, Monica Di Paola, Lisa Rizzetto, Irene Stefanini. Formal analysis: Francesco Vitali, Irene Stefanini, Matteo Ramazzotti, Monica Di Paola. Investigation: Monica Di Paola, Carlotta De Filippo, Lisa Rizzetto, Irene Stefanini, Luigina Romani, Cristina Massi-Benedetti, Noemi Tocci. Resources: Duccio Cavalieri, Carlotta De Filippo, Paolo Lionetti, Luigina Romani. Data Curation: Irene Stefanini, Francesco Vitali, Matteo Ramazzotti. Writing - Original Draft: Monica Di Paola, Duccio Cavalieri, Irene Stefanini, Lisa Rizzetto, Carlotta De Filippo.Writing - Review & Editing: Duccio Cavalieri, Monica Di Paola, Irene Stefanini.
Authorship’ contributions
DC, CDF, MDP, PL participated in study planning and designed the experiments.
FV, MDP, IS, MR carried out the metabarcoding, statistical and data analyses.
LiRi, MDP, LuRo, carried out in vitro and in vivo experiments.
NT, performed chemical analysis.
MDP, DC, IS, LiRi, wrote the manuscript.
All authors read and approved the final manuscript.
Funding sources
This project was supported by the Seventh Framework Programme [FP7/2007–2013] Integrative Project SYBARIS (Grant Number: HEALTH-2010-242,220) by the Ministero dell’ Istruzione, dell’ Università e della Ricerca, Italy Grant PRIN 2007-N352CP_001, by University of Florence, by AOU Meyer Children Hospital (Florence), AMICI onlus, by funding from Autonomous Province of Trento to Fondazione E. Mach (Accordo di Programma) and DT-SFS-26-2019-Food Cloud demonstrators- Food Nutrition Security Cloud- Horizon 2020.
Declaration of competing interest
The authors declare financial and non-financial competing interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jtauto.2020.100036.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Sokol H., Pigneur B., Watterlot L., Lakhdari O., Bermudez-Humaran L.G., Gratadoux J.J. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. U.S.A. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Willing B.P., Gill N., Finlay B.B. The role of the immune system in regulating the microbiota. Gut Microb. 2010;1:213–223. doi: 10.4161/gmic.1.4.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Joossens M., Huys G., Cnockaert M., De Preter V., Verbeke K., Rutgeerts P. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut. 2011;60:631–637. doi: 10.1136/gut.2010.223263. [DOI] [PubMed] [Google Scholar]
- 4.Seksik P., Rigottier-Gois L., Gramet G., Sutren M., Pochart P., Marteau P. Alterations of the dominant faecal bacterial groups in patients with Crohn’s disease of the colon. Gut. 2003;52:237–242. doi: 10.1136/gut.52.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frank D.N., St Amand A.L., Feldman R.A., Boedeker E.C., Harpaz N., Pace N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U.S.A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sokol H., Cosnes J., Chazouilleres O., Beaugerie L., Tiret E., Poupon R. Disease activity and cancer risk in inflammatory bowel disease associated with primary sclerosing cholangitis. World J. Gastroenterol. 2008;14:3497–3503. doi: 10.3748/wjg.14.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gevers D., Kugathasan S., Denson L.A., Vazquez-Baeza Y., Van Treuren W., Ren B. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miyoshi J., Sofia M.A., Pierre J.F. The evidence for fungus in Crohn’s disease pathogenesis. Clinical journal of gastroenterology. 2018;11:449–456. doi: 10.1007/s12328-018-0886-9. [DOI] [PubMed] [Google Scholar]
- 9.Main J., McKenzie H., Yeaman G.R., Kerr M.A., Robson D., Pennington C.R. Antibody to Saccharomyces cerevisiae (bakers’ yeast) in Crohn’s disease. Br. Med. J. 1988;297:1105–1106. doi: 10.1136/bmj.297.6656.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sendid B., Quinton J.F., Charrier G., Goulet O., Cortot A., Grandbastien B. Anti-Saccharomyces cerevisiae mannan antibodies in familial Crohn’s disease. Am. J. Gastroenterol. 1998;93:1306–1310. doi: 10.1111/j.1572-0241.1998.00415.x. [DOI] [PubMed] [Google Scholar]
- 11.McKenzie H., Main J., Pennington C.R., Parratt D. Antibody to selected strains of Saccharomyces cerevisiae (baker’s and brewer’s yeast) and Candida albicans in Crohn’s disease. Gut. 1990;31:536–538. doi: 10.1136/gut.31.5.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Standaert-Vitse A., Jouault T., Vandewalle P., Mille C., Seddik M., Sendid B. Candida albicans is an immunogen for anti-Saccharomyces cerevisiae antibody markers of Crohn’s disease. Gastroenterology. 2006;130:1764–1775. doi: 10.1053/j.gastro.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 13.Richard M.L., Lamas B., Liguori G., Hoffmann T.W., Sokol H. Gut fungal microbiota: the Yin and Yang of inflammatory bowel disease. Inflamm. Bowel Dis. 2015;21:656–665. doi: 10.1097/MIB.0000000000000261. [DOI] [PubMed] [Google Scholar]
- 14.Underhill D.M., Iliev I.D. The mycobiota: interactions between commensal fungi and the host immune system. Nat. Rev. Immunol. 2014;14:405–416. doi: 10.1038/nri3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iliev I.D., Leonardi I. Fungal dysbiosis: immunity and interactions at mucosal barriers. Nat. Rev. Immunol. 2017;17:635–646. doi: 10.1038/nri.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang T.T., Shao T.Y., Ang W.X.G., Kinder J.M., Turner L.H., Pham G. Commensal fungi recapitulate the protective benefits of intestinal bacteria. Cell Host Microbe. 2017;22:809–816 e4. doi: 10.1016/j.chom.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardison S.E., Brown G.D. C-type lectin receptors orchestrate antifungal immunity. Nat. Immunol. 2012;13:817–822. doi: 10.1038/ni.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sancho D., Reis e Sousa C. Signaling by myeloid C-type lectin receptors in immunity and homeostasis. Annu. Rev. Immunol. 2012;30:491–529. doi: 10.1146/annurev-immunol-031210-101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen L.D., Viscogliosi E., Delhaes L. The lung mycobiome: an emerging field of the human respiratory microbiome. Front. Microbiol. 2015;6:89. doi: 10.3389/fmicb.2015.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romani L., Zelante T., Palmieri M., Napolioni V., Picciolini M., Velardi A. The cross-talk between opportunistic fungi and the mammalian host via microbiota’s metabolism. Semin. Immunopathol. 2015;37:163–171. doi: 10.1007/s00281-014-0464-2. [DOI] [PubMed] [Google Scholar]
- 21.Jawhara S., Poulain D. Saccharomyces boulardii decreases inflammation and intestinal colonization by Candida albicans in a mouse model of chemically-induced colitis. Med. Mycol. 2007;45:691–700. doi: 10.1080/13693780701523013. [DOI] [PubMed] [Google Scholar]
- 22.Iliev I.D., Funari V.A., Taylor K.D., Nguyen Q., Reyes C.N., Strom S.P. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336:1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sokol H., Leducq V., Aschard H., Pham H.P., Jegou S., Landman C. Fungal microbiota dysbiosis in IBD. Gut. Jun 2017;66(6):1039–1048. doi: 10.1136/gutjnl-2015-310746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Terciolo C., Dapoigny M., Andre F. Beneficial effects of Saccharomyces boulardii CNCM I-745 on clinical disorders associated with intestinal barrier disruption. Clin. Exp. Gastroenterol. 2019;12:67–82. doi: 10.2147/CEG.S181590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leech B., McIntyre E., Steel A., Sibbritt D. Risk factors associated with intestinal permeability in an adult population: a systematic review. Int. J. Clin. Pract. 2019:e13385. doi: 10.1111/ijcp.13385. [DOI] [PubMed] [Google Scholar]
- 26.Li Q., Wang C., Tang C., He Q., Li N., Li J. Dysbiosis of gut fungal microbiota is associated with mucosal inflammation in Crohn’s disease. J. Clin. Gastroenterol. 2014;48:513–523. doi: 10.1097/MCG.0000000000000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liguori G., Lamas B., Richard M.L., Brandi G., da Costa G., Hoffmann T.W. Fungal dysbiosis in mucosa-associated microbiota of Crohn’s disease patients. J Crohns Colitis. 2016;10:296–305. doi: 10.1093/ecco-jcc/jjv209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiaro T.R., Soto R., Zac Stephens W., Kubinak J.L., Petersen C., Gogokhia L. A member of the gut mycobiota modulates host purine metabolism exacerbating colitis in mice. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aaf9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foligne B., Dewulf J., Vandekerckove P., Pignede G., Pot B. Probiotic yeasts: anti-inflammatory potential of various non-pathogenic strains in experimental colitis in mice. World J. Gastroenterol. 2010;16:2134–2145. doi: 10.3748/wjg.v16.i17.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cross M.L., Ganner A., Teilab D., Fray L.M. Patterns of cytokine induction by gram-positive and gram-negative probiotic bacteria. FEMS Immunol. Med. Microbiol. 2004;42:173–180. doi: 10.1016/j.femsim.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 36.Martins F.S., Nardi R.M., Arantes R.M., Rosa C.A., Neves M.J., Nicoli J.R. Screening of yeasts as probiotic based on capacities to colonize the gastrointestinal tract and to protect against enteropathogen challenge in mice. J. Gen. Appl. Microbiol. 2005;51:83–92. doi: 10.2323/jgam.51.83. [DOI] [PubMed] [Google Scholar]
- 38.Liti G., Carter D.M., Moses A.M., Warringer J., Parts L., James S.A. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liti G. The fascinating and secret wild life of the budding yeast S. cerevisiae. eLife. 2015;4 doi: 10.7554/eLife.05835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goddard M.R., Greig D. Saccharomyces cerevisiae: a nomadic yeast with no niche? FEMS Yeast Res. 2015;15 doi: 10.1093/femsyr/fov009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramazzotti M., Stefanini I., Di Paola M., De Filippo C., Rizzetto L., Berna L. Population genomics reveals evolution and variation of Saccharomyces cerevisiae in the human and insects gut. Environ. Microbiol. 2018 doi: 10.1111/1462-2920.14422. [DOI] [PubMed] [Google Scholar]
- 42.Aomatsu T., Yoden A., Matsumoto K., Kimura E., Inoue K., Andoh A. Fecal calprotectin is a useful marker for disease activity in pediatric patients with inflammatory bowel disease. Dig. Dis. Sci. 2011;56:2372–2377. doi: 10.1007/s10620-011-1633-y. [DOI] [PubMed] [Google Scholar]
- 43.Sebastiani F., Barberio C., Casalone E., Cavalieri D., Polsinelli M. Crosses between Saccharomyces cerevisiae and Saccharomyces bayanus generate fertile hybrids. Res. Microbiol. 2002;153:53–58. doi: 10.1016/s0923-2508(01)01286-4. [DOI] [PubMed] [Google Scholar]
- 44.Gagliardi M.C., Teloni R., Mariotti S., Bromuro C., Chiani P., Romagnoli G. Endogenous PGE2 promotes the induction of human Th17 responses by fungal ss-glucan. J. Leukoc. Biol. 2010;88:947–954. doi: 10.1189/jlb.0310139. [DOI] [PubMed] [Google Scholar]
- 45.Dallies N., Francois J., Paquet V. A new method for quantitative determination of polysaccharides in the yeast cell wall. Application to the cell wall defective mutants of Saccharomyces cerevisiae. Yeast. 1998;14:1297–1306. doi: 10.1002/(SICI)1097-0061(1998100)14:14<1297::AID-YEA310>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 46.De Filippo C., Cavalieri D., Di Paola M., Ramazzotti M., Poullet J.B., Massart S. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. U.S.A. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andersson A.F., Lindberg M., Jakobsson H., Backhed F., Nyren P., Engstrand L. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PloS One. 2008;3 doi: 10.1371/journal.pone.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Albanese D., Fontana P., De Filippo C., Cavalieri D., Donati C. MICCA: a complete and accurate software for taxonomic profiling of metagenomic data. Sci. Rep. 2015;5:9743. doi: 10.1038/srep09743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Caporaso J.G., Bittinger K., Bushman F.D., DeSantis T.Z., Andersen G.L., Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bazan S.B., Walch-Ruckheim B., Schmitt M.J., Breinig F. Maturation and cytokine pattern of human dendritic cells in response to different yeasts. Med. Microbiol. Immunol. 2018;207:75–81. doi: 10.1007/s00430-017-0528-8. [DOI] [PubMed] [Google Scholar]
- 54.Brenden R.A., Miller M.A., Janda J.M. Clinical disease spectrum and pathogenic factors associated with Plesiomonas shigelloides infections in humans. Rev. Infect. Dis. 1988;10:303–316. doi: 10.1093/clinids/10.2.303. [DOI] [PubMed] [Google Scholar]
- 55.Raimondi S., Amaretti A., Gozzoli C., Simone M., Righini L., Candeliere F. Longitudinal survey of fungi in the human gut: ITS profiling, phenotyping, and colonization. Front. Microbiol. 2019;10:1575. doi: 10.3389/fmicb.2019.01575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thompson A., Orr S.J. Emerging IL-12 family cytokines in the fight against fungal infections. Cytokine. 2018;111:398–407. doi: 10.1016/j.cyto.2018.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Strati F., Di Paola M., Stefanini I., Albanese D., Rizzetto L., Lionetti P. Age and gender affect the composition of fungal population of the human gastrointestinal tract. Front. Microbiol. 2016;7:1227. doi: 10.3389/fmicb.2016.01227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rizzetto L., Ifrim D.C., Moretti S., Tocci N., Cheng S.C., Quintin J. Fungal chitin induces trained immunity in human monocytes during cross-talk of the host with Saccharomyces cerevisiae. J. Biol. Chem. 2016;291:7961–7972. doi: 10.1074/jbc.M115.699645. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequences are available in the European Nucleotide Archive (ENA) with accession number PRJEB22036 and PRJEB22343 (http://www.ebi.ac.uk/ena/data/view/PRJEB22036 and http://www.ebi.ac.uk/ena/data/view/PRJEB22343).