Abstract
Systemic lupus erythematosus (SLE) is a typical autoimmune disease characterized by chronic inflammation and pathogenic auto-antibodies. Apart from B cells, dysregulation of other immune cells also plays an essential role in the pathogenesis and development of the disease including CD4+T cells, dendritic cells, macrophages and neutrophils. Since metabolic programs control immune cell fate and function, they are critical checkpoints in an effective immune response and are involved in the etiology of autoimmune disease. In addition, mitochondria and oxidative stress are both involved in cellular metabolism and is also essential in immune response. In this review, apart from the disturbed immune system, we will discuss mitochondrial dysfunction, oxidative stress, abnormal metabolism (including glucose, lipid and amino acid metabolism) of immune cells as well as epigenetic control of metabolism reprogramming to elucidate the underlying pathogenic mechanisms of systemic lupus erythematosus.
Keywords: Systemic lupus erythematosus (SLE), Metabolic programs, Immune response, Pathogenesis
Highlights
-
•
Mitochondria plays a vital role in cellular metabolism and is involved in immune response.
-
•
There are alterations in glucose, lipid and amino acid metabolism of various immune cells in SLE patients.
-
•
Epigenetic status is influenced by the presence of metabolic intermediates and certain autoimmunity-related genes are hypomethylated in CD4+T cells, CD19+ B cells as well as CD14+ monocytes of SLE.
1. Introduction
Systemic lupus erythematosus (SLE) is a typical autoimmune disease characterized by chronic inflammation, with involvement of various organs and diverse clinical manifestations such as thrombocytopenia, rash, vasculitis, arthritis, nephritis and even neuropsychopathy [1]. Autoantibodies secreted from B cells is the main factor that contribute to the disease and cause tissue damage. However, the aberrant immune system is not limited to B cells, other immune cells, such as T cells, neutrophils, plasmacytoid dendritic cells (pDC), and macrophages, are reported to be involved in SLE pathogenesis [2,3].
Immune cells take advantage of various metabolic pathways to provide energy for cell survival and synthesize numerous effector molecules for cellular growth, proliferation and differentiation [4]. Metabolic reprograming takes place when immune cells are activated by the stimulation of intrinsic or extrinsic signals, shifting from time-consuming oxidative phosphorylation (OXPHOS) to rapid aerobic glycolysis [5]. Since immune cell function is closely associated with its intracellular metabolic pathways, the imbalanced immune system in SLE patients and lupus mouse models may present metabolic abnormalities. Previous reports have demonstrated that T cell mitochondrial dysfunction was associated with SLE disease progression [6]. Nevertheless, the metabolic abnormalities of other immune cells are less understood in SLE. Increased occurrences of metabolic syndrome are observed among lupus patients, which is closely related to both atherosclerosis and multiple organ injury [[7], [8], [9], [10]]. Metabolomics demonstrate that intermediates related to main metabolic pathways were altered in patients with SLE by analyzing blood and urine samples [[11], [12], [13]]. This review aims to elucidate main metabolism pathways as well as epigenetic regulation of metabolic reprogramming involved in lupus, addressing the pathogenesis of SLE from the perspective of immunometabolism.
2. Disturbed immune system in SLE
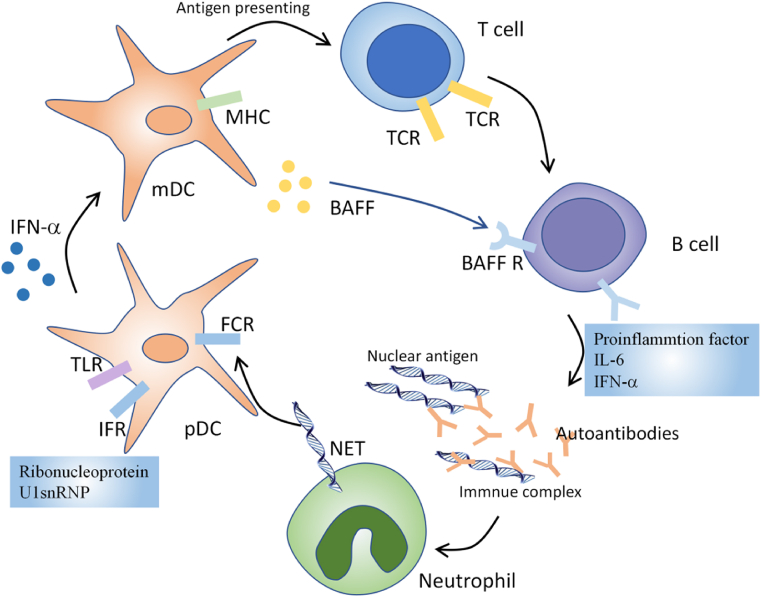
SLE is characterized by immune system activation, including autoantibody synthesis, immune complex accumulation and infiltration of proinflammatory cells [14]. Diverse immune cells and inflammatory mediators have been proven to be deleterious players in the pathogenesis of SLE (Fig. 1). Increased apoptosis and defective clearance are observed in SLE patients. It contributes to self-DNA and nuclear antigens exposition and promotes activation of multiple innate immune cells. Nuclear particles mimic viral particles and activate Toll like receptors (TLR) on antigen-presenting cells, mainly dendritic cells and promote their maturation [[15], [16], [17]]. Persistent activation of dendritic cells by lupus autoantigens induces T cell activation and proliferation. Activated T cells then lead to mature autoreactive B cells [18,19]. Besides, B cell activating factor (BAFF) and its homolog, a proliferation-inducing ligand (APRIL), can support B cell differentiation, plasma cell survival and regulate immunoglobulin class switching [20,21]. Furthermore, ribonucleoprotein and U1snRNP can induce type I IFN secretion by pDCs in SLE [22]. IFN-α upregulates TLR7 and IRF7 expression in pDC, mDC and monocytes, thus enhancing the immune response to nucleic-acid-containing immune complexes [23]. Besides, IFN-α also contributes to the maturation of mDC [24]. BlyS/BAFF can also be induced by IFN-α and promotes peripheral mature B cells survival. IFN-α can also promote the differentiation of activated B cells into plasmablasts. With the help of IL-6, IFN-α enables plasmablasts to develop into antibody-secreting plasma cells [25]. It is noteworthy that autoantibodies can bind to nuclear antigens, form immune complex and activate innate immune cells, which is a positive feedback loop and amplifies the pathogenic processes in SLE.
Fig. 1.
Disturbed immune system in SLE. Nuclear particles activate Toll like receptors (TLR) on antigen-presenting cells, mainly dendritic cells and promote their maturation. Persistent activation of dendritic cells induces T cell activation and proliferation. Activated T cells then lead to mature autoreactive B cells. Furthermore, ribonucleoprotein and U1snRNP can induce type I IFN secretion by pDCs in SLE, which promotes the differentiation of activated B cells into plasmablasts and antibody-secreting plasma cells. Autoantibodies can bind to nuclear antigens, form immune complex and activate innate immune cells, which is a positive feedback loop and amplifies the pathogenic processes in SLE.
3. Mitochondrial dysfunction, oxidative stress, and hypoxia
Mitochondria plays a vital role in cellular metabolism and is reported to be essential in immune response. It not only acts as an energy machinery but also is a signal-transducing organelle [[26], [27], [28]]. Mitochondrial hyperpolarization and reactive oxygen intermediates production were detected in peripheral blood T lymphocytes from SLE patients, together with diminished levels of intracellular ATP, all of which indicated a dysfunction in T cell mitochondria in lupus patients [29]. CD4+T cells from SLE exhibit an increased mitochondrial mass and size due to increased mitochondrial biogenesis and defective mitophagy [30]. Mitochondrial remodeling determines metabolic alterations and status of T cells. For instance, switch from oxidative phosphorylation to aerobic glycolysis is simultaneously accompanied by change from mitochondrial fussion to fission [31]. Surface glycoprotein CD3ζ chain is degraded and replaced by FcεRIγ chain in SLE T cells due to its oxidative stress. The homologous FcεRIγ can promote tyrosine-protein kinase SYK recruitment and enhance signaling upon T cell receptor activation [1]. There is a therapeutic effect with the treatment of N-acetylcysteine which protect against the oxidative stress in the mitochondria by elevating levels of glutathione and NADPH in T cells of SLE [32].
Sle1c2, a lupus susceptibility locus in mice, is associated with a decreased level of ESRRG (mitochondrial metabolism regulator) and mitochondrial dysfunction [33]. UCP2, a gene involved in both mitochondrial ATP production and reactive oxygen species (ROS) generation, is closely associated with SLE [34]. Neutrophil extracellular trap (NET) was first described by Brinkmann as neutrophil-derived extracellular structures [35]. The enhanced NETosis of low density granulocytes as well as impaired removal of NET have been reported in SLE [36,37]. Ribonucleoprotein immune complex induce mitochondrial membrane hyperpolarization and ROS generation, resulting in both NET formation and oxidation of mitochondrial DNA (mtDNA). Extracellular oxidized mtDNA is a potent proinflammatory mediator in vitro and induces type-I interferon (IFN) signaling pathway in mice models. On the contrary, mitochondrial ROS inhibition in vivo reduces disease severity and attenuates type-I IFN responses in a lupus mouse model. These facts have emphasized a role of mitochondria involvement in the pathogenesis of SLE. Accordingly, decreased spontaneous NETosis and reduced disease activity was reported in MRL/lpr mice by treatment with a mitochondrial - ROS scavenger [38,39]. DCs contribute to the SLE pathogenesis through indirect impacts on T cells. In DCs, mTORC1 activation accelerated their maturation by a Myc-dependent metabolic signal pathway, which is associated with increased ROS production. The impaired metabolism of DCs promotes their maturation and accelerates T cell activation in SLE, thus influencing disease progress and severity [[40], [41], [42]].
Hypoxia regulates immunometabolism in multiple ways which are dependent on the transcription factor HIF-1α [43,44]. Under hypoxic conditions, HIF-1α was accumulated due to inactivation of prolyl hydroxylases, an enzyme responsible for HIF-1α ubiquitylation and proteosomal degradation. HIF-1 increases levels of multiple genes involved in cell adaptations to hypoxia. HIF-1α can be upregulated via mTOR at the protein level or via STAT3 and NF-κB signaling at the mRNA level. HIF-1α has been demonstrated to increase the rate of glycolysis by upregulating glycolytic gene expression and is required for Th1 and Th17 cell differentiation [45,46]. Nevertheless, HIF-1α exerts both positive and negative effects on Treg cell differentiation [47,48]. ROS have been demonstrated to modulate the HIF pathway although the exact mechanism remains unclear [49].
4. Metabolism in immune cells in SLE
4.1. Glucose metabolism in immune cells in SLE
Glucose constitutes the fundamental energy source for most cells and is closely related to cell proliferation, growth and survival. Activated T cells enhances glucose metabolism dramatically to generate enough energy and synthesize intermediate materials to meet the requirement of cell proliferation and differentiation [50]. Glucose deprivation leads to decreased cellular ATP levels and the serine/threonine kinase AMPK activation [51]. AMPK activation has a positive regulatory effect on signaling pathways which compensate for cellular ATP. For instance, AMPK activation enhances both Glut 4 transcription and its translocation, and promotes glucose intake. In addition, it also accelerates catabolism such as fatty acid oxidation and glycolysis through ACC inhibition and PFK2 activation. AMPK negatively modulates certain key proteins in ATP-consuming reactions such as mTORC2, glycogen synthase, Sterol regulatory element binding protein 1 (SREBP-1) and TSC2 (tuberous sclerosis 2), leading to inhibition of gluconeogenesis as well as glycogen, lipid, and protein synthesis [[52], [53], [54], [55]].
However, inhibition of AMPK and the downstream mTORC1 activation by Roquin-1 promotes T helper follicular cell differentiation [56] and a lupus-prone phenotype [57]. mTOR 1 activation can be triggered not only by mitochondrial dysfunction, but also the PPP overactivation, which is correlated with the metabolic need of activating T cells [58]. mTOR is an essential metabolic sensor that regulates cell growth and energy utilization and is required for polarization into Th1 and Th17 subsets [59].
Chronically activated CD4+T cells from healthy individuals, CD4+T cells from SLE patients or lupus-like mice model all exhibit high levels of oxygen consumption. Nevertheless, acutely activated T cells utilize glycolysis as their main metabolic pathway [60]. These results imply that the chronic stimulation by autoantigens in lupus rely on OXPHOS, whereas the acute activation of T cells by foreign antigens or the in vitro TCR stimulation is supported by the aerobic glycolysis. Previous studies showed that elevated glucose metabolism and mitochondrial respiration was observed in effector memory (EM) CD4+T cells from healthy controls for cell survival, differentiation, proliferation as well as IFNγ production. Consistently, EM CD4+T cell subsets are featured by both glycolysis and OXPHOS [61]. Previous reports have demonstrated that the proportion of EM CD4+T cells is expanded in SLE patients [62], which may account for the resemblance between the metabolism of normal EM CD4+T cells and that of SLE CD4+T cells. It is noteworthy that naïve CD4+T cells in lupus-prone mice also had enhanced glycolysis and OXPHOS. These results indicate that an altered intrinsic metabolism reprograms exist in SLE T cells, including increased glycolysis and OXPHOS. SLE T cells share the EM metabolic characteristics, which may contribute to their hyperactive status [63].
Additionally, glucose transporters are expressed on T cells surface. TCR and CD28 stimulation induces GLUT1 expression, which is associated with increased glucose uptake and glycolysis [64]. GLUT1 overexpression has not been observed in human SLE and lupus mice model while GLUT1 is linked to activated CD4+T cells accumulation and antibodies production [65,66]. Additionally, GLUT1 overexpression in CD4+T cells caused effector T cells expansion, whereas AMPK activation reduces Glut1 levels and increases Treg cells. This has revealed that there is a difference in glucose metabolism for effector and regulatory T cells [67]. HIF1α not only controls the cellular response to hypoxia, but also induces GLUT1 expression which is essential for Th17 differentiation [68].
Currently, glucose metabolic in SLE immune cells are mainly focused on T cells. In fact, glucose signaling pathway is also critical to other immune cells (Fig. 2). Just as CD4+T cells, the majority of activated B cells are glycolytic [69], but the detailed mechanism is still poorly understood. It was confirmed that the enforced expression of mTORC1 can lead to plasma cell differentiation. mTORC1 is activated in the B cells of lupus-prone mice and rapamycin can inhibit B lymphocyte proliferation and survival [70,71], indicating that mTOR is associated with the pathogenic autoantibody production. Overexpression of B cell activating factor (BAFF) increase the lupus-like autoantibodies in a transgenetic mouse model, and B cells in this mouse model exhibit highly glycolytic phenotype [72].
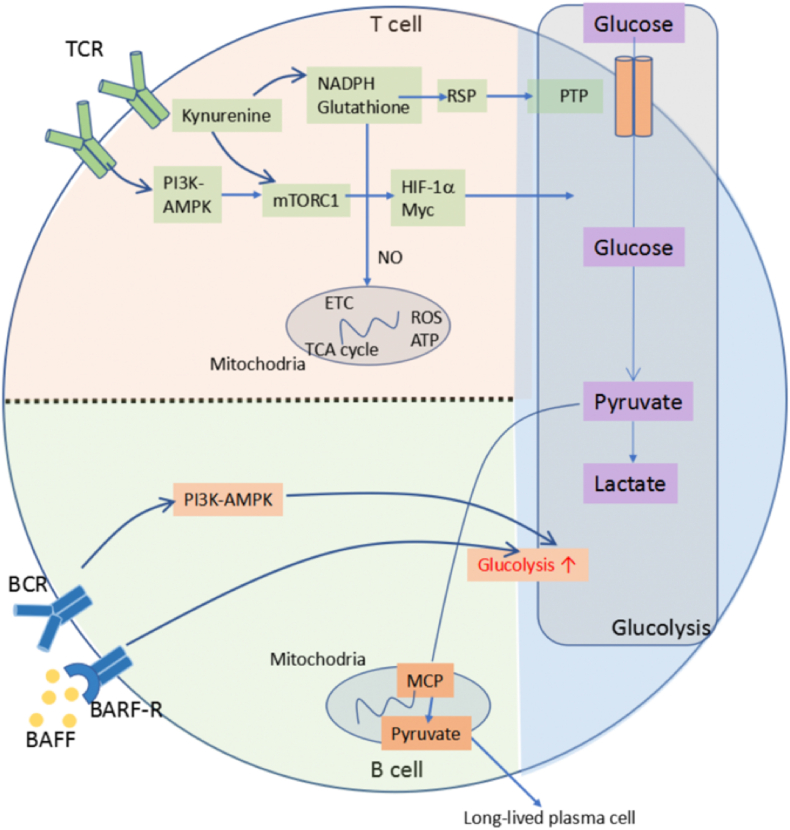
Fig. 2.
Glucose metabolic pathways in immune cells. The glucose metabolic pathway includes both glycolysis and oxidative phosphorylation. T cell receptor (TCR) stimulation activates mechanistic target of rapamycin complex 1 (mTORC1) through PI3K-AMPK pathway. Low levels of NADPH and glutathione leads to increased levels of mitochondrial reactive oxygen species (ROS) and decreased levels of ATP. It also contributes to mTORC1 activation, directly or through elevated levels of kynurenine. mTORC1 activation facilitates glucose metabolism through hypoxia-inducible factor 1α (HIF-1α) and Myc. In B cells, B cell activating factor (BAFF) and B cell receptor (BCR) signals increase glucose metabolism and glycolysis. This promotes pyruvate influx into the mitochondria, which is essential for the survival of long-lived plasma cells.
4.2. Lipid metabolism in immune cells in SLE
Latest evidences suggest that mice with high-fat diet culminate in cholesteral accumulation in spleens and lymph nodes as well as auto-antibody production [73]. It indicates that lipid metabolism also exerts a fundamental role in the immune responses and pathogenesis of autoimmunity (Fig. 3). Cholesterol and glycosphingolipids are significant constituents of lipid rafts of the cell plasma membrane and are aggregated in T cell from SLE patients [74]. Actually, SLE T cells are featured by increased glycosphingolipid synthesis, which has a close association with TCR activation. Inhibiting glycosphingolipid synthesis not only reduces T cell activation in vitro, but also decreases anti-dsDNA antibody titres in SLE patients [75]. Notably, there is disturbed glycosphingolipid metabolism in the renal specimen of MRL/lpr mice and SLE patients due to over-expression of two enzymes, β-1,4-galactosyltransferase 5 (β4GalT-5) and neuraminidase 1 (NEU1) [76]. As a nuclear receptor which regulates cellular lipid metabolism and trafficking, the oxysterols receptor LXR is responsible for glycosphingolipids accumulation in T cells from SLE patients [75]. Since LXR exerts both proinflammatory and anti-inflammatory functions [77], the role of LXR remains elusive in SLE development. It remains to be explored that whether LXR signaling drives disturbed GSL homeostasis or LXR is activated as a compensatory mechanism for the dysregulated cholesterol metabolism in autoimmune T cells.
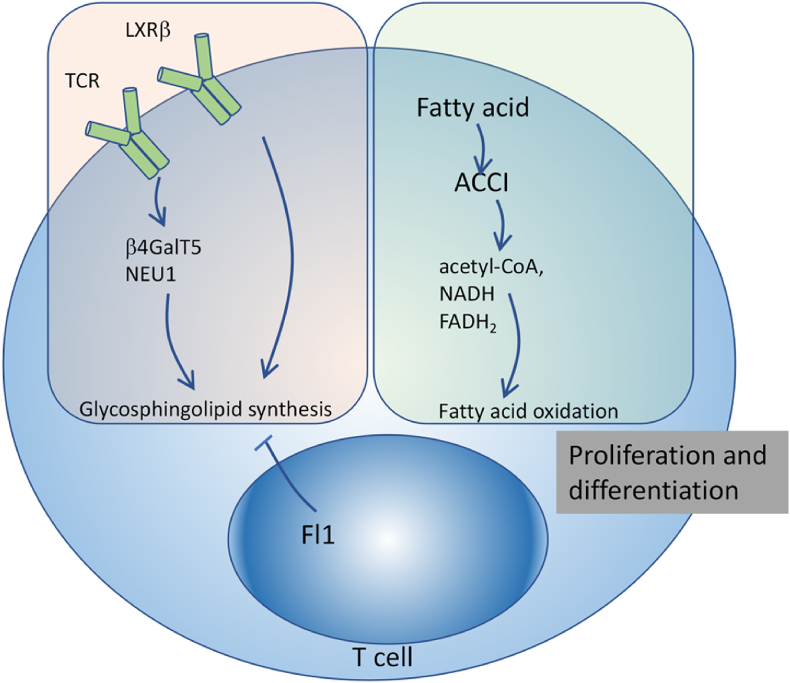
Fig. 3.
Lipid metabolic pathways in immune cells. Fatty acid oxidation pathway converts fatty acids into multiple intermediates (including acetyl-CoA, NADH and FADH2) for energy generation. Fatty acid synthesis is essential for activation-induced proliferation and differentiation of effector T cells.
FLI1 is a transcription factor expressed in T cells that regulates glycosphingolipids synthesis. It has been reported that an alteration in FLI1 promoter region resulted in elevated FLI1 levels and predisposition to SLE. Accordingly, FLI1-haplodeficiency reduces disease severity in MRL/lpr mice, which is accompanied by decreased T cell activation [78,79]. Sterol, a specific type of fatty acid, has the capacity to regulate Th17 cell differentiation via RORγ activation [80]. In clinical settings, A reduced Th17 polarization and a increased Treg cell expansion are observed in multiple sclerosis and RA patients respectively, who have received treatment with statins [81,82]. Besides, statins have proved to not only decrease cardiovascular morbidity but also stabilize renal function in SLE patients [83].
It is konwn that fatty acid oxidation pathway converts fatty acids into multiple intermediates (including acetyl-CoA, NADH and FADH2) for energy generation while fatty acid synthesis pathway produces lipids for cellular growth and proliferation [4]. In addition, fatty acid oxidation and synthesis also exert opposite roles in immune system. Fatty acid oxidation is favorably utilized by non-inflammatory and tolerogenic immune cells while fatty acid synthesis is featured by inflammatory responses [84,85].
Fatty acid synthesis is essential for activation-induced proliferation and differentiation of effector T cells, which is determined by acetyl-CoA carboxylase I (ACCI). ACCI-konck out mice are immune from autoimmune encephalitis, a model of multiple sclerosis with dominance of Th17 cells [86,87]. In light of Th17 involvement in SLE, it is worthwhile to investigate fatty acid production in SLE T cells. Fatty acid oxidation provides large energy for Treg cells and memory CD8+T cells [88]. Furthermore, Fatty acid oxidation has been reported to regulate the inflammatory functions of macrophages and macrophage differentiation [89]. The abnormal deposition of fatty acids and their derivative lipoproteins in macrophages correlate well with foam cell synthesis and pathologic inflammation [90]. It has been demonstrated that the elevated intracellular levels of unsaturated fatty acids such as oleic acid, linoleic acid and arachidonic acid, induces IL-1α secretion in foam cells, leading to aberrant inflammation in vivo [91].
Macrophages, specialized phagocytic cells, are able to uptake various kinds of lipids (LDL, VLDL, and oxidized lipoproteins) through processes such as phagocytosis, macropinocytosis, and scavenger receptor-mediated pathways [92]. Macrophages from SLE patients are reported to have impaired phagocytic ability. There is a feed-forward loop between NETs and macrophages in SLE patients. NETs, together with its constituent peptide LL-37, activates the inflammasome and induces IL-18 and IL-1β secretion. The released cytokines can in turn stimulate neutrophils to undergo NETosis and amplify the loop, thus producing multiple proinflammatory cytokines [93].
4.3. Amino acids metabolism in immune cells in SLE
Amino acids and their metabolism play a vital role in immune function (Fig. 4). Particularly, glutamine catabolism regulates immune cell function in various aspects [94]. Adequate amounts of glutamine have been demonstrated to be necessary for IL-1 induction by macrophages upon LPS stimulation [95]. Interestingly, recent reports has shown that most glutamine entered into the TCA cycle and the hexosamine pathway and induces M2 macrophage polarization upon IL-4 stimulation. Nevertheless, glutamine is not a requisite for the development of LPS-stimulated M1 macrophages [96].
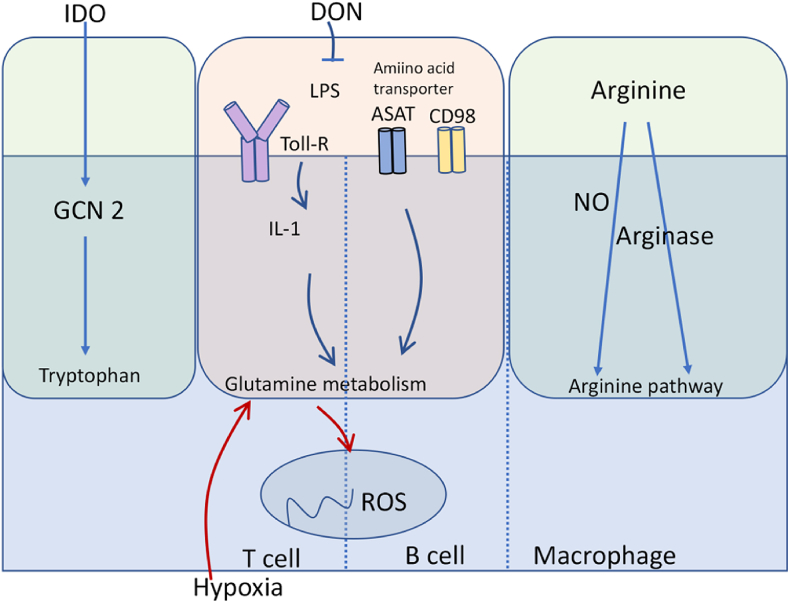
Fig. 4.
Amino acid metabolic pathways in immune cells. Amino acids and their metabolism play a vital role in immune function. Glutamine has been demonstrated to be necessary for IL-1 induction upon LPS stimulation. Indoleamine-2,3-dioxygenase (IDO), is responsible for tryptophan catabolism through General control nonderepressible 2 (GCN2). The arginine pathway is mainly modulated by Arg-degrading enzymes such as NO synthase.
Glutamine metabolism also modulates immune responses of both T cells and B cells. Besides, both T cell and B cell activation involves large glutamine consumption and requires glutamine in response to antigen receptor stimulation [97,98]. In regard to T cells, heterozygous knockout of glutaminase leads to increased ROS levels which are increased upon hypoxia. This implicates that glutamine metabolism is helpful in controlling ROS stress [99]. Amino acid transporters are fundamental for effector T cell differentiation and function, such as ASCT for glutamine and CD98 for branched amino acids [100]. Glutaminolysis is indispensable for maintaining T cell activation and proliferation. Blockage of glutamine with the 6-diazo-5-oxo-l-norleucine (DON) inhibits activation-induced proliferation in vitro [101]. Enzymes involved in glutaminolysis are significantly elevated in CD4+T cells from lupus-prone TC mice, suggesting that it leads to increased OXPHOS in these cells and that DON treatment may also be therapeutic for SLE T cells [63].
Tryptophan is another amino acid with significant role in sustaining immune function. Previous studies have demonstrated that high levels of exogenous tryptophan led to an autoimmune phenotype characterized by eosinophil dysfunction in animal models [102,103]. Indoleamine-2,3-dioxygenase (IDO), is a rate-limiting enzyme responsible for tryptophan catabolism. General control nonderepressible 2 (GCN2) is a downstream effector of IDO. It is a metabolic-stress sensing kinase eIF-2α-kinase and is essential to protect from autoimmunity through regulation of T cell responses [104]. The protective role of GCN2 against glomerular inflammation has also been reported in kidneys of nephritis mice induced by immune complex [105]. Indeed, preliminary investigations suggest that halofuginone, a GCN2 agonist, could suppress systemic autoimmunity in animal models [106]. Increased IDO activity and elevated Trp degradation are observed in SLE patients [107].
Macrophages utilize arginine in two main metabolic pathways, the nitric oxide synthesis pathway through classical activation and the arginase pathway through alternative activation [108]. The nitric oxide synthesis pathway is associated with an inflammatory M1 phenotype and inducible nitric oxide synthase (iNOS) mediates this process [109]. The production of α-ketoglutarate (αKG) via glutaminolysis is essential for M2 macrophages polarization [110]. Metabolically, M1 macrophages exhibit a glycolytic phenotype. Nevertheless, M2 macrophages employ fatty acid oxidation and mitochondrial respiration to satisfy their functional needs. Consequently, M2 macrophages have higher basal mitochondrial oxygen consumption rates [111,112]. However, whether altered metabolic profile is involved in the pathogenesis of SLE remains elusive.
Myeloid derived suppressor cells (MDSCs) are a heterogeneous cell population involved in cancer, inflammation and infection, with a distinct capacity to suppress T-cell responses [113]. An obvious increased percentage of peripheral MDSCs is observed in active SLE patients, which has a positive correlation with serum arginase-1 (Arg-1) activity, Th17 differentiation and disease severity [114]. Moreover, MDSCs are necessary for the induction of Th17 responses and are related to renal injuries in an Arg-1-dependent fashion. This has suggested an Arg-1-dependent effect of MDSCs and its pathogenic function in human SLE. Thus targeting MDSCs or Arg-1 could be a promising therapy for SLE. In another study, administration of MDSCs from healthy mice into roquinsan/san mice, a lupus mice model, led to reduced levels of serum anti-ds-DNA antibodies and decreased proteinuria. In addition, expansion of regulatory B cells and decreased follicular helper T cells, Th1 cells, and Th17 cells were also observed simultaneously [115]. In this case, the therapeutic effects were inducible NOS (iNOS)-dependent and Arg1-independent. There might be important differences between human SLE and experimental models. Endothelial nitric oxide synthase (eNOS) deficiency in MRL/lpr mice aggravates renal lesions [116]. Patients with lupus nephritis are featured by increased levels of iNOS and decreased levels of eNOS [117,118].
5. Epigenetic control of metabolism reprogramming in SLE
Gene expression can be altered by epigenetic modifications, such as methylation/demethylation of DNA and histones and acetylation/deacetylation of histone and nonhistone proteins, thus regulating immune response in lupus. However, these epigenetic regulations are reversible and are influenced by the presence of metabolic intermediates [[119], [120], [121]].
Certain autoimmunity-related genes are hypomethylated in CD4+ T cells, CD19+ B cells, and CD14+ monocytes in SLE [[122], [123], [124]]. MX1, IFI44L, NLRC5 and PLSCR1 genes were confirmed to be hypomethylated by microarray techniques. These genes were overexpressed in the type I interferon signaling pathway, which is relevant to the pathogenesis of SLE [125]. Particularly, abnormal DNA hypomethylation in T cells is an obvious epigenetic hallmark in SLE. Richardson had discovered that inhibition of DNA methylation by 5-azacytidine (5-azaC) induced autoreactive CD4+T cells and autoimmune syndrome [126]. Methylation sensitive genes include CD11a (ITGAL), perforin (PRF1), CD70 (TNFSF7) and CD40 ligand (TNFSF5) in lupus T cells. Various mechanisms may account for DNA hypomethylation in lupus T cells, such as certain miRNAs, RFX1, defective ERK pathway signaling, Gadd45α and DNA hydroxymethylation. For instance, increased miR-126 levels contributes to decreased DNA methyltransferases (DNMT1) expression in lupus CD4+T cells; Recruitment of less DNMT1 to the promoter regions of certain methylation-sensitive genes is associated with diminished activity and amount of the transcription factor RFX1; Defective ERK pathway signaling leads to reduced DNMT1 expression in lupus CD4+T cells; Gadd45α mainly acts as a DNA demethylator in lupus [123].
Oxidative stress is, to some extent, responsible for the impaired activity of DNA methyltransferases (DNMTs) in SLE and it is also essential in the mitochondrial disorder in SLE T cells [127]. The regulation of DNA or histone methylation in lupus is determined by the linkage between dynamically altered methylation/demethylation and metabolic intermediates. Sera of SLE patients have exhibited decreased levels of metabolites derived from methyl group donors, indicating that DNA hypomethylation might be due to defective S-adenosyl-l-methionine (SAM) cycle [128]. Studies have also shown that there is a remarkable amelioration of splenomegaly, lymphadenopathy, autoantibody titers, as well as renal IgG accumulation and inflammatory cell infiltration in lupus mouse model upon 5′-Deoxy-5-methylthioadenosine (MTA) treatment [129].
The JMJC domain-containing histone demethylases are capable to remove histone lysine methylation and therefore regulate gene expression [130,131]. Iron Fe(II) and α-ketoglutarate (αKG) are indispensable cofactors which are required for the oxidative demethylation reaction via hydroxymethyl lysine. ROS can oxidize Fe(II) to Fe(III) and decrease JMJC domain-containing histone demethylases activity [132,133]. This leads to the enhanced H3K27me3 levels of a kinase promoter, resulting in activated T cell and B cell in SLE patients [134]. Previous studies have demonstrated that mTOR influences histone demethylase activity through modulation of HIF1 expression, which enhances JMJC demethylase activity [135]. mTOR signaling is affected by metabolites and microenvironment. It is a master regulator that senses and integrates diverse nutritional and environmental signals, including growth factors, amino acids, energy levels as well as cellular stress [136]. mTOR is sensitive to hyperglycaemia and amino acids and therefore enhances HIF1 transcription [137]. Conversely, HIF1 expression is inhibited by EgIN prolyl hydroxylases [138], which can be induced by α-KG, but suppressed by succinate and fumarate, all of which are products of the TCA cycle [139]. In conclusion, demethylase enzyme activity is regulated by these metabolic intermediates, which implies the significance of mitochondrial dysfunction in SLE [140].
Apart from DNA methylation, acetylation of histone and non-histone proteins also has a close relationship with the development of lupus. Enhanced oxidative metabolism and increased levels of acetyl-CoA have been detected in SLE patients [141]. Acetyl-CoA is proved to affect both p300 acetyltransferase activity and p300 structure [142,143]. P300 acetyltransferase is important for sustaining self-tolerant B lymphocytes in mice models. Conditional deletion of p300 in B cells induces the presence of a lupus-like syndrome in mice, with elevated autoantibody levels and typical autoimmune-related phenotypes. This implies acetyl-CoA metabolites may contribute to lupus pathogenesis by modulating p300.
6. Interconnection of the metabolic pathways in SLE
The metabolic processes of glucose, fatty acid and amino acid are interlinked and can be co-regulated. mTOR is a target of interest which regulates both glycolysis and fatty acid synthesis in activated immune cells, mainly T cells and B cells [60,69]. Additionally, mTOR can sense amino acids and growth factors, promote mRNA translation and lipid synthesis to support cellular growth. Disturbed tryptophan metabolism could enhance CD4+ T cell activation, since kynurenine, a tryptophan metabolite, could activate mTORC1 in CD4+ T cells [144]. The elevated level of kynurenine in SLE patients is due to impaired degradation of kynurenine by NADPH-dependent kynurenine hydrolase [145]. In support of this mechanism, N-acetylcysteine treatment, which restores NADPH levels, significantly decreased kynurenine levels in peripheral blood lymphocytes [145]. In addition, mitochondrial dysfunction, over-reactivity of the pentose phosphate pathway (PPP) and transaldolase activity, as well as accumulation of kynurenine, can lead to mTORC1 activation in T cells of SLE patients [146]. Ribose-5-phosphate, produced from an over-reactive PPP in SLE patients is preferentially metabolized into ribose 1,5-bisphosphate instead of phosphoribosyl pyrophosphate (PRPP), which leads to reduced biosynthesis of amino acids, pyrimidines and purines [147]. In summary, metabolic changes are key to cell immune function.
7. Conclusion
Apart from the disturbed immune system and mitochondrial dysfunction, metabolism (including glucose, lipid and amino acid metabolism) of immune cells, as well as epigenetic, control of metabolism reprogramming is also abnormal in SLE patients. With the studies of SLE patients and mouse models, various cell types function through different metabolic ways, which indicates that cellular metabolism is, to some extent, a cell-intrinsic process. Novel drugs that modulate cell metabolic processes might ameliorate the aberrant immune response and be used to treat SLE patients. Therapy targeting mTOR activation with rapamycin or N-acetylcysteine could be a promising way to reduce the disease severity in SLE patients [32]. Regulation of the fatty acid pathways, such as glucocorticoid treatment, has been directly linked to reduce leptin levels through inhibition of mTOR in SLE patients [18]. In recent studies, DON treatment, targeting MDSCs or Arg-1 might be promising therapies for SLE [63,101,113,114]. In conclusion, metabolic pathways are potential targets for therapy in SLE patients. A comprehensive understanding of each metabolic pathway will facilitate and benefit personalized therapeutics in SLE and other autoimmune diseases.
Funding
There is no funding source for this review.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Tsokos G.C. Systemic lupus erythematosus. N. Engl. J. Med. 2011;365:2110–2121. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 2.Sang A., Zheng Y.Y., Morel L. Contributions of B cells to lupus pathogenesis. Mol. Immunol. 2014;62:329–338. doi: 10.1016/j.molimm.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herrada A.A., Escobedo N., Iruretagoyena M., Valenzuela R.A., Burgos P.I., Cuitino L., Llanos C. Innate immune cells contribution to systemic lupus erythematosus. Front. Immunol. 2019;10:772. doi: 10.3389/fimmu.2019.00772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Neill L.A., Kishton R.J., Rathmell J. A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 2016;16:553–565. doi: 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pearce E.L., Poffenberger M.C., Chang C.H., Jones R.G. Fueling immunity: insights into metabolism and lymphocyte function. Science. 2013;342:1242454. doi: 10.1126/science.1242454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee H.T., Wu T.H., Lin C.S., Lee C.S., Wei Y.H., Tsai C.Y., Chang D.M. The pathogenesis of systemic lupus erythematosus - from the viewpoint of oxidative stress and mitochondrial dysfunction. Mitochondrion. 2016;30:1–7. doi: 10.1016/j.mito.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 7.Demir S., Artim-Esen B., Şahinkaya Y., Pehlivan Ö., Alpay-Kanıtez N., Omma A., Erer B., Kamalı S., Gül A., Aral O., Öcal L., İnanç M. Metabolic syndrome is not only a risk factor for cardiovascular diseases in systemic lupus erythematosus but is also associated with cumulative organ damage: a cross-sectional analysis of 311 patients. Lupus. 2016;25:177–184. doi: 10.1177/0961203315603140. [DOI] [PubMed] [Google Scholar]
- 8.Mok C.C., Tse S.M., Chan K.L., Ho L.Y. Effect of the metabolic syndrome on organ damage and mortality in patients with systemic lupus erythematosus: a longitudinal analysis. Clin. Exp. Rheumatol. 2018;36:389–395. https://ncbi.nlm.nih.gov/pubmed/29148424 [PubMed] [Google Scholar]
- 9.Mobini M., Niksolat F., Mohammadpour R.A., Dashti Dargahloo S., Marzban D. Metabolic syndrome in patients with systemic lupus erythematosus: association with disease activity, disease damage and age. Int J Rheum Dis. 2018;21:1023–1030. doi: 10.1111/1756-185x.13276. [DOI] [PubMed] [Google Scholar]
- 10.Sinicato N.A., Postal M., de Oliveira Peliçari K., Rittner L., Marini R., Appenzeller S. Prevalence and features of metabolic syndrome in childhood-onset systemic lupus erythematosus. Clin. Rheumatol. 2017;36:1527–1535. doi: 10.1007/s10067-017-3602-0. [DOI] [PubMed] [Google Scholar]
- 11.Guleria A., Pratap A., Dubey D., Rawat A., Chaurasia S., Sukesh E., Phatak S., Ajmani S., Kumar U., Khetrapal C.L., Bacon P., Misra R., Kumar D. NMR based serum metabolomics reveals a distinctive signature in patients with Lupus Nephritis. Sci. Rep. 2016;6:35309. doi: 10.1038/srep35309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu T., Xie C., Han J., Ye Y., Weiel J., Li Q., Blanco I., Ahn C., Olsen N., Putterman C., Saxena R., Mohan C. Metabolic disturbances associated with systemic lupus erythematosus. PLoS One. 2012;7 doi: 10.1371/journal.pone.0037210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicolaou Orthodoxia. Andreas Kousios, Andreas Hadjisavvas, Bernard Lauwerys, Kleitos Sokratous, Kyriacos Kyriacou, Biomarkers of systemic lupus erythematosus identified using mass spectrometry-based proteomics: a systematic review. J. Cell Mol. Med. 2017;21:993–1012. doi: 10.1111/jcmm.13031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crispín J.C., Liossis S.N., Kis-Toth K., Lieberman L.A., Kyttaris V.C., Juang Y.T., Tsokos G.C. Pathogenesis of human systemic lupus erythematosus: recent advances. Trends Mol. Med. 2010;16:47–57. doi: 10.1016/j.molmed.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mistry Pragnesh, Kaplan Mariana J. Cell death in the pathogenesis of systemic lupus erythematosus and lupus nephritis. Clin. Immunol. 2017;185:59–73. doi: 10.1016/j.clim.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lech Maciej, Anders Hans-Joachim. The pathogenesis of lupus nephritis. J. Am. Soc. Nephrol. 2013;24:1357–1366. doi: 10.1681/ASN.2013010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mackern-Oberti Juan P., Llanos Carolina, Riedel Claudia A., Bueno Susan M., Kalergis Alexis M. Contribution of dendritic cells to the autoimmune pathology of systemic lupus erythematosus. Immunology. 2015;146:497–507. doi: 10.1111/imm.12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guiducci C., Gong M., Xu Z., Gill M., Chaussabel D., Meeker T., Chan J.H., Wright T., Punaro M., Bolland S., Soumelis V., Banchereau J., Coffman R.L., Pascual V., Barrat F.J. TLR recognition of self nucleic acids hampers glucocorticoid activity in lupus. Nature. 2010;465:937–941. doi: 10.1038/nature09102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parker D.C. T cell-dependent B cell activation. Annu. Rev. Immunol. 1993;11:331–360. doi: 10.1146/annurev.iy.11.040193.001555. [DOI] [PubMed] [Google Scholar]
- 20.Zheng L., Anne D. BAFF and selection of autoreactive B cells. Trends Immunol. 2011;32:388–394. doi: 10.1016/j.it.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gayed M., Gordon C. Novel treatments for systemic lupus erythematosus. Curr. Opin. Invest. Drugs. 2010;11:1256–1264. https://ncbi.nlm.nih.gov/pubmed/21157645 [PubMed] [Google Scholar]
- 22.Liu Z., Davidson A. Taming lupus-a new understanding of pathogenesis is leading to clinical advances. Nat. Med. 2012;18:871–882. doi: 10.1038/nm.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ganguly D., Chamilos G., Lande R., Gregorio J., Meller S., Facchinetti V., Homey B., Barrat F.J., Zal T., Gilliet M. Self-RNA-antimicrobial peptide complexes activate human dendritic cells through TLR7 and TLR8. J. Exp. Med. 2009;206:1983–1994. doi: 10.1084/jem.20090480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blanco P., Palucka A.K., Gill M., Pascual V., Banchereau J. Induction of dendritic cell differentiation by IFN-α in systemic lupus erythematosus. Science. 2001;294:1540–1543. doi: 10.1126/science.1064890. [DOI] [PubMed] [Google Scholar]
- 25.Jego G., Palucka A.K., Blanck J.P., Chalouni C., Pascual V., Banchereau J. Plasmacytoid dendritic cells induce plasma cell differentiation through type I interferon and interleukin 6. Immunity. 2003;19:225–234. doi: 10.1016/s1074-7613(03)00208-5. [DOI] [PubMed] [Google Scholar]
- 26.West A.P., Shadel G.S., Ghosh S. Mitochondria in innate immune responses. Nat. Rev. Immunol. 2011;11:389–402. doi: 10.1038/nri2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monlun M., Hyernard C., Blanco P., Lartigue L., Faustin B. Mitochondria as molecular platforms integrating multiple innate immune signalings. J. Mol. Biol. 2017;429:1–13. doi: 10.1016/j.jmb.2016.10.028. [DOI] [PubMed] [Google Scholar]
- 28.Weinberg S.E., Sena L.A., Chandel N.S. Mitochondria in the regulation of innate and adaptive immunity. Immunity. 2015;42:406–417. doi: 10.1016/j.immuni.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gergely P., Jr., Grossman C., Niland B., Puskas F., Neupane H., Allam F., Banki K., Phillips P.E., Perl A. Mitochondrial hyperpolarization and ATP depletion in patients with systemic lupus erythematosus. Arthritis Rheum. 2002;46:175–190. doi: 10.1002/1529-0131(200201)46:1<175::AID-ART10015>3.0.CO;2-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caza T.N., Talaber G., Perl A. Metabolic regulation of organelle homeostasis in lupus T cells. Clin. Immunol. 2012;144:200–213. doi: 10.1016/j.clim.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buck M.D., O’Sullivan D., Klein Geltink R.I., Curtis J.D., Chang C.H., Sanin D.E., Qiu J., Kretz O., Braas D., van der Windt G.J., Chen Q., Huang S.C., O’Neill C.M., Edelson B.T., Pearce E.J., Sesaki H., Huber T.B., Rambold A.S., Pearce E.L. Mitochondrial dynamics controls T cell fate through metabolic programming. Cell. 2016;166:63–76. doi: 10.1016/j.cell.2016.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai Z.W., Hanczko R., Bonilla E., Caza T.N., Clair B., Bartos A., Miklossy G., Jimah J., Doherty E., Tily H., Francis L., Garcia R., Dawood M., Yu J., Ramos I., Coman I., Faraone S.V., Phillips P.E., Perl A. N-acetylcysteine reduces disease activity by blocking mammalian target of rapamycin in T cells from systemic lupus erythematosus patients: a randomized, double-blind, placebo-controlled trial. Arthritis Rheum. 2012;64:2937–2946. doi: 10.1002/art.34502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huss J.M., Garbacz W.G., Xie W. Constitutive activities of estrogen-related receptors: transcriptional regulation of metabolism by the ERR pathways in health and disease. Biochim. Biophys. Acta. 2015;1852:1912–1927. doi: 10.1016/j.bbadis.2015.06.016. [DOI] [PubMed] [Google Scholar]
- 34.Yu X., Wieczorek S., Franke A., Yin H., Pierer M., Sina C., Karlsen T.H., Boberg K.M., Bergquist A., Kunz M., Witte T., Gross W.L., Epplen J.T., Alarcón-Riquelme M.E., Schreiber S., Ibrahim S.M. Association of UCP2 -866 G/A polymorphism with chronic inflammatory diseases. Gene Immun. 2009;10:601–605. doi: 10.1038/gene.2009.29. [DOI] [PubMed] [Google Scholar]
- 35.Brinkmann V., Reichard U., Goosmann C., Fauler B., Uhlemann Y., Weiss D.S., Weinrauch Y., Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 36.Knight J.S., Kaplan M.J. Lupus neutrophils: ’NET’ gain in understanding lupus pathogenesis. Curr. Opin. Rheumatol. 2012;24:441–450. doi: 10.1097/BOR.0b013e3283546703. [DOI] [PubMed] [Google Scholar]
- 37.Kegerreis B.J., Catalina M.D., Geraci N.S., Bachali P., Lipsky P.E., Grammer A.C. Genomic identification of low-density granulocytes and analysis of their role in the pathogenesis of systemic lupus erythematosus. J. Immunol. 2019;202:3309–3317. doi: 10.4049/jimmunol.1801512. [DOI] [PubMed] [Google Scholar]
- 38.Lood C., Blanco L.P., Purmalek M.M., Carmona-Rivera C., De Ravin S.S., Smith C.K., Malech H.L., Ledbetter J.A., Elkon K.B., Kaplan M.J. Neutrophil extracellular traps enriched in oxidized mitochondrial DNA are interferogenic and contribute to lupus-like disease. Nat. Med. 2016;22:146–153. doi: 10.1038/nm.4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caielli S., Athale S., Domic B., Murat E., Chandra M., Banchereau R., Baisch J., Phelps K., Clayton S., Gong M., Wright T., Punaro M., Palucka K., Guiducci C., Banchereau J., Pascual V. Oxidized mitochondrial nucleoids released by neutrophils drive type I interferon production in human lupus. J. Exp. Med. 2016;213:697–713. doi: 10.1084/jem.20151876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sukhbaatar N., Hengstschläger M., Weichhart T. mTOR-mediated regulation of dendritic cell differentiation and function. Trends Immunol. 2016;37:778–789. doi: 10.1016/j.it.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Linke M., Fritsch S.D., Sukhbaatar N., Hengstschläger M., Weichhart T. mTORC1 and mTORC2 as regulators of cell metabolism in immunity. FEBS Lett. 2017;591:3089–3103. doi: 10.1002/1873-3468.12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y., Huang G., Zeng H., Yang K., Lamb R.F., Chi H. Tuberous sclerosis 1 (Tsc1)-dependent metabolic checkpoint controls development of dendritic cells. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E4894–E4903. doi: 10.1073/pnas.1308905110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Halligan D.N., Murphy S.J., Taylor C.T. The hypoxia-inducible factor (HIF) couples immunity with metabolism. Semin. Immunol. 2016;28:469–477. doi: 10.1016/j.smim.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 44.Eckardt K.U. Immunometabolism: oxygen sensing and cell metabolism in inflammation. Nat. Rev. Nephrol. 2017;13:727–728. doi: 10.1038/nrneph.2017.145. [DOI] [PubMed] [Google Scholar]
- 45.Cheng S.C., Quintin J., Cramer R.A., Shepardson K.M., Saeed S., Kumar V., Giamarellos-Bourboulis E.J., Martens J.H., Rao N.A., Aghajanirefah A., Manjeri G.R., Li Y., Ifrim D.C., Arts R.J., van der Veer B.M., Deen P.M., Logie C., O’Neill L.A., Willems P., van de Veerdonk F.L., van der Meer J.W., Ng A., Joosten L.A., Wijmenga C., Stunnenberg H.G., Xavier R.J., Netea M.G. mTOR- and HIF-1α-mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 2014;345:1250684. doi: 10.1126/science.1250684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choudhry H., Harris A.L. Advances in hypoxia-inducible factor biology. Cell Metabol. 2018;27:281–298. doi: 10.1016/j.cmet.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 47.Feldhoff L.M., Rueda C.M., Moreno-Fernandez M.E., Sauer J., Jackson C.M., Chougnet C.A., Rupp J. IL-1β induced HIF-1α inhibits the differentiation of human FOXP3+ T cells. Sci. Rep. 2017;7:465. doi: 10.1038/s41598-017-00508-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Westendorf A.M., Skibbe K., Adamczyk A., Buer J., Geffers R., Hansen W., Pastille E., Jendrossek V. Hypoxia enhances immunosuppression by inhibiting CD4+ effector T cell function and promoting Treg activity. Cell. Physiol. Biochem. 2017;41:1271–1284. doi: 10.1159/000464429. [DOI] [PubMed] [Google Scholar]
- 49.Movafagh S., Crook S., Vo K. Regulation of hypoxia-inducible factor-1a by reactive oxygen species: new developments in an old debate. J. Cell. Biochem. 2015;116:696–703. doi: 10.1002/jcb.25074. [DOI] [PubMed] [Google Scholar]
- 50.Maciolek J.A., Pasternak J.A., Wilson H.L. Metabolism of activated T lymphocytes. Curr. Opin. Immunol. 2014;27:60–74. doi: 10.1016/j.coi.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 51.Lin S.C., Hardie D.G., AMPK Sensing glucose as well as cellular energy status. Cell Metabol. 2018;27:299–313. doi: 10.1016/j.cmet.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 52.Cantó C., Auwerx J. AMP-activated protein kinase and its downstream transcriptional pathways. Cell. Mol. Life Sci. 2010;67:3407–3423. doi: 10.1007/s00018-010-0454-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carling D., Mayer F.V., Sanders M.J., Gamblin S.J. AMP-activated protein kinase: nature’s energy sensor. Nat. Chem. Biol. 2011;7:512–518. doi: 10.1038/nchembio.610. [DOI] [PubMed] [Google Scholar]
- 54.Hardie D.G. AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes Dev. 2011;25:1895–1908. doi: 10.1101/gad.17420111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mihaylova M.M., Shaw R.J. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell Biol. 2011;13:1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramiscal R.R., Parish I.A., Lee-Young R.S., Babon J.J., Blagih J., Pratama A., Martin J., Hawley N., Cappello J.Y., Nieto P.F., Ellyard J.I., Kershaw N.J., Sweet R.A., Goodnow C.C., Jones R.G., Febbraio M.A., Vinuesa C.G., Athanasopoulos V. Attenuation of AMPK signaling by ROQUIN promotes T follicular helper cell formation. Elife. 2015;4 doi: 10.7554/eLife.08698. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pratama A., Srivastava M., Williams N.J., Papa I., Lee S.K., Dinh X.T., Hutloff A., Jordan M.A., Zhao J.L., Casellas R., Athanasopoulos V., Vinuesa C.G. MicroRNA-146a regulates ICOS-ICOSL signalling to limit accumulation of T follicular helper cells and germinal centres. Nat. Commun. 2015;6:6436. doi: 10.1038/ncomms7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fernandez D.R., Telarico T., Bonilla E., Li Q., Banerjee S., Middleton F.A., Phillips P.E., Crow M.K., Oess S., Muller-Esterl W., Perl A. Activation of mammalian target of rapamycin controls the loss of TCRζ in lupus T cells through HRES-1/Rab4-regulated lysosomal degradation. J. Immunol. 2009;182:2063–2073. doi: 10.4049/jimmunol.0803600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Delgoffe G.M., Pollizzi K.N., Waickman A.T., Heikamp E., Meyers D.J., Horton M.R., Xiao B., Worley P.F., Powell J.D. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat. Immunol. 2011;12:295–303. doi: 10.1038/ni.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wahl D.R., Petersen B., Warner R., Richardson B.C., Glick G.D., Opipari A.W. Characterization of the metabolic phenotype of chronically activated lymphocytes. Lupus. 2010;19:1492–1501. doi: 10.1177/0961203310373109. [DOI] [PubMed] [Google Scholar]
- 61.Dimeloe S., Mehling M., Frick C., Loeliger J., Bantug G.R., Sauder U., Fischer M., Belle R., Develioglu L., Tay S., Langenkamp A., Hess C. The immune-metabolic basis of effector memory CD4+ T cell function under hypoxic conditions. J. Immunol. 2016;196:106–114. doi: 10.4049/jimmunol.1501766. [DOI] [PubMed] [Google Scholar]
- 62.Sobel E.S., Brusko T.M., Butfiloski E.J., Hou W., Li S., Cuda C.M., Abid A.N., Reeves W.H., Morel L. Defective response of CD4(+) T cells to retinoic acid and TGFβ in systemic lupus erythematosus. Arthritis Res. Ther. 2011;13:R106. doi: 10.1186/ar3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yin Y., Choi S.C., Xu Z., Perry D.J., Seay H., Croker B.P., Sobel E.S., Brusko T.M., Morel L. Normalization of CD4+ T cell metabolism reverses lupus. Sci. Transl. Med. 2015;7:274ra18. doi: 10.1126/scitranslmed.aaa0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Macintyre A.N., Gerriets V.A., Nichols A.G., Michalek R.D., Rudolph M.C., Deoliveira D., Anderson S.M., Abel E.D., Chen B.J., Hale L.P., Rathmell J.C. The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function. Cell Metabol. 2014;20:61–72. doi: 10.1016/j.cmet.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cretenet G., Clerc I., Matias M., Loisel S., Craveiro M., Oburoglu L., Kinet S., Mongellaz C., Dardalhon V., Taylor N. Cell surface Glut1 levels distinguish human CD4 and CD8 T lymphocyte subsets with distinct effector functions. Sci. Rep. 2016;6:24129. doi: 10.1038/srep24129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ricciardi S., Manfrini N., Alfieri R., Calamita P., Crosti M.C., Gallo S., Müller R., Pagani M., Abrignani S., Biffo S. The translational machinery of human CD4 T cells is poised for activation and controls the switch from quiescence to metabolic remodeling. Cell Metabol. 2018;28:1–12. doi: 10.1016/j.cmet.2018.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Michalek R.D., Gerriets V.A., Jacobs S.R., Macintyre A.N., MacIver N.J., Mason E.F., Sullivan S.A., Nichols A.G., Rathmell J.C. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J. Immunol. 2011;186:3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shi L.Z., Wang R., Huang G., Vogel P., Neale G., Green D.R., Chi H. HIF1α-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J. Exp. Med. 2011;208:1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Murray P.J., Rathmell J., Pearce E. SnapShot: immunometabolism. Cell Metabol. 2015;22:190–190.e1. doi: 10.1016/j.cmet.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 70.Wu T., Qin X., Kurepa Z., Kumar K.R., Liu K., Kanta H., Zhou X.J., Satterthwaite A.B., Davis L.S., Mohan C. Shared signaling networks active in B cells isolated from genetically distinct mouse models of lupus. J. Clin. Invest. 2007;117:2186–2196. doi: 10.1172/jci30398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zeng Q., Zhang H., Qin J., Xu Z., Gui L., Liu B., Liu C., Xu C., Liu W., Zhang S., Huang S., Chen L. Rapamycin inhibits BAFF-stimulated cell proliferation and survival by suppressing mTOR-mediated PP2A-Erk 1/2 signaling pathway in normal and neoplastic B-lymphoid cells. Cell. Mol. Life Sci. 2015;72:4867–4884. doi: 10.1007/s00018-015-1976-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Caro-Maldonado A., Wang R., Nichols A.G., Kuraoka M., Milasta S., Sun L.D., Gavin A.L., Abel E.D., Kelsoe G., Green D.R., Rathmell J.C. Metabolic reprogramming is required for antibody production that is suppressed in anergic but exaggerated in chronically BAFF-exposed B cells. J. Immunol. 2014;192:3626–3636. doi: 10.4049/jimmunol.1302062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chiurchiù V., Leuti A., Maccarrone Bioactive lipids and chronic inflammation: managing the fire within. Front. Immunol. 2018;9:38. doi: 10.3389/fimmu.2018.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Krishnan S., Nambiar M.P., Warke V.G., Fisher C.U., Mitchell J., Delaney N., Tsokos G.C. Alterations in lipid raft composition and dynamics contribute to abnormal T cell responses in systemic lupus erythematosus. J. Immunol. 2004;172:7821–7831. doi: 10.4049/jimmunol.172.12.7821. [DOI] [PubMed] [Google Scholar]
- 75.McDonald G., Deepak S., Miguel L., Hall C.J., Isenberg D.A., Magee A.I., Butters T., Jury E.C. Normalizing glycosphingolipids restores function in CD4+ T cells from lupus patients. J. Clin. Invest. 2014;124:712–724. doi: 10.1172/jci69571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nowling T.K., Mather A.R., Thiyagarajan T., Hernández-Corbacho M.J., Powers T.W., Jones E.E., Snider A.J., Oates J.C., Drake R.R., Siskind L.J. Renal glycosphingolipid metabolism is dysfunctional in lupus nephritis. J. Am. Soc. Nephrol. 2015;26:1402–1413. doi: 10.1681/asn.2014050508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Waddington K.E., Jury E.C., Pineda-Torra I. Liver X receptors in immune cell function in humans. Biochem. Soc. Trans. 2015;43:752–757. doi: 10.1042/bst20150112. [DOI] [PubMed] [Google Scholar]
- 78.Richard E.M., Thiyagarajan T., Bunni M.A., Basher F., Roddy P.O., Siskind L.J., Nietert P.J., Nowling T.K. Reducing FLI1 levels in the MRL/lpr lupus mouse model impacts T cell function by modulating glycosphingolipid metabolism. PLoS One. 2013;8 doi: 10.1371/journal.pone.0075175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Morris E.E., Amria M.Y., Kistner-Griffin E., Svenson J.L., Kamen D.L., Gilkeson G.S., Nowling T.K. A GA microsatellite in the Fli1 promoter modulates gene expression and is associated with systemic lupus erythematosus patients without nephritis. Arthritis Res. Ther. 2010;12:R212. doi: 10.1186/ar3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hu X., Wang Y., Hao L.Y., Liu X., Lesch C.A., Sanchez B.M., Wendling J.M., Morgan R.W., Aicher T.D., Carter L.L., Toogood P.L., Glick G.D. Sterol metabolism controls T(H)17 differentiation by generating endogenous RORγ agonists. Nat. Chem. Biol. 2015;11:141–147. doi: 10.1038/nchembio.1714. [DOI] [PubMed] [Google Scholar]
- 81.Zhang X., Tao Y., Troiani L., Markovic-Plese S. Simvastatin inhibits IFN regulatory factor 4 expression and Th17 cell differentiation in CD4+ T cells derived from patients with multiple sclerosis. J. Immunol. 2011;187:3431–3437. doi: 10.4049/jimmunol.1100580. [DOI] [PubMed] [Google Scholar]
- 82.Tang T.T., Song Y., Ding Y.J., Liao Y.H., Yu X., Du R., Xiao H., Yuan J., Zhou Z.H., Liao M.Y., Yao R., Jevallee H., Shi G.P., Cheng X. Atorvastatin upregulates regulatory T cells and reduces clinical disease activity in patients with rheumatoid arthritis. J. Lipid Res. 2011;52:1023–1032. doi: 10.1194/jlr.m010876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Abud-Mendoza C., de la Fuente H., Cuevas-Orta E., Baranda L., Cruz-Rizo J., González-Amaro R. Therapy with statins in patients with refractory rheumatic diseases: a preliminary study. Lupus. 2003;12:607–611. doi: 10.1191/0961203303lu429oa. [DOI] [PubMed] [Google Scholar]
- 84.Qian X., Yang Z., Mao E., Chen E. Regulation of fatty acid synthesis in immune cells. Scand. J. Immunol. 2018;88 doi: 10.1111/sji.12713. [DOI] [PubMed] [Google Scholar]
- 85.Van den Bossche J., van der Windt G.J.W. Fatty acid oxidation in macrophages and T cells: time for reassessment. Cell Metabol. 2018;28:538–540. doi: 10.1016/j.cmet.2018.09.018. [DOI] [PubMed] [Google Scholar]
- 86.Berod L., Friedrich C., Nandan A., Freitag J., Hagemann S., Harmrolfs K., Sandouk A., Hesse C., Castro C.N., Bähre H., Tschirner S.K., Gorinski N., Gohmert M., Mayer C.T., Huehn J., Ponimaskin E., Abraham W.R., Müller R., Lochner M., Sparwasser T. De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nat. Med. 2014;20:1327–1333. doi: 10.1038/nm.3704. [DOI] [PubMed] [Google Scholar]
- 87.Young K.E., Flaherty S., Woodman K.M., Sharma-Walia N., Reynolds J.M. Fatty acid synthase regulates the pathogenicity of Th17 cells. J. Leukoc. Biol. 2017;102:1229–1235. doi: 10.1189/jlb.3ab0417-159rr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.O’Sullivan D., van der Windt G.J., Huang S.C., Curtis J.D., Chang C.H., Buck M.D., Qiu J., Smith A.M., Lam W.Y., DiPlato L.M., Hsu F.F., Birnbaum M.J., Pearce E.J., Pearce E.L. Memory CD8(+) T cells use cell-intrinsic lipolysis to support the metabolic programming necessary for development. Immunity. 2014;41:75–88. doi: 10.1016/j.immuni.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nomura M., Liu J., Rovira, Gonzalez-Hurtado E., Lee J., Wolfgang M.J., Finkel T. Fatty acid oxidation in macrophage polarization. Nat. Immunol. 2016;17:216–217. doi: 10.1038/ni.3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xu W., Wei Z., Dong J., Duan F., Chen K., Chen C., Liu J., Yang X., Chen L., Xiao H., Liu A. Global metabolomics reveals the metabolic dysfunction in ox-LDL induced macrophage-derived foam cells. Front. Pharmacol. 2017;8:586. doi: 10.3389/fphar.2017.00586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Freigang S., Ampenberger F., Weiss A., Kanneganti T.D., Iwakura Y., Hersberger M., Kopf M. Fatty acid-induced mitochondrial uncoupling elicits inflammasome-independent IL-1α and sterile vascular inflammation in atherosclerosis. Nat. Immunol. 2013;14:1045–1053. doi: 10.1038/ni.2704. [DOI] [PubMed] [Google Scholar]
- 92.Serbulea V., DeWeese D., Leitinger N. The effect of oxidized phospholipids on phenotypic polarization and function of macrophages. Free Radic. Biol. Med. 2017;111:156–168. doi: 10.1016/j.freeradbiomed.2017.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kahlenberg J.M., Carmona-Rivera C., Smith C.K., Kaplan M.J. Neutrophil extracellular trap-associated protein activation of the NLRP3 inflammasome is enhanced in lupus macrophages. J. Immunol. 2013;190:1217–1226. doi: 10.4049/jimmunol.1202388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cruzat V., Macedo Rogero M., Noel Keane K., Curi R., Newsholme P. Glutamine: metabolism and immune function, supplementation and clinical translation. Nutrients. 2018;10 doi: 10.3390/nu10111564. pii: E1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wallace C., Keast D. Glutamine and macrophage function. Metabolism. 1992;41:1016–1020. doi: 10.1016/0026-0495(92)90130-3. [DOI] [PubMed] [Google Scholar]
- 96.Jha A.K., Huang S.C., Sergushichev A., Lampropoulou V., Ivanova Y., Loginicheva E., Chmielewski K., Stewart K.M., Ashall J., Everts B., Pearce E.J., Driggers E.M., Artyomov M.N. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity. 2015;42:419–430. doi: 10.1016/j.immuni.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 97.Carr E.L., Kelman A., Wu G.S., Gopaul R., Senkevitch E., Aghvanyan A., Turay A.M., Frauwirth K.A. Glutamine uptake and metabolism are coordinately regulated by ERK/MAPK during T lymphocyte activation. J. Immunol. 2010;185:1037–1044. doi: 10.4049/jimmunol.0903586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Crawford J., Cohen H.J. The essential role of L-glutamine in lymphocyte differentiation in vitro. J. Cell. Physiol. 1985;124:275–282. doi: 10.1002/jcp.1041240216. [DOI] [PubMed] [Google Scholar]
- 99.Le A., Lane A.N., Hamaker M., Bose S., Gouw A., Barbi J., Tsukamoto T., Rojas C.J., Slusher B.S., Zhang H., Zimmerman L.J., Liebler D.C., Slebos R.J., Lorkiewicz P.K., Higashi R.M., Fan T.W., Dang C.V. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metabol. 2012;15:110–121. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Park B.V., Pan F. Metabolic regulation of T cell differentiation and function. Mol. Immunol. 2015;68:497–506. doi: 10.1016/j.molimm.2015.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang R., Dillon C.P., Shi L.Z., Milasta S., Carter R., Finkelstein D., McCormick L.L., Fitzgerald P., Chi H., Munger J., Green D.R. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Silver R.M., Heyes M.P., Maize J.C., Quearry B., Vionnet-Fuasset M., Sternberg E.M. Scleroderma, fasciitis, and eosinophilia associated with the ingestion of tryptophan. N. Engl. J. Med. 1990;322:874–881. doi: 10.1056/nejm199003293221302. [DOI] [PubMed] [Google Scholar]
- 103.Stahl J.L., Cook E.B., Pariza M.A., Cook M.E., Graziano F.M. Effect of L-tryptophan supplementation on eosinophils and eotaxin in Guinea pigs. Exp. Biol. Med. 2001;226:177–184. doi: 10.1177/153537020122600304. [DOI] [PubMed] [Google Scholar]
- 104.Munn D.H., Sharma M.D., Baban B., Harding H.P., Zhang Y., Ron D., Mellor A.L. GCN2 kinase in T cells mediates proliferative arrest and anergy induction in response to indoleamine 2,3-dioxygenase. Immunity. 2005;22:633–642. doi: 10.1016/j.immuni.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 105.Chaudhary K., Shinde R., Liu H., Gnana-Prakasam J.P., Veeranan-Karmegam R., Huang L., Ravishankar B., Bradley J., Kvirkvelia N., McMenamin M., Xiao W., Kleven D., Mellor A.L., Madaio M.P., McGaha T.L. Amino acid metabolism inhibits antibody-driven kidney injury by inducing autophagy. J. Immunol. 2015;194:5713–5724. doi: 10.4049/jimmunol.1500277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ravishankar B., Liu H., Shinde R., Chaudhary K., Xiao W., Bradley J., Koritzinsky M., Madaio M.P., McGaha T.L. The amino acid sensor GCN2 inhibits inflammatory responses to apoptotic cells promoting tolerance and suppressing systemic autoimmunity. Proc. Natl. Acad. Sci. U. S. A. 2015;112:10774–10779. doi: 10.1073/pnas.1504276112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pertovaara M., Hasan T., Raitala A., Oja S.S., Yli-Kerttula U., Korpela M., Hurme M. Indoleamine 2,3-dioxygenase activity is increased in patients with systemic lupus erythematosus and predicts disease activation in the sunny season. Clin. Exp. Immunol. 2007;150:274–278. doi: 10.1111/j.1365-2249.2007.03480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pekarova M., Lojek A. The crucial role of l-arginine in macrophage activation: what you need to know about it. Life Sci. 2015;137:44–48. doi: 10.1016/j.lfs.2015.07.012. [DOI] [PubMed] [Google Scholar]
- 109.MacMicking J., Xie Q.W., Nathan C. Nitric oxide and macrophage function. Annu. Rev. Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 110.Liu P.S., Wang H., Li X., Chao T., Teav T., Christen S., Di Conza G., Cheng W.C., Chou C.H., Vavakova M., Muret C., Debackere K., Mazzone M., Huang H.D., Fendt S.M., Ivanisevic J., Ho P.C. α-ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat. Immunol. 2017;18:985–994. doi: 10.1038/ni.3796. [DOI] [PubMed] [Google Scholar]
- 111.O’Neill L.A., Pearce E.J. Immunometabolism governs dendritic cell and macrophage function. J. Exp. Med. 2016;213:15–23. doi: 10.1084/jem.20151570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Galván-Peña S., O’Neill L.A. Metabolic reprograming in macrophage polarization. Front. Immunol. 2014;5:420. doi: 10.3389/fimmu.2014.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bronte V., Brandau S., Chen S.H., Colombo M.P., Frey A.B., Greten T.F., Mandruzzato S., Murray P.J., Ochoa A., Ostrand-Rosenberg S., Rodriguez P.C., Sica A., Umansky V., Vonderheide R.H., Gabrilovich D.I. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat. Commun. 2016;7:12150. doi: 10.1038/ncomms12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wu H., Zhen Y., Ma Z., Li H., Yu J., Xu Z.G., Wang X.Y., Yi H., Yang Y.G. Arginase-1-dependent promotion of TH17 differentiation and disease progression by MDSCs in systemic lupus erythematosus. Sci. Transl. Med. 2016;8:331ra40. doi: 10.1126/scitranslmed.aae0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Park M.J., Lee S.H., Kim E.K., Lee E.J., Park S.H., Kwok S.K., Cho M.L. Myeloid-derived suppressor cells induce the expansion of regulatory B cells and ameliorate autoimmunity in the sanroque mouse model of systemic lupus erythematosus. Arthritis Rheum. 2016;68:2717–2727. doi: 10.1002/art.39767. [DOI] [PubMed] [Google Scholar]
- 116.Gilkeson G.S., Mashmoushi A.K., Ruiz P., Caza T.N., Perl A., Oates J.C. Endothelial nitric oxide synthase reduces crescentic and necrotic glomerular lesions, reactive oxygen production, and MCP1 production in murine lupus nephritis. PLoS One. 2013;8 doi: 10.1371/journal.pone.0064650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Furusu A., Miyazaki M., Abe K., Tsukasaki S., Shioshita K., Sasaki O., Miyazaki K., Ozono Y., Koji T., Harada T., Sakai H., Kohno S. Expression of endothelial and inducible nitric oxide synthase in human glomerulonephritis. Kidney Int. 1998;53:1760–1768. doi: 10.1046/j.1523-1755.1998.00907.x. [DOI] [PubMed] [Google Scholar]
- 118.Bollain-y-Goytia J.J., Ramírez-Sandoval R., Daza L., Esparza E., Barbosa O., Ramirez D., Pacheco-Tovar G., Avalos-Diaz E., Rodríguez-Padilla C., Herrera-Esparza R. Widespread expression of inducible NOS and citrulline in lupus nephritis tissues. Inflamm. Res. 2009;58:61–66. doi: 10.1007/s00011-009-7215-1. [DOI] [PubMed] [Google Scholar]
- 119.Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 120.Reid M.A., Dai Z., Locasale J.W. The impact of cellular metabolism on chromatin dynamics and epigenetics. Nat. Cell Biol. 2017;19:1298–1306. doi: 10.1038/ncb3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Katada S., Imhof A., Sassone-Corsi P. Connecting threads: epigenetics and metabolism. Cell. 2012;148:24–28. doi: 10.1016/j.cell.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 122.Ballestar E., Esteller M., Richardson B.C. The epigenetic face of systemic lupus erythematosus. J. Immunol. 2006;176:7143–7147. doi: 10.4049/jimmunol.176.12.7143. [DOI] [PubMed] [Google Scholar]
- 123.Zhang Y., Zhao M., Sawalha A.H., Richardson B., Lu Q. Impaired DNA methylation and its mechanisms in CD4(+)T cells of systemic lupus erythematosus. J. Autoimmun. 2013;41:92–99. doi: 10.1016/j.jaut.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 124.Absher D.M., Li X., Waite L.L., Gibson A., Roberts K., Edberg J., Chatham W.W., Kimberly R.P. Genome-wide DNA methylation analysis of systemic lupus erythematosus reveals persistent hypomethylation of interferon genes and compositional changes to CD4+ T-cell populations. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yeung K.S., Chung B.H., Choufani S., Mok M.Y., Wong W.L., Mak C.C., Yang W., Lee P.P., Wong W.H., Chen Y.A., Grafodatskaya D., Wong R.W., Lau C.S., Chan D.T., Weksberg R., Lau Y.L. Genome-Wide DNA methylation analysis of Chinese patients with systemic lupus erythematosus identified hypomethylation in genes related to the type I interferon pathway. PLoS One. 2017;12 doi: 10.1371/journal.pone.0169553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Richardson B. Effect of an inhibitor of DNA methylation on T cells. II. 5-Azacytidine induces self-reactivity in antigen-specific T4+ cells. Hum. Immunol. 1986;17:456–470. doi: 10.1016/0198-8859(86)90304-6. [DOI] [PubMed] [Google Scholar]
- 127.Oaks Z., Perl A. Metabolic control of the epigenome in systemic Lupus erythematosus. Autoimmunity. 2014;47:256–264. doi: 10.3109/08916934.2013.834495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wu T., Xie C., Han J., Ye Y., Weiel J., Li Q., Blanco I., Ahn C., Olsen N., Putterman C., Saxena R., Mohan C. Metabolic disturbances associated with systemic lupus erythematosus. PLoS One. 2012;7 doi: 10.1371/journal.pone.0037210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yang M.L., Gee A.J., Gee R.J., Zurita-Lopez C.I., Khare S., Clarke S.G., Mamula M.J. Lupus autoimmunity altered by cellular methylation metabolism. Autoimmunity. 2013;46:21–31. doi: 10.3109/08916934.2012.732133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tsukada Y., Fang J., Erdjument-Bromage H., Warren M.E., Borchers C.H., Tempst P., Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 131.Klose R.J., Kallin E.M., Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 132.Guengerich F.P. Introduction: metals in biology: α-ketoglutarate/iron-dependent dioxygenases. J. Biol. Chem. 2015;290:20700–20701. doi: 10.1074/jbc.r115.675652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Niu Y., DesMarais T.L., Tong Z., Yao Y., Costa M. Oxidative stress alters global histone modification and DNA methylation. Free Radic. Biol. Med. 2015;82:22–28. doi: 10.1016/j.freeradbiomed.2015.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhang Q., Long H., Liao J., Zhao M., Liang G., Wu X., Zhang P., Ding S., Luo S., Lu Q. Inhibited expression of hematopoietic progenitor kinase 1 associated with loss of jumonji domain containing 3 promoter binding contributes to autoimmunity in systemic lupus erythematosus. J. Autoimmun. 2011;37:180–189. doi: 10.1016/j.jaut.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 135.Cheng S.C., Quintin J., Cramer R.A., Shepardson K.M., Saeed S., Kumar V., Giamarellos-Bourboulis E.J., Martens J.H., Rao N.A., Aghajanirefah A., Manjeri G.R., Li Y., Ifrim D.C., Arts R.J., van der Veer B.M., Deen P.M., Logie C., O’Neill L.A., Willems P., van de Veerdonk F.L., van der Meer J.W., Ng A., Joosten L.A., Wijmenga C., Stunnenberg H.G., Xavier R.J., Netea M.G. mTOR- and HIF-1α-mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 2014;345:1250684. doi: 10.1126/science.1250684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cobbold S.P. The mTOR pathway and integrating immune regulation. Immunology. 2013;140:391–398. doi: 10.1111/imm.12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hudson C.C., Liu M., Chiang G.G., Otterness D.M., Loomis D.C., Kaper F., Giaccia A.J., Abraham R.T. Regulation of hypoxia-inducible factor 1 alpha expression and function by the mammalian target of rapamycin. Mol. Cell Biol. 2002;22:7004–7014. doi: 10.1128/mcb.22.20.7004-7014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Keating S.T., El-Osta A. Epigenetics and metabolism. Circ. Res. 2015;116:715–736. doi: 10.1161/circresaha.116.303936. [DOI] [PubMed] [Google Scholar]
- 139.Friso S., Udali S., De Santis D., Choi S.W. One-carbon metabolism and epigenetics. Mol. Aspect. Med. 2017;54:28–36. doi: 10.1016/j.mam.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 140.Lee H.T., Wu T.H., Lin C.S., Lee C.S., Wei Y.H., Tsai C.Y., Chang D.M. The pathogenesis of systemic lupus erythematosus - from the viewpoint of oxidative stress and mitochondrial dysfunction. Mitochondrion. 2016;30:1–7. doi: 10.1016/j.mito.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 141.Wahl D.R., Petersen B., Warner R., Richardson B.C., Glick G.D., Opipari A.W. Characterization of the metabolic phenotype of chronically activated lymphocytes. Lupus. 2010;19:1492–1501. doi: 10.1177/0961203310373109. [DOI] [PubMed] [Google Scholar]
- 142.Henry R.A., Kuo Y.M., Bhattacharjee V., Yen T.J., Andrews A.J. Changing the selectivity of p300 by acetyl-CoA modulation of histone acetylation. ACS Chem. Biol. 2015;10:146–156. doi: 10.1021/cb500726b. https://pubs.acs.org/doi/10.1021/cb500726b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Maksimoska J., Segura-Peña D., Cole P.A., Marmorstein R. Structure of the p300 histone acetyltransferase bound to acetyl-coenzyme A and its analogues. Biochemistry. 2014;53:3415–3422. doi: 10.1021/bi500380f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Herbel C., Patsoukis N., Bardhan K., Seth P., Weaver J.D., Boussiotis V.A. Clinical significance of T cell metabolic reprogramming in cancer. Clin. Transl. Med. 2016;5:29. doi: 10.1186/s40169-016-0110-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Perl A., Hanczko R., Lai Z.W., Okas Z., Kelly R., Borsuk R., Asara J.M., Phillips E.P. Comprehensive metabolome analyses reveal N-acetylcysteine-responsive accumulation of kynurenine in systemic lupus erythematosus: implications for activation of the mechanistic target of rapamycin. Metabolomics. 2015;11:1157–1174. doi: 10.1007/s11306-015-0772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Morel L. Immunometabolism in systemic lupus erythematosus. Nat. Rev. Rheumatol. 2017;13:280–290. doi: 10.1038/nrrheum.2017.43. [DOI] [PubMed] [Google Scholar]
- 147.Rojo D., Hevia A., Bargiela R., López P., Cuervo A., González S., Suárez A., Sánchez B., Martínez-Martínez M., Milani C., Ventura M., Barbas C., Moya A., Suárez A., Margolles A., Ferrer M. Ranking the impact of human health disorders on gut metabolism: systemic lupus erythematosus and obesity as study cases. Sci. Rep. 2015;5:8310. doi: 10.1038/srep08310. [DOI] [PMC free article] [PubMed] [Google Scholar]