Abstract
Temozolomide (TMZ) resistance is a complication of treatment of glioma, and new strategies are urgently required to overcome chemoresistance in glioma cells. In the present study, it was demonstrated that tripartite motif-containing 31 (TRIM31) was abnormally upregulated in glioma tissues and cell lines compared with normal samples. Furthermore, the role of TRIM31 was assessed by overexpressing and knocking down its expression. Overexpression of TRIM31 increased cell viability, increased TMZ IC50 values and inhibited apoptosis in A172 and U251 cells; whereas overexpression of TRIM31 decreased the expression of the apoptosis-associated protein p53. Knockdown of TRIM31 increased apoptosis in cells treated with TMZ. Additionally, the mechanisms by which TRIM31 affected glioma cells treated with TMZ were determined. Overexpression of TRIM31 increased phosphorylation of AKT and inhibiting the PI3K/AKT signaling pathway abolished the increase in cell viability and decreased phospho-Akt protein expression in TRIM31 overexpressing A172 cells treated with TMZ. Together, the findings suggest that TRIM31 may be a potentially novel target for glioma chemotherapy.
Keywords: glioblastoma, tripartite motif-containing 31, chemoresistance, PI3K/Akt, apoptosis
Introduction
Malignant gliomas are the most common primary tumors observed in the central nervous system with a median survival time of 12-15 months (1-3). At present, the available treatment options are surgical resection and adjuvant temozolomide (TMZ)-based chemotherapy combined with radiotherapy (4,5). TMZ is a DNA-alkylating drug which has improved overall survival in patients (6,7). However, its efficacy is limited by the development of chemotherapeutic resistance in tumors (8,9). Therefore, identifying novel molecular mechanisms underlying TMZ resistance may improve outcomes for patients.
The tripartite motif-containing (TRIM) family of proteins contains more than 70 members and each member consists of a conserved RING finger, B-box, and coiled-coil domains (10), These members are primarily involved in important cellular biological processes (11,12). TRIM31 is one member of the TRIM family which has been reported to be involved in innate immunity and cancer development (13-20), and TRIM31 expression was found to be upregulated in pancreatic cancer tissues (20). Mechanistically, TRIM31 confers gemcitabine resistance in pancreatic cancer cells by activating the NF-κB signaling pathway (20). However, the role of TRIM31 in governing TMZ resistance remains unclear and the exact mechanism underlying its effects are unknown.
In the present study, TRIM31 was analyzed in glioma tissues and we clarified the relationship between TRIM31 upregulation and certain clinicopathological features including age, sex and WHO grade. Upregulation of TRIM31 enhanced TMZ resistance in glioma cells in vitro. Mechanistically, the PI3K/Akt pathway was determined to be involved in TRIM31-mediated chemoresistance in glioma cells. The present study demonstrated that overexpression of TRIM31 resulted in TMZ chemoresistance in glioma and it may be possible to improve the chemosensitivity to TMZ in patients with glioma by downregulating the expression of or inhibiting TRIM31.
Materials and methods
Tissue samples
A total of 40 tissue samples and 8 cases of adjacent normal brain tissues were collected at Department of Neurosurgery, The Second Hospital of Shandong University (Shandong, China) between June 2016 and March 2019. The samples ranged from 5 years old to 72 years old, 27 males and 13 females with an average age of 45.3 years old. All samples were histopathologically and clinically diagnosed and none of the patients received chemotherapy or radiotherapy prior to obtaining the tissue samples. All samples were frozen in liquid nitrogen and stored at -80˚C until further use. Patient characteristics are presented in Table I. All patients provided written informed consent and the study was approved by the Medical Ethics Committee of the Second Hospital of Shandong University.
Table I.
Correlations between TRIM31 expression and the clinicopathological characteristics of patients with glioma.
| TRIM31 expression | ||||
|---|---|---|---|---|
| Clinicopathological characteristics | Number of patients (n=40) | Low | High | P-value |
| Age | 0.7462 | |||
| <55 | 26 | 15 | 11 | |
| ≥55 | 14 | 9 | 4 | |
| Sex | 0.5106 | |||
| Male | 27 | 13 | 14 | |
| Female | 13 | 8 | 5 | |
| WHO grade | 0.007 | |||
| I-II | 14 | 12 | 2 | |
| III-IV | 26 | 10 | 16 | |
TRIM31, tripartite motif-containing 31.
Cell culture and treatments
Normal human astrocytes (NHA) were obtained from ScienCell Research Laboratories, Inc. LN229 cells were obtained from American Type Culture Collection. U87, U251 and A172 cell lines were purchased from The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences. U87 cells are a glioblastoma of unknown origin which was authenticated by STR profiling, Cells were cultured in DMEM (Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Biological Industries). The cells were cultured at 37˚C with 5% CO2. TMZ (cat. no. T2577; Sigma-Aldrich; Merck KGaA) was dissolved in DMSO (Sigma-Aldrich; Merck KGaA) at a stock concentration of 200 mM and stored at -20˚C. A 5 mM solution of LY294002 was purchased from MedChemExpress (cat. no. HY-10108), after 10 h transfection, the glioma cells were incubated with LY294002 for 12 h. Cells were harvested for further analysis.
Plasmid construction and cell transfection
TRIM31 expression plasmids were kindly provided by Professor Chengjiang Gao and Professor Lihui Han (Shandong University School of Medicine, Shandong, China). TRIM31 small interfering (si)RNAs were synthesized by Shanghai GenePharma Co., Ltd. And the sequences were as follows: siTRIM31, 5'-GGACCACAAAUCCCAUAAU-3'; and si-negative control (NC), 5'-UUCUCCGAACGUGUCACGU-3'. A172 and U251 cells were transfected using Lipofectamine® 2000 (cat .no. 11668019, Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. A total of 3x105 A172 or U251 cells/well were seeded in 6-well plates and cultured overnight. Lipofectamine® 2000 and opti-MEM (cat. no. 31985; Gibco; Thermo Fisher Scientific, Inc.) were used for transient transfection with 3 µg plasmid or 100 pmol siRNA. Subsequent experiments were performed after 24 or 48 h of transfection. Transfection efficiency was determined using reverse transcription-quantitative (RT-q)PCR.
RT-qPCR
Total RNA was extracted using a RNeasy kit (cat. no. DP430; Tiangen Biotech, Co., Ltd.) and 1 µg RNA was reverse transcribed into cDNA using a First-strand Synthesis kit (cat. no. KR116; Tiangen Biotech, Co., Ltd.). qPCR was performed on a Mastercycler ep realplex (Eppendorf) with SuperReal Premix Plus (cat. no. FP205; Tiangen Biotech, Co., Ltd.). The reverse transcription temperature protocol was: 42˚C for 15 min and reverse transcriptase inactivation 95˚C for 3 min. qPCR was performed using 10 µl 2x SuperReal Premix Plus, 20 ng cDNA template, 10 µM each forward and reverse primers, and ddH2O. The thermocycling conditions were: 95˚C for 15 min; followed by 40 cycles of denaturation at 95˚C for 10 sec, annealing at 60˚C for 25 sec and extension at 72˚C for 20 sec. The data were analyzed using the 2-ΔΔCq method (21) and normalized to the internal control. The sequence of the primers were: TRIM31 forward, 5'-GGCAGATTCAAGAGCAG-3' and reverse, 5'-TCAGTGGAGGCAACATAG-3'; and β-actin forward, 5'-GGAAATCGTGCGTGACATTAA-3' and reverse, 5'-AGGAAGGAAGGCTGGAAGAG-3'.
Western blotting
Cells were lysed with 1 ml RIPA buffer (Beijing Solarbio Science & Technology Co., Ltd.) supplemented with a protease inhibitor (Beijing Solarbio Science & Technology Co., Ltd.) and centrifuged at 4˚C 12,000 x g for 15 min, and the supernatant was collected. The protein concentration was measured using a bicinchoninic acid protein assay kit (ABP Biosciences) according to the manufacturer's protocol. Protein samples (20 µg) were loaded on a 12% SDS gel and resolved using SDS-PAGE. Resolved proteins were transferred onto PVDF membranes (EMD Millipore). The PVDF membranes were blocked in 5% nonfat milk at room temperature for 1 h and subsequently incubated with the following primary antibodies at 4˚C overnight: anti-TRIM31 (cat. no. 12543-1-AP; 1:500; ProteinTech Group, Inc.), anti-p-AKT (cat. no. 4060, 1:1,000), AKT (cat. no. 4685; 1:1,000), anti-p53 (cat. no. 2524; 1:1,000) and GADPH (cat. no. 2118s; 1:1,000) all from Cell Signaling Technology, Inc. The membranes were washed three times in PBS-Tween (PBST) for 6 min. Membranes were incubated with a secondary goat anti-rabbit immunoglobulin G (IgG) antibody (1:5,000, OriGene Technologies, Inc.) at room temperature for 1 h. Membranes were washed with PBST three times for 10 min. Signals were visualized using a chemiluminescence fluorescent detection kit (EMD Millipore) in the dark. Membranes were exposed in a gel imaging system. The ratios of gray values of each band was semi-quantified using Alpha Imager version 2011 (ProteinSimple). All experiments were performed in triplicate.
Apoptosis assay
An AnnexinV/propidium iodide (PI) double staining kit (cat. no. BB-4101; Bestbio) was used to measure apoptosis. Cells were harvested using 0.25% trypsin following TMZ treatment, washed twice with pre-cooled PBS and cells were resuspended in binding buffer. 5 µl Annexin V/fluorescein isothiocyanate and 5 µl PI was added. After incubation in the dark for 15 min at 4˚C, cells were analyzed using BD flow cytometry. All experiments were performed in triplicate.
Cell viability assay
Cell viability was measured using an MTT assay. Cells were seeded in a 96-well plate at a density of 1x103 cells/well in 100 µl of culture medium and incubated at 37˚C with 5% CO2 for 24 h. After treatment with different concentrations of TMZ for 48 h, 5 mg/ml MTT solution (Beijing Solarbio Science & Technology Co., Ltd.) was added to each group at the appropriate time and incubated for a further 4 h at 37˚C in the dark, after which 200 µl DMSO was added to dissolve the purple crystals. Cell viability was detected at 450 nm using a microplate reader (Infinite M200 Pro; Tecan Group). IC50 was calculated using GraphPad Prism version 5.01 (GraphPad Software, Inc.) by plotting a nonlinear regression curve fit using log (inhibitor) vs. normalized response (variable slope). Experiments were performed in triplicate.
Colony formation assay
A172 or U251 cells were plated in a 6-well plate at a density of 100 cells/well and cultured in DMEM supplemented with 10% FBS. Cells were treated with the indicated agents for 10-14 days. Medium was replaced every 3 days. Cell colonies were gently washed with PBS and fixed with 4% paraformaldehyde for 15 min at room temperature, and subsequently stained with 0.1% crystal violet for 20 min at room temperature. The stain was carefully washed using running water and dried. The number of colonies with >50 cells were counted under a NikonTS-100F microscope light. Three independent assays were performed for each condition.
Immunohistochemistry
Immunohistochemistry staining was performed using a kit (cat. no. PV-9001; OriGene Technologies, Inc.). The sections were deparaffinized with xylene, rehydrated and boiled in 0.01 M citrate buffer (pH 6.0) for antigen retrieval. Hydrogen peroxide was added to block endogenous peroxide activity, and the sections were washed three times, and subsequently incubated with TRIM31 antibody (1:50) overnight at 4˚C. Tissues were washed three times in PBS for 3 min each, after which the sections were incubated with a secondary goat anti-rabbit IgG antibody at room temperature for 20 min. Sections were colored using a 3,3'-diaminobenzidine (cat. no. ZLI-9018; OriGene Technologies, Inc.) at room temperature for 40 sec. Nuclei were counterstained with hematoxylin at room temperature for 3-5 min. The intensity and the ratio of positive cells in at least five separate fields were evaluated at x400 magnification under a Nikon90i microscope light. The intensity of staining was scored between 1 and 4: 1, No staining; 2, weak staining; 3, moderate staining; and 4, strong staining. The proportion of cells stained were scored as follows: 1, 0-5%; 2, 6-50%; 3, 51-75%; and 4, 76-100%. The scores of the proportion of cells stained and the intensity scores were multiplied to give a final score between 1 and 16 and classed as follows: Negative, 1-4; weakly positive, 5-8; moderately positive, 9-12; or strongly positive, >12. The scores were evaluated by two pathologists who were blinded to clinical data.
Statistical analysis
GraphPad Prism version 5.01 (GraphPad Software, Inc.) was used for all statistical analyses. All experiments were independently performed three times. Data are presented as the mean ± standard deviation. Comparisons between two groups were analyzed using a Student's t-test. A one-way ANOVA with post hoc Tukey's test was used to analyze differences between multiple groups. The associations between TRIM31 levels and clinicopathological features were analyzed using a Fisher's exact probability test. P<0.05 was considered to indicate a statistically significant difference.
Results
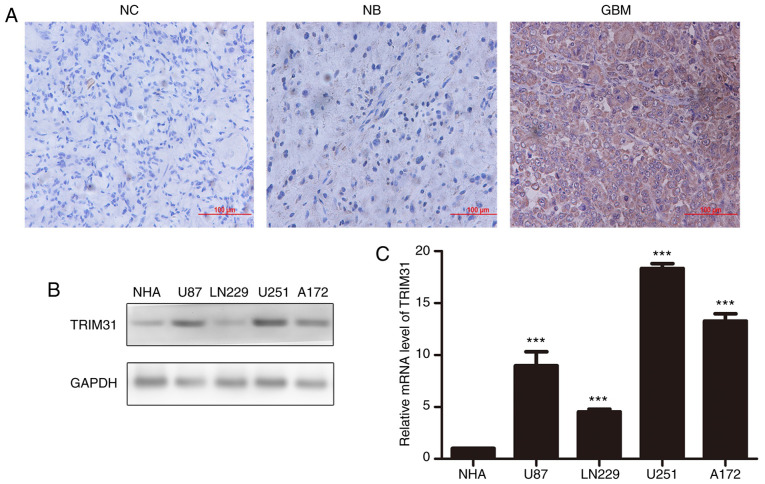
TRIM31 is upregulated in human glioma tissues
To determine the role of TRIM31 in human glioma, its expression in normal brain tissues and glioma tissues were examined by RT-qPCR and immunohistochemistry assay. The results showed that expression of TRIM31 was significantly upregulated in glioma tissues compared with normal brain tissues and it was primarily expressed in the cytoplasm of glioblastoma cells (Fig. 1A). Additionally, the expression of TRIM31 in NHA, U87, LN229 U251 and A172 cell lines was examined. TRIM31 expression was higher in U251 and A172 cells compared with the other cell lines at both the protein and mRNA levels (Fig. 1B). Therefore, the U251 and A172 cell lines were chosen for subsequent experiments. Subsequently, the association between TRIM31 expression and clinicopathological characteristics of patients with glioma were assessed. Analysis showed that TRIM31 expression was associated with World Health Organization grade. There was no significant difference between TRIM31 expression and sex or age. Taken together, these findings suggest that increased TRIM31 expression was associated with aggressive clinical features of glioma.
Figure 1.
Expression levels of TRIM31 in glioma tissues and glioma cell lines. (A) TRIM31 protein levels in glioblastoma tissues compared with normal brain tissues. Magnification, x200; scale bar, 100 µm. (B) Western blot analysis of TRIM 31 protein expression levels in four different glioma cell lines and the NHA cell line. GAPDH was used as the loading control. (C) mRNA expression levels of TRIM31 in four different glioma cell lines and the NHA cell line. ***P<0.001 NHA cell line was compared to different gliomas cell lines separately. Data are presented as the mean ± standard deviation of three independent experiments. NC, negative control; NB, normal brain tissue; GBM, glioblastoma; TRIM31, tripartite motif-containing 31; NHA, normal human astrocytes.
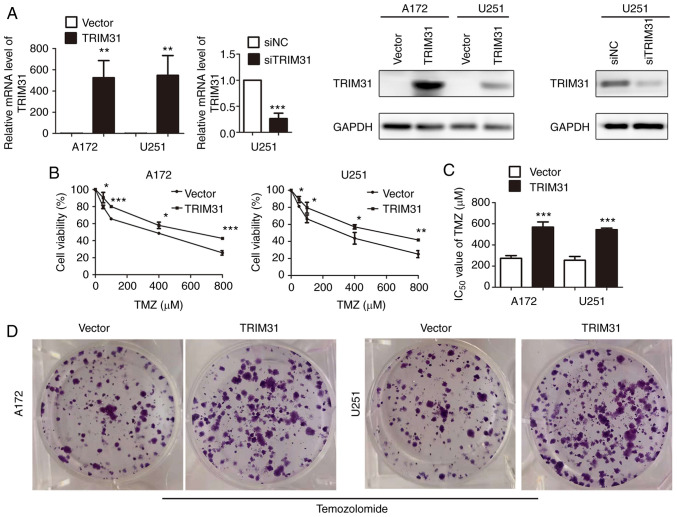
TRIM31 overexpression confers TMZ resistance in glioma cells
To examine the potential role of TRIM31 in chemoresistance in glioma cells, TRIM31 expression was knocked down using siRNA or overexpressed in A172 and U251 cells. Overexpression and knockdown efficiency were confirmed by RT-qPCR and western blotting (Fig. 2A). The effect of TRIM31 on the cell viability of A172 and U251 cells treated with different concentrations of TMZ for 48 h was subsequently determined. Cell proliferation was assessed using MTT assays and they showed that TRIM31 overexpression increased cell viability compared with the control group (Fig. 2B). The IC50 value of TMZ was significantly higher in TRIM31 overexpressing cells than transfecting blank plasmids cells (Fig. 2C). Colony formation assays showed that TRIM31 overexpression resulted in increased colony formation and exhibited greater clonogenic survival following TMZ treatment compared with the control (Fig. 2D). These data suggest that TRIM31 enhanced TMZ resistance in glioma cells in vitro.
Figure 2.
TRIM31 confers TMZ resistance in glioma cells in vitro. (A) mRNA and protein expression levels of TRIM31 in cells transfected with empty plasmid, TRIM31 overexpression plasmid, siTRIM31 and siNC. (B) Cell viability of A172 and U251 cells transfected with the indicated plasmids. (C) IC50 values of TMZ in A172 and U251 cells transfected with the empty plasmid and TRIM31 overexpression plasmid. (D) Colony formation in A172 and U251 cells transfected with empty plasmid and TRIM31 overexpression plasmid and treated with 100 µM TMZ. *P<0.05, **P<0.01, ***P<0.001. Data are presented as the mean ± standard deviation of three independent experiments. TMZ, temozolomide; si, small interfering; NC, negative control; TRIM31, tripartite motif-containing 31.
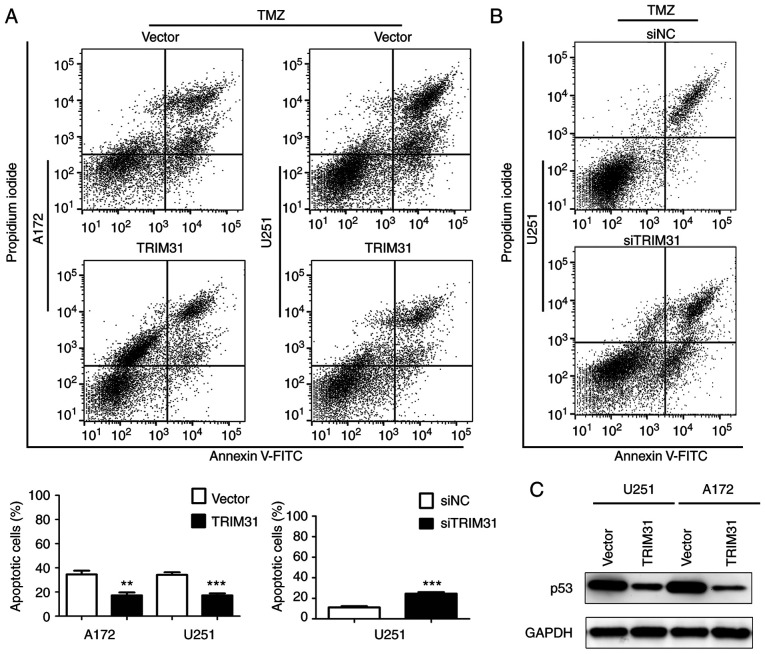
TRIM31 inhibits TMZ-induced glioma cell apoptosis
To further investigate the effect of TRIM31 on TMZ-induced apoptosis, apoptosis rates were measured by flow cytometry analysis in A172 and U251 cells treated with TMZ for 48 h. A172 and U251 cells were transfected with empty vector or TRIM31 overexpression plasmid and treated with TMZ (100 µg/ml). The results demonstrated that TRIM31 overexpression significantly decreased the apoptotic proportion of cells compared with the control cells (Fig. 3A). In U251 cells transfected with siNC or siTRIM31 and treated with TMZ (100 µg/ml), knockdown of TRIM31 increased the apoptotic proportion of cells (Fig. 3B). Subsequently, p53 protein levels were detected in U251 and A172 cells treated with TMZ, overexpressing TRIM31 decreased the expression of p53 protein (Fig. 3C). These results showed TRIM31 reduced TMZ-induced glioma cells apoptosis, which suggested that TRIM31 decreased glioma cells sensitivity to TMZ.
Figure 3.
TRIM31 decreased TMZ-induced glioma cell apoptosis. (A) Apoptotic rates of A172 and U251 cells transfected with empty plasmid and TRIM31 overexpression plasmid after 48 h of TMZ treatment (100 µM). (B) Apoptotic rate of U251 cells transfected with siTRIM31 and negative control after 48 h of TMZ treatment (100 µM). (C) Protein expression levels of p53 were analyzed by western blot in U251 and A172 cells transfected with TRIM31 plasmids and treated with TMZ. **P<0.01, ***P<0.001. Data are presented as the mean ± standard deviation of three independent experiments. Si, small interfering; TMZ, temozolomide; TRIM31, tripartite motif-containing 31.
The PI3K/Akt signaling pathway is involved in TRIM31-mediated TMZ resistance of glioma cells
Activation of PI3K/Akt signaling is the primary pathway which participates in glioblastoma growth and therapy (22-29), and activated Akt is indicative of increased signaling via this pathway. Whether PI3K/Akt pathway was involved in TRIM31-mediated chemoresistance of glioma cells was unknown. TRIM31 overexpression upregulated the phosphorylation of AKT (Fig. 4A) compared with control cells treated with TMZ. TRIM31 knockdown downregulated the phosphorylation of AKT in U251 cells compared with control cells treated with TMZ (Fig. 4B). These results suggested that TRIM31 may activate the PI3K/Akt signal pathway in glioma cells treated with TMZ.
Figure 4.
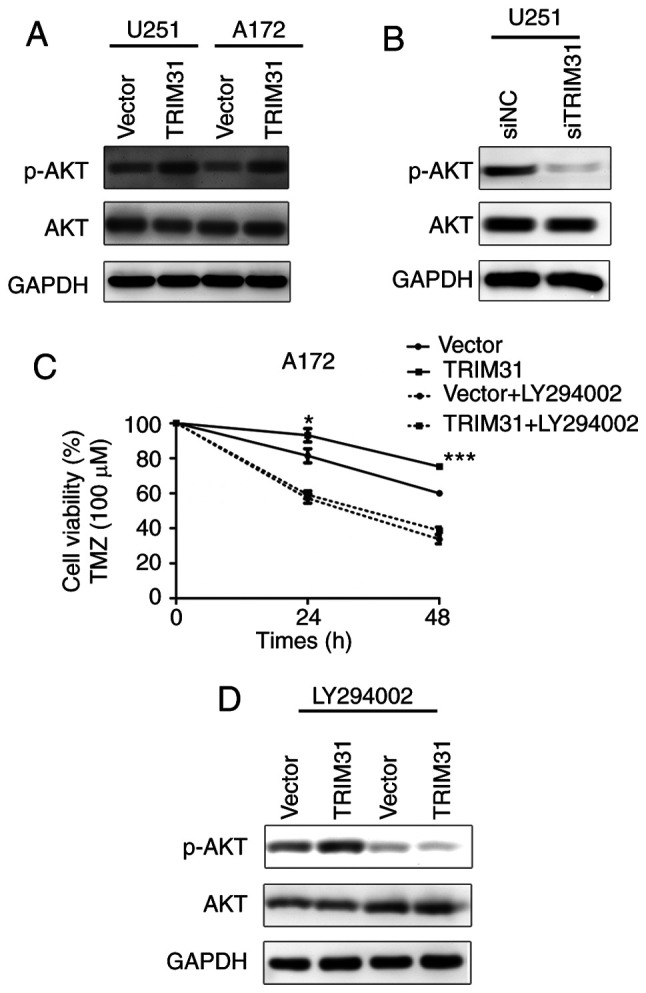
TRIM31 regulates TMZ resistance via the PI3K/Akt signaling pathway. (A) Protein expression levels of p-AKT and AKT in A172 and U251 cells transfected with the indicated plasmids and treated with TMZ. (B) Protein expression levels of p-AKT and AKT in U251 cells transfected with siTRIM31 and siNC and treated with TMZ. (C) Cell viability of A172 cells transfected with the TRIM31 and control plasmids and treated with the PI3K inhibitor, LY294002, and TMZ. (D) Protein expression levels of p-AKT and AKT in A172 cells transfected with the TRIM31 and control plasmids and treated with the PI3K inhibitor, LY294002, and TMZ. *P<0.05, ***P<0.001. Data are presented as the mean ± standard deviation of three independent experiments. TMZ, temozolomide; si, small interfering; NC, negative control; TRIM31, tripartite motif-containing 31; p, phosphorylated.
To further elucidate the mechanism by which TRIM31 induced chemoresistance in glioma cells, LY294002 was used to inhibit PI3K/Akt activation in U251 and A172 cells. Inhibition of PI3K/Akt signaling decreased glioma cell viability and abolished the effects of TRIM31 expression (Fig. 4C). LY294002 also reduced the expression of p-AKT induced by TRIM31 overexpression in A172 cells treated with TMZ (Fig. 4D). The PI3K/Akt inhibitor abolished the effects of TRIM31 on p-AKT upregulation. These data suggest that TRIM31 regulated TMZ resistance through the PI3K/Akt activation and p-AKT upregulation.
Discussion
TMZ chemoresistance is a significant challenge faced during glioma recurrence which results in treatment failure (8,9), Therefore, enhancement of sensitivity of the tumor to TMZ may improve the prognosis of patients with gliomas.
TRIM31, is a member of the TRIM family of proteins, which are been involved in various cellular processes (13-20). Recently, studies have demonstrated that TRIM31 serves an important role in the development of various types of cancer (15-20). A recent study reported that TRIM31 promoted progression of hepatocellular carcinoma through the mTORC1-HIF1α pathway by directly targeting the TSC1-TSC2 complex for degradation (19). Notably, TRIM31 has also been found to be involved in the development of chemotherapeutic resistance in cancer cells. For instance, TRIM31 overexpression conferred gemcitabine resistance in pancreatic cancer cells via the NF-κB signaling pathway (20). It has also been reported to participate in the development of drug resistance in ovarian cancer (30). In the present study, it was demonstrated that TRIM31 expression was upregulated in glioblastoma (GBM) tissues and TRIM31 expression was significantly correlated with tumor grade, indicating that TRIM31 functions as a tumor oncogene in glioma progression. The data together suggested that TRIM31 may be associated with chemoresistance in glioblastoma cells.
To clarify the biological function of TRIM31 in affecting TMZ sensitivity in glioblastoma cells, stable TRIM31 overexpression and knockdown models were created. MTT assays demonstrated that overexpression of TRIM31 increased cell viability and the effects were augmented by treatment with TMZ in a dose-dependent manner. TRIM31 overexpression combined with TMZ promoted cell growth and resulted in reduced apoptosis of glioma cells compared with untransfected cells treated with TMZ. The IC50 value was used as a measure of TMZ chemoresistance in GBM cells, thus increased IC50 generally corresponded to increased clinical chemoresistance to TMZ. TRIM31 overexpression significantly increased the IC50 values of TMZ. Furthermore, TRIM31 overexpression significantly increased colony formation ability in vitro compared with the TMZ treated untransfected cells. Similarly, flow cytometry analysis showed that combined TRIM31 overexpression and TMZ treatment resulted in significantly reduced apoptosis compared with TMZ treated untransfected cells. Conversely, TRIM31 knockdown increased apoptosis. The results of the present study suggested that overexpression of TRIM31 in glioblastoma cells induced resistance to TMZ. The tumor suppressor protein p53 serves various functional roles in the cell by regulating responses to several cellular stresses and it is inactivated in a variety of cancer cells via point mutations (31,32). p53 regulates tumor proliferation and apoptosis synergistically (33-35). p53 mimetic agents designed to stabilize the p53wt sensitize glioma cells to TMZ (36). In the present study, upregulation of TRIM31 significantly reduced the expression of p53, these results suggested that decreased p53 expression may account for the inhibitory role of TRIM31 in TMZ-induced glioma cell apoptosis.
Numerous studies have demonstrated a close association between the PI3K/Akt pathway and TMZ resistance in gliomas (37-41). Akt is a primary regulator of PI3K-initiated signaling and its activation contributes to chemoresistance (42,43). Therefore, it was determined whether the PI3K/Akt signaling pathway was involved in TRIM31 induced TMZ resistance of glioma cells. Firstly, the results showed that TRIM31 overexpression significantly increased the expression of p-Akt in glioma cells, which indicated that the biological effects of TRIM31 were partly mediated by the PI3K/Akt pathway. LY294002, a PI3K inhibitor, was used to further confirm this assumption. TRIM31 failed to upregulate cell viability when treated with LY294002, and the cell viability was reduced to levels similar to TMZ treated cells and in the TRIM31 overexpressing cells. The results indicated that PI3K inhibition decreased p-Akt protein expression and abolished the effects of TRIM31 on p-Akt upregulation, suggesting that TRIM31 regulated TMZ resistance possibly via the PI3K/Akt pathway.
In the present study, upregulation of TRIM31 resulted in an increase of p-AKT and a decrease of p53 expression, which demonstrated that TRIM31 induced chemoresistance in gliomas to TMZ via the PI3K/Akt/p53 signaling. The E3 ubiquitin ligase has been reported to catalyze the polyubiquitin of p53 and trigger the degradation of p53 in hepatocellular carcinoma cells (44). In glioma cells, whether TRIM31 directly targeted p53 for ubiquitin-mediated degradation or if TRIM31 bound adaptors involved in the PI3K/Akt pathway remains unclear.
In conclusion, the present study showed that TRIM31 mediated TMZ sensitivity via the PI3K/Akt signaling pathway in glioblastoma. The results present a possible molecular mechanism underlying TMZ resistance and proposed a novel strategy to potentially improve the therapeutic outcomes of glioblastoma treatment.
Acknowledgements
The authors would like to thank Professor Chengjiang Gao (Key Laboratory of Infection and Immunity of Shandong Province and Department of Immunology, The School of Basic Medical Sciences, Shandong University, Jinan, China) for providing valuable suggestions.
Funding
The present study was supported by grants from the Natural Science Foundation of Shandong Province (grant no. 2013ZRE27073) and China Natural Science Foundation (grant no. 81771270).
Availability of data and materials
All the datasets generated and analyzed during the present study are included in this manuscript.
Authors' contributions
MF and XZ performed the experiments. ZD, RZ and BL collected and analyzed the data. JQ and YJ conceptualized the study design and analyzed the data, drafted and reviewed the manuscript and supervised the entire study. CW and QP designed the study, revised the manuscript and provided material support. All the authors have read and approved the final manuscript.
Ethics approval and consent to participate
All patients provided written informed consent for their tissues to be used for clinical research. The present study was approved by the Medical Ethics Committee of the Second Hospital of Shandong University (Jinan, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: A clinical review. JAMA. 2013;310:1842–1850. doi: 10.1001/jama.2013.280319. [DOI] [PubMed] [Google Scholar]
- 2.Gladson CL, Prayson RA, Liu WM. The pathobiology of glioma tumors. Annu Rev Pathol. 2010;5:33–50. doi: 10.1146/annurev-pathol-121808-102109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanase C, Albulescu R, Codrici E, Popescu ID, Mihai S, Enciu AM, Cruceru ML, Popa AC, Neagu AI, Necula LG, et al. Circulating biomarker panels for targeted therapy in brain tumors. Future Oncol. 2015;11:511–524. doi: 10.2217/fon.14.238. [DOI] [PubMed] [Google Scholar]
- 4.Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: The avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60:166–193. doi: 10.3322/caac.20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 6.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 7.Minniti G, De Sanctis V, Muni R, Filippone F, Bozzao A, Valeriani M, Osti MF, De Paula U, Lanzetta G, Tombolini V, Maurizi Enrici R. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma in elderly patients. J Neurooncol. 2008;88:97–103. doi: 10.1007/s11060-008-9538-0. [DOI] [PubMed] [Google Scholar]
- 8.Chamberlain MC. Temozolomide: Therapeutic limitations in the treatment of adult high-grade gliomas. Expert Rev Neurother. 2010;10:1537–1544. doi: 10.1586/ern.10.32. [DOI] [PubMed] [Google Scholar]
- 9.Prasad G, Sottero T, Yang X, Mueller S, James CD, Weiss WA, Polley MY, Ozawa T, Berger MS, Aftab DT, et al. Inhibition of PI3K/mTOR pathways in glioblastoma and implications for combination therapy with temozolomide. Neuro Oncol. 2011;13:384–392. doi: 10.1093/neuonc/noq193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sardiello M, Cairo S, Fontanella B, Ballabio A, Meroni G. Genomic analysis of the TRIM family reveals two groups of genes with distinct evolutionary properties. BMC Evol Biol. 2008;8(225) doi: 10.1186/1471-2148-8-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimsa MW, Strzalka-Mrozik B, Kimsa MC, Mazurek U, Kruszniewska-Rajs C, Gola J, Adamska J, Twardoch M. Differential expression of tripartite motif-containing family in normal human dermal fibroblasts in response to porcine endogenous retrovirus infection. Folia Biol (Praha) 2014;60:144–151. doi: 10.14712/fb2014060030144. [DOI] [PubMed] [Google Scholar]
- 12.Hatakeyama S. TRIM family proteins: Roles in autophagy, immunity, and carcinogenesis. Trends Biochem Sci. 2017;42:297–311. doi: 10.1016/j.tibs.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Liu B, Zhang M, Chu H, Zhang H, Wu H, Song G, Wang P, Zhao K, Hou J, Wang X, et al. The ubiquitin E3 ligase TRIM31 promotes aggregation and activation of the signaling adaptor MAVS through Lys63-linked polyubiquitination. Nat Immunol. 2017;18:214–224. doi: 10.1038/ni.3641. [DOI] [PubMed] [Google Scholar]
- 14.Song H, Liu B, Huai W, Yu Z, Wang W, Zhao J, Han L, Jiang G, Zhang L, Gao C, Zhao W. The E3 ubiquitin ligase TRIM31 attenuates NLRP3 inflammasome activation by promoting proteasomal degradation of NLRP3. Nat Commun. 2016;7(13727) doi: 10.1038/ncomms13727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sugiura T, Miyamoto K. Characterization of TRIM31, upregulated in gastric adenocarcinoma, as a novel RBCC protein. J Cell Biochem. 2008;105:1081–1091. doi: 10.1002/jcb.21908. [DOI] [PubMed] [Google Scholar]
- 16.Li H, Zhang Y, Zhang Y, Bai X, Peng Y, He P. TRIM31 is downregulated in non-small cell lung cancer and serves as a potential tumor suppressor. Tumour Biol. 2014;35:5747–5752. doi: 10.1007/s13277-014-1763-x. [DOI] [PubMed] [Google Scholar]
- 17.Wang H, Yao L, Gong Y, Zhang B. TRIM31 regulates chronic inflammation via NF-κB signal pathway to promote invasion and metastasis in colorectal cancer. Am J Transl Res. 2018;10:1247–1259. [PMC free article] [PubMed] [Google Scholar]
- 18.Li H, Zhang Y, Hai J, Wang J, Zhao B, Du L, Geng X. Knockdown of TRIM31 suppresses proliferation and invasion of gallbladder cancer cells by down-regulating MMP2/9 through the PI3K/Akt signaling pathway. Biomed Pharmacother. 2018;103:1272–1278. doi: 10.1016/j.biopha.2018.04.120. [DOI] [PubMed] [Google Scholar]
- 19.Guo P, Ma X, Zhao W, Huai W, Li T, Qiu Y, Zhang Y, Han L. TRIM31 is upregulated in hepatocellular carcinoma and promotes disease progression by inducing ubiquitination of TSC1-TSC2 complex. Oncogene. 2018;37:478–488. doi: 10.1038/onc.2017.349. [DOI] [PubMed] [Google Scholar]
- 20.Yu C, Chen S, Guo Y, Sun C. Oncogenic TRIM31 confers gemcitabine resistance in pancreatic cancer via activating the NF-κB signaling pathway. Theranostics. 2018;8:3224–3236. doi: 10.7150/thno.23259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Zhang B, Liu Y, Li Y, Zhe X, Zhang S, Zhang L. Neuroglobin promotes the proliferation and suppresses the apoptosis of glioma cells by activating the PI3K/AKT pathway. Mol Med Rep. 2018;17:2757–2763. doi: 10.3892/mmr.2017.8132. [DOI] [PubMed] [Google Scholar]
- 23.Ping YF, Yao XH, Jiang JY, Zhao LT, Yu SC, Jiang T, Lin MC, Chen JH, Wang B, Zhang R, et al. The chemokine CXCL12 and its receptor CXCR4 promote glioma stem cell-mediated VEGF production and tumour angiogenesis via PI3K/AKT signalling. J Pathol. 2011;224:344–354. doi: 10.1002/path.2908. [DOI] [PubMed] [Google Scholar]
- 24.Wei Y, Jiang Y, Zou F, Liu Y, Wang S, Xu N, Xu W, Cui C, Xing Y, Liu Y, et al. Activation of PI3K/Akt pathway by CD133-p85 interaction promotes tumorigenic capacity of glioma stem cells. Proc Natl Acad Sci USA. 2013;110:6829–6834. doi: 10.1073/pnas.1217002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang P, Chen XB, Ding BQ, Liu HL, He T. Down-regulation of ABCE1 inhibits temozolomide resistance in glioma through the PI3K/Akt/NF-κB signaling pathway. Biosci Rep. 2018;38 doi: 10.1042/BSR20181711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burris HA III. Overcoming acquired resistance to anticancer therapy: Focus on the PI3K/AKT/mTOR pathway. Cancer Chemother Pharmacol. 2013;71:829–842. doi: 10.1007/s00280-012-2043-3. [DOI] [PubMed] [Google Scholar]
- 27.Li X, Wu C, Chen N, Gu H, Yen A, Cao L, Wang E, Wang L. PI3K/Akt/mTOR signaling pathway and targeted therapy for glioblastoma. Oncotarget. 2016;7:33440–33450. doi: 10.18632/oncotarget.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dimitrova V, Arcaro A. Targeting the PI3K/AKT/mTOR signaling pathway in medulloblastoma. Curr Mol Med. 2015;15:82–93. doi: 10.2174/1566524015666150114115427. [DOI] [PubMed] [Google Scholar]
- 29.Cruceru ML, Enciu AM, Popa AC, Albulescu R, Neagu M, Tanase CP, Constantinescu SN. Signal transduction molecule patterns indicating potential glioblastoma therapy approaches. Onco Targets Ther. 2013;6:1737–1749. doi: 10.2147/OTT.S52365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei Z, Liu Y, Wang Y, Zhang Y, Luo Q, Man X, Wei F, Yu X. Downregulation of Foxo3 and TRIM31 by miR-551b in side population promotes cell proliferation, invasion, and drug resistance of ovarian cancer. Med Oncol. 2016;33(126) doi: 10.1007/s12032-016-0842-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freed-Pastor WA, Prives C. Mutant p53: One name, many proteins. Genes Dev. 2012;26:1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14:359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen SR, Cai WP, Dai XJ, Guo AS, Chen HP, Lin GS, Lin RS. Research on miR-126 in glioma targeted regulation of PTEN/PI3K/Akt and MDM2-p53 pathways. Eur Rev Med Pharmacol Sci. 2019;23:3461–3470. doi: 10.26355/eurrev_201904_17711. [DOI] [PubMed] [Google Scholar]
- 34.Toledo F, Wahl GM. Regulating the p53 pathway: In vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Z, Li M, Wang H, Agrawal S, Zhang R. Antisense therapy targeting MDM2 oncogene in prostate cancer: Effects on proliferation, apoptosis, multiple gene expression, and chemotherapy. Proc Natl Acad Sci USA. 2003;100:11636–11641. doi: 10.1073/pnas.1934692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hermisson M, Klumpp A, Wick W, Wischhusen J, Nagel G, Roos W, Kaina B, Weller M. O6-methylguanine DNA methyltransferase and p53 status predict temozolomide sensitivity in human malignant glioma cells. J Neurochem. 2006;96:766–776. doi: 10.1111/j.1471-4159.2005.03583.x. [DOI] [PubMed] [Google Scholar]
- 37.Bleau AM, Hambardzumyan D, Ozawa T, Fomchenko EI, Huse JT, Brennan CW, Holland EC. PTEN/PI3K/Akt pathway regulates the side population phenotype and ABCG2 activity in glioma tumor stem-like cells. Cell Stem Cell. 2009;4:226–235. doi: 10.1016/j.stem.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li M, Liang RF, Wang X, Mao Q, Liu YH. BKM120 sensitizes C6 glioma cells to temozolomide via suppression of the PI3K/Akt/NF-κB/MGMT signaling pathway. Oncol Lett. 2017;14:6597–6603. doi: 10.3892/ol.2017.7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mueller S, Phillips J, Onar-Thomas A, Romero E, Zheng S, Wiencke JK, McBride SM, Cowdrey C, Prados MD, Weiss WA, et al. PTEN promoter methylation and activation of the PI3K/Akt/mTOR pathway in pediatric gliomas and influence on clinical outcome. Neuro Oncol. 2012;14:1146–1152. doi: 10.1093/neuonc/nos140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang BS, Luo QZ, Han Y, Huang D, Tang QP, Wu LX. MiR-223/PAX6 axis regulates glioblastoma stem cell proliferation and the chemo resistance to TMZ via regulating PI3K/Akt pathway. J Cell Biochem. 2017;118:3452–3461. doi: 10.1002/jcb.26003. [DOI] [PubMed] [Google Scholar]
- 41.Wu Y, Dong L, Bao S, Wang M, Yun Y, Zhu R. FK228 augmented temozolomide sensitivity in human glioma cells by blocking PI3K/AKT/mTOR signal pathways. Biomed Pharmacother. 2016;84:462–469. doi: 10.1016/j.biopha.2016.09.051. [DOI] [PubMed] [Google Scholar]
- 42.Cioce M, Canino C, Goparaju C, Yang H, Carbone M, Pass HI. Autocrine CSF-1R signaling drives mesothelioma chemoresistance via AKT activation. Cell Death Dis. 2014;5(e1167) doi: 10.1038/cddis.2014.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang H, He L, Kruk P, Nicosia SV, Cheng JQ. Aurora-A induces cell survival and chemoresistance by activation of Akt through a p53-dependent manner in ovarian cancer cells. Int J Cancer. 2006;119:2304–2312. doi: 10.1002/ijc.22154. [DOI] [PubMed] [Google Scholar]
- 44.Guo P, Qiu Y, Ma X, Li T, Ma X, Zhu L, Lin Y, Han L. Tripartite motif 31 promotes resistance to anoikis of hepatocarcinoma cells through regulation of p53-AMPK axis. Exp Cell Res. 2018;368:59–66. doi: 10.1016/j.yexcr.2018.04.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the datasets generated and analyzed during the present study are included in this manuscript.