Abstract
Osteosarcoma is the most common primary malignancy of bones and frequently affects young children and adolescents. There are several challenges associated with treating osteosarcoma owing to the aggressiveness of the disease, as well as the risk of chemoresistance. Numerous studies are being performed with the aim of identifying improved prognostic and therapeutic markers for this malignancy. Maternal embryonic leucine zipper kinase (MELK) is an oncogene that has been studied in several types of cancer in recent years. In the present study, the expression of MELK in osteosarcoma and normal tissue samples was examined, and the effects of MELK expression on osteosarcoma cellular proliferation, metastasis, the cell cycle and apoptosis were demonstrated using CCK-8, wound healing, migration and invasion and apoptosis assays. The role of MELK in cancer progression in osteosarcoma was determined, revealing the association between MELK expression and prognosis of osteosarcoma. It was demonstrated that knockdown of MELK resulted in reduced proliferation, migration and invasion in vitro along with potentiation of apoptosis and cell cycle arrest. Furthermore, the effect of the targeted MELK inhibitor, OTSSP167, on tumor progression of osteosarcoma in vitro and in vivo was assessed. Mechanistically, it was demonstrated that MELK promoted osteosarcoma proliferation and metastasis by regulating PCNA and MMP9 expression via the PI3K/Akt/mTOR signaling pathway. Thus, the present study revealed the oncogenic role played by MELK, and established MELK as a valuable prognostic and therapeutic marker in osteosarcoma.
Keywords: osteosarcoma, maternal embryonic lucine zipper kinase, MELK inhibitor OTSSP167, oncogene, prognostic marker
Introduction
Osteosarcoma is the most common primary malignancy of bones, and accounts for ~2% of all childhood malignancies (1–3). It is primarily diagnosed in teens and adolescents and in the elderly population over 65 years (4,5). Osteosarcoma is a markedly aggressive cancer with a high capacity to form distant metastases, and there is a high rate of resistance to the traditional therapeutic approaches (3). There have been only minimal advances made in recent decades in the treatment of osteosarcoma (5). Although multimodal therapy has revealed improved outcomes in patients with osteosarcoma, this is not the case for the patients who present with metastases at the initial diagnosis (6). Therefore, it is necessary to study the pathogenesis and metastatic mechanisms underlying the development of osteosarcoma, to identify novel therapeutic targets for improving the treatment and prognosis.
Maternal embryonic leucine zipper kinase (MELK) belongs to the Snf1/AMPK kinase family and is also referred to as murine protein K38 (MPK38) or Eg3 protein (7,8). It is highly expressed in the egg and in the pre-implanted embryo in mice, and influences embryonic development (9). MELK is a highly conserved AMP-activated Ser/Thr protein kinase (7), and its expression is upregulated in several types of cancer, including glioblastoma, melanoma, breast cancer, ovarian cancer, gastric cancer and colorectal cancer (10–15). MELK performs various functions in different types of cancer, which promote the development and progression of cancer (13,16,17). However, the role and mechanism of MELK in osteosarcoma has not been studied, to the best of our knowledge.
In the present study, the effects of MELK on crucial functions, such as proliferation, metastasis and apoptosis, which promote the progression of osteosarcoma, was studied. OTSSP167, a MELK inhibitor, is already in phase I clinical trials for the treatment of solid tumors that do not respond to any other treatment (18,19), and if successful may serve as therapy for the treatment of several different types of cancer. Therefore, the role of the novel molecular inhibitor of MELK, OTSSP167, in osteosarcoma was also explored.
The aim of the present study was to determine the roles and the underlying mechanisms of MELK in osteosarcoma. MELK expression was determined to be upregulated in osteosarcoma compared with the normal control samples, and the increased expression was associated with a less favorable prognosis. Through analysis of the clinicopathological characteristics, it was revealed that there was a positive association between MELK expression with metastasis and a poor response to chemotherapy. A series of in vitro and in vivo experiments were performed to establish the role of MELK in crucial functions such as proliferation, metastasis, cell cycle progression and apoptosis of osteosarcoma. Furthermore, the role and therapeutic effects of OTSSP167 in osteosarcoma were also studied in vitro and in vivo. The results revealed that MELK promoted osteosarcoma proliferation and metastasis by regulating PCNA and MMP9 expression through the PI3K/Akt/mTOR signaling pathway. Collectively, these results indicated that MELK may be a novel prognostic marker and therapeutic target for treatment of osteosarcoma.
Materials and methods
Analysis of public datasets
Gene expression data including 84 cases of osteosarcoma were acquired from the Therapeutically Applicable Research to Generate Effective Treatments (TARGET) program, which is accessible publicly (https://portal.gdc.cancer.gov/projects). The relative differences in MELK expression were analyzed using X-tile software (version 3.6.1; Yale University School of Medicine) to determine the thresholds for producing the most significant log-rank test P-values, which segregated the data into MELK-low and MELK-high groups. The differences in survival between the low- and high-expression groups were evaluated using Kaplan-Meier analysis.
Tissue samples and cell lines
A total of 25 osteosarcoma tissue samples that had not received any neoadjuvant chemotherapy and 5 osteoblastoma tissue samples were collected from patients who were treated at Qilu Hospital of Shandong University from January 2010 to June 2014. All patients signed informed consent to participate in the present study, and the study was approved by the Ethics Committee of Qilu Hospital of Shandong University. The small number of samples is due the fact that most osteosarcoma patients receive preoperative neoadjuvant chemotherapy. MNNG/HOS and hFOB1.19 cell lines were purchased from the American Type Culture Collection. MNNG/HOS cells were maintained in DMEM with 5% FBS (Gibco; Thermo Fisher Scientific, Inc.) and hFOB1.19 cells were maintained in a 1:1 mixture of Ham's F12 medium and DMEM supplemented with 10% FBS. All the cells were incubated at 37°C with 5% CO2. MELK inhibitor (OTSSP167; cat. no. S7159) and PI3K inhibitor (LY294002; cat. no. S1105) were purchased from Selleck Chemicals.
Immunohistochemistry
Immunohistochemical staining was performed on tissue sections (4-µm thick) of formalin-fixed at room temperature for 48 h, paraffin-embedded osteosarcoma tissue samples. Xylene and ethanol were used to deparaffinize and rehydrate the tissue slides. Antigen retrieval was performed using sodium citrate buffer solution (pH 6.0) maintained at a sub-boiling temperature for 15 min. Endogenous peroxidase and nonspecific binding were blocked using 3% hydrogen peroxide for 15 min and goat serum (SP-kit: cat. no. SP9000; Beijiing Zhongshan Golden Bridge Biotechnology) for 30 min, respectively at room temperature. Tissues were incubated with anti-MELK primary antibody (1:250; cat. no. bs-12201R; Bioss) in a humid chamber overnight at 4°C. Expression was detected using a I–View 3,3′-diaminobenzidine staining technique. To evaluate the staining intensity, semiquantitative evaluation of protein levels in tissues was performed using the H-score technique (range, 0–3) (20). The staining intensity was graded as: 0, none; 1, weak; 2, moderate; and 3, strong. The H-score was calculated using the formula: H-score=ΣPi × I; where i represents the staining intensity and Pi the percentage of cells at each level of intensity. An H-score <1.5 was classified as low, and a H-score ≥1.5 was classified as a high protein expression level.
Transient transfection
Small interfering (si)RNAs and the corresponding negative control (NC) were used to transiently knockdown expression of MELK. The siRNAs were purchased from (Shanghai GenePharma, Co., Ltd.). RNAi-mediated knockdown was performed using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The sequences of the siRNA used were: si-MELK, 5′-GACAUCCUAUCUAGCUGCA-3′ and si-NC, 5′-UUCUCCGAACGUGUCACGUTT-3′.
Reverse transcription-quantitative (RT-q)PCR
Total RNA was extracted from osteosarcoma cells using TRIzol® reagent (Ambion; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The cDNA was synthesized using a One Step PrimeScript miRNA cDNA Synthesis kit (Takara Bio, Inc.). qPCR was performed using a SYBR Green Premix Ex Taq II (Takara Bio, Inc.) with a StepOne Plus Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The reaction conditions were as follows: Denaturation at 95°C for 5 sec, followed by 40 cycles of annealing at 60°C for 10 sec and an extension at 72°C for 30 sec. GAPDH expression was used as the endogenous control for the detection of miRNA expression levels. The 2−ΔΔCq was used to calculate the relative gene expression level (21). The sequences of the primers used were: GAPDH forward, 5′-CAGGAGGCATTGCTGATGAT-3′ and reverse, 5′-GAAGGCTGGGGCTCATTT3′; and MELK, forward 5′-TCTCCCAGTAGCATTCTGCTT3′ and reverse 5′-TGATCCAGGGATGGTTCAATAGA3′. Primers were purchased from Sangon Biotech, Co., Ltd.
Western blotting
Cells and tissues were harvested and lysed using RIPA Lysis Buffer (Beyotime Institute of Biotechnology) with PMSF (1%). Protein samples were incubated for 30 min on ice and sonicated, after which the cell debris were removed by centrifugation at 12,000 × g. The protein concentration was determined using a bicinchoninic acid assay kit (Thermo Fisher Scientific, Inc.). Protein samples (30 µg) were resolved by SDS-PAGE (5% stacking gel and 10% separating gel) and transferred to a PVDF membrane (EMD Millipore). The membrane was blocked with 5% non-fat milk at room temperature for 1 h, incubated overnight at 4°C with the primary antibody, and subsequently incubated with horseradish peroxidase-conjugated anti-rabbit (dilution 1:5,000; cat. no. TA130023; OriGene Technologies, Inc.) secondary antibody for 2 h. Signals were visualized using enhanced chemiluminescence reagent and ImageQuant LAS 4000 (GE Healthcare Life Sciences). ImageJ software version 1.47v (National Institutes of Health) was used for densitometric measurement.
The antibodies used were: Phosphorylated (p)-PI3K p85 (1:1,000; product no. 4228; Cell Signaling Technology, Inc.), total PI3K p85 (1:1,000; product code ab40755; Abcam), p-Akt(S473) (1:1,000; product no. 9271S; CST Biological Reagents Co., Ltd.), total Akt (1:1,000; product no. 9272S; CST Biological Reagents, Co., Ltd.), GAPDH (1:5,000; cat. no. AF7021; Affinity Biosciences), MELK (1:1,000; product code ab108529; Abcam), mTOR (1:1,000; product no. 2972S; CST Biological Reagents, Co., Ltd.), p-mTOR (1:1,000; product no. 5536S; CST Biological Reagents, Co., Ltd.), PCNA (1:750, cat. no. AF0239) and MMP9 (1:750; cat. no. AF5228; both from Affinity Biosciences).
Cell proliferation assay
The proliferative capacity of cells was assessed using a Cell Counting Kit-8 (CCK-8) assay (BestBio). Each cell line was seeded in quintuplicate into 96-well plates (1-2×103 cells/well) for 1–5 days. At the specified time-points, 10 µl CCK-8 reagent was added to each well, and the cells were incubated for an additional 4 h at 37°C. Subsequently, the absorbance values at 450 nm were measured using a Varioskan Flash microplate reader (Thermo Fisher Scientific, Inc.). Cell proliferation was measured in si-MELK and si-NC transfected cells, and in cells treated with 40 nM OTSSP167. For the latter, cells were initially treated for 48 h with OTSSP167.
Cell viability assay
A total of 5×103 cells/well were seeded in quintuplicate into 96-well plates. The plate was incubated at 37°C for 24 h, and subsequently treated with various concentrations of OTSSP167 (0, 2, 4, 8, 16, 32 and 64 nM) for 48 h. For evaluation of cell viability, 10 µl CCK-8 reagent was added to each well and incubated at 37°C for 4 h. The cell viability was determined by measuring the absorbance at 450 nm using Varioskan microplate reader.
Migration and invasion assay
Cellular invasion and migration were analyzed using Boyden chamber-style cell culture inserts with and without Matrigel, respectively (BD Falcon; BD Biosciences). Osteosarcoma cells (1-2×105 cells) were seeded in the upper chambers of the Transwell inserts (24-wells, 8 µm pores; BD Falcon) with 200 µl serum-free media. The lower chambers were filled with 500 µl culture media supplemented with 10% FBS as a chemoattractant. After 12–48 h, cells on the lower surface of the membrane were fixed at room temperature in methanol for 20 min, stained at room temperature with 0.1% crystal violet for 30 min, and counted using a light microscope (magnification, ×200).
Wound healing assay
Cell migration was measured using a wound healing assay. MNNG/HOS cells were used for the assay at a density of 5×105/well. The scratch was generated using 200-µl pipette tip. The culture medium was replaced with serum-free DMEM after the generation of a scratch wound, and wound closure was imaged 48 h later under a light microscope (magnification, ×100). Wound closure was analyzed and is represented as percent closure. The experiments were performed in triplicate. Wound healing assays were performed on si-MELK and si-NC transfected cells, and cells treated with 10 nM OTSSP167.
Cell cycle distribution and apoptosis assay
Cell cycle synchronization was achieved by serum starvation and thymidine double blocking (22). Subsequently, the cells were harvested and stained with 5 µl propidium iodide at 4°C for 30 min in the dark according to the manufacturer's protocol (BestBio). Flow cytometry was performed using a FACScan flow cytometer (BD Biosciences).
Apoptosis was detected using an Annexin V-APC and 7-AAD kit (BioGems International, Inc.) according to the manufacturer's protocol, and flow cytometry was performed on the cells. MNNG/HOS cells were harvested 48 h after transfection or treated with OTSSP167 (10 nM) and subsequently harvested using EDTA-free trypsin, centrifuged at 800 × g for 5 min; and, after washing twice with cold PBS, the cells were resuspended at a concentration of 1×106/ml and stained with 5 µl Annexin V-APC and 5 µl 7-AAD at room temperature for 15 min in the dark. The cells were analyzed by flow cytometry (FACSCalibur) and the results were analyzed using FlowJo software (version 10; TreeStar, Inc.).
Animal experiments
Eight pathogen-free female Balb/c nude mice (weighing 10–14 g; 4 weeks old) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd and housed in standard sterilized conditions in individually ventilated cages (IVCs) with high efficiency particulate air (HEPA) filters at an ambient temperature of 30–31°C, humidity of 50–60% with 12-h light/dark cycle and adequate access to food and water. The health and behavior of the animals were monitored every 3 days. All animal experiments were approved by the Ethics Committee of Qilu Hospital, Shandong University.
For tumorigenesis assays, 1×107 MNNG/HOS cells, resuspended in 200 µl PBS, were injected subcutaneously into the armpits of the mice. The tumor volume was measured daily using a Vernier caliper, with the following formula: ½ × a × b2 where ‘a’ and ‘b’ represent the length and width of the tumor, respectively. After 7 days, when the tumor volumes reached ~100 mm3, the tumor-bearing mice were orally fed Vehicle or OTSSP167 (10 mg/kg, daily). The tumor volumes were measured daily using Vernier calipers. After 7 days, when the maximum tumor volumes reached ~1,000 mm3 the mice were euthanized using cervical dislocation method and then the growth of the tumors was examined. Following excision, the tumors were preserved at −80°C for western blot analysis.
Statistical analysis
Data were analyzed using independent samples t-test, log-rank test, or Fisher's exact test. Fisher's exact test was used to analyze the relationship between MELK expression and clinicopathological data of osteosarcoma patients. Overall survival (OS) curves were plotted using the Kaplan-Meier method and analyzed using a log-rank test. Quantitative data were analyzed using a Student's t-test. Data are expressed as the mean ± the standard deviation of three independent measurements. Statistical analysis was performed using SPSS version 18.0 (SPSS Inc.) and GraphPad Prism version 5.0 (GraphPad Software Inc.). Two-tailed P<0.05 was considered to indicate a statistically significant difference.
Results
MELK expression is upregulated in osteosarcoma and its overexpression is associated with a less favorable prognosis
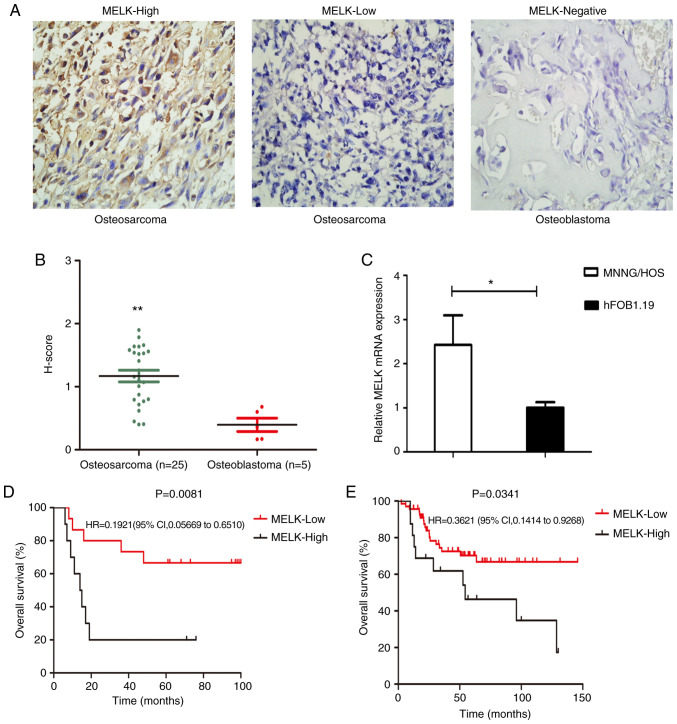
The expression levels of MELK were assessed in 25 osteosarcoma tissue samples and 5 osteoblastoma samples using immunohistochemistry (Fig. 1A). H-score evaluation revealed significantly higher protein expression levels in osteosarcoma tissues compared with the osteoblastoma tissues (Fig. 1B). Of the 25 samples, 40% exhibited high staining and 60% exhibited low staining, whereas in the osteoblastoma samples, none of the tissues showed MELK expression (Fig. 1A). MELK mRNA expression levels were also compared between MNNG/HOS and hFOB1.19 cells. MELK expression was significantly higher in the MNNG/HOS cells compared with the hFOB1.9 cells (Fig. 1C). Subsequently, the association between MELK expression and various clinicopathological characteristics of the patients were assessed. As revealed in Table I higher expression levels of MELK were associated with metastasis and the response to chemotherapy. Kaplan-Meier survival curves revealed that OS was shorter in patients with high MELK expression levels, based on the data obtained from TARGET (Fig. 1E), in agreement with the present analysis. Thus, high expression levels of MELK were associated with less favorable OS in patients (Fig. 1D).
Figure 1.
MELK expression is upregulated in osteosarcoma, and its upregulated expression is associated with a poor prognosis. (A) Immunohistochemical staining of osteosarcoma and osteoblastoma tissue samples (B) Grading of immunohistochemically-stained tissues using H-score. (C) mRNA expression levels of MELK in the MNNG/HOS and hFOB1.19 cells. (D) Survival of patients with osteosarcoma who were treated Qilu hospital with high MELK expression levels was less favorable compared with patients with low expression levels of MELK. (E) Kaplan-Meier survival curves revealed lower overall survival in the patients with high MELK expression based on data obtained from the TARGET database. *P<0.05, **P<0.01, Magnification, ×400. MELK, maternal embryonic leucine zipper kinase; TARGET, Therapeutically Applicable Research to Generate Effective Treatments.
Table I.
Association between MELK expression and clinicopathological data of osteosarcoma patients.
| MELK expression | ||||
|---|---|---|---|---|
| Parameters | N 25 | High n=10 (40%) | Low n=15 (60%) | P-value |
| Age (years) | 0.6968 | |||
| <18 | 14 | 5 | 9 | |
| ≥18 | 11 | 5 | 6 | |
| Sex | 0.2262 | |||
| Male | 13 | 7 | 6 | |
| Female | 12 | 3 | 9 | |
| Location | 0.1500 | |||
| Extremity | 23 | 8 | 15 | |
| Trunk | 2 | 2 | 0 | |
| Histological subtype | 0.8889 | |||
| Osteoblastic | 8 | 2 | 6 | |
| Fibroblastic | 4 | 3 | 1 | |
| Chondroblastic | 7 | 4 | 3 | |
| Othersa | 6 | 1 | 5 | |
| Metastasis | 0.0414b | |||
| Yes | 13 | 8 | 5 | |
| No | 12 | 2 | 10 | |
| Surgery | 0.4139 | |||
| Amputation | 14 | 7 | 7 | |
| Limb salvage | 11 | 3 | 8 | |
| Response to chemotherapy | 0.0154b | |||
| Good | 13 | 2 | 11 | |
| Poor | 12 | 8 | 4 | |
| Enneking stage | 0.472 | |||
| I | 2 | 0 | 2 | |
| II | 20 | 8 | 12 | |
| III | 3 | 2 | 1 | |
P-values were calculated by Fisher's exact test.
Others: Telangiectatic, small-cell, parosteal or undifferentiated osteosarcoma.
P<0.05. MELK, maternal embryonic leucine zipper kinase.
MELK knockdown reduces proliferation in vitro
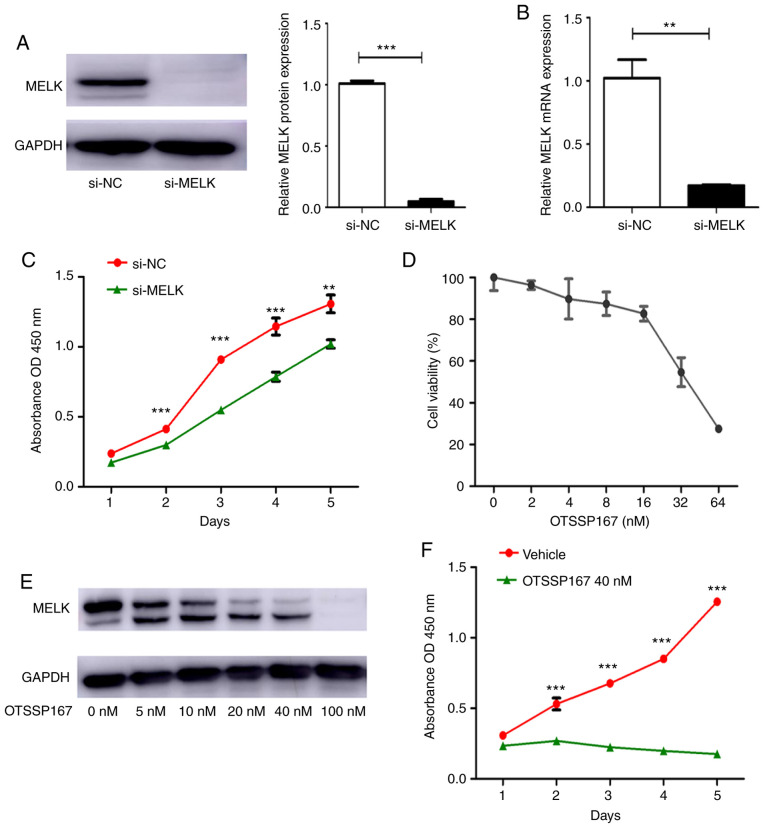
si-MELK was used to knock down the expression of MELK in MNNG/HOS cells. The mRNA and protein expression levels of MELK following knockdown are presented in Fig. 2A and B. To determine the effect of MELK on the proliferation of osteosarcoma cells, a proliferation assay was performed using MNNG/HOS cells transiently transfected with si-NC or si-MELK. As revealed in Fig. 2C, knockdown of MELK expression significantly decreased proliferation. OTSSP167, a MELK inhibitor, was used to assess its effects on proliferation. First, the IC50 of OTSSP167 was determined, which was determined to be 40 nM/ml (Fig. 2D). There was also a dose-dependent decrease in MELK expression levels following treatment with OTSSP167 (Fig. 2E). Using 40 nM OTSSP167, the proliferation of cells treated with vehicle or OTSSP167 was assessed. As revealed in Fig. 2F, there was a significant decrease in the proliferation of cells treated with OTSSP167. These in vitro experiments demonstrated that MELK serves a substantial role in the proliferation of osteosarcoma cells.
Figure 2.
MELK inhibition is associated with suppression of proliferation in vitro. (A and B) MELK expression was knocked down using si-MELK, and the efficiency of knock down was determined using western blotting and PCR in MNNG/HOS cells. (C) A CCK-8 assay revealed that proliferation was reduced following knockdown of MELK. (D) A cell viability assay was used to determine the IC50 of OTSSP167, which was determined to be 40 nM. (E) Western blots revealing the concentration-dependent inhibition of MELK by OTSSP167. (F) A CCK-8 proliferation assay revealed OTSSP167 inhibited proliferation of osteosarcoma cells compared with the vehicle control. **P<0.01, ***P<0.001. MELK, maternal embryonic leucine zipper kinase; si, small interfering; CCK-8, CELL Counting Kit-8.
OTSSP167 inhibits tumorigenesis in vivo
To determine the role of MELK in tumorigenesis, MNNG/HOS cells were subcutaneously injected into the armpits of nude mice. Tumor growth was closely monitored and tumor volume was measured daily using a Vernier caliper, using the aforementioned formula. When the tumor volume reached ~100 mm3, the mice were randomly divided into two groups; one group was orally fed OTSSP167 (10 mg/kg) daily whilst the other group was fed vehicle. After 7 days, the mice were euthanized, and the tumors were excised and analyzed. The tumor volume exhibited a gradual decrease in the group fed OTSSP167 compared with the control group fed vehicle. There was a significant difference in the tumor weight between the two groups (Fig. 3A-D). To evaluate the results, the protein expression levels were assessed between the groups, and the expression of MELK was revealed to be lower in the mice fed OTSSP167 (Fig. 3E).
Figure 3.
OTSSP167 reduces tumorigenesis in vivo. (A) Tumor xenografts were significantly smaller in mice orally fed OTSSP167 compared with the control group. (B) Time-dependent reduction in the volume of the tumor xenografts in mice orally fed OTSSP167 daily. (C) Tumor volumes and (D) tumor weights were significantly lower in the mice fed OTSSP167 compared with the vehicle control. (E) Western blots revealing reduced MELK expression in the tumors of mice fed OTSSP167. **P<0.01, ***P<0.001. MELK, maternal embryonic leucine zipper kinase; p.o, per os; q.d. quaque die.
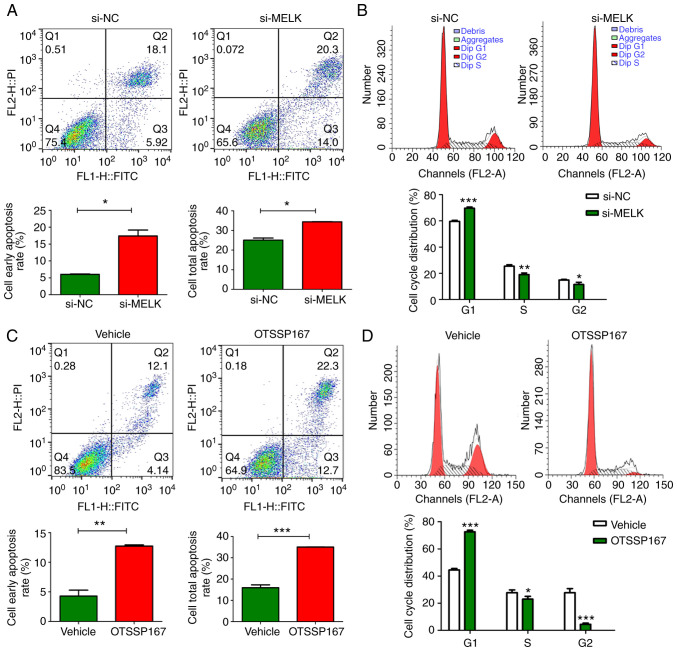
MELK reduces apoptosis and promotes cell cycle progression in osteosarcoma cells
The proportion of apoptotic MNNG/HOS cells following MELK knockdown was assessed and compared with the control cells. The results revealed that there was an increase in the total proportion of apoptotic cells and the portion of cells in early stage apoptosis when MELK expression was knocked down (Fig. 4A). Additionally, MELK knockdown resulted in inhibition of cell cycle progression in osteosarcoma cells (Fig. 4B). There was an increased proportion of cells in the G1 phase and a lower proportion of cells in the G2 and S phases in the MELK-knockdown cells compared with the control. It was hypothesized that MELK exerted its function on osteosarcoma proliferation by inhibiting apoptosis and potentiating the cell cycle. Such effects were also demonstrated when MELK was inhibited by OTSSP167. Notably, the magnitude of increase in apoptosis and cell cycle inhibition were more pronounced with OTSSP167 (Fig. 4C and D).
Figure 4.
Effect of MELK on apoptosis and cell cycle progression. (A) Knockdown of MELK using si-MELK resulted in an increase in the proportion of early apoptotic cells and in the proportion of total apoptotic cells. (B) MELK knockdown resulted in cell cycle arrest in osteosarcoma cells. (C) Potentiation of apoptosis following inhibition of MELK by OTSSP167. (D) Cell cycle arrest in osteosarcoma cells treated with OTSSP167. *P<0.05, **P<0.01, ***P<0.001. MELK, maternal embryonic leucine zipper kinase; si, small interfering.
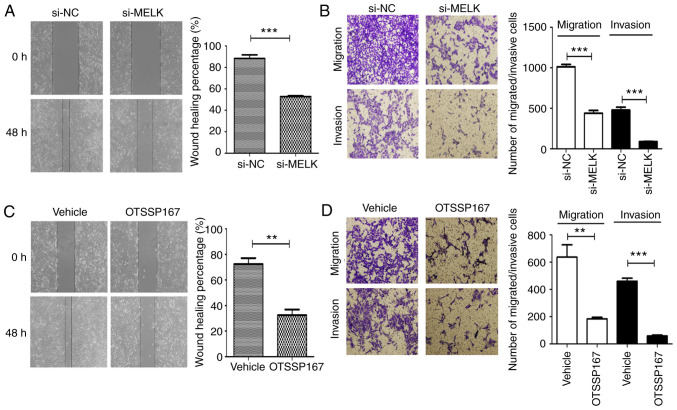
MELK knockdown decreases migration and invasion in vitro
The role of MELK on the migration and invasion of osteosarcoma cells was assessed using Transwell assays with or without Matrigel, and wound healing assays. As revealed in Fig. 5A and B, MNNG/HOS cells exhibited reduced migratory and invasive capacities following MELK knockdown. When MELK was inhibited using the inhibitor OTSSP167, similar inhibitory effects were observed on the migration and invasion abilities (Fig. 5C and D). Thus, it was hypothesized that MELK serves a role in potentiating the migratory and invasive capacities of osteosarcoma cells. The clinicopathological analysis from immunohistochemistry also revealed a significant association between upregulated MELK expression and metastasis of osteosarcoma, in agreement with the results of the in vitro experiments.
Figure 5.
MELK knockdown reduces migration and invasion in vitro. (A) Wound healing assays revealed the reduced migratory potential of cells following knockdown of MELK. (B) Transwell assays revealed reduced migratory and invasive potential of MNNG/HOS cells following knockdown of MELK. (C) Migration (wound healing assay) and (D) migration and invasion (Transwell assays) were reduced in osteosarcoma cells following treatment with OTSSP167. **P<0.01, ***P<0.001. MELK, maternal embryonic leucine zipper kinase.
MELK promotes proliferation and metastasis of osteosarcoma through the PI3K/AKT/mTOR pathway
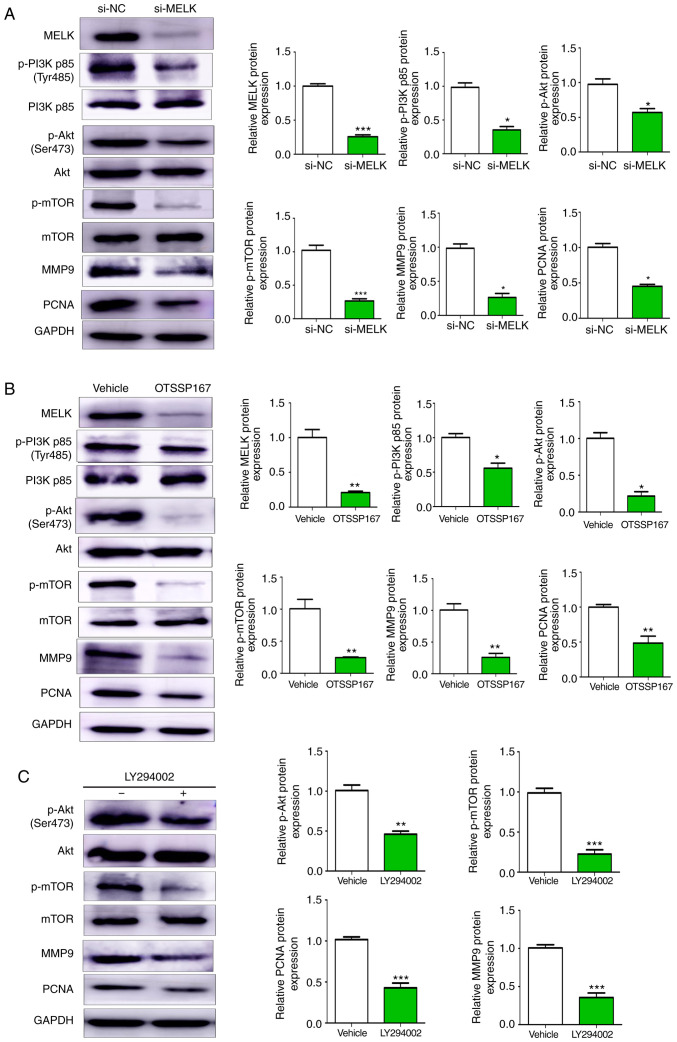
The PI3K/AKT and the mTOR signaling pathways are considered principal pathways involved in several oncogenic mechanisms. In the present study, it was revealed that knocking down MELK expression resulted in reduced phosphorylation of PI3K, AKT and mTOR, and reduced protein expression levels of MMP9 and PCNA (Fig. 6A). Similar results were also observed with the use of OTSSP167 (Fig. 6B). Furthermore, in MNNG/HOS cell treated with LY294002 (20 µM), the expression of p-AKT, p-mTOR, PCNA and MMP9 were decreased (Fig. 6C). Collectively, these results indicated that MELK promoted proliferation and metastasis of osteosarcoma cells by modulating PCNA and MMP9 expression via the PI3K/AKT/mTOR pathway.
Figure 6.
MELK promotes osteosarcoma proliferation and metastasis through the PI3K/AKT/mTOR pathway. (A) Western blot analysis of changes in protein expression following knockdown of MELK. Cells transfected with si-MELK exhibited lower expression levels of p-PI3K p85 (Tyr485), p-AKT (Ser473), p-mTOR, MMP9 and PCNA. (B) Western blotting revealed lower protein expression levels of p-PI3K, p-AKT, p-mTOR, MMP9 and PCNA in the cells treated with OTSSP167 compared with the vehicle control. (C) PI3K inhibitor LY294002 decreased the expression levels of p-AKT, p-mTOR, PCNA and MMP9. GAPDH was used as the internal control. *P<0.05, **P<0.01, ***P<0.001. MELK, maternal embryonic leucine zipper kinase; si, small interfering; p-, phospho-.
Discussion
There is an increasing body of literature showing that MELK expression is upregulated in several types of cancer (10–15), and its upregulated expression is associated with a less favorable prognosis in cancer (13,16,17) Similarly, MELK has been demonstrated to exert its oncogenic functions by affecting cellular proliferation, cell cycle, apoptosis and metastasis (23,24). The present study is the first study, to the best of our knowledge, to reveal the role of MELK in the progression and metastasis of osteosarcoma.
In the present study, MELK expression was revealed to be significantly upregulated in osteosarcoma tissues compared with normal tissues. Compared with the hFOB1.19 cell line, MELK expression levels were also upregulated in the MNNG/HOS osteosarcoma cell line. Analysis of the clinicopathological characteristics revealed that upregulated expression of MELK was closely associated with metastasis and chemotherapy response in patients with osteosarcoma. Furthermore, MELK expression was associated with the OS of the patients; patients with high MELK expression levels had a worse prognosis than patients with low expression.
MELK expression was knocked down in the MNNG/HOS osteosarcoma cell line using si-MELK or the MELK inhibitor, OTSSP167. Knockdown or inhibition of MELK activity resulted in a significant decrease in cellular proliferation and metastatic behavior. MELK has previously been revealed to regulate proliferation, migration and invasion (11–17). In agreement with the previous studies, suppressing MELK in the MNNG/HOS osteosarcoma cell line reduced proliferation, migration and invasion. There was also a significant decrease in cell cycle progression and potentiation of apoptosis due to knockdown of MELK.
In addition to identifying the role of MELK in osteosarcoma, the underlying mechanism was also identified. PCNA has been revealed to be an indicator of cell proliferation, and to serve a principal role in the regulation of cellular proliferation and cell cycle progression (25,26). Furthermore, PCNA is a known oncogene and its upregulation has been revealed to be associated with a poorer prognosis in patients with osteosarcoma (27).
In the present study, the expression levels of PCNA were revealed to be decreased following knockdown or inhibition of MELK. Thus, it was hypothesized that MELK may promote the growth of osteosarcoma by regulating PCNA expression.
MMPs are proteolytic enzymes that serve a role in extracellular matrix remodeling, affecting cell growth, differentiation, migration, invasion and angiogenesis (28). As an important member of the MMP family, MMP-9 has been revealed to affect tumor metastasis in several tumors, including osteosarcoma (29–31). The results of the present study revealed that there was a significant decrease in migration and invasion based on the Transwell and wound healing assays. Western blot analysis also revealed that MMP9 protein expression levels were lower in the cells when MELK expression was knocked down, indicating reduced invasive capacity.
The PI3K/AKT/mTOR pathway is associated with a variety of cellular functions and is known to serve a distinct role in cancer progression (32). In the present study, silencing of MELK resulted in marked suppression of p-PI3K, p-AKT, p-mTOR, PCNA and MMP9 expression. It was further revealed that a PI3K inhibitor decreased the expression of p-AKT, p-mTOR, PCNA and MMP9. Collectively, it was hypothesized that MELK affected the proliferation and migration of osteosarcoma cells by regulating PCNA and MMP9 expression via the PI3K/Akt/mTOR signaling pathway. Studies have previously revealed a link between PCNA and the PI3K/Akt/mTOR signaling pathway (33,34), and this pathway is known to be involved in regulation of migration and invasion in various types of cancer, possibly through regulation of crucial MMPs (35–37).
OTSSP167 is a potent MELK inhibitor, which has been demonstrated to possess antitumor properties in several types of cancer (15,16). OTSSP167 is a protein kinase inhibitor that prevents MELK phosphorylation and abrogates its downstream substrates (38). OTSSP167 is being developed as an anticancer drug for patients with solid tumors, which have not responded to established modes of treatment, and is emerging as a promising therapeutic for treatment of several types of solid tumors (20). A phase I clinical trial studying the efficiency of OTSSP167 was started in August of 2013 (18). In the present study, OTSSP167 served an inhibitory role in osteosarcoma progression through suppression of MELK. When xenografted tumors were produced in nude mice and were orally fed OTSSP167, there was a gradual decrease in the size of tumors over time, whereas the control group, which was fed the vehicle alone, possessed tumors which grew over time. This is in agreement with the in vitro proliferation assays and strongly supports the hypothesis that MELK is responsible for tumor progression and its knockdown or inhibition by RNAi or OTSSP167, respectively, could reduce tumor progression. In recent decades, interest in studying inhibitors of PI3K, AKT and mTOR has increased. Several of these inhibitors are in various phases of clinical trials (39). Since the MELK inhibitor OTSSP167 has been revealed to downregulate the phosphorylation of these molecules, it is hypothesized that it may be used as a substitute for these inhibitors. However, additional studies are required for verification.
In summary, MELK expression was upregulated in osteosarcoma, and this was associated with a poor prognosis. MELK increased proliferation and metastasis of osteosarcoma cells by modulating PCNA and MMP9 expression via the PI3K/AKT/mTOR pathway. The present study is the first to examine the role of MELK in osteosarcoma, the first to determine the underlying mechanism of MELK-mediated effects, and the first to determine the effect of OTSSP167 in suppressing osteosarcoma progression through inhibition of MELK. These results suggest that MELK may be used as a prognostic biomarker, and highlights its potential as a therapeutic target for the treatment of osteosarcoma.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 81672655) and the Natural Science Foundation of Shandong Province of China (grant no. ZR2017MH047).
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request. The results published here are in whole or in part are based on data generated by the Therapeutically Applicable Research to Generate Effective Treatments (https://ocg.cancer.gov/programs/target) initiative, phs000468. The data used for this analysis are available at https://portal.gdc.cancer.gov/projects.
Authors' contributions
SFAJ and XW provided substantial contributions to the conception and design of the work. SFAJ and XW were responsible for the experimental procedure and clinical studies. KL and XL conducted the literature search. QY and JL provided the data acquisition and carried out the statistical analysis. SD drafted the work, edited the manuscript and revised it critically for important intellectual content. All authors critically revised and approved the final manuscript and agree to be responsible for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
All patients signed informed consent to participate in the present study, and the study was approved by the Ethics Committee of Qilu Hospital of Shandong University. All animal experiments were approved by the Ethics Committee of Qilu Hospital, Shandong University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Biermann JS, Adkins DR, Agulnik M, Benjamin RS, Brigman B, Butrynski JE, Cheong D, Chow W, Curry WT, Frassica DA, et al. Bone cancer. J Natl Compr Canc Netw. 2013;11:688–723. doi: 10.6004/jnccn.2013.0088. [DOI] [PubMed] [Google Scholar]
- 3.Lindsey BA, Markel JE, Kleinerman ES. Osteosarcoma overview. Rheumatol Ther. 2017;4:25–43. doi: 10.1007/s40744-016-0050-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 5.Ottaviani G, Jaffe N. Pediatric and adolescent osteosarcoma. Vol. 152. US: Springer; 2009. The epidemiology of osteosarcoma; pp. 3–13. [DOI] [PubMed] [Google Scholar]
- 6.Bacci G, Longhi A, Bertoni F, Briccoli A, Versari M, Pignotti E, Picci P. Bone metastases in osteosarcoma patients treated with neoadjuvant or adjuvant chemotherapy. The Rizzoli experience in 52 patients. Acta Orthopaed. 2006;77:938–943. doi: 10.1080/17453670610013268. [DOI] [PubMed] [Google Scholar]
- 7.Heyer BS, Warsowe J, Solter D, Knowles BB, Ackerman SL. New member of the SNF1/AMPK kinase family, Melk, is expressed in the mouse egg and preimplantation embryo. Mol Reprod Dev. 1997;47:148–156. doi: 10.1002/(SICI)1098-2795(199706)47:2<148::AID-MRD4>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Zhou X, Li Y, Xu Y, Lu K, Li P, Wang X. Inhibition of maternal embryonic leucine zipper kinase with OTSSP167 displays potent anti-leukemic effects in chronic lymphocytic leukemia. Oncogene. 2018;37:5520–5533. doi: 10.1038/s41388-018-0333-x. [DOI] [PubMed] [Google Scholar]
- 9.Jiang P, Zhang D. Maternal embryonic leucine zipper kinase (MELK): A novel regulator in cell cycle control, embryonic development, and cancer. Int J Mol Sci. 2013;14:21551–21560. doi: 10.3390/ijms141121551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ganguly R, Hong CS, Smith LG, Kornblum HI, Nakano I. Maternal embryonic leucine zipper kinase: Key kinase for stem cell phenotype in glioma and other cancers. Mol Cancer Ther. 2014;13:1393–1398. doi: 10.1158/1535-7163.MCT-13-0764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janostiak R, Rauniyar N, Lam TT, Ou J, Zhu LJ, Green MR, Wajapeyee N. MELK promotes melanoma growth by stimulating the NF-κB pathway. Cell Rep. 2017;21:2829–2841. doi: 10.1016/j.celrep.2017.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li G, Yang M, Zuo L, Wang MX. MELK as a potential target to control cell proliferation in triple-negative breast cancer MDA-MB-231 cells. Oncol Lett. 2018;15:9934–9940. doi: 10.3892/ol.2018.8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohler RS, Kettelhack H, Knipprath-Mészaros AM, Fedier A, Schoetzau A, Jacob F, Heinzelmann-Schwarz V. MELK expression in ovarian cancer correlates with poor outcome and its inhibition by OTSSP167 abrogates proliferation and viability of ovarian cancer cells. Gynecol Oncol. 2017;145:159–166. doi: 10.1016/j.ygyno.2017.02.016. [DOI] [PubMed] [Google Scholar]
- 14.Li S, Li Z, Guo T, Xing XF, Cheng X, Du H, Wen XZ, Ji JF. Maternal embryonic leucine zipper kinase serves as a poor prognosis marker and therapeutic target in gastric cancer. Oncotarget. 2016;7:6266–6280. doi: 10.18632/oncotarget.6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gray D, Jubb AM, Hogue D, Dowd P, Kljavin N, Yi S, Bai W, Frantz G, Zhang Z, Koeppen H, et al. Maternal embryonic leucine Zipper Kinase/Murine protein serine-threonine kinase 38 is a promising therapeutic target for multiple cancers. Cancer Res. 2005;65:9751–9761. doi: 10.1158/0008-5472.CAN-04-4531. [DOI] [PubMed] [Google Scholar]
- 16.Wu S, Chen X, Hu C, Wang J, Shen Y, Zhong Z. Up-regulated maternal embryonic leucine zipper kinase predicts poor prognosis of hepatocellular carcinoma patients in a Chinese Han population. Med Sci Monit. 2017;23:5705–5713. doi: 10.12659/MSM.907600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pickard MR, Green AR, Ellis IO, Caldas C, Hedge VL, Mourtada-Maarabouni M, Williams GT. Dysregulated expression of Fau and MELK is associated with poor prognosis in breast cancer. Breast Cancer Res. 2009;11:R60. doi: 10.1186/bcr2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chung S, Nakamura Y. MELK inhibitor, novel molecular targeted therapeutics for human cancer stem cells. Cell Cycle. 2013;12:1655–1656. doi: 10.4161/cc.24988. [DOI] [PubMed] [Google Scholar]
- 19.Chung S, Suzuki H, Miyamoto T, Takamatsu N, Tatsuguchi A, Ueda K, Kijima K, Nakamura Y, Matsuo Y. Development of an orally-administrative MELK-targeting inhibitor that suppresses the growth of various types of human cancer. Oncotarget. 2012;3:1629–1640. doi: 10.18632/oncotarget.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu D, Zhang XX, Li MC, Cao CH, Wan DY, Xi BX, Tan JH, Wang J, Yang ZY, Feng XX, et al. C/EBPβ enhances platinum resistance of ovarian cancer cells by reprogramming H3K79 methylation. Nat Commun. 2018;9:1739. doi: 10.1038/s41467-018-03590-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Chen G, Deng X. Cell synchronization by double thymidine block. Bio Protoc. 2018;8:e2994. doi: 10.21769/BioProtoc.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ganguly R, Mohyeldin A, Thiel J, Kornblum HI, Beullens M, Nakano I. MELK-a conserved kinase: Functions, signaling, cancer, and controversy. Clin Transl Med. 2015;4:11. doi: 10.1186/s40169-014-0045-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin ML, Park JH, Nishidate T, Nakamura Y, Katagiri T. Involvement of maternal embryonic leucine zipper kinase (MELK) in mammary carcinogenesis through interaction with Bcl-G, a pro-apoptotic member of the Bcl-2 family. Breast Cancer Res. 2007;9:R17. doi: 10.1186/bcr1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strzalka W, Ziemienowicz A. Proliferating cell nuclear antigen (PCNA): A key factor in DNA replication and cell cycle regulation. Ann Bot. 2011;107:1127–1140. doi: 10.1093/aob/mcq243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Müller R, Misund K, Holien T, Bachke S, Gilljam KM, Våtsveen TK, Rø TB, Bellacchio E, Sundan A, Otterlei M. Targeting proliferating cell nuclear antigen and its protein interactions induces apoptosis in multiple myeloma cells. PLoS One. 2013;8:e70430. doi: 10.1371/journal.pone.0070430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, Wang D, Yuan N, Liu F, Wang F, Wang B, Zhou D. The prognostic value of PCNA expression in patients with osteosarcoma: A meta-analysis of 16 studies. Medicine (Baltimore) 2017;96:e8254. doi: 10.1097/MD.0000000000008254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jabłońska-Trypuć A, Matejczyk M, Rosochacki S. Matrix metalloproteinases (MMPs), the main extracellular matrix (ECM) enzymes in collagen degradation, as a target for anticancer drugs. J Enzyme Inhib Med Chem. 2016;31(Suppl 1):S177–S183. doi: 10.3109/14756366.2016.1161620. [DOI] [PubMed] [Google Scholar]
- 29.Viros D, Camacho M, Zarraonandia I, García J, Quer M, Vila L, León X. Prognostic role of MMP-9 expression in head and neck carcinoma patients treated with radiotherapy or chemoradiotherapy. Oral Oncol. 2013;49:322–325. doi: 10.1016/j.oraloncology.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 30.Vilen ST, Salo T, Sorsa T, Nyberg P. Fluctuating roles of matrix metalloproteinase-9 in oral squamous cell carcinoma. ScientificWorldJournal. 2013;2013:920595. doi: 10.1155/2013/920595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou J, Liu T, Wang W. Prognostic significance of matrix metalloproteinase 9 expression in osteosarcoma: A meta-analysis of 16 studies. Medicine (Baltimore) 2018;97:e13051. doi: 10.1097/MD.0000000000013051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou HY, Yao XM, Chen XD, Tang JM, Qiao ZG, Wu XY. Mechanism of metformin enhancing the sensitivity of human pancreatic cancer cells to gem-citabine by regulating the PI3K/Akt/mTOR signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23:10283–10289. doi: 10.26355/eurrev_201912_19666. [DOI] [PubMed] [Google Scholar]
- 33.Ou JM, Qiu MK, Dai YX, Dong Q, Shen J, Dong P, Fei ZW. Combined Blockade and Akt/mTOR pathway inhibits growth of human hemangioma via downregulation of proliferating cell nuclear antigen. Int J Immunopathol Pharmacol. 2012;25:945–953. doi: 10.1177/039463201202500412. [DOI] [PubMed] [Google Scholar]
- 34.Olaisen C, Muller R, Nedal A, Otterlei M. PCNA-interacting peptides reduce Akt phosphorylation and TLR-mediated cytokine secretion suggesting a role of PCNA in cellular signaling. Cell Signal. 2015;27:1478–1487. doi: 10.1016/j.cellsig.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 35.Dobbin ZC, Landen CN. The importance of the PI3K/AKT/mTOR pathway in the progression of ovarian cancer. Int J Mol Sci. 2013;14:8213–8227. doi: 10.3390/ijms14048213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levine DA, Bogomolniy F, Yee CJ, Lash A, Barakat RR, Borgen PI, Boyd J. Frequent mutation of the PIK3CA gene in ovarian and breast cancers. Clin Cancer Res. 2005;11:2875–2878. doi: 10.1158/1078-0432.CCR-04-2142. [DOI] [PubMed] [Google Scholar]
- 37.Matsuoka T, Yashiro M. The role of PI3K/Akt/mTOR signaling in gastric carcinoma. Cancers (Basel) 2014;6:1441–1463. doi: 10.3390/cancers6031441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ji W, Arnst C, Tipton AR, Bekier ME, II, Taylor WR, Yen TJ, Liu ST. OTSSP167 abrogates mitotic checkpoint through inhibiting multiple mitotic kinases. PLoS One. 2016;11:e0153518. doi: 10.1371/journal.pone.0153518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee JJ, Loh K, Yap YS. PI3K/Akt/mTOR inhibitors in breast cancer. Cancer Biol Med. 2015;12:342–354. doi: 10.7497/j.issn.2095-3941.2015.0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request. The results published here are in whole or in part are based on data generated by the Therapeutically Applicable Research to Generate Effective Treatments (https://ocg.cancer.gov/programs/target) initiative, phs000468. The data used for this analysis are available at https://portal.gdc.cancer.gov/projects.