Abstract
At present, a large number of exciting results have been found regarding energy metabolism within the triple-negative breast cancer (TNBC) field. Apart from aerobic glycolysis, a number of other catabolic pathways have also been demonstrated to participate in energy generation. However, the prognostic value of energy metabolism for TNBC currently remains unclear. In the present study, the association between gene expression profiles of energy metabolism and outcomes in patients with TNBC was examined using datasets obtained from the Gene Expression Omnibus and The Cancer Genome Atlas. In total, four robust TNBC subtypes were identified on the basis of negative matrix factorization clustering and gene expression patterns, which exhibited distinct immunological, molecular and prognostic (disease-free survival) features. The differentially expressed genes were subsequently identified from the subgroup that demonstrated the poorest prognosis compared with the remaining 3 subgroups, where their biological functions were assessed further by means of gene ontology enrichment analysis. Any signatures found to be associated with energy metabolism were then established using the Cox proportional hazards model to assess patient prognosis. According to results of Kaplan-Meier analysis, the constructed signature consisting of eight genes that were associated with energy metabolism distinguished patient outcomes into low- and high-risk groups. In addition, this signature, which was found to be markedly associated with the clinical characteristics of the patients, served as an independent factor in predicting TNBC patient prognosis. According to gene set enrichment analysis, the gene sets related to the high-risk group participated in the MAPK signal transduction pathway, focal adhesion and extracellular matrix receptor interaction, whilst those related to the low-risk group were revealed to be mainly associated with mismatch repair and propanoate metabolism. Findings from the present study shed new light on the role of energy metabolism within TNBC, where the eight-gene signature associated with energy metabolism constructed can be utilized as a new prognostic marker for predicting survival in patients with TNBC.
Keywords: energy metabolism, triple-negative breast cancer
Introduction
Triple-negative breast cancer (TNBC) is a type of breast cancer (BC) in which the expression of human epidermal growth factor receptor 2 (HER2), progesterone receptor and oestrogen receptor have all been deleted (1). TNBC is associated with distinct clinical characteristics and molecular complexity compared with other types of BCs (1). Typically, prognosis for women with TNBC following metastatic recurrence is poorer compared with those with other subtypes of BCs (2). Chemotherapy remains to be the preferred systemic treatment for TNBC (3). Although subpopulations showing effective response to chemotherapy can be identified, particularly in those carrying BRCA1 mutations, the effectiveness of chemotherapy is restricted in other subpopulations with TNBC (4). A number of treatment strategies have been previously developed for clinical intervention, including platinum salts, poly ADP-ribose polymerase inhibitors, immunological agents and mitogen-activated extracellular signal-regulated kinase inhibitors (5–7). Among these, although a number of predictive biomarkers, including O6-methylguanine DNA methyltransferase (8), isocitrate dehydrogenase (9), epidermal growth factor receptor (10) and phosphatase and tensin homolog (11), have been identified, though suitable molecular features for optimizing personalized treatment strategies for individual patients remain insufficient (12).
Accumulating evidence has demonstrated high heterogeneity within TNBC tumors, which is one of the main causes of recurrence and resistance (13,14). A number of previous studies have applied the use of gene expression profiles in classifying TNBCs, which can offer relevant clinical information (15,16). For example, platinum-based chemotherapy can be used for patients carrying breast cancer (BRCA) 1 and BRCA2 mutations. Harano et al (17) previously recognized six TNBC subtypes based on gene expression profiles, where the immunomodulatory subtype was frequently observed during processes involving immunocytes. In another study, Bonsang-Kitzis et al (18) classified six TNBC subgroups on the basis of gene selection driven by the biological network, where two immunity clusters were included and a gene signature was constructed based on the stromal immune module. In addition, Burstein et al (19) previously recognized four TNBC subtypes with high stability on the basis of mRNA expression and DNA genomic profile data: i) Mesenchymal; ii) luminal/androgen receptor; iii) basal-like immune activated; and iv) basal-like immune suppressed. Based on this classification, Burstein et al (19) also proposed candidate treatment targets for the aforementioned subtypes. The TNBC classification strategies aforementioned have contributed to laying the foundations for developing targeted TNBC treatments.
The reprogramming of energy metabolism can serve as a hallmark in cancer physiology, making it possible for cancer cells to produce ATP whilst maintaining redox balance and macromolecular biosynthesis, processes that are necessary for cell proliferation, growth, migration, drug resistance and angiogenesis (20). It has been previously suggested that a large number of cancer types divert their energy metabolism towards the glycolytic pathway, resulting in the excessive production of lactate even under normoxic conditions, in a phenomenon known as the Warburg effect (21,22). Compared with non-malignant cells, cancer cells generally prefer incomplete and non-oxidative glucose metabolism (22). Although glucose has been accepted as a major source of energy in tumor cells, there is growing consensus regarding the heterogeneity of metabolic phenotypes in tumors. A portion of cancer cells prefer glycolysis, whilst other tumor cell types can bias towards oxidative phosphorylation (OXPHOS) (23,24). In addition, there is an increasing number of reports revealing the presence of metabolic symbiosis between oxidative and glycolytic cancer cells. For instance, pyruvate and lactate produced by glycolysis can be transferred and utilized to become substrates of the tricarboxylic acid cycle (TCA) and for ATP production by neighboring tumor cells (25). Likewise, malignant cancer cells can absorb ketone bodies and free fatty acids generated through catabolic cells in their vicinity, thereby accelerating mitochondrial OXPHOS (26). Glutamine has also been reported to be metabolized by cancer cells through the TCA cycle for energy production under hypoxic conditions or those under glucose deficiency (27). Therefore, an extensive understanding of energy metabolism in different types of cancer cells can promote the development of novel therapeutic strategies.
Catabolic pathways have been previously reported to participate in energy metabolism in TNBC cells, which exhibit high OXPHOS rates and are not responsive to glycolytic inhibitors. Under normoxic conditions, TNBCs generally prefer OXPHOS to glycolysis, whilst under hypoxic conditions, TNBCs generally prefer glycolysis (28). Tumor stem cells do not display glycolytic phenotypes compared with those of the differentiated progeny cells for TNBC (29,30). Previous studies have indicated that fatty acids can also be utilized by TNBC cells as substrates for energy production (31,32). Inhibiting the β-oxidation of fatty acids can reduce cancer cell proliferation in myc-overexpressing TNBC (31). However, the association between the local metabolic state and its prognostic significance in patients with TNBC remain unknown at present.
The present study aimed to investigate the expression profiles of genes associated with energy metabolism, together with their clinical significance in patients with TNBC, based on data obtained from the gene expression omnibus (GEO) and The Cancer Genome Atlas (TCGA) databases. Patients were divided into four groups based on differential gene expression patterns, where these four groups exhibited significant differences in terms of prognosis, immunological and molecular features. A signature based on energy metabolism was subsequently constructed to assess TNBC patient prognosis based on the TCGA dataset, which was then verified using the GEO dataset. This signature showed close association with patient survival, suggesting that it can be used as an independent prognostic factor. These findings revealed that the energy metabolic state is closely associated with the clinical prognosis of TNBC.
Materials and methods
Pre-processing of initial sample information
In total, 1097 BC samples were collected from the TCGA database for analysis, from which 191 samples that were negative for ‘HER2’, ‘progesterone’ and ‘estrogen receptor expression’ were screened using the three key words. A total of 177 samples with disease free survival (DFS) >30 days was then obtained from the 191 samples. In addition, two GEO datasets [GSE58812 (33) and GSE21653 (34)] were extracted from NCBI database, which were then normalized according to the robust multichip average (RMA) method (35). The GSE21653 dataset included 87 TNBC cases, whilst the GSE58812 dataset included 107 TNBC cases. Of note, 2 cases were excluded due to the lack of prognostic information in the GSE21653 dataset. Following series data preprocessing aforementioned, 369 samples, including 177 from the TCGA training set and 192 from the GEO validation set, were acquired for further analysis. Using following three keywords ‘metabolism’, ‘TCA’ and ‘glycogen’, the expression profiles of 594 genes associated with energy metabolism were extracted from the Reactome database (36) based on the related metabolic pathways (Table I). Table II displays the sample clinicopathological features of the samples.
Table I.
Gene sets related to energy metabolism extracted from the Reactome database.
| Metabolic pathways from the Reactome | Pathway ID | Gene count |
|---|---|---|
| Biological oxidations | R-HSA-211859 | 221 |
| Metabolism of carbohydrates | R-HSA-71387 | 292 |
| Mitochondrial fatty acid beta-oxidation | R-HSA-77289 | 38 |
| Glycogen synthesis | R-HSA-3322077 | 16 |
| Glycogen metabolism | R-HSA-8982491 | 27 |
| Glucose metabolism | R-HSA-70326 | 92 |
| Glycogen breakdown (glycogenolysis) | R-HSA-70221 | 15 |
| Glycolysis | R-HSA-70171 | 72 |
| Pyruvate metabolism | R-HSA-70268 | 31 |
| Pyruvate metabolism and citric acid (TCA) cycle | R-HSA-71406 | 55 |
| Citric acid cycle (TCA cycle) | R-HSA-71403 | 22 |
| Sum | 881 (unique: 594) |
Table II.
Clinicopathological features of the samples after series data pre-processing.
| Characteristic | TCGA datasets (n=177) | GSE21653 (n=85) | GSE58812 (n=107) |
|---|---|---|---|
| Age (years) | |||
| ≤60 | 118 | 50 | 64 |
| >60 | 59 | 35 | 43 |
| Survival status | |||
| Living | 150 | 58 | 78 |
| Deceased | 27 | 27 | 29 |
| Gender | |||
| Female | 177 | 85 | 107 |
| Male | 0 | 0 | 0 |
| T | |||
| T1 | 45 | 16 | – |
| T2 | 110 | 45 | – |
| T3 | 14 | 23 | – |
| T4 | 7 | 0 | – |
| N | |||
| N0 | 105 | 51 | – |
| N1 | 45 | 33 | – |
| N2 | 15 | 0 | – |
| N3 | 10 | 0 | – |
| M | |||
| M0 | 154 | – | – |
| M1 | 1 | – | – |
| MX | 21 | – | – |
| Tumor stage | |||
| Stage I | 31 | – | – |
| Stage II | 109 | – | – |
| Stage III | 32 | – | – |
TCGA, the cancer genome atlas.
NMF (Negative matrix factorization) clustering
NMF clustering was performed to identify stable sample clusters based on 50 iterations according to the Brunet method (37) using genes associated with energy metabolism. In addition, the cluster number, represented by k, was set as 2–10, whilst the best cluster number was calculated based on the cluster cophenetic correlation and the observed consensus map. The mean silhouette width of the consensus membership matrix was determined using the ‘NMF’ function in the R package (version 3.5.3; http://bioconductor.org/biocLite.R). For every member, the lowest cluster number for each member was set as 10. Maftools v1.6.15 from the R package was used to analyze the gene mutation profiles within the TNBC case clusters. The tumor immune estimation resource (TIMER; http://cistrome.shinyapps.io/timer/) was applied to calculate the scores of immune cell infiltration within the TNBC clusters (38).
Identification of gene signature
Differentially expressed genes (DEGs) associated with energy metabolism in the TCGA training set (n=177) first underwent univariate survival analysis by using the ‘survival coxph function’ in the R package to investigate the genes linked to patient prognosis and survival. Typically, a difference of P<0.05 was considered to indicate a statistically significant difference. The significant genes were subsequently screened by multivariate Cox proportional hazards regression, using regional lymph nodes (N) and tumor size (T) as covariants for the model and P<0.05 as threshold for significance. Prognostic genes recognized using the glmnet of R package (family=‘cox’; nλ=100; α=1) were then refined using lasso Cox regression (39), which was appropriate for the regression analysis of high-dimensional data. By using the regularization coefficient of machine learning (λ, also known as penalty coefficient), the number of variables when the cross validation error is minimum was found. DESeq2 (40) was then used for calculating DEGs that participated in energy metabolism by applying the thresholds of false discovery rate (FDR) <0.05 and |log2 fold change|>1. The R package cluster profile (41) was subsequently used to perform enrichment analysis of the DEGs by turning them to ENTREZID. Afterwards, the ENTREZIDs were enriched using the enrich Kyoto Encyclopedia of Genes and Genomes pathways (KEGG) and the enrich gene ontology (GO) functions by applying the P<0.05 threshold. Using the enrichment map software from Cytoscape (version 3.8.0; http://cytoscape.org/) (42), a visualization of the enrichment results was then obtained. By applying the coefficients obtained from lasso Cox regression analysis and expression value of each gene, a specific risk signature was finally constructed, the formula of which was shown below:
Risk score=0.051 × IL1RL2 + 0.012 × FBLN7 + 0.01 × CA3 + 0.146 × PDE1B + 0.01 × SLURP1 + 0.002 × CILP + 0.011 × AQP7 + 0.003 × TPSB.
The risk score values of all patients in the TCGA training set were determined based on linear combination with the expression of signature-related genes weighted according to the corresponding regression coefficients. The regression coefficients from the training set were then applied into the GEO validation set for risk score calculation.
Multiple regression analyzes, gene set enrichment analysis (GSEA) and gene ontology (GO) analysis
GO analysis was performed to annotate the major functions of the DEGs using the Database for Annotation, Visualization and Integrated Discovery online tool v6.8 (version 6.8; http://david.ncifcrf.gov/). The statistically different genes identified between high risk and low risk groups, which were classified according to the median risk score values, were using the GSEA v7.1 software (MSigDB; http://software.broadinstitute.org/gsea/msigdb/index.jsp) (43). Multiple regression analyses were performed to determine the association between the clinical features and the risk score values, which were displayed as forest plots and nomograms.
Statistical analysis
All cases were classified into high-risk or low-risk groups according to the median risk score value. Differences in disease-free survival (DFS) between the two groups were evaluated using Kaplan-Meier combined with a two-sided log-rank test. Differences in the pathological characteristics between the two groups were compared using χ2 test. Multivariate and univariate cox regression analyses were performed for identifying the independent prognosis factors. Lasso regression was used for narrowing the number of genes. ROC curve was used to test the sensitivity and specificity of risk score model and to calculate the cutoff value using Youden's index (44). R or SPSS version 19.0 software (IBM Corp.) was used for all statistical analyses. P<0.05 was considered to indicate a statistically significant difference.
Results
Molecular subtypes on the basis of different genes associated with energy metabolism
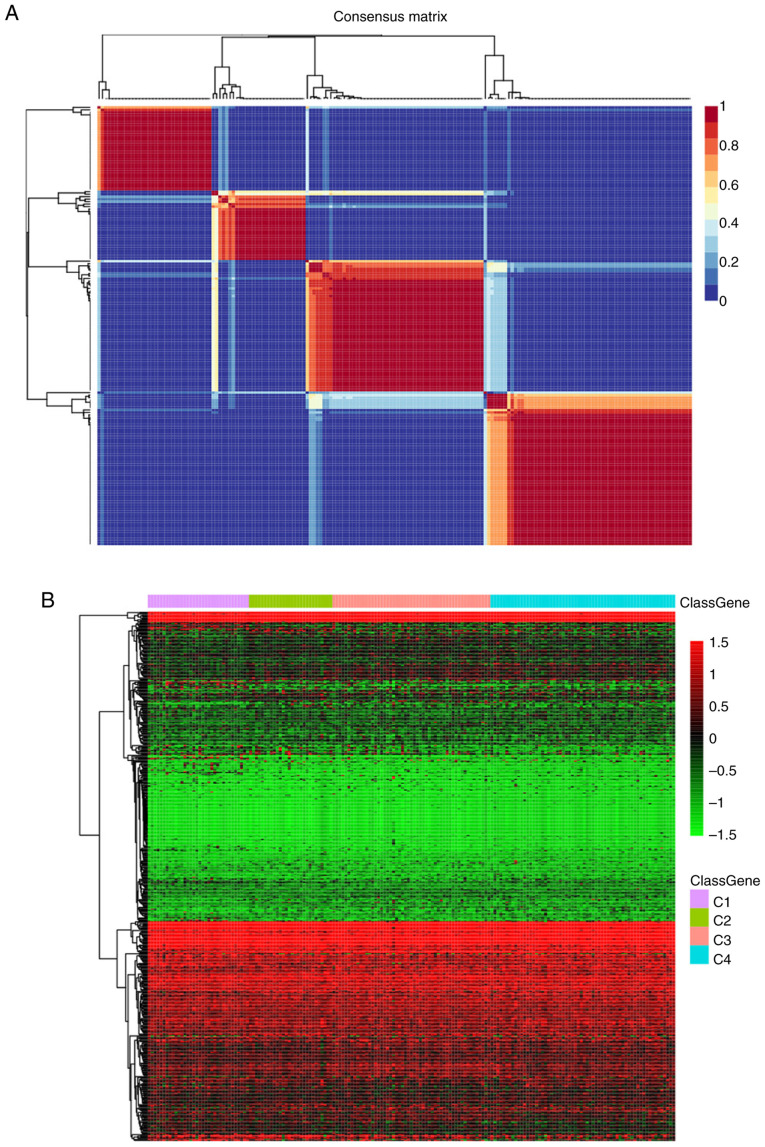
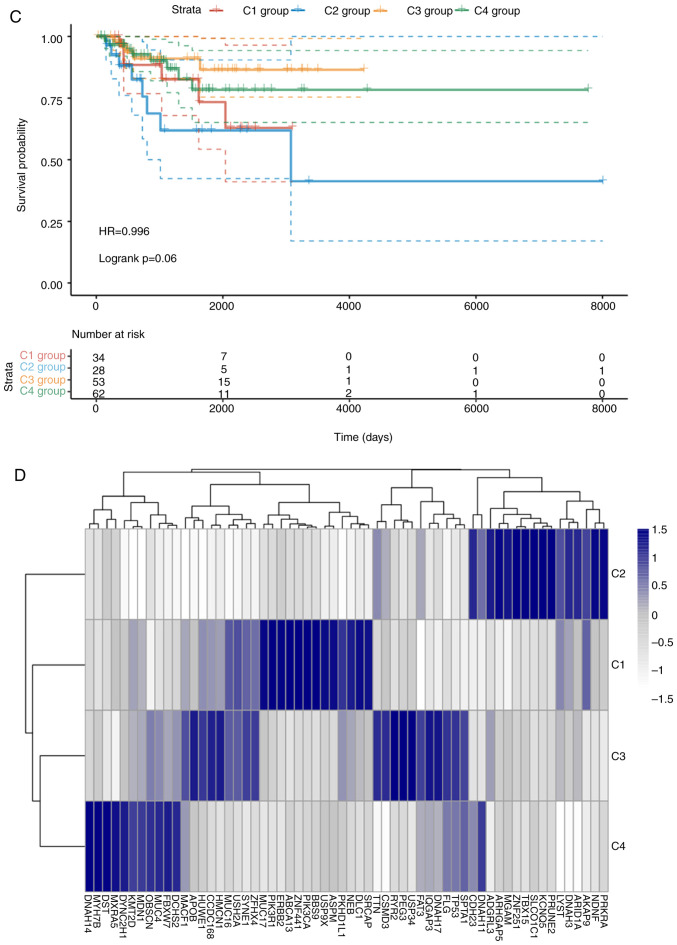
To examine the state of energy metabolism in TNBC, a total of 369 cases including available clinical data and RNA sequencing profiles were acquired from the GEO and TCGA databases. In addition, one separate gene set containing 594 genes that were associated with energy metabolism was also collected from the Reactome database. The NMF algorithm was applied for clustering the TNBC cases, where the best k number was calculated to be 4 based on silhouette, dispersion and cophenetic analyses (Fig. 1A). The expression levels of genes associated with energy metabolism across the four subtypes are displayed in Fig. 1B, where the expression profiles of some genes were shown to be heterogenous across the different subtypes. Prognosis among the patients within the four subtypes was subsequently examined. Subtype C2 exhibited the poorest prognosis, such that subtypes C1, C3 and C4 all exhibited markedly superior prognoses compared with those in the C2 subtype (log rank P=0.06; Fig. 1C).
Figure 1.
Identification of the molecular subtypes of TNBC by NMF analysis. (A) Plots showing the NMF cluster results. Colour scale represents the P-values of the cluster results, where red indicates high probability of two samples belonging to the same cluster and blue indicates low probability of two samples in the different cluster. (B) Expression levels of genes associated with energy metabolism across the four subtypes were tested. Colour scale represents the gene expression levels, where red indicates upregulation and green indicates downregulation. Identification of the molecular subtypes of TNBC by NMF analysis. (C) The difference in prognosis among the four subgroups. (D) The overall heterogeneity in mutation frequency among the four subtypes. Colour scale represents the expression levels of each gene, where blue indicates upregulation and grey indicates downregulation. NMF, non-negative factorization; TNBC, triple-negative breast cancer; HR, hazard ratio.
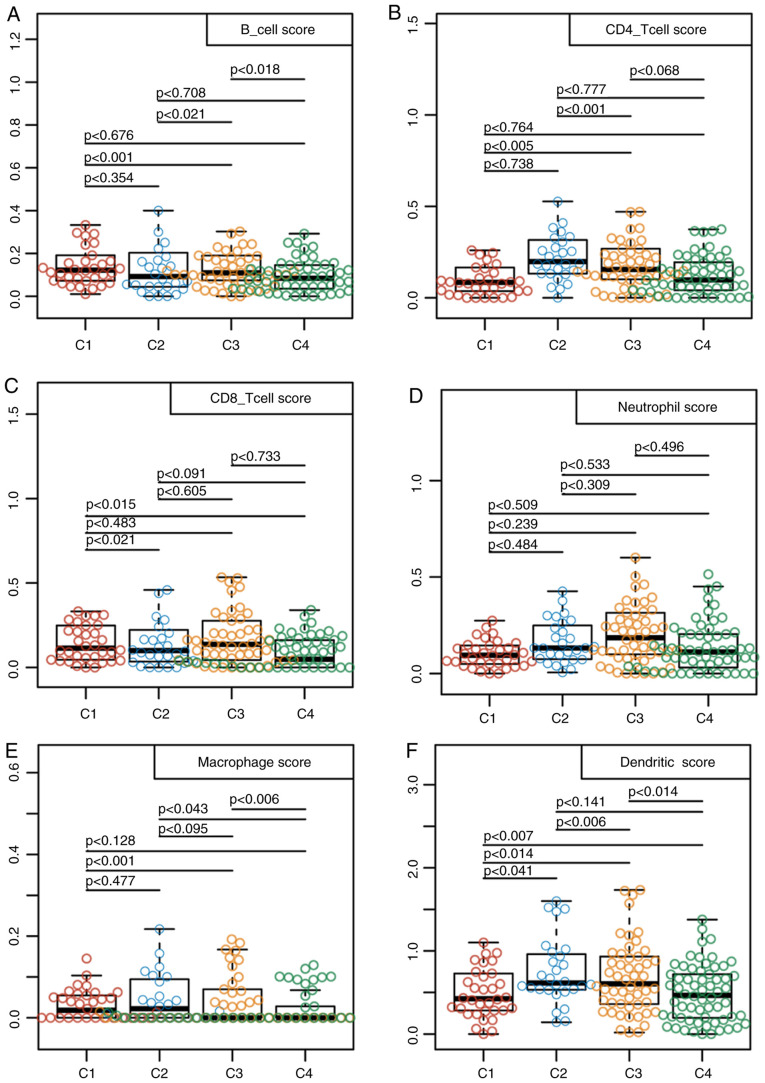
Differences in the aforementioned four TNBC case clusters was measured. Firstly, associations of the TNBC mutation profiles with each of the four sub-classifications were examined using maftools v1.6.15 from R package. After the synonymous mutations were eliminated, the missense and nonsense gene mutations were obtained. The overall heterogeneity in mutation frequency among the different subtypes was found to be of statistical significance (Fig. 1D). Subsequently, differences in the clinicopathological characteristics, including age, TNM and stage, were compared among the four subtypes. The differences in the clinicopathological features among the four TNBC subtypes were not found to be statistically significant (Table III). In addition, samples from these 4 subtypes were analyzed using the TIMER method. The scores in terms of the three types of immune cells, including dendritic cells, macrophages and CD4 T cells were found to be higher in samples from the C2 subtype compared with those from the other subtypes (Fig. 2). These findings suggested that the immune microenvironment or the degree of immunocyte infiltration in TNBC was associated with patient prognosis. In addition, the expression profiles of genes associated with energy metabolism displayed close association with the clinical characteristics and patient prognosis in TNBC.
Table III.
Clinicopathological features among the four triple-negative breast cancer subtypes.
| Clinical features | C1 | C2 | C3 | C4 | P-value |
|---|---|---|---|---|---|
| Outcome | 0.130 | ||||
| Alive | 28 | 20 | 48 | 54 | |
| Deceased | 6 | 8 | 5 | 8 | |
| T | 0.078 | ||||
| T1 | 8 | 10 | 16 | 11 | |
| T2 | 20 | 14 | 30 | 46 | |
| T3 | 3 | 1 | 6 | 4 | |
| T4 | 3 | 3 | 1 | 0 | |
| N | 0.058 | ||||
| N0 | 14 | 19 | 33 | 39 | |
| N1 | 8 | 4 | 15 | 18 | |
| N2 | 6 | 1 | 5 | 3 | |
| N3 | 4 | 4 | 0 | 2 | |
| Stage | 0.061 | ||||
| I | 5 | 8 | 10 | 8 | |
| II | 17 | 12 | 35 | 45 | |
| III | 11 | 8 | 7 | 6 | |
| Gender | – | ||||
| Female | 34 | 28 | 53 | 62 | |
| Male | 0 | 0 | 0 | 0 | |
| Age | 0.686 | ||||
| ≤60 | 20 | 19 | 35 | 44 | |
| >60 | 14 | 9 | 18 | 18 |
Figure 2.
Identification of the immune features for each triple-negative breast cancer subtype. The immune scores of samples across 4 subtypes were calculated and compared using the tumor immune estimation resource approach. The scores of (A) B cells, (B) CD4 T cells, (C) CD8 T cells, (D) neutrophils, (E) macrophages and (F) dendritic cells for each of the four subtypes were calculated.
Identification of the prognostic signature associated with energy metabolism for TNBC
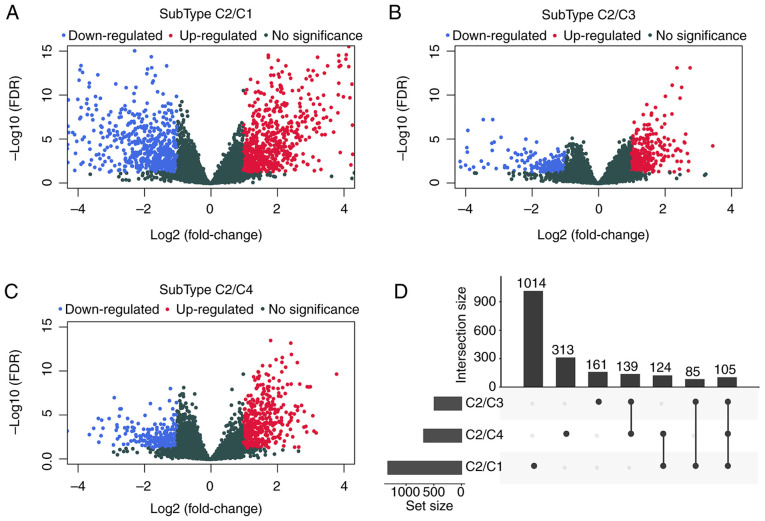
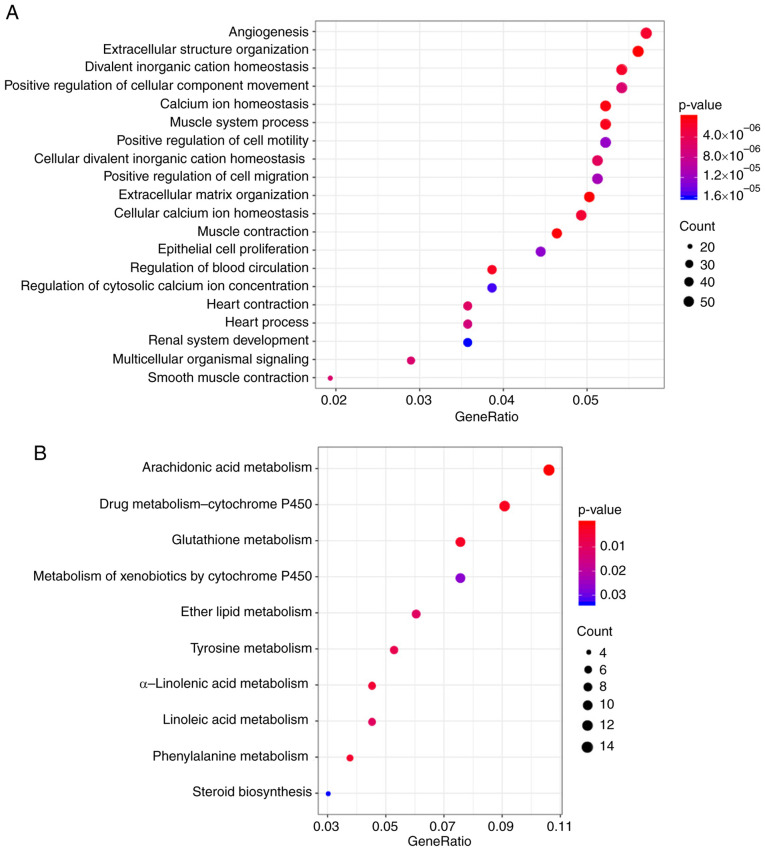
To examine the relationship between the expression profiles of genes associated with energy metabolism and the TNBC prognosis, DESeq2 was used for calculating DEGs that participated in energy metabolism between samples from the C2 subtype, which had the poorest prognosis and those in the C1, C3 or C4 subtypes, by applying the thresholds of FDR <0.05 and |log2 fold change|>1. Data shown in Fig. 3 and Table IV suggested that a total of 1941 DEGs were identified between the subtype C2 samples and those in the C1, C3 and C4 subtypes. The R package cluster profile was subsequently used to perform enrichment analysis of these 1941 DEGs. KEGG pathway enrichment and GO enrichment analysis results appeared to show that these genes were mostly enriched in biological functions and pathways associated with ‘calcium homeostasis’, ‘angiogenesis’, the metabolism of molecules including glutathione, arachidonic acid, drugs and xenobiotics and ‘positive cell migration regulation’ (Fig. 4).
Figure 3.
Calculation of DEGs associated with energy metabolism between samples of the C2 subtype and those of the C1/C3/C4 subtypes. (A) Volcano plots of DEGs comparing C1 and C2. (B) Volcano plots of DEGs comparing C3 and C2. (C) Volcano plots of DEGs comparing C4 and C2. (D) The upset intersection diagram showing the three types of DEGs between C1/C3/C4 and C2. DEGs, differentially expressed genes; FDR, false discovery rate.
Table IV.
Comparison of differentially expressed genes in samples from the C2 subtype with the C1, C3 and C4 subtypes.
| Type | C2/C1 | C2/C3 | C2/C4 |
|---|---|---|---|
| PCG_Up | 700 | 304 | 448 |
| PCG_Down | 628 | 186 | 233 |
| PCG_All | 1328 | 490 | 681 |
PCG, protein coding gene.
Figure 4.
Functional analysis of the DEGs associated with energy metabolism. (A) Top 20 gene ontology terms enriched by the 1941 DEGs. (B) Kyoto Encyclopedia of Genes and Genomes pathways enriched by the 1941 DEGs. DEGs, differentially expressed genes.
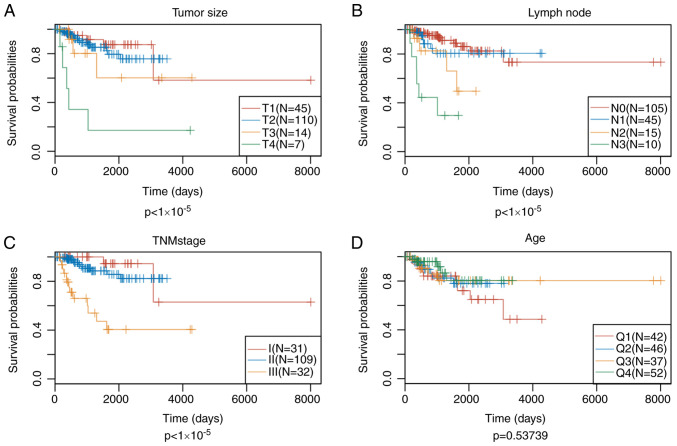
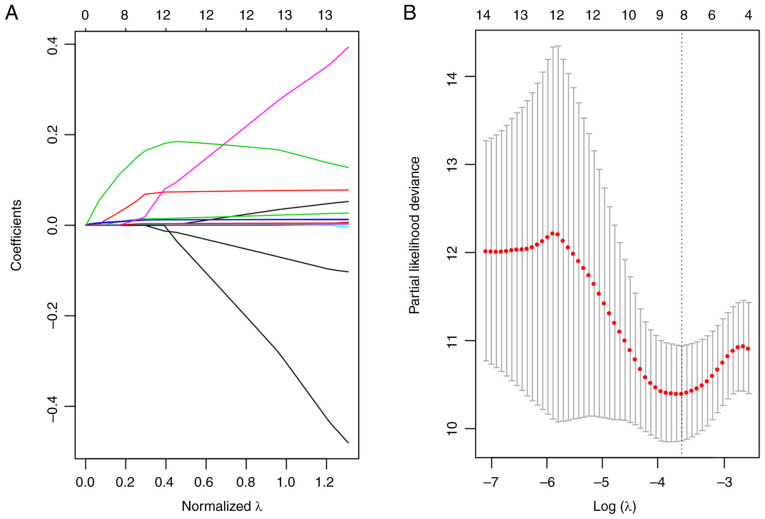
Since patient prognosis was found to be closely associated with the energy metabolism state, a signature associated with energy metabolism was therefore proposed to predict patient prognosis. Univariate Cox regression analysis suggested that 102 of these DEGs showed significant association with the DFS of patients in the training set (top 20 DEGs shown in Table V). Subsequently, the univariate Cox proportional hazards regression model was performed to analyze the age, TNM stages, stage, age and survival data. Differences in stage, lymph node and tumor size were found to be statistically significant in predicting prognosis (Fig. 5). Multivariate Cox proportional hazards regression model was utilized to select for the genes associated with energy metabolism that displayed the highest prognostic significance, using stage, N and T as covariants. A total of 14 genes associated with energy metabolism that were specific to prognosis were mined and narrowed by lasso regression to reduce the number of genes selected for the construction of the prognostic signature. Typically, the lasso algorithm uses the biased estimate to process multicollinearity data, to estimate and screen variables and to solve the multicollinearity problem encountered during regression analysis. In the present study, the glmnet function of R package was used to perform the lasso regression analysis. For every independent parameter, a variation trajectory was evaluated (Fig. 6A) The coefficients of the 14 genes were revealed to be ~0 as the λ gradually increased. Additionally, a model was constructed using a three-fold cross-validation. Fig. 6B presents the confidence interval for every λ, which suggested that λ=0.0283271 would result in the best model. Consequently, the signature with λ=0.0283271 was applied to be the eventual signature, which involves eight genes associated with energy metabolism.
Table V.
Top 20 DEGs that showed significant correlation with the disease-free survival of patients in the training set by analysis of univariate Cox regression.
| Symbol | P-value | Hazard ratio | Low 95% CI | High 95% CI |
|---|---|---|---|---|
| KRTDAP | 1.48×10−05 | 1.009 | 1.005 | 1.013 |
| PLAC9 | 4.89×10−05 | 1.021 | 1.011 | 1.031 |
| CD248 | 6.91×10−05 | 1.004 | 1.002 | 1.006 |
| SPRR2G | 2.55×10−04 | 1.035 | 1.016 | 1.055 |
| EFNB1 | 2.82×10−04 | 1.016 | 1.007 | 1.025 |
| SPON2 | 3.17×10−04 | 1.006 | 1.003 | 1.010 |
| PTGDS | 4.44×10−04 | 1.002 | 1.001 | 1.003 |
| CA3 | 5.86×10−04 | 1.010 | 1.004 | 1.015 |
| PTGER1 | 6.55×10−04 | 1.747 | 1.267 | 2.407 |
| CILP | 6.66×10−04 | 1.005 | 1.002 | 1.007 |
| COL16A1 | 7.28×10−04 | 1.009 | 1.004 | 1.014 |
| CNFN | 7.99×10−04 | 1.015 | 1.006 | 1.024 |
| PDE1B | 1.12×10−03 | 1.246 | 1.091 | 1.422 |
| ACVRL1 | 1.170×10−03 | 1.055 | 1.022 | 1.090 |
| COL5A3 | 1.39×10−03 | 1.009 | 1.003 | 1.014 |
| PYGM | 1.40×10−03 | 1.914 | 1.285 | 2.850 |
| MYL3 | 1.55×10−03 | 1.490 | 1.164 | 1.907 |
| DCSTAMP | 1.63×10−03 | 1.292 | 1.102 | 1.515 |
| RARRES2 | 1.68×10−03 | 1.003 | 1.001 | 1.005 |
| C1QTNF7 | 1.68×10−03 | 1.297 | 1.103 | 1.525 |
CI, confidence interval.
Figure 5.
Analysis of tumor size, lymph node, TNM stage and age as prognostic indicators for triple-negative breast cancer. Comparison of patient prognosis according to (A) tumor size, (B) lymph node, (C) TNM stage and (D) age.
Figure 6.
Identifying the prognosis signature associated with energy metabolism for triple-negative breast cancer using lasso. (A) Coefficients calculated for each lambda. Each line represents a gene confidence value. (B) The changing trajectory of each independent variable. The horizontal axis represents the log value of the independent variable λ, whilst the vertical axis represents the partial likelihood deviance of the log value of each independent variable λ.
The eight-gene based signature shows potential in predicting prognosis for TNBC
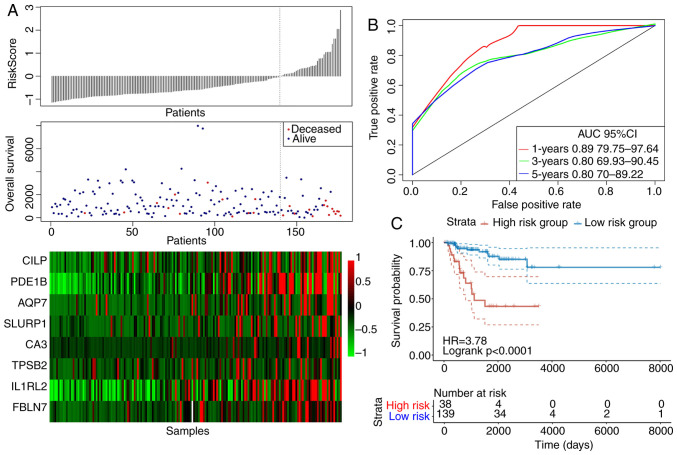
The risk score values for the samples in the training set were first calculated, following which the samples were divided into high-risk and low-risk groups according to the risk score values and set at a threshold of 1.351149 according to the 3-year area under the curve (AUC) after Youden's index analysis. Fig. 7 displays the efficiency of the constructed eight-gene signature in classifying samples in the TCGA training set. showed that, for the samples from the deceased group, the survival time was markedly reduced as the patient risk score value increased (Fig. 7A). In addition, the high-risk group was found to be associated with the increased number of deceased samples (Fig. 7A). The increased levels of the eight genes associated with energy metabolism also appeared to be risk factors, since the risk score values were found to be increased when the expression levels of these eight genes were increased (Fig. 7A). According to the ROC curves, the 1-, 3- and 5-year AUC were calculated to be 0.89, 0.80 and 0.80, respectively (Fig. 7B). After the patients were divided into low-risk and high-risk groups, the difference in DFS between the two groups was revealed to be statistically significant (HR=3.78; P<0.0001; Fig. 7C).
Figure 7.
The classification effect of the eight-gene signature associated with energy metabolism in the TCGA training set. (A) The relationship between the risk score, survival time, survival status and the expression levels of the eight genes derived from the TCGA training set. Colour scale represents the expression levels of the eight genes, where red indicates upregulation and green indicates downregulation. (B) ROC curves of the eight-gene signature in triple-negative breast cancer samples obtained from the TCGA training set. (C) Analysis of prognosis following classification into the high risk and low-risk groups according to the eight-gene signature. TCGA, The cancer genome atlas; ROC, receiver operating characteristic; AUC, area under the curve; CI, confidence interval; IL1RL2, interleukin-1 receptor-like 2; CA3, carbonic anhydrase 3; PDE1B, phosphodiesterase 1B; SLURP, secreted Ly-6/uPAR-related protein; AQP7, aquaporin 7; TPSB, tryptase-β2; FBLN7, fibulin 7; CILP, cartilage intermediate layer protein.
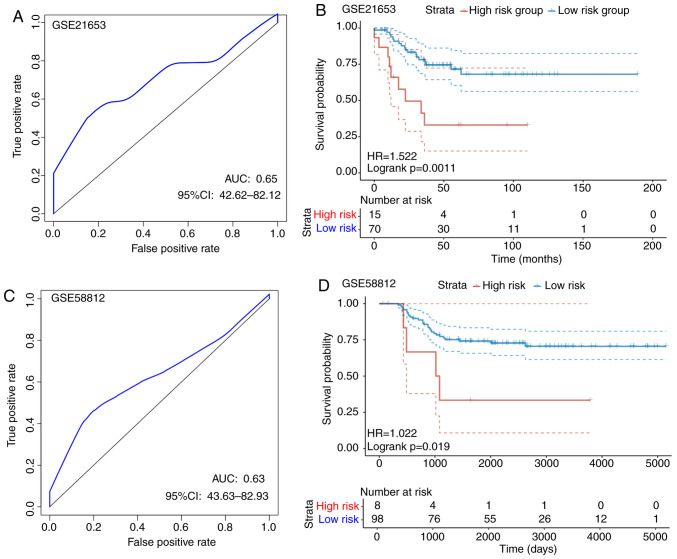
To determine the reliability of the constructed eight-gene signature, an identical model used for the TCGA training set samples was performed on the two GEO test sets. For the GSE21653 dataset, according to the ROC analysis, a 1-year AUC of 0.65 was found, although interpretation was restricted by the limited follow-up time of the samples (Fig. 8A). Subsequently, a total of 70 cases were divided into the low-risk group, whereas 15 were allocated into the high-risk group using a threshold of 1.475448 according to the 1-year AUC, where a statistically significant difference was found between the two groups (P=0.0011; Fig. 8B). For the GSE58812 dataset, ROC analysis revealed the 1-year AUC to be 0.63, but interpretation was restricted by the limited follow-up time of the samples (Fig. 8C). According to Fig. 8D, a total of 98 cases were allocated into the low-risk group, whereas 8 were assigned into the high-risk group using a threshold of 32.77999 according to the 1-year AUC, where there was a statistically significant difference between the two groups (P=0.0019). The threshold was found to be large, possibly due to the larger variation in the GSE58812 dataset. For the overall expression levels of the two GSE datasets, the calculated risk scores differed drastically. In conclusion, the signature constructed based on these eight genes showed high accuracy in predicting prognosis for patients in both internal and external TNBC datasets.
Figure 8.
The classification effect of the eight-gene signature associated with energy metabolism on the GEO test set. (A) ROC curves of the eight-gene signature in TNBC samples obtained from the GSE21653 test set. (B) Analysis of the prognosis into the high risk and low-risk groups according to the eight-gene signature in the GSE21653 test set. (C) ROC curves of the eight-gene signature in TNBC samples obtained from the GSE51882 test set. (D) Analysis of the prognosis into the high risk and low-risk groups according to the eight-gene signature in the GSE51882 test set. TNBC, triple-negative breast cancer; HR, hazard ratio; CI, confidence ratio; ROC, receiver operating characteristic; AUC, area under the curve.
Compared with recent studies that also investigated the prognostic models for TNBC (45–47), that present study also calculated the risk scores of each sample obtained from the TCGA database using the method reported in the present study and those from the previous studies, where ROC analysis was performed for each model. The samples were then divided into high-risk and low-risk groups using the median risk score, following which the difference in the prognosis of patients between the high-risk and low risk groups were compared. It was found that the signature derived from the present study was superior compared with the other models from the recent studies in terms of prognostic prediction (Fig. S1).
The signature constructed based on genes associated with energy metabolism is superior compared with other clinical features in predicting survival of patients with TNBC
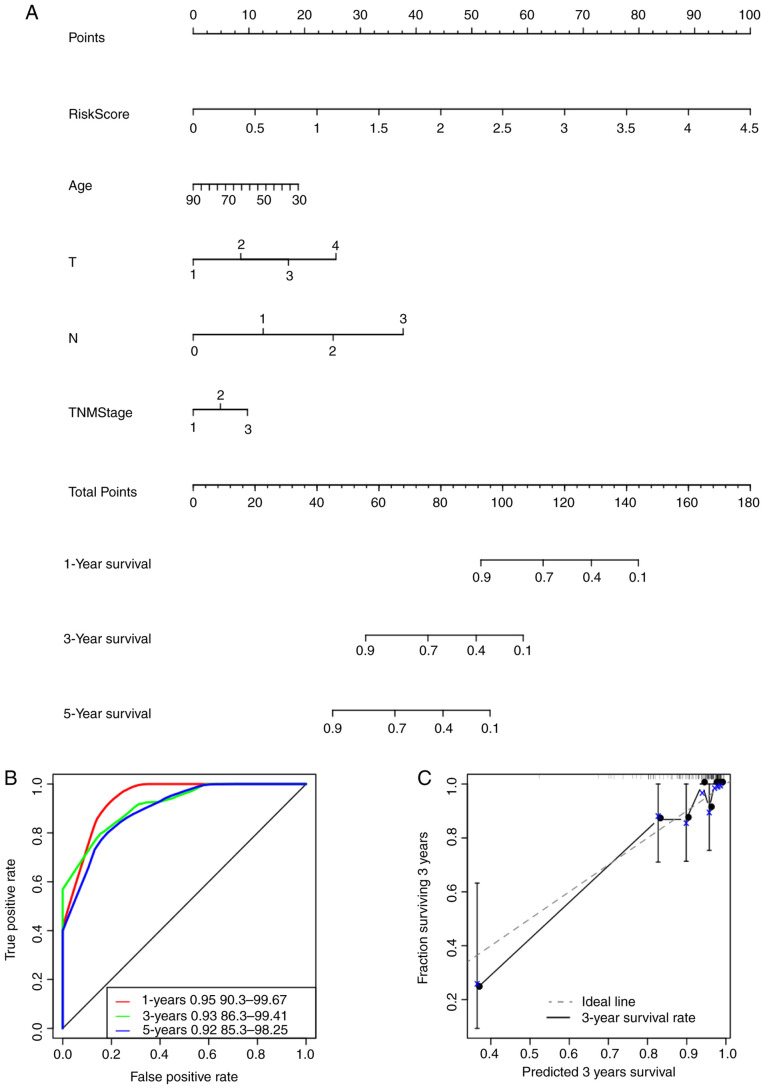
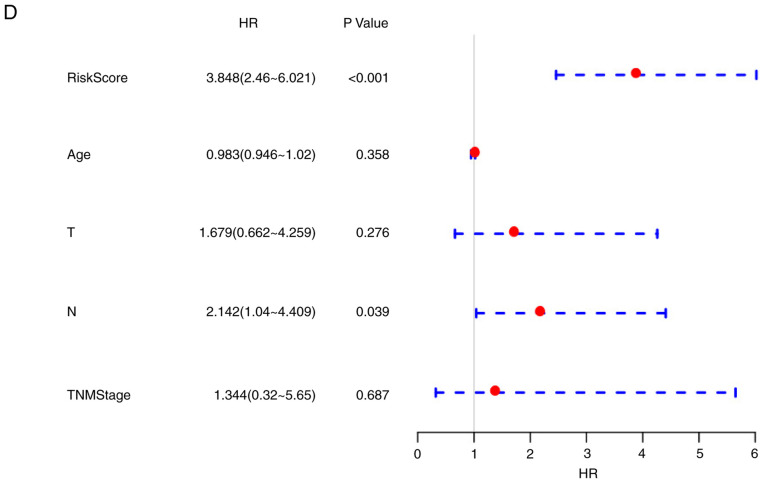
The risk score values were utilized in combination with other clinicopathological features to construct a nomogram model for the TNBC cases. Nomogram is a method for effectively and intuitively presenting the results of a risk model, which can be adopted conveniently for predicting patient outcome (48). Specifically, length of the straight line within the nomogram indicates the influence of the various parameters on the significance of patient outcome. In the present study, a nomogram was constructed by integrating age, risk scores, stage, N and T (Fig. 9A). The risk score features were revealed to be associated with survival prediction. This predictive efficiency was assessed further using the ROC analysis to calculate the AUC of the nomogram scores, which was obtained using the ‘predict’ function from the R package. According to Fig. 9B, the 1-, 3- and 5-year AUC of nomogram were calculated to be 0.95, 0.93 and 0.92, respectively. In addition, the constructed nomogram model displayed consistent predictive capacity compared with that of the ideal model (dotted line) for predicting the 3-year survival of patients with TNBC (Fig. 9C). Finally, a forest plot was constructed based on the risk score values and the clinicopathological features. The HRs of the risk score value was calculated to be ~3.848, as observed from the forest plots constructed based on risk score values, stage, N, T and age (P<0.001; Fig. 9D), suggesting that the signature constructed based on the expression profiles of the eight genes associated with energy metabolism exhibited adequate capacity in predicting prognosis in patients with TNBC.
Figure 9.
The eight-gene signature constructed based on genes associated with energy metabolism is a more accurate prognostic indicator for TNBC compared with other clinicopathological features. (A) The nomogram model constructed by combining the T, N, TNM stage, age with risk scores for TNBC samples. ‘Points’ and ‘total points’ represents the reference index for the score of nomogram. (B) ROC curves of the nomogram scores in the TNBC samples. (C) The calibration plots for predicting 3-year overall survival of patients with TNBC. nomogram-predicted probability of survival is plotted on the x-axis; actual survival is plotted on the y-axis. The eight-gene signature constructed based on genes associated with energy metabolism is a more accurate prognostic indicator for TNBC compared with other clinicopathological features. (D) The forest plot constructed based on the risk score values and the clinical features. HR, hazard ratio; TNBC, triple negative breast cancer.
Differences in pathways involved in GSEA between the low and high-risk groups
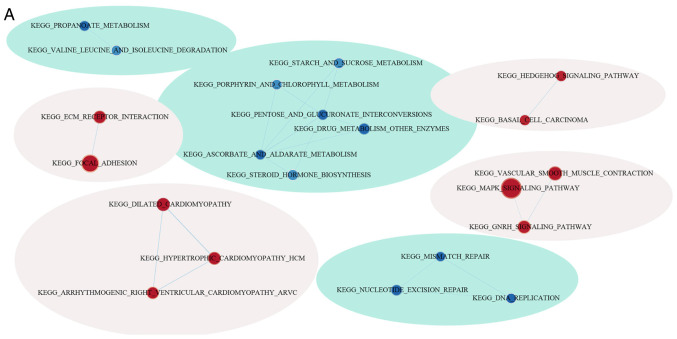
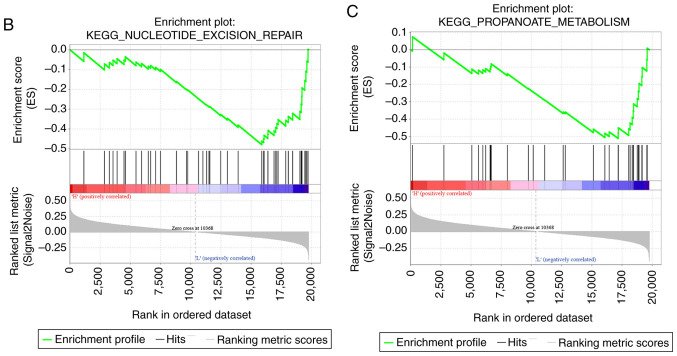
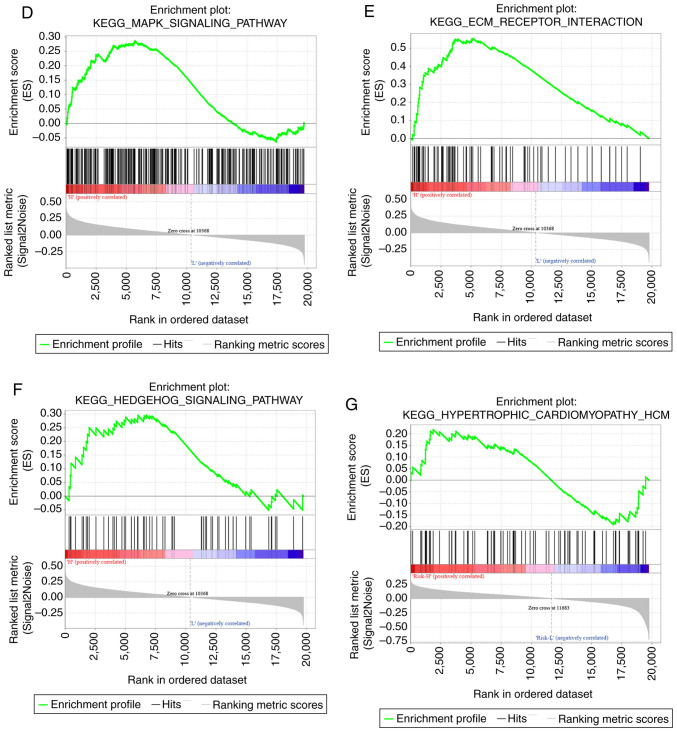
GSEA was used for analyzing the markedly enriched pathways between the low-risk and high-risk groups according to the TCGA training set samples, where 20 markedly enriched pathways were acquired in total. Genes associated with the high-risk group were particularly enriched in the ‘MAPK signal transduction pathway’, ‘focal adhesion’, ‘hedgehog signaling’, ‘cardiomyopathy’ and ‘ECM receptor interaction’, whilst those associated with the low-risk group were involved in ‘mismatch repair’ and ‘propanoate metabolism’ (Fig. 10; Table VI).
Figure 10.
Functional analysis of the eight-gene signature. (A) Visualization of the GSEA enrichment results, calculated based on the median values of the risk scores using the enrichment map software from Cytoscape. The red region represents the enriched pathways in the high-risk group, whilst the blue region represents the enriched pathways in the low-risk group. The sizes of the red/blue dots indicates the score of enrichment, the higher the score of enrichment, the larger the dot sizes. Functional analysis of the eight-gene signature. Enrichment plots for (B) nucleotide excision pair, (C) proparoate metabolism, (D) MAPK signaling pathway, (E) ECM receptor interaction, (F) hedgehog signaling pathway and (G), hypertrophic cardiomyopathy. GSEA, gene set enrichment analysis; KEGG, Kyoto Encyclopedia of Genes and Genomes pathways; ECM, extracellular matrix.
Table VI.
KEGG pathway of genes associated with the high-risk groupand those associated with the low-risk group.
| Name | Size | Esa | Nes | Nom P-value | FDR q-value |
|---|---|---|---|---|---|
| KEGG_dilated_cardiomyopathy | 90 | 0.473 | 1.816 | 0.003 | 0.475 |
| KEGG_calcium_signaling_pathway | 176 | 0.348 | 1.574 | 0.003 | 0.489 |
| KEGG_hypertrophic_cardiomyopathy_hcm | 83 | 0.448 | 1.735 | 0.008 | 0.318 |
| KEGG_ecm_receptor_interaction | 84 | 0.553 | 1.799 | 0.010 | 0.279 |
| KEGG_notch_signaling_pathway | 47 | 0.424 | 1.618 | 0.013 | 0.561 |
| KEGG_vascular_smooth_muscle_contraction | 115 | 0.379 | 1.586 | 0.023 | 0.546 |
| KEGG_mapk_signaling_pathway | 265 | 0.287 | 1.357 | 0.045 | 0.771 |
| KEGG_protein_export | 23 | −0.661 | −1.902 | 0.000 | 0.169 |
| KEGG_biosynthesis_of_unsaturated_fatty_acids | 22 | −0.643 | −1.866 | 0.002 | 0.123 |
| KEGG_n_glycan_biosynthesis | 46 | −0.504 | −1.764 | 0.006 | 0.180 |
| KEGG_steroid_biosynthesis | 16 | −0.729 | −1.804 | 0.007 | 0.163 |
| KEGG_glyoxylate_and_dicarboxylate_metabolism | 16 | −0.627 | −1.745 | 0.009 | 0.143 |
| KEGG_one_carbon_pool_by_folate | 17 | −0.568 | −1.603 | 0.012 | 0.338 |
| KEGG_terpenoid_backbone_biosynthesis | 15 | −0.663 | −1.714 | 0.013 | 0.163 |
| KEGG_pentose_and_glucuronate_interconversions | 27 | −0.609 | −1.593 | 0.026 | 0.325 |
| KEGG_proteasome | 44 | −0.617 | −1.747 | 0.034 | 0.168 |
| KEGG_nucleotide_excision_repair | 44 | −0.480 | −1.562 | 0.039 | 0.331 |
| KEGG_pantothenate_and_coa_biosynthesis | 16 | −0.552 | −1.556 | 0.040 | 0.317 |
| KEGG_propanoate_metabolism | 32 | −0.511 | −1.545 | 0.046 | 0.314 |
| KEGG_citrate_cycle_TCA_cycle | 30 | −0.564 | −1.578 | 0.049 | 0.325 |
Es>0, high risk and Es<0, low risk. KEGG, Kyoto Encyclopedia of Genes and Genomes pathways; Es, enrichment score; Nes, normalized enrichment score; Nom, normalized; FDR, false discovery rate.
Discussion
TNBC accounts for ~15-20% of all BC cases, which is particularly prevalent among younger women aged <40 years. TNBC is a complex disease with substantial heterogeneity, which is associated with inferior prognosis compared with that in other BC subtypes. Currently, treatments for TNBC mainly include surgery combined with adjuvant chemotherapy for early stage disease and chemotherapy for advanced stage disease. However, surgical resection remain ineffective due to the localized invasion of cancer cells to adjacent tissues or the development of distant metastasis (49,50). Additionally, effects exerted by chemotherapy may be diminished due to tumor heterogeneity (51,52). Therefore, identifying novel candidate biomarkers for the prognostic prediction of TNBC is important for finding novel treatment targets for this disease and to accurately manage patients potentially developing complete response from adjuvant treatment. In the present study, a gene expression signature was explored as potential approach for the prognostic prediction of TNBC.
A number of previous studies have classified TNBC subtypes using genomic profiling, which allows for the specific classification and accurate prediction of TNBC outcomes. Using the TIMER method, it was found that the scores for the three immune cell types, specifically dendritic cells, macrophages and CD4 cells, were higher in the samples of the C2 subtype compared with those of other subtypes, suggesting that the immune microenvironment or the degree of immunocyte infiltration in TNBC was associated with patient prognosis. Several molecular markers, including O6-methylguanine DNA methyltransferase, isocitrate dehydrogenase, epidermal growth factor receptor and phosphatase and tensin homolog, were previous examined among patients with TNBC, facilitating targeted and personalized anti-TNBC therapy. Fumagalli C et al (8) previously found that patients with TNBC carrying a silencing mutation in the O6-methylguanine DNA methyltransferase gene are associated with an increase in the susceptibility for alkylating agents. Wang et al (10) found that epidermal growth factor induced the expression of Fascin-1 through the activation of MAPK, which lead to poorer relapse-free survival and overall survival for patients with TNBC. However, since TNBC is associated with poor prognosis, identification of novel molecular biomarkers and development of treatment strategies are urgently sorted for unravelling the possible mechanisms of TNBC progression or improving patient prognosis.
Accumulating evidence suggested that the aberrant metabolism can serve as a hallmark for cancer cells. In recent years, significant research effort has been made on investigating the difference in energy metabolism between tumor and normal cells (53–55). Recent studies on TNBC revealed that a number of catabolic pathways can participate in energy metabolism, including fatty acid metabolism, OXPHOS and glycolysis (28,56,57). Consequently, it is of great significance to screen for new genes associated with energy metabolism that are specific to prognosis, to accurately predict the prognosis for patients, develop therapeutic strategies and identify patients with a high risk of postoperative relapse. In the present study, tumor size (T), nodal status (N), age and TNM stages were applied as the prognostic predictors to select for genes associated with energy metabolism. For TNBC, these are classical prognostic predictors that are universally acknowledged (58,59). Consistent with this, the present study found the differences in stage, N and T to be statistically significant in predicting prognosis, showing them to be good prognostic predictors.
In the present study, the relationship between energy metabolic state in the tumor microenvironment and the prognostic significance among patients with TNBC was determined based on RNA sequencing data. Use of profiling data of genes associated with energy metabolism allowed for the delineation of molecular and clinical characteristics among patients with TNBC. In addition, a signature was developed for patient stratification into low-risk or high-risk groups for predicting poor prognosis. Univariate Cox model was considered to be insufficient in selecting the variables based on dimensional data. Therefore, it was applied to first screen for the genes associated with DFS, following which an elastic net Cox regression model was used for enhancing the prognostic prediction performance of the indices. The eight genes acquired exerted cumulative effects on predicting patient survival. This signature associated with energy metabolism could be used as an effective indicator for prognosis prediction and patient stratification for future treatments targeting energy metabolism.
Based on the present study, five of the eight genes in the signature had been previously recognized to participate in TNBC pathogenesis, immune microenvironment, progression and malignant transformation, which were interleukin-1 receptor-like 2, carbonic anhydrase 3, phosphodiesterase 1B, secreted Ly-6/uPAR-related protein and aquaporin 7 (60–63). These five genes showed marked correlation with the prognosis and survival of patients. These results suggested that the bioinformatic mining results were reliable and accurate. However, it remains unclear regarding the association between the other genes and TNBC physiology, which require further study. Fibulin 7 has been previously reported to participate in the regulation of aberrant neovascularization by modulating the angiopoietin-1/angiopoietin-2-Tie2 axis (64). A previous study has shown that the cartilage intermediate layer protein gene is differentially expressed in differentiated thyroid carcinomas in comparison with normal thyroid tissues (65). By contrast, tryptase-β2 (TPSB) was documented as a major neutral protease present in mast cells that participate in tumor-associated immunity and can serve as a prognostic marker for several types of cancer (66,67). TPSB has also been demonstrated to upregulate vascular endothelial growth factor production in the tumor microenvironment via the protease-activated receptors-2, ERK1/2 and p38 MAPK signaling pathways (68).
In conclusion, the expression profiles of genes associated with energy metabolism and their prognostic significance in TNBC was revealed in the present study. A signature associated with energy metabolism was subsequently constructed, allowing for the classification of patients with TNBC into high-risk or low-risk groups in terms of survival. However, further study is required, since the predictive capacity of the eight-gene signature constructed in the present study need to be tested in a clinical setting. Results in the present study can shed new light on the relationship between the metabolic state and TNBC pathophysiology and potentially aid in the development of novel TNBC treatment strategies targeting energy metabolism.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was supported by The Natural Science Foundation of Ningbo (grant no. 2017A610163).
Availability of data and materials
The datasets generated and analyzed during the current study are available in the gene expression omnibus (https://www.ncbi.nlm.nih.gov/geo/) and The Cancer Genome Atlas (TCGA) databases (https://portal.gdc.cancer.gov/).
Authors' contributions
CLi was involved in the conception and design of the study, project administration and drafted the manuscript. XLi developed the methodology and supervised the study and critically reviewed the manuscript. GLi and LS and WZ collected data from the database and performed statistical data analysis. JJ and QG organized the figures and tables and were involved in interpretation of data for the work. All authors have read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: Challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. 2016;13:674–690. doi: 10.1038/nrclinonc.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smid M, Wang Y, Zhang Y, Sieuwerts AM, Yu J, Klijn JG, Foekens JA, Martens JW. Subtypes of breast cancer show preferential site of relapse. Cancer Res. 2008;68:3108–3114. doi: 10.1158/0008-5472.CAN-07-5644. [DOI] [PubMed] [Google Scholar]
- 3.Silver DP, Richardson AL, Eklund AC, Wang ZC, Szallasi Z, Li Q, Juul N, Leong CO, Calogrias D, Buraimoh A, et al. Efficacy of neoadjuvant Cisplatin in triple-negative breast cancer. J Clin Oncol. 2010;28:1145–1153. doi: 10.1200/JCO.2009.22.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collignon J, Lousberg L, Schroeder H, Jerusalem G. Triple-negative breast cancer: Treatment challenges and solutions. Breast Cancer (Dove Med Press. 2016;8:93–107. doi: 10.2147/BCTT.S69488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Shaughnessy J, Osborne C, Pippen JE, Yoffe M, Patt D, Rocha C, Koo IC, Sherman BM, Bradley C. Iniparib plus chemotherapy in metastatic triple-negative breast cancer. N Engl J Med. 2011;364:205–214. doi: 10.1056/NEJMoa1011418. [DOI] [PubMed] [Google Scholar]
- 6.Telli ML, Jensen KC, Vinayak S, Kurian AW, Lipson JA, Flaherty PJ, Timms K, Abkevich V, Schackmann EA, Wapnir IL, et al. Phase II study of gemcitabine, carboplatin, and iniparib as neoadjuvant therapy for triple-negative and BRCA1/2 mutation-associated breast cancer with assessment of a tumor-based measure of genomic instability: PrECOG 0105. J Clin Oncol. 2015;33:1895–1901. doi: 10.1200/JCO.2014.57.0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nanda R, Chow LQ, Dees EC, Berger R, Gupta S, Geva R, Pusztai L, Pathiraja K, Aktan G, Cheng JD, et al. Pembrolizumab in patients with advanced triple-negative breast cancer: Phase Ib KEYNOTE-012 Study. J Clin Oncol. 2016;34:2460–2467. doi: 10.1200/JCO.2015.64.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fumagalli C, Pruneri G, Possanzini P, Manzotti M, Barile M, Feroce I, Colleoni M, Bonanni B, Maisonneuve P, Radice P, et al. Methylation of O6-methylguanine-DNA methyltransferase (MGMT) promoter gene in triple-negative breast cancer patients. Breast Cancer Res Treat. 2012;134:131–137. doi: 10.1007/s10549-011-1945-9. [DOI] [PubMed] [Google Scholar]
- 9.Campone M, Valo I, Jézéquel P, Moreau M, Boissard A, Campion L, Loussouarn D, Verriele V, Coqueret O, Guette C. Prediction of recurrence and survival for triple-negative breast cancer (TNBC) by a protein signature in tissue samples. Mol Cell Proteomics. 2015;14:2936–2946. doi: 10.1074/mcp.M115.048967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang CQ, Li Y, Huang BF, Zhao YM, Yuan H, Guo D, Su CM, Hu GN, Wang Q, Long T, et al. EGFR conjunct FSCN1 as a novel therapeutic strategy in triple-negative breast cancer. Sci Rep. 2017;7:15654. doi: 10.1038/s41598-017-15939-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inanc M, Ozkan M, Karaca H, Berk V, Bozkurt O, Duran AO, Ozaslan E, Akgun H, Tekelioglu F, Elmali F. Cytokeratin 5/6, c-Met expressions, and PTEN loss prognostic indicators in triple-negative breast cancer. Med Oncol. 2014;31:801. doi: 10.1007/s12032-013-0801-7. [DOI] [PubMed] [Google Scholar]
- 12.Tutt A, Robson M, Garber JE, Domchek SM, Audeh MW, Weitzel JN, Friedlander M, Arun B, Loman N, Schmutzler RK, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: A proof-of-concept trial. Lancet. 2010;376:235–244. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- 13.Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: Challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. 2016;13:674–690. doi: 10.1038/nrclinonc.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nedeljković M, Damjanović A. Mechanisms of chemotherapy resistance in triple-negative breast cancer-how we can rise to the challenge. Cells. 2019;8:957. doi: 10.3390/cells8090957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harano K, Wang Y, Lim B, Seitz RS, Morris SW, Bailey DB, Hout DR, Skelton RL, Ring BZ, Masuda H, et al. Rates of immune cell infiltration in patients with triple-negative breast cancer by molecular subtype. PLoS One. 2018;13:e0204513. doi: 10.1371/journal.pone.0204513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonsang-Kitzis H, Sadacca B, Hamy-Petit AS, Moarii M, Pinheiro A, Laurent C, Reyal F. Biological network-driven gene selection identifies a stromal immune module as a key determinant of triple-negative breast carcinoma prognosis. Oncoimmunology. 2015;5:e1061176. doi: 10.1080/2162402X.2015.1061176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burstein MD, Tsimelzon A, Poage GM, Covington KR, Contreras A, Fuqua SA, Savage MI, Osborne CK, Hilsenbeck SG, Chang JC, et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res. 2015;21:1688–1698. doi: 10.1158/1078-0432.CCR-14-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fumarola C, Petronini PG, Alfieri R. Impairing energy metabolism in solid tumors through agents targeting oncogenic signaling pathways. Biochem Pharmacol. 2018;151:114–125. doi: 10.1016/j.bcp.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 21.Spencer NY, Stanton RC. The Warburg effect, lactate, and nearly a century of trying to cure cancer. Seminars Nephrol. 2019;39:380–393. doi: 10.1016/j.semnephrol.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 22.Schwartz L, Supuran CT, Alfarouk KO. The Warburg effect and the hallmarks of cancer. Anticancer Agents Med Chem. 2017;17:164–170. doi: 10.2174/1871520616666161031143301. [DOI] [PubMed] [Google Scholar]
- 23.Sonveaux P, Végran F, Schroeder T, Wergin MC, Verrax J, Rabbani ZN, De Saedeleer CJ, Kennedy KM, Diepart C, Jordan BF, et al. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J Clin Invest. 2008;118:3930–3942. doi: 10.1172/JCI36843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitaker-Menezes D, Martinez-Outschoorn UE, Flomenberg N, Birbe RC, Witkiewicz AK, Howell A, Pavlides S, Tsirigos A, Ertel A, Pestell RG, et al. Hyperactivation of oxidative mitochondrial metabolism in epithelial cancer cells in situ: Visualizing the therapeutic effects of metformin in tumor tissue. Cell Cycle. 2011;10:4047–4064. doi: 10.4161/cc.10.23.18151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Faubert B, Li KY, Cai L, Hensley CT, Kim J, Zacharias LG, Yang C, Do QN, Doucette S, Burguete D, et al. Lactate metabolism in human lung tumors. Cell. 2017;171:358–371.e9. doi: 10.1016/j.cell.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nieman KM, Kenny HA, Penicka CV, Ladanyi A, Buell-Gutbrod R, Zillhardt MR, Romero IL, Carey MS, Mills GB, Hotamisligil GS, et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat Med. 2011;17:1498–1503. doi: 10.1038/nm.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, Tsukamoto T, Rojas CJ, Slusher BS, Zhang H, et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012;15:110–121. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pacheco-Velazquez SC, Robledo-Cadena DX, Hernandez-Resendiz I, Gallardo-Perez JC, Moreno-Sanchez R, Rodriguez-Enriquez S. Energy metabolism drugs block triple negative breast metastatic cancer cell phenotype. Mol Pharm. 2018;15:2151–2164. doi: 10.1021/acs.molpharmaceut.8b00015. [DOI] [PubMed] [Google Scholar]
- 29.Kim S, Kim DH, Jung WH, Koo JS. Metabolic phenotypes in triple-negative breast cancer. Tumour Biol. 2013;34:1699–1712. doi: 10.1007/s13277-013-0707-1. [DOI] [PubMed] [Google Scholar]
- 30.Jeon HM, Kim DH, Jung WH, Koo JS. Expression of cell metabolism-related genes in different molecular subtypes of triple-negative breast cancer. Tumori. 2013;99:555–564. doi: 10.1177/030089161309900419. [DOI] [PubMed] [Google Scholar]
- 31.Camarda R, Zhou AY, Kohnz RA, Balakrishnan S, Mahieu C, Anderton B, Eyob H, Kajimura S, Tward A, Krings G, et al. Inhibition of fatty acid oxidation as a therapy for MYC-overexpressing triple-negative breast cancer. Nat Med. 2016;22:427–432. doi: 10.1038/nm.4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamashita Y, Nishiumi S, Kono S, Takao S, Azuma T, Yoshida M. Differences in elongation of very long chain fatty acids and fatty acid metabolism between triple-negative and hormone receptor-positive breast cancer. BMC Cancer. 2017;17:589. doi: 10.1186/s12885-017-3554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jézéquel P, Kerdraon O, Hondermarck H, Guérin-Charbonnel C, Lasla H, Gouraud W, Canon JL, Gombos A, Dalenc F, Delaloge S. Gene-expression molecular subtyping of triple-negative breast cancer tumours: Importance of immune response. Breast Cancer Res. 2015;17:43. doi: 10.1186/s13058-015-0550-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sabatier R, Finetti P, Cervera N, Lambaudie E, Esterni B, Mamessier E, Tallet A, Chabannon C, Extra JM, Jacquemier J, et al. A gene expression signature identifies two prognostic subgroups of basal breast cancer. Breast Cancer Res Treat. 2011;126:407–420. doi: 10.1007/s10549-010-0897-9. [DOI] [PubMed] [Google Scholar]
- 35.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haw R, Stein L. Using the reactome database. Curr Protoc Bioinformatics Chapter 8: Unit8.7. 2012 doi: 10.1002/0471250953.bi0807s38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brunet JP, Tamayo P, Golub TR, Mesirov JP. Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci USA. 2004;101:4164–4169. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hou JY, Wang YG, Ma SJ, Yang BY, Li QP. Identification of a prognostic 5-Gene expression signature for gastric cancer. J Cancer Res Clin Oncol. 2017;143:619–629. doi: 10.1007/s00432-016-2324-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song W, Miao DL, Chen L. Comprehensive analysis of long noncoding RNA-associated competing endogenous RNA network in cholangiocarcinoma. Biochem Biophys Res Commun. 2018;506:1004–1012. doi: 10.1016/j.bbrc.2018.10.186. [DOI] [PubMed] [Google Scholar]
- 42.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Youden WJ. Index for rating diagnostic tests. Cancer. 1950;3:32–35. doi: 10.1002/1097-0142(1950)3:1<32::AID-CNCR2820030106>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 45.Xie G, Yang H, Ma D, Sun Y, Chen H, Hu X, Jiang YZ, Shao ZM. Integration of whole-genome sequencing and functional screening identifies a prognostic signature for lung metastasis in triple-negative breast cancer. Int J Cancer. 2019;145:2850–2860. doi: 10.1002/ijc.32329. [DOI] [PubMed] [Google Scholar]
- 46.Lv X, He M, Zhao Y, Zhang L, Zhu W, Jiang L, Yan Y, Fan Y, Zhao H, Zhou S, et al. Identification of potential key genes and pathways predicting pathogenesis and prognosis for triple-negative breast cancer. Cancer Cell Int. 2019;19:172. doi: 10.1186/s12935-019-0884-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quist J, Mirza H, Cheang MCU, Telli ML, O'Shaughnessy JA, Lord CJ, Tutt ANJ, Grigoriadis A. A Four-gene decision tree signature classification of Triple-negative breast cancer: Implications for targeted therapeutics. Mol Cancer Ther. 2019;18:204–212. doi: 10.1158/1535-7163.MCT-18-0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Balachandran VP, Gonen M, Smith JJ, DeMatteo RP. Nomograms in oncology: More than meets the eye. Lancet Oncol. 2015;16:e173–e180. doi: 10.1016/S1470-2045(14)71116-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tseng LM, Hsu NC, Chen SC, Lu YS, Lin CH, Chang DY, Li H, Lin YC, Chang HK, Chao TC, et al. Distant metastasis in triple-negative breast cancer. Neoplasma. 2013;60:290–294. doi: 10.4149/neo_2013_038. [DOI] [PubMed] [Google Scholar]
- 50.Altundag K. Genomic profiling of brain metastasis and matched primary Triple-negative breast cancer. Clin Breast Cancer. 2017;17:e161. doi: 10.1016/j.clbc.2016.12.015. [DOI] [PubMed] [Google Scholar]
- 51.Abramson VG, Mayer IA. Molecular heterogeneity of triple negative breast cancer. Curr Br Cancer Rep. 2014;6:154–158. doi: 10.1007/s12609-014-0152-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chiu AM, Mitra M, Boymoushakian L, Coller HA. Integrative analysis of the inter-tumoral heterogeneity of triple-negative breast cancer. Sci Rep. 2018;8:11807. doi: 10.1038/s41598-018-29992-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hashimoto N, Nagano H, Tanaka T. The role of tumor suppressor p53 in metabolism and energy regulation, and its implication in cancer and lifestyle-related diseases. Endocr J. 2019;66:485–496. doi: 10.1507/endocrj.EJ18-0565. [DOI] [PubMed] [Google Scholar]
- 54.Zhu S, Dong Z, Ke X, Hou J, Zhao E, Zhang K, Wang F, Yang L, Xiang Z, Cui H. The roles of sirtuins family in cell metabolism during tumor development. Semin Cancer Biol. 2019;57:59–71. doi: 10.1016/j.semcancer.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 55.Lu J. The Warburg metabolism fuels tumor metastasis. Cancer Metastasis Rev. 2019;38:157–164. doi: 10.1007/s10555-019-09794-5. [DOI] [PubMed] [Google Scholar]
- 56.Urra FA, Munoz F, Cordova-Delgado M, Ramírez MP, Peña-Ahumada B, Rios M, Cruz P, Ahumada-Castro U, Bustos G, Silva-Pavez E, et al. FR58P1a; a new uncoupler of OXPHOS that inhibits migration in triple-negative breast cancer cells via Sirt1/AMPK/β1-integrin pathway. Sci Rep. 2018;8:13190. doi: 10.1038/s41598-018-31367-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li W, Tanikawa T, Kryczek I, Xia H, Li G, Wu K, Wei S, Zhao L, Vatan L, Wen B, et al. Aerobic glycolysis controls myeloid-derived suppressor cells and tumor immunity via a specific CEBPB isoform in triple-negative breast cancer. Cell Metab. 2018;28:87–103.e6. doi: 10.1016/j.cmet.2018.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cserni G, Chmielik E, Cserni B, Tot T. The new TNM-based staging of breast cancer. Virchows Arch. 2018;472:697–703. doi: 10.1007/s00428-018-2301-9. [DOI] [PubMed] [Google Scholar]
- 59.Hortobagyi GN, Edge SB, Giuliano A. New and important changes in the TNM staging system for breast cancer. Am Soc Clin Oncol Educ Book. 2018;38:457–467. doi: 10.1200/EDBK_201313. [DOI] [PubMed] [Google Scholar]
- 60.Chen YC, Gonzalez ME, Burman B, Zhao X, Anwar T, Tran M, Medhora N, Hiziroglu AB, Lee W, Cheng YH, et al. Mesenchymal Stem/Stromal cell engulfment reveals metastatic advantage in breast cancer. Cell Rep. 2019;27:3916–3926.e5. doi: 10.1016/j.celrep.2019.05.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kalantari-Dehaghi M, Parnell EA, Armand T, Bernard HU, Grando SA. The nicotinic acetylcholine receptor-mediated reciprocal effects of the tobacco nitrosamine NNK and SLURP-1 on human mammary epithelial cells. Int Immunopharmacol. 2015;29:99–104. doi: 10.1016/j.intimp.2015.04.041. [DOI] [PubMed] [Google Scholar]
- 62.Zhu L, Ma N, Wang B, Wang L, Zhou C, Yan Y, He J, Ren Y. Significant prognostic values of aquaporin mRNA expression in breast cancer. Cancer Manag Res. 2019;11:1503–1515. doi: 10.2147/CMAR.S193396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wittliff JL, Sereff SB, Daniels MW. Expression of genes for methylxanthine pathway-associated enzymes accompanied by sex steroid receptor status impacts breast carcinoma progression. Hormones Cancer. 2017;8:298–313. doi: 10.1007/s12672-017-0309-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.de Vega S, Kondo A, Suzuki M, Arai H, Jiapaer S, Sabit H, Nakada M, Ikeuchi T, Ishijima M, Arikawa-Hirasawa E, et al. Fibulin-7 is overexpressed in glioblastomas and modulates glioblastoma neovascularization through interaction with angiopoietin-1. Int J Cancer. 2019;145:2157–2169. doi: 10.1002/ijc.32306. [DOI] [PubMed] [Google Scholar]
- 65.Celestino R, Nome T, Pestana A, Hoff AM, Gonçalves AP, Pereira L, Cavadas B, Eloy C, Bjøro T, Sobrinho-Simões M, et al. CRABP1, C1QL1 and LCN2 are biomarkers of differentiated thyroid carcinoma, and predict extrathyroidal extension. BMC Cancer. 2018;18:68. doi: 10.1186/s12885-017-3948-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Atiakshin D, Buchwalow I, Samoilova V, Tiemann M. Tryptase as a polyfunctional component of mast cells. Histochem Cell Biol. 2018;149:461–477. doi: 10.1007/s00418-018-1659-8. [DOI] [PubMed] [Google Scholar]
- 67.Aponte-López A, Fuentes-Pananá EM, Cortes-Muñoz D, Muñoz-Cruz S. Mast cell, the neglected member of the tumor microenvironment: Role in breast cancer. J Immunol Res. 2018;2018:2584243. doi: 10.1155/2018/2584243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang XP, Li Y, Wang Y, Wang Y, Wang P. Beta-Tryptase up-regulates vascular endothelial growth factor expression via proteinase-activated receptor-2 and mitogen-activated protein kinase pathways in bone marrow stromal cells in acute myeloid leukemia. Leuk Lymphoma. 2010;51:1550–1558. doi: 10.3109/10428194.2010.496013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed during the current study are available in the gene expression omnibus (https://www.ncbi.nlm.nih.gov/geo/) and The Cancer Genome Atlas (TCGA) databases (https://portal.gdc.cancer.gov/).