Abstract
Non-small cell lung cancer (NSCLC) is a type of cancer that is associated with high prevalence and high mortality rates in China. Therefore, it is of importance to identify the mechanisms underlying NSCLC progression. In the present study, reverse transcription-quantitative PCR was performed to measure the expression of microRNA (miR)-365b in NSCLC cell lines. In addition, the biological roles of miR-365b and N-acetylgalactosaminyltransferase 4 (GALNT4) were investigated by manipulating the expression levels of miR-365b and GALNT4 in NSCLC cells. It was found that miR-365b expression was reduced in NSCLC tissues and cells. Overexpression of miR-365b inhibited NSCLC cell proliferation whilst promoting apoptosis, but miR-365b knockdown promoted NSCLC cell proliferation. In addition, it was demonstrated that miR-365b regulated the proliferation and apoptosis of NSCLC cells by targeting GALNT4 expression. Collectively, the present study identified a miR-365b/GALNT4 regulatory axis in NSCLC, suggesting that miR-365b may serve as a therapeutic target for NSCLC.
Keywords: microRNA-365b, N-acetylgalactosaminyltransferase 4, non-small cell lung cancer, proliferation
Introduction
The estimated cases for lung cancer in United States are ~228,820, while non-small cell lung cancer (NSCLC) represents about 85% of all lung cancer cases (1). Treatment strategies for NSCLC including surgery, chemotherapy, immunotherapy and targeted therapy (2). However, the 5-year overall survival for NSCLC remains only ~19% (1).
MicroRNAs (miRNAs or miRs) are conserved RNA molecules that are typically 18-24 nucleotides in length and do not encode proteins (3). miRNA can regulate gene expression by binding to the 3'-untranslated regions (3'-UTR) of target mRNAs, resulting in degradation or inhibition of translation (4). In addition, miRs have been reported to have the potential to be developed as diagnostic or treatment biomarkers for patients with NSCLC (5). For example, circulating serum levels of miR-590-5p have been identified as a useful biomarker for NSCLC prognosis prediction (6). It has also been previously demonstrated that miR-590-5p overexpression can inhibit NSCLC cell proliferation and metastasis, whilst miR-590-5p knockdown resulted in opposite effects being observed (6). These previous findings suggest a tumor suppressive role for miR-590-5p.
miR-365b has been reported to exert important roles in regulating disease progression (7-9). For instance, miR-365b-3p overexpression has been shown to inhibit atherosclerosis development (7). Reduced miR-365b-3p expression has been demonstrated in retinoblastoma tumor tissues, where its overexpression can inhibit cancer cell proliferation by inducing cell cycle arrest and promoting apoptosis, suggesting that miR-365b-3p has a role in suppressing tumor progression (8). However, Tian et al (9) previously showed that miR-365b expression is elevated in hepatocellular carcinoma. In addition, functional assays showed that miR-365b overexpression can promote hepatocellular carcinoma cell motility and invasion by regulating the expression of small glutamine rich tetratricopeptide repeat containing β (SGTB) proteins, conversely suggesting an oncogenic role for miR-365b (9).
N-acetylgalactosaminyltransferase 4 (GALNT4) belongs to the GALNT family of proteins that catalyze the transfer of N-acetylgalactosamine to serine or threonine residues (10). GALNT contains several isoforms, including GALNT3, GALNT5 and GALNT9, which have been reported to serve roles in cancer progression (11-13). Additionally, GALNT4 exerts crucial functions in multiple cancer types (14,15). For example, GALNT4 expression can be negatively regulated by miR-4262, resulting in the inhibition of colon cancer proliferation (14,15).
Since the reported roles of miR-365b are diverse in different types of cancer and its specific function in NSCLC remains poorly understood, the present study aimed to investigate the role of miR-365b and its associated downstream mechanism in NSCLC.
Materials and methods
Cell culture and transfection
NSCLC cell lines A549 and H1299 and the normal human bronchial epithelial cell line 16HBE were purchased from American Type Culture Collection. Cells were incubated in DMEM (Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Invitrogen; Thermo Fisher Scientific, Inc.) in a 37˚C humidified atmosphere containing 5% CO2.
miR-365b mimic (forward, 5'-AGGGACUUUCAGGGGCAGCUGU-3'; and reverse, 5'-AGCUGCCCCUGAAAGUCCCUUU-3'), inhibitor (forward, 5'-ACAGCUGCCCCUGAAAGUCCCU-3'; and reverse, 5'-GGGACUUUCAGGGGCAGCUGUUU-3') and corresponding negative controls (NC-mimic; forward, 5'-GUAUGAGCGACUAGUGGCUGCG-3'; and reverse, 5'-CAGCCACUAGUCGCUCAUACUU-3' and NC-inhibitor; forward, 5'-GACUACCGAUACACCGCCGUUC-3'; and reverse, 5'-CGGCGGUGUAUCGGUAGUCUU-3') were designed by Shanghai GeneChem Co., Ltd. Small interfering RNA targeting GALNT4 (si-GALNT4; 5'-AAGAGATCATCTTGGTGGATG-3') and corresponding negative control (NC-siR; 5'-GGTCATGATGTCGGTGAATAA-3') were also purchased from Shanghai GeneChem Co., Ltd. The pcDNA3.1 plasmid containing the coding sequence of GALNT4 (pGALNT4, Clone ID: OHu17193) was purchased from GenScript Biotech, Inc, while pcDNA3.1 was used as negative control. For transfection, cells were seeded into 6-well plates at the density of 2x103 cells/well and transfected with Lipofectamine® 2,000 (Invitrogen; Thermo Fisher Scientific, Inc.) at the concentration of miRNAs: 50 nmol/l, siRNAs: 20 nmol/l, and expression vector: 4 µg. Cells were collected for subsequent analyses 48 h after transfection.
Microarray analysis
The microarray dataset GSE53882, was downloaded from Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE53882). The dataset contained miRNA expression data from 151 paired NSCLC and healthy tissues.
Cell Counting Kit-8 (CCK-8) assay
CCK-8 assay (Beyotime Institute of Biotechnology) was used to measure cell viability according to the manufacturer's instructions. In brief, 2x103 cells were plated into 96-well plates and cultured at 37˚C for 0, 24, 48 and 72 h. 10 µl CCK-8 solution was added to each well at each of the indicated times prior to incubation at 37˚C for a further 4 h. Optical density at 450 nm was measured in each well using a microplate reader.
Reverse transcription-quantitative PCR (RT-qPCR)
RNA samples of cultured cells were extracted using the TRIzol® reagent according to manufacturer's protocol (Invitrogen; Thermo Fisher Scientific, Inc.). PrimerScript™ RT Master mix (Takara Biotechnology Co., Ltd.) was used to reverse transcribe RNA into cDNA at 37˚C for 15 min, 85˚C for 5 sec and 4˚C for 60 min according to manufacturer's protocol. qPCR was performed using SYBR® Premix Ex Taq™ II (Takara Biotechnology Co., Ltd.) in an ABI 7500 system (Applied Biosystems; Thermo Fisher Scientific, Inc.). Sequences of the primers used, which were purchased from GenScript Biotech, Inc., were as follows: miR-365b forward, 5'-TAATGCCCCTAAAAAT-3' and reverse, 5'-CCAGTGCAGGGTCCGAGGT-3'; U6 snRNA forward, 5'-TGCGGGTGCTCGCTTCGGCAGC-3' and reverse, 5'-CCAGTGCAGGGTCCGAGGT-3'; GALNT4 forward, 5'-GGCCTATATCTTCGTGGAGCTC-3' and reverse, 5'-CCTGCGGAGGCATGAAAA-5' and GAPDH forward, 5'-CTGGGCTACACTGAGCACC-3' and reverse, 5'-AAGTGGTCGTTGAGGGCAATG-3'. Thermocycling conditions were as follows: 1 cycle of at 95˚C for 3 min; followed by 40 cycles at 95˚C for 10 sec, 58˚C for 30 sec and 72˚C for 30 sec. Relative expression levels were calculated using the 2-ΔΔCq method (16).
Western blotting
Protein samples were extracted from cells using RIPA lysis buffer (Invitrogen; Thermo Fisher Scientific, Inc.) and quantified using a bicinchoninic acid protein assay kit (Beyotime Institute of Biotechnology). The samples (50 µg) were then separated at 10% SDS-PAGE and transferred to PVDF membranes. The membranes were subsequently blocked with fat-free milk at 4˚C for 2 h and incubated with anti-GALNT4 (1:5,000; cat. no. ab80676; Abcam) and anti-GAPDH (1:5,000; cat. no. ab181602; Abcam) primary antibodies 4˚C for overnight. After washing with PBS, the membranes were incubated with horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (1:1,000; cat. no. ab6721; Abcam) at room temperature for 4 h. Then, the membranes were incubated with a BeyoECL kit (Beyotime Institute of Biotechnology) to visualize the protein bands.
Colony formation assay
In total, 500 cells were seeded into 6-well plates and allowed to proliferate for 2 weeks at a 37˚C humidified atmosphere contains 5% CO2. On day 14, 95% methanol was used to fix the colonies at 37˚C for 30 min, following which 1% crystal violet was used to stain the colonies at 37˚C for 15 min. Colony numbers were then manually counted using an inverted microscope with a magnification of x200.
Cell apoptosis assay
Flow cytometry was used to measure cell apoptosis. Cells (5x105) were collected and incubated with Annexin V-FITC and propidium iodide (Beyotime Institute of Biotechnology) according to the manufacturer's protocols. Flow cytometry was performed using a BD FACSCalibur™ (BD Biosciences) and analyzed with the FlowJo v10 software (FlowJo LLC).
Bioinformatics analysis
Potential targets for miR-365b were predicted using starBase v2.0 [http://starbase.sysu.edu.cn/index.php, (17)], which is a database containing the prediction results from various prediction tools. GALNT4 was selected since it was found to contain a complete binding site for miR-365b in its 3'-UTR.
Dual-luciferase activity reporter assay
Luciferase activity constructs were established by inserting the wild-type (wt) or mutant (mt) sequences of GALNT4 3'-UTR synthesized by GenScript Biotech, Inc., into the pmirGLO vector (Promega Corporation) to generate wt-GALNT4 or mt-GALNT4 plasmids. A549 and H1299 cells were seeded into 6-well plates at the density of 2x103 cells/well and were co-transfected with the respective luciferase activity reporter vectors (4 µg) and miR-365b mimic or NC-mimic (50 nmol/l) with Lipofectamine® 2000 according to manufacturer's protocols (Invitrogen; Thermo Fisher Scientific, Inc.). Relative luciferase activity was measured 48 h after transfection using the Dual-Luciferase® Reporter Assay system (Promega Corporation) with Renilla luciferase activity serving as the internal control.
Statistical analysis
Experiments were conducted in triplicate. Data are presented as the mean ± SD. Difference between two groups were analyzed using paired Student's t-test, whilst differences among multiple groups were analyzed with ANOVA and Tukey's test. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-365b expression is reduced in NSCLC
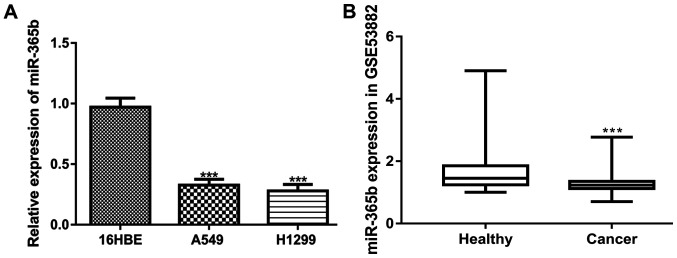
miR-365b expression was measured in two NSCLC cell lines (A549 and H1299) and a normal human bronchial epithelial cell line (16HBE). It was found that the expression of miR-365b was significantly reduced in NSCLC cells compared with that in 16HBE cells (Fig. 1A). In addition, by analyzing the GSE53882 dataset, it was demonstrated that miR-365b expression was also significantly reduced in NSCLC tissues compared with that in healthy tissues (Fig. 1B).
Figure 1.
miR-365b expression is lower in NSCLC tissues and cells compared with their non-cancerous counterparts. (A) Comparison of miR-365b expression among NSCLC and healthy cell lines. ***P<0.001 vs. 16HBE. (B) Comparison of miR-365b expression between NSCLC and healthy tissues. ***P<0.001 vs. Healthy. miR-365b, microRNA-365b; NSCLC, non-small cell lung cancer.
miR-365b knockdown promotes NSCLC cell proliferation and colony formation whilst inhibiting apoptosis
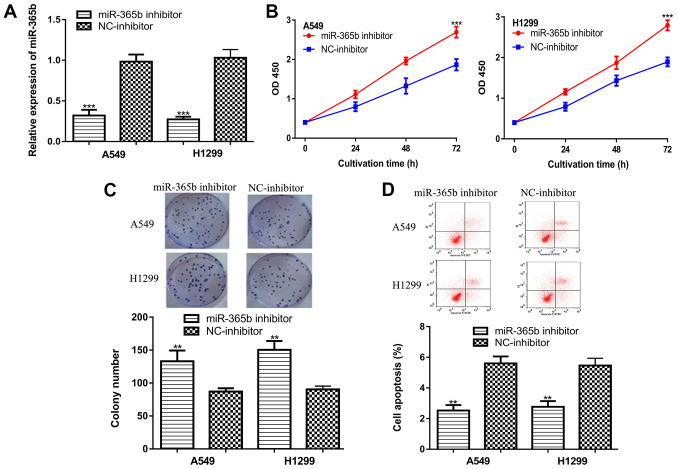
Knockdown experiments were performed to assess the functional roles of miR-365b in NSCLC cells. The efficiency of miR-365b inhibitor transfection was verified by RT-qPCR (Fig. 2A). CCK-8 assay results suggested that miR-365b knockdown significantly increased NSCLC cell viability (Fig. 2B). In addition, transfection with the miR-365b inhibitor was found to exert similar effects on NSCLC colony formation compared with the cell viability assays (Fig. 2C). In addition, flow cytometry assay results suggested that miR-365b knockdown significantly inhibited NSCLC cell apoptosis (Fig. 2D).
Figure 2.
Knockdown of miR-365b promotes NSCLC cell proliferation, promotes colony formation and inhibits apoptosis. (A) Expression of miR-365b in NSCLC cells transfected with the miR-365b inhibitor and NC-inhibitor was measured. (B) Cell Counting Kit-8 assay results indicated that miR-365b knockdown increased NSCLC cell viability. (C) Colony formation assay results suggested that miR-365b knockdown promoted NSCLC colony formation. (D) Flow cytometry assay results demonstrated that miR-365b knockdown inhibited NSCLC cell apoptosis. ***P<0.001, **P<0.01 vs. NC-inhibitor. miR-365b, microRNA-365b; NSCLC, non-small cell lung cancer; NC, negative control; OD, optical density.
Overexpression of miR-365b inhibits NSCLC cell proliferation, inhibits colony formation and promotes apoptosis
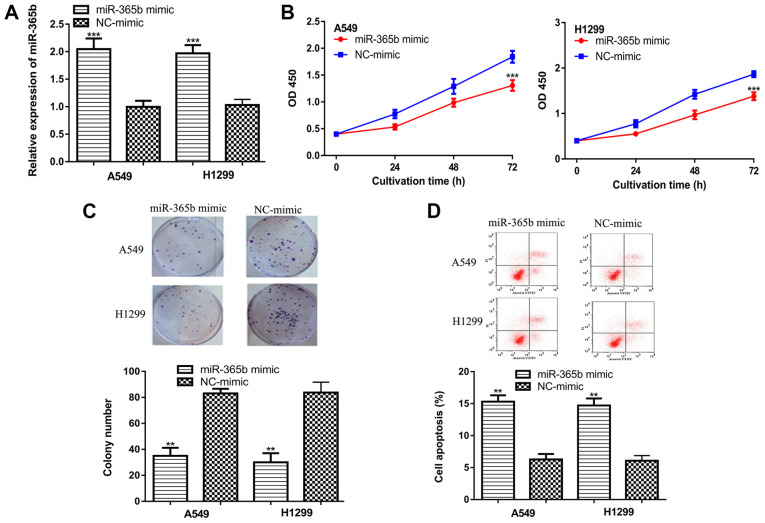
Overexpression experiments were subsequently performed to elucidate the roles of miR-365b further. It was found that transfection with the miR-365b mimic increased miR-365b expression (Fig. 3A). CCK-8 assay and colony formation assay results demonstrated that miR-365b overexpression inhibited NSCLC cell proliferation and colony formation (Fig. 3B and C). Additionally, the results indicated that the percentage of cell apoptosis was significantly increased by miR-365b overexpression (Fig. 3D).
Figure 3.
Overexpression of miR-365b inhibits NSCLC cell proliferation, inhibits colony formation and promotes apoptosis. (A) Expression of miR-365b in NSCLC cells transfected with the miR-365b mimic or NC-mimic was measured. (B) Cell Counting Kit-8 assay results indicated that miR-365b overexpression inhibited NSCLC proliferation. (C) Colony formation assay demonstrated that miR-365b overexpression inhibited NSCLC colony formation. (D) Flow cytometry assay indicated that miR-365b overexpression promoted NSCLC cell apoptosis. ***P<0.001, **P<0.01 vs. NC-mimic. miR-365b, microRNA-365b; NSCLC, non-small cell lung cancer; NC, negative control; OD, optical density.
miR-365b regulates GALNT4 expression binding to its 3'-UTR
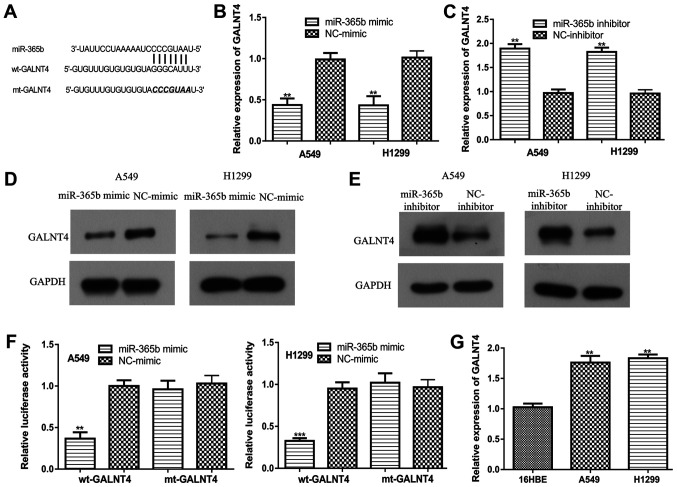
Bioinformatics analysis suggested that the 3'-UTR of GALNT4 contained a binding site for miR-365b (Fig. 4A). Overexpression of miR-365b significantly reduced GALNT4 expression in NSCLC cells (Fig. 4B), whereas miR-365b knockdown significantly increased GALNT4 expression in NSCLC cells (Fig. 4C). Western blotting results appeared to exhibit the same trends as those obtained by RT-qPCR (Fig. 4D and E). In addition, dual-luciferase activity reporter assay results suggested that miR-365b overexpression reduced luciferase activity in cells transfected with wt-GALNT4, but not in those transfected with mt-GALNT4 (Fig. 4F). It was also found that GALNT4 expression was higher in NSCLC cell lines compared with that in the non-cancerous cell line (Fig. 4G).
Figure 4.
miR-365b regulates GALNT4 expression by binding to the 3'-UTR of GALNT4. (A) Binding site between miR-365b and the 3'-UTR of GALNT4. (B) Expression of GALNT4 in NSCLC cells transfected with miR-365b mimic and NC-mimic was measured by RT-qPCR. **P<0.01 vs. NC-mimic. (C) Expression of GALNT4 in NSCLC cells transfected with miR-365b inhibitor and NC-inhibitor was measured by RT-qPCR. **P<0.01 vs. NC-inhibitor. (D) Expression of GALNT4 in NSCLC cells transfected with miR-365b mimic and NC-mimic was measured by western blotting. (E) Expression of GALNT4 in NSCLC cells transfected with miR-365b inhibitor and NC-inhibitor was measured by western blotting. (F) Relative luciferase activity as measured in NSCLC cells co-transfected with miR-365b mimic or NC-mimic and wt-GALNT4 or mt-GALNT4. **P<0.01 vs. NC-mimic. (G) GALNT4 expression in NSCLC cells and the non-cancerous cell line as measured by RT-qPCR. **P<0.01 vs. 16HBE. miR-365b, microRNA-365b; NSCLC, non-small cell lung cancer; UTR, untranslated region; GALNT4, N-acetylgalactosaminyltransferase 4; wt, wild-type; mt, mutant; NC, negative control; RT-qPCR, reverse transcription-quantitative PCR.
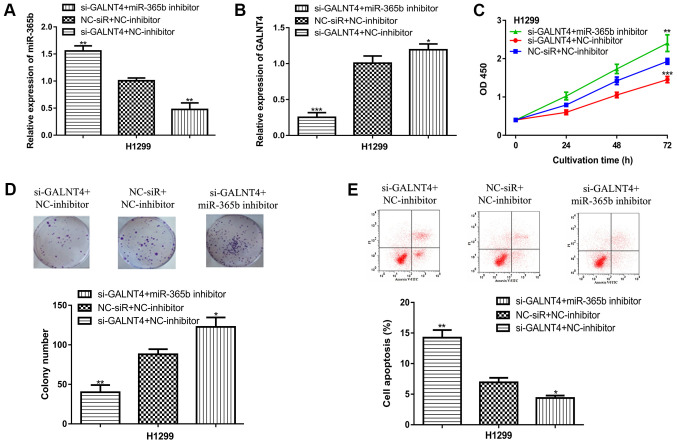
GALNT4 knockdown attenuates the effects of miR-365b inhibitors on NSCLC cell physiology
To assess whether miR-365b exerted a tumor suppressive role on NSCLC cells by regulating GALNT4 expression, si-GLANT4 and miR-365b inhibitor were co-transfected into H1299 cells. The results indicated that miR-365b expression was reduced by miR-365b inhibitor transfection, whilst si-GALNT4 transfection increased miR-365b expression slightly (Figs. 5A and S1A). In addition, it was found that GALNT4 expression was reduced by transfection with si-GALNT4 and increased by miR-365b inhibitor transfection (Figs. 5B and S1B). Functional assay results suggested that GALNT4 knockdown inhibited NSCLC cell proliferation and colony formation whilst promoting apoptosis (Fig. 5C-E). It was subsequently demonstrated that the stimulatory effects of miR-365b inhibitor transfection on NSCLC cells could be partly attenuated by si-GALNT4 co-transfection (Fig. 5C-E), whereas GALNT4 overexpression was found to increase GALNT4 expression and NSCLC cell proliferation (Fig. S2).
Figure 5.
GALNT4 knockdown attenuates the effects of miR-365b inhibitor transfection on NSCLC cells. (A) miR-365b expression in cells transfected with si-GALNT4+NC-inhibitor, NC-siR+NC-inhibitor or si-GALNT4+miR-365b inhibitor. (B) GALNT4 expression in cells transfected with si-GALNT4+NC-inhibitor, NC-siR+NC-inhibitor or si-GALNT4+miR-365b inhibitor. (C) Cell Counting Kit-8 assay results indicated that GALNT4 knockdown inhibited NSCLC proliferation and attenuated the effects of miR-365b inhibitor transfection on NSCLC cells. (D) Colony formation assay found that GALNT4 knockdown inhibited NSCLC colony formation and reversed the effects of miR-365b inhibitor transfection on NSCLC cells. (E) Flow cytometry assay suggested that GALNT4 knockdown promoted NSCLC cell apoptosis and attenuated the effects of miR-365b inhibitor transfection on NSCLC cells. ***P<0.001, **P<0.01, *P<0.05 vs. NC-siR+NC-inhibitor. miR-365b, microRNA-365b; NSCLC, non-small cell lung cancer; GALNT4, N-acetylgalactosaminyltransferase 4; si, small interfering RNA; NC, negative control.
Discussion
Results from the present study suggested that miR-365b overexpression inhibited NSCLC cell proliferation and colony formation whilst promoting apoptosis by targeting GALNT4 expression, which serve as a novel mechanism underlying the progression of NSCLC.
Dysregulated expression of miRNA has been revealed to occur in numerous types of cancer and during the carcinogenesis process (18). miRNAs have been shown to function as either tumor suppressors or oncogenes in NSCLC (19,20). For example, miR-422a has been reported to inhibit NSCLC cell proliferation, metastasis, epithelial-mesenchymal transition and tumorigenesis by regulating sulfatase 2 expression (19). By contrast, miR-671-3p can promote NSCLC cell proliferation, migration and invasion by regulating the expression of the forkhead box P2 protein (20). The present study measured the expression of miR-365b in NSCLC and non-cancerous cell lines, which found miR-365b expression to be lower in NSCLC cell lines. Cancer progression is frequently accompanied with several hallmark features, including uncontrolled cell proliferation and resistance to cell death (21). Therefore, the effects of miR-365b on NSCLC cell physiology were assessed. Knockdown of miR-365b expression was found to promote NSCLC cell proliferation and colony formation whilst inhibiting apoptosis. However, it was found that the overexpression of miR-365b using a miR-365b mimic exerted the opposite effects on NSCLC cells compared with those transfected with the miR-365b inhibitor.
Previous studies have identified a number of target genes, including ADAM metallopeptidase with thrombospondin type 1 motif 1, paired box 6 and SGTB for miR-365b in various types of malignancies (7-9). However, the relationship between miR-365b and these targets in NSCLC was not investigated in the present study. Despite this, the present study searched for potential targets for miR-365b using bioinformatics tools and identified GALNT4 to be a possible target. Subsequent luciferase activity reporter assay results demonstrated the direct interaction between miR-365b and the 3'-UTR of GALNT4. RT-qPCR and western blotting results also indicated that overexpression of miR-365b significantly reduced the expression of GALNT4 in NSCLC cells in contrast to miR-365b knockdown, which increased GALNT4 expression. Rescue experimental results suggested that GALNT4 knockdown attenuated the effects of miR-365b inhibitors on NSCLC cells. However, a limitation to this rescue experiment is that the results were only presented for the H1299 cell line. Previous studies have demonstrated that the expression of miRNAs in cancer can be regulated by long non-coding RNA, circular RNA and nucleotide polymorphism (22-24). Therefore, a limitation to the present study is that the regulatory mechanism responsible for the reduced expression of miR-365b in NSCLC was not examined, which should be investigated in future studies.
Collectively, the present results indicated that miR-365b functioned as a tumor suppressor in NSCLC. Therefore, elucidating the regulatory mechanism of GALNT4 by miR-365b may further the understanding of NSCLC carcinogenesis and facilitate the development of treatment for NSCLC.
Supplementary Material
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
LX, XDH, LC, PR and HZ performed the assay. LX, XDH, LC, PR and HZ performed the computations and data analysis. LX, XDH and HZ wrote the manuscript. LX and HZ conceived and designed the study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Watanabe SI, Nakagawa K, Suzuki K, Takamochi K, Ito H, Okami J, Aokage K, Saji H, Yoshioka H, Zenke Y, et al. Neoadjuvant and adjuvant therapy for stage III non-small cell lung cancer. Jpn J Clin Oncol. 2017;47:1112–1118. doi: 10.1093/jjco/hyx147. [DOI] [PubMed] [Google Scholar]
- 3.Acunzo M, Romano G, Wernicke D, Croce CM. MicroRNA and cancer-a brief overview. Adv Biol Regul. 2015;57:1–9. doi: 10.1016/j.jbior.2014.09.013. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.hou Q, Huang SX, Zhang F, Li SJ, Liu C, Xi YY, Wang L, Wang X, He QQ, Sun CC, et al. MicroRNAs: A novel potential biomarker for diagnosis and therapy in patients with non-small cell lung cancer. Cell Prolif. 2017;50 doi: 10.1111/cpr.12394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khandelwal A, Seam RK, Gupta M, Rana MK, Prakash H, Vasquez KM, Jain A. Circulating microRNA-590-5p acts as a liquid biopsy marker in non-small cell lung cancer. Cancer Sci. 2020;111:826–839. doi: 10.1111/cas.14199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qu Y, Zhang N. MiR-365b-3p inhibits the cell proliferation and migration of human coronary artery smooth muscle cells by directly targeting ADAMTS1 in coronary atherosclerosis. Exp Ther Med. 2018;16:4239–4245. doi: 10.3892/etm.2018.6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, Wang X, Wu G, Hou D, Hu Q. MiR-365b-3p, down-regulated in retinoblastoma, regulates cell cycle progression and apoptosis of human retinoblastoma cells by targeting PAX6. FEBS Lett. 2013;587:1779–1786. doi: 10.1016/j.febslet.2013.04.029. [DOI] [PubMed] [Google Scholar]
- 9.Tian Q, Sun HF, Wang WJ, Li Q, Ding J, Di W. MiRNA-365b promotes hepatocellular carcinoma cell migration and invasion by downregulating SGTB. Future Oncol. 2019;15:2019–2028. doi: 10.2217/fon-2018-0676. [DOI] [PubMed] [Google Scholar]
- 10.Niang B, Jin L, Chen X, Guo X, Zhang H, Wu Q, Padhiar AA, Xiao M, Fang D, Zhang J. Galnac-t4 putatively modulates the estrogen regulatory network through FOXA1 glycosylation in human breast cancer cells. Mol Cell Biochem. 2016;411:393–402. doi: 10.1007/s11010-015-2601-1. [DOI] [PubMed] [Google Scholar]
- 11.Sahasrabudhe NM, Lenos K, van der Horst JC, Rodríguez E, van Vliet SJ. Oncogenic BRAFV600E drives expression of MGL ligands in the colorectal cancer cell line HT29 through N-acetylgalactosamine-transferase 3. Biol Chem. 2018;399:649–659. doi: 10.1515/hsz-2018-0120. [DOI] [PubMed] [Google Scholar]
- 12.Detarya M, Sawanyawisuth K, Aphivatanasiri C, Chuangchaiya S, Saranaruk P, Sukprasert L, Silsirivanit A, Araki N, Wongkham S, Wongkham C. doi: 10.1093/glycob/cwz098. The O-GalNAcylating enzyme GALNT5 mediates carcinogenesis and progression of cholangiocarcinoma via activation of AKT/ERK signaling. Glycobiology: Dec 4, 2019 (Epub ahead of print). [DOI] [PubMed] [Google Scholar]
- 13.erois N, Gattolliat CH, Barrios E, Capandeguy L, Douc-Rasy S, Valteau-Couanet D, Bénard J, Osinaga E. GALNT9 gene expression is a prognostic marker in neuroblastoma patients. Clin Chem. 2013;59:225–233. doi: 10.1373/clinchem.2012.192328. [DOI] [PubMed] [Google Scholar]
- 14.Weng L, Ma J, Jia YP, Wu SQ, Liu BY, Cao Y, Yin X, Shang MY, Mao AW. MiR-4262 promotes cell apoptosis and inhibits proliferation of colon cancer cells: Involvement of GALNT4. Am J Transl Res. 2018;10:3969–3977. [PMC free article] [PubMed] [Google Scholar]
- 15.Qu JJ, Qu XY, Zhou DZ. MiR-4262 inhibits colon cancer cell proliferation via targeting of GALNT4. Mol Med Rep. 2017;16:3731–3736. doi: 10.3892/mmr.2017.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Li JH, Liu S, Zhou H, Qu LH, Yang JH. StarBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42 (Database Issue):D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 19.Li WQ, Zhang JP, Wang YY, Li XZ, Sun L. MicroRNA-422a functions as a tumor suppressor in non-small cell lung cancer through SULF2-mediated TGF-β/SMAD signaling pathway. Cell Cycle. 2019;18:1727–1744. doi: 10.1080/15384101.2019.1632135. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Li ZY, Zhang ZZ, Bi H, Zhang QD, Zhang SJ, Zhou L, Zhu XQ, Zhou J. Upregulated microRNA-671-3p promotes tumor progression by suppressing forkhead box P2 expression in non-small-cell lung cancer. Mol Med Rep. 2019;20:3149–3159. doi: 10.3892/mmr.2019.10563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 22.Yu G, Li S, Liu P, Shi Y, Liu Y, Yang Z, Fan Z, Zhu W. LncRNA TUG1 functions as a ceRNA for miR-6321 to promote endothelial progenitor cell migration and differentiation. Exp Cell Res. 2020;338(111839) doi: 10.1016/j.yexcr.2020.111839. [DOI] [PubMed] [Google Scholar]
- 23.Deng G, Mou T, He J, Chen D, Lv D, Liu H, Yu J, Wang S, Li G. Circular RNA circRHOBTB3 acts as a sponge for miR-654-3p inhibiting gastric cancer growth. J Exp Clin Cancer Res. 2020;39(1) doi: 10.1186/s13046-019-1487-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim H, Burassakarn A, Kang Y, Iizasa H, Yoshiyama H. A single nucleotide polymorphism in the BART promoter region of Epstein-Barr virus isolated from nasopharyngeal cancer cells. Biochem Biophys Res Commun. 2019;520:373–378. doi: 10.1016/j.bbrc.2019.10.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.