Abstract
Breast cancer is the most common type of malignancy in women, which remains a significant health concern worldwide. Gemcitabine is a frequently applied anticancer pharmacological agent. However, the efficacy of gemcitabine is limited by chemoresistance. In the present study, a combination of reverse transcription quantitative-PCR, cell viability, flow cytometry, luciferase reporter assay and western blot analysis were performed to elucidate the potential effects of miR-187-3p on gemcitabine sensitivity in the breast cancer cell line, MDA-MB-231. The results revealed that miR-187-3p was significantly decreased in the breast cancer tumor tissues. Moreover, the overexpression of miR-187-3p significantly inhibited cell viability and promoted apoptosis in MDA-MB-231 cells. In addition, miR-187-3p overexpression enhanced the anti-proliferative and pro-apoptotic effects of gemcitabine, indicating that miR-187-3p regulated gemcitabine sensitivity in breast cancer cells. Mechanistically, miR-187-3p negatively regulated the expression of fibroblast growth factor 9 (FGF9) by binding to its 3'-untranslated region. Overexpression of FGF9 reversed the aforementioned effects of miR-187-3p overexpression on cell viability and apoptosis in the presence of gemcitabine. In conclusion, the present study indicated that miR-187-3p increased gemcitabine sensitivity in breast cancer cells by targeting FGF9 expression.
Keywords: breast cancer, gemcitabine, microRNA-187-3p, fibroblast growth factor 9
Introduction
Breast cancer is one of the most diverse and complex types of human malignancy, which is a leading cause of mortality among women worldwide (1). In the USA, breast cancer diagnoses are particularly common, accounting for ~30% of all newly diagnosed malignancies; it was reported that breast cancer led to various malgnancies, which seriously affected patients' quality of life (1-3).
Gemcitabine (2',2'-difluorodeoxycytidine, dFdC) is a pyrimidine nucleoside analog that inhibits DNA replication and transcription, leading to cell apoptosis (4). It is frequently applied as a single agent as salvage chemotherapy following several lines of treatment (5). Considering prior treatment, dose and schedule, response rates to gemcitabine were typically 14-42% (5). The combined use of gemcitabine with capecitabine, vinorelbine, platinum and other chemotherapeutic agents can enhance the anticancer effects of the overall chemotherapy (6). Since Gemcitabine has been recommended by Chinese breast cancer diagnosis and treatment guide (2017) as an effective method in treating advanced breast cancer (7). Enhancing the therapeutic effects gemcitabine would be of great significance for the treatment of this disease.
MicroRNAs (miRNAs or miRs) are a class of evolutionarily conserved non-coding RNAs that typically consist of ~22 nucleic acids (8). They normally serve as post-transcriptional regulators of gene expression by binding to the 3'untranslated regions (3'UTRs) of target mRNAs to either inhibit translation or promote degradation (8). miRNAs have been reported to participate in a number of physiological processes, including proliferation, apoptosis, cell movement and stem cell renewal (8). Accumulating evidence has revealed that a wide variety of miRNAs are involved in oncogenesis (9). Of note, miR-187-3p has been revealed to be downregulated in colorectal cancer (10), renal cell carcinoma (11), prostate cancer (12), lung cancer (13) and breast cancer (14). In addition, miR-187-3p expression has also been previously associated with patient prognosis in breast cancer (14). However, the precise function of miR-187-3p in the pathophysiology of cancer remain controversial, as a previous study has demonstrated that miR-187 overexpression promotes lymphoma progression and enhances resistance to chemotherapy in peripheral T-cell lymphoma (15).
Fibroblast growth factor 9 (FGF9) is a member of the FGF family, which includes 23 family members each serving key functions including cellular differentiation, proliferation and tumorigenesis (16). FGFs bind to and activate FGF receptors, leading to the further activation of developmental signaling pathways that are responsible for a variety of biological functions (17). The overexpression of FGF9 has been previously identified as a novel unfavorable prognostic indicator in lung cancer (18). Intracellular signaling pathways activated by FGF serve important roles in a wide range of malignancies, including breast cancer (17,19). Fillmore et al (20) revealed that estrogen could activate FGF9/FGFR3/T box transcription factor 3 signaling to increase the numbers of breast cancer stem-like cells, whilst Yin et al (21) have previously reported that the miRNA-FGF9 pathway is important for pleuropulmonary blastoma development.
In the present study, it was revealed that the overexpression of miR-187-3p inhibited MDA-MB-231 cell proliferation, promoted apoptosis and reduced resistance to gemcitabine. Mechanistically, miR-187-3p overexpression resulted in the downregulation of FGF9 expression to regulate gemcitabine sensitivity in breast cancer cells, implicating miR-187-3p as a promising therapeutic target in the treatment of breast cancer.
Materials and methods
Clinical patient tissue samples
A total of 30 breast cancer tumor tissue samples and matched adjacent non-tumor tissue samples, 5 cm away from the tumors, were collected at Chifeng Municipal Hospital (Chifeng, China) from June 2015 to July 2017. All samples were collected from women aged between 27 and 65 years with an average age of 48±11 years. Patients who have received any chemo- or radio- therapies were excluded from the study. Written informed consent was provided by all participants prior to enrollment. The present study was approved by the Ethics Committee of Chifeng Municipal Hospital (approval no. 20150602CFMH; Chifeng, China). All tissue samples were immediately frozen in liquid nitrogen following surgery and stored in a -80˚C refrigerator prior to use.
Cell culture and reagents
MDA-MB-231 human breast cancer cell line was purchased from the American Type Culture Collection and was subsequently cultured in DMEM (Life Technologies; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (HyClone; GE Healthcare Life Sciences) and 1% penicillin-streptomycin solution (Life Technologies; Thermo Fisher Scientific, Inc.) in a humidified atmosphere at 37˚C and 5% CO2. Gemcitabine was purchased from Sigma-Aldrich (Merck KGaA).
Transient transfection
miR-187-3p mimic (50 nM, 5'-GGCCGACGUUGUGUUCUGUGCU-3') and miR-NC mimic (50 nM, 5'-UCGCUUGGUGCAGGUCGGGAA-3') were purchased from Shanghai GenePharma Co., Ltd., pcDNA3.1 (2 µg) and pcDNA-FGF9 (2 µg) were purchased from Addgene, Inc. All transfections were performed into MDA-MB-231 using Lipofectamine® 2000 transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.). After incubation for 48 h, cells were collected for the subsequent studies.
RNA extraction and reverse transcription-quantitative PCR (RT-qPCR
Total RNA was extracted from cultured MDA-MB-231 cells and tissues using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.) and cDNA synthesis was performed at 37˚C for 15 min and 85˚C for 5 sec using a PrimeScript™ RT reagent kit (Takara Bio, Inc.) according to the manufacturer's protocols. RT-qPCR was performed in triplicate using SYBR® Premix Ex Taq™ (Takara Bio, Inc.) in a Bio-Rad CFX96 Real-Time PCR System (Bio-rad Laboratories Inc.). The thermocycling conditions were as follows: 95˚C for 30 sec, followed by 35 cycles of 95˚C for 5 sec and 60˚C for 30 sec. Relative levels of miR-187-3p were normalized to that of U6 small nucleolar RNA, whereas those of FGF9 were normalized to GAPDH. The 2-ΔΔCq method was used to quantify relative gene expression (22). The primer sequences used were listed as follows: Stem loop primer, 5'-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCCGGCT-3'; miR-187-3p forward, 5'-GCCGAGTCGTGTCTTGTGTT-3' and reverse, 5'-CTCAACTGGTGTCGTGGA-3'; U6 forward, 5'-CTCAACTGGTGTCGTGGA-3' and reverse, 5'-CTCAACTGGTGTCGTGGA-3'; FGF9 forward, 5'-ATGGCTCCCTTAGGTGAAGTT-3' and reverse, 5'-CCCAGGTGGTCACTTAACAAAAC-3'; GAPDH forward, 5'-CAATGACCCCTTCATTGACC-3' and reverse, 5'-GACAAGCTTCCCGTTCTCAG-3'.
Cell viability
Cell viability was assessed by performed a cell counting kit-8 assay (CCK-8; Dojindo Molecular Technologies, Inc.) according to the manufacturer's protocol. Cells (~5x103/well) were seeded into 96-well plates. Following treatment with ascending concentrations of Gemcitabine (0.25, 0.5, 1, 2 and 4 nM) for 24 h at 37˚C and co-transfection with miR-187-3p or miR-NC mimic and pcDNA3.1-FGF9 or pcDNA3.1 plasmid for 48 h, 10 µl CCK-8 solution was added into each well and incubated at 37˚C for 2 h. Absorbance at 450 nm was subsequently measured in each well using a spectrophometer to determine cell viability.
Apoptosis assay
An Annexin-V/Dead Cell Apoptosis kit (Invitrogen; Thermo Fisher Scientific, Inc.) was used to perform cell apoptosis assay according to manufacturer's protocol. Cells were harvested and washed in cold PBS, after which they were then diluted to ~1x106 cells/ml using 1X Annexin-binding buffer in 100 µl per assay. Cells were subsequently treated with 5 µl Alexa Fluor® 488 annexin V and 1 µl 100 µg/ml PI per assay suspension. A total of 400 µl annexin-binding buffer was added and samples were incubated at room temperature for 15 min. Stained cells were analyzed using BD FACSCaliburTM flow cytometer (BD Biosciences) coupled with FlowJo software (version 10.2; FlowJo LLC).
Western blot analysis
Antibodies against FGF9 (cat. no. PA5-23719; 1:1,000) and β-catenin (cat. no. 71-2700; 1:1,000) were obtained from Thermo Fisher Scientific, Inc. Anti-c-Myc (cat. no. 5605; 1:1,000) and Cyclin D1 (cat. no. 2978; 1:1,000) antibodies were purchased from Cell Signaling Technology, Inc. GAPDH mouse monoclonal antibodies (cat. no. ab8245; 1:10,000) was obtained from Abcam. Anti-mouse (cat. no. CW0221S; 1:10,000) and anti-rabbit (cat. no. CW0234S; 1:10,000) secondary antibodies were purchased from Beijing ComWin Biotech Co., Ltd.
Following collection, cells were washed twice with cold PBS and lysed in cold RIPA buffer (Beyotime Institute of Biotechnology) supplemented with protease inhibitor cocktail (Sigma-Aldrich, Merck KGaA). Samples were then incubated on ice for 30 mins. Lysates were subsequently centrifuged at 12,000 x g at 4˚C for 15 min and protein concentration was measured using Bicinchoninic Acid Protein Assay kit (Thermo Fisher Scientific, Inc.). Equal quantities (20 µg/well) of protein were separated by 8% SDS-PAGE, transferred to polyvinylidene fluoride membranes (EMD millipore) and incubated with the respective aforementioned antibodies. Membranes were developed using SuperSignal™ West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific, Inc.) and image analysis was performed using the ImageQuant™ LAS 4000 software (GE Healthcare Life Sciences).
Dual-luciferase reporter gene assay
The TargetScan miRNA target database (http://www.targetscan.org/) predicted that a putative miR-187-3p binding site was present on the 3'-UTR of FGF9. Therefore, the FGF9 3'-UTR region was amplified from cDNA isolated from MDA-MB-231 cells and inserted into the XbaI restriction site of pGL3 Luciferase Reporter vector (Promega Corporation) with the primer pairs as listed: WT FGF9, forward, 5'-GCTCTAGACAAAGACAGTTTCTTCAC-3', reverse, 5'-GCTCTAGATTTTCAAAACTCTGTAAT-3'. In addition, two site mutations were introduced into the wild-type (WT) pGL3-FGF9 3'UTR WT to construct the mutant (Mut) FGF9 3'UTR plasmid using the Quicksite mutation kit (Agilent Technologies, Inc.) with the listed primer pairs: MT FGF9, forward, 5'-CGGAAAAAGACGGGCCACGACAGG-3', reverse, 5'-CCTGTCGTGGCCCGTCTTTTTCCG-3'. MDA-MB-231 cells were co-transfected with 100 nM FGF9 3'UTR-WT or FGF9 3'UTR-Mut plasmids and 100 nM miR-187-3p mimic or negative mimic control using Lipofectamine® 2000 transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to manufacturer's protocol for 48 h. Luciferase activity was evaluated using the Dual-Glo® Luciferase assay system according to manufacturer's protocol (Promega Corporation). The luciferase activity was normalized to Renilla luciferase activity.
Statistical analysis
All experiments in the present study were performed three times independently. GraphPad Prism 5.0 software (GraphPad Software, Inc.) was used for statistical analysis and all data were presented as mean ± standard deviation. Student's t-test was used for all comparisons between two groups whereas one-way ANOVA followed by Student-Newman-Keuls test was used for comparison of differences between ≥3 groups. Pearson's correlation analysis was performed to analyze the correlation between FGF mRNA and miR-187-3p expression in the 30 pairs of breast cancer tumor and corresponding matched adjacent non-tumor tissue samples collected from patients with breast cancer. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-187-3p expression is lower in breast cancer tumor tissues
To investigate the potential role of miR-187-3p in breast cancer, miR-187-3p expression was measured in 30 pairs of breast cancer tumor and corresponding matched adjacent non-tumor tissue samples collected from patients with breast cancer. RT-qPCR analysis revealed that miR-187-3p expression was significantly reduced in breast cancer tumor tissues compared with non-tumor tissues (Fig. 1), indicating that miR-187-3p may serve a role in the development of breast cancer.
Figure 1.
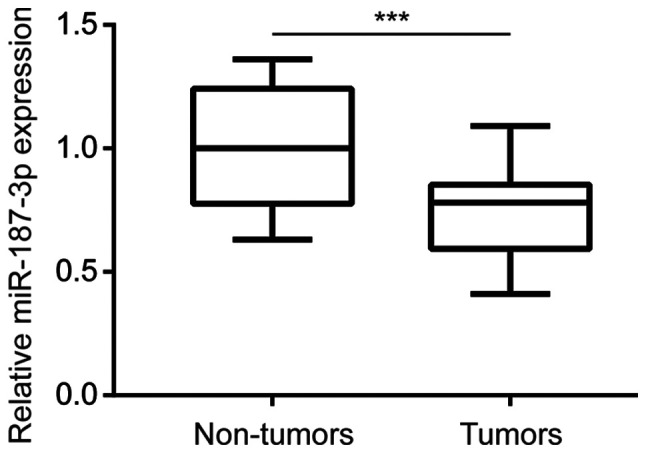
miR-187-3p expression was reduced in breast cancer tumor tissues. Compared with matched non-tumor tissues, miR-187-3p expression was significantly reduced in breast cancer tumor tissues collected from 30 patients with breast cancer. Data were presented as mean ± standard deviation. ***P<0.001 as indicated. miR, microRNA.
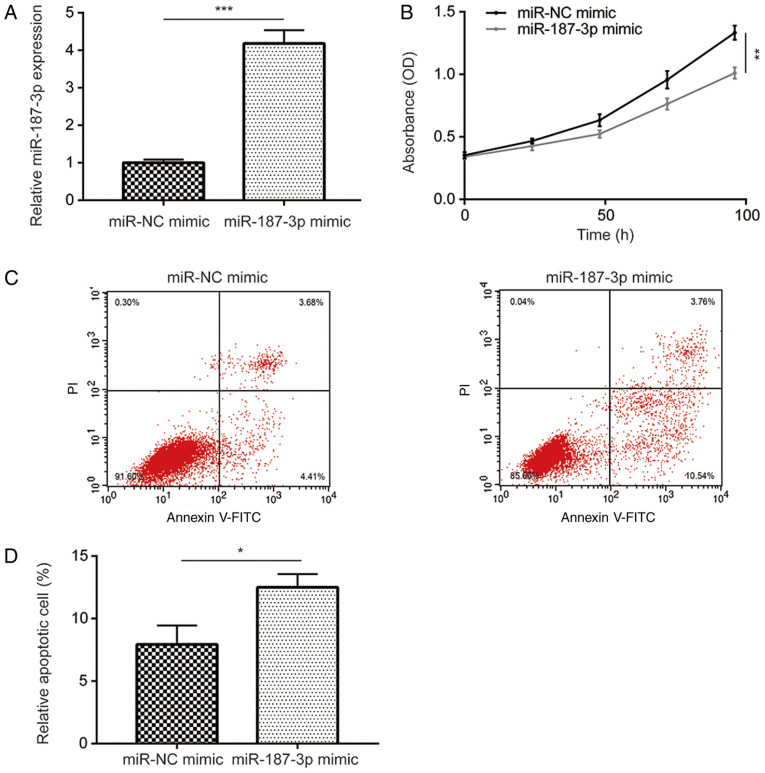
Overexpression of miR-187-3p inhibits cell viability and promotes apoptosis in MDA-MB-231 cells
The physiological function and underlying mechanism of miR-187-3p in breast cancer remain poorly understood. Therefore, MDA-MB-231 cells were successfully transfected with the miR-187-3p mimic to overexpress miR-187-3p compared with the miR-NC mimic (Fig. 2A). Compared with the miR-NC mimic, miR-187-3p overexpression was subsequently revealed to significantly inhibit MDA-MB-231 cell viability (Fig. 2B). Additionally, compared with the miR-NC mimic, overexpression of miR-187-3p significantly promoted MDA-MB-231 cell apoptosis (Fig. 2C and D). The results indicated that miR-187-3p overexpression reduced MDA-MB-231 cell viability and promoted apoptosis.
Figure 2.
miR-187-3p overexpression reduced cell viability and increased cell apoptosis in MDA-MB-231 cells. (A) Transfection with miR-187-3p mimic significantly increased miR-187-3p expression compared with those transfected with miR-NC mimic in MDA-MB-231 cells. ***P<0.001 as indicated. (B) Compared with miR-NC mimic, transfection with miR-187-3p mimic significantly reduced MDA-MB-231 cell viability and (C) increased cell apoptosis in MDA-MB-231 cells. **P<0.01 as indicated. (D) Quantified data from (C) demonstrating that transfection with miR-187-3p mimic significantly increased MDA-MB-231 cell apoptosis. Data are presented as the mean ± standard deviation. *P<0.05 as indicated. miR, microRNA; NC, negative control.
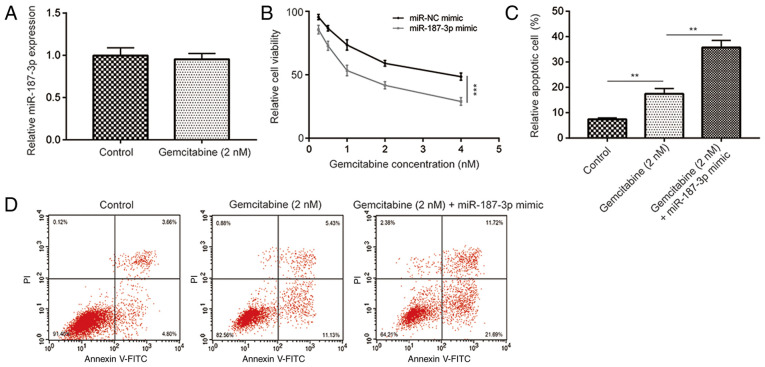
Overexpression of miR-187-3p enhances gemcitabine sensitivity in MDA-MB-231 cells
Liang et al (23) discovered that miR-187-3p was one of the most significantly downregulated miRNAs in multidrug-resistant MCF7 cells compared to parental MCF7 cells. Although gemcitabine is frequently applied as a therapeutic agent for advanced breast cancer, it remains associated with limited efficacy (7,24). In this present study, it was determined that, compared to the control, gemcitabine (2 nM) treatment did not significantly alter miR-187-3p expression in MDA-MB-231 cells (Fig. 3A). However, compared with the miR-NC mimic, overexpression of miR-187-3p significantly enhanced the gemcitabine-mediated (1, 2, 3 and 4 nM) reduction of cell viability in MDA-MB-231 cells (Fig. 3B) whilst significantly potentiating gemcitabine-induced MDA-MB-231 cell apoptosis (Fig 3C and D). These results indicated that miR-187-3p overexpression removed gemcitabine resistance in breast cancer cells.
Figure 3.
miR-187-3p enhanced the gemcitabine-mediated reduction in cell viability and induction of MDA-MB-231 cell apoptosis. (A) Gemcitabine treatment (2 nM) did not alter miR-187-3p expression in MDA-MB-231 cells. (B) Transfection with miR-187-3p mimic significantly enhanced gemcitabine-mediated reduction in MDA-MB-231 cell viability compared with those transfected with miR-NC mimic. ***P<0.001 as indicated. (C) Treatment with 2 nM gemcitabine significantly increased MDA-MB-231 cell apoptosis compared with the control, which was in turn, significantly potentiated following transfection with the miR-187-3p mimic. **P<0.01 as indicated. (D) Representative flow cytometry dot plots complementing the quantified data from (C) Data are presented as the mean ± standard deviation. miR, microRNA; NC, negative control.
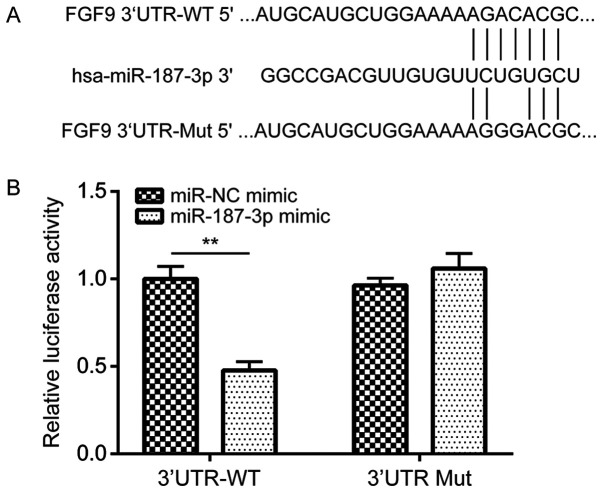
miR-187-3p regulates FGF9 expression by binding to its 3'-UTR
Following the indication that miR-187-3p enhanced the efficacy of gemcitabine, the underlying mechanism was elucidated. According to the TargetScan database, FGF9 was predicted to be a potential target gene of miR-187-3p, as a putative miR-187-3p binding site was observed on the 3'-UTR of FGF9 (Fig. 4A). WT FGF9 3'-UTR and Mut FGF9 3'-UTR luciferase reporter gene plasmids were constructed to verify this potential association. Dual-luciferase reporter gene assay data demonstrated that, compared with the miR-NC mimic, transfection with the miR-187-3p mimic significantly reduced relative luciferase activity in MDA-MB-231 cells co-transfected with the WT FGF9 3'-UTR plasmid (Fig. 4B).
Figure 4.
miR-187-3p regulated FGF9 expression by binding to its 3'-UTR. (A) Bioinformatics database prediction of the putative binding site of miR-187-3p on the 3'-UTR of FGF9. A mutant 3'UTR sequence was constructed by the introduction of two site mutations. (B) Luciferase activity was significantly decreased in MDA-MB-231 cells co-transfected with miR-187-3p mimic and FGF9 3'UTR-WT reporter plasmid compared with those transfected with the miR-NC mimic. However, luciferase activity was not altered following co-transfection with miR-187-3p mimic and FGF9 3'UTR-Mut reporter plasmid. Data are presented as the mean ± standard deviation. **P<0.01 as indicated. FGF9. Fibroblast growth factor 9; 3'UTR, 3' untranslated region; miR, microRNA; NC, negative control; WT, wild type, Mut, mutant.
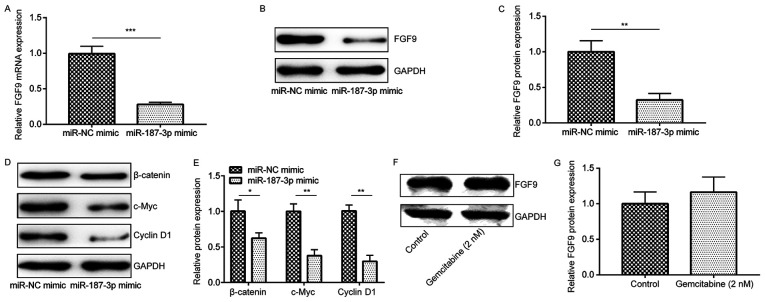
miR-187-3p overexpression negatively regulates FGF9 expression and the associated downstream Wnt pathway
To investigate the regulatory effects of miR-187-3p on FGF9, RT-qPCR and western blot analysis were performed to measure FGF9 mRNA and protein expression in MDA-MB-231 cells following miR-187-3p overexpression. It was revealed that, compared with the miR-NC mimic, miR-187-3p overexpression significantly reduced FGF9 mRNA and protein expression (Fig. 5A-C). A previous study demonstrated that FGF9 can regulate Wnt/β-catenin signaling and that deficiencies in this pathway have resulted in reduced mesenchymal proliferation (25). In addition, FGF9 has been reported to be a target for the Wnt/β-catenin pathway in ovarian endometrioid adenocarcinomas (26). Following miR-187-3p overexpression, β-catenin, c-Myc and Cyclin D1 protein expression were revealed to be significantly reduced compared with the miR-NC mimic (Fig. 5D and E). In addition, compared with the control, gemcitabine treatment did not significantly alter FGF9 expression in MDA-MB-231 cells (Fig. 5F and G). The results indicated that miR-187-3p regulated breast cancer progression by negatively regulating FGF9 expression, which in turn inactivated the Wnt signaling pathway.
Figure 5.
miR-187-3p negatively regulated FGF9 expression and the Wnt signaling pathway downstream. (A) Compared with those transfected with the miR-NC mimic, miR-187-3p mimic transfection significantly reduced FGF9 mRNA expression in MDA-MB-231 cells. ***P<0.001 as indicated. (B) Compared with those transfected with the miR-NC mimic, miR-187-3p mimic transfection reduced FGF9 protein expression in MDA-MB-231 cells. (C) Quantified data from (B) demonstrating that miR-187-3p mimic transfection significantly reduced FGF9 protein expression in MDA-MB-231 breast cancer cells. (D) β-catenin, c-Myc and Cyclin D1 protein expression were all lower in MDA-MB-231 cells transfected with miR-187-3p mimic compared with those transfected with the miR-NC mimic. (E) Quantified data of (D) demonstrating that miR-187-3p mimic transfection significantly reduced β-catenin, c-Myc and Cyclin D1 expression in MDA-MB-231 breast cancer cells. (F) Gemcitabine treatment did not alter FGF9 protein expression in MDA-MB-231 cells. (G) Quantitative analysis of FGF9 protein expression displayed in (F) Data are presented as the mean ± standard deviation. *P<0.05, **P<0.01, ***P<0.001. FGF9. fibroblast growth factor 9; miR, microRNA; NC, negative control.
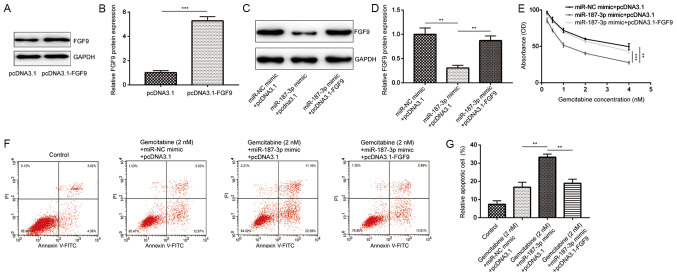
Overexpression of FGF9 reverses gemcitabine- and miR-187-3p-mediated cell viability inhibition and promotion of MDA-MB-231 cell apoptosis
To explore the functional association of miR-187-3p and FGF9 on gemcitabine sensitivity in breast cancer cells, a pcDNA3.1-FGF9 recombinant plasmid was constructed, which was co-transfected with the miR-187-3p mimic into MDA-MB-231 cells. Compared with the pcDNA3.1, transfection with the pcDNA3.1-FGF9 plasmid increased FGF9 expression in MDA-MB-231 cells (Fig. 6A and B). Compared with the miR-NC mimic and pcDNA3.1, overexpression of miR-187-3p significantly reduced the FGF9 protein expression, which was significantly reversed by co-transfection with the FGF9 plasmid (Fig. 6C and D). Cell viability and apoptosis assays were performed to measure the effect of miR-187-3p and FGF9 overexpression on gemcitabine sensitivity in breast cancer cells. In the presence of gemcitabine, co-transfection with the FGF9 plasmid significantly reversed miR-187-3p overexpression-mediated reduction in cell viability (Fig. 6E) and promotion of apoptosis (Fig. 6F and G). Taken together, the results indicated that overexpression of miR-187-3p increased gemcitabine sensitivity in MDA-MB-231 cells by suppressing FGF9 expression.
Figure 6.
Overexpression of FGF9 reversed the inhibitory effects of gemcitabine and miR-187-3p on MDA-MB-231 cells. (A) Representative western blotting images revealed that compared with pcDNA3.1, transfection with pcDNA3.1-FGF9 increased FGF9 expression in MDA-MB-231 cells. (B) Quantified data from (A) demonstrating that compared with pcDNA3.1, pcDNA3.1-FGF9 transfection significantly increased FGF9 protein expression. (C) Compared with cells co-transfected with miR-NC mimic and pcDNA3.1 plasmid, cells co-transfected with miR-187-3p mimic and pcDNA3.1 plasmid exhibited reduced FGF9 protein expression, which was reversed by co-transfection with miR-187-3p mimic and pcDNA3.1-FGF9 plasmid. (D) Quantified data from (C) demonstrating that the differences observed in (C) were statistically significant. (E) Compared with the miR-NC mimic and pcDNA3.1 group, co-transfection with the miR-187-3p mimic and pcDNA3.1 plasmid significantly reduced MDA-MB-231 cell viability at each concentration of gemcitabine (0.25, 0.5, 1, 2 and 4 nM), which was reversed by co-transfection with the recombinant FGF9 plasmid. (F) Compared with the control group, co-transfection with the miR-187-3p mimic and pcDNA3.1 plasmid significantly enhanced gemcitabine (2 nM)-induced MDA-MB-231 cell apoptosis, which was reversed by co-transfection with the recombinant FGF9 plasmid. (G) Quantified data from (F) demonstrating that the differences observed in (F) were statistically significant. Data are presented as the mean ± standard deviation. **P<0.01, ***P<0.001 as indicated. FGF9. Fibroblast growth factor 9; miR, microRNA; NC, negative control.
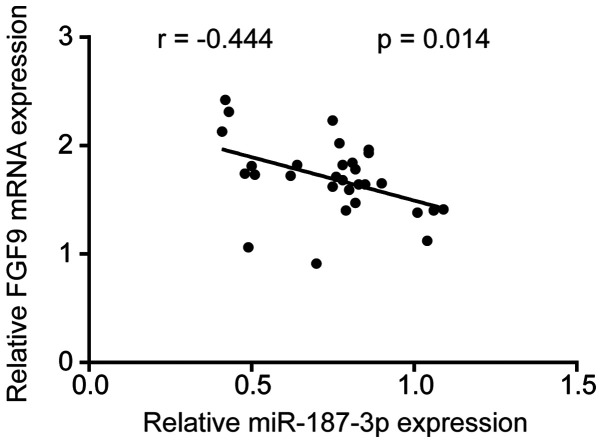
miR-187-3p expression negatively correlates with that of FGF9 mRNA in breast tumor tissues
Following observations that miR-187-3p negatively regulates FGF9 in breast cancer cells, the potential correlation between miR-187-3p and FGF9 mRNA expression was determined in 30 breast cancer tumor and their matched adjacent non-tumor tissue samples. The results revealed that FGF9 mRNA expression was negatively correlated with miR-187-3p expression (Fig. 7), indicating that miR-187-3p may regulate FGF9 during breast cancer pathogenesis.
Figure 7.
miR-187-3p expression negatively correlated with FGF9 mRNA expression in breast cancer tissues. FGF9 expression was measured via reverse transcription-quantitative PCR. Pearson correlation analysis determined that FGF9 mRNA expression negatively correlated with miR-187-3p expression in tumor tissues obtained from 30 patients with breast cancer. r=-0.444 and P=0.014. FGF9, fibroblast growth factor 9; miR, microRNA.
Discussion
Breast cancer is the most commonly diagnosed type of cancer in women, with its incidence ranking the highest among all malignancies (27). Accumulating evidence has demonstrated that targeted therapies offer superior therapeutic efficacy compared with conventional approaches (28). Recently, miRNAs have been widely demonstrated to be associated with a variety of diseases including breast cancer (29). Gasparini et al (30) reported that the downregulation of miR-27a and miR-30e in patients with breast cancer correlated with poor patient outcomes, whereas the overexpression of miR-155 and miR-493 correlated with good patient outcomes. In addition, Kumaraswamy et al (31) revealed that miR-146a expression correlated positively with that of breast cancer gene 1, which in turn negatively regulated epidermal growth factor receptor expression in breast cancer. The miR-200 family has previously been demonstrated to serve important roles in the cell migration, metastasis, proliferation and invasion of breast cancer (32,33). miRNA expression profiles may influence responses to cancer chemotherapy (34). miRNAs regulate cancer drug resistance via a number of methods, including the expression of specific drug targets, survival and/or apoptosis signaling, cell proliferation and cell cycle progression (35). Furthermore, miR-200c was revealed to be reduced in the doxorubicin-resistant MCF7 breast cancer cell line (MCF7/ADR), where the overexpression of miR-200c increased MCF7/ADR sensitivity to epirubicin (36). In addition, the expression of miR-7, miR-326 and miR-345 were revealed to be markedly lower in the MCF7 the cisplatin-resistant cell line in comparison with the MCF7 cell line (37). The function of miR-187-3p in cancer remains controversial, as it has been previously reported to be downregulated in many diseases, including prostate cancer (38), retinal ganglion (39), type 2 diabetes (40), non-small cell lung cancer (13) and breast cancer (14). miR-187-3p has been previously associated with a number of different malignancies (13,14). Using a miRNA microarray, Liang et al (23) determined that miR-187-3p was one of the most significantly downregulated miRNAs in the chemotherapy-resistant MCF7 cells compared with parental MCF7 cells. However, the function of miR-187-3p on breast cancer drug resistance has yet to be clearly elucidated. In the present study, it was revealed that miR-187-3p expression was significantly lower in breast cancer tumor tissue compared with matched non-tumor tissues collected from patients with breast cancer. miR-187-3p overexpression significantly reduced cell viability and promoted cell apoptosis, indicating that miR-187-3p is a tumor suppressor in breast cancer. Although gemcitabine has been used for the treatment of metastatic breast cancer since 2004(41), its therapeutic use in cancer chemotherapy has been impeded at least in part by drug resistance (42). Chaudhari et al (43) performed miRNA microarray analysis, which determined that doxorubicin treatment increased miR-187-3p expression in human cardiomyocytes. In the present study, cell viability and cell apoptosis assays revealed the effect of miR-187-3p on breast cancer gemcitabine sensitivity. Data from these analyses, it was indicated that miR-187-3p overexpression increased gemcitabine sensitivity in breast cancer cells.
Although Fillmore et al (20) have previously explored the role of FGF9 signaling in breast cancer cells, the association between miRNA expression and FGF9 in breast cancer remain unclear. According to the TargetScan database, FGF9 was predicted to be a potential target gene of miR-187-3p, which was subsequently confirmed by luciferase assays and observations that miR-187-3p negatively regulated FGF9 expression.
Target genes of miR-187-3p were involved in the regulation of signaling pathways, including phosphatidylinositol 3-kinase-AKT and hypoxia-inducible factor 1 signaling pathways in epileptic rats (44). Hendrix et al (45) revealed that FGF9 was a downstream target of the Wnt signaling pathway, which may be an indirect transcription target of β-catenin in ovarian endometrioid adenocarcinomas. Abdel-Rahma et al (46) demonstrated that 80% FGF9 mutations were associated with membranous β-catenin expression in colorectal and endometrial carcinomas. In addition, FGF9 has been reported as a target for the Wnt/β-catenin pathway in ovarian endometrioid adenocarcinomas (26). Interestingly, FGF9 interacted with Wnt/β-catenin signaling in a feed-forward loop in lung development, as determined by Yin et al (25). Wnt signaling is associated with drug resistance in cancer (47). In addition, Shi et al (48) elucidated that LGR5 suppressed docetaxel resistance in breast cancer through the inactivation of the Wnt/β-catenin signaling. However, information on the association between miR-187-3p and Wnt/β-catenin in breast cancer pathophysiology remains unclear. In the current study, overexpression of miR-187-3p in MDA-MB-231 cells significantly reduced the protein expression of β-catenin, c-Myc and Cyclin D1. In conclusion, miR-187-3p negatively regulated FGF9 expression, which inactivated Wnt/β-catenin signaling and increased breast cancer sensitivity to gemcitabine-mediated toxicity.
Taken together, the results of the present study suggested that miR-187-3p increased gemcitabine sensitivity by targeting FGF9 expression in breast cancer cells, indicating that miR-187-3p may be a therapeutic target for patients with breast cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
YW, YQ, WL, HY and XY carried out experiments and analyzed the data. LT and JL collected the clinical samples and analyzed the data. LL designed the study, analyzed the data and prepared the manuscript. All author read and approved the final version of the manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethic Committee of Chifeng Municipal Hospital. Written consent was provided by all participants prior to the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Feng Y, Spezia M, Huang S, Yuan C, Zeng Z, Zhang L, Ji X, Liu W, Huang B, Luo W, et al. Breast cancer development and progression: Risk factors, cancer stem cells, signaling pathways, genomics, and molecular pathogenesis. Genes Dis. 2018;5:77–106. doi: 10.1016/j.gendis.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strollo SE, Fallon EA, Gapstur SM, Smith TG. Cancer-related problems, sleep quality, and sleep disturbance among long-term cancer survivors at 9-years post diagnosis. Sleep Med. 2020;65:177–185. doi: 10.1016/j.sleep.2019.10.008. [DOI] [PubMed] [Google Scholar]
- 4.Tripathy D. Overview: Gemcitabine as single agent therapy for advanced breast cancer. Clin Breast Cancer. 2002;3 (Suppl 1):8–11. doi: 10.3816/cbc.2002.s.002. [DOI] [PubMed] [Google Scholar]
- 5.Heinemann V. Role of gemcitabine in the treatment of advanced and metastatic breast cancer. Oncology. 2003;64:191–206. doi: 10.1159/000069315. [DOI] [PubMed] [Google Scholar]
- 6.Hernandez-Aya LF, Ma CX. Chemotherapy principles of managing stage IV breast cancer in the United States. Chin Clin Oncol. 2016;5:1–15. doi: 10.21037/cco.2016.04.01. [DOI] [PubMed] [Google Scholar]
- 7.Blackstein M, Vogel CL, Ambinder R, Cowan J, Iglesias J, Melemed A. Gemcitabin as first line therapy in patient with metastatic breast cancer: A phase II trail. Oncology. 2002;62:2–8. doi: 10.1159/000048240. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Lorio MV, Croce CM. MicroRNAs in cancer: Small molecules with a huge impact. J Clin Oncol. 2009;27:5848–5856. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang F, Luo Y, Shao Z, Xu L, Liu X, Niu Y, Shi J, Sun X, Liu Y, Ding Y, Zhao L. MicroRNA-187, a downstream effector of TGF beta pathway, suppresses Smad-mediated epithelial-mesenchymal transition in colorectal cancer. Cancer Lett. 2016;373:203–213. doi: 10.1016/j.canlet.2016.01.037. [DOI] [PubMed] [Google Scholar]
- 11.Zhao J, Lei T, Xu C, Li H, Ma W, Yang Y, Fan S, Liu Y. MicroRNA-187, down-regulated in clear cell renal cell carcinoma and associated with lower survival, inhibits cell growth and migration though targeting B7-H3. Biochem Biophys Res Commun. 2013;438:439–444. doi: 10.1016/j.bbrc.2013.07.095. [DOI] [PubMed] [Google Scholar]
- 12.Casanova-Salas I, Masiá E, Armiñán A, Calatrava A, Mancarella C, Rubio-Briones J, Scotlandi K, Vicent MJ, López-Guerrero JA. MiR-187 targets the androgen-regulated gene ALDH1A3 in prostate cancer. PLoS One. 2015;10(e0125576) doi: 10.1371/journal.pone.0125576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun C, Li S, Yang C, Xi Y, Wang L, Zhang F, Li D. MicroRNA-187-3p mitigates non-small cell lung cancer (NSCLC) development through down-regulation of BCL6. Biochem Biophys Res Commun. 2016;471:82–88. doi: 10.1016/j.bbrc.2016.01.175. [DOI] [PubMed] [Google Scholar]
- 14.Mulrane L, Madden SF, Brennan DJ, Gremel G, McGee SF, McNally S, Martin F, Crown JP, Jirström K, Higgins DG, et al. Mir-187 is an independent prognostic factor in breast cancer and confers increased invasive potential in vitro. Clin Cancer Res. 2012;18:6702–6713. doi: 10.1158/1078-0432.CCR-12-1420. [DOI] [PubMed] [Google Scholar]
- 15.Yan ZX, Wu LL, Xue K, Zhang QL, Guo Y, Romero M, Leboeuf C, Janin A, Chen SJ, Wang L, Zhao WL. MicroRNA187 overexpression is related to tumor progression and determines sensitivity to bortezomib in peripheral T-cell lymphoma. Leukemia. 2014;28:880–887. doi: 10.1038/leu.2013.291. [DOI] [PubMed] [Google Scholar]
- 16.Turner N, Grose R. Fibroblast growth factor signaling: From development to cancer. Nat Rev Cancer. 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 17.Grose R, Dickson C. Fibroblast growth factor signaling in tumorigenesis. Cytokine Growth Factor Rev. 2005;16:179–186. doi: 10.1016/j.cytogfr.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 18.Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, et al. Relation between microRNA expression and progression and prognosis of gastric cancer: A microRNA expression analysis. Lancet Oncol. 2010;11:136–146. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ornitz DM, Itoh N. Fibroblast growth factors. Genome Biol. 2001;2(REVIEWS 3005) doi: 10.1186/gb-2001-2-3-reviews3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fillmore CM, Gupta PB, Rudnick JA, Caballero S, Keller PJ, Lander ES, Kuperwasser C. Estrogen expands breast cancer stem-like cells through paracrine FGF/Tbx3 signaling. Proc Natl Acad Sci USA. 2010;107:21737–21742. doi: 10.1073/pnas.1007863107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin Y, Castro AM, Hoekstra M, Yan TJ, Kanakamedala AC, Dehner LP, Hill DA, Ornitz DM. Fibroblast growth factor 9 regulation by MicroRNAs controls lung development and links DICER1 loss to the pathogenesis of pleuropulmonary blastoma. PLoS Genet. 2015;11(e1005242) doi: 10.1371/journal.pgen.1005242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Liang Z, Wu H, Xia J, Li Y, Zhang Y, Huang K, Wagar N, Yoon Y, Cho HT, Scala S, Shim H. Involvement of miR-326 in chemotherapy resistance of breast cancer through modulating expression of multidrug resistance-associated protein 1. Biochem Pharmacol. 2010;79:817–824. doi: 10.1016/j.bcp.2009.10.017. [DOI] [PubMed] [Google Scholar]
- 24.Modi S, Seidman AD. Single-agent gemcitabine in the treatment of advanced breast cancer. Clin Breast Cancer. 2004;4 (Suppl 3):S101–S106. doi: 10.3816/cbc.2004.s.002. [DOI] [PubMed] [Google Scholar]
- 25.Yin Y, Wang F, Ornitz DM. Mesothelial- and epithelial-derived FGF9 have distinct functions in the regulation of lung development. Development. 2011;138:3169–3177. doi: 10.1242/dev.065110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwartz DR, Wu R, Kardia SL, Levin AM, Huang CC, Shedden KA, Kuick R, Misek DE, Hanash SM, Taylor JM, et al. Novel candidate targets of beta-catenin/T-cell factor signaling identified by gene expression profiling of ovarian endometrioid adenocarcinomas. Cancer Res. 2003;63:2913–2922. [PubMed] [Google Scholar]
- 27.Parvizpour S, Razmara J, Omidi Y. Breast cancer vaccination comes to age: Impacts of bioinformatics. Bioimpacts. 2018;3:223–235. doi: 10.15171/bi.2018.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shabnam B, Padmavathi G, Banik K, Girisa S, Monisha J, Sethi G, Fan L, Wang L, Mao X, Kunnumakkara AB. Sorcin a potential molecular target for cancer therapy. Transl Oncol. 2018;11:1379–1389. doi: 10.1016/j.tranon.2018.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Temian DC, Pop LA, Irimie AI, Berindan-Neagoe I. The epigenetics of triple-negative and basal-like breast cancer: Current knowledge. J Breast Cancer. 2018;21:233–243. doi: 10.4048/jbc.2018.21.e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gasparini P, Cascione L, Fassan M, Lovat F, Guler G, Balci S, Irkkan C, Morrison C, Croce CM, Shapiro CL, Huebner K. MicroRNA expression profiling identifies a four microRNA signature as a novel diagnostic and prognostic biomarker in triple negative breast cancers. Oncotarget. 2014;5:1174–1184. doi: 10.18632/oncotarget.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumaraswamy E, Wendt KL, Augustine LA, Stecklein SR, Sibala EC, Li D, Gunewardena S, Jensen RA. BRCA1 regulation of epidermal growth factor receptor (EGFR) expression in human breast cancer cells involves microRNA-146a and is critical for its tumor suppressor function. Oncogene. 2015;34:4333–4346. doi: 10.1038/onc.2014.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Humphries B, Wang Z, Oom AL, Fisher T, Tan D, Cui Y, Jiang Y, Yang C. MicroRNA-200b targets protein kinase Cɑ and suppresses triple-negative breast cancer metastasis. Carcinogenesis. 2014;35:2254–2263. doi: 10.1093/carcin/bgu133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsouko E, Wang J, Frigo DE, Aydoğdu E, Williams C. MiR-200a inhibits migration of triple-negative breast cancer cells through direct repression of the EPHA2 oncogene. Carcinogenesis. 2015;36:1051–1060. doi: 10.1093/carcin/bgv087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blower PE, Chung JH, Verducci JS, Lin S, Park JK, Dai Z, Liu CG, Schmittgen TD, Reinhold WC, Croce CM, et al. MicroRNAs modulate the chemosensitivity of tumor cells. Mol Cancer Ther. 2008;7:1–9. doi: 10.1158/1535-7163.MCT-07-0573. [DOI] [PubMed] [Google Scholar]
- 35.Gottesman MM, Lav O, Hall MD, Gillet JP. Toward a better understanding of the complexity of cancer drug resistance. Annu Rev Pharmacol Toxicol. 2016;56:85–102. doi: 10.1146/annurev-pharmtox-010715-103111. [DOI] [PubMed] [Google Scholar]
- 36.Chen J, Tian W, Cai H, He H, Deng Y. Down-regulation of microRNA-200c is associated with drug resistance in human breast cancer. Med Oncol. 2012;29:2527–2534. doi: 10.1007/s12032-011-0117-4. [DOI] [PubMed] [Google Scholar]
- 37.Pogribny IP, Filkowski JN, Tryndyak VP, Golubov A, Shpyleva SI, Kovalchuk O. Alterations of microRNAs and their targets are associated with acquired resistance of MCF-7 breast cancer cells to cisplatin. Int J Cancer. 2010;127:1785–1794. doi: 10.1002/ijc.25191. [DOI] [PubMed] [Google Scholar]
- 38.Casanova-Salas I, Rubio-Briones J, Calatrava A, Mancarella C, Masiá E, Casanova J, Fernández-Serra A, Rubio L, Ramírez-Backhaus M, Armiñán A, et al. Identification of miR-187 and miR-182 as biomarkers of early diagnosis and prognosis in patients with prostate cancer treated with radical prostatectomy. J Urol. 2014;192:252–259. doi: 10.1016/j.juro.2014.01.107. [DOI] [PubMed] [Google Scholar]
- 39.Zhang QL, Wang W, Li J, Tian SY, Zhang TZ. Decreased miR-187 induces retinal ganglion cell apoptosis through upregulating SMAD7 in glaucoma. Biomed Pharm Ther. 2015;75:19–25. doi: 10.1016/j.biopha.2015.08.028. [DOI] [PubMed] [Google Scholar]
- 40.Locke JM, da Silva Xavier G, Dawe HR, Rutter GA, Harries LW. Increased expression of miR-187 in human islets from individuals with type 2 diabetes is associated with reduced glucose-stimulated insulin secretion. Diabetologia. 2014;57:122–128. doi: 10.1007/s00125-013-3089-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barton-Burke M. Gemcitabine: A pharmacologic and clinical overview. Cancer Nurs. 1999;22:176–183. doi: 10.1097/00002820-199904000-00011. [DOI] [PubMed] [Google Scholar]
- 42.Dyawanapelly S, Kumar A, Chourasia MK. Lessons learned from gemcitabine: Impact of therapeutic carrier systems and gemcitabine's drug conjugates on cancer therapy. Crit Rev Ther Drug Carrier Syst. 2017;1:63–96. doi: 10.1615/CritRevTherDrugCarrierSyst.2017017912. [DOI] [PubMed] [Google Scholar]
- 43.Chaudhari U, Nemade H, Gaspar JA, Hescheler J, Hengstler JG, Sachinidis A. MicroRNAs as early toxicity signatures of doxorubicin in human-induced pluripotent stem cell-derived cardiomyocytes. Arch Toxicol. 2016;90:3087–3098. doi: 10.1007/s00204-016-1668-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang S, Kou Y, Hu C, Han Y. MicroRNA profiling in the dentate gyrus in epileptic rats: The role of miR-187-3p. Medicine (Baltimore) 2017;96(e6744) doi: 10.1097/MD.0000000000006744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hendrix ND, Wu R, Kuick R, Schwartz DR, Fearon ER, Cho KR. Fibroblast growth factor 9 has oncogenic activity and is a downstream target of wnt signaling in ovarian endometrioid adenocarcinomas. Cancer Res. 2006;66:1354–1362. doi: 10.1158/0008-5472.CAN-05-3694. [DOI] [PubMed] [Google Scholar]
- 46.Abdel-Rahman WM, Kalinina J, Shoman S, Eissa S, Ollikainen M, Elomaa O, Eliseenkova AV, Bützow R, Mohammadi M, Peltomäki P. Somatic FGF9 mutations in colorectal and endometrial carcinomas associated with membranous β-catenin. Hum Mutat. 2008;29:390–397. doi: 10.1002/humu.20653. [DOI] [PubMed] [Google Scholar]
- 47.Galluzzi L, Spranger S, Fuchs E, López-Soto A. WNT signaling in cancer immunosurveillance. Trends Cell Biol. 2018;18:30143–30150. doi: 10.1016/j.tcb.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi S, Chen X, Liu H, Yu K, Bao Y, Chai J, Gao H, Zou L. LGR5 acts as a target of miR-340-5p in the suppression of cell progression and drug resistance in breast cancer via Wnt/β-catenin pathway. Gene. 2018;18:31039–31044. doi: 10.1016/j.gene.2018.10.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.