Abstract
DNA methylation is known to regulate the expression of numerous genes but its role in the pathogenesis of thoracic aortic dissection (TAD) has remained largely elusive. In the present study, the DNA methylome of patients with TAD was analyzed using a methylation microarray and bisulfite pyrosequencing was used to determine whether the hypermethylation of matrix metalloproteinase 2 (MMP2) specifically is associated with TAD. Chip-based whole-DNA methylome analysis was performed on 4 male patients with TAD and 4 male healthy controls using an Illumina HumanMethylation EPIC 850K BeadChip. The resulting data were analyzed by clustering and principal component analysis, and Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed on the differentially methylated genes to interrogate their biological functions. Compared to the healthy controls, 3,362 loci were differentially methylated in the patients with TAD with a statistical significance of P<0.05, while 1,223 loci had a significance of P<0.01. Among these loci, 2,019 were hypermethylated and 1,343 were hypomethylated. From GO analysis within the biological process category, the MMP2, MMP14 and WNT2B genes were identified. enrichment was observed for loci involved in cellular component organization, enzyme-linked receptor protein signaling pathways (potentially having a key role in the development of cardiopulmonary function disorders) and vascular reconstruction. Bisulfite pyrosequencing of plasma samples indicated significantly increased methylation (P<0.01) of the CpG site at position 2 in the promoter of MMP2 in the TAD group relative to the healthy controls, and the mean methylation level of four CpG sites on the MMP2 gene in the TAD group was slightly higher than that in the control group, but not significantly. Hypermethylation of the MMP2 promoter may be a promising novel diagnostic and prognostic biomarker for TAD. Future studies on the epigenetics of biomarkers linked to vascular reconstruction and immune function may provide further insight into the pathogenesis and progression of TAD.
Keywords: thoracic aortic dissection, DNA methylation, genome-wide, microarray, matrix metalloproteinase 2, hypermethylation
Introduction
Aortic dissection (AD) is a deadly condition in which the mouth of the aortic wall intima is torn, allowing pulsatile blood flow through the tear, thereby causing the layers of the aorta to separate (1). A dissection may significantly reduce vascular wall strength, finally resulting in vascular expansion or rupture (2). This is associated with the high mortality rate identified in recent years for this condition: The global fatality rate from AD and aortic aneurysms combined has increased from 2.49 per 100,000 in 1999 to 2.78 per 100,000 inhabitants in 2010, representing an increasing global health burden (3). Investigators have recently begun to develop methods for the diagnosis and treatment of thoracic AD (TAD) at the gene and protein levels. One of the characteristic features of aortic aneurysm and dissection is the disruption and degradation of structural extracellular matrix (ECM) proteins by the matrix metalloproteinases (MMPs), particularly MMP2 and MMP9. Therefore, ECM-degrading proteins within the MMP family may serve as promising biomarkers for the diagnosis and treatment of AD (4-6). DNA methylation has a key role in embryonic development and in the pathogenesis of numerous diseases by regulating gene transcription, imprinting and immune defense against the invasion of cells by exogenous genetic material. It was reported that DNA methylation involves heritable but reversible methylation or demethylation of nucleotides and has an important role in gene and RNA transcriptional regulation in cardiovascular diseases (7-9).
In the past 5 years of methylome research, enhancer regions in particular have been identified as critical sites for differential methylation in projects including ENCODE and FANTOM5 (10,11). Epigenome-wide association studies using DNA methylation arrays have become an important tool in clinical research for discovering differentially methylated regions (DMRs) associated with phenotypes of interest. For instance, Illumina's Infinium Beadchips provide unparalleled genome-wide coverage, allowing the detection of DMRs of the DNA that are likely to influence transcriptional gene activity and, thus, the regulation of metabolic processes (12,13). Indeed, by far the most widely used technology in this field is the Illumina Methylation EPIC 850K BeadChip, which may be used to compare the methylation levels of 850,000 cytosines in the context of CpG dinucleotides between patients and a control group (14).
Changes in DNA methylation may affect the expression of numerous genes (15,16). In particular, methylation frequently occurs at the 5'position of the cytosine ring in CpG dinucleotides, where it has a role in regulating gene expression (17). In a previous study, differences in the methylation level of promoters of the MMP gene family between healthy controls and patients with acute AD were identified. Therefore, it was hypothesized that DNA methylation may have an important role in the development and progression of TAD. To date, however, only few studies have been performed on the epigenetic mechanisms involved in TAD, and no study has focused on DNA methylation specifically. Thus, the aim of the present study was to identify and characterize the DMRs and associated genes in TAD in comparison with healthy controls. The DNA methylation level of MMP promoters in the peripheral blood was also quantified to determine whether altered MMP promoter methylation specifically may be associated with TAD. The present study may help to identify novel DNA methylation markers and enhance the current understanding and treatment of TAD.
Materials and methods
Patients
Subjects treated at the inpatient clinic of Ningbo Medical Center, Li Huili Hospital (Ningbo, China) between May 2016 and December 2016 were selected. The inclusion criteria for the clinical diagnosis of AD were as follows: i) AD originating in the descending thoracic aorta and not involving the ascending aorta (known as type B); ii) proximal landing zone of at least 2 cm; iii) distance between entry tear and brachiocephalic trunk of at least 0.5 cm; iv) no signs of cardiac tamponade or severe aortic regurgitation; and v) no sign of aortic branch ischemia. Patients with cardiac revascularization from the ascending aorta were excluded. Clinical information regarding the 25 patients with TAD and 27 healthy controls, from which samples were taken for methylation analysis, is provided in Table I. All participants were recruited from Li Huili Hospital (Ningbo, China).
Table I.
Clinical characteristics of 25 cases with thoracic aortic dissection and 27 healthy controls.
| Illumina Human Methylation 850K array (n=8) | MMP2 gene methylation by pyrosequencing (n=44) | |||
|---|---|---|---|---|
| Variable | Controls (n=4) | Cases (n=4) | Controls (n=23) | Cases (n=21) |
| Age (years) | 60.25±5.62 (59-64) | 61.00±2.16 (53-66) | 59.83±5.96 (47-61) | 63.8±14.72 (37-90) |
| Sex | ||||
| Male | 4 | 4 | 16 | 19 |
| Female | 0 | 0 | 7 | 2 |
| Cigarette smoking | 2 | 0 | 6 | 15 |
| Hypertension | 0 | 4 | 2 | 21 |
Values are expressed as n (%) or mean (range). MMP, matrix metalloproteinase.
Sample collection
A total of 52 blood samples were collected in EDTA tubes from patients with TAD (25 samples) and healthy controls (27 samples). The samples were centrifuged for 15 min at 936 x g to obtain the plasma, which was then stored at -80˚C. The 52 samples were divided into four groups: i) 4 samples from the patients with TAD for array analysis, ii) 4 samples from the healthy controls for array analysis, iii) 21 samples from the patients with TAD for bisulfite pyrosequencing and iv) 23 samples from the healthy controls for bisulfite pyrosequencing. All of the blood samples were collected by the same investigator (Xiuying Zhu).
Infinium HumanMethylationEPIC 850 BeadChip assay
The Infinium MethylationEPIC 850K BeadChip assay (Illumina) was performed in accordance with Illumina's standard protocol. Bisulfite-converted DNA was amplified followed by enzymatic end-point fragmentation, precipitation and resuspension. Sample labeling, hybridization to chips and image scanning were performed and all 8 samples were processed on the same chipset to avoid a batch effect. Principal component analysis was performed between the TAD cases and the controls through PCA analysis. The distribution of samples was investigated, the rationality of experimental design was verified and the homogeneity of biological duplicate samples was demonstrated by two-dimensional graph. The closer the two-dimensional spatial distribution of the same group of samples was, the more representative the selection of these samples was, as well as the better the biological duplicate.
Bisulfite treatment
Genomic DNA was extracted from plasma using a QIAamp Mini kit (Qiagen). Finally, the QIAamp Mini spin column was placed in a new 1.5-ml microcentrifuge tube, 200 µl Buffer AE or distilled water was added and the column was incubated at room temperature for 1 min. Subsequently, the column was centrifuged at 6,654 x g for 1 min to elute the DNA. Bisulfite treatment was performed with the EpiTect DNA Bisulfite kit (Qiagen), in accordance with the manufacturer's protocol, using 500 ng genomic DNA isolated from each sample.
Pyrosequencing to determine MMP2 methylation levels
PCR amplification of bisulfite-treated DNA was performed in a total volume of 38 µl of premix, using 50 pmol of each primer and 100 ng of bisulfite-treated DNA. A total of 40 cycles of PCR were performed using the PCR conditions described in a previous study by our group (18). PCR products were visualized on 1.5% agarose gels. Pyrosequencing was performed in accordance with the manufacturer's protocol on a PyroMark Q96 ID System (Qiagen). A total of 4 CpG sites in the promoter of the MMP2 gene were analyzed with the Pyro Q96-CpG software 2.5.8 (Qiagen). The mean percentage of methylation of these 11 CpG sites analyzed was calculated. The primers were as follows: MMP2 sequencing primer, 5'-ACTACCAACTCTTTATCC-3'; forward primer, 5'-TTTGTTTTTTGGGTTGTTTGTTGA-3' and reverse primer, 5'-CTCACCACTACCAACTCTTTATC-3'.
Quality control of methylation data and data analysis
DNA quality was controlled using BiQ Analyzer software. Pathway and gene ontology (GO) analyses were performed. Statistical analyses were performed using R software (version 2.1.1). The quality of sequencing data was controlled using GraphPad Prism 8 software (GraphPad Software, Inc.). Values are expressed as the mean ± standard deviation and P<0.05 was considered to indicate statistical significance. Student's t-test was used to compare differences in methylation levels between the patients with TAD and the controls. Analysis of variance was used for comparing positions 1-4 in the TAD group.
Results
Participant characteristics
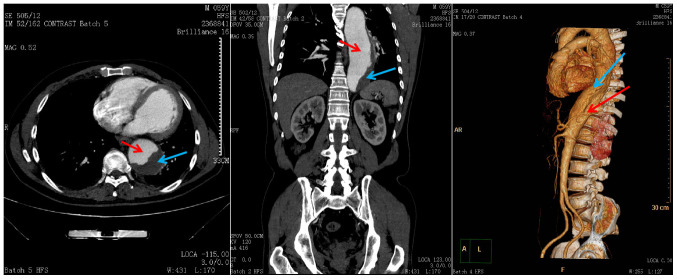
The present study aimed to compare the DMRs in cases with TAD vs. healthy controls. The clinical and pathological characteristics of the subjects are provided in Table I, with 27 controls (20 of which were male) and 25 cases (23 of which were male). A total of 4 male patients with TAD (mean age, 61±2.16 years) and 4 male healthy donors (mean age, 60.25±5.62 years) were recruited. A total of 4 patients in each group were used for the Illumina Human Methylation 850K array. Pre-operative aortography of one of the patients with TAD (Fig. 1) revealed that the true lumen was narrow in the middle of the descending aorta. The remaining 21 patients with TAD (mean age, 63.8±14.72) and 23 healthy controls (mean age, 59.83±5.96) were used to assess MMP2 gene methylation by pyrosequencing.
Figure 1.
Pre-operative aortography of a representative case (male, age 59) with thoracic aortic dissection indicated that the true lumen was narrow in the middle of the descending aorta. Red arrows point to the area of the true lumen, while blue arrows point to the area of the false lumen. Left, middle and right panels indicate the axial section, the coronal plane and 3D restruction, respectively.
Quality control of the methylation array data
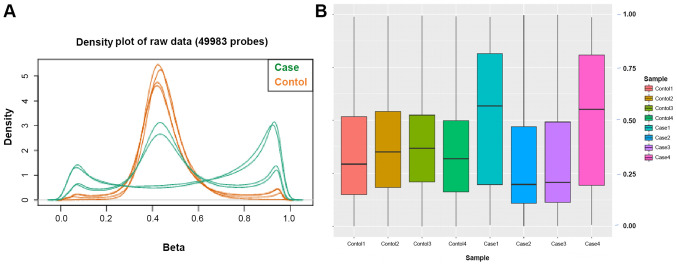
Genome-wide DNA methylation profiles of the 8 participants were generated using the Illumina Human Methylation EPIC 850 BeadChip. Methylation at each locus was measured using β-values, which were generated using the Illumina GenomeStudio software 1.8 (Illumina) based on the intensity of the methylated and unmethylated probes. The workflow of the procedure used to identify the DMRs by array analysis is presented in Fig. S1. The density distribution of the β-values indicated a typical bimodal distribution (Fig. 2A), in which the first peak corresponded to low or unmethylated probes with a β-value close to 0, while the second peak corresponded to highly or fully methylated probes with a β-value close to 1. Furthermore, boxplots of the β-value distributions of the 8 participants were presented (Fig. 2B). Using the average β-values of the two data groups, the overall distribution and concentration trend of the two groups of data in AD group and normal group were evaluated in a homogeneous distribution.
Figure 2.
Quality control of the methylation array data. (A) β-value distributions of the data from the case and control groups (patients with thoracic aortic dissection; orange: Healthy controls). (B) Boxplots of the β-values from the eight samples.
Comparison of differential methylation between patients with TAD and healthy controls
Using the BeadChip, 3,362 DMRs were identified with a significance of P<0.05, while 1,223 were identified with a significance of P<0.01. Among these, 2,019 CpG sites were hypermethylated and 1,343 were hypomethylated in the TAD group relative to the healthy controls. A volcano plot was used to graphically represent the distribution of significant CpG sites from the site-level test sorted by the mean β-differences and P-values (Fig. 3A). Fig. 3B presents the differentially methylated CpG sites between the TAD cases and the controls. Fig. 3C provides the results of the principal component analysis of the two sets of samples.
Figure 3.
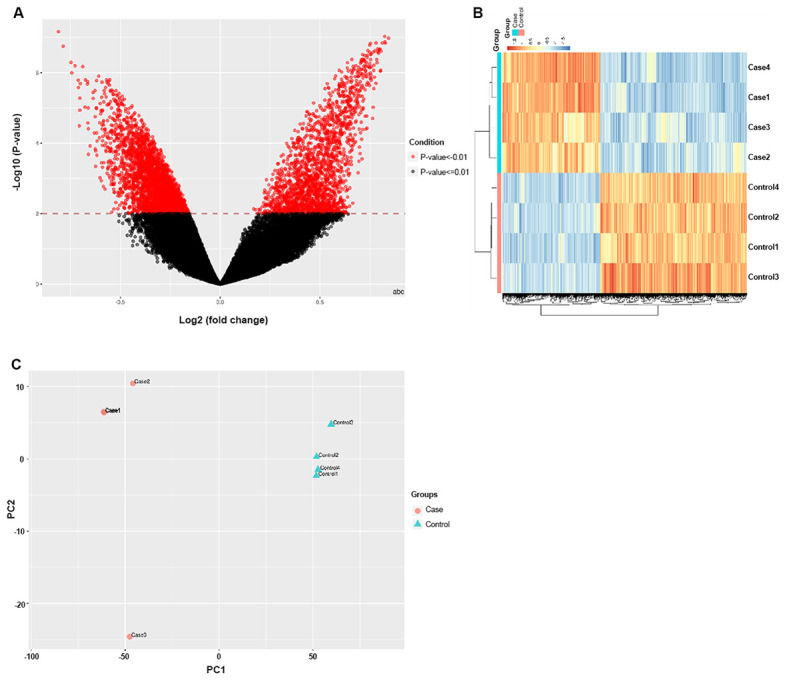
Volcano, heat and PCA maps of the differences between the healthy control samples and the thoracic aortic dissection case samples. (A) Volcano plot of the methylome data indicating the differentially methylated regions between the cases and the controls. (B) Heat map of the differentially methylated CpG sites between the cases and the controls. The DNA methylation level is color-coded. Blue, CpGs with the lowest methylation levels; red, CpGs with the highest methylation levels. (C) Results of the PCA analysis of the two sets of samples. PCA, principal component analysis.
GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses
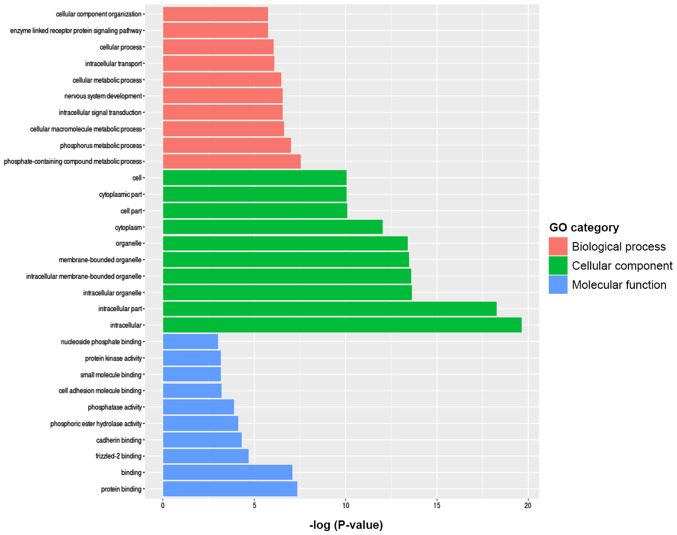
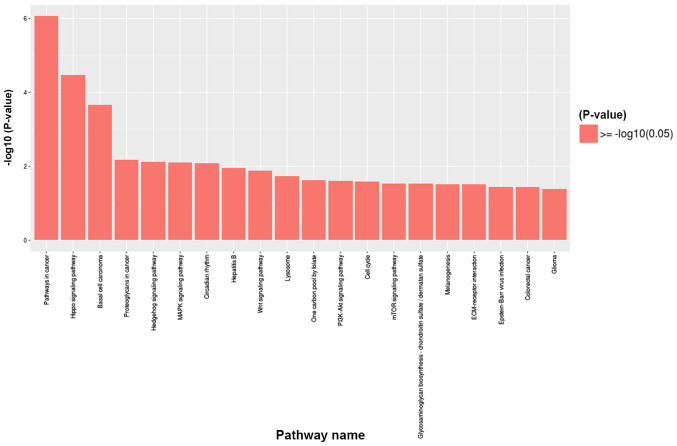
To further investigate the signaling pathways associated with the differentially methylated genes between the TAD and control groups, KEGG signaling pathway analysis was used. According to the criteria of P<0.05 and false discovery rate <0.05, the 20 most significant major signaling pathways were identified. GO functional enrichment was also analyzed according to the differentially methylated sites and the results were annotated and classified (Fig. 4). Kyoto Encyclopedia of Genes and Genomes pathway analyses of the differentially methylated genes are presented in Fig. 5. The key pathways and genes associated with TAD were identified and also listed in Table II. These results indicate that changes in the immune system may occur during the pathogenesis of TAD. The results of the KEGG analysis corresponded well with the results of the GO analysis.
Figure 4.
GO categories including biological process, cellular component, and molecular function associated with thoracic aortic dissection as identified by bioinformatics analysis. GO, Gene Ontology.
Figure 5.
Kyoto Encyclopedia of Genes and Genomes pathway analyses of the differentially methylated genes associated with thoracic aortic dissection. ECM, extracellular matrix.
Table II.
Pathway analysis of the differentially methylated regions and their associated genes in the thoracic aortic dissection vs. control group based on Kyoto Encyclopedia of Genes and Genomes pathway analysis.
| Pathway_id | Pathway_name | Pathway_class | Genes_in_list |
|---|---|---|---|
| hsa05200 | Pathways in cancer | Cancers: Overview | MMP2, WNT1, WNT2B, IGF1R, PDGFB, BCL2, PGF |
| hsa04151 | PI3K-Akt signaling pathway | Signal transduction | FGFR2, IFNAR1, PPP2R2C, IGF1R, MDM2, TNXB, RPTOR |
| hsa04010 | MAPK signaling pathway | Signal transduction | FGFR2, DUSP10, ECSIT, RASGRF1, GRB2, PDGFB, MAPT, NFATC2 |
| hsa05205 | Proteoglycans in cancer | Cancers: Overview | MMP2, WNT1, WNT2B, IGF1R, MDM2, ITPR3, GRB2, FGF12 |
| hsa04390 | Hippo signaling pathway | Signal transduction | WNT1, WNT2B, GLI2, WNT3A, APC, TCF7L1, PPP2R2B, WNT3, MPP5 |
| hsa05169 | Epstein-Barr virus infection | Infectious diseases: Viral | PSMD2, MDM2, PSMD7, MAP2K6, BCL2, HDAC4, FCER2, CCNA1 |
| hsa05161 | Hepatitis B | Infectious diseases: Viral | IFNAR1, TICAM1, NFATC3, GRB2, NFATC2, BCL2,TLR3 |
| hsa04310 | Wnt signaling pathway | Signal transduction | WNT1, WNT2B, CTBP2, NFATC3, FBXW11, NFATC2, SFRP2, RAC1 |
| hsa04142 | Lysosome | Transport and catabolism | MFSD8, AP3D1, LAMP1, GBA, NAGPA, CTSA, ARSG, PPT1, CTNS |
| hsa04110 | Cell cycle | Cell growth and death | MDM2, TFDP2, MAD1L1, MCM5, CCNA1, E2F2, TGFB1, TFDP1, MCM7 |
| hsa04916 | Melanogenesis | Endocrine system | WNT1, WNT2B, ADCY7, ADCY9, PRKCA, PRKCG, WNT5B, WNT5A |
| hsa05217 | Basal cell carcinoma | Cancers: Specific types | WNT1, WNT2B, GLI2, SUFU, APC2, WNT5B, WNT5A, FZD3, WNT3A |
| hsa04512 | ECM-receptor interaction | Signaling molecules and interaction | TNXB, COL4A1, ITGB5, LAMA3, COL4A2, ITGB1, CD44, COL1A |
| hsa04340 | Hedgehog signaling pathway | Signal transduction | WNT1, WNT2B, GLI2, SUFU, FBXW11, WNT5B, WNT5A, WNT3A |
| hsa04150 | mTOR signaling pathway | Signal transduction | RPTOR, PRKCA, PRKCG, RPS6KA2, IKBKB, AKT1S1, MTOR, TSC2 |
| hsa05210 | Colorectal cancer | Cancers: Specific types | MLH1, BCL2, RAC1, APC2, TGFB1, BIRC5, APC, TCF7L1, AKT1 |
| hsa05214 | Glioma | Cancers: Specific types | IGF1R, MDM2, GRB2, PDGFB, PRKCA, PRKCG, E2F2, MTOR, AKT1 |
| hsa04710 | Circadian rhythm | Environmental adaptation | PRKAG2, FBXW11, NR1D1, BHLHE41, RORA, RORB, PRKAA1, SKP1 |
hsa, Homo sapiens; ECM, extracellular matrix; MAPK, mitogen-activated protein kinase.
Comparison of differential methylation between patients with TAD and healthy controls with mapping to the MMP2 gene
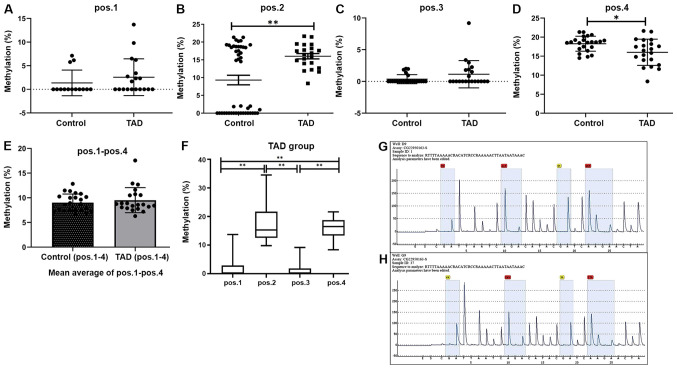
Bisulfite pyrosequencing was then performed for a fragment of the promoter region of the MMP2 gene and the results are provided in Fig. 6. This fragment contained 4 CpG sites that may be measured to evaluate the methylation levels of the MMP2 promoter (Fig. 6G and H). This analysis revealed significantly increased methylation levels at position 2 within this fragment in the TAD cases compared with the controls (P<0.01; Fig. 6B), while decreased methylation levels were present at position 4 (P<0.05; Fig. 6D). However, no significant differences in methylation levels were identified at position 1 (Fig. 6A) and position 3 (Fig. 6C). In summary, the mean methylation level of the 4 CpG sites on the MMP2 gene in the TAD group was higher than that in the control group (9.51±0.51 vs. 9.00±0.65%; Fig. 6E). However, this difference was not significant (P>0.05). Therefore, hypermethylation of the MMP2 gene may be associated with TAD. The CpG sites that were hypomethylated and hypermethylated in the TAD group compared with the healthy controls were subjected to KEGG pathway enrichment analysis (Table II). The genes identified to be associated with TAD included MMP2, Wnt family member 2B (WNT2B) and insulin-like growth factor (IGF), and these genes are actively involved in carcinogenesis, the PI3K/Akt signaling pathway, the mitogen-activated protein kinase (MAPK) signaling pathway, proteoglycans in cancer, ECM-receptor interactions and the circadian rhythm, which may all have key roles in the development of cardiopulmonary dysfunction.
Figure 6.
Methylation levels (%) at 4 different sites within the MMP2 promoter. (A) Methylation level of CpG at position 1. (B) Methylation level of CpG at position 2. (C) The methylation level of CpG at position 3. (D) Methylation level of CpG at position 4. (E) The mean average methylation level of CpG from position 1-4. (F) Mean methylation levels of the 4 CpG sites from positions 1-4 in the TAD group. (G) Methylation levels of the 4 CpG sites on the MMP2 gene body of one control sample. (H) Methylation levels of the 4 CpG sites on the MMP2 gene body of one TAD sample. TAD group: n=21; control group: n=23; **P<0.01, *P<0.05. TAD, thoracic aortic dissection; pos, position; MMP, matrix metalloproteinase.
Discussion
Aortic aneurysm is a severe vascular disease involving apoptosis of the vascular smooth muscle cells in the aortic media, endothelial cell injury, and degradation and pathological remodeling of the ECM (4,19). The etiology and pathogenesis of aortic aneurysms have remained to be comprehensively characterized. Characterization of the genetic landscape of TAD is important to identify potential targets for personalized therapeutic regimens (20). In the present study, a high-throughput array platform (Illumina Human Methylation EPIC 850K BeadChip) was used to explore the genome-scale diversity of DNA methylation in patients with AD in the Han Chinese population. The incidence of TAD in males is significantly higher than that in females, and in China, 85% of patients with TAD are male (21). In the present study, 4 samples from male patients with TAD were selected for analysis with the Methylation EPIC 850K BeadChip.
Compared with existing tools, DiMmeR may complete all data analysis steps within only a few minutes, being guided by an interactive user interface. DiMmeR is the first one-step assay for the sophisticated identification of statistically robust differentially methylated CpGs and regions in modern Illumina chip data. To date, several methylation changes in specific genes have been reported for TAD and DMRs mainly linked to the inflammatory/defense response have been detected in aortic aneurysm (4,22). In the present study, 2,019 hypermethylated and 1,343 hypomethylated CpG sites in the TAD group compared with the healthy control group were identified. Furthermore, subsequent GO pathway analysis suggested that, in the biological process category, the MMP2, WNT2B and IGF genes were particularly affected. These genes may have key roles in the development of cardiopulmonary function disorders. Furthermore, several key signaling pathways were also identified, including the MAPK signaling pathway. Further comprehensive studies including whole-genome and detailed epigenetic analyses are warranted to fully elucidate the role that these genes have in the pathogenesis of TAD.
Altered DNA methylation in genes including MMP2, MMP14, WNT1 and BCL2 may mediate the involvement of vascular smooth muscle cells and inflammatory cells in the pathogenesis and progression of aortic aneurysm. Therefore, the knowledge obtained from epigenetic studies of rheumatology may also be applicable to TAD. For instance, hypermethylation of immune system-associated genes, including MMP2, WNT1, WNT2B, IGF1 receptor (IGF1R), platelet-derived growth factor subunit B, BCL2 and placental growth factor, were identified among the TAD cases. Of these candidate genes, 4 (MMP2, WNT1, IGF1R and BCL2) are potentially involved in the pathogenesis of TAD. Aortic aneurysm, which refers to the focal dilation of the aorta, results from impaired integrity of the aortic ECM (23,24). MMPs, traditionally known as ECM-degrading enzymes, have been demonstrated to have an important role in the myocardial remodeling process; for instance, these enzymes are active in dilated failing hearts (25). One of the common CpG probes indicated a mapped gene in the present study (MMP2), which was selected for further analysis, and genomic comparison of patients with TAD and healthy controls at this locus suggested that differentially methylated MMP2 may contribute toward the pathogenesis of TAD. MMP2 has previously been associated with aneurysm in patients and in animal models (26). It may be speculated that MMP2 has a pathogenic role during matrix remodeling, which appears to be important in regulating cardiac function following stress or injury. Combining the results from the DNA methylation array and bisulfite pyrosequencing, a significant difference was observed between patients with TAD and healthy controls in terms of methylation within the MMP2 promoter, but the in-depth mechanisms require further study.
In conclusion, a total of 2,019 hypermethylated and 1,343 hypomethylated CpG sites were identified in the TAD group compared with the healthy controls. Subsequent GO and KEGG pathway analyses suggested that the MMP2, MMP14, WNT2B and IGF genes in the biological process category are actively involved in TAD. One of the genes identified, MMP2, was confirmed to be hypermethylated, which may be associated with an increased risk of TAD. The epigenetics of vascular reconstruction and immune-based biomarkers may not only provide novel insight into the pathogenesis and progression of TAD but may also assist in identifying novel therapeutic targets.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the Natural Science Foundation of Ningbo (grant no. 2016A610197), the Zhejiang Province Medical and Health Project (grant no. 2017RC026), the Ningbo Public Welfare Project (grant no. 2019C50069), the Advanced Key Scientific and Technological Programs of Ningbo (grant no. 2012C5017) and the Ningbo Health Branding Subject Fund (grant no. PPXK2018-01).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
NL, HL, JG, GS and LS conceived the study, participated in its design and coordination, and helped draft the manuscript. NL, HS and DZ performed the experiments. NL, HL, HZ and GX analyzed the data. JG participated in methylation analysis. XZ participated in data collection and analysis of clinical samples. NL, LS and GS wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The Ethics Committee of Li Huili Hospital approved the present study. Written informed consent was obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ohle R. Diagnosing acute aortic dissection: Both an artery and a science. Acad Emerg Med. 2018;25(1186) doi: 10.1111/acem.13500. [DOI] [PubMed] [Google Scholar]
- 2.Zhao H, Wen D, Duan W, An R, Li J, Zheng M. Identification of CTA-Based predictive findings for temporary and permanent neurological dysfunction after repair in acute type a aortic dissection. Sci Rep. 2018;8(9740) doi: 10.1038/s41598-018-28152-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strzyz P. A sugar rush of DNA methylation. Nat Rev Mol Cell Biol. 2018;19(617) doi: 10.1038/s41580-018-0051-9. [DOI] [PubMed] [Google Scholar]
- 4.Pan S, Lai H, Shen Y, Breeze C, Beck S, Hong T, Wang C, Teschendorff AE. DNA methylome analysis reveals distinct epigenetic patterns of ascending aortic dissection and bicuspid aortic valve. Cardiovasc Res. 2017;113:692–704. doi: 10.1093/cvr/cvx050. [DOI] [PubMed] [Google Scholar]
- 5.Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, Klotzle B, Bibikova M, Fan JB, Gao Y, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49:359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Longo GM, Xiong W, Greiner TC, Zhao Y, Fiotti N, Baxter BT. Matrix metalloproteinases 2 and 9 work in concert to produce aortic aneurysms. J Clin Invest. 2002;110:625–632. doi: 10.1172/JCI15334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moran S, Arribas C, Esteller M. Validation of a DNA methylation microarray for 850,000 CpG sites of the human genome enriched in enhancer sequences. Epigenomics. 2016;8:389–399. doi: 10.2217/epi.15.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nasu T, Satoh M, Ohmomo H, Shiwa Y, Komaki S, Ono K, Shimizu A, Taguchi S, Takahashi Y, Osaki T, et al. Epigenome-wide association study identifies a novel DNA methylation in patients with severe aortic valve stenosis. Circ Genom Precis Med. 2020;13(e002649) doi: 10.1161/CIRCGEN.119.002649. [DOI] [PubMed] [Google Scholar]
- 9.Xu H, Du S, Fang B, Li C, Jia X, Zheng S, Wang S, Li Q, Su W, Wang N, et al. VSMC-specific EP4 deletion exacerbates angiotensin II-induced aortic dissection by increasing vascular inflammation and blood pressure. Proc Natl Acad Sci USA. 2019;116(8457) doi: 10.1073/pnas.1902119116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu L, Zheng D, Wang L, Jiang D, Liu H, Xu L, Liao Q, Zhang L, Liu P, Shi X, et al. GCK gene-body hypomethylation is associated with the risk of coronary heart disease. Biomed Res Int. 2014;2014(151723) doi: 10.1155/2014/151723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pidsley R, Zotenko E, Peters TJ, Lawrence MG, Risbridger GP, Molloy P, Van Djik S, Muhlhausler B, Stirzaker C, Clark SJ. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol. 2016;17(208) doi: 10.1186/s13059-016-1066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirby MK, Ramaker RC, Roberts BS, Lasseigne BN, Gunther DS, Burwell TC, Davis NS, Gulzar ZG, Absher DM, Cooper SJ, et al. Genome-wide DNA methylation measurements in prostate tissues uncovers novel prostate cancer diagnostic biomarkers and transcription factor binding patterns. BMC Cancer. 2017;17(273) doi: 10.1186/s12885-017-3252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silva-Martinez GA, Zaina S, Lund G. Array probe density and pathobiological relevant CpG calling bias in human disease and physiological DNA methylation profiling. Brief Funct Genomics. 2018;17:42–48. doi: 10.1093/bfgp/elx017. [DOI] [PubMed] [Google Scholar]
- 14.Taylor JY, Wright ML, Crusto CA, Sun YV. The intergenerational impact of genetic and psychological factors on blood pressure (InterGEN) study: Design and methods for complex DNA analysis. Biol Res Nurs. 2016;18:521–530. doi: 10.1177/1099800416645399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng D, Chen X, Li N, Sun L, Zhou Q, Shi H, Xu G, Liu J, Xu L, Duan S, Shao G. Differentially methylated regions in patients with rheumatic heart disease and secondary pulmonary arterial hypertension. Exp Ther Med. 2017;14:1367–1372. doi: 10.3892/etm.2017.4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagata H, Kozaki KI, Muramatsu T, Hiramoto H, Tanimoto K, Fujiwara N, Imoto S, Ichikawa D, Otsuji E, Miyano S, et al. Genome-wide screening of DNA methylation associated with lymph node metastasis in esophageal squamous cell carcinoma. Oncotarget. 2017;8:37740–37750. doi: 10.18632/oncotarget.17147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen X, Shen LH, Gui LX, Yang F, Li J, Cao SZ, Zuo ZC, Ma XP, Deng JL, Ren ZH, et al. Genome-wide DNA methylation profile of prepubertal porcine testis. Reprod Fertil Dev. 2018;30:349–358. doi: 10.1071/RD17067. [DOI] [PubMed] [Google Scholar]
- 18.Liu O, Xie W, Qin Y, Jia L, Zhang J, Xin Y, Guan X, Li H, Gong M, Liu Y, et al. MMP-2 gene polymorphisms are associated with type A aortic dissection and aortic diameters in patients. Medicine (Baltimore) 2016;95(e5175) doi: 10.1097/MD.0000000000005175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shalata A, Mahroom M, Milewicz DM, Limin G, Kassum F, Badarna K, Tarabeih N, Assy N, Fell R, Cohen H, et al. Fatal thoracic aortic aneurysm and dissection in a large family with a novel MYLK gene mutation: Delineation of the clinical phenotype. Orphanet J Rare Dis. 2018;13(41) doi: 10.1186/s13023-018-0769-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang W, Yao L, Hoogeveen RC, Alonso A, Couper DJ, Lutsey PL, Steenson CC, Guan W, Hunter DW, Lederle FA, Folsom AR. The association of biomarkers of inflammation and extracellular matrix degradation with the risk of abdominal aortic aneurysm: The ARIC Study. Angiology. 2019;70:130–140. doi: 10.1177/0003319718785278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Chan YC, Cheng SW. Contemporary endovascular treatment of type B aortic dissection in China. Asian Cardiovasc Thorac Ann. 2016;24:739–749. doi: 10.1177/0218492316657464. [DOI] [PubMed] [Google Scholar]
- 22.Bhandari R, Aatre RD, Kanthi Y. Diagnostic approach and management of genetic aortopathies. Vasc Med. 2020;25:63–77. doi: 10.1177/1358863X19886361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao M, Zou S, Bao Y, Jin J, Yang J, Liu Y, Green M, Yang F, Qu L. Matrix metalloproteinases are regulated by MicroRNA 320 in macrophages and are associated with aortic dissection. Exp Cell Res. 2018;370:98–102. doi: 10.1016/j.yexcr.2018.06.011. [DOI] [PubMed] [Google Scholar]
- 24.Li T, Lv Z, Jing JJ, Yang J, Yuan Y. Matrix metalloproteinase family polymorphisms and the risk of aortic aneurysmal diseases: A systematic review and meta-analysis. Clin Genet. 2018;93:15–32. doi: 10.1111/cge.13050. [DOI] [PubMed] [Google Scholar]
- 25.Lin X, Tan JYL, The AL, Lim IY, Liew SJ, MacIsaac JL, Chong YS, Gluckman PD, Kobor MS, Cheong CY, Karnani N. Cell type-specific DNA methylation in neonatal cord tissue and cord blood: A 850K-reference panel and comparison of cell-types. Epigenetics. 2018;13:941–958. doi: 10.1080/15592294.2018.1522929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li YH, Li XM, Lu MS, LV MF, JIN X. The expression of the BRM and MMP2 genes in thoracic aortic aneurysm and aortic dissection. Eur Rev Med Pharmacol Sci. 2017;21:2743–2748. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.