Abstract
Here we generate FRET biosensors for guanine exchange factors (GEFs) by inserting a fluorescent protein pair in a structural “hinge” common to many GEFs. Fluorescent biosensors can map the activation of signaling molecules in space and time, but it has not been possible to quantify how different activation events affect one another or contribute to a specific cell behavior. By imaging the GEF biosensors in the same cells as red-shifted biosensors of Rho GTPases, we can apply partial correlation analysis to parse out the extent to which each GEF contributes to the activation of a specific GTPase in regulating cell movement. Through analysis of spontaneous cell protrusion events we identify when and where the GEF Asef regulates the GTPases Cdc42 and Rac1 to control cell edge dynamics. This approach exemplifies a powerful means to elucidate the real-time connectivity of signal transduction networks.
Introduction
The cytoskeleton is regulated by dynamic, multi-layered signaling networks that interconnect Rho family small GTPases with exquisite spatiotemporal precision1. Understanding the organization of these networks is challenging, as protein activation and interaction occur transiently and with precise subcellular localization. We and others have used fluorescent biosensors in living cells to map the activation patterns of the Rho family small GTPases relative to the changes in cell edge dynamics that they control2–4.
In these studies we could determine the relative timing and location of Rac1, Cdc42, RhoA, and RhoC activation by first using fluorescent biosensors to correlate the activation of each GTPase with cell edge movements, and then using cell edge movement as a common fiduciary to relate the different GTPases to one another3, 5, 6. This allowed us to predict the relative spatio-temporal dynamics of the four signaling activities with exquisite precision, but we could not predict whether any of the signals were directly or indirectly coupled, how specific source signals contributed to the total modulation of each target signal, and the effects of specific couplings on downstream events.
The Rho GTPases are controlled by the localized activity of numerous Rho guanine nucleotide exchange factors (RhoGEFs) with overlapping GTPase specificity7, 8. This produces complex relationships between RhoGEFs and Rho GTPases, where a single GTPase may be activated by multiple GEFs, even in the same location, or one GEF may activate multiple GTPases. Here we show that statistical approaches, coupled with multiplexed biosensor imaging, can be used to ascertain the component of a GTPase signal that results from a particular GEF, or the relative influence a multi-specific GEF has on its different downstream GTPases. Furthermore, to appreciate the functional diversity of a GEF in controlling e.g. cell motility vs other downstream functions, it is possible to statistically determine the extent to which cell edge dynamics results from a particular GEF-GTPase interaction. Below we describe an approach to generate fluorescent biosensors for GEFs, multiplexed imaging of these biosensors together with red-shifted GTPase biosensors, and correlation analysis to dissect out GEF/GTPase relationships. Applying this approach to the GEFs Asef and Vav2, and the GTPases Rac1 and Cdc42, we find that Vav2 activity is weakly coupled to cell protrusion dynamics, but Asef strongly activates Cdc42 signals that relate to cell edge movement. In contrast, the interaction between Asef and Rac1 is weaker and probably indirect.
Results
Design and testing of new RhoGEF biosensors
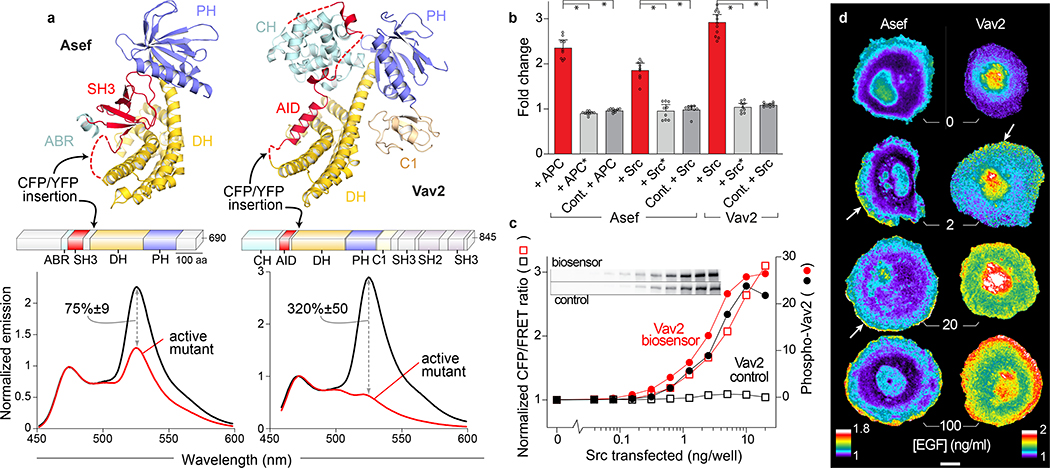
To perform these statistical analyses, it was first necessary to produce biosensors that report the activity of GEFs in living cells (Figure 1). Many Dbl family RhoGEFs are regulated through occlusion of the GTPase binding interface by an autoinhibitory domain (AID)7, providing a route to Dbl family biosensors. We first targeted GEFs with well understood AID interactions and published crystal structures. In the RhoGEF Asef, an SH3 domain acts as the AID; it undergoes a conformational rearrangement when adenomatous polyposis coli (APC) binds to the upstream ABR (APC binding region), leading to Asef activation9 (Figure 1a). We produced an Asef analog that reports these activating conformational changes by inserting two fluorescent proteins into the flexible linker between the SH3 AID and the catalytic DH domain. The activating conformational changes affected FRET between the fluorescent proteins by altering their distance or relative orientation. We used high-throughput microscopy10 to test insertion of Cerulean11 and Venus12 fluorescent proteins at a series of positions between the AID and DH domain, optimizing FRET intensity and the activation-dependent difference in the donor/FRET emission ratio (Supplementary Figure 1a). We then screened a small library combining different fluorophore pairs (Cerulean313, TagCFP14, or mTFP15; combined with YPet16 or a series of YPet circular permutations) (Supplementary Figure 1a). This led to a biosensor with 75%+/−9% difference in donor/FRET ratio for wild type versus constitutively active Asef (Figure 1a). Similar changes were seen upon co-expression of known activating proteins, a fragment of APC9 or constitutively active Src17 (Figure 1b). No ratio change was observed when kinase-dead Src or a non-binding APC mutant were used, or when the FRET pair was moved to a site in the RhoGEF that does not undergo a conformational change (Figure 1b).
Figure 1. Biosensors for Asef and Vav2.
(a) Autoinhibited structures of Asef and Vav (PDB entries 2PZ1 and 3KY9, respectively, top) and domain architecture of full length Asef and Vav2 (ABR - APC binding region, AID - autoinhibitory domain, C1 - C1 domain, CH - calponin homology, DH - Dbl homology, PH - pleckstrin homology, SH2 - Src homology 2, SH3 - Src homology 3, middle) with insertion sites of FRET pair cassettes indicated. Emission spectra (λex = 430 nm) of wild type and active forms (Asef - V252E, Vav2 - Y142E:Y159E:Y172E:Δ860) expressed in HEK293t cells (bottom). Change in donor/FRET ratio upon activation is indicated (n=9 independent transfections, +/− 95% C.I.). (b) Stimulation of Vav2 and Asef biosensors using Src or APC in HEK293t cells. Src* is kinase dead, APC* does not bind to Asef. In biosensor control (Cont.) the FRET pair is moved to the C-terminus. Ratio normalized to empty vector control. Bars represent mean of 9 independent transfections across multiple experiments. Error bars are 95% C.I. *p<0.0001 using two-tailed Welch’s t-test. (c) Effect of increasing Src levels on donor/FRET emission ratio (open squares) and phospho-Tyr172 levels (solid circles) of Vav2 biosensor (red) and control (black). Normalized to empty vector transfection control. Inset – phospho-Vav blots (Full blots see Supplementary Figure 2e). Representative of 3 independent experiments. (d) A431 cells expressing Asef (left) and Vav2 (right) biosensors after stimulation with increasing amounts of EGF. Arrows point to high ratio values at the edge of cell protrusions. Ratios in pseudocolor normalized so lowest value = 1. Scale bar 10μm. Images representative of 3 independent experiments.
We hypothesized that this approach could be applied to other RhoGEFs that undergo a conformational change upon release of autoinhibition, so we tested Vav2, where the DH domain is blocked by an upstream helical AID18 (Figure 1a). Autoinhibition is relieved when Src and other kinases phosphorylate tyrosines in the autoinhibitory regions19–21. As with Asef, we optimized the site of fluorophore insertion between the AID and DH domains, and screened fluorophore combinations (Supplementary Figure 1a). In addition, we tested fluorophore-fluorophore connectors of varying length and rigidity to impose constraints on the conformation of the inserted segment (Supplementary Figure 1b). This led to three biosensors, whose fluorescence ratio changed 330±70% (TagCFP donor), 320±50% (mCerulean3 donor), and 130±20% (mTFP donor) upon activation (Figure 1a, Supplementary Figure 1c). The detectability of GEF activity was a function of both the extent of fluorescence change and the brightness of the fluorophores. In the imaging studies below, we used the brightest donor (mTFP) even though it produced less change (Supplementary Figure 1c).
To test whether the Vav2 biosensor could report activating conformational changes in living cells, we compared the mCerulean3 biosensor with a non-responsive control biosensor in HEK293 cells, examining response to increasing amounts of constitutively active Src. Dose-dependent phosphorylation of the biosensor and the control biosensor were equivalent, as shown by blotting with a phospho-Vav antibody, but only the real biosensor showed an increase in fluorescence emission ratio (Figure 1c). As with Asef, this ratio change was not seen with inactive Src (Figure 1b). Using the optimized Vav2 biosensor as a template we also produced biosensors for other Vav family members simply through limited screening of the insertion site (Supplementary Figure 1d).
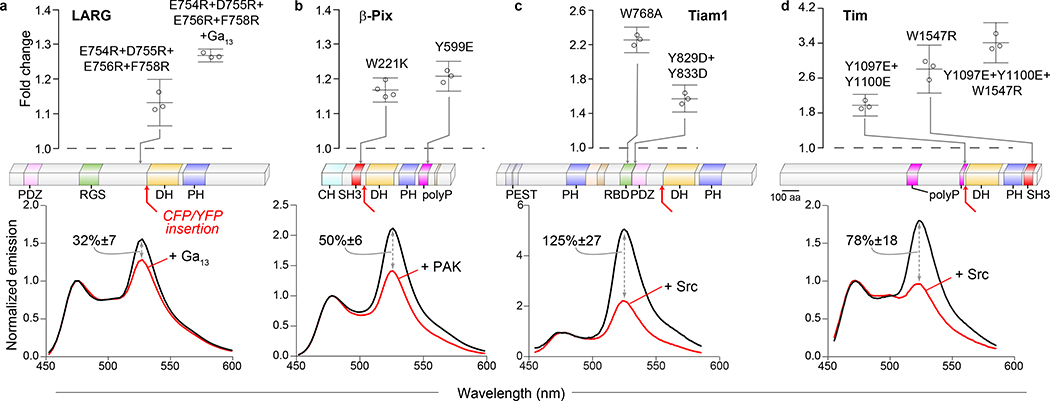
For Asef and Vav2, high-resolution crystal structures were available to identify AID interactions. We next attempted to make biosensors for RhoGEFs proposed to have autoinhibitory regulation, but where structural information was limited. The RhoGEF Tim contains a putative helical region that is thought to directly interact with the DH domain22, equivalent to the Vav2 AID, and autoinhibition is maintained by polyproline and SH3 domains that flank the DH domain23: For the GEFs Tiam1 (T-cell lymphoma invasion and metastasis 1) and LARG (Leukemia-associated RhoGEF), small-angle X-ray scattering suggests that an N-terminal domain folds over the DH domain24, 25, and binding sites for known regulators lie upstream of the DH domain (Ras26/tyrosine kinases27–29 for Tiam1, and Gα1330 for LARG). The RhoGEF β-Pix has a polyproline-SH3 domain pair flanking the DH domain and multiple sites downstream that are proposed to regulate activity31. For each of these RhoGEFs we screened donor/acceptor insertion sites directly upstream of the DH domain, and optimized the response as before (Supplementary Figure 1a). The final set of biosensors had dynamic ranges of 32% for LARG (Figure 2a), 50% for β-Pix (Figure 2b), 125% for Tiam1 (Figure 2c), and 78% for Tim (Figure 2d). For Tim, mutations within the autoinhibitory helix that mimic Src phosphorylation, and mutations within the SH3 domain that prevent polyproline binding, caused changes in the donor/FRET ratio as large as those produced by Src co-expression. The combined mutations had an additive effect (Figure 2d).
Figure 2. Biosensors for RhoGEFs without high resolution structures.
Domain structures (middle) show site of FRET pair insertion in full length RhoGEFs LARG (a), β-Pix (b), Tiam1 (c) and Tim (d). Lower graphs show emission spectra of wild type biosensors +/− indicated activators (λex = 430 nm), change in donor/FRET ratio upon activation is indicated (a, n= 6; b, n= 8; c, n=9; d, n=6; +/− 95% C.I.). Upper graphs show ratio change caused by indicated mutations as compared to wild type (n=3). Bars represent mean of n independent transfections across multiple experiments. Error bars are 95% C.I. (PDZ – PSD95/Dlg1/zo-1; RGS – regulator of G protein signaling; polyP – polyproline; PEST – Sequence rich in amino acids P, E, S and T; RBD – Ras-binding domain; other domains are as listed above).
The new LARG, Tiam1 and β-Pix biosensors provided insight into potential regulatory mechanisms. Specific residues within the RBD (Ras binding domain) of Tiam1 caused large FRET changes, suggesting that this domain has an autoinhibitory role (Figure 2c). Mutations of negatively charged residues in LARG32 and phosphorylation sites in β-Pix that were thought to be involved in RhoGEF regulation induced modest FRET changes, and mutation of the SH3 domain in β-Pix also affected FRET, suggesting that there may be an autoinhibitory role for the polyproline-SH3 domain pair flanking the DH domain, as there is for Tim (Figure 2a, b).
We validated the new biosensors by examining their response to known stimuli in living cells. In response to EGF, Vav2 and Tiam1 showed widespread activation (Figure 1d, Supplementary Figure 2a) while Asef was activated only within 2μm of the cell edge (Figure 1d). Ratio imaging was used to minimize effects of nonuniform biosensor distribution. Comparison of biosensor and activity distribution indicated that activity patterns were not simply the result of variations in cell thickness (Supplementary Figure 3a). More detailed studies of Vav2 showed that its response was dose dependent and reversible, with activation within 30 seconds (Supplementary Figure 2b). LARG and β-Pix showed activation concentrated at the leading edge of randomly migrating cells, with LARG concentrated right at the edge and β-Pix 1–2μm away from the edge (Supplementary Figure 2c).
GEF activity relative to cell edge dynamics
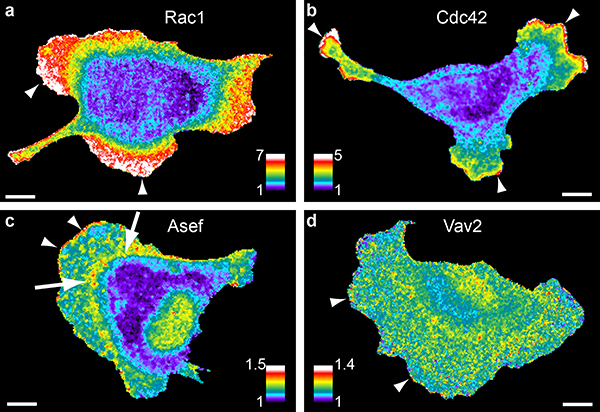
For studies of GEF-GTPase circuitry, we focused on GEFs that were stimulated by EGF, to examine protrusion/retraction cycles during EGF-induced chemokinesis. Biosensors for Asef, Vav2, and improved versions of our previously published biosensors for Rac13 and Cdc4233, 34 (Methods and Supplementary Figure 4a) were stably expressed in MDA-MB-231 cells and studied in EGF-containing medium. Biosensors were kept below expression levels that perturbed motility behaviors (Supplementary Figure 5a, b). As previously described, Rac1 and Cdc42 (Figure 3a–b, Supplementary Movie 1) showed broad gradients of activity dropping from the cell edge to 5 – 8 μm behind the edge, consistent with activation in protruding lamella (Supplementary Figure 6a, b). The new biosensor showed that Asef activation was generally restricted to a narrow band at the leading edge of protrusions (resembling that produced by acute EGF stimulation of A431 cells, Figure 1d), together with a broader region 5–10 microns back from the edge, at the base of lamella, (Figure 3c; Supplementary Figure 6a, b; Supplementary Movie 2). The latter activation occurred in the same cellular region as Rab7-mScarlet, suggesting activated Asef interacts with a sorting compartment (Supplementary Figure 6c). When we moved the FRET pair in the Asef biosensor to a region that should not undergo conformational change, activation at both cellular locations was eliminated (Supplementary Figure 6d). In contrast, Vav2 showed diffuse activation throughout the cell, with only a few ‘hot spots’ adjacent to the edge (Figure 3d, Supplementary Movie 2). Comparison of biosensor localization with activity showed that this activity pattern was not caused by variations in cell thickness (Supplementary Figure 7a).
Figure 3. RhoGEF and GTPase activity in living cells.
Rac1 (a), Cdc42 (b), Asef (c), and Vav2 (d) activation reported by biosensors in MDA-MB-231 cells undergoing random edge motion. Pseudocolor as in Fig. 1. Scale bars 10μm. Images are representative of at least 3 independent experiments.
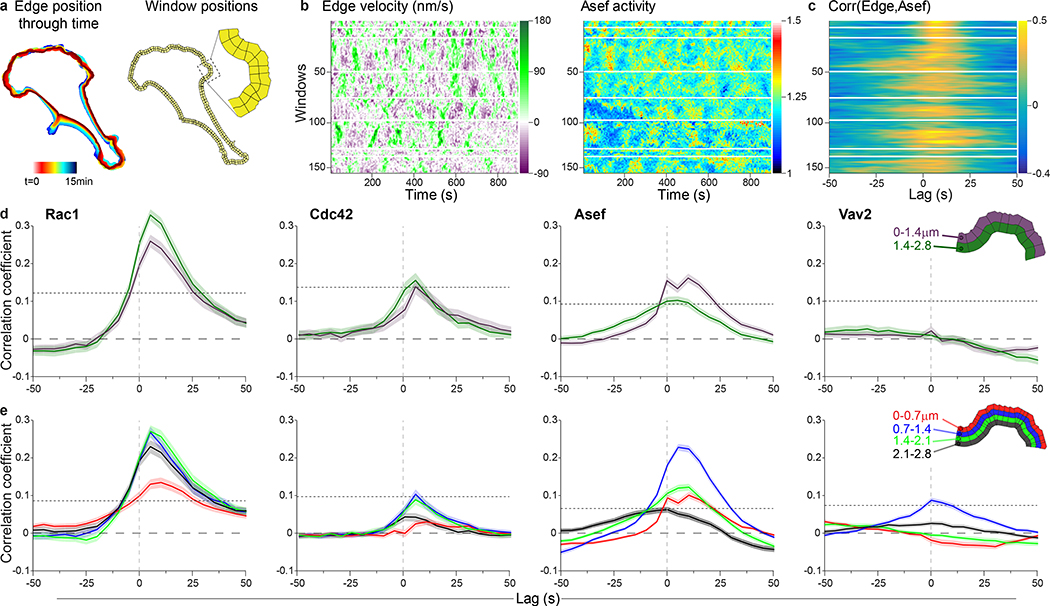
To quantify the relationship between biosensor activity and edge displacement, we used previously described local cross-correlation analysis3, which integrates data from multiple cells to assess the significance of coupling between the two cues despite heterogeneous signaling and motion along the cell periphery. Specifically, we tracked the cell edge over time and then divided the perimeter of the cell into small edge sectors and associated sampling windows (Figure 4a). Sampling windows were placed in layers of increasing but constant distance from the edge. As the cell boundary moved, the windows were rearranged such that they maintained a stable one-to-one relationship with the associated sector. Thus, we could sample for each edge sector the local, instantaneous velocity and the corresponding biosensor activity at any layer (Figure 4b). These cell shape- and motion-invariant data representations then permitted a straightforward analysis of the relationship between edge motion and signaling activity using Pearson’s correlation (Figure 4c).
Figure 4. Correlation of protein activities with edge motion.
(a) Evolution of cell edge positions (color-encoded from red (early) to blue (late) time points, left). Two rows of sampling windows, each 1.4 μm deep placed at the cell edge. (b) Maps of edge velocity (left) and Asef biosensor activities (right) along the edge. Green regions are protruding, purple regions retracting. Red/yellow regions have high activity, blue regions have low activity. Each column is a single time point. White horizontal bars show quiescent regions that are excluded. Scale bars are shown to right of each map. (c) Cross correlation coefficients between edge velocity and Asef activity. Gold shows high correlation and blue negative correlation. Each row is a single position along the edge corresponding to the activity maps. (d) Average cross-correlation functions for each biosensor. Analysis using 1.4 μm window size. (n = cells, m = windows); Rac1 (n=9, m=259); Cdc42 (n=6, m=204); Asef (n=8, m=448); Vav2 (n=6, m=360). (e) Average cross-correlation functions for each biosensor. Analysis using 0.7 μm window size. (n = cells, m = windows); Rac1 (n=9, m=518); Cdc42 (n=6, m=408); Asef (n=8, m=896); Vav2 (n=6, m=720). Inset shows window size and color key. For (d) and (e) dotted lines show the correlation coefficient above which the coupling between two variables is considered significant with 95% confidence. This depends on the number of windows (see Methods). Shading represents 95% C.I. about the mean correlation computed from m windows. The width of this interval depends on the consistency of the correlations across windows and cells (see Methods).
Improvements in instrumentation since our first published correlation analyses of biosensor fluctuations enabled us to enhance spatial resolution by sampling 0.7 × 0.7 micron windows rather than the 1.4 × 1.4 micron windows used previously. Consistent with the earlier work in fibroblasts, Rac1 correlated best with edge motion in the two layers between 0.7–2.1 μm (Figure 4d, e). Also consistent with the earlier studies, correlation was strongest when we incorporated a lag between edge movement and Rac1 activity, but the lag seen in MDA-MB-231 cells was considerably shorter than in fibroblasts (5s instead of ~40s, Figure 4d, e). The shortest time lag to protrusion/retraction cycles was in these two middle layers, suggesting that Rac1 molecules are activated in this zone and then diffuse or are transported to the cell front3. The correlation of Cdc42 with edge motion also peaked at a distance 1 – 2 μm from the cell edge, and with similar time lag, but with overall weaker significance than Rac1 (Figure 4d, e), suggesting that Rac1 is the more dominant GTPase in the regulation of MDA-MB-231 protrusion and retraction. We suggest that the delays of both Rac1 and Cdc42 relate to the upregulation of Arp2/3-mediated dendritic network growth after protrusion onset, which is required for persistent edge protrusion against increasing membrane tension35–37. Arp2/3 is the primary nucleator of the branched actin filament network and is an effector of the Rac1-WAVE and Cdc42-N-Wasp pathways.
Asef had a maximum correlation in the first layer, immediately adjacent to the edge (0–1.4 μm, Figure 4d). Remarkably, and in contrast to the GTPases, we observed a near twofold increase in the correlation peak magnitude when we switched to analyzing correlation with the new, smaller windows, despite the decreased signal/noise that they produced (compare Figures 4d and 4e). This indicated that the portion of the Asef signal related to cell motion was activated in a highly confined zone 0.7–1.4 μm behind the cell edge, which corresponds to the zone of cell adhesion formation (Supplementary Figure 6b). Like Rac1 and Cdc42, the Asef activation was slightly delayed relative to protrusion-retraction cycles. Together, these results indicate that spatially precise RhoGEF activity produces more diffuse effector activation, probably due to diffusion of the effector from the point of activation. Unlike Asef, Vav2 showed no significant correlation with edge motion at the coarser analysis resolution (Figure 4d), and only a weak correlation in finer analyses using the 0.7–1.4 μm layer (Figure 4e). This indicates that Vav2 plays no major role in coordinating signals that specifically regulate cell protrusion and retraction events in unstimulated migration. Given the strong response of Vav2 to acute EGF stimulation (Figure 1d), we conjecture that Vav2 could be important in translating directional cues to the cell protrusion machinery, or may be disconnected from the pathways that promote constitutive protrusion-retraction cycles.
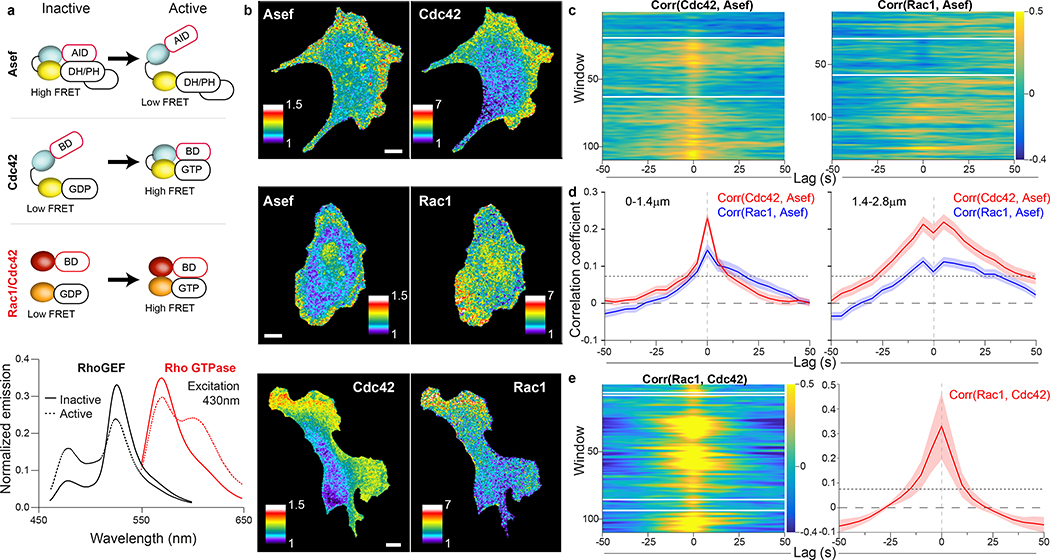
Simultaneous imaging of RhoGEF and Rho GTPase activities
In view of the significant correlation between Asef activity and cell edge movement we decided to focus on the interactions of Asef with the GTPases Rac1 and Cdc42. To complete our statistical analysis, we needed to correlate the activation of Asef and each GTPase, so we produced red shifted GTPase biosensors that could be imaged in the same cell as the Asef biosensor. We modified our published GTPase biosensors3 by changing the fluorescent proteins to LSSmOrange38 and mCherry39 (Figure 5a, b). Because of LSSmOrange’s long Stokes shift, we could capture emission from both biosensors at once using a single excitation, for rapid imaging and reduced photo-toxicity (Figure 5a, b). The new biosensors responded correctly to RhoGEFs and RhoGAPs (Supplementary Figure 4), and their correlation analysis showed GTPase/edge coordination consistent with previous findings (Supplementary Figure 8). The GTPase biosensors were transfected into cells stably expressing Asef biosensor, at levels that minimized perturbation of cell motility (Supplementary Figure 5c, d). Cells with two biosensors showed activation patterns like those of single biosensor cells (by correlation analysis and by visual inspection, Figure 5b, Supplementary Movie 3, Supplementary Figure 7) but did show some increase in edge velocity (Supplementary Figure 5). With two biosensors in the same cell we could employ the dynamic grid of sampling windows to extract the local correlation between Asef and each GTPase (Figure 5c, d). Asef activity correlated with both Rac1 and Cdc42, in both the 0–1.4 μm and 1.4–2.8 μm layers. Overall, both correlation peaks were significantly higher for Cdc42 than for Rac1. This is consistent with biochemical data that Asef directly interacts with Cdc42, but can activate Rac1 only indirectly9, 40, although our imaging data now shows that all three signals are active in the same zone. Any delay between activation of Asef and Cdc42 was too short to be captured by the 5s sampling intervals we needed when imaging both biosensors together. This means that the communication between GEF and GTPase as resolved by these sensors and the affordable image sampling is quasi-instantaneous. We noted that the overall positive correlation lobes in the second layer dip at t=0. Thorough investigation of the causes unveiled a systematic amplification of subpixel errors in cell edge segmentation and sampling window positioning that depresses the correlation values specifically at zero time lags (Supplementary Figure 9). Taken together, this analysis revealed specific regions and times when activation of Asef is correlated with Cdc42 and Rac1 activation during cell edge movement. During protrusion, the GTPases Rac1 and Cdc42 are co-regulated by multiple RhoGEFs, which our data show includes Asef but not Vav2. Using a previously published Cdc42 biosensor41 combined with our new red-shifted Rac1 biosensor, we tested whether Cdc42 and Rac1 correlate directly. Indeed, this was the case, again with a time lag of zero (Figure 5b, e; Supplementary Movie 4), suggesting that at the time scale of our sampling, the two GTPases are modulated concurrently.
Figure 5. Multiplex imaging of two protein activities.
(a) Biosensor designs and fluorescent proteins used for multiplexing (upper). Emission spectra of representative RhoGEF and Rho GTPase biosensors (lower). Inactive biosensors in solid lines, activated forms in dotted line. The two spectra were obtained using the same excitation wavelength (430 nm). (b) RhoGEF and Rho GTPase biosensors imaged simultaneously in MDA-MB-231 cells undergoing constitutive edge motion. Pseudocolor as in Fig. 1. Scale bars 10μm. Images are representative of at least 3 independent experiments. (c) Cross correlation coefficients between Rho GTPase activity and Asef activity in the 1.4–2.8 μm layer. Gold shows high correlation, blue negative correlation. (d) Average cross-correlation functions for each biosensor combination, 0–1.4 μm layer – left, 1.4–2.8 μm layer – right; (n = cells, m = windows); Cdc42 (n=5, m=729); Rac1, (n=6, m=719). Dotted lines show the correlation coefficient above which the coupling between two variables is considered significant with 95% confidence. This depends on the number of windows (see Methods). Shading represents 95% C.I. about the mean correlation computed from m windows. The width of this interval depends on the consistency of the correlations across windows and cells (see Methods). (e) Single cell cross correlation coefficients (left) and average cross-correlation functions (right) between Rac1 activity and Cdc42 activity in the 1.4–2.8 μm layer (left). (n=7, m=684)
Partial correlation analysis of RhoGEF and Rho GTPase activities at the edge
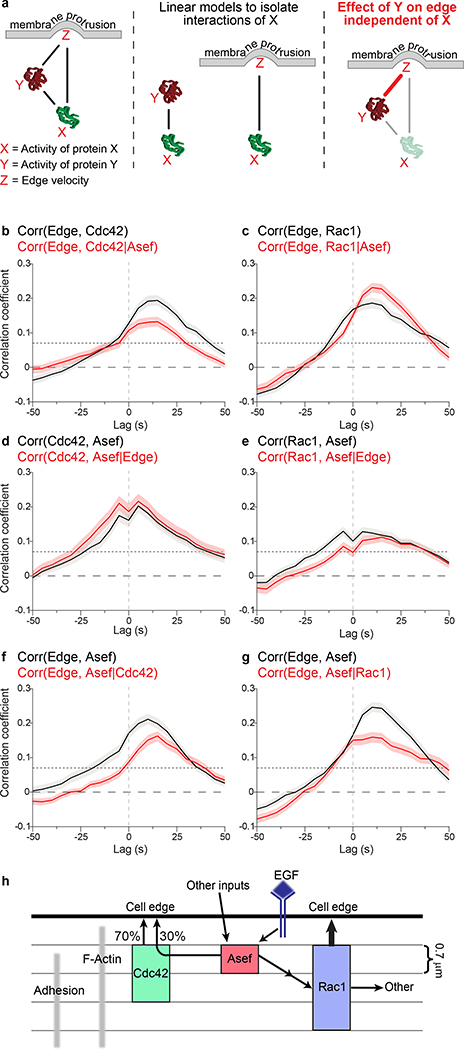
A critical question is why a RhoGEF activates multiple GTPases, and how much interaction with each one contributes to the downstream effector responses that actually produce edge motion. In concrete terms here, how much of the Cdc42 activation that is produced specifically by Asef contributes to the modulation of edge motion, and how much is contributed by Asef’s indirect activation of a Rac1 signal? By imaging all possible combinations of three variables (GEF/GTPase, GEF/edge, and GTPase/edge), we could address this question using partial correlation analysis.
Given three co-fluctuating variables (X,Y,Z), partial correlation analysis can quantify how much the fluctuations in Y and Z remain coupled after removing the effects of a third variable X, which controls both Y and Z (Figure 6a). Using this approach, we first computed the correlation between Cdc42 activation and edge motion after eliminating the influence of Asef on Cdc42 and edge motion (i.e. a “numerical knockout” of Asef). We focused on the layer 1.4 – 2.8 μm from the edge, where Asef displayed significant co-fluctuation with edge motion and Cdc42 (Figures 4e, 5d). Compared to the direct correlation between Cdc42 and edge motion, the peak value was about a third smaller (0.2 versus 0.13) (Figure 6b), indicating that 36% of the Cdc42 signal that influences edge motion is triggered by Asef. Using shRNA to reduce Asef levels experimentally also produced a reduction in the correlation between Cdc42 and edge motion (Supplementary Figure 8b). The difference between the 36% reduction seen with partial correlation analysis and the 25% reduction in the shAsef experiment may be marginally significant, but it should be noted that reducing the concentration of one GEF can be compensated by other GEFs. The numerical analysis by partial correlation is not subject to compensation by other GEFs. Thus, it is reasonable for the partial correlation analysis to report stronger differences. In stark contrast, for Rac1 the correlation with edge motion increased after eliminating the contribution of Asef (Figure 6c). This implies that the Asef to Rac1 signaling link desynchronizes the coupling of Rac1 and edge velocity. Such a conclusion can only be reached using numerical analysis. An actual perturbation of Asef would have effects on cell edge velocity that preclude analysis of the changes in the relationship between Rac1 and velocity, e.g. inducing shifts in Cdc42 signalling.
Figure 6. Partial correlation analysis.
(a) Model for partial correlation analysis using three variables, X, Y, and Z, which are all connected by pair-wise relationships (solid lines, left). Correlations describing specifically the relationships between X and Y, as well as X and Z (middle). The contribution of X to both Y and Z is then subtracted from each (grey arrows), and the relationship between the residual Y and Z is calculated, giving the partial correlation (red line, right). (b-g) Comparison between total and partial correlations for all combinations of Asef, Rho GTPase, and edge velocity (total correlation, black line; partial correlation, red line ((n = cells, m = windows); Cdc42 (n=5, m=729); Rac1, (n=6, m=719)). Edge to GTPase correlations, controlled for RhoGEF input (b, c). GTPase to RhoGEF correlations, controlled for edge input (d, e). Edge to RhoGEF correlations, controlled for GTPase input (f, g). Dotted lines show the correlation coefficient above which the coupling between two variables is considered significant with 95% confidence. This depends on the number of windows (see Methods). Shading represents 95% C.I. about the mean correlation computed from m windows. The width of this interval depends on the consistency of the correlations across windows and cells (see Methods). (h) Spatiotemporal integration of Asef-, Cdc42- and Rac1- signaling in the control of constitutive cell protrusion-retraction cycles. Asef directly activates Cdc42 to control ~30% of the effect of Cdc42 on edge motion. Asef also contributes indirectly to the activation of Rac1, but this contribution does not play a role in controlling the edge. Instead, other GEFs must be responsible for the motion-relevant signaling of Rac1, which is stronger than Cdc42’s contribution. The relative localizations of each protein are shown. Note that activation of Asef is tightly localized compared to Rac1 and Cdc42, supporting localized GTPase activation by Asef followed by GTPase diffusion or transport to the edge.
This surprising finding is consistent with an indirect activation of Rac1 by Asef, potentially via cross-talk between Cdc42 and Rac140. Alternatively, interactions between Asef and Rac1 mediated without Cdc42 may be elicited for cell functions other than protrusion, again with an adversarial effect on the interactions between Rac1 and edge motion. Distinguishing these two explanations will require concurrent imaging of Asef, Cdc42, and Rac1, which is impossible at this point. Nonetheless, these findings reveal the limitations of multi-functional signaling networks, where signaling for one purpose may impair the precision in signaling for another purpose.
Previous studies suggest that expansion of the plasma membrane during protrusion produces mechanical feedback on GEF-GTPase interactions, especially in the adhesion-rich zone near the cell edge36, 42. We therefore applied partial correlation analysis to assess the influence of cell edge motion on Asef/Cdc42 and Asef/Rac1 relationships. The correlations between GEF and GTPase fluctuations remained unchanged upon controlling for common fluctuations associated with edge motion. This shows that both interactions are nearly independent of edge motion (Figure 6d, e). Hence, Asef is unlikely to be a mediator of mechanical feedback to Rac1 and Cdc42.
Finally, we asked how eliminating the influence of the GTPases would affect the correlation between Asef and edge motion. For both Cdc42 (Figure 6f) and Rac1 (Figure 6g) the correlation curves substantially decreased, confirming that the interaction between Asef and motion is indeed mediated by Rac1 and Cdc42. Intriguingly, with Cdc42 effects removed, the Asef/motion correlation was markedly reduced at negative time lags and between 0 and 25 s (Figure 6f), whereas removal of Rac1 inputs reduced the Asef/motion correlation at lags between 0 and 50s (Figure 6g). This suggests that coupling of Asef to edge motion early in protrusion is dependent on Cdc42, while later coupling depends more on Rac1. Again, this finding is consistent with the interpretation that Asef activates Cdc42 directly, but Rac1 is coupled to Asef only via an indirect connection. Together, these analyses and conclusions indicate how direct multiplexed imaging of an activator and receiver signal in the context of a dynamic cell function allows statistical dissection of very complex signaling relations with unprecedented precision.
Discussion
The study of RhoGEFs has been limited to deciphering biochemical interactions in vitro, and to molecular perturbation in vivo43, 44. However, due to the complexity of RhoGEF-GTPase interaction networks, which contain numerous feedbacks, crosstalks and redundancies, an understanding of GEF function requires new methods to determine connectivity and function in the context of space and time. To unravel the effects of RhoGEFs on multiple GTPase targets, we first developed a generalizable approach to Dbl family GEF biosensors. The method has been applied to the GEFs described here and to GEF-H145. It is applicable to diverse structures comprising 19 of the 70 Dbl family RhoGEFs (Supplementary Figure 2d), and potentially to 10 GEFs that are similarly regulated. This enabled correlation of GEF activity and GTPase activity with the motions of the cell edge. We then modified our existing GTPase biosensors for simultaneous imaging of GEF and GTPase activity, providing the ability to directly correlate their spatio-temporal dynamics in living cells. With pairwise correlation of edge motion, GEF activity and GTPase activity in hand, we could apply partial correlation analysis to locally sampled time series and thereby determine that the RhoGEF Asef has stronger interactions with Cdc42 than with Rac1, albeit both interactions are statistically significant. We could show that specific, spatially localized Asef activations contribute to specific Cdc42 activation events, and determine to what extent the Cdc42 activation promoting cell edge activity was due to Asef (Figure 6h). For Asef activation of Rac1, several observations (timing, diffuse response, and weaker coupling between Asef and Rac1) indicated that Rac1 is activated via indirect interactions, consistent with published biochemical studies. Moreover, while Asef activity clearly has an activating effect on Rac1 signaling, our partial correlation analysis indicates that the Asef-triggered Rac1 activity does not contribute to the modulation of edge protrusion – this despite the fact that the correlation between Rac1 and edge motion is substantially stronger than the correlation between Cdc42 and motion (Figure 4d, e). Rac1 may be more involved in the regulation of constitutive protrusion-retraction cycles. This highlights that each signal can best be understood through in situ analysis in the context of a cell function.
The GEF biosensor design used here exemplifies a valuable alternative to current biosensor approaches. It can produce protein analogs that report conformational changes while maintaining upstream and downstream interactions. This contrasts with biosensors whose components bind selectively to the activated state of the target protein, thereby generating dominant negative effects. Unlike biosensors based on protein substrates, the new design will be less prone to false negative readouts where substrate cannot reach the target. The GEF biosensors described here could potentially be expressed by replacing the genes of their endogenous counterparts. Like other biosensors, in the current study we had to carefully control expression level to minimize overexpression artifacts, forcing us in some cases to work at concentrations near the limit of signal/noise for robust image analysis.
Building on multiplexed imaging of biosensors, future correlation analysis can assign functions to the perplexing multitude of RhoGEF-GTPase interactions, in the context of diverse cellular behaviors. This can ultimately be extended to a host of other complex signaling circuits.
Online Methods
RhoGEF biosensor design
The biosensors were optimized and tested in a sequential fashion. Initially, a cassette comprising mCerulean or mCerulean3, a flexible linker5, and mVenus or YPet was inserted into a series of sites between the AID and DH domains of full-length RhoGEFs and emission was measured using fluorometry for Vav2 or high-throughput microscopy for the other RhoGEFs (see detailed procedures below). For Vav2 we compared wild type to constitutively active (Y140E:Y159E:Y172E) mutants, and for the other RhoGEFs we used co-expression of an activator. For Vav2 we also tested linker variants. These comprised a flexible unit46 and a structured helical unit47 combined in different topologies and repeat numbers (2, 3, and 4 repeats for short, medium and long respectively). Once we had identified the best insertion site and linker combination we combinatorially combined donor proteins (mCerulean3, TagCFP and mTFP) with acceptor proteins (YPet, along with circular permutations of YPet) to form a library of variants of each biosensor that were screened using high-throughput microscopy for the effects of activation. The biosensor constructs were inserted into a tet-off inducible retroviral expression system and stable lines were produced in tet-off MDA-MB-231 cells (Johnson lab, UNC-CH). Cells were maintained in DMEM (Cellgro) with 10% FBS (Hyclone) and 0.2 μg/ml doxycycline to repress biosensor expression. Biosensors were named FLARE.a as part of a nomenclature system based on the biosensor design (eg Asef FLARE.a)
Rho GTPase biosensors
The Rac1 FLARE.dc1g biosensor is a modification of our previously reported dual chain biosensor design3. To improve brightness and dynamic range we used Turquoise fluorescent protein13 rather than CyPet. In brief, YPet16 is fused upstream of residues 60 −145 of human PAK1, and Turquoise is fused to the N-terminus of full-length Rac1. The two biosensor chains were expressed on one open reading frame with two consecutive 2A viral peptide sequences from Porcine teschovirus-1 (P2A) and Thosea asigna virus (T2A) inserted between them, leading to expression of the two separate biosensor chains48. The Cdc42 and RhoA FLARE.dc1g biosensors use an identical topology, wherein YPet is fused upstream of N-WASP (200–293) or Rhotekin (6–98), for Cdc42 and RhoA respectively, and mCerulean3 is fused to the N-terminus of the GTPase, with the two biosensor chains separated by the 2A sequences. The biosensor constructs were inserted into a tet-off inducible retroviral expression system and stable lines were produced in tet-off MDA-MB-231 cells (Johnson Lab, UNC-CH). Cells were maintained in DMEM (Cellgro) with 10% FBS (Hyclone) and 0.2 μg/ml doxycycline to repress biosensor expression
For the red shifted versions of the GTPase biosensors, YPet was exchanged for mCherry and the donor was exchanged for LSSmOrange. Both donor and acceptor contain the R125I mutation to increase intramolecular FRET49. For dual biosensor experiments, these red-shifted GTPase biosensors were transfected into the RhoGEF biosensor-expressing stable cell lines. For experiments with two GTPase biosensors in the same cell, the Cdc42 biosensor41 was transfected into cells stably expressing the red-shifted Rac1 biosensor.
Spectral analysis of biosensors
Emission spectra of biosensors were obtained using a Fluorolog fluorometer (Horiba). HEK-293t cells grown in 6-well plates (Nunc) were transfected with biosensor DNA plus regulator if required. After 24 h cells were detached by trypsinization (Cellgro) and resuspended in cold PBS (Sigma) + 1%FBS (Hyclone), washed and then resuspended in cold PBS. Samples were excited at 430nm and spectra obtained from 460 to 600nm for biosensors with Cerulean3/TagCFP/mTFP, and 550 to 650nm for LSSmOrange biosensors. Dual chain biosensors were corrected for acceptor bleedthrough.
High-throughput microscopy screening
Using a modification of our published procedure10, HEK-293t cells were plated in 96-well plates with μ-clear plastic bottoms (Greiner bio-one) coated with poly-l-lysine (Sigma). Cells were transfected in triplicate with a library of biosensor DNA plus regulator if required and imaged after 24 h. Growth media was replaced with HBSS (Sigma) with 1%FBS and 10mM HEPES (Gibco) prior to imaging. Cells were imaged using a 10X, 0.4 NA objective on an Olympus IX-81 inverted microscope and using Metamorph screen acquisition software (Molecular Devices) and mercury arc lamp illumination. Filters used were Ex - ET436/20X, Em; donor- ET470/24M, FRET - ET535/30M and a 445/505/580 ET dichroic mirror. Images were obtained on a Flash4 sCMOS camera (Hamamatsu). Images were analyzed using MATLAB (Mathworks). Briefly, 4 fields were taken of each well and the intensity was summed for each channel. These were then background subtracted using values from wells that were mock transfected, and ratios obtained from these background-subtracted values. For regulator titration experiments, regulator DNA was first titrated against the biosensor DNA and then transfection complexes formed in 96-well plates prior to transfection. Regulators used: APC (309–798), Src (FL, Y529F), Gα13 (FL, Q226L), PAK (FL D389R:S422D:T423E), Dbl (495–826), Vav2 (191–573), Asef (FL), Tiam1 (C1199), p115RhoGEF (FL), RhoGDI (FL), p50RhoGAP (FL), RacGAP1 (FL).
Src stimulation of Vav2 biosensors
Cells were transfected and imaged as above. After lysis, triplicate samples were pooled and Western blotted for phospho-Vav levels using a phospho-specific Tyr-172 antibody (Abcam). Samples were normalized to biosensor expression using an anti-GFP antibody (Clontech).
Stimulation experiments
A431 cells (ATCC) were plated in 6-well plates 24hr prior to transfection. Cells were incubated with transfection complexes for 5–6 hours and then replated onto #1.5 coverslips (Warner Instruments) coated with Collagen IV (Gibco). Cells were allowed to spread in DMEM/10% FBS. After 2–3 h the media was replaced with DMEM/0.5% delipidated BSA and cells were starved overnight prior to imaging. Cells were then imaged in Hams/F12 (Caisson Labs) with 0.5%BSA, 10mM HEPES (Gibco), 100 μm Trolox (Sigma), and 0.5mM Ascorbate (Sigma). Cells were imaged using a 40× 1.3NA Silicon oil objective on an Olympus IX-81 inverted microscope using Metamorph software and 100 W Hg arc lamp illumination. Excitation filters used were FF-434/17 and FF-510/10 combined with a FF462/523 dichroic mirror. Donor and FRET images were simultaneously captured using a TuCam system (Andor) fitted with FF-482/35 and FF-550/49 and an imaging flat FF509-FDi01 dichroic, together with two Flash4 sCMOS cameras (Hamamatsu). To visualize endogenous Vav2 in these cells, mock-transfected A431 cells were stimulated in exactly the same way, then fixed in 4% formaldehyde (EMS microscopy supplies), 0.1 M Pipes, 1 mM EGTA and 1 mM MgSO4 (all Sigma). They were washed in PHEM buffer (60 mM Pipes, 25 mM Hepes, 2 mM MgSO4, 10 mM EGTA, all Sigma) then blocked with BSA. The cells were incubated with anti-Vav2 antibodies (Epitomics) in BSA/PHEM buffer, washed with PHEM buffer and then visualized using Alexa-633 labelled secondary antibodies.
Constitutive migration experiments
Biosensor expression was induced 48 hr prior to imaging through trypsinization and culturing without doxycycline. On the day of imaging, cells were replated using Accumax (Innovative Cell Technologies) onto coverslips coated with collagen I (10μg/ml 37C overnight) and allowed to attach in DMEM /10%FBS. After 2 hrs the media was replaced with Hams/F12 with 0.2% BSA, 10ng/μl EGF (R and D systems) 10mM HEPES, 100 μm Trolox, and 0.5mM Ascorbate and cells were allowed to equilibrate. After a further 2–4 hrs, cells were imaged in a closed chamber with media treated with Oxyfluor (1/100). For single biosensor experiments, cells were imaged using the filters listed above. For dual biosensor experiments, the excitation filters used were FF-434/17 for Cerulean3/mTFP and LSSmOrange, and FF-546/6 for Cherry (Semrock) combined with a custom zt440/545 dichroic (Chroma). For emission, a TuCam was fitted with a FF560-FDi01 imaging flat dichroic and a Gemini dual view (Hamamatsu) was added to each emission port. For the short wavelength Gemini, the filters used were donor- FF-482/35, FRET - FF-520/15 and a FF509-FDi01 imaging flat dichroic mirror. For the Red-shifted Gemini, the filters used were Orange – FF01–575/15, FRET/mCherry – FF01–647/57 and a FF580-FDi01 imaging flat dichroic. For the shRNA experiments, multiple shRNA constructs using microRNA backbones(mir-E)50 were tested for knock-down using qRT-PCR. The most efficient was inserted downstream of iRFP-67051 in a tet-off inducible retroviral expression system and stable lines were produced. When imaging, cells expressing the highest levels of iRFP670 were chosen to ensure knockdown.
Image Processing and Analysis
Biosensor activation levels were measured in living cells by monitoring the ratio of FRET to donor emission on a pixel by pixel basis. Donor and FRET images were aligned using fluorescent beads as fiduciaries to produce a transformation matrix using the Matlab function “cp2tform” (Matlab, The Mathworks Inc.). This was then applied to the Donor image using the Matlab function “imtransform”. The camera dark current was determined by obtaining images for each camera without excitation, and the dark current was subtracted from all images. Images were corrected for shading due to uneven illumination by taking images of a uniform dye solution under conditions used for each wavelength, normalizing this image to an average intensity of 1 to produce a reference image for each wavelength, and then dividing the images corrected for dark current by the shading correction reference image. Background fluorescence was removed by subtracting, at each frame, the intensity of a region containing no cells or debris. Images were segmented into binary masks separating cell and non-cell regions using the segmentation package “MovThresh”, which is based on the Otsu algorithm. The Donor channel was used for segmentation, as it had the highest signal to noise, particularly at the cell edge. The masks were then applied to all channels, setting non-cell regions to zero intensity. For dual biosensor imaging these masked images were then corrected for bleedthough of Cerulean3/mTFP and YPet into the orange/red channels.
For RhoGEF biosensors, activation maps were obtained by dividing the corrected donor image by the FRET image. For the GTPase biosensors, the images were corrected for bleed-through and ratios were obtained using the following equation (using data from control cells expressing donor or acceptor alone to obtain the bleed-through coefficients α and β): R= (FRET – α(Donor) – β(Acceptor))/donor where R is the Ratio, FRET is the total FRET intensity as measured, α is the bleed-through of the donor into the FRET signal, β is the bleed-through of acceptor into the FRET signal, and Donor and Acceptor are the donor and acceptor intensities as measured through direct excitation. These ratio images were then corrected for photobleaching. For stimulation experiments, the ratios were divided by a reference curve derived from mock-stimulation experiments. For constitutive migration the whole cell average was fitted to a double exponential curve and this curve was used to normalise. Pseudocolor scales were produced without considering the lowest and highest 5% of ratio values to eliminate spurious pixels, and normalizing so the lowest value is 1. For the dual chain biosensors, there is ideally no FRET in the off state, so dynamic range cannot be defined as FRETon / FREToff. The lowest ratio values are in fact determined by the level of noise in the image and any binding in the off state.
Cell windowing analysis
After the corrections described above, the cell images of the ratiometric biosensor activity were compartmentalized into layers of rectangular windows along the entire cell edge. To construct the sampling windows at a constant distance from the cell edge we computed a distance map (Matlab function: bwdist) to the segmented cell edge. The distance map yielded equidistant contours at either 0.7μm (2 pixels) or 1.4μm (4 pixels) from the cell edge. At the cell edge the first contour was divided into segments of 1.8 – 3 μm width (Figure 4a). Sampling windows located at the cell edge derived a time course of edge velocity and biosensor intensity as the windows tracked the morphological changes of the cell over time. For windows placed in layer 2 and higher only the biosensor intensities were sampled. However, each of these windows maintained unambiguous correspondence to a window at the cell edge, allowing correlation of biosensor intensity fluctuations inside the cell with cell edge movements. Importantly, the ability to maintain unique correspondences depended on the cell morphology. Windows for which the correspondence was lost at one or several time points of a movie because of particularly strong cell morphological changes were eliminated from the analysis.
This in silico compartmentalization of the cell allowed us to represent the biosensor activity into a cell-shape invariant space. For each frame of the movies, the biosensor activity was averaged within the area of each sampling window, resulting in a set of matrices representing the biosensor activity of a layer of windows with a fixed distance from the cell edge. Rows correspond to windows and columns to time (Figure 4b). This method has shown to be an efficient way to spatiotemporally sample the activity of sensors expressed by the cell. For more information, see52.
Windows selection
Migrating cells usually display regions along the cell edge that are active with protrusion/retraction cycles, but they also exhibit quiescent regions with little morphodynamic activity. We implemented window selection criteria based on the autocorrelation function of the protrusion/retraction speed estimated by the windowing algorithm described above. The autocorrelation function of a random variable X can be described as the cross-correlation between X and its time-delayed version X (lag) where lag is the duration of the delay. It can measure an average duration of “memory” in the signal that can be described as the maximum time spacing between samples that still exhibit a linear association. No significant linear correlation can be measured when taking samples further than this duration apart. Consequently, the autocorrelation function of a signal with structure has a much slower decay compared with the autocorrelation function of a signal with samples independently drawn from a uniform random distribution. We used the full width at half maximum (fwhm) of the window speed autocorrelation as a measure of information in the signal that our analysis methods can make use of. Only windows with autocorrelation fwhm equal to or higher than 0.6 frames were chosen for analysis, i.e. the speed time series in such windows showed on average some temporal coherence in motion. For reference, a white noise time series with no coherence has a fwhm < 0.5 frames. The effect of this strategy was the elimination of quiescent regions. For the example in Figure 2, all windows are shown in Supplementary Figure 7.
A second method of window selection was implemented in order to avoid a phenomenon we observed when cells were expressing two biosensors. This problem arose due to a sub-pixel segmentation error that appears when two biosensors have opposing gradients. See Supplementary Figure 8. This transient error causes a strong negative cross-correlation between the activities of the two imaged biosensors. In order to minimize the impact of this artifact on our final analysis, we excluded windows that presented a sharp negative cross-correlation between two biosensors at lag zero. It is worth mentioning that windows with a negative cross-correlation trend that resulted in a negative score at lag zero were not excluded from analysis. The selection algorithm starts by decomposing the cross-correlation between the biosensors for a given window using empirical mode decomposition – EMD53. This decomposition technique recursively extracts components of the signal from the fastest, or higher frequency content, to slower component of the signal. The fastest component or first intrinsic mode function (IMF) absolves all fast variations present in the signal. The window is excluded from analysis if after its cross-correlation decomposition, the amplitude of the first IMF at lag zero exceeds a threshold and shows change in derivate for the lag zero neighborhood. The threshold is estimated as three standard deviations away from the mean using the first IMF points to build the distribution.
In our further analysis of the red-shifted biosensors, we utilized the Ljung-Box statistical test to exclude the quiescent subcellular regions automatically. The test provides a p-value where the time series of interest, i.e. here cell edged motion, can be considered as a white noise. For the implementation of the method, we used the matlab function lbqtest() and checked the statistical significance of auto-correlations up to 20 lags of the velocity time series for each window. Using a significance level of 0.05, certain subsets of the windows were identified as white noise signals, equivalent to our previous quiescent velocities. The selected windows were further excluded from our analysis.
Pearson correlation analysis
The main aim of the Pearson correlation analysis is to determine the strength of the linear relationship between two random variables. For instance, it can be applied to find time-dependent linear relationship of two measured parameters, which is the case in this work. The correlation value ranges from [−1,1] where score value 1 represents perfect linear correlation, 0 represents no correlation and −1 perfect anti-correlation. Additionally, one time series can be time-shifted in relation to the second by a value usually referred to as lag. This lag analysis can be useful when two variables are correlated with a time delay. That means there will a value of lag where the correlation score is maximum. The lag at peak correlation can then be interpreted as the time needed for one variable to process upstream information or as the time taken for information to be transferred or a combination of both. Because correlation analysis requires a stationary signal, few pre-processing steps are required before the correlation coefficient is calculated. All time series data used to calculate a correlation score in this work were mean subtracted and linear trend removed.
Pearson’s correlation coefficient ρ(a1(t),a2(t))τ between two activity time courses a1(t) and a2(t) was computed as a function of the time lag τ using the Matlab function (xcov). This function implements the mean corrected and normalized correlation functions as follows:
The operators Cov(.) and Var(.) denote the covariance and the variance of the mean corrected time courses, respectively. The variables and denote the mean value of the respective time course. Correlation functions were first calculated for pairs of time courses per window (either between edge velocity and biosensor intensity, or between two biosensor intensities in the case of simultaneous imaging GEF and GTPase activity). For each correlation curve we computed the level, which is exceeded by no more than 5% of correlations between two random time series. This level (~0.1 for all our data) depended on the duration of the movies and the number of sampling windows. Regions of the correlation curve that extend above this confidence interval are therefore considered significant with a confidence of 95%. In addition, through bootstrap analysis of the variation among all correlation curves, we computed confidence intervals for the mean cross-correlation. This reflected the number and consistency of sampling windows across experiments. This computation is performed by applying the Fisher transformation to the correlation values with a hyperbolic tangent and bringing back the distribution parameters with the hyperbolic tangent inverse54.
Partial correlation analysis
Although Pearson correlation analysis has been shown to be useful when only two variables are at hand, it can result in misleading outcomes when trying to identify linear relationships between random variables where the data generating system has confounding structure55. A simple example of this case is when a random variable A drives two other unrelated random variables B and C. There is a flow of information going from A to B and A to C but no exchange of information between B and C. A simple correlation analysis would identify the confounding information from A shared by B and C as a direct connection between B and C. However, the partial correlation between B and C control for A, or partial(B,C|A), would statistically return zero. For this simple three variable example, the partial correlation can be computed as:
The time dependency of the random variables was omitted for simplicity. The equation above shows that partial correlation removes the influence of the variable A from B and C and renormalizes it such the final value lies within the [−1,1] interval. Although simple, using the inverse of the correlation matrix is a more efficient way to calculate the partial correlation when more than 3 variables are present55.
Because of the stationary requirement, all time series were mean subtracted and linear trend corrected as was done for the simple correlation analysis. Confidence intervals and levels were calculated in the same fashion as the Pearson’s correlation.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. Figures 4, 5, and 6 have associated raw data used to produce the correlation plots.
Code availability statement
All code used for the data analysis was written in Matlab 2014b. All the code can be downloaded from the Danuser Lab github: https://github.com/DanuserLab under Biosensor, Windowing-Protrusion and Time-Series-Tools repositories
Supplementary Material
Acknowledgements
R01-GM062299 (JS), R01 GM071868 (GD), and R35GM122596 (KMH) supported this work. DM was supported by funding from the Leukemia and Lymphoma Society (LLS). JH was supported by a Human Frontiers in Sciences Program. GG was supported by the “Integrated Training in Cancer Model Systems” Training Program, T32CA009156. The authors would like to thank the Calabrese lab and F. Pimenta for the shRNA expression vector.
Footnotes
Competing Financial Interests Statement
We declare that none of the authors have competing financial or non-financial interests as defined by Nature Research, or other interests that might be perceived to influence the results and/or discussion reported in this paper.
References
- 1.Devreotes P & Horwitz AR Signaling networks that regulate cell migration. Cold Spring Harb Perspect Biol 7, a005959 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Komatsu N et al. Development of an optimized backbone of FRET biosensors for kinases and GTPases. Mol Biol Cell 22, 4647–4656 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Machacek M et al. Coordination of Rho GTPase activities during cell protrusion. Nature 461, 99–103 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang HW, Collins SR & Meyer T Locally excitable Cdc42 signals steer cells during chemotaxis. Nat Cell Biol 18, 191–201 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zawistowski JS, Sabouri-Ghomi M, Danuser G, Hahn KM & Hodgson L A RhoC biosensor reveals differences in the activation kinetics of RhoA and RhoC in migrating cells. PLoS One 8, e79877 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hodgson L et al. FRET binding antenna reports spatiotemporal dynamics of GDI-Cdc42 GTPase interactions. Nat Chem Biol 12, 802–809 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rossman KL, Der CJ & Sondek J GEF means go: turning on Rho GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol 6, 167–180 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Hall A Rho family GTPases. Biochem Soc Trans 40, 1378–1382 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Mitin N et al. Release of autoinhibition of ASEF by APC leads to Cdc42 activation and tumor suppression. Nat Struct Mol Biol 14, 814–823 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slattery SD & Hahn KM A High-Content Assay for Biosensor Validation and for Examining Stimuli that Affect Biosensor Activity. Curr Protoc Cell Biol 65, 14 15 11–31 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rizzo MA, Springer GH, Granada B & Piston DW An improved cyan fluorescent protein variant useful for FRET. Nat Biotechnol 22, 445–449 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Nagai T et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol 20, 87–90 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Markwardt ML et al. An improved cerulean fluorescent protein with enhanced brightness and reduced reversible photoswitching. PLoS One 6, e17896 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xia NS et al. Bioluminescence of Aequorea macrodactyla, a common jellyfish species in the East China Sea. Mar Biotechnol (NY) 4, 155–162 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Ai HW, Henderson JN, Remington SJ & Campbell RE Directed evolution of a monomeric, bright and photostable version of Clavularia cyan fluorescent protein: structural characterization and applications in fluorescence imaging. Biochem J 400, 531–540 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen AW & Daugherty PS Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat Biotechnol 23, 355–360 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Itoh RE et al. Phosphorylation and activation of the Rac1 and Cdc42 GEF Asef in A431 cells stimulated by EGF. J Cell Sci 121, 2635–2642 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Yu B et al. Structural and energetic mechanisms of cooperative autoinhibition and activation of Vav1. Cell 140, 246–256 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aghazadeh B, Lowry WE, Huang XY & Rosen MK Structural basis for relief of autoinhibition of the Dbl homology domain of proto-oncogene Vav by tyrosine phosphorylation. Cell 102, 625–633 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Crespo P, Schuebel KE, Ostrom AA, Gutkind JS & Bustelo XR Phosphotyrosine-dependent activation of Rac-1 GDP/GTP exchange by the vav proto-oncogene product. Nature 385, 169–172 (1997). [DOI] [PubMed] [Google Scholar]
- 21.Barreira M et al. The C-terminal SH3 domain contributes to the intramolecular inhibition of Vav family proteins. Sci Signal 7, ra35 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Yohe ME et al. Auto-inhibition of the Dbl family protein Tim by an N-terminal helical motif. J Biol Chem 282, 13813–13823 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Yohe ME, Rossman K & Sondek J Role of the C-terminal SH3 domain and N-terminal tyrosine phosphorylation in regulation of Tim and related Dbl-family proteins. Biochemistry 47, 6827–6839 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Z, Gakhar L, Bain FE, Spies M & Fuentes EJ The Tiam1 guanine nucleotide exchange factor is auto-inhibited by its pleckstrin homology coiled-coil extension domain. J Biol Chem 292, 17777–17793 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Z, Guo L, Sprang SR & Sternweis PC Modulation of a GEF switch: autoinhibition of the intrinsic guanine nucleotide exchange activity of p115-RhoGEF. Protein Sci 20, 107–117 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lambert JM et al. Tiam1 mediates Ras activation of Rac by a PI(3)K-independent mechanism. Nat Cell Biol 4, 621–625 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Miyamoto Y, Yamauchi J, Tanoue A, Wu C & Mobley WC TrkB binds and tyrosine-phosphorylates Tiam1, leading to activation of Rac1 and induction of changes in cellular morphology. Proc Natl Acad Sci U S A 103, 10444–10449 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Servitja JM, Marinissen MJ, Sodhi A, Bustelo XR & Gutkind JS Rac1 function is required for Src-induced transformation. Evidence of a role for Tiam1 and Vav2 in Rac activation by Src. J Biol Chem 278, 34339–34346 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Tolias KF et al. The Rac1 guanine nucleotide exchange factor Tiam1 mediates EphB receptor-dependent dendritic spine development. Proc Natl Acad Sci U S A 104, 7265–7270 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki N, Nakamura S, Mano H & Kozasa T Galpha 12 activates Rho GTPase through tyrosine-phosphorylated leukemia-associated RhoGEF. Proc Natl Acad Sci U S A 100, 733–738 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng Q et al. Cool-1 functions as an essential regulatory node for EGF receptor- and Src-mediated cell growth. Nat Cell Biol 8, 945–956 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Jaiswal M et al. Mechanistic Insights into Specificity, Activity, and Regulatory Elements of the Regulator of G-protein Signaling (RGS)-containing Rho-specific Guanine Nucleotide Exchange Factors (GEFs) p115, PDZ-RhoGEF (PRG), and Leukemia-associated RhoGEF (LARG). J Biol Chem 286, 18202–18212 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nalbant P, Hodgson L, Kraynov V, Toutchkine A & Hahn KM Activation of endogenous Cdc42 visualized in living cells. Science 305, 1615–1619 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Herrington KA et al. Spatial analysis of Cdc42 activity reveals a role for plasma membrane-associated Cdc42 in centrosome regulation. Mol Biol Cell 28, 2135–2145 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mendoza MC, Vilela M, Juarez JE, Blenis J & Danuser G ERK reinforces actin polymerization to power persistent edge protrusion during motility. Sci Signal 8, ra47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee K et al. Functional hierarchy of redundant actin assembly factors revealed by fine-grained registration of intrinsic image fluctuations. Cell Syst 1, 37–50 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ji L, Lim J & Danuser G Fluctuations of intracellular forces during cell protrusion. Nat Cell Biol 10, 1393–1400 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shcherbakova DM, Hink MA, Joosen L, Gadella TW & Verkhusha VV An orange fluorescent protein with a large Stokes shift for single-excitation multicolor FCCS and FRET imaging. J Am Chem Soc 134, 7913–7923 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaner NC et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22, 1567–1572 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Nishimura T et al. PAR-6-PAR-3 mediates Cdc42-induced Rac activation through the Rac GEFs STEF/Tiam1. Nat Cell Biol 7, 270–277 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Martin K et al. Spatio-temporal co-ordination of RhoA, Rac1 and Cdc42 activation during prototypical edge protrusion and retraction dynamics. Sci Rep 6, 21901 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuo JC, Han X, Hsiao CT, Yates JR 3rd & Waterman CM Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for beta-Pix in negative regulation of focal adhesion maturation. Nat Cell Biol 13, 383–393 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goicoechea SM, Awadia S & Garcia-Mata R I’m coming to GEF you: Regulation of RhoGEFs during cell migration. Cell Adh Migr 8, 535–549 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lawson CD & Ridley AJ Rho GTPase signaling complexes in cell migration and invasion. J Cell Biol 217, 447–457 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Azoitei ML et al. Spatiotemporal dynamics of GEF-H1 activation controlled by microtubule- and Src-mediated pathways. J Cell Biol 218, 3077–3097 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whitlow M et al. An improved linker for single-chain Fv with reduced aggregation and enhanced proteolytic stability. Protein engineering 6, 989–995 (1993). [DOI] [PubMed] [Google Scholar]
- 47.Kuhlman B, Yang HY, Boice JA, Fairman R & Raleigh DP An exceptionally stable helix from the ribosomal protein L9: implications for protein folding and stability. Journal of molecular biology 270, 640–647 (1997). [DOI] [PubMed] [Google Scholar]
- 48.Kim JH et al. High Cleavage Efficiency of a 2A Peptide Derived from Porcine Teschovirus-1 in Human Cell Lines, Zebrafish and Mice. Plos One 6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lindenburg LH, Hessels AM, Ebberink EH, Arts R & Merkx M Robust red FRET sensors using self-associating fluorescent domains. ACS Chem Biol 8, 2133–2139 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fellmann C et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep 5, 1704–1713 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Shcherbakova DM & Verkhusha VV Near-infrared fluorescent proteins for multicolor in vivo imaging. Nat Methods 10, 751–754 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vilela M et al. Fluctuation analysis of activity biosensor images for the study of information flow in signaling pathways. Methods Enzymol 519, 253–276 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang NE et al. The empirical mode decomposition and the Hilbert spectrum for nonlinear and non-stationary time series analysis. P Roy Soc a-Math Phy 454, 903–995 (1998). [Google Scholar]
- 54.Zoubir AM & Iskander DR Bootstrap techniques for signal processing. (Cambridge, Cambridge, England; New York; 2004). [Google Scholar]
- 55.Bailey NT J. Statistical methods in biology, Edn. 3rd. (Cambridge University Press, Cambridge England; New York; 1995). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. Figures 4, 5, and 6 have associated raw data used to produce the correlation plots.