Abstract
Curcumin, a polyphenol, is the main bioactive compound in dietary spice turmeric curcuma longa. It possesses anti-inflammatory, anti-oxidant and anti-neoplastic properties and shows potentials in treating or preventing particular diseases such as oxidative and inflammatory conditions, metabolic syndrome, arthritis, anxiety, hyperlipidemia and cancers. The diverse range and potential health beneficial effects has generated enthusiasm leading to intensive investigation into the phytochemical. However, a concern has been also raised if curcumin has a promiscuous bioassay profile and is a Pan-Assay INterference compound (PAINS). Here we present evidence indicating that curcumin is not a PAINS, but an inhibitor to APE1 redox function that affects many genes and pathways. This discovery explains the wide range of effects of curcumin on diverse human diseases and predicts a potential application in treatment of viral infection and virus-associated cancer. As a proof-of-concept, we demonstrated that curcumin is able to efficiently block Kaposi’s sarcoma-associated herpesvirus replication and inhibit the pathogenic processes of angiogenesis and cell invasion.
1. Introduction
Curcumin is the main bioactive compound in turmeric, one of the most effective nutritional supplements and traditional medicine (Schraufstatter and Bernt, 1949). Studies demonstrated that curcumin has anti-inflammatory, anti-oxidant and anti-neoplastic properties. Previous literature has described potential roles of this phytochemical in the treatment and prevention of particular diseases such as oxidative and inflammatory conditions, metabolic syndrome, arthritis, anxiety, hyperlipidemia and cancers (Gupta et al., 2013; Strimpakos and Sharma, 2008). Turmeric is generally Recognized As Safe (GRAS) by the US FDA and curcumin has been granted an acceptable daily intake (ADI) level of 3 mg/kg-BW by the joint FAO and WHO Expert Committee on Food Additives in 1996 (Clinical Development Plan: Curcumin, 1996).
Despite enthusiasm for the potential value of curcumin on human health that has led to more than 120 clinical trial of curcuminoids, efforts in curcumin-based drug development has been hampered by certain obstacles including its poor bioavailability, which is primarily due to poor absorption and metabolic instability, and enigmatic diverse effects (or promiscuous bioassay profile) that leads to a speculation of curcumin being a Pan-Assay INterference compound (PAINS) (Nelson et al., 2017). The poor bioavailability issue has led to numerous efforts with several strategies to improve the bioavailability such as modulation of route and medium of curcumin administration, blocking of metabolic pathways by concomitant administration with other agents, and conjugation and structural modifications of curcumin (Prasad et al., 2014). Since many human diseases including cancers result from multiple steps and involve multiple genes, the diverse effects of curcumin may not be a drawback but an advantage by targeting multiple genes simultaneously. However, the success in utilization of the multiple targeting property of this phytochemical in drug design heavily relies on thorough comprehension of the molecular mechanism underlying curcumin action of each biological activity and its pharmacological properties. Although many genes and several signaling pathways have been reported to be affected by curcumin, the molecular targets and biochemical mechanisms are largely unknown. To understand the diverse effect property of curcumin, we reviewed the spectrum of the genes and pathways that were affected by curcumin and found these curcumin-affected genes and pathways are largely overlapped with the pathways controlled by the redox reaction of APE1 (Apurinic/apyrimidinic endonuclease 1). Therefore, we asked if curcumin is an APE1 inhibitor and inhibition of APE1 redox activity contributes to the diverse biological activities of curcumin.
APE1, also termed Redox factor-1 (Ref-1), is a multifunctional protein. Its N-domain carries a redox activity controlling expression of the genes for cell survival pathways and its C-domain has a DNA base excision function acting as apurinic/apyrimidinic endonuclease (Tell et al., 2009). The redox function of APE1 converts its substrate proteins from oxidized inactive form to reduced active form and enhances their transcriptional activities. APE1 substrates include AP-1 (Xanthoudakis and Curran, 1992), NF-κB (Nishi et al., 2002), Egr-1 (Huang and Adamson, 1993), HIF-1α (Huang et al., 1996), p53 (Gaiddon et al., 1999), Pax protein (Tell et al., 1998) and COX-2 (Nagoya et al., 2014). It was demonstrated that the redox function of APE1 is associated with many malignancies, such as pancreatic cancer, ovarian cancer and glioblastoma (Fishel et al., 2008, 2011), and inhibition of APE1 redox function decreases cell proliferation, prevents the angiogenesis progress (Bapat et al., 2010), and blocks the differentiation of endothelial precursor cells (Zou et al., 2009).
In the current study, we investigated the possibility of curcumin being an APE1 redox inhibitor and results indicate that curcumin indeed inhibits enzymatic reaction of APE1 redox activity. Through blocking APE1-mediated redox function, curcumin effectively inhibits Kaposi’s sarcoma-associated herpesvirus (KSHV) replication and virus-associated pathogenic properties, suggesting that curcumin is an antiviral agent and has potential to be used in treatment of viral infection and associated diseases.
2. Result
2.1. Curcumin is an inhibitor to APE1 redox function
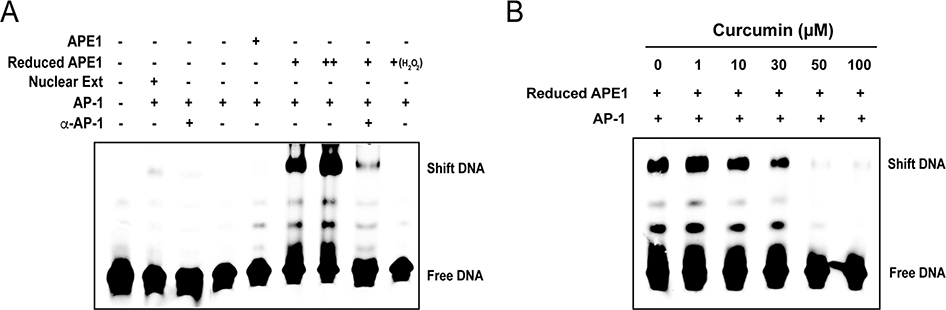
In order to determine if curcumin is an APE1 redox inhibitor, an APE1 redox enzymatic assay was established using AP-1 (c-jun/c-fos) as a substrate. Based on the fact that the ability of AP-1 binding to its DNA binding sites requires APE1 redox regulation (Xanthoudakis and Curran, 1992), electrophoresis mobility shift assay (EMSA) was employed to assess APE1 redox activity by measuring the binding ability of AP-1 to its DNA target sequence. Although the double-stranded DNA fragment containing AP-1 binding motif could be shifted when incubated with a nuclear extract, E. coli-expressed AP-1 protein complex (c-jun/c-fos heterodimer) failed to bind to the DNA fragment in the absence of APE1 (Fig. 1A). In accordance with redox regulation of APE1, the binding of AP-1 heterodimer to the double-stranded AP-1 oligonucleotides occurred only in the presence of the reduced form of APE1 (Fig. 1A). Untreated APE1 (oxidized form) showed near undetectable activity to promote AP-1 DNA binding, verifying that the AP-1 DNA binding activity correlates with the redox function of APE1 in this system. With this assay system, we examined if curcumin can inhibit APE1 redox activity and, as a consequence, block AP-1 DNA binding activity. The EMSA experiment was performed with increasing concentrations of curcumin. As shown in Fig. 1B, a dose-dependent inhibitory response by curcumin was established. This experiment clearly demonstrated that curcumin is an inhibitor to APE1 redox function.
Fig. 1. Curcumin inhibits the redox activity of APE1.
(A) An EMSA-based assay for APE1 redox activity was established with purified AP-1 (c-Jun and c-Fos) and APE-1 proteins. A double-stranded DNA carrying an AP-1 binding motif was mixed with either nuclear extract or purified c-jun/c-fos (1:1 ratio). Purified APE1 or reduced APE1 (treated with 0.25 mM DTT) were added to the reaction. AP-1-DNA binding was examined by EMSA. Nuclear extract and reduced APE1 resulted in shifted bands while oxidized APE1 (treated with H2O2) failed to support AP-1 DNA binding. (B) Increasing concentrations of curcumin were added into the reaction and the effects of them on redox activities were measured through EMSA.
2.2. Curcumin inhibits transcriptional activities by APE1 substrates AP-1 and NF-κB
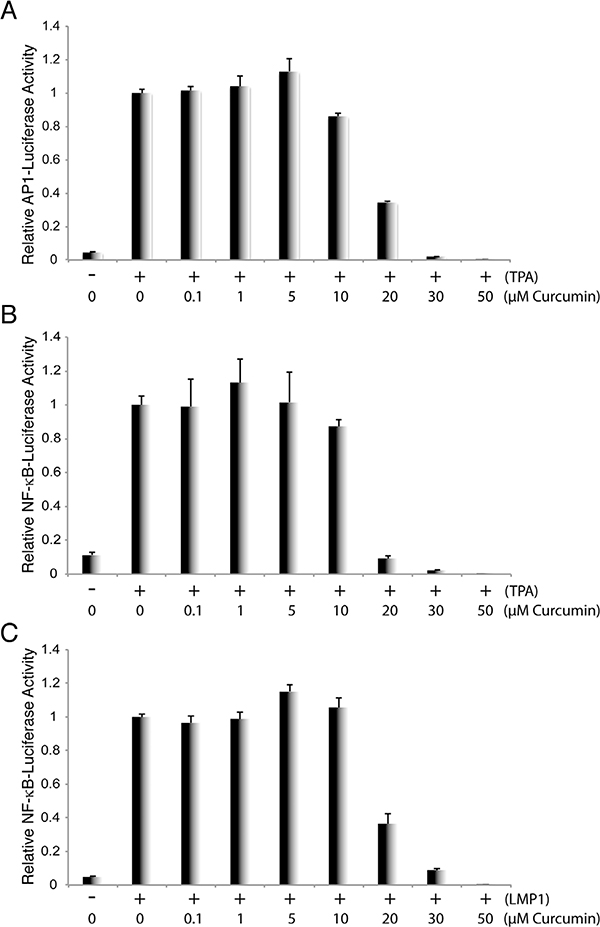
The binding of AP-1 and other APE1-regulated transcriptional factors to their specific promoter DNA leads to activation of these promoters. Therefore, we further examined the effect of curcumin on the transcription activities governed by APE1 substrates, AP-1 and NF-κB. We employed promoter-reporter assays to study the effects of curcumin on the activation of the AP-1 and NF-κB-activated promoter by these APE1-regulated transcriptional activators. 293 T cells were transfected with the AP-1 and NF-κB promoter-luciferase reporter plasmids, respectively. For the AP-1 promoter assay, cells were treated with TPA 24 h post-transfection to activate AP-1 signaling. Curcumin was added into the culture media in varying concentrations. The activation of the AP-1 promoter in the absence and presence of curcumin was measured through luciferase activity 36 h post-induction. Result shows that curcumin is able to block TPA-induced AP-1 promoter in a dose-dependent manner (Fig. 2A). In the NF-κB promoter assay, NF-κB signaling was activated by two means: treating cells with TPA and stimulating cells with EBV LMP1. The effect of curcumin on NF-κB-mediated transcriptional activities was examined using luciferase assays and results indicate that curcumin blocks NF-κB-mediated transcriptional activities in both systems (Fig. 2B and C).
Fig. 2. Curcumin blocks AP-1 and NF-κb-mediated transcription.
293 T cells were transfected with AP-1 promoter-luciferase (A) or NF-κB promoter-luciferase reporter plasmids (B), followed by induction with TPA. Curcumin in increasing concentrations were added 6 h post-induction. The promoter activities were measured through luciferase assay after 24 h (C) 293 T cells were also co-transfected with NF-κB promoter-luciferase reporter and EBV LMP1 expression vector. Curcumin in different concentrations were added 6h post-transfection and luciferase assays were performed through luciferase assay after 24 h.
2.3. Curcumin inhibits KSHV lytic replication
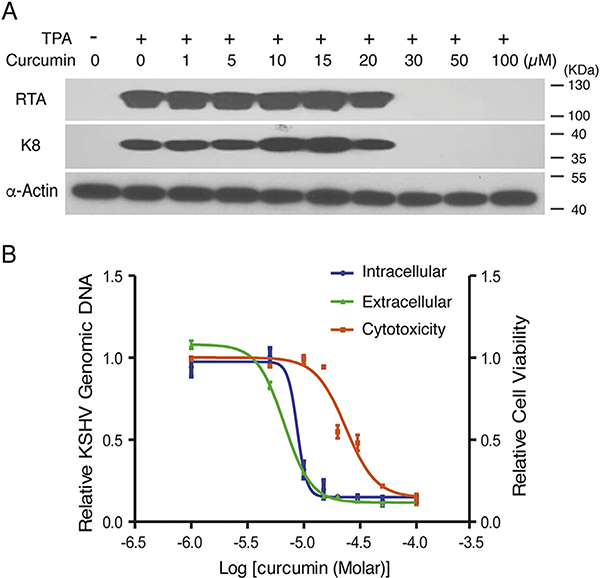
We reported previously that KSHV lytic replication is dependent on the redox function of APE1 (Zhong et al., 2017). Therefore, we wondered if the APE1 inhibitor curcumin can block KSHV lytic replication and acts as an antiviral agent. KSHV, as well as other gamma-herpesviruses including EBV, establish latency by default. Latent KSHV can be reactivated through certain signaling pathways that are under the control of APE1 redox activity (Zhong et al., 2017). For example, KSHV reactivation cascade can be initiated through activating protein kinase C (PKC), leading to stimulation of the mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK) pathway. As a consequence of the activation of the pathway, c-Fos is accumulated and c-Jun is phosphorylated, leading to the formation of an active AP-1 complex and activation of KSHV immediate-early transcriptional activator RTA and the lytic cascade of KSHV (Reviewed in Aneja and Yuan, 2017). We first determined if curcumin is able to block the expression of the KSHV switch gene RTA in a viral reactivation process as well as RTA-mediated downstream gene transcription. KSHV-carrying primary effusive lymphoma (PEL) BCBL-1 cells were treated with curcumin in a wide range of concentration 3 h after induction with TPA. The effect of curcumin on expression of RTA was analyzed using Western blot. Results showed that RTA expression in response to TPA was efficiently blocked by curcumin in a dose-dependent manner (Fig. 3A). As a consequence, the expression of delayed-early gene K8 was also inhibited in the presence of curcumin (Fig. 3A).
Fig. 3. Effect of Curcumin on KSHV lytic replication.
BCBL-1 cells were treated with a wide range of concentrations of Curcumin 3 h after being induced by TPA for viral lytic replication. (A) Whole cell extracts were prepared 48 h post-induction and subjected to Western blot analysis with anti-RTA and anti-K8 monoclonal antibodies. (B) Total DNA was purified 48-h post-induction and intracellular KSHV DNA replication was determined using real-time quantitative PCR (blue). Five days post-induction, cell culture media were collected and extracellular virions (green) were determined as described in Materials and Methods (green). Uninduced BCBL-1 cells were treated with curcumin for 120 h and cell cytotoxicity (Orange) were assessed by Trypan blue-staining procedure using a Countstar instrument. The mean values from three independent experiments and standard deviations are presented on the y-axis of dose-response curves. IC50, EC50 and CC50 are calculated using GraphPad Prism software.
Next, we analyzed the effect of curcumin on KSHV lytic DNA replication and virion production. TPA-induced BCBL-1 cells were exposed to curcumin in a wide range of concentrations. The changes of intracellular viral genomic DNA in response to curcumin treatments were determined 48 h post-induction using real-time PCR. KSHV virion production was assessed by measuring encapsidated viral DNA in the media 5 days post-induction. The half-maximal DNA replication inhibitory concentration (IC50) and half-maximal antiviral effective concentration (EC50) of curcumin were determined from the dose-response curves of the intracellular DNA and extracellular virion to be 8.76 and 6.68 μM, respectively (Fig. 3B). The cytotoxicity of curcumin was assessed for BCBL-1 using trypan blue exclusion method for cell viability in 5 days exposure to the phytochemical and the 50% cytotoxic concentration (CC50) was determined to be 23.56 μM. Based on these results, we conclude that APE-1 redox inhibitor curcumin blocks KSHV lytic replication through inhibiting TPA-induced AP-1 pathway and RTA expression.
2.4. The effect of curcumin on KSHV-mediated pathogenesis
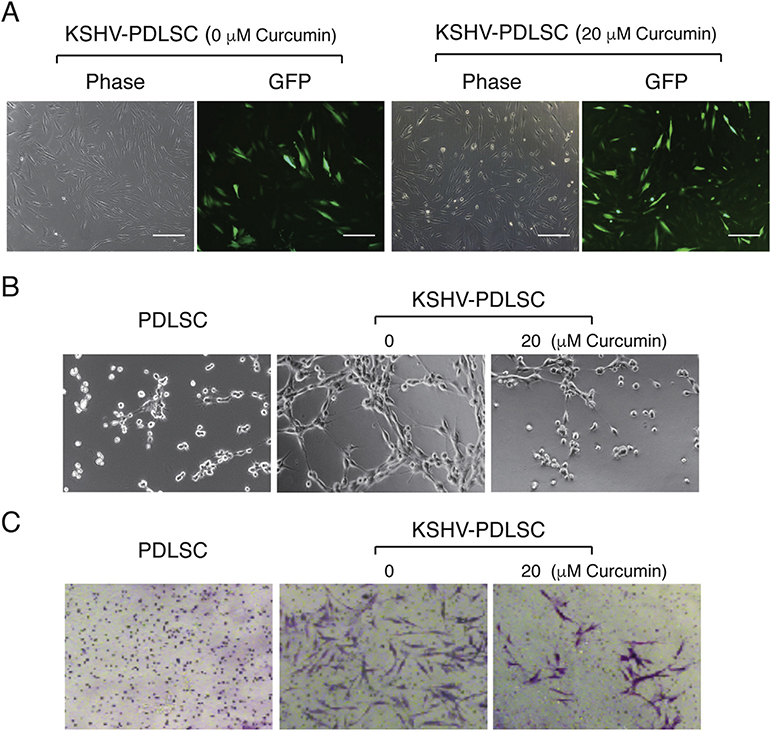
Our previous study revealed that APE1 also plays a role in KSHV-mediated oncogenesis and metastasis (Zhong et al., 2017). KS is an angiogenic and invasive tumor and abnormal neovascular channel is a pathological feature of KS. KSHV infection of mesenchymal stem cells (MSCs) confers the cells with certain KS features including angiogenic, invasive and transformation phenotypes (Zhong et al., 2017; Lee et al., 2016). When human MSCs from periodontal ligament (PDLSC) were infected with KSHV, increased angiogenesis activity was shown in an in vitro Matrigel tubulogenesis assay (Fig. 4B). We previously showed that KSHV-mediated angiogenesis depends on the maintenance of the redox status of AP-1, HIF-1α and their respective DNA binding activities, silencing APE1 expression with a specific shRNA efficiently reduced the angiogenic activity of MSCs (Zhong et al., 2017). We reasoned that curcumin, as an APE1 redox inhibitor, could inhibit KSHV-elaborated neoangiogenesis. To this question, PDLSCs were infected with KSHV in an MOI of 50 (KSHV genomic DNA equivalent) in the absence and presence of curcumin and subjected to an in vitro Matrigel tube formation assay. The presence of curcumin at 20 μM did not affect the susceptibility of PDLSC to KSHV infection (Fig. 4A), but greatly reduced the ability of KSHV-infected MSCs to form capillary-like tubules (Fig. 4B).
Fig. 4. Effect of Curcumin on KSHV-mediated angiogenesis and cell invasion of MSCs.
(A) Periodontal ligament stem cells (PDLSCs) were infected with KSHV at an MOI of 50 (viral genomic DNA equivalent) in the absence and presence of curcumin as indicated. Infectivity was shown to be 92% and 97%, respectively by GFP expression. (B) Uninfected and KSHV-infected PDLSCs were applied on Matrigel to examine the ability for tubule formation. KSHV-infected PDLSCs were placed on Matrigel in the presence of 20 μM curcumin. Tubulogenesis was examined under a microscope and quantified by measuring the total tube length using the Image J software. (C) PDLSCs or KSHV-PDLSCs (1.5 × 104 cells/well) were seeded in the upper chamber of Transwell with a layer of Matrigel. Cells migrated to the lower chamber were stained with crystal violet. KSHV-infected PDLSCs were treated with 20 μM curcumin and effect of curcumin on cell invasion were assessed by counting migrated cells in the lower chamber. Quantification of transwell-invasion assay was performed by Image J software by counting from multiple randomly selected microscopic visual fields.
Cell invasion is an important feature of KS and KSHV infection of human primary endothelial cells was reported to promote cell migration and invasion (Qian et al., 2007; Dai et al., 2012). We have shown that shRNA-mediated knock down of APE1 led to a significant decrease in cell invasion ability of KSHV-infected MSCs (PDLSCs) and an APE1 redox inhibitor also reduced the invasiveness of the KSHV-infected cells (Zhong et al., 2017). Using a Matrigel-Transwell assay, we tested if curcumin can inhibit cell invasion of KSHV-infected MSCs. Cells were stimulated for chemotaxis in a-MEM containing 10% FBS and seeded in a Matrigel on the upper chamber of a Transwell containing a-MEM free of FBS. The cells that migrated through the membrane were visualized by crystal violet staining. KSHV-infected MSC showed increased migration and invasion activity. In the presence of 20 μM of curcumin, cell invasion of KSHV-PDLSC was dramatically reduced (Fig. 4C), indicating that APE1 is required for KSHV-mediated cell invasion and APE1 redox inhibitor curcumin can efficiently block this phenotype.
3. Discussion
In the current study, we found that the high profile phytochemical curcumin is an APE1 redox inhibitor that efficiently inhibits KSHV replication, virion production and viral pathogenicity. The salient features of the results and their significance are as follows.
The finding that curcumin serves as an APE1 redox inhibitor provides explanation for the diverse-effect property of curcumin. APE1 is a multifunctional protein with DNA base excision activity in its C-terminal region and redox activity in its N-terminal domain. Both functions of APE1 have been intensively explored for therapeutic purposes. Selective inhibitors against APE1 base excision repair activity have been explored for application in combination with DNA-interactive anticancer drug to enhance the efficacy of chemotherapy (Fishel and Kelley, 2007; Wilson and Simeonov, 2010). The APE1 redox regulation of a class of transcription factors affecting cancer survival and growth makes the protein attractive to be used as a target for cancer therapeutic strategy (Tell et al., 2009; Thakur et al., 2014). Redox regulates gene expression by altering the DNA binding activity of transcription factors (Bhakat et al., 2009). The transcription factors that are affected by APE1 redox function include AP-1 (Xanthoudakis and Curran, 1992), NF-κB (Nishi et al., 2002), Egr-1 (Huang and Adamson, 1993), HIF-1α (Huang et al., 1996), HLF (Ema et al., 1999), p53 (Gaiddon et al., 1999), Pax protein (Tell et al., 1998), ATF/CREB, Myb (Xanthoudakis et al., 1992) and COX-2 (Nagoya et al., 2014). As consequences, through its redox function, APE1 affects a number of signaling pathways involved in modulating oxidative stress, cellular survival, inflammation, and cell proliferation in response to a variety of mitogenic and nonmitogenic stimuli (Tell et al., 2009; O’Hara et al., 2006).
Curcumin has been presumed to affect APE1 signaling (Bhakat et al., 2009). It was reported that curcumin decreases APE1 levels in cancer patients and treatment of follicular lymphoma patients with curcumin, in combination with epigallocatechin gallate (EGCG) from green tea, significantly lowered the levels of APE1 and substrate transcription factors (Bassiouny et al., 2011). In the current study, we found that curcumin can directly inhibit APE1 redox enzymatic activity and, as a result, reduce the transcription activities of AP-1 and NF-κB.
As an inhibitor to APE1 redox function, curcumin is predicted to be able to block APE1 redox activity in modulating pro-survival, pro-proliferation pathways and oxidative stress. Indeed, curcumin has been known to have a wide range of biological effects including anti-inflammatory, anti-oxidant and anti-neoplastic properties (Aggarwal and Harikumar, 2009). Therefore, it is possible that many of the curcumin properties, if not all, may attribute to its APE1 redox inhibition. Furthermore, clinical trials of curcumin in cancer patients revealed that treatment with curcumin led to down-regulation of certain genes and pathways, such as AP-1, NF-κB, COX-2, IL-1, IL-6, IL-8 and IL-10 (Dhillon et al., 2008; Vadhan-Raj et al., 2007). Interestingly many of them are known to be substrates of APE1 redox enzyme or their downstream genes (Tell et al., 2009; Thakur et al., 2014). Taken together, curcumin may not be a promiscuous compound, but an intracellular signaling tool involved in modulating oxidative stress and several cell functional changes by targeting one regulatory protein – APE1. In consideration of the nature of cancers or other complex diseases that always display multiple abnormalities and involve a sophisticated network system, the multiple-effect property of curcumin may not be a drawback but an advantage or even a virtue because it provides health beneficial effects by balancing a biological network system rather than shutting down a single gene.
We have previously demonstrated that APE1 redox function has critical roles in KSHV replication and pathogenic phenotypes (Zhong et al., 2017). Thus, as an APE1 redox inhibitor, curcumin is expected to possess an antiviral property against KSHV. We examined the effect of curcumin on the KSHV lytic DNA replication and progeny virion production, results showed that curcumin indeed efficiently inhibits KSHV lytic replication and blocks virion production and release. In addition, as APE1 also plays a critical role in KSHV oncogenesis including neoangiogenesis and invasion (Zhong et al., 2017), curcumin has demonstrated its efficacy in inhibiting these KSHV-associated pathogenic properties. Taken together, curcumin is a promising phytochemical with potential human health benefits. Therefore, further investigation to explore its therapeutic value and to improve its bioavailability is warranted.
4. Experimental procedures
4.1. Cells and reagents
BCBL-1 cells (obtained from the National Institutes of Health AIDS Research and Reference Reagent Program), an effusion lymphoma cell line that latently infected with KSHV, were grown in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum (FBS) and penicillin-streptomycin (50 units/ml) and 100 mg/ml amphotericin B sodium deoxycholate. Human embryonic kidney HEK293T cells were purchased from American Type Culture Collection (ATCC) and cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS and antibiotics. Periodontal ligament stem cells (PDLSCs) were isolated from the periodontal ligament tissues (n = 5 referred to 5 independent cultures from different individuals) and maintained in alpha minimal essential medium (a-MEM, GIBCO Life Technologies) containing 10% FBS, 200 mM L-glutamine and antibiotics. Curcumin [(E,E)-1,7-bis(4-Hydroxy-3-methoxyphenyl)-1,6-heptadiene-3,5-dione,Diferuloylmethane, Diferulylmethane, > 98% purity] is purchased from Fisher Scientific.
4.2. Expression and purification of APE1, c-jun and c-fos proteins in E. coli
APE1 and c-fos cDNAs were cloned into the pET-28a vector with a hexahistidine (6xHis) tagged at the N-terminus. c-jun was cloned into pGEX-4T-1 vector with a GST tag. Roseta E. coli transformed with each of the plasmids, were grown in LB media and induced with 1 mM isopropylb-D-thiogalactoside (IPTG) when the culture reached the density of 0.6 OD. The bacterial culture grew at 37 °C for 4 h for APE1 and at 25 °C for 4 h for c-jun and c-fos. Pelleted cells were resuspended and sonicated in lysis buffer containing PMSF with (for GST-tag) or without DTT (for His-tag). His-tagged APE1 and c-fos were purified with Ni2+ - NTA-resin and eluted with buffer containing imidazole. GST-tagged c-jun was purified using glutathione beads and eluted with reduced glutathione. Protein concentrations were determined using the BCA protein assay kit (Thermo Scientific).
4.3. Electrophoretic mobility shift assay (EMSA) for APE1 redox activity
An EMSA-based assay was established previously (Zhong et al., 2017) and used to measure APE1 redox activity following the APE-1-dependent AP-1 DNA binding ability. APE1 was reduced by incubating in 0.25 mM DTT over night at final concentration of 1 μM. Reduced APE1 (2 μL, the final concentration was 0.1 μM) were incubated with purified c-jun/c-fos (1:1 ratio) in EMSA reaction buffer (10 mM Tris-HCl pH 7.5, 50 mM NaCl, 5 mM MgCl2) for 20 min at 37 °C. cy5.5-labelled double-stranded DNA was added and incubated for another 20 min. The samples were resolved on 5% nondenaturing polyacrylamide gels (TBE-PAGE) at 4 °C, 100 V for 1 h and then scanned with an Odyssey imager (LI-COR). Antibodies against c-jun and c-fos (Abclone) were included in the EMSA for supershift of specific band.
4.4. Reporter plasmids and luciferase assay
The promoter-reporter plasmids pAP-1-luc and pNF-κB-luc were provided by Dr. Ersheng Kuang at Sun Yat-sen University. Subconfluent 293 T cells grown in 48-well plates were co-transfected with 50 ng of pAP1-luc or pNF-κB-luc and 5 ng of pRL-TK in aid of lipofectamine 2000 reagent (Life Technologies). Twenty-four hours after transfection, cells were treated with 12-O-Tetradecanoyl-phorbol-13-acetate (TPA). The pRL-TK plasmid expresses Renilla luciferase and was used as an internal control. For LMP1 induced pNF-κB promoter assay, 293 T cells were co-transfected with pNF-κB -luc, pRL-TK and pCR3.1-LMP1. Thirty-six hour post-induction, the luciferase assay was performed with Promega’s Dual-luciferase assay kit. Each sample was duplicated and each experiment was repeated at least three times.
4.5. Analysis of intracellular and extracellular KSHV genomic DNA content and chemical effects
BCBL-1 cells were treated with TPA to induce KSHV lytic replication. Three hour post-induction, curcumin in a wide range of concentration were added into the culture medium. Total DNA was purified using a DNasey kit (Magen) 48 h post-induction. KSHV genomic DNA copy number was quantified by real time PCR on an Applied Biosystems QuantStudio 5 instrument with primers specified for LANA (ORF73) and normalized to GAPDH. The half-maximal inhibitory concentration (IC50) values of the compound was determined from a dose-response curve of KSHV DNA content values from TPA-induced and chemical-treated cells. The viral DNA contents with those of uninduced cells subtracted were divided by those of the control cells with no drug treatment and then represented on the y-axes of dose-response curves: y-axis value = (TPAX – no TPAX)/(TPA0 – no TPA0), where X is any concentration of the drug and 0 represents nondrug treatment. The IC50 on viral DNA synthesis for the compound was calculated using GraphPad Prism software.
Extracellular virion numbers were estimated by determining encapsidated viral genomic DNA. Five days post-induction with TPA, BCBL-1 culture media were collected and virion particles were cleared by passing through 0.45-μm filters. Virions were pelleted from the medium supernatant. The virion preparations were treated with Turbo DNase I (Takara) at 37 °C for 1 h followed by proteinase K digestion. Encapsidated viral DNA was extracted with phenol-chloroform. Extracted DNA was precipitated with ice-cold ethanol, and the final DNA pellet was dissolved in TE buffer. The KSHV genomic DNA in virions was measured by real-time PCR with primers directed to LANA as described above. Virion DNA copy numbers were calculated from a standard curve established using rKSHV.219. KSHV virion numbers were presented as the copy numbers of viral genomic DNA per milliliter of culture supernatant.
4.6. Cytotoxicity assay
BCBL-1 cells were treated with curcumin in a wide range of concentration for 5 days. The viability of cells was assessed by counting Trypan blue-stained cells using a Countstar instrument. The half-maximal cytotoxic concentration (CC50) was calculated from dose-response curves with Graph-Pad Prism software.
4.7. In vitro tube formation assay
Forty eight-well plates were coated with Matrigel (100μl/well) and incubated at 37 °C for 1 h to allow gelation to occur and avoid bubble. PDLSC or KSHV-PDLSC were resuspended in 200 μl a-MEM without FBS and placed on the top of the gel. The cells were incubated at 37 °C with 5% CO2 for 8 h, and images of tube formation were captured using a ZEISS fluorescence microscope. The quantification of the tube was using the software ImageJ to measure the total length of tube in the image. The average value was used for the histogram.
4.8. Cell invasion assay
PDLSCs or KSHV-PDLSCs (1.5 × 104 cells/well) were seeded on the top of the matrigel in the upper chamber of Transwell in serum-free a-MEM. The lower chamber contained a-MEM containing 10% FBS which was used as a stimulus for chemotaxis. Cells migrated through matrigel to the lower chamber were stained with crystal violet. KSHV-infected PDLSCs were treated with curcumin (20 μM) and assessed by Transwell invasion assay. Quantification of transwell-invasion assay was performed by Image J software by counting from multiple randomly selected microscopic visual fields.
Acknowledgement
We thank members of Yuan Lab, especially Lorenzo Gonzalez-Molleda and Louis Taylor, for their support and discussion. We are grateful to Dr. Manunya Nuth at the University of Pennsylvania for technical help and constructive discussion. This work was supported by National Institutes of Health grants P01CA174439 and R01DE027901. HL is supported by a China Scholarship Council scholar project.
Footnotes
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
References
- Aggarwal BB, Harikumar KB, 2009. Potential therapeutic effects of curcumin, the anti-inflammatory agent, against neurodegenerative, cardiovascular, pulmonary, metabolic, autoimmune and neoplastic diseases. Int. J. Biochem. Cell Biol 41, 40–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aneja KK, Yuan Y, 2017. Reactivation and lytic replication of Kaposi’s sarcoma-associated herpesvirus: an update. Front. Microbiol 8, 613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bapat A, Glass LS, Luo M, Fishel ML, Long EC, et al. , 2010. Novel small-molecule inhibitor of apurinic/apyrimidinic endonuclease 1 blocks proliferation and reduces viability of glioblastoma cells. J. Pharmacol. Exp. Ther 334, 988–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhakat KK, Mantha AK, Mitra S, 2009. Transcriptional regulatory functions of mammalian AP-endonuclease (APE1/Ref-1), an essential multifunctional protein. Antioxidants Redox Signal. 11, 621–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassiouny AR, Atteya MA, El-Rashidy FH, Neenaa HM, 2011. Curcumin and EGCG suppress apurinic/apyrimidinic endonuclease 1 and induce complete remission in B-cell non-Hodgkin’s lymphoma patients. Funct. Foods Health Dis 1, 525–544. [Google Scholar]
- Clinical development plan: curcumin. J. Cell. Biochem. Suppl 1996, 26, 72–85. [PubMed] [Google Scholar]
- Dai L, Bratoeva M, Toole BP, Qin Z, Parsons C, 2012. KSHV activation of VEGF secretion and invasion for endothelial cells is mediated through viral upregulation of emmprin-induced signal transduction. Int. J. Cancer 131, 834–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon N, Aggarwal BB, Newman RA, Wolff RA, Kunnumakkara AB, Abbruzzese JL, et al. , 2008. Phase II trial of curcumin in patients with advanced pancreatic cancer. Clin. Cancer Res 14, 4491–4499. [DOI] [PubMed] [Google Scholar]
- Ema M, Hirota K, Mimura J, Abe H, Yodoi J, Sogawa K, Poellinger L, Fujii-Kuriyama Y, 1999. Molecular mechanisms of transcription activation by HLF and HIF1alpha in response to hypoxia: their stabilization and redox signal-induced interaction with CBP/p300. EMBO J. 18, 1905–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel ML, He Y, Reed AM, Chin-Sinex H, Hutchins GD, et al. , 2008. Knockdown of the DNA repair and redox signaling protein Ape1/Ref-1 blocks ovarian cancer cell and tumor growth. DNA Repair (Amst) 7, 177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel ML, Jiang Y, Rajeshkumar NV, Scandura G, Sinn AL, et al. , 2011. Impact of APE1/Ref-1 redox inhibition on pancreatic tumor growth. Mol. Cancer Therapeut 10, 1698–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel ML, Kelley MR, 2007. The DNA base excision repair protein Ape1/Ref-1 as a therapeutic and chemopreventive target. Mol. Aspect. Med 28, 375–395. [DOI] [PubMed] [Google Scholar]
- Gaiddon C, Moorthy NC, Prives C, 1999. Ref-1 regulates the transactivation and proapoptotic functions of p53 in vivo. EMBO J. 18, 5609–5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta SC, Patchva S, Aggarwal BB, 2013. Therapeutic roles of curcumin: lessons learned from clinical trials. AAPS J. 15, 195–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RP, Adamson ED, 1993. Characterization of the DNA-binding properties of the early growth response-1 (Egr-1) transcription factor: evidence for modulation by a redox mechanism. DNA Cell Biol. 12, 265–273. [DOI] [PubMed] [Google Scholar]
- Huang LE, Arany Z, Livingston DM, Bunn HF, 1996. Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its alpha subunit. J. Biol. Chem 271, 32253–32259. [DOI] [PubMed] [Google Scholar]
- Lee MS, Yuan H, Jeon H, Zhu Y, Yoo S, et al. , 2016. Human mesenchymal stem cells of diverse origins support persistent infection with Kaposi’s sarcoma-associated herpesvirus and manifest distinct angiogenic, invasive, and transforming phenotypes. mBio 7, e02109–02115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson KM, Dahlin JL, Bisson J, Graham J, Pauli GF, Walters MA, 2017. The essential medicinal chemistry of curcumin. J. Med. Chem 60, 1620–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi T, Shimizu N, Hiramoto M, Sato I, Yamaguchi Y, et al. , 2002. Spatial redox regulation of a critical cysteine residue of NF-kappa B in vivo. J. Biol. Chem 277, 44548–44556. [DOI] [PubMed] [Google Scholar]
- Nagoya H, Futagami S, Shimpuku M, Tatsuguchi A, Wakabayashi T, et al. , 2014. Apurinic/apyrimidinic endonuclease-1 is associated with angiogenesis and VEGF production via upregulation of COX-2 expression in esophageal cancer tissues. Am. J. Physiol. Gastrointest. Liver Physiol 306, G183–G190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara AM, Bhattacharyya A, Mifflin RC, Smith MF, Ryan KA, Scott KG, Naganuma M, Casola A, Izumi T, Mitra S, Ernst PB, Crowe SE, 2006. Interleukin-8 induction by Helicobacter pylori in gastric epithelial cells is dependent on apurinic/apyrimidinic endonuclease-1/redox factor-1. J. Immunol 177, 7990–7999. [DOI] [PubMed] [Google Scholar]
- Prasad S, Gupta SC, Tyagi AK, Aggarwal BB, 2014. Curcumin, a component of golden spice: from bedside to bench and back. Biotechnol. Adv 32, 1053–1064. [DOI] [PubMed] [Google Scholar]
- Qian LW, Xie J, Ye F, Gao SJ, 2007. Kaposi’s sarcoma-associated herpesvirus infection promotes invasion of primary human umbilical vein endothelial cells by inducing matrix metalloproteinases. J. Virol 81, 7001–7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schraufstatter E, Bernt H, 1949. Antibacterial action of curcumin and related compounds. Nature 164, 456. [DOI] [PubMed] [Google Scholar]
- Strimpakos AS, Sharma RA, 2008. Curcumin: preventive and therapeutic properties in laboratory studies and clinical trials. Antioxidants Redox Signal. 10, 511–545. [DOI] [PubMed] [Google Scholar]
- Tell G, Quadrifoglio F, Tiribelli C, Kelley MR, 2009. The many functions of APE1/Ref-1: not only a DNA repair enzyme. Antioxidants Redox Signal. 11, 601–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tell G, Pellizzari L, Cimarosti D, Pucillo C, Damante G, 1998. Ref-1 controls pax-8 DNA-binding activity. Biochem. Biophys. Res. Commun 252, 178–183. [DOI] [PubMed] [Google Scholar]
- Thakur S, Sarkarm B, Choliam RP, Gautam N, Dhiman M, et al. , 2014. APE1/Ref-1 as an emerging therapeutic target for various human diseases: phytochemical modulation of its functions. Exp. Mol. Med 46, e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadhan-Raj S, Weber D, Wang M, Giralt S, Alexanian R, Thomas S, et al. , 2007. Curcumin downregulates NF-κB and related genes in patients with multiple myeloma: results of a phase 1/2 study. Blood 110, 357. [Google Scholar]
- Wilson DM 3rd, Simeonov A, 2010. Small molecule inhibitors of DNA repair nuclease activities of APE1. Cell. Mol. Life Sci 67, 3621–3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xanthoudakis S, Miao G, Wang F, Pan YC, Curran T, 1992. Redox activation of Fos-Jun DNA binding activity is mediated by a DNA repair enzyme. EMBO J. 11, 3323–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xanthoudakis S, Curran T, 1992. Identification and characterization of Ref-1, a nuclear protein that facilitates AP-1 DNA-binding activity. EMBO J. 11, 653–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong C, Xu M, Wang Y, Xu J, Yuan Y, 2017. An APE1 inhibitor reveals critical roles of the redox function of APE1 in KSHV replication and pathogenic phenotypes. PLoS Pathog. 13, e1006289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou GM, Karikari C, Kabe Y, Handa H, Anders RA, et al. , 2009. The Ape-1/Ref-1 redox antagonist E3330 inhibits the growth of tumor endothelium and endothelial progenitor cells: therapeutic implications in tumor angiogenesis. J. Cell. Physiol 219, 209–218. [DOI] [PubMed] [Google Scholar]