Abstract
The nociceptive transient receptor potential vanilloid 1 (TRPV1) ion channel is a polymodal receptor for multiple painful stimuli, hence actively pursued as a target for analgesic drugs. We identified a small peptide toxin RhTx2 from the Chinese red-headed centipede that strongly modulates TRPV1 activities. RhTx2, a 31-amino-acid peptide, is similar to a TRPV1-activating toxin RhTx we have previously discovered but with four extra amino acids at the N terminus. We observed that, like RhTx, RhTx2 activated TRPV1, but RhTx2 rapidly desensitized the channel upon prolonged exposure. Desensitization was achieved by reducing both the open probability and the single-channel conductance. RhTx2 is not only a tool to study the desensitization mechanism of TRPV1, but also a promising starting molecule for developing novel analgesics.
Keywords: Peptide toxins, TRPV1, Nociception, Desensitization, Gating mechanisms
1. Introduction
TRPV1 ion channel is a non-selective cation channel highly expressed in dorsal root ganglion nociceptive neurons (Caterina et al., 1997). It has been shown that, when TRPV1 was inhibited by antagonists or genetically knocked out, pain sensation was largely attenuated in animal models, validating TRPV1 as a potential target for analgesic drugs (Caterina et al., 2000). As a polymodal receptor, TRPV1 is activated by a plethora of physical and chemical stimuli such as noxious heat above 40 °C, capsaicin in chili peppers, extracellular acidification and divalent cations, as well as peptide toxins (Tominaga et al., 1998; Zheng, 2013; Yang et al., 2010; Yang et al., 2015; Yang and Zheng, 2017; Yang et al., 2018; Cao et al., 2014; Yang et al., 2014). In particular, the sting or bite by venomous animals such as scorpions and centipedes elicits excruciating pain due to TRPV1-activating toxins in their venoms (Siemens et al., 2006; Yang et al., 2015; Yang et al., 2017). Previously we have identified a peptide toxin, RhTx, from the venom of Chinese red-headed centipede that strongly activates TRPV1 (Yang et al., 2015). RhTx is a 27-amino-acid short peptide containing two pairs of disulfide bonds. Its activation of TRPV1 can explain the burning pain caused by centipede bites.
Peptide toxins have been valuable tools for the study of ion channels (Zheng and Trudeau, 2015). For example, scorpion toxins like CTX were used to probe the pore of potassium channels (Hidalgo and MacKinnon, 1995). For structural study of ion channels, spider toxin DkTx was employed to stabilize and acquire the open state structure of TRPV1 channel (Cao et al., 2013; Bohlen et al., 2010). ProTx II and HwTx IV assisted the structure determination of voltage-gated sodium channels (Shen et al., 2018). Using RhTx, we have previously demonstrated that the outer pore domain of TRPV1 is critically involved in its heat activation process. We also observed that the channel desensitization induced by RhTx is mechanistically similar to the heat desensitization (Yang et al., 2015; Luo et al., 2019).
Besides being tools for scientific research, peptide toxins have been developed as drugs because of their high affinity and selectivity for targets (Robinson et al., 2017; Ombati et al., 2018). For instance, the conotoxin ziconotide is an analgesic drug targeting voltage-gated calcium channels (Safavi-Hemami et al., 2019). TRPV1 is also a validated target for analgesics (Julius, 2013). However, as TRPV1 is critically involved in body temperature regulation (Gavva et al., 2007), small molecule blockers of this channel failed in clinical trials as they often caused severe side effects such as altered body temperature and heat sensation in patients (Gavva, 2009; Garami et al., 2018). Therefore, to develop novel molecules targeting TRPV1 as analgesics, alternative strategies that can inhibit TRPV1 activities are required.
In this study, we have identified a variant of RhTx from the venom of the Chinese red-headed centipede Scolopendra subspinipes mutilans, which contains four more residues at the N terminus. We named this 31-residue peptide toxin RhTx2. Like RhTx, RhTx2 activated TRPV1; however, RhTx2 desensitized the channel with significantly faster kinetics. We explored the structural mechanisms underlying toxin binding and desensitization with computational modeling and docking in combination of functional analyses. Our results suggest that a distinct structural mechanism underlies RhTx2 interaction with TRPV1.
2. Results
2.1. Identification of RhTx2
From the venom of the Chinese red-headed centipede (Fig. 1A), we previously found a 27-residue peptide toxin RhTx (Yang et al., 2015). The gene encoding RhTx translates into a 69-residue peptide (Fig. 1B), which is much larger than RhTx. We hypothesized that the 69-residue peptide undergoes post-translational modification, which likely yields multiple matured peptides including RhTx. We further purified the venom fraction containing RhTx with molecular weight from 2000 to 8000 Da (dashed box, Fig. 1B) using a C18 RP-HPLC (Waters, Milford, CT, USA) column. We identified a peptide, termed RhTx2, from the fraction indicated by an arrow in red (Fig. 1C) that is identical to RhTx except for four additional residues (NSKY) at its N terminus (Fig. 1D). We determined the molecular weight of native RhTx2 to be 3458.8 Da by matrix-assisted laser desorption/ionization time-of-flight (MALDI--TOF) mass spectrometry (Bruker Daltonik GmbH, Leipzig, Germany) (Fig. 1E), which is consistent with its amino acid sequence (Fig. 1D, highlighted in yellow). To facilitate functional studies of RhTx2, we performed refolding of chemically synthesized RhTx2 peptide. The molecular weight of our refolded RhTx2 is 4 Da less than that of the linear peptide (Fig. 1F and G, respectively), supporting the formation of two disulfide bonds. Co-elution of the native and refolded RhTx2 exhibited only one peak in HPLC, which is distinct from that of linear RhTx2 peptide (Fig. 1H). These results demonstrated that the conformation of synthesized and refolded RhTx2 is similar to that of the native toxin with two disulfide bonds formed between Cys9-Cys20 and Cys14-Cys27, respectively. Therefore, we used the synthesized and refolded RhTx2 in all functional studies.
Fig. 1. Identification and purification of RhTx2 from centipede venom.
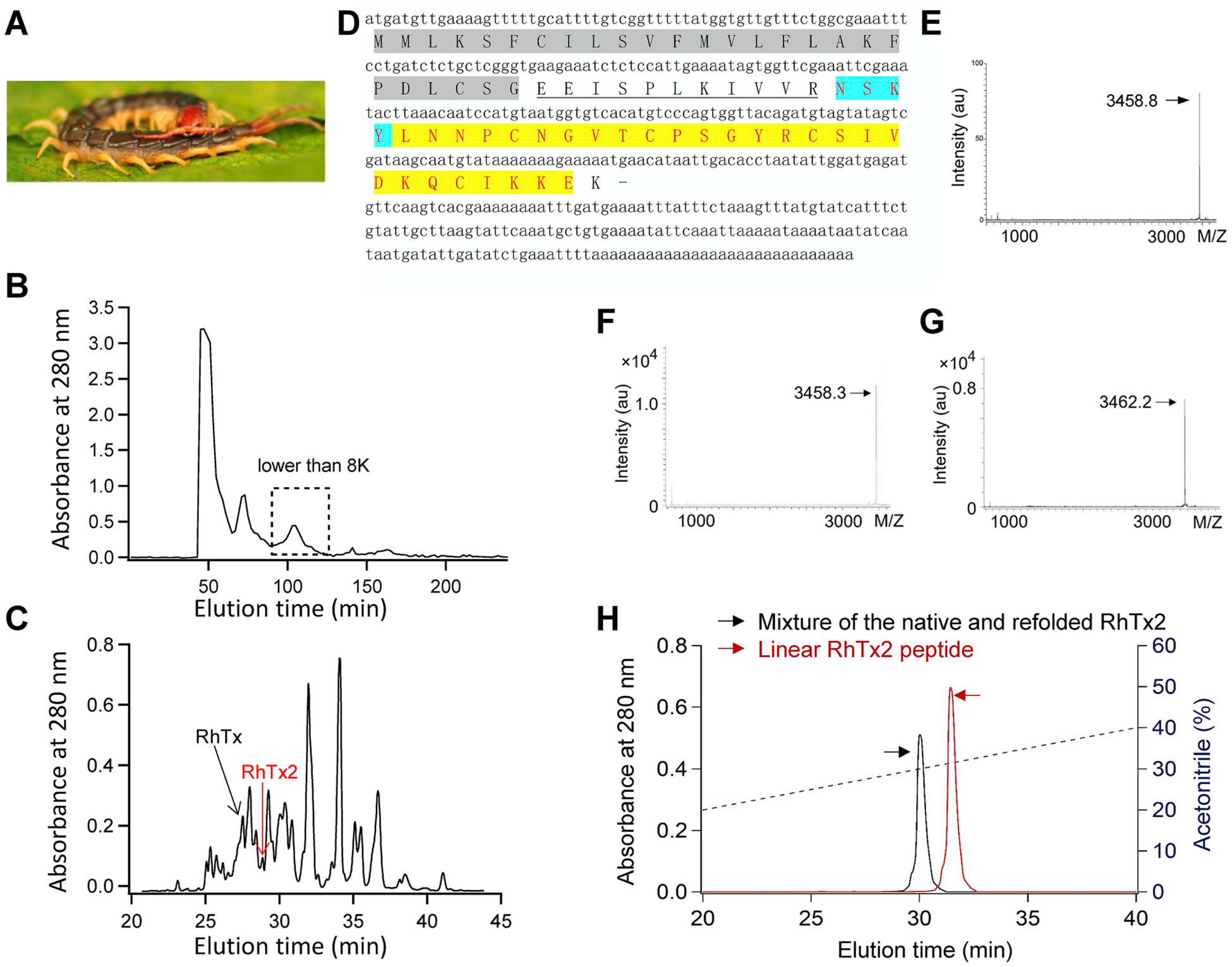
(A) The Chinese red-headed centipede Scolopendra subspinipes. (B) Centipede venom was fractionated using Sephadex G-50 gel filtration. (C) Fraction with molecular weight from 2000 to 8000 Da (dashed box in (B)) was further fractionated by an analytical C18 RP-HPLC column. (D) The cDNA and primary protein sequence of RhTx2. The signaling peptide was shaded and the propeptide was underlined, while the matured peptide sequence was highlighted in yellow. (E) The molecular weight of the purified native RhTx2 was determined by MALDI-TOF to be 3458.8 Da. (F and G) The molecular weight of the linear and refolded RhTx2 peptide was determined by MALDI-TOF to be 3458.3 Da and 3462.2 Da, respectively. (H) Co-elution of native and synthesized and refolded RhTx2 in HPLC. The elution profile of the mixture of native and refolded RhTx2, and the linear peptide of RhTx2 are colored in black and red, respectively.
We have previously determined the structure of RhTx with NMR (Yang et al., 2015), which forms a compact structure stabilized by two pairs of disulfide bonds (Fig. 2A, PDB ID: 2MVA). We have computationally modeled the structure of RhTx2 with four more residues. After three rounds of loop modeling using the Rosetta computational structural biology suite (Leaver-Fay et al., 2011), the model with the most favorable energy suggested that the N terminal NSKY residues adopted a helical conformation (dashed box in red, Fig. 2B and E). As compared to RhTx (Fig. 2A and C), RhTx2 exhibited a large patch of positively charged electrostatic surface (blue, Fig. 2D). Such a patch is formed by three residues: K3, R19 and K30. The orientation of the side chain in the extra residue K3 is similar to that of R19, so they form a large patch together. The N1 residue likely forms two hydrogen bonds with the last residue E31 (Fig. 2F, dotted lines in blue), which is expected to stabilize the conformation of the N terminus in RhTx2. Because the extracellular side of plasma membrane is negatively charged, these four extra residues are expected to impose a profound impact on the function of RhTx2.
Fig. 2. Structural modeling and characterization of RhTx2.
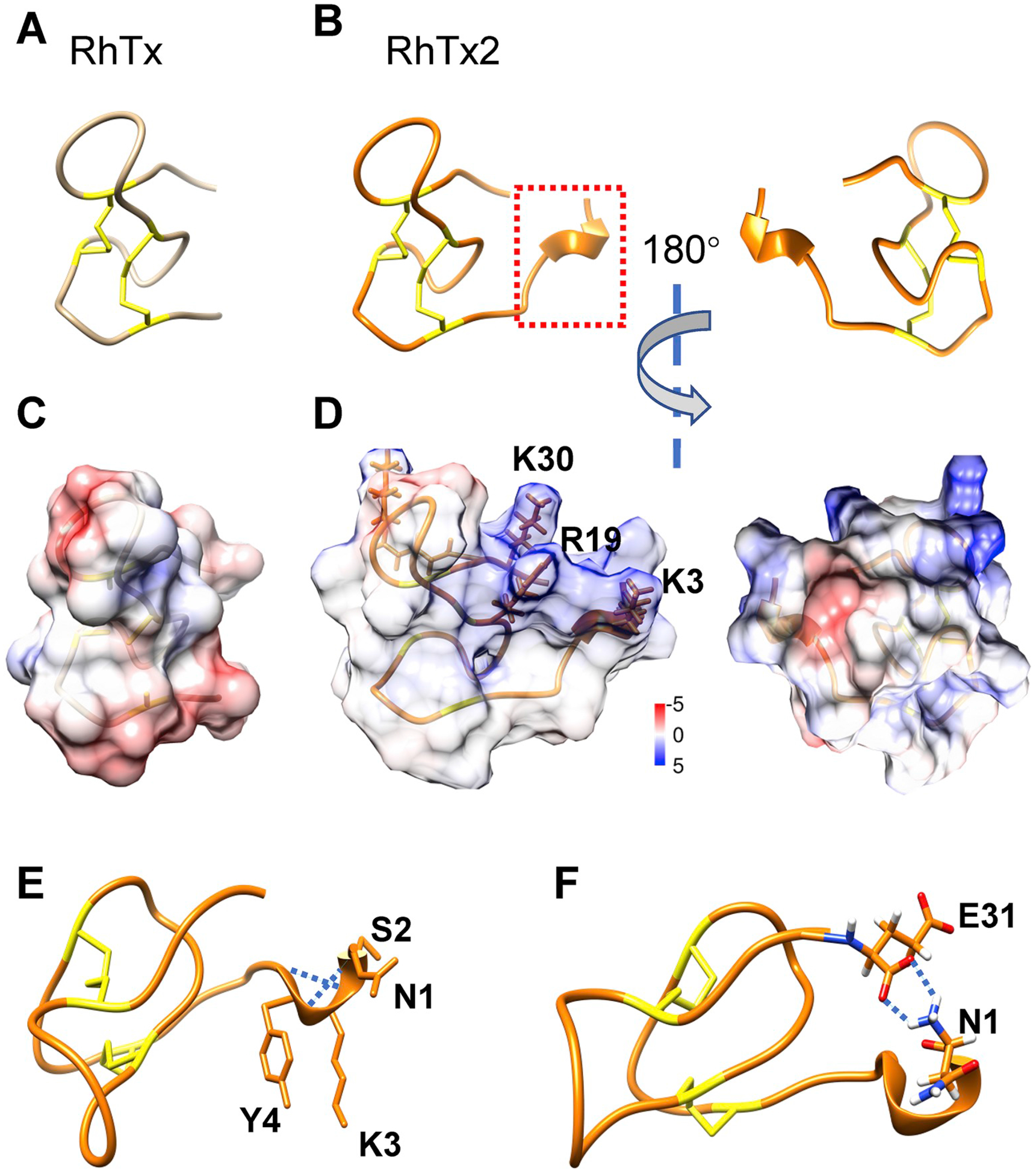
(A) NMR structure of RhTx (PDB ID: 2MVA). Two disulfide bonds are colored in yellow. (B) Rosetta model of RhTx2, with the extra four residues at the N terminus marked with a dashed box in red. (C and D) Distribution of electrostatic potential on the surface of RhTx (C) and RhTx2 (D), respectively. Coulombic electrostatic potential, in unit of kcal/mol/e, was calculated in the UCSF Chimera software, with positive and negative potential colored in blue and red, respectively. (E) The residues in the N terminus of RhTx2 forms a helical structure, which is stabilized by hydrogen bonds (dotted lines in blue). (F) The N1 residue forms two hydrogen bonds (dotted lines in blue) with the E31 residue, which stabilizes the N terminus conformation.
2.2. RhTx2 activates and desensitizes TRPV1 channel
To characterize the function of RhTx2, we first tested whether it activates TRPV1 like RhTx. With patch-clamp recording, we observed that when RhTx2 was perfused from extracellular side, it robustly activated TRPV1 expressed in HEK293T cells (Fig. 3A). RhTx2 applied to the intracellular side failed to open TRPV1 (Fig. 3B and C), indicating that RhTx2 also binds to the outer pore region of TRPV1 like RhTx (Yang et al., 2015). Consistently, RhTx2 activation cannot be blocked by capsazepine as the mixture of RhTx2 and capsazepine (289 μM and 300 μM, respectively) still activated TRPV1 (Fig. 3D). Capsazepine is a competitive inhibitor to TRPV1 agoinist capsaicin, which binds to the transmembrane domains of TRPV1(Cao et al., 2013; Yang et al., 2015). However, the concentration-response curve of RhTx2 was rightward shifted, showing that the EC50 value was increased from 0.47 ± 0.03 μM of RhTx to 38.35 ± 4.61 μM for RhTx2 (Fig. 3E). The Hill coefficients of RhTx and RhTx2 were similar (1.02 ± 0.06 and 0.79 ± 0.08, respectively). This nearly 100-fold change in EC50 suggests that apparent binding affinity of RhTx2 to TRPV1 was much lower than RhTx. As compared to TRPV1, RhTx2 induced current activation in the closely related TRPV2 and TRPV3 channels was much smaller (Fig. 4A–C), indicting the toxin is selective for TRPV1 channel.
Fig. 3. Activation and desensitization of TRPV1 by RhTx2.
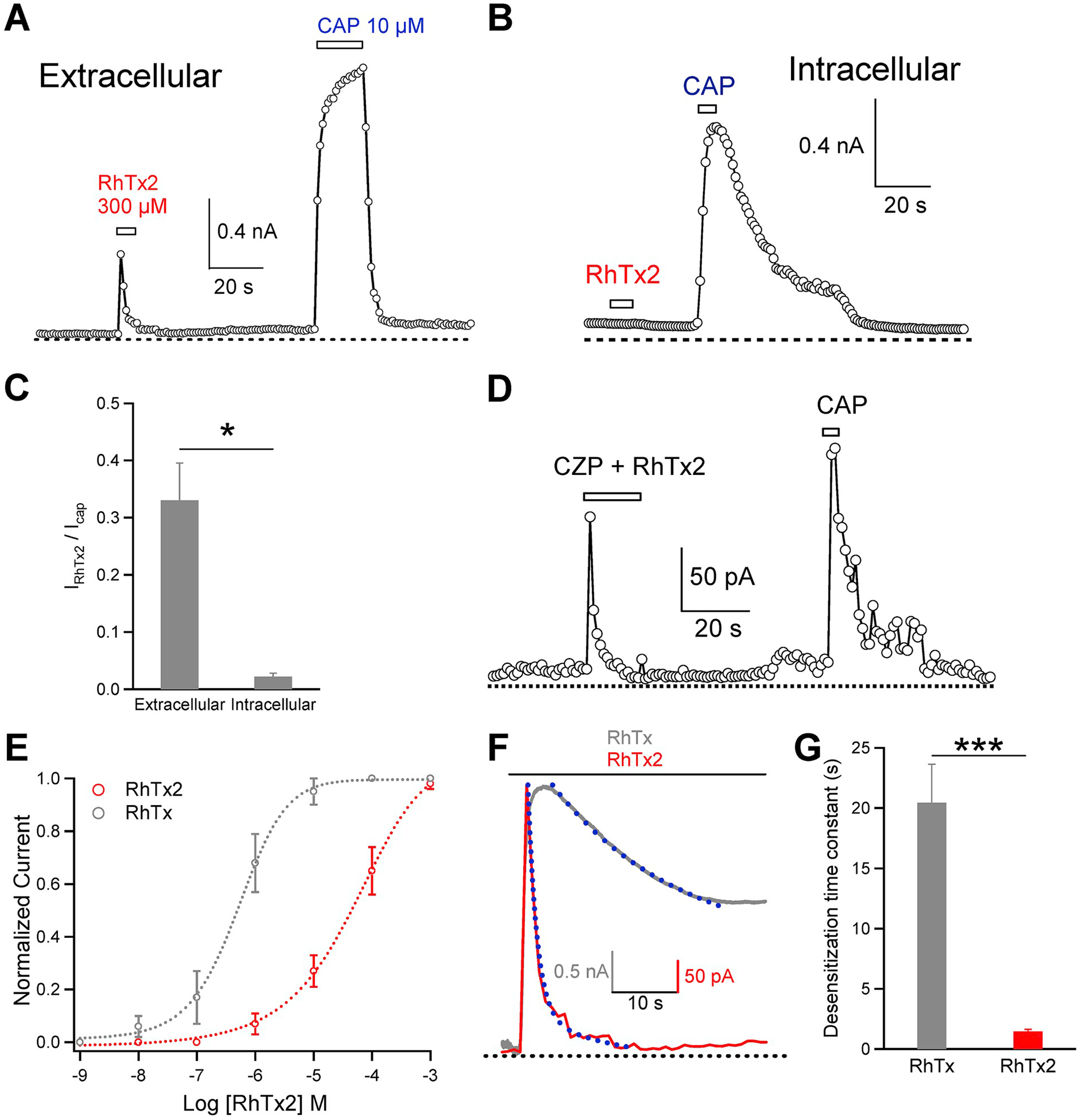
(A) RhTx2 perfused from the extracellular side activated TRPV1 channels in a representative whole-cell recording. Due to rapid desentitization, RhTx2-induced current was smaller in amplitude than capsaicin-induced current. (B) RhTx2 perfused to the intracellular side failed to activate TRPV1 channels in a representative inside-out recording, whereas these channels were robustly activated by capsaicin. (C) Comparison of TRPV1 current elicited by either extracellular or intracellular perfusion of RhTx2 at saturating concentration. (D) Represenstative outside-out recording of TRPV1 activated by extracellular perfusion of a mixture of RhTx2 and capsazepine (289 μM and 300 μM, respectively). (E) Concentration-response curves of TRPV1 activation by RhTx and RhTx2, respectively (n = 3-to-5). (F) Comparison of the kinetics in TRPV1 desensitization by RhTx and RhTx2 (10 μM and 300 μM, respectively). Superimposed are exponential fits of the current time course. (G) Time constant of TRPV1 desensitization by RhTx and RhTx2 (n = 4-to-6). The time constant was determined by fitting an exponential function to the current desensitization time course. All the electrophysiological experiments are done with the synthetic and refolded toxin.
Fig. 4. Selectivity of RhTx2.
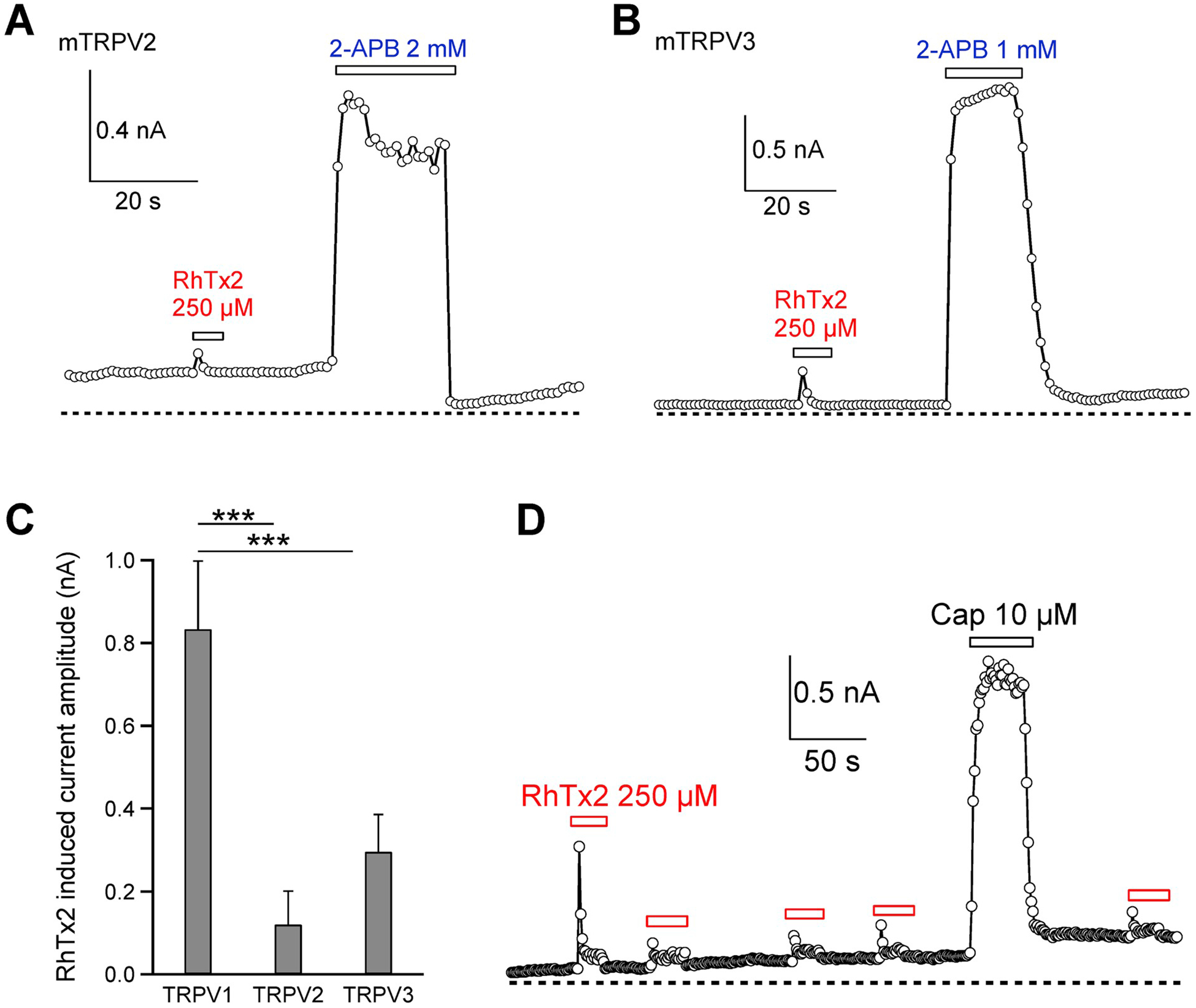
(A and B) Representative whole-cell current recordings of RhTx2 activation of TRPV2 and TRPV3, respectively. (C) Comparison of current activation by RhTx2 in TRPV channel. For each channel, n = 4. (D) Representative whole-cell current recordings of TRPV1 activation by RhTx2 and potential recovery from desensitization. All the electrophysiological experiments are done with the synthetic and refolded toxin.
Besides changes in EC50, another striking feature of RhTx2 was that it induced rapid TRPV1 desensitization upon prolonged application (Fig. 3A). Though RhTx also desensitized TRPV1 with prolonged application (Luo et al., 2019) (Fig. 3F), desensitization by RhTx2 was significantly faster (Fig. 3F and G). Moreover, with RhTx2 the steady state open probability in the desensitized state was also significantly lower than that of RhTx (Fig. 3F), suggesting that RhTx2 leads to more complete TRPV1 desensitization. We further measured the recovery of TRPV1 from RhTx2 induced desensitization (Fig. 4D). We found that upon desensitization, little recovery was observed in about 6 min, indicating the recovery is a fairly slow process. Desensitization serves as an important way to regulate ion channel functions. RhTx2 with a fast desensitization kinetics is a promising starting point for understanding the mechanisms of channel desensitization as well as for developing an inhibitor of TRPV1 ion channel.
2.3. Mechanisms of TRPV1 desensitization by RhTx2
We explored the mechanisms of RhTx2-induced desensitization with patch-clamp. With single-channel recordings, we observed that on the same membrane patch of TRPV1 channels, perfusion of 300 μM RhTx2 first induced strong channel opening (as reflected in current levels from O1 to O5, Fig. 5A). As the channels started to desensitize, both their open probabilities and single-channel currents were reduced (Fig. 5A and B). In fact, the single-channel conductance was significantly reduced from 154.4 ± 10.8 pS to 65.8 ± 6.7 pS (Fig. 5C). Therefore, desensitization of TRPV1 current involves two mechanisms: decrease of open probability and decrease in single-channel conductance upon prolonged RhTx2 application.
Fig. 5. Mechanisms of RhTx2-induced desensitization.
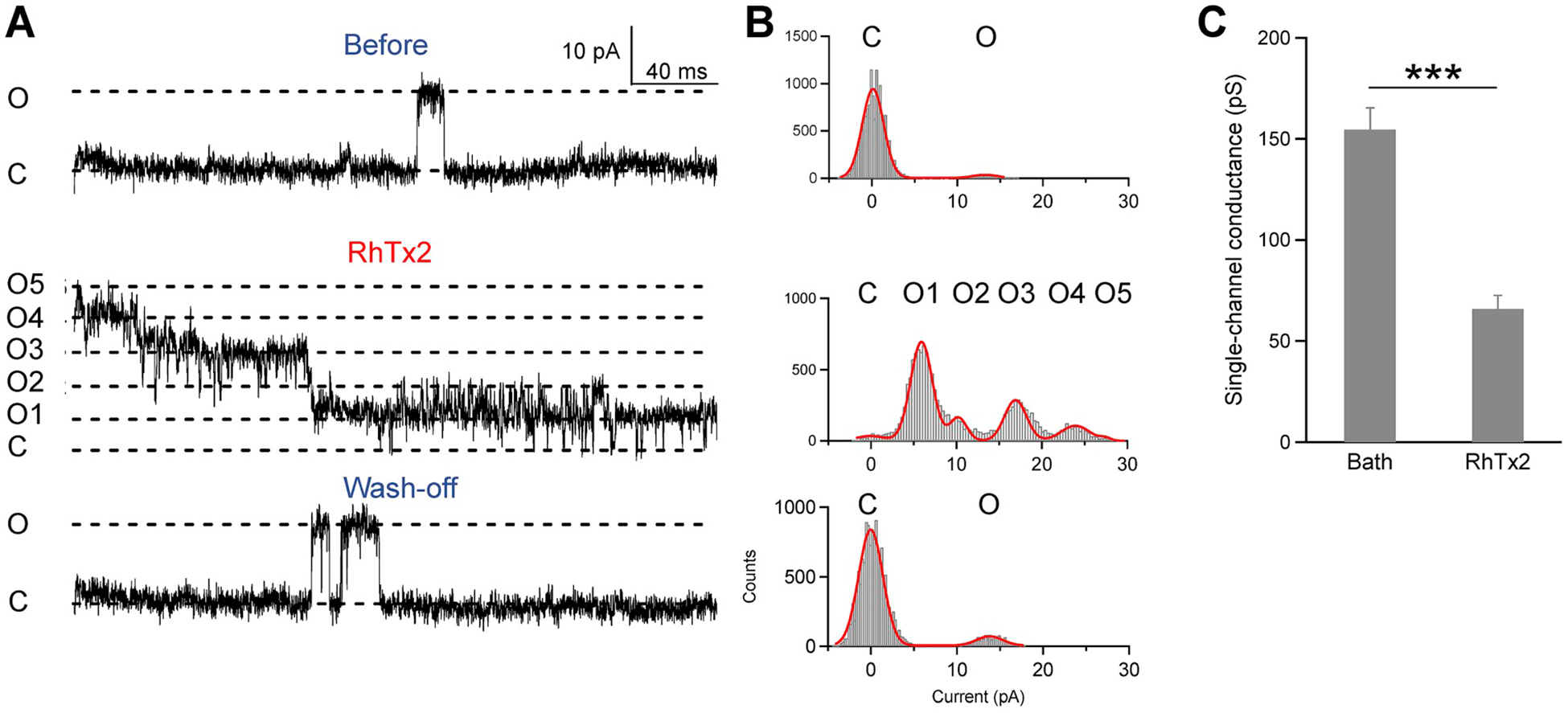
(A) From the same outside-out patch recording of TRPV1 channels, single-channel opening events were rare before RhTx2 application (top) or after wash-off (bottom). When RhTx2 was perfused onto this membrane patch, current from five channels (O1 to O5) was immediately observed (middle). The current started to desensitize as both channel open probability and single-channel current amplitude decreased over time. (B) All-point histograms of the representative single-channel recordings shown in (A). Histograms were fitted to a sum of Gauss functions. (C) Single-channel conductance was significantly reduced by RhTx2 (n = 4-to-6). All the electrophysiological experiments are done with the synthetic and refolded toxin.
2.4. Potential structural mechanisms underlying toxin binding and channel desensitization
Since RhTx binds to the outer pore region above the pore helix to activate TRPV1 channel (Yang et al., 2015), we docked the RhTx molecule to both the closed and open state structures of TRPV1 (Fig. 6A–D). From docking we observed that in both states, RhTx binds to similar out pore region of TRPV1, though the relative orientation of the RhTx molecule is different in these two states. Given that the outer pore region undergoes substantial conformational changes as revealed by the cryo-EM structures (Cao et al., 2013; Liao et al., 2013) (PDB ID: 3J5P and 3J5Q for the closed and open states, respectively) as well as FRET imaging in live cells (Yang et al. 2010, 2014), it is reasonable that the binding configurations of RhTx molecule are different. Furthermore, we reasoned that the structurally similar RhTx2 may also bind to the vicinity of pore helix to alter both the open probability and single-channel conductance. To investigate how RhTx2 binds and desensitizes TRPV1 channel, we computationally docked RhTx2 to the cryo-EM structure of TRPV1 channel in the closed state and toxin-activated state (PDB ID: 3J5P and 3J5Q, respectively) (Cao et al., 2013; Liao et al., 2013). We observed that the docking configurations of RhTx2 to the closed state of TRPV1 is indeed similar to that of RhTx, where they resided above the pore helix in the outer pore region (Fig. 6E and F). With positively charged Lys in the N terminus of RhTx2 (Fig. 2D), this helical domain pointed downward to the negatively charged cell membrane surface (dashed box in red and shaded green, respectively. Fig. 6F).
Fig. 6. Docking of RhTx and RhTx2 to TRPV1 channel. (A and B) Top and side view of RhTx (colored in magenta) docked to TRPV1 in the apo and closed state (PDB ID: 3J5P), respectively. The position of cell membrane is marked by a shaded box.
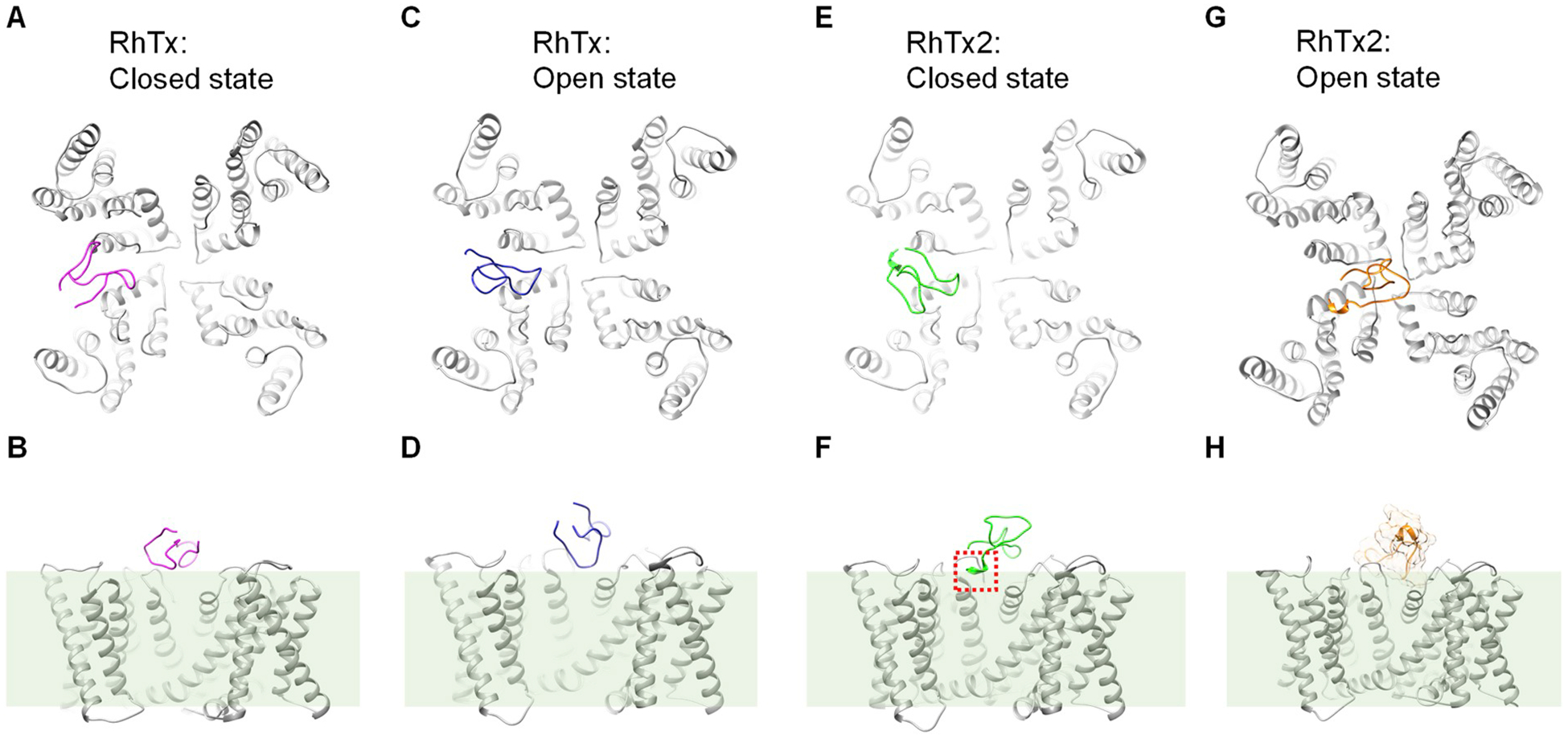
(C and D) Top and side view of RhTx (colored in blue) docked to TRPV1 in the toxin-activated open state (PDB ID: 3J5Q), respectively. (E and F) Top and side view of RhTx2 (colored in green) docked to TRPV1 in the apo and closed state (PDB ID: 3J5P), respectively. The extra four residues at the N terminus of RhTx2 is boxed in red. (G and H) Top and side view of RhTx2 (colored in orange) docked to TRPV1 in the toxin-activated open state (PDB ID: 3J5Q), respectively. The surface of RhTx2 molecule was shown in (H), where only the transmembrane domains of TRPV1 was displayed.
In single-channel recordings, we observed that when RhTx2 desensitized TRPV1 channel, the single-channel conductance was significantly reduced (Fig. 5). Consistent with this observation, when RhTx2 was docked to the open state of TRPV1, it bound preferentially right above the entrance of selectivity filter (Fig. 6G), where it may sterically hinder the permeation of ions through the selectivity filter (Fig. 6H). Therefore, our docking experiments suggested that when RhTx2 initially contacts TRPV1 channel which is still in the closed state, it binds to the periphery of the selectivity filter to first open the channel. As TRPV1 transits from the closed state to the open state, the binding configuration of RhTx2 changes to a central position above the selectivity filer, where it hinders ion permeation through the channel pore.
3. Discussion
In this study we have identified a peptide toxin RhTx2 from the venom of Chinese red-headed centipede (Fig. 1). Like RhTx (Yang et al., 2015), RhTx2 also activates the nociceptive TRPV1 channel, but prolonged perfusion of RhTx2 rapidly desensitizes TRPV1 by reducing both the channel open probability and single-channel conductance (Fig. 5). We reasoned that such differences in the functional properties of RhTx2 is largely attributed to the extra four residues NSKY at its N terminus as compared to RhTx. Our previous NMR study has revealed that the conformation of the N terminus of RhTx is highly flexible (Yang et al., 2015). In RhTx2, the four extra residues likely formed a helical structure based on our modeling studies (Fig. 2E). Moreover, with the addition of the four extra residues in RhTx2, in our structural model of RhTx2 the N1 residue likely forms two hydrogen bonds with the last residue E31 (Fig. 2F, dotted lines in blue), which is expected to stabilize the conformation of the N terminus in RhTx2. As the positively charged K brings the N terminus of RhTx2 towards the cell membrane, like a lever it alters the binding conformation of other parts of the toxins, which further may lead to sterical hinderence of the binding of RhTx2 and changes in binding affinity. Indeed, the concentration-response curve of TRPV1 activation of RhTx2 was largely right-shifted (Fig. 3D).
In addition, the K3 within the extra four residues introduced a positive charge that drastically changes surface electrostatic potential (Fig. 2D). Electrostatic interactions are one of the major forces that determine protein folding and interaction. In cation channels like voltage-gated potassium, sodium and calcium channels as well as TRP channels, there are many negatively charged residues in the outer pore region that attract and concentrate cations (Doyle et al., 1998; Wu et al., 2015; Shen et al., 2018; Liao et al., 2013). Moreover, the surface of the cell membrane is also negatively charged because of the phosphate group within the phospholipids. Therefore, it is not surprising for the positively charged N terminus of RhTx2 to be in the proximity of cell membrane as suggested by our docking experiments (Fig. 6G and H). Likely though this mechanism, the binding configuration of RhTx2 is altered as compared to that of RhTx, leading to changes in functional properties of RhTx2. Such a mechanism of toxin binding has been previously exploited to optimize existing toxins. For instance, the binding configuration of scorpion toxin BmKTX on Kv1.3 channel was changed by introducing a single negatively charged point mutation, which increased the selectivity of the toxin to Kv1.3 channel (Chen et al. 2014, 2015).
Peptide toxins are a promising starting point for the development of biologic drugs due to relatively small size, high affinity and high selectivity (Robinson et al., 2017; Ombati et al., 2018). Specifically for the analgesic target TRPV1 ion channel, direct inhibition of the channel failed in clinical trials because of severe side effects (Gavva, 2009). For this reason, alternative strategies to modulate TRPV1 channel have been actively pursued. For instance, a small molecule MRS1477 can positively and allosterically modulate the ligand activation of TRPV1, leading to analgesic effects without side effects such as hyperthermia in animal studies (Kaszas et al., 2012; Lebovitz et al., 2012). Interestingly, peptide toxins APHC1 and APHC3 from sea anemone Heteractis crispa have been reported to antagonize TRPV1 to achieve analgesia without changing body temperature (Andreev et al., 2013). Under physiological conditions with the presence of extracellular calcium, calcium ions permeate through activated TRPV1 to bind with intracellular calmodulins, which further bind to the intracellular domains to cause calcium dependent desensitization (Lishko et al., 2007; Lau et al., 2012). RhTx2 rapidly desensitizes TRPV1 activation by binding to the outer pore region, so it employs distinct mechanisms to induce desensitization. In this sense, RhTx2 and calcium ions may work synergistically to desensitize the TRPV1 channel, so it is promising to examine whether RhTx2 exerts analgesic effects in animal models in future.
4. Materials and methods
4.1. Venom collection, toxin purification and protein sequencing
Adult Scolopendra subspinipes L. Koch (both sexes, n = 1000) were purchased from Jiangsu Province, China. As previously reported (Yang et al., 2012; Yang et al., 2015), venom was collected manually by stimulating the venom glands with a 3-V alternating current. Unique peptide toxins were purified from the raw venom using a combination of a Sephadex G-50 gel filtration column and reverse-phase (RP) HPLC. The purity of target peptides was analyzed using a matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF). Lyophilized HPLC fractions were dissolved in 0.1% (v/v) trifluoroacetic acid/water, from which 0.5 μl was spotted onto a MALDI-TOF plate with 0.5 μl α-cya-no-4-hydroxycinnamic acid matrix (10 mg/ml in 60% acetonitrile). Spots were analyzed by an UltraFlex I mass spectrometer (Bruker Daltonics) in a positive ion mode. Peptides with purity over 99.5% were collected and stored at −20 °C until further use. A Shimadzu protein sequencer (PPSQ-31A, Shimadzu, Japan) was used for the determination of primary sequence of peptides.
4.2. cDNA library and cloning
The venom-gland cDNA library was prepared as previously described (Li et al., 2007). Briefly, the total RNAs were extracted from the venom gland of 20 centipedes using TRIzol (Life Technologies Ltd.) and used to prepare the cDNA library using a SMART™ PCR cDNA synthesis kit (Clontech, Palo Alto, CA). The first strand was synthesized by using the 3′ SMART CDS Primer II A (5′ AAGCAGTGGTATCAACGCAGAGTACT (30)N–1N 3′, where N = A, C, G, or T and N−1 = A, G, or C) and SMART II A oligonucleotide, (5′ AAGCAGTGGTATCAACGCAGAGTACGCGGG 3′). The 5′ PCR primer II A (5′ AAGCAGTGGTATCAACGCAGAGT 3′) provided by the kit was used to synthesize the second strand using Advantage polymerase (Clontech, Palo Alto, CA). RACE (Rapid Amplification of cDNA ends) was used to clone transcripts encoding RhTx from the venom-gland cDNA library.
For cloning, the sense-direction primers were designed according to the amino acid sequences determined by Edman degradation. These primers were used in conjunction with an antisense SMART™ II A primer II in PCR reactions to screen for transcripts encoding neurotoxins. PCR was performed using Advantage polymerase (Clontech) under the following conditions: 2 min at 94 °C, followed by 30 cycles of 10 s at 92 °C, 30 s at 50 °C, and 40 s at 72 °C. Finally, the PCR products were cloned into pGEM®-T Easy vector (Promega, Madison, WI). DNA sequencing was performed on an ABI PRISM 377 DNA sequencer (Applied Biosystems).
4.3. Toxins synthesis, refolding and purification
Linear RhTx and RhTx2 peptides were synthesized by GL Biochem (Shanghai) Ltd. Crude reduced peptides were further purified by RP-HPLC. Once the purity of a peptide of interest was determined to be higher than 95% by MALDI-TOF mass spectrometry and HPLC techniques, the peaks were pooled and lyophilized. The linear reduced peptide was dissolved in 0.1 M Tris-HCl buffer (pH 7.0) at a final concentration of 30 μM glutathione containing 5 mM reduced glutathione and 0.5 mM oxidized glutathione. Oxidization and folding were performed at room temperature and monitored at 280 nm by analytical RP-HPLC and MALDI-TOF mass spectrometry.
4.4. Molecular biology and cell transfection
cDNA of Murine TRPV1 (a gift from Dr. Michael X. Zhu, University of Texas Health Science Center at Houston) was used. The construct used in this study also carries a point mutation E571A to increase protein expression and facilitate patch-clamp recordings; functional characterizations of E571A were reported in our previous study (Yang et al., 2015). To facilitate identification of channel-expressing cells, eYFP was fused to the C-terminus of TRPV1. Our previous study has shown that tagging of the fluorescence protein did not alter channel functions (Cheng et al., 2007).
HEK293T cells, purchased from and authenticated by American Type Culture Collection (ATCC), were cultured in DMEM medium with 10% FBS, 100 u/ml penicillin and 100 mg/ml streptomycin at 37 °C with 5% CO2. Cells were plated on glass coverslips 24 h before transfection. Transient transfection was conducted by adding 4 μl Lipo-fectamine 3000 (Invitrogen) and 4 μg plasmid DNA into opti-MEM and then stored for 20 min. The mixer was added to a 35 mm cell culture dish. Electrophysiological experiments and fluorescence imaging recordings were performed between 24 h and 48 h after transfection.
4.5. Chemicals
All chemicals such as capsaicin were purchased from Sigma-Aldrich.
4.6. Electrophysiology
Patch-clamp recordings were performed with a HEKA EPC10 amplifier with PatchMaster software (HEKA) in the outside-out or whole-cell configuration. Patch pipettes were prepared from borosilicate glass and fire-polished to a resistance of ~4 MΩ. For whole-cell recordings, serial resistance was compensated by 60%. For single-channel recordings, patch pipettes were fire-polished to a higher resistance of 6-to-10 MΩ. A normal solution containing 130 mM NaCl, 10 mM glucose, 0.2 mM EDTA and 3 mM HEPES (pH 7.2) was used in both bath and pipette. HEPES was used as the pH buffer. Current signal was sampled at 10 kHz and filtered at 2.9 kHz. All recordings were performed at room temperature (22 °C) with the maximum variation of 1 °C.
To apply solutions containing capsaicin or other reagents during patch-clamp recording, a rapid solution changer with a gravity-driven perfusion system was used (RSC-200, Bio-Logic). Each solution was delivered through a separate tube so that there was no mixing of solutions. Pipette tip with a membrane patch was placed directly in front of the perfusion outlet during recording.
4.7. Molecular docking
To model the RhTx2 with four more residues (NSKY) than RhTx, we first generated a fragment file of the RhTx2 amino acid sequence with the Robetta server (Kim et al., 2004). Then the conformation of the N terminus in RhTx2 was modeled de novo with cyclic coordinate descent loop modeling protocol (Wang et al., 2007). In each round of loop modeling, 10,000 to 20,000 models were generated. The top 20 models by energy were selected as the input for the next round of loop modeling. After three rounds of loop modeling, the top five models converged well. The model with the lowest energy was finally selected as the model of RhTx2.
To prepare RhTx (PDB ID: 2mva), RhTx2 and TRPV1 (apo and closed state, 3J5P; toxin-activated open state, 3J5Q) structure models for molecular docking, they were first relaxed in Rosetta version 3.5 (Leaver-Fay et al., 2011), respectively. For each structure 10,000 decoys were generated. The top 10 lowest energy decoys converged well, so the lowest energy decoy among them was picked for docking. To dock the toxin, membrane environment was first setup on the channel model using RosettaScripts (Fleishman et al., 2011; Yarov-Yarovoy et al., 2006). A total of 20,000 docking decoys were generated with Rosetta-Scripts. The top 10 decoys with largest binding energy were selected from top 1000 total energy score decoys. Among these 10 decoys, the one satisfies best with experimental findings was chosen as the final docking model.
4.8. Statistics
All experiments have been independently repeated for at least three times. All statistical data are given as mean ± SEM. Two-tailedStudent’s t-test was applied to examine the statistical significance. N.S. indicates no significance. ***, p < 0.001.
Acknowledgements
We are grateful to our lab members for assistance. This work was supported by funding from the National Science Foundation of China (31640071 and 31770835), Chinese Academy of Sciences (Youth Innovation Promotion Association and “Light of West China” Program), and Yunnan Province (2017FB037 and 2018FA003) to S.Y.; from National Science Foundation of China (31741067 and 31800990) and Zhejiang Provincial Natural Science Foundation of China (LR20C050002) to F.Y; and from NIH (R01GM132110) to J.Z. This work was also supported by the Core Facilities in Zhejiang University School of Medicine, including the Bioinformatics Computation Platform and Dr. Cheng Ma at the Protein Facility.
Footnotes
Declaration of competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data availability
All data needed to evaluate the conclusions in the report are present in the paper. Additional data are available from authors upon request.
Ethics statement
No vertebrate was used in this study. All the animal experiments were performed in accordance with recommendations in the Guide for the Care and Use of Laboratory Animals of Kunming Institute of Zoology, Chinese Academy of Sciences. Experimental protocols using animals in this study were approved by the Institutional Animal Care and Use Committees at Kunming Institute of Zoology, Chinese Academy of Sciences (approval ID: SMKXLLWYH20120520–01).
References
- Andreev YA, Kozlov SA, Korolkova YV, Dyachenko IA, Bondarenko DA, Skobtsov DI, Murashev AN, Kotova PD, Rogachevskaja OA, Kabanova NV, Kolesnikov SS, Grishin EV, 2013. Polypeptide modulators of TRPV1 produce analgesia without hyperthermia. Mar. Drugs 11, 5100–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlen CJ, Priel A, Zhou S, King D, Siemens J, Julius D, 2010. A bivalent tarantula toxin activates the capsaicin receptor, TRPV1, by targeting the outer pore domain. Cell 141, 834–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao E, Liao M, Cheng Y, Julius D, 2013. TRPV1 structures in distinct conformations reveal activation mechanisms. Nature 504, 113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Ma L, Yang F, Wang K, Zheng J, 2014. Divalent cations potentiate TRPV1 channel by lowering the heat activation threshold. J. Gen. Physiol 143, 75–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caterina MJ, Leffler A, Malmberg AB, Martin WJ, Trafton J, Petersen-Zeitz KR, Koltzenburg M, Basbaum AI, Julius D, 2000. Impaired nociception and pain sensation in mice lacking the capsaicin receptor. Science 288, 306–313. [DOI] [PubMed] [Google Scholar]
- Caterina MJ, Schumacher MA, Tominaga M, Rosen TA, Levine JD, Julius D, 1997. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature 389, 816–824. [DOI] [PubMed] [Google Scholar]
- Chen Z, Hu Y, Hong J, Hu J, Yang W, Xiang F, Yang F, Xie Z, Cao Z, Li W, Lin D, Wu Y, 2015. Toxin acidic residue evolutionary function-guided design of de novo peptide drugs for the immunotherapeutic target, the Kv1.3 channel. Sci. Rep 5, 9881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Hu Y, Hu J, Yang W, Sabatier JM, De Waard M, Cao Z, Li W, Han S, Wu Y, 2014. Unusual binding mode of scorpion toxin BmKTX onto potassium channels relies on its distribution of acidic residues. Biochem. Biophys. Res. Commun 447, 70–76. [DOI] [PubMed] [Google Scholar]
- Cheng W, Yang F, Takanishi CL, Zheng J, 2007. Thermosensitive TRPV channel subunits coassemble into heteromeric channels with intermediate conductance and gating properties. J. Gen. Physiol 129, 191–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R, 1998. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science 280, 69–77. [DOI] [PubMed] [Google Scholar]
- Fleishman SJ, Leaver-Fay A, Corn JE, Strauch EM, Khare SD, Koga N, Ashworth J, Murphy P, Richter F, Lemmon G, Meiler J, Baker D, 2011. RosettaScripts: a scripting language interface to the Rosetta macromolecular modeling suite. PloS One 6, e20161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garami A, Pakai E, McDonald HA, Reilly RM, Gomtsyan A, Corrigan JJ, Pinter E, Zhu DXD, Lehto SG, Gavva NR, Kym PR, Romanovsky AA, 2018. TRPV1 antagonists that cause hypothermia, instead of hyperthermia, in rodents: compounds’ pharmacological profiles, in vivo targets, thermoeffectors recruited and implications for drug development. Acta Physiol. 223, e13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavva NR, 2009. Setbacks in the clinical development of TRPV1 antagonists. Open Drug Discov. J 1, 1–35. [Google Scholar]
- Gavva NR, Bannon AW, Surapaneni S, Hovland DN Jr., Lehto SG, Gore A, Juan T, Deng H, Han B, Klionsky L, Kuang R, Le A, Tamir R, Wang J, Youngblood B, Zhu D, Norman MH, Magal E, Treanor JJ, Louis JC, 2007. The vanilloid receptor TRPV1 is tonically activated in vivo and involved in body temperature regulation. J. Neurosci 27, 3366–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo P, MacKinnon R, 1995. Revealing the architecture of a K+ channel pore through mutant cycles with a peptide inhibitor. Science 268, 307–310. [DOI] [PubMed] [Google Scholar]
- Julius D, 2013. TRP channels and pain. Annu. Rev. Cell Dev. Biol 29, 355–384. [DOI] [PubMed] [Google Scholar]
- Kaszas K, Keller JM, Coddou C, Mishra SK, Hoon MA, Stojilkovic S, Jacobson KA, Iadarola MJ, 2012. Small molecule positive allosteric modulation of TRPV1 activation by vanilloids and acidic pH. J. Pharmacol. Exp. Therapeut 340, 152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DE, Chivian D, Baker D, 2004. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 32, W526–W531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau SY, Procko E, Gaudet R, 2012. Distinct properties of Ca2+-calmodulin binding to N- and C-terminal regulatory regions of the TRPV1 channel. J. Gen. Physiol 140, 541–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman K, Renfrew PD, Smith CA, Sheffler W, Davis IW, Cooper S, Treuille A, Mandell DJ, Richter F, Ban YE, Fleishman SJ, Corn JE, Kim DE, Lyskov S, Berrondo M, Mentzer S, Popovic Z, Havranek JJ, Karanicolas J, Das R, Meiler J, Kortemme T, Gray JJ, Kuhlman B, Baker D, Bradley P, 2011. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 487, 545–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebovitz EE, Keller JM, Kominsky H, Kaszas K, Maric D, Iadarola MJ, 2012. Positive allosteric modulation of TRPV1 as a novel analgesic mechanism. Mol. Pain 8, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Xu X, Xu C, Zhou W, Zhang K, Yu H, Zhang Y, Zheng Y, Rees HH, Lai R, Yang D, Wu J, 2007. Anti-infection peptidomics of amphibian skin. Mol. Cell. Proteomics 6, 882–894. [DOI] [PubMed] [Google Scholar]
- Liao M, Cao E, Julius D, Cheng Y, 2013. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature 504, 107–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lishko PV, Procko E, Jin X, Phelps CB, Gaudet R, 2007. The ankyrin repeats of TRPV1 bind multiple ligands and modulate channel sensitivity. Neuron 54, 905–918. [DOI] [PubMed] [Google Scholar]
- Luo L, Wang Y, Li B, Xu L, Kamau PM, Zheng J, Yang F, Yang S, Lai R, 2019. Molecular basis for heat desensitization of TRPV1 ion channels. Nat. Commun 10, 2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ombati R, Luo L, Yang S, Lai R, 2018. Centipede envenomation: clinical importance and the underlying molecular mechanisms. Toxicon 154, 60–68. [DOI] [PubMed] [Google Scholar]
- Robinson SD, Undheim EAB, Ueberheide B, King GF, 2017. Venom peptides as therapeutics: advances, challenges and the future of venom-peptide discovery. Expert Rev. Proteomics 14, 931–939. [DOI] [PubMed] [Google Scholar]
- Safavi-Hemami H, Brogan SE, Olivera BM, 2019. Pain therapeutics from cone snail venoms: from Ziconotide to novel non-opioid pathways. J Proteomics 190, 12–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Li Z, Jiang Y, Pan X, Wu J, Cristofori-Armstrong B, Smith JJ, Chin YKY, Lei J, Zhou Q, King GF, Yan N, 2018. Structural basis for the modulation of voltage-gated sodium channels by animal toxins. Science 362. [DOI] [PubMed] [Google Scholar]
- Siemens J, Zhou S, Piskorowski R, Nikai T, Lumpkin EA, Basbaum AI, King D, Julius D, 2006. Spider toxins activate the capsaicin receptor to produce inflammatory pain. Nature 444, 208–212. [DOI] [PubMed] [Google Scholar]
- Tominaga M, Caterina MJ, Malmberg AB, Rosen TA, Gilbert H, Skinner K, Raumann BE, Basbaum AI, Julius D, 1998. The cloned capsaicin receptor integrates multiple pain-producing stimuli. Neuron 21, 531–543. [DOI] [PubMed] [Google Scholar]
- Wang C, Bradley P, Baker D, 2007. Protein-protein docking with backbone flexibility. J. Mol. Biol 373, 503–519. [DOI] [PubMed] [Google Scholar]
- Wu J, Yan Z, Li Z, Yan C, Lu S, Dong M, Yan N, 2015. Structure of the voltage-gated calcium channel Cav1.1 complex. Science 350, aad2395. [DOI] [PubMed] [Google Scholar]
- Yang F, Cui Y, Wang K, Zheng J, 2010. Thermosensitive TRP channel pore turret is part of the temperature activation pathway. Proc. Natl. Acad. Sci. U. S. A 107, 7083–7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Ma L, Cao X, Wang K, Zheng J, 2014. Divalent cations activate TRPV1 through promoting conformational change of the extracellular region. J. Gen. Physiol 143, 91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Xiao X, Cheng W, Yang W, Yu P, Song Z, Yarov-Yarovoy V, Zheng J, 2015. Structural mechanism underlying capsaicin binding and activation of the TRPV1 ion channel. Nat. Chem. Biol 11, 518–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Zheng J, 2017. Understand spiciness: mechanism of TRPV1 channel activation by capsaicin. Protein Cell 8, 169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Fan, Xiao Xian, Lee Hyun, Bo Simon, Vu Yang, Wei Yarov Yarovoy, Vladimir Zheng Jie, 2018. The conformational wave in capsaicin activation of transient receptor potential vanilloid 1 ion channel. Nat. Commun 9, 2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Liu Z, Xiao Y, Li Y, Rong M, Liang S, Zhang Z, Yu H, King GF, Lai R, 2012. Chemical punch packed in venoms makes centipedes excellent predators. Mol. Cell. Proteomics 11, 640–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Yang F, Wei N, Hong J, Li B, Luo L, Rong M, Yarov-Yarovoy V, Zheng J, Wang K, Lai R, 2015. A pain-inducing centipede toxin targets the heat activation machinery of nociceptor TRPV1. Nat. Commun 6, 8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Yang F, Zhang B, Lee BH, Li B, Luo L, Zheng J, Lai R, 2017. A bimodal activation mechanism underlies scorpion toxin-induced pain. Sci Adv 3, e1700810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarov-Yarovoy V, Schonbrun J, Baker D, 2006. Multipass membrane protein structure prediction using Rosetta. Proteins 62, 1010–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, 2013. Molecular mechanism of TRP channels. Comp. Physiol 3, 221–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Jie, Trudeau Matthew C., 2015. Handbook of Ion Channels. CRC Press, Boca Raton, FL. [Google Scholar]


