Abstract
Our understanding of repair outcomes after Cas9-induced DNA cleavage is still limited, especially in primary human cells. We sequence repair outcomes at 1,656 on-target genomic sites in primary human T cells and use these data to train a machine learning model, CRISPR Repair OUTcome (SPROUT). SPROUT accurately predicts the length, probability, and sequence of nucleotide insertions and deletions and will facilitate SpCas9 guide RNA design in therapeutically-important primary human cells.
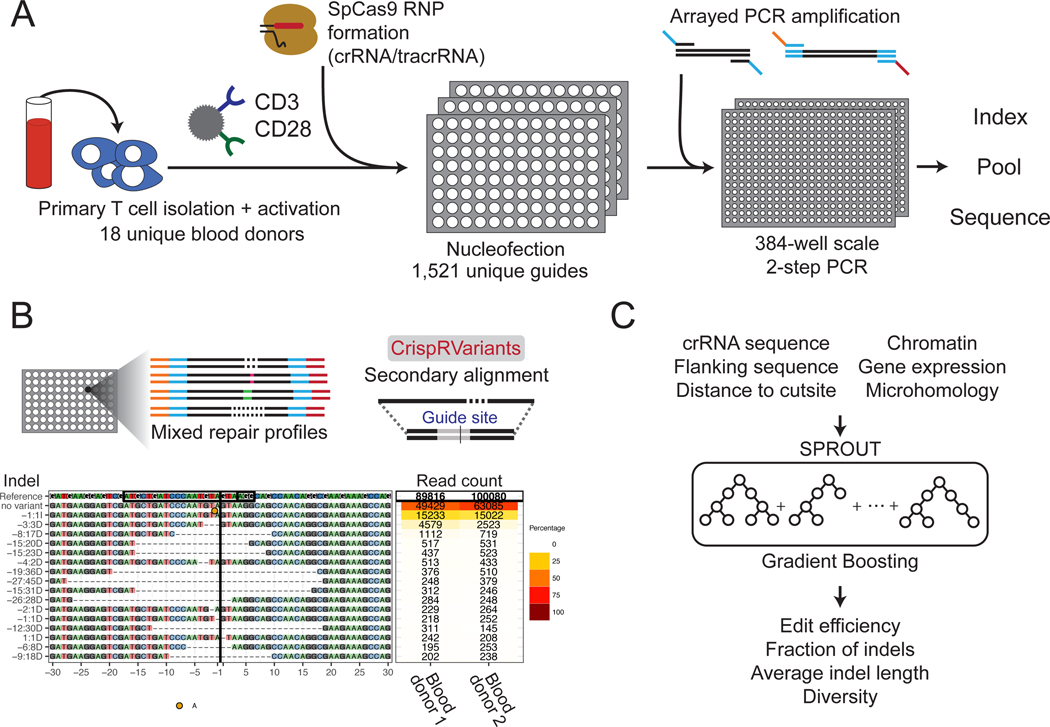
Primary T cells are a promising cell type for therapeutic genome editing, as they can be engineered efficiently ex vivo and adoptively transferred to patients1. However there lacks detailed information about the genomic outcomes of Cas9-dependent editing in primary human cells. Here, we systematically characterize Streptococcus pyogenes Cas9 (SpCas9) repair outcomes in primary T cells from 18 healthy blood donors (Supplementary Fig. 1).
Targeted sequencing was applied to 1,656 unique genomic locations within 559 genes in primary CD4+ T cells. Guide RNAs were combined with SpCas9 to assemble ribonucleoprotein complexes (RNPs) and electroporated into T cells2,3. DNA was isolated from cells after 6 days of recovery and expansion, and a 180–260 base pair (bp) region around each site was PCR amplified and sequenced (Fig. 1). We quantified the distribution of repair outcomes at each target site from the generated amplicon library using CrispRVariants4 (Fig. 1). In total, 31% of reads contained deletions centered around the cut site with an average deletion length of 13 bps. We also found that 20% of the reads had insertions at the cut site, and 95% of these insertions were of exactly one nucleotide (Supplementary Fig. 2). Only 0.008% of reads contained both an insertion and deletion.
Figure 1.
Overview of the method. (A) Primary T cells were isolated, activated, and electroporated with Cas9/crRNA/tracrRNA RNPs in 96-well plates. After 6 days of expansion, genomic DNA was isolated from each well, amplified and sequenced. (B) The CrispRVariants R package4 was used to quantify each SpCas9 RNP knockout. An example alignment is plotted here, with quantification shown for two blood donors. Each site has this same unique plot, all of which can be found on figshare. (C) A gradient boosting machine learning algorithm was trained to predict multiple DNA repair outcomes given the guide RNA sequence, flanking nucleotides and additional features.
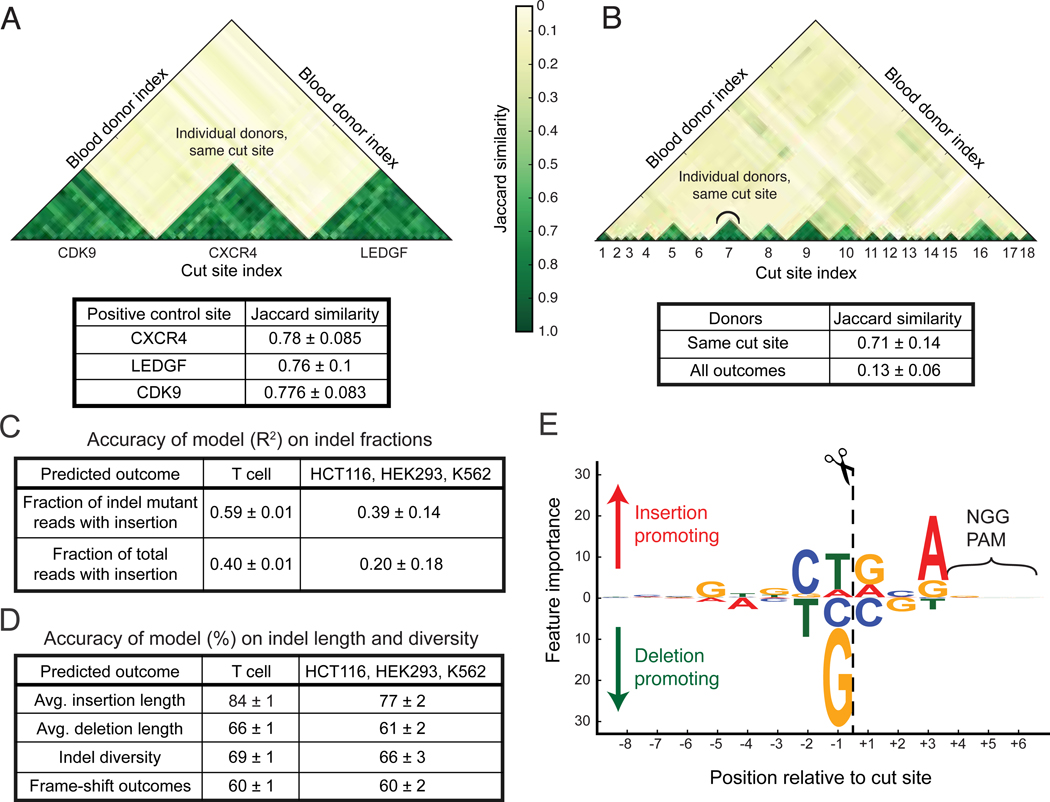
There was an average of 98 discrete repair outcomes per target site that were observed at a frequency greater than 1 in 1000 reads, and different sites were highly variable in the proportion and length distribution of insertions and deletions. The repair outcomes from each target site were similar between donors, but very different across target sites (Fig. 2A). Comparisons of repair outcomes between all sites showed that replicate editing experiments from individual target sites were significantly more similar to each other than to outcomes from different sites (Fig. 2B, Supplementary Fig. 3).
Figure 2.
SPROUT predicts DNA repair outcomes. (A) The DNA repair outcomes resulting from RNP activity in T cells derived from different blood donors were compared for control guides targeting CDK9, CXCR4, and LEDGF, analyzing the top 20 indels at each site. These guides were used in every blood donor. Jaccard similarity is calculated for each guide site across donors. (B) Jaccard similarity of DNA repair outcomes for 18 randomly chosen guides, again using the top 20 indels. Jaccard coefficients are plotted comparing outcomes from different guide RNAs and between blood donors. (C) The trained model was used to predict DNA repair indel fractions in a hold-out (un-seen) portion of the T cell dataset. The model was also evaluated on previously published data5 obtained from immortalized cell lines to test generalization performance for other cell types and experimental conditions. (D) Accuracy of the trained model in predicting the average insertion and deletion length, indel diversity and whether a target has high, medium or low fraction of frame-shift outcomes on both T cells and previously published data5. (E) The importance that SPROUT assigns to nucleotides at each position relative to the cut site. Larger text indicates that the presence of a particular nucleotide at a position has greater importance in determining the likelihood of insertion versus deletion. Bootstrap mean and standard deviation are shown in each table. This study assayed 1,656 genomic sites in T cells.
We hypothesized that the variation in repair outcomes across cut sites was largely due to sequence variation near the cut site5–7. To test this, we developed a machine learning model, SPROUT, to predict SpCas9 repair outcomes (Fig. 1). The model takes as input the 20 nucleotides of the spacer sequence plus the PAM, and it uses gradient boosting to train an ensemble of decision trees over the nucleotides. At each target site, the model predicted the fraction of indel mutant reads with an insertion (Fig. 2C) and deletion (1 - fraction of insertions) and the average length of insertions and deletions (Fig. 2D). We included fraction of both indel mutant reads and total reads in order to separate the dependency on the edit efficiency. On an independent set of 304 target sites in primary T cells, SPROUT was able to accurately predict the fraction of indel mutant reads with an insertion (R2 = 0.59, Spearman Rank = 0.81) and the fraction of total reads with an insertion (R2 = 0.40, Spearman Rank = 0.68, Fig. 2C, Supplementary Fig. 4, Supplementary Fig. 5). SPROUT was also able to predict if a target has high (greater than 60%), medium (40%−60%) or low (less than 40%) fraction of frame-shift repair outcomes with accuracy of 0.6 (Fig. 2D).
SPROUT can also be used for in silico guide design. For each of the 532 genes with multiple guides, we used SPROUT’s predictions to rank the targets in a gene from the most likely to have frame-shift repair outcomes to the least likely. SPROUT correctly identified the best performing frame-shift guide in 54% of the genes, and it correctly predicted the complete ranking in 38% of the genes (Supplementary Fig. 6). We further investigated whether SPROUT could correctly select which SpCas9 target site in a gene was the most likely to have an enrichment of insertions over deletions. For each gene, we used SPROUT’s prediction to rank the target sites by their predicted fraction of indel mutant reads with an insertion. For 73% of the genes, SPROUT correctly chose the top sgRNA, and for 60% of genes it correctly predicted the complete ranking of all the candidate guides by their insertion proportion, significantly above random guessing (Supplementary Fig. 6, p < 10−10).
The prediction signal was primarily localized in the three nucleotides immediately to the left and right of the cut site. The −1 position (immediately to the 5’ end of the cleavage site) was the most influential (Fig. 2E and Supplementary Fig. 7). This is consistent with previous observations where this nucleotide is duplicated at many cut sites, which has been suggested to be the result of repair of single-base overhangs generated by Cas97. The presence of a G or C nucleotide at this position decreased the insertion proportion: 7% and 10% of indel mutant reads were insertions, respectively. Comparatively, the presence of A or T nucleotide at this position increased this proportion to 23% and 26%, respectively. The +3 position is also important in determining the proportion of outcomes as insertions or deletions (Supplementary Fig. 7). A or G nucleotides at this position increase the insertion proportion to 25% and 23% respectively, compared with 16% and 15% for C and T. The presence of homopolymers (a run of two or more identical nucleotides) adjacent to the cut site increased the proportion of deletions (p < 0.02). For example, targets with G homopolymers abutting the cut site have deletions in 92% of the indel mutant reads, compared to 77% deletions when there is no homopolymer at the cut site (Supplementary Fig. 8), which could be a reflection of microhomology mediated end joining8.
Next, we assessed the robustness of the algorithm to sequence- and cell-specific features by using the SPROUT model trained on the T cell data to predict SpCas9 repair outcomes in other human cell types. We re-analyzed published targeted sequencing data from 96 unique target sites tested in HEK293, K562, and HCT116 cells5. These 96 targets were distinct from the 1,521 sites that were used to train SPROUT, and hence constitute new test data. SPROUT achieved an accuracy of R2 = 0.40 in predicting the fraction of indel mutant reads with an insertion and an R2 = 0.23 in predicting the fraction of total reads with an insertion. The relatively high cross-cell-type performance of SPROUT further suggests that the primary factor influencing the repair outcomes after SpCas9 cleavage within dividing cells is the nucleotide sequence context near the cut site.
We systematically compared SPROUT with two recently developed methods for predicting SpCas9 repair outcomes, inDelphi and FORECasT9,10. The methods were compared on tasks that all three algorithms perform—predicting the fraction of repair outcomes with frameshift, the repair precision (defined as one minus the indel diversity), and the fraction of indel reads that are insertions. To rigorously compare the algorithms, we generated three new SpCas9 repair datasets collected after all three models have been trained. We collected two new primary T cell SpCas9 repair outcome data: first for 32 sites tiled across the CXCR4 gene and next for 182 unique sites from 91 immune related genes. Each site was replicated across multiple donors. These sites are distinct from the T cell sites used to train SPROUT, and hence act as independent validation. At these sites, SPROUT substantially outperforms both InDelphi and FORECasT in the repair prediction tasks (Supplementary Fig. 9, p < 0.01). We also collected repair data from the same CXCR4 sites when edited in human induced pluripotent stem cells (iPSC). SPROUT was not trained on these sites nor had it seen data from iPSC, and this constitutes a strong test in another therapeutically-relevant cell type. Again SPROUT was more accurate than inDelphi and FORECasT on the iPSC data for each of the three prediction tasks (p < 0.05, Supplementary Fig. 9). These results demonstrate that SPROUT is state-of-the-art in predicting SpCas9 editing outcomes in both T cells and iPSCs, two cell types in which concerted efforts are underway to harness CRISPR for engineered cellular therapies.
In 90% of the T-cell SpCas9 target locations we discovered long (>25 base pairs) DNA insertions in the repair outcome sequencing data. Across sites, 40% of the long insertions aligned to the human genome, and they correspond to 0.07% of indel-containing reads. Among the aligned long insertions, 36% aligned to the same chromosome as the SpCas9 target site, with 27% aligning to within 1kb of the target (Supplementary Fig. 10). The remaining insertions aligned to locations on different chromosomes which are enriched for HiC interaction with the target sites (p < 10−5, Supplementary Figs. 11–13). These findings suggest a possible model whereby genomic regions physically proximal to the cut site could be inserted during the DNA repair process. Recent reports have indicated that cells may undergo genomic rearrangements in response to SpCas9 cleavage11,12, although these should be interpreted cautiously given the cell types used and other variables. The potential therapeutic applications of CRISPR in primary T cells and other human cells motivate further investigations into the mechanisms and prevalence of insertions and other rearrangements during genome editing.
Online Methods
T cell Editing
Lyophilized crRNA and tracrRNA (Dharmacon) was resuspended at a concentration of 160 μM in 10 mM Tris-HCL (7.4 pH) with 150 mM KCl. Cas9 ribonucleoproteins (RNPs) were made as previously described by combining 5μL of 160μM crRNA with 5μL of 160μM tracrRNA for 30 min at 37°C, followed by incubation of this 80μM gRNA product with 10μL of 40μM Cas9 (UC Macrolab) to form RNPs at 20μM13. Five 3.5μL aliquots were frozen in lo-bind 96-well V-bottom plates (E&K Scientific) at −80°C until used. All crRNA guide sequences were designed by Dharmacon for gene knockout.
T cell editing was conducted according to published protocols14. Briefly, peripheral blood mononuclear cells (PBMC) were isolated from whole blood (numeric donors, under a protocol approved by UCSF Committee on Human Research, CHR #13–11950) or de-identified residuals from leukoreduction chambers after Trima Apheresis (alphabetic donors, from Blood Centers of the Pacific) from healthy human donors by Ficoll centrifugation with SepMate tubes (STEMCELL, per manufacturer’s instructions). CD4+ T cells were then isolated from PBMCs with magnetic negative selection (STEMCELL), cultured at 1 million cells/mL in complete RPMI (RPMI-1640 with 20 IU/mL IL2, 10% FBS, 50 μg/mL Pen/Strep and 5mM HEPES) and activated with plate-bound anti-CD3 (OKT3) and anti-CD28 (CD28.2) antibodies.
After three days of culture on stimulating antibodies at 37°C / 5% CO2, cells were resuspended and counted before editing. Approximately 3.5 × 105 cells were edited per blood donor per guide. Immediately before electroporation, cells were centrifuged at 400xg for 5 minutes, supernatant was aspirated, and the pellet resuspended in 20 μL of room-temperature Lonza electroporation buffer P3 (Lonza). The cell suspension was then gently mixed with thawed RNP and carefully aliquoted into 96-well electroporation cuvette for nucleofection with the 4D 96 well shuttle unit (Lonza) using code EH-115. Immediately after electroporation, 80 μL of pre-warmed media without IL2 were added to each well and cells were allowed to rest for at least one hour in a 37°C cell culture incubator. Subsequently cells were moved to 96-well flat-bottomed culture plates pre-filled with 100 μL warm complete media with IL2 at 40 IU/mL (for a final concentration of 20 IU/mL) and anti-CD3/anti-CD2/anti-CD28 beads (T cell Activation and Stimulation Kit, Miltenyi Biotec) or anti-CD3/anti-CD28 dynabeads (ThermoFisher) at 1:1 bead:to:cell ratio.
Cells were then cultured at 37°C / 5% CO2 in a dark cell culture incubator for a further 6 days, and were supplemented with IL2-containing complete media on days 3 and 5 of culture. On day 6 of culture, one eighth of each culture, approximately 35 μL, was reserved for genomic DNA analysis by 1:1 mixing with QuickExtract buffer (EpiCentre) in a 96-well plate, sealing carefully with foil and heating to 65°C for 20 min followed by heating to 98°C for 5 minutes on a thermocycler. Genomic DNA extracts were stored at −20°C until use.
For the validation set across 91 immune genes (New T Cells II, Supplementary Fig. 9), editing was conducted in a similar manner with the following exceptions: bulk T cells were isolated instead of CD4+ T cells (STEMCELL, magnetic negative selection per manufacturer’s instructions.) These cells were edited after two, not three, days of stimulation with anti-CD3/anti-CD28 beads (ThermoFisher) and were not given additional stimulation beads.
RNP editing for iPSC cells was performed in a very similar manner. AltR guides (IDT) suspended in IDT nuclease free TE were incubated at 37oC for 15 minutes to form crRNA trRNA complex (2=1:1 ratio). The crRNA:trRNA complex was then incubated with spCas9-NLS (UC-Berkeley MacroLab) at 37oC for 15 minutes in a 2:1 ratio, forming a final RNP concentration of 10 μM. iPSCs were treated with ROCK inhibitor Y-276932 at 10 μM (STEMCELL Technologies CN# NC0791122) for 2 hours prior to nucleofection and were dissociated with Accutase (STEMCELL Technologies CN# 07920) to single cells suspension prior to nucleofection. 200K cells were nucleofected in the Amaxa 96 well shuttle using 18 μL P3 buffer and 2 μl RNP (1μM final concentration), using nucleofection code DS-138. Cells were rescued into 96 well culture plates and maintained on growth factor-reduced Matrigel (Corning Life Sciences CN# CB-40230C) in feeder-free media conditions (Gibco Essential 8 Flex Media) supplemented with ROCK inhibitor Y-276932 (10 μM) for 72 hours before harvesting gDNA for analysis with QuickExtract (Lucigen).
PCR amplification of cut sites
PCR primers were designed using an in-house Python wrapper around Primer3 (github.com/czbiohub/Primer3Wrapper)15. Primers were designed to amplify a 180 to 260 nucleotide region, ensuring that the cut site was at least 50 nucleotides from the end of each primer, as well as 15 nucleotides from the center of the read to ensure there was enough sequence to accurately quantify larger indels. Sequencing adapters (Forward: 5’-CTCTTTCCCTACACGACGCTCTTCCGATCT-3’ and Reverse 5’-CTGGAGTTCAGACGTGTGCTCTTCCGATCT-3’) were appended to the designed primers, and a homodimer and heterodimer filter was applied to ensure no secondary structure existed between primers. Sites were amplified using between 4,000 and 10,000 genomic copies, 0.5 μM of each primer, and Q5 hot start high-fidelity 2x master mix (NEB). PCR was performed using the standard protocol: 98°C for 30 seconds; then 35 cycles of 98°C for 10 seconds, 60°C for 30 seconds, and 72°C for 30 seconds; followed by a final extension at 72°C for 2 minutes (NEB). Samples were diluted 1:100 and individually indexed in a second, 12-cycle PCR using index primers containing Illumina sequencing adapters and 8 base barcodes, under the same conditions as the first PCR. After the second PCR, indexed samples were pooled and purified using a 0.7x SPRIselect purification and sequenced on an Illumina NextSeq 500.
Repair outcome pre-processing pipeline
Fastq sequencing files were first merged using FLASH16, then subjected to adapter and quality trimming with trimmomatic17. These merged reads were then initially aligned to the hg38 genomic contig using bwa mem18, creating individual .bam files. Each sample was individually analyzed using the CrispRVariants bioconductor package in R4, which performs a secondary alignment and quantifies each unique insertion and deletion per sequencing read. Repair outcomes were then further parsed using embedded CrispRVariants packages to quantify individual DNA repair outcomes, the insertion sequences, mutation efficiencies, and SNVs. Sites where the total number of reads was less than 1,000 were considered dropouts and filtered from all analysis. There were 1,521 unique target sites from 549 genes that passed this filtering, and 1,361 of these sites were replicated in two or more donors. The subsequent analyses focus on these 1,521 sites. The average number of reads per site, after filtering, is approximately 59,000. Fewer than 1% of the reads contained single nucleotide variation (SNV) but no indel, some of which may be attributable to sequencing error, and we chose to focus our analysis on reads containing at least one insertion or deletion.
T cell and iPSC data summary
This study involved 3,989 DNA repair profiles from T cells isolated from 18 patients. These outcomes targeted 1,521 unique sites within 549 genes in the human genome. Guides were chosen targeting genes encoding for HIV-interacting proteins. The top three guides from the Dharmacon Edit-R Predesigned knockout library were selected for each gene. Three distinct non-targeting controls chosen from the Edit-R library were included on every plate, as well as three validated, custom-designed guides known to knock out the genes CXCR4, CDK9 and LEDGF with high efficiency. The RNP knockouts were repeated on average 2 times, each across unique primary T cells from different blood donors (Supplementary Fig. 1). The repair outcomes were averaged over the repeats across the blood donors, and DNA repair outcome data from each target site has been deposited on figshare.
Two additional validation datasets were generated by designing guides tiling along the CXCR4 gene, which were repeated in biological triplicate in both new primary T cell and iPSC donors. After filtering for quality, repair outcome sequencing data were analyzed for 32 new guides in primary T cells and 30 new guides in iPSC using the same process as was done for the original T cell data. A third additional validation dataset was generated on primary T cells using guides targeting 182 distinct loci close to the start codons in 91 immune related genes. Each guide was tested on 6 unique donors. All of the target sites in these new validation experiments are distinct from the sites used to train SPROUT.
HCT116, HEK293, and K562 data summary
Published sequencing data7 from three other cell types (HEK293, K562, and HCT116; BioProject PRJNA326019) were analyzed according to the same procedure as the T cell data and used for validation of the machine learning model. The dataset we used from the manuscript comprised the RNP knockouts, after 48 hours, from 96 unique cut sites on the human genome.
Statistical analysis
We use the gradient boosting algorithm to train SPROUT. Gradient boosting is an aggregation model which iteratively learns a weighted ensemble of base classifiers. SPROUT uses decision trees as the base classifiers. The depth and number of the trees are hyperparameters of the algorithm which we set by cross validation. In SPROUT, typically 20 to 200 decision trees are utilized each of which are 3 to 20 layers deep depending on the prediction task. A complete list of all features that were assessed for inclusion in SPROUT can be found in Supplementary Fig. 14.
We used five-fold cross-validation to train SPROUT. We randomly split the unique cut sites in T cells (a total of 1,521) into 5 folds and trained SPROUT on four of the five folds. We then tested the performance of SPROUT on the remaining unseen fifth fold (304 cut sites). We repeated the random data split procedure 10 times and report the average and standard deviation of the prediction performance over the 10 random repeats. We performed the training using varying sizes of the training set and the performance of SPROUT appears to saturate with our current data size on T cells (Supplementary Fig. 15). We evaluated the prediction performance of regression tasks, i.e., predicting the fraction of total or indel mutant reads with insertion or deletion and the edit efficiency, using the coefficient of determination (R2). We also evaluated the prediction performance of classification tasks, i.e., predicting if the average insertion or deletion length or the diversity is larger/smaller than the median of the distribution, using the accuracy of the classifier. Repair diversity is defined as the entropy of the distribution of repair outcomes in the reads. High diversity suggest that the repair outcome at the site is more variable. A naive (or random) guess would be 50% accurate in predicting the correct output labels.
For the models evaluated on three other cell types (HCT116, HEK293, and K562), we trained SPROUT on the full T cell data (1,521 cut sites) and tested the performance of the model on the other cell types. We did not fine-tune SPROUT using features specific to these other cell types in order to quantify the robustness of the model. For the classification tasks, we used the median of the cell type distributions to set the threshold. Additional detail is in Supplementary Note.
Comparison to InDelphi and FORECasT
We compared SPROUT with InDelphi and FORECast on four benchmark datasets: 1) held-out T cell test data that was not used during SPROUT training; 2) a primary T cell data set of 32 SpCas9 target sites tiled across CXCR4; 3) a primary T cell data set of 182 SpCas9 target sites in 91 immune related genes; and 4) a new data set from iPSC detailed in the main text. The data sets 2, 3 and 4 were collected after SPROUT had been developed, and they consist of new genomic loci and new donors that were not seen during SPROUT training.
We used the trained inDelphi model provided at the website http://indelphi.giffordlab.mit.edu/ to test the performance of this method on the benchmark datasets. U2OS was set as the input cell type (the closest outcome found to T cells among the provide cell types). Frameshift and precision was directly downloaded from the website graphical interface. We used the definition of the repair precision proposed in the inDelphi paper, which is one minus the entropy of the distribution of the deletion lengths frequency. We downloaded the repair outcome for each experiment and used a script to find the fraction of reads with an insertion. We used the website https://partslab.sanger.ac.uk/FORECasT to evaluate the frameshifts in the FORECasT method. To measure precision and fraction of reads with insertions we used the batch mode of the trained model provided at https://github.com/felicityallen/SelfTarget and a post processing script. For frameshift and precision we thresholded the predicted values and binned them into “high” and “low” categories and reported the percentage of the method predicted the categories correctly. For fraction of insertion we reported the R2 value.
Nucleotide feature interpretations extracted from SPROUT
To measure the importance of individual features in the gradient boosting model, the information gain concept was used. The information gain associated to a feature measures the decrease in entropy after a dataset is split based on that particular feature. A higher information gain corresponds to a more predictive feature. We also determined the influence of each feature (enrichment or depletion) from the sign of the coefficients of a linear regression model trained on the data. Note that the algorithm was completely blind to the actual location of the cut site. Additionally, the feature importance for nucleotides (e.g., ‘G’) showed an alternating pattern. We speculate that one reason for the enrichment of alternating pattern for an insertion outcome and thus depletion for a deletion outcome is the homopolymer effect. It has been observed that homopolymers – the repetition of one base creating long runs of the same nucleotide – favor a deletion outcome5,8.
Ranking guides based on a desired repair outcome
We evaluated SPROUT in ranking the guides based on fitness to produce a desired repair outcome. Two outputs were used to train the regression: the fractions of indel reads, and the fractions of total reads. After training on 400 genes, the model was used to predict the fraction of insertions and deletion of a hold-out set of guides targeting 149 different genes. We assessed the ranking performance of the guides on only the genes that have more than one guide in our datasets (142 test genes out of 149 genes total). The guides were then ranked within each gene based on the insertion and deletion fractions, and the rank correlation between the observed result and predicted ranking was evaluated.
The performance was measured using Kendall’s tau ranking coefficient and the percentage of completely correct predictions. Kendall’s tau ranking coefficient measures the difference between the observed result and the predicted rankings. The Kendall’s tau coefficient is a ranking measure between −1 and 1, where 1 indicates that rankings match exactly, 0 means that there is no ranking correlation, and −1 means that there is complete reverse ranking correlation. Supplementary Fig. 6 summarizes the ranking results for guides in hold-out genes from T cells and guides from the three other validation cell types (HCT116, HEK293, and K562).
Extracting and aligning long insertion data from the repair outcomes
To obtain the insertion data, the repair outcomes of all 1,521 cut sites were parsed and reads with an inserted sequence of length at least 25 bp were selected, totaling 22,495 unique insertions which centered on the cleavage site. All insertions were aligned to the human genome with the BLAST algorithm (blastn command, https://blast.ncbi.nlm.nih.gov/Blast.cgi) under default conditions and input parameters. For the cases with more than one alignment, the site with the highest alignment score was selected. A total of 8,946 unique insertions aligned to the human genome.
Supplementary Material
Acknowledgments
This work was supported by the Chan Zuckerberg Biohub. J.Z. was supported by a Chan-Zuckerberg Investigator grant and by National Science Foundation grant CRII 1657155. A.M. was supported by NIH/NIDA Avenir New Innovator Award (DP2DA042423, A.M.), NIH/NIGMS funding for the HIV Accessory & Regulatory Complexes (HARC) Center (P50 GM082250, A.M. and N.J.K.), and gifts from J. Aronov, B. Bakar, K. Jordan, F. Caufield, and D. Wolkoff. A.M. holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund, has received funding from the Innovative Genomics Institute (IGI) and the Parker Institute for Cancer Immunotherapy (PICI), and is an investigator at the Chan Zuckerberg BioHub. A. A. was supported by National Institute of Health grant 7R01HG008164–04 and the Stanford data science initiative. J.H. was supported by the UCSF Medical Scientist Training Program. We would like to thank N. Neff and R. Sit for assistance collecting sequence data and A. Sellas for lab support. We also thank J. Palacios for statistical discussions.
Footnotes
Competing interests
A.M. is a co-founder of Spotlight Therapeutics. A.M. has served as an advisor to Juno Therapeutics and is a member of the scientific advisory board at PACT Pharma and an advisor to Sonoma Biotherapeutics. The Marson laboratory has received sponsored research support from Juno Therapeutics, Epinomics, Sanofi and a gift from Gilead. A.M. and T.L.R. are co-founders of Arsenal Biosciences and T.L.R. is chief scientific officer of the company.
Data availability
All the raw data and analyses are openly available through SRA (BioProject: PRJNA486372) and figshare (https://bit.ly/2PoccsB), respectively.
Code availability
The SPROUT software is publicly available at https://zou-group.github.io/SPROUT.
References
- 1.Fischbach MA, Bluestone JA & Lim WA Cell-Based Therapeutics: The Next Pillar of Medicine. Sci. Transl. Med. 5, 179ps7–179ps7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simeonov D et al. A large CRISPR-induced bystander mutation causes immune dysregulation. Commun. Biol. 2(1), 70 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hultquist JF et al. CRISPR-Cas9 genome engineering of primary CD4+ T cells for the interrogation of HIV-host factor interactions. Nat. Protoc. 14, 1–27 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindsay H et al. CrispRVariants charts the mutation spectrum of genome engineering experiments. Nat. Biotechnol. 34, 701 (2016). [DOI] [PubMed] [Google Scholar]
- 5.van Overbeek M et al. DNA Repair Profiling Reveals Nonrandom Outcomes at Cas9-Mediated Breaks. Mol. Cell 63, 633–646 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Brinkman EK et al. Kinetics and Fidelity of the Repair of Cas9-Induced Double-Strand DNA Breaks. Mol. Cell 70, 801–813.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lemos BR et al. CRISPR/Cas9 cleavages in budding yeast reveal templated insertions and strand-specific insertion/deletion profiles. Proc. Natl. Acad. Sci. U. S. A. 115, E2040–E2047 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deriano L & Roth DB Modernizing the nonhomologous end-joining repertoire: alternative and classical NHEJ share the stage. Annu. Rev. Genet. 47, 433–455 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Shen MW et al. Predictable and precise template-free CRISPR editing of pathogenic variants. Nature 563, 646–651 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allen F et al. Predicting the mutations generated by repair of Cas9-induced double-strand breaks. Nat. Biotechnol. (2018). doi: 10.1038/nbt.4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shin HY et al. CRISPR/Cas9 targeting events cause complex deletions and insertions at 17 sites in the mouse genome. Nat. Commun. 8, 15464 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kosicki M, Tomberg K & Bradley A Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 36, 765 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roth TL et al. Reprogramming human T cell function and specificity with non-viral genome targeting. Nature 559, 405–409 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simeonov D & Marson A CRISPR-Based Tools in Immunity. Annu. Rev. Immunol. 37 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Untergasser A et al. Primer3--new capabilities and interfaces. Nucleic Acids Res. 40, e115 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Magoč T & Salzberg SL FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bolger AM, Lohse M & Usadel B Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H & Durbin R Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.