Abstract
MicroRNAs (miRNAs) are known to have regulatory roles in the osteogenic differentiation of various mesenchymal stem cells (MSCs), although their regulatory role on human adipose-derived mesenchymal stem cells (hADSCs) remains unclear. The aim of the present study was to investigate the biological function and underlying molecular mechanism of miRNAs in regulating the osteogenic differentiation of hADSCs using microarray assay. hADSCs differentiated into osteoblasts under culture with osteogenic medium, with an increase observed in calcium deposits and alkaline phosphatase activity. The mRNA levels of bone sialoprotein, osteopontin and osteocalcin increased, whereas Runt-related transcription factor-2 expression decreased during osteogenic differentiation. In addition, miR-143 was markedly downregulated during osteogenic differentiation, while miR-143 overexpression inhibited and miR-143 knockdown enhanced this process. miR-143 overexpression also blocked extracellular signal-regulated kinase 1/2 (ERK1/2) pathway activation, while miR-143 inhibition enhanced it. The promoting effects of miR-143 knockdown on the osteogenic differentiation of hADSCs were partly diminished by the mitogen-activated protein kinase (MEK) inhibitors U0126 and PD98059. Bioinformatics analysis further revealed that miR-143 targets k-Ras and directly binds to the 3′-untranslated region of its mRNA. Inhibition of miR-143 enhanced the activation of the k-Ras/MEK/ERK pathway during osteogenic differentiation, whereas miR-143 overexpression had the opposite effect. Collectively, these results demonstrated that miR-143 negatively regulates the osteogenic differentiation of hADSCs through the k-Ras/MEK/ERK pathway, providing further insight into the underlying molecular mechanisms.
Keywords: microRNA-143, human adipose-derived mesenchymal stem cells, osteogenic differentiation, k-Ras/MEK/ERK signaling pathway
Introduction
Human adipose-derived mesenchymal stem cells (hADSCs) are a type of adult mesenchymal stem cells (MSCs) that have the plasticity to differentiate into osteoblasts, chondrocytes and adipocytes in response to the appropriate conditions (1-3). In recent studies, hADSCs have been successfully induced into an osteogenic lineage in vitro and have been used as seed cells to effectively repair bone defects (4,5). However, the molecular mechanisms governing the osteogenic differentiation of hADSCs are not fully elucidated, and thus investigating the potential mechanisms is of great importance.
MicroRNAs (miRNAs) are small (18-25 nucleotides in length), single-stranded noncoding RNAs that mediate gene suppression by binding to the 3′-untranslated region (3′UTR) of target mRNAs by promoting degradation or inhibiting the translation of target mRNAs (6,7). Recently, several studies have revealed that miRNAs serve critical roles in the regulation of MSC osteogenic differentiation. For instance, miR-145 was decreased during osteogenic differentiation of C2C12 and MC3T3-E1 cells, and may suppress their osteogenic differentiation potential by targeting Sp7 (8). Wang et al (9) further revealed that miR-193a served a suppressive role in the osteogenic differentiation of human bone marrow-derived stromal cells (hBMSCs) via targeting high mobility group box 1 (HMGB1). In addition, Li et al have indicated that miR-23a suppressed the osteogenic differentiation of hBMSCs by possibly targeting low-density lipoprotein receptor-related protein 5 (10). However, the roles of miRNAs in regulating the osteogenic differentiation of hADSCs remain largely unknown.
Osteogenic differentiation is a complex process governed by the interplay of multiple signaling pathways, such as bone morphogenetic protein (11), Wnt (12) and mitogen-activated protein kinase (MAPK) signaling pathways (13,14). These pathways are often constitutively activated during osteogenic differentiation of MSCs. It has been reported that MAPK signaling components, including extracellular-signal regulated kinase 1/2 (ERK1/2), strongly increased the expression of Runt-related transcription factor-2 (Runx2) protein, which is one of several key transcriptional factors in osteogenesis (15). Furthermore, several studies revealed that the ERK signaling pathway is closely associated with the osteogenic differentiation of rat and human MSCs (16,17). For example, Ye et al (18) demonstrated that knockdown of forkhead box protein A2 enhanced the osteogenic differentiation of BMSCs partly via activation of the ERK signaling pathway. Wang et al (19) further reported that naringin, a traditional Chinese medicine, enhanced the BMSC osteogenic differentiation through the activation of ERK signaling. Each of these studies has led us to speculate that ERK signaling may serve an important role in the differentiation of hADSCs into an osteogenic lineage.
In the present study, the expression profiles of miRNAs during osteogenic differentiation of hADSCs were analyzed using miRNA microarray, and revealed that miR-143 was significantly downregulated in this process. The study then investigated the underlying mechanisms involved in the regulatory role of miR-143 on hADSC osteogenic differentiation in order to identify a potential molecular therapeutic strategy for bone regeneration.
Materials and methods
Cell culture
All protocols involving human subjects were approved by the Ethics Committee of Minhang Hospital, Fudan University (Shanghai, China). Adipose tissue specimens were obtained from five healthy donors undergoing tumescence liposuction (age range, 32-53 years; median age, 41 years; 2 males and 3 females) who underwent surgery at Minhang Hospital, Fudan University between April 2017 and April 2018. Clinical and biochemical examinations confirmed that these subjects did not have acute inflammation, cancer, endocrine diseases or infectious diseases. The inclusion criteria were as follows: i) Patients who were willing to participate in the study; and ii) clinical and biochemical examinations confirmed that these subjects did not have acute inflammation, cancer, endocrine diseases or infectious diseases. Patients who received chemotherapy prior to the study were excluded from the present study. Written informed consent for participation in the study was obtained from all patients. The hADSCs were isolated from the adipose tissues according to a previously described method (20).
Following isolation, hADSCs were cultured in basal MSC culture medium (bM), containing Dulbecco's modified Eagle medium (DMEM), 10% fetal calf serum, 1% antibiotics (100 U/ml penicillin and 100 mg/ml streptomycin; Thermo Fisher Scientific, Inc.) and 1% L-glutamine (200 mM; Lonza) at 37°C with 5% CO2.
Adipogenic, osteogenic and chondrogenic differentiation
For adipogenic differentiation, hADSCs (3×105 cells/well) were seeded into 6-well culture plates and cultured for 21 days with adipogenic differentiation medium (Cyagen Bioscience, Inc.), and the medium was replaced every 3 days. Following the culture, the cells were washed with PBS and then fixed for 30 min at room temperature with 4% paraformaldehyde solution. Next, 0.6% Oil Red O (Sigma-Aldrich; Merck KGaA) solution was used to stain the fixed cells for 1 h at room temperature, and the cells were then observed using a phase contrast microscope (Motic).
For osteogenic differentiation, hADSCs (3×105 cells/well) were seeded into 6-well culture plates containing 3 ml bM. On the following day, the osteogenic differentiation was initiated by replacing the bM with osteogenic differentiation medium (odM), which consisted of DMEM, 100 nM dexamethasone, L-glutamine, 50 mM ascorbate, 1% antibiotics, mesangial cell growth supplement and 10 mM β-glycerophosphate (Lonza). The medium was replaced with fresh odM every 2-3 days thereafter. After the cells were cultured with odM for a maximum of 21 days, Alizarin Red S staining was performed to assess the osteogenic differentiation potential of hADSCs. For this, the cells were fixed in 4% paraformaldehyde solution for 15 min, followed by staining with 2% Alizarin Red S (Sigma-Aldrich; Merck KGaA) solution for 40 min at room temperature. Images were captured under a light microscope (DM-1000 Microscope; Leica Microsystems). Subsequently, the stained cells were incubated with 10% acetic acid for 30 min and collected with a cell scraper and vortexed. Samples were then heated at 85°C for 10 min, cooled down, and centrifuged at 20,000 × g for 15 min. The supernatant was neutralized with 10% ammonium hydroxide. The absorbance was measured at 405 nm as previously described (21).
For the differentiation of hADSCs into chondrocytes, an MSC Chondrogenic Differentiation kit (Cyagen Bioscience, Inc.) was used according to the manufacturer's protocol. Next, Alcian Blue staining was performed to assess the chondrogenic differentiation potential of hADSCs. Briefly, the cells were fixed with 4% paraformaldehyde solution for 30 min, washed three times with PBS, stained with 1% Alcian Blue (Solarbio Science & Technology Co., Ltd.) solution, and finally observed under a microscope (Olympus Corp.).
Alkaline phosphatase (ALP) activity
ALP activity was determined using an Alkaline Phosphatase Diethanolamine Activity Detection kit (cat. no. AP0100-1KT; Sigma-Aldrich; Merck KGaA) according to the manufacturer's protocol.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total cellular RNAs were isolated 7, 14 and 21 days after osteogenic differentiation using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. For miR-143 detection, RT was performed using the miScript II RT kit (Qiagen), while for the detection of Runx2, bone sialoprotein (BSP), osteopontin (OPN), osteocalcin (OCN) and k-Ras mRNA levels, RT was performed using the High Capacity cDNA reverse transcription kit (Invitrogen; Thermo Fisher Scientific, Inc.). miR-143 were measured using the miScript SYBR® Green PCR kit (Exiqon; Qiagen), while Runx2, OCN, BSP, OPN and k-Ras expression levels were measured using a Power SYBR Green PCR Master mix (Roche Diagnostics), on a Light Cycler instrument (Bio-Rad Laboratories, Inc.). The primers for qPCR analysis were as follows: miR-143 forward, 5′-ACA CTC CAG CTG GGG GTG CAG TGC TGC ATC -3′, and reverse, 5′-CTC AAC TGG TGT CGT GGA GTC GGC AAT TCA GTT GAG ACC AGA -3′; U6 forward, 5′-GCT TCG GCA GCA CAT ATA CTA AAA T-3′, and reverse, 5′-CGC TTC ACG AAT TTG CGT GTC AT-3′; Runx2 forward, 5′-GTC TCA CTG CCT CTC ACT TG-3′, and reverse, 5′-CAC ACA TCT CCT CCC TTC TG-3′; OCN forward, 5′-ACA GAC AAG TCC CAC ACA GCA GC-3′, and reverse, 5′-TGA AGG CTT TGT CAG ACT CAG GGC-3′; BSP forward, 5′-GCC AGA GGA GCA ATC ACC AA-3′, and reverse, 5′-CAG GCT GGA GGT TCA CTG GT-3′; OPN forward, 5′-TTG GCT TTG CAG TCT CCT GCG G-3′, and reverse, 5′-AGG CAA GGC CGA ACA GGC AAA-3′; k-Ras forward, 5′-ACT GAA TAT AAA CCT TGT GGT AG-3′, and reverse, 5′-TCA AAG AAT GGT CCT GGA CC-3′; GAPDH forward, 5′-CGA GCC ACA TCG CTC AGA CA-3′, and reverse, 5′-GTC TCA CTG CCT CTC ACT TG-3′. The thermocycling were as follows: 95°C for 1 min; and 40 cycles of 95°C for 30 sec, 58°C for 30 sec and 68°C for 3 min/kb; followed by 68°C for 10 min. The relative expression of each gene was calculated using the 2−∆∆Cq method (22).
Microarray assay
Total cellular RNA were extracted 14 days after osteoinduction using miRNeasy mini kit (Qiagen). Next, the samples were assessed using the miRCURY LNA™ Array kit, version 18.0 (Exiqon; Qiagen). The procedure and imaging processes were performed as previously described (23).
Cell transfection
When hADSCs in a 6-well plate were grown to ~80% confluence, miR-143 mimics, mimics negative control (NC), miR-143 inhibitor and inhibitor NC (Shanghai GenePharma Co., Ltd.) were transfected into the cells using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol.
In addition, transfection of cultured hADSCs with agomir-143, agomir NC, antagomir-143 and antagomir-NC (Shanghai GenePharma Co., Ltd.) was performed at a final concentration of 100 nM in bM. The medium was replaced with osteogenic medium after 16 h, and hADSCs were continuously cultured in odM for 14 days and then cells were harvested for RT-qPCR and western blot analyses.
MEK/ERK inhibition
To block the activity of the ERK1/2 signaling pathway, hADSCs were treated with the pathway inhibitors U0126 (10 µmol/l; cat no. 662009; Sigma-Aldrich; Merck KGaA) or PD98059 (10 µmol/l; cat no. P215; Sigma-Aldrich; Merck KGaA) at 37°C for 15 min.
Dual-luciferase reporter assay
miRNA target prediction tools, including Miranda (http://miranda.org.uk) and TargetScan v7.0 (http://targetscan.org/) were used to search for the putative targets of miR-143. Dual-Luciferase Reporter Assay system (Promega Corporation) was used to analyze the double luciferase activities according to the manufacturer's protocol. In brief, a sequence containing miR-143-predicted target within the k-Ras 3′ untranslated region (UTR) or a mutant sequence lacking any complementarity with miR-143 seed sequence were cloned in the 3′ UTR of the luciferase gene, generating the reporter vector wild-type-pGL3-k-Ras or mut-type-pGL3-k-Ras, respectively. When hADSCs grew to 60-70% confluence in 12-well plates, the cells were co-transfected with 50 nM miR-143 mimics, miR-143 inhibitor and 2 µg luciferase reporter plasmids using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Luciferase activity was determined 48 h after transfection. Normalization of firefly luciferase activity to Renilla luciferase activity was performed.
Western blot analysis
Western blot analysis was performed as previously described (19). Briefly, cells were lysed with RIPA buffer (cat. no. P0013C; Beyotime Institute of Biotechnology), and the total protein concentration was determined with using a bicinchoninic acid assay (cat. no. P0012; Beyotime Institute of Biotechnology). Subsequently, 40 µg protein samples were separated by 12% SDS-PAGE, and then transferred onto a polyvinylidene difluoride (Millipore) membrane, and this membrane was then blocked with 5% skim milk for 2 h at 4°C overnight. Next, the samples were probed with primary antibodies at 37°C for 2 h as follows: k-Ras (1:1,000; ab180772), p-p38 (1:1,000; ab31828) and c-Raf/1 (1:1,000; cat. no. ab137435), purchased from Abcam; p-c-Raf/1 (1:1,000; cat. no. sc-81513), obtained from Santa Cruz Biotechnology, Inc., mitogen-activated protein kinase 1/2 (MEK1/2; 1:1,000; cat. no. 9122), p-MEK1/2 (1:1,000; cat. no. 9121), ERK1/2 (1:2,000; cat. no. 4695), p-ERK1/2 (1:2,000; cat. no. 4370), c-Jun N-terminal kinase (JNK; 1:1,000; cat. no. 9258), p-JNK (1:1,000; cat. no. 4671) and p38 (1:2,000; cat. no. 9212), purchased from Cell Signaling Technology, Inc.; and β-actin (1:1,000; bs-0061R) from Beijing Biosynthesis Biotechnology Co., Ltd. Membranes were subsequently incubated with horseradish peroxidase-conjugated anti-rabbit IgG secondary antibody (1:2,000; cat. no. 7074; Cell Signaling Technology, Inc.) for 1 h at room temperature. Protein bands were visual-ized using an enhanced chemiluminescence detection system (GE Healthcare Life Sciences), and the blot bands were then quantified with ImageJ software (version 1.46; National Institutes of Health).
Statistical analysis
The difference between means was analyzed using Student's t-test and analysis of variance using SPSS software package, version 13.0 (SPSS, Inc.). All data are showed as the mean ± standard deviation (SD). A P-value of <0.05 was considered to denote a statistically significant difference.
Results
Aberrant expression of miRNAs during osteogenic differentiation
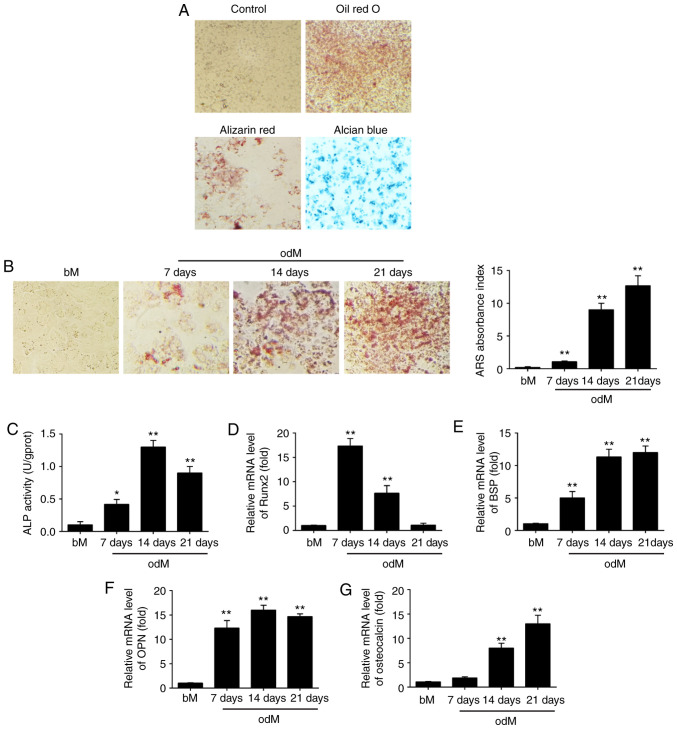
After hADSCs were isolated, the cells were cultured to induce adipogenic, osteogenic and chondrogenic differentiation to assess their multilineage differentiation potential. As shown in Fig. 1A, hADSCs were able to differentiate into adipocytes (Oil Red O staining), osteogenic cells (Alizarin Red S staining) and chondrocytes (Alcian Blue staining), which suggest that the isolated hADSCs exhibit the typical characteristics of MSCs. Next, Alizarin Red S staining was conducted to observe the changes of the extracellular matrix mineralization and the results showed that extracellular matrix mineralization was significantly increased under osteogenic differentiation conditions in a time-dependent manner (Fig. 1B). In addition, ALP activity was further measured by ALP assay. As shown in Fig. 1C, ALP activity increased with a peak at 14 days and then decreased, typical of the well-established program of osteoblast differentiation.
Figure 1.
miR-143 was downregulated during hADSC differentiation. (A) hADSCs were seeded in 6-well plates, incubated in normal, adipogenic, osteogenic or chondrogenic differentiation medium for 21 days, and then stained with Oil Red O, Alizarin Red S or Alcian Blue, respectively. Representative images are shown. The control group was stained by Alizarin Red S. Magnification, ×200. (B) Alizarin Red S staining to examine the mineralization of hADSCs cultured in odM or bM at 7, 14 and 21 days. A representative example of three independent experiments is shown. Magnification, ×200. (C) ALP activity was assessed by colorimetric assay. (D) Runx2, (E) BSP, (F) OPN and (G) OCN mRNA levels were assessed by RT-qPCR analysis on days 7, 14 and 21 post-induction of osteogenic differentiation. Data are represented as the mean ± standard deviation of three independent experiments. *P<0.05, **P<0.01 vs. bM group. miR, microRNA; hADSCs, human adipose-derived mesenchymal stem cells; odM, osteogenic differentiation medium; bM, basal medium; ALP, alkaline phosphatase; Runx2, Runt-related transcription factor-2; BSP, bone sialoprotein; OPN, osteopontin; OCN, osteocalcin; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Multiple transcription factors drive the control of osteogenesis, such as Runx2, BSP, OPN and OCN. Runx2 is a key transcriptional regulator that promotes differentiation by inducing the expression of osteo-specific genes during the early stages of differentiation (24), whereas OCN is one of the late-onset genes associated with osteogenesis (25). BSP and OPN are non-collagenous phosphoproteins proven to be involved in the mineralization of bone (26). In the present study, RT-qPCR was subsequently used to analyze the expression levels of these osteogenic specific genes. The results demonstrated that the expression of Runx2 was time-dependently decreased under osteogenic differentiation conditions, while the expression levels of BSP, OPN and OCN were significantly increased (Fig. 1D-G). These data suggested that osteogenic differentiation of hADSCs was successfully induced using an osteogenic induction medium.
miR-143 expression was downregulated during the osteogenic differentiation of hADSCs
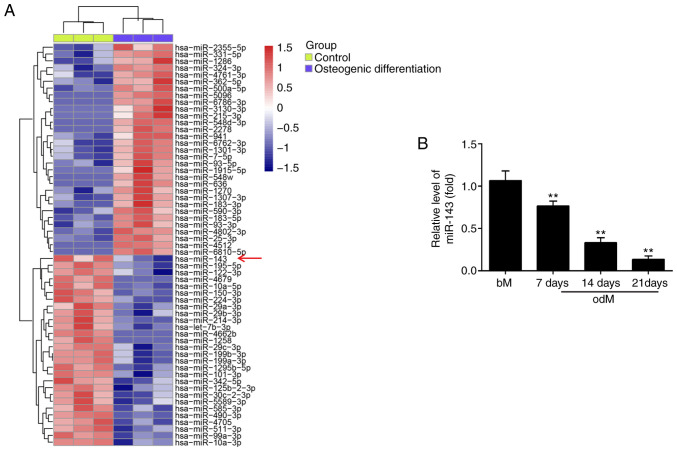
To screen the expression patterns of miRNAs involved in the osteogenic differentiation of hADSCs, a microarray analysis was performed. Compared with the control group, 31 miRNAs were upregulated and 28 miRNAs were downregulated following osteogenic differentiation (Fig. 2A). Among the aberrantly expressed miRNAs, miR-143 was selected for further investigation in the present study, since its expression level was the most significantly downregulated in the osteogenic differentiation group. Previous studies have reported the suppressive functions of miR-143 in different MSCs (18,27); however, whether miR-143 has a similar role in the osteogenic differentiation of hADSCs remains unknown. To further verify the microarray analysis results on miR-143 expression, the miR-143 levels were detected at different time points during the osteogenesis process. As shown in Fig. 2B, miR-143 expression was significantly decreased during this process in a time-dependent manner, compared with the control group. These results implied that miR-143 may be involved in the osteogenic differentiation of hADSCs.
Figure 2.
miR-143 expression is downregulated during the osteogenic differentiation of hADSCs. (A) Heat map of differentially expressed miRNAs between differentiated and non-differentiated hADSCs. Green indicates low expression levels, and red indicates high expression levels. (B) RT-qPCR analysis was used to assess the expression levels of miR-143 on days 7, 14 and 21 post-induction of osteogenic differentiation. Data are represented as the mean ± standard deviation of three independent experiments. **P<0.01 vs. bM group. miR, microRNA; hADSCs, human adipose-derived mesenchymal stem cells; odM, osteogenic differentiation medium; bM, basal medium; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
miR-143 negatively regulates the osteogenic differentiation of hADSCs
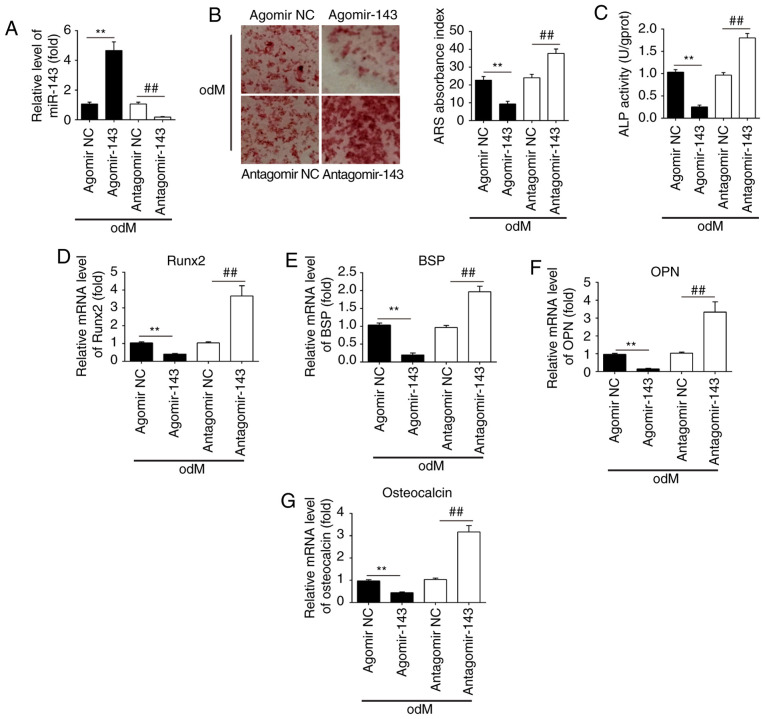
To investigate the role of miR-143 in the osteogenic differentiation of hADSCs, the cells were transfected with agomir-143 and antagomir-143, and the efficiency of transfection was determined by qPCR on day 21 of osteogenic differentiation of hADSCs. The results of RT-qPCR (Fig. 3A) revealed that miR-143 was notably upregulated and downregulated following agomir-143 and antagomir-143 transfection, respectively. Subsequently, Alizarin Red S and ALP staining were performed to evaluate the effects of miR-143 on calcium deposits and ALP activity during the osteogenic differentiation of hADSCs. The results demonstrated that agomir-143 treatment led to a significant decrease in both calcium deposits and ALP activity compared with those in the agomir-NC group, while antagomir-143 transfection markedly enhanced these processes (Fig. 3B and C). RT-qPCR was also conducted to assess the mRNA expression levels of the bone markers Runx2, OPN, BSP and OCN. The results demonstrated that agomir-143 significantly reduced the expression levels of these markers, whereas antagomir-143 significantly promoted these levels compared with those in the corresponding NC groups (Fig. 3D-G). Collectively, these data suggested that miR-143 was a negative regulator of hADSC osteogenesis.
Figure 3.
miR-143 negatively regulated the osteogenic differentiation of hADSCs. hADSCs were transfected with agomir-143, antagomir-143 or the corresponding NC (100 nM) for 16 h. The medium was then replaced with odM, and hADSCs were continuously cultured for 14 days. (A) miR-143 expression was assessed by RT-qPCR. (B) Osteogenic differentiation was determined by Alizarin Red S staining and (C) ALP activity was assessed by colorimetric assay on day 14 post-induction. Magnification, ×200. (D) Runx2, (E) BSP, (F) OPN and (G) osteocalcin mRNA levels were assessed by RT-qPCR analysis on day 14 post-induction. Data represent the mean ± standard deviation of three independent experiments. **P<0.01 and ##P<0.01. miR, microRNA; hADSCs, human adipose-derived mesenchymal stem cells; NC, negative control; odM, osteogenic differentiation medium; ALP, alkaline phosphatase; Runx2, Runt-related transcription factor-2; BSP, bone sialoprotein; OPN, osteopontin; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
miR-143 negatively regulates the ERK1/2 signaling pathway during the osteogenic differentiation of hADSCs
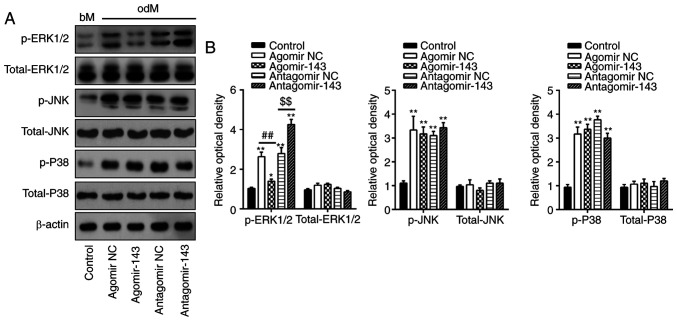
Previous studies have reported that MAPKs, including ERK, JNK and p38-MAPK, are important signal transducers during osteogenic differentiation of MSCs (28-30). Therefore, the present study attempted to investigate whether miR-143 can influence the MAPK (ERK1/2, JNK, and p38) signaling pathways during hADSC osteogenic differentiation. Consistent with the findings of previous studies (31,32), the levels of p-ERK1/2, p-JNK and p-p38 were significantly increased during osteogenic differentiation compared with undifferentiated control groups (Fig. 4A and B). Notably, agomir-143 and antagomir-143 transfection resulted in a decrease and increase, respectively, only in the expression of p-ERK1/2, whereas the expression levels of p-JNK or p-p38 were not markedly affected (Fig. 4A and B). Taken together, these results suggested that miR-143 was a negative regulator of the ERK1/2 signaling pathway during the osteogenic differentiation of hADSCs.
Figure 4.
miR-143 negatively regulated the ERK1/2 signaling pathway. hADSCs were transfected with agomir-143, antagomir-143 or the corresponding NC (100 nM) for 16 h. The medium was replaced with osteogenic medium, and hADSCs were continuously cultured for 14 days. (A) Western blot analysis was used to assess the protein expression levels of p-ERK1/2, ERK1/2, p-JNK, JNK, p-P38 and P38, with β-actin used as an internal control. (B) The bands were semi-quantitatively analyzed using ImageJ software and normalized to β-actin density. Data represent the mean ± standard deviation of three independent experiments. *P<0.05 and **P<0.01, vs. control group; ##P<0.01 and $$P<0.01. miR, microRNA; hADSCs, human adipose-derived mesenchymal stem cells; NC, negative control; ERK1/2, extracellular-signal regulated kinase 1/2; JNK, c-Jun N-terminal kinase.
miR-143 downregulation enhances osteogenic differentiation of hADSCs through activation of the ERK1/2 signaling pathway
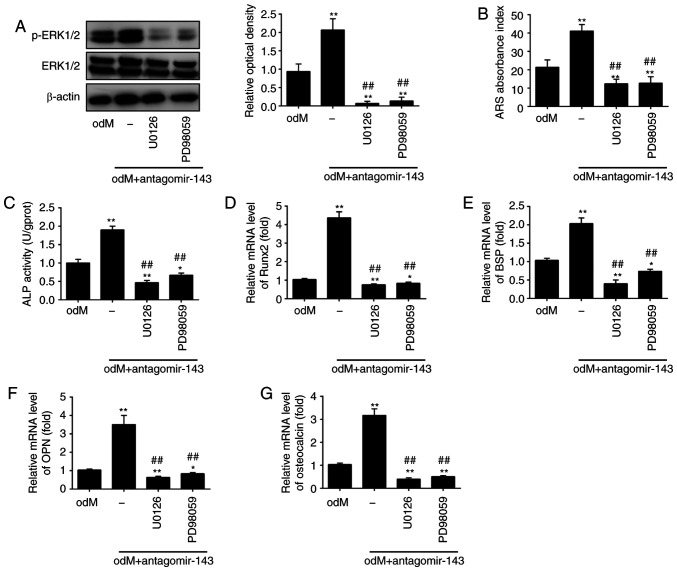
To investigate whether the ERK1/2 signaling pathway is critically involved in the positive effect of miR-143 on the osteogenic differentiation of hADSCs, the activity of the ERK1/2 signaling pathway was blocked using the pathway inhibitors U0126 and PD98059 (33). The results revealed that the level of p-ERK1/2 was markedly increased in the odM + antagomir-143 group compared with that in odM group; however, the protein levels of p-ERK1/2 were significantly decreased upon U0126 or PD98059 treatment (Fig. 5A). Next, Alizarin Red S and ALP staining were conducted to measure the calcium deposits and ALP activity, respectively. The results indicated that both U0126 and PD98059 markedly reversed the antagomir-143-induced increase in calcium deposits and ALP activity (Fig. 5B and C). In addition, the upregulation of Runx2, OPN, BSP and OCN levels induced by antagomir-143 was also abrogated by U0126 and PD98059 treatment (Fig. 5D-G). These results indicated the significance of the ERK1/2 signaling pathway in the osteogenic differentiation of hADSCs mediated by antagomir-143.
Figure 5.
miR-143 downregulation enhanced the osteogenic differentiation of human adipose-derived mesenchymal stem cells through activation of the ERK1/2 signaling pathway. Cells were treated with antagomir-143 or antagomir NC, with or without 10 µmol/l U0126 or PD98059 for 15 min. (A) Protein expression levels of p-ERK1/2 and ERK1/2, detected by western blot analysis and quantified using ImageJ software. (B) Graphical representation of the optical density following Alizarin Red S staining at day 21 of culture. (C) ALP activity assessed by colorimetric assay. (D) Runx2, (E) BSP, (F) OPN and (G) osteo-calcin mRNA levels were assessed by reverse transcription-quantitative polymerase chain reaction analysis. Data represent the mean ± standard deviation of three independent experiments. *P<0.05 and **P<0.01, vs. odM group; ##P<0.01 vs. antagomir-143 + odM group. miR, microRNA; ERK1/2, extracellular-signal regulated kinase 1/2; NC, negative control; ALP, alkaline phosphatase; Runx2, Runt-related transcription factor-2; BSP, bone sialoprotein; OPN, osteopontin; odM, osteogenic differentiation medium.
k-Ras is a direct target of miR-143
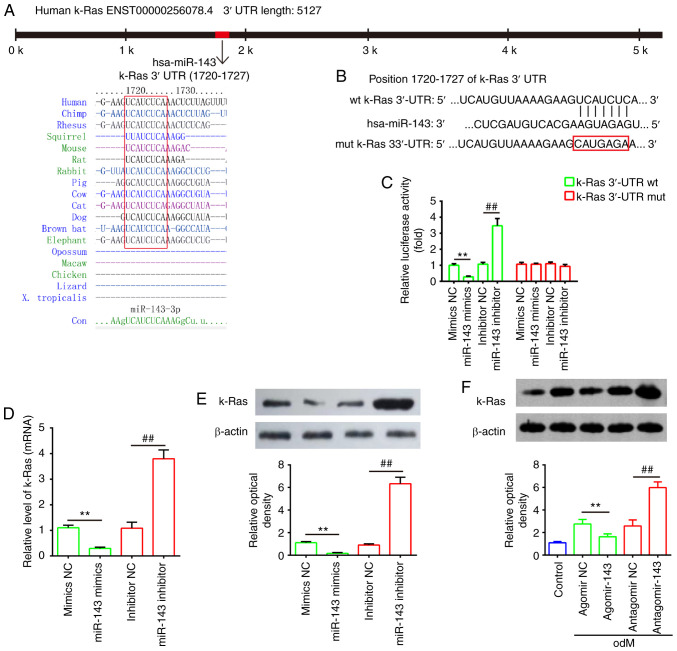
Through bioinformatics prediction using TargetScan 7.0 and miRanda, a putative target site of miR-143 was identified in the 3′-UTR of k-Ras mRNA (Fig. 6A and B). To further validate that miR-143 targeted k-Ras, a dual-luciferase reporter assay was conducted. The results demonstrated that miR-143 mimics markedly inhibited the luciferase activity of the k-Ras-3′UTR WT reporter, whereas co-transfection with the miR-143 inhibitor and WT reporter resulted in increased luciferase activity; however, no evident changes were observed following co-transfection of k-Ras 3′-UTR-Mut with miR-143 mimics or inhibitor (Fig. 6C). Furthermore, to determine whether miR-143 regulates k-Ras, the mRNA and protein levels of k-Ras were measured by RT-qPCR and western blot analysis, respectively. As shown in Fig. 6D and E, k-Ras expression was markedly decreased following miR-143 mimics transfection, while its expression was increased by miR-143 inhibitor in hADSCs at the mRNA and protein levels. It was also observed that k-Ras levels were markedly downregulated in hADSCs transfected with agomir-143 during osteogenic differentiation, but upregulated in cells transfected with antagomir-143 (Fig. 6F). These data indicated that miR-143 directly targeted k-Ras during the osteogenic differentiation of hADSCs.
Figure 6.
k-Ras was a direct target of miR-143. (A) Sequences of miR-143 binding sites are highly conserved among different species. (B) Putative binding site of miR-143 and k-Ras. (C) Luciferase activity of hADSCs co-transfected with luciferase reporter constructs containing WT or Mut k-Ras 3′-UTR, and with miR-143 mimics, miR-143 inhibitor or NC (n=3). (D) Reverse transcription-quantitative polymerase chain reaction and (E) western blot analysis of mRNA and protein expression levels of k-Ras in hADSCs transfected with miR-143 mimics, miR-143 inhibitor or NC miRNA (n=3). (F) Western blot analysis of k-Ras protein expression in hADSCs transfected with agomir-143, antagomir-143 and the corresponding NC. The medium was replaced with odM after 16 h, and hADSCs were continuously cultured for 14 days prior to protein level assessment. Data represent the mean ± standard deviation of three independent experiments. **P<0.01 and ##P<0.01. miR, microRNA; hADSCs, human adipose-derived mesenchymal stem cells; WT, wild-type; Mut, mutant; NC, negative control; odM, osteogenic differentiation medium.
miR-143 negatively regulates the k-Ras/MEK/ERK signaling pathway during osteogenic differentiation of hADSCs
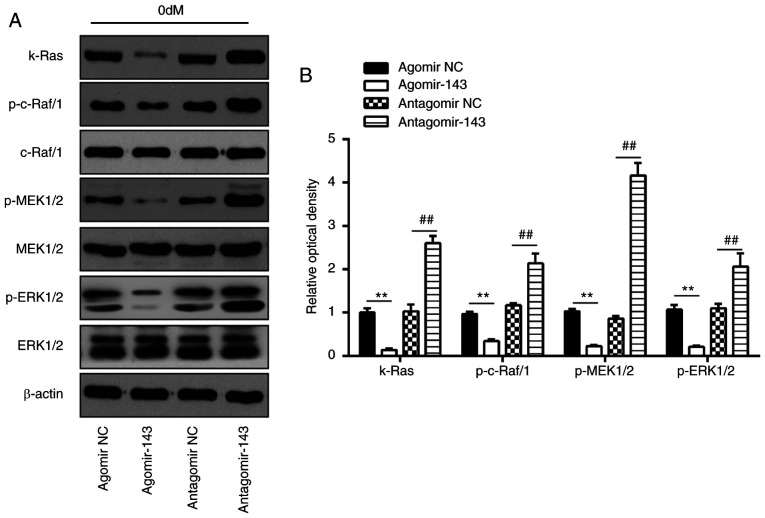
It has been reported that k-Ras functions as an important activator of the ERK signaling pathway, which has been implicated in the regulation of the osteogenic differentiation of MSCs (34). Given this, in the present study, it was speculated that miR-143 may modulate the ERK signaling pathway via targeting k-Ras during osteogenic differentiation of hADSCs. As shown in Fig. 7A and B, miR-143 overexpression significantly decreased the expression levels of k-Ras, p-c-Raf/1, p-MEK1/2 and p-ERK1/2, as compared with those in the agomir NC group. By contrast, inhibition of miR-143 enhanced the expression levels of these proteins when compared with those in antagomir-NC group. These findings indicated that inhibition of miR-143 may promote the osteogenic differentiation of hADSCs via activating the k-Ras/MEK/ERK signaling pathway.
Figure 7.
miR-143 negatively regulated k-Ras/MEK/ERK signaling pathway during osteogenic differentiation of hADSCs. hADSCs were transfected with agomir-143, antagomir-143 or the corresponding NC, and osteogenic differentiation medium was added. (A) Western blot analysis was used to assess the protein expression levels of k-Ras, p-Raf/1, Raf/1, p-MEK1/2, MEK1/2, p-ERK1/2 and ERK1/2, with β-actin used as an internal control. (B) The bands were semi-quantitatively analyzed using ImageJ software and normalized to β-actin density. Data represent the mean ± standard deviation of three independent experiments. **P<0.01 and ##P<0.01. miR, microRNA; hADSCs, human adipose-derived mesenchymal stem cells; MEK, mitogen-activated protein kinase; ERK, extracellular-signal regulated kinase; NC, negative control.
Discussion
In the present study, the data revealed that miR-143 was down-regulated during the osteogenic differentiation of hADSCs. Functional analyses indicated that miR-143 overexpression suppressed the osteogenic potential of hADSCs, whereas miR-143 inhibition enhanced this potential. Notably, the current study data revealed that miR-143 knockdown promoted the osteogenic differentiation of these cells by activating the k-Ras/MEK/ERK signaling pathway. These findings suggested that miR-143 serves a potentially crucial role in the osteogenic differentiation of hADSCs.
Mounting evidence has indicated the vital roles of several other miRNAs in the osteogenic differentiation of hADSCs. For instance, Kim et al (35) reported that miR-196a is a key modulator of this process in hADSCs, and lentiviral overexpression of this miRNA enhanced osteogenic differentiation by targeting HOXC8. Luzi et al (36) observed that miR-26a expression increased during hADSC differentiation, while inhibition of miR-26a modulated late osteogenic differentiation by targeting the SMAD1 transcription factor. Zeng et al (37) also identified miR-100 as a negative regulator of the osteogenic differentiation of hADSCs, and overexpression of miR-100 inhibited this differentiation. In the present study, using an miRNA microarray, several miRNAs were found to be differently expressed during hADSC osteogenic differentiation. In particular, the study focused on miR-143, since its expression was decreased the most during this process. It has previously been reported that miR-143 was downregulated in MSC osteogenic differentiation and negatively regulated this process by targeting different genes (27,38). However, little is known regarding the effect of miR-143 on the differentiation of hADSCs. To the best of our knowledge, the present study investigated is the first to investigate the role of miR-143 in the osteogenic differentiation of hADSCs. The data revealed that overexpression of miR-143 significantly inhibited osteogenic differentiation, while inhibition of miR-143 enhanced this process, suggesting the important role of this miRNA in this cellular change.
Activation of the MAPK signaling pathway, particularly of ERK1/2, has been implicated in the process of osteogenic differentiation of hADSCs (18) and human periodontal ligament stem cells (hPDLSCs) (33). For instance, activation of the ERK1/2 signaling pathway has been demonstrated to mediate the promoting effect of fibroblast growth factor-7 on the osteogenic differentiation of embryonic stem cells (39) and the naringin-induced osteogenic differentiation of immortalized hPDLSCs (33). Notably, previous studies have reported that miRNAs are important in the regulation of osteogenic differentiation of MSCs through activating the ERK1/2 signaling pathway. For example, Mei et al (40) revealed that miR-21 modulated ERK-MAPK signaling activity by repressing SPRY2 expression to affect the human MSC differentiation. Of note, Dong and Hu (41) demonstrated that overexpression of miR-143 suppressed the phosphorylation of ERK1/2 in non-small-cell lung cancer PC9/GR cells. To further elucidate the mechanisms by which miR-143 promotes the osteogenic differentiation of hADSCs, the effects of miR-143 downregulation on key kinases in the MAPK signaling pathway were examined in the present study. The data revealed that only the ERK1/2 signaling pathway was regulated by miR-143, whereas the p38 and JNK pathways did not appear to be involved. Further assessments indicated that the promoting effect of antagomir-143 on the osteogenic differentiation of hADSCs was markedly reversed by treatment with MEK-specific inhibitors, U0126 and PD98059, which suggested that the promoting effects of miR-143 on the osteogenic differentiation of hADSCs may be mediated by the ERK1/2 signaling pathway.
It is well-known that k-Ras is an upstream regulator of the ERK1/2 signaling pathway (42). Under physiological conditions, k-Ras stimulates the transformation of Ras-GTP to Ras-GDP and promotes the activation of downstream of MEK, which in turn activates the ERK1/2 pathway (43,44). The Ras/Raf/MEK/ERK signaling pathway has been reported to be closely associated with osteogenic differentiation of MSCs. A previous study by Feng et al (45) indicated that the HMGB1 promoted the secretion of multiple cytokines and potentiated the osteogenic differentiation of MSCs through promoting the activation of the Ras/MAPK signaling pathway. Another study also revealed that strontium enhanced the MSC osteo-genic differentiation by activating the Ras/MAPK signaling pathway (46). It has previously been reported that miR-143 directly targets k-Ras in human cancer (47,48); however, whether k-Ras is a functional target of miR-143 during hADSC osteogenic differentiation was unclear. In the present study, k-Ras was confirmed as a direct target of miR-143 in hADSCs. Furthermore, it was observed that miR-143 blocked the activation of k-Ras/c-Raf/MEK/ERK signaling, whereas inhibition of miR-143 enhanced this signaling. Collectively, these findings indicated that miR-143 may regulate the osteogenic differentiation of hADSCs through the k-Ras/MEK/ERK signaling pathway.
To date, the majority of cell-based therapies are conducted using the well-characterized BMSCs. The osteogenic process of BMSCs is a critical step for bone formation, and it has been already used in clinical trials for bone damage treatment (49-51). Compared with the BMSCs, hADSCs displayed a significantly higher proliferating ability and differentiation potential, thus confirming that hADSCs are a better candidate for clinical application (52,53). In addition, hADSC harvesting involves less pain and ethical issues (54). Since BMSCs have been involved for some time in clinical protocols of cell therapy, we will further verify the results of the present study on BMSCs in the future.
In conclusion, the data demonstrated that miR-143 was the most significantly downregulated miRNA during osteogenesis in hADSC. Furthermore, miR-143 inhibition promoted the osteogenic differentiation of hADSCs by activating k-Ras/c-Raf/MEK/ERK pathway. The present study may provide a theoretical basis for the development of stem cell-based therapy of bone fractures in the future.
Acknowledgments
Not applicable.
Funding
This study was supported by the Shanghai Municipal Planning Commission of Science and Research Fund (grant no. 201640219).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
YZ, KZ, LW, HG and ZH performed the experiments, analysed the data and wrote the paper. JX conceptualized the study design, and contributed to data analysis and experimental materials. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All individuals provided informed consent for the use of human specimens for clinical research. The present study was approved by the Ethics Committee of Minhang Hospital, Fudan University (Shanghai, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Locke M, Feisst V, Dunbar PR. Concise review: Human adipose-derived stem cells: Separating promise from clinical need. Stem Cells. 2011;29:404–411. doi: 10.1002/stem.593. [DOI] [PubMed] [Google Scholar]
- 2.Wang S, Qu X, Zhao RC. Mesenchymal stem cells hold promise for regenerative medicine. Front Med. 2011;5:372–378. doi: 10.1007/s11684-011-0164-4. [DOI] [PubMed] [Google Scholar]
- 3.Gimble JM, Guilak F. Differentiation potential of adipose derived adult stem (ADAS) cells. Curr Top Dev Biol. 2003;58:137–160. doi: 10.1016/S0070-2153(03)58005-X. [DOI] [PubMed] [Google Scholar]
- 4.Alvira-González J, Sánchez-Garcés MÀ, Cairó JR, Del Pozo MR, Sánchez CM, Gay-Escoda C. Assessment of bone regeneration using adipose-derived stem cells in critical-size alveolar ridge defects: An experimental study in a dog model. Int J Oral Maxillofac Implants. 2016;31:196–203. doi: 10.11607/jomi.4190. [DOI] [PubMed] [Google Scholar]
- 5.Sánchez-Garcés MÀ, Alvira-González J, Sánchez CM, Barbany Cairó JR, Del Pozo MR, Gay-Escoda C. Bone regeneration using adipose-derived stem cells with fibronectin in dehiscence-type defects associated with dental implants: An experimental study in a dog model. Int J Oral Maxillofac Implants. 2017;32:e97–e106. doi: 10.11607/jomi.5169. [DOI] [PubMed] [Google Scholar]
- 6.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Jia J, Tian Q, Ling S, Liu Y, Yang S, Shao Z. miR-145 suppresses osteogenic differentiation by targeting Sp7. FEBS Lett. 2013;587:3027–3031. doi: 10.1016/j.febslet.2013.07.030. [DOI] [PubMed] [Google Scholar]
- 9.Wang SN, Zhao XQ, Yu B, Wang BW. miR-193a inhibits osteogenic differentiation of bone marrow-derived stroma cell via targeting HMGB1. Biochem Biophys Res Commun. 2018;503:536–543. doi: 10.1016/j.bbrc.2018.05.132. [DOI] [PubMed] [Google Scholar]
- 10.Li C, Yang D, Cao X, Wang F, Jiang H, Guo H, Du L, Guo Q, Yin X. LFG-500, a newly synthesized flavonoid, attenuates lipopolysaccharide-induced acute lung injury and inflammation in mice. Biochem Pharmacol. 2016;113:57–69. doi: 10.1016/j.bcp.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 11.Afzal F, Pratap J, Ito K, Ito Y, Stein JL, van Wijnen AJ, Stein GS, Lian JB, Javed A. Smad function and intranuclear targeting share a Runx2 motif required for osteogenic lineage induction and BMP2 responsive transcription. J Cell Physiol. 2005;204:63–72. doi: 10.1002/jcp.20258. [DOI] [PubMed] [Google Scholar]
- 12.Da Forno PD, Pringle JH, Hutchinson P, Osborn J, Huang Q, Potter L, Hancox RA, Fletcher A, Saldanha GS. WNT5A expression increases during melanoma progression and correlates with outcome. Clin Cancer Res. 2008;14:5825–5832. doi: 10.1158/1078-0432.CCR-07-5104. [DOI] [PubMed] [Google Scholar]
- 13.Corrêa SA, Eales KL. The Role of p38 MAPK and its substrates in neuronal plasticity and neurodegenerative disease. J Signal Transduct. 2012;2012:649079. doi: 10.1155/2012/649079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thouverey C, Caverzasio J. The p38alpha MAPK positively regulates osteoblast function and postnatal bone acquisition. Cell Mol Life Sci. 2012;69:3115–3125. doi: 10.1007/s00018-012-0983-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ying X, Cheng S, Wang W, Lin Z, Chen Q, Zhang W, Kou D, Shen Y, Cheng X, Peng L, Zi Xu H, Zhu Lu C. Effect of lactoferrin on osteogenic differentiation of human adipose stem cells. Int Orthop. 2012;36:647–653. doi: 10.1007/s00264-011-1303-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y, Wang J, Chen G, Feng S, Wang P, Zhu X, Zhang R. Quercetin promotes the osteogenic differentiation of rat mesen-chymal stem cells via mitogen-activated protein kinase signaling. Exp Ther Med. 2015;9:2072–2080. doi: 10.3892/etm.2015.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou C, Zhang X, Xu L, Wu T, Cui L, Xu D. Taurine promotes human mesenchymal stem cells to differentiate into osteoblast through the ERK pathway. Amino Acids. 2014;46:1673–1680. doi: 10.1007/s00726-014-1729-8. [DOI] [PubMed] [Google Scholar]
- 18.Ye C, Chen M, Chen E, Li W, Wang S, Ding Q, Wang C, Zhou C, Tang L, Hou W, et al. Knockdown of FOXA2 enhances the osteogenic differentiation of bone marrow-derived mesenchymal stem cells partly via activation of the ERK signalling pathway. Cell Death Dis. 2018;9:836. doi: 10.1038/s41419-018-0857-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H, Li C, Li J, Zhu Y, Jia Y, Zhang Y, Zhang X, Li W, Cui L, Li W, Liu Y. Naringin enhances osteogenic differentiation through the activation of ERK signaling in human bone marrow mesenchymal stem cells. Iran J Basic Med Sci. 2017;20:408–414. doi: 10.22038/IJBMS.2017.8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boquest AC, Shahdadfar A, Brinchmann JE, Collas P. Isolation of stromal stem cells from human adipose tissue. Methods Mol Biol. 2006;325:35–46. doi: 10.1385/1-59745-005-7:35. [DOI] [PubMed] [Google Scholar]
- 21.Li B, Qu C, Chen C, Liu Y, Akiyama K, Yang R, Chen F, Zhao Y, Shi S. Basic fibroblast growth factor inhibits osteogenic differentiation of stem cells from human exfoliated deciduous teeth through ERK signaling. Oral Dis. 2012;18:285–292. doi: 10.1111/j.1601-0825.2011.01878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Mei LL, Wang WJ, Qiu YT, Xie XF, Bai J, Shi ZZ. miR-125b-5p functions as a tumor suppressor gene partially by regulating HMGA2 in esophageal squamous cell carcinoma. PLoS One. 2017;12:e0185636. doi: 10.1371/journal.pone.0185636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu TM, Lee EH. Transcriptional regulatory cascades in Runx2-dependent bone development. Tissue Eng Part B Rev. 2013;19:254–263. doi: 10.1089/ten.teb.2012.0527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee MJ, Chen HT, Ho ML, Chen CH, Chuang SC, Huang SC, Fu YC, Wang GJ, Kang L, Chang JK. PPARγ silencing enhances osteogenic differentiation of human adipose-derived mesenchymal stem cells. J Cell Mol Med. 2013;17:1188–1193. doi: 10.1111/jcmm.12098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ying X, Chen X, Cheng S, Guo X, Chen H, Xu HZ. Phosphoserine promotes osteogenic differentiation of human adipose stromal cells through bone morphogenetic protein signalling. Cell Biol Int. 2014;38:309–317. doi: 10.1002/cbin.10203. [DOI] [PubMed] [Google Scholar]
- 27.Li E, Zhang J, Yuan T, Ma B. MiR-143 suppresses osteogenic differentiation by targeting Osterix. Mol Cell Biochem. 2014;390:69–74. doi: 10.1007/s11010-013-1957-3. [DOI] [PubMed] [Google Scholar]
- 28.Ge C, Xiao G, Jiang D, Franceschi RT. Critical role of the extracellular signal-regulated kinase-MAPK pathway in osteoblast differentiation and skeletal development. J Cell Biol. 2007;176:709–718. doi: 10.1083/jcb.200610046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greenblatt MB, Shim JH, Glimcher LH. Mitogen-activated protein kinase pathways in osteoblasts. Annu Rev Cell Dev Biol. 2013;29:63–79. doi: 10.1146/annurev-cellbio-101512-122347. [DOI] [PubMed] [Google Scholar]
- 30.Artigas N, Ureña C, Rodriguez-Carballo E, Rosa JL, Ventura F. Mitogen-activated protein kinase (MAPK)-regulated interactions between Osterix and Runx2 are critical for the transcriptional osteogenic program. J Biol Chem. 2014;289:27105–27117. doi: 10.1074/jbc.M114.576793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park S, Arai Y, Kim BJ, Bello A, Ashraf S, Park H, Park KS, Lee SH. SPRY4 suppression facilitates osteogenic differentiation in adipose-derived mesenchymal stem cells. Tissue Eng Part A. 2019;25:1645–1657. doi: 10.1089/ten.tea.2019.0056. [DOI] [PubMed] [Google Scholar]
- 32.Yang X, Yang Y, Zhou S, Gong X, Dai Q, Zhang P, Jiang L. Puerarin stimulates osteogenic differentiation and bone formation through the ERK1/2 and p38-MAPK signaling pathways. Curr Mol Med. 2018;17:488–496. doi: 10.2174/1566524018666171219101142. [DOI] [PubMed] [Google Scholar]
- 33.Wei K, Xie Y, Chen T, Fu B, Cui S, Wang Y, Cai G, Chen X. ERK1/2 signaling mediated naringin-induced osteogenic differentiation of immortalized human periodontal ligament stem cells. Biochem Biophys Res Commun. 2017;489:319–325. doi: 10.1016/j.bbrc.2017.05.130. [DOI] [PubMed] [Google Scholar]
- 34.Wei F, Liu Y, Bellail AC, Olson JJ, Sun SY, Lu G, Ding L, Yuan C, Wang G, Hao C. K-Ras mutation-mediated IGF-1-induced feedback ERK activation contributes to the rapalog resistance in pancreatic ductal adenocarcinomas. Cancer Lett. 2012;322:58–69. doi: 10.1016/j.canlet.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim YJ, Bae SW, Yu SS, Bae YC, Jung JS. miR-196a regulates proliferation and osteogenic differentiation in mesenchymal stem cells derived from human adipose tissue. J Bone Miner Res. 2009;24:816–825. doi: 10.1359/jbmr.081230. [DOI] [PubMed] [Google Scholar]
- 36.Luzi E, Marini F, Sala SC, Tognarini I, Galli G, Brandi ML. Osteogenic differentiation of human adipose tissue-derived stem cells is modulated by the miR-26a targeting of the SMAD1 transcription factor. J Bone Miner Res. 2008;23:287–295. doi: 10.1359/jbmr.071011. [DOI] [PubMed] [Google Scholar]
- 37.Zeng Y, Qu X, Li H, Huang S, Wang S, Xu Q, Lin R, Han Q, Li J, Zhao RC. MicroRNA-100 regulates osteogenic differentiation of human adipose-derived mesenchymal stem cells by targeting BMPR2. FEBS Lett. 2012;586:2375–2381. doi: 10.1016/j.febslet.2012.05.049. [DOI] [PubMed] [Google Scholar]
- 38.Zhang P, Yang W, Wang G, Li Y. miR-143 suppresses the osteogenic differentiation of dental pulp stem cells by inactivation of NF-κB signaling pathway via targeting TNF-α. Arch Oral Biol. 2018;87:172–179. doi: 10.1016/j.archoralbio.2017.12.031. [DOI] [PubMed] [Google Scholar]
- 39.Jeon YM, Kook SH, Rho SJ, Lim SS, Choi KC, Kim HS, Kim JG, Lee JC. Fibroblast growth factor-7 facilitates osteogenic differentiation of embryonic stem cells through the activation of ERK/Runx2 signaling. Mol Cell Biochem. 2013;382:37–45. doi: 10.1007/s11010-013-1716-5. [DOI] [PubMed] [Google Scholar]
- 40.Mei Y, Bian C, Li J, Du Z, Zhou H, Yang Z, Zhao RC. miR-21 modulates the ERK-MAPK signaling pathway by regulating SPRY2 expression during human mesenchymal stem cell differentiation. J Cell Biochem. 2013;114:1374–1384. doi: 10.1002/jcb.24479. [DOI] [PubMed] [Google Scholar]
- 41.Dong YZ, Hu T. Effects of miR-143 overexpression on proliferation, apoptosis, EGFR and downstream signaling pathways in PC9/GR cell line. Eur Rev Med Pharmacol Sci. 2018;22:1709–1716. doi: 10.26355/eurrev_201803_14584. [DOI] [PubMed] [Google Scholar]
- 42.Castellano E, Santos E. Functional specificity of ras isoforms: So similar but so different. Genes Cancer. 2011;2:216–231. doi: 10.1177/1947601911408081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng D, Zhao L, Xu Y, Ou R, Li G, Yang H, Li W. K-Ras promotes the non-small lung cancer cells survival by cooperating with sirtuin 1 and p27 under ROS stimulation. Tumour Biol. 2015;36:7221–7232. doi: 10.1007/s13277-015-3429-8. [DOI] [PubMed] [Google Scholar]
- 44.Georgieva M, Krasteva M, Angelova E, Ralchev K, Dimitrov V, Bozhimirov S, Georgieva E, Berger MR. Analysis of the K-ras/B-raf/Erk signal cascade, p53 and CMAP as markers for tumor progression in colorectal cancer patients. Oncol Rep. 2008;20:3–11. [PubMed] [Google Scholar]
- 45.Feng L, Xue D, Chen E, Zhang W, Gao X, Yu J, Feng Y, Pan Z. HMGB1 promotes the secretion of multiple cytokines and potentiates the osteogenic differentiation of mesenchymal stem cells through the Ras/MAPK signaling pathway. Exp Ther Med. 2016;12:3941–3947. doi: 10.3892/etm.2016.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peng S, Zhou G, Luk KD, Cheung KM, Li Z, Lam WM, Zhou Z, Lu WW. Strontium promotes osteogenic differentiation of mesenchymal stem cells through the Ras/MAPK signaling pathway. Cell Physiol Biochem. 2009;23:165–174. doi: 10.1159/000204105. [DOI] [PubMed] [Google Scholar]
- 47.Qin HX, Cui HK, Pan Y, Hu RL, Zhu LH, Wang SJ. miR-143 inhibits cell proliferation through targeted regulating the expression of K-ras gene in HeLa cells. Zhonghua Zhong Liu Za Zhi. 2016;38:893–897. doi: 10.3760/cma.j.issn.0253-3766.2016.12.003. In Chinese. [DOI] [PubMed] [Google Scholar]
- 48.Takai T, Tsujino T, Yoshikawa Y, Inamoto T, Sugito N, Kuranaga Y, Heishima K, Soga T, Hayashi K, Miyata K, et al. Synthetic miR-143 exhibited an anti-cancer effect via the down-regulation of K-RAS networks of renal cell cancer cells in vitro and in vivo. Mol Ther. 2019;27:1017–1027. doi: 10.1016/j.ymthe.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caplan AI. Review: Mesenchymal stem cells: Cell-based reconstructive therapy in orthopedics. Tissue Eng. 2005;11:1198–1211. doi: 10.1089/ten.2005.11.1198. [DOI] [PubMed] [Google Scholar]
- 50.Arinzeh TL. Mesenchymal stem cells for bone repair: Preclinical studies and potential orthopedic applications. Foot Ankle Clin. 2005;10:651–665. viii. doi: 10.1016/j.fcl.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 51.Bhansali S, Dutta P, Kumar V, Yadav MK, Jain A, Mudaliar S, Bhansali S, Sharma RR, Jha V, Marwaha N, et al. Efficacy of autologous bone marrow-derived mesenchymal stem cell and mononuclear cell transplantation in type 2 diabetes mellitus: A randomized, placebo-controlled comparative study. Stem Cells Dev. 2017;26:471–481. doi: 10.1089/scd.2016.0275. [DOI] [PubMed] [Google Scholar]
- 52.Zuk PA, Zhu M, Mizuno H, Huang J, Futrell JW, Katz AJ, Benhaim P, Lorenz HP, Hedrick MH. Multilineage cells from human adipose tissue: Implications for cell-based therapies. Tissue Eng. 2001;7:211–228. doi: 10.1089/107632701300062859. [DOI] [PubMed] [Google Scholar]
- 53.Tobita M, Orbay H, Mizuno H. Adipose-derived stem cells: Current findings and future perspectives. Discov Med. 2011;11:160–170. [PubMed] [Google Scholar]
- 54.Sempere JM, Martinez-Peinado P, Arribas MI, Reig JA, De La Sen ML, Zubcoff JJ, Fraga MF, Fernández AF, Santana A, Roche E. Single cell-derived clones from human adipose stem cells present different immunomodulatory properties. Clin Exp Immunol. 2014;176:255–265. doi: 10.1111/cei.12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.