Abstract
The role of T cells in the resolution or exacerbation of COVID-19, as well as their potential to provide long-term protection from reinfection with SARS-CoV-2, remains debated. Nevertheless, recent studies have highlighted various aspects of T cell responses to SARS-CoV-2 infection that are starting to enable some general concepts to emerge.
Subject terms: SARS-CoV-2, Cellular immunity, T cells
In this Progress article, Zeyu Chen and E. John Wherry summarize early reports of the T cell responses observed in patients with COVID-19, emphasizing how different immune response characteristics in different patients may reflect a spectrum of disease phenotypes.
Introduction
COVID-19 caused by SARS-CoV-2 infection is a global pandemic, with more than 15.8 million infections and more than 641,000 deaths as of 25 July 2020, according to the COVID-19 Map of the Johns Hopkins Coronavirus Resource Center. SARS-CoV-2 infection can result in a range of clinical manifestations, from asymptomatic or mild infection to severe COVID-19 that requires hospitalization. Patients who are hospitalized often progress to severe pneumonia and acute respiratory distress syndrome (ARDS)1–3. Although relatively little is known about the immunology of individuals who are asymptomatic or individuals with mild disease who do not require hospitalization, recent studies have revealed important insights into the immune responses of patients who are hospitalized. Similar to other respiratory viral infections, adaptive immune responses4–9, particularly of T cells6,8,10, have a prominent role in SARS-CoV-2 infection. However, it remains unclear whether T cell responses are helpful or harmful in COVID-19, and whether T cell responses are suboptimal and dysfunctional or excessive, with evidence having been provided for both ends of the spectrum.
Here, we summarize some of the recent data on conventional T cell responses in COVID-19, noting, however, that emerging data also highlight impacts on other lymphocyte populations, including B cells4,8,11, innate lymphoid cells4, natural killer cells4,11, mucosa-associated invariant T cells4 and γδT cells11. We highlight some of the key observations made for conventional αβ CD8+ and CD4+ T cells in COVID-19, including the prominent lymphopenia observed in patients with severe disease, relevant features of CD8+ versus CD4+ T cell responses in patients who are hospitalized, features of T cell differentiation that may be altered and current data on whether the overall magnitude of the T cell response in patients with COVID-19 is insufficient or excessive and how these features may relate to disease. Finally, we discuss emerging data on SARS-CoV-2-specific T cells found in patients who have recovered from COVID-19 and the implications for T cell memory. In this rapidly emerging field, we summarize the most recent studies that have addressed T cell responses in COVID-19. Some of these are as yet only available on preprint servers and conclusions from non-peer-reviewed data should be treated with caution. A summary of the data sets that are published versus those available as preprints is provided in Table 1 to aid the reader in establishing the weight of evidence for particular features.
Table 1.
Studies reporting T cell analysis in patients with COVID-19
| Donor cohort | Sample origin | Profiling technology used | Major conclusions for αβT cells | Refs |
|---|---|---|---|---|
| 3 healthy, 3 mild/moderate disease, 6 severe disease | Bronchoalveolar lavage fluid | 10x Genomics scRNA-seq, 10× Genomics scTCR-seq | Greater clonal expansion of T cells in moderate disease than severe disease; T cells in moderate disease have stronger signatures of tissue residency | 5 |
| 8 moderate disease, 11 severe disease | Nasopharyngeal and bronchial samples | 10x Genomics scRNA-seq | Fewer CTLs in severe disease than moderate disease; hyperactivation of CTLs in the respiratory tract, with a signature of interacting with epithelial cells and other immune cell types | 9 |
| 5 healthy, 5 early-recovered, 5 late-recovered | PBMCs | 10x Genomics scRNA-seq, 10x Genomics scTCR-seq | Greater clonal expansion of T cells in late-recovered than in early-recovered patients; fewer CD8+ T cells but greater cytotoxic signatures in early-recovered than in late-recovered patients | 28 |
| 6 healthy, 3 non-ventilated, 4 with ARDS | PBMCs | Seq-Well scRNA-seq | Heterogeneity of immune responses, including of interferon-stimulated genes; no transcriptional signature of exhaustion; features of T cell hyperactivation in some of the patients with ARDS | 7 |
| 3 healthy, 6 mild/moderate disease, 4 severe disease | PBMCs | 10x Genomics scRNA-seq, flow cytometry | Strong T cell lymphopenia in severe disease with potential systemic adaptive immune dysregulation; altered T cell differentiation and a hyperactivation stage in severe disease; thymosin α1 can expand the memory-like T cell population and prevent T cell hyperactivation | 25 (Preprint) |
| 15 healthy, 79 COVID-19 (15 with ARDS), 26 influenza (7 with ARDS) | PBMCs | 10x Genomics scRNA-seq (3 influenza and 4 COVID-19), flow cytometry | Similar total and activated T cell counts for influenza and COVID-19 groups; higher IFNα-responding and IFNγ-responding signatures in the severe influenza groups than in the COVID-19 groups | 40 (Preprint) |
| 28 with ARDS, 26 non-ARDS, other infection controls | PBMCs | Flow cytometry | Stronger T cell lymphopenia in more severe COVID-19; lower CD4+ T cell counts in COVID-19 (n = 17) than in influenza (H1N1; n = 4) | 6 |
| 12 healthy, 7 recovered, 7 moderate disease, 27 severe disease | PBMCs | High-dimensional flow cytometry | Stronger T cell lymphopenia in more severe disease; heterogeneity of T cell responses related to activation and cytotoxicity signatures; T cells express more markers of terminal differentiation or exhaustion in severe disease | 4 |
| 60 healthy, 36 recovered, 125 hospitalized patients (NIH ordinal score 2–5) | PBMCs | High-dimensional flow cytometry | Stronger T cell lymphopenia in severe disease, with a bias towards CD8+ T cells; heterogeneity of T cell responses based on high-dimensional immune profiling, with three potential immune subtypes; T cells more activated but also express more markers of terminal differentiation and exhaustion in patients with COVID-19 than in individuals who are healthy or who recovered | 8 |
| 40 healthy, 522 with varying disease severity | PBMCs | Flow cytometry | Stronger T cell lymphopenia in ICU patients, elderly patients and severe disease, for both CD4+ and CD8+ T cells; IL-6, IL-10 and TNF levels negatively correlate with lymphocyte count; T cells express higher levels of PD1 and TIM3 in ICU patients than in non-ICU individuals | 10 |
| 55 healthy, 6 mild disease, 26 moderate disease, 31 severe disease | PBMCs | High-dimensional flow cytometry | Stronger T cell lymphopenia in severe disease; increased number of hyperactivated proliferating CD4+ and CD8+ T cells in severe disease; increased markers of terminal differentiation or exhaustion in severe disease compared with milder disease | 11 (Preprint) |
| 10 moderate disease, 11 severe disease | PBMCs | Flow cytometry | Higher lymphocyte counts in moderate disease than in severe disease, for both CD4+ and CD8+ T cells; more IFNγ-producing T cells in moderate disease than severe disease | 13 |
| 20 healthy, 30 with varying disease severity | PBMCs | Flow cytometry | T cell lymphopenia in patients compared with healthy controls, with an increased ratio of CD4+ T cells to CD8+ T cells; increased proportion of terminally differentiated or senescent CD8+ T cells in patients, with a reduced proportion of IFNγ-producing cells; tocilizumab improves lymphocyte counts (n = 5) | 14 |
| 30 healthy, 55 mild disease, 13 severe disease | PBMCs | Flow cytometry | Stronger T cell lymphopenia in severe disease than in mild disease or healthy controls, with recovery of T cell numbers in convalescent individuals; reduced levels of multiple cytokines in CD8+ T cells in disease groups, with higher levels of NKG2A expression than healthy controls | 23 |
| 6 healthy, 10 mild disease, 6 severe disease | PBMCs | Flow cytometry | Increased cytotoxicity but decreased cytokine secretion of T cells, particularly CD8+ T cells, in severe disease compared with mild disease; CD8+ T cells in severe disease express more inhibitory receptors | 24 |
| Case report, multiple time points | PBMCs | Flow cytometry | ICOS+PD1+ circulating TFH cells increase during recovery; activated CD4+ and CD8+ T cells peak at day 9 post disease onset and decline after recovery | 26 |
| 20 healthy, 20 convalescent, other common cold coronaviruses | PBMCs | Flow cytometry | CD4+ and CD8+ T cells from convalescent patients respond to SARS-CoV-2 epitopes, including S, M and N proteins and other ORFs; T cell reactivity to SARS-CoV-2 also detected in non-exposed donors, with potential cross-reactivity to other common cold coronaviruses | 29 |
| 14 convalescent | PBMCs | Flow cytometry | CD4+ and CD8+ T cells from convalescent patients respond to SARS-CoV-2 epitopes | 30 |
| 16 healthy, 28 recovered from mild disease, 14 recovered from severe disease | PBMCs | Flow cytometry | T cells from convalescent patients with mild or severe disease respond to SARS-CoV-2 epitopes; convalescent patients with mild disease have a better memory CD8+ T cell response than convalescent patients with severe disease | 31 (Preprint) |
| 8 healthy, 8 with varying disease severity | PBMCs | Flow cytometry | T cell lymphopenia in COVID-19 compared with healthy controls, with an increase of T cell activation phenotypes; SARS-CoV-2-specific T cells mainly produce TH1-type cytokines | 35 |
| 10 healthy, 21 non-ICU, 12 in ICU | PBMCs | Flow cytometry | CD4+ T cells in ICU patients produce more GM-CSF and IL-6 than non-ICU individuals and healthy controls | 37 |
| 245 healthy, 19 mild disease, 41 severe disease | PBMCs | Flow cytometry | Stronger T cell lymphopenia in severe disease compared with mild disease and healthy controls; IL-6 levels negatively correlate with lymphocyte count; patients who respond to treatment recover lymphocyte numbers | 38 |
| 15 male and 29 female healthy, 17 male and 21 female with COVID-19 | PBMCs | High-dimensional flow cytometry | Male and female patients have T cell lymphopenia; female patients have more T cell activation than male patients; male patients with severe disease have greater reduction of T cell activation and loss of IFNγ-producers than those with stable disease | 60 (Preprint) |
ARDS, acute respiratory distress syndrome; CTL, cytotoxic T lymphocyte; GM-CSF, granulocyte–macrophage colony-stimulating factor; ICOS, inducible co-stimulator; ICU, intensive care unit; IFNγ, interferon-γ; ORF, open reading frame; PBMC, peripheral blood mononuclear cell; PD1, programmed cell death protein 1; scRNA-seq, single-cell RNA sequencing; scTCR-seq, single-cell T cell receptor sequencing; TFH cell, T follicular helper cell; TH1 cell, T helper 1 cell; TIM3, T cell immunoglobulin and mucin domain-containing protein 3; TNF, tumour necrosis factor.
Lymphopenia in COVID-19
One prominent feature of SARS-CoV-2 infection is lymphopenia1,6,12,13. Lymphopenia is associated with severe disease10,12,13 but is reversed when patients recover4,12. In some patients, lymphopenia has been reported to affect CD4+ T cells, CD8+ T cells, B cells and natural killer cells4,6, whereas other data suggest that SARS-CoV-2 infection has a preferential impact on CD8+ T cells8,9,14. Transient lymphopenia is a common feature of many respiratory viral infections, such as with influenza A H3N2 virus, human rhinovirus or respiratory syncytial virus, but lymphopenia in these other infections typically occurs for only 2–4 days around symptom onset and rapidly recovers15. By contrast, COVID-19-associated lymphopenia may be more severe or persistent than in these other infections and seems to be more selective for T cell lineages. It is possible that the peripheral lymphopenia observed in patients with COVID-19 reflects recruitment of lymphocytes to the respiratory tract or adhesion to inflamed respiratory vascular endothelium. However, although autopsy studies of patients’ lungs and single-cell RNA sequencing (scRNA-seq) of bronchoalveolar lavage fluid do identify the presence of lymphocytes, the lymphocytic infiltration is not excessive5,16. In addition, a recent scRNA-seq analysis of the upper respiratory tract from patients with COVID-19 showed that there was a markedly decreased contribution of cytotoxic T lymphocytes in patients with severe disease compared with those with moderate disease9. In severe disease, lymphopenia may be associated with high levels of IL-6, IL-10 or tumour necrosis factor (TNF)6,10,14, potentially through a direct effect of these cytokines on T cell populations17,18 and/or indirect effects via other cell types, such as dendritic cells19 and neutrophils20,21. Hyperactivation of T cells or high levels of expression of pro-apoptotic molecules, such as FAS (also known as CD95)8, TRAIL or caspase 3 (ref.11), could also contribute to T cell depletion.
Thus, although the mechanisms of lymphopenia in COVID-19 remain incompletely understood, the reduction in the number of T cells, in particular, in the periphery is a prominent feature of many individuals with severe disease. It remains unclear why the lymphopenia is T cell biased and perhaps specifically CD8+ T cell biased. In animal models, lymphopenia can augment T cell activation and proliferation22. Therefore, determining how lymphopenia in patients with COVID-19 might impact T cell hyperactivation and, potentially, immunopathology is an important future goal, as therapeutics such as IL-7 could be beneficial in this regard.
CD8+ T cell responses in COVID-19
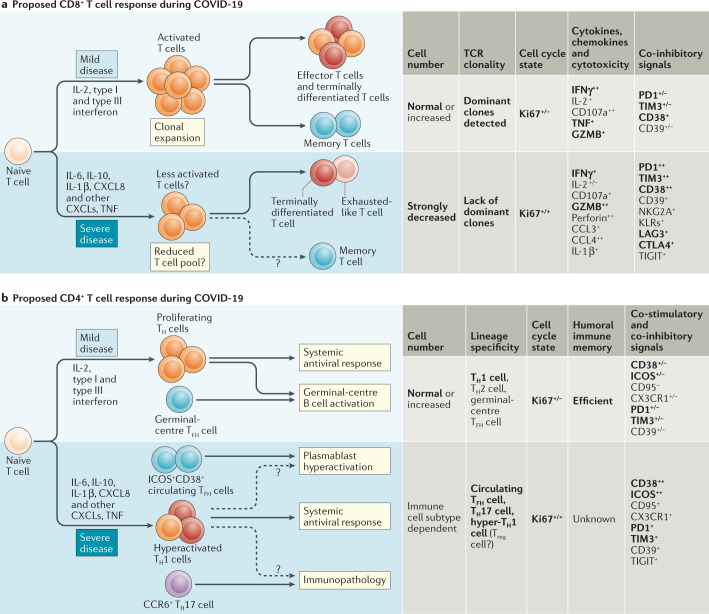
Early studies of small numbers of patients, or sometimes even a single patient, reported alterations in the activation and/or differentiation status of CD8+ T cells in severe COVID-19 (Fig. 1a). For example, there is evidence of terminally differentiated T cells or possibly exhausted T cells in severe disease, with reported increases in expression levels of the inhibitory receptors PD1, TIM3, LAG3, CTLA4, NKG2A and CD39 (refs4,8,10,11,23,24). However, expression of these receptors could also reflect recent activation, and it is not clear whether the T cells in patients with COVID-19 are exhausted or just highly activated. By contrast, at least one blood scRNA-seq study has reported limited expression of inhibitory receptors by CD8+ T cells in patients with COVID-19 compared with healthy controls7. In one report, CD8+ T cells from patients with severe COVID-19 had reduced cytokine production upon stimulation23. Alternatively, other data suggest that CD8+ T cells might have a hyperactivation signature, including high levels of expression of natural killer cell-related markers and increased cytotoxicity4,23,25. Some of these studies imply the presence of an overaggressive CD8+ T cell response or a hyperactive state in patients with COVID-19 (ref.25). Increased numbers of CD38+HLA-DR+ activated CD8+ T cells or Ki67+ proliferating CD8+ T cells were also observed in many, but not all, patients4,8,11,26. In other settings, CD38+HLA-DR+ or Ki67+ CD8+ T cells that are present in the blood during the acute phase of a viral infection or after live-attenuated vaccination contain virus-specific T cells27, which indicates that there might be virus-specific CD8+ T cell responses in those patients with COVID-19 who have robust CD38+HLA-DR+ or Ki67+ CD8+ T cell responses. However, not all patients have this T cell activation phenotype and current data point to potentially diverse patterns of CD8+ T cell responses in patients with COVID-19 (ref.8).
Fig. 1. Potential model of T cell responses during COVID-19 progression.
A proposed model of CD8+ T cell responses (a) and CD4+ T cell responses (b) during COVID-19 progression in mild versus severe disease. Tables show the immune parameters that have been reported to differ between mild and severe COVID-19. Phenotype data are collated from the references cited in this Progress article. Results that have been confirmed by multiple studies are indicated in bold type. CCL, CC-chemokine ligand; CCR6, CC-chemokine receptor 6; CTLA4, cytotoxic T lymphocyte antigen 4; CX3CR1, CX3C-chemokine receptor 1; CXCL, CXC-chemokine ligand; GZMB, granzyme B; ICOS, inducible co-stimulator; IFNγ, interferon-γ; KLR, killer cell lectin-like receptor; LAG3, lymphocyte activation gene 3; TCR, T cell receptor; TFH cell, T follicular helper cell; TH1 cell, T helper 1 cell; TH2 cell, T helper 2 cell; TH17 cell, T helper 17 cell; TIGIT, T cell immunoreceptor with immunoglobulin and ITIM domains; TIM3, T cell immunoglobulin and mucin domain-containing protein 3; TNF, tumour necrosis factor; Treg cell, regulatory T cell.
Several important considerations arise from these studies: how does heterogeneity of the CD8+ T cell response relate to disease; how do T cell responses in peripheral blood reflect events in the respiratory tract; and are the CD8+ T cell responses antigen specific? First, multiple studies have indicated heterogeneity in immune responses to SARS-CoV-2, including in CD8+ T cells, and emerging data identify potentially distinct patient immunotypes6,8 that might, in some cases, be related to disease features. Second, autopsy studies and scRNA-seq data from bronchoalveolar lavage fluid suggest the importance of respiratory CD8+ T cell responses5,16, and recent scRNA-seq data from the upper respiratory tract imply that interactions occur there between epithelial cells and cytotoxic T lymphocytes, in particular through an interferon-γ (IFNγ) axis9. Indeed, more robust clonal expansion of CD8+ T cells in peripheral blood28 or bronchoalveolar lavage fluid5 may be associated with milder disease or recovery, although it is not clear whether this CD8+ T cell clonal expansion is the cause or consequence of the disease recovery. Last, there is evidence of SARS-CoV-2-specific CD8+ T cells in patients who have recovered, which provides evidence not only of virus-specific CD8+ T cell responses but also of CD8+ T cell memory in many convalescent patients29–32. The precise role of these virus-specific CD8+ T cells in control of initial acute SARS-CoV-2 infection and their capacity to protect from future infection remain to be determined.
CD4+ T cell responses in COVID-19
Similar to CD8+ T cells, there is evidence of functional impairment and increased expression of activation and/or exhaustion markers by CD4+ T cells in patients with COVID-19 (refs10,14) (Fig. 1b). Case reports have suggested that CD8+ T cell activation might be greater than CD4+ T cell activation, as defined by activation markers such as CD38 and HLA-DR26,33,34. However, another study identified a subset of patients with higher levels of CD4+ T cell activation who possibly do worse clinically8. One report has described a higher proportion of IFNγ-producing T helper 1 (TH1)-like cells in patients with moderate disease than in patients with severe disease13. Moreover, CD4+ T cells specific for the SARS-CoV-2 spike protein have been identified in acute infection and have a TH1 cell cytokine profile35. A role for T helper 2 (TH2) cell-type responses in severe COVID-19 is unclear, although patients with mild disease may have a normal TH2 cell response11. Given the prominent role of TH2 cell responses in other lung diseases, this is an important area for further study. Two individual reports have also described a strong CCR6+CD4+ T cell signature in severe COVID-19 (refs2,34), which indicates a potential role for T helper 17 (TH17) cell-mediated immunopathology, whereas another study identified possible increases in the number of CD4+ T cells producing transforming growth factor-β (TGFβ)36. A population of CD4+ T cells producing granulocyte–macrophage colony-stimulating factor (GM-CSF) or IL-6 has also been reported in patients with COVID-19 (ref.37), although T cells do not typically produce IL-6. Regulatory T cells36 and ICOS+CD38+ circulating T follicular helper (activated cTFH) cells8 may also be altered in patients with COVID-19, with increased numbers of activated cTFH cells in some settings perhaps connected to a reported increase in the number of circulating plasmablasts4.
Lymphopenia also affects CD4+ T cells, although some studies suggest that the impact is less than that for CD8+ T cells8,38. It remains to be determined how lymphopenia might relate to CD4+ T cell activation and/or dysfunction. Moreover, in addition to memory CD8+ T cells, patients who have recovered from SARS-CoV-2 infection have virus-specific memory CD4+ T cells29–32, which bodes well for the possibility of T cell memory and, perhaps, protective immunity. In individuals who recovered from mild COVID-19, CD4+ T cells gained a typical memory phenotype with high levels of expression of IL-7Rα (ref.32). Interestingly, cross-reactive memory CD4+ T cells are also found in subjects who have never been exposed to SARS-CoV-2 (ref.29). It is unclear how these pre-existing memory CD4+ T cells, which are presumably generated in response to human endemic coronaviruses, affect immunity or pathology upon SARS-CoV-2 infection. There are other settings in which the presence of memory CD4+ T cells in the absence of effective CD8+ T cells or antibodies can cause pathology39. Thus, CD4+ T cell responses are present in patients with COVID-19 and perhaps form memory following resolution of the disease. Whether CD4+ T cells responding in acute infection with SARS-CoV-2 are functionally impaired or hyperactivated, and how these responses relate to disease outcome, remain important unanswered questions.
T cell differentiation in COVID-19
Most acute viral infections in humans induce the activation and proliferation of both CD4+ T cells and CD8+ T cells, so SARS-CoV-2 infection may not be unique in this regard. However, hyperactivation or hypoactivation of T cells, or skewing towards an ineffective differentiation state (for example, TH17 cells, exhausted T cells or terminally differentiated T cells), might not be optimal in some patients with COVID-19. Several features that have been described in patients with severe COVID-19 — including high levels of systemic cytokines or chemokines, most notably IL-6, CXCL8, CXCL9 and CXCL10 (refs8,11,40), or delayed or defective type I interferon responses41 — could potentially skew T cell responses. The potential type I interferon deficiency described in patients with COVID-19 is consistent with a recent observation of reduced numbers of plasmacytoid dendritic cells11. This observation, however, contrasts with the robust and consistent upregulation of the interferon-inducible chemokine CXCL10 observed in one patient cohort11, perhaps owing to non-interferon inducers of expression of this chemokine.
Another key gap in our understanding is the relationship between viral load and the immune response. Viral load could have a major impact on the magnitude and quality of the T cell response, and future studies that quantify virus replication should provide context for understanding evolving T cell responses during SARS-CoV-2 infection. A key feature of patients with COVID-19 is the enrichment for co-morbidities such as hypertension and diabetes42 (see next section), but it remains to be defined how an individual patient’s background ‘immune health’ landscape shapes responses to SARS-CoV-2 infection. Although lymphopenia is not a unique feature of SARS-CoV-2 infection43, it may be more prolonged in patients with COVID-19, and lymphopenia-induced proliferation can clearly affect T cell differentiation and activation; however, the contribution of lymphopenia to T cell differentiation in patients with COVID-19 has not yet been investigated.
Co-morbidities and COVID-19
Co-morbidities such as cardiovascular disease44,45, diabetes42 and obesity46 are some of the most common underlying conditions associated with worse clinical outcome and severe COVID-19. Moreover, older patients experience greater clinical severity of COVID-19 (refs1,47), males may experience more severe disease than females1,47 and genetic variations, including the ABO blood type8,48,49, have been reported to affect the clinical outcomes for patients with COVID-19. However, how these co-morbidities are associated with T cell responses during COVID-19 remains largely unknown.
Advanced age is a common co-morbidity for severity of disease during respiratory viral infections, and disease severity may be associated with altered T cell responses50,51. Ageing affects T cell repertoire diversity52, both for CD4+ T cells53 and for CD8+ T cells54. This age-related reduction in T cell clonal diversity is associated with impaired responses to viral infections such as influenza55. Advanced age can also be associated with T cell senescence, which perhaps contributes to ineffective responses to infections56,57. However, senescent T cells may also paradoxically be pro-inflammatory and therefore perhaps contribute to immunopathology. Older patients experience more severe lymphopenia during COVID-19 (ref.10), although it is not clear whether ageing-associated lymphopenia is causal to disease severity or vice versa.
Men with COVID-19 have higher rates of hospitalization and mortality than women58, and among severe cases of disease, men have more severe lymphopenia59. There may also be a bias to stronger CD4+ and CD8+ T cell activation in women with COVID-19 (ref.60). It is unclear whether these sex biases relate to X chromosome-encoded immune genes58 and/or the role of sex hormones in regulating immune responses61. Nevertheless, these data highlight the importance of accounting for sex in ongoing clinical trials.
A very high proportion of patients with COVID-19 who are hospitalized have one or more underlying cardiovascular co-morbidities44,45,62–64. Moreover, many of the manifestations of severe disease relate to coagulopathies and vascular complications62,64. Even the newly recognized SARS-CoV-2-associated multisystem inflammatory syndrome in children has similarities to Kawasaki disease, which often involves inflammation of the coronary artery65,66. Other viral infections, including Ebola virus and virulent lymphocytic choriomeningitis virus in mice, can cause damage to the vascular endothelium. These viruses infect the vascular endothelium, and T cells can then cause vascular damage by attacking the virus-infected cells67–69. However, disease presentation in these settings is distinct from that for COVID-19, and extensive SARS-CoV-2 infection of the vascular endothelium is unlikely in most patients.
Obesity also contributes to more severe COVID-19 and higher rates of hospitalization46,70, which are perhaps associated with the role of body mass index in cardiovascular health. However, obesity can also directly affect T cell responses to influenza vaccination71 or infection72, and a role for obesity in altering T cell differentiation has been observed in asthma73 and chronic inflammation74. Nevertheless, the precise role of T cells in driving the coagulopathies or inflammation that are characteristic of severe COVID-19 in patients with cardiovascular co-morbidities is unclear.
Finally, a recent genetic association study compared patients with severe COVID-19 and ARDS with healthy controls49. Several chemokine receptor genes, including CCR9, CXCR6 and XCR1, and the locus controlling the ABO blood type were identified as being associated with severe disease. Whether these genes are directly or indirectly related to T cell responses in COVID-19, however, remains unknown and needs further investigation.
Thus, although severe COVID-19 is partially characterized by patients with co-morbidities, how these co-morbidities relate to T cell responses remains poorly understood. Causal associations may exist, perhaps connected to some of the underlying genetics. However, it is also possible that certain co-morbidities or pre-existing conditions make patients with severe disease less able to tolerate the severe virus-mediated and immune-mediated pathology associated with SARS-CoV-2 infection.
Strength of COVID-19 T cell response
The published data discussed here indicate that patients with severe COVID-19 can have either insufficient or excessive T cell responses. It is possible, therefore, that disease might occur in different patients at either end of this immune response spectrum, in one case from virus-mediated pathology and in the other case from T cell-driven immunopathology4. However, it is unclear why some patients respond too little and some patients too much, and whether the strength of the T cell response in the peripheral blood reflects the T cell response intensity in the respiratory tract and other SARS-CoV-2-infected organs. Some information exists on SARS-CoV-2-specific T cells in patients who are hospitalized, and approaches using activation markers (such as Ki67) likely capture virus-specific T cell responses; recent studies are beginning to fill in this picture by identifying virus-specific CD4+ T cells and CD8+ T cells in acute disease5,29,30. However, one missing set of information is a careful immune analysis of outpatients who, during acute infection with SARS-CoV-2, are undergoing a ‘successful’ immune response. Moreover, it will be crucial to incorporate into any model of COVID-19 how the magnitude and quality of T cell responses relate to signals from other parts of the immune system, such as innate cytokines, antigen presentation and myeloid cells. Lastly, the strength of the T cell response may also relate to the stage of disease or infection. Accurately assessing the different ‘phases’ of acute SARS-CoV-2 infection may also be valuable in interpreting T cell responses.
Thus, future studies should provide insights into how to identify patients at different stages of disease or with different immune response characteristics and then to match them with treatment strategies. Such a precision medicine strategy based on analysis of ‘immune health’ could be used to better tailor the use of immunostimulatory strategies such as thymosin α1 (ref.25) or type I interferon75 versus immunosuppressive drugs such as tocilizumab76–78, ruxolitinib79 or dexamethasone80 to patients who will receive most benefit. These efforts should be aided by the development of better preclinical models, including human ACE2-expressing mice81–85. It will be interesting in the future to use such models to address key questions including: what is an ‘appropriate’ profile of T cell activation and differentiation for a successful antiviral response to SARS-CoV-2 without triggering severe pathology; how does viral load or initial inoculum of SARS-CoV-2 impact T cell responses and pathology; how do T cell responses in the blood relate to lung-infiltrating T cells, considering that many respiratory viruses are controlled by tissue-resident immune cells that may not be present in the blood; and what are the molecular mechanisms that support a successful T cell response during SARS-CoV-2 infection?
T cell memory in COVID-19
A key question is whether protective T cell memory can form following either SARS-CoV-2 infection or vaccination. Although data on vaccination will await trial results, early data from patients with COVID-19 who have recovered are promising. Memory CD4+ T cells and CD8+ T cells were detected in 100% and 70% of patients who recovered, respectively29. Moreover, memory T cell responses were detected for multiple SARS-CoV-2 proteins, including not only spike protein but also nucleoprotein and membrane protein31. Whether these T cells provide protective immunity is unclear, and addressing this question for T cells alone will be confounded by the presence of SARS-CoV-2-specific antibodies in patients who recover. Nevertheless, ongoing analysis of patients who have recovered should provide insights into the protective capacity of immune memory, including humoral and cellular memory, as these individuals are potentially re-exposed to SARS-CoV-2 in the community. Further studies should also define the differentiation state and durability of T cell memory. Moreover, defining how T cell memory forms in patients who experience mild symptoms of COVID-19 versus severe disease will be important. Although evidence of T cell memory to other coronaviruses is encouraging, such immune natural history studies for SARS-CoV-2 will likely be valuable for examining vaccine-induced T cell responses.
Conclusions
The accumulating evidence supports a role for T cells in COVID-19 and probably in the immunological memory that forms following recovery from SARS-CoV-2 infection. Most, although not all, patients who are hospitalized seem to mount both CD8+ and CD4+ T cell responses, and evidence points to possible suboptimal, excessive or otherwise inappropriate T cell responses associated with severe disease. In fact, multiple distinct patterns of T cell response may exist in different patients, which suggests the possibility of distinct clinical approaches tailored to the particular immunotype of a specific patient. Much of the available data on T cell responses in patients with COVID-19 who are hospitalized is still available only in preprint form. This format has been essential for rapid dissemination of information in the setting of pandemic urgency. However, as these studies move through peer review, we should gain increasing confidence and clarity about the nature of T cell responses to SARS-CoV-2 infection. More carefully defining distinct classes of T cell response types in COVID-19 and delineating how pre-existing conditions, co-morbidities, race, ‘immune health’ status and other variables influence T cell responses should reveal novel opportunities for treatment and prevention.
Acknowledgements
Z.C. is supported by grant CA234842 from the National Institutes of Health (NIH). Work in the Wherry laboratory is supported by NIH grant AI082630, the Allen Institute for Immunology and the Parker Institute for Cancer Immunotherapy.
Author contributions
The authors contributed equally to all aspects of the article.
Competing interests
E.J.W. has consulting agreements with and/or is a scientific advisor for Merck, Roche, Pieris, Elstar and Surface Oncology. E.J.W. has a patent licensing agreement on the PD1 pathway with Roche/Genentech. E.J.W. is a founder of Arsenal Biosciences. Z.C. declares no competing interests.
Footnotes
Peer review information
Nature Reviews Immunology thanks Susan L. Swain and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
COVID-19 Map of the Johns Hopkins Coronavirus Resource Center: https://coronavirus.jhu.edu/map.html
References
- 1.Huang C, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu Z, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Resp. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matthay MA, Aldrich JM, Gotts JE. Treatment for severe acute respiratory distress syndrome from COVID-19. Lancet Resp. Med. 2020;8:433–434. doi: 10.1016/S2213-2600(20)30127-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuri-Cervantes L, et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci. Immunol. 2020;5:eabd7114. doi: 10.1126/sciimmunol.abd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liao M, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;8:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 6.Giamarellos-Bourboulis EJ, et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27:992–1000. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilk AJ, et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mathew D, et al. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020 doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chua RL, et al. COVID-19 severity correlates with airway epithelium–immune cell interactions identified by single-cell analysis. Nat. Biotech. 2020 doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- 10.Diao B, et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front. Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laing AG, et al. A consensus Covid-19 immune signature combines immuno-protection with discrete sepsis-like traits associated with poor prognosis. medRxiv. 2020 doi: 10.1101/2020.06.08.20125112. [DOI] [Google Scholar]
- 12.Tan L, et al. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal. Transduct. Target. Ther. 2020;5:33. doi: 10.1038/s41392-020-0148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen G, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mazzoni A, et al. Impaired immune cell cytotoxicity in severe COVID-19 is IL-6 dependent. J. Clin. Invest. 2020 doi: 10.1172/JCI138554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McClain MT, et al. Longitudinal analysis of leukocyte differentials in peripheral blood of patients with acute respiratory viral infections. J. Clin. Virol. 2013;58:689–695. doi: 10.1016/j.jcv.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 16.Wichmann D, et al. Autopsy findings and venous thromboembolism in patients with COVID-19: a prospective cohort study. Ann. Intern. Med. 2020 doi: 10.7326/M20-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taga K, Tosato G. IL-10 inhibits human T cell proliferation and IL-2 production. J. Immunol. 1992;148:1143–1148. [PubMed] [Google Scholar]
- 18.Böttcher JP, et al. IL-6 trans-signaling-dependent rapid development of cytotoxic CD8+ T cell function. Cell Rep. 2014;8:1318–1327. doi: 10.1016/j.celrep.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 19.Malefyt R, et al. Interleukin 10 (IL-10) and viral IL-10 strongly reduce antigen-specific human T cell proliferation by diminishing the antigen-presenting capacity of monocytes via downregulation of class II major histocompatibility complex expression. J. Exp. Med. 1991;174:915–924. doi: 10.1084/jem.174.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, et al. Neutrophil-to-lymphocyte ratio as an independent risk factor for mortality in hospitalized patients with COVID-19. J. Infect. 2020;81:e6–e12. doi: 10.1016/j.jinf.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma Y, et al. Predictive value of the neutrophil-to-lymphocyte ratio (NLR) for diagnosis and worse clinical course of the COVID-19: findings from ten provinces in China. SSRN. 2020 doi: 10.2139/ssrn.3569838. [DOI] [Google Scholar]
- 22.Surh CD, Sprent J. Homeostasis of naive and memory T cells. Immunity. 2008;29:848–862. doi: 10.1016/j.immuni.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 23.Zheng M, et al. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol. 2020;17:533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng H-Y, et al. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell. Mol. Immunol. 2020;17:541–543. doi: 10.1038/s41423-020-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu K, et al. Thymosin α-1 protected T cells from excessive activation in severe COVID-19. Research Square. 2020 doi: 10.21203/rs.3.rs-25869/v1. [DOI] [Google Scholar]
- 26.Thevarajan I, et al. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat. Med. 2020;26:453–455. doi: 10.1038/s41591-020-0819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller JD, et al. Human effector and memory CD8+ T cell responses to smallpox and yellow fever vaccines. Immunity. 2008;28:710–722. doi: 10.1016/j.immuni.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 28.Wen W, et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 2020;6:31. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grifoni A, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ni L, et al. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity. 2020;52:971–977. doi: 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong T, et al. Broad and strong memory CD4 and CD8 T cells induced by SARS-CoV-2 in UK convalescent COVID-19 patients. bioRxiv. 2020 doi: 10.1101/2020.06.05.134551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neidleman J, et al. SARS-CoV-2-specific T cells exhibit unique features characterized by robust helper function, lack of terminal differentiation, and high proliferative potential. bioRxiv. 2020 doi: 10.1101/2020.06.08.138826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vabret N, et al. Immunology of COVID-19: current state of the science. Immunity. 2020;52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qin C, et al. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weiskopf D, et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020;5:eabd2071. doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang W, et al. High-dimensional immune profiling by mass cytometry revealed immunosuppression and dysfunction of immunity in COVID-19 patients. Cell. Mol. Immunol. 2020;17:650–652. doi: 10.1038/s41423-020-0447-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou Y, et al. Pathogenic T-cells and inflammatory monocytes incite inflammatory storms in severe COVID-19 patients. Natl Sci. Rev. 2020;7:998–1002. doi: 10.1093/nsr/nwaa041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang F, et al. Characteristics of peripheral lymphocyte subset alteration in COVID-19 pneumonia. J. Infect. Dis. 2020;221:1762–1769. doi: 10.1093/infdis/jiaa150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Penaloza-MacMaster P, et al. Vaccine-elicited CD4 T cells induce immunopathology after chronic LCMV infection. Science. 2015;347:278–282. doi: 10.1126/science.aaa2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mudd PA, et al. Targeted immunosuppression distinguishes COVID-19 from influenza in moderate and severe disease. medRxiv. 2020 doi: 10.1101/2020.05.28.20115667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blanco-Melo D, et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fang L, Karakiulakis G, Roth M. Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection? Lancet Resp. Med. 2020;8:e21. doi: 10.1016/S2213-2600(20)30116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Panesar NS. Lymphopenia in SARS. Lancet. 2003;361:1985. doi: 10.1016/S0140-6736(03)13557-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li B, et al. Prevalence and impact of cardiovascular metabolic diseases on COVID-19 in China. Clin. Res. Cardiol. 2020;109:531–538. doi: 10.1007/s00392-020-01626-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo T, et al. Cardiovascular implications of fatal outcomes of patients with coronavirus disease 2019 (COVID-19) JAMA Cardiol. 2020;5:811–818. doi: 10.1001/jamacardio.2020.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lighter J, et al. Obesity in patients younger than 60 years is a risk factor for COVID-19 hospital admission. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richardson S, et al. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City area. JAMA. 2020;323:2052–2059. doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao J, et al. Relationship between the ABO blood group and the COVID-19 susceptibility. medRxiv. 2020 doi: 10.1101/2020.03.11.20031096. [DOI] [Google Scholar]
- 49.Ellinghaus D, et al. Genomewide association study of severe Covid-19 with respiratory failure. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee N, Shin MS, Kang I. T-cell biology in aging, with a focus on lung disease. J. Gerontol. A Biol. Sci. Med. Sci. 2012;67:254–263. doi: 10.1093/gerona/glr237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goronzy JJ, Weyand CM. Successful and maladaptive T cell aging. Immunity. 2017;46:364–378. doi: 10.1016/j.immuni.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Naylor K, et al. The influence of age on T cell generation and TCR diversity. J. Immunol. 2005;174:7446–7452. doi: 10.4049/jimmunol.174.11.7446. [DOI] [PubMed] [Google Scholar]
- 53.Pourgheysari B, et al. The cytomegalovirus-specific CD4+ T-cell response expands with age and markedly alters the CD4+ T-cell repertoire. J. Virol. 2007;81:7759–7765. doi: 10.1128/JVI.01262-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Messaoudi I, Lemaoult J, Guevara-Patino JA, Metzner BM, Nikolich-Zugich J. Age-related CD8 T cell clonal expansions constrict CD8 T cell repertoire and have the potential to impair immune defense. J. Exp. Med. 2004;200:1347–1358. doi: 10.1084/jem.20040437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yager EJ, et al. Age-associated decline in T cell repertoire diversity leads to holes in the repertoire and impaired immunity to influenza virus. J. Exp. Med. 2008;205:711–723. doi: 10.1084/jem.20071140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goronzy JJ, Li G, Yu M, Weyand CM. Signaling pathways in aged T cells—a reflection of T cell differentiation, cell senescence and host environment. Semin. Immunol. 2012;24:365–372. doi: 10.1016/j.smim.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu HT, Park S, Shin E-C, Lee W-W. T cell senescence and cardiovascular diseases. Clin. Exp. Med. 2016;16:257–263. doi: 10.1007/s10238-015-0376-z. [DOI] [PubMed] [Google Scholar]
- 58.Scully EP, Haverfield J, Ursin RL, Tannenbaum C, Klein SL. Considering how biological sex impacts immune responses and COVID-19 outcomes. Nat. Rev. Immunol. 2020;20:442–447. doi: 10.1038/s41577-020-0348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meng Y, et al. Sex-specific clinical characteristics and prognosis of coronavirus disease-19 infection in Wuhan, China: a retrospective study of 168 severe patients. PLoS Pathog. 2020;16:e1008520. doi: 10.1371/journal.ppat.1008520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takahashi T, et al. Sex differences in immune responses to SARS-CoV-2 that underlie disease outcomes. medRxiv. 2020 doi: 10.1101/2020.06.06.20123414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bouman A, Heineman MJ, Faas MM. Sex hormones and the immune response in humans. Hum. Reprod. Update. 2005;11:411–423. doi: 10.1093/humupd/dmi008. [DOI] [PubMed] [Google Scholar]
- 62.Zheng Y-Y, Ma Y-T, Zhang J-Y, Xie X. COVID-19 and the cardiovascular system. Nat. Rev. Cardiol. 2020;17:259–260. doi: 10.1038/s41569-020-0360-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Magro C, et al. Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID-19 infection: a report of five cases. Transl Res. 2020;220:1–13. doi: 10.1016/j.trsl.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Klok FA, et al. Incidence of thrombotic complications in critically ill ICU patients with COVID-19. Thromb. Res. 2020;191:145–147. doi: 10.1016/j.thromres.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Viner RM, Whittaker E. Kawasaki-like disease: emerging complication during the COVID-19 pandemic. Lancet. 2020;395:1741–1743. doi: 10.1016/S0140-6736(20)31129-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Belhadjer Z, et al. Acute heart failure in multisystem inflammatory syndrome in children (MIS-C) in the context of global SARS-CoV-2 pandemic. Circulation. 2020 doi: 10.1161/CIRCULATIONAHA.120.048360. [DOI] [PubMed] [Google Scholar]
- 67.Frebel H, et al. Programmed death 1 protects from fatal circulatory failure during systemic virus infection of mice. J. Exp. Med. 2012;209:2485–2499. doi: 10.1084/jem.20121015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang ZY, et al. Identification of the Ebola virus glycoprotein as the main viral determinant of vascular cell cytotoxicity and injury. Nat. Med. 2000;6:886–889. doi: 10.1038/78645. [DOI] [PubMed] [Google Scholar]
- 69.Wolf T, et al. Severe Ebola virus disease with vascular leakage and multiorgan failure: treatment of a patient in intensive care. Lancet. 2015;385:1428–1435. doi: 10.1016/S0140-6736(14)62384-9. [DOI] [PubMed] [Google Scholar]
- 70.Stefan N, Birkenfeld AL, Schulze MB, Ludwig DS. Obesity and impaired metabolic health in patients with COVID-19. Nat. Rev. Endocrinol. 2020;16:341–342. doi: 10.1038/s41574-020-0364-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sheridan PA, et al. Obesity is associated with impaired immune response to influenza vaccination in humans. Int. J. Obes. 2012;36:1072–1077. doi: 10.1038/ijo.2011.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Paich HA, et al. Overweight and obese adult humans have a defective cellular immune response to pandemic H1N1 influenza A virus. Obesity. 2013;21:2377–2386. doi: 10.1002/oby.20383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Leiria LOS, Martins MA, Saad MJA. Obesity and asthma: beyond TH2 inflammation. Metabolism. 2015;64:172–181. doi: 10.1016/j.metabol.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 74.DeFuria J, et al. B cells promote inflammation in obesity and type 2 diabetes through regulation of T-cell function and an inflammatory cytokine profile. Proc. Natl Acad. Sci. USA. 2013;110:5133–5138. doi: 10.1073/pnas.1215840110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sallard E, Lescure F-X, Yazdanpanah Y, Mentre F, Peiffer-Smadja N. Type 1 interferons as a potential treatment against COVID-19. Antivir. Res. 2020;178:104791. doi: 10.1016/j.antiviral.2020.104791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu X, et al. Effective treatment of severe COVID-19 patients with tocilizumab. Proc. Natl Acad. Sci. USA. 2020;117:10970–10975. doi: 10.1073/pnas.2005615117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Luo P, et al. Tocilizumab treatment in COVID-19: a single center experience. J. Med. Virol. 2020;92:814–818. doi: 10.1002/jmv.25801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guo C, et al. Tocilizumab treatment in severe COVID-19 patients attenuates the inflammatory storm incited by monocyte centric immune interactions revealed by single-cell analysis. bioRxiv. 2020 doi: 10.1101/2020.04.08.029769. [DOI] [Google Scholar]
- 79.Cao Y, et al. Ruxolitinib in treatment of severe coronavirus disease 2019 (COVID-19): a multicenter, single-blind, randomized controlled trial. J. Allergy Clin. Immunol. 2020;146:137–146. doi: 10.1016/j.jaci.2020.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Abdolahi N, et al. Letter to the editor: efficacy of different methods of combination regimen administrations including dexamethasone, intravenous immunoglobulin, and interferon-β to treat critically ill COVID-19 patients: a structured summary of a study protocol for a randomized controlled trial. Trials. 2020;21:549. doi: 10.1186/s13063-020-04499-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Goldman-Israelow B, Song E, Mao T, Lu P, Meir A. Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. bioRxiv. 2020 doi: 10.1101/2020.05.27.118893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sun J, et al. Generation of a broadly useful model for COVID-19 pathogenesis, vaccination, and treatment. Cell. 2020 doi: 10.1016/j.cell.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hassan AO, et al. A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell. 2020 doi: 10.1016/j.cell.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bao L, et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020 doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 85.Jiang R-D, et al. Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell. 2020;182:50–58. doi: 10.1016/j.cell.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]