Abstract
The Varkud Satellite (VS) ribozyme catalyzes site-specific RNA cleavage and ligation, and serves as an important model system to understand RNA catalysis. Here we combine stereospecific phosphorothioate substitution, precision nucleobase mutation and linear free energy relationship measurements with molecular dynamics, molecular solvation theory, and ab initio quantum mechanical/molecular mechanical free energy simulations to gain insight into catalysis. Through this confluence of theory and experiment, we unify the existing body of structural and functional data to unveil the catalytic mechanism in unprecedented detail, including the degree of proton transfers in the transition state. Further, we provide evidence for a critical Mg2+ ion in the active site that interacts with the scissile phosphate and anchors the general base guanine in position for nucleophile activation. This novel role for Mg2+ adds to the diversity of known catalytic RNA strategies and unifies functional features observed in the VS, hairpin, and hammerhead ribozyme classes.
The remarkable ability of RNA molecules to catalyze complex chemical transformations has profoundly influenced our understanding of the role of RNA in biology, the design of new biotechnology, and the formulation of theories into the origin of life itself.1,2 From a chemistry perspective, it is of fundamental interest to understand how RNA, with its limited repertoire of fairly inert functional groups, can achieve rate enhancements typically up to 6 orders of magnitude or more,3 in some cases comparable to the intrinsic rates of protein enzymes.4 A predictive understanding of catalytic RNA mechanisms may enable general principles to emerge that are transferable to the design of synthetic systems, such as the recently reported Hachimoji DNA/RNA,5 with great promise for new biotechnological applications. In-depth studies of self-cleaving ribozymes, of which there are currently nine known classes, have contributed substantially to our understanding of these mechanisms.6,7
The Varkud Satellite (VS) ribozyme is the largest known self-cleaving ribozyme, and among the largest endonucleolytic ribozymes (only certain RNase P constructs are comparable). It is found in the mitochondria of natural isolates of Neurospora and performs site-specific scission of multimeric VS RNAs into linear monomers.8,9 Since its discovery nearly three decades ago, the VS ribozyme has been the subject of numerous experimental studies that provide tremendous insight into its structural and functional properties. The ribozyme cleavage proceeds in a fashion similar to that observed in other self-cleaving ribozymes - through nucleophilic attack of a 2’-hydroxyl on the scissile phosphate to form 2’,3’-cyclic phosphate and 5’-hydroxyl termini. Electrophoretic mobility and Förster resonance energy transfer (FRET) studies10,11 have led to models for the VS tertiary structure, and mutagenesis and chemical probing studies, along with pH-rate measurements,11–14 have identified residues critical for activity. A significant body of functional data suggests that two critical active site nucleobases, G638 and A756, act as general base and acid, respectively, in the reaction (Figure 1).11–18 It has been established that Mg2+ ions stabilize the VS ribozyme structure, but the specific catalytic role of Mg2+ in the active site has not previously been determined.
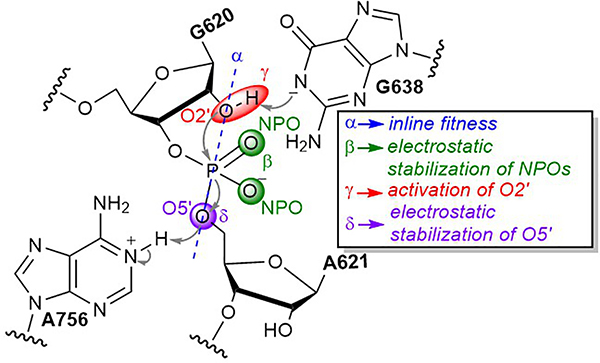
Figure 1. Schematic illustration of the VS ribozyme catalytic reaction.
The ribozyme catalyzes site-specific phosphodiester cleavage by using a guanine, G638, and an adenine, A756, as a general base and acid, respectively. The key catalytic strategies employed by ribozymes23 are highlighted: α, acquirement of in-line nucleophilic attack (blue), β neutralization of negative charge on NPOs (green), γ deprotonation of the 2’-hydroxyl group to activate the nucleophile (red), and δ neutralization of negative charge on the 5’O leaving group (purple).
Despite the abundance of functional data available for the VS ribozyme, in the absence of an atomic-resolution structure, its active site architecture and key catalytic interactions remained elusive. Recently, we reported the first pre-catalytic high-resolution crystal structures of the VS ribozyme, the G638A (PDB ID 4R4V) and A756G (PDB ID 4R4P) mutants, by introducing a C634G mutation that facilitated remodeling and binding of the substrate helix.19 In a follow up study, we crystallized the G638A mutant (PDB ID 5V3I) without the C634G mutation and demonstrated that the substrate helix can remodel itself naturally during the course of binding.20 These structures revealed the complex interlacing of the different helices that gave rise to the dimeric, domain swapped structure of the ribozyme, and were largely consistent with the existing body of VS ribozyme literature.21,22 While these structures provide meaningful insight into the global architecture of the ribozyme, they represent only average structures of artificially deactivated catalysts in a crystal, which may not be representative of the dynamic active state in solution. Moreover, these structures were obtained in high ammonium ion concentrations, which may preclude divalent metal ion binding. As a consequence, major gaps exist in our understanding of the active site architecture and the catalytic mechanism. Here, we take an integrated experimental and computational approach to establish a dynamical model of VS catalysis that extends and unifies the existing body of experimental data, defines the catalytic configuration, and reveals the modes of transition state stabilization unambiguously. These findings broaden our mechanistic understanding of the diverse array of catalytic strategies available to RNA.
In this work, we perform a series of computational and experimental investigations on the VS ribozyme in order to characterize its functionally active state, identify its key catalytic strategies, and unify the existing body of structural and functional data on this important system. We adopt a recently developed ontology for discussion of RNA cleavage reactions,23 extended from the original framework introduced by Breaker24, specifically, α catalysis, acquirement of in-line nucleophilic attack; β catalysis, stabilization of negative charge on a non-bridge phosphoryl oxygens (NPOs); ɣ catalysis, activation (deprotonation) of the 2’-hydroxyl group to generate the nucleophile; and δ catalysis, stabilization of negative charge on the 5’-oxygen of the leaving group (Figure 1). The results are divided into eight sections. In the first section, mutational analysis of the catalytic nucleobases A756 and G638 in the background of oxo and phosphorothioate substrates reveals a stereospecific functional linkage between the exocyclic amines of both residues and the pro-RP NPO of the scissile phosphate. In section 2, molecular dynamics (MD) simulations departing independently from each of the three existing crystal structures (none of which contained an Mg2+ ion in the active site) collectively converge to an ensemble that is reflective of a catalytically inactive state of the ribozyme. In section 3, MD simulations and molecular solvation theory-based calculations predict a Mg2+ ion binding pocket at the pro-SP NPO position at the scissile phosphate (not yet observed crystallographically). In section 4, the predicted Mg2+-bound model is verified by stereospecific thio/metal ion rescue effect experiments. In section 5, MD simulations with a Mg2+ ion at the pro-SP position converge to an ensemble that is reflective of the active state of the ribozyme and illustrate that the metal ion organizes the active site for catalysis in a way that appears to be unique to VS. In section 6, chemically precise mutational experiments further validate the Mg2+ bound model and probe interactions in the metal ion binding pocket. In section 7, linear free energy analysis provides insight into the nature of the rate-controlling transition state, and in particular, the degree of proton transfer for the general base and acid. In section 8, ab initio quantum mechanical/molecular mechanical (QM/MM) free energy simulations departing from the Mg2+-bound model report a concerted mechanism for RNA backbone cleavage with a late transition state for the cleavage reaction with extensive/complete proton transfer from the 2’-hydroxyl to G638, but only partial proton transfer from A756 to the 5’O leaving group, consistent with LFER analysis. Taken together, the integrated experimental and theoretical data provide detailed atomic-level insight into the VS ribozyme catalytic mechanism. These results establish new functional connections between VS and both the hammerhead and hairpin ribozymes, and identify a novel role for a divalent metal ion in organizing the active site by anchoring the general base guanine and tuning its pKa to promote general base (ɣ) catalysis.
Results
Phosphorothioate substitution and mutational analysis of A756 and G638 reveal catalytic interactions in the active site
In this section, we use stereospecific phosphorothioate substitution and chemically precise nucleobase modifications to pinpoint key functional interactions between the pro-RP position of the substrate scissile phosphate and A756 and G638 in the active site. Results are summarized in Table 1 (details are provided in Supplementary Information). Building on earlier work by others,25,26 our stereospecific phosphorothioate substitution experiments indicated that under standard conditions (10mM MgCl2, 25mM KCl, 2mM spermidine, pH = 8), the wild type ribozyme cleaved with kobs = 0.32 min−1, whereas the A621 RP and SP thio substrates cleaved with rates below the limit of detection (here, 10−4 min−1) corresponding to thio effects of at least 3000-fold. These data suggest that critical catalytic interactions are present at both NPOs. Later, we show that the SP thio substrate exhibits metal ion rescue effects, whereas the RP thio substrate does not.
Table 1. kobs for cleavage of VS substrates by ribozyme variants under various ionic conditions.
k/k′ corresponds to the ratio , where the subscript o corresponds to the oxo substrate, and superscripts Mut, WT correspond to a ribozyme mutant and wildtype, respectively. Metal ion rescue is calculated according to the ratio of the ratios (ko/ks)Mg2+/(ko/ks)Cd2+, where the subscript s corresponds to the thio substrate, superscripts Mg2+ and Cd2+ correspond to experiments in presence of Mg2+ alone, and experiments in presence of both Mg2+ and Cd2+, respectively. Mutational rescue corresponds to the ratio of the ratios (ko/ks)WT/(ko/ks)Mut. The error of kobs measurements is the standard deviation of three or more independent measurements and errors for thio effect, metal ion rescue, and mutational rescue are the propagated errors of relative errors. Reactions that proceed with slower than the limit of detection (<5% cleavage after 24 hours) have been assigned a rate of 10−4 min−1 and cannot be assigned an error (“n.a.”). Propagated errors as well as ratios based on these measurements are also “n.a.”
| variant | ionic conditions | substrate | kobs (min−1) | k/k′ | ko/ks | metal ion rescue | mutational rescue |
|---|---|---|---|---|---|---|---|
| Wild Type | 10mM Mg2+ | oxo | 0.32 ± 0.01 | ||||
| A621 SP | < 0.0001 | > 3.2 × 103 | |||||
| A621 RP | < 0.0001 | > 3.2 × 103 | |||||
| A622 SP | 0.0294 ± 0.0008 | 11 ± 0.3 | |||||
| A622 RP | < 0.0001 | > 3.2 × 103 | |||||
| 10mM Mg2+ | oxo | 0.283 ± 0.005 | |||||
| and | A621 SP | 0.0053 ± 0.0001 | 53 ± 1.4 | > 60 ± n.a. | |||
| 20μM Cd2+ | A621 RP | < 0.0001 | > 2.8 × 103 | n.a. | |||
| A622 SP | 0.0328 ± 0.003 | 8.6 ± 0.8 | 1.3 ± 0.1 | ||||
| A622 RP | 0.0190 ± 0.007 | 15 ± 5.5 | > 213 ± n.a. | ||||
| G638I | 10mM Mg2+ | oxo | 0.0135 ± 0.0006 | 24 ± 1.3 | |||
| A621 SP | < 0.0001 | > 130 | n.a. | ||||
| A621 RP | 0.0020 ± 0.0001 | 6.8 ± 0.5 | > 470 ± n.a. | ||||
| 10mM Mg2+ | oxo | 0.0141 ± 0.0005 | 21 ± 0.8 | ||||
| and | A621 SP | 0.0009 ± 0.0001 | 16 ± 1.8 | > 8.1 ± n.a. | 3.3 ± 0.1 | ||
| 20μM Cd2+ | A621 RP | 0.0023 ± 0.0001 | 6.1 ± 0.3 | 1.1 ± 0.1 | > 460 ± n.a. | ||
| A756(3cP) | 10mM Mg2+ | oxo | 0.0336 ± 0.0002 | 9.5 ± 0.3 | |||
| A621 SP | < 0.0001 | > 330 | n.a. | ||||
| A621 RP | 0.0041 ± 0.0001 | 8.2 ± 0.2 | > 390 ± n.a. | ||||
| 10mM Mg2+ | oxo | 0.035 ± 0.002 | 8.1 ± 0.5 | ||||
| and | A621 SP | 0.0016 ± 0.0001 | 22 ± 1.9 | > 15 ± n.a. | 2.4 ± 0.2 | ||
| 20μM Cd2+ | A621 RP | 0.0033 ± 0.0001 | 11 ± 0.7 | 0.8 ± 0.1 | > 250 ± n.a | ||
| G638I | 10mM Mg2+ | oxo | 0.0023 ± 0.0004 | 140 ± 7.4 | |||
| and | A621 SP | < 0.0001 | > 23 | n.a. | |||
| A756(3cP) | A621 RP | 0.0140 ± 0.0006 | 0.2 ± 0.1 | > 16 × 103 ± n.a. | |||
| 10mM Mg2+ | oxo | 0.0018 ± 0.0001 | 160 ± 9.2 | ||||
| and | A621 SP | 0.0009 ± 0.0005 | 2 ± 1.1 | > 12 ± n.a. | 27 ± 15 | ||
| 20μM Cd2+ | A621 RP | 0.011 ± 0.003 | 0.2 ± 0.1 | 1.0 ± 0.5 | > 14 × 103 ± n.a. | ||
Inspection of the crystal structures shows that the exocyclic amines of G638 and A756 reside spatially close to the scissile phosphate NPOs, and mutational studies11,14 of these nucleobases have hinted at the importance of their exocyclic amines. To identify potential interactions of the G638 and A756 exocyclic amines to the NPOs, we observed cleavage for ribozymes with the G638 mutated to inosine (G638I), A756 mutated to 3-deazapurine (A756(3cP)), or both (referred to here as “double mutant”) in oxo and stereospecific phosphorothioate backgrounds. The use of 3-deazapurine provides a strategy to test removal of A’s exocyclic amine without major perturbation to the N1 pKa.
Both the G638I and A756(3cP) ribozymes exhibit mutational rescues (Table 1) of at least 470 and 390, respectively, suggesting that the exocyclic amines of both G638 and A756 form catalytically important interactions with the pro-RP NPO of the scissile phosphate (Figure 1). Interestingly, the double mutant cleaves the RP thio substrate with a rate of 0.014 min−1 corresponding to an inverse thio effect of 6 and a mutational rescue of nearly 16,000. The inverse thio effect is unexpected but not without precedent. A similar effect was observed in the glmS aporibozyme where, in the absence of cofactor, the 2’-hydroxyl at N-1 engaged in an unproductive interaction with pro-RP NPO, which was eliminated in the RP thio background.27 A similar mechanism could explain the inverse thio effect observed here; in the absence G638:N2 and A756:N6 the pro-RP NPO might engage the 2’-hydroxyl in an unproductive interaction that would deter the reaction center from achieving an in-line conformation. In the RP thio substrate, the sulfur atom would be expected to disrupt this unproductive interaction, resulting in a modest (6-fold) rate enhancement.
To summarize, the loss in reactivity due to a sulfur substitution at the pro-RP NPO position of the WT ribozyme was recovered to some extent by removing the exocyclic amines of either or both A756 and G638. These observations strongly suggest that the active site is organized by interactions between G638:N2, A756:N6 and the pro-RP NPO, and the disruption of either or both of these interactions substantially reduces the rate of cleavage.
Independent MD simulations departing from different crystal structures converge on a functionally inactive state of the ribozyme in the absence of Mg2+
In an effort to unify the VS ribozyme crystal structures with the existing body of solution data, we tested whether molecular motions from the configurations around the scissile phosphate observed in the static crystal structures could plausibly generate active sites consistent with a consensus picture of VS catalysis. We define such active sites as those with in-line fitness of the nucleophile (τ >140°) consistent with α catalysis, and G638 (deprotonated at N1) and A756 (protonated at N1) positioned to act in general base (γ) and acid (δ) catalysis, respectively, while having their exocyclic amines interacting with the pro-RP NPO of the scissile phosphate (β catalysis) as established above. To that end, we carried out MD simulations of the WT ribozyme using all three available crystal structures as departure points so as to ensure predictions were independent of starting state.19,20 Results from these simulations are summarized in Figure 2[a–b].
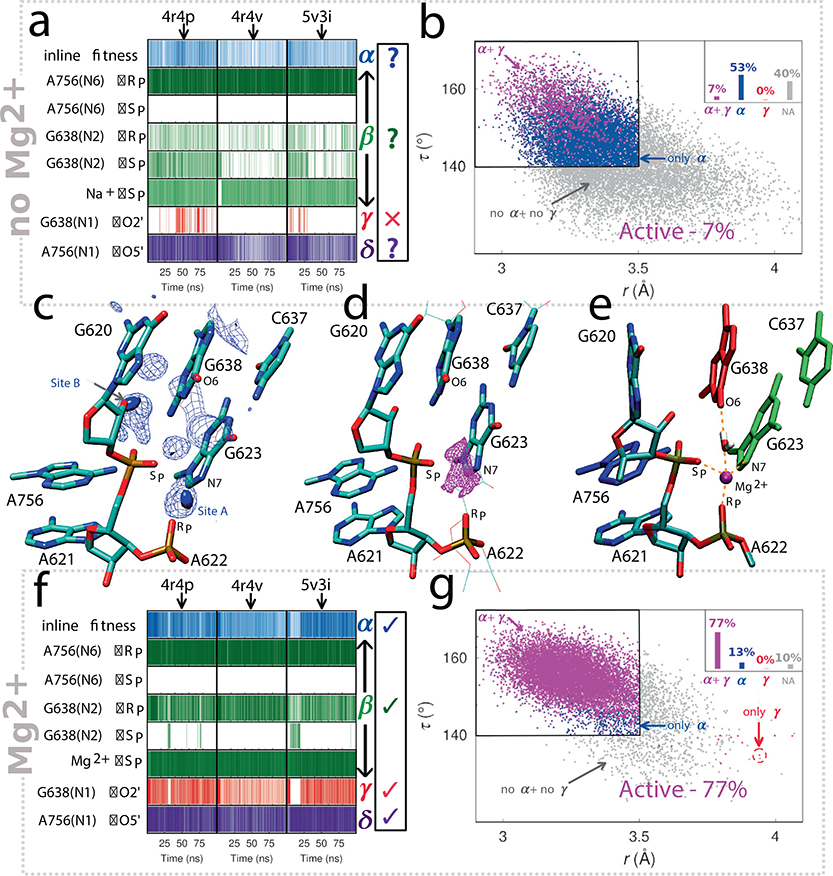
Figure 2. Computational investigation of catalytic strategies adopted by the VS ribozyme.
Panels (a) and (b) summarize results from MD simulations based on the existing crystal structures as is. Panel (a) illustrates the persistence of active site interactions during the various MD simulations. The three columns correspond to simulations departing from the 4R4P, 4R4V, and 5V3I crystal structures, while the rows correspond to interactions related to different catalytic strategies (α-δ). The first row refers to in-line fitness, which is dictated by the τ (O2’-P-O5’) angle ≥ 140° and the O2’-P distance ≤ 3.5 Å. Hydrogen bond and metal-ion interaction distance thresholds (β-δ) were chosen to ≤ 2 Å and ≤ 2.5 Å, respectively. Panel (b) is 2D scatter plot of in-line attack angle τ and O2’-P distance (r), and include data from all MD simulations. Data points are classified into 4 categories – configurations that exhibit both α and γ catalysis (magenta), only α catalysis (blue), only γ catalysis (red), and configurations that do not exhibit either α or γ catalysis (non-active, gray). Datapoints corresponding to the different categories are indicated by arrows and the black rectangle outlines the region that exhibits α catalysis. Panel (c) illustrates Na+ ion charge isodensity plots in the active site region calculated from MD simulations. To construct these isodensity plots, a 3D cubic grid having an edge length of 12 Å and spacing of 1 Å was centered on the scissile phosphate phosphorus atom. For a given MD trajectory, the total charge per grid cell was determined by summing over the charges of all ions in that particular cell. The blue solid surface and the mesh represent the top 30% and top 70% of the charge density of the entire grid, respectively. Panel (d) illustrates Mg2+ ion binding site predicted by 3D-RISM calculations. The magenta surface represents the top 10% of Mg2+ ion isodensity. Panel (e) depicts the Mg2+ bound active site model obtained from MD simulations. In this model, the Mg2+ ion forms inner sphere contacts with the A621 pro-SP NPO, A622 pro-RP NPO and G623:N7, and interacts with G638:O6 via a water molecule. Panels (f) and (g) are analogous to panels (a) and (b), respectively, and summarize results from MD simulations based on the crystal structures but with an added Mg2+ ion bound at the pro-SP NPO of the scissile phosphate.
Figure 2a depicts the important active site interactions along the various MD trajectories in the form of interaction maps. These maps report on the probability of observing specific active site interactions identified from functional data that support each of the catalytic strategies (α-δ). In the simulations, the putative general base G638 and general acid A756 were kept deprotonated and protonated, respectively, at their N1 positions, assumptions supported by linear free energy relationships described below. According to Figure 2a, the simulations based on the different crystal structures paint a consistent dynamical picture of the ribozyme active site – one in which each of the key elements of catalysis (α, β, γ, and δ) are either absent or not present in full strength to support efficient catalysis and hence does not reflect an active state in solution. Most concerning in the simulations was the lack of formation of a catalytically active state characterized by in-line fitness and positioning of the O2’ nucleophile for activation (α and γ catalysis, respectively), which accounted for only a small fraction (7%) of all configurations (Figure 2b). Taken together the results from these MD simulations suggest that the active site ensemble emerging from simulations based on the ribozyme crystal structures as is, does not reflect the catalytically active state of the ribozyme in solution under standard conditions. Detailed discussion of Figure 2[a–b] is provided in Supplementary Information.
MD simulations and molecular solvation theory calculations predict a Mg2+ binding site at the active site
No divalent metal ions were observed at the active site in the VS ribozyme crystal structures, possibly because they were obtained in the presence of high ammonium ion concentrations. However, spectroscopic data28–32 suggest that VS activity depends on multiple localized divalent ions. VS cleavage typically requires Mg2+ ions for activity, except in very high concentrations of monovalent ions.33 The ribozyme is not active in cobalt hexammine alone, but in the presence of small amounts of Mg2+, cobalt hexammine enhances catalysis.34 The Mg2+ ion dependence of the ribozyme folding, inferred from hydroxyl radical footprinting, was estimated to be an order of magnitude lower concentration than that required for catalysis.35 Taken together these studies indicate that Mg2+ ions are not only important for global folding but are more directly involved in interactions (either in organization or transition state stabilization) at the active site that affect catalysis.
Analysis of the MD trajectories revealed two sites in the active site where Na+ ions bind with high occupancy (Figure 2c). Site A is at the pro-SP NPO of the scissile phosphate, and site B is between the 2’O nucleophile and the pro-RP NPO of the scissile phosphate. To identify probable Mg2+ binding sites, we carried out 3D-RISM calculations36,37 on the different crystal structures as well as configurations obtained from simulations. Results from the 3D-RISM calculations predict high probability of Mg2+ ion binding at site A in the active site (Figure 2d). A Mg2+ ion at this position is consistent with the previously reported thio effects25 observed for a mixture of diastereoisomers for the scissile phosphate (although this was not rescued in the presence of Mn2+) and at the phosphate of the N+1 nucleotide (A622) downstream of the scissile phosphate. Thus, analysis of Na+ ion binding at the active site and results from 3D-RISM calculations jointly suggest that site A, the position coordinating the pro-SP NPO of A621 (the scissile phosphate), and close to the pro-RP NPO of A622 and the N7 of G623 is a plausible Mg2+ binding site.
Metal ion rescue experiments confirm a Mg2+ ion is bound stereospecifically at the pro-SP non-bridging position
Given the large thio effects observed and the computational prediction that a divalent metal ion likely occupies a position near the active site, we tested the rate of cleavage of A621 RP and SP thio substrates in the presence of a thiophilic metal ion. Two prior studies had investigated metal ion rescue for the scissile phosphate and did not observe any rescue. However, as mentioned previously, those studies were complicated by the use of a mixture of diastereomeric substrates25 or observation of the reverse reaction rather than cleavage26. Furthermore, both studies tested for rescue in the presence of Mn2+ rather than the more thiophilic Cd2+. We investigated metal ion rescue with Cd2+ in a background of 10mM Mg2+ in an effort to identify the putative metal ion binding site. The results are summarized in Table 1. In the oxo background, Cd2+ has a minimal impact on the rate of cleavage at low concentration, but inhibits cleavage when the concentration of Cd2+ approaches the concentration of Mg2+. The presence of 20μM Cd2+ stimulated the rate of cleavage for the SP thio substrate to 0.0053 min−1, corresponding to at least a 53-fold enhancement over rate of cleavage in the absence of Cd2+ and a metal ion rescue of at least 60. Note, the actual metal ion rescue is somewhat higher, but since the actual rate in the absence of Cd2+ is less than the limit of detection, we assume a rate of 10−4 min-1. In contrast to the SP thio substrate, the RP thio substrate exhibits no enhanced cleavage in the presence of Cd2+ corresponding to no metal ion rescue, indicating that the stimulatory effect of the thiophilic metal is site specific. Additionally, we tested the effect of metal ion rescue in the G638I, A756(3cP) and double mutant ribozyme backgrounds and observed metal ion rescues of at least 8, 15 and 12, respectively, for the SP thio substrate, and no metal ion rescue for the RP thio substrate for each variant. These data provide support for a mechanism wherein a divalent metal ion specifically coordinates to the pro-SP NPO in the ribozyme active site.
Simulations with an active site Mg2+ ion converge on a functionally active state of the ribozyme
Motivated by the results from the metal ion rescue experiments, we sought to investigate the ramifications of a Mg2+ ion for the ribozyme active site dynamics. We re-propagated MD trajectories, departing from each of the different crystal structures and having G638 and A756 in their active protonation states as previously, but with a Mg2+ ion bound at the A621 pro-SP NPO, as well as close to the pro-RP NPO of A622 and the N7 of G623 as predicted above (site A in the active site, Figure 2[c–e]). The results of these simulations are summarized in Figure 2[f–g]. Interestingly, as illustrated by the interaction maps (Figure 2f), these independent trajectories converge to an active site ensemble that agrees with experimental data and exhibits all the key catalytic strategies.
For the simulations in the presence of a Mg2+ ion bound at site A in the active site, the majority of the configurations (77%) sampled a catalytically active state (exhibiting both α and γ catalysis), compared with only 7% of configurations in the simulations without Mg2+. This eleven-fold increase in active configurations observed in MD simulations in presence of the Mg2+ ion at site A suggests that the metal ion enhances catalysis, at least in part, by organizing the active site.
The Mg-bound model of the active site emerging from these MD simulations is shown in Figure 2e and Supplementary Figure 3. The Mg2+ ion remains mostly localized in the simulations, and apart from the scissile phosphate A621 pro-SP NPO, maintains an inner sphere contact with the N7 of G623. In two of the three simulations, the metal ion also maintains an inner sphere contact with the A622 pro-RP NPO and interacts with G638:O6 via an water molecule, while in one simulation, the metal ion moves up in the active site to form an inner sphere contact with the G638:O6 and interacts with the A622 pro-RP NPO via a water molecule. In each of the simulations, two other water molecules complete the hexacoordination of the ion.
Mutation experiments further validate the Mg2+ bound model
We sought to validate the Mg-bound model (Figure 2e), first, by testing for metal ion rescue at the downstream phosphate, A622. Previous studies showed a substantial thio effect at the phosphorothioate downstream of the scissile phosphate rescuable with Mn2+,25,38 but lack clarity on the stereochemistry of the putative metal ion–phosphate interaction. In presence of 10mM Mg2+, the A622:SP thio substrate cleaves with a rate of 0.029 min−1 corresponding to a modest thio effect of about 11. The A622:RP thio substrate, on the other hand, cleaves with a rate below the limit of detection corresponding to a thio effect of at least 3000. In the presence of 20μM Cd2+, cleavage of the A622:SP thio substrate is virtually unaffected but the A622:RP thio substrate is accelerated at least 190-fold to a rate of 0.019 min−1 corresponding to metal rescue of 213. These data strongly suggest that the pro-RP NPO of the phosphate downstream of the cleavage center coordinates a divalent metal ion, consistent with the computational prediction. We also tested cleavage with a G623(7cG) ribozyme variant, which eliminates the putative G623:N7–Mg2+ interaction. This ribozyme failed to cleave the phosphate substrate with a detectable rate, implying that G623:N7 plays an important role in catalysis. Regarding the predicted outer sphere Mg2+ interaction with G638:O6, the unique configuration of 7-deaza-5-azaguanosine (a member of the Hachimoji alphabet,5,39–41 and referred to herein as “P*”) The “P*” nucleobase enables a strategy to test for a possible role of the O6 keto group. “P*” ionizes with the same pKa as 2-aminopurine riboside (2-amP) and differs from 2-aminopurine on the Watson-Crick face only by the presence of the keto group (Supplementary Figure 5). In contexts where N7 makes no interactions (as in G638 of the VS ribozyme), comparisons of RNAs bearing P* versus 2-amP can reveal the functional significance of a G residue’s O6 keto group. At all pH values tested, a substrate containing the G638P* mutation reacts significantly faster than a substrate containing G638(2amP), consistent with an interaction involving the O6 keto group. In summary, the phosphorothioate and subsequent metal ion rescue effect observed at A622 pro-RP NPO, inactivity of the G623(7cG) ribozyme variant, and faster reactivity of the G638P* variant compared to the G638(2amP) (the latter does not have the O6 keto group at position 638), suggest that the Mg2+ ion interacts with the A622 pro-RP NPO, G623:N7, and G638:O6 (through a coordinated water molecule), respectively, and provide crucial evidence in support of our proposed metal binding site.
LFERs suggest proton transfer is nearly complete for the general base and partial for the general acid in the transition state
The consensus view of VS catalysis holds that G638 and A756 function as general base and general acid, respectively.22,42 Although evidence exists to support these hypotheses,11,14–18 significant gaps in our understanding remain. For instance, phosphorothiolate rescue experiments17 demonstrate a functional linkage between A756 and the 5ʹO-leaving group, but those data leave open the possibility of catalysis through some mechanism other than proton transfer to the leaving group. Also, although pH-rate profiles implicate titration of G638, no data to-date establish a functional linkage between G638 and the 2ʹO-nucleophile. Moreover, nothing is known about the nature of the transition state in terms of extents of proton transfer from the general acid and to the general base. To shed light on these aspects of VS ribozyme catalysis, we measured Brønsted coefficients for a series of ribozymes with A and G analogs (see Supplementary Information for details).
To test the extent of proton transfer between the general acid, A756, and the 5ʹO-leaving group, we observed the rate of cleavage over a range of pH values for ribozymes with adenosine (WT), 3-deazaadenosine (3cA), 7-deazaadenosine (7cA), 8-azaadenosine (8nA), 8-aza-7-deazaadenosine (8n7cA), and 3-deazapurine (3cP) at the 756 position. The observed linear relationship observed in the Brønsted plot for these A analogs, excluding 3cP, (Figure 3a) strongly suggests that A756 is involved in proton transfer and the expected deviation from linearity observed for the A756(3cP) ribozyme results from the deletion of A756:N6, which coordinates the pro-RP NPO of the scissile phosphate. The small Brønsted coefficient observed (α = 0.23) indicates that a) proton transfer from A756 occurs in the rate-controlling step and not in a pre-equilibrium step and b) the extent of proton transfer is only partial in the transition state. In contrast, proton transfer to the leaving group in the HDV ribozyme appears to be significantly more advanced.43
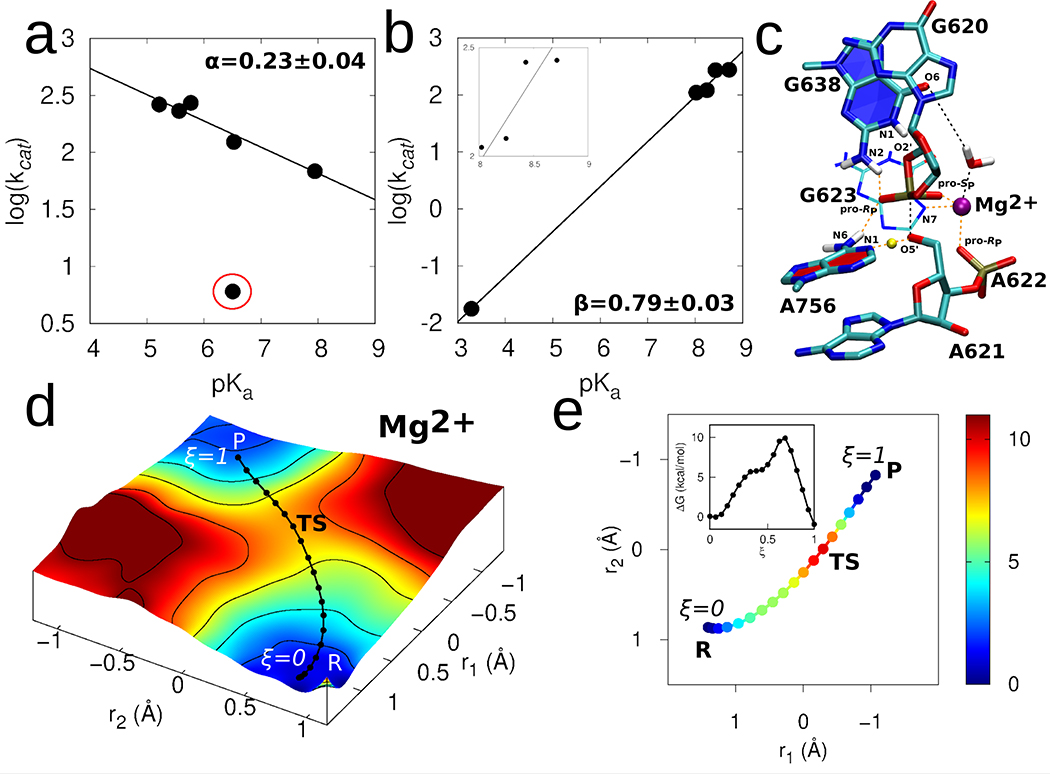
Figure 3. Catalytic mechanism of the VS ribozyme.
(a) Brønsted plot for VS ribozymes with A756 analogs. kcat and pKa values were determined through fitting pH-profiles (Supplementary Figure 6 to a model for double ionization. For all A analogs, the lower pKa was assumed to correspond to the general acid (A756 in wild type ribozyme). The data (excluding the outlier 3cP mutant) were fit linearly to reveal a slope = 0.23 ± 0.04. (b) Brønsted plot for VS ribozymes with G638 analogs. kcat and pKa values were determined through fitting pH-profiles (Supplementary Figure 7) to a model for double ionization with the exception of the G638P* substrate, which was fit to a model for single ionization. For all G analogs, the higher pKa was assumed to correspond to the general base (G638 in wild type ribozyme). The pKa of P* (3.3) was inferred from titration of the nucleoside in solution. The linear fitting of the resulting plot revealed a slope of 0.79 ± 0.03. The inset in (b) zooms into the datapoints that falls in the pKa range 8–9. The linear fitting of only these datapoints and excluding the datapoint corresponding to P* (at 3.3) results in a slope of 0.67 ± 0.04. (c) 3D structure of representative transition state geometry along the reaction pathway obtained in presence of the active site Mg2+ ion. (d) The 2D free energy surface underlying the catalytic reaction obtained using ab initio QM/MM umbrella sampling simulations. The solid black line corresponds to the converged pathway obtained using FTS simulations. (e) Converged pathway obtained from the FTS simulations in presence of active site Mg2+ ion. In (d) and (e), reaction coordinate r1 corresponds to the phosphorus bond breaking/forming coordinate represented by the difference between the A621:P-G620:O2’ and A621:P-A621:O5’ distances, and r2 corresponds to the general acid proton transfer coordinate represented by the difference between the A756:N1-A756:H1 and A621:O5’-A756:H1 distances. The color scale denotes relative free energy values in units of kcal/mol. Inset in (e) shows the 1D free energy profile of the pathway obtained in presence of the active site Mg2+ ion.
We repeated the same procedure to investigate proton transfer to the putative general base, G638, with a series of substrates containing guanosine (WT), 7-deazaguanosine (7cG), 8-azaguanosine (8nG), 6-thio-guanosine (6sG), or 7-deaza-5-azaguanosine (P*) at the 638 position. The Brønsted plot constructed for the G analogs reveal a linear dependence on pKa of the G analogs with a slope near 1 (β = 0.79±0.03) (Figure 3b). Considering the uncertainty of the data point at pKa 3.3, due to the need to estimate the reaction pKa from the nucleobase solution pKa, the derived β slope of 0.79 should be viewed with caution and considered a rough estimate for the degree of proton transfer to the nucleobase. Omission of the datapoint at pKa 3.3, which limits the dataset to a relatively narrow pKa range (8–9), yields a modestly attenuated slope (β value) of 0.67±0.04. Despite these limits to their quantitative interpretation, the data qualitatively support a mechanism involving substantial proton transfer from the nucleophile to the general base in the transition state.
Taken together, the LFER data strongly support the roles of the catalytic nucleobases A756 and G638 in general acid-base catalysis established previously by Lilley and co-workers,11,12,14,15,17,22 and provide additional information about the nature of the transition state involving each of the proton transfer steps. Proton transfer between A756 and the 5ʹO-leaving group occurs in the rate-controlling step and the extent of transfer is partial in the transition state. Brønsted analysis also implicates G638 in proton transfer. The data suggest that proton transfer from the nucleophile has advanced significantly in the transition state but lack the resolution to distinguish between pre-equilibrium or concerted proton transfer for the catalytic guanine. Computational results presented below provide predictive insight to resolve this ambiguity.
QM/MM free energy simulations departing from the Mg2+ bound model predict a catalytic mechanism that is consistent with LFER data
We calculated the free energy surface and the minimum free energy pathway associated with the catalytic reaction by combining semi-empirical finite temperature string (FTS) simulations 44,45 and ab initio 1D and 2D QM/MM umbrella sampling (US) simulations (details provided in Supplementary Information). The FTS simulations suggested a sequential catalytic pathway in which the 2’O nucleophile deprotonation by G638 occurs initially, as a separate step, followed by concerted nucleophilic attack and leaving group departure; the associated free energy profile along the pathway suggested the latter to be rate-limiting (Supplementary Figure 9). This result is consistent with Brønsted analysis of the G638 analogs, and together support a mechanism where the proton transfer from the 2’O-hydroxyl nucleophile occurs in a pre-equilibrium step.
The chemical step of the reaction was studied in more detail using ab initio 2D QM/MM umbrella sampling (US) simulations. The 2D free energy surface calculated from these ab initio simulations (Figure 3d) is in excellent agreement with the converged pathway obtained from the semi-empirical FTS simulations (see Supplementary Figure 10 for direct comparison) and suggest that the P-O2’/P-O5’ bond formation/cleavage and general acid proton transfer to the leaving group occur in a concerted fashion. In order to obtain better resolution of free energy along the reaction pathway, we performed additional ab initio 1D umbrella US along the converged pathway obtained from the FTS simulations, the results of which are shown in Figures 3[d–e]. The free energy barrier of the reaction obtained from these simulations is ~10 kcal/mol, which, using the Arrhenius rate model, corresponds to an intrinsic rate constant of ~104 s−1 (assuming a prefactor of 1 ps−1). This calculated rate constant is consistent with the intrinsic rates obtained for certain fast-cleaving VS constructs (1.4×103 s−1),18,46 but is somewhat higher than the rates observed in this work. Presumably, fast dynamical equilibria reduce the observed rate of cleavage, especially in the trans-constructs employed in this study that require substrate docking; these processes are not accounted for in the QM/MM calculations. Thus, the estimated rate constant obtained from the simulations represents an upper limit of plausible intrinsic rate constants. The transition state obtained from these simulations (Figure 3c) is a “late” transition state, in which the P-O2’ bond is almost fully formed, the P-O5’ bond is mostly broken, and the proton from A756 is partly transferred to the 5’O leaving group, consistent with the LFER data, and further suggesting that the proton transfer from G638 likely occurs in a pre-equilibrium step prior to arriving at the rate-controlling transition state.
To summarize, the QM/MM simulations departing from the Mg2+ bound model suggest a mechanism in which the 2’O nucleophile activation by the general base occurs in a pre-equilibrium proton transfer step; the rate-controlling step consists of the P-O2’/P-O5’ bond formation/cleavage and general acid proton transfer to the 5’O leaving group. The free energy barrier for this step is consistent with the ribozyme intrinsic reaction rates and passes through a “late” transition state in which the P-O2’ bond is mostly formed, the P-O5’ bond is mostly broken, and the proton from the general acid has partially transferred to the 5’O leaving group, in agreement with the LFER data.
Discussion
Ribozymes use two main chemical moieties for catalysis: metal ions and nucleobases. From biophysical studies spanning nearly three decades, the consensus view held that the VS catalytic mechanism depended entirely on nucleobase catalysis and the contributions of metal ions were restricted to structural organization.12,22 Herein, we presented a combination of computational and experimental data that converge on a configuration of interactions within the active site that imparts catalysis (Figure 4), including a newly identified divalent metal ion that interacts with the pro-SP oxygen of the scissile phosphate and helps to organize the active site through additional inner sphere coordination to the pro-RP oxygen of the downstream phosphate (A622) and G623:N7 and an outer sphere coordination to G638:O6.
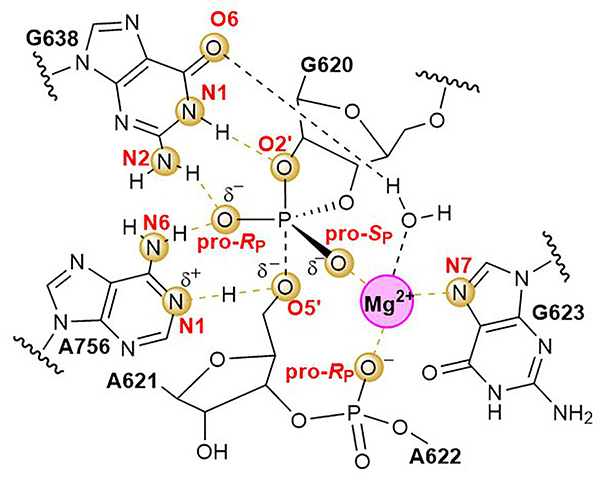
Figure 4. An elaborate network of interactions configures the VS ribozyme for proton transfer and transition state stabilization.
2D schematic of the transition state configuration developed in this work. The transition state is a “late” transition state, in which the P-O2’ bond is almost fully formed, the P-O5’ bond is mostly broken, and the proton from A756 is partly transferred to the 5’O leaving group, consistent with the LFER data, and suggesting that the proton transfer from G638 likely occurs in a pre-equilibrium step prior to arriving at the rate-controlling transition state. Atoms and dashed line connections highlighted in gold indicate functional linkages established in this study. Dotted line connections indicate forming and breaking O-P bonds and H atom in flight from A756 to O5’.
Prior to this work, the VS and hairpin ribozymes were speculated to be mechanistically nearly identical, having topologically similar active sites and employing exclusively nucleobase catalysis in their cleavage mechanisms.12 In our previous study,19 based on the then-recent VS structural data, we took the parallels in these ribozymes a step further by illustrating that both VS and hairpin ribozymes, along with the hammerhead ribozyme, share a common active site scaffold, referred to as the L-platform; an L-shaped motif in which the nucleotide 3’ to the general base makes the corner of the L, the N-1 nucleotide forming the short arm of the L, and the nucleotide 5’ to the general base forming the long arm of the L with the general base (Figure 5). The presence of this L-shaped motif in three distinct ribozyme classes implicates the L-platform motif as a functional strategy to position the N-1 nucleotide and direct the general base towards the 2’-hydroxyl group.
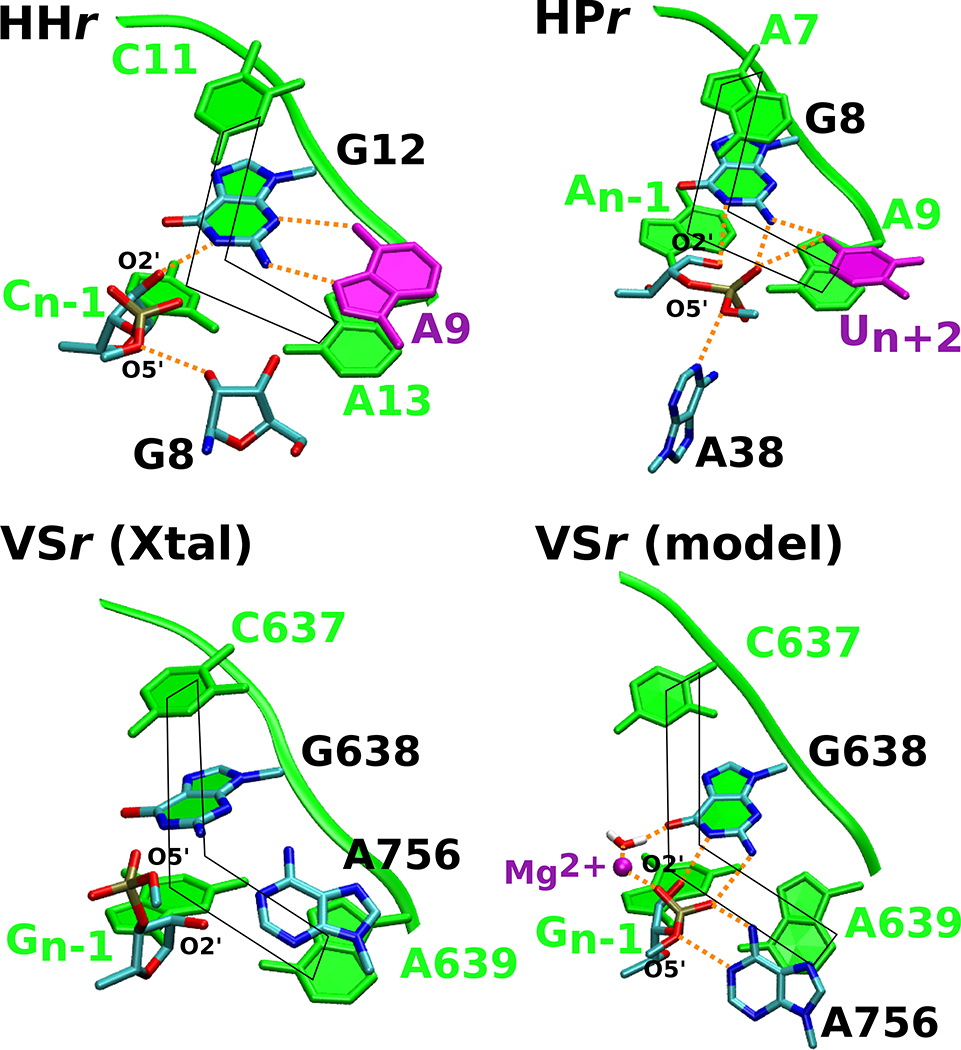
Figure 5. The hammerhead, hairpin and VS ribozymes use distinct strategies to anchor the L-platform.
Active sites in the crystal structures of the three ribozymes are compared to the Mg2+-bound VS ribozyme model proposed in this work to highlight common features. Each of the ribozyme active sites exhibit the L-platform motif (shown in fade green) formed by the N-1 nucleotide and nucleotide 3’ to the general base as the base of the L, and the nucleotide 3’ to the general base, general base itself, and the nucleotide 5’ to the general base as the long arm of the L. Both the hammerhead and hairpin contain a L-anchor residue (shown in purple) that engages with the general base and presumably holds it in position; this active site feature is lacking in the VS ribozyme crystal structures. The Mg2+-bound model defined in this study suggests that the metal ion steps into the role of the L-anchor, interacting with G638 and positioning the base for catalysis. In this figure, the hairpin, hammerhead, and VS ribozyme active sites are based on PDB IDs 1M5O,50 3ZP8,51 and 5V3I,20 respectively.
Herein, our joint experimental and computational efforts illustrate that despite having the L-platform, the VS ribozyme fails to exhibit the key catalytic interactions in the absence of the active site Mg2+ ion, implying that the presence of the L-platform alone may not provide sufficient positioning for the general base. Considering this new data, we went back to the hairpin and hammerhead crystal structures, as well as the subsequently determined twister ribozyme structure, and found that, instead of a Mg2+ ion as in the VS ribozyme, these ribozymes have an active site nucleotide, referred to here as the L-anchor, that interacts with the general base and presumably contributes to its positioning for nucleophile activation. In hammerhead, a conserved adenine (A21) serves as the L-anchor, whereas in hairpin an invariant uracil (U+2) serves this role. In the currently available structural data, the VS ribozyme lacks a nucleobase that could plausibly serve the role of the L-anchor. However, inspection of the model of the VS ribozyme defined in this work suggests that the newly identified Mg2+ ion may serve as the L-anchor, fulfilling the role served by a nucleobase in the hairpin and hammerhead ribozymes. The metal ion assists in organizing the active site through direct stereospecific interactions with the A621 and A622 phosphates and G623:N7, and a water-mediated interaction with G638:O6. The outer sphere interaction between the Mg2+ and G638:O6, supported functionally by the faster reaction rates of the G638P mutant as compared to G638(2amP) mutant, may also assist with shifting the G638 pKa to its apparent value of 8.4. Similar pKa tuning of the general base caused by divalent metal ion binding has been observed in X-ray structures47,48 of the hammerhead ribozyme at elevated pH and have been subsequently interpreted mechanistically in recent computational work.49
Thus, while our studies corroborate the existing notion that the VS ribozyme bears mechanistic similarity to the hairpin ribozyme12, they reveal previously unknown facets of the VS ribozyme active site architecture and elicit mechanistic similarities to the hammerhead ribozyme. In one view, VS is hairpin-like in that it utilizes the same nucleobase residues to carry out general acid-base catalysis. However, the two ribozymes differ in terms of how the catalytic nucleobases utilize their exocyclic amines and by the presence of an active site Mg2+ ion that the VS requires for catalysis. The role of Mg2+ in VS is both organizational as an anchoring construct, and electrostatic in tuning the pKa of the general base and presumably in stabilizing the transition state through interaction with the scissile phosphate. Like the VS ribozyme, the hammerhead ribozyme uses a guanine as a general base and requires a conserved adenine and Mg2+ ion for catalysis. However, the latter roles are reversed in the two ribozymes: in VS A756 provides general acid catalysis, and Mg2+ serves as the L-anchor, whereas in hammerhead the Mg2+ assists in acid catalysis56,58–60, and A9 serves as the L-anchor. Taken together, these observations reveal new functional features shared by the hammerhead, hairpin and VS ribozymes.
Methods
Methods used in this study and associated references are described in the Supplementary Information.
Supplementary Material
Acknowledgements
We thank Dr. Saurja Dasgupta for valuable discussion. A.G., B.W., J.A.P. and D.M.Y. are grateful for financial support provided by the National Institutes of Health (Grant GM62248 to D.M.Y. and Grant GM131568 to J.A.P.). B.W. acknowledges support from the Predoctoral Training Program in Chemistry and Biology (T32-GM008720). Computational resources were provided by the National Institutes of Health under Grant No. S10OD012346, the Office of Advanced Research Computing (OARC) at Rutgers, the State University of New Jersey, Rutgers Discovery Information Institute (RDI2), the State University of New Jersey, and by the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation Grant No. OCI-1053575 (Project No. TG-MCB110101). This research is also part of the Blue Waters sustained-petascale computing project, which is supported by the National Science Foundation (awards OCI-0725070 and ACI-1238993) and the state of Illinois. Blue Waters is a joint effort of the University of Illinois at Urbana-Champaign and its National Center for Supercomputing Applications.
Footnotes
Competing Interests
The authors declare no competing interests.
Data Availability
The data to support the findings of this study are available in the Supplementary Information file and from the corresponding authors upon request.
Code Availability
Simulation software are available in the latest release of AMBER18. Example input files, representative structures, animation of the active site in presence and absence of the Mg2+ ion derived from the MD simulations, and an animation of the catalytic reaction derived from the simulations are provided online free to download: http://theory.rutgers.edu.
REFERENCES
- 1.Abelson J The discovery of catalytic RNA. Nat Rev Mol Cell Biol 18, 653, doi: 10.1038/nrm.2017.105 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Symons RH SMALL CATALYTIC RNAs. Ann Rev Biochem 61, 641–671, doi: 10.1146/annurev.bi.61.070192.003233 (1992). [DOI] [PubMed] [Google Scholar]
- 3.Herschlag D & Cech TR Catalysis of RNA cleavage by the Tetrahymena thermophila ribozyme. 1. Kinetic description of the reaction of an RNA substrate complementary to the active site. Biochemistry 29, 10159–10171, doi: 10.1021/bi00496a003 (1990). [DOI] [PubMed] [Google Scholar]
- 4.Narlikar GJ & Herschlag D MECHANISTIC ASPECTS OF ENZYMATIC CATALYSIS: Lessons from Comparison of RNA and Protein Enzymes. Ann Rev Biochem 66, 19–59, doi: 10.1146/annurev.biochem.66.1.19 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Hoshika S et al. Hachimoji DNA and RNA: A genetic system with eight building blocks. Science 363, 884, doi: 10.1126/science.aat0971 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jimenez RM, Polanco JA & Lupták A Chemistry and Biology of Self-Cleaving Ribozymes. Trends Biochem Sci 40, 648–661, doi: 10.1016/j.tibs.2015.09.001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson TJ, Liu Y & Lilley DMJ Ribozymes and the mechanisms that underlie RNA catalysis. Front. Chem. Sci. Eng 10, 178–185 (2016). [Google Scholar]
- 8.Kennell JC et al. The VS catalytic RNA replicates by reverse transcription as a satellite of a retroplasmid. Gene Dev 9, 294–303, doi:DOI 10.1101/gad.9.3.294 (1995). [DOI] [PubMed] [Google Scholar]
- 9.Saville BJ & Collins RA A site-specific self-cleavage reaction performed by a novel RNA in neurospora mitochondria. Cell 61, 685–696, doi:Doi 10.1016/0092-8674(90)90480-3 (1990). [DOI] [PubMed] [Google Scholar]
- 10.Lafontaine DA, Norman DG & Lilley DMJ The global structure of the VS ribozyme. The EMBO journal 21, 2461–2471, doi: 10.1093/emboj/21.10.2461 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lafontaine DA, Wilson TJ, Zhao ZY & Lilley DMJ Functional group requirements in the probable active site of the VS ribozyme. J Mol Biol 323, 23–34, doi: 10.1016/S0022-2836(02)00910-5 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Wilson TJ & Lilley DMJ Do the hairpin and VS ribozymes share a common catalytic mechanism based on general acid-base catalysis? A critical assessment of available experimental data. RNA 17, 213–221, doi: 10.1261/rna.2473711 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiley SL, Sood VD, Fan J & Collins RA 4-thio-U cross-linking identifies the active site of the VS ribozyme. The EMBO journal 21, 4691–4698, doi: 10.1093/emboj/cdf462 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson TJ, McLeod AC & Lilley DMJ A guanine nucleobase important for catalysis by the VS ribozyme. Embo J 26, 2489–2500, doi: 10.1038/sj.emboj.7601698 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lafontaine DA, Wilson TJ, Norman DG & Lilley DMJ The A730 loop is an important component of the active site of the VS ribozyme. J Mol Biol 312, 663–674, doi:DOI 10.1006/jmbi.2001.4996 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Jaikaran D, Smith MD, Mehdizadeh R, Olive J & Collins RA An important role of G638 in the cis-cleavage reaction of the Neurospora VS ribozyme revealed by a novel nucleotide analog incorporation method. RNA 14, 938–949, doi: 10.1261/rna.936508 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson TJ et al. Nucleobase-mediated general acid-base catalysis in the Varkud satellite ribozyme. Proc Natl Acad Sci USA 107, 11751–11756, doi: 10.1073/pnas.1004255107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith MD & Collins RA Evidence for proton transfer in the rate-limiting step of a fast-cleaving Varkud satellite ribozyme. Proc Natl Acad Sci USA 104, 5818–5823, doi: 10.1073/pnas.0608864104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suslov NB et al. Crystal structure of the Varkud satellite ribozyme. Nat Chem Biol 11, 840–846, doi: 10.1038/nchembio.1929 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DasGupta S, Suslov NB & Piccirilli JA Structural Basis for Substrate Helix Remodeling and Cleavage Loop Activation in the Varkud Satellite Ribozyme. J Am Chem Soc 139, 9591–9597, doi: 10.1021/jacs.7b03655 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collins RA The Neurospora Varkud satellite ribozyme. Biochem Soc Trans 30, 1122, doi: 10.1042/bst0301122 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Lilley DMJ The Varkud satellite ribozyme. RNA 10, 151–158, doi: 10.1261/rna.5217104 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bevilacqua PC et al. An Ontology for Facilitating Discussion of Catalytic Strategies of RNA-Cleaving Enzymes. ACS Chemical Biology 14, 1068–1076, doi: 10.1021/acschembio.9b00202 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emilsson GM, Nakamura S, Roth A & Breaker RR Ribozyme speed limits. RNA 9, 907–918, doi: 10.1261/rna.5680603 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kovacheva YS, Tzokov SB, Murray IA & Grasby JA The role of phosphate groups in the VS ribozyme-substrate interaction. Nucleic Acids Res 32, 6240–6250, doi: 10.1093/nar/gkh957 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zamel R & Collins RA Rearrangement of substrate secondary structure facilitates binding to the Neurospora VS ribozyme. J Mol Biol 324, 903–915, doi: 10.1016/S0022-2836(02)01151-8 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Bingaman JL et al. The GlcN6P cofactor plays multiple catalytic roles in the glmS ribozyme. Nat Chem Biol 13, 439–445 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell DO & Legault P Nuclear magnetic resonance structure of the varkud satellite ribozyme stem-loop V RNA and magnesium-ion binding from chemical-shift mapping. Biochemistry 44, 4157–4170, doi:DOI 10.1021/bi047963l (2005). [DOI] [PubMed] [Google Scholar]
- 29.Campbell DO, Bouchard P, Desjardins G & Legault P NMR structure of Varkud satellite ribozyme stem-loop V in the presence of magnesium ions and localization of metal-binding sites. Biochemistry 45, 10591–10605, doi: 10.1021/bi0607150 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Bonneau E & Legault P Nuclear magnetic resonance structure of the III-IV-V three-way junction from the varkud satellite ribozyme and identification of magnesium-binding sites using paramagnetic relaxation Enhancement. Biochemistry 53, 6264–6275, doi: 10.1021/bi500826n (2014). [DOI] [PubMed] [Google Scholar]
- 31.Bonneau E & Legault P NMR Localization of Divalent Cations at the Active Site of the Neurospora VS Ribozyme Provides Insights into RNA-Metal-Ion Interactions. Biochemistry 53, 579–590, doi: 10.1021/bi401484a (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dagenais P, Girard N, Bonneau E & Legault P Insights into RNA structure and dynamics from recent NMR and X-ray studies of the Neurospora Varkud satellite ribozyme. Wires RNA 8, doi:ARTN e142110.1002/wrna.1421 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murray JB, Seyhan AA, Walter NG, Burke JM & Scott WG The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem Biol 5, 587–595, doi:Doi 10.1016/S1074-5521(98)90116-8 (1998). [DOI] [PubMed] [Google Scholar]
- 34.Maguire JL & Collins RA Effects of cobalt hexammine on folding and self-cleavage of the Neurospora VS ribozyme. J Mol Biol 309, 45–56, doi:DOI 10.1006/jmbi.2001.4625 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Tzokov SB, Murray IA & Grasby JA The role of magnesium ions and 2 ‘-hydroxyl groups in the VS ribozyme-substrate interaction. J Mol Biol 324, 215–226, doi: 10.1016/S0022-2836(02)01063-X (2002). [DOI] [PubMed] [Google Scholar]
- 36.Luchko T et al. Three-dimensional molecular theory of solvation coupled with molecular dynamics in Amber. J Chem Theory Comput 6, 607–624, doi: 10.1021/ct900460m (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Genheden S, Luchko T, Gusarov S, Kovalenko A & Ryde U An MM/3D-RISM approach for ligand binding affinities. J Phys Chem B 114, 8505–8516, doi: 10.1021/jp101461s (2010). [DOI] [PubMed] [Google Scholar]
- 38.Sood VD, Beattie TL & Collins RA Identification of phosphate groups involved in metal binding and tertiary interactions in the core of the Neurospora VS ribozyme. J Mol Biol 282, 741–750, doi:DOI 10.1006/jmbi.1998.2049 (1998). [DOI] [PubMed] [Google Scholar]
- 39.Kim SH, Bartholomew DG, Allen LB, Robins RK & Revankar GR Imidazo[1,2-a]-s-triazine nucleosides. Synthesis and antiviral activity of the N-bridgehead guanine, guanosine, and guanosine monophosphate analogues of imidazo[1,2-a]-s-triazine. J Med Chem 21, 883–889 (1978). [DOI] [PubMed] [Google Scholar]
- 40.Krauch T et al. New Base-Pairs for DNA and RNA. Abstracts of Papers at the 196th ACS National Meeting of the American Chemical Society, Los Angeles, CA, Sep. 25–30 (1988). [Google Scholar]
- 41.Georgiadis MM et al. Structural basis for a six nucleotide genetic alphabet. J Am Chem Soc 137, 6947–6955, doi: 10.1021/jacs.5b03482 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jimenez RM, Polanco JA & Luptak A Chemistry and Biology of Self-Cleaving Ribozymes. Trends Biochem Sci 40, 648–661, doi: 10.1016/j.tibs.2015.09.001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koo SC et al. Transition State Features in the Hepatitis Delta Virus Ribozyme Reaction Revealed by Atomic Perturbations. J Am Chem Soc 137, 8973–8982, doi: 10.1021/jacs.5b01189 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.E, W., Liu D & Vanden-Eijnden E Nested stochastic simulation algorithm for chemical kinetic systems with disparate rates. J Chem Phys 123, 194107, doi: 10.1063/1.2109987 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Vanden-Eijnden E & Venturoli M Revisiting the finite temperature string method for the calculation of reaction tubes and free energies. J Chem Phys 130, 194103, doi: 10.1063/1.3130083 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Zamel R et al. Exceptionally fast self-cleavage by a Neurospora Varkud satellite ribozyme. Proc Natl Acad Sci USA 101, 1467–1472, doi: 10.1073/pnas.0305753101 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mir A et al. Two Divalent Metal Ions and Conformational Changes Play Roles in the Hammerhead Ribozyme Cleavage Reaction. Biochemistry 54, 6369–6381, doi: 10.1021/acs.biochem.5b00824 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mir A & Golden BL Two Active Site Divalent Ions in the Crystal Structure of the Hammerhead Ribozyme Bound to a Transition State Analogue. Biochemistry 55, 633–636, doi: 10.1021/acs.biochem.5b01139 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen H, Giese TJ, Golden BL & York DM Divalent Metal Ion Activation of a Guanine General Base in the Hammerhead Ribozyme: Insights from Molecular Simulations. Biochemistry 56, 2985–2994, doi: 10.1021/acs.biochem.6b01192 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rupert PB, Massey AP, Sigurdsson ST & Ferre-D’Amare AR Transition state stabilization by a catalytic RNA. Science 298, 1421–1424, doi: 10.1126/science.1076093 (2002). [DOI] [PubMed] [Google Scholar]
- 51.Anderson M, Schultz EP, Martick M & Scott WG Active-site monovalent cations revealed in a 1.55-Å-resolution hammerhead ribozyme structure. J Mol Biol 425, 3790–3798, doi: 10.1016/j.jmb.2013.05.017 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.