Abstract
The main characteristic of biofilm formation is extracellular matrix (ECM) production. The cells within the biofilm are surrounded by ECM which provides structural integrity and protection. During an infection, this protection is mainly against cells of the immune system and antifungal drugs. A. fumigatus forms biofilms during static growth on a solid substratum and in chronic aspergillosis infections. It is important to understand how, and which, A. fumigatus signal transduction pathways are important for the adhesion and biofilm formation in a host during infection. Here we investigated the role of MAP kinases and protein phosphatases in biofilm formation. The loss of the MAP kinases MpkA, MpkC and SakA had an impact on the cell surface and the ECM during biofilm formation and reduced the adherence of A. fumigatus to polystyrene and fibronectin-coated plates. The phosphatase null mutants ΔsitA and ΔptcB, involved in regulation of MpkA and SakA phosphorylation, influenced cell wall carbohydrate exposure. Moreover, we characterized the A. fumigatus protein phosphatase PphA. The ΔpphA strain was more sensitive to cell wall-damaging agents, had increased β-(1,3)-glucan and reduced chitin, decreased conidia phagocytosis by Dictyostelium discoideum and reduced adhesion and biofilm formation. Finally, ΔpphA strain was avirulent in a murine model of invasive pulmonary aspergillosis and increased the released of tumor necrosis factor alpha (TNF-α) from bone marrow derived macrophages (BMDMs). These results show that MAP kinases and phosphatases play an important role in signaling pathways that regulate the composition of the cell wall, extracellular matrix production as well as adhesion and biofilm formation in A. fumigatus.
Keywords: Aspergillus fumigatus, Polysaccharide, Extracellular matrix, Biofilm, Mitogen activated protein kinases, Phosphatases
Introduction
Biofilms are composed by microorganisms attaching to each other, often associated with a surface, and typically encased within an extracellular matrix (ECM) complex polysaccharide. The ECM produces a structural scaffold for cohesion between cells, and to surfaces (Flemming and Wingender, 2010, O’Toole, 2003). In addition, the ECM can contribute to the retention of nutrients and water and also provide structural integrity and protection to the biofilm cells. In a human host this protection is mainly against cells of the immune system, and often has a role in resistance to antifungal therapies (Costerton et al., 1999, Donlan, 2001, Flemming and Wingender, 2010). The ECM of fungal biofilms is composed of different classes of macromolecules, such as proteins, carbohydrates, lipids and nucleic acids (Cegelski, 2015, Martins et al., 2010, Reichhardt et al., 2015, Zarnowski et al., 2014).
The fungal pathogen Candida albicans frequently forms biofilms on implanted medical devices and the environmental circumstances strongly affect ECM production (Hawser et al., 1998). C. albicans biofilm development involves a series of sequential events. In the adherence step, the yeast cells adhere to a substrate. Then, the cells proliferate to establish microcolonies and produce elongated projections that yield the filamentous forms. In the maturation step occurs the accumulation of the ECM, biofilm expansion and increased drug resistance (Finkel & Mitchell, 2011). Initial investigations identified proteins, hexosamine, carbohydrates, phosphorus, uronic acid and extracellular DNA (eDNA) as components of the C. albicans ECM (Al-Fattani and Douglas, 2006, Baillie and Douglas, 2000). Several groups have since performed comprehensive analysis of the composition and relative abundance of each C. albicans ECM component. Unexpectedly, the β-1,3 glucan, an earlier identified sugar with important role in the ECM, was much less abundant when compared to β-1,6 glucan and α-1,6 mannan (Al-Fattani and Douglas, 2006, Faria-Oliveira et al., 2014, Thomas et al., 2006, Zarnowski et al., 2014). However, increased susceptibility to fluconazole treatments, both in vivo and in vitro, was observed when biofilms were treated with β-1,3 glucanase. Further studies have shown a role for β-1,3 glucan in the resistance to other antimicrobial agents in C. tropicalis, C. parapsilosis, and C. glabrata (Fernandes et al., 2015, Mitchell et al., 2013, Nett et al., 2010, Yi et al., 2011). Although there are similar polysaccharides found in the ECM and cell wall of C. albicans, several studies suggest that synthesis of the ECM polysaccharides is regulated by pathways distinct of those involved in cell wall synthesis (Chaffin, 2008). In C. albicans, ECM synthesis and biofilm production are regulated by specific transcription factors that govern morphological changes, adhesion and ECM production. The two main transcription factors are Bcr1p, a regulator that positively affect the expression of many adhesins during biofilm formation (Nobile and Mitchell, 2005, Srikantha et al., 2013), and Zap1p, which negatively regulates the expression of genes required for β-1,3 glucan production (Nobile et al., 2009). Fks1p, a β-1,3 glucan synthase, is involved directly in production of β-1,3 glucan and this enzyme controls the level of this ECM polysaccharide (Nett et al., 2010, Taff et al., 2012). The deletion of SMI1 or RLM1, members of kinase C (PKC) pathway, resulted in lower levels of β-1,3 glucan in the ECM and in azole susceptibility (Nett et al., 2011). These phenotypes were repaired by FKS1 overexpression in the deletion mutants. However, no effect was observed when upstream components of the PKC pathway were deleted (Nett et al., 2011). These results clearly demonstrate that the PKC pathway regulates in a distinct manner the mechanisms involved in biofilm ECM production compared to general cell wall integrity.
Aspergillus fumigatus, the principal airborne fungal pathogen, grows as a typical biofilm with hyphae surrounded by an ECM (Beauvais et al., 2007, Beauvais et al., 2014, Loussert et al., 2010). A. fumigatus biofilms can be produced during chronic pulmonary infections such as asthma, cystic fibrosis, obstructive pulmonary disease and allergic bronchopulmonary aspergillosis (Taylor et al., 2014, Xu et al., 2013). On rare occasions, A. fumigatus biofilms are found on medical devices for healthcare (Loussert et al., 2010, Ramage et al., 2011). The initial studies of ECM components of A. fumigatus biofilms showed the presence of galactomannan, galactosaminogalactan (GAG), α-1,3 glucan, proteins, polyols and melanin. Furthermore, eDNA has been detected in biofilms produced by A. fumigatus. Immunolabeling analysis of the in vitro secreted proteins in A. fumigatus ECM showed the presence of two major antigens, dipeptidylpeptidase V (DPPV) and catalase B (CatB), and one allergen, AspF1 (Rajendran et al., 2013). Several studies have demonstrated that the presence of GAG in the A. fumigatus ECM is an important virulence factor that permits the fungus to avoid the host innate immune system (Fontaine et al., 2011, Gravelat et al., 2013, Gresnigt et al., 2014, Robinet et al., 2014). Microarrays and RNA sequencing (RNAseq) analysis of A. fumigatus biofilms showed transcriptional up-regulation of hydrophobins and some important genes involved in biosynthesis of secondary metabolites. As expected, cell surface proteins and some proteins involved in adhesion, such as glycophosphatidylinositol (GPI)- anchored cell wall were found to be up-regulated in A. fumigatus biofilms (Gibbons et al., 2012, Muszkieta et al., 2013). It has been reported that A. fumigatus biofilm production increased resistance to azoles, echinocandins and polyenes (Beauvais et al., 2007, Mowat et al., 2007, Mowat et al., 2008, Seidler et al., 2008). The mechanisms involved in the resistance of A. fumigatus biofilms to antifungal drugs include the increased expression of multidrug resistance (MDR) pumps (Muszkieta et al., 2013, Ramage et al., 2012), antioxidant defenses (Bruns et al., 2010, Delattin et al., 2014), Hsp90 (Robbins et al., 2011), and the presence of persister cells (Beauvais & Müller, 2009).
Despite these findings, the regulation of biofilm formation in A. fumigatus is relatively unknown. Some work showed that production of GAG is dependent on proteins that regulate development, including StuA (Gravelat, et al., 2013), MedA (Al Abdallah et al., 2012, Gravelat et al., 2010, Gravelat et al., 2013), SomA (Lin et al., 2015), and more recently PtaB, a lim-domain binding protein that regulates biofilm formation (Zhang et al., 2017). Recent studies by our group showed that SitA and PtcB phosphatases are important for ECM production and biofilm formation in A. fumigatus (Bom et al., 2015, Winkelströter et al., 2015a). Biofilms have the capacity to resist commonly used doses of antimicrobial agents, and this characteristic has become an important clinical issue. Thus, the characterization of proteins and pathways involved in biofilm production is essential to understand the genetic control of fungal biofilm development and the establishment of a biofilm in a host during A. fumigatus infection.
This work aimed to investigate the role of MAP kinases and protein phosphatases in biofilm formation. Here we presented evidence which support the role of Aspergillus fumigatus MAP kinases and phosphatases in controlling biofilm formation, ECM production, adhesion, cell wall composition as well as pathogenicity and virulence.
Material and methods
Ethics statement
The principles that guide our studies are based on the Declaration of Animal Rights ratified by the UNESCO in January 27, 1978 in its 8th and 14th articles. All protocols used in this study were approved by the local ethics committee for animal experiments from the Campus of Ribeirão Preto, Universidade de São Paulo (Permit Number: 08.1.1277.53.6; Studies on the interaction of Aspergillus fumigatus with animals). All animals were housed in groups of five within individually ventilated cages and were cared for in strict accordance with the principles outlined by the Brazilian College of Animal Experimentation (Princípios Éticos na Experimentação Animal – Colégio Brasileiro de Experimentação Animal, COBEA) and Guiding Principles for Research Involving Animals and Human Beings, American Physiological Society. All efforts were made to minimize suffering. Animals were clinically monitored at least twice daily and humanely sacrificed if moribund (defined by lethargy, dyspnoea, hypothermia and weight loss). All stressed animals were sacrificed by cervical dislocation.
Strains and media
The A. fumigatus strains used in this study are described in Table 1. Media were of two basic types. A complete medium with three variants: (2% (w/v) glucose, 0.5% (w/v) yeast extract, 2% (w/v) agar, trace elements), YUU (YAG supplemented with 1.2 g/L each of uracil and uridine) and liquid YG or YUU medium of the same composition (but without agar). A modified minimal medium (MM: 1% (w/v) glucose, original high nitrate salts, trace elements, 2% (w/v) agar, pH 6.5) was also applied. Trace elements, vitamins, and nitrate salts were described by (Käfer, 1977).
Table 1.
Strains used in this work.
| Strain | Genotype | Source |
|---|---|---|
| Afs35 | Wild-type strain | FGSC A1159 |
| CEA17 | Wild-type strain | Our laboratory |
| Ku80 | Δku80:: pyrGAF | FGSCA1151 |
| Ku80pyrG | Δku80:: pyrGAF pyrG- | Our laboratory |
| ΔmpkC | ΔmpkC::prtA | Hagiwara et al. (2014) |
| ΔsakA | ΔsakA::hph | Hagiwara et al. (2014) |
| ΔmpkC ΔsakA | ΔmpkC::prtA ΔsakA::hph | Bruder Nascimento et al. (2016) |
| ΔmpkC::mpkC+ | ΔmpkC::mpkC+::prtA | Bruder Nascimento et al. (2016) |
| ΔsakA::sakA+ | ΔsakA::sakA+::prtA | Bruder Nascimento et al. (2016) |
| ΔmpkA | ΔmpkA::hph | Valiante et al. (2009) |
| ΔmpkA::mpkA+ | ΔmpkA:: sakA+::prtA | Valiante et al. (2009) |
| ΔsitA | ΔsitA::pyrG+ | Bom et al., 2015 |
| ΔsitA::sitA+ | ΔsitA::sitA+::hph | Bom et al., 2015 |
| ΔptcB | ΔptcB::pyrG+ | Winkelströter et al. (2015a) |
| ΔptcB::ptcB+ | ΔptcB::ptcB+::hph | Winkelströter et al. (2015a) |
| ΔpphA | ΔpphA::pyrG+ | Winkelströter et al. (2015b) |
| ΔpphA::pphA+ | ΔpphA::pphA+::hph | Winkelströter et al. (2015b) |
Phenotypic assays
The phenotypes of the deletion mutants were evaluated by radial growth at different temperatures, in the presence or absence of stressing agents. Five µl of a concentration of 2 × 107 for the wild-type and mutant strains were spotted on different growth media and grown for 96 h at 37 °C.
The adherence to fibronectin was assessed according to Gravelat et al. (2010) with modifications. Briefly, wells from a sterile 96-well flat-bottom polystyrene plate were previously coated with 250 µL of fibronectin 0.01 mg/mL (Sigma–Aldrich) diluted in PBS. The plates were incubated at 37 °C for 16 h and washed twice with PBS. Further, 1 x 104 conidia were inoculated into 200 µL of liquid MM in each well, incubated for additional 24 h at 37 °C and washed 3 times with sterile PBS (NaCl, 137 mM; KCl, 2.7 mM; Na2HPO4, 10 mM; and KH2PO4, 1.8 mM). The adherent cells were stained with 200 µL of 0.5% crystal violet solution for 5 min at room temperature, exhaustively washed with PBS and air dried. Finally, the crystal violet was eluted from the wells using 200 µL of 100% ethanol, and the absorbance was measured at 590 nm. The capacity of adhesion to plastic surface was performed similarly to described above except by the fact the wells were not covered with fibronectin (Mowat et al., 2007). All the experiments were performed at least three times.
Staining for dectin-1, chitin, and other carbohydrates
This procedure was performed as described by Graham et al., 2006, Winkelströter et al., 2015a, Winkelströter et al., 2015b. Briefly, A. fumigatus conidia (2 × 103) were grown in 200 µL of MM for 16 h at 37 °C. The culture medium was removed and the germlings were UV-irradiated (600,000 µJ). The germlings were washed with PBS and 200 µL blocking solution (goat serum 2% (w/v), BSA 1% (w/v), 0.1% (v/v) Triton X-100, 0.05% (v/v) Tween 20, 0.05% (v/v) sodium azide and 0.01 M PBS) were added and incubated for 30 min at room temperature. For dectin staining, 0.2 µg/mL of Fc-h-dectin-hFc were added and incubated for 1 h at room temperature, followed by adding 1:1000 DyLight 594-conjugated, goat anti-human IgG1 and incubated for 1 h at room temperature (Graham et al., 2006). Germlings were washed with PBS and fluorescence was read at 587 nm excitation and 615 nm emission. For chitin staining, 200 µL of a PBS solution with 10 µg/mL of CFW were added to UV-irradiated germlings, incubated for 5 min at room temperature, washed three times with PBS and fluorescence was read at 380 nm excitation and 450 nm emission. For GAG, GlcN, and mannose staining, 200 µL of PBS with 0.1 mg/ml of SBA-FITC (Glycine max Soybean lectin SBA-FITC bioworld cat# 21761024-2), WGA (Lectin-FITC L4895, Sigma) or ConA (Concanavalin A C7642, Sigma), respectively, were added to UV-irradiated germlings. The germlings were incubated for 1 h at room temperature, washed with PBS, and fluorescence was read at 492 nm excitation and 517 nm emission. All the experiments were performed with 12 repetitions and fluorescence was read in a microtiter plate reader SpectraMax i3 (Molecular Devices).
SYTO9 fluorescence assay
Adhesion of A. fumigatus conidia to polystyrene surface was assessed by SYTO9 fluorescence assay (Honraet et al., 2005). Briefly, A. fumigatus conidia (1 × 106) were grown in RPMI media in black 96 well polystyrene plate for 90 min at 37 °C. The culture medium was removed and any non-adherent cells were washed off with distilled water. Subsequently, adherent cells were stained with 100 μL of SYTO 9 dye (10 μM) for 15 min at 37 °C. Fluorescence was then measured using a microtiter plate reader (FLUOstar omega, BMG Labtech) at an excitation and emission wavelength of 485 nm and 505 nm, respectively.
Scanning electron microscopy (SEM)
Standardized conidia of ΔpphA and its parental strain were inoculated in RPMI medium onto Thermanox coverslips (13 mm) within a 24-well tissue culture plate. After 24 h incubation at 37 °C, biofilms were processed and imaged as previously described (Erlandsen et al., 2004a, Erlandsen et al., 2004b). Briefly, the biofilms were washed in PBS and fixed in 2% paraformaldehyde, 2% glutaraldehyde, and 0.15% (w/v) alcian blue in 0.15 M sodium cacodylate (pH 7.4). The biofilms were sputter coated with gold and viewed under a JEOL JSM-6400 scanning electron microscope in high vacuum mode at 10 kV.
Transmission electron microscopy (TEM)
Preparation of samples was as previously described (Walker et al., 2008) with the following modifications. Briefly, cells were collected and the pellets were fixed with 2.5% (v/v) glutaraldehyde in 0.1 M sodium phosphate buffer (pH 7.3) for 24 h at 4 °C. Samples were encapsulated in 3% (w/v) low melting point agarose prior to processing to Spurr resin following a 24 h schedule on a Lynx tissue processor (secondary 1% OsO4 fixation, 1% Uranyl acetate contrasting, ethanol dehydration and infiltration with acetone/Spurr resin). Additional infiltration was provided under vacuum at 60 °C before embedding in TAAB capsules and polymerising at 60 °C for 48 h. Semi-thin (0.5 µm) survey sections were stained with toluidine blue to identify areas of best cell density. Ultrathin sections (60 nm) were prepared using a Diatome diamond knife on a Leica UC6 ultramicrotome, and stained with uranyl acetate and lead citrate for examination with a JEM-1400Plus transmission electron microscope (JEOL (U.K.) LTD, Hertfordshire, UK) and imaging with an AMT UltraVUE camera and AMT Image Capture Engine V602 software (Deben UK Limited, Suffolk, UK).
Dictyostelium discoideum phagocytosis assay
D. discoideum was grown in Petri dishes for 16 h at 22 °C in HL5 (axenic medium) supplemented with streptomycin sulfate (300 μg/mL). Subsequently, the plates were washed and the supernatant was centrifuged for 5 min at 1500 rpm, the supernatant was discarded, and the pelleted cells were resuspended in 3 mL of HL5 medium. The density of D. discoideum cells was determined by counting the cells in a hemocytometer as described at http://dictybase.org/. As described by Hillmann et al. (2015), 10 conidia per cell of D. discoideum (different multiplicities of infection, MOIs) were used for the co-incubation. D. discoideum (105) cells and A. fumigatus conidia (106) were added to a six-well plate and the plate was incubated at 22 °C for 5 h, after which time the 50 D. discoideum cells were counted and the number of phagocytosed conidia evaluated.
Murine model of pulmonary aspergillosis
The murine model of pulmonary aspergillosis was performed according to Dinamarco et al. (2012). Outbred female mice (BALB/c strain; body weight, 20–22 g) were housed in vented cages containing 5 animals. Mice were immunosuppressed with cyclophosphamide at a concentration of 150 mg per kg of body weight, which was administered intraperitoneally on days −4, −1, and 2 prior to and post infection (day 0). Hydrocortisone acetate (200 mg/kg) was injected subcutaneously on day −3. The A. fumigatus conidia used for inoculation were grown on Aspergillus complete YAG for 2 days prior to infection. Fresh conidia were harvested in PBS and filtered through a Miracloth (Calbiochem). Conidial suspensions were spun for 5 min at 3000×g, washed three times with PBS, counted using a hemocytometer, then resuspended at a concentration of 5.0 × 106 conidia/mL. Viability counts for the administered inoculum were determined, following serial dilution, and plating on Aspergillus YGA, and the conidia were grown at 37 °C. Mice were anesthetized by halothane inhalation and infected by intranasal instillation of 1.0 × 105 conidia in 20 μL of PBS. As a negative control, a group of 5 mice received PBS only. Mice were weighed every 24 h from the day of infection and visually inspected twice daily. In the majority of cases, the endpoint for survival experimentation was identified when a 20% reduction in body weight was recorded, at which time the mice were sacrificed. The statistical significance of comparative survival values was calculated using log rank analysis using the Prism statistical analysis package. Additionally, at 3 days post infection, 2 mice per strain were sacrificed and the lungs were removed, fixed, and processed for histological analysis.
Lung histopathology and fungal burden
After sacrifice, the mice lungs were removed and fixed for 24 h in 3.7% (v/v) formaldehyde–PBS. Samples were washed several times in 70% (v/v) alcohol before dehydration in a series of alcohol solutions of increasing concentrations. Finally, the samples were diafanized in xylol and embedded in paraffin. For each sample, sequential 5-µm-thick sections were collected on glass slides and stained with Gomori methenamine silver (GMS) or hematoxylin and eosin (HE) stain following standard protocols (Greenberger, 2002). Briefly, sections were deparaffinized, oxidized with 4% chromic acid, stained with methenamine silver solution, and counterstained with hematoxylin. Tissue sections were also stained with hematoxylin and eosin for histological examination to determine lung damage. All stained slides were immediately washed, preserved in mounting medium, and sealed with a coverslip. Microscopic analyses were performed using an Axioplan 2 imaging microscope (Carl Zeiss) at the stated magnifications under bright-field conditions.
To investigate fungal burden in the lungs, mice were infected as described previously, but with a higher inoculum of 1 × 106 conidia/20 µL. A higher inoculum, in comparison to the survival experiments, was used to increase fungal DNA detection. Animals were sacrificed 72 h post infection, and both lungs were harvested and immediately frozen in liquid nitrogen. Samples were homogenized by vortexing with glass beads for 10 min, and DNA was extracted via the phenol–chloroform method. DNA quantity and quality were assessed using a NanoDrop 2000 spectrophotometer (Thermo Scientific). At least 500 ng of total DNA from each sample was used for quantitative real-time PCR. A primer and a Lux probe (Invitrogen) were used to amplify the 18S rRNA region of A. fumigatus and an intronic region of mouse GAPDH (glyceraldehyde-3-phosphate dehydrogenase). Six-point standard curves were calculated using serial dilutions of gDNA from all the A. fumigatus strains used and the uninfected mouse lung. Fungal and mouse DNA quantities were obtained from the threshold cycle (CT) values from an appropriate standard curve. Fungal burden was determined as the ratio between picograms of fungal and micrograms of mouse DNA.
Determination of TNF-α levels
For cytokine determination, bone marrow derived macrophages (BMDMs) from C57BL/6 mice were prepared as previously described (Marim et al., 2010). Briefly, bone marrow cells from femurs of adult mice were cultured for 6 days in RPMI 1640, containing 20% (v/v) fetal bovine serum (FBS) and 30% (v/v) L-929 cell conditioned media (LCCM). Macrophages (5.0 × 105) were plated in 48-well plates for 16 h at 37 °C, 5% (v/v) CO2 in RPMI 140 media containing 10% (v/v) FBS and 5% (v/v) of LCCM. For fungal infection, strains were cultured for 18 h up to a hyphal stage at a density of 2 × 104 per well, UV-irradiated and used to stimulate the macrophages. The cells were centrifuged to synchronize the infection and allowed to infect for 18 h. The supernatant was collected and the cytokine was measured by enzyme-linked immunosorbent assay (ELISA) with a mouse TNF-α kit (R&D Quantikine ELISA) according to the manufacturer's instructions.
Statistical analysis
All data, except the survival curves, were analyzed using t-test (Prism, GraphPad) with significance levels of ∗∗p < 0.01 and ∗∗∗p < 0.001.The statistical significance of comparative survival values was calculated using log rank analysis using the Prism statistical analysis package.
Results
MAPKs are important for biofilm formation
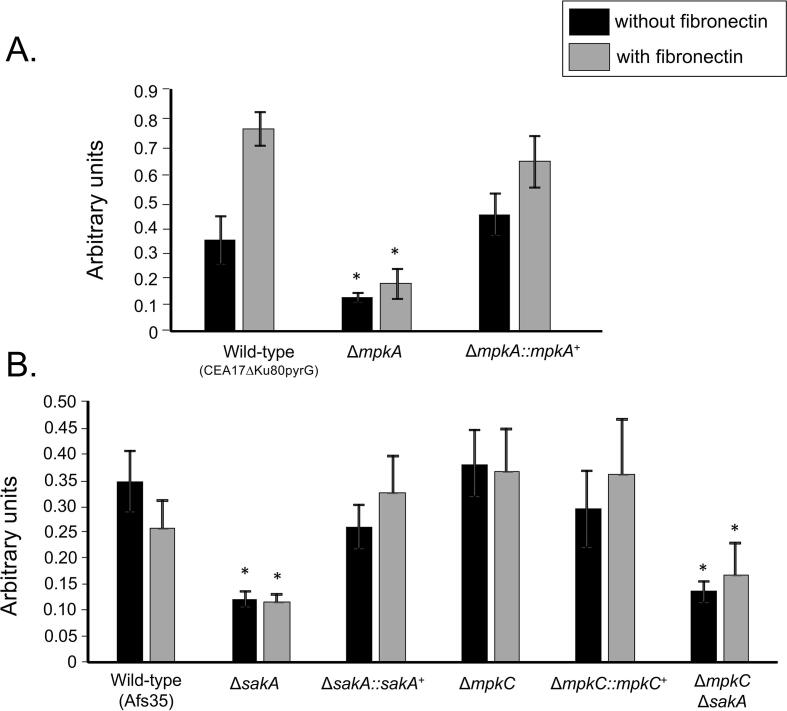
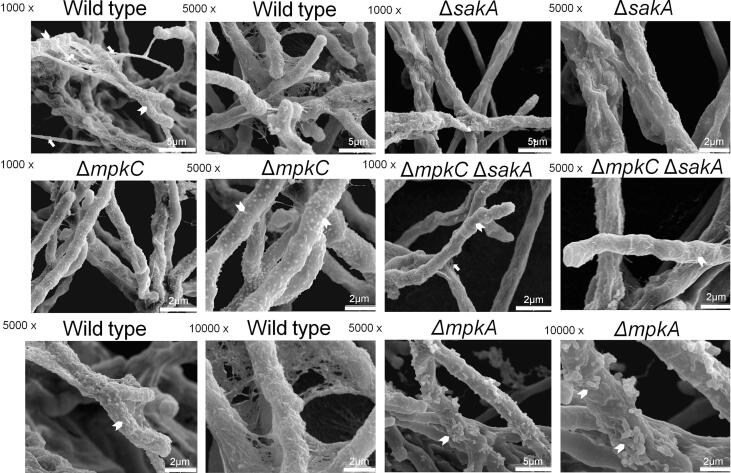
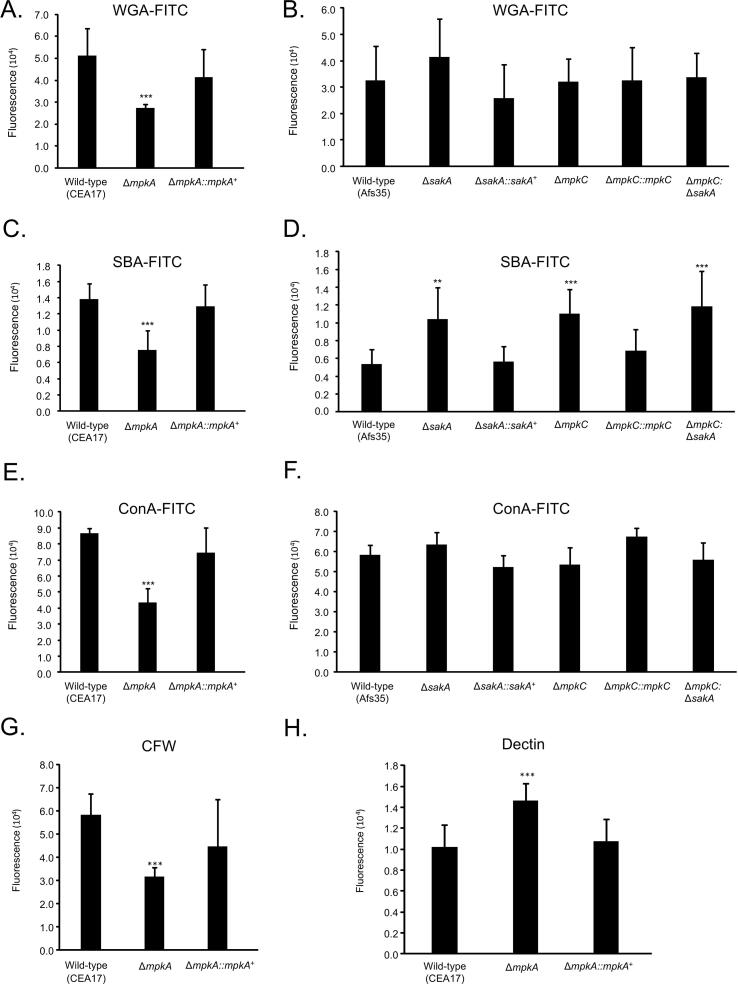
Initially, we assessed the adhesive capacity of the respective mutants, which is the initial step in the foundation of biofilm formation, to assess the impact of loss of the MAP kinases on the adherence to plastic or fibronectin. Germlings of the ΔmpkA, ΔsakA, and ΔmpkC ΔsakA mutants displayed reduced adherence to polystyrene plates ranging from 70 to 75% when compared to the wild type. The adherence to fibronectin-coated plates was reduced ranging from 50 to 60% when comparing the mutants ΔmpkA, ΔsakA, and ΔmpkC ΔsakA to the wild type strain (Fig. 1A and B). Scanning electron microscopy (SEM) of the mycelia revealed that ΔsakA, ΔmpkC, ΔmpkC ΔsakA, and ΔmpkA influenced to different extents the cell surface and the ECM during biofilm formation (Fig. 2). The surface of the MAP kinase null mutants appeared to have reduced ECM, which was in stark contrast to the wild-type strain (Fig. 2). This decreased adhesion and ECM formation could be due to alterations in the composition of the carbohydrates present in the cell wall. Thus, cell wall stains and lectins were used to identify differences in the content or exposure of different carbohydrates on the surface of the fungal cell wall, in both the wild-type and MAP kinase mutants. These included: (i) SBA (Soybean Agglutinin)-FITC (preferentially binds to oligosaccharide structures with terminal α- or β-linked N-acetylgalactosamine (GalNAc), and to a lesser extent, galactose residues – important for recognizing galactosaminogalactan, GAG), (ii) WGA (Wheat Germ Agglutinin)-FITC [recognizing surface exposed N-acetyl glucosamine (GlcNAc)], (iii) ConA (Concanavalin A)-FITC (recognizes α-linked mannose), (iv) soluble dectin-1 staining (recognizing β-glucans), and (v) CFW (recognizing chitin). When the mutants were compared with the wild-type and the corresponding complemented strains (Fig. 3A–H), for ΔmpkA we observed 50% less N-acetyl glucosamine (Fig. 3A), 50% less GAG (Fig. 3C), 50% less mannose (Fig. 3E), 40% less chitin (Fig. 3G), and 40% more β-glucan (Fig. 3H). In contrast there was 50 % more GAG in the ΔmpkC, ΔsakA, ΔmpkC ΔsakA mutants (Fig. 3D). Previously, we have shown that the intensity of CFW staining per fungal area was 30% higher in ΔmpkC mutant than wild-type and complemented strains, while ΔsakA and ΔmpkC ΔsakA were 20% lower than wild-type and complemented strains (Bruder Nascimento et al., 2016). Moreover, the ΔmpkC, ΔsakA and ΔmpkC ΔsakA mutant strains were shown to have more abundant β-glucans than the wild-type and complemented strains. The intensity of dectin-1 staining per fungal area was 50, 40, and 30% higher in the ΔmpkC, ΔsakA and ΔmpkC ΔsakA mutants, respectively compared with wild-type strain (Bruder Nascimento et al., 2016).
Fig. 1.
The adhesion of A. fumigatus wild-type and MAP kinase mutants. (A and B) Fungal adhesion to plastic or fibronectin was measured by crystal violet (CV) assay. The results are the average of three experiments ± standard deviation (* = p < 0.001 by t-tests).
Fig. 2.
Scanning Electron Microscopy for the wild–type and MAP kinase null mutant strains. In the upper, middle, and lower panels 1000, 5000, and 10,000 times magnification were used respectively. The arrow heads indicate the hypha surface.
Fig. 3.
Carbohydrate content and exposure on the surface of the wild-type and MAP kinase mutants. (A and B) WGA-FITC staining, (C and D) SBA-FITC staining, (E and F) ConA-FITC staining, (G) CFW staining, and (H) Dectin staining. The results are the average of twelve repetitions ± standard deviation (*** = p < 0.001 and ** = p < 0.01 by t-tests).
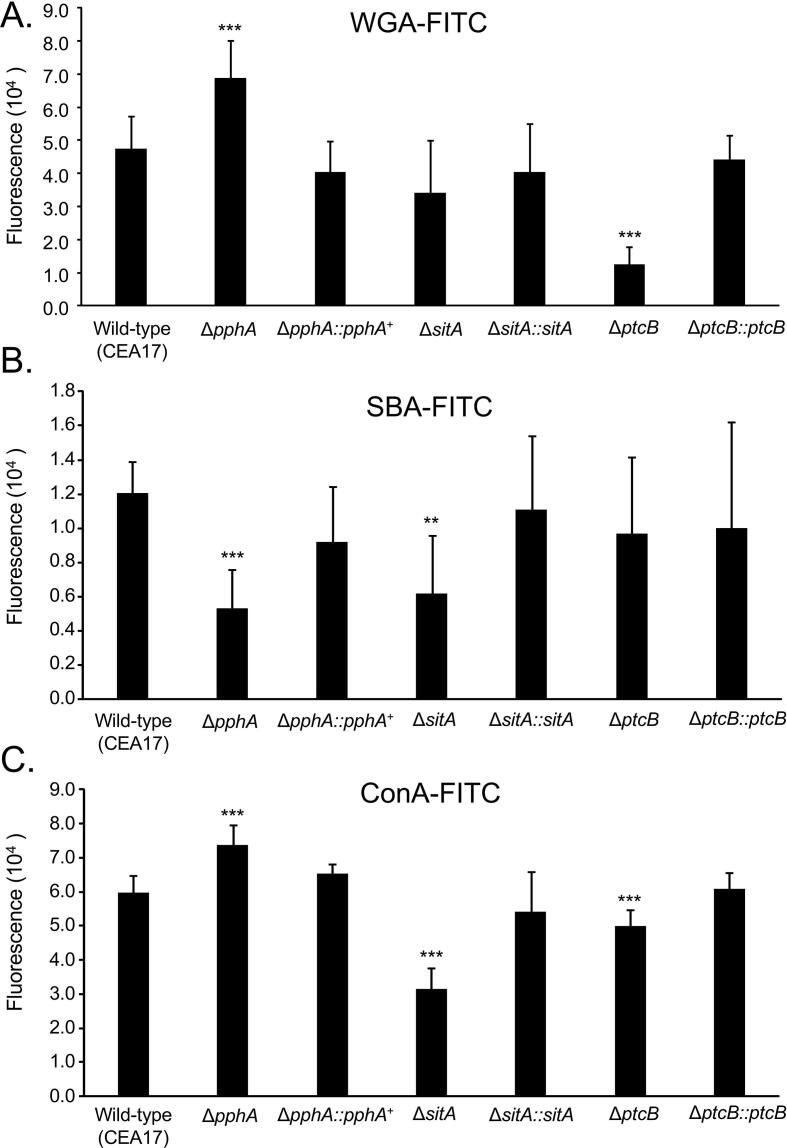
Next step, we characterized the distribution of WGA-FITC, SBA-FITC, and ConA-FITC binding (Fig. 4A–C) to phosphatase null mutants previously shown to be involved in modulating MpkA or SakA phosphorylation, ΔsitA, ΔptcB, and ΔpphA (Winkelströter et al., 2015a, Winkelströter et al., 2015b, Bom et al., 2015). A number of differences were observed when the mutants were compared with the wild-type and the corresponding complemented strains (Fig. 4A–H). We have observed 40% more WGA-FITC binding to ΔpphA and fourfold less to ΔptcB (Fig. 4A), 50–60% less SBA-FITC binding to ΔpphA and ΔsitA (Fig. 4B). In addition there was 20% more mannose in ΔpphA and 50% and 10% less mannose in ΔsitA and ΔptcB, respectively (Fig. 4C). Again, we have previously shown the intensity of dectin-1 and CFW staining per fungal area was twofold and 25% higher in the ΔptcB mutant compared to wild–type strain (Winkelströter et al., 2015a). The intensity of dectin-1 and CFW staining per fungal area was 40% and 20% higher in ΔsitA mutant than wild–type strain (Bom et al., 2015). All these modifications in the cell wall increase the MAP kinase, ΔptcB, and ΔsitA mutants conidial phagocytosis by Dictyostelium discoideum in about two to threefold when compared to the wild-type and complemented strains (Fig. 5A–F).
Fig. 4.
Carbohydrate content and exposure on the surface of the wild-type and phosphatase mutants. (A) WGA-FITC staining, (B) SBA-FITC staining, and (C) ConA-FITC staining. The results are the average of twelve repetitions ± standard deviation (*** = p < 0.001 and ** = p < 0.01 by t-tests).
Fig. 5.
Phagocytosis of MAP kinase and phosphatase null mutants by Dictyostelium discoideum. (A–F) Conidia from the wild-type and MAP kinase and phosphatase null mutants were exposed to D. discoideum and the total number of phagocytosed conidia evaluated. The results are the average of three repetitions with ±standard deviation. The asterisks indicate p < 0.001.
Collectively, our results demonstrate that MAP kinases and phosphatases that affect MpkA and SakA phosphorylation, influence the carbohydrate content and exposure at the cell wall, thus impacting adhesion, biofilm formation and phagocytosis by Dictyostelium.
PphA is a novel phosphatase that affects the CWI pathway
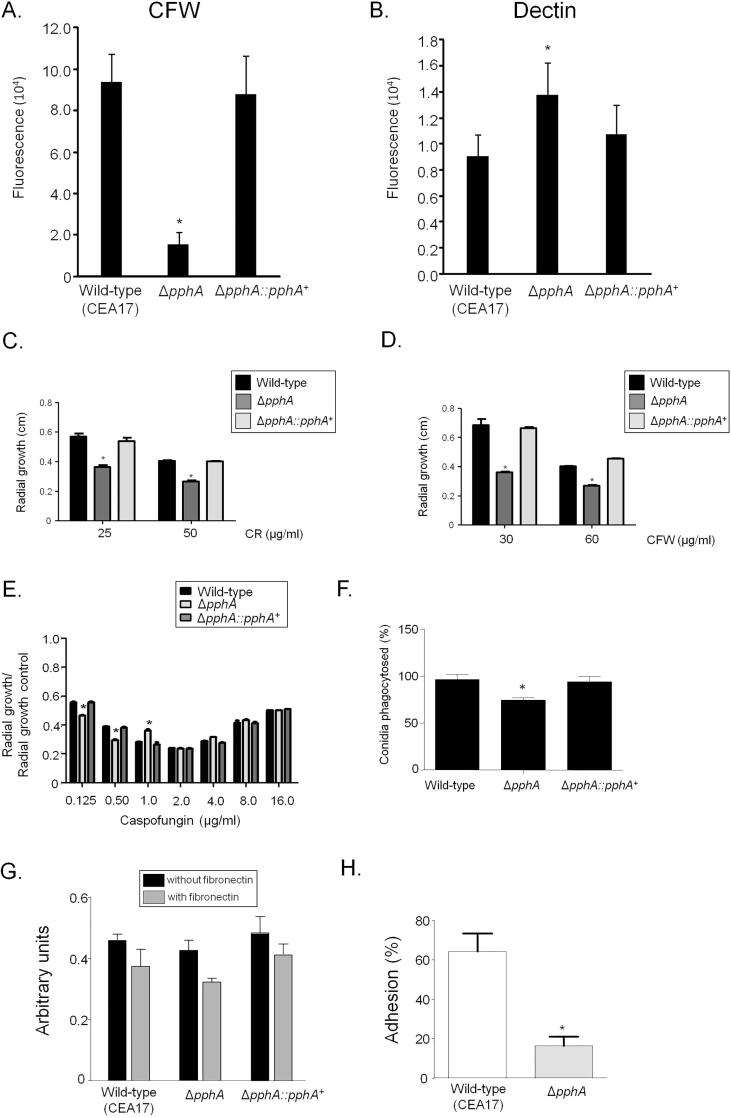
We decided to investigate in more detail the ΔpphA phosphatase mutant. The ΔpphA mutant has less chitin and more β-1-3 glucan than the wild-type and complemented strains. The intensity of CFW and dectin-1 staining were 80% lower and 30% higher in the ΔpphA mutant, respectively compared with wild-type strain (Fig. 6A and B). This mutant is more sensitive to Congo Red (CR) and CFW than the wild-type and complemented strains (Fig. 6C and D). It shows sensitivity to lower caspofungin concentrations such as 0.125 and 0.5 µg/ml, but it preserved the caspofungin paradoxical effect or trailing, (i.e. the escape of A. fumigatus from caspofungin inhibition at concentrations above the minimal inhibitory concentration; Chen et al., 2011; Fig. 6E). Possible modifications in the cell wall decrease ΔpphA conidia phagocytosis by about 30% when compared to the wild-type and complemented strains (Fig. 6F). Germlings of ΔpphA showed comparable % adherence to polystyrene plates and fibronectin-coated plates (Fig. 6G). Furthermore, a sensitive SYTO 9 fluorescence based assay showed a significant (p < 0.05) reduction of about 4-fold in adherence with pphA compared to the wild type strain (Fig. 6H).
Fig. 6.
The PphA null mutant has impaired cell wall integrity. (A and B) The A. fumigatus ΔpphA mutant has decreased chitin and increased β-1,3-glucan. The results are the average of twelve repetitions ± standard deviation (* = p < 0.001 by t-tests). (C–E) The wild-type, ΔpphA, and ΔpphA::pphA+ strains were grown for 120 h at 37 °C in MM in the presence or absence of different concentrations of Congo Red (CR), Calcofluor White (CFW) and Caspofungin. These experiments were performed by measuring radial diameter in triplicate, and the results are displayed as mean values with standard errors (* = p < 0.001 by t-tests). (F) Phagocytosis of A. fumigatus conidia by Dictyostelium discoideum. A ratio of 106 amoeba cells to 107 conidia was used in each experiment and they were incubated for 5 h at 22 °C. Fifty amoeba cells were evaluated for phagocytosis. The results are the average of three independent experiments ± standard deviation. (G) The adhesion of A. fumigatus wild-type, ΔpphA, and ΔpphA::pphA+ mutants. Fungal adhesion as measured by crystal violet (CV) assay on the adherence to plastic or fibronectin was examined. (H) SYTO9 adhesion assay with the wild-type and ΔpphA mutant strains. The results are displayed as mean values with standard errors (* = p < 0.001 by t-tests).
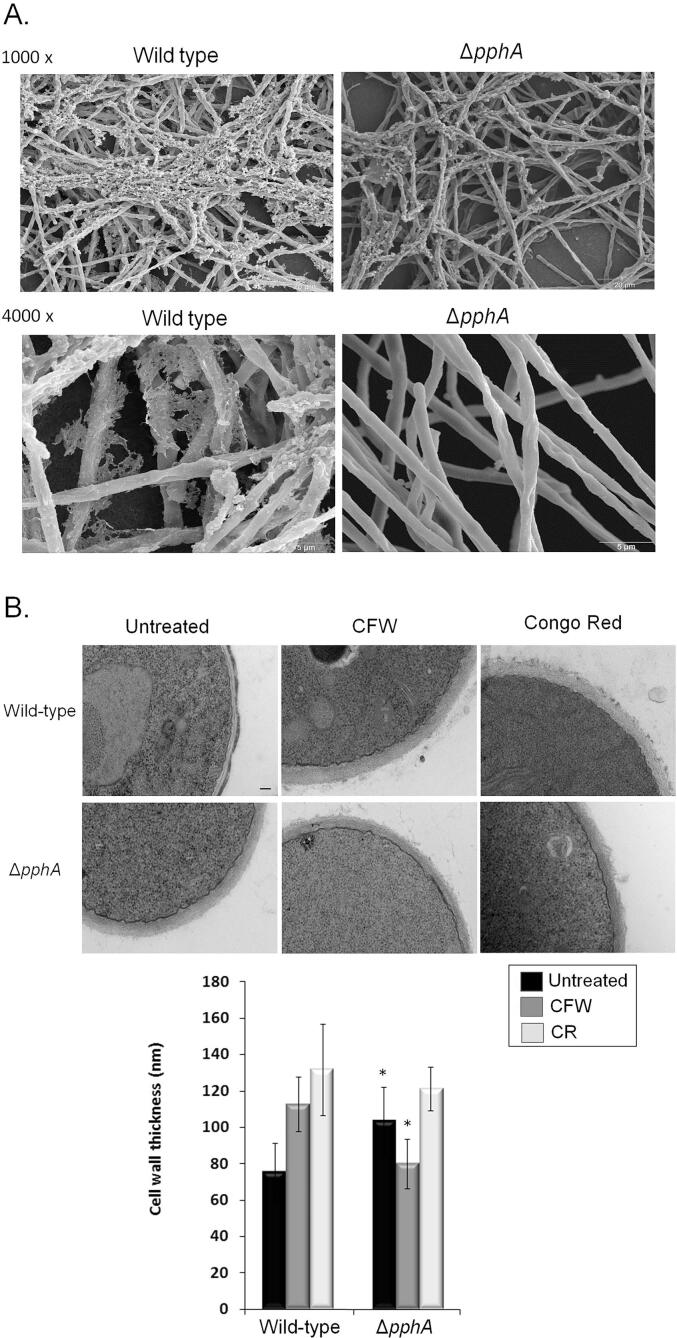
Additional evidence for a role of PphA in biofilm formation and in the organization of the cell wall was provided by SEM and TEM (Transmission Electron Microscopy) analyses (Fig. 7A). SEM of the mycelia revealed that PphA influenced the cell surface and the ECM during biofilm formation (Fig. 7A). The surface of the ΔpphA appeared smooth, which was in stark contrast to the wild-type strain (Fig. 7A). TEM analysis showed that untreated ΔpphA germlings grown in MM have about 25% thicker cell walls compared to the wild-type (Fig. 7B). Further exposure of the ΔpphA germlings to CFW decreased the cell wall thickness about 30% (Fig. 7B). In contrast, when wild-type strain was exposed to CFW and CR, it had about 50% increased cell wall thickness (Fig. 7B).
Fig. 7.
PphA plays a role in biofilm formation and in the organization of the cell wall as demonstrated by SEM and TEM analyses. (A) SEM for the wild–type and ΔpphA mutant strains. The magnification is 1000 and 4000 times. (B) TEM for A. fumigatus wild-type and ΔpphA mutant strains. Germlings were grown in the absence or presence of CFW (Calcofluor White) or Congo Red. TEM analysis (bars, 100 nm). Cell wall thickness (µm) of fifty sections of different germlings were measured. The results are expressed as average ± standard deviation (* = p < 0.001 by t-tests).
We also investigated if PphA was involved in the MpkA and/or SakA pathways in A. fumigatus, by determining the phosphorylation state of MpkA and SakA in the presence and absence of CR and sorbitol stresses. In both stressing conditions, the phosphorylation levels of MpkA and SakA were similar in both the wild-type and ΔpphA mutant strains (Supplementary Fig. S1). Calphostin C, chelerythrine, and cercosporamide have been used as protein kinase C inhibitors in mammals and in fungi (Arpaia et al., 1999, da Rocha et al., 2002, Herbert et al., 1990, Jarvis and Grant, 1999, Juvvadi et al., 2007, Sussman et al., 2004). We hypothesize if ΔpphA is modulating the protein kinase C activity, the susceptibility status of ΔpphA to these inhibitors would be altered. The ΔpphA and wild-type growth was similarly inhibited by calphostin C, cercosporamide, and chelerythrine (Supplementary Fig. S2).
Taken together these results strongly indicate that PphA is important for the A. fumigatus cell wall integrity pathway. However, PphA is not modulating either MpkA, SakA or Protein kinase C activity.
PphA is important for A. fumigatus virulence
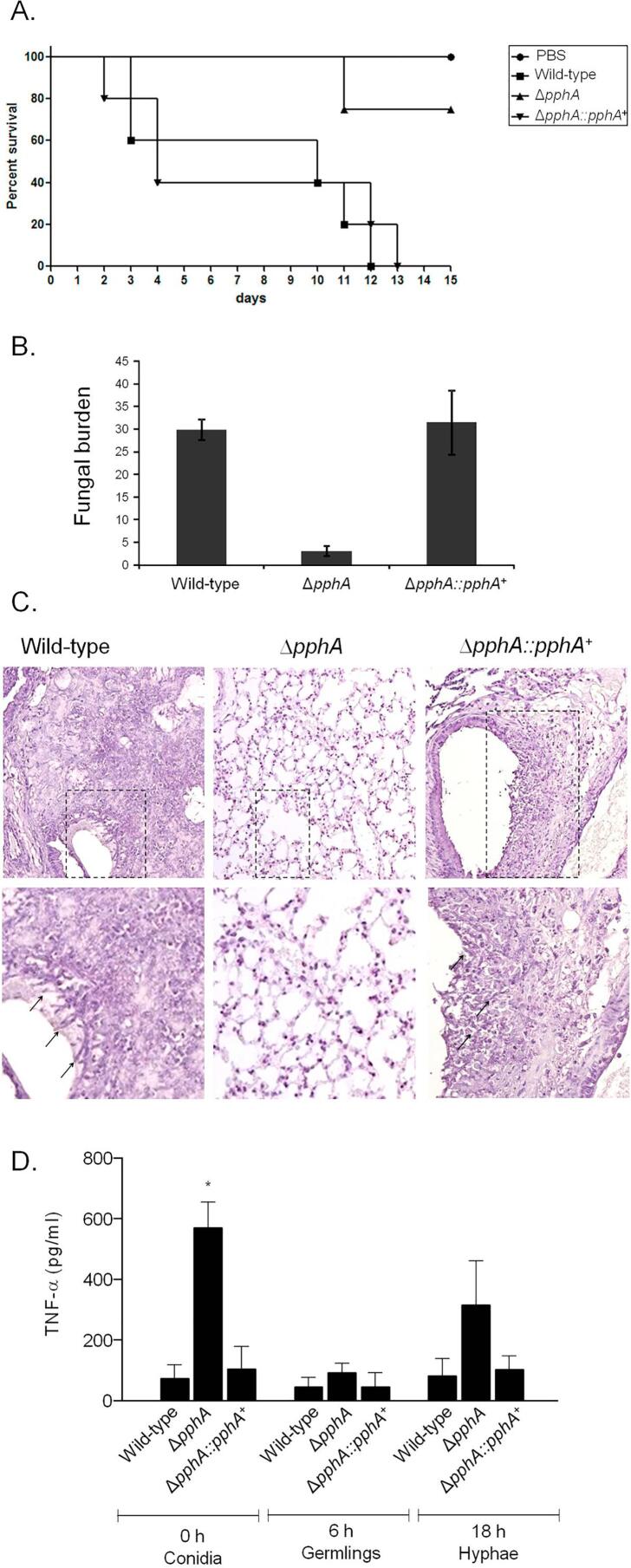
The importance of PphA for A. fumigatus pathogenicity was evaluated in a neutropenic murine model of invasive pulmonary aspergillosis (Fig. 8A). There are no statistical differences among the wild-type and ΔpphA::pphA+ strains (using two different statistical tests, Log–rank Mantel–Cox and Gehan–Breslow–Wilcoxon). Wild-type infection resulted in 100% mortality 12 days post-infection, while ΔpphA::pphA+ infection resulted in a significant mortality rate of 100% after 13 days post-infection (Fig. 8A). The ΔpphA infection resulted in the greatest reduction in mortality rate, with only 25% of mice dying after 15 days post-infection (Fig. 8A, p < 0.0025 and p < 0.0041 for the comparison between the wild-type and the ΔpphA mutant, Log–rank Mantel–Cox and Gehan–Breslow–Wilcoxon tests, respectively).
Fig. 8.
A. fumigatus PphA contributes to virulence in neutropenic mice. (A) Comparative analysis of wild-type and mutant strains in a neutropenic murine model of pulmonary aspergillosis. Mice in groups of 10 per strain were infected intranasally with a 20 μl suspension of conidiospores at a dose of 105. (B) Fungal burden results, determined by qPCR 72 h post-infection, were expressed based on 18S rRNA gene of A. fumigatus divided by the results of an intronic region of the mouse GAPDH gene. (C) Histological analysis of infection murine lung was performed 72 h after infection. Lower panels show increased magnification of the lined area from upper figure. Arrows indicate germlings. (D) TNF–α secretion from bone marrow-derived macrophages (BMDMs). The ΔpphA conidia triggered significantly increased release of TNF–α from BMDMs compared to wild–type and the reconstituted strain at 0 h time point. BMDMs from C57BL/6 mice were infected with A. fumigatus conidia, germlings, or hyphae for up to 18 h and the supernatant of cells was collected to measure the TNF-α levels by ELISA. Data show average ± standard deviation and *p ≤ 0.005 compared to the wild–type and the complemented strain.
Fungal burden was measured by qPCR, ΔpphA had only 15% of the fungal burden of the wild type and complemented strains (Fig. 8B). Histopathological examination revealed that at 72 h post-infection the lungs of mice infected with the wild-type or complemented strains contained multiple foci of invasive hyphal growth; the majority of the disease seems peri-bronchiole with some extension into the lung parenchyma (Fig. 8C). In contrast, ΔpphA infections revealed inflammatory infiltrates in bronchioles, with some containing poorly germinated or non-germinated conidia (Fig. 8C). Taken together, these results clearly demonstrate that PphA plays an important role in A. fumigatus virulence.
The impaired ΔpphA CWI together with the dramatic attenuation in virulence could contribute to an altered immune response. Subsequently, the cytokine Tumour Necrosis Factor alpha (TNF–α) levels released from bone marrow derived macrophages (BMDMs) after co-incubation with A. fumigatus conidia, germlings, and hyphae were investigated. TNF–α is an important inflammatory mediator secreted by macrophages when exposed to A. fumigatus (Hayashi et al., 2005, Taramelli et al., 1996) and it has shown to be an important signal in the initiation and maintenance of innate immunity in different animal models of pneumonia (Gosselin et al., 1995, Laichalk et al., 1996). The innate immunity represents the principal pathway by which A. fumigatus is cleared from the lung. This innate response consists specially by macrophages, which represent the first line of defense against conidia entering the alveolus, and recruited neutrophils (Schaffner et al., 1982). Bone Marrow Derived Macrophages (BMDMs) co-cultured with wild-type, ΔpphA ΔpphA::pphA+ showed a comparable TNF–α production when exposed to germlings and hypha (Fig. 8D). However, the ΔpphA conidia showed about 7-fold TNF-α induction than conidia from the wild–type or complemented strains (Fig. 8D). These results suggest that the effect caused by the absence of PphA on conidia is important for macrophage recognition and inducing inflammatory responses.
Discussion
A. fumigatus represents the most common airborne human pathogenic fungus in the world. Humans inhale hundreds of A. fumigatus asexual conidia daily and they cause a number of diseases, ranging from simple allergies to lethal invasive aspergillosis (IA) in immunocompromised patients (Latgé & Steinbach, 2009). Although A. fumigatus virulence has been investigated in many studies in recent years (Carberry et al., 2012, Chotirmall et al., 2014, Ding et al., 2014, Grahl et al., 2012, Haas, 2014, Hartmann et al., 2011, Moore, 2013, Scharf et al., 2012, Schrettl and Haas, 2011, Wezensky and Cramer, 2011), the signal transduction pathways that are activated during the different clinical forms of the disease are still unclear. Recently, aiming to investigate the signal transduction pathways that make A. fumigatus a successful pathogen, we have characterized the role of Mitogen-Activated Protein (MAP)-kinases and phosphatases in the adaptive response of this fungus to different types of stresses (Bom et al., 2015, Bruder Nascimento et al., 2016, Winkelströter et al., 2015a, Winkelströter et al., 2015b). A. fumigatus has four MAPKs: MpkA, which regulates the cell wall integrity (CWI) signaling and also has a role in the oxidative stress response (Valiante et al., 2015), MpkC and SakA, the mitogen-activated protein kinases of the high-osmolarity glycerol (HOG) pathway, that are involved in response to osmotic and oxidative stress, play a role in carbon source utilization and caspofungin adaptation (Altwasser et al., 2015, Bruder Nascimento et al., 2016, Du et al., 2006, Reyes et al., 2006, Posas et al., 1996), and MpkB, homologous to yeast Fus3, still uncharacterized (Elion et al., 1990). Here, we explored the impact of loss of the MAP kinases and phosphatases on the adhesion properties and biofilm formation in A. fumigatus. Such A. fumigatus biofilms are formed in vitro during static growth on a solid substratum (Beauvais et al., 2007) and in mouse lung with IA and in human lung aspergilloma (Loussert et al., 2010). Germlings of the ΔmpkA, ΔsakA, and the double ΔmpkC ΔsakA showed reduced adherence. Scanning electron microscopy for the wild-type and the mutant strains demonstrated that all the MAP kinase null mutants displayed altered cell surfaces and have altered ECM production during biofilm formation. In C. albicans, the CWI MAP kinase Mkc1p, the homologue of the A. fumigatus MpkA, is activated in a contact-dependent manner and it was shown that this kinase is necessary for normal biofilm development and invasive hyphal growth (Kumamoto, 2005). Another C. albicans kinase, Pkh3, is crucial for cell substrate adherence (Fanning et al., 2012), an essential stage of biofilm establishment (Finkel & Mitchell, 2011). Six master transcriptional regulators control C. albicans biofilm development at the genetic level. These six proteins (Efg1, Tec1, Bcr1, Ndt80, Brg1, and Rob1) are required for biofilm development both in vivo and in vitro (Nobile et al., 2012) and regulate the expression of around 1000 target genes. Bcr1, a transcriptional master regulator of biofilm production, and its cell wall protein targets Hwp1, Als1 and Als3, are needed for adherence during biofilm establishment (Chandra et al., 2001, Nobile et al., 2006a, Nobile et al., 2006b, Nobile et al., 2008, Nobile and Mitchell, 2005, Zhao et al., 2006). The distinct phases of biofilm production in C. albicans are regulated by different signaling pathways: yeast to hyphae transition involves Ras/cAMP/PKA (Hogan & Sundstrom, 2009); the induction of pseudohyphal growth is controlled by Cek1 MAPK (Srinivasa et al., 2012); and the calcium signaling pathways are important for morphogenesis (Sanglard et al., 2003). For example, the inactivation of the transcription regulator Efg1p (Ras/cAMP/PKA pathway) or Cph1p (MAP kinase pathway) can block hyphal growth and biofilm formation (Lo et al., 1997). In addition, it was demonstrated that filamentous growth was inhibited in cells deficient in Cdc35p showing the role of the adenylyl cyclase pathway in hyphal development (Rocha et al., 2001). A recent study involving a collection of 63 C. albicans protein kinase mutants showed that 38 displayed some degree of biofilm defect. Network analysis demonstrated functional redundancy of the proteins involved in some processes, including MAP kinases, since individual components are not required for biofilm formation (Konstantinidou & Morrissey, 2015). In this work we presented that the single deletion of Map kinases in A. fumigatus caused impaired biofilm formation. However, it is important to note that some changes in growth conditions for biofilm formation, such as surface support and characteristics of the medium, may produce different results.
We decided to investigate if this decreased biofilm formation could be due to alterations in the exposure of the polysaccharides present in the cell wall. The A. fumigatus cell wall contains different types of polysaccharides, such as: β-(1,3)-/β-(1,4)-glucan, chitin and galactomannan, which are covalently bound to branched β-(1,3)-glucan and α-(1,3)-glucan, which maintain the stability of the cell wall through its adhesive properties (Beauvais et al., 2013). The cell wall of the conidia that are inhaled are different compared to vegetative mycelium. The conidial cell wall has an outer layer composed of melanin and rodlets. These components display hydrophobic properties and prevent immune recognition of airborne fungal spores (Aimanianda et al., 2009, Aimanianda and Latgé, 2010, Carrion et al., 2013). In contrast to this, the external layer of the hypha presents galactosaminogalactan (GAG), an important A. fumigatus virulence factor (Beauvais et al., 2014). The A. fumigatus biofilm ECM is mainly composed of galactomannan, GAG, α-1,3 glucan, proteins, polyols, melanin and eDNA (Rajendran et al., 2013). A. fumigatus ΔmpkA germlings have reduced exposure of N-acetyl glucosamine, GAG, mannose and chitin, and increased β-1-3-glucan. The cell wall of A. fumigatus has pathogen-associated molecular patterns (PAMPs) that are identified by pattern recognition receptors (PRRs) on innate immune cells of the host. β-1-3-glucan acts as a PAMP and is recognized by the C–type lectin receptor (CLR) dectin 1 and this process is essential for initial host defense mechanisms in the respiratory tract (Brown and Gordon, 2001, Gessner et al., 2012, Werner et al., 2009). The cell wall component GAG, which is secreted during hyphal growth and is present in the A. fumigatus biofilm, mediates adherence and has a role attenuating the host immune response through inducing interleukin-1 receptor antagonist and also by masking β-1,3 glucan (Gravelat et al., 2013, Gresnigt et al., 2014). Thus, our results show that the ΔmpkA mutant displays changes in exposure of β-1,3-glucan and GAG, the two most important polysaccharides involved in the recognition of the A. fumigatus during host infection.
Recently our group showed that the ΔsakA and the double ΔmpkC ΔsakA mutants were more sensitive to cell wall damaging agents (Bruder Nascimento et al., 2016). The distribution of β-1,3-glucan and chitin content on the cell surface of these mutants was altered when compared to the wild type. The ΔsakA, ΔmpkC and ΔmpkC ΔsakA mutant strains have more abundant β-glucans than the wild-type, and the chitin content was higher in the ΔmpkC and lower in ΔsakA and ΔmpkC ΔsakA. Moreover, the single mutants ΔsakA and ΔmpkC were virulent in a low dose murine infection model and the double ΔmpkC ΔsakA had highly attenuated virulence in the same model of infection (Bruder Nascimento et al., 2016). In this work we showed that ΔsakA, ΔmpkC and ΔmpkC ΔsakA displayed reduced exposure of GAG in the cell wall. All these results suggest that the mitogen-activated protein kinases of the HOG pathway play an important role in the CWI pathway and biofilm formation in A. fumigatus. However, which transcriptional factors are activated downstream of the SakA and MpkC during the cell wall stress response and biofilm formation is still unknown. Previously, our group showed that deletion of the SitA and PtcB phosphatases activated the cell wall integrity pathway in A. fumigatus and this event was linked to suppression of ECM production and biofilm formation (Bom et al., 2015, Winkelströter et al., 2015a). PtcB is involved in the HOG pathway regulating the expression of osmo-dependent genes and phosphorylation of SakA. Moreover, ΔsitA and ΔptcB had increased exposure of β-1-3 glucan and chitin in the cell wall and both these mutant strains were avirulent in a murine model of invasive pulmonary aspergillosis. The results presented here showed that ΔsitA and ΔptcB have reduced exposure of mannose content in the cell wall and chitin and GAG were less abundant in the ΔptcB and ΔsitA, respectively. All these modifications in the cell wall increase the MAP kinase, ptcB, and sitA mutant conidia phagocytosis by D. discoideum.
Here we investigated in detail the phosphatase-encoding gene PphA. The A. fumigatus gene pphA codes for a Ser/Thr protein phosphatase PphA member of the PPP4 phosphatase family, which is homologous to yeast PPH3. In S. cerevisiae, PPH3 forms a phosphatase complex Pph3/Psy2 and it is involved in the regulation of Non-Homologous End-Joining (NHEJ) Pathway. Pph3/Psy2 complex controls the cell cycle progression dephosphorylating the key protein Rad53p and allowing cell cycle to continue (Omidi et al., 2014). Furthermore, Pph3/Psy2 targets the glucose signal protein Mth1 and this activity is required for the dephosphorylation of the downstream transcriptional repressor Rgt1 under glucose removal, an essential event in the repression of HXT genes, which encode glucose transporters. Mth1 and its associated corepressor Rgt1 are dephosphorylated by Pph3/Psy2, but this complex does not dephosphorylate Rgt1 at sites correlated with inactivation. The protein kinase A (PKA), the most important protein kinase activated in response to glucose, phosphorylates Mth1 via putative Pph3/Psy2 dephosphorylation sites in vivo. S. cerevisiae Pph3/Psy2 phosphatase negatively regulates Mth1 phosphorylation by protein kinase A (PKA) (Ma et al., 2013). In C. albicans, Pph3 dephosphorylates Rad53, a DNA checkpoint kinase that regulates effectors witch execute the stress response to DNA damage (Omidi et al., 2014). The A. fumigatus PphA phosphatase has a notable influence on the composition of the carbohydrates present in the cell wall. ΔpphA germlings have increased exposure of β-1,3 glucan, mannose and N-acetyl glucosamine, and the GAG and chitin content are reduced. Unexpectedly, when confronted with D. discoideum, the ΔpphA deletion mutant conidia were less frequently phagocytosed than those of the parental wild-type strain.
The A. fumigatus ΔpphA strain was more sensitive to Congo Red (CR) and CFW than the wild type strain, and we also have observed its sensitivity to lower caspofungin concentrations. PphA also had an important impact on the cell surface, conidia adhesion, and biofilm formation, features usually associated with the CWI pathway. Germlings of ΔpphA showed similar adherence to polystyrene and fibronectin-coated plates compared to the wild type. However, adhesion assay with conidia displayed significant reduction in adherence. Again, these results show that conidia and hyphae may exhibit differences in surface carbohydrates that result in alterations in important mechanisms during the host infection process, including adhesion properties. Immunoblot analysis showed that MpkA and SakA phosphorylation in response to CR and osmotic stresses are not affected by PphA. Moreover, our results demonstrated that ΔpphA and wild-type showed similar growth in presence of protein kinase C inhibitors. The activation of the CWI in fungi is dependent of the protein kinase C-mediated mitogen-activated protein kinase (PKC1-MAPK) pathway (Monge et al., 2006, Levin, 2005). Thus, more studies are required to understand which signaling pathways are being regulated by this phosphatase under cell wall stresses.
In this work, we demonstrated that PphA contributes to virulence of A. fumigatus. Comparative analysis of wild-type and the mutant strain in a neutropenic murine model of pulmonary aspergillosis showed that the ΔpphA is avirulent. The reduction of GAG and increased β-(1,3)-glucan in the ΔpphA cell wall can contribute to recognition of this strain by the host immune system and to induction of inflammatory responses. Corroborating this hypothesis, we observed that the cytokine Tumour Necrosis Factor alpha (TNF–α), released from macrophages in response to A. fumigatus exposure (Taramelli et al., 1996, Hayashi et al., 2005), was increased in presence of the ΔpphA conidia.
In summary, this study shows that the loss of specific MAP kinases and phosphatases had striking effects on the cell surface, adhesion of the conidia and germlings, and biofilm formation in A. fumigatus. However, how the upstream sensors trigger the MAP kinase cascades and which transcriptional regulators are being activated during the biofilm formation need to be elucidated.
Acknowledgments
We would like to thank the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) for providing financial support. LW and CM would like to acknowledge support from the University of Aberdeen and the MRC Centre for Medical Mycology (MR/N006364/1). All authors declare no financial conflict of interest.
Footnotes
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.tcsw.2018.03.002.
Appendix A. Supplementary data
References
- Aimanianda V., Bayry J., Bozza S., Kniemeyer O., Perruccio K., Elluru S.R., Latgé J.P. Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature. 2009;460:1117–1121. doi: 10.1038/nature08264. [DOI] [PubMed] [Google Scholar]
- Aimanianda V., Latgé J.P. Fungal hydrophobins form a sheath preventing immune recognition of airborne conidia. Virulence. 2010;1:185–187. doi: 10.4161/viru.1.3.11317. [DOI] [PubMed] [Google Scholar]
- Al Abdallah Q., Choe S.I., Campoli P., Baptista S., Gravelat F.N., Lee M.J., Sheppard D.C. A conserved C-terminal domain of the Aspergillus fumigatus developmental regulator MedA is required for nuclear localization, adhesion and virulence. PLoS One. 2012;7:e49959. doi: 10.1371/journal.pone.0049959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Fattani M.A., Douglas L.J. Biofilm ECM of Candida albicans and Candida tropicalis: chemical composition and role in drug resistance. J. Med. Microbiol. 2006;55(Pt 8):999–1008. doi: 10.1099/jmm.0.46569-0. [DOI] [PubMed] [Google Scholar]
- Altwasser R., Baldin C., Weber J., Guthke R., Kniemeyer O., Brakhage A.A., Valiante V. Network modeling reveals cross talk of MAP Kinases during adaptation to Caspofungin stress in Aspergillus fumigatus. PLoS One. 2015;10:e0136932. doi: 10.1371/journal.pone.0136932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia G., Cerri F., Baima S., Macino G. Involvement of protein kinase C in the response of Neurospora crassa to blue light. Mol. Gen. Genet. 1999;262:314–322. doi: 10.1007/s004380051089. [DOI] [PubMed] [Google Scholar]
- Baillie G.S., Douglas L.J. ECM polymers of Candida biofilms and their possible role in biofilm resistance to antifungal agents. J. Antimicrob. Chemother. 2000;46(3):397–403. doi: 10.1093/jac/46.3.397. [DOI] [PubMed] [Google Scholar]
- Beauvais A., Schmidt C., Guadagnini S., Roux P., Perret E., Henry C., Latge J.P. An ECM glues together the aerial-grown hyphae of Aspergillus fumigatus. Cell Microbiol. 2007;9(6):1588–1600. doi: 10.1111/j.1462-5822.2007.00895.x. [DOI] [PubMed] [Google Scholar]
- Beauvais A., Müller F.M. Biofilm formation in Aspergillus fumigatus. In: Latgé J.P., Steinbach W.J., editors. Aspergillus fumigatus and Aspergillosis. ASM Press; Washington DC: 2009. pp. 149–158. [Google Scholar]
- Beauvais A., Bozza S., Kniemeyer O., Formosa C., Balloy V., Henry C., Latgé J.P. Deletion of the alpha-(1,3)-glucan synthase genes induces a restructuring of the conidial cell wall responsible for the avirulence of Aspergillus fumigatus. PLoS Pathog. 2013;9:e1003716. doi: 10.1371/journal.ppat.1003716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauvais A., Fontaine T., Aimanianda V., Latge J.P. Aspergillus cell wall and biofilm. Mycopathologia. 2014;178(5–6):371–377. doi: 10.1007/s11046-014-9766-0. [DOI] [PubMed] [Google Scholar]
- Bom V.L., de Castro P.A., Winkelströter L.K., Marine M., Hori J.I., Ramalho L.N., Goldman G.H. The Aspergillus fumigatus sitA phosphatase homologue is important for adhesion, cell wall integrity, biofilm formation, and virulence. Eukaryot. Cell. 2015;14:728–744. doi: 10.1128/EC.00008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown G.D., Gordon S. Immune, recognition A new receptor for β-glucans. Nature. 2001;413:36–37. doi: 10.1038/35092620. [DOI] [PubMed] [Google Scholar]
- Bruder Nascimento A.C., Dos Reis T.F., de Castro P.A., Hori J.I., Bom V.L., de Assis L.J., Goldman G.H. Mitogen activated protein kinases SakA (HOG1) and MpkC collaborate for Aspergillus fumigatus virulence. Mol. Microbiol. 2016;100:841–859. doi: 10.1111/mmi.13354. [DOI] [PubMed] [Google Scholar]
- Bruns S., Seidler M., Albrecht D., Salvenmoser S., Remme N., Hertweck C., Muller F.M. Functional genomic profiling of Aspergillus fumigatus biofilm reveals enhanced production of the mycotoxin gliotoxin. Proteomics. 2010;10:3097–3107. doi: 10.1002/pmic.201000129. [DOI] [PubMed] [Google Scholar]
- Carberry S., Molloy E., Hammel S., O’Keeffe G., Jones G.W., Kavanagh K., Doyle S. Gliotoxin effects on fungal growth: mechanisms and exploitation. Fungal Genet. Biol. 2012;49:302–312. doi: 10.1016/j.fgb.2012.02.003. [DOI] [PubMed] [Google Scholar]
- Carrion S.D.J., Leal S.M., Jr, Ghannoum M.A., Aimanianda V., Latgé J.P., Pearlman E. The RodA hydrophobin on Aspergillus fumigatus spores masks dectin-1- and dectin-2-dependent responses and enhances fungal survival in vivo. J. Immunol. 2013;191:2581–2588. doi: 10.4049/jimmunol.1300748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cegelski L. Bottom-up and top-down solid-state NMR approaches for bacterial biofilm ECM composition. J. Magn. Reson. 2015;253:91–97. doi: 10.1016/j.jmr.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra J., Kuhn D.M., Mukherjee P.K., Hoyer L.L., McCormick T., Ghannoum M.A. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J. Bacteriol. 2001;183:5385–5394. doi: 10.1128/JB.183.18.5385-5394.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffin W.L. Candida albicans cell wall proteins. Microbiol. Mol. Biol. Rev. 2008;72(3):495–544. doi: 10.1128/MMBR.00032-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.C., Slavin M.A., Sorrell T.C. Echinocandin antifungal drugs in fungal infections: a comparison. Drugs. 2011;71:11–41. doi: 10.2165/11585270-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Chotirmall S.H., Mirkovic B., Lavelle G.M., McElvaney N.G. Immunoevasive Aspergillus virulence factors. Mycopathologia. 2014;178:363–370. doi: 10.1007/s11046-014-9768-y. [DOI] [PubMed] [Google Scholar]
- Costerton J.W., Stewart P.S., Greenberg E.P. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284(5418):318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- da Rocha A.B., Mans D.R., Regner A., Schwartsmann G. Targeting protein kinase C: new therapeutic opportunities against high-grade malignant gliomas? Oncologist. 2002;7:17–33. doi: 10.1634/theoncologist.7-1-17. [DOI] [PubMed] [Google Scholar]
- Delattin N., Cammue B.P., Thevissen K. Reactive oxygen species inducing antifungal agents and their activity against fungal biofilms. Future Med. Chem. 2014;6:77–90. doi: 10.4155/fmc.13.189. [DOI] [PubMed] [Google Scholar]
- Dinamarco T.M., Almeida R.S., de Castro P.A., Brown N.A., dos Reis T.F., Ramalho L.N., Goldman G.H. Molecular characterization of the putative transcription factor SebA involved in virulence in Aspergillus fumigatus. Eukaryot. Cell. 2012;11:518–531. doi: 10.1128/EC.00016-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding C., Festa R.A., Sun T.S., Wang Z.Y. Iron and copper as virulence modulators in human fungal pathogens. Mol. Microbiol. 2014;93:10–23. doi: 10.1111/mmi.12653. [DOI] [PubMed] [Google Scholar]
- Donlan R.M. Biofilm formation: a clinically relevant microbiological process. Clin. Infect. Dis. 2001;33(8):1387–1392. doi: 10.1086/322972. [DOI] [PubMed] [Google Scholar]
- Du C., Sarfati J., Latge J.P., Calderone R. The role of the sakA (Hog1) and tcsB (sln1) genes in the oxidant adaptation of Aspergillus fumigatus. Med. Mycol. 2006;44:211–218. doi: 10.1080/13693780500338886. [DOI] [PubMed] [Google Scholar]
- Erlandsen S.L., Kristich C.J., Dunny G.M., Wells C.L. High-resolution visualization of the microbial glycocalyx with low-voltage scanning electron microscopy: dependence on cationic dyes. J. Histochem. Cytochem. 2004;52:1427–1435. doi: 10.1369/jhc.4A6428.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlandsen S.L., Kristich C.J., Dunny G.M., Wells C.L. High-resolution visualization of the microbial glycocalyx with low-voltage scanning electron microscopy: dependence on cationic dyes. J. Histochem. Cytochem. 2004;5:1427–1435. doi: 10.1369/jhc.4A6428.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion E.A., Grisafi P.L., Fink G.R. FUS3 encodes a cdc2+/CDC28-related kinase required for the transition from mitosis into conjugation. Cell. 1990;60:649–664. doi: 10.1016/0092-8674(90)90668-5. [DOI] [PubMed] [Google Scholar]
- Fanning S., Xu W., Beaurepaire C., Suhan J.P., Nantel A., Mitchell A.P. Functional control of the Candida albicans cell wall by catalytic protein kinase A subunit Tpk1. Mol. Microbiol. 2012;86:284–302. doi: 10.1111/j.1365-2958.2012.08193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria-Oliveira F., Carvalho J., Belmiro C.L., Martinez- Gomariz M., Hernaez M.L., Pavao M., Ferreira C. Methodologies to generate, extract, purify and fractionate yeast ECM for analytical use in proteomics and glycomics. BMC Microbiol. 2014;14:244. doi: 10.1186/s12866-014-0244-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes T., Silva S., Henriques M. Candida tropicalis biofilm’s ECM-involvement on its resistance to amphotericin B. Diagn. Microbiol. Infect. Dis. 2015;83(2):165–169. doi: 10.1016/j.diagmicrobio.2015.06.015. [DOI] [PubMed] [Google Scholar]
- Finkel J.S., Mitchell A.P. Genetic control of Candida albicans biofilm development. Nat. Rev. Microbiol. 2011;9:109–118. doi: 10.1038/nrmicro2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine T., Delangle A., Simenel C., Coddeville B., van Vliet S.J., van Kooyk Y., Latgé J.P. Galactosaminogalactan, a new immunosuppressive polysaccharide of Aspergillus fumigatus. PLoS Pathog. 2011;7:e1002372. doi: 10.1371/journal.ppat.1002372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming H.C., Wingender J. The biofilm ECM. Nat. Rev. Microbiol. 2010;8(9):623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- Gessner M.A., Werner J.L., Lilly L.M., Nelson M.P., Metz A.E., Dunaway C.W., Steele C. Dectin–1–dependent interleukin–22 contributes to early innate lung defense against Aspergillus fumigatus. Infect. Immun. 2012;80:410–417. doi: 10.1128/IAI.05939-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons J.G., Beauvais A., Beau R., McGary K.L., Latgé J.P., Rokas A. Global transcriptome changes underlying colony growth in the opportunistic human pathogen Aspergillus fumigatus. Eukaryot. Cell. 2012;11:68–78. doi: 10.1128/EC.05102-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin D., DeSanctis J., Boulé M., Skamene E., Matouk C., Radzioch D. Role of tumor necrosis factor alpha in innate resistance to mouse pulmonary infection with Pseudomonas aeruginosa. Infect. Immun. 1995;63(9):3272–3278. doi: 10.1128/iai.63.9.3272-3278.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham L.M., Tsoni S.V., Willment J.A., Williams D.L., Taylor P.R. Soluble Dectin-1 as a tool to detect beta-glucans. J. Immunol. Methods. 2006;314:164–169. doi: 10.1016/j.jim.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Grahl N., Shepardson K.M., Chung D., Jr. R.A., Cramer Hypoxia and fungal pathogenesis: to air or not to air? Eukaryot. Cell. 2012;11:560–570. doi: 10.1128/EC.00031-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravelat F.N., Ejzykowicz D.E., Chiang L.Y., Chabot J.C., Urb M., Macdonald K.D., Sheppard D.C. Aspergillus fumigatus MedA governs adherence, host cell interactions and virulence. Cell Microbiol. 2010;12:473–488. doi: 10.1111/j.1462-5822.2009.01408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravelat F.N., Beauvais A., Liu H., Lee M.J., Snarr B.D., Chen D., Sheppard D.C. Aspergillus galactosaminogalactan mediates adherence to host constituents and conceals hyphal beta-glucan from the immune system. PLoS Pathog. 2013;9:e1003575. doi: 10.1371/journal.ppat.1003575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberger P.A. Allergic bronchopulmonary aspergillosis. J. Allergy Clin. Immunol. 2002;110:685–692. doi: 10.1067/mai.2002.130179. [DOI] [PubMed] [Google Scholar]
- Gresnigt M.S., Bozza S., Becker K.L., Joosten L.A., Abdollahi-Roodsaz S., van der Berg W.B., van de Veerdonk F.L. A polysaccharide virulence factor from Aspergillus fumigatus elicits anti-inflammatory effects through induction of interleukin-1 receptor antagonist. PLoS Pathog. 2014;10:e1003936. doi: 10.1371/journal.ppat.1003936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas H. Fungal siderophore metabolism with a focus on Aspergillus fumigatus. Nat. Prod. Rep. 2014;31:1266–1276. doi: 10.1039/c4np00071d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara D., Suzuki S., Kamei K., Gonoi T., Kawamoto S. The role of AtfA and HOG MAPK pathway in stress tolerance in conidia of Aspergillus fumigatus. Fungal Genet. Biol. 2014;73:138–149. doi: 10.1016/j.fgb.2014.10.011. [DOI] [PubMed] [Google Scholar]
- Hartmann T., Sasse C., Schedler A., Hasenberg M., Gunzer M., Krappmann S. Shaping the fungal adaptome-stress responses of Aspergillus fumigatus. Int. J. Med. Microbiol. 2011;301:408–416. doi: 10.1016/j.ijmm.2011.04.008. [DOI] [PubMed] [Google Scholar]
- Hayashi N., Nomura T., Sakumoto N., Mukai Y., Kaneko Y., Harashima S., Murakami S. The SIT4 gene, which encodes protein phosphatase 2A, is required for telomere function in Saccharomyces cerevisiae. Curr. Genet. 2005;47:359–367. doi: 10.1007/s00294-005-0577-1. [DOI] [PubMed] [Google Scholar]
- Hawser S.P., Baillie G.S., Douglas L.J. Production of ECM by Candida albicans biofilms. J. Med. Microbiol. 1998;47(3):253–256. doi: 10.1099/00222615-47-3-253. [DOI] [PubMed] [Google Scholar]
- Herbert J.M., Augereau J.M., Gleye J., Maffrand J.P. Chelerythrine is a potent and specific inhibitor of protein kinase C. Biochem. Biophys. Res. Commun. 1990;172:993–999. doi: 10.1016/0006-291x(90)91544-3. [DOI] [PubMed] [Google Scholar]
- Hillmann F., Novohradská S., Mattern D.J., Forberger T., Heinekamp T., Westermann M., Brakhage A.A. Virulence determinants of the human pathogenic fungus Aspergillus fumigatus protect against soil amoeba predation. Environ. Microbiol. 2015;17(8):2858–2869. doi: 10.1111/1462-2920.12808. [DOI] [PubMed] [Google Scholar]
- Hogan D.A., Sundstrom P. The Ras/cAMP/PKA signaling pathway and virulence in Candida albicans. Future Microbiol. 2009;4:1263–1270. doi: 10.2217/fmb.09.106. [DOI] [PubMed] [Google Scholar]
- Honraet K., Goetghebeur E., Nelis H.J. Comparison of three assays for the quantification of Candida biomass in suspension and CDC reactor grown biofilms. J. Microbiol. Methods. 2005;63(3):287–295. doi: 10.1016/j.mimet.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Jarvis W.D., Grant S. Protein kinase C targeting in antineoplastic treatment strategies. Invest. New Drugs. 1999;17:227–240. doi: 10.1023/a:1006328303451. [DOI] [PubMed] [Google Scholar]
- Juvvadi P.R., Maruyama J., Kitamoto K. Phosphorylation of the Aspergillus oryzae Woronin body protein, AoHex1, by protein kinase C: evidence for its role in the multimerization and proper localization of the Woronin body protein. Biochem. J. 2007;405:533–540. doi: 10.1042/BJ20061678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käfer E. The anthranilate synthetase enzyme complex and the trifunctional trpC gene of Aspergillus. Can. J. Genet. Cytol. 1977;19:723–738. doi: 10.1139/g77-079. [DOI] [PubMed] [Google Scholar]
- Konstantinidou N., Morrissey J.P. Co-occurence of filamentation defects and impaired biofilms in Candida albicans protein kinase mutants. FEMS Yeast Res. 2015;15(8) doi: 10.1093/femsyr/fov092. [DOI] [PubMed] [Google Scholar]
- Kumamoto C.A. A contact-activated kinase signals Candida albicans invasive growth and biofilm development. Proc. Natl. Acad. Sci. U.S.A. 2005;102(15):5576–5581. doi: 10.1073/pnas.0407097102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laichalk L.L., Kunkel S.L., Strieter R.M., Danforth J.M., Bailie M.B., Standiford T.J. Tumor necrosis factor mediates lung antibacterial host defense in murine Klebsiella pneumonia. Infect. Immun. 1996;64:5211. doi: 10.1128/iai.64.12.5211-5218.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latgé J.P., Steinbach W.J. A perspective view of Aspergillus fumigatus research for the next ten years. In: Latge J.P., Steinbach W.J., editors. Aspergillus fumigatus and Aspergillosis. ASM Press; Washington, DC: 2009. pp. 549–558. [Google Scholar]
- Levin D., E. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2005;69:262–291. doi: 10.1128/MMBR.69.2.262-291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.J., Sasse C., Gerke J., Valerius O., Irmer H., Frauendorf H., Braus G.H. Transcription factor SomA is required for adhesion, development and virulence of the human pathogen Aspergillus fumigatus. PLoS Pathog. 2015;11:e1005205. doi: 10.1371/journal.ppat.1005205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo H.J., Kohler J.R., DiDomenico B., Loebenberg D., Cacciapuoti A., Fink G.R. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- Loussert C., Schmitt C., Prevost M.C., Balloy V., Fadel E., Philippe B., Beauvais A. In vivo biofilm composition of Aspergillus fumigatus. Cell Microbiol. 2010;12(3):405–410. doi: 10.1111/j.1462-5822.2009.01409.x. [DOI] [PubMed] [Google Scholar]
- Ma H., Han B.K., Guaderrama M., Aslanian A., Yates J.R., 3rd., Hunter T., Wittenberg C. Psy2 targets the PP4 family phosphatase Pph3 to dephosphorylate Mth1 and repress glucose transporter gene expression. Mol. Cell Biol. 2013;34(3):452–463. doi: 10.1128/MCB.00279-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marim F.M., Silveira T.N., Lima D.S., Jr, Zamboni D.S. A method for generation of bone marrow-derived macrophages from cryopreserved mouse bone marrow cells. PLoS One. 2010;5:e15263. doi: 10.1371/journal.pone.0015263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins M., Uppuluri P., Thomas D.P., Cleary I.A., Henriques M., Lopez-Ribot J.L., Oliveira R. Presence of extracellular DNA in the Candida albicans biofilm ECM and its contribution to biofilms. Mycopathologia. 2010;169(5):323–331. doi: 10.1007/s11046-009-9264-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell K.F., Taff H.T., Cuevas M.A., Reinicke E.L., Sanchez H., Andes D.R. Role of ECM beta- 1,3 Glucan in antifungal resistance of non-albicans Candida Biofilms. Antimicrob. Agents Chemother. 2013;57(4):1918–1920. doi: 10.1128/AAC.02378-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monge R.A., Román E., Nombela C., Pla J. The MAP kinase signal transduction network in Candida albicans. Microbiology. 2006;152:905–912. doi: 10.1099/mic.0.28616-0. [DOI] [PubMed] [Google Scholar]
- Moore M.M. The crucial role of iron uptake in Aspergillus fumigatus virulence. Curr. Opin. Microbiol. 2013;16:692–699. doi: 10.1016/j.mib.2013.07.012. [DOI] [PubMed] [Google Scholar]
- Mowat E., Butcher J., Lang S., Williams C., Ramage G. Development of a simple model for studying the effects of antifungal agents on multicellular communities of Aspergillus fumigatus. J. Med. Microbiol. 2007;56:1205–1212. doi: 10.1099/jmm.0.47247-0. [DOI] [PubMed] [Google Scholar]
- Mowat E., Lang S., Williams C., McCulloch E., Jones B., Ramage G. Phase-dependent antifungal activity against Aspergillus fumigatus developing multicellular filamentous biofilms. J. Antimicrob. Chemother. 2008;62:1281–1284. doi: 10.1093/jac/dkn402. [DOI] [PubMed] [Google Scholar]
- Muszkieta L., Beauvais A., Pahtz V., Gibbons J.G., Anton Leberre V., Beau R., Latgé J.P. Investigation of Aspergillus fumigatus biofilm formation by various “omics” approaches. Front. Microbiol. 2013;4:13. doi: 10.3389/fmicb.2013.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett J.E., Crawford K., Marchillo K., Andes D.R. Role of Fks1p and ECM glucan in Candida albicans biofilm resistance to an echinocandin, pyrimidine, and polyene. Antimicrob. Agents Chemother. 2010;54(8):3505–3508. doi: 10.1128/AAC.00227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett J.E., Sanchez H., Cain M.T., Ross K.M., Andes D.R. Interface of Candida albicans biofilm matrix-associated drug resistance and cell wall integrity regulation. Eukaryot. Cell. 2011;10(12):1660–1669. doi: 10.1128/EC.05126-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile C.J., Mitchell A.P. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr. Biol. 2005;15:1150–1155. doi: 10.1016/j.cub.2005.05.047. [DOI] [PubMed] [Google Scholar]
- Nobile C.J., Andes D.R., Nett J.E., Smith F.J., Yue F., Phan Q.T., Mitchell A.P. Critical role of Bcr1-dependent adhesions in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2006;2:e63. doi: 10.1371/journal.ppat.0020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile C.J., Nett J.E., Andes D.R., Mitchell A.P. Function of Candida albicans adhesin Hwp1 in biofilm formation. Eukaryot. Cell. 2006;5:1604–1610. doi: 10.1128/EC.00194-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile C.J., Schneider H.A., Nett J.E., Sheppard D.C., Filler S.G., Andes D.R., Mitchell A.P. Complementary adhesin function in C. albicans biofilm formation. Curr. Biol. 2008;18:1017–1024. doi: 10.1016/j.cub.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile C.J., Nett J.E., Hernday A.D., Homann O.R., Deneault J.S., Nantel A., Mitchell A.P. Biofilm ECM regulation by Candida albicans Zap1. PLoS Biol. 2009;7(6):e1000133. doi: 10.1371/journal.pbio.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile C.J., Fox E.P., Nett J.E., Sorrells T.R., Mitrovich Q.M., Hernday A.D., Johnson A.D. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell. 2012;148:126–138. doi: 10.1016/j.cell.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omidi K., Hooshyar M., Jessulat M., Samanfar B., Sanders M., Burnside D., Golshani A. Phosphatase complex Pph3/Psy2 is involved in regulation of efficient non-homologous end-joining pathway in the yeast Saccharomyces cerevisiae. PLoS One. 2014;9(1):e87248. doi: 10.1371/journal.pone.0087248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole G.A. To build a biofilm. J. Bacteriol. 2003;185(9):2687–2689. doi: 10.1128/JB.185.9.2687-2689.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas F., Wurgler-Murphy S.M., Maeda T., Witten E.A., Thai T.C., Saito H. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- Rajendran R., Williams C., Lappin D.F., Millington O., Martins M., Ramage G. Extracellular DNA release acts as an antifungal resistance mechanism in mature Aspergillus fumigatus biofilms. Eukaryot. Cell. 2013;12(3):420–429. doi: 10.1128/EC.00287-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage G., Rajendran R., Gutierrez-Correa M., Jones B., Williams C. Aspergillus biofilms: clinical and industrial significance. FEMS Microbiol. Lett. 2011;324(2):89–97. doi: 10.1111/j.1574-6968.2011.02381.x. [DOI] [PubMed] [Google Scholar]
- Ramage G., Rajendran R., Sherry L., Williams C. Fungal biofilm resistance. Int. J. Microbiol. 2012:528521. doi: 10.1155/2012/528521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichhardt C., Ferreira J.A., Joubert L.M., Clemons K.V., Stevens D.A., Cegelski L. Analysis of the Aspergillus fumigatus biofilm ECM by solid-state nuclear magnetic resonance spectroscopy. Eukaryot. Cell. 2015;14(11):1064–1072. doi: 10.1128/EC.00050-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes G., Romans A., Nguyen C.K., May G.S. Novel mitogen activated protein kinase MpkC of Aspergillus fumigatus is required for utilization of polyalcohol sugars. Eukaryot. Cell. 2006;5:1934–1940. doi: 10.1128/EC.00178-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinet P., Baychelier F., Fontaine T., Picard C., Debre P., Vieillard V., Elbim C. A polysaccharide virulence factor of a human fungal pathogen induces neutrophil apoptosis via NK cells. J. Immunol. 2014;192:5332–5342. doi: 10.4049/jimmunol.1303180. [DOI] [PubMed] [Google Scholar]
- Robbins N., Uppuluri P., Nett J., Rajendran R., Ramage G., Lopez-Ribot J.L., Cowen L.E. Hsp90 governs dispersion and drug resistance of fungal biofilms. PLoS Pathog. 2011;7:e1002257. doi: 10.1371/journal.ppat.1002257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha C.R., Schröppel K., Harcus D., Marcil A., Dignard D., Taylor B.N., Leberer E. Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol. Biol. Cell. 2001;12:3631–3643. doi: 10.1091/mbc.12.11.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanglard D., Ischer F., Marchetti O., Entenza J., Bille J. Calcineurin A of Candida albicans: involvement in antifungal tolerance, cell morphogenesis and virulence. Mol. Microbiol. 2003;48:959–976. doi: 10.1046/j.1365-2958.2003.03495.x. [DOI] [PubMed] [Google Scholar]
- Schaffner A., Douglas H., Braude A. Selective protection against conidia by mononuclear and against mycelia by polymorphonuclear phagocytes in resistance to Aspergillus: observations on these two lines of defense in vivo and in vitro with human and mouse phagocytes. J. Clin. Invest. 1982;69:617. doi: 10.1172/JCI110489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidler M.J., Salvenmoser S., Muller F.M. Aspergillus fumigatus forms biofilms with reduced antifungal drug susceptibility on bronchial epithelial cells. Antimicrob. Agents Chemother. 2008;52:4130–4136. doi: 10.1128/AAC.00234-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf D.H., Heinekamp T., Remme N., Hortschansky P., Brakhage A.A., Hertweck C. Biosynthesis and function of gliotoxin in Aspergillus fumigatus. Appl. Microbiol. Biotechnol. 2012;93:467–472. doi: 10.1007/s00253-011-3689-1. [DOI] [PubMed] [Google Scholar]
- Schrettl M., Haas H. Iron homeostasis-Achilles’ heel of Aspergillus fumigatus? Curr. Opin. Microbiol. 2011;14:400–405. doi: 10.1016/j.mib.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikantha T., Daniels K.J., Pujol C., Kim E., Soll D.R. Identification of genes upregulated by the transcription factor Bcr1 that are involved in impermeability, impenetrability and drug-resistance of Candida albicans a/alpha biofilms. Eukaryot. Cell. 2013;12(6):875–888. doi: 10.1128/EC.00071-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasa K., Kim J., Yee S., Kim W., Choi W. A MAP kinase pathway is implicated in the pseudohyphal induction by hydrogen peroxide in Candica albicans. Mol. Cells. 2012;33:183–193. doi: 10.1007/s10059-012-2244-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman A., Hus K., Chio L.C., Heidler S., Shaw M., Ma D., Ye X.S. Discovery of cercosporamide, a known antifungal natural product, as a selective Pkc1 kinase inhibitor through high-throughput screening. Eukaryot. Cell. 2004;3:932–943. doi: 10.1128/EC.3.4.932-943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taff H.T., Nett J.E., Zarnowski R., Ross K.M., Sanchez H., Cain M.T., Andes D.R. A Candida biofilm-induced pathway for ECM glucan delivery: implications for drug resistance. PLoS Pathog. 2012;8(8):e1002848. doi: 10.1371/journal.ppat.1002848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taramelli D., Malabarba M.G., Sala G., Basilico N., Cocuzza G. Production of cytokines by alveolar and peritoneal macrophages stimulated by Aspergillus fumigatus conidia or hyphae. J. Med. Vet. Mycol. 1996;34:49–56. [PubMed] [Google Scholar]
- Taylor P.R., Leal S.M., Jr, Sun Y., Pearlman E. Aspergillus and Fusarium corneal infections are regulated by Th17 cells and IL-17-producing neutrophils. J. Immunol. 2014;192:3319–3327. doi: 10.4049/jimmunol.1302235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D.P., Bachmann S.P., Lopez-Ribot J.L. Proteomics for the analysis of the Candida albicans biofilm lifestyle. Proteomics. 2006;6(21):5795–5804. doi: 10.1002/pmic.200600332. [DOI] [PubMed] [Google Scholar]
- Valiante V., Jain R., Heinekamp T., Brakhage A.A. The MpkA MAP kinase module regulates cell wall integrity signaling and pyomelanin formation in Aspergillus fumigatus. Fungal Genet. Biol. 2009;46(12):909–918. doi: 10.1016/j.fgb.2009.08.005. [DOI] [PubMed] [Google Scholar]
- Valiante V., Macheleidt J., Foge M., Brakhage A.A. The Aspergillus fumigatus cell wall integrity signaling pathway: drug target, compensatory pathways, and virulence. Front. Microbiol. 2015;6:325. doi: 10.3389/fmicb.2015.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L.A., Munro C.A., de Bruijn I., Lenardon M.D., McKinnon A., Gow N.A. Stimulation of chitin synthesis rescues Candida albicans from echinocandins. PLoS Pathog. 2008;4:e1000040. doi: 10.1371/journal.ppat.1000040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner J.L., Metz A.E., Horn D., Schoeb T.R., Hewitt M.M., Schwiebert L.M., Steele C. Requisite role for the dectin–1 β-glucan receptor in pulmonary defense against Aspergillus fumigatus. J. Immunol. 2009;182:4938–4946. doi: 10.4049/jimmunol.0804250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wezensky S.J., Cramer R.A., Jr. Implications of hypoxic microenvironments during invasive aspergillosis. Med. Mycol. 2011;49(Suppl. 1):S120–S124. doi: 10.3109/13693786.2010.495139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkelströter L.K., Bom V.L., de Castro P.A., Ramalho L.N., Goldman M.H., Brown N.A., Goldman G.H. High osmolarity glycerol response PtcB phosphatase is important for Aspergillus fumigatus virulence. Mol. Microbiol. 2015;96:42–54. doi: 10.1111/mmi.12919. [DOI] [PubMed] [Google Scholar]
- Winkelströter L.K., Dolan S.K., Fernanda Dos Reis T., Bom V.L., Alves de Castro P., Hagiwara D., Goldman G.H. Systematic global analysis of 24 genes encoding protein phosphatases in Aspergillus fumigatus. G3 (Bethesda) 2015;5:1525–1539. doi: 10.1534/g3.115.016766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Gao C., Li X., He Y., Zhou L., Pang G., Sun S. In vitro antifungal activity of silver nanoparticles against ocular pathogenic filamentous fungi. J. Ocul. Pharmacol. Ther. 2013;29:270–274. doi: 10.1089/jop.2012.0155. [DOI] [PubMed] [Google Scholar]
- Yi S., Sahni N., Daniels K.J., Lu K.L., Srikantha T., Huang G., Soll D.R. Alternative mating type configurations (a/alpha versus a/a or alpha/alpha) of Candida albicans result in alternative biofilms regulated by different pathways. PLoS Biol. 2011;9(8):e1001117. doi: 10.1371/journal.pbio.1001117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnowski R., Westler W.M., Lacmbouh G.A., Marita J.M., Bothe J.R., Bernhardt J., Andes D.R. Novel entries in a fungal biofilm ECM encyclopedia. mBio. 2014;5(4) doi: 10.1128/mBio.01333-14. e01333-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Chen Y., Ma Z., Chen Q., Ostapska H., Gravelat F.N., Sheppard D.C. PtaB, a lim-domain binding protein in Aspergillus fumigatus regulates biofilm formation and conidiation through distinct pathways. Cell Microbiol. 2017;20(1) doi: 10.1111/cmi.12799. [DOI] [PubMed] [Google Scholar]
- Zhao X., Daniels K.J., Oh S.H., Green C.B., Yeater K.M., Soll D.R., Hoyer L.L. Candida albicans Als3p is required for wild-type biofilm formation on silicone elastomer surfaces. Microbiology. 2006;152:2287–2299. doi: 10.1099/mic.0.28959-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.