Abstract
Background
Identification and characterization of key enzymes associated with cell wall biosynthesis and modification is fundamental to gain insights into cell wall dynamics. However, it is a challenge that activity assays of glycosyltransferases are very low throughput and acceptor substrates are generally not available.
Results
We optimized and validated microscale thermophoresis (MST) to achieve high throughput screening for glycosyltransferase substrates. MST is a powerful method for the quantitative analysis of protein–ligand interactions with low sample consumption. The technique is based on the motion of molecules along local temperature gradients, measured by fluorescence changes. We expressed glycosyltransferases as YFP-fusion proteins in tobacco and optimized the MST method to allow the determination of substrate binding affinity without purification of the target protein from the cell lysate. The application of this MST method to the β-1,4-galactosyltransferase AtGALS1 validated the capability to screen both nucleotide-sugar donor substrates and acceptor substrates. We also expanded the application to members of glycosyltransferase family GT61 in sorghum for substrate screening and function prediction.
Conclusions
This method is rapid and sensitive to allow determination of both donor and acceptor substrates of glycosyltransferases. MST enables high throughput screening of glycosyltransferases for likely substrates, which will narrow down their in vivo function and help to select candidates for further studies. Additionally, this method gives insight into biochemical mechanism of glycosyltransferase function.
Keywords: Plant cell wall, Glycosyltransferase, AtGALS1, Microscale thermophoresis, Sorghum, GT61
Background
Glycosylation is a fundamental process in biology, conserved across all domains of life. Proteins, lipids, and small molecules are frequently glycosylated, and polysaccharides are ubiquitous to all organisms [1]. The principal enzymes responsible for glycosylation are glycosyltransferases (GTs). GTs use activated sugars, usually nucleotide sugars (NDP-sugars), as donor substrate [2]. There are about 10–15 common NDP-sugars used by GTs whereas potential acceptor substrates are unlimited [3]. While GTs can often be recognized by their primary structures their specificity cannot currently be predicted, and elucidating GT function has been excruciatingly slow. In plants less than 10% have a known function [4]. Unique assays must be developed for every glycosylated product studied. Determining the function of a GT currently requires correct identification of both donor and acceptor substrates in addition to the development of an assay for product identification. Some commercial (UDP/CMP/GDP-Glo™, Promega) and reported glycosylation assays are based on detection of UDP or GDP released during the reaction through coupling to another reaction [5, 6] making them more suitable for high throughput studies, but they still depend on acceptor substrates. Often acceptor substrates are not only unknown, but candidate substrates are generally not available from commercial and other sources.
A large proportion of the GTs in plants are known or likely to be involved in cell wall biosynthesis. The plant cell wall, an extracellular matrix of polysaccharides and glycoproteins, is crucial for plant growth, morphology, integrity, and biomass recalcitrance [7, 8]. Dynamic structural change in the plant cell wall promotes plant development through deposition, crosslinking, remodeling, and degradation [9, 10]. Polysaccharides of the plant cell wall comprises cellulose synthesized at the plasma membrane and matrix polysaccharides, which are synthesized in the Golgi apparatus and transported by endo-membrane vesicles within the secretory pathway for deposition [11]. During this process, biosynthetic GTs are the main enzymes required for the synthesis of the polysaccharides. In order to gain insights into cell wall biology, it is fundamental to identify and characterize key GTs associated with cell wall biosynthesis and modification.
However, long-standing challenges to characterizing these Golgi-localized GTs exist. First, protein purification for biochemical activity assay has only been accomplished in few cases due to the labile nature and low abundance of these enzymes [12, 13]. Heterologous expression in E. coli has been largely ineffective, likely limited by insufficient folding and post-translational modification in bacterial systems [14–16]. Heterologous expression in eukaryotic systems has been reported, but no generally successful system has been found. Second, activity assays are often difficult since assays must use both nucleotide-sugar donor and acceptor substrates, which are generally not available. Finally, based on sequence homology, GTs have been classified into 110 families in the Carbohydrate Active enZyme database (CAZy, http://www.cazy.org) [17]. For each GT family at least one member was characterized biochemically, whereas most of the other family members have been assigned based on their sequence. Substrate specificity, however, is very difficult to predict from the sequence. Many GT families contain enzymes with widely different activities despite the sequence similarity. When there is no clear hypothesis for the function of a GT it is essentially impossible to determine both the sugar donor and acceptor. Even when mutant studies or sequence similarity lead to hypothesis of potential activity, it can be difficult to know if inability to detect activity is due to the wrong substrates being tested or a problem with the activity of the heterologously expressed enzyme. Indeed, some members of GT families may not have GT activity in vivo but could have other roles, e.g. as part of GT complexes [18, 19]. As a result, there are significantly fewer well characterized GTs compared to the large number predicted by bioinformatic methods. However, the nucleotide sugar donor substrates constitute a limited set of about thirteen common ones in plants (UDP-α-d-Glc, UDP-α-d-Xyl, UDP-β-l-Araf, UDP-β-l-Arap, UDP-α-d-Gal, UDP-β-l-Rha, UDP-α-d-GlcNAc, GDP-α-l-Gal, GDP-β-l-Fuc, GDP-α-d-Man, GDP-α-d-Glc, UDP-α-d-GalA, UDP-α-d-GlcA). Less common nucleotide sugars in plants include CMP-β-d-Kdo, UDP-α-d-Api and the unknown precursors of Dha (2-keto-3-deoxy-d-lyxo-heptulosaric acid) and Ace (3-C-carboxy-5-deoxy-l-xylose). Recent evidence indicates the presence of UDP-α-d-Fuc at least in some plants [20], but d-Fuc is not known to be part of any cell wall polysaccharide. A method to screen for putative donor substrate in the absence of acceptor would therefore be extremely useful in initial testing of putative biochemical function and in validating that a heterologously expressed enzyme is likely to be active. Acceptor substrates are much more diverse, but a method to screen putative acceptor substrates in the absence of donor would likewise be extremely useful.
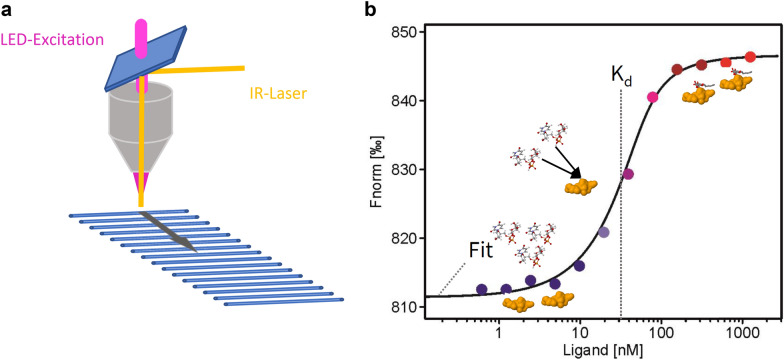
An emerging and sensitive method for studying molecular interactions is microscale thermophoresis (MST). The technique is based on the motion of molecules along local temperature gradients, measured by fluorescence changes [21, 22]. A spatial temperature difference (ΔT) leads to a mass flow of molecules in the temperature-elevated region. The Soret coefficient, also known as thermophoresis, was defined by ST: chot = ccold * exp(− ST ΔT ) and quantified the relation of temperature gradient and the molecule flow [23, 24]. The thermophoresis of a protein typically differs significantly from the thermophoresis of a protein–ligand complex due to binding-induced changes in size, charge, and hydration shell [24]. Figure 1 shows the setup of the instrument for MST.
Fig. 1.
Setup of the apparatus to determine the thermophoresis of biomolecules. a An infrared (IR) laser heats the aqueous sample filled in a thin glass capillary and generates a localized microscopic temperature gradient in the range of 2–6 °C. Protein complexes with interaction partners demonstrate slower movement through the temperature gradient compared with free molecules. This movement is monitored via fluorescence of the target protein. Rapid scanning of 16 capillaries loaded with fluorescent target protein at a constant concentration and substrates in increasing concentration gradients enables the determination of equilibrium binding constants. b A binding curve can be calculated from the gradual difference of thermophoresis between the fluorescent molecules of both unbound and bound states, which is plotted as Fnorm, defined as Fhot/Fcold against the ligand concentration. The binding constant Kd can be then derived from the binding curve
With no size limitations of the interacting molecules, this technique makes it possible to study interactions between GTs and their substrates without requiring much sample volume. Here we optimized and validated MST to facilitate high throughput screening for GT substrates. We expressed GTs as YFP-fusion proteins in tobacco (Nicotiana benthamiana) and optimized the MST method to allow the determination of substrate binding affinity, without purification of the target protein from the cell lysate. The application of this MST method to the β-1,4-galactosyltransferase AtGALS1 validated the capability to screen nucleotide-sugar substrates as well as acceptor substrates. We also expanded the application to the identification of GTs in sorghum, which will enable selection of candidates for further studies and engineering.
Results
MST assay optimization of reaction buffer formulation
To determine the best buffer formulation for analyzing GT-substrate interaction, several buffers and non-ionic detergents were pre-tested based on the produced florescence counts and thermograph profiles during thermophoresis. The well-characterized Arabidopsis thaliana β‐1,4‐galactan synthase 1 (AtGALS1), which catalyzes the elongation of pectic β‐1,4‐galactan chains using UDP-Gal as donor substrate [25], was used to test the buffer formulation and conditions. While N-terminal tags may in some cases interfere with targeting, YFP-AtGALS1 is known to be efficiently targeted to Golgi and we therefore used this well-characterized construct [25]. Since divalent cations are commonly required for GT function, and are known to be essential for AtGALS1 activity [26], all the buffers tested contained 5 mM Mn2+. Microsomes extracted from N. benthamiana leaves transiently expressing YFP-AtGALS1 were used as a source of AtGALS1 protein. Extracted microsomes were diluted to reach an optimal fluorescence level between 200 and 1600 units of fluorescence (F1 units). The final concentration of the total microsomal protein was kept constant at 1.5 mg/ml. The concentration of AtGALS1 is not known, but assuming that it may constitute 1% of total microsomal protein, the final concentration would be in the order of 250 nM. As shown for YFP-AtGALS1, MES or HEPES buffer with 1% Triton X100 provided the highest fluorescent counts (Table 1), allowing further dilution of the AtGALS1-containing microsomes if necessary. The MES buffer also presented a smooth curve with no indication of protein aggregation that can hinder the interpretation of measurements (Table 1, Additional file 1: Figure S1). A buffer with Tween20 with concentrations ranging from 0.05 to 2% all yielded much lower fluorescence counts and signs of aggregation compared to that using a Triton X100-based buffer, suggesting Triton X100 serves as better detergent in this case (Table 1, Additional file 1: Figure S1). Since aggregation can also be caused by too low of a salt concentration and resulting charge-to-charge interaction of biomolecules, adding 125 mM KCl to the MES buffer was also tested. However, this also resulted in thermograph profile suggesting protein aggregation (Table 1, Additional file 1: Figure S1).
Table 1.
Fluorescence counts and aggregation status of YFP-AtGALS1 microsomes in different incubation buffers
| Buffer composition | Fluorescence counts | Aggregation observed |
|---|---|---|
| HEPES pH 7.0, 50 mM NaCl, 5 mM MnCl2, 1% Tx100 | 1247 | (+) |
| PBS pH 7.4, 5 mM MnCl2, 0.05% Tween | 479 | − |
| MES pH 6.5, 5 mM MnCl2, 125 mM KCl, 1% Tx100 | 974 | + |
| MES pH 6.5, 5 mM MnCl2, 1% Tx100 | 1263 | − |
| MES pH 6.5, 5 mM MnCl2, 0.05% Tween | 101 | ++ |
| MES pH 6.5, 5 mM MnCl2, 0.5% Tween | 174 | + |
| MES pH 6.5, 5 mM MnCl2, 2% Tween | 272 | − |
MES buffer with 1% Triton X-100 provides highest fluorescence counts and did not produce bumpy thermograph curve as indication of protein aggregation. Tween as detergent with concentration from 0.05 to 2% did not work well with MES. Thermographs corresponding to the different conditions are shown in Additional file 1: Figure S1
Method validation with YFP-AtGALS1 binding its substrate UDP-Gal
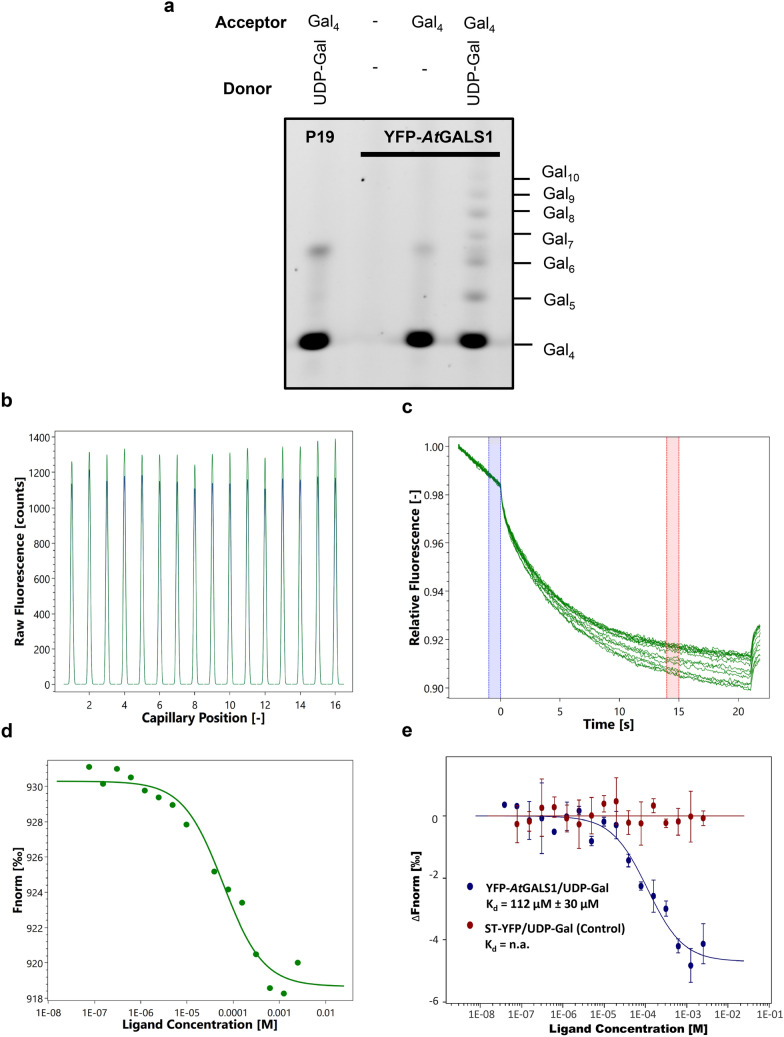
To analyze GT-substrate interaction using MST, AtGALS1 was tested for binding to its substrate UDP-Gal. To first validate that YFP-AtGALS1-containing microsomes retained the native protein confirmation and activity of AtGALS1, we employed Polysaccharide Analysis using Carbohydrate gel Electrophoresis (PACE) [27] to detect the enzymatic activity. A PACE assay using Gal4 substrate conjugated with the fluorophore ANTS (8-amino-naphthalene-1,3,6-trisulfonic acid) [28] in the presence of 200 μM UDP‐Gal as donor sugar displayed elongation activity, and produced a galactan chain with maximum detected degree of polymerization (DP) of approximately 11 (Fig. 2a). A control with microsomes from leaves expressing p19 alone revealed no activity.
Fig. 2.
Substrate binding assay of AtGALS1. a Activity analysis of AtGASL1 using carbohydrate gel electrophoresis (PACE). Incubation of YFP-AtGALS1 microsomes and ANTS-labeled β-1,4-galactotetraose (Gal4) in the presence and absence of 200 μM UDP-Gal displayed activity of catalyzing the elongation of galactan backbone. No activity was observed in the p19 control. b Initial fluorescence counts of YFP-AtGALS1 microsomes in MES buffer at different concentrations of UDP-Gal. The variation in fluorescence across the concentration gradients is within the tolerance range (± 10%). c Thermographs of YFP-AtGALS1 binding to UDP-Gal provide well-defined curves. The blue region at 0 s indicates cold spot before the temperature gradient was applied, and the red region at 15 s shows the hot spot during the thermophoresis. d Dose–response curve for the binding interaction between YFP-AtGALS1 and UDP-Gal by plotting Fnorm against the ligand concentration. The binding curve yields a Kd of 101 μM. e Normalized binding curve of YFP-AtGALS1 and ST-YFP control in presence of UDP-Gal. The binding curve yields a Kd of 112 ± 30 μM. Concentration of YFP-AtGALS1 or ST-YFP were kept constant while the UDP-Gal concentration varied from 2.5 mM to 76.3 nM. Data were fitted to Kd model and confidence interval of the Kd is calculated from the variance of the fitted parameter, derived from Levenberg–Marquardt fit-algorithm. Error Bar: SD, n = 3. Values are the average ± 68% confidence
With an activity-validated microsome preparation, we proceeded to use MST to detect the interaction of AtGALS1 with UDP-Gal. Based on the initial pretest of optimal buffers, MES buffer with 1% Triton X100 was selected to perform the binding affinity analysis. The measurements were performed at 22 °C in standard capillaries with 2% excitation power and 40% MST power. The initial fluorescence was uniform among the sixteen capillaries within a 10% tolerance threshold from the average (Fig. 2b). The thermographs showed no signs of aggregation or molecule adsorption to the capillary (Fig. 2c). The smooth binding curve relating the substrate concentration to the normalized fluorescence could be fitted based on a Kd model (Fig. 2d). Each point represents the mean of three sets of measurements. The binding of YFP-AtGALS1 with UDP-Gal in MES buffer had an affinity of 112 ± 30 μM (± 68% confidence) (Fig. 2e), compared with data using Golgi-associated rat sialyltransferase ST-YFP [29], which served as a control protein and showed no significant binding with UDP-Gal. This data is in good agreement with the Km of 142 ± 30 μM (± SE) measured by activity assay [28]. Km and Kd are related as Km = Kd + kcat/k1, based on standard Michaelis–Menten reactions, and often Km is close to Kd because kcat is low compared to k-1 [30].
Analysis of substrates specificity of AtGAL1
AtGALS1 is reported as bifunctional, catalyzing the addition of galactose with UDP-Gal as substrate as well as termination of galactan chains by transferring arabinopyranose from UDP‐β‐l‐Arap. However, UDP‐Gal is the preferred substrate with a tenfold lower Km [28]. To further screen the substrate specificity of AtGALS1, we applied the optimized protocol to detect interaction of AtGALS1 with nucleotide moiety and multiple nucleotide sugar donors. The interaction of AtGALS1 with UDP‐Arap resulted in a calculated Kd of 0.96 ± 0.51 mM (± 68% confidence) (Fig. 3a), consistent with the Km of 1.1 ± 0.2 mM previously reported [28]. Interestingly, UDP interacted with AtGALS1 with a Kd of 165 ± 30 μM (± 68% confidence) (Fig. 3b). Since UDP is a product of the enzymatic reaction, it would appear that the reaction would produce a competitive inhibitor. However, in vivo the UDP concentration is kept low by the action of nucleoside diphosphatases [31]. No binding was detected with UDP-Araf or UDP-Xyl, indicating they cannot serve as substrates for AtGALS1 (Additional file 1: Figure S2). These results underline the ability to screen the donor substrates of GTs in complex samples such as crude microsomes without purification.
Fig. 3.
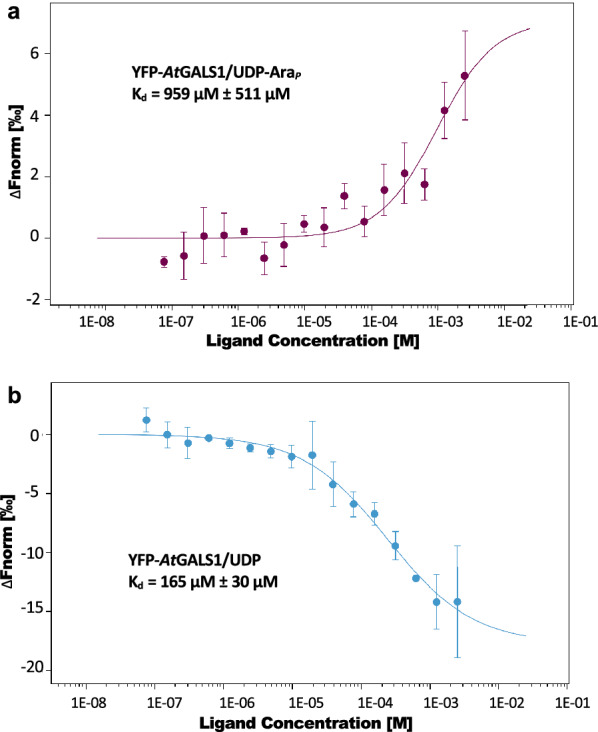
Substrate specificity assay of AtGALS1. Dose–response curve for the binding interaction between YFP-AtGALS1 and a UDP-Araf, or b UDP. Concentration of YFP-AtGALS1 was kept constant, while the ligand varied from 2.5 mM to 76.3 nM. Data were fitted to Kd model and confidence interval of the Kd is calculated from the variance of the fitted parameter, derived from Levenberg–Marquardt fit-algorithm. Error Bar: SD, n = 3. Values are the average ± 68% confidence
Acceptor binding preference of AtGALS1
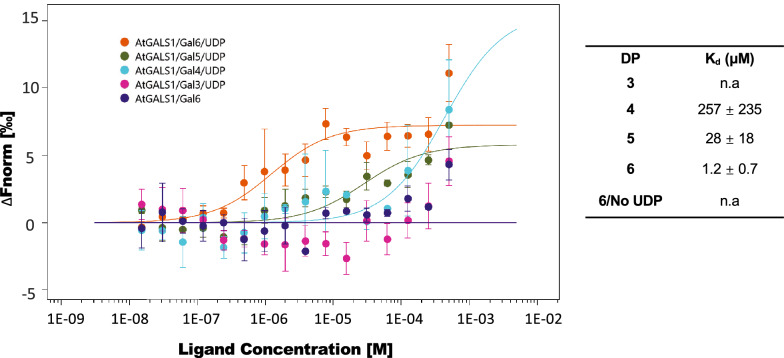
To test whether MST can also be used to detect acceptor binding of GTs, galactan substrates of different DP in the presence or absence of UDP were used for studying interaction with AtGALS1. As evidenced in the results shown before (Fig. 3b), UDP can bind to the GT, and this would be expected to induce a conformational change that enables subsequent binding of the acceptor substrate [32]. The MST results showed that galactooligosaccharides longer than three residues can bind to AtGALS1, and longer acceptors up to six residues have higher preferences, with higher binding affinities (Fig. 4). This result is consistent with the activity preference of AtGALS1 for galactooligosaccharide substrates with a minimum of four galactose units and increasing activity with longer substrates (Gal4‐6) [28]. No binding of Gal6 could be detected in the absence of UDP.
Fig. 4.
Acceptor binding assay of AtGASL1 with β-1,4-galactooligosaccharides Gal3-6. AtGALS1 expressed in N. benthamiana showed increasing binding affinity from DP4 to DP6 upon addition of UDP and no binding was detected without UDP. Gal3 did not bind
Substrate screening with selected sorghum GT61
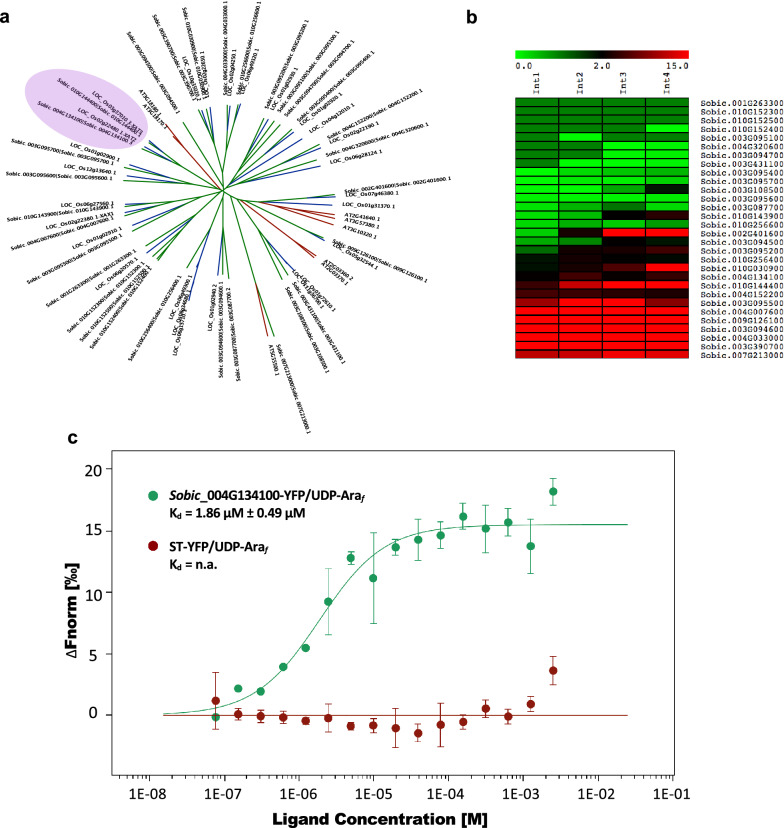
Sorghum (Sorghum bicolor) has emerged as a promising target for lignocellulosic biofuel production, but very little is known about the identities and functions of sorghum GTs. With a bioinformatics approach, we performed genome-wide identification of GT genes in sorghum based on the presence of the corresponding Pfam domains. The GT61 family is very large in grasses compared to dicots, and is known to include several members involved in decorating the xylan backbone [33, 34]. Since xylan is the predominant non-cellulosic polysaccharide in biomass, we were particularly interested in the GT61 family. To facilitate downstream biochemical and functional analysis, phylogenomic analysis was performed to prioritize candidates for in-depth characterization. In Fig. 5a, the phylogenetic clade highlighted in purple showed sorghum orthologs of rice OsXAT2 which have potential roles in arabinoxylan biosynthesis [34]. The expression patterns of these genes also suggested their roles in cell wall biosynthesis (Fig. 5b). Heterologous expression of OsXAT2 in Arabidopsis leads to arabinosylation of xylan, suggesting gain-of-function evidence for α-(1,3)-arabinosyltransferase activity [34]. However, direct biochemical evidence of the activity is still lacking. Here we selected one representative sorghum ortholog of OsXAT2, Sobic_004G134100, for substrate screening using MST. A binding curve of Sobic_004G134100-YFP with potential substrate UDP-Araf yielded a Kd of 1.86 μM ± 0.49 μM (± 68% confidence) while the control ST-YFP exhibited no detectable binding. The binding of UDP-Araf is much stronger than the binding of UDP-Gal to AtGALS1 as would be expected since the cellular concentrations of UDP-Araf in leaves is about 30-fold lower than that of UDP-Gal [35]. Arabidopsis UDP-Araf transporters that transport UDP-Araf from the cytosolic compartment to the Golgi lumen have Kms in the 7-10 µM range [35]. UDP-Xyl, UDP-Arap and UDP-GlcA did not exhibit significant binding (Additional file 1: Figures S3 and S4). UDP sugar preparations often contain some free UDP and the very weak binding of UDP-GlcA (Kd > 0.1 M) could indicate binding of a small amount of free UDP in the sample rather than binding of UDP-GlcA. These results provide supporting evidence to predict the sorghum ortholog of OsXAT2 as an arabinofuranosyltransferase.
Fig. 5.
Substrate testing of sorghum GT61. a Combined phylogenetic tree of rice, A. thaliana and sorghum GT61 proteins. Purple circle highlights the clade containing OsXAT orthologs in sorghum. b Heatmap showing expression patterns of sorghum GT61 genes in growing internodes of sorghum. c Dose–response curve for the binding interaction between sorghum Sobic_004g134100 and UDP-Araf. Error Bar: SD, n = 3. Values are the average ± 68% confidence
Discussion
The identification and characterization of key enzymes associated with cell wall biosynthesis and modification is fundamental to gaining insights into cell wall dynamics. However, it is challenging that activity assays of GTs are very low throughput and acceptor substrates are generally not available. In this work we optimized and validated MST using the well-characterized enzyme AtGASL1, and we also developed an analysis flow that uses MST-based strategies to screen potential substrates for novel GTs. We confirmed that in the presence of the appropriate nucleoside diphosphate, acceptor substrates can be screened in a similar way to donor substrates. In contrast to activity screens which must use two substrates, MST allows individual substrate screening to determine likely donors/acceptors of GTs. Furthermore, the detection of binding is a universal method, unlike activity assays which generally require different methods to detect activity depending on the product formed. Compared to other competitive techniques, such as saturation transfer difference nuclear magnetic resonance (NMR) [36], mass spectrometry (MS) [37] and isothermal titration calorimetry (ITC), MST is more straightforward, less time-consuming, and possesses high-throughput potential. A drawback is the relatively high cost of capillaries of about $12 per assay with 16 capillaries. Furthermore, in this assay, we expressed GTs as YFP-fusion proteins in N. benthamiana and optimized the MST method to allow the determination of substrate binding affinity without purification of the target protein from the cell lysate or heterologous expression system. This overcomes the obstacles of low quantities of native GTs in plants or inappropriate protein modification in a heterologous expression system, and at the same time causes no loss of enzymatic activity.
Thanks to the availability of both bioinformatics tools and full sequences of the sorghum genome, potential gene candidates that encode putative GTs for wall polysaccharide biosynthesis have been identified. As a proof of concept for the MST method we show that a sorghum GT from family GT61 can be identified as a likely arabinofuranosyltransferase, probably involved in xylan biosynthesis.
Conclusions
In summary, our study optimized and validated MST to facilitate high throughput screening for GT substrates. A transient system in N. benthamiana was used which enables easy expression of fluorescently labeled protein. The filter combinations of the MST instrument determine which fluorophore is most suitable. The blue filter we used is suitable for GFP and YFP, and the green filter would be suitable for red fluorescing fluorophores. Expression in a plant system is an advantage since any necessary modification, e.g. N- and O-glycosylation, is likely to take place normally. The MST method was optimized to allow the determination of substrate binding affinity with crude microsomes, without purification of the target protein from the cell lysate. The buffer conditions we recommend should work well for Golgi-localized GTs. The pH of 6.5 of the recommended buffer is close to the pH of the Golgi in plants, and should also work for cytoplasm, ER, and plastid stroma, although a pH closer to 7 might be more optimal for those compartments [38]. We have only tested the method with several Golgi-localized GTs from plants, but we find it likely that good MST signal should be achievable for most GTs without further optimization. The MST method validated the interaction of UDP-Gal to the β-1,4-galactosyltransferase AtGALS1, and showed consistent binding affinity as reported from activity assays. MST is also sensitive to capture the rare substrate of AtGALS1 as UDP-Arap which is needed for termination of galactan and showed a tenfold lower affinity than UDP-Gal. The method of substrate screening we developed can be adapted to any plant species and is obviously not limited to GTs involved in cell wall biosynthesis. Obviously, binding of a nucleotide sugar does not prove that it is a substrate, but strong binding of non-substrates is unlikely to be common since that would cause non-productive competitive inhibition. Some GTs may not be entirely specific, as seen for example for AtGALS1, which has low but biologically relevant activity with UDP-Arap and binds this substrate at relatively high but still physiologically meaningful concentrations. Therefore, binding data should be interpreted with caution, but a nucleotide sugar that does not bind at a physiologically relevant concentration is highly unlikely to be a substrate in vivo.
MST, as a rapid and sensitive method to screen the substrates of GTs, will greatly facilitate selection of candidates for further studies and engineering. The method will also be powerful in studying how interaction with other proteins, posttranslational modifications, allosteric regulators, or competitive inhibitors can affect the enzyme activity through altered substrate binding.
Methods
Chemicals
UDP-Gal and UDP-Xyl were of analytical grade and purchased from Sigma‐Aldrich. UDP‐β‐l‐Arap was purchased from Carbosource Service (Complex Carbohydrate Research Center, Athens, GA, USA) and and UDP‐β‐l‐Araf was purchased from Peptides International (Louisville, KY, USA). Galactan substrates Galn (n = 3–6) were chemically synthesized as previously described [39].
Plasmid construction
cDNA of Sobic_004G134100 was amplified by PCR using primers as follows: F: 5′-CACCATGAAGGCGGTGGAG-3′ and R: 5′-TTGGTTCAATTGATCAAGAGCC-3′. A purified fragment was cloned into pENTR/D/TOPO entry vectors through GATEWAY LR reactions (Invitrogen), following the manufacturer’s protocol. AtGALS1 and rat ST expression constructs were described previously [25, 40].
To generate transient protein expression constructs in N. benthamiana, the pENTR/D vector containing coding sequence of Sobic_004G134100 were recombined into pEarleyGate 101 [41] using LR Clonase II (Invitrogen). The binary vectors were electro-transformed into Agrobacterium tumefaciens strain GV3101::pMP90 for tobacco leaf infiltration.
Transient expression in Nicotiana benthamiana
Agrobacterium tumefaciens strains harboring binary vectors were cultured in 10 ml LB medium overnight and harvested at 4500 rpm for 10 min. Concentrated cells were then washed by 10 ml of 10 mM MgCl2 twice and left at room temperature for 3 h prior to infiltration. Equal volumes of cell culture for expression of the gene of interest and cell culture expressing the p19 protein, which suppresses gene silencing [42] were mixed with an OD600 = 0.5 and infiltrated into the 4-week-old tobacco expanding leaves. The leaves were harvested 4 days after the infiltration for microsomal membrane preparation.
Microsome extraction
Microsomal membranes were prepared according to the protocol previously described [43]. The N. benthamiana leaves transiently expressing AtGASL1 or Sobic_004G134100 were ground in microsomal extraction buffer (50 mM HEPES‐KOH pH 7.0, 400 mM sucrose, 20 mM sodium ascorbate, 1 mM phenylmethylsulfonyl fluoride, 1% w/v polyvinylpolypyrrolidone). The Miracloth-filtered suspension was centrifugated at 3000×g for 10 min to remove cell debris. The supernatant was collected and centrifugated at 100,000×g for 1 h to isolate membranes. The precipitated pellet was then resuspended in buffer (50 mM HEPES‐KOH pH 7.0, 400 mM sucrose) and aliquots were flash‐frozen in liquid nitrogen and stored at − 80 °C until use. All above steps were performed at 4 °C. Total protein was measured by Coomassie Blue (Bio-Rad) assay based on bovine serum albumin standard.
ANTS‐labeling and analysis of labeled galactan substrates
Each galactan oligosaccharide (200 μg) was reductively aminated with ANTS (Invitrogen) as follows: speedvac-dried oligosaccharides were resuspended in 5 μl of 0.2 M ANTS solution (in H2O:acetic acid = 17:3), 10 μl of dimethylsulfoxide (DMSO) buffer (30 μl acetic acid, 170 μl H2O, 200 μl DMSO) and 5 μl of 0.2 M 2‐picoline borane (in DMSO). After overnight incubation at 37 °C, samples were dried in a speedvac and resuspended in 100 μl H2O, as described in Mortimer et al. [44].
PACE
Reactions were performed in a total of 25 μl containing MnCl2 (10 mm), Triton X‐100 (1% v/v) in buffer (50 mm MES, pH 6.5), 2 μg galactooligosaccharide substrate, 200 µM UDP‐Gal and microsomal membranes (50 μg total protein), as previously described [28]. Reactions were incubated at 30 °C for 2 h and then terminated by heating at 100 °C for 5 min, followed by centrifugation at 10,000×g for 10 min. Supernatants (15 µl) were mixed with 15 μl 3 m urea, and 5 μl samples were loaded on large format Tris‐borate acrylamide gel prepared as described previously [27], and electrophoresed at 200 V for 30 min followed by 1000 V for 1.5 h. The PACE gels were visualized with Syngene G‐box at Tumi-wavelength with a UV detection filter and long‐wave UV tubes (365 nm emission).
MST buffer optimization
Various buffer solutions were tested to determine the best buffer composition based on stability and homogeneity within capillaries: HEPES buffer (50 mM HEPES pH 7.0; 5 mM MnCl2; 50 mM NaCl, 1% v/v Triton X-100), MES buffer (20 mM MES pH 6.5; 5 mM MnCl2, 1% v/v Triton X-100), MES buffer with extra KCl (20 mM MES pH 6.5; 5 mM MnCl2; 125 mM KCl, 1% v/v Triton X-100), MES buffer with Tween (20 mM MES pH 6.5; 5 mM MnCl2; 125 mM KCl, 0.05–2% v/v Tween 20) and PBS buffer (pH 7.4; 137 mM NaCl; 2.7 mM KCl; 10 mM Na2HPO4; 1.8 mM K2HPO4; 5 mM MnCl2; 0.05% Tween 20).
MST experiments were performed on a NanoTemper ® Monolith NT.115 (NanoTemper Technologies, Germany) with blue/red filters. Samples were diluted 200X in different buffers listed above, and final concentration yield detectable fluorescent signals, between 200 and 1600 units of fluorescence (FI units). Prepared samples were loaded into standard treated capillaries for measurements using 40% MST power with laser off/on times of 0 s and 20 s, respectively, at 22 °C.
Binding assay
To test the binding affinity between AtGASL1 and UDP-Gal, microsomes containing YFP-AtGASL1 were diluted in MES buffer (1% Tx100) to a final concentration of 1.5 mg/ml total protein. Diluted microsomes were centrifuged at 20,000×g for 10 min to remove potential protein aggregates. 10 µl of 5 mM UDP-Gal solution was diluted 1:1 in 10 µl buffer to make a 16-sample serial dilution from 2.5 mM to 76.3 nM. 10 µl of prepared microsome was then added to 10 µl of each ligand solution and incubated at room temperature for 10 min. For the acceptor binding between YFP-AtGASL1 and Gal3-6, similar serial dilution of galactan substrates was prepared using a 1:1 dilution and the final concentration range from 0.5 mM to 15.3 nM, with the other parameters remaining the same. 2% excitation power and 40% MST power with laser off/on times of 0 s and 20 s were used in all MST experiments. All experiments were repeated three times for each measurement. Data analyses were performed using the MO.Affinity Analysis software (version 2.3, NanoTemper Technologies).
The binding constant Kd was calculated according to the protocol of NanoTemper Technologies. The fluorescence change in MST signal is normalized (Fnorm), defined as Fhot/Fcold (Fhot as the hot region at 20 s after IR laser heating and Fcold as the cold region at 0 s). A dose–response curve is plotted as Fnorm against the ligand concentration. The Kd constants between a protein and its substrate was calculated using the saturation binding curve at equilibrium [45]. The fitting function is derived from the law of mass action:
| 1 |
unbound is the response value of unbound state; bound is the response value of bound state; [Ptot] is the total protein concentration; [Ltot] is the total ligand concentration; Kd is the dissociation constant
Sequence retrieval and phylogenetic analysis
The Hidden Markov Model (HMM) profile for GT61 proteins (PF04577) was downloaded from Pfam Database (http://pfam.xfam.org/) and used as a query to search against sorghum proteome available at Phytozome (http://www.phytozome.net/) using HMMER V3.1b1 (http://hmmer.org/) with default parameters. The information about GT61 proteins of rice and Arabidopsis was obtained from the previously published studies [33, 46]. Corresponding rice and Arabidopsis protein sequences were retrieved from rice genome annotation project database (http://rice.plantbiology.msu.edu/) and The Arabidopsis Information Resource (TAIR) database (https://www.arabidopsis.org/), respectively. GT61 proteins of sorghum, rice and Arabidopsis were aligned using ClustalX [47] and a neighbor-joining phylogenetic tree was generated using default parameters.
Expression analysis
For expression analysis of GT61 genes, we used publicly available RNA Seq-based expression data corresponding to four internodes of Sorghum [48]. The normalized expression was downloaded from NCBI-GEO (accession number GSE98817) and expression values for GT61 genes in sorghum was extracted. The expression heatmap was generated using MeV microarray data analysis platform (http://www.tm4.org/mev/).
Supplementary information
Additional file 1: Figure S1. Thermographs corresponding to the different buffer conditions shown in Table 1. Figure S2.AtGALS1 expressed in Nicotiana benthamiana has no detectable binding of UDP-Araf. Figure S3.Sobic_004G134100 AtGALS1 has no detectable binding of UDP-Arap or UDP-Xyl. Figure S4.AtGALS1 has very weak interaction with UDP-GlcA at physiologically irrelevant concentrations.
Acknowledgements
The authors thank Ms. Nurgul Kaplan at the Joint BioEnergy Institute for help in applying automated liquid handling technique for sample preparations, Dr. Mi Yeon Lee for help with growing plants, Dr. Venkataramana Pidatala for valuable instructions on ANTS-labeling and PACE, and Ms. Leah Sloan for help in editing the manuscript.
Abbreviations
- GT
Glycosyltransferase
- MST
Microscale thermophoresis
- DP
Degree of polymerization
- ANTS
8-Amino-naphthalene-1,3,6-trisulfonic acid
Authors’ contributions
WS designed and performed experiments, analyzed data, prepared figures, and cowrote the manuscript. RS performed bioinformatic analysis of sorghum glycosyltransferase family GT61. MHC synthesized the galactan substrates Galn (n = 3–7) for acceptor binding assay. HVS designed experiments, supervised the study, provided financial support, and cowrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was funded by the DOE Joint BioEnergy Institute (http://www.jbei.org) supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research through Contract DEAC0205CH11231 between Lawrence Berkeley National Laboratory and the U.S. Department of Energy. RS was supported by a GETin fellowship from Indo-US Science and Technology Forum.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its additional files, or are available from the corresponding authors per request. All constructs are available via the JBEI registry (https://registry.jbei.org/).
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13007-020-00641-1.
References
- 1.Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, et al., editors. Essentials of glycobiology. 2. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2009. [PubMed] [Google Scholar]
- 2.Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 3.Thibodeaux CJ, Melançon CE, Liu H. Natural-product sugar biosynthesis and enzymatic glycodiversification. Angew Chem Int Ed Engl. 2008;47:9814–9859. doi: 10.1002/anie.200801204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hansen SF, Harholt J, Oikawa A, Scheller HV. Plant glycosyltransferases beyond cazy: a perspective on DUF families. Front Plant Sci. 2012;3:59. doi: 10.3389/fpls.2012.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu ZL, Ethen CM, Prather B, Machacek M, Jiang W. Universal phosphatase-coupled glycosyltransferase assay. Glycobiology. 2011;21:727–733. doi: 10.1093/glycob/cwq187. [DOI] [PubMed] [Google Scholar]
- 6.McGraphery K, Schwab W. Comparative analysis of high-throughput assays of family-1 plant glycosyltransferases. Int J Mol Sci. 2020 doi: 10.3390/ijms21062208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voxeur A, Höfte H. Cell wall integrity signaling in plants: “to grow or not to grow that’s the question”. Glycobiology. 2016;26:950–960. doi: 10.1093/glycob/cww029. [DOI] [PubMed] [Google Scholar]
- 8.Cosgrove DJ. Plant cell wall extensibility: connecting plant cell growth with cell wall structure, mechanics, and the action of wall-modifying enzymes. J Exp Bot. 2016;67:463–476. doi: 10.1093/jxb/erv511. [DOI] [PubMed] [Google Scholar]
- 9.Drakakaki G. Polysaccharide deposition during cytokinesis: challenges and future perspectives. Plant Sci. 2015;236:177–184. doi: 10.1016/j.plantsci.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 10.Cosgrove DJ. Catalysts of plant cell wall loosening. [version 1; peer review: 2 approved] F1000Res. 2016 doi: 10.12688/f1000research.7180.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim S-J, Brandizzi F. The plant secretory pathway: an essential factory for building the plant cell wall. Plant Cell Physiol. 2014;55:687–693. doi: 10.1093/pcp/pct197. [DOI] [PubMed] [Google Scholar]
- 12.Sandhu APS, Randhawa GS, Dhugga KS. Plant cell wall matrix polysaccharide biosynthesis. Mol Plant. 2009;2:840–850. doi: 10.1093/mp/ssp056. [DOI] [PubMed] [Google Scholar]
- 13.Fukuda M, Bierhuizen MFA, Nakayama J. Expression cloning of glycosyltransferases. Glycobiology. 1996;6:683–689. doi: 10.1093/glycob/6.7.683. [DOI] [PubMed] [Google Scholar]
- 14.Welner DH, Shin D, Tomaleri GP, De Giovanni AM, Tsai AYL, Tran HM, et al. Plant cell wall glycosyltransferases: high-throughput recombinant expression screening and general requirements for these challenging enzymes. PLoS ONE. 2017;12:e0177591. doi: 10.1371/journal.pone.0177591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Culbertson AT, Chou Y-H, Smith AL, Young ZT, Tietze AA, Cottaz S, et al. Enzymatic activity of xyloglucan xylosyltransferase 5. Plant Physiol. 2016;171:1893–1904. doi: 10.1104/pp.16.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaur J, Kumar A, Kaur J. Strategies for optimization of heterologous protein expression in E. coli: roadblocks and reinforcements. Int J Biol Macromol. 2018;106:803–822. doi: 10.1016/j.ijbiomac.2017.08.080. [DOI] [PubMed] [Google Scholar]
- 17.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42(Database issue):D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Atmodjo MA, Sakuragi Y, Zhu X, Burrell AJ, Mohanty SS, Atwood JA, et al. Galacturonosyltransferase (GAUT)1 and GAUT7 are the core of a plant cell wall pectin biosynthetic homogalacturonan:galacturonosyltransferase complex. Proc Natl Acad Sci USA. 2011;108:20225–20230. doi: 10.1073/pnas.1112816108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ren Y, Hansen SF, Ebert B, Lau J, Scheller HV. Site-directed mutagenesis of IRX9, IRX9L and IRX14 proteins involved in xylan biosynthesis: glycosyltransferase activity is not required for IRX9 function in Arabidopsis. PLoS ONE. 2014;9:e105014. doi: 10.1371/journal.pone.0105014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jozwiak A, Sonawane PD, Panda S, Garagounis C, Papadopoulou KK, Abebie B, et al. Plant terpenoid metabolism co-opts a component of the cell wall biosynthesis machinery. Nat Chem Biol. 2020 doi: 10.1038/s41589-020-0541-x. [DOI] [PubMed] [Google Scholar]
- 21.Seidel SAI, Dijkman PM, Lea WA, van den Bogaart G, Jerabek-Willemsen M, Lazic A, et al. Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods. 2013;59:301–315. doi: 10.1016/j.ymeth.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jerabek-Willemsen M, Wienken CJ, Braun D, Baaske P, Duhr S. Molecular interaction studies using microscale thermophoresis. Assay Drug Dev Technol. 2011;9:342–353. doi: 10.1089/adt.2011.0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jung H, Gusev VE, Baek H, Wang Y, Diebold GJ. Ludwig-Soret effect in a linear temperature field: theory and experiments for steady state distributions. Phys Lett A. 2011;375:1917–1920. doi: 10.1016/j.physleta.2011.03.047. [DOI] [Google Scholar]
- 24.Duhr S, Braun D. Why molecules move along a temperature gradient. Proc Natl Acad Sci USA. 2006;103:19678–19682. doi: 10.1073/pnas.0603873103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liwanag AJM, Ebert B, Verhertbruggen Y, Rennie EA, Rautengarten C, Oikawa A, et al. Pectin biosynthesis: GALS1 in Arabidopsis thaliana is a β-1,4-galactan β-1,4-galactosyltransferase. Plant Cell. 2012;24:5024–5036. doi: 10.1105/tpc.112.106625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ebert B, Birdseye D, Liwanag AJM, Laursen T, Rennie EA, Guo X, et al. The three members of the arabidopsis glycosyltransferase family 92 are functional β-1,4-galactan synthases. Plant Cell Physiol. 2018;59:2624–2636. doi: 10.1093/pcp/pcy180. [DOI] [PubMed] [Google Scholar]
- 27.Goubet F, Jackson P, Deery MJ, Dupree P. Polysaccharide analysis using carbohydrate gel electrophoresis: a method to study plant cell wall polysaccharides and polysaccharide hydrolases. Anal Biochem. 2002;300:53–68. doi: 10.1006/abio.2001.5444. [DOI] [PubMed] [Google Scholar]
- 28.Laursen T, Stonebloom SH, Pidatala VR, Birdseye DS, Clausen MH, Mortimer JC, et al. Bifunctional glycosyltransferases catalyze both extension and termination of pectic galactan oligosaccharides. Plant J. 2018;94:340–351. doi: 10.1111/tpj.13860. [DOI] [PubMed] [Google Scholar]
- 29.Teh O-K, Moore I. An ARF-GEF acting at the Golgi and in selective endocytosis in polarized plant cells. Nature. 2007;448:493–496. doi: 10.1038/nature06023. [DOI] [PubMed] [Google Scholar]
- 30.Robinson PK. Enzymes: principles and biotechnological applications. Essays Biochem. 2015;59:1–41. doi: 10.1042/bse0590001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiu T-Y, Christiansen K, Moreno I, Lao J, Loqué D, Orellana A, et al. AtAPY1 and AtAPY2 function as Golgi-localized nucleoside diphosphatases in Arabidopsis thaliana. Plant Cell Physiol. 2012;53:1913–1925. doi: 10.1093/pcp/pcs131. [DOI] [PubMed] [Google Scholar]
- 32.Albesa-Jové D, Guerin ME. The conformational plasticity of glycosyltransferases. Curr Opin Struct Biol. 2016;40:23–32. doi: 10.1016/j.sbi.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 33.Chiniquy D, Sharma V, Schultink A, Baidoo EE, Rautengarten C, Cheng K, et al. XAX1 from glycosyltransferase family 61 mediates xylosyltransfer to rice xylan. Proc Natl Acad Sci USA. 2012;109:17117–17122. doi: 10.1073/pnas.1202079109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anders N, Wilkinson MD, Lovegrove A, Freeman J, Tryfona T, Pellny TK, et al. Glycosyl transferases in family 61 mediate arabinofuranosyl transfer onto xylan in grasses. Proc Natl Acad Sci USA. 2012;109:989–993. doi: 10.1073/pnas.1115858109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rautengarten C, Ebert B, Moreno I, Temple H, Herter T, Link B, et al. The Golgi localized bifunctional UDP-rhamnose/UDP-galactose transporter family of Arabidopsis. Proc Natl Acad Sci USA. 2014;111:11563–11568. doi: 10.1073/pnas.1406073111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blume A, Angulo J, Biet T, Peters H, Benie AJ, Palcic M, et al. Fragment-based screening of the donor substrate specificity of human blood group B galactosyltransferase using saturation transfer difference NMR. J Biol Chem. 2006;281:32728–32740. doi: 10.1074/jbc.M600424200. [DOI] [PubMed] [Google Scholar]
- 37.Ishii K, Noda M, Uchiyama S. Mass spectrometric analysis of protein-ligand interactions. Biophys Physicobiol. 2016;13:87–95. doi: 10.2142/biophysico.13.0_87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen J, Zeng Y, Zhuang X, Sun L, Yao X, Pimpl P, et al. Organelle pH in the Arabidopsis endomembrane system. Mol Plant. 2013;6:1419–1437. doi: 10.1093/mp/sst079. [DOI] [PubMed] [Google Scholar]
- 39.Andersen MCF, Kračun SK, Rydahl MG, Willats WGT, Clausen MH. Synthesis of β-1,4-Linked Galactan Side-Chains of Rhamnogalacturonan I. Chem Eur J. 2016;22:11543–11548. doi: 10.1002/chem.201602197. [DOI] [PubMed] [Google Scholar]
- 40.Brandizzi F, Snapp EL, Roberts AG, Lippincott-Schwartz J, Hawes C. Membrane protein transport between the endoplasmic reticulum and the Golgi in tobacco leaves is energy dependent but cytoskeleton independent: evidence from selective photobleaching. Plant Cell. 2002;14:1293–1309. doi: 10.1105/tpc.001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 42.Voinnet O, Rivas S, Mestre P, Baulcombe D. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 2003;33:949–956. doi: 10.1046/j.1365-313x.2003.01676.x. [DOI] [PubMed] [Google Scholar]
- 43.Nunan KJ, Scheller HV. Solubilization of an arabinan arabinosyltransferase activity from mung bean hypocotyls. Plant Physiol. 2003;132:331–342. doi: 10.1104/pp.102.019406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mortimer JC, Faria-Blanc N, Yu X, Tryfona T, Sorieul M, Ng YZ, et al. An unusual xylan in Arabidopsis primary cell walls is synthesised by GUX3, IRX9L, IRX10L and IRX14. Plant J. 2015;83:413–426. doi: 10.1111/tpj.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swillens S. Interpretation of binding curves obtained with high receptor concentrations: practical aid for computer analysis. Mol Pharmacol. 1995;47:1197–1203. [PubMed] [Google Scholar]
- 46.Cao P-J, Bartley LE, Jung K-H, Ronald PC. Construction of a rice glycosyltransferase phylogenomic database and identification of rice-diverged glycosyltransferases. Mol Plant. 2008;1:858–877. doi: 10.1093/mp/ssn052. [DOI] [PubMed] [Google Scholar]
- 47.Kebrom TH, McKinley B, Mullet JE. Dynamics of gene expression during development and expansion of vegetative stem internodes of bioenergy sorghum. Biotechnol Biofuels. 2017;10:159. doi: 10.1186/s13068-017-0848-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim T, Joo H. ClustalXeed: a GUI-based grid computation version for high performance and terabyte size multiple sequence alignment. BMC Bioinform. 2010;11:467. doi: 10.1186/1471-2105-11-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Thermographs corresponding to the different buffer conditions shown in Table 1. Figure S2.AtGALS1 expressed in Nicotiana benthamiana has no detectable binding of UDP-Araf. Figure S3.Sobic_004G134100 AtGALS1 has no detectable binding of UDP-Arap or UDP-Xyl. Figure S4.AtGALS1 has very weak interaction with UDP-GlcA at physiologically irrelevant concentrations.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its additional files, or are available from the corresponding authors per request. All constructs are available via the JBEI registry (https://registry.jbei.org/).