Abstract
The severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is responsible for the current COVID-19 pandemic. An unbalanced immune response, characterized by a weak production of type I interferons (IFN-Is) and an exacerbated release of proinflammatory cytokines, contributes to the severe forms of the disease. SARS-CoV-2 is genetically related to SARS-CoV and Middle East respiratory syndrome-related coronavirus (MERS-CoV), which caused outbreaks in 2003 and 2013, respectively. Although IFN treatment gave some encouraging results against SARS-CoV and MERS-CoV in animal models, its potential as a therapeutic against COVID-19 awaits validation. Here, we describe our current knowledge of the complex interplay between SARS-CoV-2 infection and the IFN system, highlighting some of the gaps that need to be filled for a better understanding of the underlying molecular mechanisms. In addition to the conserved IFN evasion strategies that are likely shared with SARS-CoV and MERS-CoV, novel counteraction mechanisms are being discovered in SARS-CoV-2–infected cells. Since the last coronavirus epidemic, we have made considerable progress in understanding the IFN-I response, including its spatiotemporal regulation and the prominent role of plasmacytoid dendritic cells (pDCs), which are the main IFN-I–producing cells. While awaiting the results of the many clinical trials that are evaluating the efficacy of IFN-I alone or in combination with antiviral molecules, we discuss the potential benefits of a well-timed IFN-I treatment and propose strategies to boost pDC-mediated IFN responses during the early stages of viral infection.
Introduction
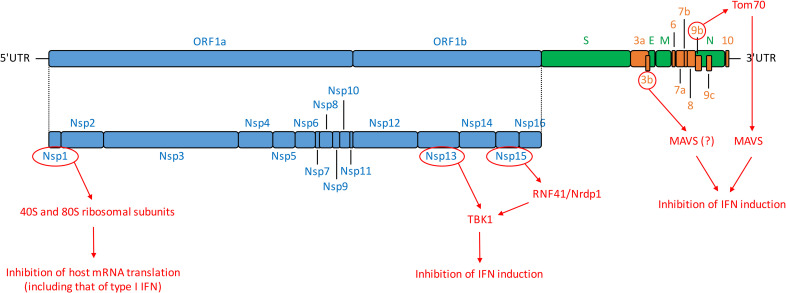
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a beta-coronavirus that emerged at the end of 2019 in China and rapidly spread around the world, causing a pandemic [1, 2]. SARS-CoV-2 infection is responsible for COVID-19, a disease associated with mild symptoms in the majority of cases but that can progress to an acute respiratory distress syndrome [1, 3]. So far (July 16th, 2020), the virus has infected more than 13 million people and caused more than 500,000 deaths worldwide. SARS-CoV-2 is genetically related to other beta-coronaviruses that have caused epidemics: SARS-CoV and MERS-CoV (for Middle East respiratory syndrome-related coronavirus), in 2003 and 2013, respectively. Beta-coronaviruses are enveloped positive-sense single-stranded RNA viruses. The 30-kb genome of SARS-CoV-2 has 82% nucleotide identity with SARS-CoV and contains at least 14 open reading frames (ORFs) [4, 5] (Fig 1). It comprises a 5′-untranslated region (5′-UTR); ORF1a/b, encoding a polyprotein proteolytically processed into 16 nonstructural proteins (Nsp1–16); structural proteins including spike (S), envelope (E), membrane (M), and nucleocapsid (N); 9 accessory proteins (ORF3a, 3b, 6, 7a, 7b, 8, 9b, 9c, and 10); and a 3′-UTR [4, 5] (Fig 1).
Fig 1. SARS-CoV-2 genomic organization and encoded proteins.
ORF1a/1b encode a polyprotein, which is proteolytically processed into Nsp1–16, represented in blue. Structural proteins, including S, E, M, and N proteins are in green. Accessory proteins encoded at the 3′ end of the viral genome comprise ORF3a, 3b, 6, 7a, 7b, 8, 9b, 9c, and 10 and are colored in orange. Untranslated extremities of the genome (5′-UTR and 3′-UTR) are also represented. In red are depicted SARS-CoV-2 proteins that interfere with IFN induction pathway as well as their known or hypothetic target [5, 37, 147]. E, envelope; IFN, interferon; M, membrane; MAVS, mitochondrial antiviral-signaling protein; N, nucleocapsid; Nrdp1, neuregulin receptor degradation protein-1; Nsp, nonstructural protein; ORF, open reading frame; RNF41, ring finger protein 41; S, spike; SARS-CoV-2, severe acute respiratory syndrome coronavirus-2; TANK, TRAF family member-associated NF-κB activator; TBK1, TANK-binding kinase 1; Tom70, translocase of outer mitochondrial membrane 70; UTR, untranslated region.
Type I interferon (IFN-I) response is critical for providing an efficient protection against viral infections. IFN-I production is rapidly triggered by the recognition by host sensors of pathogen-associated molecular patterns (PAMPs), such as viral nucleic acids [6]. IFN-I–induced signaling converges on transcription factors, which rapidly induces the expression of hundreds of genes called interferon-stimulated genes (ISGs) (reviewed in [7, 8]). This antiviral signaling cascade occurs in virtually all cell types exposed to IFN-I. ISGs, along with other downstream molecules controlled by IFN-I (including proinflammatory cytokines), have diverse functions, ranging from direct inhibition of viral replication to the recruitment and activation of various immune cells [9, 10]. A robust, well-timed, and localized IFN-I response is thus required as a first line of defense against viral infection because it promotes virus clearance, induces tissue repair, and triggers a prolonged adaptive immune response against viruses.
Like most, if not all, RNA viruses, coronavirus RNA is detected by cytosolic sensors including retinoic acid-inducible gene 1 (RIG-I/DExD/H-box helicase 58 [DDX58]) and melanoma differentiation-associated gene 5 (MDA5/IFN induced with helicase C domain 1 [IFIH1]) [11, 12]. Upon activation, RIG-I and MDA5 interact with the downstream adaptor, the mitochondrial antiviral signaling protein (MAVS, also known as IFN-B promoter stimulator 1 [IPS-1], CARD adaptor inducing IFN-beta [CARDIF], or virus-induced signaling adaptor [VISA]) on mitochondria. MAVS activation leads, via the recruitment of tumor necrosis factor receptor-associated factor 3 (TRAF3), TRAF family member-associated NF-κB activator (TANK)-binding kinase 1 (TBK1) and inhibitor of nuclear factor κB (IκB) kinase-ε (IKKε), to the phosphorylation of the IFN gene “master regulators” IFN regulatory factor (IRF)3 and IRF7. Upon phosphorylation, IRF3 and/or IRF7 dimerize and translocate into the nucleus, where they induce the expression of IFN-I and a subset of ISGs referred to as early ISGs (reviewed in [13]). Secreted IFN-I then bind to the interferon alpha and beta receptor (IFNAR, composed of the IFNAR1 and IFNAR2 subunits), leading to the activation of the Jak tyrosine kinases tyrosine kinase 2 (Tyk2) and Janus kinase 1 (JAK1), which in turn phosphorylate the signal transducer and activator of transcription (STAT)1 and STAT2 [14, 15]. Phosphorylated STATs heterodimerize and associate with the DNA binding protein IRF9 to form a complex known as IFN-stimulated growth factor 3 (ISGF3). The ISGF3 complex translocates into the nucleus and binds to interferon-stimulated response elements (ISREs) in ISG promoters, thus inducing the expression of hundreds of ISG products that establish the antiviral state at the site of viral infection [15]. The antiviral response is intensified by various signaling factors, including sensors and transcriptional regulators, which are themselves ISGs induced by ISGF3 and/or directly by the IRF3/IRF7 transcriptional activators. Aside from the IFN-I response, the recognition of double-stranded viral RNA elements by the protein kinase receptor (PKR) triggers a translational arrest in infected cells (reviewed in [8, 16, 17]). This host response is highly connected to the IFN-I response because PKR is also an ISG (reviewed in [16, 18]).
IFN-I response requires fine-tuning because its overactivation is deleterious to the host. Notably, some ISGs are involved in the regulation of cell metabolism, intracellular RNA degradation, translation arrest, and cell death, for which changes can be potentially detrimental to the host. IFN-I also potentiates the recruitment and activation of various immune cells. Thus, although a robust IFN-I response is required as a first line of defense against viral infections, systemic/uncontrolled or prolonged IFN-I production can lead to inflammatory diseases. For example, an exacerbated IFN-I response contributes to the development of autoimmune diseases [19]. COVID-19 is no exception to the rule, and it is therefore critical to understand the regulation of the IFN-I response upon infection.
SARS-CoV-2 and IFN-I response
SARS-Cov-2 is a poor inducer of IFN-I response in vitro and in animal models as compared with other respiratory RNA viruses [20, 21]. IFN-I levels in the serum of infected patients are below the detection levels of commonly used assays, yet ISG expression is detected [4, 22], thus suggesting that a limited IFN-I production could be sufficient to induce ISGs. Alternatively, IFN-I production could be restricted to specific immune cells, such as plasmacytoid dendritic cells (pDCs). Despite a more efficient replication in human lung tissues, SARS-CoV-2 induced even less IFN-I than SARS-CoV [4], which is itself a weak inducer in human cells [23–25]. An ineffective IFN-I response seems to be a hallmark of other coronavirus infections, as observed with MERS-CoV in ex vivo respiratory tissue cultures [26] and with animal coronaviruses such as the porcine epidemic diarrhea virus (PEDV) or the mouse hepatitis virus (MHV), which are alpha- and beta-coronaviruses, respectively [27, 28]. Indeed, coronaviruses have developed multiple strategies to escape and counteract innate sensing and IFN-I production.
Inhibition of IFN-I induction and signaling by SARS-CoV-2
SARS-CoV encodes at least 10 proteins that allow the virus to either escape or counteract the induction and antiviral action of IFN (Fig 2 and Table 1) [29–48]. Initial observations already suggest that the SARS-CoV-2 anti-IFN arsenal is at least as efficient as that of SARS-CoV [4, 20, 22], although detailed mechanistic studies are required to determine whether the IFN antagonists identified in other coronaviruses have equivalently competent counterparts in SARS-CoV-2. A virus–cell protein interaction map performed with 26 of the 29 SARS-CoV-2 proteins expressed in human embryonic kidney (HEK)293T identified several innate immune signaling proteins as partners of viral proteins cells (Fig 1) [5]. SARS-CoV-2 ORF9b, like SARS-CoV ORF9b, interacts with MAVS through its association with Tom70, thus suggesting a conserved mechanism of IFN-I evasion [5, 40] (Fig 1). Furthermore, SARS-CoV-2 Nsp13 and Nsp15 were found to interact with TBK1 and the TBK1 activator ring finger protein 41 (RNF41)/Nrdp1, respectively [5] (Fig 1). Nsp15, which is a highly conserved endoribonuclease encoded by various coronaviruses, including SARS-CoV [30, 49, 50], antagonizes the induction of IFN-I by cleaving the 5′-polyuridines of the negative-sense viral RNA, as demonstrated for MHV and PEDV in various cellular models [31, 32, 50] (Table 1 and Fig 2). If further validated, the interaction between SARS-CoV-2 nsp15 and TBK1 may reveal that the viral endoribonuclease antagonizes IFN induction via at least 2 mechanisms. SARS-CoV ORF3b was reported to inhibit IFN induction and to act either on IRF3 or possibly on MAVS because it translocates to mitochondria when overexpressed in Vero cells [36, 42]. Despite the fact it encodes a shorter protein than SARS-CoV, SARS-CoV-2 ORF3b was recently found to suppress IFN induction even more efficiently [51]. By screening 15,000 SARS-CoV-2 sequences, the authors identified a natural variant encoding a longer ORF3b and displaying an even greater inhibitory activity [51]. Finally, SARS-CoV-2 Nsp1 was also recently found to bind 40S ribosomal subunits (Fig 1), thus inhibiting host mRNA translation, including that of IFN-I [52], a feature that was previously demonstrated for other coronavirus-encoded Nsp1, including SARS-CoV [43, 45] (Fig 2 and Table 1).
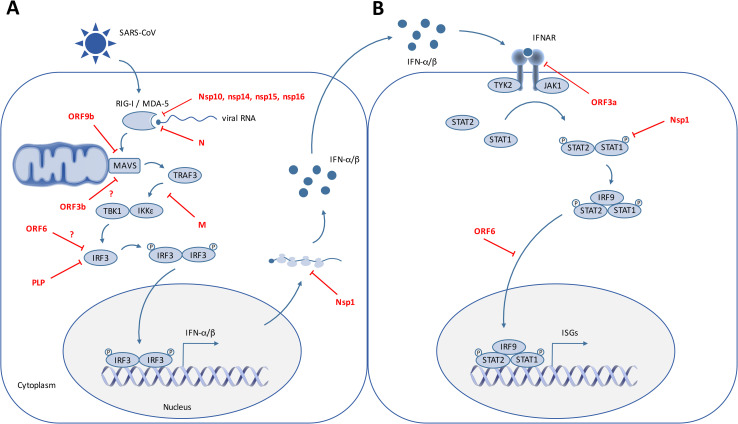
Fig 2. SARS-CoV interfering with IFN induction and signaling.
On this cartoon are schematically represented the signaling pathways triggered by SARS-CoV RNA recognition by the cytoplasmic RNA sensors RIG-I and MDA5, which leads to IFN induction (A) and subsequent IFN signaling in surrounding cells, resulting in the expression of ISGs (B). SARS-CoV proteins that have been reported to interfere with these pathways are indicated. IFN, interferon; IFNAR, interferon alpha and beta receptor; IκB, inhibitor of nuclear factor κB; IKKε, IκB kinase-ε; IRF, IFN regulatory factor; ISG, IFN-stimulated gene; JAK, Janus kinase; M, membrane; MAVS, mitochondrial antiviral signaling protein; MDA5, melanoma differentiation-associated gene 5; N, nucleocapsid; Nsp, nonstructural protein; ORF, open reading frame; P, phosphate; PLP, papain-like protease; RIG-I, retinoic acid-inducible gene 1; SARS-CoV, severe acute respiratory syndrome coronavirus; STAT, signal transducer and activator of transcription; TANK, TRAF family member associated NF-κB activator; TBK1, TANK-binding kinase 1; TRAF3, tumor necrosis factor receptor-associated factor 3; TYK2, tyrosine kinase 2.
Table 1. SARS-CoV proteins interfering with IFN-I induction and signaling.
| Protein | Mechanism | Affected Step | Experimental Approach | Cellular Model | References |
|---|---|---|---|---|---|
| Inhibition of IFN-I Induction | |||||
| Nsp14 | Guanine-N7-methyltransferase activity—methylates capped RNA transcripts | Sensing | Yeast genetic system—validated using a SARS-CoV replicon | Yeast | [29] |
| Nsp15 (EndoU) | Cleaves the 5′-polyuridines from negative-sense viral RNA | Sensing | Protein overexpression (mutants for other coronaviruses) |
MA104 cells | [30–32] |
| Nsp16 | 2′-O-methyltransferase activity involved in viral RNA capping |
Sensing | SARS-CoV mutants | Vero, Calu-3, and HAE cells; mice | [33] |
| Nsp10 | Cofactor of Nsp16, required for RNA cap methylation | Sensing | In vitro reconstitution of SARS-CoV mRNA cap methylation | In vitro | [34] |
| N | Inhibits TRIM25-mediated RIG-I ubiquitination | Sensing | Protein overexpression | HEK293T and A549 cells | [35–37] |
| Nsp3 (PLP) | Deubiquitinates cellular substrates, possibly including RIG-I | Sensing | Protein overexpression | HEK293 cells | [38] |
| Inhibits IRF3 phosphorylation | IRF3 activation | Protein overexpression | HEK293, HeLa, and MA104 cells | [30, 39] | |
| ORF9b | Targets MAVS, TRAF3, and TRAF6 to degradation | Downstream signaling | Protein overexpression | HEK293 and A549 cells | [40] |
| M | Impedes the formation of TRAF3/TBK1/IKKε complex | Downstream signaling | Protein overexpression; also observed in infected cells | HEK293 and HeLa cells | [41] |
| ORF6 | Unknown | Downstream signaling | Protein overexpression | HEK293T and A549 cells | [36] |
| ORF3b | Mechanism unclear; may target MAVS | Downstream signaling | Protein overexpression | HEK293T and A549 cells | [36, 42] |
| Nsp1 | Promotes cellular mRNA degradation and blocks host mRNA translation | Expression of IFN-I | Protein overexpression—validated using a SARS-CoV mutant virus | HEK293 and Vero cells | [43–45] |
| Inhibition of IFN-I Signaling | |||||
| ORF3a | Promotes IFNAR1 degradation | IFN binding to its receptor | Protein overexpression | Huh7 cells | [46] |
| Nsp1 | Decreases STAT1 phosphorylation | STAT1 activation | Protein overexpression—validated using a SARS-CoV mutant virus | HEK293T and Vero cells | [47] |
| ORF6 | Sequesters karyopherin alpha 2 and beta 1 | Nuclear translocation of STAT1 | Protein overexpression—Validated using a SARS-CoV mutant virus | HEK293(T), A549, and Vero cells | [36, 48] |
Abbreviations: EndoU, endoribonuclease; HAE, human airway epithelial; HEK, human embryonic kidney; IFN, interferon; IFNAR, interferon alpha and beta receptor; IFN-I, type I IFN; IκB, inhibitor of nuclear factor κB; IKKε, IκB kinase-ε; IRF, IFN regulatory factor; M, membrane; MAVS, mitochondrial antiviral signaling protein; Nsp, nonstructural protein; ORF, open reading frame; PLP, papain-like protease; RIG-I, retinoic acid-inducible gene 1; SARS-CoV, severe acute respiratory syndrome coronavirus; STAT, signal transducer and activator of transcription; TANK, TRAF family member-associated NF-κB activator; TBK1, TANK-binding kinase 1; TRAF, tumor necrosis factor receptor-associated factor; TRIM25, tripartite motif containing 25.
Another viral strategy to inhibit IFN-I signaling is to enhance the host retrocontrol of this pathway. Several ISGs are themselves repressors of the IFN-I response, and their regulatory functions operate at the viral and host mRNA transcription and translation steps, acting via a wide-range of mechanisms (reviewed in [7, 53]). For example, the inducible negative regulators such as the suppressor of cytokine signaling (SOCS1 and SOCS3) act at various levels of the Jak–STAT pathway or by targeting IRF7 for degradation [54]. In the context of SARS-CoV, the S protein induces the expression of SOCS3 expression in B cells [55]. Induction of SOCS1/3 expression is also detected in SARS-CoV–infected cells, albeit to a lower extent as compared with other respiratory viruses [56]. Recent genomic screen approaches identified a set of repressors of the IFN-I response depending on the cell type and activation pathway involved [57–59]. Hence, one might anticipate that distinct repressors of the IFN-I response are induced depending on the cell type targeted by SARS-CoV-2, the level of replication, and the microenvironment. For example, in the context of coronaviruses, an inefficient detection of MHV infection likely results from an inhibition of the basal levels of sensors mRNA expression in several cell types [60]. It is conceivable that this inhibition might involve negative regulators such as the IFN-inducible RNF125, which targets signaling components such as RIG-I, MDA5, and MAVS for degradation [61].
Interplay between host translation and the IFN-I response
Inhibition of protein synthesis is a conserved host response to prevent viral infections. The host translation is dynamically regulated by PKR, activated via recognition of viral RNA (reviewed by [62]). Activated PKR inhibits the eukaryotic initiation factor 2 (eIF2α), a major regulator of the initiation phase of mRNA translation, by phosphorylating its α subunit. The PKR-induced translational arrest shuts down the negative feedback on the IFN-I response, which can thus result in a prolonged and/or amplified IFN-I response [63]. Because PKR is an ISG, the translational arrest is, in turn, potentiated by the IFN-I response (reviewed in [64]). This highlights a paradoxical situation in which translation arrest prevents viral replication but also set a threshold of viral detection to commensurate the host transcriptional antiviral response to the level of infection [63].
Whether the PKR pathway is modulated by SARS-CoV2 is unknown, yet different coronaviruses regulate PKR-eIF2α axis and host translation. For example, the MERS-CoV protein 4a (p4a) accessory protein impedes PKR activation [65, 66]. Future studies are needed to further uncover the relationship between IFN-I response and host translation and their dynamics in the context of SARS-CoV2 infection.
Dynamics of the IFN-I response define severity of infection
Dynamics of the IFN-I response at the levels of individual cells and cell population
The IFN-I response varies among different cell types and within different microenvironments. Studies at the single-cell level suggest that the amplitude and kinetic of the response is also heterogeneous for a given cell type. Mathematic modeling revealed that IFN-I response is, at least in part, stochastic because only a fraction of cells are able to produce IFN-I upon activation by agonists of the sensors and are sensitive to the paracrine stimulation by IFN-I [67–71]. The heterogeneity of the IFN-I response can be imprinted by the state of the cell at the activation time, including its global translation activity, metabolism, expression levels of signaling molecules (sensors, adaptors, and receptors) [67–70]. Additionally, the distinct onsets of the IFN-I induction depend on the rapidity and amplitude of viral replication. This heterogeneous responsiveness at the individual cell level consequently shapes the dynamics of the host antiviral response at the whole population level [69–71]. This model of the IFN-I response dynamics yielded in the context of various RNA viruses provides a framework likely at play for coronavirus infections. A delayed induction of the ISG expression via virus-induced modulation of the basal activity of transcriptional activity of STAT1 and PKR pathways leads to a peak of coronavirus replication preceding the ISG response [72]. Additionally, in vivo study of the dynamic of MHV infection showed that a fast and robust IFN-I production by pDCs down-regulate the IFN-I response by other cells [73]. This suggests that the IFN-I response at play in different cell types might drive the control of coronavirus infection and potentially contribute to the progression of the disease.
Impaired or delayed IFN-I response contributes to the disease
As mentioned above, coronaviruses possess various mechanisms to defeat the IFN-I response within infected cells, and this inhibition ability is associated with clinical severity (reviewed in [74]). Clinical studies showed that coronaviruses evade innate immunity during the first 10 days of infection, which corresponds to a period of widespread inflammation and steadily increasing viral load [75, 76]. Elevated virus replication eventually leads to inflammation and hypercytokinemia, referred to as a “cytokine storm” [77–80] (Fig 3). The delayed IFN-I response indeed promotes the accumulation of pathogenic monocyte–macrophages [77, 81]. This cell infiltrate results in lung immunopathology, vascular leakage, and suboptimal T cell response [77, 81]. Immune phenotypic profiling in peripheral blood mononuclear cells (PBMCs) of COVID-19 patients similarly revealed that high viremia is associated with an exacerbated IFN-I response, an aggravated cytokine secretion, and inflammation, driving clinical severity [22]. Although the IFNAR signaling pathway was up-regulated at an earlier disease stage, down-regulation of ISGs, together with exacerbated NF-κB activation, promotes a cytokine storm and hyperinflammation, found in critically ill patients [22].
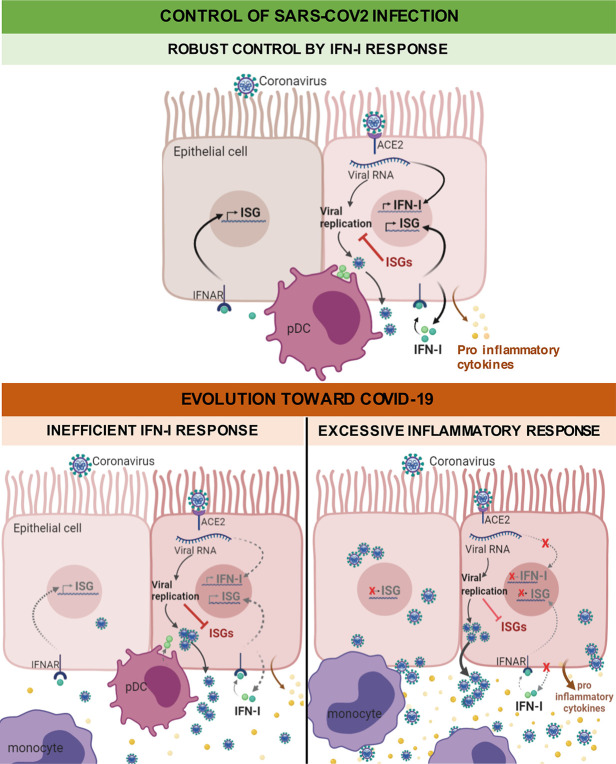
Fig 3. Working model of the failure of IFN-I to control of SARS-CoV-2 infection, leading to COVID-19.
While IFN-I can control viral infection (upper panel), IFN-I deficiency is believed to play a key role in SARS-CoV-2 pathogenesis (lower panel). As shown for related coronaviruses, a delayed IFN-I signaling is associated with robust virus replication and severe complications, i.e., inflammation and “cytokine storm,” notably via the accumulation of monocytes resulting in lung immunopathology, vascular leakage, and suboptimal T cell response. ACE2, angiotensin I converting enzyme 2; IFNAR, interferon alpha and beta receptor; IFN-I, type I interferon; ISG, IFN-stimulated gene; pDC, plasmacytoid dendritic cell; SARS-CoV-2, severe acute respiratory syndrome coronavirus-2.
Collectively, these findings highlight the negative impact of a delayed IFN-I response on viral control and disease severity. However, the underlining mechanisms that drive the temporal control of the IFN-I response in patients are still elusive. In particular, the host and viral determinants driving the on/off switch of the IFN-I response in infected cells, noninfected cells, and/or stimulated immune cells need to be investigated. Such studies will certainly benefit from longitudinal studies of immune profiling in SARS-CoV2 infected patients at the single-cell level and in combination with the clinical data.
Robust and localized IFN-I response by pDCs
By producing 1,000-fold more IFN-I than any other cell types, pDCs are at the heart of the antiviral IFN-I response [82, 83]. They also produce other proinflammatory cytokines, which contribute to modulate the function of several immune cells, such as the mobilization of natural killer (NK) cells or the licensing of virus-specific T cell responses [82–85]. Because pDCs are refractory to most viral replication, their antiviral response cannot be inhibited by viral proteins [82, 86]. This unopposed response likely contributes to the exceptional magnitude of pDC IFN-I production [82, 87–89]. pDCs are circulating immune cells; nonetheless, their response is mostly localized at the infection site because their activation requires physical contact with infected cells [82, 83, 90]. The contact site between pDCs and infected cells, which we named the interferogenic synapse, is a specialized platform for PAMP transfer from infected cell to the Toll-like receptor 7 (TLR7) sensor in pDC, leading to an antiviral response [91].
Previous studies on SARS- and MERS-CoV demonstrated that the rapid production of IFN-I by pDCs is essential for the control of potentially lethal coronavirus infections in mouse models [77, 92]. pDCs migrate into the lungs at the early phase of infection (i.e., pDC number peaks at day 2), temporally coinciding with the peak of IFNα production [92, 93]. The pDC number was found to be reduced in blood of COVID-19 patients as compared with control patients [22], potentially resulting from a prior response followed by a vanished number of circulating pDCs and/or their mobilization to the infected site. Future studies are needed to address how pDC responsiveness evolves in the course of SARS-CoV-2 infection and how pDCs respond to contact with coronavirus-infected cells.
Are IFNs protective of or detrimental to SARS-CoV-2?
Despite the abovementioned viral inhibitory mechanisms of IFN-I response (Table 1), exogenous IFN-I in cell cultures efficiently inhibit SARS-CoV, SARS-CoV-2, and MERS-CoV spread [5, 20, 26, 51, 94–101]. Consistently, IFN-I was shown to have a protective effect against SARS-CoV and MERS-CoV, alone or in combination with other antivirals, in various animal models including mice, marmosets, and macaques [97, 102, 103]. IFN-I and III interfere with virus infection by inducing the expression of several hundred ISGs [7]. Numerous well-described ISGs exhibit direct antiviral activities by targeting specific stages of the viral life cycle, including entry into host cells, replication, protein translation, and assembly of new virus particles. As mentioned above, many signaling regulators are themselves ISGs, thus leading to amplification of the antiviral IFN-I pathway.
Detrimental effects of IFNs on SARS-CoV-2 replication
As a first step towards identifying ISGs able to restrict SARS-CoV-2 replication, transcriptomic responses to infection have been analyzed in different cellular models, including primary cells, organoids, and clinical samples [20, 104–106], as summarized in Table 2. These studies demonstrate that, despite triggering very little to no IFN expression (Table 2), SARS-CoV-2 replication induces moderate levels of a limited number of ISGs. A small subset of infected cells may be refractory to the antagonistic mechanisms of SARS-CoV-2, producing minute but sufficient amounts of IFNs to trigger ISG induction in larger population of cells. Alternatively, ISGs may be up-regulated in noninfected cells, which were analyzed together with infected ones. Indeed, interpretation of genome-wide investigations of virus–pathogen interactions are often obscured by analyses of mixed populations of infected and uninfected cells [107].
Table 2. ISGs and IFN signature of SARS-CoV-2–infected samples.
| Infection (Viral Dose and Time Point) | Cellular Model | ISGs and IFN Signatures (Top Up-Regulated ISGs) |
References |
|---|---|---|---|
| MOI 0.2 24 h |
A549 lung alveolar cells expressing ACE2 | Moderate up-regulation of a subset of ISGs; no IFN-I/III | [20] |
| MOI 2 24 h |
A549 lung alveolar cells expressing ACE2 | High induction of ISGs (OASL, IFIT1, IFIT2, IFIT3, OAS2) and IFNs | [20] |
| MOI 2 24 h |
Lung Calu-3 cells | High induction of ISGs (TRIM22, MX2, IFIT2, IFIT3, and RSAD2/viperin) and IFNs | [20] |
| MOI 2 24 h |
NHBE cells | A small subset of ISGs induced (IFI27, MX1, OAS1, MX2, and IFITM1); undetectable level of IFN | [20] |
| N/A | Postmortem lung samples from 2 COVID-19 patients | A subset of ISGs (IFI6, IFIT1, OAS1, IFITM2, and IFIT3); undetectable level of IFN-I/III | [20] |
| N/A | BALF of 8 COVID-19 patients | IFIT family members and IFITM family members, as well as ISG15 and RSAD2/viperin | [105] |
| MOI 1 60 h |
Expending human intestinal organoids | Broader response than in lung cells, ISGs (IFI6, IFI27, IFITM3, MX1, and RN7SK); low expression of IFN-I/III | [106] |
Abbreviations: ACE2, angiotensin I converting enzyme 2; BALF, bronchoalveolar lavage fluid; IFI, interferon-inducible protein; IFIT, interferon-induced protein with tetratricopeptide repeats; IFN, interferon; IFN-I/III, type I/III IFN; ISG, IFN-stimulated gene; MOI, multiplicity of infection; MX, myxovirus resistance protein; N/A, not applicable; NHBE, normal human bronchial epithelial; OAS, 2′-5′-oligoadenylate synthetase; RSAD2, Radical S-Adenosyl Methionine Domain Containing 2; RN7SK, RNA component of 7SK nuclear ribonucleoprotein; SARS-CoV-2, severe acute respiratory syndrome coronavirus-2; TRIM22, tripartite motif containing 22.
Of note, by contrast to low-multiplicity of infection (MOI) infection of A549 cells expressing angiotensin I converting enzyme 2 (ACE2), normal human bronchial epithelial (NHBE) cells, and patient samples, high-MOI infections of A549-ACE2 and Calu-3 cells led to the high induction of IFNs and ISGs, including ISGs with broad antiviral activities [20] (Table 2). This discrepancy of IFN production/signaling between the levels of viral replication and/or proportion of infected cells might reflect that the counteraction measures employed by SARS-CoV-2 are less potent at high MOI. Alternatively, as suggested by Blanco-Melo and colleagues, high-MOI infections in cell culture may generate more PAMPs, such as defective noninfectious viral particles, than low-MOI infections [108].
Despite being expressed at moderate levels in vitro and in vivo, several up-regulated ISGs identified by these transcriptomic studies (Table 2) exhibit well-characterized broad-spectrum antiviral activities and could thus have additive restrictive effects on SARS-CoV-2 replication. For instance, the 3 members of the interferon-induced protein with tetratricopeptide repeats (IFITM) family, known to inhibit entry of numerous enveloped RNA viruses [109], similarly restrict entry of SARS-CoV, MERS-CoV, and the globally circulating human coronaviruses 229E and NL63 in 293T and A549 cell lines [110, 111]. OAS1 and mycovirus resistance protein (Mx)A could also contribute to the IFN-I–mediated inhibitory effect on SARS-CoV-2 because a clinical study revealed that single nucleic polymorphisms in the OAS1 3′-UTR and MxA promoter region appear associated with host susceptibility to SARS-CoV in the Chinese Han population [112]. Moreover, the fact that MERS-CoV nonstructural protein 4B (NS4b) is a 2′-5′-oligoadenylate synthetase (OAS)-RNase L antagonist [113] suggests that the OAS pathway contributes to the antiviral effects of IFNs on coronavirus replication. ISGs positively potentiating IFN signaling, such as IFIH1/MDA5, TANK, IRF7, and STAT1, were also increased in the bronchoalveolar lavage fluid (BALF) of COVID-19 patients as compared with healthy controls [105] and could potentially contribute to the amplification of IFN-I response against SARS-CoV-2 replication.
Zinc finger antiviral protein (ZAP), which is encoded by an ISG, contributes to the anti-SARS-CoV-2 effect of IFNs in human lung Calu-3 cells [114]. ZAP is known for restricting the replication of numerous viruses such as retroviruses and filoviruses [115]. The protein recruits the cellular mRNA degradation machinery to viral RNA via 5′-C-phosphate-G-3′ (CpG) dinucleotide recognition [115].
To further determine which individual ISG or combination of ISGs mainly restricts SARS-CoV-2 replication in vitro, several previously established approaches could be used, such as, for example, screening for single or combined ISG activity using a lentiviral vector-based library, as successfully performed by Schoggins and colleagues for other viral infections [116–119]. Indeed, this library of around 380 human ISGs was recently screened in human hepatoma cells for antiviral activity against HCoV-229E [120]. The screen identified IFN-inducible lymphocyte antigen 6 complex, locus E (LY6E) as a potent inhibitor of the replication of multiple coronaviruses, including SARS-CoV, SARS-CoV-2, and MERS-CoV, by blocking fusion of viral and cellular membranes [120]. Mice studies revealed that LY6E directly protects primary B cells and dendritic cells from murine coronavirus infection [120]. Pursuing the identification and characterization of IFN effectors with potent anti-SARS-CoV-2 activities will reveal weakness points in the life cycle of SARS-CoV-2 and may lead to the design of drugs that activate antiviral ISGs or either mimic or amplify their action.
Beneficial effects of IFN on SARS-CoV-2 replication
Recent advances in systematic screening strategies have revealed the existence of a small subset of ISGs exhibiting proviral activities [116, 119]. These proviral ISGs act either by exhibiting direct proviral activities such as facilitating viral entry [119] or via their abilities to negatively regulate IFN signaling and facilitate the return to cellular homeostasis. The receptor tyrosine kinase AXL is a well-characterized example of an ISG that is used by enveloped virus for cellular internalization [121–123]. Alternatively, ISGs that possess antiviral activities against a viral family can be hijacked by unrelated viruses to favor infection. This is the case for IFITM2 and IFITM3, which potently block entry of a broad range of enveloped viruses [109] while promoting entry step of human coronavirus OC43 (HCoV-OC43) in human cells [124].
SARS-CoV-2 uses ACE2 and transmembrane serine protease 2 (TMPRSS2) to enter cells [125]. Viral tropism is thus largely dictated by ACE2 and TMPRSS2 coexpression. Analysis of human, nonhuman primate, and mouse single-cell RNA-sequencing (scRNA-seq) data sets generated from healthy or diseased individuals revealed that expression of ACE2 is primarily restricted to type II pneumocytes in the lung, absorptive enterocytes within the gut, and goblet secretory cells of the nasal mucosa [126]. Interestingly, this meta-analysis identified an association between ACE2 expression and canonical ISGs or components of the IFN-signaling pathway in different tissues. Independent analyzes of publicly available data sets concluded that ACE2 expression pattern is similar to ISGs [127]. In vitro validations were performed by treating primary human upper airway cells with numerous inflammatory cytokines. IFNα2, and to some extent IFNy, led to greater and more significant ACE2 up-regulation compared with all other tested cytokines [126]. Substantial up-regulation of ACE2 was also observed in primary skin and primary bronchial cells treated with either IFN-I or IFN-II. Moreover, ACE2 expression was also up-regulated upon ex vivo influenza A infection in human lung explants isolated following surgical resection [126]. Because the majority of cells robustly up-regulating ACE2 were epithelial, this observation potentially explains why previous analyses to define canonical ISGs within immune populations did not identify ACE2 as an induced gene [116]. Finally, STAT1, STAT3, IRF8, and IRF1 binding sites were identified within −1,500 to +500 bp of the transcription start site of ACE2 [126]. Despite need for direct evidence that IFNs up-regulate ACE2 in target cells in vivo, altogether these studies suggest that ACE2 could be an ISG that enhances SARS-CoV-2 internalization in human epithelial cells [125, 126].
Elucidating tissue and cell type specificity of ISGs, as well as their mechanisms of action, is essential for understanding the potential dual role of IFNs during human SARS-CoV-2 infection. It may also guide the use of IFNs in clinical trials.
Clinical implications of the dual role of IFN on SARS-CoV-2 replication
Although IFN-I treatment gave some encouraging results against SARS-CoV and MERS-CoV in vitro and in animal models, including mice, marmosets, and macaques [97, 102, 103, 128, 129], additional knowledge to optimize its therapeutic efficiency in humans is required [130–134]. Previous information yielded from these animal studies provided guidance for treating the current pandemic virus. First, it became clear from these former studies that IFNβ is a more potent inhibitor than IFNα as shown both in vitro and in patients [129, 132]. Second, the timing of IFN-I treatment seems determinant for infection outcomes. Indeed, as shown in mice and in macaques, IFN-I is protective when administered prior to SARS-CoV or MERS-CoV infection or early in the course of infection, whereas late administration could be either ineffective or detrimental [77, 135]. In humans as well, IFN-I–based therapies were not beneficial to critically ill patients with multiple comorbidities and who were diagnosed late with MERS-CoV, thus pointing out that IFN-I has to be administered early after infection [134, 136].
The first clinical trials using IFN-I alone or in combination with other antivirals are currently carried out in COVID-19 patients in several countries. For instance, the multicenter, adaptive, randomized, open clinical trial DisCoVeRy evaluates, among other treatment, the efficacy of IFNβ as a treatment for COVID-19 in hospitalized adults in Europe. A recent open-label, randomized, phase 2 trial performed in adults with COVID-19 in Hong Kong showed that the triple combination of IFNβ-1b, lopinavir–ritonavir, and ribavirin was safe and superior to lopinavir–ritonavir alone in alleviating symptoms and shortening the duration of viral shedding and hospital stay in patients with mild to moderate COVID-19 [137]. It has to be noted that the patients were treated in the early stages of the disease because the median number of days from symptom onset to start of study treatment was 5 days, further reinforcing the fact that the timing of IFN-I treatment is key [137].
Other therapeutic approaches are under investigation to avoid the adverse effects of IFN-I therapy and/or its potential inefficacity when administrated too late postinfection. One strategy is to use aerosol formulations of recombinant IFN to deliver the cytokine directly inside the lung [138, 139]. This approach has several benefits because it is a noninvasive route of administration, and the local concentration reached in the tissue can be higher than through systemic injection and is thus expected to minimize the adverse effects of IFN. Nebulized IFNα-2b was used on COVID-19 patients in Wuhan, alone or in combination with arbidol [140]. The study, performed on 77 adults, showed a significant reduction of the duration of detectable virus in the upper respiratory tract in IFNα-2b–treated patients, with or without arbidol [140]. Another study currently ongoing in Beijing aims at evaluating the efficacy and safety of recombinant human IFNα spray in preventing SARS-CoV-2 infection in highly exposed medical staffs (ChiCTR2000030013).
Type III IFNs (IFNλs or IFN-III) are gaining an increased interest in antiviral therapies [141–143]. Like IFN-I, they activate the JAK–STAT signaling pathway. They do so via a receptor that is largely restricted to cells of epithelial origin, including respiratory epithelial cells (reviewed in [144]). IFN-IIIs are induced upon viral infections, and they are growing evidence that they provide important first-line defense against viral infections of the respiratory and gastrointestinal tracts [145–147]. In mice, IFN-III was shown to protect epithelial cells of the respiratory and tract from infections with several respiratory viruses, including MERS-CoV [147]. A study investigating SARS-CoV-2 infection of intestinal epithelial cells, using both colon-derived cell lines and primary colon organoids, showed that IFN-III response was more efficient than IFN-I at controlling viral replication [148]. However, IFN-IIIs produced by dendritic cells in the lung were recently shown to cause barrier damage and to compromise host tissue tolerance and predispose to lethal bacterial superinfections [149]. Therefore, although the antiviral properties are promising, the benefit of IFN-III to treat COVID-19 patients awaits careful evaluation. The first clinical trials using IFN-III are ongoing, including one launched at the Massachusetts General Hospital to evaluate the safety and efficacy of pegylated IFNλ on a small number of COVID-19 patients (NCT04343976).
Besides the use of recombinant IFN as a therapeutic treatment, one interesting alternative strategy would be to boost the natural innate immune defenses of COVD-19 patients at early stages of the disease. Because pDCs are seemingly crucial to control coronavirus infections [77, 92], a possibility would be to either amplify or prolong their activation to make them produce more IFN-I and IFN-III. A number of negative feedback loops prevent an exacerbated activation of pDCs, which can be deleterious for the organism in the long term. Thus, transitorily inhibiting these negative retrocontrols may increase the antiviral activity of pDCs. For instance, the bone marrow stromal cell antigen 2 (BST2) is an ISG that activates the immunoglobulin-like transcript 7 (ILT7) inhibitory receptor expressed by pDCs to interrupt the IFN-I response [150]. The blockade of this interaction using either antibodies or inhibitory molecules should thus increase the duration of pDC activation. One could also envisage to take advantage of viral proteins that counteract the antiviral activity of BST2, such as HIV-1 viral protein U (vpu) [151]. Other pDC inhibitory molecules include natural monamines such as histamine, dopamine, or serotonin, which bind to the C-X-C motif chemokine receptor 4 (CXCR4) at the surface of pDCs [152]. Because the CXCR4 antagonist AMD3100 (also known as plerixafor) blocks the binding of monoamines to pDCs, it can prevent the amine-dependent inhibition of pDC activation [152]. AMD3100 is already used in clinics as an immunostimulatory molecule able to mobilize hematopoietic stem cells in cancer patients [153]. Finally, we recently reported that the peptidyl-prolyl isomerase peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (Pin1) switches off the IFN-I expression by pDCs by inducing IRF7 degradation [154]. A number of Pin1 inhibitors have been developed and could be tested for their potential activity on human pDCs [155] and could represent another possible therapeutic strategy to boost pDC-mediated IFN-I production.
Conclusions and perspectives
SARS-CoV-2 emerged in the human population around 7 months ago, yet it seems well adapted to avoid and inhibit the IFN-I response in its new host. Such efficient strategies allow the virus to replicate and disseminate in infected individuals without encountering the initial host defense. This modest IFN response could explain why viremia peaks at early stages of the disease, at the time of symptoms appearance, and not around 7 to 10 days following symptoms, like during SARS-CoV and MERS-CoV infections. IFNβ treatment would be expected to improve the antiviral response of patients at the early stage of COVID-19 and, if possible, at the site of infection. Indeed, IFNβ appeared to be pivotal to improve patient states in a combined therapy regiment of IFNβ, lopinavir–ritonavir, and ribavirin [137]. Nonetheless, IFN-resistant viral mutants may arise and be able to control IFN even more efficiently than parental viruses.
The exacerbated production of proinflammatory cytokines observed at later stage of COVID-19 might challenge the efficiency of an IFNβ treatment administrated after appearance of symptoms. There is indeed an increasing appreciation of the detrimental effects of inappropriate, excessive, or mistimed IFN-I responses in viral infections [156]. The underlying mechanisms by which IFN-I promote disease severity likely include immunopathology due to excessive inflammation and direct tissue damage. At the late stages of COVID-19, immunomodulatory drugs that diminish inflammation may benefit patients. The therapeutic benefits of such treatments have been demonstrated in the context of influenza infection in clinical trials [157] and in mice models [158]. Discovery of host markers associated with disease progression will be instrumental to defining the appropriate treatment and time of administration.
Luckily, most COVID-19 patients develop no or mild symptoms. In these patients, the virus ends up being cleared by the immune system, possibly through a partial protection conferred by cross-reactive CD4+ T cells that have been found in between 40% and 60% of unexposed individuals [159]. It is therefore probable that a viral replication that is under control thanks to an efficient adaptive immunity prevents a systemic viral spread and the subsequent cytokine storm. Prior and coinfections along with age, gender, immunological state, and comorbidities also likely play a key role in the ability of the patients to efficiently respond to SARS-CoV-2 infection.
The current studies that aim to better understand the mechanisms that render some patients particularly sensitive to SARS-CoV-2 infections raise hope for the possibility of treating patients with drugs that either enhance the IFN response at the early stage of the disease or dampen it at later stages.
Acknowledgments
We thank Nathalie J. Arhel for helpful discussion.
Funding Statement
Research in the laboratory of NJ is funded by the CNRS, the Institut Pasteur, the European Molecular Biology Organisation (EMBO) Young Investigator Program, and the Agence Nationale de la Recherche Scientifique (ANR 16 CE15 0025 01 VIRO-STORM). Research related to the topic of this review in the team led by MD is supported by the Agence Nationale de la Recherche (ANR Flash COVID-19 and ANR 19-CE15-0025-01 JCJC iSYN). Research related to the topic of this review in the team led by SN is supported by the Labex EpiGenMed, an Investissements d’avenir program (ANR-10-LABX-12-01), the Région Occitanie and the Agence Nationale de la Recherche (ANR Flash COVID-19). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet. 2020;395(10223):470–3. 10.1016/S0140-6736(20)30185-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–33. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–9. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan JF, Kok KH, Zhu Z, Chu H, To KK, Yuan S, et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9(1):221–36. 10.1080/22221751.2020.1719902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583: 459–468. 10.1038/s41586-020-2286-9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Streicher F, Jouvenet N. Stimulation of Innate Immunity by Host and Viral RNAs. Trends Immunol. 2019;40(12):1134–48. 10.1016/j.it.2019.10.009 . [DOI] [PubMed] [Google Scholar]
- 7.Schoggins JW. Interferon-Stimulated Genes: What Do They All Do? Annu Rev Virol. 2019;6(1):567–84. 10.1146/annurev-virology-092818-015756 . [DOI] [PubMed] [Google Scholar]
- 8.Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol. 2014;32:513–45. 10.1146/annurev-immunol-032713-120231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crouse J, Kalinke U, Oxenius A. Regulation of antiviral T cell responses by type I interferons. Nat Rev Immunol. 2015;15(4):231–42. 10.1038/nri3806 . [DOI] [PubMed] [Google Scholar]
- 10.Makris S, Paulsen M, Johansson C. Type I Interferons as Regulators of Lung Inflammation. Front Immunol. 2017;8: 259 10.3389/fimmu.2017.00259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, Liu Y, Zhang X. Murine coronavirus induces type I interferon in oligodendrocytes through recognition by RIG-I and MDA5. J Virol. 2010;84(13): 6472–82. 10.1128/JVI.00016-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zalinger ZB, Elliott R, Rose KM, Weiss SR. MDA5 Is Critical to Host Defense during Infection with Murine Coronavirus. J Virol. 2015;89(24):12330–40. 10.1128/JVI.01470-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loo YM, Gale M Jr. Immune signaling by RIG-I-like receptors. Immunity. 2011;34(5):680–92. 10.1016/j.immuni.2011.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darnell JE Jr., Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264(5164):1415–21. 10.1126/science.8197455 . [DOI] [PubMed] [Google Scholar]
- 15.Schindler C, Levy DE, Decker T. JAK-STAT signaling: from interferons to cytokines. J Biol Chem. 2007;282(28):20059–63. 10.1074/jbc.R700016200 . [DOI] [PubMed] [Google Scholar]
- 16.Dabo S, Meurs EF. dsRNA-dependent protein kinase PKR and its role in stress, signaling and HCV infection. Viruses. 2012;4(11):2598–635. 10.3390/v4112598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14(1):36–49. 10.1038/nri3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCormick C, Khaperskyy DA. Translation inhibition and stress granules in the antiviral immune response. Nat Rev Immunol. 2017;17(10):647–60. 10.1038/nri.2017.63 . [DOI] [PubMed] [Google Scholar]
- 19.Uggenti C, Lepelley A, Crow YJ. Self-Awareness: Nucleic Acid-Driven Inflammation and the Type I Interferonopathies. Annu Rev Immunol. 2019;37:247–67. 10.1146/annurev-immunol-042718-041257 . [DOI] [PubMed] [Google Scholar]
- 20.Blanco-Melo D, Nilsson-Payant BE, Liu WC, Uhl S, Hoagland D, Moller R, et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell. 2020;181(5):1036–45 e9. 10.1016/j.cell.2020.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chu H, Chan JF, Wang Y, Yuen TT, Chai Y, Hou Y, et al. Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: an ex vivo study with implications for the pathogenesis of COVID-19. Clin Infect Dis. 2020: ciaa410 10.1093/cid/ciaa410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Smith N, et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020: eabc6027 10.1126/science.abc6027 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spiegel M, Pichlmair A, Martinez-Sobrido L, Cros J, Garcia-Sastre A, Haller O, et al. Inhibition of Beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J Virol. 2005;79(4):2079–86. 10.1128/JVI.79.4.2079-2086.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheung CY, Poon LL, Ng IH, Luk W, Sia SF, Wu MH, et al. Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: possible relevance to pathogenesis. J Virol. 2005;79(12):7819–26. 10.1128/JVI.79.12.7819-7826.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ziegler T, Matikainen S, Ronkko E, Osterlund P, Sillanpaa M, Siren J, et al. Severe acute respiratory syndrome coronavirus fails to activate cytokine-mediated innate immune responses in cultured human monocyte-derived dendritic cells. J Virol. 2005;79(21):13800–5. 10.1128/JVI.79.21.13800-13805.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan RW, Chan MC, Agnihothram S, Chan LL, Kuok DI, Fong JH, et al. Tropism of and innate immune responses to the novel human betacoronavirus lineage C virus in human ex vivo respiratory organ cultures. J Virol. 2013;87(12):6604–14. 10.1128/JVI.00009-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Q, Shi K, Yoo D. Suppression of type I interferon production by porcine epidemic diarrhea virus and degradation of CREB-binding protein by nsp1. Virology. 2016;489:252–68. 10.1016/j.virol.2015.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roth-Cross JK, Martinez-Sobrido L, Scott EP, Garcia-Sastre A, Weiss SR. Inhibition of the alpha/beta interferon response by mouse hepatitis virus at multiple levels. J Virol. 2007;81(13):7189–99. 10.1128/JVI.00013-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y, Cai H, Pan J, Xiang N, Tien P, Ahola T, et al. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc Natl Acad Sci U S A. 2009;106(9):3484–9. 10.1073/pnas.0808790106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frieman M, Ratia K, Johnston RE, Mesecar AD, Baric RS. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-kappaB signaling. J Virol. 2009;83(13):6689–705. 10.1128/JVI.02220-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng X, Hackbart M, Mettelman RC, O'Brien A, Mielech AM, Yi G, et al. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc Natl Acad Sci U S A. 2017;114(21):E4251–E60. 10.1073/pnas.1618310114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hackbart M, Deng X, Baker SC. Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. Proc Natl Acad Sci U S A. 2020;117(14):8094–103. 10.1073/pnas.1921485117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Menachery VD, Yount BL Jr., Josset L, Gralinski LE, Scobey T, Agnihothram S, et al. Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2'-o-methyltransferase activity. J Virol. 2014;88(8):4251–64. 10.1128/JVI.03571-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bouvet M, Debarnot C, Imbert I, Selisko B, Snijder EJ, Canard B, et al. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 2010;6(4):e1000863 10.1371/journal.ppat.1000863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu X, Pan J, Tao J, Guo D. SARS-CoV nucleocapsid protein antagonizes IFN-beta response by targeting initial step of IFN-beta induction pathway, and its C-terminal region is critical for the antagonism. Virus Genes. 2011;42(1):37–45. 10.1007/s11262-010-0544-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kopecky-Bromberg SA, Martinez-Sobrido L, Frieman M, Baric RA, Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J Virol. 2007;81(2):548–57. 10.1128/JVI.01782-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu Y, Li W, Gao T, Cui Y, Jin Y, Li P, et al. The Severe Acute Respiratory Syndrome Coronavirus Nucleocapsid Inhibits Type I Interferon Production by Interfering with TRIM25-Mediated RIG-I Ubiquitination. J Virol. 2017;91(8): e02143–16. 10.1128/JVI.02143-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clementz MA, Chen Z, Banach BS, Wang Y, Sun L, Ratia K, et al. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J Virol. 2010;84(9):4619–29. 10.1128/JVI.02406-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Devaraj SG, Wang N, Chen Z, Chen Z, Tseng M, Barretto N, et al. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J Biol Chem. 2007;282(44):32208–21. 10.1074/jbc.M704870200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi CS, Qi HY, Boularan C, Huang NN, Abu-Asab M, Shelhamer JH, et al. SARS-coronavirus open reading frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J Immunol. 2014;193(6):3080–9. 10.4049/jimmunol.1303196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siu KL, Kok KH, Ng MH, Poon VK, Yuen KY, Zheng BJ, et al. Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3.TANK.TBK1/IKKepsilon complex. J Biol Chem. 2009;284(24):16202–9. 10.1074/jbc.M109.008227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Freundt EC, Yu L, Park E, Lenardo MJ, Xu XN. Molecular determinants for subcellular localization of the severe acute respiratory syndrome coronavirus open reading frame 3b protein. J Virol. 2009;83(13):6631–40. 10.1128/JVI.00367-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Narayanan K, Huang C, Lokugamage K, Kamitani W, Ikegami T, Tseng CT, et al. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J Virol. 2008;82(9):4471–9. 10.1128/JVI.02472-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kamitani W, Narayanan K, Huang C, Lokugamage K, Ikegami T, Ito N, et al. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc Natl Acad Sci U S A. 2006;103(34):12885–90. 10.1073/pnas.0603144103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lokugamage KG, Narayanan K, Huang C, Makino S. Severe acute respiratory syndrome coronavirus protein nsp1 is a novel eukaryotic translation inhibitor that represses multiple steps of translation initiation. J Virol. 2012;86(24):13598–608. 10.1128/JVI.01958-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Minakshi R, Padhan K, Rani M, Khan N, Ahmad F, Jameel S. The SARS Coronavirus 3a protein causes endoplasmic reticulum stress and induces ligand-independent downregulation of the type 1 interferon receptor. PLoS ONE. 2009;4(12):e8342 10.1371/journal.pone.0008342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wathelet MG, Orr M, Frieman MB, Baric RS. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J Virol. 2007;81(21):11620–33. 10.1128/JVI.00702-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frieman M, Yount B, Heise M, Kopecky-Bromberg SA, Palese P, Baric RS. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J Virol. 2007;81(18):9812–24. 10.1128/JVI.01012-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ivanov KA, Hertzig T, Rozanov M, Bayer S, Thiel V, Gorbalenya AE, et al. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc Natl Acad Sci U S A. 2004;101(34):12694–9. 10.1073/pnas.0403127101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deng X, Baker SC. An "Old" protein with a new story: Coronavirus endoribonuclease is important for evading host antiviral defenses. Virology. 2018;517:157–63. 10.1016/j.virol.2017.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Konno Y, Kimura I, Uriu K, Fukushi M, Irie T, Koyanagi Y, et al. SARS-CoV-2 ORF3b is a potent interferon antagonist whose activity is further increased by a naturally occurring elongation variant. bioRxiv 088179 [Preprint]. 2020. [cited 2020 May 15]. Available from: https://www.biorxiv.org/content/10.1101/2020.05.11.088179v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thoms M, Buschauer R, Ameismeier M, Koepke L, Denk T, Hirschenberger M, et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. bioRxiv 102467 [Preprint]. 2020. [cited 2020 May 15]. Available from: https://www.biorxiv.org/content/10.1101/2020.05.18.102467v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li MM, MacDonald MR, Rice CM. To translate, or not to translate: viral and host mRNA regulation by interferon-stimulated genes. Trends Cell Biol. 2015;25(6):320–9. 10.1016/j.tcb.2015.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu CF, Peng WM, Schlee M, Barchet W, Eis-Hubinger AM, Kolanus W, et al. SOCS1 and SOCS3 Target IRF7 Degradation To Suppress TLR7-Mediated Type I IFN Production of Human Plasmacytoid Dendritic Cells. J Immunol. 2018;200(12):4024–35. 10.4049/jimmunol.1700510 [pii]. . [DOI] [PubMed] [Google Scholar]
- 55.Chiang SF, Lin TY, Chow KC, Chiou SH. SARS spike protein induces phenotypic conversion of human B cells to macrophage-like cells. Mol Immunol. 2010;47(16):2575–86. 10.1016/j.molimm.2010.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Okabayashi T, Kariwa H, Yokota S, Iki S, Indoh T, Yokosawa N, et al. Cytokine regulation in SARS coronavirus infection compared to other respiratory virus infections. J Med Virol. 2006;78(4):417–24. 10.1002/jmv.20556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lumb JH, Li Q, Popov LM, Ding S, Keith MT, Merrill BD, et al. DDX6 Represses Aberrant Activation of Interferon-Stimulated Genes. Cell Rep. 2017;20(4):819–31. 10.1016/j.celrep.2017.06.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li J, Ding SC, Cho H, Chung BC, Gale M, Chanda SK, et al. A short hairpin RNA screen of interferon-stimulated genes identifies a novel negative regulator of the cellular antiviral response. mBio. 2013;4(3):e00385–13. 10.1128/mBio.00385-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu S, Jiang M, Wang W, Liu W, Song X, Ma Z, et al. Nuclear RNF2 inhibits interferon function by promoting K33-linked STAT1 disassociation from DNA. Nat Immunol. 2018;19(1):41–52. 10.1038/s41590-017-0003-0 . [DOI] [PubMed] [Google Scholar]
- 60.Rose KM, Weiss SR. Murine Coronavirus Cell Type Dependent Interaction with the Type I Interferon Response. Viruses. 2009;1(3):689–712. 10.3390/v1030689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arimoto K, Takahashi H, Hishiki T, Konishi H, Fujita T, Shimotohno K. Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc Natl Acad Sci U S A. 2007;104(18):7500–5. 10.1073/pnas.0611551104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gal-Ben-Ari S, Barrera I, Ehrlich M, Rosenblum K. PKR: A Kinase to Remember. Front Mol Neurosci. 2019;11:480 10.3389/fnmol.2018.00480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dalet A, Arguello RJ, Combes A, Spinelli L, Jaeger S, Fallet M, et al. Protein synthesis inhibition and GADD34 control IFN-beta heterogeneous expression in response to dsRNA. EMBO J. 2017;36(6):761–82. 10.15252/embj.201695000 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dalet A, Gatti E, Pierre P. Integration of PKR-dependent translation inhibition with innate immunity is required for a coordinated anti-viral response. FEBS Lett. 2015;589(14):1539–45. 10.1016/j.febslet.2015.05.006 [pii]. . [DOI] [PubMed] [Google Scholar]
- 65.Rabouw HH, Langereis MA, Knaap RC, Dalebout TJ, Canton J, Sola I, et al. Middle East Respiratory Coronavirus Accessory Protein 4a Inhibits PKR-Mediated Antiviral Stress Responses. PLoS Pathog. 2016;12(10):e1005982 10.1371/journal.ppat.1005982 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nakagawa K, Narayanan K, Wada M, Makino S. Inhibition of Stress Granule Formation by Middle East Respiratory Syndrome Coronavirus 4a Accessory Protein Facilitates Viral Translation, Leading to Efficient Virus Replication. J Virol. 2018;92(20): e00902–18. 10.1128/JVI.00902-18 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wimmers F, Subedi N, van Buuringen N, Heister D, Vivie J, Beeren-Reinieren I, et al. Single-cell analysis reveals that stochasticity and paracrine signaling control interferon-alpha production by plasmacytoid dendritic cells. Nat Commun. 2018;9(1):3317 10.1038/s41467-018-05784-3 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pervolaraki K, Rastgou Talemi S, Albrecht D, Bormann F, Bamford C, Mendoza JL, et al. Differential induction of interferon stimulated genes between type I and type III interferons is independent of interferon receptor abundance. PLoS Pathog. 2018;14(11):e1007420 10.1371/journal.ppat.1007420 PPATHOGENS-D-18-02029 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Patil S, Fribourg M, Ge Y, Batish M, Tyagi S, Hayot F, et al. Single-cell analysis shows that paracrine signaling by first responder cells shapes the interferon-beta response to viral infection. Sci Signal. 2015;8(363):ra16 10.1126/scisignal.2005728 [pii]. . [DOI] [PubMed] [Google Scholar]
- 70.Talemi SR, Hofer T. Antiviral interferon response at single-cell resolution. Immunol Rev. 2018;285(1):72–80. 10.1111/imr.12699 . [DOI] [PubMed] [Google Scholar]
- 71.Rand U, Rinas M, Schwerk J, Nohren G, Linnes M, Kroger A, et al. Multi-layered stochasticity and paracrine signal propagation shape the type-I interferon response. Mol Syst Biol. 2012;8:584 10.1038/msb.2012.17 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Menachery VD, Eisfeld AJ, Schafer A, Josset L, Sims AC, Proll S, et al. Pathogenic influenza viruses and coronaviruses utilize similar and contrasting approaches to control interferon-stimulated gene responses. mBio. 2014;5(3):e01174–14. 10.1128/mBio.01174-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bocharov G, Zust R, Cervantes-Barragan L, Luzyanina T, Chiglintsev E, Chereshnev VA, et al. A systems immunology approach to plasmacytoid dendritic cell function in cytopathic virus infections. PLoS Pathog. 2010;6(7):e1001017 10.1371/journal.ppat.1001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14(8):523–34. 10.1038/nrmicro.2016.81 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Peiris JS, Chu CM, Cheng VC, Chan KS, Hung IF, Poon LL, et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361(9371):1767–72. 10.1016/s0140-6736(03)13412-5 [pii]. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wong RS, Wu A, To KF, Lee N, Lam CW, Wong CK, et al. Haematological manifestations in patients with severe acute respiratory syndrome: retrospective analysis. BMJ. 2003;326(7403):1358–62. 10.1136/bmj.326.7403.1358 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Channappanavar R, Fehr AR, Vijay R, Mack M, Zhao J, Meyerholz DK, et al. Dysregulated Type I Interferon and Inflammatory Monocyte-Macrophage Responses Cause Lethal Pneumonia in SARS-CoV-Infected Mice. Cell Host Microbe. 2016;19(2):181–93. 10.1016/j.chom.2016.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lau YL, Peiris JS. Pathogenesis of severe acute respiratory syndrome. Curr Opin Immunol. 2005;17(4):404–10. 10.1016/j.coi.2005.05.009 [pii]. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nicholls JM, Poon LL, Lee KC, Ng WF, Lai ST, Leung CY, et al. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361(9371):1773–8. 10.1016/s0140-6736(03)13413-7 [pii]. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lo AW, Tang NL, To KF. How the SARS coronavirus causes disease: host or organism? J Pathol. 2006;208(2):142–51. Epub 2005 Dec 17. 10.1002/path.1897 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Channappanavar R, Fehr AR, Zheng J, Wohlford-Lenane C, Abrahante JE, Mack M, et al. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J Clin Invest. 2019;130:3625–39. 10.1172/JCI126363.126363 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Reizis B. Plasmacytoid Dendritic Cells: Development, Regulation, and Function. Immunity. 2019;50(1):37–50. 10.1016/j.immuni.2018.12.027 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Webster B, Assil S, Dreux M. Cell-Cell Sensing of Viral Infection by Plasmacytoid Dendritic Cells. J Virol. 2016;90(22):10050–3. 10.1128/JVI.01692-16 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cervantes-Barragan L, Lewis KL, Firner S, Thiel V, Hugues S, Reith W, et al. Plasmacytoid dendritic cells control T-cell response to chronic viral infection. Proc Natl Acad Sci U S A. 2012;109(8):3012–7. 10.1073/pnas.1117359109 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Swiecki M, Gilfillan S, Vermi W, Wang Y, Colonna M. Plasmacytoid dendritic cell ablation impacts early interferon responses and antiviral NK and CD8(+) T cell accrual. Immunity. 2010;33(6):955–66. 10.1016/j.immuni.2010.11.020 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Decembre E, Assil S, Hillaire ML, Dejnirattisai W, Mongkolsapaya J, Screaton GR, et al. Sensing of immature particles produced by dengue virus infected cells induces an antiviral response by plasmacytoid dendritic cells. PLoS Pathog. 2014;10(10):e1004434 10.1371/journal.ppat.1004434 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Esashi E, Bao M, Wang YH, Cao W, Liu YJ. PACSIN1 regulates the TLR7/9-mediated type I interferon response in plasmacytoid dendritic cells. Eur J Immunol. 2012;42(3):573–9. 10.1002/eji.201142045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Blasius AL, Arnold CN, Georgel P, Rutschmann S, Xia Y, Lin P, et al. Slc15a4, AP-3, and Hermansky-Pudlak syndrome proteins are required for Toll-like receptor signaling in plasmacytoid dendritic cells. Proc Natl Acad Sci U S A. 2010;107(46):19973–8. 10.1073/pnas.1014051107 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sasai M, Linehan MM, Iwasaki A. Bifurcation of Toll-like receptor 9 signaling by adaptor protein 3. Science. 2010;329(5998):1530–4. 10.1126/science.1187029 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Webster B, Werneke SW, Zafirova B, This S, Coleon S, Decembre E, et al. Plasmacytoid dendritic cells control dengue and Chikungunya virus infections via IRF7-regulated interferon responses. Elife. 2018;7: e34273 10.7554/eLife.34273 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Assil S, Coleon S, Dong C, Decembre E, Sherry L, Allatif O, et al. Plasmacytoid Dendritic Cells and Infected Cells Form an Interferogenic Synapse Required for Antiviral Responses. Cell Host Microbe. 2019;25(5):730–45 e6. 10.1016/j.chom.2019.03.005 [pii]. . [DOI] [PubMed] [Google Scholar]
- 92.Cervantes-Barragan L, Zust R, Weber F, Spiegel M, Lang KS, Akira S, et al. Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood. 2007;109(3):1131–7. 10.1182/blood-2006-05-023770 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen J, Lau YF, Lamirande EW, Paddock CD, Bartlett JH, Zaki SR, et al. Cellular immune responses to severe acute respiratory syndrome coronavirus (SARS-CoV) infection in senescent BALB/c mice: CD4+ T cells are important in control of SARS-CoV infection. J Virol. 2010;84(3):1289–301. Epub 2009 Nov 11. 10.1128/JVI.01281-09 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cinatl J, Morgenstern B, Bauer G, Chandra P, Rabenau H, Doerr HW. Treatment of SARS with human interferons. Lancet. 2003;362(9380):293–4. 10.1016/s0140-6736(03)13973-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hensley LE, Fritz LE, Jahrling PB, Karp CL, Huggins JW, Geisbert TW. Interferon-beta 1a and SARS coronavirus replication. Emerg Infect Dis. 2004;10(2):317–9. 10.3201/eid1002.030482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stroher U, DiCaro A, Li Y, Strong JE, Aoki F, Plummer F, et al. Severe acute respiratory syndrome-related coronavirus is inhibited by interferon- alpha. J Infect Dis. 2004;189(7):1164–7. 10.1086/382597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Falzarano D, de Wit E, Martellaro C, Callison J, Munster VJ, Feldmann H. Inhibition of novel beta coronavirus replication by a combination of interferon-alpha2b and ribavirin. Sci Rep. 2013;3:1686 10.1038/srep01686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.de Wilde AH, Raj VS, Oudshoorn D, Bestebroer TM, van Nieuwkoop S, Limpens R, et al. MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-alpha treatment. J Gen Virol. 2013;94(Pt 8):1749–60. 10.1099/vir.0.052910-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lokugamage KG, Hage A, Schindewolf C, Rajsbaum R, Menachery VD. SARS-CoV-2 is sensitive to type I interferon pretreatment. bioRxiv 982264 [Preprint]. 2020. [cited 2020 May 15]. Available from: https://www.biorxiv.org/content/10.1101/2020.03.07.982264v3 [Google Scholar]
- 100.Mantlo E, Bukreyeva N, Maruyama J, Paessler S, Huang C. Antiviral activities of type I interferons to SARS-CoV-2 infection. Antiviral Res. 2020;179:104811 10.1016/j.antiviral.2020.104811 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xie X, Muruato A, Lokugamage KG, Narayanan K, Zhang X, Zou J, et al. An Infectious cDNA Clone of SARS-CoV-2. Cell Host Microbe. 2020;27(5):841–8 e3. 10.1016/j.chom.2020.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chan JF, Yao Y, Yeung ML, Deng W, Bao L, Jia L, et al. Treatment With Lopinavir/Ritonavir or Interferon-beta1b Improves Outcome of MERS-CoV Infection in a Nonhuman Primate Model of Common Marmoset. J Infect Dis. 2015;212(12):1904–13. 10.1093/infdis/jiv392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sheahan TP, Sims AC, Leist SR, Schafer A, Won J, Brown AJ, et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat Commun. 2020;11(1):222 10.1038/s41467-019-13940-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Xiong Y, Liu Y, Cao L, Wang D, Guo M, Jiang A, et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microbes Infect. 2020;9(1):761–70. 10.1080/22221751.2020.1747363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhou Z, Ren L, Zhang L, Zhong J, Xiao Y, Jia Z, et al. Heightened Innate Immune Responses in the Respiratory Tract of COVID-19 Patients. Cell Host Microbe. 2020;27(6):883–90 e2. 10.1016/j.chom.2020.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lamers MM, Beumer J, van der Vaart J, Knoops K, Puschhof J, Breugem TI, et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020: eabc1669 10.1126/science.abc1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Carlin AF, Vizcarra EA, Branche E, Viramontes KM, Suarez-Amaran L, Ley K, et al. Deconvolution of pro- and antiviral genomic responses in Zika virus-infected and bystander macrophages. Proc Natl Acad Sci U S A. 2018;115(39):E9172–E81. 10.1073/pnas.1807690115 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rezelj VV, Levi LI, Vignuzzi M. The defective component of viral populations. Curr Opin Virol. 2018;33:74–80. 10.1016/j.coviro.2018.07.014 [pii]. . [DOI] [PubMed] [Google Scholar]
- 109.Bailey CC, Zhong G, Huang IC, Farzan M. IFITM-Family Proteins: The Cell's First Line of Antiviral Defense. Annu Rev Virol. 2014;1:261–83. 10.1146/annurev-virology-031413-085537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wrensch F, Winkler M, Pohlmann S. IFITM proteins inhibit entry driven by the MERS-coronavirus spike protein: evidence for cholesterol-independent mechanisms. Viruses. 2014;6(9):3683–98. 10.3390/v6093683 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Huang IC, Bailey CC, Weyer JL, Radoshitzky SR, Becker MM, Chiang JJ, et al. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011;7(1):e1001258 10.1371/journal.ppat.1001258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.He J, Feng D, de Vlas SJ, Wang H, Fontanet A, Zhang P, et al. Association of SARS susceptibility with single nucleic acid polymorphisms of OAS1 and MxA genes: a case-control study. BMC Infect Dis. 2006;6:106 10.1186/1471-2334-6-106 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Comar CE, Goldstein SA, Li Y, Yount B, Baric RS, Weiss SR. Antagonism of dsRNA-Induced Innate Immune Pathways by NS4a and NS4b Accessory Proteins during MERS Coronavirus Infection. mBio. 2019;10(2): e00319–19. 10.1128/mBio.00319-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nchioua R, Kmiec D, Müller J, Conzelmann C, Groß R, Swanson C, et al. The Zinc Finger Antiviral Protein restricts SARS-CoV-2. bioRxiv 134379 [Preprint]. 2020. [cited 2020 May 15]. Available from: https://www.biorxiv.org/content/10.1101/2020.06.04.134379v2 [Google Scholar]
- 115.Ghimire D, Rai M, Gaur R. Novel host restriction factors implicated in HIV-1 replication. J Gen Virol. 2018;99(4):435–46. 10.1099/jgv.0.001026 . [DOI] [PubMed] [Google Scholar]
- 116.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472(7344):481–5. 10.1038/nature09907 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wilson SJ, Schoggins JW, Zang T, Kutluay SB, Jouvenet N, Alim MA, et al. Inhibition of HIV-1 particle assembly by 2',3'-cyclic-nucleotide 3'-phosphodiesterase. Cell Host Microbe. 2012;12(4):585–97. 10.1016/j.chom.2012.08.012 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;505(7485):691–5. 10.1038/nature12862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mar KB, Rinkenberger NR, Boys IN, Eitson JL, McDougal MB, Richardson RB, et al. LY6E mediates an evolutionarily conserved enhancement of virus infection by targeting a late entry step. Nat Commun. 2018;9(1):3603 10.1038/s41467-018-06000-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pfaender S, Mar KB, Michailidis E, Kratzel A, Hirt D, V'Kovski P, et al. LY6E impairs coronavirus fusion and confers immune control of viral disease. bioRxiv 979260 [Preprint]. 2020. [cited 2020 May 15]. Available from: https://www.biorxiv.org/content/10.1101/2020.03.05.979260v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bhattacharyya S, Zagorska A, Lew ED, Shrestha B, Rothlin CV, Naughton J, et al. Enveloped viruses disable innate immune responses in dendritic cells by direct activation of TAM receptors. Cell Host Microbe. 2013;14(2):136–47. 10.1016/j.chom.2013.07.005 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Monel B, Compton AA, Bruel T, Amraoui S, Burlaud-Gaillard J, Roy N, et al. Zika virus induces massive cytoplasmic vacuolization and paraptosis-like death in infected cells. EMBO J. 2017;36(12):1653–68. 10.15252/embj.201695597 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Meertens L, Carnec X, Lecoin MP, Ramdasi R, Guivel-Benhassine F, Lew E, et al. The TIM and TAM families of phosphatidylserine receptors mediate dengue virus entry. Cell Host Microbe. 2012;12(4):544–57. 10.1016/j.chom.2012.08.009 [pii]. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhao X, Guo F, Liu F, Cuconati A, Chang J, Block TM, et al. Interferon induction of IFITM proteins promotes infection by human coronavirus OC43. Proc Natl Acad Sci U S A. 2014;111(18):6756–61. 10.1073/pnas.1320856111 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181(2):271–80 e8. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, et al. SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell. 2020;181(5):1016–35 e19. 10.1016/j.cell.2020.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang P-H, Cheng Y. Increasing Host Cellular Receptor—Angiotensin-Converting Enzyme 2 (ACE2) Expression by Coronavirus may Facilitate 2019-nCoV Infection. bioRxiv 963348 [Preprint]. 2020. [cited 2020 May 15]. Available from: https://www.biorxiv.org/content/10.1101/2020.02.24.963348v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Morgenstern B, Michaelis M, Baer PC, Doerr HW, Cinatl J Jr. Ribavirin and interferon-beta synergistically inhibit SARS-associated coronavirus replication in animal and human cell lines. Biochem Biophys Res Commun. 2005;326(4):905–8. 10.1016/j.bbrc.2004.11.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Scagnolari C, Vicenzi E, Bellomi F, Stillitano MG, Pinna D, Poli G, et al. Increased sensitivity of SARS-coronavirus to a combination of human type I and type II interferons. Antivir Ther. 2004;9(6):1003–11. . [PubMed] [Google Scholar]
- 130.Arabi YM, Shalhoub S, Mandourah Y, Al-Hameed F, Al-Omari A, Al Qasim E, et al. Ribavirin and Interferon Therapy for Critically Ill Patients With Middle East Respiratory Syndrome: A Multicenter Observational Study. Clin Infect Dis. 2020;70(9):1837–44. 10.1093/cid/ciz544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Omrani AS, Saad MM, Baig K, Bahloul A, Abdul-Matin M, Alaidaroos AY, et al. Ribavirin and interferon alfa-2a for severe Middle East respiratory syndrome coronavirus infection: a retrospective cohort study. Lancet Infect Dis. 2014;14(11):1090–5. 10.1016/S1473-3099(14)70920-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Stockman LJ, Bellamy R, Garner P. SARS: systematic review of treatment effects. PLoS Med. 2006;3(9):e343 10.1371/journal.pmed.0030343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Loutfy MR, Blatt LM, Siminovitch KA, Ward S, Wolff B, Lho H, et al. Interferon alfacon-1 plus corticosteroids in severe acute respiratory syndrome: a preliminary study. JAMA. 2003;290(24):3222–8. 10.1001/jama.290.24.3222 . [DOI] [PubMed] [Google Scholar]
- 134.Shalhoub S, Farahat F, Al-Jiffri A, Simhairi R, Shamma O, Siddiqi N, et al. IFN-alpha2a or IFN-beta1a in combination with ribavirin to treat Middle East respiratory syndrome coronavirus pneumonia: a retrospective study. J Antimicrob Chemother. 2015;70(7):2129–32. 10.1093/jac/dkv085 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Haagmans BL, Kuiken T, Martina BE, Fouchier RA, Rimmelzwaan GF, van Amerongen G, et al. Pegylated interferon-alpha protects type 1 pneumocytes against SARS coronavirus infection in macaques. Nat Med. 2004;10(3):290–3. 10.1038/nm1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Al-Tawfiq JA, Momattin H, Dib J, Memish ZA. Ribavirin and interferon therapy in patients infected with the Middle East respiratory syndrome coronavirus: an observational study. Int J Infect Dis. 2014;20:42–6. 10.1016/j.ijid.2013.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hung IF, Lung KC, Tso EY, Liu R, Chung TW, Chu MY, et al. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020;395(10238):1695–704. 10.1016/S0140-6736(20)31042-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dhanani J, Fraser JF, Chan HK, Rello J, Cohen J, Roberts JA. Fundamentals of aerosol therapy in critical care. Crit Care. 2016;20(1):269 10.1186/s13054-016-1448-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Laube BL. The expanding role of aerosols in systemic drug delivery, gene therapy and vaccination: an update. Transl Respir Med. 2014;2:3 10.1186/2213-0802-2-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhou Q, Chen V, Shannon CP, Wei X-S, Xiang X, Wang X, et al. Interferon-α2b Treatment for COVID-19. Frontiers in Immunology. 2020;11(11):1061 10.3389/fimmu.2020.01061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Andreakos E, Tsiodras S. COVID-19: lambda interferon against viral load and hyperinflammation. EMBO Mol Med. 2020;12(6):e12465 10.15252/emmm.202012465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.O'Brien TR, Thomas DL, Jackson SS, Prokunina-Olsson L, Donnelly RP, Hartmann R. Weak Induction of Interferon Expression by SARS-CoV-2 Supports Clinical Trials of Interferon Lambda to Treat Early COVID-19. Clin Infect Dis. 2020: ciaa453 10.1093/cid/ciaa453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Prokunina-Olsson L, Alphonse N, Dickenson RE, Durbin JE, Glenn JS, Hartmann R, et al. COVID-19 and emerging viral infections: The case for interferon lambda. J Exp Med. 2020;217(5): e20200653 10.1084/jem.20200653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Broggi A, Granucci F, Zanoni I. Type III interferons: Balancing tissue tolerance and resistance to pathogen invasion. J Exp Med. 2020;217(1): e20190295 10.1084/jem.20190295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Galani IE, Triantafyllia V, Eleminiadou EE, Koltsida O, Stavropoulos A, Manioudaki M, et al. Interferon-lambda Mediates Non-redundant Front-Line Antiviral Protection against Influenza Virus Infection without Compromising Host Fitness. Immunity. 2017;46(5):875–90 e6. 10.1016/j.immuni.2017.04.025 . [DOI] [PubMed] [Google Scholar]
- 146.Andreakos E, Salagianni M, Galani IE, Koltsida O. Interferon-lambdas: Front-Line Guardians of Immunity and Homeostasis in the Respiratory Tract. Front Immunol. 2017;8: 1232 10.3389/fimmu.2017.01232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mordstein M, Neugebauer E, Ditt V, Jessen B, Rieger T, Falcone V, et al. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J Virol. 2010;84(11):5670–7. 10.1128/JVI.00272-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Stanifer ML, Kee C, Cortese M, Triana S, Mukenhirn M, Kraeusslich H-G, et al. Critical role of type III interferon in controlling SARS-CoV-2 infection, replication and spread in primary human intestinal epithelial cells. bioRxiv 059667 [Preprint]. 2020. [cited 2020 May 15]. Available from: https://www.biorxiv.org/content/10.1101/2020.04.24.059667v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Broggi A, Ghosh S, Sposito B, Spreafico R, Balzarini F, Lo Cascio A, et al. Type III interferons disrupt the lung epithelial barrier upon viral recognition. Science. 2020: eabc3545 10.1126/science.abc3545 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Cao W, Bover L, Cho M, Wen X, Hanabuchi S, Bao M, et al. Regulation of TLR7/9 responses in plasmacytoid dendritic cells by BST2 and ILT7 receptor interaction. J Exp Med. 2009;206(7):1603–14. 10.1084/jem.20090547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.O'Neil JD, Owen TJ, Wood VH, Date KL, Valentine R, Chukwuma MB, et al. Epstein-Barr virus-encoded EBNA1 modulates the AP-1 transcription factor pathway in nasopharyngeal carcinoma cells and enhances angiogenesis in vitro. J Gen Virol. 2008;89(Pt 11):2833–42. 10.1099/vir.0.2008/003392-0 [pii]. . [DOI] [PubMed] [Google Scholar]
- 152.Smith N, Pietrancosta N, Davidson S, Dutrieux J, Chauveau L, Cutolo P, et al. Natural amines inhibit activation of human plasmacytoid dendritic cells through CXCR4 engagement. Nat Commun. 2017;8: 14253 10.1038/ncomms14253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.De Clercq E. Mozobil(R) (Plerixafor, AMD3100), 10 years after its approval by the US Food and Drug Administration. Antivir Chem Chemother. 2019;27:2040206619829382 10.1177/2040206619829382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Maarifi G, Smith N, Maillet S, Moncorge O, Chamontin C, Edouard J, et al. TRIM8 is required for virus-induced IFN response in human plasmacytoid dendritic cells. Sci Adv. 2019;5(11): eaax3511 10.1126/sciadv.aax3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Nakatsu Y, Matsunaga Y, Ueda K, Yamamotoya T, Inoue Y, Inoue MK, et al. Development of Pin1 inhibitors and their potential as therapeutic agents. Curr Med Chem. 2020;27(20): 3314–3329. 10.2174/0929867325666181105120911 . [DOI] [PubMed] [Google Scholar]
- 156.Davidson S, Maini MK, Wack A. Disease-promoting effects of type I interferons in viral, bacterial, and coinfections. J Interferon Cytokine Res. 2015;35(4):252–64. 10.1089/jir.2014.0227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Marsolais D, Hahm B, Walsh KB, Edelmann KH, McGavern D, Hatta Y, et al. A critical role for the sphingosine analog AAL-R in dampening the cytokine response during influenza virus infection. Proc Natl Acad Sci U S A. 2009;106(5):1560–5. 10.1073/pnas.0812689106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Teijaro JR, Walsh KB, Cahalan S, Fremgen DM, Roberts E, Scott F, et al. Endothelial cells are central orchestrators of cytokine amplification during influenza virus infection. Cell. 2011;146(6):980–91. 10.1016/j.cell.2011.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181(7):1489–1501.e15. 10.1016/j.cell.2020.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]