Abstract
Despite the important roles of freshwater gastropods in aquatic ecosystems, the taxonomic status of many taxa is unclear, which is compounded by a lack of information on species population genetic structuring, distribution, and dispersal patterns. The objective of this study was to address the biogeography of the freshwater snail Planorbella trivolvis (Gastropoda: Planorbidae) in the western United States. We amplified two genetic markers (16S, COI) from individuals belonging to western USA populations and downloaded genetic data from GenBank. We utilized minimum spanning networks to assess the genetic patterns and performed Analysis of Molecular Variance and linear regression analyses to determine how geographic distance and watershed identity contributed to the observed genetic structuring. For both markers, we found that the majority of genetic variation was associated within and among populations, rather than among watersheds. Correspondingly, there was no significant effect of geographic distance on genetic distance, suggesting that long-distance dispersal was promoting gene flow between populations. The genetic similarity could reflect avian-mediated dispersal of snails along the Pacific Flyway, a major waterfowl migratory corridor. Further analysis of the population structuring across North America revealed East-West genetic structuring, suggesting that across longitudinal gradients P. trivolvis experiences significant genetic isolation.
1 Introduction
Freshwater snails play important roles in complex ecological interactions, acting as bioindicators of water quality, invasive species, and intermediate hosts for multi-host parasites [1–5]. However, freshwater mollusks also represent one of the most threatened animal groups on Earth [6, 7]. Despite the important roles freshwater gastropods play in aquatic ecology, climatology, conservation, and epidemiology, their basic biology and ecology are still understudied [2]. The taxonomic status of many taxa is uncertain, which is further impaired by a lack of information on species distribution, dispersal patterns and population structure [7]. If fundamental questions regarding freshwater gastropod ecosystem functions are to be answered, a thorough understanding of their biogeography is essential [8, 9].
One of the major factors that determines biogeographical distribution of freshwater invertebrates is their dispersal mode and ability [10, 11]. The ability to move between often discrete habitats has significant consequences for gene flow and population structuring. While many freshwater invertebrates are believed to readily disperse, analyses of their population structure show high levels of genetic differentiation [12]. Although animal vectors, wind or other mechanisms enable long-range dispersal to new water bodies, other factors may restrict gene flow among populations, including founder and priority effects [11, 12]. This discrepancy has been the focus of a larger discussion about dispersal, gene flow, and genetic differentiation in aquatic organisms [13]. Among the freshwater invertebrate taxa where information regarding their dispersal patterns and associated mechanisms remains unresolved are the freshwater gastropods.
Aquatic snails can move through the environment by both active and passive dispersal mechanisms. Active dispersal, such as crawling, is considered important at a local scale [14]. Freshwater snails mostly employ passive dispersal mechanisms for large-scale, rapid dispersal. Within water bodies, many biotic and abiotic factors, such as transport via fish and through currents, influence their passive dispersal ability [14]. For example, freshwater fishes in the Salmonidae family have been known to successfully transport several freshwater gastropod taxa including species in the families Valvatidae, Lymnaeidae and Tateidae [15–17]. Passive drifting downstream via currents is considered the most common dispersal mechanism in freshwater mollusks [14]. Freshwater snails within the families Tateidae and Hydrobiidae often drift downstream in the water column [18, 19]. However, the above mechanisms only apply to hydrologically connected water bodies. Dispersal of freshwater gastropods across non-connected habitats requires assistance from other vectors.
Extra-aquatic dispersal can occur via abiotic vectors such as flooding events or tornadoes; and biotic vectors such as mammals (including humans), insects, and birds [14]. Migratory waterfowl have long been proposed as long-distance dispersers of freshwater snails [20–26]. Gastropod taxa in the families Physidae, Planorbidae and Lymnaeidae have been found attached to feathers of the White-Faced Ibis (Plegadis chihi) (Vieillot) [21] and experimentally on the Tundra Swan (Cygnus columbianus) (Ord) [24]. This suggests that waterfowl-mediated dispersal promotes ongoing gene flow over large geographic ranges [27, 28] and could strongly influence freshwater gastropod distribution as well [29, 30].
Planorbidae is one of the most abundant and widespread freshwater gastropod families in the world [31, 32]. Among the species restricted to North America, one abundant genus is Planorbella (formerly Helisoma) [31]. Compared to other common freshwater snails, Planorbella is generally considered to be a less effective disperser. Species, such as Physella acuta (Draparnaud) (Family Physidae), are known to have higher fecundity, a shorter hatching time, and significant reproductive plasticity, which all aid stochastic dispersal [33–37]. While Planorbella is hermaphroditic and able to reproduce via self-fertilization, studies indicate this genus has lower viability through self-fertilization and reproduces primarily through outcrossing [38–42]. Low reproductive success through self-fertilization would reduce the probability of colonization of a new environment by a single individual [43]. In addition to lacking opportunistic life history characteristics, Planorbella is less effective at utilizing passive dispersal mechanisms. For instance, Planorbella spp. have been found attached to feathers of waterfowl, but experimental flight simulations have shown they are less likely to remain attached to a feather for any length of time [21, 24]. While it is known that other freshwater snails can be transported via the intestinal tract of birds, this method of dispersal was not found for Planorbella [22, 23]. Cumulatively, the results suggest that Planorbella is less likely to disperse long distances compared to other common freshwater snails.
Like many freshwater gastropod families, Planorbidae snails exhibit considerable diversity in shell morphology within species and share extremely homogenous anatomical traits between species [44–47], often leading to unclear species identifications [48, 49]. The variations in the diagnostic shell morphology has resulted in considerable uncertainty about Planorbella classification [50]. Planorbella was raised from a subgenus of Helisoma to the genus level by Burch [51], as was initially suggested by Taylor [52]. Of the previously 17 described species of Helisoma, 16 were moved to Planorbella based on differences in shell coiling, leaving only Helisoma anceps. However, previous work concluded that classification in Planorbella based on shell alone is not sufficient in distinguishing species [31]. Therefore, genetic identification based on mitochondrial markers is necessary to advance our understanding of Planorbidae taxonomy [44, 53]. Furthermore, there is a significant knowledge gap regarding the population genetic structuring of Planorbella in North America. To date, studies focus predominantly on Planorbidae species at global scales, with considerably less sampling within the United States [8]. Overall, a lack of information on the genetic diversity and population structuring of these species in their native range has limited opportunities to make meaningful comparisons and precluded an understanding of the family-level global genetic diversity.
The objective of the current study was to investigate the biogeographic genetic structuring of P. trivolvis in the western United States. Specifically, we opportunistically collected 60 samples from 22 ponds and reservoirs in the western USA and sequenced the 16S and COI mitochondrial loci. We created haplotype networks and calculated molecular diversity indexes and Fst values to describe levels of gene flow between the populations. An AMOVA was conducted to characterize the genetic variance within populations, among populations and across watersheds in the West Coast. By building a statistical model, we tested the relative influence of geographic distance on population genetic structuring. We further compared our findings with samples from GenBank to describe population structuring across North America. These results aim to provide insights into the genetic variation of this ecologically important group of gastropods while concurrently testing the influence of alternative hypothesized explanatory variables on population genetic structure.
2 Methods
2.1 Sampling
Planorbella trivolvis snails were opportunistically collected by hand or by dipnet from 22 ponds and reservoirs in California, Oregon, and Washington States (Fig 1). Samples were collected under the Scientific Take Permit (permit numbers: 029-18 and 101-19) issued by the Oregon Department of Fish and Wildlife and by the California Department of Fish and Wildlife (permit number: SC-3683). Localities included 13 sites in California, eight sites in Oregon, and one site in Washington. To assess genetic structuring, the 22 localities were grouped according to watershed identity (Fig 1), as determined using United States Geological Survey delineation maps and the 4-digit Hydrologic Unit Code (HUC) (https://water.usgs.gov/wsc/map_index.html). The localities spanned six different watersheds along the West Coast of the United States (Lower Columbia (HUC 1708), Middle Columbia (HUC 1707), Oregon-Washington Coastal (HUC 1710), Willamette (HUC 1709), Sacramento River (HUC 1802), and San Francisco Bay (HUC 1805)). Detailed locality information can be found in S1 Appendix.
Fig 1. Map of the West Coast of the United States (Washington, Oregon, California) showing the 22 study sampling locations.
The locality site colors correspond to each specific sampling location. The localities are grouped according to state and watershed (designated by the USGS). Detailed locality information can be found in S1 Appendix.
At each of the 22 sites, between one and seven snails were opportunistically collected for a total of 60 specimens. The snails were identified by the collectors as H. trivolvis, based on diagnostic shell attributes originally described in Hubendick & Rees [54]. Both Helisoma and Planorbella are genus names used today to refer to P. trivovlis specimens [42, 55]. In this paper, we use the accepted name listed by the Integrated Taxonomic Information System (https://www.itis.gov/), Planorbella trivolvis, as described in Burch [51]. Species identities were later confirmed with genetic data (see below). Upon collection, some of the specimens were euthanized and stored in 95% EtOH. Others were kept alive, temporarily stored in the refrigerator, and subsequently transferred to 95% EtOH for DNA extraction.
2.2 DNA amplification
Genomic DNA was extracted from the snail foot tissue utilizing the E.Z.N.A Mollusc DNA Kit (Omega Biotek). Two mitochondrial DNA loci were amplified and sequenced (16S and COI) as both have been used successfully to recover population genetic variation in freshwater snails [9, 53, 56].
The 16S mitochondrial gene was amplified using primer sets 16Sar 5’-CGCCTGTTTATCAAAAACAT-3’ and 16Sbr 5’-CCGGTCTGAACTCAGATCACGT-3’ [57]. The COI mitochondrial gene was amplified using primer sets LCOI490 5’-GGTCAACAAATCATAAAGATATTGG-3’ and HCO2198 5’-TAAACTTCAGGGTGACCAAAAAATC-3’ [58]. PCR amplifications were performed in a total reaction volume of 12.5 μL with 6.25 μL GoTaq Green Master Mix (Promega), 3.75 μL of nuclease-free water, 1 μL of each primer and 0.5 μL of the DNA template.
The PCR protocol for 16S included an initial denaturation at 94 °C for 60 s, 35 cycles of 94 °C for 30 s, 55 °C for 50 s and 72 °C for 60 s, and a final extension at 72 °C for 10 minutes [59]. The PCR protocol for COI included an initial denaturation at 96 °C for 2 minutes, 9 cycles of 96 °C for 40 s, 55 °C for 60 s and 72 °C for 60 s, 30 cycles of 96 °C for 40 s, 46 °C for 60 s and 72 °C for 60 s, and a final extension at 72 °C for 7 minutes. PCR products were assessed through gel electrophoresis.
Amplified products were sequenced by Sanger sequencing at Quintara Biosciences (California). Raw sequences were processed in CodonCode Aligner 3.1.7 to remove primer sequences and manually correct for low quality read. The final size of the 16S gene segment was 456 bp. The final size of COI gene segment was 606 bp. All Planorbella specimens sampled were confirmed to be P. trivolvis based on BLAST match with existing sequences in GenBank (>97% similarity with known P. trivolvis and H. trivolvis sequences).
In addition, all available COI sequences for P. trivolvis (including the one named H. trivolvis) were downloaded from GenBank, resulting in 38 COI sequences for P. trivolvis. 23 of the 38 COI sequences for P. trivolvis are West Coast samples and the remaining 15 are from other North American samples, including Maryland, New Mexico and Canadian localities. GenBank accession numbers and associated studies can be found in S1 Appendix.
2.3 Population structure
Amplified sequences were aligned using the ClustalW algorithm [60] implemented in CodonCode Aligner 3.1.7 and corrected by eye. Low quality regions at the beginning and end of the alignments were removed and not included in the final analysis. The final size of the 16S gene segment was 454 bp. The final alignment size of the COI segment was 400 bp.
Minimum Spanning Networks (MSN) were constructed with PopARt v. 1.7 [61] (http://popart.otago.ac.nz) for both gene fragments to view the connections among haplotypes. First, we constructed MSN networks for P. trivolvis in their native range along the West Coast of the United States (Fig 2).
Fig 2. Haplotype networks for Planorbella trivolvis in the West Coast.
(a) The 16S network of 16 haplotypes. (b) The COI network of 7 haplotypes. Each circle represents a unique haplotype. The size of the circle is proportionate to the frequency of that haplotype in the population. Each hashmark represents one base pair change. Snail haplotypes are color-coded according to their locality.
To assess the genetic structure in the West Coast, we conducted an Analysis of Molecular Variance (AMOVA) [62]. The AMOVA was performed using Arlequin v. 3.5 [63] on the 16S and COI datasets to evaluate degrees of genetic variation within populations, among populations within a watershed and among the six West Coast watersheds. Φ statistics and the variance components were calculated for within populations, among populations within watersheds, and among watersheds. ΦST estimates the genetic variation within populations; ΦSC estimates the variation among populations within the watersheds; and ΦCT estimates variation among all of the watersheds sampled. In order to further examine the population structure of the species, pairwise ΦST between the watersheds were calculated from the 16S and COI dataset in Arlequin v. 3.5. Significance of ΦST were determined through 10,000 random permutations (p < 0.05).
Genetic diversity summary statistics were calculated for each gene segment in Arlequin v. 3.5. Summary statistics were estimated for each watershed and across the entire sampling range. The molecular diversity indexes for each watershed included number of haplotypes (h), haplotype diversity (Hd), and nucleotide diversity. The indexes estimated for across the entire sampling range include gene diversity, mean number of pairwise differences and nucleotide diversity.
To further assess whether geographic distance is associated with P. trivolvis population structuring, general linear models were built using the lm function in R version 3.4.2 [64] to test for a relationship between geographic distance and genetic distance utilizing the package ggplot2. Genetic distance was the dependent variable, and geographic distance was the predictor variable. Pairwise genetic distances between West Coast localities were obtained in Arlequin v. 3.5. Very small negative values of genetic distance known to be statistical noise were rounded to 0. Geographic distance was calculated as the shortest distance between two localities on the surface of a sphere, known as the orthodromic distance.
To assess the structuring of P.trivolvis across North America, we aligned the newly amplified sequences and the ones downloaded from GenBank using the ClustalW algorithm [60] implemented in CodonCode Aligner 3.1.7. Low quality regions were removed for the final analysis. We constructed a Minimum Spanning Network that included all available COI data for P. trivolvis (Fig 3). The average genetic distances between the sequences were calculated in MEGA X [65]. A phylogenetic reconstruction using the North American COI dataset was also conducted to compliment the network analyses. Biomphalaria straminea was used as an outgroup (GenBank ID KY697249, KY697248, KY697200). A maximum-likelihood reconstruction was conducted using RAxML 8.2.11 [66] with the GTR+G model and 100 bootstrap replicates. The best scoring tree is used to represent the phylogeny. Although occurrences of P. trivolvis have been documented globally [67], there is currently no available genetic information for us to assess P. trivolvis population genetics outside of North America.
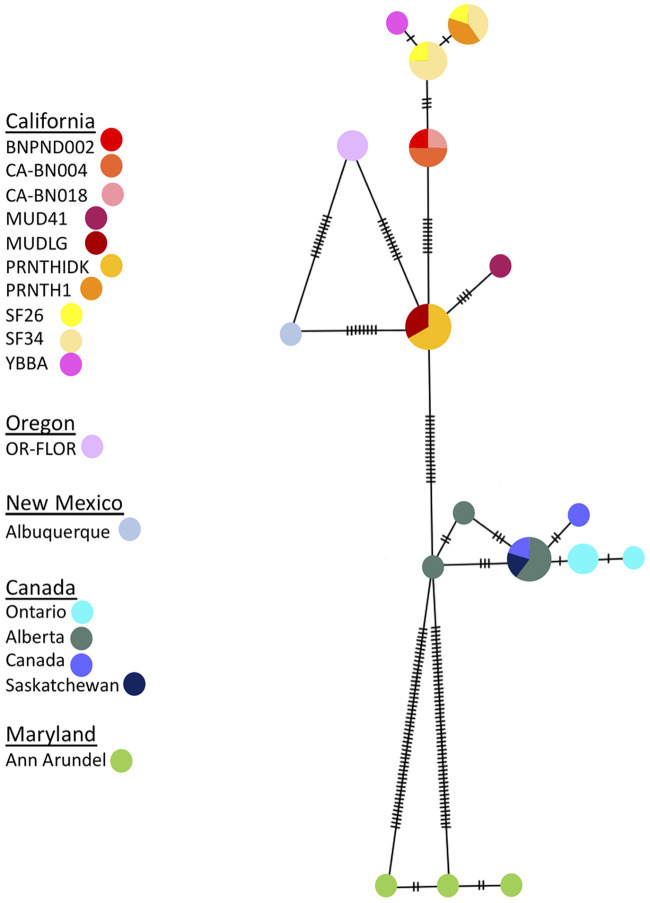
Fig 3. COI haplotype network for Planorbella trivolvis in North America.
Each circle represents a unique haplotype. The size of the circle is proportionate to the frequency of that haplotype in the population. Each hashmark represents one base pair change. Snail haplotypes are color-coded according to their locality.
3 Results
3.1 West Coast
A total of 57 out of 60 P. trivolvis individuals were successfully amplified for the 16S gene fragment. The genotyped individuals yielded 16 unique haplotypes, and seven singleton haplotypes are present in the populations (Fig 2). Two haplotypes were identified as the most common (H1 and H9). H1 is shared by 14 individuals from California and Oregon representing two watersheds, San Francisco and Willamette. H9 is shared by 16 individuals from California and Oregon representing three watersheds, San Francisco, Willamette and Sacramento River. Results from the AMOVA are shown in Table 1. Within population variance (ΦST) accounts for 71.84% of the total genetic variation (p < 0.05), among populations within a watershed and among watershed account for 12.31% and 15.85% (p < 0.05) respectively.
Table 1. Analysis of Molecular Variance (AMOVA) results to test for subdivision in Planorbella trivolvis populations in the West Coast.
| 16s (N = 57) | ||||
| Source of Variation | d.f. | % Variation | Fixation Indices | p-value |
| Among Watershed | 4 | 15.85 | ΦCT = 0.1585 | 0.0301 |
| Among Populations within Watersheds | 16 | 12.31 | ΦSC = 0.1463 | 0.1605 |
| Within Populations | 35 | 71.84 | ΦST = 0.2816 | 0.0111 |
| COI (N = 23) | ||||
| Source of Variation | d.f. | % Variation | Fixation Indices | p-value |
| Among Watershed | 1 | 20.91 | ΦCT = 0.2091 | <0.001 |
| Among Populations within Watersheds | 9 | 77.59 | ΦSC = 0.9811 | 0.0949 |
| Within Populations | 12 | 1.50 | ΦST = 0.9850 | <0.001 |
For the COI gene fragment, a total of 23 of 60 P. trivolvis individuals were successfully amplified. 20 of the individuals were also analyzed for the 16S gene fragment. The low amplification success for the COI gene fragment results in lower statistical power compared to the 16S gene fragment. Seven unique haplotypes were obtained as a result, and three singletons are present in the populations (Fig 2). The AMOVA suggest a significant ΦSC and ΦST (p<0.001 and p<0.001, respectively). Among populations within a watershed accounts for 77.59% of the genetic variation, within population and among watershed account for 1.5% and 20.91% respectively (Table 1).
Tables 2 and 3 summarize the molecular diversity indexes for each watershed and across the entire West Coast sampling range. Haplotype diversity was high for almost every sampled watershed, even reaching 100% (Hd = 1) in some watersheds. Low haplotype diversity (Hd = 0) within certain watersheds is mostly attributed to low sample sizes. The haplotype diversity across watersheds was 0.839 for the 16S dataset and 0.85 for the COI dataset. Gene diversity for 16S was 0.8509 and for COI was 0.8498; and nucleotide diversity was 0.0414 in the 16S dataset and 0.0360 in the COI dataset.
Table 2. Population statistics and molecular diversity indexes for Planorbella trivolvis populations in the West Coast, separated by gene and watershed.
| Gene | Watershed | N | H | HD | Nucleotide |
|---|---|---|---|---|---|
| COI | San Francisco | 21 | 6 | 0.82 | 0.03 ± 0.02 |
| Willamette | 2 | 1 | 0 | 0 | |
| Total | 23 | 7 | 0.85 | ||
| 16S | Lower Columbia | 2 | 1 | 0 | 0 |
| San Francisco | 42 | 11 | 0.72 | 0.04 ± 0.02 | |
| Middle Columbia | 2 | 2 | 1 | 0.05 ± 0.05 | |
| Willamette | 8 | 7 | 0.99 | 0.06 ± 0.03 | |
| Coastal | 1 | 1 | 1 | 0 | |
| Sacramento | 2 | 1 | 0 | 0 | |
| Total | 57 | 16 | 0.839 |
Population statistic abbreviations: N, number of individuals sampled, H, number of haplotypes, Hd, haplotype diversity, and nucleotide diversity
Table 3. Overall molecular diversity indexes for Planorbella trivolvis in the West Coast, by gene.
| COI | 16S | |
|---|---|---|
| Gene diversity | 0.85 ± 0.04 | 0.85 ± 0.03 |
| Mean number of pairwise differences | 14.40 ± 6.70 | 18.79 ± 8.45 |
| Nucleotide diversity (average over loci) | 0.04 ± 0.02 | 0.04 ± 0.02 |
In order to further assess the population structuring of P. trivolvis, pairwise ΦST values were calculated between watersheds for each gene segment (Table 4). Only one marginally significant population genetic divergence was found, between the San Francisco and Willamette watersheds (p<0.05), among the six watersheds sampled. This finding supports the AMOVA results that only small amounts of variation can be attributed to the among watershed level.
Table 4. Population ΦST values for Planorbella trivolvis populations between watersheds in the West Coast, separated by gene.
| Gene | 1 | 2 | 3 | 4 | 5 | 6 | |
|---|---|---|---|---|---|---|---|
| COI | 1) San Francisco | - | - | - | - | - | - |
| 2) Willamette | 0.41 | - | - | - | - | - | |
| 16S | 1) San Francisco | - | - | - | - | - | - |
| 2) Willamette | 0.14 | - | - | - | - | - | |
| 3) Sacramento | 0.02 | 0.07 | - | - | - | - | |
| 4) Lower Columbia | 0.19 | 0.22 | 1.0 | - | - | - | |
| 5) Middle Columbia | 0.02 | -0.20 | 0.41 | 0.12 | - | - | |
| 6) Coastal | 0.54 | 0.46 | 1.0 | 1.0 | 0.45 | - |
Values with a p-value <0.05 are bolded.
To determine whether geographic distance was associated with genetic distance, linear regressions models were produced. Within both the 16S and COI dataset, orthodromic geographic distance did not predict genetic distance, indicating there is not a significant isolation-by-distance pattern (16S: R2 = 0.002, p = 0.2273; COI: R2 = -0.006, p = 0.417) (Fig 4).
Fig 4. Effect of geographic distance (km) on pairwise genetic distance in Planorbella trivolvis.
(a) The relationship between geographic distance to pairwise genetic distance in the 16S dataset. (b) The relationship between geographic distance to pairwise genetic distance in the COI dataset. Geographic distance did not determine the pairwise genetic distance (p>0.05) in either gene.
3.2 North America
GenBank sequences were included in this study in order to assess the overall genetic diversity of P. trivolvis across their entire native range. A lack of available genetic data meant we were only able to access the genetic structure of P. trivolvis within North America, despite its scattered global appearance [67]. Even within North America, there is only limited existing genetic data from many parts of the United States and Canada. Nonetheless, when the 15 additional GenBank COI sequences were added to create a range-wide network, the result suggested significant East-West structuring across North America (Fig 3).
The added GenBank samples from Canada, Maryland and New Mexico did not fall into the existing haplogroups from the West Coast but formed their own distinct clusters. The average genetic distances among the Canada haplotypes and Maryland haplotypes to the West Coast populations are 8% and 16%, respectively. The average genetic distance between the Canada haplotypes and Maryland haplotypes is 14%. These results are consistent with previous studies that show a higher similarity between freshwater organism populations in a North-South than an East-West direction in North America [27, 28]. The COI gene tree analysis of P. trivolvis highlights phylogenetic structuring and the presence of five well supported clades (Fig 5). Clade A includes all samples from Maryland and Canada and clades B-E include all western United States samples. Clade A contains samples as far west as Alberta, Canada where the observed East-West structuring begins. The clades largely correspond to the haplotype groups recovered in the MSN network.
Fig 5. Maximum likelihood tree based on the COI dataset for Planorbella trivolvis samples in North America.
Bootstrap values for nodes are provided when ≥83%. The tree is shaded to depict the five clades. Tips are color-coded based on sample locality.
4 Discussion
4.1 West Coast
The objective of this study was to address the genetic structuring of the freshwater snail P. trivolvis in the western United States. The results of the AMOVA provided insight into the population structuring of P. trivolvis. Within the 16S dataset, most of the genetic variation derived from within-population differentiation. Little variation was detected within or among watersheds, indicating overall panmixia. Within the COI dataset, most of the genetic diversity was attributed to among populations within a watershed, indicating that the species may exhibit moderate levels of population segregation within watershed. The lack of variation among watersheds is consistent with other studies examining the population structuring of freshwater gastropods, including P. acuta and Taylorconcha serpenticola (Hershler) [9, 19].
Differences in the results between the two genes could be a consequence of smaller sample sizes in the COI dataset or from the differences between the two mitochondrial genes. The COI gene used in this study is considered a universal marker because of its ability to successfully recover gene fragments and rapid rate of evolution in diverse invertebrates [58, 68, 69]. COI has been shown to evolve at a rate that is three times faster than that of 16S in certain groups [69]. The differences in the source of variation in the COI gene segment may result from the faster evolution of the COI gene that is not detected by the 16S gene. Additionally, 16S has been used to assess intrafamilial differentiation within mollusks [70]. Therefore, it may not exhibit enough variations to reveal fine scale genetic structuring compared to COI.
Another goal of this study was to determine the degree to which watershed identity (which helps reflect hydrological connectivity and passive dispersal opportunity) or geographic distance contributed to the observed genetic structuring. We found that watersheds were not a significant factor contributing to the observed population structure. Similarly, we discovered that geographic distance was unrelated to genetic distance, at least based on our measurements from 16S and COI. This result corroborates prior research, which shows that geographic distance is not strongly correlated to the genetic distance between populations of freshwater organisms [28, 71, 72]. It is noteworthy, however, in that freshwater snails generally and Planorbella spp. in particular are often considered to be dispersal-limited owing to the apparent dependence on hydrological connectivity for direct movement [14].
One explanation for the lack of association between watersheds, geographic distance and genetic distance is that dispersal via waterfowl is promoting gene flow across the watersheds. The Pacific Flyway, the westernmost of the four primary bird corridors in North America [73], is a major pathway for migratory birds along the Pacific Coast from Alaska to Mexico. As the waterfowl travel along this flyway, they are likely transporting the snails between sites. Indeed, avian-mediated dispersal has been identified as a potential vector for promoting long-distance gene flow in freshwater snails [21–26, 29, 30]. The occasional dispersal by waterfowl may help explain the North-South genetic similarity in this species. These findings parallel other genetically-based biogeographic assessments of freshwater gastropods which demonstrate population structuring that correlates with dispersal, specifically similar genetic patterns mapping along the Pacific Flyway [19, 74].
Importantly, a lack of geographically isolated populations suggests that despite the limited sampling of this study, most of the shared common haplotypes among populations have been recovered. If the populations were truly isolated, and thus do not share a lot of common haplotypes, the limited sampling would have retrieved a higher number of unique haplotypes. Additionally, if waterfowl were only transporting a small number of snails that successfully establish new populations, genetic bottlenecks are more likely to occur and result in more genetically diverse populations. This suggests that dispersal among populations is relatively frequent. Therefore, although a more comprehensive sampling is needed to further explore P. trivolvis geography, it will likely strengthen the conclusion of this study. There is still a possibility that, by chance, we sampled more common haplotypes than what is actually present in the populations. This can also be evaluated with expanded sampling efforts in the future.
At the regional scale, watersheds are not major barriers to the distribution of P. trivolvis. These results contradict the historical view that Planorbella would be a less effective long-distance disperser when compared to other freshwater gastropods [22–24]. Their generally larger size suggests that they would be less likely to remain attached the waterfowl feathers for any length of time [24]. Remaining attached to waterfowl for long periods of time is only important if dispersal occurs over a smaller number of long-distance dispersal events. Alternatively, their migration along the Pacific Flyway may involve many small, gradual dispersal events, which is consistent with bird migration behavior along the Pacific Flyway where waterfowl utilize waterbodies as stopover points [73]. Additionally, the final size of the adult shell may not dictate their dispersal ability. First, younger snails, whether as egg-masses, newly hatched individuals or sub-adults, tend to be dispersed most often by waterfowl [75]. Second, research has shown that each species has a size at which they disperse that maximizes their success [24]. The final adult size of each species can then be considered less indicative of their dispersal ability.
Future studies are needed to examine the role of waterfowl in freshwater gastropod dispersal. The emphasis should be on smaller, more abundant birds, such as ducks and waders, that are likely important dispersers, but have received less scientific attention relative to larger birds, such as geese and swans [25]. At this time, other long-distance dispersal vectors cannot be ruled out. Anthropogenic-mediated dispersal is also unlikely to cause the observed population structuring pattern because the collection localities are isolated ponds or reservoirs not used for recreation, although the potential role of aquarium releases and the spread of invasive aquatic macrophytes—including snail “hitch-hikers”—have the potential to be influential in certain areas [76, 77].
4.2 North America
Results of our efforts to compile genetic sequences of P. trivolvis populations emphasize the degree to which this taxon is grossly under sampled for molecular analyses of population structure. Efforts to understand the population structuring are complicated by lack of available genetic data, unawareness of their global occurrence and the historic methods of identification based on morphology. This study represents the first published effort to characterize the population genetic structure of P. trivolvis.
The COI range-wide haplogroup network suggests that P. trivolvis displays strong East-West genetic structuring in their native range, despite their continuous distribution throughout North America (https://www.gbif.org/). These results imply that P. trivolvis experiences a high degree of genetic isolation across North America, even if such isolation was not observed along the West Coast specifically. The structuring observed in the haplotype network is further corroborated by the COI gene tree. The phylogenetic analysis supports the presence of five clades, where the Maryland and Canadian samples form their own clade (A) and exhibits long genetic distance from the western US clades B-E. The East-West genetic structuring coincides with the Rocky Mountains. Specimens collected east of the Rocky Mountains fall within clade A, and samples collected west of the Rocky Mountains fall within clades B-E. This pattern has been seen in other freshwater gastropods, such as P. acuta [9]. The isolation could be caused by insurmountable biogeographical barriers, a lack of dispersal vectors that traverse longitudinally across the continent, or both if the barriers also prevent dispersal of the vectors. Alternatively, the significant structuring across the United States may indicate the presence of cryptic species, as the COI genetic distances among these populations are relatively high. These genetic distances are higher than the standard 5% divergence in mtDNA between mollusk species, as seen in studies of Rhagada [78], Eobania [79], Iberus [80], Helixena [81], and Goniobasis species [82]. Other studies assessing the biogeographic patterns of freshwater snails have used mtDNA genetic distances of 1.3% in Pyrgulopsis [74] and 5% in Physella [9] to distinguish between species. A taxonomic study of freshwater Pleuroceridae snails found that interspecific variation in mtDNA ranged from 3.1-5.1% in Pleurocera, 2.2-22.6% in Leptoxis and up to 7.9% in Elimia [83]. The sequence divergences among the samples might suggest that they are distinct species. Further genetic sampling and morphological studies are needed in order to fill in the geographic gaps and determine the true taxonomic identity of Planorbella species in their native range.
The current global distribution of Planorbella is arguably unknown. Small, scattered populations of P. trivolvis have been identified outside North America, but their general abundance and distribution levels have yet to be elucidated [84, 85]. Despite populations of P. trivolvis existing outside their native range, the species is not considered invasive. One area of study relevant to their global distribution is their dispersal ability. As evidenced by its current proposed distribution, the ability of P. trivolvis to disperse effectively at a regional scale does not translate to the global scale. A possible explanation is that cross-continental dispersal, primarily mediated by humans, may be a less effective dispersal mechanism for P. trivolvis. Anthropogenic transport may occur infrequently, or the snails are not able to survive the journey. Additionally, waterfowl species that effectively disperse P. trivolvis in their native range may not be present in other parts of the globe, limiting P. trivolvis ability to successfully colonize new habitats. Alternatively, the life history traits of Planorbella could play a significant role in their limited global distribution. For example, once P. trivolvis reaches a new environment, it may be less likely to successfully establish a population. This could be a result of the species preferential outcrossing mating system, where more than just one specimen is required to start a population [43]. Ecological factors in the new environments also may prevent P. trivolvis from successfully establishing a population. The aquatic environments may lack suitable habitat or resources, support effective predators and competitors, or have physiochemical properties or pollution levels that P. trivolvis cannot tolerate [86].
The lack of knowledge about the global distribution of Planorbella is compounded by their phenotypic plasticity. There is a large gap in the understanding of many freshwater snails due to the traditional methods in snail identification, which rely almost exclusively on shell morphology and internal anatomy. The morphological similarity of Planorbella to other planorbid snail species has impeded the overall understanding of their distribution [87]. Improving their systematics will resolve current species distribution confusion and inform the study of their ecology.
A scarcity of information on the population structuring of P. trivolvis in their native range has prevented an understanding of their role in freshwater ecological interactions. Our results suggest that long-distance dispersal helps shape and maintain the structure of P. trivolvis, which can influence their ecosystem interactions both in their native and global range. Their effective dispersal has important consequences for their role as intermediate hosts to trematode parasites and conservation. Mechanical barriers, such as dams, are often proposed as a way to stop the spread of parasites in freshwater habitats by preventing the spread of their gastropod hosts [88]. Our results suggest that mechanical barriers would mitigate only local dispersal. The snails would still overcome mechanical barriers via bird-mediated dispersal; and that the parasites may also experience similar, if not more, levels of dispersal and gene flow. Additionally, this study suggests that the sustainable population growth of P. trivolvis relies on, in part, the frequency and abundance of contact with waterfowl. These important interactions with waterfowl depend on the availability of suitable habitat. Freshwater habitat modification threatens both aquatic and avian biodiversity by impairing the ability of freshwater snails to disperse locally and impeding contact with waterfowl that may be required for long-distance dispersal [7, 19]. Importantly, our data cannot fully address these questions and future work is necessary to understand how the dispersal patterns of P. trivolvis influence its ecological interactions.
5 Conclusions
This study addressed the population structuring of the freshwater snail P. trivolvis in the western United States and across North America. The observed genetic patterns were not explained by watersheds. Additionally, genetic distance was unrelated to geographic distance. The North-South genetic similarity reflects the Pacific Flyway, which suggests that long-distance dispersal vectors, such as waterfowl, are promoting ongoing gene flow over large geographic ranges and that the population structure of P. trivolvis may be strongly influenced by long-distance dispersal in the West Coast. Cumulatively, these results indicate that, at a regional scale, their distribution is not impeded by dispersal.
The efforts to compare the range-wide structuring of the species was complicated by lack of available information on P. trivolvis. This study is the first to examine the population structure of P. trivolvis. Our results showed that P. trivolvis exhibits significant East-West genetic structuring across their native range. Overall, this paper provides an initial framework for continued biogeographical analysis of Planorbella in their native range.
Supporting information
(CSV)
Acknowledgments
We would like to thank the editor and two reviewers for their excellent comments and suggestions which improved the quality of this manuscript. We would also like to thank Travis McDevitt-Galles, Wynne Moss, Dana Calhoun, and Laura Guderyahn for facilitating snail collection and processing, Dr. Leanne Elder for help with sample curation and Bridget Chalifour for providing comments on the drafts of this manuscript.
Data Availability
All sequences are available from the GenBank database. Accession numbers are listed in the Supporting Information files.
Funding Statement
This study was financed in part by the David and Lucile Packard Foundation (1149308) and the National Science Foundation (1754171) to PTJJ. The Museum Student Research Fund and Collie Fund, both awarded by The University of Colorado Boulder Museum of Natural History, provided some financial support to KRM. A grant from the Oregon Community Foundation to the Bend Science Station provided publishing cost assistance to JB. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Mahmoud KM, Taleb HMA. Fresh water snails as bioindicator for some heavy metals in the aquatic environment. Afr J Ecol. 2013;51:193–8. [Google Scholar]
- 2. Adema CM, Bayne CJ, Knight M, Loker ES, Yoshino TP, Zhang SM. Will All Scientists Working on Snails and the Diseases They Transmit Please Stand Up? PLoS Negl Trop Dis. 2012;6(12):e1835 10.1371/journal.pntd.0001835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cowie RH. Apple snails (Ampullariidae) as agricultural pests: Their biology, impacts and management In: Barker GM, editor. Molluscs as crop pests. Wallingford: CABI Publishing; 2002. p. 145–192. [Google Scholar]
- 4. Carlsson NOL, Bronmark C, Hansson LA. Invading herbivory: The golden apple snail alters ecosystem functioning in Asian wetlands. Ecology. 2004;85:1575–80. [Google Scholar]
- 5. Burlakova LE, Karatayev AY, Padilla DK, Cartwright LD, Hollas DN. Wetland Restoration and Invasive Species: Apple snail (Pomacea insularum) Feeding on Native and Invasive Aquatic Plants. Restor Ecol. 2009;17:433–40. [Google Scholar]
- 6. Lydeard C, Cowie RH, Ponder WF, Bogan AE, Bouchet P, Clark SA, et al. The global decline of nonmarine mollusks. Bioscience. 2004;54:321–30. [Google Scholar]
- 7. Strong EE, Gargominy O, Ponder WF, Bouchet P. Global diversity of gastropods (Gastropoda; Mollusca) in freshwater. Hydrobiologia. 2008;595:149–66. [Google Scholar]
- 8. Morgan JAT, DeJong RJ, Jung YH, Khallaayoune K, Kock S, Mkoji GM, et al. A phylogeny of planorbid snails, with implications for the evolution of Schistosoma parasites. Mol Phylogenet Evol. 2002;25:477–88. [DOI] [PubMed] [Google Scholar]
- 9. Ebbs ET, Loker ES, Brant SV. Phylogeography and genetics of the globally invasive snail physa acuta Draparnaud 1805, and its potential to serve as an intermediate host to larval digenetic trematodes. BMC Evol Biol. 2018;18:103 10.1186/s12862-018-1208-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brown KM, Lydeard C. Mollusca: Gastropoda In: Thorp JH, Covich AP, editors. Ecology and classification of North American freshwater invertebrates. San Diego: Academic Press; 2010. p. 277–306. [Google Scholar]
- 11. Bilton DT, Freeland JR, Okamura B. Dispersal in freshwater invertebrates. Annu Rev Ecol Evol Syst. 2001;32:159–181. [Google Scholar]
- 12. Meester LD, Gómez A, Okamura B, Schwenk K. The Monopolization Hypothesis and the dispersal–gene flow paradox in aquatic organisms. Acta Oecol. 2002;23:121–135. [Google Scholar]
- 13. Bohonak AJ, Jenkins DG. Ecological and evolutionary significance of dispersal by freshwater invertebrates. Ecol Lett. 2003;6:783–796. [Google Scholar]
- 14. Kappes H, Haase P. Slow, but steady: dispersal of freshwater molluscs. Aquat Sci. 2012;74:1–14. [Google Scholar]
- 15. Bondesen P, Kaiser EW. Hydrobia (Potamopyrgus) jenkinsi Smith in Denmark illustrated by its ecology. Oikos. 1949;1:252–81. [Google Scholar]
- 16. Haynes A, Taylor BJR, Varley ME. The influence of the mobility of Potamopyrgus jenkinsi (Smith, E.A.) (Prosobranchia, Hydrobiidae) on its spread. Arch Hydrobiol. 1985;103:497–508. [Google Scholar]
- 17. Brown RJ. Freshwater mollusks survive fish gut passage. Arctic. 2007;60:124–8. [Google Scholar]
- 18. Ribi G. Within-lake dispersal of the prosobranch snails, Viviparus ater and Potamopyrgus jenkinsi. Oecologia. 1986;69:60–3. [DOI] [PubMed] [Google Scholar]
- 19. Liu HP, Hershler R. Genetic diversity and population structure of the threatened Bliss Rapids snail (Taylorconcha serpenticola). Freshw Biol. 2009;54:1285–99. [Google Scholar]
- 20. Darwin C. The origin of species by means of natural selection: or, the preservation of favored races in the struggle for life. Harmondsworth: Penguin; 1959. [Google Scholar]
- 21. Roscoe EJ. Aquatic snails found attached to feathers of white-faced glossy ibis. Wilson Bull. 1955;67:66–7. [Google Scholar]
- 22.Malone CR. Dispersal of fresh-water gastropods by water birds [M.S. Thesis]. Texas Technological College; 1965.
- 23. Malone CR. Dispersal of aquatic gastropods via the intestinal tract of water birds. Nautilus. 1965;78:135–9. [Google Scholar]
- 24. Boag DA. Dispersal in Pond Snails—Potential Role of Waterfowl. Can J Zool. 1986;64:904–9. [Google Scholar]
- 25. Figuerola J, Green AJ. Dispersal of aquatic organisms by waterbirds: a review of past research and priorities for future studies. Freshw Biol. 2002;47:483–94. [Google Scholar]
- 26. Green AJ, Figuerola J. Recent advances in the study of long-distance dispersal of aquatic invertebrates via birds. Divers Distrib. 2005;11:149–56. [Google Scholar]
- 27. Taylor DJ, Finston TL, Hebert PDN. Biogeography of a widespread freshwater crustacean: Pseudocongruence and cryptic endemism in the North American Daphnia laevis complex. Evolution. 1998;52:1648–70. [DOI] [PubMed] [Google Scholar]
- 28. Freeland JR, Romualdi C, Okamura B. Gene flow and genetic diversity: a comparison of freshwater bryozoan populations in Europe and North America. Heredity. 2000;85:498–508. [DOI] [PubMed] [Google Scholar]
- 29. Leeuwen CHV, Huig N, der Velde GV, Alen TAV, Wagemaker CA, Sherman CD, et al. How did this snail get here? Several dispersal vectors inferred for an aquatic invasive species. Freshw Biol. 2013;58:88–99. [Google Scholar]
- 30. Saito T, Hirano T, Prozorova L, Tu DV, Sulikowska-Drozd A, Sitnikova T, et al. Phylogeography of freshwater planorbid snails reveals diversification patterns in Eurasian continental islands. BMC Evol Biol. 2018;18(164). 10.1186/s12862-018-1273-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Baker FC, Cleave HJV. Molluscan family Planorbidae. Ubana: University of Illinois press; 1945. [Google Scholar]
- 32. Burch JB. Freshwater snails (Mollusca: Gastropoda) of North America. Washington D.C.: Environmental Protection Agency; 1982. [Google Scholar]
- 33. Clampitt PT. Comparative ecology of the snails Physa gyrina and Physa intgra (Basommatophora: Physidae). Malacologia. 1970;10:113–51. [Google Scholar]
- 34. Brackenbury TD, Appleton CC. Effect of controlled temperatures on gametogenesis in the gastropods Physa Acuta (Physidae) and Bulinus Tropicus (Planorbidae). J Molluscan Stud. 1991;57:461–9. [Google Scholar]
- 35. Monsutti A, Perrin N. Dinucleotide microsatellite loci reveal a high selfing rate in the freshwater snail Physa acuta. Mol Ecol. 1999;8:1076–8. [Google Scholar]
- 36. Gittenberger E, Janssen AW, Kuijper WJ, Kuiper JG, Meijer T, der Velde GV, et al. De Nederlandse Zoetwatermollusken: Recente en fossiele weekdieren uit zoet en brak water. 2nd ed Utrecht: KNNV Publishing; 2004. [Google Scholar]
- 37. Zukowski S, Walker KF. Freshwater snails in competition: alien Physa acuta (Physidae) and native Glyptophysa gibbosa (Planorbidae) in the River Murray, South Australia. Mar Freshw Res. 2009;60:999–1005. [Google Scholar]
- 38. DeWitt RM, Sloan WC. Reproduction in Physa pomilia and Helisoma duryi. Anim Behav. 1959;7:81–4. [Google Scholar]
- 39. Paraense WL, Correa LR. Self-fertilization in the freshwater snails Helisoma duryi and Helisoma trivolvis. Memórias do Instituto Oswaldo Cruz. 1988;83:405–10. [DOI] [PubMed] [Google Scholar]
- 40. Jarne P, Vianeyliaud M, Delay B. Selfing and outcrossing in hermaphrodite freshwater gastropods (Basommatophora)—where, when and why. Biol J Linn Soc Lond. 1993;49:99–125. [Google Scholar]
- 41. Escobar JS, Auld JR, Correa AC, Alonso JM, Bony YK, Coutellec MA, et al. Patterns of mating-system evolution in hermaphroditic animals: Correlations among selfing rate, inbreeding depression, and the timing of reproduction. Evolution. 2011;65:1233–53. [DOI] [PubMed] [Google Scholar]
- 42. Norton CG, Johnson AF, Nelson BM. The genetic basis of albinism in the hermaphroditic freshwater snail Planorbella trivolvis. Am Malacol Bull. 2018;36:153–7. [Google Scholar]
- 43. Bousset L, Henry PY, Jarne P. Population biology of the invasive freshwater snail Physa acuta approached through genetic markers, ecological characterization and demography. Mol Ecol. 2004;13:2023–36. [DOI] [PubMed] [Google Scholar]
- 44. Pfenninger M, Cordellier M, Streit B. Comparing the efficacy of morphologic and DNA-based taxonomy in the freshwater gastropod genus Radix (Basommatophora, Pulmonata). BMC Evol Biol. 2006;6(100). 10.1186/1471-2148-6-100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fiorentino V, Salomone N, Manganelli G, Giusti F. Phylogeography and morphological variability in land snails: the Sicilian Marmorana (Pulmonata, Helicidae). Biol J Linn Soc Lond. 2008;94:809–23. [Google Scholar]
- 46. Schniebs K, Gloeer P, Vinarski MV, Beran L, Hundsdoerfer AK. Intraspecific morphological and genetic variability in the palaearctic freshwater snail Radix ampla (Hartmann, 1821) (Gastropoda: Basommatophora: Lymnaeidae). J Conchol. 2019;43:245–67. [Google Scholar]
- 47. Whelan NV, Galaska MP, Sipley BN, Weber JM, Johnson PD, Halanych KM, et al. Riverscape genetic variation, migration patterns, and morphological variation of the threatened Round Rocksnail, Leptoxis ampla. Mol Ecol. 2019;28:1593–610. [DOI] [PubMed] [Google Scholar]
- 48. Minton RL, Norwood AP, Hayes DM. Quantifying phenotypic gradients in freshwater snails: a case study in Lithasia (Gastropoda: Pleuroceridae). Hydrobiologia. 2008;605:173–82. [Google Scholar]
- 49. Perez KE, Minton RL. Practical applications for systematics and taxonomy in North American freshwater gastropod conservation. J North Am Benthol Soc. 2008;27:471–83. [Google Scholar]
- 50. Dillon RT Jr. The classification of the Planorbidae In: the Freshwater Gastropods of North America. vol. 2, Essays on the Pulmonates. Charleston: FWGNA Press; 2019. p. 127–135. [Google Scholar]
- 51. Burch JB. North American freshwater snails. Hamburg: Malacological Publications; 1989. [Google Scholar]
- 52. Taylor DW. Summary of North American Blancan nonmarine mollusks. Malacologia. 1966;4:1–172. [Google Scholar]
- 53. Wethington AR, Lydeard C. A molecular phylogeny of Physidae (Gastropoda: Basommatophora) based on mitochondrial DNA sequences. J Molluscan Stud. 2007;73:241–57. [Google Scholar]
- 54. Hubendick B, Rees WJ. Phylogeny in the Planorbidae. Trans Zool Soc Lond. 1955;28:453–542. [Google Scholar]
- 55. Wang SY, Tattersall GJ, Koprivnikar J. Trematode Parasite Infection Affects Temperature Selection in Aquatic Host Snails. Physiol Biochem Zool. 2019;92:71–9. [DOI] [PubMed] [Google Scholar]
- 56. Wethington AR, Guralnick R. Are populations of physids from different hot springs distinctive lineages? Am Malacol Bull. 2004;19:135–44. [Google Scholar]
- 57. Paraense WL, Correa LR. Self-fertilization in the freshwater snails Helisoma duryi and Helisoma trivolvis. Memórias do Instituto Oswaldo Cruz. 1996;83:405–10. [DOI] [PubMed] [Google Scholar]
- 58. Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–9. [PubMed] [Google Scholar]
- 59. Collado GA, Mendez MA. Phylogenetic relationships and taxonomy of Altiplano populations of Biomphalaria (Gastropoda: Planorbidae): inference from a multilocus approach. Zool J Linn Soc. 2012;165(4):795–808. [Google Scholar]
- 60. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–8. [DOI] [PubMed] [Google Scholar]
- 61. Leigh JW, Bryant D. POPART: full-feature software for haplotype network construction. Methods in Ecol Evol. 2015;6:1100–6. [Google Scholar]
- 62. Excoffier L, Smouse PE, Quattro JM. Analysis of Molecular Variance Inferred from Metric Distances Among DNA Haplotypes—Application to Human Mitochondrial-DNA Restriction Data. Genetics. 1992;131:479–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–7. [DOI] [PubMed] [Google Scholar]
- 64. Team RC. R: a language and environment for statistical computing; 2018. [Google Scholar]
- 65. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–9. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Appleton CC. Alien and invasive fresh water Gastropoda in South Africa. Afr J Aquat Sci. 2003;28:669–81. [Google Scholar]
- 68. Zhang DX, Hewitt GM. Assessment of the universality and utility of a set of conserved mitochondrial COI primers in insects. Insect Mol Biol. 1997;6:143–50. [DOI] [PubMed] [Google Scholar]
- 69. Knowlton N, Weigt LA. New dates and new rates for divergence across the Isthmus of Panama. Pro Biol Sci. 1998;265:2257–63. [Google Scholar]
- 70. Bonnaud L, Boucherrodoni R, Monnerot M. Phylogeny of Decapod Cephalopods based on partial 16S rDNA nucleotide-sequences. Comptes Rendus de l’Académie des Sciences—Series III—Sciences de la Vie. 1994;317:581–8. [PubMed] [Google Scholar]
- 71. Hebert PDN, Finston TL. Genetic differentiation in Daphnia obtusa: A continental perspective. Freshw Biol. 1996;35:311–21. [Google Scholar]
- 72. Vanoverbeke J, Meester LD. Among-populational genetic differentiation in the cyclical parthenogen Daphnia magna (Crustacea, Anomopoda) and its relation to geographic distance and clonal diversity. Hydrobiologia. 1997;360:135–42. [Google Scholar]
- 73. Wilson RM. Seeking Refuge: Birds and Landscapes of the Pacific Flyway. Seattle: University of Washington Press; 2010. [Google Scholar]
- 74. Liu HP, Hershler R, Clift K. Mitochondrial DNA sequences reveal extensive cryptic diversity within a western American springsnail. Mol Ecol. 2003;12:2771–82. [DOI] [PubMed] [Google Scholar]
- 75. Boss KJ. Pulmonates. vol. 2A New York: Academic Press; 1978. [Google Scholar]
- 76. Madsen H, Frandsen F. The spread of freshwater snails including those of medical and veterinary importance. Acta Trop. 1989;46:139–46. [DOI] [PubMed] [Google Scholar]
- 77. Beran L. Unintentional introduction of aquatic molluscs from Poland to Prague (Czech Republic). Malacol Bohemoslov. 2006;5:6–9. [Google Scholar]
- 78. Johnson MS, Stankowski S, Whisson CS, Teale RJ, Hamilton ZR. Camaenid land snails on Barrow Island: distributions, molecular phylogenetics and taxonomic revision. Rec West Aust Mus. 2013;83:159–71. [Google Scholar]
- 79. Desouky MM, Busais S. Phylogenetic relationships of the land snail; Eobania vermiculata (Müller, 1774) from Egypt and Saudi Arabia. A combined morphological and molecular analysis. J Basic Appl Zool. 2012;65:144–51. [Google Scholar]
- 80. Elejalde MA, Madeira MJ, Arrebola JR, Munoz B, Gomez-Moliner BJ. Molecular phylogeny, taxonomy and evolution of the land snail genus Iberus (Pulmonata: Helicidae). J Zool Syst Evol Res. 2008;46(3):193–202. [Google Scholar]
- 81. Riel PV, Jordaens K, Houtte NV, Martins AMF, Verhagen R, Backeljau T. Molecular systematics of the endemic Leptaxini (Gastropoda: Pulmonata) on the Azores islands. Mol Phylogenet Evol. 2005;37:132–43. [DOI] [PubMed] [Google Scholar]
- 82. Dillon RT, Frankis RC. High levels of mitochondrial DNA sequence divergence in isolated populations of freshwater snails of the genus Goniobasis Lea, 1862. Am Malacol Bull. 2004;19:69–77. [Google Scholar]
- 83. Lydeard C, Holznagel WE, Garner J, Hartfield P, Pierson JM. Molecular phylogeny of mobile river drainage basin pleurocerid snails (Caenogastropoda: Cerithioidea). Mol Phylogenet Evol. 1997;7:117–28. [DOI] [PubMed] [Google Scholar]
- 84. Frandsen F, Madsen H. Review of Helisoma duryi in biological control. Acta Trop. 1979;36:67–84. [PubMed] [Google Scholar]
- 85. Cianfanelli S, Lori E, Bodon M. Non-indigenous freshwater molluscs and their distribution in Italy In: Gherardi G, editor. Biological invaders in inland waters: Profiles, distribution, and threats. Dordrecht: Springer; 2007. p. 103–21. [Google Scholar]
- 86. Fernandez MA, Thiengo SC, Bezerra FSM, Alencar LMS. Current distribution of the exotic freshwater snail Helisoma duryi (Gastropoda: Planorbidae) in Brazil. Nautilus. 2010;124:44–50. [Google Scholar]
- 87. Jelnes JE. Enzyme analyses on 7 laboratory stocks and 2 natural populations of Helisoma duryi—electrophoretic patterns of 8 enzymes with genetic information on 4 polymorphic enzymes. Hereditas. 1982;97:9–15. [Google Scholar]
- 88. Clennon JA, King CH, Muchiri EM, Kitron U. Hydrological modelling of snail dispersal patterns in Msambweni, Kenya and potential resurgence of Schistosoma haematobium transmission. Parasitology. 2006;134:683–693. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(CSV)
Data Availability Statement
All sequences are available from the GenBank database. Accession numbers are listed in the Supporting Information files.