Abstract
The arbovirus vectors Aedes aegypti (yellow fever mosquito) and Ae. albopictus (Asian tiger mosquito) are both common throughout the Indo-Pacific region, where 70% of global dengue transmission occurs. For Ae. aegypti all Indo-Pacific populations are invasive, having spread from an initial native range of Africa, while for Ae. albopictus the Indo-Pacific includes invasive populations and those from the native range: putatively, India to Japan to Southeast Asia. This study analyses the population genomics of 480 of these mosquitoes sampled from 27 locations in the Indo-Pacific. We investigated patterns of genome-wide genetic differentiation to compare pathways of invasion and ongoing gene flow in both species, and to compare invasive and native-range populations of Ae. albopictus. We also tested landscape genomic hypotheses that genetic differentiation would increase with geographical distance and be lower between locations with high connectivity to human transportation routes, the primary means of dispersal at these scales. We found that genetic distances were generally higher in Ae. aegypti, with Pacific populations the most highly differentiated. The most differentiated Ae. albopictus populations were in Vanuatu, Indonesia and Sri Lanka, the latter two representing potential native-range populations and potential cryptic subspeciation respectively. Genetic distances in Ae. aegypti increased with geographical distance, while in Ae. albopictus they decreased with higher connectivity to human transportation routes. Contrary to the situation in Ae. aegypti, we found evidence of long-distance Ae. albopictus colonisation events, including colonisation of Mauritius from East Asia and of Fiji from Southeast Asia. These direct genomic comparisons indicate likely differences in dispersal ecology in these species, despite their broadly sympatric distributions and similar use of human transport to disperse. Our findings will assist biosecurity operations to trace the source of invasive material and for biocontrol operations that benefit from matching genetic backgrounds of released and local populations.
Author summary
The mosquitoes Ae. aegypti and Ae. albopictus are highly invasive and transmit dengue and other arboviruses. This study investigates the genetics of these mosquitoes in the Indo-Pacific region, where 70% of global dengue transmission occurs and where both species have established widespread invasions by hitch-hiking on human transport vessels. We compared patterns of genetic differentiation to determine the pathways these species have taken while spreading through the Indo-Pacific, and to better understand how they disperse. We sequenced DNA from 480 mosquitoes sampled from 27 locations in the Indo-Pacific, and found many genetic differences between the two species. Populations of Ae. aegypti, which is not native to the region, tended to be genetically different from each other, and populations in the Pacific Ocean were particularly divergent. Aedes albopictus populations were generally more similar to each other, though genetically different populations in Sri Lanka and Indonesia point to these having a different history to other populations. Genetic differences between Ae. aegypti populations were larger when populations were geographically distant, while differences between Ae. albopictus populations were larger when populations likely had limited access to human transportation. These results will help improve strategies for controlling these species and stopping their spread around the world.
Introduction
The Indo-Pacific region, here defined as encompassing the Indian Ocean, the western and central Pacific Ocean, and the coastal territories therein, is the site of 70% of global dengue transmission [1]. Infections are vectored by two mosquito species, Aedes aegypti (yellow fever mosquito) and Ae. albopictus (Asian tiger mosquito) [2,3], both of which have established widespread invasions in the region. Aedes aegypti, the primary regional vector, has a putative native range of West Africa [4] and is thought to have invaded the Indian Ocean via the Mediterranean [5], while its invasion history in the western Pacific remains unclear but likely happened in the ~18-19th c. [6]. Aedes albopictus has a hypothesised native range that includes Japan, China, northern India, and parts of Southeast Asia [7]. The southern extent of this range remains unclear and may extend as far as Indonesia and New Guinea [7–9]. Regional expansion of Ae. albopictus is thought to have occurred more recently than Ae. aegypti, with colonisation likely involving successive waves [8,10].
Like many invasive taxa, Ae. aegypti and Ae. albopictus (hereafter Aedes spp.) exhibit stratified dispersal. This consists of two distinct processes: short-range, active dispersal and long-range, passive dispersal [11]. Active dispersal of Aedes aegypti is generally highly localised, as measured by both traditional mark-release-recapture methods (8–199 m/gen [12]) or current methods using genomic estimates of relatedness (33–131 m/gen [13]). Similar observations have been made of Ae. albopictus using mark-release-recapture [14]. At the metropolitan scale (< 50 km), passive dispersal in Aedes spp. has been observed directly [15] or inferred from the spatial distribution of close kin [16,17]. Dispersal at broader scales such as between cities is almost exclusively by passive transportation on ships, aircraft and land vehicles [18–21].
The unrestricted spread of Aedes spp. has many adverse consequences. Both species are presently continuing to colonise new tropical, subtropical and, for Ae. albopictus, temperate regions [22], and future expansions are expected in the face of increased urbanisation and climate change [23]. In addition to range expansions, dispersal between Aedes spp. populations can spread advantageous alleles that reduce the efficacy of local control programs [24]. Incursions of these species are frequently linked to vessels such as ships and planes [19,20,25], and when source locations can be identified they are often far from the point of collection [20,25].
Understanding the forces that influence dispersal, gene flow, and population structure in Aedes spp. can help efforts to prevent, control, and prepare for future threats from these species. As well as being a critical focus for dengue control efforts, the Indo-Pacific presents a useful location to investigate these processes due to the broad sympatry of Aedes spp. within the region. Investigating the genetics of sympatric taxa can help link patterns of genetic differentiation to the forces that shape population structure [26,27]. For instance, when taxa share a common area but have contrasting patterns of differentiation, this may indicate intrinsic differences in how they disperse [28–30], while similar dispersal behaviour can produce similar patterns of differentiation [31].
Here we investigate these processes in Aedes spp. from the Indo-Pacific region at spatial scales from ~20 km (inter-city) to ~16,000 km (trans-oceanic). Dispersal at these scales largely proceeds along the network of shipping routes that link each inhabited coastal location with every other location (Fig 1). As Aedes spp. are thought to make use of the same vessel types for transportation, their dispersal may take place along similar pathways, which may produce similar patterns of genetic structure [31,32]. However, there are several reasons why different patterns might be observed. Aedes albopictus may be able to survive transportation over longer distances than Ae. aegypti by undergoing quiescence and diapause [33–35] which confers resistance to cold and desiccation [36]. Aedes aegypti does not undergo diapause but can make use of egg quiescence to survive adverse conditions [35]. Heterogeneous histories of colonisation may also produce differences in structure. At broad scales, this relates to differences between the two species in the timing and direction of invasions. At finer scales, stochastic processes during colonisation can produce rapid change in allele frequencies [37]. For any of the above processes, genetic structure can be resistant to ‘erosion’ via gene flow if local population densities are high [38] as they generally are for Aedes spp.
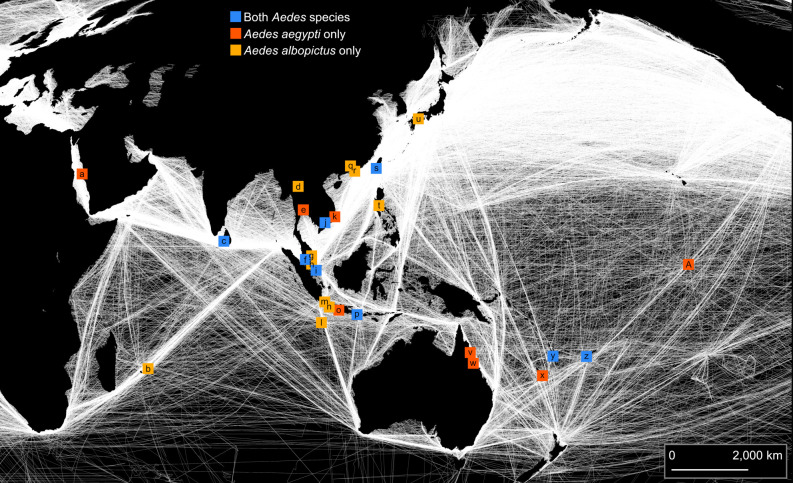
Fig 1. Sampling locations of Aedes aegypti and Ae. albopictus.
White lines indicate density of shipping routes. The map uses a Mollweide projection with a central meridian of 120°E. The map was produced in arcmap 10.5.1 using shipping route data made available from Halpern et al. [39].
This study uses high-resolution markers to compare patterns of broad-scale genetic structure in Aedes spp.. Few such comparative studies have been conducted [40,41], and none have used high-resolution molecular markers, which have greater demonstrated power to distinguish between Aedes populations [42]. We also test landscape genomic hypotheses of isolation by distance [43] and gene flow via connectivity to human transportation routes. These analyses provide a means of disentangling the putative influences of human transportation routes which may direct gene flow, high local population density following invasion which may limit the effects of gene flow, and diapause in Ae. albopictus which may permit gene flow between more distant populations than for Ae. aegypti. Recently, single-species studies using high-resolution molecular markers have reported strong divergence among Ae. aegypti populations [5,42] and weaker divergence among Ae. albopictus populations [44,45], though highly-differentiated Ae. albopictus populations have been recorded [46]. As broad-scale studies have typically included a greater number of samples from European, African and American Aedes spp. populations than from Indo-Pacific populations [5,44,45,47], this study also fills an important geographical gap in the population genomics literature on Aedes spp..
We found clear differences between the spatial genetic structure of Ae. aegypti and Ae. albopictus, and our landscape genomic analyses indicated that Ae. aegypti populations were generally structured by geographical distance, and Ae. albopictus populations by connectivity to human transport routes. These results may reflect important differences in the capacity of Aedes spp. to disperse and to invade new regions, and may also reflect the limited power of gene flow to erode existing structure in species with high census sizes. The findings of this study will assist biosecurity operations that aim to trace the source of invasive material [20,48] and for biocontrol operations that benefit from matching genetic backgrounds of released and local populations [49].
Materials and methods
Sample collection, genotyping, processing and subsampling
Aedes aegypti were sampled from 16 locations in the Indo-Pacific region, across a ~16,000 km range from Jeddah, Saudi Arabia to Kiribati (Fig 1, Table 1). Aedes albopictus were sampled from 19 locations ranging from Mauritius to Fiji to Matsuyama, Japan (Fig 1, Table 2). We considered mosquitoes collected within the same city to be from the same population, as structuring within these scales is typically weak (e.g. [17]). The smallest distance between two distinct populations was ~20 km, between Singapore and Johor, Malaysia. Among populations within the same country, the smallest distance was ~130 km, between Guangzhou and Hong Kong in China.
Table 1. Details of Aedes aegypti sampled from 16 populations.
See Fig 1 for map id locations. See Materials and Methods for details regarding filtering and calculation of connectivity indices.
| map id (see Fig 1) | sample | location(s) | country | year(s) collected | no. genotypes after filtering | connectivity (marine) | connectivity (aerial) |
|---|---|---|---|---|---|---|---|
| a | Jeddah | Various locations in Jeddah | Saudi Arabia | 2018 | 18 | 7.92 | 194 |
| c | Colombo | Kiribathgoda | Sri Lanka | 2017 | 12 | 9.62 | 101 |
| e | Bangkok | Seven locations in Bangkok | Thailand | 2016 | 18 | 0.17 | 326 |
| f | Kuala Lumpur | Bandar Sunway/UKM Specialist Centre | Malaysia | 2015/2017 | 18 | 3 | 257 |
| i | Singapore | Various locations in Singapore | Singapore | 2015 | 15 | 2.64 | 408 |
| j | Ho Chi Minh City | Various locations in Ho Chi Minh City | Vietnam | 2014 | 18 | 0.43 | 109 |
| k | Nha Trang | Hon Mieu Island | Vietnam | 2016 | 11 | 5.26 | 7 |
| o | Yogyakarta | Various locations in Yogyakarta, Java | Indonesia | 2014 | 13 | 0.44 | 29 |
| p | Bali | Near Denpasar Airport, Bali | Indonesia | 2016/2017 | 18 | 1.24 | 100 |
| s | Taiwan | Various locations in Southwest Taiwan | Republic of China | 2016 | 8 | 7.25 | 65 |
| v | Cairns | Six locations in urban Cairns | Australia | 2015 | 18 | 1.86 | 56 |
| w | Townsville | Near Townsville Airport | Australia | 2014 | 10 | 2.25 | 16 |
| x | New Caledonia | Various locations in Nouméa | New Caledonia | 2018 | 13 | 1.24 | 13 |
| y | Vanuatu | Various locations in Efate | Vanuatu | 2018 | 14 | 0.71 | 28 |
| z | Fiji | Various locations in Nadi | Fiji | 2018 | 11 | 0.85 | 43 |
| A | Kiribati | Various locations in Kiribati | Kiribati | 2018 | 9 | 0.8 | 3 |
Table 2. Details of Aedes albopictus sampled from 20 populations.
See Fig 1 for map id locations. See Materials and Methods for details regarding filtering and calculation of connectivity indices. Colombo-1 was treated as a distinct population due to divergences (see text), and omitted (*) from landscape genomic analyses.
| map id (see Fig 1) | sample name | location(s) | country | year(s) collected | no. genotypes after filtering | connectivity (marine) | connectivity (aerial) |
|---|---|---|---|---|---|---|---|
| b | Mauritius | Near Chamarel village | Mauritius | 2017 | 7 | 2.68 | 41 |
| c | Colombo-1 | Delgoda | Sri Lanka | 2017 | 7 | * | * |
| c | Colombo-2 | Delgoda and Kiribathgoda | Sri Lanka | 2017 | 8 | 9.62 | 101 |
| d | Chiang Mai | Near Wachirathan Waterfall | Thailand | 2017 | 5 | 0 | 48 |
| f | Selangor | Kuala Lumpur and Gombak, Selangor | Malaysia | 2016 | 11 | 3 | 257 |
| g | Pahang | Kuantan | Malaysia | 2015 | 18 | 3.2 | 5 |
| h | Johor | Johor Baru | Malaysia | 2016 | 18 | 3.28 | 18 |
| i | Singapore | Near Bukit Timah Nature Reserve | Singapore | 2017 | 14 | 2.07 | 408 |
| j | Ho Chi Minh City | Various locations in Ho Chi Minh City | Vietnam | 2017 | 18 | 0.44 | 109 |
| l | Christmas Island | Near Christmas Island International Airport | Australia | 2017 | 7 | 1.91 | 2 |
| m | Jakarta | Various locations in Jakarta, Java | Indonesia | 2016 | 17 | 1.09 | 185 |
| n | Bandung | Various locations in Bandung, Java | Indonesia | 2017 | 14 | 0.53 | 21 |
| p | Bali | Various locations in Bali | Indonesia | 2016/2017 | 17 | 1.24 | 100 |
| q | Guangzhou | 13 locations in Guangzhou | China | 2015 | 16 | 0.24 | 338 |
| r | Hong Kong | Various locations in Hong Kong | China | 2016 | 16 | 3.87 | 355 |
| s | Taiwan | Various locations in Southwest Taiwan | Republic of China | 2016 | 15 | 7.24 | 65 |
| t | Manila | Manila, Quezon City and Makati City | Philippines | 2017 | 6 | 1.56 | 193 |
| u | Matsuyama | Near Ehime University | Japan | 2017 | 11 | 1.97 | 12 |
| y | Vanuatu | Various locations in Efate | Vanuatu | 2018 | 15 | 0.71 | 28 |
| z | Fiji | Various locations in and near to Nadi | Fiji | 2018 | 16 | 0.84 | 43 |
Mosquitoes were genotyped using a pipeline that has been described elsewhere [20] and is described here in S1 Text. Briefly, we used the double-digest restriction site-associated (ddRAD) sequencing protocol for Ae. aegypti [42] to obtain sequence reads, which were processed in Stacks v2.0 [50]. We used Bowtie v2.0 [51] to align reads to the Ae. aegypti [52] and Ae. albopictus [53] mitochondrial genome (mtDNA) assemblies. Reads that did not align to the mtDNA assemblies were aligned to the nuclear assemblies AaegL5 [54] and AaloF1 [55] to obtain nuclear genotypes for Ae. aegypti and Ae. albopictus respectively. This ensured that the nuclear genotypes would not include transpositions of mitochondrial DNA into the nuclear genome (NUMTs), a common occurrence in Aedes spp. [56]. As some population samples contained more close kin than others, we removed individuals in order of missing data so that no first order kin pairs remained and no population had greater than 18 genotypes, following the procedure described in S1 Text.
We retained 224 Ae. aegypti and 256 Ae. albopictus genotypes. We used the Stacks program ref_map to build catalogs containing all mosquito genotypes from each species, from which we called single nucleotide polymorphism (SNP) genotypes at RAD stacks using default Stacks settings. Only SNPs scored in at least 75% of genotypes from each population were used in analyses.
Individual-level genetic structure
We investigated genetic structure among individual mosquito genotypes using three methods: discriminant analysis of principal components (DAPC [57]); sparse non-negative matrix factorisation (sNMF [58]); and PCA-UMAP [59], which combines principal components analysis (PCA) with uniform manifold approximation and projection (UMAP [60]). For these analyses, we filtered the 224 Ae. aegypti and 256 Ae. albopictus genotypes to retain biallelic SNPs with < 20% missing data, with minimum depth of coverage of 3, and maximum depth of coverage of 45. Genotypes were phased and missing data imputed in Beagle v4.1 [61] using default settings and 10 iterations.
DAPC was run in the R package adegenet [62]. In DAPC, each genotype is assigned to one of K genetic clusters. We ran the ‘find.clusters’ algorithm to determine the optimal K for 1 ≤ K ≤ N, with N equal to the number of populations (16 for Ae. aegypti, 20 for Ae. albopictus). We ran 1x109 iterations and selected the value of K with the lowest Bayesian information criterion (BIC). We then repeated the above analysis, selecting the value of K with the lowest Akaike information criterion (AIC), a less conservative measure than BIC.
We ran sNMF in the R package LEA [63]. This analysis estimates individual ancestry coefficients, assuming that individual genotypes are produced from the admixture of K ancestral lineages, where K is unknown a priori. To determine which value of K was optimal for summarising the variation in each species, we set 1 ≤ K ≤ N and ran 100 iterations of the sNMF algorithm for each K, selecting the K with the lowest cross-entropy across all runs. For the chosen K we selected the iteration with the lowest cross-entropy for visual presentation.
To generate PCA-UMAPs, we adapted code provided in Diaz-Papkovich et al. [59]. UMAP provides a means of projecting high-dimensional data onto two-dimensional space, and has advantages over other dimensionality reduction techniques in that it better preserves the global data structure between clusters in reduced dimensions. Combining UMAP and PCA has been shown to produce optimum definition of population clusters [59]. PCA-UMAPs used the first 5 principal components of the PCA, which were projected in two dimensions via UMAP using 50 neighbours and a minimum distance of 0.5.
Population-level genetic structure
For investigations of population-level genetic structure, we were cautious to process and filter the data so that uneven population sample size (n) would not bias analyses, while still including as many genotypes as possible. To balance these aims, we subsampled each dataset ten times, sampling (with replacement) from each population a number of genotypes (five) equal to the minimum n among all populations. For each subsample we used the Stacks program populations to calculate FST’ [64] between each pair of populations, and to calculate mean observed heterozygosity (HO), nucleotide diversity (π), number of private alleles and proportion of missing data for each population. We used the results from the ten subsamples to calculate the mean and 95% confidence intervals of each estimated parameter.
For genome-wide SNP datasets, genetic differentiation can be estimated with five genotypes [65]. To determine whether using only five genotypes in each subsample affected our FST’ estimates, we took ten subsamples of 16 individuals from the Ae. aegypti populations in Bali, Bangkok, Jeddah and Kuala Lumpur, and calculated FST’ means and 95% confidence intervals as above. We compared these results with those calculated with five genotypes.
Analysis of the data indicated a deep genetic division within the Sri Lankan Ae. albopictus sample (see Fig 2ii and Results: Individual-level genetic structure). Seven of the 15 genotypes formed a group highly divergent from all others, including the other Sri Lankan genotypes sampled from the same region of Colombo. This did not appear to be an artefact of sampling close kin; only a single half-sib dyad was present in the 7 genotypes, a similar incidence as in other populations. We therefore treated the Sri Lankan Ae. albopictus sample as two separate populations, denoted as Colombo-1 (highly divergent) and Colombo-2. We investigated this divergence further by calculating the folded allele frequency spectrum for each Ae. albopictus population in VCFtools [66], using the ten subsamples of five genotypes to calculate means and 95% confidence intervals.
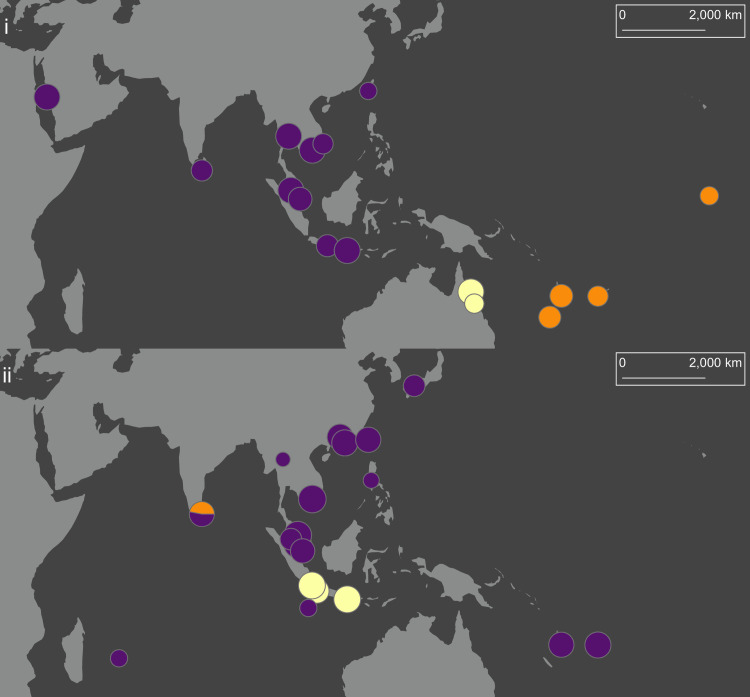
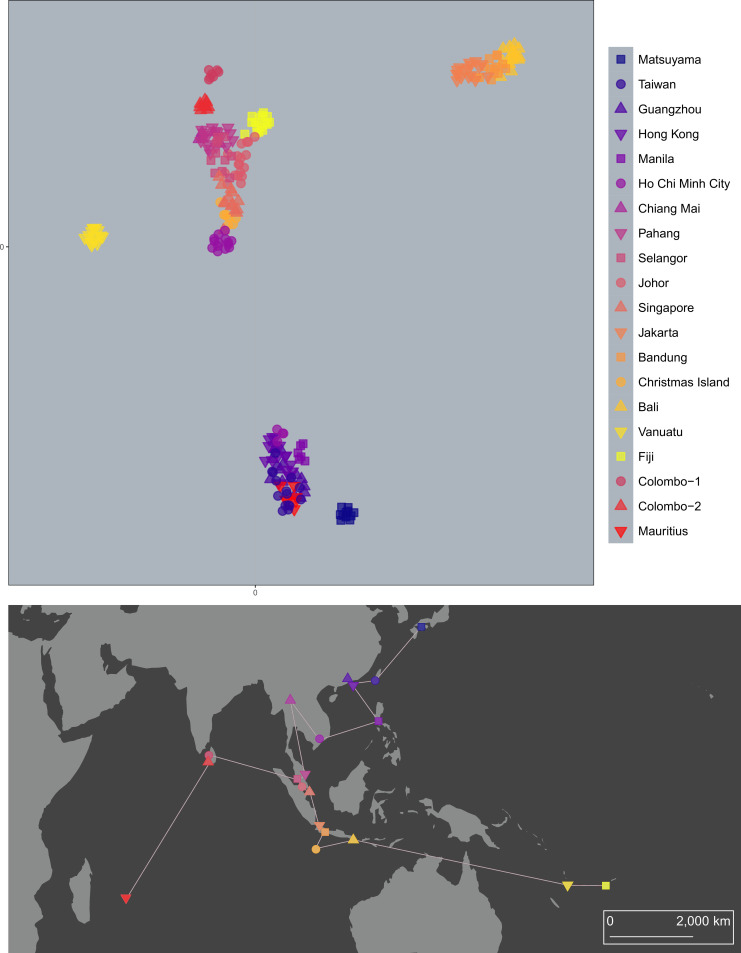
Fig 2. DAPCs of Ae. aegypti (i) and Ae. albopictus (ii) using lowest BIC (K = 3).
Colours indicate cluster membership. Circles are sized relative to population sample size. The map uses a Mollweide projection with a central meridian of 120°E. The map was produced in arcmap 10.5.1, using shapefiles made available by www.naturalearthdata.com.
Landscape genomics
We investigated the geographical structuring of genetic variation in Aedes spp. using distance-based redundancy analysis (dbRDA), performed in the R package vegan [67]. These analyses tested two hypotheses: that genetic differentiation would increase with geographical distance (i.e. isolation by distance), and that genetic differentiation would decrease with greater connectivity to shipping or aerial transportation routes. We omitted the outlier Ae. albopictus subpopulation Colombo-1 from analyses (see Results: Individual-level genetic structure). Mean FST’ estimates among the 16 Ae. aegypti and 19 Ae. albopictus populations were used as a measure of genetic differentiation.
Our measure for connectivity to shipping routes was derived from previously published raster data of 799,853 commercial shipping tracks from October 2004 to October 2005 (Halpern et al., 2008; see Supporting Online Material therein for full description). We used arcmap 10.5.1 [68] to calculate the average density of shipping tracks within a 200 km radius from each population sampling point, so that the inland site at Chiang Mai scored 0 while Jeddah, Colombo-2, and Taiwan received the highest scores (see Fig 1). For connectivity to aerial transportation routes, we used data from the OpenFlights Routes Database (https://openflights.org/data.html#route), which lists all airline routes active at June 2014. For each sampled population, we used the total number of active flight routes at the largest international airport of that city or town. This gave values of between 2 (Christmas Island) to 408 (Singapore). Shipping and aerial connectivity scores for each population are listed in Tables 1 and 2. No correlation between shipping and aerial connectivity scores were observed for either Ae. aegypti (R2 = 0.003, P = 0.838) or Ae. albopictus (R2 < 0.001, P = 0.938) populations.
The connectivity of each population pair to the shipping and aerial networks was calculated as the average of their connectivity scores for that network type. These average scores were used to construct pairwise connectivity matrices of each transportation type for each species. As independent variables in a dbRDA must be rectangular, these matrices were transformed into principle coordinates using the function pcnm. We likewise calculated geographical distance matrices for each species, and transformed these into principle coordinates.
To test for isolation by distance in each species, we built dbRDAs using the scaled vectors of geographical distance as the independent variable and FST’ as the dependent variable. To test for effects of connectivity to transportation networks, we first determined scaled geographical distance vectors with which to condition the model, by building dbRDAs where each vector was treated as a separate independent variable, and selecting vectors with P < 0.01. We assessed shipping and aerial connectivity separately. We built dbRDAs using the scaled vectors of connectivity as the independent variable and FST’ as the dependent variable, with the model conditioned using the significant vectors for geographical distance. All dbRDAs were built using the function capscale. Marginal significance of variables was assessed using anova.cca with 9,999 permutations.
Once transformed into principal coordinates, geographical and connectivity variables ceased to indicate directionality, and thus dbRDAs could only ascertain significance of associations and not the direction of these associations. To determine directionality, we ran linear regressions on the normalised pairwise scores of these variables against normalised FST’ estimates, and used the regression coefficients as indicators of directionality. These analyses were not used to determine significance of variables due to non-independence among pairwise terms.
Results
Individual-level genetic structure
We retained 54,296 SNPs for analysis of the 224 Ae. aegypti (Table 1) and 40,016 SNPs for analysis of the 256 Ae. albopictus (Table 2), with mean read depths of coverage of 17.67 (s.d. 5.31) and 25.23 (s.d. 7.49) respectively. The three analyses of genetic structure among individual genotypes, DAPC, sNMF, and PCA-UMAP, were consistent in their findings, and indicated several broad differences in genetic structure among Aedes spp. in the Indo-Pacific.
DAPC determined that K = 3 was the optimal number of clusters for both Ae. aegypti and Ae. albopictus when lowest BIC (Fig 2) was used for cluster detection. For Ae. aegypti, DAPC partitioned genotypes into three spatial groups representing the Pacific Islands (orange), Australia (cream), and all central and Western populations (purple) (Fig 2i). DAPC of Ae. albopictus (Fig 2ii) showed a very different pattern, with a single cluster (purple) present across the entire sampling range. One other cluster (cream) was found in Indonesia, indicating deep differences between these populations and nearby populations like Christmas Island, despite separation distances of only ~450 km. The final Ae. albopictus cluster was found within the Colombo population, consisting of a subset of seven genotypes that were deeply divergent from the other eight Colombo genotypes. In all further analyses the Colombo genotypes were treated as two populations, denoted as Colombo-1 (highly divergent) and Colombo-2. As BIC is a conservative measure, these results represent broad genetic groupings, with more detailed variation described in sNMFs (Fig 3), PCA-UMAPs (Figs 4 and 5), and DAPC using lowest AIC (S1 Fig).
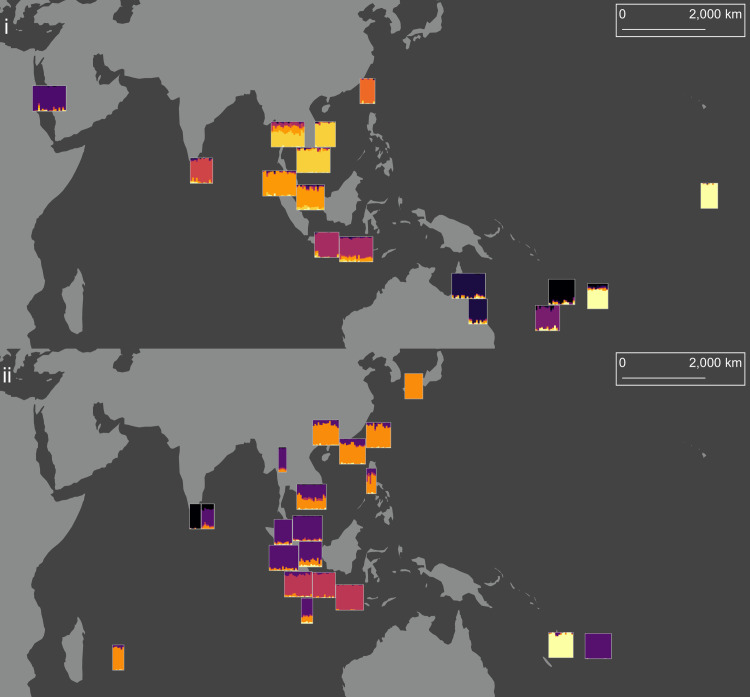
Fig 3. sNMFs of Ae. aegypti (i) and Ae. albopictus (ii).
Number of ancestral lineages (K) was 10 for Ae. aegypti and 5 for Ae. albopictus. Each population is a rectangle, with each genotype a vertical line made of between 1 and K colours. Colours indicate ancestral lineages. The map uses a Mollweide projection with a central meridian of 120°E. The map was produced in arcmap 10.5.1, using shapefiles made available by www.naturalearthdata.com.
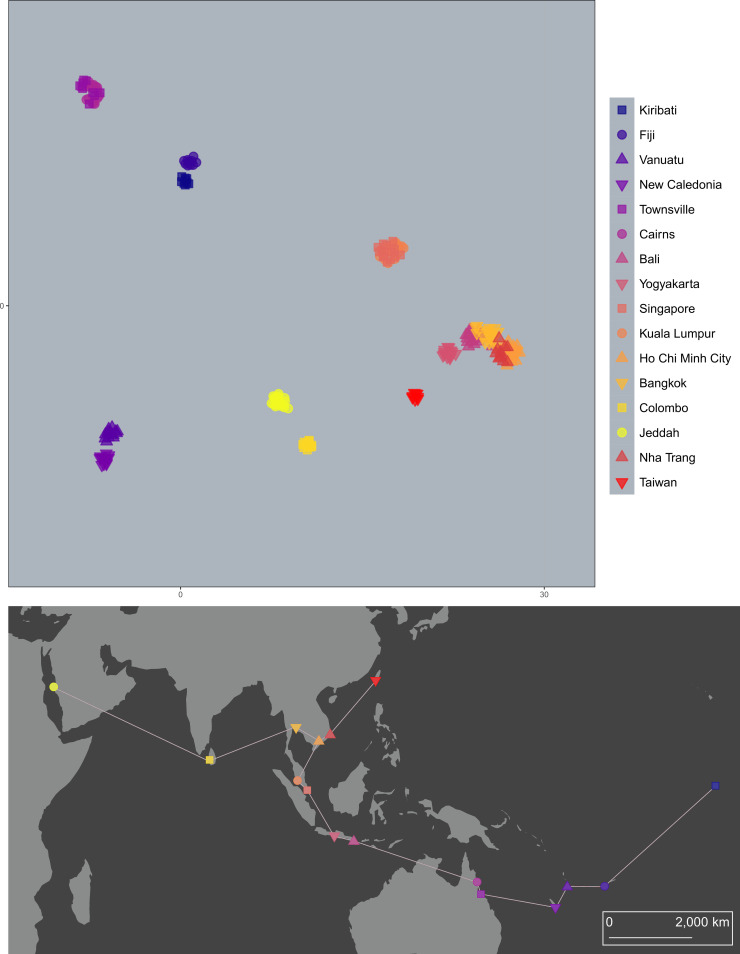
Fig 4. PCA-UMAP of 224 Ae. aegypti genotypes.
Genotypes are coloured by population, with similar colours given to geographically proximate populations.
Fig 5. PCA-UMAP of 256 Ae. albopictus genotypes.
Genotypes are coloured by population, with similar colours given to geographically proximate populations.
The sNMF of Ae. aegypti (Fig 3i) found K = 10 was an optimal number of ancestral lineages for summarising genetic variation among genotypes. This high number relative to the number of populations (16) largely reflected the high level of structuring between populations, with five of the ten ancestral lineages almost wholly confined to single locations: Jeddah, Colombo, Taiwan, New Caledonia and Vanuatu. Proximate population pairs such as Cairns and Townsville, Kuala Lumpur and Singapore, and Ho Chi Minh City and Nha Trang each had common ancestral lineages, as did Fiji and Kiribati, despite their distance of separation. The most putatively admixed population was Bangkok, which showed genetic similarities with the other Southeast Asian populations, but not with either Taiwan or Colombo.
These results contrasted sharply with those of Ae. albopictus (Fig 3ii), for which K = 5 was optimal. Fewer ancestral lineages indicated greater genetic similarity among Ae. albopictus populations. The interpretation of the spatial distribution of ancestral lineages among Ae. albopictus requires reference to the native and invasive ranges of this species. The northernmost native range lineage, in orange, indicated a common heritage among the East Asian populations. This lineage was also dominant in Mauritius. A second native range lineage, in purple, indicated common heritage among Southeast Asian populations from Chiang Mai to Singapore, and was also found in Christmas Island and Fiji, but not Vanuatu. The population at Ho Chi Minh City was made up of roughly even contributions from the two lineages. The Indonesian Ae. albopictus genotypes (Fig 3ii, red) of Jakarta, Bandung and Bali were genetically distinct from the East and Southeast Asian native range genotypes.
Treating Colombo Ae. albopictus as two separate populations revealed that genotypes from the highly-divergent Colombo-1 population were almost entirely of one ancestral lineage (Fig 3ii, black) found only in Colombo. Colombo-2 was a composite of lineages, including the unique lineage from Colombo and the purple native range lineage.
PCA-UMAP of Ae. aegypti (Fig 4) showed genotypes grouping clearly by spatial location, echoing the sNMF results but providing more detailed information on differentiation between similar populations. Genotypes clustered first by population, then by region, with Pacific and Australian genotypes the most distinct. PCA-UMAP indicated similarities between genotypes from Fiji and Kiribati, Vanuatu and New Caledonia, and Singapore and Kuala Lumpur. The westernmost samples from Jeddah and Colombo also showed evidence of similarity. Despite Taiwan’s proximity to Southeast Asia, Taiwanese Ae. aegypti were clearly distinct from all others.
PCA-UMAP of Ae. albopictus (Fig 5) likewise gave results similar to the sNMF but with more detail. The Matsuyama population was revealed as distinct from others in East Asia, and there was evidence that the Fijian genotypes were most closely related to Johor, Malaysia, and those of Christmas Island were most closely related to Singapore. The Ho Chi Minh City genotypes were split into two clusters, four grouping with the East Asian genotypes and 14 with the Southeast Asian genotypes. This likely reflects Ho Chi Minh City as having ancestry from both of these groups (Fig 3ii). A corresponding split was observed in the DAPC results using lowest AIC (S1 Fig).
Population-level genetic structure
Numbers of nuclear SNPs retained for population-level analyses had ranges of 46,151–54,647 (Ae. aegypti) and 35,984–57,639 (Ae. albopictus). Pairwise FST’ estimates for Ae. aegypti and Ae. albopictus are listed in S1A Table and S1B Table respectively. Mean estimates are given with 95% confidence ranges, which show that pairwise FST’ estimates were generally consistent across the ten subsamples. FST’ confidence intervals in Ae. aegypti were on average only 5.3% the size of the mean; for Ae. albopictus, they were only 6.4% the size of the mean. Mean HO and π estimates were also mostly consistent among subsamples within populations, and are listed in S1C Table and S1D Table along with the number of private alleles and proportion of missing data among genotypes in each population.
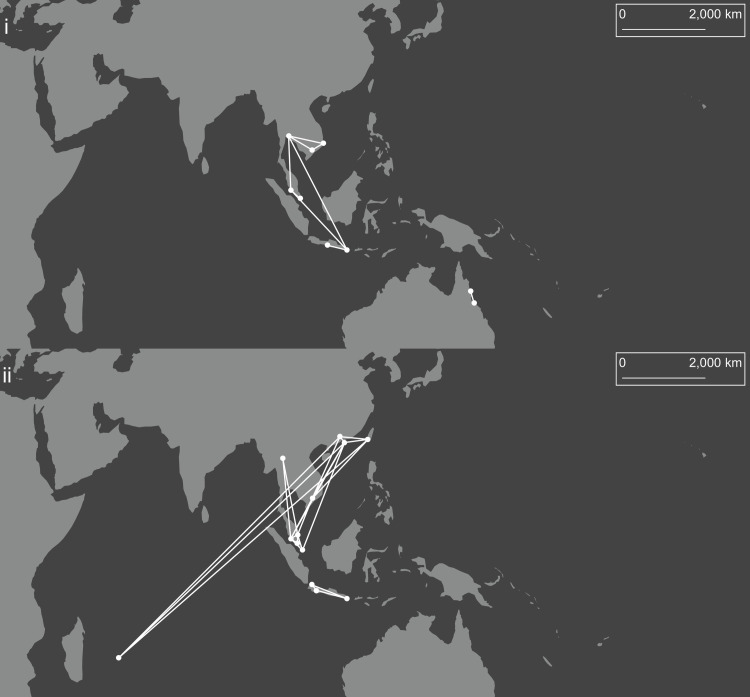
Fig 6 shows all population pairs with mean pairwise FST’ < 0.03 (a value selected to help visualise patterns), indicating high genetic similarity. Australian Ae. aegypti populations were genetically very similar, and there was a network of similarity among Southeast Asian populations (Fig 6i). Indonesian Ae. albopictus populations were genetically similar, as were a network of populations covering parts of the native range in East and Southeast Asia as well as Mauritius (Fig 6ii). Taiwan Ae. aegypti had the highest mean FST’ from all other populations ( = 0.093; s.d. = 0.016). Colombo-1 Ae. albopictus had a particularly high mean FST’ when compared to all other populations ( = 0.153; s.d. = 0.018), including substantial differentiation from Colombo-2 ( = 0.113; s.d. = 0.008). Colombo-2 had a much lower mean FST’ when compared to all other populations ( = 0.070; s.d. = 0.020).
Fig 6. Population pairs of Ae. aegypti (i) and Ae. albopictus (ii) with mean pairwise FST’ < 0.03.
Circles connected by lines indicate populations with mean pairwise FST’ < 0.03. The map uses a Mollweide projection with a central meridian of 120°E. The map was produced in arcmap 10.5.1, using shapefiles made available by www.naturalearthdata.com.
When 16 genotypes were used to calculate FST’, results were similar to when 5 genotypes were used (S1E Table). Using five genotypes tended to slightly overestimate FST’, particularly for highly differentiated populations, though FST’ scores were consistent relative to one another. Likewise, while using only five genotypes will lead to lower estimates of HO and π than when estimated with more genotypes, HO and π can be compared across populations and species. Both HO and π were much higher in Ae. aegypti (S1C Table) than in Ae. albopictus (S1D Table).
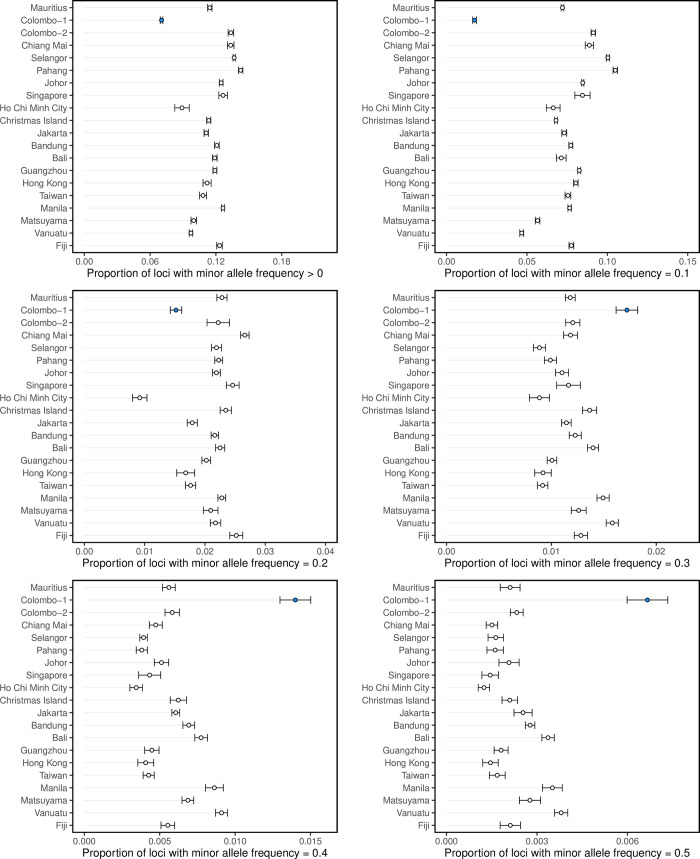
Analysis of the folded allele frequency spectrum in Ae. albopictus showed that Colombo-1 had lower frequencies of rare alleles and higher frequencies of common alleles compared with other populations (Fig 7). The proportion of loci in Colombo-1 with a minor allele frequency of 0.1 was only 0.23 times the size of the mean for all populations. The proportions of loci in Colombo-1 with minor allele frequencies of 0.4 and 0.5 were 2.32 and 2.75 times higher respectively than the mean for all populations. Neither Colombo-2 nor any of the other populations consistently differed from other populations in minor allele frequency. Colombo-1 had the largest proportion of monomorphic loci out of all populations.
Fig 7. Minor allele frequencies of Ae. albopictus populations.
Circles represent the mean frequency from the 10 subsamples, with 95% confidence intervals. The Colombo-1 population is indicated in blue.
Landscape genomics
The dbRDAs assessing isolation by distance indicated that geographical distance was significantly associated with FST’ in Ae. aegypti (Table 3: F9 = 2.136, P = 0.002) but not in Ae. albopictus (Table 4: F9 = 0.965, P = 0.534). Linear regression indicated that this relationship was positive (regression coefficient = 0.52). When each scaled vector for geographical distance was treated as an independent variable, one was significant at P < 0.01 in Ae. aegypti and none were significant in Ae. albopictus.
Table 3. Results of dbRDAs assessing landscape genomic hypotheses in Ae. aegypti.
All geographical distances and connectivity indices were transformed into principal coordinates (PCs) for analysis. The single PC with P < 0.01 when testing for isolation by distance was used to condition the models assessing connectivity to transportation networks.
| Df | Sum of Squares | F | P | |
|---|---|---|---|---|
| Geographical distance | 9 | 0.026 | 2.136 | 0.002 |
| Residual | 6 | 0.008 | ||
| Marine connectivity | 12 | 0.022 | 0.811 | 0.727 |
| Geographical distance PC1 | 1 | 0.006 | 2.659 | 0.041 |
| Residual | 2 | 0.004 | ||
| Aerial connectivity | 11 | 0.021 | 1.075 | 0.467 |
| Geographical distance PC1 | 1 | 0.003 | 1.692 | 0.131 |
| Residual | 3 | 0.005 |
Table 4. Results of dbRDAs assessing landscape genomic hypotheses in Ae. albopictus.
All geographical distances and connectivity indices were transformed into principal coordinates (PCs) for analysis.
| Df | Sum of Squares | F | P | |
|---|---|---|---|---|
| Geographical distance | 9 | 0.018 | 0.965 | 0.534 |
| Residual | 9 | 0.019 | ||
| Marine connectivity | 15 | 0.034 | 2.411 | 0.034 |
| Residual | 3 | 0.003 | ||
| Aerial connectivity | 13 | 0.032 | 2.65 | 0.006 |
| Residual | 5 | 0.005 |
The dbRDAs assessing connectivity to transport networks indicated that FST’ in Ae. albopictus was significantly associated with both shipping (F15 = 2.411, P = 0.034) and aerial (F13 = 2.650, P = 0.006) transportation routes (Table 4) with negative associations being detected (regression coefficients: shipping = -0.20; aerial = -0.24). Neither transport network was significantly associated with FST’ in Ae. aegypti (Table 3), irrespective of whether the significant geographical distance vector was used to condition the models.
Discussion
Here we present the first direct comparison of genome-wide genetic structure in the globally-invasive, dengue-vectoring mosquitoes Ae. aegypti and Ae. albopictus. Our study region, the Indo-Pacific, is the site of 70% of global dengue transmission [1], and this study contributes important knowledge to the understanding of invasion histories and ongoing gene flow within this region. While both species are already widespread in the Indo-Pacific [22], increased urbanisation and climate change may allow them to invade new regions in the coming decades [23]. Furthermore, the same dispersal processes facilitating invasions into new regions can also lead to genetic invasions of advantageous alleles into established populations, as observed recently in this region [24]. Our comparative approach indicated different genetic structure patterns in these species at each level of analysis: individual, population, and landscape genomic. For the most part, Ae. aegypti populations were genetically differentiated from each other, and differentiation increased with geographical distance. Aedes albopictus populations in the native range were split between three main regions of genetic similarity, one in East Asia, one in Southeast Asia (excluding Indonesia), and one in Indonesia. Certain populations outside the native range showed clear signs of recent invasion from these regions; specifically, Mauritius from East Asia, and Fiji and Christmas Island from Southeast Asia. Aedes albopictus populations in Indonesia were highly differentiated from all others, despite their proximity to other Southeast Asian populations. In Colombo, we observed two distinct Ae. albopictus subpopulations, one of which may represent an ancestral native-range population or cryptic subspecies. Overall these results indicate that Aedes spp. have established invasions in the Indo-Pacific along different pathways, and that recent gene flow patterns are different between the two species. Our findings also point to various regions of interest in the Ae. albopictus native range that require further investigation.
The patterns of divergence observed in Ae. aegypti fit with the hypothesis that Ae. aegypti began its invasion of the Indian Ocean region from regions to the west [5,6]. Putatively, the Indian Ocean regions and Australia were invaded from the Mediterranean, though there is some uncertainty over the timing and direction of this invasion [69,70], and the strong genetic differentiation we observed in Australian populations confounds this somewhat. Interestingly, the most highly differentiated Ae. aegypti populations were in the western Pacific populations of Australia, the Pacific Islands and Taiwan, suggesting that these regions do not share an invasion history with Indian Ocean regions. Separate invasions of Australia and the Pacific Islands also seem likely, based on the strong differentiation between Australian Ae. aegypti and those of nearby Pacific Islands such as Vanuatu and New Caledonia.
The high differentiation of western Pacific Ae. aegypti populations may reflect an earlier invasion of these regions, possibly from the Americas. However, these divergences could instead be due to a lack of gene flow into these populations following invasion, with strong genetic differentiation produced by the effects of drift following founder events [37]. Different structural patterns in these populations may be reinforced by high local population densities, reflecting a “founder takes all” [38] system following colonisation. Considering the strong isolation by distance observed in this species (Table 3), the relative remoteness of the Pacific Islands suggests that populations from these islands experience limited gene flow, leading to pronounced differences among them. By contrast, transport connectivity was an important influence on Ae. albopictus genetic structure (Table 4), and there were signs of recent dispersal into Fiji from Southeast Asia (likely Malaysia); a non-Indonesian Southeast Asian population is thus a likely source of the Fijian invasion of Ae. albopictus in 1989 [71]. These results require cautious interpretation, however, as most Ae. albopictus invasions of the Pacific are more recent than those of Ae. aegypti, and thus low differentiation among populations may reflect recent invasion rather than ongoing gene flow. Nevertheless, genetic similarities among geographically distant populations were more apparent in Ae. albopictus than in Ae. aegypti, and the isolation by distance patterns observed in Ae. aegypti indicate that gene flow in this species is more common between geographically proximate populations.
Our landscape genomics analyses point to different processes structuring Aedes spp. populations in the Indo-Pacific region. For Ae. aegypti, population structuring by geographical distance but not transportation routes accords with a pre-modern invasion timeline. Aedes aegypti is thought to have begun its invasion of the region in ~18-19th c. [6], a time when shipping routes likely differed from the 21st century routes used in our analyses. In contrast, the more recent expansion of Ae. albopictus occurred at a time when international shipping routes more closely resembled the 2004 routes analysed here. Although dispersal by aircraft may be less common than by shipping in this species [25], it is worth noting that much of the material used in this study was collected near to international airports, which may exaggerate the influence of the aerial dispersal network. Also, as the metric for aerial connectivity was derived from the activity of nearby airports, aerial connectivity scores may correlate closely with other untested variables that could influence regional genetic structure, such as economic activity.
Although the observation of isolation by distance in Ae. aegypti but not in Ae. albopictus may reflect the greater impact of long-distance transportation in modern times, the capacity of Ae. albopictus to undergo diapause as well as quiescence [35] could be important, as mosquitoes must survive long-distance transportation for gene flow to occur. A comparison of incursion pathways of Aedes spp. species into Australia indicated that not only was each species dispersing from a different set of locations, but also that each species was likely transported through different vessel types, with Ae. albopictus found more frequently at seaports and Ae. aegypti at airports [25]. The most common source of Ae. albopictus incursions was southern China, where diapause has been observed in field populations [72]. Many other factors may also influence long-distance dispersal outcomes, such as differences in behaviour when boarding and travelling on vessels and the relative abundance of each species around airports and seaports. Better survivorship on long ship journeys may allow Ae. albopictus to colonise distant locations without first colonising geographically intermediate ‘stepping stones’, which accords with much of the broad-scale literature for this species [44,73]. For taxa capable of long-distance dispersal, long-range colonisation becomes more probable than stepping-stone colonisation as the required number of successful colonisations is minimised [74].
The investigation of Ae. albopictus in its native range revealed a pair of native-range clades covering East and Southeast Asia, with both clades found in Ho Chi Minh City (Fig 3ii). Differentiation was low between these clades (Fig 6ii). A third potential native-range clade was found in Indonesia, where populations were strongly differentiated from all other clades, confirming previous results from allozyme data [75]. Indonesia could represent the southern border of the native range, and whereas the East and Southeast Asian populations have become largely homogenised through gene flow, the Indonesian populations have remained distinct from these. Alternatively, Indonesia may have been invaded earlier than other regions. A possible ancient invasion source is India, which had considerable connectivity with Java and Bali in antiquity [76].
The Ae. albopictus of Colombo, Sri Lanka, appeared to represent two sympatric subpopulations: Colombo-1, a highly-divergent, ancestral subpopulation of Ae. albopictus, possibly a cryptic subspecies; and Colombo-2, a subpopulation seemingly produced by admixture between the ancestral Colombo-1 lineage and the Southeast Asian native-range lineage (Fig 3ii). The folded allele frequency spectrum of Colombo-1 was unlike other populations, having greater proportions of high-frequency minor alleles and fewer rare alleles (Fig 7). Several potential cryptic species closely related to Ae. albopictus have been identified in its Chinese native range [77], and the western extent of its range in India is not well-studied genetically. It is possible that Colombo is also within the native range of Ae. albopictus, and that Colombo-1 is an ancestral, native-range subpopulation. Alternatively, Colombo may have been invaded by an ancestral, native-range population from India or elsewhere. The putatively admixed Colombo-2 subpopulation is evidence of recent incursions into Colombo from the native range.
The different patterns of population genetic structure in the Aedes spp. described here has implications for biosecurity and biocontrol. Recently, genomics has been used to identify source locations of Aedes incursions into Australia and New Zealand [20,25]; these incursions threaten not only the arrival of species themselves but also the introduction of insecticide resistance alleles [24]. The genomic patterns identified here can help in identifying biosecurity threats in both species, both by revealing likely pathways of gene flow and demonstrating that long-distance marine incursions may be more likely in Ae. albopictus than in Ae. aegypti. The identification of the highly-differentiated Ae. albopictus clusters in Indonesia and Sri Lanka may also indicate regions where vector competence is higher or lower. For instance, Ae. albopictus from three populations putatively invaded from East Asia were highly susceptible to DENV-2 [78], while Ae. albopictus from the Torres Strait Islands, putatively invaded from Indonesia [79], had low susceptibility [80]. Genetically differentiated populations may also have differential Wolbachia infection status, as found in cryptic Ae. albopictus subspecies in China [77]. Variation in Wolbachia infection status may be exploitable by future dengue control efforts that involve the release of this bacterium into mosquito populations [81]. Likewise, patterns of population genetic structure can indicate areas where mosquito genetic backgrounds are likely to be similar or different to those of target populations–an important consideration for widespread species such as Aedes spp. that can be locally adapted to different conditions [49].
Supporting information
(DOCX)
These tables show means and upper and lower confidence ranges of: (S1A Table) pairwise FST' for Ae. aegypti in the Indo-Pacific region; (S1B Table) pairwise FST' for Ae. albopictus in the Indo-Pacific region; (S1C Table) heterozygosity and nucleotide diversity for Ae. aegypti in the Indo-Pacific region; (S1D Table) heterozygosity and nucleotide diversity for Ae. albopictus in the Indo-Pacific region; and (S1E Table) FST when calculated with five genotypes compared with when calculated with 16 genotypes.
(XLSX)
Colours indicate cluster membership. Circles are sized relative to population sample size. For Ae. aegypti, K = 15 was used, while K = 11 was used for Ae. albopictus. The map uses a Mollweide projection with a central meridian of 120°E. The map was produced in arcmap 10.5.1, using shapefiles made available by www.naturalearthdata.com.
(TIF)
Acknowledgments
We thank Tim Hurst, Samia Elfekih, Dayanath Meegoda, Hadian Sasmita, Nazni Wa, Pattamaporn Kittayapong, Lia Faridah, Kozo Watanabe, Thaddeus Carvajal, Chi Yung Jim, Gwendolin Wong, Ashley Callahan, Jason Axford, Stephen Doggett, Elizabeth Valerie, Craig Williams, Joe Davis and other researchers for samples. We also thank Moshe Jasper, Anthony van Rooyen, Nancy Endersby-Harshman and Nick Bell for sample processing. This research was facilitated by use of the Nectar Research Cloud, a collaborative Australian research platform supported by the Australian Research Data Commons (ARDC) and National Collaborative Research Infrastructure Strategy (NCRIS).
Data Availability
Aligned .bam files for 224 Ae. aegypti and 256 Ae. albopictus are available at the NCBI Sequence Read Archive under project accession number PRJNA641723.
Funding Statement
AAH was supported by Program and Fellowship grants from the National Health and Medical Research Council (NHMRC), no. 1037003 (https://www.nhmrc.gov.au). TLS and AAH were also supported by the Wellcome Trust UK, no. 108508 (https://wellcome.ac.uk). A-CH was funded the German Science Foundation (HO 5981/1-1) and the Swiss Tropical and Public Health Institute. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, et al. The global distribution and burden of dengue. Nature. 2013;496(7446):504–7. 10.1038/nature12060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gubler DJ. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev. 1998;11(3):480–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu J-Y, Lun Z-R, James AA, Chen X-G. Dengue Fever in Mainland China. Am J Trop Med Hyg. 2010;83(3):664–71. 10.4269/ajtmh.2010.09-0755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crawford JE, Alves JM, Palmer WJ, Day JP, Sylla M, Ramasamy R, et al. Population genomics reveals that an anthropophilic population of Aedes aegypti mosquitoes in West Africa recently gave rise to American and Asian populations of this major disease vector. BMC Biol. 2017;15(1):16 10.1186/s12915-017-0351-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kotsakiozi P, Gloria-Soria A, Schaffner F, Robert V, Powell JR. Aedes aegypti in the Black Sea: recent introduction or ancient remnant? Parasit Vectors. 2018;11(1):396 10.1186/s13071-018-2933-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Powell JR, Gloria-Soria A, Kotsakiozi P. Recent History of Aedes aegypti: Vector Genomics and Epidemiology Records. Bioscience. 2018;68(11):854–60. 10.1093/biosci/biy119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hawley WA. The biology of Aedes albopictus. J Am Mosq Control Assoc Suppl. 1988;1:1–39. [PubMed] [Google Scholar]
- 8.Goubert C, Minard G, Vieira C, Boulesteix M. Population genetics of the Asian tiger mosquito Aedes albopictus, an invasive vector of human diseases. Heredity (Edinb). 2016;117(3):125–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Porretta D, Mastrantonio V, Bellini R, Somboon P, Urbanelli S. Glacial History of a Modern Invader: Phylogeography and Species Distribution Modelling of the Asian Tiger Mosquito Aedes albopictus. Moreira LA, editor. PLoS One. 2012;7(9):e44515 10.1371/journal.pone.0044515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonizzoni M, Gasperi G, Chen X, James AA. The invasive mosquito species Aedes albopictus: Current knowledge and future perspectives. Trends Parasitol. 2013;29(9):460–8. 10.1016/j.pt.2013.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hengeveld R. Dynamics of biological invasions. London: Chapman & Hall; 1989. [Google Scholar]
- 12.Harrington LC, Scott TW, Lerdthusnee K, Coleman RC, Costero A, Clark GG, et al. Dispersal of the dengue vector Aedes aegypti within and between rural communities. Am J Trop Med Hyg. 2005;72(2):209–20. [PubMed] [Google Scholar]
- 13.Jasper M, Schmidt TL, Ahmad NW, Sinkins SP, Hoffmann AA. A genomic approach to inferring kinship reveals limited intergenerational dispersal in the yellow fever mosquito. Mol Ecol Resour. 2019;19(5):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marini F, Caputo B, Pombi M, Tarsitani G, Della Torre A. Study of Aedes albopictus dispersal in Rome, Italy, using sticky traps in mark-release-recapture experiments. Med Vet Entomol. 2010;24(4):361–8. 10.1111/j.1365-2915.2010.00898.x [DOI] [PubMed] [Google Scholar]
- 15.Eritja R, Palmer JRB, Roiz D, Sanpera-Calbet I, Bartumeus F. Direct Evidence of Adult Aedes albopictus Dispersal by Car. Sci Rep. 2017;7(1):14399–413. 10.1038/s41598-017-12652-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt TL, Filipović I, Hoffmann AA, Rašić G. Fine-scale landscape genomics helps explain the slow spatial spread of Wolbachia through the Aedes aegypti population in Cairns, Australia. Heredity (Edinb). 2018;120(5):386–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt TLTL, Rašić G, Zhang D, Zheng X, Xi Z, Hoffmann AAA. Genome-wide SNPs reveal the drivers of gene flow in an urban population of the Asian Tiger Mosquito, Aedes albopictus. Lenhart A, editor. PLoS Negl Trop Dis. 2017;11(10):e0006009 10.1371/journal.pntd.0006009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tatem AJ, Hay SI, Rogers DJ. Global traffic and disease vector dispersal. Proc Natl Acad Sci U S A. 2006;103(16):6242–7. 10.1073/pnas.0508391103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ammar SE, Mclntyre M, Swan T, Kasper J, Derraik JGB, Baker MG, et al. Intercepted Mosquitoes at New Zealand’s Ports of Entry, 2001 to 2018: Current Status and Future Concerns. Trop Med Infect Dis. 2019;4(3):101–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt TL, van Rooyen AR, Chung J, Endersby‐Harshman NM, Griffin PC, Sly A, et al. Tracking genetic invasions: Genome‐wide single nucleotide polymorphisms reveal the source of pyrethroid‐resistant Aedes aegypti (yellow fever mosquito) incursions at international ports. Evol Appl. 2019;12(6):1136–46. 10.1111/eva.12787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gloria-Soria A, Ayala D, Bheecarry A, Calderon-Arguedas O, Chadee DD, Chiappero M, et al. Global genetic diversity of Aedes aegypti. Mol Ecol. 2016;25(21):5377–95. 10.1111/mec.13866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kraemer MUG, Sinka ME, Duda KA, Mylne AQN, Shearer FM, Barker CM, et al. The global distribution of the arbovirus vectors Aedes aegypti and Ae. albopictus. Elife. 2015;4:e08347 10.7554/eLife.08347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kraemer MUG, Reiner RC, Brady OJ, Messina JP, Gilbert M, Pigott DM, et al. Past and future spread of the arbovirus vectors Aedes aegypti and Aedes albopictus. Nat Microbiol. 2019;4(5):854–63. 10.1038/s41564-019-0376-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Endersby-Harshman NM, Schmidt TL, Chung J, van Rooyen A, Weeks AR, Hoffmann AA. Heterogeneous genetic invasions of three insecticide resistance mutations in Indo-Pacific populations of Aedes aegypti (L.). Mol Ecol. 2020;29(9):1628–41. 10.1111/mec.15430 [DOI] [PubMed] [Google Scholar]
- 25.Schmidt TL, Chung J, van Rooyen A, Sly A, Weeks AR, Hoffmann AA. Incursion pathways of the Asian tiger mosquito (Aedes albopictus) into Australia contrast sharply with those of the yellow fever mosquito (Aedes aegypti). Pest Manag Sci. 2020. 10.1002/ps.5977 [DOI] [PubMed] [Google Scholar]
- 26.McMillen-Jackson AL, Bert TM. Disparate patterns of population genetic structure and population history in two sympatric penaeid shrimp species (Farfantepenaeus aztecus and Litopenaeus setiferus) in the eastern United States. Mol Ecol. 2003;12(11):2895–905. 10.1046/j.1365-294x.2003.01955.x [DOI] [PubMed] [Google Scholar]
- 27.Avise JC. The history and purview of phylogeography: A personal reflection. Mol Ecol. 1998;7(4):371–9. [Google Scholar]
- 28.Joseph L, Moritz C, Hugall A. Molecular support for vicariance as a source of diversity in rainforest. Proc R Soc London Ser B Biol Sci. 1995;260(1358):177–82. [DOI] [PubMed] [Google Scholar]
- 29.Trewick SA, Wallis GP. Bridging the “Beech-gap”: New Zealand invertebrate phylogeography implicates Pleistocene glaciation and Pliocene isolation. Evolution (N Y). 2001;55(11):2170–80. [DOI] [PubMed] [Google Scholar]
- 30.Wagner CE, McCune AR. Contrasting patterns of spatial genetic structure in sympatric rock-dwelling cichlid fishes. Evolution (N Y). 2009;63(5):1312–26. [DOI] [PubMed] [Google Scholar]
- 31.Dawson MN. Parallel phylogeographic structure in ecologically similar sympatric sister taxa. Mol Ecol. 2012;21(4):987–1004. 10.1111/j.1365-294X.2011.05417.x [DOI] [PubMed] [Google Scholar]
- 32.Slatkin M. Gene Flow and the Geographic Structure of Natural Populations. Source Sci New Ser. 1987;236(4803):787–92. [DOI] [PubMed] [Google Scholar]
- 33.Armbruster PA. Photoperiodic Diapause and the Establishment of Aedes albopictus (Diptera: Culicidae) in North America. J Med Entomol. 2016;53(5):1013–23. 10.1093/jme/tjw037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Medley KA, Westby KM, Jenkins DG. Rapid local adaptation to northern winters in the invasive Asian tiger mosquito Aedes albopictus: A moving target. Bauer S, editor. J Appl Ecol. 2019;56(11):2518–27. [Google Scholar]
- 35.Diniz DFA, De Albuquerque CMR, Oliva LO, De Melo-Santos MAV, Ayres CFJ. Diapause and quiescence: Dormancy mechanisms that contribute to the geographical expansion of mosquitoes and their evolutionary success. Parasit Vectors. 2017;10(1):310–22. 10.1186/s13071-017-2235-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Urbanski JM, Benoit JB, Michaud MR, Denlinger DL, Armbruster P. The molecular physiology of increased egg desiccation resistance during diapause in the invasive mosquito, Aedes albopictus. Proc R Soc B Biol Sci. 2010;277(1694):2683–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Excoffier L, Ray N. Surfing during population expansions promotes genetic revolutions and structuration. Trends Ecol Evol. 2008;23(7):347–51. 10.1016/j.tree.2008.04.004 [DOI] [PubMed] [Google Scholar]
- 38.Waters JM, Fraser CI, Hewitt GM. Founder takes all: density-dependent processes structure biodiversity. Trends Ecol Evol. 2013;28:78–85. 10.1016/j.tree.2012.08.024 [DOI] [PubMed] [Google Scholar]
- 39.Halpern BS, Walbridge S, Selkoe KA, Kappel C V, Micheli F, D’Agrosa C, et al. A global map of human impact on marine ecosystems. Science. 2008;319(5865):948–52. 10.1126/science.1149345 [DOI] [PubMed] [Google Scholar]
- 40.Mousson L, Dauga C, Garrigues T, Schaffner F, Vazeille M, Failloux AB. Phylogeography of Aedes (Stegomyia) aegypti (L.) and Aedes (Stegomyia) albopictus (Skuse) (Diptera: Culicidae) based on mitochondrial DNA variations. Genet Res. 2005;86(1):1–11. 10.1017/S0016672305007627 [DOI] [PubMed] [Google Scholar]
- 41.Eskildsen GA, Rovira JR, Smith O, Miller MJ, Bennett KL, McMillan WO, et al. Maternal invasion history of Aedes aegypti and Aedes albopictus into the Isthmus of Panama: implications for the control of emergent viral disease agents. PLoS One. 2018;13(3):e0194874 10.1371/journal.pone.0194874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rašić G, Filipović I, Weeks AR, Hoffmann AA. Genome-wide SNPs lead to strong signals of geographic structure and relatedness patterns in the major arbovirus vector, Aedes aegypti. BMC Genomics. 2014;15(1):275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wright S. Isolation by Distance. Genetics. 1943;28(2):114–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kotsakiozi P, Richardson JB, Pichler V, Favia G, Martins AJ, Urbanelli S, et al. Population genomics of the Asian tiger mosquito, Aedes albopictus: insights into the recent worldwide invasion. Ecol Evol. 2017;7(23):10143–57. 10.1002/ece3.3514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pichler V, Kotsakiozi P, Caputo B, Serini P, Caccone A, Torre A Della. Complex interplay of evolutionary forces shaping population genomic structure of invasive Aedes albopictus in southern Europe. PLoS Negl Trop Dis. 2019;13(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sherpa S, Rioux D, Pougnet-Lagarde C, Després L. Genetic diversity and distribution differ between long-established and recently introduced populations in the invasive mosquito Aedes albopictus. Infect Genet Evol. 2018;58:145–56. 10.1016/j.meegid.2017.12.018 [DOI] [PubMed] [Google Scholar]
- 47.Sherpa S, Blum MGB, Capblancq T, Cumer T, Rioux D, Després L. Unravelling the invasion history of the Asian tiger mosquito in Europe. Mol Ecol. 2019;28(9):2360–77. 10.1111/mec.15071 [DOI] [PubMed] [Google Scholar]
- 48.Hamelin RC, Roe AD. Genomic biosurveillance of forest invasive alien enemies: a story written in code. Evol Appl. 2019;eva.12853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garcia G de A, Sylvestre G, Aguiar R, da Costa GB, Martins AJ, Lima JBP, et al. Matching the genetics of released and local Aedes aegypti populations is critical to assure Wolbachia invasion. Kittayapong P, editor. PLoS Negl Trop Dis. 2019;13(1):e0007023 10.1371/journal.pntd.0007023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA. Stacks: an analysis tool set for population genomics. Mol Ecol. 2013;22(11):3124–40. 10.1111/mec.12354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–9. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Behura SK, Lobo NF, Haas B, DeBruyn B, Lovin DD, Shumway MF, et al. Complete sequences of mitochondria genomes of Aedes aegypti and Culex quinquefasciatus and comparative analysis of mitochondrial DNA fragments inserted in the nuclear genomes. Insect Biochem Mol Biol. 2011;41(10):770–7. 10.1016/j.ibmb.2011.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang H, Xing D, Wang G, Li C, Zhao T. Sequencing and analysis of the complete mitochondrial genome of Aedes albopictus (Diptera: Culicidae) in China. Mitochondrial DNA. 2016;27(4):2787–8. 10.3109/19401736.2015.1053067 [DOI] [PubMed] [Google Scholar]
- 54.Matthews BJ, Dudchenko O, Kingan SB, Koren S, Antoshechkin I, Crawford JE, et al. Improved reference genome of Aedes aegypti informs arbovirus vector control. Nature. 2018;1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen X-G, Jiang X, Gu J, Xu M, Wu Y, Deng Y, et al. Genome sequence of the Asian Tiger mosquito, Aedes albopictus, reveals insights into its biology, genetics, and evolution. Proc Natl Acad Sci. 2015;112(44):E5907–15. 10.1073/pnas.1516410112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hlaing T, Tun-Lin W, Somboon P, Socheat D, Setha T, Min S, et al. Mitochondrial pseudogenes in the nuclear genome of Aedes aegypti mosquitoes: Implications for past and future population genetic studies. BMC Genet. 2009;10(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jombart T, Devillard S, Balloux F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 2010;11(1):94–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Frichot E, Mathieu F, Trouillon T, Bouchard G, François O. Fast and efficient estimation of individual ancestry coefficients. Genetics. 2014;196(4):973–83. 10.1534/genetics.113.160572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Diaz-Papkovich A, Anderson-Trocmé L, Ben-Eghan C, Gravel S. UMAP reveals cryptic population structure and phenotype heterogeneity in large genomic cohorts. PLOS Genetics. 2019;15(11):e1008432 10.1371/journal.pgen.1008432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McInnes L, Healy J, Melville J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv. 2018;1802.03426. [Google Scholar]
- 61.Browning BL, Browning SR. Genotype Imputation with Millions of Reference Samples. Am J Hum Genet. 2016;98(1):116–26. 10.1016/j.ajhg.2015.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jombart T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24(11):1403–5. 10.1093/bioinformatics/btn129 [DOI] [PubMed] [Google Scholar]
- 63.Frichot E, François O. LEA: An R package for landscape and ecological association studies. O’Meara B, editor. Methods Ecol Evol. 2015;6(8):925–9. [Google Scholar]
- 64.Meirmans PG. Using the AMOVA framework to estimate a standardized genetic differentiation measure. Evolution (N Y). 2006;60(11):2399–402. [PubMed] [Google Scholar]
- 65.Willing E-M, Dreyer C, van Oosterhout C. Estimates of Genetic Differentiation Measured by FST Do Not Necessarily Require Large Sample Sizes When Using Many SNP Markers. Chave J, editor. PLoS One. 2012;7(8):e42649 10.1371/journal.pone.0042649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, et al. The variant call format and VCFtools. Bioinformatics. 2011;27(15):2156–8. 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, et al. vegan: Community Ecology Package [Internet]. 2019. p. 2395–6. [Google Scholar]
- 68.ESRI. ArcGIS. Redlands, CA: Environmental Systems Research Institute; 2017. [Google Scholar]
- 69.Kuno G. Letter to the Editor. Bioscience. 2019;69(3):161–161. [Google Scholar]
- 70.Powell JR, Gloria-Soria A, Kotsakiozi P. Response to Kuno. Bioscience. 2019;69(3):161–2. [Google Scholar]
- 71.Guillaumot L, Ofanoa R, Swillen L, Singh N, Bossin HC, Schaffner F. Distribution of Aedes albopictus (Diptera, Culicidae) in southwestern Pacific countries, with a first report from the Kingdom of Tonga. Parasites and Vectors. 2012;5(1):247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xia D, Guo X, Hu T, Li L, Teng PY, Yin QQ, et al. Photoperiodic diapause in a subtropical population of Aedes albopictus in Guangzhou, China: Optimized field-laboratory-based study and statistical models for comprehensive characterization. Infect Dis Poverty. 2018;7(1):89 10.1186/s40249-018-0466-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Manni M, Guglielmino CR, Scolari F, Vega-Rúa A, Failloux AB, Somboon P, et al. Genetic evidence for a worldwide chaotic dispersion pattern of the arbovirus vector, Aedes albopictus. Apperson C, editor. PLoS Negl Trop Dis. 2017;11(1):e0005332 10.1371/journal.pntd.0005332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gillespie RG, Baldwin BG, Waters JM, Fraser CI, Nikula R, Roderick GK. Long-distance dispersal: a framework for hypothesis testing. Trends Ecol Evol. 2012;27(1):47–56. 10.1016/j.tree.2011.08.009 [DOI] [PubMed] [Google Scholar]
- 75.Urbanelli S, Bellini R, Carrieri M, Sallicandro P, Celli G. Population structure of Aedes albopictus (Skuse): The mosquito which is colonizing Mediterranean countries. Heredity (Edinb). 2000;84(3):331–7. [DOI] [PubMed] [Google Scholar]
- 76.Ardika IW, Bellwood P. Sembiran: the beginnings of Indian contact with Bali. Antiquity. 1991;65(247):221–32. [Google Scholar]
- 77.Guo Y, Song Z, Luo L, Wang Q, Zhou G, Yang D, et al. Molecular evidence for new sympatric cryptic species of Aedes albopictus (Diptera: Culicidae) in China: A new threat from Aedes albopictus subgroup? Parasit Vectors. 2018;11(1):228–41. 10.1186/s13071-018-2814-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lourenço de Oliveira R, Vazeille M, de Filippis AMB, Failloux AB. Large genetic differentiation and low variation in vector competence for dengue and yellow fever viruses of Aedes albopictus from Brazil, the United States, and the Cayman Islands. Am J Trop Med Hyg. 2003;69(1):105–14. [PubMed] [Google Scholar]
- 79.Maynard AJ, Ambrose L, Cooper RD, Chow WK, Davis JB, Muzari MO, et al. Tiger on the prowl: Invasion history and spatio-temporal genetic structure of the Asian tiger mosquito Aedes albopictus (Skuse 1894) in the Indo-Pacific. Vasilakis N, editor. PLoS Negl Trop Dis. 2017;11(4):e0005546 10.1371/journal.pntd.0005546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Moore PR, Johnson PH, Smith GA, Ritchie SA, Van Den Hurk AF. Infection and dissemination of dengue virus type 2 in Aedes aegypti, Aedes albopictus, and Aedes scutellaris from the Torres Strait, Australia. J Am Mosq Control Assoc. 2007;23(4):383–8. 10.2987/5598.1 [DOI] [PubMed] [Google Scholar]
- 81.Zheng X, Zhang D, Li Y, Yang C, Wu Y, Liang X, et al. Incompatible and sterile insect techniques combined eliminate mosquitoes. Nature. 2019;572(7767):56–61. 10.1038/s41586-019-1407-9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
These tables show means and upper and lower confidence ranges of: (S1A Table) pairwise FST' for Ae. aegypti in the Indo-Pacific region; (S1B Table) pairwise FST' for Ae. albopictus in the Indo-Pacific region; (S1C Table) heterozygosity and nucleotide diversity for Ae. aegypti in the Indo-Pacific region; (S1D Table) heterozygosity and nucleotide diversity for Ae. albopictus in the Indo-Pacific region; and (S1E Table) FST when calculated with five genotypes compared with when calculated with 16 genotypes.
(XLSX)
Colours indicate cluster membership. Circles are sized relative to population sample size. For Ae. aegypti, K = 15 was used, while K = 11 was used for Ae. albopictus. The map uses a Mollweide projection with a central meridian of 120°E. The map was produced in arcmap 10.5.1, using shapefiles made available by www.naturalearthdata.com.
(TIF)
Data Availability Statement
Aligned .bam files for 224 Ae. aegypti and 256 Ae. albopictus are available at the NCBI Sequence Read Archive under project accession number PRJNA641723.