Abstract
Epizoic diatoms form an important part of micro-epibiota of marine vertebrates such as whales and sea turtles. The present study explores and compares the diversity and biogeography of diatom communities growing on the skin and shell of loggerhead sea turtles (Caretta caretta) from four different localities: Adriatic Sea (Croatia), Ionian Sea (Greece), South Africa and Florida Bay (USA) using both light and scanning electron microscopy. We observed almost 400 diatom taxa belonging to more than 100 genera. Diatom communities from Greece and Croatia showed the highest similarity and were statistically different from those recorded from South Africa and Florida. Part of this variation could be attributed to differences in sampling techniques; however, we believe that geography had an important role. In general, contrary to several previous observations from sea turtles, the presumably exclusively epizoic diatoms contributed less than common benthic taxa to the total diatom flora, which might have been related to the loggerhead feeding behavior. Moreover, skin samples differed from carapace samples in having a distinct diatom composition with a higher proportion of the putative true epizoonts. Our results indicate that epizoic diatom communities differ according to loggerhead geographical location and substrate (skin vs. carapace). The relative abundances of common benthic diatoms and putative exclusive epizoic taxa may inform about sea turtle habitat use or behavior though detailed comparisons among different host species have yet to be performed.
Introduction
Diatoms (Bacillariophyceae) are unicellular eukaryotic microalgae characterized by a silica outer shell (frustule). The number of diatom species worldwide is estimated between 30 000 and 100 000 [1]. Of these, around 55 000 are estimated to exist in marine habitats. To date, however, less than 5 000 of marine diatoms have been described [2]. Diatoms occur wherever water is available, including terrestrial, freshwater and marine habitats. They are part of the phytobenthos attached to humid or submerged surfaces or thrive as free-floating phytoplankton in open water bodies [3]. Attached diatom communities can be classified by the substratum they live on. For instance, epipsammic diatoms are attached to sand grains, epilithic diatoms grow on rocks, epiphytic diatoms live on plants and epizoic diatoms grow on animals, the latter two categories commonly called epibionts [3].
The external surfaces of large marine vertebrates, such as whales, sea turtles and manatees, provide suitable hard substrata for the development of rich microbial biofilms. In these biofilms, composed of, among others, bacteria, fungi, cyanobacteria, and micro- and macroalgae, diatoms are often one of the main components, with densities sometimes exceeding those known from other marine substrata [4].
Several presumably exclusively epizoic diatom genera including Bennettella, Epipellis, Epiphalaina, Plumosigma, and Tursiocola have been described from the skin of cetaceans [5–9]. More recently, epizoic diatoms, including novel species, were described from freshwater turtles in the Rio Negro, Brazil [10–11]. Since 2015, there is a growing literature on epizoic diatoms observed on the carapaces and skin of all known sea turtle species [12–15].
Exclusive epibiosis is still debated as a lot of diatom taxa can be found on both animal and non-animal surfaces, and occur only haphazardly on marine turtles as a result of the physical contact with a variety of immersed substrata during the animal feeding and grooming activities [14]. However, currently, several sea turtle-associated genera are considered strictly epizoic. Chelonicola and Poulinea have so far been found on the carapaces of olive ridleys (Lepidochelys olivacea) [13, 14], and later on, green turtles (Chelonia mydas) [16–18], flatbacks (Natator depressus) [14], hawksbills (Eretmochelys imbricata) [14], loggerheads (Caretta caretta) [19], and Kemp’s ridleys (Lepidochelys kempii) [20, 21], whereas Medlinella is known only from the skin of loggerheads [12]. Additionally, several new species belonging to non-strictly epizoic genera were described in the recent past from the carapaces of sea turtles. Examples include Achnanthes elongata Majewska & Van de Vijver and A. squaliformis Majewska & Van de Vijver, found on the carapaces of olive ridleys [17], Kemp’s ridleys, loggerheads, and green turtles [20], Labellicula lecohuiana Majewska & Van de Vijver, living on the carapaces of green turtles [22, 23], and five species of Proschkinia, found associated with different sea turtle species [19].
The present research was conducted on loggerhead sea turtles, named after their large head and jaws. These middle-sized sea turtles (60–200 kg) are characterized in having a yellow coloured plastron and a dark brown carapace. Loggerheads are widely distributed in the subtropical and temperate regions of the Atlantic, Indian and Pacific Ocean and the Mediterranean Sea [24]. They can occur in both deeper areas and shallow river estuaries [25] and are highly migratory. Wallace et al. [24] proposed to subdivide the world loggerhead population into several Regional Management Units (RMUs) that enables the identification of important geographic areas for different subpopulations in terms of their presence, density and richness, including for example Northeast Atlantic, Northwest Atlantic, Mediterranean and Southwest Indian RMU. The present study reports on the diatom communities growing on loggerhead sea turtles from four distinct geographical localities (Adriatic Sea, Ionian Sea (both Mediterranean population), South Africa (Southwest Indian population) and Florida Bay (Northwest Atlantic population) with the objective to provide the baseline data on their diversity, species composition, and biogeography. Additionally, differences between communities living on the various sea turtle body parts (skin versus carapace) are explored.
Material and methods
Study area
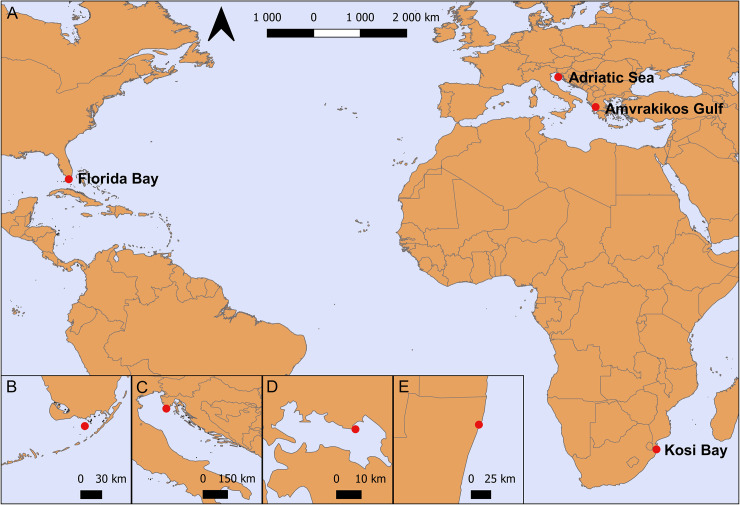
Samples used in this study were collected from loggerheads found in four different localities: northeastern Adriatic Sea (Croatia), Amvrakikos Gulf (Greece), Kosi Bay (South Africa) and Florida Bay (USA) (Fig 1).
Fig 1. The sampling areas of loggerhead sea turtles.
(A) Red dots indicate locations of sampled loggerheads. Inserts show details of the sampling locations: (B) Amvrakikos Gulf, Greece; (C) Adriatic Sea, Croatia; (D) Florida Bay, USA; (E) Kosi Bay, South Africa. The maps were made with Natural Earth. Free vector and raster map data @ naturalearthdata.com.
The Adriatic Sea, connected to the Mediterranean Sea via the Otranto Strait, is one of the most important foraging areas for juvenile and adult loggerhead turtles in the Mediterranean Basin [26]. Samples from Adriatic Sea loggerheads were obtained from animals brought into the Marine Turtle Rescue Centre in Aquarium Pula (Croatia) for rehabilitation in 2016 and 2017. A second Mediterranean loggerhead turtle population was sampled in Amvrakikos Gulf (Ionian Sea, Greece), an important foraging area with a very high density of loggerheads [27]. Diatom samples were collected in 2018 in the framework of the research and conservation activities conducted by ARCHELON in Amvrakikos Gulf by capturing them with the rodeo technique [28]. The rodeo technique was also used to capture and sample loggerheads in 2015 during an annual survey of sea turtle populations in Florida Bay, a shallow lagoon. The South African turtle population was sampled from the beaches in Kosi Bay (northeastern South Africa), an important nesting area for Indian Ocean loggerheads and leatherbacks. Samples were taken in 2018 from nesting loggerheads.
Sample collection and processing
From each subpopulation, five loggerheads were arbitrarily selected for diatom sampling. Basic information about each turtle, such as carapace length and weight, was also collected at the time of sampling (Table 1). The material collection was performed by researchers licenced for animal handling and well-informed volunteers following institutional guidelines for the care and use of animals. All the procedures involved respecting the ethical standards in the Helsinki Declaration of 1975, as revised in 2013, as well as the applicable national laws. All sampling activities performed in the iSimangaliso Wetland Park (South Africa) were carried out under research permits issued by the South African Department of Environmental Affairs (RES2017/73). Sampling activities in Croatia were done in accordance with the authorization of the Marine Turtle Rescue Centre by the Ministry of Environment and Energy of the Republic of Croatia. Sampling activities in Greece were carried out with permission from the Hellenic Ministry of Agriculture and Environment.
Table 1. List of samples and information on loggerhead sea turtles.
| Sample code | Body part | ID Tag / Turtle name | Sampling Date | Sex | Weight (kg) | SCL (cm) | CCL (cm) | SCW (cm) | CCW (cm) |
|---|---|---|---|---|---|---|---|---|---|
| Sampling location: Amvrakikos Gulf (Greece) 39° 1' 29" N - 39° 1' 44" N; 21° 3' 36" E - 21° 4' 19" E | |||||||||
| GRE-01 | carapace | Y6343- Y6344 | 8/1/2018 | 75.4 | 78.6 | 55.0 | 68.5 | ||
| GRE-02 | skin | Y6343- Y6344 | |||||||
| GRE-03 | carapace | Y6366- Y6367 | 8/1/2018 | 47.4 | 51.0 | 36.3 | 46.8 | ||
| GRE-04 | skin | Y6366- Y6367 | |||||||
| GRE-05 | carapace | Y6368- Y6369 | 8/1/2018 | 66.1 | 69.6 | 51.5 | 63.8 | ||
| GRE-06 | skin | Y6368- Y6369 | |||||||
| GRE-07 | carapace | Y6370- Y6371 | 8/1/2018 | 54.5 | 58.5 | 43.7 | 55.2 | ||
| GRE-08 | skin | Y6370- Y6371 | |||||||
| GRE-09 | carapace | M9123- M9124 | 8/1/2018 | 50.4 | 53.2 | 37.5 | 47.5 | ||
| GRE-10 | skin | M9123- M9124 | |||||||
| Sampling location: Florida Bay (USA) 24° 55’18” N; 80° 48’ 28” W | |||||||||
| FLO-U8 | carapace | HA5053 - HA5054 | 6/24/2015 | F | 58.5 | 74.1 | |||
| FLO-U9 | carapace | HB5559 - HB5560 | 6/24/2015 | 48.0 | 66.9 | ||||
| FLO-U10 | carapace | HB5668 - X7596 | 6/24/2015 | M | 89.0 | 87.3 | |||
| FLO-U11 | carapace | W1924 - W2176 | 6/24/2015 | F | 75.7 | 79.5 | |||
| FLO-U12 | carapace | HB5581 - HB5582 | 6/25/2015 | 71.2 | 78.5 | ||||
| Sampling location: Kosi Bay (South Africa) 26° 59' 38.9" S; 32° 51' 59.8" E | |||||||||
| SA-33 | carapace | ZA0447D - ZA0427D | 1/16/2018 | F | 86.4 | 83.6 | |||
| SA-37 | carapace | ZA0829D - ZA0828D | 1/11/2018 | F | 80.2 | 85.7 | 62.0 | 78.8 | |
| SA-45 | carapace | ZA0924D - ZA0186D | 1/11/2018 | F | 73.9 | 80.2 | 58.2 | 74.8 | |
CCL = curved carapace length, CCW = curved carapace width, SCL = straight carapace length, SCW = straight carapace width.
The sampling method differed between sampling events. Carapace samples from Greece and South Africa were collected by scrubbing the carapace with a single-use toothbrush on at least three arbitrarily chosen areas of the carapace, ensuring a scraped surface of at least 60 cm2. Samples were stored in plastic vials filled with at least 70% ethanol for fixation. Carapace samples from Croatia were scraped off with a curette and stored in plastic vials (100 mL) filled with seawater and fixed with formaldehyde at a final concentration of 4%. Samples from the carapaces of Florida loggerheads were collected using cotton-tipped applicators to rub diatoms from the carapace and onto the cotton tips. The cotton tips were removed from the applicators and stored in sealed plastic bags on ice until further processing. Additionally, diatoms from the skin of loggerheads from Greece were collected by gently scrubbing the dorsal area of the neck and/or the upper side of the flippers with a single-use toothbrush, and then rinsing the toothbrush head into a 50 ml Falcon tube filled with 96% ethanol. In total, we collected 25 samples of which 20 from loggerhead carapace and for five of these animals we were also able to simultaneously sample their skin.
Samples were processed following the methods described by Hasle and Syvertsen [29] for South African samples and van der Werff [30]. In most cases, portions of the biofilm were cleaned by adding 37% H2O2 and heating to 80°C for about 1h. The reaction was completed by the addition of KMnO4 [30]. South African samples were digested with boiling concentrated acids (HNO3 and H2SO4) [29]. Following digestion and centrifugation (three times 10 minutes at 3 500 rpm, Phoenix Instruments, Clinical Centrifuge CD-0412), cleaned material was diluted with distilled water to avoid excessive concentrations of diatom valves on the slides. Samples from Florida were prepared by removing in the laboratory the cotton tips of the applicators using a razor blade and then oxidizing the tips for diatom examination by boiling the cotton fibers of the applicator tip and epizoic organic material in 100 ml of 30% nitric acid followed by addition of potassium dichromate when 50 ml of acid remained. Cleaned diatoms were settled from the mixture for a minimum of 6 h and the remaining acid solution decanted. Settled diatoms were rinsed with deionized water. The rinsing/settling/decanting process was repeated six times until the solution reached a neutral pH. All slides were prepared using Naphrax mounting medium and analyzed using an Olympus BX53 microscope equipped with differential interference contrast (Nomarski) optics and the Olympus UC30 Imaging System. For scanning electron microscope (SEM) analyses, parts of the oxidized suspensions were filtered through a 2 μm Isopore™ polycarbonate membrane filter (Merck Millipore). The filters were mounted on stubs and sputter-coated with 10 nm of platinum or 20 nm of gold-palladium. The stubs were analyzed at Meise Botanic Garden using a JSM-7100F Jeol Field Emission Scanning Electron Microscope at 2 kV and with a working distance of 4 mm. For a more detailed analysis of very finely structured species, some samples were studied using a ZEISS Ultra Scanning Electron Microscope at 3 kV in the Natural History Museum London, UK. Samples and slides are stored at the BR collection (Meise Botanic Garden, Belgium).
In each slide, 400 diatom valves were counted and identified in random transects to estimate the species richness and composition in the samples. After counting, a complete slide was examined to record all occurring taxa in a sample. Extensive literature including both monographs [31–34] and other taxonomic publications were used to identify the observed taxa listed in S1 Table.
Data analyses
To make the pair-wise comparison between geographic localities we used the Sørenson similarity index [35]. This index uses presence/absence data, and the following formula 2c/(a + b + 2c), where a and b are the numbers of taxa exclusively observed in each of the two populations and c is the number of taxa shared by these populations. The Shannon-Wiener diversity index (ln-based) was calculated using the Multivariate Statistical Package (MVSP) [36]. Abundance data were square-root transformed to downweight dominant taxa. Only taxa with a total abundance of at least 2% in one sample were included in all further statistical analyses to avoid excessive noise in the dataset. Two-dimensional non-metric multidimensional scaling (NMDS) was used based on Bray-Curtis similarity matrix to reveal the patterns in taxa composition between different localities and turtle body parts. Analysis of similarity percentages (SIMPER) was performed to detect taxa that were responsible for most of the dissimilarity observed between different loggerhead localities and body parts [37]. Two sampling designs, one using four distinct loggerhead subpopulations and the second using two body parts (skin and carapace) were used to perform distance-based permutational multivariate analysis of variance (PERMANOVA) [38]. The PERMANOVA pairwise test was performed on the matrix of square root transformed data calculated on Bray Curtis similarity, using Type III Sums of Squares (i.e. partial sums of squares), with fixed effects and unrestricted permutation of raw data (9999 permutations). All multivariate analyses were performed using the software packages PRIMER v6 and v7 [39], including the add-on package v6 PERMANOVA+.
Results
Taxonomic composition and diversity
A total of 183 diatom taxa (including species, varieties and forms) belonging to 56 genera were identified during the counts. One carapace sample from Florida (sample FLO-U8) did not contain a sufficient number of diatom valves and was therefore removed from further analyses. An additional 214 taxa were observed outside the count procedures, bringing the total number of recorded taxa to 397 (S2 Table). Several common diatoms found on both carapace and skin of loggerheads from all four investigated localities are illustrated in Fig 2. Only 41% (166 taxa) of all taxa could be identified to the species level. An additional 14% (56) were given provisional names as ‘cf.’. In the Florida and Greek samples, more taxa were identified at the species level (57% and 52%, respectively) compared to the Croatian and South African samples (40% and 37%, respectively).
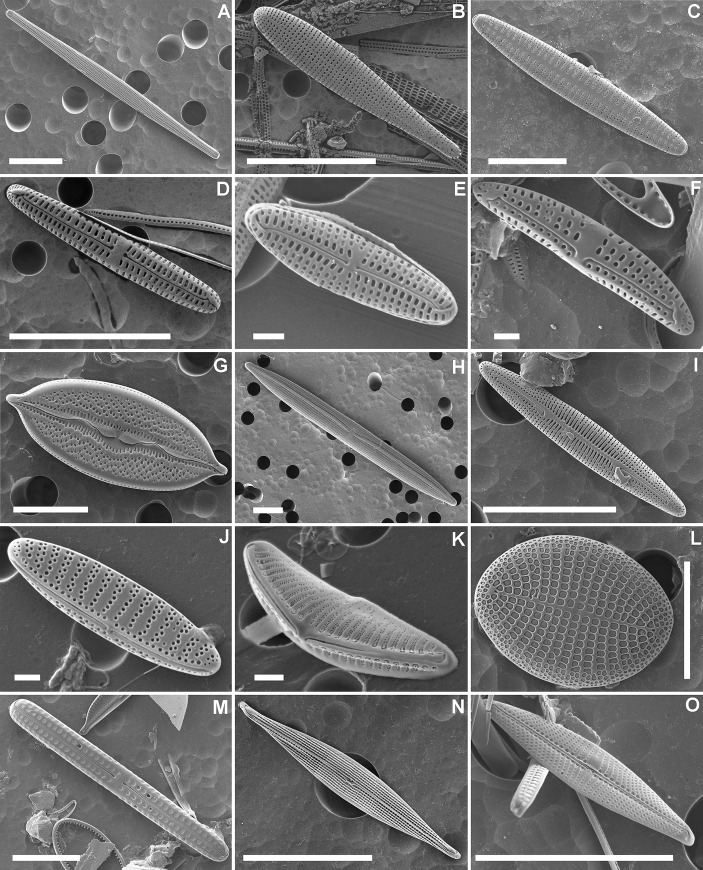
Fig 2. Scanning electron micrographs of diatom taxa associated with loggerhead sea turtles.
(A) Hyalosynedra laevigata (FLO). (B) Licmophora debilis (GRE). (C) Tabularia cf. investiens (FLO). (D) Poulinea lepidochelicola (CRO). (E) Chelonicola sp. (SA) (F) Medlinella amphoroidea (GRE). (G) Mastogloia cf. corsicana (FLO). (H) Nitzschia cf. scalpelliformis (FLO). (I) Berkeleya fennica (CRO). (J) Nitzschia cf. inconspicua (CRO). (K) Bifibulatia sp. (CRO) (L) Cocconeis scutellum (GRE). (M) Achnanthes elongata (GRE). (N) Proschkinia sulcata (GRE) (O) Proschkinia vergostriata (GRE). Scale bars represent 10 μm, except for E, F, J & K where scale bar = 1 μm. CRO–Croatia, Adriatic Sea; GRE = Greece, Ionian Sea; FLO = Florida Bay, USA; SA–South Africa; Kosi Bay.
The most taxon-rich genera found in all samples included Mastogloia (42 taxa), Navicula (32 taxa), Amphora (30 taxa) and Nitzschia (30 taxa) (Table 2). Diatom genus composition differed among the carapace samples from different locations. The carapace flora on loggerheads sampled in Croatia contained mostly Nitzschia (13 taxa) and Mastogloia (11 taxa) whereas in Greek samples Navicula (13 taxa) and Mastogloia (12 taxa), in Florida samples Mastogloia (26 taxa) and the South African samples Cocconeis (16 taxa) and Licmophora (15 taxa) were the most species-rich genera.
Table 2. The number of diatom taxa in the most diverse genera in samples from different localities.
| Diatom genera | Overall | Croatia | Greece | South Africa | Florida |
|---|---|---|---|---|---|
| Mastogloia | 42 | 11 | 12 | 12 | 26 |
| Navicula | 32 | 11 | 13 | 11 | 6 |
| Amphora | 30 | 4 | 9 | 12 | 7 |
| Nitzschia | 30 | 13 | 10 | 12 | 8 |
| Cocconeis | 26 | 8 | 6 | 16 | 2 |
| Licmophora | 20 | 5 | 5 | 15 | 0 |
| Diploneis | 20 | 10 | 5 | 6 | 2 |
| Seminavis | 12 | 2 | 5 | 5 | 3 |
| Achnanthes | 10 | 4 | 2 | 9 | 1 |
| Tryblionella | 8 | 2 | 0 | 6 | 0 |
| other genera | 167 | 57 | 71 | 71 | 32 |
Diatom counts indicated that the most frequently occurring species in all (carapace + skin) samples were Nitzschia CRO sp.2 (present 83.3% of all samples), Amphora crenulata Wachn. & E.E.Gaiser (70.8%), Cocconeis lineata Ehrenb. (70.8%), Nitzschia cf. inconspicua (62.5%) and Poulinea CRO sp.1 (54.2%). Of all counted valves, N. cf. inconspicua contributed to 16.6%, Hyalosynedra laevigata (Grunow) D.M.Williams & Round to 13.1%, Nitzschia CRO sp.2 (12.2%), Chelonicola SA sp.1 (9.9%), and Poulinea CRO sp.1 (4.4%). Altogether, only ten taxa contributed more than 71% of all counted valves, whereas 17 taxa together account for 1% of the total number of valves.
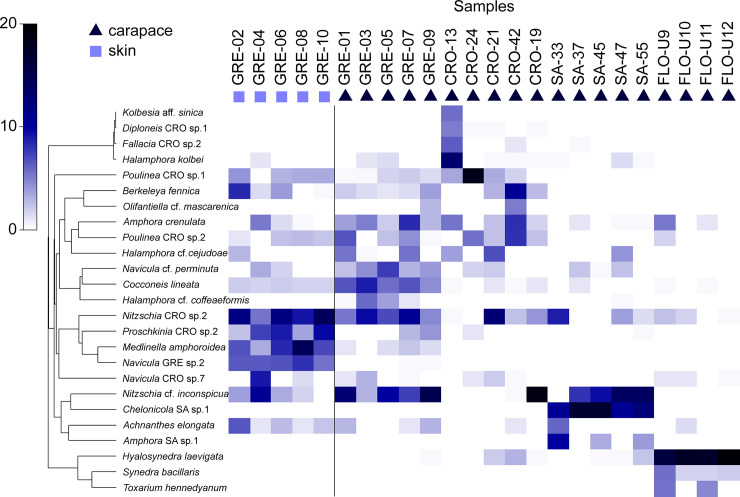
Although most taxa occurred in only one investigated locality, the Greek and Croatian samples shared 45 taxa, a relatively large number (Fig 3, S2 Table). Several taxa, such as Nitzschia cf. inconspicua and Nitzschia CRO sp. 2, appeared abundantly in almost all groups except the Florida samples. In the Florida samples, taxa such as Hyalosynedra laevigata, Synedra bacillaris (Grunow) Hust. and Toxarium hennedyanum (W.Greg.) Pelletan reached the highest relative abundances (Fig 3). All South African samples were dominated by Chelonicola SA sp.1, whereas those collected from the Meditteranean region (the Croatian-Greek group) by Poulinea CRO sp.1 and sp. 2., Amphora crenulata, Berkeleya fennica Juhl.-Dannf., Cocconeis lineata, and Navicula cf. perminuta Grunow (Fig 3). One-fifth of all counted valves belonged to the presumed exclusively-epizoic taxa such as Achnanthes elongata Majewska & Van de Vijver, Medlinella amphoroidea Frankovich et al., Poulinea spp., and Chelonicola sp. (S2 Table). The total relative abundance of these species varied strongly among the populations (Fig 3), with the lowest values (0.3%) recorded from the Florida samples and highest from the South African ones (49.1%). Moreover, a significant difference was observed between the carapace and skin samples from Greece, where the relative abundance of the presumably epizoic taxa reached 25.7% and 5.3%, respectively.
Fig 3. The most abundant diatom taxa associated with loggerhead sea turtles.
Shade plot illustrating the 25 most abundant taxa recorded on loggerhead carapaces (triangle) and skins (square) from investigated localities based on square root-transformed abundance data. The white cells represent the absence of the taxa and the darkest cells the largest abundances. Taxa are ordered by a hierarchical cluster analysis of their mutual associations across samples based on Index of Association calculated on the standardized counts. CRO = Croatia, Adriatic Sea; FLO = Florida Bay, USA; GRE = Greece, Amvrakikos Gulf; SA = South Africa, Kosi Bay.
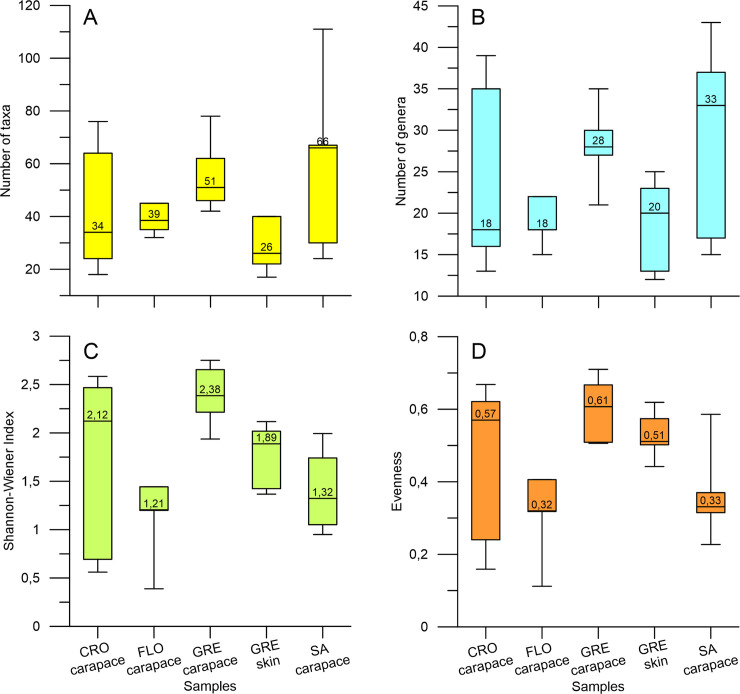
Species number in a single sample varied between 11 and 111 taxa (including taxa observed outside the counts) (Fig 4A), and the median from the same area was generally lower in the skin samples (26) than in the carapace samples (51). Among carapace samples, the South African samples were the most taxa-abundant, whereas the lowest number of taxa characterized Florida and some of the Croatian samples (Fig 4A). Likewise, the number of genera differed among the populations (Fig 4B), being highest for the South African material. The carapace samples from Greece showed the highest diversity (median value 2,38) and evenness (0.61), while the samples from Florida had the lowest diversity (1,21) and evenness (0,32) (Fig 4C and 4D).
Fig 4. Box and whisker plots of diatom community diversity across localities.
The diatom community diversity for loggerhead carapace samples from every locality, and the skin samples from Greece. (A) The number of taxa. (B) genera, (C) the Shannon-Wiener diversity index and D) evenness. Whiskers indicate maximum and minimum, the median value is denoted within the box. CRO = Croatia, Adriatic Sea; FLO = Florida Bay, USA; GRE = Greece, Amvrakikos Gulf; SA = South Africa, Kosi Bay.
Comparative analyses
The Sørenson similarity index showed that the Croatian and Greek samples were the most similar both at infrageneric (species level and below) and genus level, 35% and 62%, respectively. The lowest similarity on infrageneric level (almost 16%) is noted between the Florida and South Africa samples and on genus level between Florida and Greece (42.5%; Table 3).
Table 3. Sørensen-similarity index of the carapace samples between the different localities.
| Taxon level | Croatia | Florida | Greece |
| Florida | 20.37 | ||
| Greece | 35.02 | 21.20 | |
| South Africa | 25.57 | 15.85 | 22.22 |
| Genus level | Croatia | Florida | Greece |
| Florida | 46.91 | ||
| Greece | 62.14 | 42.50 | |
| South Africa | 54.05 | 45.45 | 49.09 |
The index was calculated at both species and genus level, expressed as percentages.
According to the SIMPER analysis, Croatian samples had the lowest within-site similarity (average similarity 21,1%), followed by South African and Greek samples (49,3% and 57,8%, respectively), whereas Florida samples were the most homogenous ones (60,4%; S3 Table).
In general, the most abundant taxa in each sample group were also the ones contributing the most to the within-group similarity such as Poulinea CRO sp.1 and sp.2 for Croatia, Nitzschia cf. insconspicua and Nitzschia CRO sp.2 for Greece, Chelonicola SA sp.1. for South Africa and Hyalosynedra laevigata for Florida (S3 Table). SIMPER dissimilarity analysis (Table 4) showed that ten taxa contributed approx. 50% to the total differences observed between Greek and Croatian samples, with Nitzschia cf. inconspicua and Cocconeis lineata having the highest contributions. Samples from Florida differed from those from other locations mainly due to Hyalosynedra laevigata with 20.6%, 18.3%, and 21.7% contributions to the total dissimilarity observed between Florida and Croatia, Florida and Greece, and Florida and South Africa, respectively. South African samples were characterized by high abundances of Chelonicola SA sp. 1 that contributed 15.86%, 17,34%, and 15.67% to the total dissimilarity between South Africa and Croatia, South Africa and Florida, and South Africa and Greece. Medlinella amphoroides, Nitzschia cf. inconspicua, Navicula GRE sp. 2, and Proschkinia CRO sp. 2 were responsible for most of the differences between the skin and carapace diatom communities from Greece (Table 4).
Table 4. Contribution of species to dissimilarities between epizoic diatom assemblages of loggerhead populations–discriminating species.
| Species | Average Abundance | Average Abundance2 | Average Dissimilarity | Dissimilarity/SD | Contribution % | Cumulative contribution % |
|---|---|---|---|---|---|---|
| Croatia & Florida | ||||||
| CRO | FLO | |||||
| Hyalosynedra laevigata | 1.04 | 17.56 | 19.05 | 3.41 | 20.61 | 20.61 |
| Poulinea CRO sp. 1 | 5.37 | 0.00 | 6.61 | 0.70 | 7.15 | 27.76 |
| Nitzschia cf. inconspicua | 3.83 | 0.00 | 5.29 | 0.49 | 5.72 | 33.48 |
| Poulinea CRO sp. 2 | 3.71 | 0.43 | 3.88 | 1.28 | 4.20 | 37.67 |
| Nitzschia CRO sp.2 | 3.89 | 1.10 | 3.78 | 0.92 | 4.09 | 41.76 |
| Amphora crenulata | 3.54 | 1.55 | 3.43 | 1.30 | 3.71 | 45.47 |
| Berkeleya fennica | 3.24 | 0.00 | 3.29 | 0.87 | 3.56 | 49.02 |
| Synedra bacillaris | 0.00 | 2.78 | 3.05 | 1.77 | 3.30 | 52.33 |
| Croatia & Greece | ||||||
| CRO | FLO | |||||
| Nitzschia cf. inconspicua | 3.83 | 9.61 | 8.05 | 2.13 | 10.82 | 10.82 |
| Cocconeis lineata | 0.49 | 6.45 | 5.00 | 3.65 | 6.72 | 17.54 |
| Nitzschia CRO sp.2 | 3.89 | 7.38 | 4.68 | 1.91 | 6.29 | 23.83 |
| Poulinea CRO sp. 1 | 5.37 | 1.12 | 4.33 | 0.69 | 5.81 | 29.64 |
| Navicula cf. pavillardii | 0.00 | 4.29 | 3.62 | 4.65 | 4.87 | 34.51 |
| Navicula cf. perminuta | 0.75 | 4.29 | 2.97 | 1.73 | 3.99 | 38.49 |
| Poulinea CRO sp. 2 | 3.71 | 2.73 | 2.59 | 1.37 | 3.49 | 41.98 |
| Amphora crenulata | 3.54 | 4.36 | 2.57 | 1.31 | 3.45 | 45.43 |
| Berkeleya fennica | 3.24 | 1.91 | 2.38 | 1.10 | 3.20 | 48.62 |
| Halamphora kolbei | 2.74 | 0.48 | 2.37 | 0.59 | 3.19 | 51.82 |
| Florida & Greece | ||||||
| FLO | GRE | |||||
| Hyalosynedra laevigata | 17.56 | 0.04 | 17.08 | 6.51 | 18.26 | 18.26 |
| Nitzschia cf. inconspicua | 0.00 | 9.61 | 9.44 | 2.13 | 10.09 | 28.34 |
| Cocconeis lineata | 0.11 | 6.45 | 6.13 | 4.28 | 6.55 | 34.89 |
| Nitzschia CRO sp.2 | 1.10 | 7.38 | 6.09 | 2.39 | 6.50 | 41.40 |
| Navicula cf. pavillardii | 0.00 | 4.29 | 4.17 | 5.22 | 4.45 | 45.85 |
| Navicula cf. perminuta | 0.00 | 4.29 | 4.13 | 2.43 | 4.41 | 50.26 |
| Croatia & South Africa | ||||||
| CRO | SA | |||||
| Chelonicola SA sp. 1 | 0.00 | 13.46 | 13.97 | 2.96 | 15.86 | 15.86 |
| Nitzschia cf. inconspicua | 3.83 | 9.05 | 9.49 | 1.76 | 10.77 | 26.64 |
| Poulinea CRO sp. 1 | 5.37 | 0.00 | 5.91 | 0.72 | 6.71 | 33.35 |
| Nitzschia CRO sp.2 | 3.89 | 2.77 | 4.07 | 1.12 | 4.62 | 37.97 |
| Poulinea CRO sp. 2 | 3.71 | 0.00 | 3.74 | 1.35 | 4.25 | 42.22 |
| Amphora SA sp. 1 | 0.00 | 3.44 | 3.38 | 0.97 | 3.84 | 46.05 |
| Amphora crenulata | 3.54 | 0.29 | 3.13 | 1.28 | 3.56 | 49.61 |
| Berkeleya fennica | 3.24 | 0.00 | 2.99 | 0.87 | 3.40 | 53.01 |
| Florida & South Africa | ||||||
| FLO | SA | |||||
| Hyalosynedra laevigata | 17.56 | 0.54 | 20.77 | 4.90 | 21.66 | 21.66 |
| Chelonicola SA sp. 1 | 0.00 | 13.46 | 16.62 | 3.08 | 17.34 | 39.00 |
| Nitzschia cf. incospicua | 0.00 | 9.05 | 11.17 | 1.76 | 11.65 | 50.65 |
| Greece & South Africa | ||||||
| GRE | SA | |||||
| Chelonicola SA sp. 1 | 0.00 | 13.46 | 12.12 | 3.52 | 15.67 | 15.67 |
| Cocconeis lineata | 6.45 | 0.20 | 5.52 | 4.28 | 7.14 | 22.81 |
| Nitzschia CRO sp.2 | 7.38 | 2.77 | 4.84 | 1.72 | 6.26 | 29.07 |
| Nitzschia cf. inconspicua | 9.61 | 9.05 | 4.56 | 1.36 | 5.89 | 34.96 |
| Navicula cf. pavillardii | 4.29 | 0.04 | 3.78 | 5.30 | 4.88 | 39.85 |
| Amphora crenulata | 4.36 | 0.29 | 3.61 | 1.81 | 4.66 | 44.51 |
| Amphora SA sp. 1 | 0.00 | 3.44 | 2.95 | 0.98 | 3.82 | 48.33 |
| Navicula cf. perminuta | 4.29 | 0.98 | 2.92 | 1.57 | 3.77 | 52.10 |
| Greece carapace & skin | ||||||
| Carapace | Skin | |||||
| Medlinella amphoroides | 1.21 | 8.00 | 5.70 | 1.67 | 9.22 | 9.22 |
| Nitzschia cf. inconspicua | 9.61 | 3.90 | 5.70 | 1.48 | 9.21 | 18.43 |
| Navicula GRE sp.2 | 0.09 | 6.64 | 5.40 | 6.46 | 8.73 | 27.16 |
| Proschkinia CRO sp.2 | 1.39 | 6.42 | 4.31 | 1.65 | 6.97 | 34.13 |
| Cocconeis lineata | 6.45 | 1.84 | 3.78 | 3.21 | 6.11 | 40.25 |
| Nitzschia CRO sp.2 | 7.38 | 10.46 | 3.43 | 1.44 | 5.55 | 45.79 |
| Amphora crenulata | 4.36 | 1.37 | 2.97 | 1.55 | 4.81 | 50.60 |
Summary of SIMPER analysis of carapace and skin data based on Bray-Curtis dissimilarity, 70% cut off, taxa cumulatively contributing to the dissimilarity over 50% are shown. Croatia (CRO), Greece (GRE), South Africa (SA), Florida (FLO).
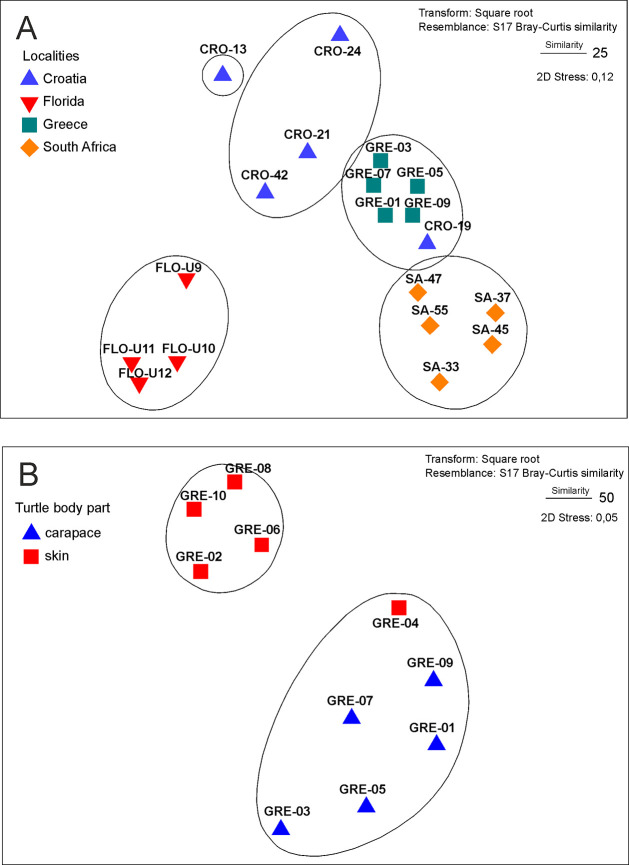
Non-metric multidimensional scaling based on carapace diatom abundance data separated samples into five distinct groups (Fig 5A). The Florida cluster was the most distant from all remaining groups, while the South African, Greek, and Croatian samples were placed closer to each other, but in general, maintaining a good separation among the different localities. Croatian samples showed the lowest group homogeneity, with the main group of three samples and a single group comprised of one sample(CRO-13) and one sample placed in a different cluster (CRO 19). An additional nMDS analysis performed only on the Greek carapace and skin samples showed good separation of the two groups (Fig 5B).
Fig 5. Non-metric Multi-Dimensional Scaling (nMDS) plots of diatom assemblages on loggerhead turtles.
(A) Carapace samples from four localities. (B) Skin and carapace samples from Greece. The overlayed cluster analysis indicates grouping based on sample similarity of 25 and 50 in (A) and (B), respectively. CRO = Croatia, Adriatic Sea; FLO = Florida Bay, USA; GRE = Greece, Amvrakikos Gulf; SA = South Africa, Kosi Bay.
The PERMANOVA pair-wise test confirmed the significant effect of both the location (p<0,01) and the sea turtle body part (p = 0.008) on the associated diatoms (Table 5).
Table 5. PERMANOVA analyses based on pairwise tests on square-root transformed data Sums of squares type: Type III (partial), fixed effects sum to zero for mixed terms.
| Groups | df | t | P(perm) | Unique perms |
| Croatia, Florida | 7 | 2.3624 | 0.0078 | 126 |
| Croatia, Greece | 8 | 1.7282 | 0.0078 | 125 |
| Croatia, South Africa | 8 | 2.1666 | 0.0105 | 126 |
| Florida, Greece | 7 | 4.3461 | 0.0066 | 126 |
| Florida, South Africa | 7 | 3.8197 | 0.0083 | 126 |
| Greece, South Africa | 8 | 3.0402 | 0.0095 | 126 |
| Greece carapace & skin | 8 | 2.7412 | 0.008 | 126 |
| Average similarity between/within groups | ||||
| Croatia | Florida | Greece | South Africa | |
| Croatia | 21.046 | |||
| Florida | 7.5637 | 60.355 | ||
| Greece | 25.578 | 6.4227 | 57.828 | |
| South Africa | 11.931 | 4.1172 | 22.668 | 49.322 |
| carapace | skin | |||
| carapace | 57.828 | |||
| skin | 38.183 | 62.108 |
Discussion
Loggerheads from the analyzed populations harbour a very diverse diatom flora with almost 400 taxa belonging to more than 100 genera. This number is most likely an underestimation of the exact taxon richness as a sampling of a limited number of turtle individuals may limit the number of diatom taxa found. Additionally, several taxa mostly belonging to the genera Amphora, Navicula and Nitzschia were grouped under a common name and detailed SEM and molecular analysis would be necessary to clarify their correct taxonomic identity. That would most likely result in the increase of the true taxon diversity. A clear example is Nitzschia cf. inconspicua, most likely representing a group of taxa difficult to disentangle rather than one single species. In the past, the N. frustulum-inconspicua group has been the subject of several taxonomic and molecular revisions resulting in the description of several new species and a better characterization of others such as N. frustulum (Kütz.) Grunow [40, 41] based on small morphological differences. Nitzschia cf. inconspicua was found in other epibiont diatom communities, for instance living predominantly on olive ridley turtles in Costa Rica [4]. Rivera et al. [23] applied both molecular and microscopic analyses of carapace samples from green turtles in the Marine Nature Park of Mayotte (Indian Ocean) and found N. inconspicua to be one of the most abundant taxa observed with a homogenous morphology across all seven investigated sea turtles. DNA analysis, on the contrary, indicated the presence of tens of OTU’s (Operational Taxonomical Units), resulting in four groups implying a high (pseudo)cryptic diversity in N. inconspicua.
The observed taxon richness is clearly higher than currently observed from any other sea turtle species sampled so far. Majewska et al. [4] recorded only 21 taxa in 38 carapace samples from olive ridley sea turtles in Costa Rica whereas, in another study, Majewska et al. [16] reported 26 taxa belonging to 20 genera in 76 carapace samples from green turtles in Costa Rica and Iran. It is possible that the applied sampling technique in the latter two studies (i.e. use of razor blade or scalpel on a limited surface of the carapace) in contrast with the application of a toothbrush brushing off a larger surface influenced the observed taxon richness. Rivera et al. [23] used the toothbrush method to sample seven juvenile green turtles from Mayotte and observed 57 taxa. Our results also indicate a certain influence of the sampling technique. The Florida samples, collected with a cotton-tipped applicator, were the least diverse of all carapace samples. This method may have been too gentle to remove firmly attached, adnate diatom taxa, such as taxa from the genera Cocconeis and Amphora from the carapace, compared to the toothbrush and/or curette methods applied to sample the other populations. The dominating genera in the Florida samples, Hyalosynedra, Synedra and Toxarium, are all large, erect diatom genera [3, 33], only attached by their apices to the surface and therefore more easily removed when using a cotton-tipped applicator. Brushing the surface with a hard toothbrush removes more efficiently the well-attached, adnate diatom taxa from the hard carapaces. Recently this method was designated as the standard sampling method for epizoic diatom communities [42].
Despite the high taxon richness, the percentage of the presumably truly epizoic taxa is rather low, although, we cannot be certain of an exact number of taxa that belong to that group. Several taxa were recently described from loggerhead samples from this dataset such as Catenula exigua K.Robert et al., Planothidium kaetherobertianum Van de Vijver & Bosak, and Lucanicum ashworthianum Majewska et al [21, 43, 44]. These taxa have not yet been found in epizoic samples from other localities and substrata. Similarly, the newly described Proschkinia species such as P. vergostriata Frankovich et al. and P. sulcata Majewska et al. have so far only been found on turtle carapaces and skin [19]. Thus, more sampling and analyses of marine benthic diatom communities from both biotic (including marine animals) and abiotic substrata will be necessary to determine the exact habitat preferences of these diatoms.
For turtles sampled in Greece, we sampled both skin and carapace. Interestingly, a large difference in the relative abundance of the presumably strictly epizoic taxa was observed. Skin communities were dominated by Medlinella amphoroidea, Poulinea spp, and Achnanthes elongata, all currently known only from sea turtles [12, 16, 21] whereas taxa belonging to common epiphytic and epipelic diatom genera, such as Amphora, Halamphora, Berkeleya, and Cocconeis, were more abundant in carapace samples. Skin sample GRE-04 and the matching carapace sample GRE-03 were collected from the same turtle. The high abundances of Nitzschia cf. inconspicua and Navicula sp.7 (Fig 3) present in the above-mentioned skin sample resulted in its grouping with carapace samples.
Many of the observed diatom taxa are probably ‘ecological hitchhikers’ using the animal surface as yet another hard substratum suitable for their development [45–48]. On the other hand, some species in common benthic genera may as well be the obligately epizoic taxa. This seems to be true for the two Achnanthes species that are regularly found in high abundances on various sea turtles from different oceans [16, 17, 20, pers.observations]. Moreover, several Proschkinia and Craspedostauros species described from the sea turtle carapaces and skin occur frequently on the animal substratum and, so far, have never been recorded from a non-animal habitat [19, 20, pers. observations].
The current results indicate that sea turtle skin is likely a much more specific substratum for diatom growth than the carapace, the latter sharing more similarities with other biotic (e.g. shells of snails and molluscs, barnacles) or abiotic surfaces (e.g. rocks). Strictly epizoic diatom taxa develop well on the physiologically active substratum, whereas opportunistic benthic species, lacking some vital adaptations, may attach to the skin only temporarily when the external conditions are favourable. One of the striking examples is Medlinella amphoroidea, described from the skin of loggerheads in Florida [12]. In the present study, Medlinella was almost exclusively found in several skin samples but almost entirely absent from the carapace samples (only 10 valves found on the carapaces). Numerous opportunistic diatom taxa may end up on the carapaces of the loggerheads due to the foraging behavior of this turtle species [49, 50]. Other sea turtle species such as olive ridley and green turtle show a different feeding behavior and have a different diet [25], which may influence the epizoic diatom species composition. Robinson et al. [51] observed that the macro-epibiont diversity of nesting sea turtles is partially linked to the diversity of their foraging habitats. Thus, sea turtle species with more diverse foraging areas should have more diverse epibiont communities. Fuller et al. [52] reported that loggerheads host more macro-epibiotic species, such as barnacles, than green turtles. The authors of this study also suggest that the differences in epibiont communities between the two sea turtle species could be attributed to the difference in feeding behavior and diet, as adult loggerheads are benthic foragers, feeding by infaunal mining [53] and green turtles are herbivores, grazing on seagrass with little sediment disturbance [54]. Loggerheads often develop a rich macro-algal flora composed of a large number of filamentous algal taxa such as Polysiphonia carettia Hollenberg [55] or Ectocarpus fasciculatus Harvey. Epiphytic diatoms on these macroalgae, such as various species of Cocconeis or Amphora, although not directly attached to the animal body, may therefore further enrich the sea turtle-associated diatom community composition. As biofilm accumulates, the available and uncolonized substratum surface on the carapace decreases and so there will be also a decline in the relative abundance of strictly epizoic diatom taxa [16].
Thus, the behavior of the turtles and its impact on the attached diatom flora may explain why clear bioregionalism was found in the present study. Loggerhead samples from the Mediterranean localities (i.e. neighbouring Adriatic Sea and Amvrakikos Gulf), were found to be the most similar and distinct from both Southwest Indian (South Africa) and Northwest Atlantic (Florida) samples. Amvrakikos Gulf (Greek samples) is connected with the Adriatic Sea (Croatian samples) via the Ionian Sea. Satellite tracking has revealed that loggerhead turtles in Amvrakikos Gulf generally remain resident in this area but do occasionally venture to the northern Adriatic to forage l [56]. Sample CRO-13 differed significantly from other Croatian diatom communities, as indicated by the nMDS plot. The sample was taken eight months after the injured turtle arrived in the rescue center and after it was cleaned from its original epizoic biofilm due to standard procedures applied at the facility. The observed diatom flora showed a remarkable similarity to the diatom flora that was growing on the walls of the plastic housing tank in which the turtle was undergoing rehabilitation for several months (Bosak, pers. observation). This may reflect a rather easy transfer of diatom taxa present on the objects within the enclosure to the carapace surface of captive turtles, especially if the new environment restrains the animal from exhibiting its natural behaviour (e.g. feeding by diving, fast-swimming). As already proposed by Holmes et al. [8], and later by Wetzel et al. [11] and Majewska et al. [16], transfer of surface-associated diatoms between different animals occurs likely through body-to-body contact. It is plausible that physical contact will also be required for a diatom transfer between the animal host and inanimate objects.
A considerable part of the observed diatom taxa in the samples from Croatia and Greece was illustrated previously in the monograph of Álvarez-Blanco & Blanco [34] on the benthic diatom flora of the Mediterranean coasts. On the other hand, the dominating genera in the samples from Florida include Hyalosynedra, Synedra, Toxarium and Mastogloia, the latter present with a fairly large number of species, are often reported from the Florida Bay region [33, 57]. These observations seem to support the previously suggested hypothesis [14] that diatom composition may serve as a biogeographical indicator of the whereabouts of sea turtles, especially loggerheads that host particularly diverse diatom communities. By comparing the diatom flora on the sea turtle with known marine benthic diatom floras worldwide, it may be possible to detect where the loggerhead has been residing. Studies on epiphytic diatoms show that the epibiotic diatom communities may vary greatly depending on geographical locality and external environmental conditions [4, 16]. A follow-up study should explore both the epibiotic loggerhead flora and the local benthic (including diverse abiotic substrata and hard-surfaced animals) diatom communities. Additionally, a study combining the analysis of the epibiotic diatom flora and satellite tracking may be an interesting research venue.
Conclusion
The diatom flora on the carapaces and skin of loggerhead sea turtles from geographically distinct locations shows a remarkable diversity and a generally low similarity. Loggerheads from the same location share a common pool of diatoms, showing clear bioregionalism, and diatom communities on sea turtles from more distant regions show less similarity between each other than those from neighbouring areas.
In many cases, the presumably truly epizoic species were outnumbered by the local benthic taxa and had only a minor contribution to the sea turtle-associated diatom floras. This may be partially explained by the frequent physical contact with a variety of substrata occurring during the specific foraging activities of loggerheads. Although species-rich diatom communities are found on both the sea turtle carapace and skin, those associated with the latter appear to be less diverse with a higher abundance of the presumably exclusively epizoic taxa.
Loggerheads serve as reservoirs and probable vectors for diverse and often unique diatom communities. This ecological role of sea turtles is still poorly understood and rarely discussed, and future studies are required to throw more light on the sea turtle contribution to the benthic diatom dispersal and their modern biogeography.
Supporting information
(PDF)
Presumably exclusively epizoic taxa are indicated in bold.
(PDF)
SIMPER analysis was based on Bray-Curtis similarity, 70% cut off, taxa cumulatively contributing to the similarity over 70% are shown. Croatia (CRO), Greece (GRE), South Africa (SA), Florida (FLO).
(PDF)
Acknowledgments
Ronel Nel and Diane Z. M. Le Gouvello du Timat (Nelson Mandela University, South Africa) are thanked for their help during the material collection in South Africa and obtaining the necessary sampling permits. We thank Brian Stacy of the US National Marine Fisheries Service for the collection of diatom samples and Allen Foley of the Florida Fish and Wildlife Conservation Commission and Jennifer Keene of the University Of Florida College of Veterinary Medicine for allowing us to receive samples from captured loggerhead turtles during the annual Florida Bay sea turtle survey. For the Croatian samples, we are thankful to Milena Mičić and Karin Gobić Medica as well the rest of the staff from Marine Turtle Rescue Centre, Aquarium Pula. ARCHELON volunteers are thanked for their help during Amvrakikos Gulf sampling activities. Mrs Myriam de Haan is thanked for preparing the samples for LM and SEM analysis.This is a contribution 188 from the Division of Coastlines and Oceans of the Institute of Environment at Florida International University.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
SB was funded by Croatian Science Fund (HRZZ) UIP-2017-05-5635, RM was funded by the Systematics Association (UK) through the Systematics Research Fund Award (R. Majewska/2017). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Mann DG, Vanormelingen P (2013) An inordinate fondness? The number, distributions, and origins of diatom species. J Eukaryot Microbiol 60: 414–420. 10.1111/jeu.12047 [DOI] [PubMed] [Google Scholar]
- 2.Appeltans W, Ahyong ST, Anderson G, Angel MV, Artois T, Bailly N, et al. (2012) The magnitude of global marine species diversity. Curr Biol 22: 2189–2202. 10.1016/j.cub.2012.09.036 [DOI] [PubMed] [Google Scholar]
- 3.Round FE, Crawford RM, Mann DG (1990) The diatoms. Biology & morphology of the genera. Cambridge: Cambridge University Press. [Google Scholar]
- 4.Majewska R, Santoro M, Bolaños F, Chaves G, De Stefano M (2015. a) Diatoms and other epibionts associated with olive ridley (Lepidochelys olivacea) sea turtles from the Pacific coast of Costa Rica. PLoS ONE 10: e0130351 10.1371/journal.pone.0130351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennett AG (1920) On the occurrence of diatoms on the skin of whales. Proc Roy Soc London, Ser B 91: 352–357 [Google Scholar]
- 6.Nemoto T (1956) On the diatoms of the skin film of whales in the northern Pacific. Sci Rep Whales Res Inst Tokyo 11: 97–132 [Google Scholar]
- 7.Holmes RW (1985) The morphology of diatoms epizoic on cetaceans and their transfer from Cocconeis to two new genera, Bennettella and Epipellis. Brit Phycol J 20: 43–57 [Google Scholar]
- 8.Holmes RW, Nagasawa S, Takano H (1993. a) The morphology and geographic distribution of epidermal diatoms of the Dall’s porpoise (Phocoenoides dalli True) in the Northern Pacific Ocean. Bull Nat Sci Mus, Ser B (Botany) 19: 1–18 [Google Scholar]
- 9.Denys L (1997) Morphology and taxonomy of epizoic diatoms (Epiphalaina and Tursiocola) on a sperm whale (Physeter macrocephalus) stranded on the coast of Belgium. Diatom Res 12: 1–18. 10.1080/0269249X.1997.9705398 [DOI] [Google Scholar]
- 10.Wetzel C, Van de Vijver B, Ector L (2010) Luticola deniseae sp. nov. A new epizoic diatom from the Rio Negro (Amazon hydrographic Basin). Vie et Milieu 60: 177–184 [Google Scholar]
- 11.Wetzel CE, Van de Vijver B, Cox EJ, Bicudo D de C, Ector L (2012) Tursiocola podocnemicola sp. nov., a new epizoic freshwater diatom species from the Rio Negro in the Brazilian Amazon Basin. Diatom Res 27: 1–8. 10.1080/0269249X.2011.642498 [DOI] [Google Scholar]
- 12.Frankovich T, Ashworth MP, Sullivan MJ, Veselá J, Stacy NI (2016) Medlinella amphoroidea gen. et sp. nov. (Bacillariophyta) from the neck skin of loggerhead sea turtles (Caretta caretta). Phytotaxa 272: 101–114. 10.11646/phytotaxa.272.2.1 [DOI] [Google Scholar]
- 13.Majewska R, Kociolek P, Thomas E, De Stefano M, Santoro M, Bolaños F, et al. (2015. b) Chelonicola and Poulinea, two new gomphonemoid diatom genera (Bacillariophyta) living on marine turtles from Costa Rica. Phytotaxa 233: 236–250. 10.11646/phytotaxa.233.3.2 [DOI] [Google Scholar]
- 14.Robinson NJ, Majewska R, Lazo-Wasem EA, Nel R, Paladino FV, Rojas L, et al. (2016) Epibiotic diatoms are universally present on all sea turtle species. PLoS ONE 11: e0157011 10.1371/journal.pone.0157011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaleli A, Krzywda M, Witkowski A, Riaux-Gobin C, Solak CN, Zgłobicka I, et al. (2018) A new sediment dwelling and epizoic species of Olifantiella (Bacillariophyceae), with an account on the genus ultrastructure based on Focused Ion Beam nanocuts. Fottea 18: 212–226. 10.5507/fot.2018.007 [DOI] [Google Scholar]
- 16.Majewska R, Van de Vijver B, Nasrolahi A, Ehsanpour M, Afkhami M, Bolaños F, et al. (2017. a) Shared epizoic taxa and differences in diatom community structure between green turtles (Chelonia mydas) from distant habitats. Microbial Ecol 74: 969–978. 10.1007/s00248-017-0987-x [DOI] [PubMed] [Google Scholar]
- 17.Majewska R, De Stefano M, Ector L, Bolanos F, Frankovich TA, Sullivan MJ, et al. (2017b) Two new epizoic Achnanthes species (Bacillariophyta) living on marine turtles from Costa Rica. Bot Mar 60: 303–318. 10.1515/bot-2016-0114 [DOI] [Google Scholar]
- 18.Riaux-Gobin C, Witkowski A, Kociolek JP, Ector L, Chevallier D, Compere P (2017) New epizoic diatom (Bacillariophyta) species from sea turtles in the Eastern Caribbean and South Pacific. Diat Res 32: 109–125. 10.1080/0269249X.2017.1299042 [DOI] [Google Scholar]
- 19.Majewska R, Bosak S, Frankovich TA, Ashworth MP, Sullivan MJ, Robinson NJ, et al. (2019a) Six new epibiotic Proschkinia (Bacillariophyta) species and new insights into the genus phylogeny. Eur J Phycol 54: 609–631. 10.1080/09670262.2019.1628307 [DOI] [Google Scholar]
- 20.Majewska R, Ashworth MP, Lazo-Wasem E, Robinson NJ, Rojas L, Van de Vijver B, et al. (2018) Craspedostauros alatus sp. nov., a new diatom (Bacillariophyta) species found on museum sea turtle specimens. Diat Res 33:229–40. 10.1080/0269249X.2018.1491426 [DOI] [Google Scholar]
- 21.Majewska R, Robert K, Van de Vijver B, Nel R (2019b) A new species of Lucanicum (Cyclophorales, Bacillariophyta) associated with loggerhead sea turtles from South Africa. Bot Letters. 10.1080/23818107.2019.1691648 [DOI] [Google Scholar]
- 22.Majewska R, De Stefano M, Van de Vijver B (2017c) Labellicula lecohuiana, a new epizoic diatom species living on green turtles in Costa Rica. Nova Hedwigia 146: 23–31. 10.1127/1438-9134/2017/023 [DOI] [Google Scholar]
- 23.Rivera SF, Vasselon V, Ballorain K, Carpentier A, Wetzel CE, Ector L, et al. (2018) DNA metabarcoding and microscopic analyses of sea turtles biofilms: complementary to understand turtle behavior. PLoS ONE 13: e0195770 10.1371/journal.pone.0195770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wallace BP, DiMatteo AD, Hurley BJ, Finkbeiner EM, Bolten AB, Chaloupka MY, et al. (2010) Regional management units for marine turtles: a novel framework for prioritizing conservation and research across multiple scales. PLoS ONE 5: e15465 10.1371/journal.pone.0015465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson NJ, Paladino FV (2013) Sea Turtles. Ref Mod Earth Syst Environ Sci 1–13. [DOI] [Google Scholar]
- 26.Haywood JC, Casale P, Freggi D, Fuller WJ, Godley BJ, Lazar B, et al. (2020) Foraging ecology of Mediterranean juvenile loggerhead turtles: insights from C and N stable isotope ratios. Mar Biol 167: 28 10.1007/s00227-020-3647-5 [DOI] [Google Scholar]
- 27.Rees AF, Margaritoulis D, Newman R, Riggall TE, Tsaros P, Zbinden JA, et al. (2013) Ecology of loggerhead marine turtles Caretta in a neritic foraging habitat: movements, sex ratios and growth rates. Mar Biol 160: 519–529. 10.1007/s00227-012-2107-2 [DOI] [Google Scholar]
- 28.Ehrhart LM, Ogren LH (1999) Studies in foraging habitats: capturing and handling turtles In Research and management techniques for the conservation of sea turtles In: Eckert KL, Bjorndal KA, Abreu-Grobois FA, Donnelly M 4, 61–64 Washington, DC: IUCN/SSC Marine Turtle Specialist Group [Google Scholar]
- 29.Hasle GR, Syvertsen EE, 1997. Marine diatoms. In Tomas CR: Identifying marine phytoplankton. Academic Press, San Diego. [Google Scholar]
- 30.van der Werf A (1955) A new method of concentrating and cleaning diatoms and other organisms. Proc Int Assoc Theor Appl Limnol 7: 276–277 [Google Scholar]
- 31.Foged N (1975) Some Littoral Diatoms from the Coast of Tanzania. Bibl Phycol 16: 1–127 [Google Scholar]
- 32.Witkowski A, Lange-Bertalot H, Metzeltin D (2000) Diatom Flore of Marine Coasts I. Icon Diatomol 7: 1–925 [Google Scholar]
- 33.Hein MK, Winsborough BM, Sullivan MJ (2008) Bacillariophyta (diatoms) of the Bahamas. Icon Diatom 19: 1–303 [Google Scholar]
- 34.Álvarez-Blanco I, Blanco S (2014) Benthic diatoms from Mediterranean coasts. Bibl Diatomol 60: 1–409 [Google Scholar]
- 35.Sørenson T (1948) A method of establishing groups of equal amplitudes in plant sociology based on similarity of species content and its application to analyses of the vegetation on Danish commons. Kong Danske Vid Selsk, Biol Skrif 5: 1–34 [Google Scholar]
- 36.Kovach Computing Services (1993) Multivariate statistical package version 21, users’ manual Pentraeth, Kovach Computing Services
- 37.Clarke KR, Warwick RM (2001) Change in Marine Communities: An Approach to Statistical Analysis and Interpretation. 2nd Edition, PRIMER-E, Ltd, Plymouth Marine Laboratory, Plymouth [Google Scholar]
- 38.Anderson MJ (2001) A new method for non‐parametric multivariate analysis of variance. Austral Ecol 26: 32–4627 [Google Scholar]
- 39.Clarke KR, Gorley RN, Somerfield PJ, Warwick RM.(2014) Change in marine communities: an approach to statistical analysis and interpretation. Primer-E Ltd Plymouth Marine Laboratory, Plymouth [Google Scholar]
- 40.Trobajo R, Cox E, Quintana X (2004) The effects of some environmental variables on the morphology of Nitzschia frustulum (Bacillariophyta), in relation its use as a bioindicator. Nova Hedwigia 79: 433–445. 10.1127/0029-5035/2004/0079-0433 [DOI] [Google Scholar]
- 41.Trobajo R, Rovira L, Ector L, Wetzel CE, Kelly M, Mann D (2013) Morphology and identity of some ecologically important small Nitzschia species Diatom Res 28: 37–59. 10.1080/0269249X.2012.734531 [DOI] [Google Scholar]
- 42.Pinou T, Domenech F, Lazo-Wasem EA, Majewska R, Pfaller JB, Zardus JD, et al. (2019) Standardizing sea turtle epibiont sampling: outcomes of the epibiont workshop at the 37th International Sea Turtle Symposium. Mar Turtle Newsletter 157:22–32 [Google Scholar]
- 43.Robert K, Bosak S, Van de Vijver B (2019) Catenula exigua sp nov, a new marine diatom (Bacillariophyta) species from the Adriatic Sea. Phytotaxa 414 113–118 [Google Scholar]
- 44.Van de Vijver B & Bosak S (2019) Planothidium kaetherobertianum, a new marine diatom (Bacillariophyta) species from the Adriatic Sea. Phytotaxa 425: 105–112 [Google Scholar]
- 45.Belando MD, Jimenez JF, Marín A, Aboal M (2018) Morphology and molecular phylogeny of Hyalosynedra lanceolata sp nov and an extended description of Hyalosynedra (Bacillariophyta). Eur J Phycol 53: 208–218 [Google Scholar]
- 46.Car A, Witkowski A, Dobosz S, Burfeind DD, Meinesz A, Jasprica N, et al. (2012): Description of a new marine diatom, Cocconeis caulerpacola sp nov (Bacillariophyceae), epiphytic on invasive Caulerpa species. Eur J Phycol 47: 433–448 [Google Scholar]
- 47.Haiying J, Chao-Qun H, Hai-Peng J, Lv-Ping Z, Peng-Fei P, Peng L, et al. (2015) Morphology and phylogeny of Halamphora yongxingensis sp nov (Bacillariophyta), a new marine benthic diatom isolated from Yongxing Island, South China Sea. Phytotaxa 195: 53–64 [Google Scholar]
- 48.Park J, Koh C-H (2012) Taxonomic studies on Korean marine benthic diatoms—LM and SEM observations of the diatom genus Amphora (Bacillariophyceae) from Korean tidal flats with the first recordings of A arenicola, A beaufortiana and A maletractata var constricta. Ocean Sci J 47: 101–112 [Google Scholar]
- 49.Casale P, Broderick AC, Camiñas JA, Cardona L, Carreras C, Demetropoulos A, et al. (2018) Mediterranean sea turtles: current knowledge and priorities for conservation and research. Endanger Species Res 36: 229–267. 10.3354/esr00901 [DOI] [Google Scholar]
- 50.Schofield G, Katselidis KA, Dimopoulos P, Pantis JD, Hays GC (2006) Behavior analysis of the loggerhead sea turtle Caretta from direct in-water observation. Endanger Species Res 2: 71–79 [Google Scholar]
- 51.Robinson NJ, Lazo-Wasem EA, Paladino FV, Zardus JD, Pinou T (2017) Assortative epibiosis of leatherback, olive ridley and green sea turtles in the Eastern Tropical Pacific. J Mar Biol Assoc UK 97: 1233–1240. 10.1017/S0025315416000734 [DOI] [Google Scholar]
- 52.Fuller WJ, Broderick AC, Enever R, Thorne P, Godley BJ (2010) Motile homes: A comparison of the spatial distribution of epibiont communities on Mediterranean sea turtles. J Nat Hist 44: 25–28. 10.1080/00222931003624820 [DOI] [Google Scholar]
- 53.Preen AR (1996) Infaunal mining: a novel foraging method of loggerhead turtles. J Herpetol 3: 94–96. 10.2307/1564718 [DOI] [Google Scholar]
- 54.Bjorndal KA (1980) Nutrition and grazing behavior of the green turtle Chelonia mydas. Marine Biology 56: 147–154 10.1007/BF00397131 [DOI] [Google Scholar]
- 55.Battelli C, Rindi F (2016) First report of the epizoic red alga Polysiphonia carettia (Hollenberg, 1971) on the loggerhead turtle Caretta in the Adriatic Sea. Acta Adriat 57: 173–178. [Google Scholar]
- 56.Rees AF, Carreras C, Broderick AC, Margaritoulis D, Stringell TB, Godley BJ (2017) Linking loggerhead locations: using multiple methods to determine the origin of sea turtles in feeding grounds. Marine Biol 164: 30 10.1007/s00227-016-3055-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frankovich TA, Wachnicka A (2015) Epiphytic diatoms along phosphorus and salinity gradients in Florida Bay (Florida, USA), an illustrated guide and annotated checklist. In: Entry JA, Gottlieb AD, Jayachandran K, Ogram A(eds) Microbiology of the Everglades Ecosystem, CRC Press, Boca Raton, pp. 239–286 [Google Scholar]