ABSTRACT
Peptide therapeutics, unlike small-molecule drugs, display crucial advantages of target specificity and the ability to block large interacting interfaces, such as those of transcription factors. The transcription co-factor of the Hippo pathway, YAP/Yorkie (Yki), has been implicated in many cancers, and is dependent on its interaction with the DNA-binding TEAD/Sd proteins via a large Ω-loop. In addition, the mammalian vestigial-like (VGLL) proteins, specifically their TONDU domain, competitively inhibit YAP-TEAD interaction, resulting in arrest of tumor growth. Here, we show that overexpression of the TONDU peptide or its oral uptake leads to suppression of Yki-driven intestinal stem cell tumors in the adult Drosophila midgut. In addition, comparative proteomic analyses of peptide-treated and untreated tumors, together with chromatin immunoprecipitation analysis, reveal that integrin pathway members are part of the Yki-oncogenic network. Collectively, our findings establish Drosophila as a reliable in vivo platform to screen for cancer oral therapeutic peptides and reveal a tumor suppressive role for integrins in Yki-driven tumors.
This article has an associated First Person interview with the first author of the paper.
KEY WORDS: Drosophila, Peptide therapeutic, Yki, Intestinal stem cells, Integrin signaling
Summary: We demonstrate that, when fed to flies, TONDU peptide suppresses Yki-driven intestinal stem cell (ISC) tumors, and identify integrins as essential components of ISC tumorigenesis.
INTRODUCTION
Drosophila has emerged as an effective tumor model for the screening of small-molecule therapeutics (Dar et al., 2012; Khoo et al., 2013; Markstein et al., 2014; Bangi et al., 2016). Whereas cancer-promoting misregulated kinases are amenable to inhibition by small molecules, others, such as transcription factors (TFs) and co-factors, are largely considered undruggable (Bhagwat and Vakoc, 2015; Lambert et al., 2018). In this regard, peptides are particularly attractive as therapeutic molecules (Lau and Dunn, 2018; Drucker, 2019) because of their high selectivity, improved tolerance and ability to target large interacting interfaces (Furet et al., 2019). While most peptide therapeutics require parenteral injection, their oral delivery is highly desirable; indeed, currently a few orally derived therapeutic peptides are being tested in clinical trials (Drucker, 2019).
The proto-oncogene Yes-associated protein [YAP; Yorkie (Yki) in Drosophila] – the transcription co-factor of the Hippo pathway – interacts with its DNA-binding partner, transcriptional enhanced associate domain 1-4 [TEAD1-4; Scalloped (Sd) in Drosophila] (Wu et al., 2008), and is implicated in cancers (Zanconato et al., 2016). YAP binds to TEAD via an unusually large interface, the Ω-loop (Pobbati et al., 2012; Furet et al., 2019), which lacks a defined binding pocket, making it an unlikely target of inhibition by small molecules. TEAD proteins also bind to other transcriptional co-factors, such as the vestigial-like (VGLL1-4) proteins that display a highly conserved 26 amino acid TONDU domain (Pobbati et al., 2012; Koontz et al., 2013). VGLL4 competitively inhibits binding of YAP and TEAD, thereby acting as a tumor suppressor (Zhang et al., 2014). Interestingly, a synthetic peptide analog of the TONDU domain of VGLL4 was found to inhibit gastric cancer growth (Jiao et al., 2014) in a mouse xenograft model.
Similar to the mammalian VGLL-TEAD-YAP partnership, Drosophila TONDU-containing proteins, such as Vestigial (Vg) and Tondu-domain-containing Growth Inhibitor (Tgi) interact with Sd and Yki (Guo et al., 2013; Koontz et al., 2013). Sd, when not bound to Yki, interacts with the ubiquitously expressed Tgi via the Tgi TONDU domain. The conserved interaction between Vg/Tgi with Sd-Yki in Drosophila therefore makes the fly a relevant platform to screen for large-molecule inhibitors of YAP-TEAD interaction. Here, we used the adult Drosophila gut – which displays Sd-dependent Yki activity for intestinal stem cell (ISC) homeostasis (Jin et al., 2013) – to test whether a TONDU peptide can suppress ISC tumors triggered by gain of an activated form of Yki (Kwon et al., 2015; Song et al., 2019). We show that ISC tumors in the adult midgut induced by gain of activated Yki are suppressed by feeding TONDU peptide-supplemented food. Further, comparative proteome analysis and genetic tests reveal that integrin pathway members are part of the Yki-oncogenic network. Altogether, our results establish our Drosophila ISC tumor model as a reliable platform for screening therapeutic peptides with the added advantage of rapid resolution of the mechanistic underpinning of tumor suppression.
RESULTS
Genetic suppression of Yki-driven ISC tumor growth by the TONDU peptide
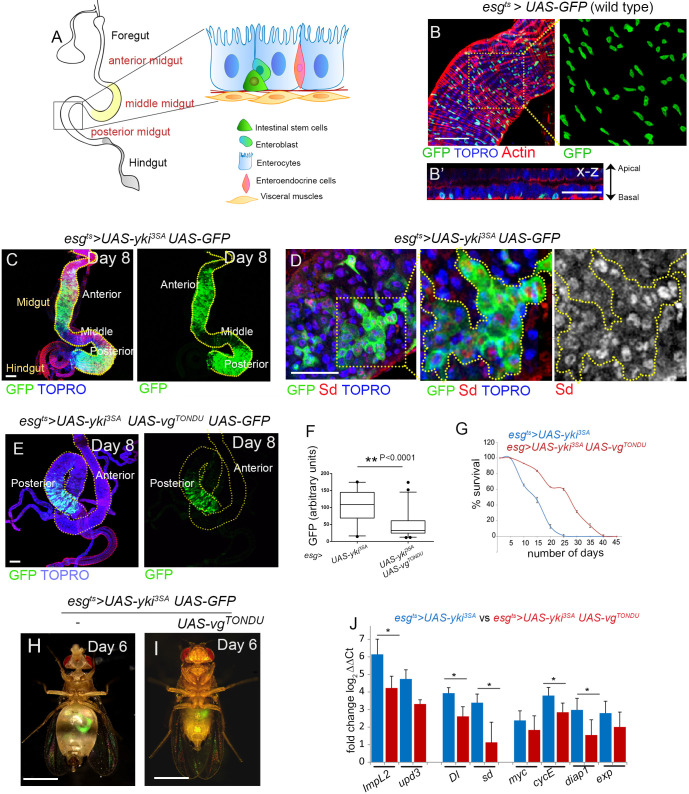
The Drosophila gut closely resembles the mammalian gut and is divided into the foregut, midgut and hindgut (Guo et al., 2016). The midgut makes up most of the gut and contains three cell types: differentiated enterocytes, entero-endocrine cells and ISCs (Fig. 1A,B). Expression of a phosphorylation-defective and therefore constitutively active form of Yki in the ISCs (esg-Gal4 Gal80ts>UAS-yki3SA, referred to as esgts>yki3SA) results in gut stem cell tumors (Kwon et al., 2015) (Fig. 1C; Fig. S1A-D). Yki gut tumor-bearing flies display a systemic wasting syndrome (Kwon et al., 2015) (Fig. 1H; Fig. S2A) and display elevated levels of the insulin antagonist ImpL2 (Fig. 1J) (Kwon et al., 2015), in addition to canonical Yki targets (Fig. 1J) that include Sd (Fig. 1D,J), the DNA-binding partner of Yki (Wu et al., 2008).
Fig. 1.
Expression of the TONDU peptide inhibits Yki-driven ISC tumors. (A) Schematic representation depicting the different cell types in the adult Drosophila gut. (B,B′) esgts>UAS-GFP labels ISCs in the Drosophila midgut. (B) ISCs (marked by GFP) are interspersed throughout the gut. Overlying muscles are marked with F-Actin (red). (B′) X-Z section displaying basally located ISCs (GFP). (C) esgts>yki3SAUAS-GFP gut shows an increase in ISC numbers. (D) esgts>yki3SAUAS-GFP tumors show increase in Sd level. (E) Decrease in ISCs (marked by GFP) in the anterior and posterior midgut of esgts>yki3SA UAS-vgTONDU flies that co-express the TONDU peptide. (F) Quantification of GFP in TONDU-expressing and non-expressing esgts>yki3SAguts. Box plots indicate the median (horizontal lines), 25th and 75th percentiles (box), and 2.5 to 97.5 percentile range (whiskers). Outliers are displayed as filled circles. Significance displayed as P-value, for unpaired Student's t-test. (G) Increase in survival of esgts>yki3SAUAS-vgTONDU flies compared to esgts>yki3SA (n=50 each genotype). (H) Abdominal bloating in esgts>yki3SAUAS-GFP flies as seen on day 6 after tumor induction (n=19/25 are bloated). (I) esgts>yki3SA UAS-vgTONDU UAS-GFP flies display delay in bloating (n=14/25 are not bloated) as seen on day 6. (J) qPCR displaying the decrease in mRNA levels of candidate genes in TONDU-expressing flies. Data presented as mean±s.e.; *P≤0.025 for Student's t-test. Scale bars: 100 µm (B,C,E), 50 µm (B′,D), 1 mm (H,I).
Since the TONDU-containing proteins Vg (Khan et al., 2013) and Tgi (Guo et al., 2013; Koontz et al., 2013) can inhibit Yki-regulated growth by competing for Sd, we tested whether co-expression of TONDU peptide alone (CVVFTNYSGDTASQVDEHFSRALNY) in ISCs with gain of Yki (esgts>yki3SA UAS-vgTONDU) would inhibit Yki-driven ISC tumor growth. Indeed, a striking inhibition in ISC tumors (Fig. 1E,F), with an accompanying loss of proliferation (Fig. S2C-E), was seen under this condition. In addition, these flies showed improved life span (Fig. 1G) and a delay in the onset of tumor-associated wasting phenotypes (Fig. 1H,I; Fig. S2B,F,G), with a concomitant decrease in expression of Impl2 (Fig. 1J), a hallmark of esgts>yki3SA tumors (Kwon et al., 2015). By contrast, overexpression of the TONDU peptide alone in ISCs (esgts>vgTONDU) did not affect the number of ISCs (Fig. S2H,I). Altogether, these results reveal that Yki-driven ISC tumors are suppressed upon co-expression of the TONDU peptide, with an accompanying delay in the onset of tumor-associated syndromes.
Oral uptake of synthetic TONDU peptide inhibits Yki-driven ISC tumors
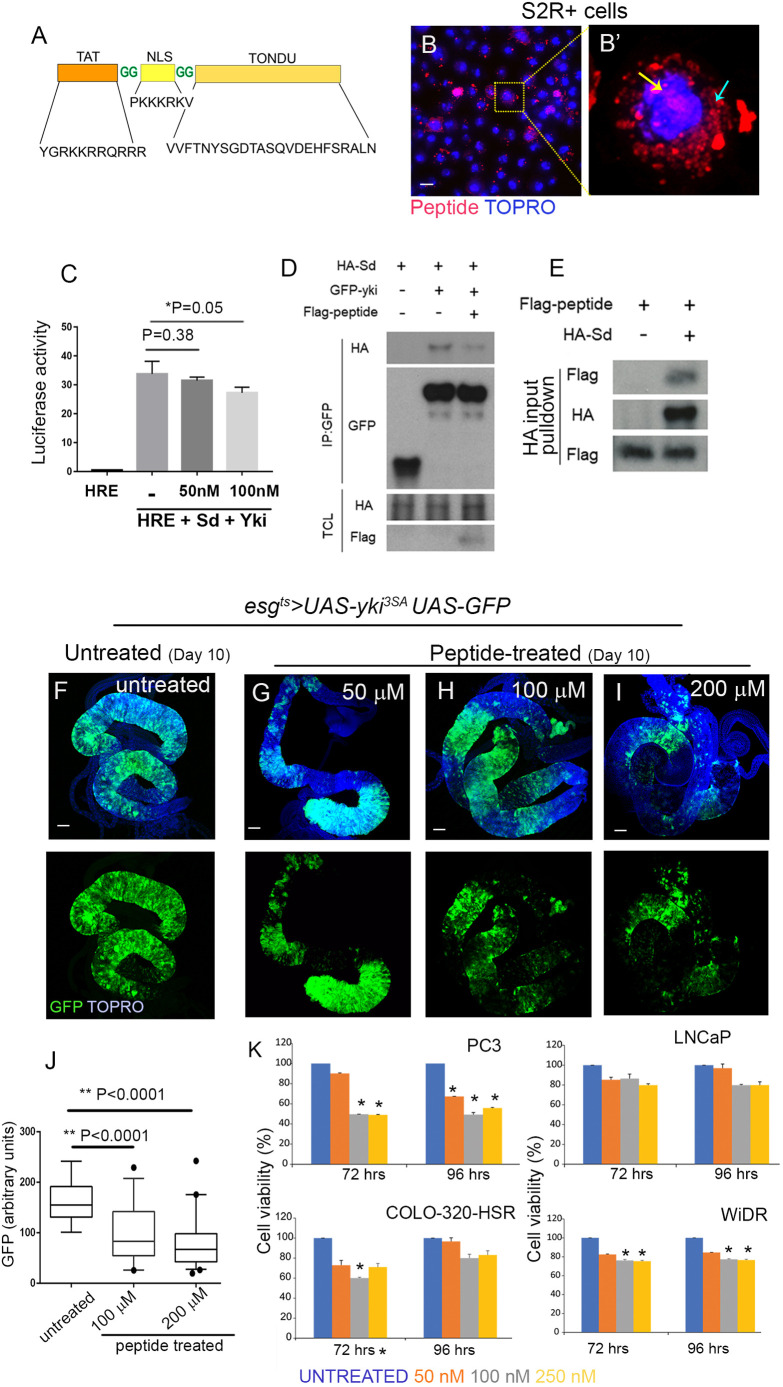
Next, we asked whether feeding a synthetic TONDU peptide could inhibit Yki-driven ISC tumors comparable to its overexpression in ISCs (Fig. 1). We designed a synthetic peptide (Fig. 2A) derived from the TONDU domain of Vg that retained the conserved TEAD/Sd-interacting interfaces I and II containing the critical VXXHF motif (Pobbati et al., 2012). Further, since we aimed to administer the peptide orally to tumor-bearing flies, unlike in a previous study that involved tail vein injection of VGLL4-derived peptide (Jiao et al., 2014), we tagged the TONDU peptide with an HIV-TAT motif (RKKRRQRRR) and a nuclear localizing signal (NLS) (PKKKRKV) to facilitate cellular uptake (Wadia and Dowdy, 2005) and nuclear localization, respectively. Prior to oral administration of the peptide to adult flies, we first tested cellular uptake of a fluorescent-labeled TONDU peptide in S2R+ cultured cells, and observed its cytoplasmic and nuclear localization (Fig. 2B).
Fig. 2.
Synthetic TONDU peptide inhibits Yki-driven ISC tumors. (A) Representation of the synthetic TONDU peptide. (B,B′) Nuclear localization of fluorescent-tagged (red) TONDU peptide in S2R+ cells. (B′) Magnified view of the boxed area in B. TONDU peptide (red) in the nucleus (yellow arrow) and cytoplasm (cyan arrow). (C) HRE-luciferase reporter activity in S2R+ cells when treated with TONDU peptide. (D) Immunoblots showing competitive binding of TONDU peptide to Yki-Sd complex. IP, immunoprecipitation; TCL, total cell lysate. (E) Binding of TONDU peptide to Sd. (F-I) Guts from esgts>yki3SA flies fed TONDU peptide: (F) unfed (control), (G) 50 μM (n=10), (H) 100 μM (n=12) and (I) 200 μM (n=10). (J) Quantification of GFP in TONDU peptide-fed and -unfed esgts>yki3SA flies. Box plots indicate the median (horizontal lines), 25th and 75th percentiles (box), and 2.5 to 97.5 percentile range (whiskers). Outliers are displayed as filled circles. P-values for Student's t-test are displayed. (K) Viability of cancer cells on treatment with TONDU peptide, as estimated using the resazurin cell viability assay. Data presented as mean±s.e.; *P≤0.001 for Student's t-test. Scale bars: 10 µm (B), 100 µm (F-I).
Further, to test whether the TONDU peptide can inhibit Yki-Sd complex formation, we used the Hippo response element (HRE)-luciferase reporter as a readout for Yki-Sd transcriptional activity (Wu et al., 2008). Specifically, we co-transfected S2R+ cells with the HRE-luciferase reporter along with Yki- and Sd-expressing vectors, treated the cells with 100 nM of TONDU peptide, and observed a moderate but consistent decrease in luciferase activity (Fig. 2C). Next, to confirm binding of the synthetic TONDU peptide to Sd and subsequent inhibition of Yki-Sd interaction, we carried out co-immunoprecipitation studies using S2R+ cells transfected with HA-Sd and GFP-Yki in the presence of the FLAG-tagged TONDU peptide. Indeed, we found that the TONDU peptide competitively inhibits binding of Sd to Yki (Fig. 2D). Finally, when purified HA-Sd from S2R+ cells was incubated with FLAG-tagged TONDU peptide, we observed binding with Sd, as revealed by immunoblots using anti-FLAG antibody (Fig. 2E). These observations are in agreement with previous studies (Guo et al., 2013; Koontz et al., 2013) that displayed binding of TONDU-containing protein Tgi to Sd via the TONDU domain. Together, these results indicate that TONDU peptide disrupts the Sd-Yki interaction by binding to Sd.
Next, we tested whether oral uptake of TONDU peptide inhibits esgts>yki3SA ISC tumors. To first estimate the maximum tolerated dose, we examined the viability of esgts>GFP flies when continuously fed different concentrations (25, 50, 100, 200 and 400 µM) of the TONDU peptide in standard fly food for 6-10 days at 29°C and scored their survival soon after. We observed that at 400 µM concentration of the TONDU peptide, only 55% (n=50) of the peptide-fed esgts>GFP flies survived on day 6, whereas approximately 97%, 98%, 93% and 91% (n=50 in all cases) of flies survived at 25, 50, 100 and 200 µM of the peptide, respectively. We therefore fed esgts>yki3SA flies, 24 h post-eclosion, 50, 100 or 200 µM TONDU peptide-supplemented food continuously for 10 days. Remarkably, we noted a progressive reduction in tumor load (Fig. 2F-I) with increasing concentration of TONDU peptide, as seen from a decrease in the numbers of GFP-marked ISCs (Fig. 2J). By contrast, tumor load was only moderately reduced when esgts>yki3SA flies were fed food supplemented with sequence-scrambled TONDU peptide (Fig. S3A) at comparable concentrations (Fig. S3B-E); the residual inhibition of tumor growth observed with scrambled peptide could presumably be due to a partial retention of the secondary structure (Pobbati et al., 2012) of the TONDU peptide in the scrambled version as revealed by its predicted structure (Fig. S3F). We observed that, compared to poor survival (65.6% on day 10, Fig. 1G) of untreated esgts>yki3SA flies, TONDU peptide-fed esgts>yki3SA flies displayed a consistent increase in survival (68.2%, 74.3% and 79.7%) accompanying lowering of the tumor burden (Fig. 2G-I). We note, however, that peptide-fed flies continued to display some mortality, which we believe could be attributed to residual tumor load and/or off-target toxicity by the TONDU peptide in vivo due to perturbations in levels of a number of proteins in peptide-treated fly gut tumors (discussed below). Further, to confirm cellular uptake of TONDU peptide by the gut epithelia, we fed FLAG-tagged TONDU peptide (at a final concentration of 200 µM) to esgts>yki3SA flies, and detected its cellular uptake in gut lysates by immunoblotting using an anti-FLAG antibody (Fig. S3G). In parallel, we also noted that feeding TONDU (at 200 µM) did not affect the numbers of ISCs in control guts (esgts>GFP) (Fig. S3H).
In addition, we tested the tumor-inhibitory property of Drosophila TONDU peptide on human cancer cells. We observed that cell lines derived from human tumors with elevated YAP1 levels (Fig. S3I), such as PC3 (prostate cancer), COLO-320 and WiDR (colorectal cancer), displayed growth arrest to varying extents upon uptake of TONDU peptide (Fig. 2K). On the other hand, the prostate cancer line LNCaP, which displayed negligible levels of YAP1 (Fig. S3I), was not significantly inhibited by the TONDU peptide (Fig. 2K), even at higher concentrations, thereby revealing specificity of the TONDU peptide to inhibit YAP-mediated tumor growth and presumably low off-target toxicity. Altogether, these results suggest that TONDU is therapeutically relevant in YAP-driven tumors and can effectively inhibit cancers of different tissues of origin.
Yki-driven tumor proteome reveals enrichment in integrin pathway components
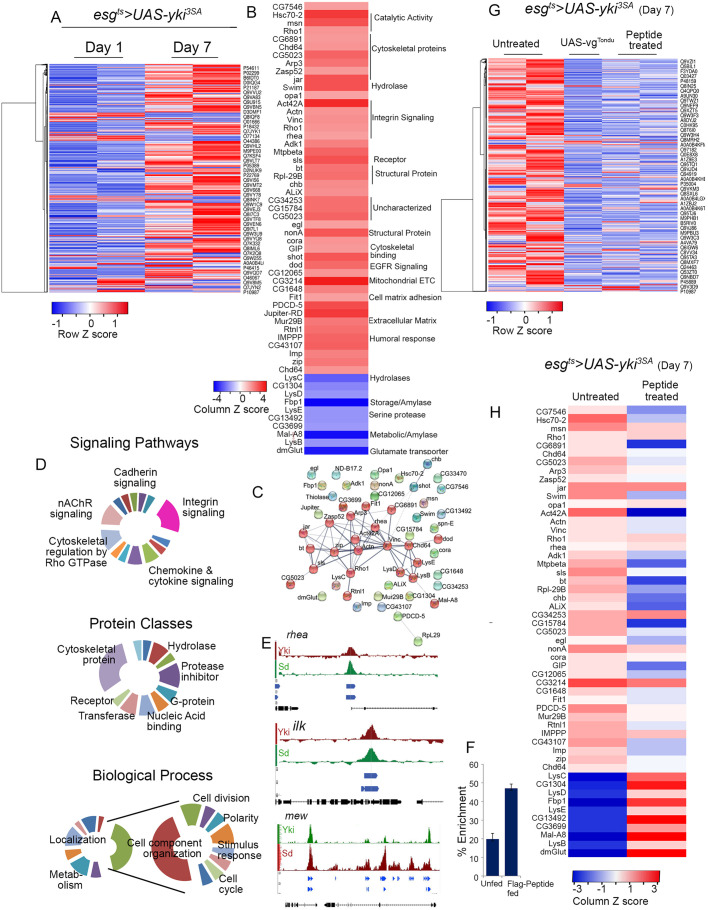
We reasoned that significantly perturbed proteins in esgts>yki3SA tumors, which are restored to normal levels following TONDU feeding, are likely to represent Yki-Sd targets that are crucial to ISC tumorigenesis, and, therefore, could be therapeutically relevant. Thus, we carried out a proteome analysis using unlabeled liquid chromatography–tandem mass spectrometry (LC-MS/MS) of esgts>yki3SA tumors on day 1 and day 7 of tumor induction, with or without TONDU peptide-supplemented food. Altogether, we identified 1219 proteins (including isoforms), corresponding to 2771 unique UniProt IDs at a false discovery rate (FDR) cutoff of q<0.05 (Fig. 3A; Table S1) present in both day 1 and day 7 esgts>yki3SA tumors. We next compared the proteomes of day 7 to day 1 esgts>yki3SA tumors, and prioritized proteins that displayed at least a log2 ±2 fold change (at a P-value <0.05) for further analysis. Fold change was derived from the abundance measure of peptides (for a given protein) in day 7 versus day 1 esgts>yki3SA tumors (see Materials and Methods). We identified 127 proteins (corresponding to 144 unique UniProt IDs, including isoforms) that were differentially expressed in day 7 esgts>yki3SA tumors, and these matched to 55 unique genes (Fig. 3B; Table S2). Forty-five of these genes showed a greater than 2-fold (log2) increase and ten displayed a greater than 2-fold (log2) decrease in protein levels in day 7 compared to day 1 tumors (Table S2).
Fig. 3.
Comparative proteomic analysis of Yki-driven ISC tumors and tumors inhibited by the TONDU peptide. (A) Heat map displaying changes in protein levels in day 7 and day 1 esgts>yki3SA tumors. (B) 55 differentially (>±2 log2fold, P=0.05) expressed proteins in day 7 esgts>yki3SA tumors. (C) Protein-protein interaction (PPI) network of enriched proteins (>log2 2 fold) in esgts>yki3SA tumors generated with STRING (Szklarczyk et al., 2019), representing 55 nodes and 63 edges (PPI enrichment P<0.0001). (D) Different GO classes identified by PANTHER (Mi et al., 2007) in differentially expressed proteins between esgts>yki3SA day 7 versus day 1 tumor proteome. (E) Sd and Yki binding sites in the regulatory regions of select integrin pathway members as determined in Nagaraj et al. (2012). (F) Percentage enrichment for Sd binding upstream of mew (αPS1) inferred by ChIP with anti-FLAG antibody. (G) Heat map displaying the effect of TONDU peptide on esgts>yki3SA tumor proteome. (H) Heat map displaying change in levels of protein (>±2 fold in day 7 tumors) upon TONDU peptide treatment.
To further examine whether the proteins enriched in the ISC tumors are biologically relevant, we performed a protein-protein interaction (PPI) network analysis using STRING (Szklarczyk et al., 2019). We noted significant (P<0.001) interaction among some of the enriched proteins (Fig. 3C), suggesting that these were not random. Furthermore, we noted that the enriched gene set included known members of the Hippo protein-protein interaction network (Kwon et al., 2013), including junction proteins Coracle, Jar and Misshapen (Table S3). We also observed an increase in protein levels of the secreted Wg transporter Swim (Mulligan et al., 2012). Furthermore, comparison of the day 7 proteome of esgts>yki3SA tumors with a recently published transcriptome of esgts>yki3SA tumors (Song et al., 2019) revealed a close correlation between changes in proteins and their respective transcript levels (r=0.548) (Fig. S4A).
Signaling pathways perturbed in tumors are often causally linked to tumor progression (Khan et al., 2013; Bajpai and Sinha, 2020). We thus undertook a gene ontology (GO) classification of the proteins found enriched in ISC tumors to identify critical signaling pathways. GO classification using PANTHER (Mi et al., 2007) revealed perturbations in several signaling pathways and protein classes (Fig. 3D; Table S4). In particular, we observed an increase in protein levels of key members of the integrin signaling pathway, including Talin (2.39-fold; all fold changes have been mentioned at log2 conversion) and the Talin-interacting adaptor proteins Vinculin (2.4-fold) and Paxillin (6.05-fold). Other members, such as αPS3 and Integrin-linked kinase (Ilk), also displayed ∼2-fold change, albeit at P>0.05 (Table S5). Consistent with these findings, we noted transcriptional upregulation of the genes encoding the integrin members in the transcriptome of comparatively aged esgts>yki3SA tumors (Table S5; also see Song et al., 2019), including mew (αPS1), scb (αPS3), mys (βPS), and integrin-binding ligands such as LanA and LanB (Table S5) (Song et al., 2019), which were otherwise not detected in the tumor proteome (Fig. 3B). It is likely that some integrin pathway components went undetected in proteomes owing to the limitation of unlabeled LC-MS/MS, such as failure to detect some proteins due to poor yield of their trypsinized products (Bantscheff et al., 2007). Significantly, we also observed an increase in levels of polarity proteins such as tight junction protein, Ferritin, Fit1 and the apical protein Shot (Fig. 3B), which are known to be regulated by the integrin pathway in the gut epithelium (Chen et al., 2018).
These observations suggest that genes encoding proteins enriched in esgts>yki3SA tumors could be Yki-Sd transcriptional targets. To further examine this possibility, we searched for putative Yki-Sd binding sites from studies on genome-wide binding of Yki (Nagaraj et al., 2012; Oh et al., 2013) and Sd (Nagaraj et al., 2012). We noted that ∼51% (23 of 45) of the genes for which protein levels were increased in esgts>yki3SA tumors displayed putative Sd and Yki binding sites in their upstream regulatory regions (Table S6). These included several key members of the integrin pathway, including mew, vinculin, paxillin, and integrin-linked kinase and rhea (Fig. 3E; Table S6). The integrin pathway has been reported to be essential for maintenance of both ISCs (Lin et al., 2013) and enterocytes (Chen et al., 2018); further, since αPS1 (encoded by mew) is particularly enriched in ISCs and critical for ISC maintenance (Lin et al., 2013), we sought to examine the role of the integrin αPS1 and the critical integrin-interacting protein Talin in esgts>yki3SA tumors, despite αPS1 not being identified in the tumor proteome (Fig. 3B). Thus, we examined the binding of Sd to the upstream regulatory region of mew, and, given that TONDU peptide binds to Sd (Fig. 2D), we further reasoned that chromatin immunoprecipitation (ChIP) – using anti-FLAG antibody – on gut lysate of esgts>yki3SA flies fed FLAG-tagged TONDU peptide could reveal binding of Sd. Indeed, we observed a significant enrichment of Sd binding in the upstream regulatory region of mew, in vivo in the guts of flies fed FLAG-tagged TONDU peptide (47.04±2.3%), compared to unfed control (19.8±3.0%) (Fig. 3F), suggesting that TONDU peptide binds to Sd and could therefore interfere with transcriptional regulation of mew by the Yki-Sd complex in esgts>yki3SA tumors in vivo. By extension, it is likely that the TONDU peptide can also affect the expression of other transcriptional targets of Sd-Yki (Tables S2 and S6), including other members of the integrin pathway.
Next, to assess the impact of TONDU peptide on the tumor proteome, we compared the proteome of esgts>yki3SA tumors with the proteome of esgts>yki3SAUAS-vgTONDU and tumors from flies fed 200 µM of the peptide. Our earlier observation of comparable phenotypic suppression of esgts>yki3SA tumors by either overexpression of TONDU peptide or its oral uptake, was supported by a strong correlation between their proteomes (Fig. S4B,C). Thus, we combined these two datasets for a robust representation of TONDU peptide-treated tumor proteome and compared it with that of untreated esgts>yki3SA tumors. We observed an overall decrease in levels of proteins in peptide-treated ISC tumors compared to those of esgts>yki3SA tumors from unfed controls (Fig. 3G). In addition, we noted that peptide-treated tumors displayed a significant decrease in protein levels (Fig. 3H), including the levels of critical members of the integrin pathway, such as Paxillin (−1.9-fold), Vinculin (−1.3-fold) and Talin (−1.2-fold) (Table S7).
Other notable perturbations included proteins involved in RNA processing, such as Pre-RNA processing factor 19 (Prp19) (−2.16-fold) (Guilgur et al., 2014) and Rumpelstiltskin (Rump) (−3.15-fold). Furthermore, decrease in Chromosome bows (Chb) (−2.16-fold), which is involved in mitotic spindle assembly (Reis et al., 2009), could presumably contribute to the lowering of cell proliferation of peptide-treated tumors. We also noted a decrease (−2.3-fold) in mitochondrial trifunctional protein β (Mtp-β), which catalyzes oxidation of long-chain fatty acids (Biswas et al., 2012), and could limit the energy source for peptide-treated tumors (Koundouros and Poulogiannis, 2020). Interestingly the tumor proteome revealed some novel and yet uncharacterized candidates that could be of functional significance. For instance, we noted a significant decrease in levels of proteins encoded by genes CG15784 (−2.65-fold) and CG7546 (−1.96-fold) in ISC tumors upon peptide treatment. Interestingly, these uncharacterized proteins interact with members of the Insulin/Akt (Vinayagam et al., 2016) and Hippo (Kwon et al., 2013) pathways, respectively, and could thus represent novel nodes connecting the Yki-oncogenic network with metabolic networks in the ISCs.
Taken together, our proteomic analyses reveal that TONDU peptide treatment perturbs a host of Yki-Sd targets that impinge upon cellular processes such as growth, proliferation and survival of tumor cells. It is likely that inhibition of esgts>yki3SA tumors by the TONDU peptide could be a cumulative effect of suppression of multiple Yki-Sd targets affecting more than one signaling pathway or cellular process. We chose, however, to further examine the role of integrin signaling pathway since it plays a critical role in ISC maintenance (Lin et al., 2013).
Genetic suppression of integrin signaling phenocopies TONDU-mediated suppression of Yki-driven ISC tumors
Integrins form an essential component of the Drosophila gut epithelia, including the basally located ISCs (Lin et al., 2013; Chen et al., 2018). Integrin, such as αPS1, is found specifically enriched in the ISCs (Lin et al., 2013) (also see Fig. 4A,B) and has been proposed to be important in the anchorage of the ISCs to the basement membrane and in their proliferation (Lin et al., 2013). Consistent with the enrichment of integrin pathway members in the esgts>yki3SA proteome, we observed an overall increase in levels of integrin αPS1 (Fig. 4C) and Talin (Fig. 4D) in esgts>yki3SA tumors. This observation, together with suppression of integrin pathway members such as Talin in TONDU peptide-fed flies suggests that integrin downregulation is likely to be causally linked to TONDU peptide-mediated inhibition of esgts>yki3SA tumors. To test this possibility, we downregulated rhea or mew in the ISCs of esgts>yki3SA tumors (Fig. 4E-G); indeed downregulation of rhea (esgts>yki3SAUAS-rhea-RNAi, Fig. 4G) or mew (esgts>yki3SAUAS-mew-RNAi, Fig. 4F) resulted in a marked reduction in ISC numbers (Fig. 4H), which was most obvious in the anterior midgut (Fig. 4F,G) when compared to similarly aged esgts>yki3SA tumors (Fig. 4E; Fig. S5A). Moreover, examination of early (day 3) esgts>yki3SAUAS-mew-RNAi guts revealed poor growth of ISC tumors. In particular, most of the ISC tumors were made up of small clusters of three to four cells (Fig. S5B,C), suggesting a strong decrease in tumor growth. These results are consistent with the observation that integrin signaling is required for ISC homeostasis (Lin et al., 2013). Interestingly, we note that gain of integrin alone, using a constitutively active form of the βPS integrin (Martin-Bermudo et al., 1999) in the ISCs (esgts>torsoD/βCyt) failed to trigger ISC proliferation (Fig. S5D). These observations suggest that although gain of integrin signaling alone per se does not transform ISCs, it is an essential partner for the progression of Yki-driven ISC tumors.
Fig. 4.
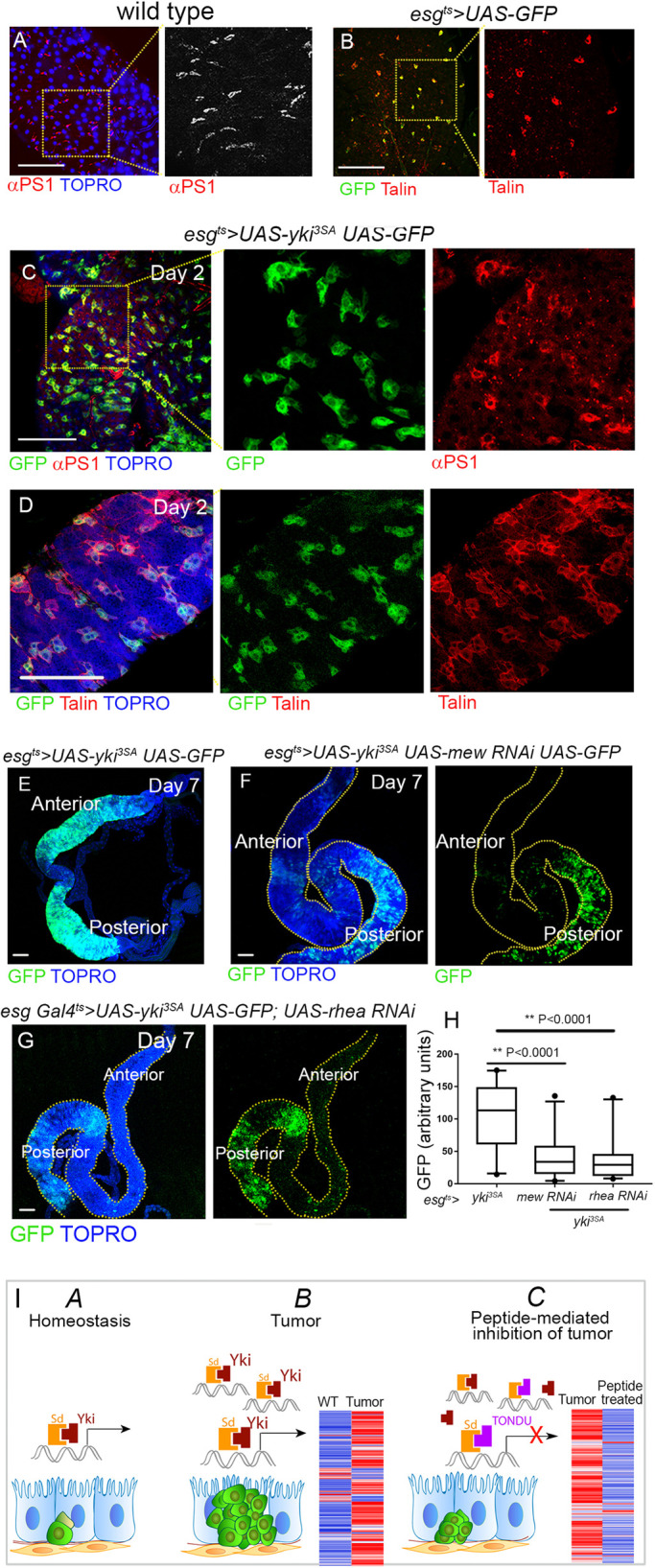
Loss of integrin signaling inhibits growth of Yki-driven ISC tumors. (A,B) αPS1 (A) and Talin (B) staining in esgts>UAS-GFP marked ISCs. (C,D) Overall increase in αPS1 (C) and Talin (D) in esgts>yki3SA tumors. (E-G) Inhibition of Yki-driven tumors upon simultaneous downregulation of αPS1 (esgts>yki3SA UAS-mew-RNAi, n=9; F) or Talin (esgts>yki3SA UAS-rhea-RNAi, n=9; G), when compared to similarly aged esgts>yki3SA tumors (E and Fig. S5A, respectively). (H) Quantification of GFP from E, F and G. Box plots indicate the median (horizontal lines), 25th and 75th percentiles (box), and 2.5 to 97.5 percentile range (whiskers). Outliers are displayed as filled circles. Significance displayed as P-values, for unpaired Student's t-test. (I) Schematic of Yki-Sd mediated transcription in wild-type (WT) guts (A), in Yki-driven tumor (B) and in Yki-driven tumor in the presence of the TONDU peptide (C). Scale bars: 100 µm.
DISCUSSION
TFs can be potent cancer drivers: suppression of TFs therefore constitutes a tumor inhibitory mechanism. The promise of targeting TFs for tumor therapy (Bhagwat and Vakoc, 2015; Lambert et al., 2018) is limited by the fact that small molecules often fail to target the large interacting surfaces associated with TFs (Lau and Dunn, 2018). Peptides, by contrast, have proven to be effective at interacting with large surfaces. Nevertheless, although peptides targeting extracellular receptors (Arosio et al., 2017) or intracellular inhibitors (Chang et al., 2013) have been explored, targeting of nuclear bound TFs with peptides remains poorly explored. The TEA/ATTS domain-containing TEAD proteins are a class of TFs that regulate YAP-induced proliferation and drive differentiation programs of VGLLs on the other (Gibault et al., 2018; Huh et al., 2019). In this study, using Yki-driven ISC tumors, we document in vivo inhibition of Yki-driven ISC tumor progression (Fig. 4I) by oral uptake of a Drosophila Vg-derived TONDU peptide. Remarkably, we observe a marked decrease in tumor load in TONDU peptide-fed flies, similar to what is observed upon ectopic expression of the peptide. Furthermore, a comparative proteomic analysis of ISC tumors in control and TONDU peptide-fed flies suggested a potential causal association between tumor suppression and downregulation of the integrin signaling pathway, a key player implicated in ISC homeostasis (Lin et al., 2013). Our study strongly suggests a critical role of integrin pathway in Yki-driven tumorigenesis; however, a mechanistic understanding of how these regulate each other in ISC tumorigenesis remains to be further elucidated.
TONDU peptide derived from VGLL4 was earlier shown to inhibit gastric tumors in mouse xenograft models (Jiao et al., 2014). Here, we further show the ability of TONDU peptide to inhibit proliferation of prostate and colon cancer cell lines with elevated YAP1 levels. It is likely that other solid tumors with activated YAP/TAZ (Zanconato et al., 2016) and/or TEAD proteins (Gibault et al., 2018; Huh et al., 2019) could be sensitive to inhibition by the TONDU peptide. Moreover, tumors that display loss of tumor suppressor VGLLs (Deng and Fang, 2018), and thereby activated TEAD protein, are likely targets of TONDU peptide-mediated inhibition. Further, our finding that inhibition of integrins suppresses Yki-driven tumors offers integrins as an alternative therapeutic target, which, being cell membrane localized, could be readily accessed (Ley et al., 2016). Finally, cross-species conservation of integrin signaling pathways (Cooper and Giancotti, 2019) and YAP/TAZ activity (Sebé-Pedrós et al., 2012) makes Drosophila tumor models ideal for exploring peptide and combinatorial therapeutic strategies for YAP-driven cancers.
It is noteworthy that peptide therapy in Drosophila has been successfully used to test therapeutic peptides that inhibit aggregate formation such as in Huntington's disease (Kazantsev et al., 2002) and Alzheimer's disease (Popiel et al., 2007), as well as peptides that exhibit immuno-modulatory roles (Pal et al., 2007). In most of these studies, however, peptides were injected into adult flies rather than administered orally. By contrast, oral administration of therapeutic peptides for treatment of human diseases, in general, carries the advantages of ease of administration, high patient compliance and, often, low production costs (Renukuntla et al., 2013). As is true for the development of small-molecule therapeutics, an in vivo Drosophila platform offers multiple advantages for peptide therapeutics, including scalability, genetic tractability and rapid elucidation of the mechanistic underpinning of peptide-based tumor suppression. Drosophila has emerged as a powerful model system to design and screen novel small-molecule drugs (Dar et al., 2012; Markstein et al., 2014; Bangi et al., 2016) as potential treatments for diverse diseases, including cancers. Our study expands the repertoire of Drosophila model-based screening options to include peptides.
Extrapolation of the TONDU peptide as a therapeutic for intestinal cancer is not without caveats. In mammals, intestinal cancers arise from multiple cell types: intestinal crypt stem cells (Barker et al., 2009), crypt progenitors or transit amplifying cells and, occasionally, via reprogramming of differentiated intestinal cells (Sadanandam et al., 2013). In a subset (Lgr5+) of crypt stem cells, gain of YAP surprisingly displays a tumor-inhibitory role via its cytoplasmic sequestration of disheveled 2 (DVL 2) (Barry et al., 2013) or by inhibiting the activity of the TCF transcriptional complex (Li et al., 2020). However, in intestinal crypt cells, activation of YAP drives their unrestricted proliferation (Camargo et al., 2007; Zhou et al., 2011; Li et al., 2020), resulting in intestinal adenomas; this pro-tumorigenic property of YAP is TEAD dependent (Li et al., 2020). Given the dual role of YAP – tumor suppression versus tumor promotion – in a cell type-specific manner, TONDU peptide-mediated therapeutic strategy may hold promise only in intestinal cancers that are mediated by the pro-tumorigenic YAP-TEAD complex. Indeed, a number of inhibitors targeting YAP/TAZ-TEAD complexes have now shown therapeutic promises in arresting growth of cancers, particularly those that display TEAD dependencies (for review, see Pobbati and Hong, 2020).
Caveats and future directions
A major drawback of peptide therapeutics is the short half-life and poor bioavailability of the peptides. Use of non-natural amino acids (Verdurmen et al., 2011) and chemical modifications to stabilize the peptide backbone could help overcome these disadvantages (Furet et al., 2019). Moreover, oral administration of peptides presents additional challenges, including a need to survive harsh digestive milieu of the gastrointestinal tract and their enzymatic degradation (Renukuntla et al., 2013). Furthermore, the intestinal mucosa is found to act as a barrier to peptide absorption. Indeed addition of TAT domains that facilitate cellular uptake (Wang et al., 2017) to the TONDU peptide in our study could have contributed to the success of the oral TONDU peptide. Further improvement to stabilize therapeutic peptide to enhance bioavailability is a challenge for future work.
An additional challenge is that TFs can have multiple binding partners, such that targeting of a TF might result in off-target activities. For instance, with regard to the TONDU peptide, whereas Drosophila has a single TEAD protein, mammals have multiple TEAD proteins (TEAD1-4) that share the TONDU-interacting TEA/ATTS motif (Holden and Cunningham, 2018). This could lead to possible off-target activity of the TONDU peptide and consequent side effects. Further, since TEAD4 also binds to co-factors other than YAP and VGLLs such as the p160 nuclear receptors (Belandia and Parker, 2000), TONDU peptide administration might disrupt regulation by p160 of target genes, which include chromatic modifiers and epigenetic regulators. Identifying and limiting off-target activity of TONDU peptide therefore presents future goals essential for its therapeutic use.
MATERIALS AND METHODS
Fly lines, antibodies and primer sequences
Details on fly lines, antibodies and primer sequences used in the study are provided in Tables S8-S10.
Genotypes of the flies used in the study
Genotypes of the flies used in the study are listed in Table S11.
Induction of Yki-driven ISC tumors
We used the UAS-Gal4 system (Brand and Perrimon, 1993) to drive constitutively active Yki (UAS-ykiS111A.S168A.S250A) in which three Serine phosphorylation sites are mutated (Oh and Irvine, 2009; Kwon et al., 2015), in the ISCs, using an ISC-specific Gal4 driver (esg-Gal4) under control of temperature-sensitive tub-Gal80ts (Kwon et al., 2015). Flies were mated and maintained at 18°C until eclosion of the F1 generation. Freshly eclosed F1 flies of the genotype esg>Gal4, tub-Gal80tsUAS-yki3SA were shifted to 29°C and maintained until dissection.
Generation of UAS-vgTONDU fly line
We synthesized an oligonucleotide coding for the Drosophila TONDU domain (CVVFTNYSGDTASQVDEHFSRALNY) (Pobbati and Hong, 2013). We introduced a start (ATG) and stop (TAA) codon flanking the nucleotide sequences, and inserted a 5′EcoR1 and 3′ Xba1 endonuclease restriction enzyme site on either side to allow directional cloning into pUASt vector (Addgene). We replaced Cytosine on position one and Alanine on position 22 with Serine (SVVFTNYSGDTASQVDEHFSRSLNY) to make the encoded peptide more polar and therefore improve its solubility. Substitution of terminal Cysteine would also reduce chances of aberrant dimer formation. The VXXHF domain of the TONDU domain, which is essential for interaction with TEAD/Sd (Pobbati et al., 2012), was left unchanged. The synthesized oligonucleotide was cloned into pUAST vector carrying mini white, and injected into Canton S embryos at the Center for Cellular and Molecular Platforms (C-CAMP), National Centre for Biological Sciences (NCBS), Bangalore, India. Adults were screened for insertion of the vector into the third chromosome.
Design and synthesis of the TONDU peptide and its variants
TONDU peptide
We synthesized a peptide corresponding to the TONDU domain with certain modifications. The basic peptide is a 46-amino-acid peptide (YGRKKRRQRRRGGPKKKRKVGG [VVFTNYSGDTASQVDEHFSRALNY]) comprised of 24 amino acids of the TONDU domain (VVFTNYSGDTASQVDEHFSRALNY) preceded by the conserved SV40 T-antigen nuclear localizing signal (PKKKRKV) (Lanford et al., 1986) and a cell-penetrating peptide (YGRKKRRQRRR) derived from human immunodeficiency virus (HIV) (Vivès et al., 1997); the NLS sequence was flanked by a di-glycine (GG) spacer to avoid any steric hindrance between the tag and the rest of the peptide. The first Cytosine on the TONDU domain was removed to prevent dimerization of the peptide.
FLAG-tagged TONDU peptide
To test for binding partners to TONDU peptide, we added a FLAG tag (DYKDDDDK) at its C-terminus (YGRKKRRQRRRGGPKKKRKVGG-VVFTNYSGDTASQVDEHFSRALNYDYKDDDDK) to allow protein immunoprecipitation using an anti-FLAG antibody.
Fluorescent-tagged TONDU peptide
To track uptake of the peptide and facilitate its cellular localization, we added 5-TAMARA, a fluorescent tag, to the C-terminus of the TAT-NLS-TONDU peptide. The peptides were synthesized at GL Biochem (Shanghai, China).
Administration of TONDU peptide
Lyophylized TONDU peptide was dissolved in water to a final concentration of 1 mM (used as stock), which was then used to prepare 50, 100, 200 or 400 µM of working stock. Then, 100 µl of each was sprayed over freshly cooled standard fly food (not containing any anti-fungal or anti-bacterial agent), on which flies were reared. The flies were transferred into fresh vials (containing TONDU peptide) every 24 h for 10 days.
Immunostaining of Drosophila adult midguts
Prior to dissection, female flies of desired genotype were starved briefly and fed water for 2 h to flush out food from the gut. Midguts were dissected in 1× PBS and fixed in 4% paraformaldehyde in PBS containing 0.2% Triton X-100 for 30 min at room temperature, followed by washing in PBS containing 0.2% Triton X-100 for 15 min. The guts were then incubated in primary antibody at 4°C overnight, followed by blocking with 0.1% bovine serum albumin (BSA) for 1 h and incubation with secondary antibody (Alexa Fluor 555 against mouse or rabbit) for 4 h at room temperature. Next, guts were washed in 1× PBS and counterstained for nuclei using TO-PRO-3 (Invitrogen, S33025) or F-actin using Alexa Fluor Phalloidin-633 (Invitrogen, A22284; 1:100), followed by mounting in an anti-fade mounting medium, Vectashield (Sigma-Aldrich).
Microscopy and image processing
Images were acquired using a Leica SP5 confocal microscope and processed using the Leica application software and Adobe Photoshop CS5.
Measurement of GFP from confocal images
GFP was quantitated from full projections of images acquired using confocal microscopy. GFP intensity in gray scale from regions of interest (ROIs) covering the entire gut was acquired using the Leica-LSM proprietary software. GFP intensity was normalized to the area of each ROI. Student’s t-test was performed using MS Excel to look for statistical significance in GFP variation.
EdU cell proliferation assay
Cell proliferation was detected by 5-ethynyl-2 deoxyuridine (EdU) uptake using a Click-iT Alexa-Fluor-555 290 kit by Invitrogen. Briefly, unfixed guts from female esgts>UAS-yki3SA flies were incubated with 100 μM EdU in Schneider's insect medium, for 1 h at room temperature. Tissue was then fixed in 4% paraformaldehyde and incubated in secondary buffer containing fluorescent-tagged dye (following the manufacturer's instructions) for 1 h at room temperature and subsequently washed in PBS, counter-stained with TO-PRO-3 (Invitrogen, S33025) and mounted using an anti-fade mounting medium (Invitrogen).
Quantitative RT-PCR
Real-time PCR (RT-PCR) was performed using SYBR green (Applied Biosystems) on ABI7 900 HT. Prior to dissection, esgts>UAS-yki3SA females were starved briefly and fed water for 2 h to flush out food from the gut. Total RNA from 20 midguts was isolated using Qiagen RNeasy columns. For human cancer cells, total RNA was extracted using TRIzol (Ambion). RNA was treated with RNase-free DNase (Roche) to get rid of any traces of DNA before converting RNA to complementary DNA (cDNA) using a cDNA preparation kit (Invitrogen). The resulting cDNA was used as substrate for relative quantitation using SYBR green on ABI7 900 HT. β-Tubulin was used as an endogenous control. Genes were assayed from four biological replicates for each condition. Quantitative PCR (qPCR) was performed using the following conditions: DNA polymerase activation for 10 min at 95°C, followed by 40 cycles of duplex melting for 15 s at 95°C and a combined annealing and extension step for 1 min at 60°C. The threshold-cycle (Ct) values were generated automatically. The relative expression value of each gene in the two conditions was calculated using the 2−ΔΔCt method.
Cancer cell line and cell culture conditions
The prostate (PC3 and LNCaP) and colorectal (COLO 320-HSR) cancer cell lines were obtained from American Type Cell Culture (ATCC; Manassas, VA, USA). The colorectal cancer cell line WiDr was a kind gift from Dr Eric R. Fearon, University of Michigan, Ann Arbor, MI, USA. All of the cell lines were cultured as per ATCC guidelines in a CO2 incubator (Thermo Fisher Scientific) supplied with 5% CO2 at 37°C. Cell line authentication was done via short tandem repeats (STR) profiling at Lifecode Technologies Private Limited (Bangalore, India) and DNA Forensics Laboratory (New Delhi, India). Routine check for mycoplasma contamination of all cell lines was carried out using a PlasmoTest mycoplasma detection kit (InvivoGen).
Cell viability assay of human cancer cell lines
To determine the effect of TONDU peptides on the cell viability of prostate cancer (PC3 and LNCaP) and colorectal cancer (COLO320 and WiDR) cells, ∼3000 cells were plated in each well of a 96-well plate. After 24 h, TONDU peptide was added to the cultured cells at three different concentrations: 50 nM, 100 nM and 250 nM. No peptide was added in the control group. After 72 h and 96 h of peptide treatment, cell viability was determined using resazurin sodium salt solution (R7107, Sigma-Aldrich). Briefly, resazurin (0.02 mg/ml; w/v) diluted in culture medium was added to the cells and incubated for 4 h in the dark at 37°C. Fluorescence was measured at 530/590 nm (excitation/emission) using a Synergy™ H4 Hybrid Microplate Reader (BioTek, Winooski, VT, USA). Three biologically independent samples were used in each experiment; data represent mean±s.e.m. Statistical significance was determined using two-tailed unpaired Student's t-test.
Immunoprecipitation studies to determine binding of TONDU peptide to Sd
Drosophila S2R+ cells (sex: male) were cultured in Schneider's medium supplemented with 10% fetal bovine serum (FBS) at 25°C. Full-length Sd (GEO03367) and Yki (GEO02945) cDNAs from the Drosophila Genomics Resource Center were cloned into the Drosophila Gateway vector pAWH and pAWG, respectively. GFP was cloned into pAWM as a control.
Immunoprecipitation and immunoblotting were performed as previously described (Tang et al., 2018). In brief, DNA was transfected into S2R+ using Effectene transfection reagent (Qiagen, 301427). After 2 days of incubation, cells were incubated with or without 1 µM TONDU peptide for 24 h and then lysed with lysis buffer (Pierce, 87788) containing a protease and phosphatase inhibitor cocktail (Pierce, 78440). Lysate was incubated with Chromotek-GFP-Trap (Bulldog Biotechnology, gta-20) for 2 h at 4°C to precipitate the proteins. Beads were washed three to four times with 1 ml lysis buffer and then boiled in SDS sample buffer, run on a 4-20% polyacrylamide gel (Bio-Rad, 4561096), and transferred to an Immobilon-P polyvinylidene fluoride (PVDF) membrane (Millipore). The membrane was blocked by 5% BSA in TBST (Tris-buffered saline with 0.1% Tween 20) at room temperature for 1 h and then probed with anti-GFP (Molecular Probes, A6455), anti-hemagglutinin (HA) (Covance/BioLegend, MMS-101P) or anti-FLAG (Sigma-Aldrich, F3165) antibody in 1× TBST with 5% BSA overnight, followed by horseradish peroxidase (HRP)-conjugated secondary antibody, and signal was detected by enhanced chemiluminescence (Amersham, RPN2209; Pierce, 34095).
For the TONDU-Sd binding assay, HA-Sd was expressed in S2R+ cells and purified through immunoprecipitation with RIPA buffer (Pierce, 89901) and anti-HA agarose (Sigma-Aldrich, A2095). Purified HA-Sd proteins were incubated with 1 µM TONDU peptide directly. The sample was then washed and subjected to immunoblotting.
Quantitation of the effect of the TONDU peptide on Yki-Sd-driven transcription using HRE
Luciferase reporter
Drosophila S2R+ cells were maintained at 25°C in Schneider's medium (GIBCO) with 10% heat-inactivated FBS (Sigma-Aldrich) and 5% Pen-Strep (GIBCO). Experiments were run on 24-well plates, with three replicates per condition. Cells were co-transfected with 100 ng each of HRE-luciferase reporter [containing two copies of an HRE cloned upstream of an hsp70 basal promoter in pGL3 basic vector (Wu et al., 2008)], along with Sd- or Yki-expressing pAc5.1/V5-HisB plasmids (Wu et al., 2008) (gift from Duojia Pan, UT Southwestern Medical Center, Dallas, TX, USA); 10 ng Act-Renilla was used for transfection control. Transfection was carried out using Effectene (Qiagen), as per the manufacturer's recommended protocol. Then, 24 h after transfection, 50 nM or 100 nM of the TONDU peptide was added to wells in triplicate, and 48 h after addition of the TONDU peptide, cells were harvested and luciferase activity was measured using Dual Glo (Promega) as per the kit instructions, measured using a Spectramax Luminescence plate reader.
Detection of fluorescent-labeled TONDU peptide in S2R+ cells
Drosophila S2R+ cells were grown to confluence in Schneider's medium (GIBCO) supplemented with 10% heat-inactivated FBS (Sigma-Aldrich) and 5% Pen-Strep (GIBCO) at 25°C in 24-well plates. TAMARA-tagged TONDU peptide was added to the medium to a final concentration of 100 nM and cells were incubated for 6 h. Next, the medium was discarded and cells were washed three times with 1× PBS. Cells were then added to lysine-coated slides, fixed with 4% formaldehyde in 1× PBS and counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Cells were imaged with a Nikon Ti, CSU-X1 spinning disk confocal microscope and the images were processed using Fiji image processing software (https://imagej.net/Fiji).
ChIP to determine binding of TONDU peptide to Sd in the upstream regulatory region of gene mew
ChIP was performed using LowCell# ChIP kit protein A (Diagenode, C01010072) according to the manufacturer's instructions. Briefly, midguts from 35 adult esgts>UAS-yki3SA females (pre-starved for 1 h) were dissected in ice-cold 1× PBS and crosslinked in 1% formaldehyde (Sigma-Aldrich) for 15 min at 37°C. Crosslinking was quenched with 125 mM glycine. The guts were washed with PBS and precipitated by centrifugation at 3500 g for 5 min. The pellet was lysed in 250 ml Buffer B (LowCell# ChIP kit) supplemented with complete protease inhibitor (Roche) and phenylmethylsulfonyl fluoride (PMSF; Sigma-Aldrich). Lysed chromatin (130 µl) was sheared using a Bioruptor (Diagenode) at high frequency for 15 cycles of 30 s ON, 30 s OFF. Then, 870 µl of Buffer A (LowCell# ChIP kit) supplemented with complete protease inhibitor (Roche) and PMSF (Sigma-Aldrich) was added to the shared chromatin, and 8 µl of the chromatin solution was saved as an input control. Magnetic beads (11 µl) were washed twice with Buffer A (LowCell# ChIP kit) and resuspended in 800 µl Buffer A. Anti-FLAG antibody (2 µg; Sigma-Aldrich, F1804) was then added to the washed beads and gently agitated at 4°C for 4 h. The beads-antibody complex was precipitated with a magnet and the supernatant was removed, and 800 µl shared chromatin was added to the beads-antibody complex and rotated at 4°C overnight. The immobilized chromatin was then washed with Buffer A three times and Buffer C once, and eluted in 100 µl elution buffer (1% SDS, 0.1 M sodium bicarbonate with proteinase K and RNaseA). The chromatin was subjected to either phenol-chloroform extraction for DNA purification and subsequent qPCR analysis, or the protein was extracted by heating the washed beads at 95°C in 20 μl SDS loading dye (4×) for 10 min and centrifuged at 15,000 g for 10 min. The supernatant was collected and used for dot blot analysis.
Protein dot blot
TONDU peptide (1 mM) was serially diluted (10−1, 10−2, 10−3) and blotted using a narrow-mouth pipette tip, and 7.5 µl peptide or enriched protein fraction from ChIP was applied slowly onto the nitrocellulose membrane (Thermo Fisher Scientific, 0.2 μm pore size). The membrane was air dried and then blocked in 5% BSA in TBST for 2 h at room temperature, then incubated for 3 h with a secondary antibody conjugated to HRP (Jackson ImmunoResearch, 711035152), washed three times with TBST, detected with chemiluminescent substrate (Thermo Fisher Scientific, 34080) and visualized on X-ray film (Fuji, Super HR-t).
Proteomics of Yki-driven ISC tumors
Protein extraction from fly guts for LC-MS/MS analysis
Prior to dissection, female esgts>UAS-yki3SA flies were briefly starved and fed water for 2 h to clear the gut. Adult guts were dissected in cold 1× PBS from 20 flies. The fore- and hindguts were removed, and the midguts were put in 100 µl extraction buffer (6 M GnHCl in 50 mM Tris-HCl pH 7.4, 65 mM dithiothreitol) with 50 mM sodium acetate and protease inhibitors (1× protease inhibitor cocktail with 0.2 mM PMSF) was added to the sample. The guts were sonicated with a Bioruptor (Diagenode) using the following settings: sonication cycle: 30 s ON and 30 s OFF for 5 cycles at 4°C. Cell debris was removed by centrifuging at 6000 g for 3 min; then the supernatant was transferred to a new tube. The protein concentration was determined spectrophotometrically using Nanodrop and BCA protein assay (Thermo Fisher Scientific), following the manufacturer's protocol. Five micrograms of the protein were used for LC-MS-MS analysis, and we made certain that the tissue was processed within 30 min of dissection.
Sample preparation for LC-MS/MS
Five micrograms of the protein samples were reduced with 5 mM tris(2-carboxyethyl)phosphine (TCEP), further alkylated with 50 mM iodoacetamide and digested with Trypsin (1:50, Trypsin/lysate ratio) for 16 h at 37°C. Digests were cleaned using a C18 silica cartridge to remove the salt and dried using a speed vac. The dried pellet was resuspended in 5% acetonitrile, 0.1% formic acid (Buffer A).
Mass spectrometric analysis of peptide mixtures
The experiment was performed using an EASY-nLC 1000 system (Thermo Fisher Scientific) coupled to a Thermo Fisher-Orbitrap Fusion mass spectrometer equipped with a nanoelectrospray ion source. One microgram of the peptide mixture was resolved using a 25 cm Thermo Easy-spray PepMap C18 column. The peptides were loaded with Buffer A and eluted with a 0-40% gradient of Buffer B (95% acetonitrile, 0.1% formic acid) at a flow rate of 300 nl/min for 60 min. Mass spectrometry (MS) data were acquired using a data-dependent top 20 method, dynamically choosing the most abundant precursor ions from the survey scan. The LC-MS/MS RAW files have been submitted to MassIVE repository (https://massive.ucsd.edu) and can be accessed using MSV000084841.
Data processing
All samples were processed and the eight RAW files generated were analyzed with Proteome Discoverer (v2.2) against the UniProt Drosophila melanogaster reference proteome database. For Sequest search, the precursor and fragment mass tolerances were set at 10 ppm and 0.5 Da, respectively. The protease used to generate peptides, i.e. enzyme specificity, was set for trypsin/P (cleavage at the C terminus of ‘K/R: unless followed by P’) along with a maximum missed cleavages value of two. Carbamidomethyl on cysteine as fixed modification and oxidation of methionine and N-terminal acetylation were considered as variable modifications for database search. Both the peptide spectrum match and the protein FDR were set to 0.01 FDR.
Proteome data analysis
To identify biologically relevant protein signatures in esgts>yki3SA tumors and characterize their status in the presence of the TONDU peptide, we calculated the log2 abundance ratios, using mean abundance values for individual UniProt IDs of esgts>UAS-yki3SA day 7 versus day 1 proteome. Only those with combined FDR confidence <0.05 (medium) or <0.01 (high) were taken into consideration; those with combined FDR >0.05 (low) were discarded. We further filtered out peptides that were not detected in either MS or MS/MS spectra, depending on the peak calling. We noted that the number of peptides that matched each UniProt ID, ranged from 1 to 67. To ascertain statistically significant calls, we applied Student’s t-test on replicate readings for the individual UniProt IDs and only those with P<0.05 were considered. We first calculated log2 abundance ratio of proteins in day 7 with day 1 of esgts>yki3SA tumors, and considered only gene products for which log2 fold change was ≥2. Next, we examined the status of tumor proteins from TONDU peptide-fed flies. Since the TONDU-peptide treated tumors phenocopied the tumor suppression seen by overexpression of the TONDU peptide (compare Fig. 1 with Fig. 2), and their protein profiles displayed closed correlation (Fig. S4), we chose to combine these two data sets. We therefore calculated log2 abundance ratio of individual proteins in untreated day 7 esgts>yki3SA tumors, to that of TONDU peptide-treated and to TONDU-peptide expressed (UAS-vgTONDU) ISC tumors. We applied the Student's t-test to look for statistical significance for each log2 fold change and considered only those with P<0.05.
GO analysis
To identify the biological function of genes and look for enrichment of functional classes, we undertook GO analysis using the Protein ANalysisTHroughEvolutionaryRelationships (PANTHER) classification system (http://www.pantherdb.org; Mi et al., 2007). Protein functions were inferred by classification of genes into one or more groups, depending on: (1) molecular function, (2) biological process, (3) protein class, (4) pathways and (5) cellular component.
Heat maps
Heat maps were generated using Heatmapper (http://heatmapper.ca/). For the heat map in Fig. 3A, raw abundance values for individual UniProt IDs were subjected to row scaling, and clustered using average linkage clustering with Euclidean method for distance measure.
Supplementary Material
Acknowledgements
We thank Jose F. de Celis for the antibody against Sd, Duojia Pan for HRE-luciferase reporter vectors, Nick Brown for the UAS-torsoD/βCyt fly line and Stephanie Mohr for comments on the manuscript. We also acknowledge the fly transgenic facility at C-CAMP (NCBS, Bangalore, India) and proteomics service by Valerian Chem (Gurgaon, India).
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: A.B., P.S.; Methodology: A.B., P.S.; Validation: A.T., B.A., N.P.; Formal analysis: T.A.Q.; Investigation: A.B., T.A.Q., H.-W.T., N.M., V.S.; Resources: A.T., B.A., N.P., P.S.; Data curation: A.B.; Writing - original draft: A.B., P.S.; Writing - review & editing: N.P.; Visualization: A.B., P.S.; Supervision: A.B., P.S.; Funding acquisition: A.B.
Funding
This study was supported by the Wellcome Trust DBT India Alliance (IA/E/13/1/501271 to A.B.). Work in the N.P. laboratory is supported by the Howard Hughes Medical Institute.
Data availability
The LC-MS/MS reads for unlabeled proteomics generated in this study are available at the MassIVE repository (https://massive.ucsd.edu) and can be accessed using MSV000084841.
Supplementary information
Supplementary information available online at https://dmm.biologists.org/lookup/doi/10.1242/dmm.044420.supplemental
References
- Arosio D., Manzoni L., Corno C. and Perego P. (2017). Integrin-targeted peptide- and peptidomimetic-drug conjugates for the treatment of tumors. Recent Pat. Anticancer Drug Discov. 12, 148-168. 10.2174/1574892812666170203151930 [DOI] [PubMed] [Google Scholar]
- Bajpai A. and Sinha P. (2020). Hh signaling from de novo organizers drive lgl neoplasia in Drosophila epithelium. Dev. Biol. 457, 1-8. 10.1016/j.ydbio.2019.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangi E., Murgia C., Teague A. G. S., Sansom O. J. and Cagan R. L. (2016). Functional exploration of colorectal cancer genomes using Drosophila. Nat. Commun. 7, 13615 10.1038/ncomms13615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantscheff M., Schirle M., Sweetman G., Rick J. and Kuster B. (2007). Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem. 389, 1017-1031. 10.1007/s00216-007-1486-6 [DOI] [PubMed] [Google Scholar]
- Barker N., Ridgway R. A., van Es J. H., van de Wetering M., Begthel H., van den Born M., Danenberg E., Clarke A. R., Sansom O. J. and Clevers H. (2009). Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 457, 608-611. 10.1038/nature07602 [DOI] [PubMed] [Google Scholar]
- Barry E. R., Morikawa T., Butler B. L., Shrestha K., de la Rosa R., Yan K. S., Fuchs C. S., Magness S. T., Smits R., Ogino S. et al. (2013). Restriction of intestinal stem cell expansion and the regenerative response by YAP. Nature 493, 106-110. 10.1038/nature11693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belandia B. and Parker M. G. (2000). Functional interaction between the p160 coactivator proteins and the transcriptional enhancer factor family of transcription factors. J. Biol. Chem. 275, 30801-30805. 10.1074/jbc.C000484200 [DOI] [PubMed] [Google Scholar]
- Bhagwat A. S. and Vakoc C. R. (2015). Targeting transcription factors in cancer. Trends Cancer 1, 53-65. 10.1016/j.trecan.2015.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S., Lunec J. and Bartlett K. (2012). Non-glucose metabolism in cancer cells--is it all in the fat? Cancer Metastasis Rev. 31, 689-698. 10.1007/s10555-012-9384-6 [DOI] [PubMed] [Google Scholar]
- Brand A. H. and Perrimon N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401-415. [DOI] [PubMed] [Google Scholar]
- Camargo F. D., Gokhale S., Johnnidis J. B., Fu D., Bell G. W., Jaenisch R. and Brummelkamp T. R. (2007). YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Biol. 17, 2054-2060. 10.1016/j.cub.2007.10.039 [DOI] [PubMed] [Google Scholar]
- Chang Y. S., Graves B., Guerlavais V., Tovar C., Packman K., To K.-H., Olson K. A., Kesavan K., Gangurde P., Mukherjee A. et al. (2013). Stapled alpha-helical peptide drug development: a potent dual inhibitor of MDM2 and MDMX for p53-dependent cancer therapy. Proc. Natl. Acad. Sci. USA 110, E3445-E3454. 10.1073/pnas.1303002110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Sayadian A.-C., Lowe N., Lovegrove H. E. and St Johnston D. (2018). An alternative mode of epithelial polarity in the Drosophila midgut. PLoS Biol. 16, e3000041 10.1371/journal.pbio.3000041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J. and Giancotti F. G. (2019). Integrin signaling in cancer: mechanotransduction, stemness, epithelial plasticity, and therapeutic resistance. Cancer Cell 35, 347-367. 10.1016/j.ccell.2019.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar A. C., Das T. K., Shokat K. M. and Cagan R. L. (2012). Chemical genetic discovery of targets and anti-targets for cancer polypharmacology. Nature 486, 80-84. 10.1038/nature11127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X. and Fang L. (2018). VGLL4 is a transcriptional cofactor acting as a novel tumor suppressor via interacting with TEADs. Am. J. Cancer Res. 8, 932-943. [PMC free article] [PubMed] [Google Scholar]
- Drucker D. J. (2019). Advances in oral peptide therapeutics. Nat. Rev. Drug Discov. 19, 277-289. 10.1038/s41573-019-0053-0 [DOI] [PubMed] [Google Scholar]
- Furet P., Salem B., Mesrouze Y., Schmelzle T., Lewis I., Kallen J. and Chène P. (2019). Structure-based design of potent linear peptide inhibitors of the YAP-TEAD protein-protein interaction derived from the YAP omega-loop sequence. Bioorg. Med. Chem. Lett. 29, 2316-2319. 10.1016/j.bmcl.2019.06.022 [DOI] [PubMed] [Google Scholar]
- Gibault F., Sturbaut M., Bailly F., Melnyk P. and Cotelle P. (2018). Targeting Transcriptional Enhanced Associate Domains (TEADs). J. Med. Chem. 61, 5057-5072. 10.1021/acs.jmedchem.7b00879 [DOI] [PubMed] [Google Scholar]
- Guilgur L. G., Prudêncio P., Sobral D., Liszekova D., Rosa A. and Martinho R. G. (2014). Requirement for highly efficient pre-mRNA splicing during Drosophila early embryonic development. eLife 3, e02181 10.7554/eLife.02181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T., Lu Y., Li P., Yin M.-X., Lv D., Zhang W., Wang H., Zhou Z., Ji H., Zhao Y. et al. (2013). A novel partner of Scalloped regulates Hippo signaling via antagonizing Scalloped-Yorkie activity. Cell Res. 23, 1201-1214. 10.1038/cr.2013.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Lucchetta E., Rafel N. and Ohlstein B. (2016). Maintenance of the adult Drosophila intestine: all roads lead to homeostasis. Curr. Opin. Genet. Dev. 40, 81-86. 10.1016/j.gde.2016.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden J. K. and Cunningham C. N. (2018). Targeting the Hippo pathway and cancer through the TEAD family of transcription factors. Cancers 10, 81 10.3390/cancers10030081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh H. D., Kim D. H., Jeong H.-S. and Park H. W. (2019). Regulation of TEAD transcription factors in cancer biology. Cells 8, 600 10.3390/cells8060600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao S., Wang H., Shi Z., Dong A., Zhang W., Song X., He F., Wang Y., Zhang Z., Wang W. et al. (2014). A peptide mimicking VGLL4 function acts as a YAP antagonist therapy against gastric cancer. Cancer Cell 25, 166-180. 10.1016/j.ccr.2014.01.010 [DOI] [PubMed] [Google Scholar]
- Jin Y., Xu J., Yin M.-X., Lu Y., Hu L., Li P., Zhang P., Yuan Z., Ho M. S., Ji H. et al. (2013). Brahma is essential for Drosophila intestinal stem cell proliferation and regulated by Hippo signaling. eLife 2, e00999 10.7554/eLife.00999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantsev A., Walker H. A., Slepko N., Bear J. E., Preisinger E., Steffan J. S., Zhu Y.-Z., Gertler F. B., Housman D. E., Marsh J. L. et al. (2002). A bivalent Huntingtin binding peptide suppresses polyglutamine aggregation and pathogenesis in Drosophila. Nat. Genet. 30, 367-376. 10.1038/ng864 [DOI] [PubMed] [Google Scholar]
- Khan S. J., Bajpai A., Alam M. A., Gupta R. P., Harsh S., Pandey R. K., Goel-Bhattacharya S., Nigam A., Mishra A. and Sinha P. (2013). Epithelial neoplasia in Drosophila entails switch to primitive cell states. Proc. Natl. Acad. Sci. USA 110, E2163-E2172. 10.1073/pnas.1212513110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo P., Allan K., Willoughby L., Brumby A. M. and Richardson H. E. (2013). In Drosophila, RhoGEF2 cooperates with activated Ras in tumorigenesis through a pathway involving Rho1-Rok-Myosin-II and JNK signalling. Dis. Model. Mech. 6, 661-678. 10.1242/dmm.010066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koontz L. M., Liu-Chittenden Y., Yin F., Zheng Y., Yu J., Huang B., Chen Q., Wu S. and Pan D. (2013). The Hippo effector Yorkie controls normal tissue growth by antagonizing scalloped-mediated default repression. Dev. Cell 25, 388-401. 10.1016/j.devcel.2013.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koundouros N. and Poulogiannis G. (2020). Reprogramming of fatty acid metabolism in cancer. Br. J. Cancer 122, 4-22. 10.1038/s41416-019-0650-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y., Vinayagam A., Sun X., Dephoure N., Gygi S. P., Hong P. and Perrimon N. (2013). The Hippo signaling pathway interactome. Science 342, 737-740. 10.1126/science.1243971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y., Song W., Droujinine I. A., Hu Y., Asara J. M. and Perrimon N. (2015). Systemic organ wasting induced by localized expression of the secreted insulin/IGF antagonist ImpL2. Dev. Cell 33, 36-46. 10.1016/j.devcel.2015.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert M., Jambon S., Depauw S. and David-Cordonnier M.-H. (2018). Targeting transcription factors for cancer treatment. Molecules 23, 1479 10.3390/molecules23061479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanford R. E., Kanda P. and Kennedy R. C. (1986). Induction of nuclear transport with a synthetic peptide homologous to the SV40 T antigen transport signal. Cell 46, 575-582. 10.1016/0092-8674(86)90883-4 [DOI] [PubMed] [Google Scholar]
- Lau J. L. and Dunn M. K. (2018). Therapeutic peptides: historical perspectives, current development trends, and future directions. Bioorg. Med. Chem. 26, 2700-2707. 10.1016/j.bmc.2017.06.052 [DOI] [PubMed] [Google Scholar]
- Ley K., Rivera-Nieves J., Sandborn W. J. and Shattil S. (2016). Integrin-based therapeutics: biological basis, clinical use and new drugs. Nat. Rev. Drug Discov. 15, 173-183. 10.1038/nrd.2015.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Sun Y., Jarugumilli G. K., Liu S., Dang K., Cotton J. L., Xiol J., Chan P. Y., DeRan M., Ma L. et al. (2020). Lats1/2 sustain intestinal stem cells and Wnt activation through TEAD-dependent and independent transcription. Cell Stem Cell 26, 675-692.e8. 10.1016/j.stem.2020.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G., Zhang X., Ren J., Pang Z., Wang C., Xu N. and Xi R. (2013). Integrin signaling is required for maintenance and proliferation of intestinal stem cells in Drosophila. Dev. Biol. 377, 177-187. 10.1016/j.ydbio.2013.01.032 [DOI] [PubMed] [Google Scholar]
- Markstein M., Dettorre S., Cho J., Neumuller R. A., Craig-Muller S. and Perrimon N. (2014). Systematic screen of chemotherapeutics in Drosophila stem cell tumors. Proc. Natl. Acad. Sci. USA 111, 4530-4535. 10.1073/pnas.1401160111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Bermudo M. D., Alvarez-Garcia I. and Brown N. H. (1999). Migration of the Drosophila primordial midgut cells requires coordination of diverse PS integrin functions. Development 126, 5161-5169. [DOI] [PubMed] [Google Scholar]
- Mi H., Guo N., Kejariwal A. and Thomas P. D. (2007). PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 35, D247-D252. 10.1093/nar/gkl869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan K. A., Fuerer C., Ching W., Fish M., Willert K. and Nusse R. (2012). Secreted Wingless-interacting molecule (Swim) promotes long-range signaling by maintaining Wingless solubility. Proc. Natl. Acad. Sci. USA 109, 370-377. 10.1073/pnas.1119197109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj R., Gururaja-Rao S., Jones K. T., Slattery M., Negre N., Braas D., Christofk H., White K. P., Mann R. and Banerjee U. (2012). Control of mitochondrial structure and function by the Yorkie/YAP oncogenic pathway. Genes Dev. 26, 2027-2037. 10.1101/gad.183061.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H. and Irvine K. D. (2009). In vivo analysis of Yorkie phosphorylation sites. Oncogene 28, 1916-1927. 10.1038/onc.2009.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H., Slattery M., Ma L., Crofts A., White K. P., Mann R. S. and Irvine K. D. (2013). Genome-wide association of Yorkie with chromatin and chromatin-remodeling complexes. Cell Rep. 3, 309-318. 10.1016/j.celrep.2013.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S., St Leger R. J. and Wu L. P. (2007). Fungal peptide Destruxin A plays a specific role in suppressing the innate immune response in Drosophila melanogaster. J. Biol. Chem. 282, 8969-8977. 10.1074/jbc.M605927200 [DOI] [PubMed] [Google Scholar]
- Pobbati A. V. and Hong W. (2013). Emerging roles of TEAD transcription factors and its coactivators in cancers. Cancer Biol. Ther. 14, 390-398. 10.4161/cbt.23788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pobbati A. V. and Hong W. (2020). A combat with the YAP/TAZ-TEAD oncoproteins for cancer therapy. Theranostics 10, 3622-3635. 10.7150/thno.40889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pobbati A. V., Chan S. W., Lee I., Song H. and Hong W. (2012). Structural and functional similarity between the Vgll1-TEAD and the YAP-TEAD complexes. Structure 20, 1135-1140. 10.1016/j.str.2012.04.004 [DOI] [PubMed] [Google Scholar]
- Popiel H. A., Nagai Y., Fujikake N. and Toda T. (2007). Protein transduction domain-mediated delivery of QBP1 suppresses polyglutamine-induced neurodegeneration in vivo. Mol. Ther. 15, 303-309. 10.1038/sj.mt.6300045 [DOI] [PubMed] [Google Scholar]
- Reis R., Feijao T., Gouveia S., Pereira A. J., Matos I., Sampaio P., Maiato H. and Sunkel C. E. (2009). Dynein and mast/orbit/CLASP have antagonistic roles in regulating kinetochore-microtubule plus-end dynamics. J. Cell Sci. 122, 2543-2553. 10.1242/jcs.044818 [DOI] [PubMed] [Google Scholar]
- Renukuntla J., Vadlapudi A. D., Patel A., Boddu S. H. S. and Mitra A. K. (2013). Approaches for enhancing oral bioavailability of peptides and proteins. Int. J. Pharm. 447, 75-93. 10.1016/j.ijpharm.2013.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadanandam A., Lyssiotis C. A., Homicsko K., Collisson E. A., Gibb W. J., Wullschleger S., Ostos L. C. G., Lannon W. A., Grotzinger C., Del Rio M. et al. (2013). A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat. Med. 19, 619-625. 10.1038/nm.3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebé-Pedrós A., Zheng Y., Ruiz-Trillo I. and Pan D. (2012). Premetazoan origin of the hippo signaling pathway. Cell Rep. 1, 13-20. 10.1016/j.celrep.2011.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Kir S., Hong S., Hu Y., Wang X., Binari R., Tang H.-W., Chung V., Banks A. S., Spiegelman B. et al. (2019). Tumor-derived ligands trigger tumor growth and host wasting via differential MEK activation. Dev. Cell 48, 277-286; e6. 10.1016/j.devcel.2018.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Gable A. L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N. T., Morris J. H., Bork P. et al. (2019). STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607-D613. 10.1093/nar/gky1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H.-W., Hu Y., Chen C.-L., Xia B., Zirin J., Yuan M., Asara J. M., Rabinow L. and Perrimon N. (2018). The TORC1-regulated CPA complex rewires an RNA processing network to drive autophagy and metabolic reprogramming. Cell Metab. 27, 1040-1054; e8. 10.1016/j.cmet.2018.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdurmen W. P. R., Bovee-Geurts P. H., Wadhwani P., Ulrich A. S., Hallbrink M., van Kuppevelt T. H. and Brock R. (2011). Preferential uptake of L- versus D-amino acid cell-penetrating peptides in a cell type-dependent manner. Chem. Biol. 18, 1000-1010. 10.1016/j.chembiol.2011.06.006 [DOI] [PubMed] [Google Scholar]
- Vinayagam A., Kulkarni M. M., Sopko R., Sun X., Hu Y., Nand A., Villalta C., Moghimi A., Yang X., Mohr S. E. et al. (2016). An integrative analysis of the InR/PI3K/Akt network identifies the dynamic response to insulin signaling. Cell Rep. 16, 3062-3074. 10.1016/j.celrep.2016.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivès E., Brodin P. and Lebleu B. (1997). A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J. Biol. Chem. 272, 16010-16017. 10.1074/jbc.272.25.16010 [DOI] [PubMed] [Google Scholar]
- Wadia J. S. and Dowdy S. F. (2005). Transmembrane delivery of protein and peptide drugs by TAT-mediated transduction in the treatment of cancer. Adv. Drug Deliv. Rev. 57, 579-596. 10.1016/j.addr.2004.10.005 [DOI] [PubMed] [Google Scholar]
- Wang X., Qiao Y., Asangani I. A., Ateeq B., Poliakov A., Cieślik M., Pitchiaya S., Chakravarthi B. V. S. K., Cao X., Jing X. et al. (2017). Development of Peptidomimetic Inhibitors of the ERG Gene Fusion Product in Prostate Cancer. Cancer Cell 31, 532-548.e7. 10.1016/j.ccell.2017.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Liu Y., Zheng Y., Dong J. and Pan D. (2008). The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev. Cell 14, 388-398. 10.1016/j.devcel.2008.01.007 [DOI] [PubMed] [Google Scholar]
- Zanconato F., Cordenonsi M. and Piccolo S. (2016). YAP/TAZ at the roots of cancer. Cancer Cell 29, 783-803. 10.1016/j.ccell.2016.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Gao Y., Li P., Shi Z., Guo T., Li F., Han X., Feng Y., Zheng C., Wang Z. et al. (2014). VGLL4 functions as a new tumor suppressor in lung cancer by negatively regulating the YAP-TEAD transcriptional complex. Cell Res. 24, 331-343. 10.1038/cr.2014.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Zhang Y., Wu H., Barry E., Yin Y., Lawrence E., Dawson D., Willis J. E., Markowitz S. D., Camargo F. D. et al. (2011). Mst1 and Mst2 protein kinases restrain intestinal stem cell proliferation and colonic tumorigenesis by inhibition of Yes-associated protein (Yap) overabundance. Proc. Natl. Acad. Sci. USA 108, E1312-E1320. 10.1073/pnas.1110428108 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.