Objectives
This is a protocol for a Cochrane Review (diagnostic). The objectives are as follows:
To determine the accuracy of Xpert MTB/RIF and Xpert Ultra for screening for tuberculosis in adults irrespective of signs or symptoms of pulmonary tuberculosis in the general population (i.e. low‐risk population).
To determine the accuracy of Xpert MTB/RIF and Xpert Ultra for screening of pulmonary tuberculosis in adults in the following high‐risk groups.
People living with HIV.
Household contacts of people with tuberculosis.
Patients residing in high‐tuberculosis‐burden settings attending primary health facilities.
Homeless people.
Miners.
People with diabetes mellitus.
People who abuse alcohol.
Smokers.
People residing in prisons.
Healthcare workers.
To determine the accuracy of Xpert MTB/RIF and Xpert Ultra for the detection of rifampicin resistance in the general population and in the high‐risk groups and settings described above.
Secondary objectives
To compare the accuracy of Xpert MTB/RIF and Xpert Ultra in the above high‐risk groups and settings.
To investigate potential sources of heterogeneity in accuracy estimates, including the percentage of participants with tuberculosis symptoms, tuberculosis burden, tuberculosis/HIV burden, and MDR‐TB burden.
Background
Tuberculosis is the world’s leading cause of infectious disease‐related death and is one of the top 10 causes of death worldwide (WHO Global TB Report 2019). In 2018, an estimated 10 million people developed tuberculosis disease, a 2% decline from 2017 (MacNeil 2020; WHO Global TB Report 2019).
Among all tuberculosis cases, about 9% were in people living with HIV (WHO Global TB Report 2019). The risk of developing tuberculosis is much higher in people living with HIV, estimated to be 20 to 37 times higher in HIV‐positive individuals than in HIV‐negative individuals (Getahun 2010). Signs and symptoms of tuberculosis in people living with HIV vary, which makes it challenging to determine when to consider a diagnosis of tuberculosis ‐ tuberculosis is the leading cause of hospitalization and death in people with HIV worldwide (Ford 2016). In addition, there were around 500,000 new cases of rifampicin‐resistant tuberculosis, of which 78% had multidrug‐resistant tuberculosis (tuberculosis that is resistant to both rifampicin and isoniazid, the two most essential anti‐tuberculosis drugs) (WHO Global TB Report 2019). When tuberculosis is detected early and is effectively treated, the disease is largely curable. Ending the tuberculosis epidemic by 2030 is among the health‐related targets described in United Nations Sustainable Development Goal 3 (WHO END TB 2015). The United Nations Sustainable Development Goals represent a collective plan to end poverty, decrease inequality, and protect the planet from degradation by 2030 (UN Sustainable Development Goals 2030).
The World Health Organization (WHO) recommends the use of specific molecular tests, including Xpert MTB/RIF or Xpert Ultra, the newest version of the assay, as the initial diagnostic tests for the detection of tuberculosis and rifampicin resistance in people with signs and symptoms of tuberculosis (WHO Xpert MTB/RIF 2013; WHO Xpert Ultra 2017; WHO Rapid Communication 2020a). However, the WHO estimates that nearly one‐third of all active tuberculosis cases go undiagnosed and unreported (WHO Global TB Report 2019). In an effort to close this diagnostic gap, the WHO is seeking evidence to recommend case‐finding approaches and strategies to improve tuberculosis case detection of the 'missing millions'. In particular, the WHO is interested in case‐finding approaches in high‐risk populations and settings, such as people living with HIV, people with diabetes mellitus, and people residing in prisons. Stated another way, the WHO is interested in the best ways to find the so‐called ‘missing millions’.
Tuberculosis screening is a term that has been used differently in the literature depending on the context. We use tuberculosis screening as defined by the WHO: the "systematic identification of people with suspected active TB [tuberculosis], in a predetermined target group, using tests, examinations or other procedures that can be applied rapidly." Further, we define intensified case‐finding as tuberculosis screening activities set in health facilities, and active case‐finding as tuberculosis screening activities set in the community, including household‐based or residence‐based screening activities (WHO Tuberculosis screening 2013). The End‐TB strategy emphasizes early diagnosis of tuberculosis, including universal drug susceptibility testing, and systematic screening of contacts and high‐risk groups (WHO Global TB Report 2019).
Current screening approaches for active tuberculosis typically recommend initial screening for four cardinal signs and symptoms of tuberculosis: cough, fever, weight loss, and night sweats. People with a positive symptom screen then may go on to receive additional screening with a chest X‐ray and diagnostic testing using sputum Xpert MTB/RIF or Xpert Ultra as recommended. Concerning people living with HIV, a recent systematic review found that the four‐symptom screen had lower sensitivity and specificity for active tuberculosis in HIV‐positive people on antiretroviral therapy (ART) than in HIV‐positive people not taking ART (Hamada 2018). Compared to Xpert MTB/RIF, Xpert Ultra has shown increased sensitivity for tuberculosis in HIV‐positive people (Dorman 2018). WHO Tuberculosis Standard 8 states, "For persons living with HIV, the Xpert MTB/RIF Ultra assay should be used as an initial diagnostic test" (WHO Compendium of WHO guidelines 2018).
Several Cochrane Reviews have been published or are in process to assess the diagnostic accuracy of Xpert MTB/RIF and Xpert Ultra for different target conditions and in various populations. Of relevance to the current review, a recent Cochrane Review found Xpert MTB/RIF and Xpert Ultra to be highly sensitive and specific for pulmonary tuberculosis and rifampicin resistance in adults with signs and symptoms of tuberculosis (Horne 2019). The current review will determine the accuracy of Xpert MTB/RIF and Xpert Ultra for tuberculosis and rifampicin resistance in adults irrespective of signs and symptoms of tuberculosis, that is, when used as a screening test.
Target condition being diagnosed
Tuberculosis is caused by the bacterium Mycobacterium tuberculosis (M tuberculosis) and is spread from person to person through the air. Tuberculosis most commonly affects the lungs (pulmonary tuberculosis), but may affect any organ or tissue outside of the lungs (extrapulmonary tuberculosis). Signs and symptoms of pulmonary tuberculosis include cough, fever, chills, night sweats, weight loss, haemoptysis (coughing up blood), and fatigue. Signs and symptoms of extrapulmonary tuberculosis depend on the site of disease. Tuberculosis treatment regimens must contain multiple drugs, to which the organisms are sensitive, to cure tuberculosis and avoid selection for drug resistance. The treatment of multidrug‐resistant TB (MDR‐TB) is complex, historically requiring two years or more of therapy, although the WHO conditionally recommended a regimen of nine to 12 months in 2016 (WHO Guidelines 2016). The drugs used to treat MDR‐TB are less potent and more toxic than the drugs used to treat drug‐susceptible tuberculosis. Based on new evidence on the management of drug‐resistant tuberculosis, the WHO recently released a rapid communication stating that, "All patients with MDR‐TB or rifampicin‐resistant tuberculosis, including those with additional resistance to fluoroquinolones, stand to benefit from effective all‐oral treatment regimens, either shorter or longer, implemented under programmatic conditions" (WHO Rapid Communication 2020b).
Index test(s)
Xpert MTB/RIF is an automated polymerase chain reaction (PCR) test (molecular test) using the GeneXpert platform (Cepheid 2009). Xpert MTB/RIF is a single test that can detect both M tuberculosis complex and rifampicin resistance within two hours after starting the test, with minimal hands‐on technical time. Unlike conventional nucleic acid amplification (NAA) tests, Xpert MTB/RIF is unique because sample processing and PCR amplification and detection are integrated into a single, self‐enclosed test unit, the GeneXpert cartridge. Following sample loading, all steps in the assay are completely automated and self‐contained. In addition, the assay’s sample reagent, used to liquefy sputum, has potent tuberculocidal (the ability to kill tuberculosis bacteria) properties and so largely eliminates biosafety concerns during the test procedure (Banada 2010). Xpert MTB/RIF requires an uninterrupted and stable electrical power supply, temperature control, and yearly calibration of the cartridge modules (Global Laboratory Initiative 2019).
Since Xpert MTB/RIF was released, there have been four generations of the test (G1, G2, G3, and G4), involving different software and cartridge combinations. G4 contains modifications that improved determination of rifampicin resistance detection as previous Xpert MTB/RIF versions had found that some rifampicin susceptibility results were falsely resistant. In order to improve on Xpert MTB/RIF sensitivity, Cepheid developed Xpert MTB/RIF Ultra (hereafter referred to as Xpert Ultra), a re‐engineered assay that uses a newly developed cartridge but may be run on the same device after a software upgrade. Xpert Ultra incorporates two different multi‐copy amplification targets and a larger DNA reaction chamber than Xpert MTB/RIF (WHO Xpert Ultra 2017). A laboratory study reported that the limit of detection using Xpert Ultra improved to 15.6 CFU/mL of sputum compared to 112.6 CFU/mL for Xpert MTB/RIF (Chakravorty 2017). Of note, Xpert Ultra has added a new result category, ‘trace call', that corresponds to the lowest bacillary burden for M tuberculosis detection (WHO Xpert Ultra 2017). Although no result for rifampicin resistance will be available for people with trace results, a trace‐positive result is sufficient to initiate anti‐tuberculosis therapy in children or HIV‐positive people, according to the WHO report. Xpert Ultra is available for clinical use and several countries have moved from using Xpert MTB/RIF to using Xpert Ultra instead. In this Cochrane Review, we will include studies that used any generation of the index tests.
Clinical pathway
There are two complementary approaches to detection of active tuberculosis, Figure 1. The first is the patient‐initiated pathway, also known as passive case finding. The second is the provider‐initiated screening pathway, which represents the analytic framework for this review (WHO Systematic screening 2015). The index test, either Xpert MTB/RIF or Xpert Ultra, would be performed as the only test for pulmonary tuberculosis and rifampicin resistance in adults, irrespective of signs or symptoms of pulmonary tuberculosis, in high‐risk groups and in primary health facilities or community settings.
1.
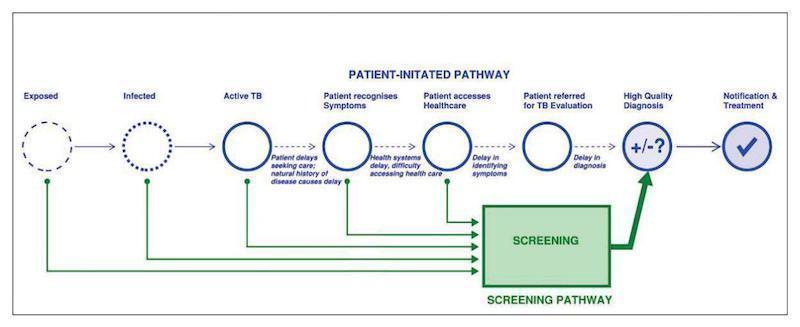
There are two complementary approaches to detection of active tuberculosis. The first is the patient‐initiated pathway, also known as passive case finding. The second is the provider‐initiated screening pathway (WHO Systematic screening 2015), which represents the analytic framework for this review. In the latter pathway, the index test would be applied as the only test, to adults, irrespective of signs and symptoms of tuberculosis, in high‐risk groups and in primary health facilities or community settings.
The purpose of the index tests is screening.
The role of the index tests is replacement for usual practice. This may include replacement for the WHO four‐question symptom screen.
The downstream consequences of screening include the following.
True‐positive (TP): patients would benefit from rapid diagnosis and initiation of appropriate treatment.
True‐negative (TN): patients would be spared unnecessary treatment and would benefit from reassurance, pursuit of an alternative diagnosis if they have symptoms, and determination of eligibility for tuberculosis preventive therapy if indicated.
False‐positive (FP): patients would probably experience anxiety and morbidity caused by additional testing, unnecessary treatment, and possible adverse events; possible stigma associated with a tuberculosis or MDR‐TB diagnosis; and the chance that a false‐positive result may halt further diagnostic evaluation of the true underlying condition.
False‐negative (FN): patients would experience an increased risk of morbidity and mortality, and delayed or inappropriate treatment initiation; there would be risk of ongoing tuberculosis transmission.
Alternative test(s)
Alternative screening tests for tuberculosis include no screening (or passive case‐finding), and one or more of symptom screening (such as the WHO four‐question symptom screen) and chest X‐ray, which must be further confirmed with a diagnostic test. Other tools that may be useful in screening include urine lipoarabinomannan (LAM) testing and smear microscopy, which require additional definitive drug resistance testing even if used as simultaneous screening and diagnostic tests. We have previously described selected alternative tests for detection of pulmonary tuberculosis and rifampicin resistance (Horne 2019; Lewinsohn 2017; Unitaid 2017). Recently, we published a Special Collection, curated by Cochrane contributors, that includes Cochrane Reviews from Cochrane Infectious Diseases and other systematic reviews from other international teams. The Special Collection describes key WHO guidelines on tuberculosis diagnostics, and their underpinning systematic reviews (Cochrane Special Collection 2019). Below we review screening tools and highlight several recent developments in tuberculosis diagnostics.
Numerous symptoms, singly and in combination, have been proposed to screen for tuberculosis in different settings. A healthcare or community worker asks the person being screened if they are experiencing any of the selected symptoms, and those who report symptoms according to local criteria go on to receive additional testing such as chest X‐ray or diagnostic testing. The most commonly assessed symptoms are cough (varying duration), fever, weight loss, drenching night sweats, loss of appetite, haemoptysis, and fatigue. Single symptoms have modest to low sensitivity; defining a positive screen as any one or more of multiple symptoms improves sensitivity but reduces specificity, consequently increasing the number of diagnostic confirmatory tests. Accuracy of symptom screening varies with the HIV status of the people screened. One study found that any one of cough of any duration, fever of any duration, or night sweats lasting three or more weeks was the most sensitive combination of symptoms for identification of tuberculosis in people living with HIV (93% sensitivity, 36% specificity; Cain 2010). In mixed HIV‐positive and HIV‐negative populations, a single symptom of cough of greater than two weeks' duration identified 35% (95% confidence interval (CI) 24 to 46) of adults with culture‐positive pulmonary tuberculosis in one systematic review and modelling analysis; any one of a list of tuberculosis symptoms had 70% sensitivity and 61% specificity for pulmonary tuberculosis in low‐HIV‐prevalence settings (van't Hoog 2013).
Chest X‐ray can involve posterior‐anterior, anterior‐posterior, or lateral recording, or a combination of two or all of these. Major types of chest X‐ray include conventional chest X‐ray (producing 36 cm x 43 cm film), digital radiography, and computed radiography. Chest X‐ray findings including hilar lymphadenopathy, cavitary lesions, and evidence of granulomas can all suggest pulmonary tuberculosis, but are also nonspecific and must be confirmed with additional testing. Accurate interpretation of pulmonary tuberculosis findings on chest X‐ray are dependent on the ability of the individual interpreting the chest X‐ray, and wide inter‐observer variation has been reported (Zellweger 2006). Computer‐aided interpretation of chest X‐ray for pulmonary tuberculosis is a promising new technology, especially for resource‐limited settings where expertise in chest X‐ray interpretation is limited (Harris 2019).
Smear microscopy is the examination of smears for acid‐fast bacilli (tuberculosis bacteria) under a microscope. The examination may be performed by light microscopy (Ziehl‐Neelsen), fluorescence microscopy, or light‐emitting diode (LED) fluorescence microscopy. Microscopy cannot distinguish between drug‐susceptible tuberculosis and drug‐resistant tuberculosis. The WHO recommends that microscopy, as the initial diagnostic test, should be replaced with WHO‐recommended rapid tests that can simultaneously detect tuberculosis and tuberculosis drug resistance (WHO Compendium of WHO guidelines 2018).
Nucleic acid amplification (NAA) tests are molecular systems that can detect small quantities of genetic material (DNA or RNA) from micro‐organisms, such as M tuberculosis. The key advantage of NAA tests is that they are rapid diagnostic tests, potentially providing results in a few hours. Several new commercial NAA tests are in the diagnostic pipeline or have recently come to market (e.g. Truenat MTB, Truenat MTBplus, and Truenat MTB‐RIF Dx, Molbio Diagnostics, India). Truenat MTB, MTB Plus and MTB‐RIF Dx assays show comparable accuracy with Xpert MTB/RIF and Xpert Ultra for detection of tuberculosis (Truenat MTB and Truenat MTB Plus), and for sequential detection of rifampicin resistance (Truenat MTB‐Rif Dx). The WHO recommends these tests as initial tests for the diagnosis of tuberculosis and rifampicin resistance (WHO Rapid Communication 2020).
Alere Determine TB LAM Ag (AlereLAM, Alere Inc, Waltham, USA) is a commercially available, point‐of‐care test for tuberculosis disease (pulmonary and extrapulmonary tuberculosis). The test detects lipoarabinomannan (LAM), a component of the bacterial cell wall, which is present in the urine of some people with tuberculosis. AlereLAM is performed by placing urine on one end of a test strip, with results appearing as a band on the strip if tuberculosis is present. The test is simple, requires no special equipment, and shows results in 25 minutes (Bjerrum 2019). In two randomized trials, the use of Alere LAM in HIV‐positive inpatients has been shown to reduce mortality (Gupta‐Wright 2018; Peter 2016). Based on evidence from the randomized trials and a Cochrane Review (Bjerrum 2019), the WHO recommends that AlereLAM should be used to assist in the diagnosis of active tuberculosis in HIV‐positive adults, adolescents and children. The full recommendations, which differ for inpatients and outpatients, are described here: WHO Lateral flow LAM 2019.
Fujifilm SILVAMP TB LAM (FuijiLAM, co‐developed by Foundation for Innovative New Diagnostics (FIND), Geneva, Switzerland and Fujifilm, Tokyo, Japan) is a new, urine‐based, point‐of‐care test for tuberculosis diagnosis in people living with HIV. In an individual participant data meta‐analysis that included five cohorts of people living with HIV, FujiLAM was found to have superior sensitivity, 70.7% (95% CI 59.0% to 80.8%), compared to AlereLAM sensitivity of 42.3% (31.7% to 51.8%), against a microbiological reference standard; FujiLAM had lower specificity, 90.9% (87.2% to 93.7%), compared to AlereLAM specificity of 95.3% (92.2% to 97.7%) (Broger 2020).
Alternative molecular methods for drug susceptibility testing include the commercial line probe assays, GenoType MTBDRplus assay (MTBDRplus, Hain LifeScience, Nehren, Germany), and the Nipro NTM+MDRTB detection kit 2 (Nipro, Tokyo, Japan), which detect the presence of mutations associated with drug resistance to isoniazid and rifampicin (WHO LPA 2016). Advantages of line probe assays are that they can provide a result for detection of tuberculosis and drug resistance in one to two days. Drawbacks are that line probe assays are expensive and need to be used in intermediate and central laboratories (Unitaid 2017).
Rationale
Xpert MTB/RIF and Xpert Ultra provide obvious benefits for patients (earlier diagnosis and the opportunity to begin earlier, appropriate treatment), and for public health (opportunities to interrupt tuberculosis transmission), especially in countries with a high tuberculosis burden.
Since 2010, the WHO has recommended the use of Xpert MTB/RIF as the preferred initial diagnostic test for people thought to have MDR‐TB or HIV‐associated tuberculosis (WHO 2011). In 2013, the WHO expanded the recommendations, stating that Xpert MTB/RIF may be used rather than conventional microscopy and culture as the initial diagnostic test in all adults suspected of having tuberculosis (conditional recommendation acknowledging resource implications, high‐quality evidence; WHO Xpert MTB/RIF 2013). In addition, the WHO recommended that following an Xpert MTB/RIF test that demonstrates rifampicin resistance, subsequent drug susceptibility testing (e.g. using a line probe assay for second‐line drugs) remains essential to detect resistance to drugs other than rifampicin (WHO Xpert MTB/RIF 2013). In 2017, based on a non‐inferiority analysis of Xpert Ultra compared with Xpert MTB/RIF (Dorman 2018), the WHO stated that recommendations on the use of Xpert MTB/RIF also apply to the use of Xpert Ultra as the initial diagnostic test for all adults and children with signs and symptoms of tuberculosis (WHO Xpert Ultra 2017).
We are interested in whether a single test, Xpert MTB/RIF or Xpert Ultra, can be useful to identify people with active pulmonary tuberculosis in high‐risk populations in community settings or attending healthcare settings for reasons unrelated to tuberculosis. This is a different approach than diagnosing active tuberculosis in people with signs and symptoms of tuberculosis who seek care in health facilities. We performed this Cochrane Review to inform an updated WHO policy review on tuberculosis screening, 2020 Revision of the Guidelines for Systematic Screening for Active Tuberculosis: Updated and Consolidated Recommendations and Implementation Guidance. The 2020 WHO guidelines will also include Cochrane and non‐Cochrane systematic reviews on symptom screening, chest radiography, and other tests and strategies for screening for tuberculosis in adults and children.
Objectives
To determine the accuracy of Xpert MTB/RIF and Xpert Ultra for screening for tuberculosis in adults irrespective of signs or symptoms of pulmonary tuberculosis in the general population (i.e. low‐risk population).
To determine the accuracy of Xpert MTB/RIF and Xpert Ultra for screening of pulmonary tuberculosis in adults in the following high‐risk groups.
People living with HIV.
Household contacts of people with tuberculosis.
Patients residing in high‐tuberculosis‐burden settings attending primary health facilities.
Homeless people.
Miners.
People with diabetes mellitus.
People who abuse alcohol.
Smokers.
People residing in prisons.
Healthcare workers.
To determine the accuracy of Xpert MTB/RIF and Xpert Ultra for the detection of rifampicin resistance in the general population and in the high‐risk groups and settings described above.
Secondary objectives
To compare the accuracy of Xpert MTB/RIF and Xpert Ultra in the above high‐risk groups and settings.
To investigate potential sources of heterogeneity in accuracy estimates, including the percentage of participants with tuberculosis symptoms, tuberculosis burden, tuberculosis/HIV burden, and MDR‐TB burden.
Methods
Criteria for considering studies for this review
Types of studies
We will include cross‐sectional studies and cohort studies that assessed the accuracy of one or both index tests for both pulmonary tuberculosis and rifampicin resistance or pulmonary tuberculosis alone. We will use abstracts to identify published studies and include the full publications when they meet our inclusion criteria. We will only include studies that reported data comparing the index test(s) to an acceptable reference standard from which we could extract true positive (TP), true negative (TN), false positive (FP), and false negative (FN) values. The index tests could be assessed alone or together with other tests. We will include studies designed to find people with active tuberculosis in community settings.
We will exclude case reports and studies with a case‐control design, the latter because these types of studies are prone to bias, in particular, studies enrolling participants with severe disease and healthy participants without disease. We will exclude drug resistance surveys.
Participants
Adults, defined as 15 years of age and older, irrespective of signs or symptoms of pulmonary tuberculosis in the general population, and adults in high‐risk populations, including the following populations.
People living with HIV.
Household contacts of people with tuberculosis.
Patients attending primary health facilities.
Homeless people.
Miners.
People with diabetes mellitus.
People who abuse alcohol.
Smokers.
People residing in prisons.
Healthcare workers.
The settings of interest are primary healthcare facilities and other community settings.
We will exclude studies that selected participants for enrolment based on the results of prior tuberculosis testing, such as symptom screening or chest radiography.
Index tests
Sputum Xpert MTB/RIF and sputum Xpert Ultra
Index test results are automatically generated (i.e. there is a single threshold), and the user is provided with a printable test result as follows.
MTB (M tuberculosis) DETECTED.
Rif (rifampicin) resistance DETECTED.
MTB DETECTED; Rif resistance NOT DETECTED.
MTB DETECTED; Rif resistance INDETERMINATE.
MTB NOT DETECTED.
INVALID (the presence or absence of MTB cannot be determined).
ERROR (the presence or absence of MTB cannot be determined).
NO RESULT (the presence or absence of MTB cannot be determined)
Xpert Ultra incorporates a semi‐quantitative classification for results: trace, very low, low, moderate, and high. ‘Trace' corresponds to the lowest bacterial burden for detection of M tuberculosis (Chakravorty 2017). We will consider a trace result to mean MTB detected.
Target conditions
The target conditions are active pulmonary tuberculosis and rifampicin resistance.
Reference standards
For tuberculosis, the reference standards are solid culture or automated liquid culture.
For rifampicin resistance, the reference standards are culture‐based drug susceptibility testing (DST) and line probe assays (WHO LPA 2016). Acceptable methods for DST are the proportion method, performed on solid media, such as Lowenstein‐Jensen, and use of a commercial liquid culture system, such as Mycobacteria Growth Indicator Tube (MGIT) 960 automated mycobacterial detection system (BD, USA).
Search methods for identification of studies
We will attempt to identify all relevant studies regardless of language or publication status (published, unpublished, in press, and ongoing).
Electronic searches
We will search the following databases without language restriction, using the search terms and strategy described in Appendix 1:
Cochrane Infectious Diseases Specialized Register.
MEDLINE (OVID, from 1966).
Embase (OVID, from 1974).
Science Citation Index ‐ Expanded (from 1900), Conference Proceedings Citation Index ‐ Science (CPCI‐S, from 1990), and BIOSIS Previews (from 1926); all three from the Web of Science.
Scopus (Elsevier, from 1970).
Latin American Caribbean Health Sciences Literature (LILACS) (BIREME, from 1982).
We will also search ClinicalTrials.gov, the WHO International Clinical Trials Registry Platform (ICTRP; www.who.int/trialsearch), and the International Standard Randomized Controlled Trials Number (ISRCTN) registry (www.isrctn.com/), for trials in progress, and ProQuest Dissertations & Theses A&I (from 1990) for dissertations.
Searching other resources
We will review reference lists of included articles and any relevant review articles identified through the above methods. We will also contact researchers at the Foundation for Innovative New Diagnostics (FIND), the WHO Global Tuberculosis Programme, and other experts in the field of tuberculosis diagnostics for information on ongoing and unpublished studies.
Data collection and analysis
Selection of studies
We will use Covidence to manage the selection of studies (Covidence). Two review authors will independently and in parallel scrutinize titles and abstracts identified from literature searching to identify potentially eligible studies. We will retrieve the article of any citation, identified by any review author, for full‐text review. Then, two review authors will independently and in parallel assess articles for inclusion using the predefined selection criteria. We will resolve any discrepancies by discussion or with a third review author. We will record all studies excluded after full‐text assessment, along with our reasons for their exclusion in the Characteristics of excluded studies table and illustrate the study selection process in a PRISMA diagram (Moher 2009).
Data extraction and management
We will extract data on the following characteristics.
Author, publication year, study design, country where study was located, clinical setting.
Population characteristics: age, gender, smear status, HIV status.
Index test(s), Xpert MTB/RIF or Xpert Ultra.
Reference standard.
Quality Assessment of Studies of Diagnostic Accuracy ‐ Revised (QUADAS‐2) items (Whiting 2011).
Number of TP, FP, FN, and TN (i.e. true positives, false positives, false negatives, and true negatives, with respect to culture).
Number of uninterpretable results for detection of pulmonary tuberculosis.
Number of indeterminate results for detection of rifampicin resistance.
We will classify country income status as either low‐ and middle‐income or high‐income, according to the World Bank List of Economies (World Bank 2019). In addition, we will classify ‘country' as being high burden or not high burden for tuberculosis, TB/HIV, or MDR‐TB, according to the classification by the WHO (WHO Global TB Report 2019).
Assessment of methodological quality
We will use the QUADAS‐2 tool, tailored to this review, to assess the quality of the included studies (Whiting 2011; Appendix 2). QUADAS‐2 consists of four domains: patient selection, index test, reference standard, and flow and timing. We will assess all domains for risk of bias and the first three domains for concerns regarding applicability. We will present the results of this quality assessment in text, tables, and graphs.
Statistical analysis and data synthesis
We will perform descriptive analyses for the results of the included studies using Stata 15 (Stata). We will determine sensitivity and specificity estimates and 95% confidence intervals (CIs) for individual studies and generate forest plots using Review Manager 5 (Review Manager 2020).
When possible, we will carry out meta‐analyses to estimate the pooled sensitivity and specificity of the index tests separately for tuberculosis detection and rifampicin resistance detection. We will determine pooled accuracy estimates using an adaptation of the bivariate random‐effects model of Reitsma 2005, which uses the exact binomial likelihood for the observed proportions (Chu 2006). The bivariate random‐effects approach will allow us to calculate the pooled estimates of sensitivity and specificity while accounting for
variation in sensitivity and specificity estimates within individual studies;
correlation between sensitivity and specificity across studies; and
variation in sensitivity and specificity between studies.
For analysis of Xpert MTB/RIF or Xpert Ultra accuracy for detection of rifampicin resistance, we will include participants who
were culture‐positive;
had a valid phenotypic drug susceptibility test (DST) or line probe assay (LPA) result;
were Xpert MTB/RIF or Xpert Ultra tuberculosis‐positive; and
had a valid Xpert MTB/RIF or Xpert Ultra result for rifampicin resistance, detected or not detected (susceptible).
Sensitivity = Xpert MTB/RIF (or Xpert Ultra) rifampicin resistance detected/phenotypic DST or LPA rifampicin‐resistant
Specificity = Xpert MTB/RIF (or Xpert Ultra) rifampicin resistance not detected/phenotypic DST or LPA rifampicin‐susceptible
We will estimate all models using a Bayesian approach, with low‐information prior distributions, using OpenBUGS software (Version 3.2.3; Lunn 2009), along with R (Version 3.3.2; R Core Team 2019). Under the Bayesian approach, all unknown parameters must be provided a prior distribution that defines the range of possible values of the parameter and the likelihood of each of those values based on information external to the data. In order to let the observed data determine the final results, we will choose to use low‐information prior distributions over the pooled sensitivity and specificity parameters and their between‐study standard deviation parameters.
It is known that meta‐analysis models can be sensitive to the choice of prior distributions over between‐study standard deviation parameters. We will therefore carry out sensitivity analyses and consider alternative prior distributions that are less informative, allowing a wider range of possible values. To study the sensitivity of all results to the choice of prior distributions, we will consider alternative prior distributions that are less informative, allowing a wider range of possible values. We will include information from the prior distribution in combination with the observed data in accordance with Bayes' theorem to obtain a posterior distribution for each unknown parameter.
Using a sample from the posterior distribution, we can obtain various descriptive statistics of interest. We will estimate the median pooled sensitivity and specificity and their 95% credible intervals (CrIs). The median or the 50% quantile is the value below which lies 50% of the posterior sample. We will report the median because the posterior distributions of some parameters may be skewed and the median would be considered a better point estimate of the unknown parameter than the mean in such cases. The 95% CrI is the Bayesian equivalent of the classical (frequentist) 95% CI. (We will indicate 95% CI for individual study estimates and 95% CrI for pooled study estimates, as appropriate.) The 95% CrI may be interpreted as an interval that has a 95% probability of capturing the true value of the unknown parameter, given the observed data and the prior information.
We will also estimate the ‘predicted' sensitivity and specificity in a future study together with their 95% CrIs. The predicted estimate is our best guess for the estimate in a future study and is the same as the pooled estimate. The CrIs, however, may be different. These values are derived from the predicted region typically reported in a bivariate meta‐analysis plot. If there is no heterogeneity at all between studies, the CI (or CrI) around the predicted estimate will be the same as the CI around the pooled estimate. On the other hand, if there is considerable heterogeneity between studies, the CI around the predicted estimate will be much wider than the CI around the pooled estimate. We will generate bivariate plots of the credible and prediction regions in the receiver operating characteristic (ROC) space using R (version 3.3.2; R Core Team 2019).
As we anticipate finding few if any studies that compare the accuracy of Xpert MTB/RIF and Xpert Ultra in the selected high‐risk groups and settings, we plan to analyse the accuracy estimates descriptively in text, tables, and forest plots.
Approach to uninterpretable index test results
The index tests report an uninterpretable test result for unexpected results with any of the internal control measures of the assay.
The uninterpretable rate for detection of pulmonary tuberculosis is the number of tests classified as ‘invalid', ‘error', or ‘no result' divided by the total number of index tests performed.
The uninterpretable rate for detection of rifampicin resistance (referred to as indeterminate rate) is the number of tests classified as ‘MTB detected; Rif (rifampicin) resistance INDETERMINATE‘ divided by the total number of index test‐positive results.
In previous reviews, we found very few uninterpretable results reported, and chose to exclude them from the bivariate meta‐analyses (Horne 2019). Instead, we used a Bayesian hierarchical model for a single proportion to estimate the pooled proportion of uninterpretable index test results and anticipate that we will do the same in this review.
Investigations of heterogeneity
We will visually inspect forest plots and the summary receiver operating characteristic (SROC) plots for heterogeneity. If the data allow, we will investigate sources of heterogeneity using bivariate meta‐regression analyses. We will consider each source of heterogeneity to be a single covariate affecting the pooled sensitivity or the specificity, or both, in a bivariate meta‐analysis model. We plan to assess the following as categorical study‐level covariates.
Detection of pulmonary tuberculosis
Percentage of participants with tuberculosis symptoms, 50% or more with tuberculosis symptoms and less than 50% tuberculosis symptoms
High tuberculosis burden, yes or no
High TB/HIV burden, yes or no
Detection of rifampicin resistance
High MDR‐TB burden, yes or no
Sensitivity analyses
If there are sufficient data, we will perform sensitivity analyses to explore the effect of risk of bias, population characteristics, and other factors on the accuracy of Xpert MTB/RIF and Xpert Ultra. Specifically, we will limit inclusion in the meta‐analyses to the following:
studies that explicitly represented the use of the index tests for the screening of individuals irrespective of signs and symptoms of tuberculosis;
studies that used liquid culture as the reference standard;
studies where a consecutive or random sample of participants were enrolled. We will exclude studies where we answered no or unclear to the QUADAS‐2 patient selection signalling question: "Could the selection of patients have introduced bias?";
studies where the reference standard was blinded. We will exclude studies where we answered no or unclear to the QUADAS‐2 Reference standard signalling question: "Were the reference standard results interpreted without knowledge of the results of the index test?";
studies that accounted for all participants in the analysis. We will exclude studies where we answered ‘no' or ‘unclear' to the QUADAS‐2 flow and timing signalling question: "Were all patients included in the analysis?".
Assessment of reporting bias
We will not formally assess reporting bias using funnel plots or regression tests as these have not been reported as helpful for diagnostic test accuracy studies (Macaskill 2010).
Summary of findings and assessment of the certainty of the evidence
We will assess the certainty of the evidence using the GRADE approach (Balshem 2011; Schünemann 2008; Schünemann 2016), and GRADEpro GDT 2015 software. In the context of a systematic review, ratings of the certainty of the evidence reflect the extent of our confidence that the estimates of effect (including test accuracy and associations) are correct. As recommended, we will rate the certainty of the evidence as either high (not downgraded), moderate (downgraded by one level), low (downgraded by two levels), or very low (downgraded by more than two levels) for five domains: risk of bias, indirectness, inconsistency, imprecision, and publication bias.
For each outcome, we will consider the certainty of the evidence to begin as high when high‐quality observational studies (cross‐sectional or cohort studies) enrolled participants with diagnostic uncertainty. If we have a reason for downgrading, we will use our judgement to classify the reason as serious (downgraded by one level) or very serious (downgraded by two levels). We will summarize this information in the ‘Summary of findings' tables.
As recommended, we will apply GRADE in the following ways (Schünemann 2020a; Schünemann 2020b).
Risk of bias: we will use QUADAS‐2 to assess risk of bias.
Indirectness: we will assess indirectness in relation to the population (including disease spectrum), setting, interventions, and outcomes (accuracy measures). For example, we will note whether the population was the same in the studies compared to the question asked. We will also use prevalence as a guide to whether there was indirectness in the population.
Inconsistency: GRADE recommends downgrading for unexplained inconsistency in sensitivity and specificity estimates. We will carry out prespecified analyses and downgrade only when we cannot explain inconsistency in the accuracy estimates.
Imprecision: we will consider a precise estimate to be one that would allow a clinically meaningful decision. We will consider the width of the CrI and ask ourselves, ‘Would we make a different decision if the lower or upper boundary of the CrI represented the truth?’ In addition, we will determine projected ranges for true positives (TP), false negatives (FN), true negatives (TN), and false positives (FP) for a given prevalence of tuberculosis and make judgements on imprecision from these calculations.
Publication bias: we will consider the comprehensiveness of the literature search and outreach to researchers in tuberculosis, the presence of only studies that produce precise estimates of high accuracy despite small sample size, and knowledge about studies that were conducted, but are not published.
History
Protocol first published: Issue 7, 2020
Acknowledgements
The Academic Editor is Professor Yemisi Takwoingi.
We are grateful to Vittoria Lutje, Cochrane Infectious Diseases' Information Specialist, for help with the search strategy.
The editorial base of Cochrane Infectious Diseases is funded by UK aid from the UK government for the benefit of low‐ and middle‐income countries (project number 300342‐104). The views expressed do not necessarily reflect the UK government’s official policies.
Appendices
Appendix 1. Search strategy
MEDLINE (PubMed)
| Search | Query |
| #1 | Search Tuberculosis or MDR‐TB or XDR‐TB or tuberculous Field: Title/Abstract |
| #2 | Search “Mycobacterium tuberculosis” [Mesh] |
| #3 | Search "Tuberculosis"[Mesh] or ("Tuberculosis, Multidrug‐Resistant"[Mesh]) OR "Extensively Drug‐Resistant Tuberculosis"[Mesh] |
| #4 | Search ((#3) OR #2) OR #1 |
| #5 | Search Xpert* or GeneXpert or Ultra or cepheid Field: Title/Abstract |
| #6 | Search "near* patient*" or near‐patient Field: Title/Abstract |
| #7 | Search (#6) OR #5 |
| #8 | Search "active case" Field: Title/Abstract |
| #9 | Search "case finding" Field: Title/Abstract |
| #10 | Search prevalence Field: Title/Abstract |
| #11 | Search Asymptomatic Field: Title/Abstract |
| #12 | Search comorbidity or co‐morbidity Field: Title/Abstract |
| #13 | Search screening Field: Title/Abstract |
| #14 | Search Detect* or missed or undetect* or undiagnosed Field: Title/Abstract |
| #15 | Search ((((((#14) OR #13) OR #12) OR #11) OR #10) OR #9) OR #8 |
| #16 | Search (#4) AND #7 AND #15 |
Database: Embase 1947‐present, updated daily
Search Strategy: ‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐ 1 (tuberculosis or TB).mp. 2 Tuberculosis, Multidrug‐Resistant/ or Extensively Drug‐Resistant Tuberculosis/ or Tuberculosis/ or tuberculosis.mp. or Mycobacterium tuberculosis/ 3 (MDR‐TB or XDR‐TB).mp. 4 1 or 2 or 3 5 Xpert* MTB RIF.ti. or Xpert* MTB RIF.ab. 6 (Xpert* or GeneXpert or cepheid).mp. 7 (near* patient or near‐patient).ti. or (near* patient or near‐patient).ab. 8 5 or 6 or 7 9 4 and 8 10 detection.mp. 11 diagnostic error/ or missed.mp. 12 (undetected or undiagnosed).mp. 13 asymptomatic.mp. 14 comorbidity.mp. or comorbidity/ 15 prevalence/ 16 active case finding.mp. or case finding/ 17 10 or 11 or 12 or 13 or 14 or 15 or 16 18 9 and 17
Web of Science
| # | 3 | #2 AND #1 Indexes=SCI‐EXPANDED, SSCI, CPCI‐S, CPCI‐SSH Timespan=All years |
| # | 2 |
TOPIC: (asymptomatic or undetected or undiagnosed) ORTOPIC: ("case finding" or prevalence or comorbidity) Indexes=SCI‐EXPANDED, SSCI, CPCI‐S, CPCI‐SSH Timespan=All years |
| # | 1 |
TOPIC: (tuberculosis OR tb OR mycobacterium) ANDTOPIC: (xpert* OR genexpert OR cepheid) Indexes=SCI‐EXPANDED, SSCI, CPCI‐S, CPCI‐SSH Timespan=All years |
Scopus
( TITLE‐ABS‐KEY ( tuberculosis OR tb OR mycobacterium ) AND TITLE‐ABS‐KEY ( ( xpert* OR genexpert OR cepheid ) ) AND TITLE‐ABS‐KEY ( asymptomatic OR undetected OR undiagnosed OR "case finding" OR prevalence OR comorbidity ) )
LILAC
(tuberculosis OR TB OR mycobacterium) (Words) AND (xpert OR Genexpert OR Cepheid) (Words)
Appendix 2. QUADAS‐2
In QUADAS‐2, we will assess methodological quality separately for each of the objectives, Xpert for pulmonary tuberculosis detection and Xpert for rifampicin resistance detection.
Domain 1: patient selection
Xpert MTB/RIF and Xpert Ultra for detection of pulmonary tuberculosis
Risk of bias: could the selection of patients have introduced bias?
Signalling question 1: was a consecutive or random sample of patients enrolled? We will answer ‘yes' if the study enrolled a consecutive or random sample of eligible patients; ‘no' if the study selected patients by convenience; and ‘unclear' if the study did not report the manner of patient selection or we cannot tell.
Signalling question 2: did the study avoid inappropriate exclusions? We will answer ‘yes' if the study included all individuals in the general population or the high‐risk group considered for tuberculosis screening. We will answer ‘no' if the study primarily or exclusively included individuals with a history of tuberculosis; individuals who had undergone previous treatment (retreatment patients); or those with signs and symptoms of tuberculosis. We will answer ‘unclear' if we cannot tell.
Applicability: are there concerns that the included patients and setting do not match the review question?
We are interested in how Xpert MTB/RIF or Xpert Ultra perform in patients who were evaluated as they would be in the settings of intended use. We will answer ‘low concern' if the study population resembled a population that was selected for tuberculosis screening in community settings or primary care centres. We will answer ‘high concern' if the study population does not resemble a population that was selected for tuberculosis screening in a community setting. We will answer ‘unclear concern' if there was insufficient information to make a decision.
Xpert MTB/RIF and Xpert Ultra for Xpert MTB/RIF or Xpert Ultra rifampicin resistance
Domain 1: patient selection is the same as for MTB/RIF or Xpert Ultra for pulmonary tuberculosis.
Domain 2: index test
Xpert MTB/RIF and Xpert Ultra for detection of pulmonary tuberculosis
Risk of bias: could the conduct or interpretation of the index test have introduced bias?
Signalling question 1: were the index test results interpreted without knowledge of the results of the reference standard? We will answer this question ‘yes' for all studies because Xpert test results were automatically generated and the user was provided with printable test results. Thus, there is no room for subjective interpretation of test results.
Signalling question 2: if a threshold was used, was it prespecified? The threshold was prespecified in all versions of Xpert. We will answer this question ‘yes' for all studies.
For risk of bias, we judge ‘low concern' for all studies.
Applicability: are there concerns that the index test, its conduct, or its interpretation differ from the review question? Variations in test technology, execution, or interpretation may affect estimates of the diagnostic accuracy of a test. All steps in the Xpert MTB/RIF and Xpert Ultra assays are completely automated and self‐contained following sample loading. We will answer ‘low concern' if the index test was performed as recommended by the manufacturer, which we anticipate will be true for most studies. We will answer ‘unclear concern' if the ratio of the Xpert MTB/RIF or Xpert Ultra sample reagent: specimen volume was not 2:1 for a raw specimen or 3:1 for a sediment, as recommended by the manufacturer.
Xpert MTB/RIF and Xpert Ultra for detection of rifampicin resistance
Domain 2: index test is the same as for MTB/RIF or Xpert Ultra for pulmonary tuberculosis.
Domain 3: reference standard
Xpert MTB/RIF and Xpert Ultra for detection of pulmonary tuberculosis
Risk of bias: could the reference standard, its conduct, or its interpretation have introduced bias?
Signalling question 1: is the reference standard likely to correctly classify the target condition?
We will answer ‘yes' for all studies, since culture as a reference standard was a criterion for inclusion in the review.
Signalling question 2: were the reference standard results interpreted without knowledge of the results of the index test?
We will answer ‘yes' if the reference test provided an automated result (for example, MGIT 960), blinding was explicitly stated, or it was clear that the reference standard was performed at a separate laboratory and/or performed by different people. We will answer ‘no' if the study stated that the reference standard result was interpreted with knowledge of the Xpert MTB/RIF or Xpert Ultra test result. We will answer ‘unclear' if we cannot tell.
Applicability: are there concerns that the target condition as defined by the reference standard does not match the question? We will answer ‘high concern' if included studies did not speciate mycobacteria isolated in culture; ‘low concern' if speciation was performed; and ‘unclear concern' if we cannot tell.
Xpert MTB/RIF and Xpert Ultra for detection of rifampicin resistance
Risk of bias: could the selection of patients have introduced bias?
Signalling question 1: is the reference standard likely to correctly classify the target condition?
We will answer ‘yes' if either culture‐based drug susceptibility testing (DST) or line probe assay (such as MTBDRplus) was used. These are the criteria for inclusion for this objective of the review.
Signalling question 2: were the reference standard results interpreted without knowledge of the results of the index test?
We will answer ‘yes' if the reference test provided an automated result (for example, MGIT 960), blinding was explicitly stated, or it was clear that the reference standard was performed at a separate laboratory and/or performed by different people. We will answer ‘no' if the study stated that the reference standard result was interpreted with knowledge of the Xpert MTB/RIF or Xpert Ultra test result. We will answer ‘unclear' if we cannot tell.
Applicability: are there concerns that the target condition as defined by the reference standard does not match the question? We will judge applicability to be of ‘low concern' for those studies evaluating Xpert MTB/RIF or Xpert Ultra for rifampicin resistance because these specimens had already been identified as Mycobacterium tuberculosis positive.
Domain 4: flow and timing
Xpert MTB/RIF and Xpert Ultra for detection of pulmonary tuberculosis detection
Risk of bias: could the patient flow have introduced bias?
Signalling question 1: was there an appropriate interval between the index test and reference standard? In most included studies, we expect that specimens for Xpert MTB/RIF or Xpert Ultra and culture would be obtained at the same time, when patients were screened. However, even if there were a delay of several days between index test and reference standard, tuberculosis is a chronic disease and we considered misclassification of disease status to be unlikely, as long as treatment was not initiated in the interim. We will answer ‘yes' if the index test and reference standard were performed at the same time or if the time interval was less than or equal to seven days, ‘no' if the time interval is greater than seven days, and ‘unclear' if we cannot tell.
Signalling question 2: did all patients receive the same reference standard? We will answer this question ‘yes' for all studies as an acceptable reference standard (either solid or liquid culture) was specified as a criterion for inclusion in the review. However, we acknowledge that it is possible that some specimens could undergo solid culture and others liquid culture. This could potentially result in variations in accuracy, but we think the variation would be minimal.
Signalling question 3: were all patients included in the analysis? We will determine the answer to this question by comparing the number of patients enrolled with the number of patients included in the 2 x 2 tables. We will answer ‘yes' if the numbers matched and ‘no' if there were patients enrolled in the study that were not included in the analysis. We will answer ‘unclear' if we cannot tell.
Xpert MTB/RIF and Xpert Ultra for detection of rifampicin resistance
Domain 4: flow and timing is the same as for Xpert MTB/RIF or Xpert Ultra for pulmonary tuberculosis.
Judgements for ‘Risk of bias' assessments for a given domain
If we answered all signalling questions for a domain ‘yes', then we will judge risk of bias as ‘low'.
If we answered all or most signalling questions for a domain ‘no', then we will judge risk of bias as ‘high'.
If we answered only one signalling question for a domain ‘no', we will discuss further the risk of bias judgement.
If we answered all or most signalling questions for a domain ‘unclear', then we will judge risk of bias as ‘unclear'.
If we answered only one signalling question for a domain ‘unclear', we will discuss further the risk of bias judgement for the domain.
Contributions of authors
AES, JMR, MK, KRS, and DJH drafted the manuscript. ND and IS wrote the statistical analysis section. All review authors read and approved the final manuscript draft.
Sources of support
Internal sources
Liverpool School of Tropical Medicine, UK
External sources
-
Department for International Development, UK
Project number 300342‐104
-
United States Agency for International Development (USAID), USA
Development of this project was in part made possible with financial support from USAID administered by the World Health Organization Global TB Programme
Declarations of interest
AES received funding from USAID, administered by the World Health Organization Global Tuberculosis Programme, Switzerland. She has received salary compensation from the University of Washington, where she is an Acting Assistant Professor in Global Health and Medicine/Infectious Diseases. A portion of her salary derives from NIH grants and from grants from the Bill & Melinda Gates Foundation.
JMR received funding from USAID, administered by the World Health Organization Global Tuberculosis Programme, Switzerland. JMR has grants/grants pending to her host institution from US National Institutes of Health, KNCV TB Foundation, and The Global Fund to Fight AIDS, TB, and Malaria, The Firland Foundation.
IS has no known conflicts of interest to declare.
MK has received funding from USAID, administered by the World Health Organization Global Tuberculosis Programme, Switzerland for related systematic reviews.
ND has no known conflicts of interest to declare.
KRS has received financial support from Cochrane Infectious Diseases, UK, McGill University, Canada, and USAID, USA, administered by the World Health Organization (WHO) Global Tuberculosis Programme, Switzerland, for the preparation of systematic reviews and educational materials, consultancy fees from Foundation for Innovative New Diagnostics (FIND), Switzerland (for the preparation of systematic reviews and GRADE tables), honoraria, and travel support to attend WHO guideline meetings.
DJH has received funding from USAID, administered by the World Health Organization Global Tuberculosis Programme, Switzerland for related systematic reviews.
New
References
Additional references
Balshem 2011
- Balshem H, Helfand M, Schünemann HJ, Oxman AD, Kunz R, Brozek J, et al. GRADE guidelines: 3. Rating the quality of evidence. Journal of Clinical Epidemiology 2011;64(4):401-6. [DOI] [PubMed] [Google Scholar]
Banada 2010
- Banada PP, Sivasubramani SK, Blakemore R, Boehme C, Perkins MD, Fennelly K, et al. Containment of bioaerosol infection risk by the Xpert MTB/RIF assay and its applicability to point-of-care settings. Journal of Clinical Microbiology 2010;48(10):3551-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Bjerrum 2019
- Bjerrum S, Schiller I, Dendukuri N, Kohli M, Nathavitharana RR, Zwerling AA, et al. Lateral flow urine lipoarabinomannan assay for detecting active tuberculosis in people living with HIV. Cochrane Database of Systematic Reviews 2019, Issue 10. Art. No: CD011420. [DOI: 10.1002/14651858.CD011420.pub3] [DOI] [PMC free article] [PubMed] [Google Scholar]
Broger 2020
- Broger T, Nicol MP, Székely R, Bjerrum S, Sossen B, Schutz C, et al. Diagnostic accuracy of a novel tuberculosis point-of-care urine lipoarabinomannan assay for people living with HIV: A meta-analysis of individual in- and outpatient data. PLoS Medicine 2020;17(5):e1003113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Cain 2010
- Cain KP, McCarthy KD, Heilig CM, Monkongdee P, Tasaneeyapan T, Kanara N, et al. An algorithm for tuberculosis screening and diagnosis in people with HIV. New England Journal of Medicine 2010;362:707-16. [DOI] [PubMed] [Google Scholar]
Cepheid 2009
- Cepheid. Brochure: Xpert®MTB/RIF. Two-hour detection of MTB and resistance to rifampicin. cepheid.com/administrator/components/com_productcatalog/library-files/8dbd1dd8b83a9780dd88a0d9852ffd98-a464e0bea6122c5c648ccdf617ecbd0c-Xpert-MTBRIF-Brochure-EU-0089-02-LOR.pdf (accessed 16 May 2019).
Chakravorty 2017
- Chakravorty S, Simmons AM, Rowneki M, Parmar H, Cao Y, Ryan J, et al. The new Xpert MTB/RIF Ultra: improving detection of Mycobacterium tuberculosis and resistance to rifampin in an assay suitable for point-of-care testing. mBio 2017;8(4):1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Chu 2006
- Chu H, Cole SR. Bivariate meta-analysis of sensitivity and specificity with sparse data: a generalized linear mixed model approach. Journal of Clinical Epidemiology 2006;59(12):1331-2. [DOI] [PubMed] [Google Scholar]
Cochrane Special Collection 2019
- Cochrane Infectious Diseases Group. Diagnosing tuberculosis. www.cochranelibrary.com/collections/doi/SC000034/full (accessed 7 January 2020).
Covidence [Computer program]
- Veritas Health Innovation Covidence. Version accessed after 12 June 2020. Melbourne, Australia: Veritas Health Innovation. Available at covidence.org.
Dorman 2018
- Dorman SE, Schumacher SG, Alland D, Nabeta P, Armstrong DT, King B, et al. Xpert MTB/RIF Ultra for detection of Mycobacterium tuberculosis and rifampicin resistance: a prospective multicentre diagnostic accuracy study. Lancet Infectious Diseases 2018;18(1):76-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
Ford 2016
- Ford N, Matteelli A, Shubber Z, Hermans S, Meintjes G, Grinsztejn B, et al. TB as a cause of hospitalization and in-hospital mortality among people living with HIV worldwide: a systematic review and meta-analysis. Journal of the International AIDS Society 2016;19(1):20714. [DOI] [PMC free article] [PubMed] [Google Scholar]
Getahun 2010
- Getahun H, Gunneberg C, Granich R, Nunn P. HIV infection-associated tuberculosis: the epidemiology and the response. Clinical Infectious Diseases 2010;50 Suppl 3:S201–7. [DOI] [PubMed] [Google Scholar]
Global Laboratory Initiative 2019
- Global Laboratory Intiaitive. Practical guide to implementing a quality assurance system for Xpert MTB/RIF testing. www.stoptb.org/wg/gli/assets/documents/Xpert-QA-guide-2019.pdf (accessed 7 January 2020).
GRADEpro GDT 2015 [Computer program]
- McMaster University (developed by Evidence Prime) GRADEpro GDT. Version accessed after 12 June 2020. Hamilton (ON): McMaster University (developed by Evidence Prime), 2015. Available at gradepro.org.
Gupta‐Wright 2018
- Gupta-Wright A, Corbett EL, Van Oosterhout JJ, Wilson D, Grint D, Alufandika-Moyo M, et al. Rapid urine-based screening for tuberculosis in HIV-positive patients admitted to hospital in Africa (STAMP): a pragmatic, multicentre, parallel-group, double-blind, randomised controlled trial. Lancet 2018;392:292–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
Hamada 2018
- Hamada Y, Lujan J, Schenkel K, Ford N, Getahun H. Sensitivity and specificity of WHO's recommended four-symptom screening rule for tuberculosis in people living with HIV: a systematic review and meta-analysis. Lancet HIV 2018;5(9):e515-23. [DOI] [PubMed] [Google Scholar]
Harris 2019
- Harris M, Qi A, Jeagal L, Torabi N, Menzies D, Korobitsyn A, et al. A systematic review of the diagnostic accuracy of artificial intelligence-based computer programs to analyze chest X-rays for pulmonary tuberculosis. PLoS One 2019;14(9):e0221339. [DOI] [PMC free article] [PubMed] [Google Scholar]
Horne 2019
- Horne DJ, Kohli M, Zifodya JS, Schiller I, Dendukuri N, Tollefson D, et al. Xpert MTB/RIF and Xpert MTB/RIF Ultra for pulmonary tuberculosis and rifampicin resistance in adults. Cochrane Database of Systematic Reviews 2019, Issue 6. Art. No: CD009593. [DOI: 10.1002/14651858.CD009593.pub4] [DOI] [PMC free article] [PubMed] [Google Scholar]
Lewinsohn 2017
- Lewinsohn DM, Leonard MK, LoBue PA, Cohn DL, Daley CL, Desmond E, et al. Official American Thoracic Society/Infectious Diseases Society of America/Centers for Disease Control and Prevention Clinical Practice Guidelines: Diagnosis of tuberculosis in adults and children. Clinical Infectious Diseases 2017;64(2):e1-e33. [PMID: ] [DOI] [PubMed] [Google Scholar]
Lunn 2009
- Lunn D, Spiegelhalter D, Thomas A, Best N. The BUGS project: evolution, critique, and future directions. Statistics in Medicine 2009;28(25):3049-67. [DOI] [PubMed] [Google Scholar]
Macaskill 2010
- Macaskill P, Gatsonis C, Deeks JJ, Harbord RM, Takwoingi Y. Chapter 10: Analysing and presenting results. In: Deeks JJ, Bossuyt PM, Gatsonis C, editors, Cochrane Handbook for Systematic Reviews of Diagnostic Test Accuracy Version 1.0. The Cochrane Collaboration, 2010. Available from srdta.cochrane.org.
MacNeil 2020
- MacNeil A, Glaziou P, Sismanidis C, Date A, Maloney S, Floyd K. Global epidemiology of tuberculosis and progress toward meeting global targets — worldwide, 2018. Morbidity and Mortality Weekly Report 2020;69:281–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Moher 2009
- Moher D, Liberati A, Tetzlaff J, Altman DG, The PRISMA Group (2009). Preferred reporting items for systematic reviews and meta-analyses: the PRISMA Statement. PLoS Medicine 6;7:e1000097. [DOI: 10.1371/journal.pmed1000097] [DOI] [PMC free article] [PubMed] [Google Scholar]
Peter 2016
- Peter JG, Zijenah LS, Chanda D, Clowes P, Lesosky M, Gina P, et al. Effect on mortality of point-of-care, urine-based lipoarabinomannan testing to guide tuberculosis treatment initiation in HIV-positive hospital inpatients: a pragmatic, parallel-group, multicountry, open-label, randomised controlled trial. Lancet 2016;387(10024):1187-97. [DOI] [PubMed] [Google Scholar]
R Core Team 2019 [Computer program]
- R Foundation for Statistical Computing R Core Team (2019). R: A language and environment for statistical computing. Version accessed after 12 June 2020. Vienna, Austria: R Foundation for Statistical Computing, 2019. Available at www.R-project.org.
Reitsma 2005
- Reitsma JB, Glas AS, Rutjes AW, Scholten RJ, Bossuyt PM, Zwinderman AH. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. Journal of Clinical Epidemiology 2005;58(10):982-90. [DOI] [PubMed] [Google Scholar]
Review Manager 2020 [Computer program]
- Nordic Cochrane Centre, The Cochrane Collaboration Review Manager 5 (RevMan 5). Version 5.4. Copenhagen: Nordic Cochrane Centre, The Cochrane Collaboration, 2020.
Schünemann 2008
- Schünemann HJ, Oxman AD, Brozek J, Glasziou P, Jaeschke R, Vist GE, et al. Grading quality of evidence and strength of recommendations for diagnostic tests and strategies. BMJ 2008;336(7653):1106-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Schünemann 2016
- Schünemann HJ, Mustafa R, Brozek J, Santesso N, Alonso-Coello P, Guyatt G, et al. GRADE Working Group. GRADE Guidelines: 16. GRADE evidence to decision frameworks for tests in clinical practice and public health. Journal of Clinical Epidemiology 2016;76:89-98. [DOI: 10.1016/j.jclinepi.2016.01.032] [DOI] [PubMed] [Google Scholar]
Schünemann 2020a
- Schünemann HJ, Mustafa R, Brozek J, Steingart KR, Leeflang M, Murad MH, et al. GRADE guidelines: 21 part 1. Study design, risk of bias and indirectness in rating the certainty across a body of evidence for test accuracy. Journal of Clinical Epidemiology 2020;122:129-41. [DOI: 10.1016/j.jclinepi.2019.12.020] [DOI] [PubMed] [Google Scholar]
Schünemann 2020b
- Schünemann HJ, Mustafa R, Brozek J, Steingart KR, Leeflang M, Murad MH, et al. GRADE guidelines: 21 part 2. Inconsistency, Imprecision, publication bias and other domains for rating the certainty of evidence for test accuracy and presenting it in evidence profiles and summary of findings tables. Journal of Clinical Epidemiology 2020;122:142-52. [DOI: 10.1016/j.jclinepi.2019.12.021] [DOI] [PubMed] [Google Scholar]
Stata [Computer program]
- Stata. Version 15. College Station, TX, USA: StataCorp, 2017. Available at www.stata.com.
UN Sustainable Development Goals 2030
- United Nations General Assembly. Transforming our world: the 2030 agenda for sustainable development. Resolution adopted by the General Assembly on 25 September 2015. https://sustainabledevelopment.un.org/post2015/transformingourworld/publication (accessed 22 July 2020).
Unitaid 2017
- Boyle D. Tuberculosis Diagnostics Technology and Market Landscape. 5th edition. Vernier: World Health Organization Unitaid Secretariat, 2017. [Google Scholar]
van't Hoog 2013
- van't Hoog AH, Langendam MW, Mitchell E, Cobelens FG, Sinclair D, Leeflang MM, et al. A systematic review of the sensitivity and specificity of symptom- and chest-radiography screening for active pulmonary tuberculosis in HIV-negative persons and persons with unknown HIV status. Report to the World Health Organization. 2013. who.int/tb/Review2Accuracyofscreeningtests.pdf?ua=1 (accessed 23 June 2020).
Whiting 2011
- Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Annals of Internal Medicine 2011;155(8):529-36. [DOI] [PubMed] [Google Scholar]
WHO 2011
- World Health Organization. Automated real-time nucleic acid amplification technology for rapid and simultaneous detection of tuberculosis and rifampicin resistance: Xpert MTB/RIF system for the diagnosis of pulmonary and extrapulmonary TB in adults and children. apps.who.int/iris/handle/10665/112472 (accessed prior to 29 April 2019). [PubMed]
WHO Compendium of WHO guidelines 2018
- World Health Organanization. Compendium of WHO guidelines and associated standards: ensuring optimum delivery of the cascade of care for patients with tuberculosis. 2nd edition. Geneva: World Health Organization, 2018. [Google Scholar]
WHO END TB 2015
- World Health Organization. Implementing the End TB Strategy: the essentials. WHO/HTM/TB/2015.31. who.int/tb/publications/2015/The_Essentials_to_End_TB/en/ (accessed 23 June 2020).
WHO Global TB Report 2019
- World Health Organization. Global Tuberculosis Report 2019. Geneva: World Health Organization, 2019. Licence: CC BY-NC-SA 3.0 IGO. [Google Scholar]
WHO Guidelines 2016
- World Health Organization. WHO treatment guidelines for drug-resistant tuberculosis, 2016 update. October 2016 revision. Geneva: World Health Organization, 2016. WHO/HTM/TB/2016.04. [Google Scholar]
WHO Lateral flow LAM 2019
- World Health Organization. Lateral flow urine lipoarabinomannan assay (LF-LAM) for the diagnosis of active tuberculosis in people living with HIV. Policy update 2019. Geneva: World Health Organization, 2019. Licence: CC BY-NC-SA 3.0 IGO. [Google Scholar]
WHO LPA 2016
- World Health Organization. The use of molecular line probe assays for the detection of resistance to isoniazid and rifampicin: policy update. WHO/HTM/TB/2016.12. Geneva: World Health Organization 2016.
WHO Rapid Communication 2020a
- World Health Organization. World Health Organization. WHO consolidated guidelines on tuberculosis. Module 3: diagnosis – rapid diagnostics for tuberculosis detection; June 2020. who.int/publications/i/item/who-consolidated-guidelines-on-tuberculosis-module-3-diagnosis---rapid-diagnostics-for-tuberculosis-detection (accessed 1 July 2020).
WHO Rapid Communication 2020b
- World Health Organization. WHO consolidated guidelines on tuberculosis. Module 4: treatment - drug-resistant tuberculosis treatment; June 2020. who.int/publications/i/item/9789240007048 (accessed 1 July 2020).
WHO Systematic screening 2015
- World Health Organization. Systematic screening for active tuberculosis: an operational guide WHO/HTM/TB/2015.16. who.int/tb/publications/systematic_screening/en/ (accessed 23 June 2020).
WHO Tuberculosis screening 2013
- World Health Organization. Systematic screening for active tuberculosis: principles and recommendations. WHO/HTM/TB/2013.04. Geneva: World Health Organization, 2013. [PubMed] [Google Scholar]
WHO Xpert MTB/RIF 2013
- World Health Organization. Automated real-time nucleic acid amplification technology for rapid and simultaneous detection of tuberculosis and rifampicin resistance: Xpert MTB/RIF assay for the diagnosis of pulmonary and extrapulmonary TB in adults and children. https://apps.who.int/iris/handle/10665/112472 (accessed 23 June 2020). [PubMed]
WHO Xpert Ultra 2017
- World Health Organization. WHO meeting report of a technical expert consultation: non-inferiority analysis of Xpert MTB/RIF Ultra compared to Xpert MTB/RIF. WHO/HTM/TB/2017.04. Geneva: WHO 2017.
World Bank 2019
- World Bank. World Bank List of Economies. Washington (District of Columbia): World Bank, 2019. [Google Scholar]
Zellweger 2006
- Zellweger JP, Heinzer R, Touray M, Vidondo B, Altpeter E. Intra-observer and overall agreement in the radiological assessment of tuberculosis. International Journal of Tuberculosis and Lung Diseases 2006;10(10):1123-6. [PubMed] [Google Scholar]


