Abstract
Regenerative ability varies tremendously across species. A common feature of regeneration of appendages such as limbs, fins, antlers, and tails is the formation of a blastema—a transient structure that houses a pool of progenitor cells that can regenerate the missing tissue. We have identified the expression of von Willebrand factor D and EGF domains (vwde) as a common feature of blastemas capable of regenerating limbs and fins in a variety of highly regenerative species, including axolotl (Ambystoma mexicanum), lungfish (Lepidosiren paradoxa), and Polpyterus (Polypterus senegalus). Further, vwde expression is tightly linked to the ability to regenerate appendages in Xenopus laevis. Functional experiments demonstrate a requirement for vwde in regeneration and indicate that Vwde is a potent growth factor in the blastema. These data identify a key role for vwde in regenerating blastemas and underscore the power of an evolutionarily informed approach for identifying conserved genetic components of regeneration.
Keywords: axolotl, blastema, lungfish, Polypterus, regeneration, von Willebrand factor and EGF domains, Xenopus
Identification of a gene, von Willebrand Factor D and EGF Repeats (vwde), whose expression is highly enriched in axolotl limb blastemas prompted comparative experimentation in lungfish, Polypterus, and Xenopus. Vwde expression is induced in key tissues during regeneration in all four models (fins in lungfish and polypterus; tail in Xenopus embryos; limbs in axolotl). Functional experimentation in axolotl supports a role for vwde in promoting the growth of blastema as well as in patterning the regenerate limb.
Research Highlights
vwde expression is a common feature of blastemas capable of fin and limb regeneration.
vwde expression is tightly tied to regeneration‐competency.
vwde is required for axolotl limb regeneration, with transient knockdown resulting in severe endpoint phenotypes.
1. INTRODUCTION
The underlying reasons why some animals have the ability to regenerate complex structures, while others cannot, remains an important and open question. This knowledge gap has led to intense study of how regeneration‐competent species are able to perform complex multitissue regeneration, with a particular focus on the ability to regenerate paired appendages, such as limbs and fins. However, this has long been a pursuit without an understanding of whether this ability was present when paired appendages first evolved or was acquired by certain phylogenetic lineages (e.g., urodele amphibians).
Recent work regarding the evolutionary origins of regenerative capacity has indicated that the ability to regenerate paired appendages is an inherited feature of the fin‐to‐limb transition (Darnet et al., 2019; Fröbisch, Bickelmann, Olori, & Witzmann, 2015; Fröbisch, Constanze, & Florian, 2014; Nogueira et al., 2016). Evidence found in the fossil record (Fröbisch et al., 2014, 2015), functional studies across species (Darnet et al., 2019), and comparisons of gene expression profiles of regenerating tissue (Darnet et al., 2019; Nogueira et al., 2016) support the notion that paired appendage regeneration is a feature lost by certain lineages and was not a newly derived capacity in highly regenerative species. This indicates that the amniote lineage (which includes humans) has lost regenerative tendencies in appendages over evolutionary time. Therefore, the ability to stimulate regeneration in nonregenerative species, potentially in a therapeutic context, may require the reinitiation of a core, evolutionary conserved program.
All species that are able to regenerate appendages share a conserved trait: the ability to form a blastema. The blastema is the morphological structure that forms at the amputation plane and houses the progenitor cells responsible for regeneration. Recent efforts have focused on elucidating the molecular definition of the blastema, with many of these efforts aimed at the axolotl limb blastema due to the ease of tissue acquisition and the ability to perform experimentation in the lab (Bryant et al., 2017; Gerber et al., 2018; Knapp et al., 2013; Leigh et al., 2018; Monaghan et al., 2012, 2009; Stewart et al., 2013; Voss et al., 2015; Wu, Tsai, Ho, Chen, & Lee, 2013). These studies provide a wealth of information about transcriptomic changes over time, cell types, and blastema‐enriched genes. More recently, sequencing efforts of nonmodel species have allowed for comparisons to the axolotl limb blastema and indicate a core molecular signature that is shared between the blastemas of distantly related species (Darnet et al., 2019; Nogueira et al., 2016). Due to these similarities and the common evolutionary origin of limb regeneration capacity, we can use an evolutionarily informed approach to understand what constitutes a blastema and for identifying core features required for regeneration. We expect that expression of a gene within this core, evolutionarily conserved program would be present, and possibly enriched, in the blastemas capable of regenerating appendages of multiple species and be required for blastema function.
A recent approach to identify the unique gene expression in the axolotl limb blastema compared blastema gene expression to a variety of homeostatic and embryonic tissues and identified over 150 blastema‐enriched genes (Bryant et al., 2017). These blastema‐enriched genes may help to explain the unique functions of the blastema, but the question remains as to whether these genes represent a core program and are functionally required for regeneration. One of the most blastema‐enriched genes in this data set was von Willebrand factor D and EGF domains (vwde), which to date has not been functionally studied in any context.
We decided to apply an evolutionary framework to determine if vwde fit the description of an evolutionary‐conserved, blastema‐enriched gene and if such an approach may help to identify genes required for regeneration. We found that vwde expression is a common feature of both fin and limb blastemas and was highly enriched in regenerating appendages as compared with preamputation intact appendages. In addition, using the natural regeneration‐competent and regeneration‐refractory periods during Xenopus laevis development, we observed that vwde expression was tightly linked to the regeneration‐competent environment. This suggests that vwde may be a critical factor in the regenerative niche. Finally, we found that vwde is functionally required for axolotl limb regeneration, with transient knockdown of protein levels resulting in aberrant regeneration. These data suggest that an evolutionarily informed approach can help to prioritize target genes and that genes that are blastema‐enriched across different species may prove to be critical factors in the ability to regenerate appendages.
2. METHODS
2.1. Animal experimentation
All axolotl experiments were performed in accordance with Brigham and Women's Hospital Institutional Animal Care and Use Committee in line with Animal Experimentation Protocol #04160. All animals were bred in house and derived from crosses between white (d/d strain RRID:AGSC_101A) parents, but the colony was originally derived from animals obtained from Ambystoma Genetic Stock Center (AGSC; Lexington, KY, NIH grant P40‐OD019794, RRID:SCR_006372). Axolotls were housed between 20°C and 21°C in 40% Holtfreter's solution and fed pellets (ordered from AGSC) three times weekly. For amputations, animals were narcotized in 0.1% MS‐222, confirmed to be fully narcotized by pinch test, amputated mid‐zeugopod, and the bone was trimmed. Animals were allowed to recover overnight in 0.5% sulfamerazine. For all functional experiments, all four limbs were amputated and injected individually. Functional experiments were performed on animals ranging from 3.8 to 8 cm and 2–5 months of age, before knowing sex and thus contain a mixture of male and female axolotls.
Polypterus senegalus and Lepidosiren paradoxa were maintained in individual tanks in a recirculating freshwater system. Animals were anesthetized before amputations: P. senegalus in 0.1% MS‐222 (Sigma‐Aldrich) and L. paradoxa in 0.1% clove oil diluted in the system water. Experiments and animal care were performed following animal care guidelines approved by the Animal Care Committee at the Universidade Federal do Para (protocol no.: 037–2015). Pectoral fins in both species were bilaterally amputated. For L. paradoxa fins were amputated at approximately 1 cm distance from the body, and for P. senegalus, fins were amputated across the fin endoskeleton. Amputated fins (regenerating and uninjured) were used for histology, in situ hybridization and quantitative reverse transcription polymerase chain reaction (qRT‐PCR) analysis.
2.2. Electroporation
Electroporation was performed while axolotls were anesthetized in 0.1% tricaine and subsequently immersed in ice cold 1× phosphate‐buffered saline (PBS) using a NepaGene Super Electroporator NEPA21 Type II electroporator. Settings for electroporation included: three poring pulses at 150 Volts with a pulse length of 5 ms, a pulse interval of 10 ms, a decay rate of 0%, and a positive (+) polarity. Transfer pulse consisted of five pulses at 50 Volts with a pulse length of 50 ms, a pulse interval of 950 ms, a decay rate of 0%, and a positive (+) polarity.
2.3. Quantitative reverse transcription polymerase chain reaction
Total RNA from regenerating or uninjured pectoral fins was extracted using TRIzol reagent (Thermo Fisher Scientific). Residual DNA removal and RNA cleanup were performed following the RNeasy Mini Kit (Qiagen) protocol. Complementary DNA (cDNA) was synthesized from 0.5 μg RNA using the Superscript III First‐Strand Synthesis Supermix (Thermo Fisher Scientific) with oligo‐dT. For qPCR, amplification reactions (10 μl) prepared with the GoTaq Probe qPCR Master Mix (Promega) were run in a StepOnePlus Real‐Time PCR System (Applied Biosystems). Gene‐specific oligos (Table 1) for qRT‐PCR assays were designed using Primer 3.0 (http://bioinfo.ut.ee/primer3/) and used in a final concentration of 200 nM to each primer. Each qPCR determination was performed with three biological and three technical replicates. Relative messenger RNA expressions were calculated with the method (Livak & Schmittgen, 2001), using sdha (P. senegalus) or polrc1 (L. paradoxa) genes as endogenous control and the uninjured fin (mean ΔC T value of the three biological replicates) as reference sample.
Table 1.
Oligos used for qPCR, morpholinos, and ISH probe templates
| Primer | Application | Sequence (5′–3′) |
|---|---|---|
| Ax_vwde_ISH_F | Riboprobe template | TGTGGAAAGAAACTTGTGCATCA |
| Ax_vwde_ISH_R | Riboprobe template | TTTAATCTGAAAATGGACCAGTAGATT |
| vwde MO1 | Antisense morpholino | ATATCCCATACATCCTTGCGTTGGC |
| vwde MO2 | Antisense morpholino | AGAAACCATCACAGTTCCTCACAGT |
| vwde MO1 inverted (control (INV)) | Control morpholino | CGGTTGCGTTCCTACATACCCTATA |
| Standard control MO (control (SC)) | Control morpholino | CCTCTTACCTCAGTTACAATTTATA |
| Ps_Vwde‐qPCR_F | qPCR | AGAATTCCTGTGACTGTGCGA |
| Ps_Vwde‐qPCR_R | qPCR | TTCTGGTGTTGTTGGTGAGGG |
| Ps_Vwde‐ISH_F | Riboprobe template | GGCCGCGGGCATGCGGAATAATGTGTGCT |
| Ps_Vwde‐ISH_R | Riboprobe template | CCCGGGGCAGTCCAGTCTTCAGCAGTGTG |
| Lp_Vwde‐qPCR_F | qPCR | TTCTTCTTGGAGACCCCTGAT |
| Lp_Vwde‐qPCR_R | qPCR | GGTCTTGCTGGCTAGTGTCAG |
| Lp_Vwde‐ISH_F | Riboprobe template | GGCCGCGGAGCTAACAGCCTGTGCAACAT |
| Lp_Vwde‐ISH_R | Riboprobe template | CCCGGGGCATCAGGGGTCTCCAAGAAGAA |
| 3′_T7 universal | Riboprobe template, 2nd‐round PCR | AGGGATCCTAATACGACTCACTATAGGGCCCGGGGC |
| 5′_T7 universal | Riboprobe template, 2nd‐round PCR | GAGAATTCTAATACGACTCACTATAGGGCCGCGG |
Abbreviations: ISH, in situ hybridization; qPCR, quantitative polymerase chain reaction.
2.4. In situ hybridization
For in situ hybridization using axolotl samples a gene fragment from the 3′‐untranslated region was amplified from blastema cDNA and cloned into the pGEM‐T Easy vector and sequenced. Depending upon orientation, T7 or Sp6 polymerase was used to transcribe the probe. Primers for in situ probes against axolotl vwde (contig c1084387_g3_i1 from Bryant et al., 2017) can be found in Table 1. Colorimetric in situ hybridization in axolotl tissue harvested from animals with snout to tail lengths of 9.5–11.5 cm and was performed as previously described at protocols.io (https://www.protocols.io/view/rna‐in‐situ‐hybridization‐p33dqqn).
For in situ hybridizations with fish samples, fins of P. senegalus (5 days postamputation [dpa] and uninjured) and L. paradoxa (21 dpa and uninjured) were amputated, embedded in TissueTek O.C.T compound (Thermo Fisher Scientific), and maintained at −80°C until use. Frozen sections of 20 μm were obtained on a Leica CM1850 UV cryostat, positioned on slides (Color Frost Plus/Thermo Fisher Scientific) and fixed as previously described (Nogueira et al., 2016). Riboprobe templates containing a gene‐specific segment (400–500 bp) and a T7 promoter sequence were produced by a two‐round PCR strategy (primers are listed in Table 1). Riboprobes were synthesized with T7 RNA polymerase (Roche) and DIG‐labeling mix (Roche). Controls probes (sense riboprobes) were synthesized from a template containing the T7 promoter in a reverse orientation. A total of 300 ng of DIG‐labeled riboprobe was used per slide during in situ hybridization performed as previously described (Nogueira et al., 2016). Images were obtained on a Nikon Eclipse 80i microscope and processed using the NIS‐Element D4.10.1 program.
2.5. Whole mount RNA‐FISH
X. laevis eggs were obtained, fertilized, and cultured as embryos at 18°C using standard methods as in (Sive, Grainger, & Harland, 2010). All experimental procedures using X. laevis were approved by the Institutional Animal Care and Use Committee (IACUC) and Tufts University Department of Laboratory Animal Medicine under protocol number M2017‐53. Once embryos reached regeneration‐competent (Stage 40) or regeneration‐incompetent (Stage 46) stages, animals were anesthetized using 0.005% MS222 in 0.1X MMR and tails were amputated at the posterior third of the tail and allowed to regenerate for 24 hr. Embryos at both stages, which had not been amputated, were also collected as intact controls. Regenerating and intact control embryos were anesthetized in 0.005% MS222 and then fixed at 4°C, rocking overnight, in either 4% paraformaldehyde in 1× diethylpyrocarbonate (DEPC) PBS or MEMPA buffer (0.1 M MOPS (pH 7.4), 2 mM ethylene glycol‐bis(β‐aminoethyl ether)‐N,N,N′,N′‐tetraacetic acid, 1 mM MgSO4, 3.7% paraformaldehyde). We used a slightly modified whole‐mount mouse protocol (Choi et al., 2016) using hybridization chain reaction v3.0 (Choi et al., 2018). After overnight incubation, embryos were washed three times for 5 min in PBST and then taken through a methanol series on ice. This series consisted of 10 min washes on ice in ice‐cold 25%MeOH/75% PBST, 50%MeOH/50% PBST, 75%MeOH/25%PBST, 100%MeOH, and then finally stored in a fresh 100%MeOH solution. Dehydrated embryos were then stored at −20°C until use. For in situ, embryos were subsequently rehydrated via a reverse methanol series, on ice, with 10 min washes of 75% MeOH/25% PBST, 50% MeOH/50% PBST, 25% MeOH/75% PBST, 100% PBST, and another final wash in 100% PBST. Embryos were then digested with proteinase K (10 µg/ml) in DEPC PBS for 5 min at room temperature. Postfixation was then performed in 4% paraformaldehyde (PFA) in 1× DEPC PBS for 20 min at room temperature. Next, three 5 min washes with PBST at room temperature was followed by 5 min at 37°C in hybridization solution (50% formamide, 5× sodium chloride sodium citrate (SSC), 9 mM citric acid (pH 6.0), 0.1% Tween‐20, 50 µg/ml heparin, 1× Denhardt's solution, and 20% dextran sulfate). Samples were prehybridized by full immersion in hybridization solution without probes for 30 min at 37°C. Hybridization was performed overnight at 37°C with samples immersed in hybridization solution containing 20 probe pairs against vwde.L (XM_018267342.1) diluted at 1:200 of 1 µM (hybridization chain reaction v3.0 RNA fluorescent in situ probes were ordered from Molecular Instruments (https://www.molecularinstruments.com/). The following day, samples were washed four times at 37°C in probe wash buffer (50% formamide, 5× SSC, 9 mM citric acid (pH 6.0), 0.1% Tween‐20, and 50 µg/ml heparin). Samples were then washed two times in 5× SSC at room temperature. Preamplification was then performed at room temperature for 30 min in amplification buffer (5× SSC, 0.1% Tween‐20, 10% dextran sulfate). During preamplification, hairpin probes (ordered from https://www.molecularinstruments.com/) compatible with vwde.L probe pairs were heated individually at 95°C for 30 s and then snap cooled for 30 min at room temperature in the dark. After 30 min, probe pairs were added to amplification buffer at 1:50 (3 µM stock) and this probe containing buffer was subsequently added to samples, ensuring that samples were fully immersed. Incubation was performed overnight at room temperature. The next day, samples were washed for 5 min in 5× SSCT, twice for 30 min in 5× SSCT, and a 5 min wash in 5× SSCT. Samples were then stained with DAPI for 5 min in 1× PBS, washed for 5 min in 1× PBS, and then stored in 1× PBS. Samples were then mounted in low melt agarose and imaged on a Zeiss LSM 880 Upright. A median 3 × 3 filter followed by maximum projection was applied to all images.
2.6. Morpholino design and administration
Morpholinos were designed and synthesized by GeneTools. Morpholino sequences can be found in Table 1. About 1.25 µl of morpholino was injected in the blastema and electroporation was performed as described in electroporation. All morpholinos were 3′ fluorescein conjugated to allow for visualization. Morpholinos were reconstituted to 1 mM in 2× PBS and diluted to a working concentration of 500 μM in 1× PBS before injection.
2.7. 5‐Ethynyl‐2′‐deoxyuridine (EdU) staining
Stock solutions of EdU dissolved in dimethyl sulfoxide were prepared per manufacturer's instructions (Thermo Fisher Scientific). Axolotls (3–6 cm tail to snout) were narcotized in 0.1% tricaine at 7 dpa and control or vwde‐targeting morpholino was injected and subsequently electroporated as described in electroporation section of methods into the blastema. At 9 dpa, intraperitoneal injections with 400 µM EdU in 0.7× PBS at a volume of 20 µ/g were performed. Eighteen hours later blastemas were harvested, fixed for 1–2 hr in 4% PFA and then taken through a sucrose gradient to 30% sucrose in 1× PBS. Tissue was then embedded in optimal cutting temperature compound and frozen in a dry ice/ethanol bath. Sections were cut at 16 µm with a cryostat, collected on Superfrost Plus slides (Fisher), and stored at −80°C. EdU staining was performed with the Click‐iT EdU Alexa Fluor 594 Imaging Kit per manufacturer's instructions (Thermo Fisher Scientific).
2.8. Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay
TUNEL assays were performed as previously described (Bryant et al., 2017).
2.9. Skeletal preparations and scoring
Limbs were stained with Alcian blue/Alizarin red according to Whited et al. (2012). In brief, limbs were incubated with rocking overnight in 95% ethanol and then rocking overnight an acetone. Limbs were then incubated for at least 7 days in alcian blue/alizarin red at 37°C. Limbs were then cleared by incubation in 1% (wt/vol) KOH, followed by 1% (vol/vol) KOH/25% glycerol, 1% KOH/50% glycerol, and 1% KOH/75% glycerol. Limbs were imaged in 1% KOH/75% glycerol. Alcian blue stock was 0.3% alcian blue in 70% ethanol; alizarin red stock was 0.1% alizarin red 95% ethanol; the working solution was 5% alcian blue stock/5% alizarin red stock/5% glacial acetic acid/volume in 70% ethanol.
Definitions for limbs after regeneration. Normal: All digits and carpals present, zeugopod and stylopod intact. Spike: Single outgrowth from amputation plane without obvious turn at joint. Loss of distal elements: Distal elements without obvious autopod. Oligodactyly: Loss or reduction in size at least one digit. Syndactyly: Fusion of digits. Additional elements: Extra bones in stylopod or zeugopod. For statistical analysis normal was compared with all of the above listed abnormalities.
2.10. Ortholog analysis
The following proteomes were downloaded from uniprot.org, human (Homo sapiens, UP000005640, accessed May 18, 2019), zebrafish (Danio rerio, UP000000437, accessed May 18, 2019), mouse (Mus musculus, accessed May 18, 2019), amphioxus (Branchiostoma floridae, accessed May 27, 2019), chick (Gallus gallus, accessed August 27, 2019), sea squirt (Ciona intestinalis, accessed August 27, 2019), lamprey (Petromyzon marinus, accessed August 27, 2019), green anole (Anolis carolinensis, August 27, 2019), frog (X. laevis, UP000186698, accessed July 11, 2019), and frog (Xenopus tropicalis, UP000008143, accessed July 11, 2019). The South American lungfish transcriptome was downloaded from https://www.ncbi.nlm.nih.gov/Traces/wgs/?val=GEHZ01 and converted to a putative reference protein using TransDecoder (version 5.3.0) like so: “TransDecoder.LongOrfs ‐t.” The Polpyterus transcriptome can be found here: https://www.ncbi.nlm.nih.gov/bioproject/480698 and converted with TransDecoder as referenced above. The axolotl (Ambystoma mexicanum) predicted proteome was obtained from https://data.broadinstitute.org/Trinity/SalamanderWeb/Axolotl.Trinity.CellReports2017.transdecoder.pep.gz (Bryant et al., 2017). Cloning of axolotl vwde revealed a sequencing error in the axolotl transcriptome which eliminated the first ~500 bp of the sequence. We manually changed the axolotl proteome to include this corrected version of vwde (Supporting Information File 1).
To predict orthologs, we used OrthoFinder2.0 (version 2.3.3; Emms & Kelly, 2019). Orthofinder was implemented as follows:
“orthofinder ‐f/path/to/proteomes ‐M msa ‐A mafft ‐T fasttree ‐t 20 ‐o/path/to/output/directory”
Output from Orthofinder was visualized using dendroscope (Huson & Scornavacca, 2012).
The Uniprot version of mouse vwde (Uniprot ID: Q6DFV8) in the proteome used did not contain EGF‐like domains, so we manually searched UCSC genome browser to confirm this lack of EGF‐like domains. This revealed a full length Vwde (ENSMUST00000203074.2), containing EGF‐like domains.
2.11. Protein domain diagrams
The R package, drawProteins (Brennan, 2018) was used to draw protein domains for different species Vwde. For all genes contained within Uniprot, these were downloaded directly with drawProteins. For genes not available via Uniprot (https://www.uniprot.org/; UniProt Consortium, 2019; e.g., Polpyterus, axolotl, and lungfish), the amino acid sequence of the protein was queried via Interpro with default settings (https://www.ebi.ac.uk/interpro/; Hunter et al., 2009) and positions and domain annotations were extracted and made into a matrix that matched the required structure for drawProteins.
2.12. Vwde knockdown confirmation
Two separate constructs to test the target specificity of each morpholino used. GFP was removed from pCAG‐GFP (pCAG‐GFP was a gift from Connie Cepko (Addgene plasmid # 11150; http://n2t.net/addgene:11150; RRID: Addgene_11150) (Matsuda & Cepko, 2004)) and replaced with vectors containing td‐Tomato sequence and the morpholino binding site (Supporting Information File 2). To confirm knockdown we coinjected and electroporated into medium‐bud blastemas the generated constructs and the appropriate fluorescein‐conjugated morpholino. Fluorescein fluorescence was used to confirm injection efficiency and td‐tomato expression was used to measure ability to block translation.
2.13. Statistics
Nested one‐way analysis of variance was used to determine significance between blastema lengths. Each limb was considered a technical replicate within one biological (i.e., animal) replicate. Nested t tests were used to determine significance in EdU and TUNEL experiments, again treating each limb as a technical replicate and placing limbs from the same animal within one biological replicate. Fisher's exact tests (control vs. treated) were used to determine significance of outgrowth phenotypes. Significant results were considered as p < .05. All statistical analyses were performed with GraphPad Prism (RRID:SCR_002798) version 8.1.2 for Mac OS X, GraphPad Software, La Jolla California, www.graphpad.com.
3. RESULTS
With the goal of identifying genes enriched to the regenerating blastema, a tissue‐mapped axolotl transcriptome was recently published (Bryant et al., 2017). Of particular interest are blastema‐enriched genes with high expression in the blastema and relative low expression in all other tissues sampled. We found that vwde was highly enriched to the axolotl limb blastema (Figure 1a). This analysis was limited to one‐time point, the medium‐bud blastema, and analysis of publicly available data indicates that vwde is likely expressed in limbs before amputation, with expression increasing with blastema formation ((Voss et al., 2015) https://ambystoma.uky.edu/). We next sought to determine spatial information about the expression of vwde across the regenerating limb. To understand the spatial and temporal regulation of vwde, we performed RNA in situ hybridizations over a time course of axolotl limb regeneration. We found vwde expression evident upon blastema formation and expressed exclusively in the blastema and not the overlying wound epidermis (Figure 1b–e). Thus, vwde fits the description of an axolotl limb blastema‐enriched gene and we were interested in pursuing whether vwde may be a core component of blastemas able to regenerate appendages.
Figure 1.
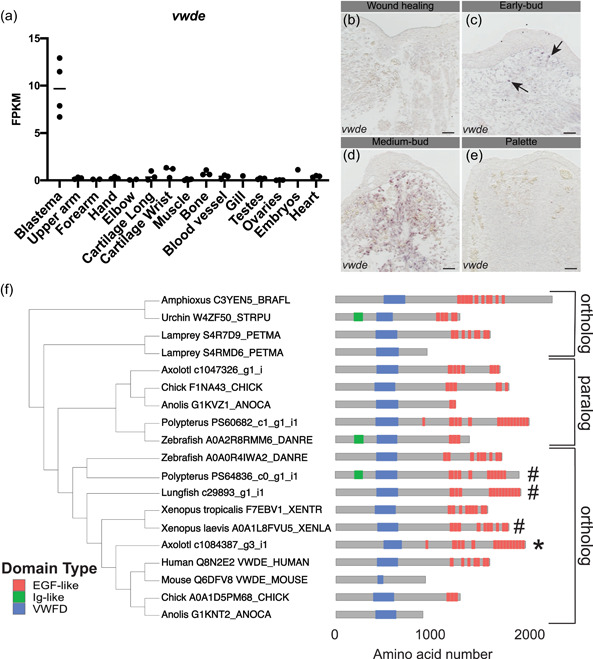
von willebrand factor D and EGF‐like domains (vwde) is a blastema‐enriched gene that is found across deuterostomes. (a) vwde (contig c1084387_g3_i1) expression in FPKM across tissues sampled from Bryant et al. (2017). Proximal and distal blastema samples are combined. (b–e) RNA in situ hybridization for vwde at (b) wound healing (3 days postamputation), (c) early‐bud blastema, (d) medium‐bud blastema, and (e) palette stage regenerating limbs. Black arrows indicate vwde expression, scale bar is 100 µm. (f) OrthoFinder 2.0 phylogeny with corresponding protein domain structure for putative Vwde orthologs. Protein domain pictures were generated with drawProteins (Brennan, 2018). Species and Uniprot ID or transcriptome contig number are included. Polypterus vwde contained multiple splice isoforms and the closest match to axolotl Vwde is shown here. Axolotl Vwde is denoted with “*” and other species Vwde that are described in this manuscript are marked with “#.” Orthologs to the Vwde studied in this study are indicated with brackets, paralog is also denoted with brackets. FPKM, fragments per kilobase of exon mapped [Color figure can be viewed at wileyonlinelibrary.com]
We next sought to determine if vwde was present in a selection of deuterostomes, including species with various regenerative abilities. Using a comparative genomics approach (Emms & Kelly, 2019) focusing only on deuterostome genomes and not protostomes, we found vwde to have orthologs across deuterostomes (though no ortholog was detected in C. intestinalis), as well as a nonblastema‐enriched paralogous gene in axolotl and other species (Figures 1f and S1). We compared axolotl Vwde to proteins from other species, and we found putative orthologs harboring predicted von Willebrand factor D domains and EGF‐like domains. The number of EGF domains may be more variable across species. However, since this gene has not yet been studied in‐depth in any species, additional experimental work may be required to fully characterize the expressed transcripts and proteins for individual species. Using these identified orthologs, we moved forward to ask whether vwde was a blastema‐enriched gene during paired fin regeneration.
We explored the possibility that vwde could be a common feature of blastemas responsible for regenerating paired fins, which share a deep homology with limbs (Shubin, Tabin, & Carroll, 1997), and likely share an inherited gene regulatory program for regeneration (Darnet et al., 2019; Nogueira et al., 2016). We chose two highly regenerative, but distantly related fish species to determine if vwde expression was a conserved feature of blastemas capable of regenerating paired appendages. These include a species in the sister group to tetrapods, the lungfish (L. paradoxa), which is a lobe‐finned fish, and P. senegalus, a ray‐finned fish that is capable of regenerating after amputation through skeletal elements that develop by endochondral ossification. We first inspected publicly available transcriptome datasets of lungfish and Polypterus regenerating fins for the vwde orthologs we previously identified (Figure 1f). The lungfish LG29893_g1_i1 contig was upregulated in blastemas 21 dpa relative to uninjured fins (Nogueira et al., 2016), and the Polypterus PS64836c0_g1_i1 contig was upregulated in 9 dpa blastemas relative to uninjured fins (Darnet et al., 2019). Assessment of expression levels via qPCR at various regeneration stages showed an upregulation of vwde coinciding with blastema formation during lungfish fin regeneration (Figure S2A). A similar pattern was seen for Polypterus fin regeneration, with expression reaching highest levels at 5 dpa (Figure S2B).
Next, we assessed the spatial pattern of vwde in histological sections of regenerating fins. lungfish 21 dpa blastemas show distal mesenchymal expression of vwde (Figure 2a). In 5 dpa Polypterus blastemas, expression is observed distal to the amputation plane in mesenchymal cells but also in the epithelium, suggesting that Polpyterus may use vwde in both compartments (Figure 2c and S2C,2D). In situ hybridization with control sense probes did not yield specific signal (Figure S2E–S2G). Histologically, these samples are similar to the medium‐bud blastema time point in which we identified vwde in the axolotl limb (Figures 2b and 2d). Together, these data indicate that vwde is expressed in regenerating fins and limbs and that vwde expression is a conserved feature of blastemas.
Figure 2.
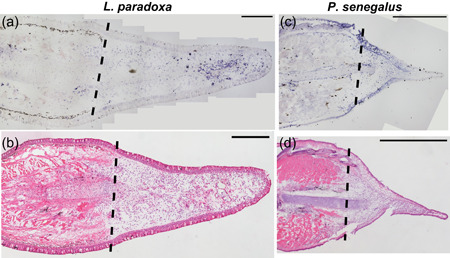
vwde is enriched in the regenerating fin of lungfish (Lepidosiren paradoxa) and Polypterus (P. senegalus). Expression pattern of vwde in the fin blastema tissues of L. paradoxa and P. senegalus. Longitudinal histological sections of fins from L. paradoxa at 21 dpa (a,b), and from P. senegalus at 5 dpa (c,d). (a,c) In situ hybridization using an antisense riboprobe to vwde. (b,d) H&E staining on sequential sections. All panels show posterior view, dorsal to the top. Dotted lines indicate amputation site (Scale bars = 1 mm in all panels). H&E, hematoxylin and eosin [Color figure can be viewed at wileyonlinelibrary.com]
Limbs are not the only appendages that highly regenerative animals are capable of regenerating. Many species, including X. laevis, can also regenerate tails at certain developmental stages (Beck, Christen, & Slack, 2003). Tails and limbs share similar tissue compositions (i.e., connective tissue and muscle), and both require the use of a blastema to regenerate lost tissue. To further investigate vwde during regeneration, we took advantage of the regeneration‐competent and regeneration‐refractory periods during X. laevis tail development (Aztekin et al., 2019; Beck et al., 2003). In X. laevis, a blastema forms in response to tail amputation during both distinct developmental stages, but only in the regeneration‐competent setting is full regeneration accomplished. This developmental feature provides an ideal situation to compare regeneration‐competent versus regeneration‐refractory environments. We reasoned that finding factors that differentiate these two contexts may provide clues for identifying the core requirements for successful regeneration. We probed for the expression of vwde during the regeneration‐competent and regeneration‐refractory periods of X. laevis tail development. Interestingly, we found that vwde expression was present in tails before amputation in both the regeneration‐competent and regeneration‐refractory setting (Figures 3a,b and 3g,h). We found robust vwde expression along the peripheral edge of the amputation plane and near the blastema in regeneration‐competent tails (Figure 3c–f). In contrast, in the regeneration‐refractory setting, vwde expression was restricted to the peripheral edges of the amputation plane and was not detected near the blastema (Figure 3i–l). This indicated a striking correlation between vwde expression and regeneration, providing evidence that vwde may be an important factor in forming a proregenerative niche. These expression data across a range of species indicate that vwde fits the profile of an evolutionarily conserved, regeneration‐enriched gene and that vwde may play an important role in the blastema niche.
Figure 3.
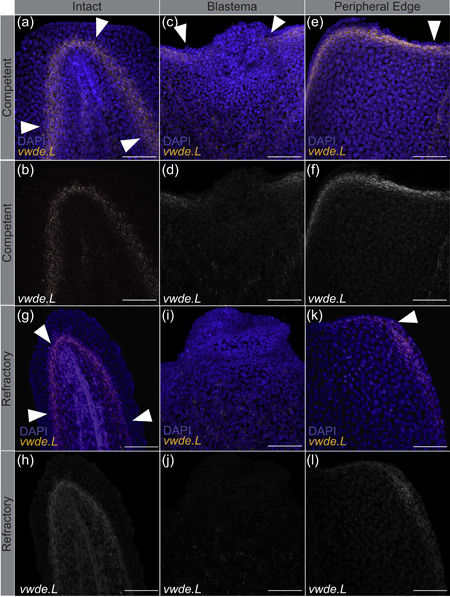
vwde expression is tightly linked with the regeneration‐competent environment. In situ hybridization chain reaction probing for vwde in (a–f) regeneration‐competent Xenopus laevis tails (a,b) before amputation, (c,d) blastema 24 hr postamputation, and (e,f) the peripheral edge of the amputation plane 24 hr postamputation. (g–l) Regeneration‐refractory tails (g,h) before amputation, (i,j) blastema 24 hr postamputation, and (k,l) the peripheral edge of the amputation plane 24 hr postamputation. White arrows indicate the location of vwde expression. Scale bars = 100 µm. DAPI, 4′,6‐diamidino‐2‐phenylindole [Color figure can be viewed at wileyonlinelibrary.com]
To investigate if vwde is required for regeneration, we performed morpholino‐mediated knockdown at its peak expression in the medium‐bud limb blastema. We found a substantial reduction in the length of the blastemas when Vwde was knocked down with two separate translation‐blocking morpholinos (Figure 4a,b). Fluorescent reporter constructs with vwde‐morpholino binding sites confirmed that both unique vwde‐targeting morpholinos were capable of blocking translation (Figure S3). Due to the dramatic reduction in blastema length, we investigated if vwde was important for blastema proliferation and/or cell survival. We found that knockdown of Vwde substantially reduced the number of EdU+ cells in the blastema (Figure 4c,d) and did not alter cell survival compared to control limbs (Figure S4). Due to the observed delay in blastema growth, we questioned whether blastemas treated with translation‐blocking morpholinos were capable of recovering from the transient knockdown of Vwde and produce fully regenerated limbs. We, therefore, performed the same Vwde morpholino‐mediated knockdown on a separate group of axolotls, and then allowed for the full course of regeneration to complete, harvesting limbs more than 8 weeks postamputation. We observed that one‐time injection of Vwde‐targeting morpholino caused substantial abnormalities in regenerated limbs, suggesting an essential role for vwde during limb regeneration (Figure 4e,g). We found defects in 4.2% (1/24) standard control (“Control (SC)”) treated limbs compared to 46% (13/28) of limbs treated with vwde MO1 and 31% (5/16) of limbs treated with vwde MO2 (Fisher's exact test p < .05; Figures 4f and S5; Table 2). A second experiment using an inverted control (“Control (INV)”) yielded similar results, with defects at endpoint in 20% (8/39) of Control (INV) treated limbs compared to 47% (18/38) of limbs treated with vwde MO1 (Fisher's exact test p < .05; Figures 4g and S6; Table 3). We found a variety of defects, some of which are reminiscent of limb development phenotypes where limited distal elements are present such as has been observed in fgf4,8‐double‐knockout mice (Mariani, Ahn, & Martin, 2008) and in the absence of sonic hedgehog (ssh) (Chiang et al., 1996). In addition, these phenotypes also resemble the defective regenerative spike characteristic of Xenopus limb regeneration (Dent, 1962). Altogether, these data highlight the functional requirement for vwde during limb regeneration.
Figure 4.
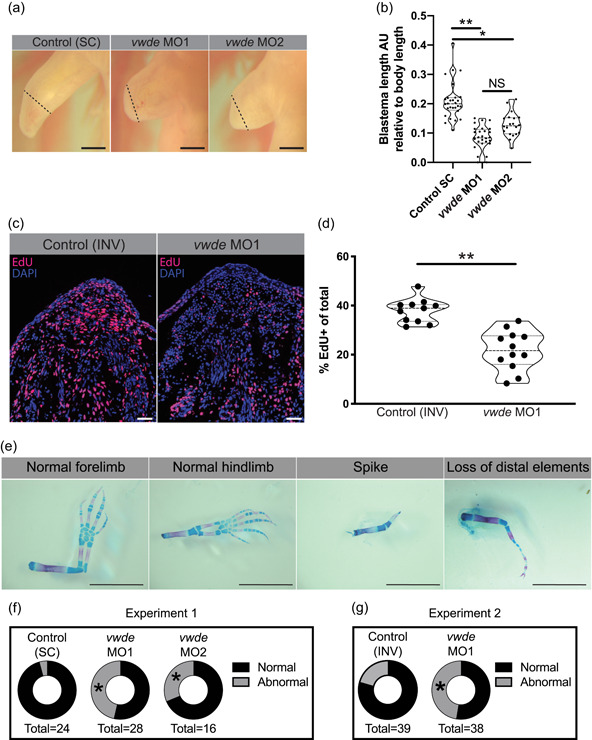
Vwde is essential for limb regeneration. (a) Representative images of blastemas 16 days postamputation (9 days postmorpholino administration) from standard control morpholino (“Control (SC)”), vwde‐targeting morpholino 1 (vwde MO1), and vwde‐targeting morpholino 2 (vwde MO2). Dotted line indicates amputation plane, blastemas are all tissue distal to amputation plane. Scale bars = 1 mm. (b) Quantification of blastema length 16 days postamputation. Median and quartiles noted with dotted lines, **p < .01, *p < .05 by nested T test. (c) Representative EdU stained sections of blastemas 10 dpa (3 days postelectroporation) of vwde morpholino 1 inverted (“Control (INV)”) and vwde‐targeting morpholino 1. Scale bars = 100 µm. (d) Quantification of percent of blastema cells positive for EdU in control (INV) and knockdown (vwde MO1). Each dot represents a limb, 4–5 animals per group. Median and quartiles noted with dotted lines, **p < .01 by nested T test. (e) Representative skeletal preparations of limbs after full regeneration after knockdown of Vwde at 7 dpa. From left to right, normal forelimb, normal hindlimb, spike, and loss of distal elements. Scale bars = 5 mm. (f) Donut plots of regenerative outcomes, pooled as abnormal versus normal from experiment with Control (SC), vwde MO1, and vwde MO2. *P < .05 by Fisher's exact test comparing Control (SC) versus vwde MO1 and Control (SC) versus vwde MO2. (g) Donut plots of regenerative outcomes, pooled as abnormal versus normal from experiment with Control (INV) and vwde MO1. *p < .05 by Fisher's exact test comparing outcomes from Control (INV) compared to vwde MO1. Edu, 5‐ethynyl‐2′‐deoxyuridine [Color figure can be viewed at wileyonlinelibrary.com]
Table 2.
Phenotypic outcomes of morpholino‐mediated knockdown in axolotl (standard control (“Control (SC)”) vs. vwde MO1, vwde MO2)
| Normal | Spike | Loss of distal elements | Oligodactyly | Polydactyly | Syndactyly | Additional elements | |
|---|---|---|---|---|---|---|---|
| Control (SC) | 23 | 0 | 0 | 0 | 0 | 1 | 0 |
| vwde MO1 | 15 | 4 | 5 | 4 | 0 | 0 | 0 |
| vwde MO2 | 11 | 0 | 0 | 3 | 1 | 1 | 0 |
Table 3.
Phenotypic outcomes of morpholino‐mediated knockdown in axolotl (Inverted control (“Control (INV)”) vs. vwde MO1)
| Normal | Spike | Loss of distal elements | Oligodactyly | Polydactyly | Syndactyly | Additional elements | |
|---|---|---|---|---|---|---|---|
| Control (INV) | 30 | 0 | 4 | 4 | 0 | 0 | 0 |
| vwde MO1 | 20 | 6 | 9 | 1 | 0 | 1 | 1 |
4. DISCUSSION
Recent work, most notably next generation sequencing, has led to a plethora of information about the genes and cells that define the blastema (Bryant et al., 2017; Darnet et al., 2019; Gerber et al., 2018; Knapp et al., 2013; Leigh et al., 2018; Monaghan et al., 2012, 2009; Nogueira et al., 2016; Stewart et al., 2013; Voss et al., 2015; Wu et al., 2013). However, it is difficult to determine which genes may have functional relevance based purely on their expression. We decided to investigate a single blastema‐enriched gene, vwde, using an evolutionarily informed approach, assuming that a gene whose expression is enriched in blastemas of multiple, distantly related, species is likely a key factor during regeneration.
The in vivo assays used here implicate Vwde as a putative growth factor during axolotl limb regeneration. Proliferation is a complex, but fundamental aspect of regeneration, as there are many different cell types and potential origins of proliferative signals. Previous work indicates that proliferative signals are produced directly following amputation independent of the nerve or wound epidermis (Johnson, Bateman, DiTommaso, Wong, & Whited, 2017; Mescher & Tassava, 1975), but are also provided by the nerve (Brockes & Kintner, 1986; Farkas, Freitas, Bryant, Whited, & Monaghan, 2016) or wound epidermis (Sugiura, Wang, Barsacchi, Simon, & Tanaka, 2016). There are thus multiple sources of proliferative signals in the regenerating limb, but it is unclear if proliferative signals from multiple tissues are required simultaneously or perhaps in a more stepwise fashion to maintain blastema proliferation. Our data indicate that Vwde may be a growth factor derived from cells residing in the blastema, which adds to the potential sources of such signals in the regenerating limb. Interestingly, the in situ profiles in axolotl appear to have rather ubiquitous expression, but due to the high cellular heterogeneity of blastema‐resident cells it will be important to determine if vwde is produced by mesenchymal progenitor cells, blastema‐resident hematopoietic cells, or both. It has been previously postulated that nerve‐derived signals are required early on during blastema formation and growth, but a fibroblast‐derived factor is required for complete regeneration (Endo, Bryant, & Gardiner, 2004). We speculate that vwde, which appears to be expressed across the majority of cells in the blastema, may provide one of the essential proliferative cues derived from blastema cells that is important after the nerve has provided sufficient input. While there is limited knowledge of blastema cell‐derived growth factors, in vitro cultures have shown that blastema protein extracts are able to drive blastema cell proliferation (Boilly & Albert, 1990). More generally, a global and temporally based view of the cellular origins of growth factors and the cell types that require these factors will provide a better understanding of what is driving proliferation during different stages of regeneration.
In addition to the dramatic reduction in number of cells undergoing DNA synthesis, we observed striking end point phenotypes after the transient knockdown of Vwde. The loss of distal elements and spike‐like phenotypes observed after Vwde knockdown suggests the possibility that Vwde could play a role in proximal‐distal determination in the regenerating limb. These phenotypes showing similarities to ssh and fgf4,8‐double‐knockout mice, suggest that Vwde may be working similarly to—or in concert with—FGFs during regeneration. In line with this notion, genes similar to human VWDE include fibulins, which in mice have previously been shown to directly interact with FGF8 (Fresco, Kern, Mohammadi, & Twal, 2016). Though many FGFs are epidermal factors during limb development in mice and chick, FGFs are expressed in the mesenchyme during axolotl limb development (Purushothaman, Elewa, & Seifert, 2019) and regeneration (Nacu, Gromberg, Oliveira, Drechsel, & Tanaka, 2016). Thus, it may be that during axolotl limb regeneration, blastema‐derived factors are primarily responsible for proximal‐distal patterning and that Vwde is working to promote the formation of distal elements. However, Vwde may not directly influence patterning and it will be important to determine if Vwde could be working with or promoting/inhibiting known regulators of skeletal patterning. Intriguingly, vwde has remained unexplored in highly studied, but less regenerative species such as mouse and human, so whether vwde plays a role in limb development in these species is unknown.
Due to the dearth of information on vwde, considering the function of protein domains or paralogs may provide insights to potential functions. The repeated EGF‐like domains, as well as the von Willebrand factor D domain and the relationship with fibulins, suggest that vwde may be an extracellular matrix (ECM) glycoprotein. The ECM plays an integral role in limb regeneration, instructing cellular behaviors such as DNA synthesis and migration (Calve, Odelberg, & Simon, 2010). Further, fibulins have been shown to be an essential component of the ECM during limb development (Debeer et al., 2002). Thus, it will be interesting to tease apart how vwde fits into the complex, proregenerative ECM and which domains are required during regeneration as EGF‐like domains have been implicated in a variety of functions required for successful regeneration including proliferation, migration, and differentiation (Singh & Harris, 2005). Previous work in newt (Wang, Marchionni, & Tassava, 2000) and axolotl (Farkas et al., 2016) with neuregulins, which feature EGF domains, have implicated EGF/EGFR in nerve‐dependent limb regeneration. Future experiments may intersect the function of Vwde with those factors described in one or more of these existing studies.
It is interesting to speculate on what has been lost in amniotes that prevents appendage regeneration. One possibility is genes that are lost in amniotes and present in anamniotes can explain differences in regenerative capacity (Korotkova et al., 2019). However, the absence of a gene in amniotes is not necessarily a prerequisite when considering which candidate genes might be responsible for high regenerative capacity. Alternative scenarios include, but are not limited to, genes that have lost ancestral proregenerative function or have altered expression domains/kinetics. Vwde may fit the paradigm of a gene that is present in both regeneration‐competent and regeneration‐incompetent species, but may exclusively be used in the blastema, a structure that cannot be produced by most regeneration‐incompetent species.
While the blastema is required for regeneration, wound healing and activation of progenitor cells required for the formation of the blastema must precede blastema formation. Based on the expression profile, we do not expect vwde to be a driver of blastema formation, but more likely a downstream effector once a blastema has been established. In most cases of amputation in less regenerative species, the blastema is not able to form, and thus we suspect that a more upstream or systemic factor may prevent blastema formation. While there may have initially been one primary cause of the loss of regenerative ability, such as the rise of adaptive immunity (Godwin, Pinto, & Rosenthal, 2017) or trade‐offs associated with endothermy (Hirose et al., 2019), it is likely that other aspects of the regenerative response have now been lost due to their lack of utility. If vwde played a relatively specialized function in the blastema and blastemas generally do not exist in less regenerative species then the use for vwde decreases. This could explain why 42.7% of human genomes have a predicted loss‐of‐function copy of VWDE, leading to speculation that VWDE is potentially drifting towards inactivation in the human population (MacArthur et al., 2012). While the blastema remains the elusive feature required for appendage regeneration, this study illustrates that taking an evolutionarily informed approach can lead to identification of functionally important genes. This also suggests that further work to understand the similarities between different species blastemas may help to elucidate the core molecular program of the blastema.
AUTHOR CONTRIBUTIONS
N. D. L.: Wrote the paper, initiation/conception, animal experiments/in situ, orthology analysis, experimental design, and manuscript editing. S. S., A. C. D., D. P. D., J. F. S., G. S. D.: Animal experiments/in situ, experimental design, manuscript editing. A. N. A., K. J., M. L.: Animal experiments/in situ and Manuscript editing. B. J. H.: Initiation/conception and Manuscript editing. I. S.: Experimental design, Manuscript editing, Contributed resources. J. L. W.: Wrote the paper, initiation/conception, corresponding author, animal experiments/in situ, experimental design, manuscript editing, and contributed resources.
Supporting information
Supporting information
Supporting information
Supporting information
ACKNOWLEDGMENTS
This study was supported by the Eunice Kennedy Shriver National Institute of Child and Human Development (R03HD083434 and R01HD095494 to Jessica L. Whited) and the Office of the Director (DP2HD087953 to Jessica L. Whited) of the National Institutes of Health. Nicholas D. Leigh was supported by Award Number F32HD092120 from the Eunice Kennedy Shriver National Institute of Child and Human Development of the National Institutes of Health. This study was supported by the Allen Discovery Center program through The Paul G. Allen Frontiers Group (12171 to Michael Levin). Portions of this study were conducted on the O2 High Performance Compute Cluster, supported by the Research Computing Group, at Harvard Medical School. See http://rc.hms.harvard.edu for more information. Support for work on lungfish and Polypterus was provided by CNPq Universal Program (Grant 403248/2016‐7) and CAPES/DAAD PROBRAL (Grant 88881.198758/2018‐01) to Igor Schneider, and a postdoctoral fellowship from CNPq to Aline C. Dragalzew. We thank William Ye, Adam Gramy, Sarah Lemire, and Bonney Couper‐Kiablick for expert animal care and other members of the Whited lab for feedback and discussions. We also thank Dr. Mansi Srivastava and Dr. Ryan Walker for their insights on orthology and protein structure, Dr. Jenny Lanni for insightful input, and Dr. James Monaghan and Timothy Duerr for sharing information on whole mount in situ protocols.
Leigh ND, Sessa S, Dragalzew AC, et al. von Willebrand factor D and EGF domains is an evolutionarily conserved and required feature of blastemas capable of multitissue appendage regeneration. Evolution & Development. 2020;22:297–311. 10.1111/ede.12332
REFERENCES
- Aztekin, C. , Hiscock, T. W. , Marioni, J. C. , Gurdon, J. B. , Simons, B. D. , & Jullien, J. (2019). Identification of a regeneration‐organizing cell in the xenopus tail. Science, 364(6441), 653–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, C. W. , Christen, B. , & Slack, J. M. W. (2003). Molecular pathways needed for regeneration of spinal cord and muscle in a vertebrate. Developmental Cell, 5(3), 429–439. [DOI] [PubMed] [Google Scholar]
- Boilly, B. , & Albert, P. (1990). In vitro control of blastema cell proliferation by extracts from epidermal cap and mesenchyme of regenerating limbs of axolotls. Roux's Archives of Developmental Biology, 198(8), 443–447. [DOI] [PubMed] [Google Scholar]
- Brennan, P. (2018). drawProteins: A Bioconductor/R package for reproducible and programmatic generation of protein schematics. F1000Research, 7(July), 1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockes, J. P. , & Kintner, C. R. (1986). Glial growth factor and nerve‐dependent proliferation in the regeneration blastema of urodele amphibians. Cell, 45(2), 301–306. [DOI] [PubMed] [Google Scholar]
- Bryant, D. M. , Johnson, K. , DiTommaso, T. , Tickle, T. , Couger, M. B. , Payzin‐Dogru, D. , … Lee, J. L. (2017). A tissue‐mapped axolotl de novo transcriptome enables identification of limb regeneration factors. Cell Reports, 18(3), 762–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calve, S. , Odelberg, S. J. , & Simon, H. G. (2010). A transitional extracellular matrix instructs cell behavior during muscle regeneration. Developmental Biology, 344(1), 259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang, C. , Litingtung, Y. , Lee, E. , Youngt, K. E. , Cordent, J. L. , Westphal, H. , & Beachyt, P. A. (1996). Cyclopia and defective axial patterning in mice lacking sonic hedgehog gene function. Nature, 383(6599), 407–413. [DOI] [PubMed] [Google Scholar]
- Choi, H. M. T. , Calvert, C. R. , Husain, N. , Huss, D. , Barsi, J. C. , Deverman, B. E. , … Hunter, N. A. (2016). Mapping a multiplexed zoo of mRNA expression. Development, 143(19), 3632–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, H. M. T. , Schwarzkopf, M. , Fornace, M. E. , Acharya, A. , Artavanis, G. , Stegmaier, J. , … Pierce, N. A. (2018). Third‐generation in situ hybridization chain reaction: Multiplexed, quantitative, sensitive, versatile, robust. Development, 145(12), dev165753. 10.1242/dev.165753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnet, S. , Dragalzew, A. C. , Amaral, D. B. , Sousa, J. F. , Thompson, A. W. , Cass, A. N. , … Lorena, I. (2019). Deep evolutionary origin of limb and fin regeneration. Proceedings of the National Academy of Sciences of the United States of America, 116(30), 15106–15115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeer, P. , Schoenmakers, E. F. P. M. , Twal, W. O. , Argraves, W. S. , De Smet, L. , Fryns, J. P. , & Van De Ven, W. J. M. (2002). The fibulin‐1 gene (FBLN1) is disrupted in a t(12;22) associated with a complex type of synpolydactyly. Journal of Medical Genetics, 39(2), 98–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent, J. N. (1962). Limb regeneration in larvae and metamorphosing individuals of the South African clawed toad. Journal of Morphology, 110(1), 61–77. [DOI] [PubMed] [Google Scholar]
- Emms, D. M. , & Kelly, S. (2019). OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biology, 20(238), 10.1186/s13059-019-1832-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo, T. , Bryant, S. V. , & Gardiner, D. M. (2004). A stepwise model system for limb regeneration. Developmental Biology, 270(1), 135–145. [DOI] [PubMed] [Google Scholar]
- Farkas, J. E. , Freitas, P. D. , Bryant, D. M. , Whited, J. L. , & Monaghan, J. R. (2016). Neuregulin‐1 signaling is essential for nerve‐dependent axolotl limb regeneration. Development, 143(15), 2724–2731. [DOI] [PubMed] [Google Scholar]
- Fresco, V. M. , Kern, C. B. , Mohammadi, M. , & Twal, W. O. (2016). Fibulin‐1 binds to fibroblast growth factor 8 with high affinity: Effects on embryo survival. The Journal of Biological Chemistry, 291(36), 18730–18739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröbisch, N. B. , Bickelmann, C. , Olori, J. C. , & Witzmann, F. (2015). Deep‐time evolution of regeneration and preaxial polarity in tetrapod limb development. Nature, 527(7577), 231–234. [DOI] [PubMed] [Google Scholar]
- Fröbisch, N. B. , Constanze, B. , & Florian, W. (2014). Early evolution of limb regeneration in tetrapods: Evidence from a 300‐million‐year‐old amphibian. Proceedings of the Royal Society B: Biological Sciences, 281(1794):20141550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber, T. , Murawala, P. , Knapp, D. , Masselink, W. , Schuez, M. , Hermann, S. , … Gac‐Santel, B. (2018). Single‐cell analysis uncovers convergence of cell identities during axolotl limb regeneration. Science, 362(6413), eaaq0681. 10.1126/science.aaq0681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godwin, J. W. , Pinto, A. R. , & Rosenthal, N. A. (2017). Chasing the recipe for a pro‐regenerative immune system. Seminars in Cell and Developmental Biology, 61(January), 71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose, K. , Payumo, A. Y. , Cutie, S. , Hoang, A. , Zhang, H. , Guyot, R. , … Lunn, G. N. (2019). Evidence for hormonal control of heart regenerative capacity during endothermy acquisition. Science, 364(6436), 184–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, S. , Apweiler, R. , Attwood, T. K. , Bairoch, A. , Bateman, A. , Binns, D. , … Bork, C. (2009). InterPro: The integrative protein signature database. Nucleic Acids Research, 37(Database issue), D211–D215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson, D. H. , & Scornavacca, C. (2012). Dendroscope 3: An interactive tool for rooted phylogenetic trees and networks. Systematic Biology, 61(6), 1061–1067. [DOI] [PubMed] [Google Scholar]
- Johnson, K. , Bateman, J. , DiTommaso, T. , Wong, A. Y. , & Whited, J. L. (2017). Systemic cell cycle activation is induced following complex tissue injury in axolotl. Developmental Biology, 433, 461–472. 10.1016/j.ydbio.2017.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp, D. , Schulz, H. , Rascon, C. A. , Volkmer, M. , Scholz, J. , Nacu, E. , … Le, E. M. (2013). Comparative transcriptional profiling of the axolotl limb identifies a tripartite regeneration‐specific gene program. PLOS One, 8(5):e61352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkova, D. D. , Lyubetsky, V. A. , Ivanova, A. S. , Rubanov, L. I. , Seliverstov, A. V. , Zverkov, O. A. , … Martynova, A. G. (2019). Bioinformatics screening of genes specific for well‐regenerating vertebrates reveals c‐answer, a regulator of brain development and regeneration. Cell Reports, 29(4), 1027–1040.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh, N. D. , Dunlap, G. S. , Johnson, K. , Mariano, R. , Oshiro, R. , Wong, A. Y. , … Bryant, J. L. (2018). Transcriptomic landscape of the blastema niche in regenerating adult axolotl limbs at single‐cell resolution. Nature Communications, 9(1), 5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K. J. , & Schmittgen, T. D. (2001). Analysis of relative gene expression data using real‐time quantitative pcr and the 2(‐delta delta C(T)) method. Methods, 25(4), 402–408. [DOI] [PubMed] [Google Scholar]
- MacArthur, D. G. , Balasubramanian, S. , Frankish, A. , Huang, N. , Morris, J. , Walter, K. , … Jostins, C. (2012). A systematic survey of loss‐of‐function variants in human protein‐coding genes. Science, 335(6070), 823–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani, F. V. , Ahn, C. P. , & Martin, G. R. (2008). Genetic evidence that FGFs have an instructive role in limb proximal‐distal patterning. Nature, 453(7193), 401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda, T. , & Cepko, C. L. (2004). Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proceedings of the National Academy of Sciences of the United States of America, 101(1), 16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mescher, A. L. , & Tassava, R. A. (1975). Denervation effects on DNA replication and mitosis during the initiation of limb regeneration in adult newts. Developmental Biology, 44(1), 187–197. [DOI] [PubMed] [Google Scholar]
- Monaghan, J. R. , Athippozhy, A. , Seifert, A. W. , Putta, S. , Stromberg, A. J. , Maden, M. , … Voss, S. R. (2012). Gene expression patterns specific to the regenerating limb of the Mexican axolotl. Biology Open, 1(10), 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan, J. R. , Epp, L. G. , Putta, S. , Page, R. B. , Walker, J. A. , Beachy, C. K. , … Zhu, S. R. (2009). Microarray and cDNA sequence analysis of transcription during nerve‐dependent limb regeneration. BMC Biology, 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacu, E. , Gromberg, E. , Oliveira, C. R. , Drechsel, D. , & Tanaka, E. M. (2016). FGF8 and SHH substitute for anterior‐posterior tissue interactions to induce limb regeneration. Nature, 533(7603), 407–410. [DOI] [PubMed] [Google Scholar]
- Nogueira, A. F. , Costa, C. M. , Lorena, J. , Moreira, R. N. , Frota‐Lima, G. N. , Furtado, C. , … Schneider, I. (2016). Tetrapod limb and sarcopterygian fin regeneration share a core genetic programme. Nature Communications, 7, 13364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purushothaman, S. , Elewa, A. , & Seifert, A. W. (2019). Fgf‐signaling is compartmentalized within the mesenchyme and controls proliferation during salamander limb development. eLife, 8(e48507), 10.7554/eLife.48507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shubin, N. , Tabin, C. , & Carroll, S. (1997). Fossils, genes and the evolution of animal limbs. Nature, 388(6643), 639–648. [DOI] [PubMed] [Google Scholar]
- Singh, A. B. , & Harris, R. C. (2005). Autocrine, paracrine and juxtacrine signaling by EGFR ligands. Cellular Signalling, 17(10), 1183–1193. [DOI] [PubMed] [Google Scholar]
- Sive, H. L. , Grainger, R. M. , & Harland, R. M. (2010, March 8). Early development of Xenopus Laevis by Hazel L Sive, Robert M Grainger | Waterstones. Retrieved from https://www.waterstones.com/book/early‐development‐of‐xenopus‐laevis/hazel‐l‐sive//9780879699420
- Stewart, R. , Rascón, C. A. , Tian, S. , Nie, J. , Barry, C. , Chu, L. F. , … Ardalani, C. N. (2013). Comparative RNA‐Seq analysis in the unsequenced axolotl: The oncogene burst highlights early gene expression in the blastema. PLOS Computational Biology, 9(3):e1002936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura, T. , Wang, H. , Barsacchi, R. , Simon, A. , & Tanaka, E. M. (2016). MARCKS‐like protein is an initiating molecule in axolotl appendage regeneration. Nature, 531(7593), 237–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium . (2019). UniProt: A worldwide hub of protein knowledge. Nucleic Acids Research, 47(D1), D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss, S. R. , Palumbo, A. , Nagarajan, R. , Gardiner, D. M. , Muneoka, K. , Stromberg, A. J. , & Athippozhy, A. T. (2015). Gene Expression during the first 28 days of axolotl limb regeneration I: Experimental design and global analysis of gene expression. Regeneration, 2(3), 120–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Marchionni, M. A. , & Tassava, R. A. (2000). Cloning and neuronal expression of a type III newt neuregulin and rescue of denervated, nerve‐dependent newt limb blastemas by rhGGF2. Journal of Neurobiology, 43(2), 150–158. [DOI] [PubMed] [Google Scholar]
- Whited, J. L. , Lehoczky, J. A. , & Tabin, C. J. (2012). Inducible genetic system for the axolotl. Proceedings of the National Academy of Sciences of the United States of America, 109(34), 13662–13667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C. H. , Tsai, M. H. , Ho, C. C. , Chen, C. Y. , & Lee, H. S. (2013). De novo transcriptome sequencing of axolotl blastema for identification of differentially expressed genes during limb regeneration. BMC Genomics, 14, 434. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Supporting information
Supporting information


