Abstract
D cyclins include three isoforms: D1, D2, and D3. D cyclins heterodimerize with cyclin-dependent kinase 4/6 (CDK4/6) to form kinase complexes that can phosphorylate and inactivate Rb. Inactivation of Rb triggers the activation of E2F transcription factors, which in turn regulate expression of genes whose products drive cell cycle progression. Because D-type cyclins function as mitogenic sensors that link growth factor signaling directly with G1 phase progression, it is not surprising that D cyclin accumulation is dysregulated in a variety of human tumors. Elevated expression of D cyclins results from gene amplification, increased gene transcription and protein translation, decreased microRNA levels, and inefficiency or loss of ubiquitylation-mediated protein degradation. This review focuses on the clinicopathological importance of D cyclins, how dysregulation of Ubiquitin-Proteasome System (UPS) contributes to the overexpression of D cyclins, and the therapeutic potential through targeting D cyclin-related machinery in human tumors.
Keywords: D cyclin, E3 Ubiquitin Ligase, Protein degradation, Cancer
1. Introduction
Cell cycle defines the process through which cells duplicate their genome and divide into two daughter cells [1]. There are four phases of cell cycle: G1, cell growth and generation of proteins required for DNA replication; S-phase, genome duplication; G2, Gap phase to monitor fidelity of DNA replication; and M-phase, cell division [2, 3]. Progression through the cell cycle is catalyzed by cyclins and cyclin-dependent kinases (CDKs) [2, 4]. Cyclins can activate their cognate CDKs through the formation of heterodimers, which ultimately catalyze the phosphorylation of downstream genes to promote cell cycle progression. Due to their significance in regulating cell cycle progression, the activities of CDKs are tightly regulated by the abundance of cyclins and CDK inhibitory machinery like INK4, CIP and KIP family proteins [2, 5].
G1 phase is unique as it regulates cell cycle re-entry in response to extrinsic factors such as growth factors, nutrients, and integrin-related adhesion signaling during G1 phase [6]. The D-type cyclins along with their cognate CDKs, CDK4/6, are exquisitely sensitive to such extrinsic signals and serve to integrate such signals with cell division. Mammalian cells encode three D-type cyclins, D1, D2, and D3, with approximately 57% identity for the coding region among different isoforms, and 78% identity for the N-terminal “cyclin box” domain [7]. Moreover, D cyclins also possess a C-terminal phospho-degron and an Rb binding motif [8]. D cyclins heterodimerize with CDK4/6 to form kinase complexes, which phosphorylate and inactivate retinoblastoma (Rb) protein, leading to the release of the E2F transcription factors, thereafter, the expression of downstream genes to drive cell cycle progression [9] (Fig. 1).
Figure 1. Schematic illustration of cell cycle regulation.
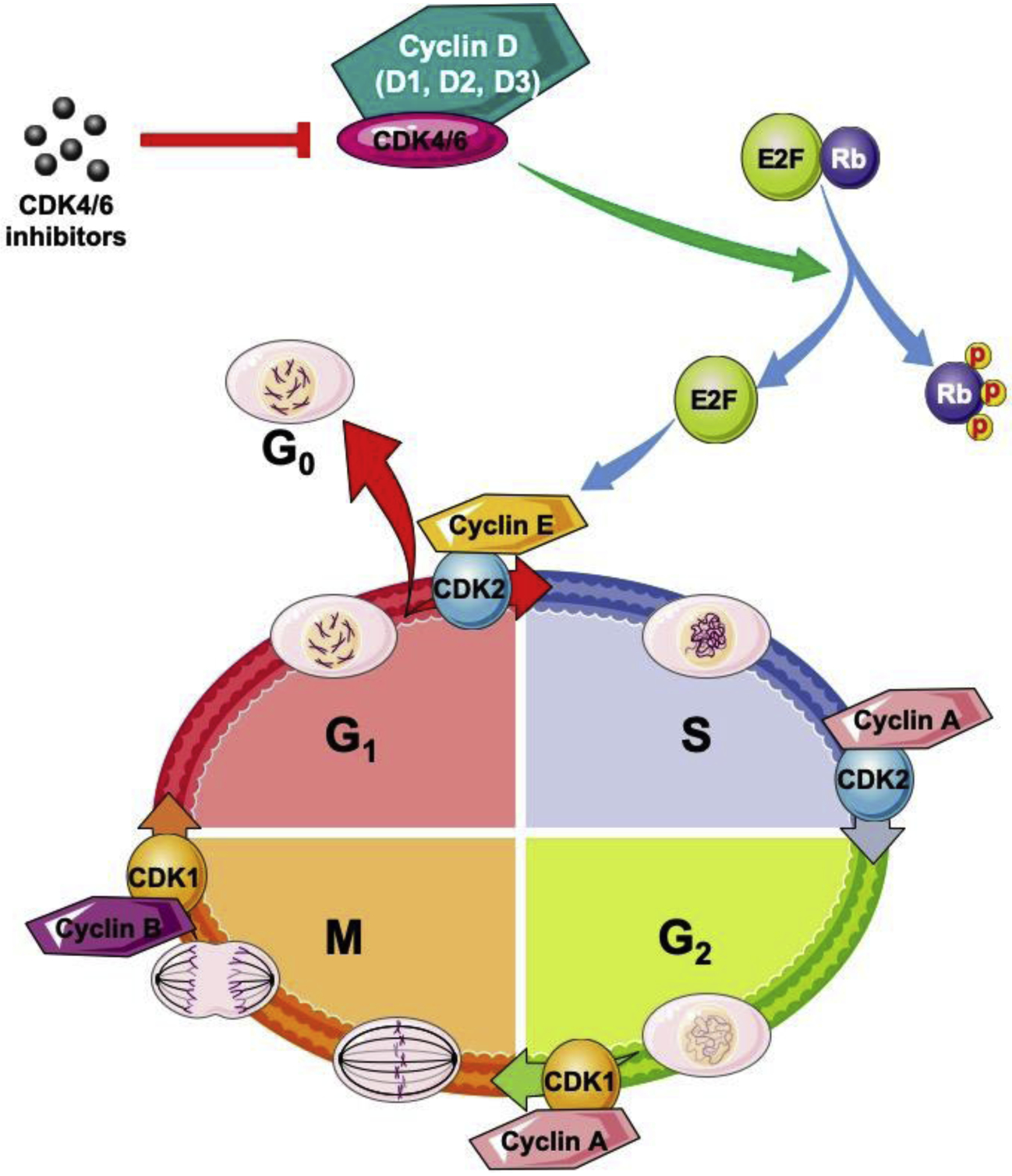
Cell cycle is divided into four phases: G1 (gap phase 1), S (DNA synthesis), G2 (gap phase 2), and M (mitosis). Cell cycle progression is regulated by cyclins and their cognate CDKs. As the important regulator of G1 phase, CDK4/6 inhibitors have been developed, for example, PD-0332991 (palbociclib), LY2835219 (abemaciclib), and LEE011 (ribociclib) that are approved by U.S. Food and Drug Administration to treat human tumors.
The Restriction Point (R), analogous to “start” in budding yeast, occurs in late G1 phase and represents an experimentally defined point where cells are committed to one round of cell division. Growth factor and nutrient signals are required for cells to reach R point. Molecularly, this reflects the assembly and activation of the D-type cyclin-CDK4/6 kinase and the initial phosphorylation-dependent inactivation of Rb. This initial Rb phosphorylation triggers an initial wave of transcription and activation of subsequent cyclin E-CDK2 and cyclin A-CDK2 kinases that drive G1 to S-phase transition at which point cells are committed to finishing one round of cell division [10, 11]. Given the central role of D-type cyclins as sensors for various growth factors, it stands to reason that accumulation and activity of cyclin D-CDK4/6 kinase is dysregulated in a large fraction of human cancers [1, 2]. Elevated expression of D cyclins reflects gene amplification, increased gene transcription and protein translation, decreased microRNA expression, and inefficiency or loss of ubiquitylation-mediated protein degradation [2, 7]. This review will focus on the clinic-pathological importance of D cyclins, how dysregulation of Ubiquitin-Proteasome System (UPS) contributes to the overexpression of D cyclins, and the therapeutic potential through targeting D cyclin-related machinery in human tumors.
2. D cyclins and human cancers
2.1. Cyclin D1
Cyclin D1 is a direct target of mitogenic signaling cascades including epidermal growth factor receptor (EGFR) [12, 13], and phosphatidylinositol 3-kinase (PI3K)-Akt [14–16], and it is sensitive to transcription factors such as β-Catenin [17, 18], and nuclear factor-kappa B (NF-κB) [19, 20]. In addition, the CCND1 gene is amplified in a variety of human cancers including pancreatic cancer [21], non-small cell lung carcinoma (NSCLC) [22–24], breast cancer [25], head and neck squamous cell carcinoma (HNSCC) [26, 27], melanoma [28], endometrial cancer [29, 30], esophageal squamous cell carcinoma (ESCC) [31, 32], and colorectal carcinoma [7] (Table 1). CCND1 is dysregulated via chromosome translocation in mantle cell lymphoma (MCL) and multiple myeloma [33, 34]. Overexpression of cyclin D1 can also be driven through mutations in the 3’-untranslated region, resulting in reduced mRNA degradation-mediated by microRNAs [35–39]. In addition, cyclin D1 levels directly correlate with tumor size, local invasion and metastasis as well as advanced clinical stages in human cancers [1, 2, 7]. These observations support cyclin D1 as an important prognostic indicator for patients with pancreatic adenocarcinoma, lung cancer, breast cancer, HNSCC, cutaneous melanoma, endometrial cancer, colorectal carcinoma, ESCC and MCL [7, 40]. Furthermore, its clinical importance highlights the therapeutic potential through targeting cyclin D1-CDK4/6 kinase complexes in human cancers [41].
Table 1.
Amplification and overexpression of D cyclins in human tumors
| D Cyclin | Enzymes | Tumors | Incidence Rate | References |
|---|---|---|---|---|
| D1 | CCND1 Amplification | ESCC | 23–57% | [31, 32, 174] |
| HNSCC | 26–39% | [27, 175] | ||
| NSCLC | 5–30% | [22, 23] | ||
| Endometrial cancer | 26% | [29, 30] | ||
| Melanoma | 0–25% | [7, 28] | ||
| Pancreatic cancer | 25% | [7, 21] | ||
| Breast cancer | 15–20% | [22, 25] | ||
| Colorectal carcinoma | 2.5% | [7, 176] | ||
| Overexpression | ESCC | 42–87% | [177, 178] | |
| HNSCC | 20–68% | [27, 175] | ||
| NSCLC | 18–76% | [24, 58, 179] | ||
| Endometrial cancer | 40–56% | [29, 30] | ||
| Melanoma | 30–65% | [7, 28] | ||
| Pancreatic cancer | 42–82% | [7, 21] | ||
| Breast cancer | 50–70% | [22, 25] | ||
| Colorectal carcinoma | 55% | [7, 180] | ||
| D2 | CCND2 Amplification | Germ cell tumor | 15.38% | [181] |
| Adrenal carcinoma | 12% | [182] | ||
| Glioma | 1.95–5% | [44, 181, 182] | ||
| ESCC | 0.58–5% | [181, 182] | ||
| Lung SCC | 1–4% | [181, 182] | ||
| Colorectal carcinoma | 0.97–3% | [181] | ||
| Breast cancer | 0.61–3% | [181, 182] | ||
| Non-Hodgkin lymphoma | 0.92% | [181] | ||
| Overexpression | Lymphoplasmacytic lymphoma | 100% | [183] | |
| B-cell chronic lymphocytic leukemia | 85% | [183] | ||
| Colon carcinoma | 57% | [184] | ||
| AML | 47% | [183] | ||
| Gastric cancer | 26.2% | [55] | ||
| Diffuse large B-cell lymphoma | 10.6% | [185] | ||
| D3 | CCND3 Amplification | Colorectal carcinoma | 0.3–21% | [181, 182, 186] |
| ESCC | 1.78–10% | [181, 182] | ||
| HNSCC | 4% | [182] | ||
| Gastric cancer | 1.96–3% | [181, 182] | ||
| Breast cancer | 0.73–3% | [181, 182] | ||
| Urothelial carcinoma | 3% | [182] | ||
| Overexpression | Thyroid carcinoma | 72% | [187] | |
| Melanoma | 20–42% | [188] | ||
| B-cell chronic lymphocytic leukemia | 20–41% | [189–191] | ||
| Oral SCC | 39% | [192] | ||
| Colon carcinoma | 21–35% | [184, 186] | ||
| Breast cancer | 30% | [193] | ||
| Non-Hodgkin lymphoma | 22% | [194] | ||
| Renal cell carcinoma | 16% | [195] |
2.2. Cyclin D2
Less is known about the contribution of cyclin D2 to tumor development and progression. Cyclin D2 is a downstream target of c-Myc and RUNX1/ETO oncogenes [42, 43]. Hyperactivation of proto-oncogenic signaling by these oncogenic factors accelerates cell cycle progression in part through increased cyclin D2 transcription. In addition, the CCND2 gene is amplified in glioblastoma and anaplastic astrocytoma, but without significant overexpression of cyclin D2 mRNA [44] (Table 1). In contrast, in gastric cancer, overexpression of cyclin D2 is the consequence of hypermethylation of CCND2 promoter [45, 46]. In addition to the aforementioned mechanisms, cyclin D2 mutations are identified in patients with acute myeloid leukemia (AML) [47]; these mutations, localize to the C-terminus, which contains sequences that direct ubiquitin-dependent degradation and thus the mutations appear to contribute directly to increased stability of cyclin D2 protein. In T-cell acute lymphoblastic leukemia (T-ALL), chromosomal translocations involving CCND2 locus and T-cell receptors have been revealed to account for the overexpression of cyclin D2 [48]. Furthermore, the stability of cyclin D2 mRNA is also regulated by microRNAs in thyroid carcinoma [49], NSCLC [50], prostate cancer [51], and oral squamous cell carcinoma [52]; insufficient expression or lack of those microRNAs also results in elevated cyclin D2 protein levels. Overexpression of cyclin D2 is associated with tumor invasion, metastasis and the prognosis in patients with colon cancer and gastric carcinoma [53–55].
2.3. Cyclin D3
Cyclin D3 is also implicated in human cancers of hematopoietic origin [56]. Genomic rearrangements of CCND3 locus are found in multiple myeloma and diffuse large cell lymphoma. In addition, several microRNAs like miR-4779, miR-212, miR-138, and miR-503 can destabilize cyclin D3 mRNA and block cell cycle progression in cell lines derived from colon cancer, T-cell leukemia/lymphoma, lung cancer, and hepatocellular carcinoma (HCC) [57–60]. Like cyclin D1, cyclin D3 protein degradation is determined by a C-terminal phospho-degron [61, 62]. However, the E3 ligase that directs phosphorylation-dependent and ubiquitylation-dependent degradation of cyclin D3 remains to be identified. Mutations at the conserved threonine (Thr283) occur at high frequency in Burkitt’s lymphoma resulting in elevated cyclin D3 protein levels [56, 63]. In addition, dysregulation of cyclin D3 also occurs in HCC, gliomas, bladder carcinoma, prostate cancer and osteosarcoma, wherein cyclin D3 expression correlates with increased cell proliferation and tumor growth [53]. Although it remains to be established whether cyclin D3 or mutant cyclin D3 possesses the tumor promoting capability comparable to cyclin D1, it has been demonstrated to be indispensable for the maintenance of T-ALL [64].
3. Ubiquitylation-medicated protein degradation
3.1. Protein degradation via the UPS
Protein abundance reflects the balance of protein synthesis versus protein degradation [65]. Ubiquitin itself is a stable and highly conserved protein with 76 amino acids [66]. Ubiquitin is initially covalently conjugated through its C-terminal carboxylate group (glycine residue) to an available amino group of a lysine residue within substrate proteins. Additional ubiquitin molecules are linked through the utilization of one of seven internal lysine residues (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48 and Lys63) [67]; the precise lysine linkage determines whether the substrate protein is destined for degradation in the 26S proteasome (e.g. Lys48) or for other fates (e.g. Lys63). Thus, different linkages and lengths of ubiquitin chains render diverse different biological functions [68, 69].
Protein ubiquitylation is catalyzed by the coordinated action of three enzymes: E1, ubiquitin-activating enzyme that produces an active ubiquitin using ATP hydrolysis to generate a thioester linkage between itself and ubiquitin molecule; E2, ubiquitin-conjugating enzyme that is both an ubiquitin receptor for E1 and an ubiquitin donor for substrates in concert with an E3; E3, ubiquitin ligase that binds to an E2 and its substrates to assist ubiquitin transfer from the E2 to a substrate or directly recognizes and conjugates ubiquitin to its targets [70–72] (Fig. 2). Currently, there are two E1s, approximately forty E2s, and more than six hundred E3s have been identified in human genome [73]. The E3 generally provides the specificity for this system, with each E3 specifically targeting a small subset of substrates. There are four groups of E3 ubiquitin ligases, including Really Interesting New Gene (RING), U-box, Homologous to E6-Associated Protein C-Terminus (HECT), and Ring-between-Ring (RBR).
Figure 2. Ubiquitylation-medicated protein degradation.
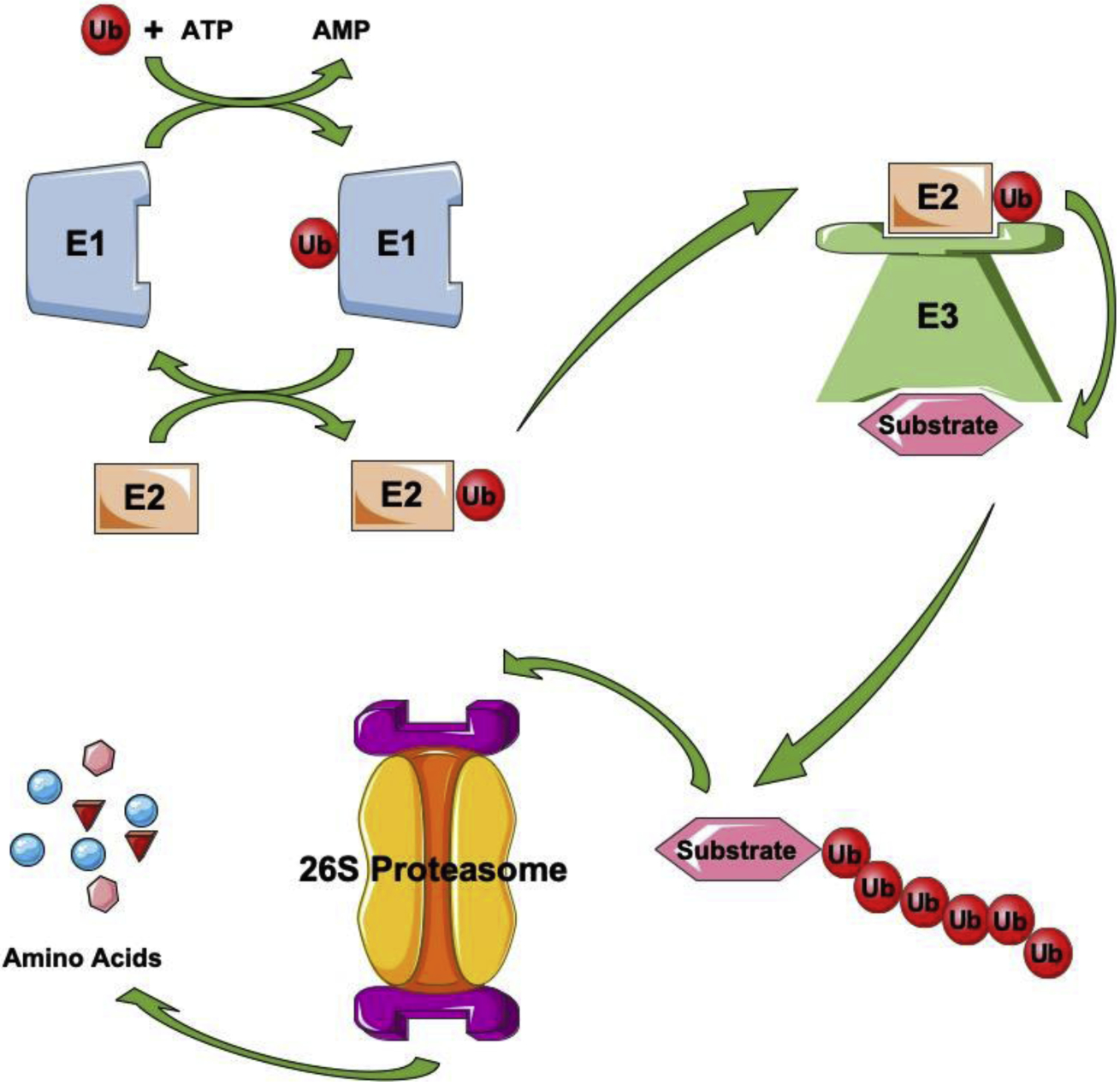
Protein ubiquitylation is catalyzed by the concerted and coordinated action of three enzymes: E1, ubiquitin-activating enzyme that produces an active ubiquitin using energy to generate a thioester linkage between itself and ubiquitin molecule; E2, ubiquitin-conjugating enzyme that is both an ubiquitin receptor for E1 and an ubiquitin donor for substrates or E3s; E3, ubiquitin ligase that binds to E2s and its substrates to assist ubiquitin transfer from E2s to substrates or directly recognizes and conjugates ubiquitin to its targets. Polyubiquitylated proteins will be degraded in proteasome, a complex intracellular structure composed of multiple enzymatic complexes.
3.2. Skp1-Cul1-F-box E3 ubiquitin ligases
RING E3 ubiquitin ligases have a conserved RING finger domain, which serves as the docking domain for E2s and facilitates the catalytic activity of ubiquitin ligases [74]. RING E3s are divided into four subgroups: Cullin RING ligases (CRL), Anaphase Promoting Complex/Cyclosome (APC/C), mono-subunit ligases, and Fanconi Anemia (FANC) E3 ligase [75–77]. G1 to S-phase transition is in part regulated by Cul1 based RING E3 ligases; Cul1 functions as a scaffold protein for large molecular complex composed of Skp1, Cul1, Rbx1 (with the Ring box domain), and an F-box protein, which functions as a substrate specific adaptor, hence termed SCF complex.
F-box proteins are a family of proteins with an F-box motif that contains 40–50 amino acids and serves as a docking domain for protein-protein interaction [78]. In human genome, there are approximately 69 F-box proteins, which are categorized into three subfamilies: Fbxw (WD40 repeats as a substrate binding domain), Fbxl (leucine-rich repeats (LRRs) as a substrate binding domain), and Fbxo (with other substrate binding domains) [79]. There are 10 members of Fbxw proteins that are mainly involved in cell cycle regulation and tumorigenesis [80]. As for Fbxl proteins, there are 22 different members with LRR domains formed by 20–29 amino acids that are crucial for substrate recognition and integration [81]. Although without clearly defined C-terminal regions, Fbxo proteins have diverse binding domains like proline-rich domains, cyclin box, zinc-finger, and so forth. Generally, different F-box proteins can target a specific group of substrates based on their unique ‘degron’ motifs.
3.3. Deubiquitinase (DUB)
Besides the enzymes catalyzing ubiquitylation, there is another group of enzymes termed DUB, which favors the recycle of ubiquitin and prevents it from 26S proteasome-mediated degradation. In human genome, there are about 100 DUBs that normally reside in proteasome [82]. The balance between ubiquitylation and deubiquitylation works in a Yin-Yang manner to control the intracellular abundance of ubiquitin and its substrates. Importantly, the recycle of ubiquitin benefits the cells through saving energy and facilitating the translocation of substrates in-and-out of narrow pores. DUBs specifically recognize their substrates based on the linkage of ubiquitin chain. Functionally, DUBs can trim or deubiquitylate the ubiquitin chains conjugated to their substrates, thereafter, change the stability, localization or functions of targeted proteins [83].
4. Ubiquitylation-mediated degradation of D cyclins
D cyclins are highly labile proteins with short half-lives ranging from 10–60 min depending on cell lines investigated [9, 14, 84–86]. As oscillating proteins, the ubiquitylation and degradation of D cyclins is cell cycle dependent, and these processes are normally regulated by the phosphorylation of D cyclins [14, 86] (Fig. 3). D cyclins are critical regulators of cell cycle progression from G1 to S phase; therefore, their degradation mostly happens in late G1 phase and early S phase [2, 87, 88]. However, some studies revealed controversial findings with cyclins D2 and D3 being ubiquitylated and degraded during G2-M transition [84, 85]; additionally, there are also controversies regarding the importance of phosphorylation as an initiating event prior to the ubiquitylation of D cyclins.
Figure 3. Illustration of phosphorylation sites of D cyclins.
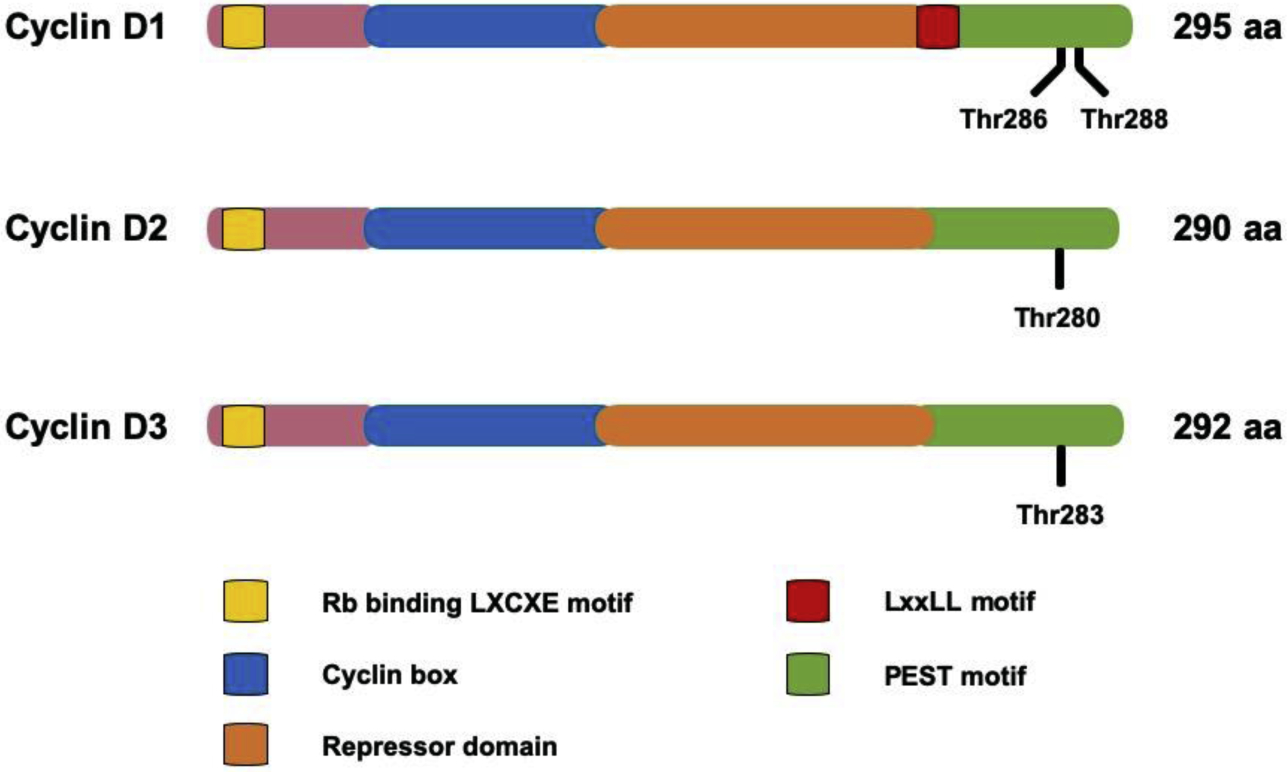
As for phosphorylation-mediated degradation, the conserved sites for Thr286 in cyclin D1 are Thr280 in cyclin D2 and Thr283 in cyclin D3, respectively.
4.1. Ubiquitylation and degradation of cyclin D1
Cyclin D1 is broadly expressed in a variety of mammalian cell types, with the notable exception in cells of hematopoietic origin. In the presence of mitogenic stimuli and abundant nutrients, cyclin D1 is synthesized; to perform its functions, cyclin D1 assembles with CDK4 or CDK6 to form a kinase complex; conversely, growth factor withdrawal or nutrient depletion compromises the formation of new cyclin D1-CDK4/6 complexes, leading to an immediate degradation of cyclin D1 in order to facilitate cell cycle exit [6]. Previous work demonstrated that cyclin D1 is phosphorylated at Thr286 during the G1-S transition, and this phosphorylation in turn directs export of cyclin D1 from the nucleus to the cytoplasm where cyclin D1 is ubiquitylated and degraded by the proteasome [14, 89]. This entire regulatory process depends exquisitely upon Thr286 phosphorylation as mutation of the threonine acceptor results in a constitutively nuclear and stable cyclin D1 [89, 90]. Critically, Thr286 is mutated in a number of cancers including uterine and endometrial cancers [89, 91], and the expression of mutant alleles is sufficient to drive spontaneous cancers in mice [92], highlighting the true oncogenic potential of cyclin D1.
Glycogen synthase kinase-3β (GSK-3β) is the primary Thr286 kinase [14]. The activity of GSK-3β is regulated by Ras via PI3K-Akt signaling. This pathway is regulated by growth factor receptors that provide a direct link from growth factor signaling to the regulation of cyclin D1 stability and accumulation [93]. Critically, any cancer derived mutations that dysregulate signaling through Ras-PI3K-Akt-GSK-3β will directly impact and dysregulate cyclin D1, contributing to excess nuclear cyclin D1-CDK4/6 activity and this is a frequent occurrence in many cancer types independent of direct genetic alterations in the CCND1 locus [94–96]. In addition to GSK-3β, other Thr-286 kinases have been proposed including p38SAPK2 and ERK2 [97, 98], and Thr288 kinase such as mirk/Dyrk1b [99].
It is not uncommon that more than one E3 ubiquitin ligase mediates the ubiquitylation and degradation of proteins like the master regulators mediating cell cycle progression. For example, c-Myc can be ubiquitylated and degraded by more than one E3 ubiquitin ligases like Fbxw7, Skp2, CHIP, Fbxo29, etc. [100–103]. Ubiquitylation of cyclin D1 has been linked with E3 ubiquitin ligases including Fbxo4, Fbxo31, β-transducin repeat-containing protein (β-TrCP) and APC/C [87, 98, 104–106] (Table 2).
Table 2.
Ubiquitylation-mediated degradation of cyclin D1
| Enzymes | Cell Lines | Cell Cycle | Phosphorylation-dependency | References |
|---|---|---|---|---|
| E3 ubiquitin ligase | ||||
| Fbxo4 | NIH3T3, MEFs, U2OS, MCF-7, TE8, TE10, TE15, 451Lu, WM793B, 1205Lu | G1-S | Phospho-Thr286 mediated by GSK-3β | [87, 94, 95, 107, 117] |
| Fbxw8 | HCT116, SW480, T98G | G1-S | Phospho-Thr286 mediated by MAPK/Erk | [98] |
| Fbxo31 | SK-MEL-28 | G1-S | Phospho-Thr286 mediated by MAPK/Erk | [104] |
| β-TrCP | LNCaP | G1-S | Phospho-Thr286 mediated by IKKα | [105] |
| APC/C | MCF-7, MEFs | — | Phospho-Thr286 mediated by GSK-3β | [106] |
| Deubiquitinase | ||||
| USP2 | HCT116, MCF-7, U2OS, PC3, 3T3-L1 | G1-S | — | [137, 139] |
| USP22 | HCT116, MCF-7, U2OS, PC3, H1299, 2091 | G1-S | Not depending on Thr286 phosphorylation | [143] |
4.1.1. Fbxo4-mediated cyclin D1 degradation
Fbxo4 belongs to the F-box protein family with a conserved F-box domain [65]. Several substrates have been identified as Fbxo4 targets such as cyclin D1, Fxr1, Trf1, Peroxisome-Proliferator-Activated Receptor gamma (PPAR-γ), Mcl-1, and ICAM-1 [65, 107–113]. Structurally, Fbxo4 has four domains: C-terminal D domain for dimerization, F-box domain for interacting with Skp1, linker region, and N-terminal region for substrate recognition and interaction [95]. Like many other E3 ubiquitin ligases such as Skp2, Fbxw7 and β-TrCP, Fbxo4 normally forms homodimers in order to perform the E3 ubiquitin ligase function [107]. The dimerization of Fbxo4 is regulated by the phosphorylation of Ser12 in D domain, which is typically catalyzed by GSK-3β [114]. The significance of Fbxo4 phosphorylation is testified by data from mammalian cells with ectopically expressing S12A mutant that loses its dimerization capacity as well as E3 ubiquitin ligase activity.
Mass spectrometry screening revealed both Fbxo4 and αB-crystallin as binding partners of cyclin D1 in NIH3T3 cells [87]. Biochemical analyses demonstrated the interaction between Fbxo4 and cyclin D1 mainly depends on the phosphorylation at Thr286 in cyclin D1 and the presence of αB-crystallin. Like introduced above, cyclin D1 is commonly phosphorylated at Thr286 during G1-S transition; not surprisingly, Ser12 phosphorylation of Fbxo4 also oscillates accordingly across the cell cycle, resulting in a prominent enrichment of the association of Fbxo4 with cyclin D1. Upon phosphorylation, cyclin D1 is transported from nucleus to cytoplasm where homodimerized SCFFbxo4 complex ubiquitylates and targets cyclin D1 to proteasome for degradation [115]. With regard to the importance of cyclin D1 in cell cycle regulation, loss of function mutations in Fbxo4 lead to the accumulation of cyclin D1, cell cycle progression, cell proliferation, and eventually tumor development and progression.
Taking advantage of available human ESCC tumor samples, we screened and identified Fbxo4 mutations in these tumors, and defined the importance of Fbxo4 in ESCC development [1, 107]. In human ESCC, the majority of Fbxo4 mutations present in the N-terminal region, for example, S8R, S12L, P13S, L23Q and G30N that undoubtedly disrupt the dimerization and activation of Fbxo4 without changing the ability for substrate recognition. In addition, it is as well revealed another mutation, P76T, which abrogates Skp1 binding and produces a dominant-negative form similar to the mutant with the deletion of F-box domain. Furthermore, the protein levels of cyclin D1 are elevated and accumulated in the nuclei in all ESCC samples harboring Fbxo4 mutations. The transformation phenotype related to cells expressing Fbxo4 mutants unquestionably supports that compromising the activity of cyclin D1 E3 ubiquitin ligase is oncogenic; therefore, Fbxo4 is defined as a tumor suppressor. However, in human HCC, it is identified a different mechanism how Fbxo4 activity is regulated. Sequencing analyses demonstrate four different Fbxo4 isoforms in HCC: α isoform with full length of amino acids, β isoform with 7 more amino acids encoded by code in intron5 and a sequence replacement of exon6, γ isoform without 168–245 nt in exon1, and δ isoform without exon6, among which only the α isoform can mediate the ubiquitylation and degradation of cyclin D1 [116].
These findings in heterozygous and knockout mice confirmed the tumor suppressor activity of Fbxo4. For example, Fbxo4−/− or Fbxo4−/+ mice can spontaneously develop diverse tumors including lymphoblastic lymphoma, extramedullary hematopoiesis/intense myeloid proliferation, hemangioma/angioinvasive tumor, dendritic cell tumor, histiocytic sarcoma, early myeloid tumor, mammary carcinoma, and uterine tumors [117]. Moreover, loss of Fbxo4 facilitates the development of N-nitrosomethylbenzylamine (NMBA)-induced papilloma in mice [118]. Importantly, loss of Fbxo4 also enhances the aggressiveness of melanoma in BrafV600E/+ mouse models, highlighting the importance of Fbxo4 as a factor related to tumor progression [95].
4.1.2. Additional F-box E3 ubiquitin ligases for cyclin D1
Fbxo31 is a senescence-related F-box protein. Under physiological conditions, Fbxo31 is highly expressed in brain and fat tissues, but its expression is low in stomach, pancreas and breast tissues as well as in bone marrow [119]. Under pathological conditions, Fbxo31 plays crucial roles in DNA damage response and tumorigenesis; however, the function of Fbxo31 varies in a context dependent manner in human cancers, for example, it has been detected that loss of heterozygosity of FBXO31 gene in HCC, prostate cancer, breast cancer and ovarian carcinoma [120]. A genome-wide RNA knockdown analysis revealed Fbxo31 is a key factor controlling BRAF-induced senescence in primary fibroblasts and melanocytes; moreover, overexpression of Fbxo31 can induce G1 cell cycle arrest in a cyclin D1-depdendent manner [104]. Biochemical analyses found Fbxo31 directly interacts with cyclin D1, ubiquitylates and targets it to proteasome for degradation that requires the phosphorylation of cyclin D1 at Thr286. While another structural analysis demonstrated that cyclin D1 phosphorylation at Thr286 is not essential for its association with Fbxo31, and thereafter ubiquitylation by SCFFbxo31 complex. Consistent with previous reports, T286A cyclin D1 mutant is mainly localized in the nucleus; however, this phospho-dead mutant can still be ubiquitylated by Fbxo31. These findings confirm that phosphorylation at Thr286 is important for nuclear exportation of cyclin D1, but it is not indispensable for Fbxo31-mediated ubiquitylation and subsequent degradation [121]. In addition, Fbxo31 can be induced by DNA-damage stresses like γ-irradiation, ultraviolet irradiation, X-ray irradiation, oxidative stress or chemotherapeutic agents (etoposide, adriamycin, cisplatin or fluorouracil); under these conditions, cyclin D1 expression is compromised through Fbxo31-mediated degradation, resulting in cell cycle arrest.
β-TrCP is a WD40-repeat containing F-box protein. It regulates both physiological and pathological processes such as cell cycle progression, cell proliferation, signal transduction, and tumor development through targeting its substrates for ubiquitylation and degradation [122]. β-TrCP recognizes its substrates in a phosphorylation-dependent manner, through which a specific binding motif known as degron is phosphorylated at Ser or Thr, for example, DSGxxS sequence or its variants like DSG/DDG/EEG/SSG/TSGxxS/E/D [123]. β-TrCP possesses both tumor-promoting and tumor-suppressing properties because it can promote the degradation of both oncoproteins (β-catenin, YAP, and WEE1) and tumor suppressors (Bim EL, IκB, PDCD4, and CDC25A) [124, 125]. STG28 is a derivative of troglitazone that compromises cyclin D1 expression in a phosphorylation dependent but GSK-3β independent manner; instead, compound screening identified IKKα involved in nuclear exportation, and thereafter the degradation of cyclin D1 [105]. STG28 treatment elevates the expression of β-TrCP, but compromises the expression of Fbxo4, Fbxw8, Fbxw7 and Skp2, suggesting β-TrCP is an E3 ubiquitin ligase targeting cyclin D1 for ubiquitylation. Further biochemical analyses revealed SCFβ-TrCP can ubiquitylate and degrade cyclin D1 after STG28 treatment. Cyclin D1 contains an unconventional recognition sequence (279EEVDLACpT286) that can be targeted by β-TrCP; this finding is convinced by mutational and modeling analyses [105]. Interestingly, besides STG28 treatment, nutrient depletion like glucose starvation also promotes cyclin D1 turnover in a β-TrCP-dependent manner, highlighting the physiological relevance of these findings.
4.1.3. APC/C-mediated cyclin D1 degradation
The CCAAT/enhancer binding protein delta (C/EBPδ) is a transcription factor that regulates many aspects of cell functions such as cell proliferation, differentiation, movement, and apoptosis [126]. The diverse functions of C/EBPδ are partially cell type and cellular context dependent. In human cancer cells, C/EBPδ compromises cyclin D1 expression in a proteasome-dependent manner [127]. Mechanistically, C/EBPδ induces the expression of Cdc27/APC3, a subunit of the APC/C complex, resulting in the ubiquitylation and degradation of cyclin D1 [106]. Previous report showed γ-irradiation can mediate the degradation of cyclin D1 depending on the RXXL motif or D-box [128]; moreover, this motif is also a conserved motif of APC/C targets, and it can be recognized by the Cdh1 coactivator. Mutational analyses verified the mutation of RXXL to QXXA successfully blocks the ubiquitylation and degradation of cyclin D1 mediated by Cdc27. Well-known APC/C targets include cyclin A and cyclin B1 that are involved in cell cycle transition from G2 to M phase [129]; while cyclin D1 is one of the critical regulators for G1-S transition; therefore, APC/C-mediated cyclin D1 degradation might not play important roles in normal cells. However, this mechanism may help maintaining mitotic viability in cells with Fbxo4 loss or mutations. Further investigations are required to dissect these potential and relative mechanisms.
4.1.4. Discrepancies in E3 ubiquitin ligases targeting cyclin D1 degradation
In addition to the above mentioned E3 ubiquitin ligases, there are additional E3 ubiquitin ligases implicated in cyclin D1 ubiquitylation and degradation. For example, Skp2 and Fbxw8. Skp2 can form a SCF complex with Cul1 and Skp1 that controls the cell cycle transition from G1 to S phase [130]. Well known substrates of Skp2 include p21 and p27 [131, 132]. Additionally, one previous study revealed that Skp2 can interact with cyclin D1 and knockdown of Skp2 leads to the accumulation of cyclin D1 [131]. Because both p21 and p27 can stabilize cyclin D1 [133], it is probable that cyclin D1 accumulation upon Skp2 knockdown is the consequence of reduced ubiquitylation and degradation of p21 and p27.
Fbxw8 is an F-box protein that utilizes Cul7. Cul7 indirectly associates with Skp1 in an Fbxw8-dependent manner, and Cul7 demonstrates a higher affinity with Fbxw8 than Cul1 [134, 135]. Biological analysis revealed similar phenotypes of placenta in Fbxw8−/− and Cul7−/− mice, suggesting Fbxw8 and Cul7 can form an E3 ubiquitin ligase complex in order to ubiquitylate and degrade their substrates. The levels of cyclin D1 are constant during S phase in normal cells, while its levels dropped dramatically during S phase. The alteration of cyclin D1 stability is mediated by Thr286 phosphorylation in a Ras-Raf-MEK-Erk signaling-dependent manner in these experiments. Biochemical analyses revealed Fbxw8 specifically associates with cyclin D1 in a phosphorylation-dependent way [98].
However, another study using transgenic mouse models introduced more challenges in E3 ubiquitin ligases that regulate cyclin D1 ubiquitylation and degradation [136]. In this study, several different transgenic mouse models were established with Fbxo4−/−, Fbxw8−/−, and Fbxo4−/− & Fbxw8−/− genotypes. However, no difference of cyclin D1 half-life was monitored in mouse embryonic fibroblasts (MEFs) derived from these transgenic mice, even with the knockdown of Fbxo31 and Skp2 in Fbxo4−/− & Fbxw8−/− MEFs. Although cyclin D1 protein levels are elevated, its regulation was proposed to be transcriptionally regulated in Fbxo31 knockdown MEFs. This is in sharp contrast to a majority of work demonstrating decreased rates of degradation of cyclin D1 in MEFs derived from Fbxo4 deficient MEFs under normal proliferating conditions as well as following a variety of cellular insults [1, 95, 117, 118]. The precise reason for these discrepancies remains to be discerned, but may reflect context-dependent and tissue-specific mechanisms.
4.2. DUBs and cyclin D1 degradation
4.2.1. Ubiquitin Specific Peptidase 2 (USP2)
USP2 has been identified as a specific DUB for cyclin D1 [137]. USP2 directly interacts with cyclin D1 to perform deubiquitylation and thereby antagonize proteasome-dependent degradation of cyclin D1. Conversely, knockdown of USP2 dramatically shortens the half-life of cyclin D1 and compromises G1-S transition and cell proliferation in human cancer cell lines. However, these effects are not significant in normal human fibroblasts or in cancer cells without cyclin D1 expression. An independent investigation demonstrated the effects of ML364, a small molecule inhibitor of USP2, on cyclin D1 expression. ML364, a reversible USP2 inhibitor with an IC50 of 1.1 μM, can effectively reduce cyclin D1 protein levels in a time- and dose-dependent manner in human cancer cells; mechanistically, ML364 promotes the ubiquitylation and the proteasome-mediated degradation of cyclin D1, leading to cell cycle arrest and reduced cell proliferation [138]. Consistently, other USP2 inhibitors like lithocholic acid (LCA) derivative, LCAHA, and 3,3’-Diindolylmethane can also promote cyclin D1 degradation and cell cycle arrest in G1 phase [139, 140].
4.2.2. USP22
USP22 is a highly conserved DUB from yeast to vertebrates. USP22 functions as a subunit of the human Spt-Ada-Gcn5-acetyltransferase (SAGA) transcriptional coactivator complex [141]. USP22 can alter chromatin structure and gene transcription through deubiquitylating histones H2A and H2B [142]. Elevated USP22 expression is correlated with aggressiveness of human cancer cells and cell cycle progression from G1 to S phase. In addition to histones, cyclin D1 is also a direct target of USP22 [143]. USP22 function reverses cyclin D1 ubiquitylation; overexpression of cyclin D1 can partially rescue cell cycle arrest in USP22 knockdown cells. Consistent with this functional relationship, CDK4/6 inhibitors inhibit proliferation of cells with ectopic expression of USP22.
5. Targeting D cyclin-related signaling and cancer treatment
5.1. Targeting D cyclin-related CDK4/6 kinases
5.1.1. CDK4/6 kinase inhibitors
D cyclins are overexpressed in a variety of human cancers and elevated D cyclin levels are associated with tumor size, invasion, and metastasis. Although this would suggest that D cyclins are potential therapeutic targets, the fact that D cyclins do not have enzymatic activity makes directly targeting D cyclins a challenge [1, 2]. In contrast, highly specific CDK4/6 inhibitors have been developed, including PD-0332991 (palbociclib), LY2835219 (abemaciclib), and LEE011 (ribociclib), all of which are approved by U.S. Food and Drug Administration to treat patients with tumors [144, 145]. These inhibitors have IC50s in the nanomolar range and are being intensively investigated in biomedical studies and clinical trials to treat human cancers like pancreatic cancer, NSCLC, breast cancer, HNSCC, melanoma, endometrial cancer, ESCC, colorectal carcinoma, MCL, and multiple myeloma [1, 146]. The high potency of CDK4/6 inhibitors sets the stage for new therapeutic options for cancers with overexpression of D-type cyclins. CDK4/6 inhibitors are generally well tolerated with minimal side effects, while the most prominent one is neutropenia [147]. However, the development of resistance to CDK4/6 inhibitors is regarded likely inevitable. While there is much to be learned, some mechanistic insights regarding resistance to CDK4/6 inhibitors have been elucidated and are discussed subsequently.
5.1.2. Mechanisms of resistance to CDK4/6 inhibitors
1). Cell cycle-related mechanisms:
Dysregulation of certain factors involved in the cell cycle machinery is a critical contributor to the development of resistance to CDK4/6 inhibitors. Loss of downstream target genes like Rb, Fizzy and Cell Division Cycle 20 Related 1 (FZR1) leads to the bypass of requirement of D cyclin-CDK4/6 for cell cycle progression [1, 148, 149]. In addition, overexpression or hyperactivation of factors directly or indirectly involved in cell cycle regulation are implicated the development of resistance to CDK4/6 inhibitors. Examples include the amplification of CDK4 [150], CDK6 [151], CCNE1/2 [152], CDK2 [153], E2F [154] CDK7, WEE1 and MDM2 [155], the loss of p16 [156], and the activation of HDAC [155].
2). Cell cycle-nonspecific mechanisms:
Hyperactivation of mitogenic signaling, such as FGFR [157], PI3K-Akt-mTOR [158], and AP-1 [159], can promote the bypass of pathways for cell cycle progression, and the development of resistance to CDK4/6 inhibitors. Hormone-mediated ER activation is a driver of D cyclin-CDK4/6 activation in breast cancer cells. Loss of ER or PR in resistant tumors in preclinical studies and clinical trials indicates the alteration of ER or PR levels is correlated with the development of resistance [151]. Other mechanisms include the suppression of Smad3, the activation of autophagy, and the change of immune response [155, 160, 161].
5.2. Therapeutic vulnerability targeting degradation machinery for D cyclins
5.2.1. Targeting glutamine metabolism in tumors with dysregulated Fbxo4-cyclin D1
The importance of glutamine as a key carbon source for many tumor types has recently been noted [162, 163]. Through a process termed glutaminolysis, glutamine is converted to glutamate, and this process is catalyzed by the glutaminase enzyme (GLS) [164, 165]. Glutamine can be utilized as an energy source, a nitrogen source, and a substrate-donor for glutathione synthesis. Recent work from a number of laboratories have demonstrated that oncogene-dependent transformation of normal cells to tumorigenic cells is associated with increased glutamine dependency. Such tumor cells require glutamine for survival and proliferation, and they are considered glutamine-addicted. Glutamine-addiction is associated with the dysregulation of tumor promoting oncoproteins such as c-Myc, N-Myc, ErbB2 or the inactivation of tumor suppressors like p53 and Rb [1, 166]. Glutamine-addiction of Rb deficient cells has broad sweeping implications given that Rb can be functionally inhibited by phosphorylation, an event mediated by cyclin D1-CDK4/6 kinases. This implies that cells with dysregulated cyclin D1-CDK4 would exhibit glutamine-dependency. Indeed, recent work revealed that oncogenic mutations in cyclin D1 driving its nuclear overexpression or inactivating mutations of the cyclin D1 E3 ubiquitin ligase, Fbxo4, can promote glutamine-addiction in mouse embryonic fibroblasts, and tumor cells derived from esophageal cancer [1].
Loss of Fbxo4 results in reduced cyclin D1 degradation and triggers a corresponding increase in CDK4/6 activity and a tumor prone phenotype in Fbxo4 deficient mice [117, 118]; for example, mice with the loss of Fbxo4 exhibit increased sensitivity to NMBA-induced gastrointestinal tumors [118]. Consistent with cyclin D1 as a critical Fbxo4 target, the NMBA-induced tumors are sensitive to treatment with palbociclib. With regard to glutamine metabolism in esophageal cancers, upregulation of GLS is observed in primary ESCC tissues, consistent with the findings that ESCC cells harboring dysregulated Fbxo4-cyclin D1 are glutamine-addicted [1]. Accordingly, the dysregulation of Fbxo4-cyclin D1 enhances the dependency of ESCC cells on glutamine in accordance with cell proliferation and growth. Furthermore, the dysregulation of Fbxo4-cyclin D1 not only drives glutamine-addiction through Rb inactivation and mTORC1 hyperactivation, but also promotes glutamine uptake as well as GLS expression [1].
Besides driving glutamine-addiction, loss of Fbxo4 or overexpression of cyclin D1 also compromises mitochondrial activity and oxidative phosphorylation, leading to deficient energy production [1]. These metabolic vulnerabilities make tumor cells with dysregulated Fbxo4-cyclin D1-CDK4/6-Rb sensitive to combined treatment with CB-839 (GLS inhibitor) and metformin, leading to energy catastrophe and apoptosis in cell culture and in mouse xenografts. This pre-clinical study provides a potential therapy for various human cancers with cyclin D1 overexpression like pancreatic cancer, NSCLC, breast cancer, HNSCC, and melanoma, or tumors with Rb loss, such as, small cell lung cancer [167], and genitourinary cancers [168–170].
5.2.2. Targeting β-TrCP mediated cyclin D1 degradation
Troglitazone, rosiglitazone, and pioglitazone are synthetic ligands of PPAR-γ, and they are approved for treating type II diabetes [171]. In addition, all three compounds exhibit antitumor effects in pre-clinical studies and favorable outcomes in some clinical trials involving in thyroid carcinoma, glioma, prostate cancer, colon cancer, lung cancer, breast cancer, sarcoma, and melanoma [172, 173]. Although the molecular mechanisms remain undefined, troglitazone at high concentration can promote the ubiquitylation and degradation of cyclin D1 in breast cancer cells. STG28, a new PPAR-γ-inactive form of troglitazone, can effectively compromise cyclin D1 expression in a Thr286 phosphorylation-dependent manner [105]. STG28 can upregulate the expression of E3 ubiquitin ligase, β-TrCP, and promote the translocation of cyclin D1 into cytoplasm, where β-TrCP interacts with cyclin D1, enhances its ubiquitylation and degradation, highlighting the therapeutic potential of STG28 to treat human tumors with cyclin D1 overexpression.
5.2.3. Targeting USP2 and promoting cyclin D1 degradation
Another dimension of new therapies is through the development of inhibitors targeting deubiquitinating enzymes (DUBs) that antagonize ubiquitylation and degradation of cyclin D1. As such, various inhibitors have been developed based on the mechanisms how specific DUB performs its functions. However, it is critical to develop inhibitors with specificity that determines the efficiency and usability of these inhibitors. Several compounds have been shown to suppress the activity of USP2, which can promote the deubiquitylation and stability of cyclin D1 [137]. For example, LCAHA, ML364, and 3,3’-diindolylmethane (DIM), all of which can successfully suppress USP2, and enhance the ubiquitylation and degradation of cyclin D1, leading to reduced cyclin D1 protein levels [138–140]. Therefore, these compounds effectively block cell cycle progression from G1 to S phase. Importantly, LCAHA can effectively antagonize the proliferative signals delivered by the presence of mitogenic stimuli in a cyclin D1 dependent manner.
6. Conclusion and future perspectives
As a central hub sensing extracellular stimuli and driving cell cycle progression, D cyclins act as a switch to integrate cell division with cell proliferation and growth. As such, D cyclins have critical roles in both normal and tumor cells. In general, the expression of D cyclins is dysregulated in various human cancers and cyclin D1 is regarded as an oncogenic driver in cancers including breast cancer, lung cancer, melanoma and HNSCC. Moreover, CCND1 gene, encoding cyclin D1, is broadly amplified with an incidence rate about 15–40% in human tumors. MCL has about ninety percent of CCND1 gene translocation with immunoglobulin heavy chain locus, while a small group of patients have the overexpression of cyclin D1 or cyclin D3. Although there are reports of CCND2 and CCND3 gene amplification, the incidence rate is dramatic low comparing to that of CCND1 gene. Interestingly, CCND2 promoter is frequently methylated, leading to the loss of cyclin D2 in pancreatic, breast and prostate cancer, raising questions regarding its function in these tumors.
Currently, the best opportunity for targeting D cyclins is through inhibition of CDK4/6. High efficacy of CDK4/6 inhibitors endows them as favorable therapies for a variety of tumors with the overexpression of D cyclins. However, the development of resistance is one major challenge that cannot be overlooked. Opportunities to overcome resistance include: 1) combined treatment with inhibitors that target signaling pathways driving resistance (inhibition of mTOR or MEK signaling); 2) identification of alternative weaknesses that develop as a consequence of CDK4/6 inhibitor resistance (e.g. GLS inhibitors in Rb deficient tumors). It is critical to ascertain the molecular basis for acquired resistance to CDK4/6 inhibitors in order to develop new therapies. Additional opportunities may pertain to the molecular machinery related to ubiquitylation-mediated degradation of D cyclins. For example, targeting DUBs that antagonize cyclin D degradation. Further studies are required to investigate the detailed molecular mechanisms how DUBs control the ubiquitylation and degradation of D cyclins, especially, the structural analyses of DUBs that will facilitate the development of inhibitors with high specificity and potency. A collective understanding of cyclin D post-translational regulation will benefit patients with tumors harboring the overexpression of D cyclins.
Acknowledgements
This work is supported by the National Institute of Health: P01CA098101 and R01CA093237 (JAD).
List of Abbreviations:
- CDK
cyclin-dependent kinase
- INK4
Inhibitors of CDK4
- CIP/KIP
CDK interacting protein/kinase inhibitor protein
- Akt
Protein Kinase B
- Myc
MYC Proto-Oncogene, BHLH Transcription Factor
- RUNX1/ETO
RUNX Family Transcription Factor 1
- Cul1
Cullin 1
- Skp1
S-phase kinase-associated protein 1
- Rbx1
Ring-Box 1
- Cdh1
Cadherin 1
- p38SAPK2
Stress-Activated Protein Kinase-2
- ERK2
Extracellular Signal-Regulated Kinase 2
- CHIP
STIP1 Homology And U-Box Containing Protein 1
- Fxr1
FMR1 Autosomal Homolog 1
- Trf1
Telomeric Repeat Binding Factor 1
- Mcl-1
Myeloid Cell Leukemia 1
- ICAM-1
Intercellular Adhesion Molecule 1
- YAP
Yes Associated Protein 1
- WEE1
WEE1 G2 Checkpoint Kinase
- Bim EL
BIM-extra long
- IκB
inhibitor of κB
- PDCD4
Programmed Cell Death 4
- CDC25A
Cell Division Cycle 25A
- IKKα
Inhibitor of κB kinase α
- ErbB2
Erb-B2 Receptor Tyrosine Kinase 2
- mTOR
Target Of Rapamycin Kinase
- MDM2
MDM2 Proto-Oncogene
- HDAC
Histone Deacetylase
- FGFR
Fibroblast Growth Factor Receptor
- AP-1
Activator protein 1
- ER
Estrogen Receptor
- PR
Progesterone Receptor
- MEK
Mitogen-activated protein kinase kinase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Competing Interest
There are no conflicts of interest to declare.
References
- [1].Qie S, Yoshida A, Parnham S, Oleinik N, Beeson GC, Beeson CC, Ogretmen B, Bass AJ, Wong KK, Rustgi AK, Diehl JA, Targeting glutamine-addiction and overcoming CDK4/6 inhibitor resistance in human esophageal squamous cell carcinoma, Nat Commun 10(1) (2019) 1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Qie S, Diehl JA, Cyclin D1, cancer progression, and opportunities in cancer treatment, J Mol Med (Berl) 94(12) (2016) 1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Sherr CJ, Cancer cell cycles, Science 274(5293) (1996) 1672–7. [DOI] [PubMed] [Google Scholar]
- [4].Zhang J, Bu X, Wang H, Zhu Y, Geng Y, Nihira NT, Tan Y, Ci Y, Wu F, Dai X, Guo J, Huang YH, Fan C, Ren S, Sun Y, Freeman GJ, Sicinski P, Wei W, Cyclin DCDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance, Nature 553(7686) (2018) 91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sherr CJ, Principles of tumor suppression, Cell 116(2) (2004) 235–46. [DOI] [PubMed] [Google Scholar]
- [6].Sherr CJ, Beach D, Shapiro GI, Targeting CDK4 and CDK6: From Discovery to Therapy, Cancer Discov 6(4) (2016) 353–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL, Cyclin D as a therapeutic target in cancer, Nat Rev Cancer 11(8) (2011) 558–72. [DOI] [PubMed] [Google Scholar]
- [8].Spinella MJ, Freemantle SJ, Sekula D, Chang JH, Christie AJ, Dmitrovsky E, Retinoic acid promotes ubiquitination and proteolysis of cyclin D1 during induced tumor cell differentiation, J Biol Chem 274(31) (1999) 22013–8. [DOI] [PubMed] [Google Scholar]
- [9].Diehl JA, Zindy F, Sherr CJ, Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway, Genes Dev 11(8) (1997) 957–72. [DOI] [PubMed] [Google Scholar]
- [10].Hinds PW, Mittnacht S, Dulic V, Arnold A, Reed SI, Weinberg RA, Regulation of retinoblastoma protein functions by ectopic expression of human cyclins, Cell 70(6) (1992) 993–1006. [DOI] [PubMed] [Google Scholar]
- [11].Weinberg RA, The retinoblastoma protein and cell cycle control, Cell 81(3) (1995) 323–30. [DOI] [PubMed] [Google Scholar]
- [12].Perry JE, Grossmann ME, Tindall DJ, Epidermal growth factor induces cyclin D1 in a human prostate cancer cell line, Prostate 35(2) (1998) 117–24. [DOI] [PubMed] [Google Scholar]
- [13].Lee RJ, Albanese C, Fu M, D’Amico M, Lin B, Watanabe G, Haines GK 3rd, Siegel PM, Hung MC, Yarden Y, Horowitz JM, Muller WJ, Pestell RG, Cyclin D1 is required for transformation by activated Neu and is induced through an E2F-dependent signaling pathway, Mol Cell Biol 20(2) (2000) 672–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Diehl JA, Cheng M, Roussel MF, Sherr CJ, Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis and subcellular localization, Genes Dev 12(22) (1998) 3499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Takuwa N, Fukui Y, Takuwa Y, Cyclin D1 expression mediated by phosphatidylinositol 3-kinase through mTOR-p70(S6K)-independent signaling in growth factor-stimulated NIH 3T3 fibroblasts, Mol Cell Biol 19(2) (1999) 1346–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Daniel P, Filiz G, Brown DV, Hollande F, Gonzales M, D’Abaco G, Papalexis N, Phillips WA, Malaterre J, Ramsay RG, Mantamadiotis T, Selective CREB-dependent cyclin expression mediated by the PI3K and MAPK pathways supports glioma cell proliferation, Oncogenesis 3 (2014) e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Shtutman M, Zhurinsky J, Simcha I, Albanese C, D’Amico M, Pestell R, Ben-Ze’ev A, The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway, Proc Natl Acad Sci U S A 96(10) (1999) 5522–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Tetsu O, McCormick F, Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells, Nature 398(6726) (1999) 422–6. [DOI] [PubMed] [Google Scholar]
- [19].Guttridge DC, Albanese C, Reuther JY, Pestell RG, Baldwin AS Jr., NF-kappaB controls cell growth and differentiation through transcriptional regulation of cyclin D1, Mol Cell Biol 19(8) (1999) 5785–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Karin M, Cao Y, Greten FR, Li ZW, NF-kappaB in cancer: from innocent bystander to major culprit, Nat Rev Cancer 2(4) (2002) 301–10. [DOI] [PubMed] [Google Scholar]
- [21].Garcea G, Neal CP, Pattenden CJ, Steward WP, Berry DP, Molecular prognostic markers in pancreatic cancer: a systematic review, Eur J Cancer 41(15) (2005) 2213–36. [DOI] [PubMed] [Google Scholar]
- [22].Santarius T, Shipley J, Brewer D, Stratton MR, Cooper CS, A census of amplified and overexpressed human cancer genes, Nat Rev Cancer 10(1) (2010) 59–64. [DOI] [PubMed] [Google Scholar]
- [23].Gautschi O, Ratschiller D, Gugger M, Betticher DC, Heighway J, Cyclin D1 in non-small cell lung cancer: a key driver of malignant transformation, Lung Cancer 55(1) (2007) 1–14. [DOI] [PubMed] [Google Scholar]
- [24].Li R, An SJ, Chen ZH, Zhang GC, Zhu JQ, Nie Q, Xie Z, Guo AL, Mok TS, Wu YL, Expression of cyclin D1 splice variants is differentially associated with outcome in non-small cell lung cancer patients, Hum Pathol 39(12) (2008) 1792–801. [DOI] [PubMed] [Google Scholar]
- [25].Arnold A, Papanikolaou A, Cyclin D1 in breast cancer pathogenesis, J Clin Oncol 23(18) (2005) 4215–24. [DOI] [PubMed] [Google Scholar]
- [26].Medvedev VN, Kostiuk ZN, Medvedeva VN, Nikolaeva NN, Bykova OF, [Diagnostic criteria of gastric secretory insufficiency and its classification], Sov Med (2) (1976) 17–21. [PubMed] [Google Scholar]
- [27].Thomas GR, Nadiminti H, Regalado J, Molecular predictors of clinical outcome in patients with head and neck squamous cell carcinoma, Int J Exp Pathol 86(6) (2005) 347–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Li W, Sanki A, Karim RZ, Thompson JF, Soon Lee C, Zhuang L, McCarthy SW, Scolyer RA, The role of cell cycle regulatory proteins in the pathogenesis of melanoma, Pathology 38(4) (2006) 287–301. [DOI] [PubMed] [Google Scholar]
- [29].Moreno-Bueno G, Rodriguez-Perales S, Sanchez-Estevez C, Marcos R, Hardisson D, Cigudosa JC, Palacios J, Molecular alterations associated with cyclin D1 overexpression in endometrial cancer, Int J Cancer 110(2) (2004) 194–200. [DOI] [PubMed] [Google Scholar]
- [30].Wu W, Slomovitz BM, Soliman PT, Schmeler KM, Celestino J, Milam MR, Lu KH, Correlation of cyclin D1 and cyclin D3 overexpression with the loss of PTEN expression in endometrial carcinoma, Int J Gynecol Cancer 16(4) (2006) 1668–72. [DOI] [PubMed] [Google Scholar]
- [31].Song Y, Li L, Ou Y, Gao Z, Li E, Li X, Zhang W, Wang J, Xu L, Zhou Y, Ma X, Liu L, Zhao Z, Huang X, Fan J, Dong L, Chen G, Ma L, Yang J, Chen L, He M, Li M, Zhuang X, Huang K, Qiu K, Yin G, Guo G, Feng Q, Chen P, Wu Z, Wu J, Ma L, Zhao J, Luo L, Fu M, Xu B, Chen B, Li Y, Tong T, Wang M, Liu Z, Lin D, Zhang X, Yang H, Wang J, Zhan Q, Identification of genomic alterations in oesophageal squamous cell cancer, Nature 509(7498) (2014) 91–5. [DOI] [PubMed] [Google Scholar]
- [32].N. Cancer Genome Atlas Research, U. Analysis Working Group: Asan, B.C.C. Agency, Brigham, H. Women’s, I. Broad, U. Brown, U. Case Western Reserve, I. Dana-Farber Cancer, U. Duke, C. Greater Poland Cancer, S. Harvard Medical, B. Institute for Systems, K.U. Leuven, C. Mayo, C. Memorial Sloan Kettering Cancer, I. National Cancer, H. Nationwide Children’s, U. Stanford, A. University of, M. University of, C. University of North, P. University of, R. University of, C. University of Southern, M.D.A.C.C. University of Texas, W. University of, I. Van Andel Research, U. Vanderbilt, U. Washington, I. Genome Sequencing Center: Broad, L. Washington University in St, B.C.C.A. Genome Characterization Centers, I. Broad, S. Harvard Medical, U. Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins, C. University of North, C. University of Southern California Epigenome, M.D.A.C.C. University of Texas, I. Van Andel Research, I. Genome Data Analysis Centers: Broad, U. Brown, S. Harvard Medical, B. Institute for Systems, C. Memorial Sloan Kettering Cancer, C. University of California Santa, M.D.A.C.C. University of Texas, C. Biospecimen Core Resource: International Genomics, H. Research Institute at Nationwide Children’s, S. Tissue Source Sites: Analytic Biologic, C. Asan Medical, B. Asterand, H. Barretos Cancer, BioreclamationIvt, C. Botkin Municipal, S. Chonnam National University Medical, S. Christiana Care Health, Cureline, U. Duke, U. Emory, U. Erasmus, M. Indiana University School of, M. Institute of Oncology of, C. International Genomics, Invidumed, H. Israelitisches Krankenhaus, M. Keimyung University School of, C. Memorial Sloan Kettering Cancer, G. National Cancer Center, B. Ontario Tumour, C. Peter MacCallum Cancer, S. Pusan National University Medical, S. Ribeirao Preto Medical, H. St. Joseph’s, C. Medical, U. St. Petersburg Academic, B. Tayside Tissue, D. University of, C. University of Kansas Medical, M. University of, H. University of North Carolina at Chapel, M. University of Pittsburgh School of, M.D.A.C.C. University of Texas, U. Disease Working Group: Duke, C. Memorial Sloan Kettering Cancer, I. National Cancer, M.D.A.C.C. University of Texas, M. Yonsei University College of, C.I. Data Coordination Center, H. Project Team: National Institutes of, Integrated genomic characterization of oesophageal carcinoma, Nature 541(7636) (2017) 169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bertoni F, Rinaldi A, Zucca E, Cavalli F, Update on the molecular biology of mantle cell lymphoma, Hematol Oncol 24(1) (2006) 22–7. [DOI] [PubMed] [Google Scholar]
- [34].Bergsagel PL, Kuehl WM, Molecular pathogenesis and a consequent classification of multiple myeloma, J Clin Oncol 23(26) (2005) 6333–8. [DOI] [PubMed] [Google Scholar]
- [35].Chen RW, Bemis LT, Amato CM, Myint H, Tran H, Birks DK, Eckhardt SG, Robinson WA, Truncation in CCND1 mRNA alters miR-16–1 regulation in mantle cell lymphoma, Blood 112(3) (2008) 822–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Abraham M, Klein S, Bulvik B, Wald H, Weiss ID, Olam D, Weiss L, Beider K, Eizenberg O, Wald O, Galun E, Avigdor A, Benjamini O, Nagler A, Pereg Y, Tavor S, Peled A, The CXCR4 inhibitor BL-8040 induces the apoptosis of AML blasts by downregulating ERK, BCL-2, MCL-1 and cyclin-D1 via altered miR-15a/16–1 expression, Leukemia 31(11) (2017) 2336–2346. [DOI] [PubMed] [Google Scholar]
- [37].Wang M, Yu W, Gao J, Ma W, Frentsch M, Thiel A, Liu M, Rahman N, Qin Z, Li X, MicroRNA-487a-3p functions as a new tumor suppressor in prostate cancer by targeting CCND1, J Cell Physiol (2019). [DOI] [PubMed] [Google Scholar]
- [38].Jiang L, Yang W, Bian W, Yang H, Wu X, Li Y, Feng W, Liu X, MicroRNA-623 Targets Cyclin D1 to Inhibit Cell Proliferation and Enhance the Chemosensitivity of Cells to 5-Fluorouracil in Gastric Cancer, Oncol Res 27(1) (2018) 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Yu Z, Wang C, Wang M, Li Z, Casimiro MC, Liu M, Wu K, Whittle J, Ju X, Hyslop T, McCue P, Pestell RG, A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation, J Cell Biol 182(3) (2008) 509–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bhalla K, Liu WJ, Thompson K, Anders L, Devarakonda S, Dewi R, Buckley S, Hwang BJ, Polster B, Dorsey SG, Sun Y, Sicinski P, Girnun GD, Cyclin D1 represses gluconeogenesis via inhibition of the transcriptional coactivator PGC1alpha, Diabetes 63(10) (2014) 3266–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Qie S, Diehl JA, Glutamine addiction: an Achilles heel in esophageal cancers with dysregulation of CDK4/6, Mol Cell Oncol 6(4) (2019) 1610257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bouchard C, Thieke K, Maier A, Saffrich R, Hanley-Hyde J, Ansorge W, Reed S, Sicinski P, Bartek J, Eilers M, Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27, EMBO J 18(19) (1999) 5321–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Martinez-Soria N, McKenzie L, Draper J, Ptasinska A, Issa H, Potluri S, Blair HJ, Pickin A, Isa A, Chin PS, Tirtakusuma R, Coleman D, Nakjang S, Assi S, Forster V, Reza M, Law E, Berry P, Mueller D, Osborne C, Elder A, Bomken SN, Pal D, Allan JM, Veal GJ, Cockerill PN, Wichmann C, Vormoor J, Lacaud G, Bonifer C, Heidenreich O, The Oncogenic Transcription Factor RUNX1/ETO Corrupts Cell Cycle Regulation to Drive Leukemic Transformation, Cancer Cell 34(4) (2018) 626–642 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Buschges R, Weber RG, Actor B, Lichter P, Collins VP, Reifenberger G, Amplification and expression of cyclin D genes (CCND1, CCND2 and CCND3) in human malignant gliomas, Brain Pathol 9(3) (1999) 435–42; discussion 432–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Yu J, Leung WK, Ebert MP, Leong RW, Tse PC, Chan MW, Bai AH, To KF, Malfertheiner P, Sung JJ, Absence of cyclin D2 expression is associated with promoter hypermethylation in gastric cancer, Br J Cancer 88(10) (2003) 1560–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Oshimo Y, Nakayama H, Ito R, Kitadai Y, Yoshida K, Chayama K, Yasui W, Promoter methylation of cyclin D2 gene in gastric carcinoma, Int J Oncol 23(6) (2003) 1663–70. [PubMed] [Google Scholar]
- [47].Eisfeld AK, Kohlschmidt J, Schwind S, Nicolet D, Blachly JS, Orwick S, Shah C, Bainazar M, Kroll KW, Walker CJ, Carroll AJ, Powell BL, Stone RM, Kolitz JE, Baer MR, de la Chapelle A, Mrozek K, Byrd JC, Bloomfield CD, Mutations in the CCND1 and CCND2 genes are frequent events in adult patients with t(8;21)(q22;q22) acute myeloid leukemia, Leukemia 31(6) (2017) 1278–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Karrman K, Andersson A, Bjorgvinsdottir H, Strombeck B, Lassen C, Olofsson T, Nguyen-Khac F, Berger R, Bernard O, Fioretos T, Johansson B, Deregulation of cyclin D2 by juxtaposition with T-cell receptor alpha/delta locus in t(12;14)(p13;q11)-positive childhood T-cell acute lymphoblastic leukemia, Eur J Haematol 77(1) (2006) 27–34. [DOI] [PubMed] [Google Scholar]
- [49].Zhao Z, Yang F, Liu Y, Fu K, Jing S, MicroRNA-409–3p suppresses cell proliferation and cell cycle progression by targeting cyclin D2 in papillary thyroid carcinoma, Oncol Lett 16(4) (2018) 5237–5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Li YL, Wang J, Zhang CY, Shen YQ, Wang HM, Ding L, Gu YC, Lou JT, Zhao XT, Ma ZL, Jin YX, MiR-146a-5p inhibits cell proliferation and cell cycle progression in NSCLC cell lines by targeting CCND1 and CCND2, Oncotarget 7(37) (2016) 59287–59298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Dong Q, Meng P, Wang T, Qin W, Qin W, Wang F, Yuan J, Chen Z, Yang A, Wang H, MicroRNA let-7a inhibits proliferation of human prostate cancer cells in vitro and in vivo by targeting E2F2 and CCND2, PLoS One 5(4) (2010) e10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Zeng Q, Tao X, Huang F, Wu T, Wang J, Jiang X, Kuang Z, Cheng B, Overexpression of miR-155 promotes the proliferation and invasion of oral squamous carcinoma cells by regulating BCL6/cyclin D2, Int J Mol Med 37(5) (2016) 1274–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Ding ZY, Li R, Zhang QJ, Wang Y, Jiang Y, Meng QY, Xi QL, Wu GH, Prognostic role of cyclin D2/D3 in multiple human malignant neoplasms: A systematic review and meta-analysis, Cancer Med 8(6) (2019) 2717–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Sarkar R, Hunter IA, Rajaganeshan R, Perry SL, Guillou P, Jayne DG, Expression of cyclin D2 is an independent predictor of the development of hepatic metastasis in colorectal cancer, Colorectal Dis 12(4) (2010) 316–23. [DOI] [PubMed] [Google Scholar]
- [55].Takano Y, Kato Y, Masuda M, Ohshima Y, Okayasu I, Cyclin D2, but not cyclin D1, overexpression closely correlates with gastric cancer progression and prognosis, J Pathol 189(2) (1999) 194–200. [DOI] [PubMed] [Google Scholar]
- [56].Schmitz R, Young RM, Ceribelli M, Jhavar S, Xiao W, Zhang M, Wright G, Shaffer AL, Hodson DJ, Buras E, Liu X, Powell J, Yang Y, Xu W, Zhao H, Kohlhammer H, Rosenwald A, Kluin P, Muller-Hermelink HK, Ott G, Gascoyne RD, Connors JM, Rimsza LM, Campo E, Jaffe ES, Delabie J, Smeland EB, Ogwang MD, Reynolds SJ, Fisher RI, Braziel RM, Tubbs RR, Cook JR, Weisenburger DD, Chan WC, Pittaluga S, Wilson W, Waldmann TA, Rowe M, Mbulaiteye SM, Rickinson AB, Staudt LM, Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics, Nature 490(7418) (2012) 116–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Koo KH, Kwon H, MicroRNA miR-4779 suppresses tumor growth by inducing apoptosis and cell cycle arrest through direct targeting of PAK2 and CCND3, Cell Death Dis 9(2) (2018) 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Wang H, Guo Q, Yang P, Long G, Restoration of microRNA-212 causes a G0/G1 cell cycle arrest and apoptosis in adult T-cell leukemia/lymphoma cells by repressing CCND3 expression, J Investig Med 65(1) (2017) 82–87. [DOI] [PubMed] [Google Scholar]
- [59].Wang W, Zhao LJ, Tan YX, Ren H, Qi ZT, MiR-138 induces cell cycle arrest by targeting cyclin D3 in hepatocellular carcinoma, Carcinogenesis 33(5) (2012) 1113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Xiao F, Zhang W, Chen L, Chen F, Xie H, Xing C, Yu X, Ding S, Chen K, Guo H, Cheng J, Zheng S, Zhou L, MicroRNA-503 inhibits the G1/S transition by downregulating cyclin D3 and E2F3 in hepatocellular carcinoma, J Transl Med 11 (2013) 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Naderi S, Gutzkow KB, Lahne HU, Lefdal S, Ryves WJ, Harwood AJ, Blomhoff HK, cAMP-induced degradation of cyclin D3 through association with GSK3beta, J Cell Sci 117(Pt 17) (2004) 3769–83. [DOI] [PubMed] [Google Scholar]
- [62].Casanovas O, Jaumot M, Paules AB, Agell N, Bachs O, P38SAPK2 phosphorylates cyclin D3 at Thr-283 and targets it for proteasomal degradation, Oncogene 23(45) (2004) 7537–44. [DOI] [PubMed] [Google Scholar]
- [63].Schmitz R, Ceribelli M, Pittaluga S, Wright G, Staudt LM, Oncogenic mechanisms in Burkitt lymphoma, Cold Spring Harb Perspect Med 4(2) (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Sawai CM, Freund J, Oh P, Ndiaye-Lobry D, Bretz JC, Strikoudis A, Genesca L, Trimarchi T, Kelliher MA, Clark M, Soulier J, Chen-Kiang S, Aifantis I, Therapeutic targeting of the cyclin D3:CDK4/6 complex in T cell leukemia, Cancer Cell 22(4) (2012) 452–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Qie S, Majumder M, Mackiewicz K, Howley BV, Peterson YK, Howe PH, Palanisamy V, Diehl JA, Fbxo4-mediated degradation of Fxr1 suppresses tumorigenesis in head and neck squamous cell carcinoma, Nat Commun 8(1) (2017) 1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Hershko A, Ubiquitin: roles in protein modification and breakdown, Cell 34(1) (1983) 11–2. [DOI] [PubMed] [Google Scholar]
- [67].Mansour MA, Ubiquitination: Friend and foe in cancer, Int J Biochem Cell Biol 101 (2018) 80–93. [DOI] [PubMed] [Google Scholar]
- [68].Yau RG, Doerner K, Castellanos ER, Haakonsen DL, Werner A, Wang N, Yang XW, Martinez-Martin N, Matsumoto ML, Dixit VM, Rape M, Assembly and Function of Heterotypic Ubiquitin Chains in Cell-Cycle and Protein Quality Control, Cell 171(4) (2017) 918–933 e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Chan CH, Li CF, Yang WL, Gao Y, Lee SW, Feng Z, Huang HY, Tsai KKC, Flores LG, Shao Y, Hazle JD, Yu D, Wei W, Sarbassov D, Hung MC, Nakayama KI, Lin HK, The Skp2-SCF E3 Ligase Regulates Akt Ubiquitination, Glycolysis, Herceptin Sensitivity, and Tumorigenesis, Cell 151(4) (2012) 913–914. [DOI] [PubMed] [Google Scholar]
- [70].Wilkinson KD, Protein ubiquitination: a regulatory post-translational modification, Anticancer Drug Des 2(2) (1987) 211–29. [PubMed] [Google Scholar]
- [71].Ernst A, Avvakumov G, Tong J, Fan Y, Zhao Y, Alberts P, Persaud A, Walker JR, Neculai AM, Neculai D, Vorobyov A, Garg P, Beatty L, Chan PK, Juang YC, Landry MC, Yeh C, Zeqiraj E, Karamboulas K, Allali-Hassani A, Vedadi M, Tyers M, Moffat J, Sicheri F, Pelletier L, Durocher D, Raught B, Rotin D, Yang J, Moran MF, Dhe-Paganon S, Sidhu SS, A strategy for modulation of enzymes in the ubiquitin system, Science 339(6119) (2013) 590–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Williams KM, Qie S, Atkison JH, Salazar-Arango S, Alan Diehl J, Olsen SK, Structural insights into E1 recognition and the ubiquitin-conjugating activity of the E2 enzyme Cdc34, Nat Commun 10(1) (2019) 3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Rape M, Ubiquitylation at the crossroads of development and disease, Nat Rev Mol Cell Biol 19(1) (2018) 59–70. [DOI] [PubMed] [Google Scholar]
- [74].Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, Conaway RC, Conaway JW, Harper JW, Pavletich NP, Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex, Nature 416(6882) (2002) 703–9. [DOI] [PubMed] [Google Scholar]
- [75].Hoeller D, Dikic I, Targeting the ubiquitin system in cancer therapy, Nature 458(7237) (2009) 438–44. [DOI] [PubMed] [Google Scholar]
- [76].Cappell SD, Mark KG, Garbett D, Pack LR, Rape M, Meyer T, EMI1 switches from being a substrate to an inhibitor of APC/C(CDH1) to start the cell cycle, Nature 558(7709) (2018) 313–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER 3rd, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D’Andrea AD, Elledge SJ, Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair, Cell 129(2) (2007) 289–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ, SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box, Cell 86(2) (1996) 263–74. [DOI] [PubMed] [Google Scholar]
- [79].Cenciarelli C, Chiaur DS, Guardavaccaro D, Parks W, Vidal M, Pagano M, Identification of a family of human F-box proteins, Curr Biol 9(20) (1999) 1177–9. [DOI] [PubMed] [Google Scholar]
- [80].Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW, F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex, Cell 91(2) (1997) 209–19. [DOI] [PubMed] [Google Scholar]
- [81].Zheng N, Zhou Q, Wang Z, Wei W, Recent advances in SCF ubiquitin ligase complex: Clinical implications, Biochim Biophys Acta 1866(1) (2016) 12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Mevissen TET, Komander D, Mechanisms of Deubiquitinase Specificity and Regulation, Annu Rev Biochem 86 (2017) 159–192. [DOI] [PubMed] [Google Scholar]
- [83].Komander D, Clague MJ, Urbe S, Breaking the chains: structure and function of the deubiquitinases, Nat Rev Mol Cell Biol 10(8) (2009) 550–63. [DOI] [PubMed] [Google Scholar]
- [84].Chen BB, Glasser JR, Coon TA, Zou C, Miller HL, Fenton M, McDyer JF, Boyiadzis M, Mallampalli RK, F-box protein FBXL2 targets cyclin D2 for ubiquitination and degradation to inhibit leukemic cell proliferation, Blood 119(13) (2012) 3132–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Chen BB, Glasser JR, Coon TA, Mallampalli RK, F-box protein FBXL2 exerts human lung tumor suppressor-like activity by ubiquitin-mediated degradation of cyclin D3 resulting in cell cycle arrest, Oncogene 31(20) (2012) 2566–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Yoshida A, Choi J, Jin H, Li Y, Bajpai S, Qie S, Diehl JA, Fbxl8 suppresses lymphoma growth and hematopoietic transformation through degradation of cyclin D3, Unpublished. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Lin DI, Barbash O, Kumar KG, Weber JD, Harper JW, Klein-Szanto AJ, Rustgi A, Fuchs SY, Diehl JA, Phosphorylation-dependent ubiquitination of cyclin D1 by the SCF(FBX4-alphaB crystallin) complex, Mol Cell 24(3) (2006) 355–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Sherr CJ, Roberts JM, CDK inhibitors: positive and negative regulators of G1-phase progression, Genes Dev 13(12) (1999) 1501–12. [DOI] [PubMed] [Google Scholar]
- [89].Benzeno S, Lu F, Guo M, Barbash O, Zhang F, Herman JG, Klein PS, Rustgi A, Diehl JA, Identification of mutations that disrupt phosphorylation-dependent nuclear export of cyclin D1, Oncogene 25(47) (2006) 6291–303. [DOI] [PubMed] [Google Scholar]
- [90].Barbash O, Egan E, Pontano LL, Kosak J, Diehl JA, Lysine 269 is essential for cyclin D1 ubiquitylation by the SCF(Fbx4/alphaB-crystallin) ligase and subsequent proteasome-dependent degradation, Oncogene 28(49) (2009) 4317–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Moreno-Bueno G, Rodriguez-Perales S, Sanchez-Estevez C, Hardisson D, Sarrio D, Prat J, Cigudosa JC, Matias-Guiu X, Palacios J, Cyclin D1 gene (CCND1) mutations in endometrial cancer, Oncogene 22(38) (2003) 6115–8. [DOI] [PubMed] [Google Scholar]
- [92].Augello MA, Berman-Booty LD, Carr R 3rd, Yoshida A, Dean JL, Schiewer MJ, Feng FY, Tomlins SA, Gao E, Koch WJ, Benovic JL, Diehl JA, Knudsen KE, Consequence of the tumor-associated conversion to cyclin D1b, EMBO Mol Med 7(5) (2015) 628–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Barbash O, Diehl JA, SCF(Fbx4/alphaB-crystallin) E3 ligase: when one is not enough, Cell Cycle 7(19) (2008) 2983–6. [DOI] [PubMed] [Google Scholar]
- [94].Barbash O, Lee EK, Diehl JA, Phosphorylation-dependent regulation of SCF(Fbx4) dimerization and activity involves a novel component, 14-3-3varepsilon, Oncogene 30(17) (2011) 1995–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Lee EK, Lian Z, D’Andrea K, Letrero R, Sheng W, Liu S, Diehl JN, Pytel D, Barbash O, Schuchter L, Amaravaradi R, Xu X, Herlyn M, Nathanson KL, Diehl JA, The FBXO4 tumor suppressor functions as a barrier to BRAFV600E-dependent metastatic melanoma, Mol Cell Biol 33(22) (2013) 4422–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Lin DI, Lessie MD, Gladden AB, Bassing CH, Wagner KU, Diehl JA, Disruption of cyclin D1 nuclear export and proteolysis accelerates mammary carcinogenesis, Oncogene 27(9) (2008) 1231–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Casanovas O, Miro F, Estanyol JM, Itarte E, Agell N, Bachs O, Osmotic stress regulates the stability of cyclin D1 in a p38SAPK2-dependent manner, J Biol Chem 275(45) (2000) 35091–7. [DOI] [PubMed] [Google Scholar]
- [98].Okabe H, Lee SH, Phuchareon J, Albertson DG, McCormick F, Tetsu O, A critical role for FBXW8 and MAPK in cyclin D1 degradation and cancer cell proliferation, PLoS One 1 (2006) e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Zou Y, Ewton DZ, Deng X, Mercer SE, Friedman E, Mirk/dyrk1B kinase destabilizes cyclin D1 by phosphorylation at threonine 288, J Biol Chem 279(26) (2004) 27790–8. [DOI] [PubMed] [Google Scholar]
- [100].Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, Ishida N, Okumura F, Nakayama K, Nakayama KI, Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7, EMBO J 23(10) (2004) 2116–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Kim SY, Herbst A, Tworkowski KA, Salghetti SE, Tansey WP, Skp2 regulates Myc protein stability and activity, Mol Cell 11(5) (2003) 1177–88. [DOI] [PubMed] [Google Scholar]
- [102].Paul I, Ahmed SF, Bhowmik A, Deb S, Ghosh MK, The ubiquitin ligase CHIP regulates c-Myc stability and transcriptional activity, Oncogene 32(10) (2013) 1284–95. [DOI] [PubMed] [Google Scholar]
- [103].Koch HB, Zhang R, Verdoodt B, Bailey A, Zhang CD, Yates JR 3rd, Menssen A, Hermeking H, Large-scale identification of c-MYC-associated proteins using a combined TAP/MudPIT approach, Cell Cycle 6(2) (2007) 205–17. [DOI] [PubMed] [Google Scholar]
- [104].Santra MK, Wajapeyee N, Green MR, F-box protein FBXO31 mediates cyclin D1 degradation to induce G1 arrest after DNA damage, Nature 459(7247) (2009) 722–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Wei S, Yang HC, Chuang HC, Yang J, Kulp SK, Lu PJ, Lai MD, Chen CS, A novel mechanism by which thiazolidinediones facilitate the proteasomal degradation of cyclin D1 in cancer cells, J Biol Chem 283(39) (2008) 26759–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Pawar SA, Sarkar TR, Balamurugan K, Sharan S, Wang J, Zhang Y, Dowdy SF, Huang AM, Sterneck E, C/EBP{delta} targets cyclin D1 for proteasome-mediated degradation via induction of CDC27/APC3 expression, Proc Natl Acad Sci U S A 107(20) (2010) 9210–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Barbash O, Zamfirova P, Lin DI, Chen X, Yang K, Nakagawa H, Lu F, Rustgi AK, Diehl JA, Mutations in Fbx4 inhibit dimerization of the SCF(Fbx4) ligase and contribute to cyclin D1 overexpression in human cancer, Cancer Cell 14(1) (2008) 68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Tan R, Nakajima S, Wang Q, Sun H, Xue J, Wu J, Hellwig S, Zeng X, Yates NA, Smithgall TE, Lei M, Jiang Y, Levine AS, Su B, Lan L, Nek7 Protects Telomeres from Oxidative DNA Damage by Phosphorylation and Stabilization of TRF1, Mol Cell 65(5) (2017) 818–831 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Lee TH, Perrem K, Harper JW, Lu KP, Zhou XZ, The F-box protein FBX4 targets PIN2/TRF1 for ubiquitin-mediated degradation and regulates telomere maintenance, J Biol Chem 281(2) (2006) 759–68. [DOI] [PubMed] [Google Scholar]
- [110].Zeng Z, Wang W, Yang Y, Chen Y, Yang X, Diehl JA, Liu X, Lei M, Structural basis of selective ubiquitination of TRF1 by SCFFbx4, Dev Cell 18(2) (2010) 214–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Peng J, Li Y, Wang X, Deng S, Holland J, Yates E, Chen J, Gu H, Essandoh K, Mu X, Wang B, McNamara RK, Peng T, Jegga AG, Liu T, Nakamura T, Huang K, Perez-Tilve D, Fan GC, An Hsp20-FBXO4 Axis Regulates Adipocyte Function through Modulating PPARgamma Ubiquitination, Cell Rep 23(12) (2018) 3607–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Feng C, Yang F, Wang J, FBXO4 inhibits lung cancer cell survival by targeting Mcl-1 for degradation, Cancer Gene Ther 24(8) (2017) 342–347. [DOI] [PubMed] [Google Scholar]
- [113].Kang JH, Choi MY, Cui YH, Kaushik N, Uddin N, Yoo KC, Kim MJ, Lee SJ, Regulation of FBXO4-mediated ICAM-1 protein stability in metastatic breast cancer, Oncotarget 8(47) (2017) 83100–83113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Pontano LL, Aggarwal P, Barbash O, Brown EJ, Bassing CH, Diehl JA, Genotoxic stress-induced cyclin D1 phosphorylation and proteolysis are required for genomic stability, Mol Cell Biol 28(23) (2008) 7245–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Pontano LL, Diehl JA, DNA damage-dependent cyclin D1 proteolysis: GSK3beta holds the smoking gun, Cell Cycle 8(6) (2009) 824–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Chu X, Zhang T, Wang J, Li M, Zhang X, Tu J, Sun S, Chen X, Lu F, Alternative splicing variants of human Fbx4 disturb cyclin D1 proteolysis in human cancer, Biochem Biophys Res Commun 447(1) (2014) 158–64. [DOI] [PubMed] [Google Scholar]
- [117].Vaites LP, Lee EK, Lian Z, Barbash O, Roy D, Wasik M, Klein-Szanto AJ, Rustgi AK, Diehl JA, The Fbx4 tumor suppressor regulates cyclin D1 accumulation and prevents neoplastic transformation, Mol Cell Biol 31(22) (2011) 4513–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Lian Z, Lee EK, Bass AJ, Wong KK, Klein-Szanto AJ, Rustgi AK, Diehl JA, FBXO4 loss facilitates carcinogen induced papilloma development in mice, Cancer Biol Ther 16(5) (2015) 750–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Choppara S, Malonia SK, Sankaran G, Green MR, Santra MK, Degradation of FBXO31 by APC/C is regulated by AKT- and ATM-mediated phosphorylation, Proc Natl Acad Sci U S A 115(5) (2018) 998–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Kumar R, Neilsen PM, Crawford J, McKirdy R, Lee J, Powell JA, Saif Z, Martin JM, Lombaerts M, Cornelisse CJ, Cleton-Jansen AM, Callen DF, FBXO31 is the chromosome 16q24.3 senescence gene, a candidate breast tumor suppressor, and a component of an SCF complex, Cancer Res 65(24) (2005) 11304–13. [DOI] [PubMed] [Google Scholar]
- [121].Li Y, Jin K, Bunker E, Zhang X, Luo X, Liu X, Hao B, Structural basis of the phosphorylation-independent recognition of cyclin D1 by the SCF(FBXO31) ubiquitin ligase, Proc Natl Acad Sci U S A 115(2) (2018) 319–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Uddin S, Bhat AA, Krishnankutty R, Mir F, Kulinski M, Mohammad RM, Involvement of F-BOX proteins in progression and development of human malignancies, Semin Cancer Biol 36 (2016) 18–32. [DOI] [PubMed] [Google Scholar]
- [123].Inuzuka H, Tseng A, Gao D, Zhai B, Zhang Q, Shaik S, Wan L, Ang XL, Mock C, Yin H, Stommel JM, Gygi S, Lahav G, Asara J, Xiao ZX, Kaelin WG Jr., Harper JW, Wei W, Phosphorylation by casein kinase I promotes the turnover of the Mdm2 oncoprotein via the SCF(beta-TRCP) ubiquitin ligase, Cancer Cell 18(2) (2010) 147–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Frescas D, Pagano M, Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer, Nat Rev Cancer 8(6) (2008) 438–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Zhao Y, Sun Y, Cullin-RING Ligases as attractive anti-cancer targets, Curr Pharm Des 19(18) (2013) 3215–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Zhang L, Pan J, Dong Y, Tweardy DJ, Dong Y, Garibotto G, Mitch WE, Stat3 activation links a C/EBPdelta to myostatin pathway to stimulate loss of muscle mass, Cell Metab 18(3) (2013) 368–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].O’Rourke JP, Newbound GC, Hutt JA, DeWille J, CCAAT/enhancer-binding protein delta regulates mammary epithelial cell G0 growth arrest and apoptosis, J Biol Chem 274(23) (1999) 16582–9. [DOI] [PubMed] [Google Scholar]
- [128].Agami R, Bernards R, Distinct initiation and maintenance mechanisms cooperate to induce G1 cell cycle arrest in response to DNA damage, Cell 102(1) (2000) 55–66. [DOI] [PubMed] [Google Scholar]
- [129].Peters JM, The anaphase promoting complex/cyclosome: a machine designed to destroy, Nat Rev Mol Cell Biol 7(9) (2006) 644–56. [DOI] [PubMed] [Google Scholar]
- [130].Latres E, Chiarle R, Schulman BA, Pavletich NP, Pellicer A, Inghirami G, Pagano M, Role of the F-box protein Skp2 in lymphomagenesis, Proc Natl Acad Sci U S A 98(5) (2001) 2515–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Carrano AC, Eytan E, Hershko A, Pagano M, SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27, Nat Cell Biol 1(4) (1999) 193–9. [DOI] [PubMed] [Google Scholar]
- [132].Bornstein G, Bloom J, Sitry-Shevah D, Nakayama K, Pagano M, Hershko A, Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase, J Biol Chem 278(28) (2003) 25752–7. [DOI] [PubMed] [Google Scholar]
- [133].Alt JR, Gladden AB, Diehl JA, p21(Cip1) Promotes cyclin D1 nuclear accumulation via direct inhibition of nuclear export, J Biol Chem 277(10) (2002) 8517–23. [DOI] [PubMed] [Google Scholar]
- [134].Kim SJ, DeStefano MA, Oh WJ, Wu CC, Vega-Cotto NM, Finlan M, Liu D, Su B, Jacinto E, mTOR complex 2 regulates proper turnover of insulin receptor substrate-1 via the ubiquitin ligase subunit Fbw8, Mol Cell 48(6) (2012) 875–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Tsunematsu R, Nishiyama M, Kotoshiba S, Saiga T, Kamura T, Nakayama KI, Fbxw8 is essential for Cul1-Cul7 complex formation and for placental development, Mol Cell Biol 26(16) (2006) 6157–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Kanie T, Onoyama I, Matsumoto A, Yamada M, Nakatsumi H, Tateishi Y, Yamamura S, Tsunematsu R, Matsumoto M, Nakayama KI, Genetic reevaluation of the role of F-box proteins in cyclin D1 degradation, Mol Cell Biol 32(3) (2012) 590–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Shan J, Zhao W, Gu W, Suppression of cancer cell growth by promoting cyclin D1 degradation, Mol Cell 36(3) (2009) 469–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Davis MI, Pragani R, Fox JT, Shen M, Parmar K, Gaudiano EF, Liu L, Tanega C, McGee L, Hall MD, McKnight C, Shinn P, Nelson H, Chattopadhyay D, D’Andrea AD, Auld DS, DeLucas LJ, Li Z, Boxer MB, Simeonov A, Small Molecule Inhibition of the Ubiquitin-specific Protease USP2 Accelerates cyclin D1 Degradation and Leads to Cell Cycle Arrest in Colorectal Cancer and Mantle Cell Lymphoma Models, J Biol Chem 291(47) (2016) 24628–24640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Magiera K, Tomala M, Kubica K, De Cesare V, Trost M, Zieba BJ, Kachamakova-Trojanowska N, Les M, Dubin G, Holak TA, Skalniak L, Lithocholic Acid Hydroxyamide Destabilizes Cyclin D1 and Induces G0/G1 Arrest by Inhibiting Deubiquitinase USP2a, Cell Chem Biol 24(4) (2017) 458–470 e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Yang H, Seo SG, Shin SH, Min S, Kang MJ, Yoo R, Kwon JY, Yue S, Kim KH, Cheng JX, Kim JR, Park JS, Kim JH, Park JHY, Lee HJ, Lee KW, 3,3’-Diindolylmethane suppresses high-fat diet-induced obesity through inhibiting adipogenesis of pre-adipocytes by targeting USP2 activity, Mol Nutr Food Res 61(10) (2017). [DOI] [PubMed] [Google Scholar]
- [141].Zhou A, Lin K, Zhang S, Chen Y, Zhang N, Xue J, Wang Z, Aldape KD, Xie K, Woodgett JR, Huang S, Nuclear GSK3beta promotes tumorigenesis by phosphorylating KDM1A and inducing its deubiquitylation by USP22, Nat Cell Biol 18(9) (2016) 954–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, Georgieva SG, Schule R, Takeyama K, Kato S, Tora L, Devys D, A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing, Mol Cell 29(1) (2008) 92–101. [DOI] [PubMed] [Google Scholar]
- [143].Gennaro VJ, Stanek TJ, Peck AR, Sun Y, Wang F, Qie S, Knudsen KE, Rui H, Butt T, Diehl JA, McMahon SB, Control of CCND1 ubiquitylation by the catalytic SAGA subunit USP22 is essential for cell cycle progression through G1 in cancer cells, Proc Natl Acad Sci U S A 115(40) (2018) E9298–E9307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Gong X, Litchfield LM, Webster Y, Chio LC, Wong SS, Stewart TR, Dowless M, Dempsey J, Zeng Y, Torres R, Boehnke K, Mur C, Marugan C, Baquero C, Yu C, Bray SM, Wulur IH, Bi C, Chu S, Qian HR, Iversen PW, Merzoug FF, Ye XS, Reinhard C, De Dios A, Du J, Caldwell CW, Lallena MJ, Beckmann RP, Buchanan SG, Genomic Aberrations that Activate D-type Cyclins Are Associated with Enhanced Sensitivity to the CDK4 and CDK6 Inhibitor Abemaciclib, Cancer Cell 32(6) (2017) 761–776 e6. [DOI] [PubMed] [Google Scholar]
- [145].Klein ME, Kovatcheva M, Davis LE, Tap WD, Koff A, CDK4/6 Inhibitors: The Mechanism of Action May Not Be as Simple as Once Thought, Cancer Cell 34(1) (2018) 9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Vilgelm AE, Saleh N, Shattuck-Brandt R, Riemenschneider K, Slesur L, Chen SC, Johnson CA, Yang J, Blevins A, Yan C, Johnson DB, Al-Rohil RN, Halilovic E, Kauffmann RM, Kelley M, Ayers GD, Richmond A, MDM2 antagonists overcome intrinsic resistance to CDK4/6 inhibition by inducing p21, Sci Transl Med 11(505) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Sammons SL, Topping DL, Blackwell KL, HR+, HER2- Advanced Breast Cancer and CDK4/6 Inhibitors: Mode of Action, Clinical Activity, and Safety Profiles, Curr Cancer Drug Targets 17(7) (2017) 637–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Yoshida A, Lee EK, Diehl JA, Induction of Therapeutic Senescence in Vemurafenib-Resistant Melanoma by Extended Inhibition of CDK4/6, Cancer Res 76(10) (2016) 2990–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].The I, Ruijtenberg S, Bouchet BP, Cristobal A, Prinsen MB, van Mourik T, Koreth J, Xu H, Heck AJ, Akhmanova A, Cuppen E, Boxem M, Munoz J, van den Heuvel S, Rb and FZR1/Cdh1 determine CDK4/6-cyclin D requirement in C. elegans and human cancer cells, Nat Commun 6 (2015) 5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Cen L, Carlson BL, Schroeder MA, Ostrem JL, Kitange GJ, Mladek AC, Fink SR, Decker PA, Wu W, Kim JS, Waldman T, Jenkins RB, Sarkaria JN, p16-Cdk4-Rb axis controls sensitivity to a cyclin-dependent kinase inhibitor PD0332991 in glioblastoma xenograft cells, Neuro Oncol 14(7) (2012) 870–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Yang C, Li Z, Bhatt T, Dickler M, Giri D, Scaltriti M, Baselga J, Rosen N, Chandarlapaty S, Acquired CDK6 amplification promotes breast cancer resistance to CDK4/6 inhibitors and loss of ER signaling and dependence, Oncogene 36(16) (2017) 2255–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Taylor-Harding B, Aspuria PJ, Agadjanian H, Cheon DJ, Mizuno T, Greenberg D, Allen JR, Spurka L, Funari V, Spiteri E, Wang Q, Orsulic S, Walsh C, Karlan BY, Wiedemeyer WR, Cyclin E1 and RTK/RAS signaling drive CDK inhibitor resistance via activation of E2F and ETS, Oncotarget 6(2) (2015) 696–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Etemadmoghadam D, Au-Yeung G, Wall M, Mitchell C, Kansara M, Loehrer E, Batzios C, George J, Ftouni S, Weir BA, Carter S, Gresshoff I, Mileshkin L, Rischin D, Hahn WC, Waring PM, Getz G, Cullinane C, Campbell LJ, Bowtell DD, Resistance to CDK2 inhibitors is associated with selection of polyploid cells in CCNE1-amplified ovarian cancer, Clin Cancer Res 19(21) (2013) 5960–71. [DOI] [PubMed] [Google Scholar]
- [154].Finn RS, Dering J, Conklin D, Kalous O, Cohen DJ, Desai AJ, Ginther C, Atefi M, Chen I, Fowst C, Los G, Slamon DJ, PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro, Breast Cancer Res 11(5) (2009) R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Pandey K, An HJ, Kim SK, Lee SA, Kim S, Lim SM, Kim GM, Sohn J, Moon YW, Molecular mechanisms of resistance to CDK4/6 inhibitors in breast cancer: A review, Int J Cancer 145(5) (2019) 1179–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Dean JL, McClendon AK, Hickey TE, Butler LM, Tilley WD, Witkiewicz AK, Knudsen ES, Therapeutic response to CDK4/6 inhibition in breast cancer defined by ex vivo analyses of human tumors, Cell Cycle 11(14) (2012) 2756–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Brooks AN, Kilgour E, Smith PD, Molecular pathways: fibroblast growth factor signaling: a new therapeutic opportunity in cancer, Clin Cancer Res 18(7) (2012) 1855–62. [DOI] [PubMed] [Google Scholar]
- [158].Jansen VM, Bhola NE, Bauer JA, Formisano L, Lee KM, Hutchinson KE, Witkiewicz AK, Moore PD, Estrada MV, Sanchez V, Ericsson PG, Sanders ME, Pohlmann PR, Pishvaian MJ, Riddle DA, Dugger TC, Wei W, Knudsen ES, Arteaga CL, Kinome-Wide RNA Interference Screen Reveals a Role for PDK1 in Acquired Resistance to CDK4/6 Inhibition in ER-Positive Breast Cancer, Cancer Res 77(9) (2017) 2488–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Smith LM, Wise SC, Hendricks DT, Sabichi AL, Bos T, Reddy P, Brown PH, Birrer MJ, cJun overexpression in MCF-7 breast cancer cells produces a tumorigenic, invasive and hormone resistant phenotype, Oncogene 18(44) (1999) 6063–70. [DOI] [PubMed] [Google Scholar]
- [160].Vijayaraghavan S, Karakas C, Doostan I, Chen X, Bui T, Yi M, Raghavendra AS, Zhao Y, Bashour SI, Ibrahim NK, Karuturi M, Wang J, Winkler JD, Amaravadi RK, Hunt KK, Tripathy D, Keyomarsi K, CDK4/6 and autophagy inhibitors synergistically induce senescence in Rb positive cytoplasmic cyclin E negative cancers, Nat Commun 8 (2017) 15916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Goel S, DeCristo MJ, Watt AC, BrinJones H, Sceneay J, Li BB, Khan N, Ubellacker JM, Xie S, Metzger-Filho O, Hoog J, Ellis MJ, Ma CX, Ramm S, Krop IE, Winer EP, Roberts TM, Kim HJ, McAllister SS, Zhao JJ, CDK4/6 inhibition triggers anti-tumour immunity, Nature 548(7668) (2017) 471–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Qie S, He D, Sang N, Overview of Glutamine Dependency and Metabolic Rescue Protocols, Methods Mol Biol 1928 (2019) 427–439. [DOI] [PubMed] [Google Scholar]
- [163].Yin C, Qie S, Sang N, Carbon source metabolism and its regulation in cancer cells, Crit Rev Eukaryot Gene Expr 22(1) (2012) 17–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Qie S, Chu C, Li W, Wang C, Sang N, ErbB2 activation upregulates glutaminase 1 expression which promotes breast cancer cell proliferation, J Cell Biochem 115(3) (2014) 498–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Dong M, Miao L, Zhang F, Li S, Han J, Yu R, Qie S, Nuclear factor-kappaB p65 regulates glutaminase 1 expression in human hepatocellular carcinoma, Onco Targets Ther 11 (2018) 3721–3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Qie S, Liang D, Yin C, Gu W, Meng M, Wang C, Sang N, Glutamine depletion and glucose depletion trigger growth inhibition via distinctive gene expression reprogramming, Cell Cycle 11(19) (2012) 3679–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Niederst MJ, Sequist LV, Poirier JT, Mermel CH, Lockerman EL, Garcia AR, Katayama R, Costa C, Ross KN, Moran T, Howe E, Fulton LE, Mulvey HE, Bernardo LA, Mohamoud F, Miyoshi N, VanderLaan PA, Costa DB, Janne PA, Borger DR, Ramaswamy S, Shioda T, Iafrate AJ, Getz G, Rudin CM, Mino-Kenudson M, Engelman JA, RB loss in resistant EGFR mutant lung adenocarcinomas that transform to small-cell lung cancer, Nat Commun 6 (2015) 6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Knudsen ES, Knudsen KE, Tailoring to RB: tumour suppressor status and therapeutic response, Nat Rev Cancer 8(9) (2008) 714–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Giacinti C, Giordano A, RB and cell cycle progression, Oncogene 25(38) (2006) 5220–7. [DOI] [PubMed] [Google Scholar]
- [170].Aparicio A, Den RB, Knudsen KE, Time to stratify? The retinoblastoma protein in castrate-resistant prostate cancer, Nat Rev Urol 8(10) (2011) 562–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Frohlich E, Wahl R, Chemotherapy and chemoprevention by thiazolidinediones, Biomed Res Int 2015 (2015) 845340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Pancione M, Sabatino L, Fucci A, Carafa V, Nebbioso A, Forte N, Febbraro A, Parente D, Ambrosino C, Normanno N, Altucci L, Colantuoni V, Epigenetic silencing of peroxisome proliferator-activated receptor gamma is a biomarker for colorectal cancer progression and adverse patients’ outcome, PLoS One 5(12) (2010) e14229. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [173].Ogino S, Shima K, Baba Y, Nosho K, Irahara N, Kure S, Chen L, Toyoda S, Kirkner GJ, Wang YL, Giovannucci EL, Fuchs CS, Colorectal cancer expression of peroxisome proliferator-activated receptor gamma (PPARG, PPARgamma) is associated with good prognosis, Gastroenterology 136(4) (2009) 1242–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Takeuchi H, Ozawa S, Ando N, Shih CH, Koyanagi K, Ueda M, Kitajima M, Altered p16/MTS1/CDKN2 and cyclin D1/PRAD-1 gene expression is associated with the prognosis of squamous cell carcinoma of the esophagus, Clin Cancer Res 3(12 Pt 1) (1997) 2229–36. [PubMed] [Google Scholar]
- [175].Hardisson D, Molecular pathogenesis of head and neck squamous cell carcinoma, Eur Arch Otorhinolaryngol 260(9) (2003) 502–8. [DOI] [PubMed] [Google Scholar]
- [176].Toncheva D, Petrova D, Tzenova V, Dimova I, Yankova R, Yordanov V, Damjanov D, Todorov T, Zaharieva B, Tissue microarray analysis of cyclin D1 gene amplification and gain in colorectal carcinomas, Tumour Biol 25(4) (2004) 157–60. [DOI] [PubMed] [Google Scholar]
- [177].Shamma A, Doki Y, Shiozaki H, Tsujinaka T, Inoue M, Yano M, Kimura Y, Yamamoto M, Monden M, Effect of cyclin D1 and associated proteins on proliferation of esophageal squamous cell carcinoma, Int J Oncol 13(3) (1998) 455–60. [DOI] [PubMed] [Google Scholar]
- [178].Dey B, Raphael V, Khonglah Y, GiriLynrah K, Expression of Cyclin D1 and P16 in Esophageal Squamous Cell Carcinoma, Middle East J Dig Dis 7(4) (2015) 220–5. [PMC free article] [PubMed] [Google Scholar]
- [179].Xu G, Jaffrey SR, The new landscape of protein ubiquitination, Nat Biotechnol 29(12) (2011) 1098–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].McKay JA, Douglas JJ, Ross VG, Curran S, Murray GI, Cassidy J, McLeod HL, Cyclin D1 protein expression and gene polymorphism in colorectal cancer. Aberdeen Colorectal Initiative, Int J Cancer 88(1) (2000) 77–81. [DOI] [PubMed] [Google Scholar]
- [181].Consortium APG, AACR Project GENIE: Powering Precision Medicine through an International Consortium, Cancer Discov 7(8) (2017) 818–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Helsten T, Kato S, Schwaederle M, Tomson BN, Buys TP, Elkin SK, Carter JL, Kurzrock R, Cell-Cycle Gene Alterations in 4,864 Tumors Analyzed by Next-Generation Sequencing: Implications for Targeted Therapeutics, Mol Cancer Ther 15(7) (2016) 1682–90. [DOI] [PubMed] [Google Scholar]
- [183].Delmer A, Ajchenbaum-Cymbalista F, Tang R, Ramond S, Faussat AM, Marie JP, Zittoun R, Overexpression of cyclin D2 in chronic B-cell malignancies, Blood 85(10) (1995) 2870–6. [PubMed] [Google Scholar]
- [184].Mermelshtein A, Gerson A, Walfisch S, Delgado B, Shechter-Maor G, Delgado J, Fich A, Gheber L, Expression of D-type cyclins in colon cancer and in cell lines from colon carcinomas, Br J Cancer 93(3) (2005) 338–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Zlamalikova L, Moulis M, Salek D, Jarkovsky J, Smarda J, Smardova J, Expression of D-type cyclins in mantle cell and diffuse large B-cell lymphomas, Oncol Rep 35(5) (2016) 2673–80. [DOI] [PubMed] [Google Scholar]
- [186].Bondi J, Husdal A, Bukholm G, Nesland JM, Bakka A, Bukholm IR, Expression and gene amplification of primary (A, B1, D1, D3, and E) and secondary (C and H) cyclins in colon adenocarcinomas and correlation with patient outcome, J Clin Pathol 58(5) (2005) 509–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Baldassarre G, Belletti B, Bruni P, Boccia A, Trapasso F, Pentimalli F, Barone MV, Chiappetta G, Vento MT, Spiezia S, Fusco A, Viglietto G, Overexpressed cyclin D3 contributes to retaining the growth inhibitor p27 in the cytoplasm of thyroid tumor cells, J Clin Invest 104(7) (1999) 865–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Florenes VA, Faye RS, Maelandsmo GM, Nesland JM, Holm R, Levels of cyclin D1 and D3 in malignant melanoma: deregulated cyclin D3 expression is associated with poor clinical outcome in superficial melanoma, Clin Cancer Res 6(9) (2000) 3614–20. [PubMed] [Google Scholar]
- [189].Metcalf RA, Zhao S, Anderson MW, Lu ZS, Galperin I, Marinelli RJ, Cherry AM, Lossos IS, Natkunam Y, Characterization of D-cyclin proteins in hematolymphoid neoplasms: lack of specificity of cyclin-D2 and D3 expression in lymphoma subtypes, Mod Pathol 23(3) (2010) 420–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Igawa T, Sato Y, Takata K, Iwaki N, Tanaka T, Asano N, Maeda Y, Orita Y, Nakamura N, Nakamura S, Yoshino T, De novo CD5-positive diffuse large B-cell lymphomas show high specificity for cyclin D2 expression, Diagn Pathol 8 (2013) 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Filipits M, Jaeger U, Pohl G, Stranzl T, Simonitsch I, Kaider A, Skrabs C, Pirker R, Cyclin D3 is a predictive and prognostic factor in diffuse large B-cell lymphoma, Clin Cancer Res 8(3) (2002) 729–33. [PubMed] [Google Scholar]
- [192].Pruneri G, Pignataro L, Valentini S, Fabris S, Maisonneuve P, Carboni N, Pece S, Capra M, Del Curto B, Neri A, Viale G, Cyclin D3 immunoreactivity is an independent predictor of survival in laryngeal squamous cell carcinoma, Clin Cancer Res 11(1) (2005) 242–8. [PubMed] [Google Scholar]
- [193].Keyomarsi K, Tucker SL, Buchholz TA, Callister M, Ding Y, Hortobagyi GN, Bedrosian I, Knickerbocker C, Toyofuku W, Lowe M, Herliczek TW, Bacus SS, Cyclin E and survival in patients with breast cancer, N Engl J Med 347(20) (2002) 1566–75. [DOI] [PubMed] [Google Scholar]
- [194].Moller MB, Nielsen O, Pedersen NT, Cyclin D3 expression in non-Hodgkin lymphoma. Correlation with other cell cycle regulators and clinical features, Am J Clin Pathol 115(3) (2001) 404–12. [DOI] [PubMed] [Google Scholar]
- [195].Hedberg Y, Roos G, Ljungberg B, Landberg G, Cyclin D3 protein content in human renal cell carcinoma in relation to cyclin D1 and clinico-pathological parameters, Acta Oncol 41(2) (2002) 175–81. [DOI] [PubMed] [Google Scholar]


