Summary
It has been known for more than 50 years that transcription and translation are physically coupled in bacteria, but whether or not this coupling may be mediated by the two-domain protein N-utilization substance (Nus) G in Escherichia coli is still heavily debated. Here, we combine integrative structural biology and functional analyses to provide conclusive evidence that NusG can physically link transcription with translation by contacting both RNA polymerase and the ribosome. We present a cryo-electron microscopy structure of a NusG:70S ribosome complex and nuclear magnetic resonance spectroscopy data revealing simultaneous binding of NusG to RNAP and the intact 70S ribosome, providing the first direct structural evidence for NusG-mediated coupling. Furthermore, in vivo reporter assays show that recruitment of NusG occurs late in transcription and strongly depends on translation. Thus, our data suggest that coupling occurs initially via direct RNAP:ribosome contacts and is then mediated by NusG.
Subject Areas: Biochemistry, Biochemistry Methods, Structural Biology, Three-Dimensional Reconstruction of Biomolecular Structures
Graphical Abstract
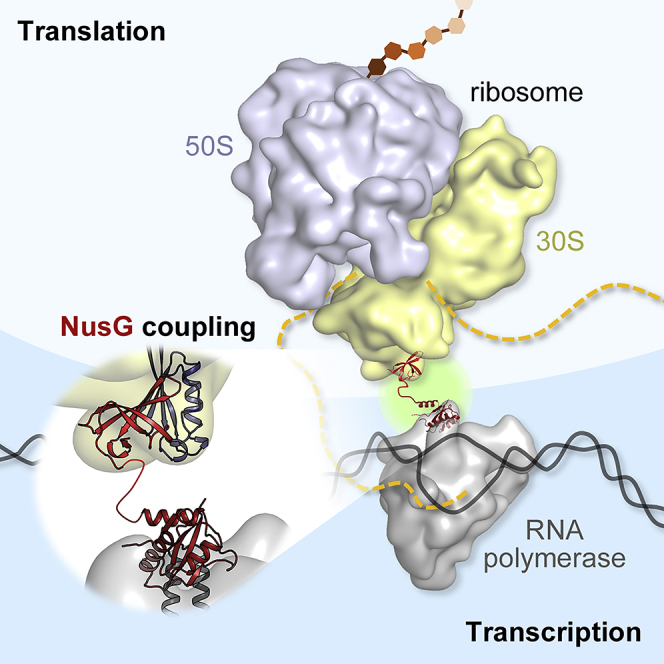
Highlights
-
•
NusG can contact RNAP and lead ribosome simultaneously
-
•
NusG recruitment occurs late during transcription and depends on translation
-
•
NusG-mediated coupling happens during late translation
Biochemistry; Biochemistry Methods; Structural Biology; Three-Dimensional Reconstruction of Biomolecular Structures
Introduction
Gene expression is a universal process in all cells and consists of transcription, i.e., the synthesis of RNA based on the DNA, and—if RNA is not the final gene product—translation, i.e., the messenger RNA (mRNA)-guided synthesis of a protein. Since the late 1960s it has been known that the rates of transcription and translation are synchronized in Escherichia coli (E. coli) so that mRNA is translated while being transcribed (Das et al., 1967; Mehdi and Yudkin, 1967; Miller et al., 1970; Proshkin et al., 2010; Vogel and Jensen, 1994, 1995). This process, called transcription:translation coupling, is possible due to the lack of a physical barrier between transcription and translation in bacteria (reviewed in Conn et al., 2019). Only recently, direct physical interactions between RNA polymerase (RNAP) and the ribosome have been demonstrated (Demo et al., 2017; Fan et al., 2017; Kohler et al., 2017), consistent with earlier observations that transcriptional events may control translation activity and vice versa (Proshkin et al., 2010). As transcription and translation are closely connected to other central processes in a bacterial cell, such as DNA repair (Pani and Nudler, 2017) and protein folding (Thommen et al., 2017), transcription:translation coupling constitutes one of the key regulatory functions in bacterial gene expression.
However, there are also indications that transcription:translation coupling may involve a member of the family of N-utilization substance (Nus) G proteins, which serves as an adapter connecting RNAP and the lead ribosome (Burmann et al., 2010, 2012; Saxena et al., 2018; Zuber et al., 2019). E. coli NusG, member and eponym of the only universally conserved class of transcription factors (Werner, 2012), consists of two domains, an N- and a C-terminal domain (NTD and CTD), respectively, connected via a flexible linker, which move independently (Burmann et al., 2011; Mooney et al., 2009a). NusG-NTD binds RNAP and accelerates transcription elongation (Burova et al., 1995; Kang et al., 2018; Mooney et al., 2009a). Structural studies demonstrate that NusG-CTD, which is a five-stranded, antiparallel β barrel with a Kyrpides-Ouzounis-Woese motif (Kyrpides et al., 1996), is a versatile interaction platform for various transcription factors. By binding to protein S10, which is part of the 30S subunit of the ribosome, NusG may link transcription and translation (Burmann et al., 2010). Saxena et al. also demonstrated specific 1:1 binding of NusG to 70S ribosomes both in vitro and in vivo (Saxena et al., 2018). S10 is identical with transcription factor NusE and forms a ribosome-free complex with NusB, NusA, and NusG which suppresses transcription termination (Dudenhoeffer et al., 2019; Huang et al., 2019; Krupp et al., 2019; Said et al., 2017; Squires et al., 1993). Finally, NusG-CTD binds to termination factor Rho and is required for most Rho activity in vivo (Burmann et al., 2010; Lawson et al., 2018; Mitra et al., 2017). Transcription:translation coupling prevents Rho factor from terminating transcription by sequestering the NusG-CTD and by blocking Rho access to RNAP via untranslated mRNA. Cryptic E. coli Rho-dependent terminators located within open reading frames (orfs) are revealed when ribosomes are released by polar nonsense mutations (Cardinale et al., 2008; Newton et al., 1965).
Nevertheless, there is evidence for intragenic uncoupling and Rho-dependent transcription termination in the absence of nonsense mutations: Washburn and Gottesman (2011) and Dutta et al. (2011) found that Rho resolves clashes between transcription and replication. Such conflicts are likely to occur within, rather than at the end of, genes. Uncoupling would allow Rho to release the stationary transcription elongation complexes (TECs).
Mutations in nusE/s10 or nusG that uncouple transcription from translation increase sensitivity to chloramphenicol (Saxena et al., 2018). This antibiotic retards translation, breaking the bond between the lead ribosome and the TEC. Consequently, the uncoupled TEC may backtrack or terminate prematurely (Dutta et al., 2011).
In this report, we present a cryo-electron microscopy (cryo-EM) structure showing NusG binding to the S10 subunit in a 70S ribosome. The NusG-CTD binding site of S10 is also target of the ribosome-release factor, transfer-messenger RNA (tmRNA), raising the possibility that tmRNA might displace NusG at rare codons, thereby uncoupling transcription from translation (Roche and Sauer, 1999). We also show by solution-state nuclear magnetic resonance (NMR) spectroscopy that NusG, once bound to RNAP, can interact with S10 or with a complete ribosome, setting the structural basis for NusG-mediated coupling.
NusG couples transcription with translation in vivo, as proposed earlier (Burmann et al., 2010). Uncoupling of RNAP from the lead ribosome is enhanced when translation is compromised. Importantly, we demonstrate that uncoupled RNAP can outpace translation, leading to Rho-dependent transcription termination. This intragenic termination explains the necessity for the apparent perfect synchronization between transcription and translation (Proshkin et al., 2010).
Results
Structural Evidence of NusG Binding to the Ribosomal Protein S10 on the 70S Ribosome
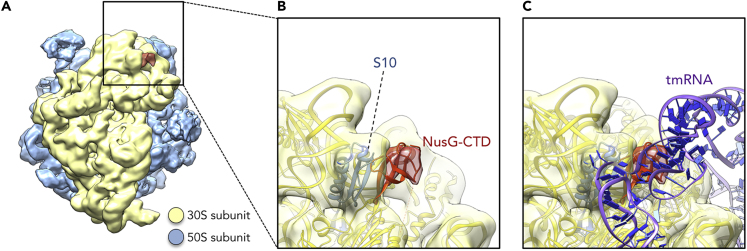
We assembled a NusG:70S complex by incubating 70S ribosomes with an excess of NusG and determined the structure of this complex by cryo-EM and single-particle reconstruction (Table S1). Overall, 188,127 particles were extracted from 1,327 images and ∼5% of these particles showed an extra mass of density attached to the mass identified as protein S10 (Figures 1A and 1B). This additional density perfectly matches the size of NusG-CTD, suggesting that NusG binds at the site predicted from the solution NMR structure of NusG-CTD bound to the free ribosomal protein S10 in a 1:1 stoichiometry (Figures 1A and 1B [Burmann et al., 2010]). The density map reconstructed from the class of NusG:70S particles was refined to an average resolution of 6.8 Å. No density could be observed for NusG-NTD, indicating that it is flexibly bound to the NusG-CTD and does not interact with the ribosome.
Figure 1.
Structure of NusG-CTD bound to 70S Ribosome
(A) Cryo-EM density of the 70S ribosome:NusG complex (see also Table S1). The density of the 50S subunit is shown in light blue, the density of the 30S subunit in yellow, the density corresponding to NusG-CTD in red.
(B) Close-up view of the region boxed in (A). 70S (yellow), S10 (blue), and NusG-CTD (red) are in ribbon representation; cryo-EM density is shown as transparencies.
(C) Superposition of the 70S:NusG complex with the 70S:tmRNA complex (tmRNA is in ribbon representation, purple and dark blue; EMD 5234, PDB: 3IZ4). 30S and NusG-CTD are displayed as in (B).
During translation ribosomes may stall on incomplete mRNAs, i.e., they reach the 3′ end of an mRNA without terminating, resulting in an unproductive translation complex. Together with the small protein B (SmpB) tmRNA can bind to these stalled ribosomes in order to rescue them and to tag the nascent polypeptide chain for degradation in a process called trans-translation (Weis et al., 2010). Interestingly, the NusG-CTD binding site overlaps with the region of S10 that is contacted by the tmRNA when it is bound to a ribosome in its resume state (Figure 1C [Burmann et al., 2010; Fu et al., 2010; Rae et al., 2019; Weis et al., 2010]). From this we conclude that NusG-CTD and tmRNA share binding sites on S10, raising the possibility that, in addition to releasing stalled ribosomes, tmRNA competes with NusG for ribosome binding, thus preventing NusG from maintaining a linkage between the lead ribosome and RNAP. In other words, tmRNA might be able to displace NusG and thereby facilitate uncoupled transcription.
Simultaneous Binding of NusG to S10 and RNAP
In the cryo-EM structure of E. coli NusG bound to a paused TEC (Kang et al., 2018) only the density of NusG-NTD was observable, indicating that NusG-CTD moves freely and does not interact with RNAP. Binding of NusG-CTD to S10 was observed both in a binary system (Burmann et al., 2010) and a λN-dependent antitermination complex (Krupp et al., 2019; Said et al., 2017).
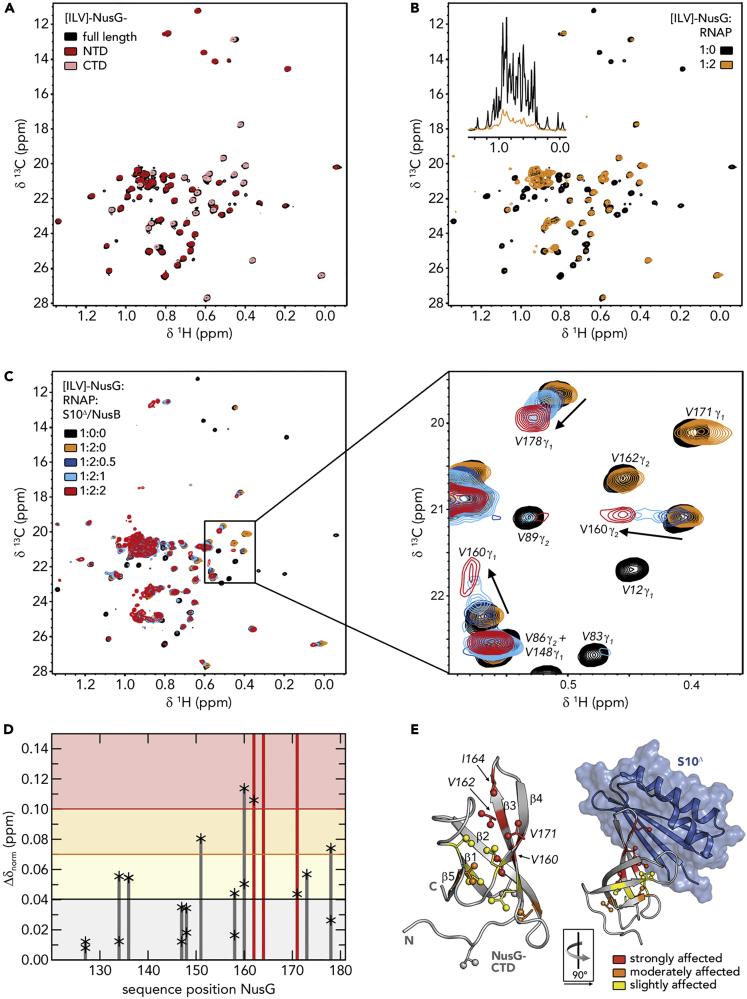
Since the NusG-CTD:S10 interaction is a prerequisite for NusG-mediated transcription:translation coupling, we probed this contact when NusG was bound to RNAP—but not in an antitermination context—by solution-state NMR spectroscopy. We employed NusG samples where [1H,13C]-labeled methyl groups of Ile, Leu, and Val residues in perdeuterated proteins served as NMR-active probes ([ILV]-NusG) to increase sensitivity, allowing us to study large systems.
In the methyl-transverse relaxation optimized spectroscopy (methyl-TROSY) spectrum of free [ILV]-NusG (Figure 2A), signals of the NusG-NTD and NusG-CTD perfectly superimpose with the signals of the isolated [ILV]-labeled protein domains, suggesting that the domains move independently, confirming a previous report stating that there are no intramolecular domain interactions (Burmann et al., 2011). Upon addition of RNAP in a two-fold molar excess, [ILV]-NusG signals were significantly decreased in the one-dimensional methyl-TROSY spectrum (Figure 2B, inset), indicating [ILV]-NusG:RNAP complex formation. Binding of RNAP increases the molecular mass of [ILV]-NusG dramatically, resulting in enhanced relaxation, which ultimately leads to drastic line broadening and a decrease in signal intensity. Interestingly, the two-dimensional spectra revealed a non-uniform signal decrease (Figure 2B), which is caused by a combination of several effects. First, there is a general loss of signal intensity due to the increase in molecular mass upon complex formation, as discussed above. Second, upon binding, methyl groups of Ile, Leu, and Val residues located in the binding surface come into close proximity of RNAP protons. Dipole-dipole interactions contribute to relaxation processes so that the signal intensity of these methyl groups is decreased more strongly than that of methyl groups located elsewhere in [ILV]-NusG. Finally, signal intensities may be affected by chemical exchange processes. We analyzed the signal intensity of [ILV]-NusG signals in the presence of RNAP quantitatively by calculating relative signal intensities, i.e., the ratio of the remaining signal intensity of [ILV]-NusG in the presence of RNAP to the signal intensity of free [ILV]-NusG (Figure S1).
Figure 2.
RNAP-Bound NusG Interacts with S10
(A) Superposition of 2D [1H, 13C]-methyl-TROSY spectra of [ILV]-NusG (black, 20 μM), [ILV]-NusG-NTD (dark red, 100 μM), and [ILV]-NusG-CTD (light red, 30 μM).
(B) 2D [1H, 13C]-methyl-TROSY spectra of [ILV]-NusG in the absence (black, 20 μM) and presence (orange, 18 μM) of two equivalents of RNAP. Inset: Normalized 1D [1H,13C]-methyl TROSY spectra, colored as 2D spectra. See also Figure S1.
(C) 2D [1H, 13C]-methyl-TROSY spectra of [ILV]-NusG alone (20 μM), in the presence of a 2-fold molar excess of RNAP (18 μM [ILV]-NusG), and upon titration of [ILV]-NusG:RNAP with 218 μM S10Δ:NusB. The molar ratio of [ILV]-NusG:RNAP:S10Δ:NusB is indicated in color. The panel on the right shows an enlargement of the boxed region. Selected signals are labeled and arrows indicate chemical shift changes upon S10Δ:NusB addition.
(D) [1H,13C]-methyl-TROSY-derived normalized chemical shift perturbations of [ILV]-NusG-CTD methyl group signals of RNAP-bound [ILV]-NusG upon complex formation with S10Δ:NusB. Asterisks mark the values of individual methyl group signals, bars represent the highest values. Red bars indicate vanishing signals. Horizontal lines are thresholds for affected methyl groups: slightly affected (0.04 ppm ≤ Δδnorm < 0.07 ppm; black), moderately affected (0.07 ppm ≤ Δδnorm < 0.1 ppm; orange), and strongly affected (Δδnorm ≥ 0.10 ppm; red).
(E) Mapping of affected methyl groups on the structure of isolated NusG-CTD (left; PDB ID: 2JVV) and NusG-CTD in complex with S10Δ (right; PDB ID 2KVQ). NusG-CTD is shown in ribbon (gray), S10Δ in ribbon and surface (blue) representation. Affected Ile, Leu, and Val residues are colored according to (D); non-affected Ile, Leu, and Val residues are gray. Side chains of Ile, Leu, and Val residues are depicted as sticks, their methyl groups as spheres. Strongly affected Ile, Leu, and Val residues are labeled. The orientation of NusG-CTD in the complex relative to the isolated state is indicated.
The average relative intensity of NusG-NTD signals was significantly lower than that of the linker or the NusG-CTD, suggesting that NusG-NTD binds to RNAP, whereas NusG-CTD remains flexible and moves independently, able to interact with other partners, as indicated by the NusG:TEC structure (Kang et al., 2018). The signal intensity of all Ile, Leu, and Val residues in the RNAP binding site of NusG was completely extinguished, confirming that NusG-NTD binds to RNAP at its known binding site (Drögemüller et al., 2015; Kang et al., 2018; Krupp et al., 2019; Said et al., 2017).
To test if NusG-CTD can bind to S10 while being tethered to RNAP via NusG-NTD, we titrated the [ILV]-NusG:RNAP complex with S10Δ (Figure 2C). In order to increase stability, we used an S10 variant lacking the ribosome binding loop in complex with NusB (Luo et al., 2008). Chemical shift changes of [ILV]-NusG-CTD signals upon titration of [ILV]-NusG:RNAP with S10Δ:NusB were determined (Figure 2D) and affected residues were mapped onto the three-dimensional structure of NusG-CTD (Figure 2E). Strongly affected residues are located in β strands 3 and 4 as well as in the connecting loop, in agreement with the binding site observed in the binary NusG-CTD:S10Δ complex (Burmann et al., 2010). The loop between β strands 1 and 2 is also part of the NusG-CTD:S10Δ binding site, but as it does not contain any Ile, Leu, or Val residues, no NMR-active probes are available in this region; nevertheless, affected residues can be found in β strand 1, directly preceding this loop. This suggests that the CTD:S10Δ binding surface in the RNAP:NusG:S10Δ:NusB complex is identical to the one determined in the binary system. Importantly, the NusG-NTD signals do not change when S10Δ is added to the NusG:RNAP complex, indicating that S10Δ binding does not release the bound RNAP.
We conclude that the S10 interaction site of NusG-CTD is accessible in the NusG:RNAP complex and thus can promote ribosome binding and formation of a ribosome:NusG:RNAP complex.
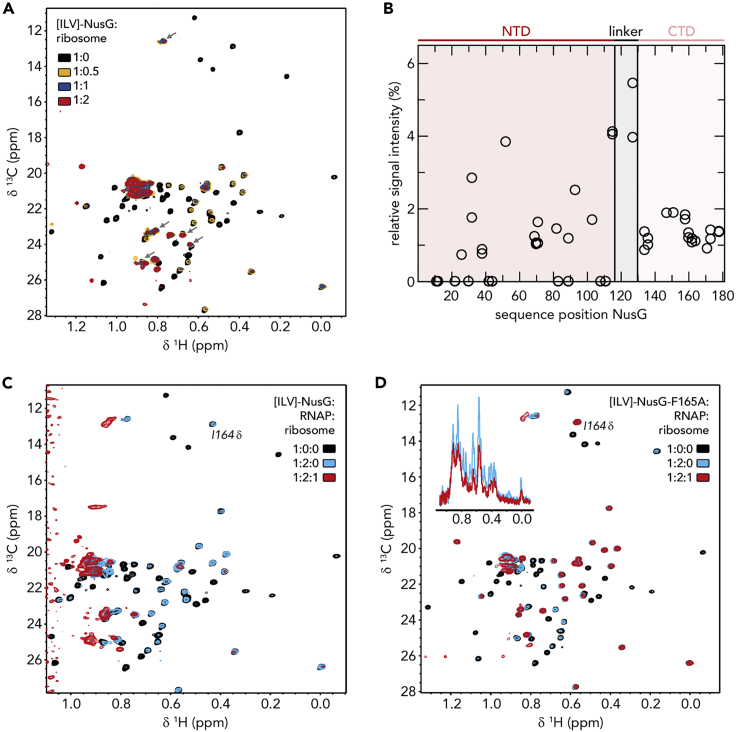
To look for a ribosome:NusG:RNAP complex, we repeated the experiment using intact 70S ribosomes instead of S10Δ:NusB (Figure 3). In a first test, we titrated [ILV]-NusG with 70S ribosomes (Figure 3A). As in the [ILV]-NusG:RNAP experiment, signal intensity of [ILV]-NusG methyl groups was significantly, but not uniformly, decreased. In the presence of a 2-fold molar excess of ribosomes some NusG-NTD signals remained visible, whereas most NusG-CTD signals were nearly completely extinguished. Quantitative analysis of the [ILV]-NusG methyl group signal intensity in the presence of 0.5 equivalents of 70S ribosomes clearly shows that the relative intensity of NusG-CTD signals was in a narrow range <2%, whereas the relative intensity of NusG-NTD signals covered values from 0%–4% and was higher on average (Figure 3B). Relative intensities of zero of NusG-NTD signals can be attributed to the fact that these signals are weak even in free NusG and can thus not be quantified upon ribosome binding. Owing to the flexibility of the linker, signals corresponding to amino acids in this region had the highest relative signal intensities. From these results we conclude that NusG binds to the ribosome via its CTD, in agreement with our cryo-EM structure (Figure 1). Owing to the drastic increase in molecular mass we were unable to determine a binding site from these experiments, but nevertheless, the pattern of intensity changes of NusG-CTD signals was similar to that resulting from the titration of RNAP-bound NusG with S10Δ, i.e., the most drastic decrease of signal intensity can be observed for residues 160–170, which are part of β strands 3 and 4 and the intervening loop. Consequently, we conclude that the ribosome binding site is identical with the binding site for isolated S10Δ.
Figure 3.
RNAP-bound NusG Interacts with the 70S Ribosome
(A and B) NusG interacts with 70S ribosome via its CTD. (A) 2D [1H, 13C]-methyl-TROSY spectra of free [ILV]-NusG (11 μM, black) and [ILV]-NusG in the presence of 70S ribosome (molar ratio [ILV]-NusG:ribosome = 1:0.5 (6.6 μM [ILV]-NusG, orange); = 1:1 (7.5 μM [ILV]-NusG, blue); = 1:2 (4 μM [ILV]-NusG, red)). Arrows indicate [ILV]-NusG-NTD signals that are well visible in the [ILV]-NusG:ribosome complex. (B) Quantitative analysis of [ILV]-NusG methyl group signal intensities in the presence of 0.5 equivalents of 70S ribosome. Relative signal intensities are plotted versus the sequence position of NusG. The domain organization of NusG is indicated above the diagram.
(C) 2D [1H, 13C]-methyl-TROSY spectra of [ILV]-NusG (11 μM, black), [ILV]-NusG in the presence of RNAP (molar ratio 1:2, 6 μM [ILV]-NusG, blue), and [ILV]-NusG in the presence of RNAP and 70S ribosome (molar ratio 1:2:1, 6 μM [ILV]-NusG, red).
(D) 2D [1H, 13C]-methyl-TROSY spectra of [ILV]-NusGF165A (20 μM, black), [ILV]-NusGF165A in the presence of RNAP (molar ratio 1:2, 6 μM [ILV]-NusGF165A, blue), and [ILV]-NusGF165A in the presence of RNAP and 70S ribosome (molar ratio 1:2:1, 6 μM [ILV]-NusGF165A, red). The inset shows the normalized 1D spectra of the corresponding titration step.
See also Figure S2.
Next, we formed a complex of [ILV]-NusG and RNAP (molar ratio 1:2). The 2D methyl-TROSY spectrum of the complex revealed a decrease of signal intensities (Figure 3C) typical for NusG binding to RNAP (see Figure 2C), i.e., primarily NusG-CTD signals remained visible. When we then added one equivalent of 70S ribosomes nearly all [ILV]-NusG signals were diminished (e.g., the signal corresponding to I164, which is in the loop responsible for ribosome binding; Figure 3C). Strikingly, the spectrum differs from the spectrum of [ILV]-NusG in the presence of 70S ribosome (Figure 3A). These results can be explained by three scenarios: (1) NusG-NTD is bound to RNAP, NusG-CTD is bound to a ribosome, and the ribosome directly interacts with RNAP; (2) NusG-NTD is bound to RNAP, NusG-CTD is bound to the ribosome, but the ribosome does not interact with RNAP; (3) NusG-NTD is bound to RNAP, the ribosome directly interacts with RNAP, and NusG-CTD is free but is in the vicinity of the ribosome. To exclude the last scenario we repeated the experiment using a NusG variant, NusGF165A, in which F165, essential for ribosome binding (Burmann et al., 2010; Knowlton et al., 2003), is substituted by an Ala. Having ensured that the amino acid substitution does not influence the structure of NusG (Figure S2A) we tested in a control experiment [ILV]-NusGF165A binding to S10Δ. Indeed, we detected no interaction (Figures S2B and S2C). When we added 70S ribosomes to a preformed [ILV]-NusGF165A:RNAP complex (molar ratio 1:2), the spectrum corresponding to the [ILV]-NusGF165A:RNAP complex did not change significantly and, in particular, NusG-CTD signals remained visible, suggesting that the ribosome was not bound (Figure 3D). However, the general decrease in signal intensity indicates a direct RNAP:ribosome interaction. Thus, we conclude that NusG can serve as physical linker between ribosome and RNAP, although it remains elusive if a direct interaction between RNAP and a ribosome occurs in this NusG-coupled complex.
Translation Promotes NusG Attachment to TEC
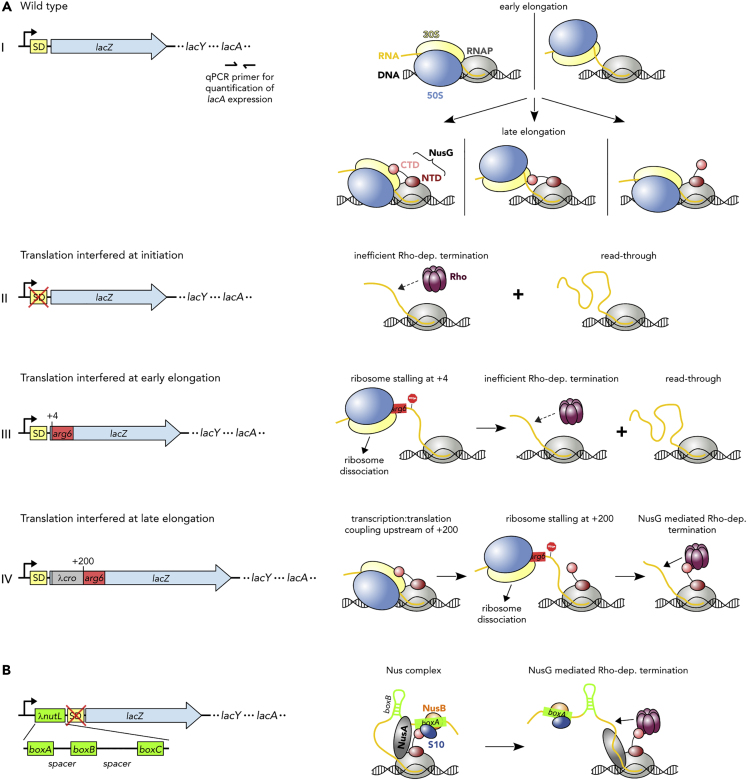
Chromatin immunoprecipitation (ChIP) analysis showed that NusG binds to TEC well after transcription and translation initiation (Mooney et al., 2009b). Thus, we asked whether translation was, in fact, required for attachment of NusG to the TEC. To approach this question, we examined the effects of translation on NusG-mediated Rho-dependent termination within the lac operon (Figure 4A, Table 1) as NusG recruitment to the TEC is necessary for efficient Rho-dependent termination. Rho-dependent termination occurs within lacZ both in vitro (Burns and Richardson, 1995) and, upon the introduction of lacZ nonsense mutations, in vivo (Adhya and Gottesman, 1978; Newton et al., 1965). Polarity was measured using a probe to lacA, comparing mRNA levels with or without treatment with the Rho inhibitor bicyclomycin (BCM). Wild-type (WT) cells revealed no detectable termination (Table 1 and Figure 4A-I), which may be attributed to (1) sequestering of NusG-CTD by the ribosome, (2) binding of the ribosome to the nascent RNA, or (3) both. In all scenarios, however, the presence of the translating ribosome prevents Rho binding. We interfered with translation initiation by mutating the ribosome-binding site, i.e., the Shine-Dalgarno (SD) sequence (Figure 4A-II), or translation elongation by introducing six successive rare arginine codons at two different locations in lacZ (Figure 4A-III and IV). Introduction of two G to A mutations in the lacZ SD sequence prohibits translation initiation of lacZ (Figure 4A-II). lacA mRNA measurements gave a readthrough of 21%, indicating that Rho-dependent termination occurs, but was inefficient in the absence of translation of lacZ mRNA. Introduction of the six in-frame rare arginine residues at the +4 position of lacZ (Figure 4A-III and Table 1) allowed 27% readthrough, i.e., Rho-dependent termination is present but still inefficient if translation of lacZ mRNA is interfered with at early elongation. In contrast, introduction of the rare arginine residues 200 nucleotides (nt) from the start site of transcription (Figure 4A-IV and Table 1) resulted in high polarity, yielding <1% readthrough. As efficient Rho-dependent termination requires NusG our results suggest that NusG binding to TEC occurs late and is dependent on translation.
Figure 4.
Translation Is Required for NusG Recruitment to the TEC
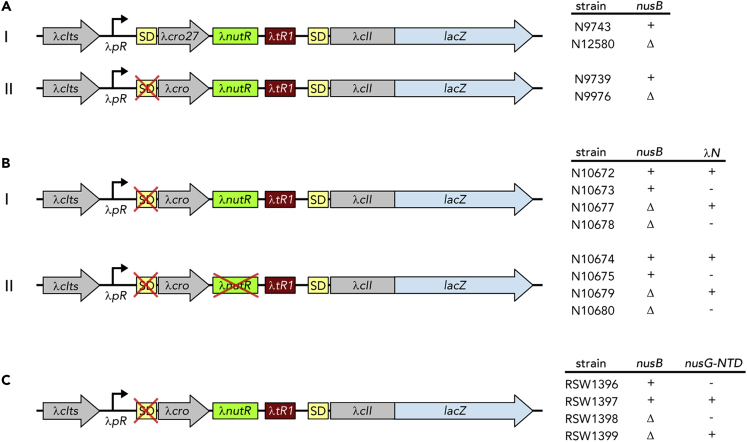
(A and B) Left: Organization of the E. coli lac operon in strains MDS42 (A-I; wild type lacZ), RSW1225 (A-II; mutant [inactive] lacZ SD sequence), RSW1245 (A-III; in-frame insertion of six rare Arg codons [arg6] at position +4 of lacZ), RSW1276 (A-IV; in-frame insertion of λcro and six rare Arg codons at position +4 of lacZ [equivalent to arg6 being at position +200 of the gene]), and RSW1297 (B; λnutL site upstream of mutant lacZ SD sequence). lacY and lacA are only indicated for clarity. qPCR primers specific to the 3′ end of lacA (position indicated in A-I) were used to measure mRNA levels and thereby readthrough of lacA (see Table 1). Right: Schemes of possible effects on transcription:translation coupling and Rho-dependent termination within lacZ. A-I, top: Ribosomes are recruited in the early elongation phase, leading to a directly coupled RNAP:ribosome complex (left) or uncoupled transcription and translation (right). A-I, bottom: NusG is recruited in late elongation, resulting in a NusG-coupled complex with (left) or without (middle) direct RNAP:ribosome contacts, or modifying the pre-existing RNAP:ribosome complex without establishing an CTD:S10 interaction (right). A-II: Failure of NusG recruitment results in inefficient Rho-dependent termination and high lacZ readthrough. A-III: arg6 stops the translating ribosome at position +4, whereas transcription elongation proceeds (left), resulting in ribosome dissociation and no NusG recruitment. Transcription proceeds and is only inefficiently terminated by Rho (right). A-IV: NusG couples transcription and translation (left) until arg6 stops the ribosome at position +200 (middle), allowing efficient, NusG-stimulated Rho-dependent termination (right). (B) λnutL recruits NusA, NusG, and the S10/NusB dimer, creating a Nus complex. NusG can thus support Rho-dependent termination.
Table 1.
NusG Couples Late after Transcription Initiation
| Strain | lacZ | nutL | Fold Increase of RNA Level (BCM−) | Fold Increase of RNA Level (BCM+) | RT (%) |
|---|---|---|---|---|---|
| MDS42 | wt | – | .25 ± 0.04 | .26 ± 0.03 | 96 ± 19 |
| RSW1225 | SD- | – | .12 ± 0.03 | .56 ± 0.10 | 21 ± 7 |
| RSW1245 | arg(6)—early | – | .13 ± 0.01 | .49 ± 0.02 | 27 ± 2 |
| RSW1276 | arg(6)—late | – | <.001 ± 0.002 | .12 ± 0.003 | <1 |
| RSW1297 | SD- | + | .01 ± 0.006 | .59 ± 0.01 | 2 ± 1 |
Expression of lacZ was induced for 20 min from the lac operon with 1 mM IPTG. Where indicated, Rho-dependent termination was inhibited by adding 100 μg/mL BCM 1 min prior to induction. Readthrough was calculated from the fold increase of lacA RNA compared with ompA RNA in the presence or absence of BCM. RNA levels were measured using qRT-PCR and the fold increase was calculated using the ΔΔCt method (Livak and Schmittgen, 2001). Values are the average of ≥3 independent experiments, each carried out in duplicate. RSW1225 carries two G to A mutations in the lacZ ribosome-binding site. RSW1245 carries an insertion of six rare arginine codons (atg-acc-atg-AGG-AGA-CGA-AGG-AGA-CGA) at the amino terminus of lacZ. RSW1276 contains six rare arginine codons 200 nt distal to the start site of translation. RSW1297 carries an insertion of λnutL immediately 5′ to the mutated ribosome binding site.
To confirm the hypothesis that NusG failed to attach to TEC in the absence of translation, we asked if a complex comprising Nus factors A, B, and E (Nus complex) assembled at a λ nutL site was able to recruit NusG so that it associates with TEC. Accordingly, we introduced the λ nutL site just upstream of the flawed lacZ SD sequence and measured lacA mRNA level (Figure 4B and Table 1). Indeed, Rho-dependent termination was highly efficient, indicating that NusG had been recruited to TEC. Thus, counterintuitively, the Nus complex, which normally suppresses transcription termination in ribosomal (rrn) operons (Dudenhoeffer et al., 2019; Huang et al., 2019; Squires et al., 1993) and, together with λN, on the phage λ chromosome, stimulates termination in this case.
We finally demonstrated that reduced termination efficiency in the mutant with the non-functional SD sequence was due to the failure of NusG recruitment to the TEC. In this assay we monitored Rho-dependent termination in a fusion construct that carries λ cro, the λ nutR site, the Rho-dependent λ tR1 terminator, and a lacZ reporter, with lacZ expression being heat-inducible (Figure 5). Termination at the λ tR1 site is poor when cro is translated, as seen with the cro ms27 fusion (Table 2 and Figure 5A-I. In the presence of an intact SD sequence we used cro ms27, where codon 27 carries a missense mutation so that the resulting protein is non-functional. The 3′ end of cro is adjacent to the λ tR1 terminator, limiting the amount of free RNA available for Rho attachment if cro mRNA is translated. When λ cro carried an SD mutation translation initiation was ablated, but nevertheless there was significant termination at λ tR1 (Table 2 and Figure 5A-II). Formation of the Nus complex at λ nutR allows NusG recruitment and efficient termination. In the absence of NusB, the complex does not assemble and there is extensive readthrough at λ tR1.
Figure 5.
NusG Can Be Recruited via a Nus Complex
Genetic constructs used to monitor NusG-mediated Rho-dependent termination are shown with the corresponding strains and their properties indicated on the right side. Transcription is started from the λpR promoter, followed by WT-λcro or λcro carrying a missense mutation at codon 27 (λcro27), a WT or mutant λnutR site, the Rho-dependent terminator λtR1, and a λcII::lacZ transcriptional fusion with a corresponding SD site. All strains encode a temperature-sensitive λcI construct (λcIts) to allow temperature-controlled induction of gene expression from the λpR promoter. (A) Nus complex formation compensates the lack of an SD sequence. (B) BoxA mutation impairs NusG recruitment (C) Uncoupling by NusG-NTD. λN+ strains listed in (B) further encode the λN protein; in (B-II) the non-functional λnutR sequence was generated by the boxA69 mutation; NusG-NTD for strains listed in (C) was supplied from plasmid pRM442. See also Tables 2, 3, and 4.
Table 2.
NusG Coupling at nutR Requires NusB
| Strain | cro | nusB | β-Galactosidase Activity (BCM−) in Miller Units | β-Galactosidase Activity (BCM+) in Miller Units | RT (%) |
|---|---|---|---|---|---|
| 9,743 | ms27 | + | 530 ± 3 | 680 ± 5 | 78 ± 0.7 |
| 12,580 | ms27 | Δ | 890 ± 11 | 1,150 ± 15 | 77 ± 1.4 |
| 9,739 | SD- | + | 141 ± 3 | 613 ± 25 | 23 ± 1.1 |
| 9,976 | SD- | Δ | 1,191 ± 17 | 1,290 ± 36 | 92 ± 3.0 |
Expression of lacZ was induced from a chromosomal cII::lacZ transcriptional fusion (λcIts-pR-cro-nutR-tR1-cII::lacZ) by incubating at 42°C for 30 min. N9743 and N12580 carry a missense mutation at cro codon 27; N9739 and 9,976 have a G to C mutation in the cro SD sequence (SD−); N12580 and N9976 are deleted for nusB. Where indicated, BCM was added to 100 μg/mL prior to induction of lacZ expression. Readthrough (RT) was calculated from the ratio of β-galactosidase activity (in Miller units) in the presence or absence of BCM (BCM+ and BCM−, respectively). Miller units from ≥3 independent experiments were averaged.
The boxA69 mutation also reduces Nus complex formation at λ nutR and, like the nusB− mutation, enhances readthrough of λ tR1 (Table 3 and Figure 5B). In this experiment, we suppressed termination at λ tR1 with λ N antitermination factor instead of BCM. Finally, we showed that expression of nusG-NTD, which competes with NusG for binding to RNAP, enhances readthrough (Table 4 and Figure 5C). Taken together, these results strongly support the idea that NusG can be supplied by the Nus complex assembled at λ nutR in the absence of translation, inducing Rho-dependent termination at λ tR1.
Table 3.
BoxA Mutations Block NusG Coupling at nutR
| Strains | boxA | nusB | β-Galactosidase Activity (λN−) in Miller Units | β-Galactosidase Activity (λN+) in Miller Units | RT (%) |
|---|---|---|---|---|---|
| 10,673; 10,672 | + | + | 125 ± 1 | 946 ± 23 | 13 ± 0.3 |
| 10,675; 10,674 | 69 | + | 1,212 ± 30 | 2,211 ± 87 | 55 ± 2.5 |
| 10,678; 10,677 | + | Δ | 2,874 ± 24 | 2,616 ± 103 | 100 ± 4.4 |
| 10,680; 10,679 | 69 | Δ | 1,896 ± 25 | 2,416 ± 80 | 78 ± 2.8 |
Expression of lacZ was induced from a chromosomal cII::lacZ transcriptional fusion (λcIts-pR-cro (SD-) -nutR-tR1-cII::lacZ) by incubation at 42°C for 30 min. Strains N10672, N10674, N10677, and N10679 express λn, which encodes the transcription termination inhibitor λΝ. boxA69 and ΔnusB strain numbers are indicated in Table 3. RT was calculated from the ratio of β-galactosidase activity in the presence or absence of λN (λN + and λN -, respectively). Miller units from ≥3 independent experiments were averaged.
Table 4.
NusG-NTD Uncouples Transcription and Translation
| Strain | nusG-NTD | nusB | β-Galactosidase Activity (BCM−) in Miller Units | β-Galactosidase Activity (BCM+) in Miller Units | RT (%) |
|---|---|---|---|---|---|
| RSW1396 | – | + | 247 ± 5 | 862 ± 2 | 29 ± 0.6 |
| RSW1397 | + | + | 944 ± 3 | 1,013 ± 7 | 93 ± 0.7 |
| RSW1398 | – | Δ | 2,013 ± 33 | 2,314 ± 55 | 87 ± 2.5 |
| RSW1399 | + | Δ | 2,360 ± 37 | 2,760 ± 150 | 86 ± 4.8 |
Expression of lacZ was induced from a chromosomal cII::lacZ transcriptional fusion (λcIts-pR-cro(SD−)-nutR-tR1-cII::lacZ) by incubating at 42°C for 30 min. nusG-NTD expression was induced from the plasmid pRM442 tac promoter with 1 mM IPTG for 10 min prior to induction of lacZ in strains RSW1397 and RSW 1399. Strains RSW1396 and RSW1398 carried an empty vector (ptrc99A) and were exposed to IPTG as above. Where indicated BCM was added to 100 μg/mL prior to induction of lacZ. RT was calculated from the ratio of β-galactosidase activity in the presence or absence of BCM (BCM+ and BCM−, respectively). Miller units from ≥3 independent experiments were averaged.
Discussion
Structural Basis of NusG-Mediated Transcription:Translation
We determined a cryo-EM structure of a NusG:70S complex showing binding of one molecule NusG per ribosome, consistent with previous results (Saxena et al., 2018). NusG binds to the S10 protein on the 30S subunit via its CTD as indicated by the study of isolated NusG-CTD and S10Δ (Burmann et al., 2010); density for NusG-NTD was not observable, suggesting that it remains flexible. We must attribute the low occupancy of the NusG-CTD on the 70S ribosome in the cryo-EM experiment to weak binding adversely affected by the conditions of sample preparation. Notably, although tmRNA contacts the ribosome at various sites, the binding of NusG-CTD and tmRNA on S10 seems to be mutually exclusive. This suggests a model in which uncoupling at rare codons, at which tmRNA releases ribosomes, is promoted by tmRNA-induced release of NusG (Roche and Sauer, 1999). The freed NusG:TEC complex exposes the NusG-CTD and is then subject to Rho-dependent transcription termination. This model, however, requires that the affinity of tmRNA for S10 is higher than for the NusG-CTD:S10 interaction. This could be the subject of further studies. Alternatively, tmRNA binding to S10 might only occur once NusG-mediated coupling has been disrupted owing to ribosome stalling, allowing transcription to continue while tmRNA rescues the ribosome.
Simultaneous binding of NusG to S10Δ and RNAP has been demonstrated by solution-state NMR studies, confirming the S10Δ binding site on NusG-CTD as identified in a binary NusG-CTD:S10Δ system (Figure 2) (Burmann et al., 2010). Moreover, we show that NusG can bind isolated RNAP and isolated 70S ribosome concurrently. Although this is not an actively transcribing and translating system, our data provide the first direct structural evidence consistent with NusG-mediated transcription:translation coupling. The flexibility of the linker between the NusG-NTD and the NusG-CTD permits these interactions.
The operon-specific E. coli NusG paralog, RfaH, likewise simultaneously binds S10Δ and RNAP in the context of a paused TEC (Burmann et al., 2012; Zuber et al., 2019). RfaH, which also comprises an NTD and a flexibly connected CTD (Belogurov et al., 2007; Burmann et al., 2012), uses the same binding sites as NusG to interact with RNAP and S10 (Burmann et al., 2010, 2012; Kang et al., 2018; Sevostyanova et al., 2011; Zuber et al., 2019). However, RfaH, unlike NusG, complexes with TEC early after transcription initiation, when TEC pauses at an operon polarity suppressor (ops) site, a representative of the E. coli consensus pause sequence (Larson et al., 2014; Vvedenskaya et al., 2014). Located in the untranslated leader region of RfaH-controlled operons, ops is responsible for sequence-specific recruitment of RfaH (Zuber et al., 2018). Importantly, RfaH-dependent operons lack a consensus SD sequence. To initiate translation, RfaH recruits a ribosome to these mRNAs, making coupling essential for translation activation and efficient gene expression (Burmann et al., 2012). The binding modes of RfaH and NusG to RNAP and S10 are very similar, indicating that coupling as observed for RfaH can also be mediated by NusG and vice versa. However, once recruited, RfaH excludes NusG (Kang et al., 2018), thus preventing intra-operon Rho-dependent transcription termination in RfaH-controlled operons (see Artsimovitch and Knauer, 2019).
Recruitment of NusG Requires Translation and Stimulates Rho-Dependent Termination
We have confirmed the results of Mooney et al. that NusG binds to TEC only after significant RNA synthesis (Mooney et al., 2009b). As postulated by these authors, binding depends on active translation of the mRNA. Thus, efficient Rho-dependent transcription termination, which requires the attachment of Rho to the NusG-CTD, does not occur at the end of an untranslated gene. We have shown that the failure of NusG to bind TEC is responsible for the absence of termination. Thus, placing a λ nut site at the start of the gene recruits NusG and restores termination. At present, it is not understood why NusG appears to be delivered to TEC by ribosomes in vivo, whereas it binds directly to RNAP in a purified system lacking ribosomes. A possible explanation would be that NusG attaches to RNAP discontinuously in an on-and-off mode in the untranslated leader region and that the NusG:RNAP interaction is only stabilized when the ribosome is coupled upon translation initiation. We should recall that NusG has two binding sites in the coupled system, which significantly increases its avidity.
Multiple Modes of Transcription:Translation Coupling
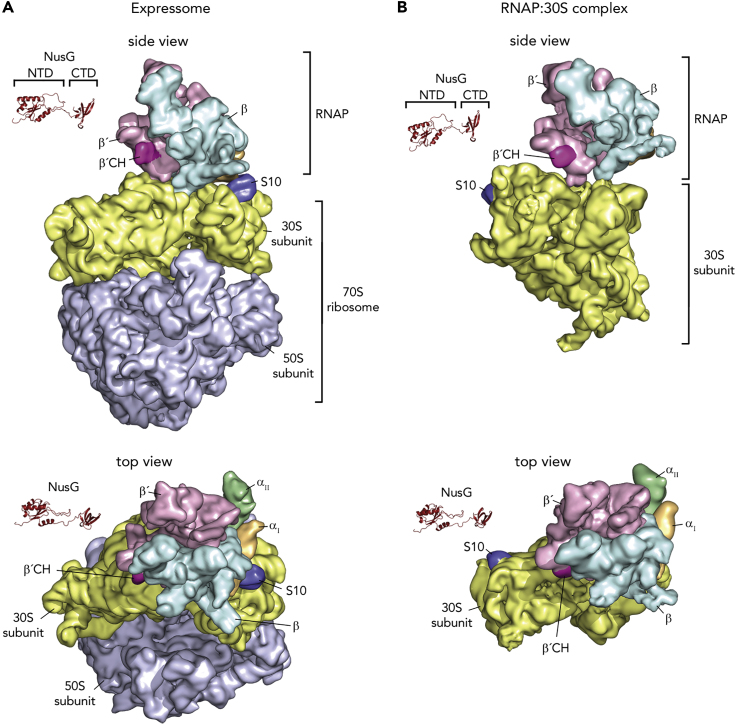
A direct connection between transcription and translation was first predicted in 1964 (Byrne et al., 1964), and later it was shown that transcription:translation coupling is necessary to coordinate gene expression as well as to maintain genome stability (McGary and Nudler, 2013). In 1970, Miller et al. performed electron microscopy analyses of lysed E. coli cells (Miller et al., 1970). They demonstrated that all mRNA molecules are connected to the E. coli genome and that the ribosome at the newly synthesized end of a polyribosome is almost always immediately adjacent to the putative RNAP molecule. Finally, they concluded that translation is completely coupled with transcription. Coupling could allow RNAP to monitor the translation rate while providing newly synthesized mRNA to the ribosome. The structural basis of this coupling is, however, still only poorly understood. Our results strongly suggest that NusG may mediate coupling (“indirect coupling”). Since NusG attaches to the TEC downstream to the translation initiation site, the coupled transcription:translation complex must initially consist of a ribosome bound directly to TEC. This “direct coupling” mode is in agreement with both structural and biochemical data (Demo et al., 2017; Fan et al., 2017; Kohler et al., 2017). A cryo-EM structure of a directly coupled complex has been published where a translating ribosome collided into a stalled RNAP, forming a so-called expressome (Figure 6A [Kohler et al., 2017]). In this complex, RNAP directly binds to the 30S subunit with the RNA exit region of RNAP docking onto the ribosome near the mRNA tunnel entry between ribosomal proteins S3, S4, and S5, allowing the mRNA exiting from RNAP to enter directly into the ribosome. Another cryo-EM structure showed an RNAP:30S complex generated by mixing 30S subunit with a 3-fold excess of RNAP (Figure 6B [Demo et al., 2017]). In this structure RNAP is bound to the 30S subunit near the mRNA binding site between the head and the platform domains, contacting ribosomal proteins S1, S2, S18, S21, and hairpin loop 40 of 16S rRNA, in agreement with cross-linking data (Fan et al., 2017). Strikingly, this position is located more than 80 Å from the binding site observed in the expressome structure, i.e., on the opposite side of the 30S head. Importantly, it ensures that RNAP interacts with the cytosolic side of the 30S ribosomal subunit so that the nascent RNA exiting from RNAP is directly guided to the entry site on the ribosome. Assuming that both the RNAP:30S complex and the expressome correspond to active coupling complexes, the structures indicate that multiple coupling modes exist, which involve massive relocalization of RNAP relative to ribosome.
Figure 6.
Structures of Coupled Complexes
Structures of the expressome (A) and an RNAP:30S complex (B) determined by cryo-EM. RNAP and ribosomal subunits are in surface representation, NusG is shown as ribbon. αI, orange; αII green; β, cyan; β′, light violet; 30S, yellow; 50S, light blue; β′CH, pink; S10, dark blue. PDB IDs: expressome, 5MY1 and 6O9J; RNAP:30S, 6AWD; NusG-NTD: 2K06; NusG-CTD: 2JVV.
Interestingly, neither the RNAP:30S nor the expressome structures allow NusG- (or RfaH-) mediated coupling: the linker of NusG/RfaH is too short (Figure 6). However, as the cryo-EM structures suggest that the position of RNAP on the 30S subunit might be flexible, these structures could be snapshots of distinct situations during translation. Thus, we suggest that, at some distance downstream of the translation initiation site, NusG recognizes and enters the coupled complex, rearranging its structure.
While our manuscript was under review, two preprints have been published reporting several structures of coupled complexes from E. coli (Wang et al., 2020; Webster et al., 2020). Overall, the structures indicate that there are indeed various types of transcription:translation coupling modes, both direct and indirect coupling. The coupling mode depends on the length of the mRNA separating the RNAP active site and the ribosomal P-site and is determined by the position of RNAP relative to the ribosome. For example, the structure of the collided complex (see above [Kohler et al., 2017]) was confirmed, but this coupling mode may be relevant only under certain conditions or when the RNA spacer is very short. In NusG-coupled complexes (Wang et al., 2020; Webster et al., 2020) NusG bridges RNAP and ribosome with NusG-NTD contacting the RNAP at the expected binding site (Kang et al., 2018) and NusG-CTD interacting with S10. This is similar to what was found for the binary NusG-CTD:S10 system (Burmann et al., 2012) and is in agreement with our data. As compared with the collided complex, RNAP is significantly rotated relative to the ribosome, but, interestingly, no stable contacts between RNAP and ribosome were observable (Webster et al., 2020). However, more coupling modes are possible, emphasizing the complexity of the interplay between transcription and translation (Wang et al., 2020; Webster et al., 2020).
Conclusion
In summary, multiple transcription:translation coupling modes exist. Based on our results, we hypothesize that direct coupling between the ribosome and TEC occurs during translation initiation and early elongation, whereas NusG-mediated coupling is established later in translation in E. coli. Notably, the transition between the different coupling modes requires significant rearrangement of the relative position of RNAP and ribosome, with NusG serving as additional anchor point to restrict the freedom of RNAP movement relative to the ribosome.
Coupling may synchronize transcription and translation; in particular, the leading ribosome may “push” RNAP to overcome transcriptional pauses while RNAP could “pull” the ribosome to prevent/escape translational pausing (Wang et al., 2020). In NusG-coupled complexes the small transcription factor may serve as cushion, conferring the system the flexibility necessary to keep transcription and translation synchronized, even if these processes are regulated differently or occur at different rates.
Limitations of the Study
-
(1)
RNA extraction efficiencies are subject to some variability.
-
(2)
Cryo-EM modeling: (a) The accuracy of the atomic model is limited by the cryo-EM resolution and the low occupancy of NusG on the ribosome. With the current resolution of the map, it is impossible to identify any rearrangement of side chains involved in interactions that stabilize the formation of the NusG:70S complex. The limited resolution also has the consequence that we had to rely on the accuracy of published structures used in the fitting and docking. (b) Furthermore, we modeled the 30S subunit structure only, based on a published atomic structure, even though the 70S-NusG complex was visualized. However, there are only minor differences between the structures of the isolated and 50S-bound 30S subunit, and the geometry of NusG binding determined by the modeling should not be affected.
-
(3)
NMR: (a) We used isolated RNAP and isolated ribosome in our experiments, and, consequently, not an actively transcribing/translating system. Thus, we provide only structural evidence that NusG can link RNAP and ribosome in the absence of nucleic acids. The situation might, theoretically, be different in vivo (although this is not very probable). (Already indicated in the section “Structural basis of NusG-mediated transcription:translation.”) (b) Although we find that NusG can serve as linker between RNAP and ribosome we cannot distinguish between two scenarios: (i) NusG links RNAP and ribosome without direct interactions between RNAP and ribosome and (ii) NusG links RNAP and ribosome, but RNAP and ribosome make direct contacts in this coupled complex (already indicated at the end of section “Simultaneous binding of NusG to S10 and RNAP”).
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Joachim Frank (jf2192@cumc.columbia.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
For the 70S:NusG complex visualized by cryo-EM, only the region of 30S:NusG-CTD was modeled for simplicity. Electron densities and the final atomic model for the 70S:NusG complex have been deposited in the Electron Microscopy Data Bank (EMDB) and the Protein Data Bank (PDB) with accession codes EMD-22143 and 6XE0.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We gratefully acknowledge the help of D. Shapoval, M. Bubunenko, N. Costantino, and D. Court, and we are deeply thankful for numerous useful discussions with P. Rösch. Supported by HHMI and NIH R01 GM29169 (to J.F.), NIH R01 GM037219 (to M.E.G.), and the German Research Foundation grant Ro617/21-1 (to P. Rösch). S.H. was supported by an Amgen Fellowship.
Author Contributions
B.S., M.S., P.K.Z., R.S.W., S.H., and Y.H. performed the experiments and processed the data. J.F., M.E.G., M.S., P.K.Z., R.S.W., S.H.K., and Y.H. designed the experiments and interpreted the results. F.J.A.R., M.S., and W.L. modeled the atomic structure of 30S:NusG-CTD. J.F., M.E.G., M.S., P.K.Z., and S.H.K. wrote the paper. This publication was funded by the German Research Foundation and the University of Bayreuth in the funding program “Open Access Publishing”.
Declaration of Interests
The authors declare no competing financial interests.
Published: August 21, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101352.
Contributor Information
Max E. Gottesman, Email: meg8@cumc.columbia.edu.
Stefan H. Knauer, Email: stefan.knauer@uni-bayreuth.de.
Joachim Frank, Email: jf2192@cumc.columbia.edu.
Supplemental Information
References
- Adhya S., Gottesman M. Control of transcription termination. Annu. Rev. Biochem. 1978;47:967–996. doi: 10.1146/annurev.bi.47.070178.004535. [DOI] [PubMed] [Google Scholar]
- Artsimovitch I., Knauer S.H. Ancient transcription factors in the news. MBio. 2019;10 doi: 10.1128/mBio.01547-18. e01547–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belogurov G.A., Vassylyeva M.N., Svetlov V., Klyuyev S., Grishin N.V., Vassylyev D.G., Artsimovitch I. Structural basis for converting a general transcription factor into an operon-specific virulence regulator. Mol. Cell. 2007;26:117–129. doi: 10.1016/j.molcel.2007.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmann B.M., Schweimer K., Luo X., Wahl M.C., Stitt B.L., Gottesman M.E., Rösch P. A NusE:NusG complex links transcription and translation. Science. 2010;328:501–504. doi: 10.1126/science.1184953. [DOI] [PubMed] [Google Scholar]
- Burmann B.M., Scheckenhofer U., Schweimer K., Rösch P. Domain interactions of the transcription-translation coupling factor Escherichia coli NusG are intermolecular and transient. Biochem. J. 2011;435:783–789. doi: 10.1042/BJ20101679. [DOI] [PubMed] [Google Scholar]
- Burmann B.M., Knauer S.H., Sevostyanova A., Schweimer K., Mooney R.A., Landick R., Artsimovitch I., Rösch P. An α helix to β barrel domain switch transforms the transcription factor RfaH into a translation factor. Cell. 2012;150:291–303. doi: 10.1016/j.cell.2012.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns C.M., Richardson J.P. NusG is required to overcome a kinetic limitation to Rho function at an intragenic terminator. Proc. Natl. Acad. Sci. U S A. 1995;92:4738–4742. doi: 10.1073/pnas.92.11.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burova E., Hung S.C., Sagitov V., Stitt B.L., Gottesman M.E. Escherichia coli NusG protein stimulates transcription elongation rates in vivo and in vitro. J. Bacteriol. 1995;177:1388–1392. doi: 10.1128/jb.177.5.1388-1392.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne R., Levin J.G., Bladen H.A., Nirenberg M.W. The in vitro formation of a DNA-ribosome complex. Proc. Natl. Acad. Sci. U S A. 1964;52:140–148. doi: 10.1073/pnas.52.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale C.J., Washburn R.S., Tadigotla V.R., Brown L.M., Gottesman M.E., Nudler E. Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science. 2008;320:935–938. doi: 10.1126/science.1152763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn A.B., Diggs S., Tam T.K., Blaha G.M. Two old dogs, one new trick: a review of RNA polymerase and ribosome interactions during transcription-translation coupling. Int. J. Mol. Sci. 2019;20:2595. doi: 10.3390/ijms20102595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das H.K., Goldstein A., Lowney L.I. Attachment of ribosomes to nascent messenger RNA in Escherichia coli. J. Mol. Biol. 1967;24:231–245. doi: 10.1016/0022-2836(67)90329-4. [DOI] [PubMed] [Google Scholar]
- Demo G., Rasouly A., Vasilyev N., Svetlov V., Loveland A.B., Diaz-Avalos R., Grigorieff N., Nudler E., Korostelev A.A. Structure of RNA polymerase bound to ribosomal 30S subunit. Elife. 2017;6:e28560. doi: 10.7554/eLife.28560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drögemüller J., Strauß M., Schweimer K., Jurk M., Rösch P., Knauer S.H. Determination of RNA polymerase binding surfaces of transcription factors by NMR spectroscopy. Sci. Rep. 2015;5:16428. doi: 10.1038/srep16428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudenhoeffer B.R., Schneider H., Schweimer K., Knauer S.H. SuhB is an integral part of the ribosomal antitermination complex and interacts with NusA. Nucleic Acids Res. 2019;47:6504–6518. doi: 10.1093/nar/gkz442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D., Shatalin K., Epshtein V., Gottesman M.E., Nudler E. Linking RNA polymerase backtracking to genome instability in E. coli. Cell. 2011;146:533–543. doi: 10.1016/j.cell.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H., Conn A.B., Williams P.B., Diggs S., Hahm J., Gamper H.B., Hou Y.-M., O’Leary S.E., Wang Y., Blaha G.M. Transcription-translation coupling: direct interactions of RNA polymerase with ribosomes and ribosomal subunits. Nucleic Acids Res. 2017;45:11043–11055. doi: 10.1093/nar/gkx719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Hashem Y., Wower I., Lei J., Liao H.Y., Zwieb C., Wower J., Frank J. Visualizing the transfer-messenger RNA as the ribosome resumes translation. EMBO J. 2010;29:3819–3825. doi: 10.1038/emboj.2010.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.-H., Said N., Loll B., Wahl M.C. Structural basis for the function of SuhB as a transcription factor in ribosomal RNA synthesis. Nucleic Acids Res. 2019;47:6488–6503. doi: 10.1093/nar/gkz290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J.Y., Mooney R.A., Nedialkov Y., Saba J., Mishanina T.V., Artsimovitch I., Landick R., Darst S.A. Structural basis for transcript elongation control by NusG family universal regulators. Cell. 2018;173:1650–1662.e14. doi: 10.1016/j.cell.2018.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowlton J.R., Bubunenko M., Andrykovitch M., Guo W., Routzahn K.M., Waugh D.S., Court D.L., Ji X. A spring-loaded state of NusG in its functional cycle is suggested by X-ray crystallography and supported by site-directed mutants. Biochemistry. 2003;42:2275–2281. doi: 10.1021/bi0272508. [DOI] [PubMed] [Google Scholar]
- Kohler R., Mooney R.A., Mills D.J., Landick R., Cramer P. Architecture of a transcribing-translating expressome. Science. 2017;356:194–197. doi: 10.1126/science.aal3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupp F., Said N., Huang Y.-H., Loll B., Bürger J., Mielke T., Spahn C.M.T., Wahl M.C. Structural basis for the action of an all-purpose transcription Anti-termination factor. Mol. Cell. 2019;74:143–157.e5. doi: 10.1016/j.molcel.2019.01.016. [DOI] [PubMed] [Google Scholar]
- Kyrpides N.C., Woese C.R., Ouzounis C.A. KOW: a novel motif linking a bacterial transcription factor with ribosomal proteins. Trends Biochem. Sci. 1996;21:425–426. doi: 10.1016/s0968-0004(96)30036-4. [DOI] [PubMed] [Google Scholar]
- Larson M.H., Mooney R.A., Peters J.M., Windgassen T., Nayak D., Gross C.A., Block S.M., Greenleaf W.J., Landick R., Weissman J.S. A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science. 2014;344:1042–1047. doi: 10.1126/science.1251871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson M.R., Ma W., Bellecourt M.J., Artsimovitch I., Martin A., Landick R., Schulten K., Berger J.M. Mechanism for the regulated control of transcription by a universal adapter protein. Mol. Cell. 2018;71:1–12. doi: 10.1016/j.molcel.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luo X., Hsiao H.-H., Bubunenko M., Weber G., Court D.L., Gottesman M.E., Urlaub H., Wahl M.C. Structural and functional analysis of the E. coli NusB-S10 transcription antitermination complex. Mol. Cell. 2008;32:791–802. doi: 10.1016/j.molcel.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGary K., Nudler E. RNA polymerase and the ribosome: the close relationship. Curr. Opin. Microbiol. 2013;16:112–117. doi: 10.1016/j.mib.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehdi Q., Yudkin M.D. Coupling of transcription to translation in the induced synthesis of beta-galactosidase. Biochim. Biophys. Acta. 1967;149:288–290. doi: 10.1016/0005-2787(67)90710-1. [DOI] [PubMed] [Google Scholar]
- Miller O.L., Hamkalo B.A., Thomas C.A. Visualization of bacterial genes in action. Science. 1970;169:392–395. doi: 10.1126/science.169.3943.392. [DOI] [PubMed] [Google Scholar]
- Mitra P., Ghosh G., Hafeezunnisa M., Sen R. Rho protein: roles and mechanisms. Annu. Rev. Microbiol. 2017;71:687–709. doi: 10.1146/annurev-micro-030117-020432. [DOI] [PubMed] [Google Scholar]
- Mooney R.A., Schweimer K., Rösch P., Gottesman M., Landick R. Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. J. Mol. Biol. 2009;391:341–358. doi: 10.1016/j.jmb.2009.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney R.A., Davis S.E., Peters J.M., Rowland J.L., Ansari A.Z., Landick R. Regulator trafficking on bacterial transcription units in vivo. Mol. Cell. 2009;33:97–108. doi: 10.1016/j.molcel.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton W.A., Beckwith J.R., Zipser D., Brenner S. Nonsense mutants and polarity in the lac operon of Escherichia coli. J. Mol. Biol. 1965;14:290–296. doi: 10.1016/s0022-2836(65)80250-9. [DOI] [PubMed] [Google Scholar]
- Pani B., Nudler E. Mechanistic insights into transcription coupled DNA repair. DNA Repair (Amst.) 2017;56:42–50. doi: 10.1016/j.dnarep.2017.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proshkin S., Rahmouni A.R., Mironov A., Nudler E. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science. 2010;328:504–508. doi: 10.1126/science.1184939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae C.D., Gordiyenko Y., Ramakrishnan V. How a circularized tmRNA moves through the ribosome. Science. 2019;363:740–744. doi: 10.1126/science.aav9370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche E.D., Sauer R.T. SsrA-mediated peptide tagging caused by rare codons and tRNA scarcity. EMBO J. 1999;18:4579–4589. doi: 10.1093/emboj/18.16.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Said N., Krupp F., Anedchenko E., Santos K.F., Dybkov O., Huang Y.-H., Lee C.-T., Loll B., Behrmann E., Bürger J. Structural basis for λN-dependent processive transcription antitermination. Nat. Microbiol. 2017;2:17062. doi: 10.1038/nmicrobiol.2017.62. [DOI] [PubMed] [Google Scholar]
- Saxena S., Myka K.K., Washburn R., Costantino N., Court D.L., Gottesman M.E. Escherichia coli transcription factor NusG binds to 70S ribosomes. Mol. Microbiol. 2018;108:495–504. doi: 10.1111/mmi.13953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevostyanova A., Belogurov G.A., Mooney R.A., Landick R., Artsimovitch I. The β subunit gate loop is required for RNA polymerase modification by RfaH and NusG. Mol. Cell. 2011;43:253–262. doi: 10.1016/j.molcel.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires C.L., Greenblatt J., Li J., Condon C., Squires C.L. Ribosomal RNA antitermination in vitro: requirement for Nus factors and one or more unidentified cellular components. Proc. Natl. Acad. Sci. U S A. 1993;90:970–974. doi: 10.1073/pnas.90.3.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thommen M., Holtkamp W., Rodnina M.V. Co-translational protein folding: progress and methods. Curr. Opin. Struct. Biol. 2017;42:83–89. doi: 10.1016/j.sbi.2016.11.020. [DOI] [PubMed] [Google Scholar]
- Vogel U., Jensen K.F. Effects of guanosine 3’,5’-bisdiphosphate (ppGpp) on rate of transcription elongation in isoleucine-starved Escherichia coli. J. Biol. Chem. 1994;269:16236–16241. [PubMed] [Google Scholar]
- Vogel U., Jensen K.F. Effects of the antiterminator BoxA on transcription elongation kinetics and ppGpp inhibition of transcription elongation in Escherichia coli. J. Biol. Chem. 1995;270:18335–18340. doi: 10.1074/jbc.270.31.18335. [DOI] [PubMed] [Google Scholar]
- Vvedenskaya I.O., Vahedian-Movahed H., Bird J.G., Knoblauch J.G., Goldman S.R., Zhang Y., Ebright R.H., Nickels B.E. Interactions between RNA polymerase and the “core recognition element” counteract pausing. Science. 2014;344:1285–1289. doi: 10.1126/science.1253458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Molodtsov V., Firlar E., Kaelber J., Blaha G., Su M., Ebright R.H. Structural basis of transcription-translation coupling. bioRxiv. 2020 doi: 10.1101/2020.03.01.972380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn R.S., Gottesman M.E. Transcription termination maintains chromosome integrity. Proc. Natl. Acad. Sci. U S A. 2011;108:792–797. doi: 10.1073/pnas.1009564108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster M.W., Takacs M., Zhu C., Vidmar V., Eduljee A., Abdelkareem M., Weixlbaumer A. Structural basis of transcription-translation coupling and collision in bacteria. bioRxiv. 2020 doi: 10.1101/2020.03.01.971028. [DOI] [PubMed] [Google Scholar]
- Weis F., Bron P., Giudice E., Rolland J.-P., Thomas D., Felden B., Gillet R. tmRNA-SmpB: a journey to the centre of the bacterial ribosome. EMBO J. 2010;29:3810–3818. doi: 10.1038/emboj.2010.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner F. A nexus for gene expression-molecular mechanisms of Spt5 and NusG in the three domains of life. J. Mol. Biol. 2012;417:13–27. doi: 10.1016/j.jmb.2012.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber P.K., Artsimovitch I., NandyMazumdar M., Liu Z., Nedialkov Y., Schweimer K., Rösch P., Knauer S.H. The universally-conserved transcription factor RfaH is recruited to a hairpin structure of the non-template DNA strand. Elife. 2018;7:e36349. doi: 10.7554/eLife.36349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber P.K., Schweimer K., Rösch P., Artsimovitch I., Knauer S.H. Reversible fold-switching controls the functional cycle of the antitermination factor RfaH. Nat. Commun. 2019;10:702. doi: 10.1038/s41467-019-08567-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
For the 70S:NusG complex visualized by cryo-EM, only the region of 30S:NusG-CTD was modeled for simplicity. Electron densities and the final atomic model for the 70S:NusG complex have been deposited in the Electron Microscopy Data Bank (EMDB) and the Protein Data Bank (PDB) with accession codes EMD-22143 and 6XE0.