Abstract
Mycoplasma hominis is an opportunistic bacterium that can cause acute and chronic infections of the urogenital tract. This bacterium, like all other Mycoplasma species, is characterized by the reduced genome size, and, consequently, reduction of the main metabolic pathways. M. hominis cells cannot effectively use glucose as a carbon and energy source. Therefore, the main pathway of energy metabolism is the arginine dihydrolase pathway. However, several bacteria can use nucleosides as the sole energy source. Biochemical studies using Salmonella typhimurium have shown that three enzymes (thymidine phosphorylase, phosphopentose mutase and deoxyribose-phosphate aldolase) are involved in the thymidine catabolic pathway. All these enzymes are present in M. hominis. For understanding changes in the energy metabolism of M. hominis we performed shotgun proteome analysis of M. hominis cells in liquid medium with arginine or thymidine as a carbon source. LC-MS analysis was performed with an Ultimate 3000 Nano LC System (Thermo Fisher Scientific) coupled to a Q Exactive HF benchtop Orbitrap mass spectrometer (Thermo Fisher Scientific) via a nanoelectrospray source (Thermo Fisher Scientific). Data are available via ProteomeXchange with identifier PXD018714 (https://www.ebi.ac.uk/pride/archive/projects/PXD018714).
Keywords: Mycoplasma hominis, Proteome, Cultivation conditions, Carbon source, Arginine, Thymidine
Specifications table
| Subject | Biology |
| Specific subject area | Proteomics |
| Type of data | LC-MS/MS data and identification data |
| How data were acquired | LC-MS analysis is performed with an Ultimate 3000 Nano LC System (Thermo Fisher Scientific) coupled to a Q Exactive HF benchtop Orbitrap mass spectrometer (Thermo Fisher Scientific) via a nanoelectrospray source (Thermo Fisher Scientific). |
| Data format | Raw and analyzed data |
| Parameters for data collection | Shotgun proteomes for M. hominis cells growing in two conditions of culturing. |
| Description of data collection | M. hominis cells growing in culture with arginine or thymidine as carbon source at log phase were collected, and their total proteomes were analyzed by shotgun proteomics in three biological replicates. |
| Data source location | Research and Clinical Center of Physical-Chemical Medicine, Moscow, Russian Federation |
| Data accessibility | Data were deposited to the PRIDE repository: Project accession: PXD018714 Project https://www.ebi.ac.uk/pride/archive/projects/PXD018714 |
Value of the data
-
•
This dataset provides proteome data for M. hominis cells growing in culture with arginine or thymidine as carbon source.
-
•
These data can be interesting for the investigation of interaction with the environment of opportunistic bacteria M. hominis.
-
•
These data can be interesting for the investigation of metabolism of M. hominis and another mycoplasma species that can be a model of a minimal cell.
1. Data description
Mycoplasma hominis is a human opportunistic bacterium that can cause acute and chronic infections of the urogenital tract [1]. Like all other Mycoplasma species, it is characterized by the reduced genome size (about 550 ORFs), and, consequently, reduction of the main metabolic pathways. M. hominis cells cannot effectively use glucose as a carbon and energy source. Therefore, the main pathway of energy metabolism is the arginine dihydrolase pathway, which includes arginine deiminase, ornithine carbamoyltransferase and carbamate kinase [2]. However, M. hominis cells can utilize nucleosides. In thymidine catabolic pathway the thymidine phosphorylase, phosphopentose mutase and deoxyribose-phosphate aldolase are involved [3].
We performed shotgun proteome analysis of M. hominis cells in liquid medium with arginine or thymidine as a carbon and energy source. LC-MS analysis was performed with an Ultimate 3000 Nano LC System (Thermo Fisher Scientific) coupled to a Q Exactive HF benchtop Orbitrap mass spectrometer (Thermo Fisher Scientific) via a nanoelectrospray source (Thermo Fisher Scientific). Protein identification and label-free quantification were made by PEAKS software. The data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the identifier PXD018714.
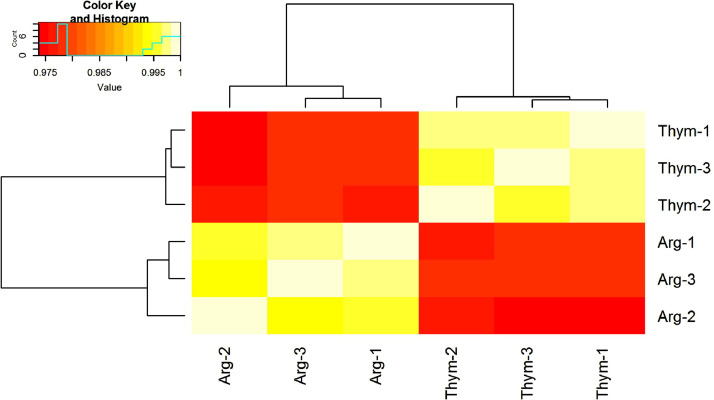
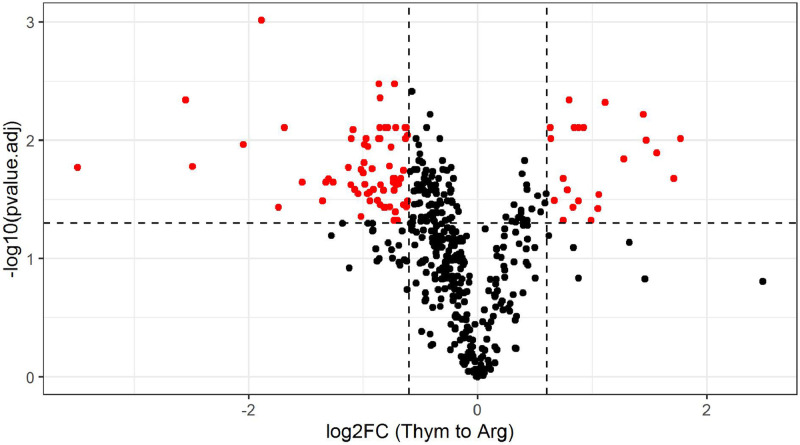
Totally, 466 proteins were identified in both cases of M. hominis culturing with arginine or thymidine as carbon source (table S1). Obtained datasets show good reproducibility between biological replicas as demonstrated by the heatmap (Fig. 1). Range of differences in the protein changes is shown on volcano plot (Fig. 2). Proteins, significantly changed between aforementioned conditions (fold change > 1.5, t-test with Benjamini–Hochberg correction, p < 0.05), are presented in Table 1. All the above-mentioned enzymes from the metabolic pathways of arginine and thymidine utilization have been identified. When growing on thymidine, the only thymidine phosphorylase abundance was increased by 1.8 times, the abundance of other enzymes did not significantly change.
Fig. 1.
Similarity of proteomic data between samples.
Fig. 2.
Different protein abundance for M. hominis growing with thymidine to growing with arginine.
Table 1.
Significantly changed proteins for M. hominis growing in different growth conditions. Log2FC – logarithm of fold change ratio for growth with thymidine to growth with arginine.
| Accession | Log2FC | P value adj. | Description |
|---|---|---|---|
| WP_012855601.1 | 1.77 | 0.010 | hypothetical protein |
| WP_012855692.1 | 1.71 | 0.021 | pyruvate kinase |
| WP_012855537.1 | 1.56 | 0.013 | hypothetical protein |
| WP_012855507.1 | 1.47 | 0.010 | transcriptional regulator MraZ |
| WP_012855710.1 | 1.44 | 0.006 | ABC transporter ATP-binding protein |
| WP_012855796.1 | 1.27 | 0.014 | 30S ribosome-binding factor RbfA |
| WP_012855529.1 | 1.11 | 0.005 | N(G) N(G)-dimethylarginine dimethylaminohydrolase |
| WP_012855730.1 | 1.06 | 0.029 | tRNA uridine-5-carboxymethylaminomethyl(34) synthesis GTPase MnmE |
| WP_012855297.1 | 1.05 | 0.038 | chaperone protein ClpB |
| WP_012855435.1 | 0.99 | 0.048 | hypothetical protein |
| WP_012855457.1 | 0.92 | 0.008 | NAD(+) synthase |
| WP_012855371.1 | 0.88 | 0.033 | hypothetical protein |
| WP_012855709.1 | 0.88 | 0.008 | hypothetical protein |
| WP_012855608.1 | 0.84 | 0.008 | thymidine phosphorylase |
| WP_012855370.1 | 0.83 | 0.037 | NUDIX domain-containing protein |
| WP_012855640.1 | 0.80 | 0.005 | DUF885 domain-containing protein |
| WP_012855422.1 | 0.78 | 0.026 | 30S ribosomal protein S12 |
| WP_012855390.1 | 0.74 | 0.048 | single-stranded DNA-binding protein |
| WP_012855783.1 | 0.74 | 0.021 | type I glyceraldehyde-3-phosphate dehydrogenase |
| WP_012855759.1 | 0.67 | 0.032 | serine/threonine-protein phosphatase |
| WP_012855649.1 | 0.64 | 0.010 | protein LemA |
| WP_012855632.1 | 0.64 | 0.008 | lactate dehydrogenase |
| WP_012855504.1 | 0.63 | 0.008 | cell division protein FtsZ |
| WP_012855582.1 | −0.61 | 0.033 | leucine–tRNA ligase |
| WP_012855330.1 | −0.61 | 0.009 | Lmp1 protein |
| WP_012855717.1 | −0.62 | 0.036 | GTPase Era |
| WP_012855408.1 | −0.62 | 0.008 | alanine–tRNA ligase |
| WP_012855532.1 | −0.63 | 0.037 | XRE family transcriptional regulator |
| WP_012855641.1 | −0.63 | 0.010 | oligoendopeptidase F |
| WP_012855592.1 | −0.64 | 0.008 | hypothetical protein |
| WP_012855418.1 | −0.65 | 0.018 | RluA family pseudouridine synthase |
| WP_012855433.1 | −0.65 | 0.035 | rRNA pseudouridine synthase |
| WP_012855356.1 | −0.67 | 0.021 | hypothetical protein |
| WP_012855477.1 | −0.69 | 0.023 | NAD-dependent DNA ligase LigA |
| WP_012855316.1 | −0.70 | 0.022 | 1-acyl-sn-glycerol-3-phosphate acyltransferase |
| WP_012855626.1 | −0.70 | 0.022 | ABC transporter ATP-binding protein |
| WP_012855757.1 | −0.70 | 0.048 | 16S rRNA (guanine(966)-N(2))-methyltransferase RsmD |
| WP_012855399.1 | −0.71 | 0.024 | TlyA family rRNA (cytidine-2′-O)-methyltransferase |
| WP_012855628.1 | −0.72 | 0.008 | 16S rRNA (guanine(527)-N(7))-methyltransferase RsmG |
| WP_012855539.1 | −0.72 | 0.040 | DegV family EDD domain-containing protein |
| WP_012855668.1 | −0.73 | 0.021 | YihA family ribosome biogenesis GTP-binding protein |
| WP_012855402.1 | −0.73 | 0.003 | DNA polymerase IV |
| WP_012855542.1 | −0.73 | 0.048 | DNA-directed RNA polymerase subunit beta' |
| WP_012855470.1 | −0.73 | 0.026 | deoxyguanosine kinase |
| WP_012855471.1 | −0.73 | 0.021 | hypoxanthine phosphoribosyltransferase |
| WP_020002555.1 | −0.74 | 0.021 | DUF402 domain-containing protein |
| WP_012855437.1 | −0.74 | 0.023 | peptide chain release factor 1 |
| WP_041359585.1 | −0.76 | 0.011 | hypothetical protein |
| WP_012855413.1 | −0.77 | 0.037 | tRNA1(Val) (adenine(37)-N6)-methyltransferase |
| WP_012855453.1 | −0.77 | 0.016 | alcohol dehydrogenase |
| WP_012855553.1 | −0.78 | 0.008 | type I methionyl aminopeptidase |
| WP_012855662.1 | −0.81 | 0.008 | RpiB/LacA/LacB family sugar-phosphate isomerase |
| WP_012855625.1 | −0.81 | 0.037 | hypothetical protein |
| WP_012855642.1 | −0.82 | 0.037 | adenine phosphoribosyltransferase |
| WP_080569061.1 | −0.82 | 0.026 | hypothetical protein |
| WP_012855743.1 | −0.85 | 0.024 | ribonuclease III |
| WP_012855497.1 | −0.85 | 0.004 | ribosome biogenesis GTPase YlqF |
| WP_012855518.1 | −0.85 | 0.035 | hypothetical protein |
| WP_012855403.1 | −0.85 | 0.010 | nicotinate (nicotinamide) nucleotide adenylyltransferase |
| WP_012855376.1 | −0.86 | 0.008 | DNA polymerase III subunit |
| WP_012855694.1 | −0.86 | 0.003 | exodeoxyribonuclease V subunit alpha |
| WP_012855699.1 | −0.87 | 0.010 | tRNA (guanosine(46)-N7)-methyltransferase TrmB |
| WP_012855514.1 | −0.88 | 0.032 | RNA methyltransferase |
| WP_012855721.1 | −0.91 | 0.026 | spermidine/putrescine ABC transporter permease |
| WP_012855655.1 | −0.92 | 0.017 | 2 3-bisphosphoglycerate-independent phosphoglycerate mutase |
| WP_012855740.1 | −0.94 | 0.033 | hypothetical protein |
| WP_012855290.1 | −0.94 | 0.028 | TatD family deoxyribonuclease |
| WP_012855419.1 | −0.96 | 0.011 | hypothetical protein |
| WP_012855591.1 | −0.96 | 0.028 | hypothetical protein |
| WP_012855323.1 | −0.98 | 0.010 | hypothetical protein |
| WP_012855798.1 | −0.99 | 0.023 | 23S rRNA (pseudouridine(1915)-N(3))-methyltransferase RlmH |
| WP_012855725.1 | −0.99 | 0.011 | RDD family protein |
| WP_080569060.1 | −0.99 | 0.015 | hypothetical protein |
| WP_012855552.1 | −1.00 | 0.019 | translation initiation factor IF-1 |
| WP_012855289.1 | −1.02 | 0.018 | ribosomal RNA small subunit methyltransferase A |
| WP_012855634.1 | −1.02 | 0.044 | hypothetical protein |
| WP_012855765.1 | −1.05 | 0.028 | hypothetical protein |
| WP_012855334.1 | −1.08 | 0.026 | hypothetical protein |
| WP_012855502.1 | −1.09 | 0.008 | 16S rRNA (uracil(1498)-N(3))-methyltransferase |
| WP_012855595.1 | −1.11 | 0.010 | putative immunoglobulin-blocking virulence protein |
| WP_041359577.1 | −1.11 | 0.024 | hypothetical protein |
| WP_012855512.1 | −1.13 | 0.017 | tRNA pseudouridine(55) synthase TruB |
| WP_012855378.1 | −1.26 | 0.023 | 30S ribosomal protein S20 |
| WP_012855580.1 | −1.30 | 0.021 | signal peptidase II |
| WP_012855576.1 | −1.33 | 0.023 | hypothetical protein |
| WP_012855409.1 | −1.36 | 0.033 | Holliday junction resolvase RuvX |
| WP_012855438.1 | −1.53 | 0.023 | peptide chain release factor N(5)-glutamine methyltransferase |
| WP_012855523.1 | −1.69 | 0.008 | hypothetical protein |
| WP_012855355.1 | −1.74 | 0.037 | hypothetical protein |
| WP_012855495.1 | −1.89 | 0.001 | signal recognition particle protein |
| WP_012855794.1 | −2.05 | 0.011 | DUF448 domain-containing protein |
| WP_012855696.1 | −2.49 | 0.017 | tRNA lysidine(34) synthetase TilS |
| WP_012855645.1 | −2.55 | 0.005 | hypothetical protein |
| WP_012855350.1 | −3.49 | 0.017 | hypothetical protein |
2. Experimental design, materials, and methods
2.1. Cell cultivation
M. hominis H34 strain was grown on Brain Heart Infusion (DIFCO, USA) supplemented with 10% horse serum (Biolot, Russia), 1% yeast extract (Helicon, Russia), penicillin (Sintez, Russia) with a final concentration 500 units/ml with the addition of 1% arginine or thymidine as a carbon source. The culture was grown at 37 °C till log-phase for 48 h with arginine or 96 h with thymidine carbon source.
2.2. Protein extraction
Aliquots (10 ml) of log-phase growing cells of M. hominis H34 were collected by centrifugation at 12,000 g at 4 °C for 10 min. Then cells were washed twice by addition of 1 ml cold PBS buffer and centrifugation at 12,000 g at 4 °C for 10 min, 10 μl of 10% sodium deoxycholate (DCNa) and 0.5 μl nuclease mix (GE Healthcare, USA) was added to the cell pellet. After incubation for 1 hour at 4 °C, the sample was resuspended in 100 µl 100 mM Tris-HCl buffer (pH 8.0) containing 0.1% DCNa, 8 M urea and 2.5 mM EDTA. After incubation for 20 min the sample was centrifuged at 16,000 g for 10 min at 4 °C to remove intact cells and debris. The supernatant was collected, and protein concentration was measured using BCA Assay Kit (Sigma-Aldrich, USA).
2.3. Protein preparation to shotgun proteomic
Disulfide bonds were reduced in supernatant (containing 200 μg of total protein) by the addition of Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) (Sigma-Aldrich, USA) to a final concentration of 5 mM and reaction was incubated for 60 min at 37 °C. To alkylate free cysteines, chloroacetamide (Sigma-Aldrich, USA) was added to a final concentration of 30 mM and placed at room temperature in the dark for 30 min. The step of adding TCEP was repeated. Then the sample was diluted 6-fold with 50 mM Tris-HCl, pH 8.0 with 0.01% DCNa. Trypsin Gold (Promega, USA) was added for a final trypsin:protein ratio of 1:50 (w/w) and incubated at 37 °C overnight. To stop trypsinolysis and degrade the acid-labile DCNa, trifluoroacetic acid (TFA) was added to the final concentration of 0.5% (v/v) (the pH should be less than 2.0), incubated at 37 °C for 45 min and the samples were centrifuged at 14,000 g for 10 min to remove the DCNa. Peptide extract was desalted using a Discovery DSC-18 Tube (Supelco, USA) according to the manufacturer protocol. Peptides were eluted with 1 ml of 75% acetonitrile in water containing 0.1% TFA, dried in an Acid-Resistant CentriVap Benchtop Vacuum concentrator (Labconco, USA) and resuspended in 3% acetonitrile in water containing 0.1% TFA to the final concentration of 5 μg/μl.
2.4. LC-MS analysis
LC-MS analysis was carried out on an Ultimate 3000 RSLC nano HPLC system connected to a QExactive Plus mass spectrometer (Thermo Fisher Scientific, USA). Samples were loaded to a home-made trap column 20×0.1 mm, packed with Inertsil ODS3 3 μm sorbent (GL Sciences, Japan), in the loading buffer (2% ACN, 98% H2O, 0.1% TFA) at 10 μl/min flow and separated at RT in a home-packed fused-silica column 500×0.1 mm packed with Reprosil PUR C18AQ 1.9 (Dr. Maisch, Germany) into the emitter prepared with P2000 Laser Puller (Sutter, USA) [4]. Samples were eluted with a linear gradient of 80% ACN, 19.9% H2O, 0.1% FA (buffer B) in 99.9% H2O, 0.1% FA (solvent A) from 4 to 36% of solvent B in 1 h at 0.44 μl/min flow at RT.
MS data were collected in DDA mode. MS1 parameters were as follows: 70 K resolution, 350–2000 scan range, max injection time 50 ms, AGC target 3 × 106. Ions were isolated with 1.4 m/z window and 0.2 m/z offset targeting 10 highest intensity peaks of +2 to +6 charge, 8 × 103 minimum AGC, preferred peptide match and isotope exclusion. Dynamic exclusion was set to 40 s. MS2 fragmentation was carried out in HCD mode at 17,5 K resolution with 27% NCE. Ions were accumulated for max 45 ms with target AGC 1 × 105.
2.5. Protein identification and quantitative analysis
Identification and label-free quantification analysis were performed with PEAKS software [5] with default settings. The data were searched against M. hominis ATCC 23,114 NCBI database and have been deposited to the ProteomeXchange Consortium via the PRIDE [6] partner repository with the dataset identifier PXD018714 and project 10.6019/PXD018714 (http://dx.doi.org/10.6019/PXD018714, https://www.ebi.ac.uk/pride/archive/projects/PXD018714). Further calculations and visualizations were made in R [7].
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships which have, or could be perceived to have, influenced the work reported in this article.
Acknowledgments
This work was supported by Russian Science Foundation 19-75-10124 «Mycoplasma hominis adaptation mechanisms to new niches in the host organism and the formation of its resistant persistent form».
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.dib.2020.106034.
Appendix. Supplementary materials
References
- 1.Waites K., Talkington D. New developments in human diseases due to mycoplasmas. In: Blanchard A., Browning G.F., editors. Mycoplasmas Molecular. 2005. pp. 289–354. [Google Scholar]
- 2.Pereyre S., Sirand-Pugnet P., Beven L., Charron A., Renaudin H. et al., Life on arginine for mycoplasma hominis: clues from its minimal genome and comparison with other human urogenital mycoplasmas. PLoS Genet. 2009;10 doi: 10.1371/journal.pgen.1000677. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robertson B.C., Jargiello P., Blank J., Hoffee P.A. Genetic regulation of ribonucleoside and deoxyribonucleoside catabolism in Salmonella typhimurium. J. Bacteriol. 1970;102:628–635. doi: 10.1128/jb.102.3.628-635.1970. https://jb.asm.org/content/102/3/628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kovalchuk S.I., Jensen O.N., Rogowska-Wrzesinska A. FlashPack: fast and simple preparation of ultrahigh-performance capillary columns for LC-MS. Mol. Cell. Proteom. 2019;18:383–390. doi: 10.1074/mcp.TIR118.000953. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.PEAKS Studio - Bioinformatics Solutions Inc.https://www.bioinfor.com/peaks-studio/, 2020(accessed 31 March 2020)
- 6.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., Pérez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz S., Tiwary S., Cox J., Audain E., Walzer M., Jarnuczak A.F., Ternent T., Brazma A., Vizcaíno J.A. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucl. Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. PubMed ID: 30395289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The R Project for Statistical Computinghttps://www.r-project.org, 2020 (accessed 31 March 2020)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.