Summary
Diet-microbe interactions play a crucial role in modulation of the early life microbiota and infant health. Bifidobacterium dominates the breast-fed infant gut and may persist in individuals during transition from a milk-based to a more diversified diet. Here, we investigated adaptation of Bifidobacterium longum to the changing nutritional environment. Genomic characterization of 75 strains isolated from nine either exclusively breast- or formula-fed (pre-weaning) infants in their first 18 months revealed subspecies- and strain-specific intra-individual genomic diversity with respect to carbohydrate metabolism, which corresponded to different dietary stages. Complementary phenotypic studies indicated strain-specific differences in utilization of human milk oligosaccharides and plant carbohydrates, whereas proteomic profiling identified gene clusters involved in metabolism of selected carbohydrates. Our results indicate a strong link between infant diet and B. longum diversity and provide additional insights into possible competitive advantage mechanisms of this Bifidobacterium species and its persistence in a single host.
Subject Areas: Dietary Supplement, Microbiology, Microbiome
Graphical Abstract
Highlights
-
•
B. longum strains can persist in individual infants despite dietary changes
-
•
New strains with additional metabolism capabilities can also be acquired
-
•
B. longum carbohydrate metabolism repertoires are individual and strain-specific
-
•
Strains at different dietary stages show genomic adaptations to specific substrates
Dietary Supplement; Microbiology; Microbiome
Introduction
Microbial colonization shortly after birth is the first step in establishment of the mutualistic relationship between the host and its microbiota (Backhed et al., 2015; Wampach et al., 2017; Lawson et al., 2020). The microbiota plays a central role in infant development by modulating immune responses, providing resistance to pathogens, and also digesting the early life diet (Heikkila and Saris, 2003; Sela et al., 2008; Marcobal and Sonnenburg, 2012; Sivan et al., 2015; de Aguero et al., 2016; Thongaram et al., 2017). Indeed, diet-microbe interactions are proposed to play a crucial role during infancy and exert health effects that extend to later life stages (Turnbaugh et al., 2006; Renz et al., 2012; Olszak et al., 2012; Feng et al., 2015; Bokulich et al., 2016; Tang et al., 2017). The gastrointestinal tract of vaginally delivered full-term healthy infants harbors a relatively simple microbiota characterized by the dominance of the genus Bifidobacterium (Dogra et al., 2015; Shao et al., 2019). In contrast, caesarean-section-born infants have disrupted transmission of maternal gastrointestinal bacteria, such as Bifidobacterium, and high levels of opportunistic hospital-associated pathogens (Shao et al., 2019).
Breast milk is considered the gold nutritional standard for infants, which also acts as an important dietary supplement for early life microbial communities, including Bifidobacterium. The strong diet-microbe association has further been supported by reports of differences in microbial composition between breast- and formula-fed infants (e.g. high versus low Bifidobacterium abundance) and related differential health outcomes between the two groups: e.g. increased instances of asthma, allergy, and obesity in formula-fed infants (Ip et al., 2007; Das, 2007; O'sullivan et al., 2015; Martin et al., 2016; Stiemsma and Michels, 2018; Ortega-Garcia et al., 2018; Forbes et al., 2018).
The high abundance of Bifidobacterium in breast-fed infants has been linked to the presence of specific carbohydrate utilization genes and gene clusters in their genomes, particularly the ones involved in the degradation of breast milk-associated human milk oligosaccharides (HMOs) (Sela et al., 2008). The presence of these genes is often species- and indeed strain-specific and has been described in Bifidobacterium breve, Bifidobacterium bifidum, Bifidobacterium longum, Bifidobacterium infantis, and more rarely in Bifidobacterium pseudocatenulatum (Sela et al., 2008; James et al., 2016; Katayama, 2016; Garrido et al., 2016). However, previous studies have indicated co-existence of Bifidobacterium species and strains in individual hosts, resulting in interaction and metabolic co-operation within a single (HMO-associated) ecosystem (Milani et al., 2015a; Lawson et al., 2020).
Transition from breastfeeding to a more diversified diet and the introduction of solid foods has been considered to initiate the development of a functionally more complex adult-like microbiome, including presence of genes responsible for degradation of plant-derived complex carbohydrates, starches, and xenobiotics, as well as production of vitamins (Koenig et al., 2011, Mckeen et al., 2019). Non-digestible complex carbohydrates such as inulin-type fructans (ITF), arabinoxylans (AX), or arabinoxylo-oligosaccharides (AXOS) in complementary foods have been proposed to potentially exert beneficial health effects through their bifidogenic and prebiotic properties and resulting modulation of the intestinal microbiota and metabolic end-products (Roberfroid, 2007; Broekaert et al., 2011; Hald et al., 2016; Riviere et al., 2016).
Despite the shift in microbiota composition during weaning, specific strains of Bifidobacterium, and B. longum in particular, have previously been shown to persist in individuals over time (Maldonado-Gomez et al., 2016; Oki et al., 2018). B. longum is currently recognized as four subspecies: longum and infantis (characteristic of the human gut microbiota) and suis and suillum (from animal hosts) (Mattarelli et al., 2008; Yanokura et al., 2015). It is considered the most common and prevalent species found in the human gut, with B. longum subsp. infantis detected in infants, and B. longum subsp. longum widely distributed in both infants and adults (Turroni et al., 2009, 2012). The differences in prevalence between the two subspecies and the ability of infant, adult, and elderly hosts to acquire new B. longum strains during a lifetime have been attributed to distinct bacterial carbohydrate utilization capabilities and the overall composition of the resident microbiota (Garrido et al., 2012; Odamaki et al., 2018).
There have been several recent studies that have explored the early life microbiota in breast- and formula-fed babies (Magne et al., 2006; Palmer et al., 2007; Roger and Mccartney, 2010; Roger et al., 2010). Strain-level metagenomic investigation of the DIABIMMUNE cohort provided insights into diet-related functional aspects of B. infantis in breast-fed infants (Vatanen et al., 2019). Longitudinal studies focusing specifically on B. longum have highlighted intraspecies diversity, colonization, and long-term persistence (years) of this species in hosts; however, there have been limited investigations into diet-related functions at early life stages (Chaplin et al., 2015; Oki et al., 2018; Odamaki et al., 2018). Furthermore, although there are studies examining B. longum strains in relation to diet, these have not been profiled over longitudinal and changing dietary periods (Arboleya et al., 2018). Hence, longitudinal assessments of B. longum strains in single hosts over time, with focus on changing dietary patterns, are lacking, and further detailed studies are required.
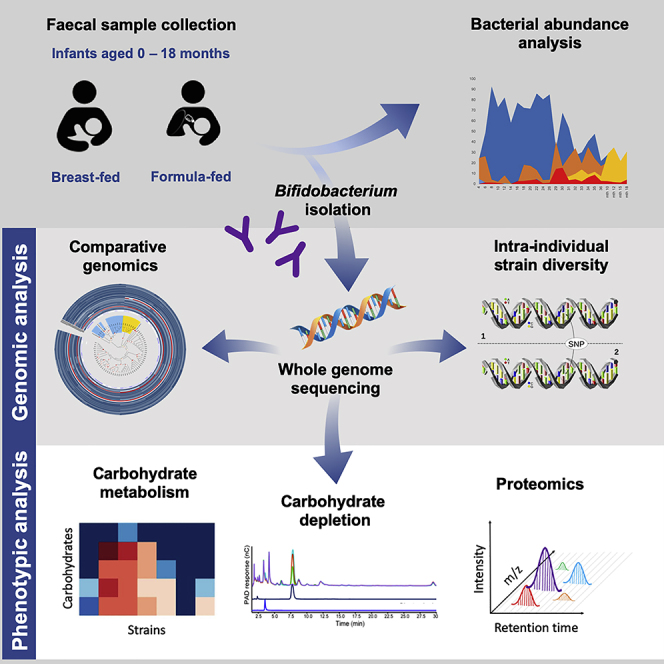
Here, we investigate the adaptations of Bifidobacterium to the changing infant diet and examine a unique collection of B. longum strains isolated from nine infants across their first 18 months, encompassing pre-weaning, weaning, and post-weaning phases. We probed the genomic and phenotypic similarities between 62 B. longum strains and 13 B. infantis strains isolated from either exclusively breast-fed or formula-fed infants (pre-weaning). Our results indicate a strong link between host diet and Bifidobacterium species/strains, which appears to correspond to the changing nutritional environment.
Results
Previous investigations into B. longum across the human lifespan have determined a broad distribution of this species, including prolonged periods of colonization (Maldonado-Gomez et al., 2016; Oki et al., 2018). To gain insight into potential mechanisms facilitating these properties during the early life window, we investigated the genotypic and phenotypic characteristics of B. longum strains within individual infant hosts in relation to diet (i.e. breast milk versus formula) and dietary stages (i.e. pre-weaning, weaning and post-weaning), following up on a longitudinal study of the infant fecal microbiota published in 2010 (Roger and Mccartney, 2010). Briefly, fecal samples from exclusively breast-fed infants and exclusively formula-fed infants were collected regularly from 1 month to 18 months of age (Roger and Mccartney, 2010). The number of samples obtained from the breast-fed infants during the pre-weaning period was higher than that obtained from the formula-fed group, which may correlate with differences in weaning age (~20.6 versus ~17 weeks old). Collected samples were subjected to quantitative analysis using fluorescence in situ hybridization (FISH) to enumerate the predominant bacterial groups (Table S1) (Roger and Mccartney, 2010). Bacterial isolation was also carried out on selected samples and the isolated colonies identified using ribosomal intergenic spacer analysis (Roger and Mccartney, 2010).
Quantitative Analysis of Microbial Communities in Breast- and Formula-Fed Infants
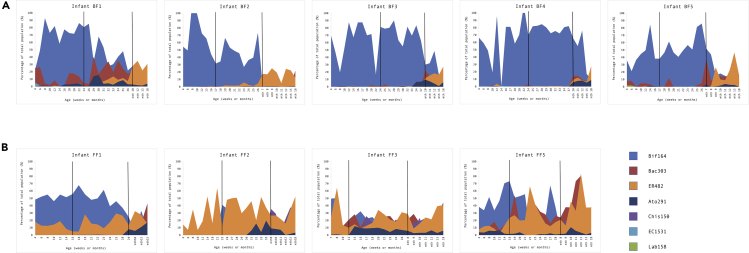
To provide context to the microbiome environment the strains selected for the present study, we reanalyzed the data originally generated by FISH (Figure 1 and Table S1) (Roger and Mccartney, 2010). Bacteria detected using probe Bif164 (bifidobacteria) proportionally constituted the predominant group in samples isolated from breast-fed infants during pre-weaning and weaning: between 16.5% and 100% of the microbiota across the study period. During post-weaning, proportions of bifidobacteria across all breast-fed samples decreased considerably and ranged from 4.6% to 12.1%. The levels of bacteria detected by ER482 (members of Clostridium cluster XIVa) started to increase during weaning, increasing to 18.2% (from 0.25% at pre-weaning). Bacteria detectable by probe Bac303 (members of genus Bacteroides, Parabacteroides and Prevotella species, Paraprevotella, Xylanibacter, Barnesiella species and Odoribacter splanchnicus) were identified in all samples throughout the study, with this bacterial group showing extensive inter-individual variation. Other microbiota members were detected in breast-fed samples at lower levels, including members of family Coriobacteriia (Ato291, mean < 2% of microbiota), Escherichia coli (EC1531, <1%), members of Clostridium clusters I and II (Chis150, <1%), and lactic acid bacteria (Lab158, mean < 1%).
Figure 1.
Proportional Representation of Bacterial Populations in the Fecal Microbiota of Infants
Based on FISH analysis in (A) breast-fed and (B) formula-fed infants. Numbers are expressed as percentage of the total bacterial population obtained using DAPI. The vertical solid black lines mark the different dietary phases in each infant (pre-weaning, weaning, and post-weaning). Oligonucleotide probes used to determine bacterial populations: Bif164—most Bifidobacterium species and Parascardovia denticolens; Bac303—most members of the genus Bacteroides, some Parabacteroides and Prevotella species, Paraprevotella, Xylanibacter, Barnesiella species, and Odoribacter splanchnicus; ER482—most members of Clostridium cluster XIVa; Ato291—Cryptobacterium curtum, Gordonibacter pamelaeae, Paraeggerthella hongkongensis, all Eggerthella, Collinsella, Olsenella and Atopobium species; Chis150—most members of Clostridium cluster I, all members of Clostridium cluster II; EC1531—Escherichia coli; Lab158—all Oenococcus, Vagococcus, Melissococcus, Tetragenococcus, Enterococcus, Catellicoccus, Paralactobacillus, Pediococcus and Lactococcus species, most Lactobacillus, Weissella, and Leuconostoc species. See also Table S1.
In contrast to the breast-fed group, no drastic shift in bacterial populations was observed in formula-fed infants throughout the study. Overall, lower levels of bifidobacteria were detected during pre-weaning and weaning, fluctuating from 0.0% to 73.3% of the microbiota at different time points. Similar to the breast-fed group, proportions of Bifidobacterium decreased during post-weaning across all formula-fed samples and ranged from 6.5% to 12% at month 18. The levels of bacteria detected by probe ER482 were overall higher in formula-fed samples throughout study duration: 19.96 ± 17.41%, 25.39 ± 14.63%, and 30.6 ± 15.92% for pre-weaning, weaning, and post-weaning phases. Similarly, proportions of bacteria detected by Bac303 during all dietary phases were higher in the formula-fed group compared with the breast-fed group. Contrastingly to the breast-fed group, levels of bacteria detected by Chis150 (Clostridium clusters I and II) started to increase during weaning in the formula-fed group and continued to increase (1.23 ± 1.28%, 7.03 ± 9.18%, and 21.72 ± 11.47% for pre-weaning, weaning, and post-weaning, respectively). Levels of bacteria identified by Ato291 and EC1531 in formula-fed samples were slightly higher than in the breast-fed group (means of <3.5% and <1.25%, respectively), whereas the mean proportion of lactic acid bacteria (Lab158) remained below 1%.
Overall, these results confirm previous studies that have indicated differences in fecal microbiota composition between breast- and formula-fed babies, particularly during the pre-weaning and weaning phases, and demonstrate the succession of bacterial species over time and in relation to diet, including Bifidobacterium.
General Features of B. longum Genomes
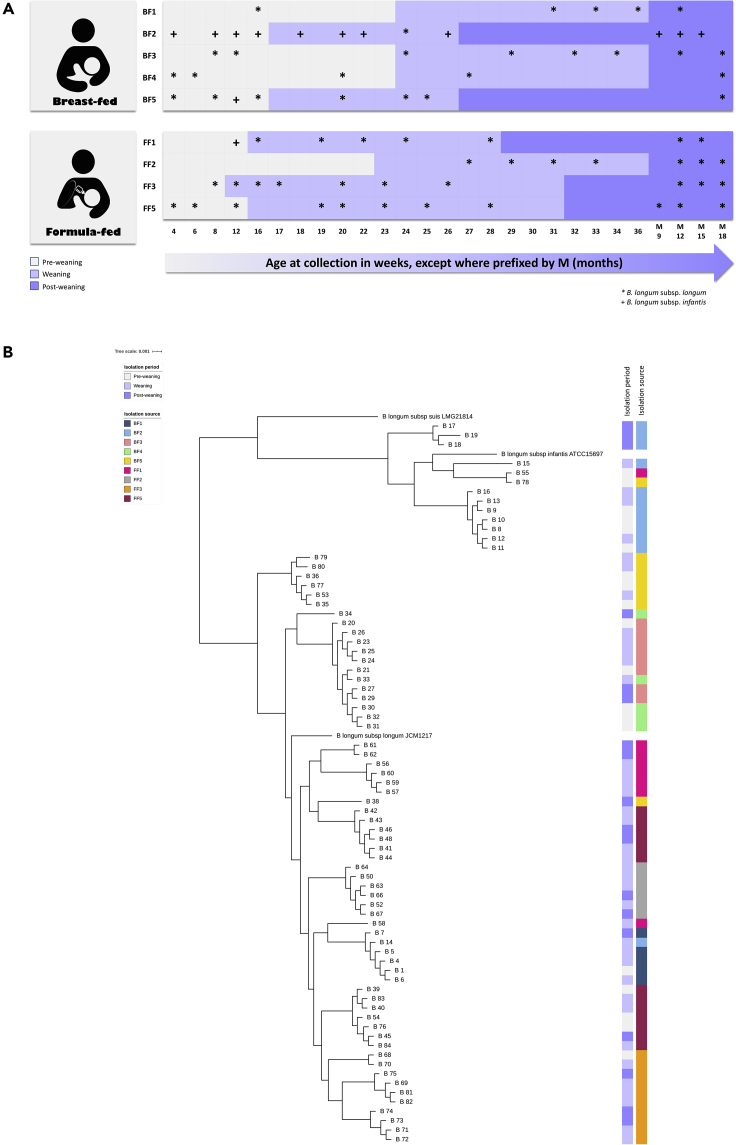
Based on the results of bacterial culture and colony identification published previously (for details, refer to (Roger et al., 2010)), 88 isolates originally identified as Bifidobacterium were selected for this study, 46 from five exclusively breast-fed infants (BF1-BF5, including identical twins BF3 and BF4) and 42 from four exclusively formula-fed infants (FF1-FF3 and FF5). Following sequencing and ANI analysis (Tables S2 and S3), 75 strains were identified as B. longum sp. and included in further analysis, with 62 strains identified as B. longum subsp. longum (B. longum) and 13 strains identified as B. longum subsp. infantis (B. infantis) (Figure 2A).
Figure 2.
Identification and Relatedness of B. longum Strains
(A) Sampling scheme and strain identification within individual breast-fed (BF1-BF5) and formula-fed (FF1-FF3 and FF5) infants based on average nucleotide identity values (ANI). The three levels of shading mark different dietary phases: pre-weaning, weaning, and post-weaning.
(B) Relatedness of B. longum strains based on core proteins. Colored strips represent isolation period (pre-weaning, weaning, and post-weaning) and isolation source (individual infants), respectively.
To determine possible genotypic factors facilitating establishment and persistence of B. longum in the changing early life environment, we assessed the genome diversity of our strains. Sequencing generated between 12 and 193 contigs for each B. longum strain, with 74/75 containing fewer than 70 contigs, yielding a mean of 66.95-fold coverage for strains (Table S2). The predicted genome size for strains identified as B. longum ranged from 2.21 Mb to 2.58 Mb, possessing an average G + C% content of 60.11%, an average predicted ORF number of 2,023, and number of tRNA genes ranging from 55–88. For strains identified as B. infantis, the predicted genome size ranged from 2.51 Mb to 2.75 Mb, with an average G + C% content of 59.69%, an average predicted ORF number of 2,280, and the number of tRNA genes ranging from 57 to 62.
Comparative Genomics
To identify B. longum strains among the sequenced isolates and assess nucleotide-level genomic differences, we performed ANI analysis. Results (Table S3) indicated that B. longum strains isolated from individual infant hosts displayed higher levels of sequence identity than strains isolated from different hosts. More specifically, pairwise identity values for strains isolated from infant BF3 showed the narrowest range (average value of 99.99 ± 3.15 × 10−5%), followed by infant FF2 strains (99.98 ± 1.12 × 10−4%), with infant BF2 strains having the broadest identity value range (averaging 99.13 ± 7.8 × 10−3%).
Next, we examined genetic diversity of newly sequenced B. longum strains and their relatedness to each other, alongside B. longum type strains. We identified a total of 1,002 core genes present in at least 99% of the analyzed B. longum subspecies genomes that allowed clear distinction between B. longum subspecies (i.e. longum versus infantis) based on the presence/absence of specific genes (Table S4). Phylogenetic analysis revealed that B. longum strains within each subspecies clustered mainly according to isolation source, i.e. individual infants, rather than dietary stage (i.e. pre-weaning, weaning and post-weaning) (Figure 2B). Interestingly, strains isolated from formula-fed baby FF5 clustered into two separate clusters, irrespective of the isolation period, suggesting presence of two highly related B. longum groups within this infant. Furthermore, strains isolated from identical twins BF3 and BF4 clustered together, indicating their close relatedness.
We next sought to identify whether specific components of the B. longum subspecies pangenome were enriched in infant hosts. Each candidate gene in the accessory genome was sequentially scored according to its apparent correlation to host diet (breast vs. formula) or dietary stage. A gene annotated as α-L-arabinofuranosidase, along with four other genes coding for hypothetical proteins, were predicted to be enriched in B. longum strains isolated from breast-fed infants. Alpha-L-arabinofuranosidases are enzymes involved in hydrolysis of terminal non-reducing α-L-arabinofuranoside residues in α-L-arabinosides and act on such carbohydrates as (arabino)xylans (Ichinose et al., 2008; Ahmed et al., 2013). In addition, two genes coding for hypothetical proteins and a gene coding for mobility protein A were overrepresented in strains isolated from formula-fed infants. No associations between genes and dietary stages in B. longum nor any associations in B. infantis were observed (Table S5).
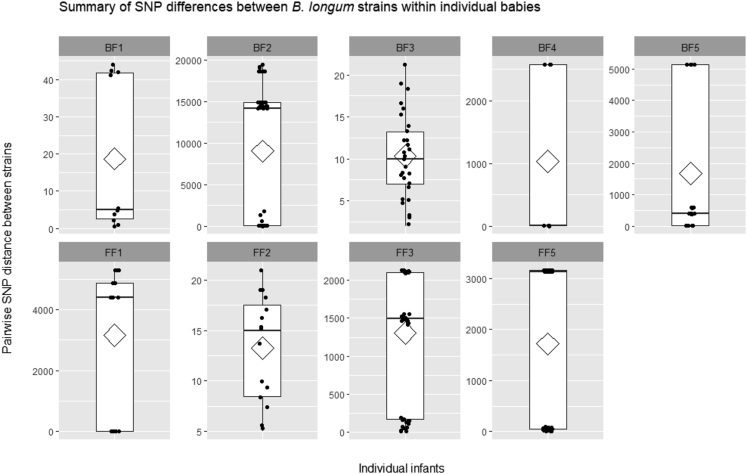
As our strains were isolated from individual infants at different time points, we next sought to determine their intra-strain diversity; for this we used the first B. longum isolate from each infant as the “reference” strain to which all other strains from the same infant were compared (Figure 3). Infants BF1, BF3, and FF2 had the lowest strain diversity, with respective mean pairwise SNP distances of 18.7 ± 20.3 SNPs (mean ± sd), 10.3 ± 5.0 SNPs, and 13.3 ± 5.3 SNPs. These results suggest strains isolated from these infants may be clonal, indicating long-term persistence despite dietary changes. Surprisingly, analysis of strains isolated from breast-fed identical twins BF3 and BF4 revealed higher strain diversity in baby BF4 (1034.5 ± 1327.1 SNPs), compared with the highly similar strains in infant BF3 (i.e. 10.3 ± 5.0 SNPs). Based on these results, we conducted SNP analysis on B. longum strains isolated from both babies and found that out of 13 strains analyzed (n = 8 from BF3 and n = 5 from BF4), 12 isolated during pre-weaning, weaning, and post-weaning appeared to be clonal (with mean pairwise SNP distance of 10.0 ± 5.5 SNPs) and one strain from baby BF4 isolated post-weaning was more distant, 2,595.4 ± 2.8 SNPs. The difference in strain diversity may relate to the fact that infant BF4 received a course of antibiotics during pre-weaning (Figure 1 and Tables S1 and S2) (Roger and Mccartney, 2010). Furthermore, the presence of clonal strains in both babies suggests vertical transmission of B. longum from mother to both infants, or potential horizontal transmission between babies, consistent with previous reports (Makino et al., 2011, 2013; Milani et al., 2015b; Odamaki et al., 2018). B. infantis strains isolated from infant BF2 showed the highest strain diversity of 9,030.9 ± 8,036.6 SNPs. Seven strains isolated during both pre-weaning and weaning periods appeared to be clonal, 6.3 ± 1.6 SNPs, whereas four strains isolated during weaning and post-weaning were more distant, with mean pairwise SNP distance of 14,983.5 ± 4,658.3 SNPs (Table S6).
Figure 3.
Pairwise SNP Distances between B. longum Strains of the Same Subspecies within Individual Infants
Individual points show data distribution, diamonds indicate the group mean, box plots show group median and interquartile range. See also Table S6.
Functional Annotation of B. longum Subspecies Genomes—Carbohydrate Utilization
To assess genomic differences between our strains at a functional level, we next assigned functional categories to ORFs of each B. longum genome. Carbohydrate transport and metabolism was identified as the second most abundant category (after unknown function), reflecting the saccharolytic lifestyle of Bifidobacterium (Figure S1) (Pokusaeva et al., 2011, Milani et al., 2015a). B. longum had a slightly higher proportion of carbohydrate metabolism and transport genes (11.39 ± 0.31%) compared with B. infantis (10.20 ± 0.60%), which is consistent with previous reports (Ventura et al., 2009; Sela and Mills, 2010). B. longum strains isolated during pre-weaning had a similar proportion of carbohydrate metabolism genes in comparison with the strains isolated post-weaning: 11.28 ± 0.23% and 11.48 ± 0.38%, respectively. Furthermore, we obtained similar results for B. longum strains isolated from breast- and formula-fed infants, with respective values of 11.41 ± 0.21% and 11.38 ± 0.38%. In contrast, B. infantis strains isolated pre-weaning had a lower proportion of carbohydrate metabolism genes in their genomes compared with the ones isolated post-weaning: 9.90 ± 0.24% and 11.20 ± 0.01%, respectively (Table S7).
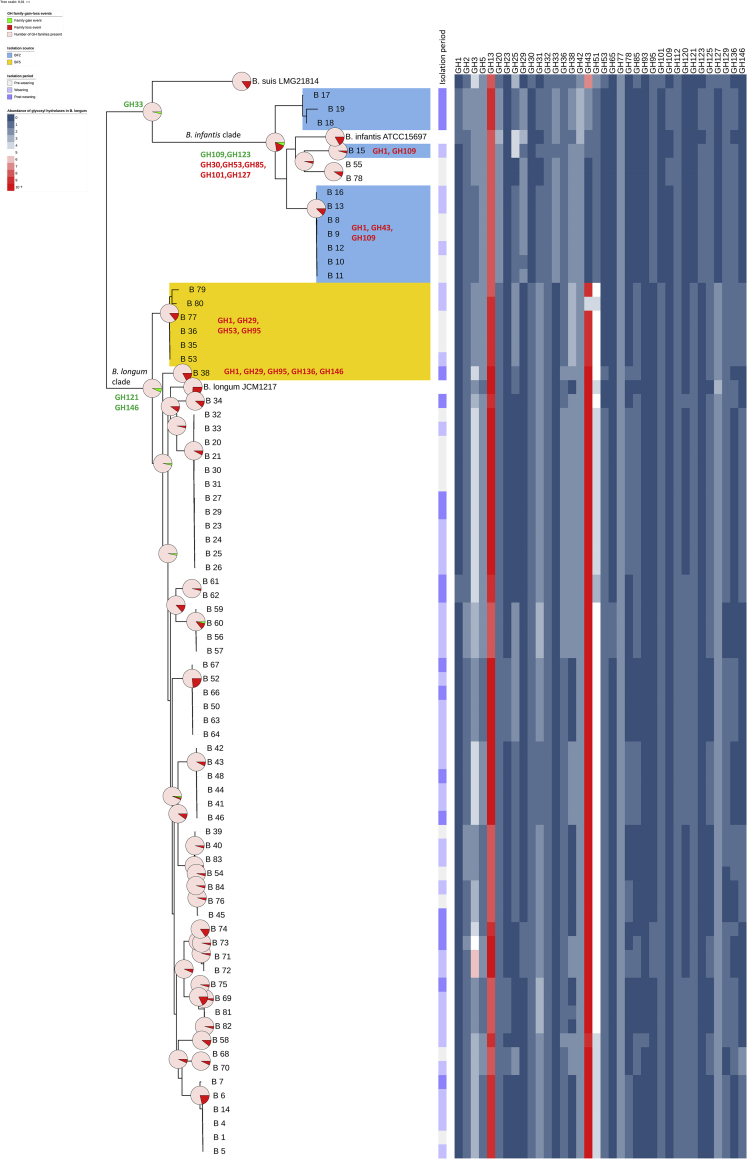
One of the major classes of carbohydrate-active enzymes comprises glycosyl hydrolases (GH), which facilitate glycan metabolism in the gastrointestinal tract. We thus sought to investigate and compare the arsenal of GHs in B. longum using dbCAN2. We identified a total of 36 different GH families in all Bifidobacterium strains. B. longum was predicted to contain 55 GH genes per genome on average (2.72% of OFRs), whereas this number was lower for B. infantis strains, ~37 GH genes per genome (1.62% of ORFs) (Figure 4 and Table S8). The predominant GH family was GH43—enzymes involved in metabolism of complex plant carbohydrates such as (arabino)xylans (Viborg et al., 2013), followed by GH13 (starch), GH51 (hemicelluloses), and GH3 (plant glycans) (Milani et al., 2015a, 2016).
Figure 4.
Gene-Loss Events and Abundance of GH Families within B. longum Subspecies
Pie charts superimposed on the whole genome SNP tree represent predicted GH family gain-loss events within B. longum and B. infantis lineages. Due to the size of the tree, examples of detailed gain loss events have been provided for main lineages, as well as baby BF2 (strains highlighted with light blue) and BF5 (strains highlighted with yellow). Heatmap represents abundance of specific GH families predicted in analyzed B. longum strains. See also Tables S8 and S9.
Within the B. longum group, strains isolated during pre-weaning had a slightly lower mean number of GH genes compared with strains isolated post-weaning (54.46 ± 2.81 vs. 56.85 ± 2.77). Moreover, strains isolated from breast-fed babies contained an average of 53.96 ± 3.82 GH genes per genome, whereas this number was slightly higher for strains isolated from formula-fed infants—56.47 ± 2.96. Further analysis revealed that these differences appeared to be intra-host-specific and diet-related. For example, strains isolated from breast-fed twins BF3 and BF4 pre-weaning had 11 GH43 genes per genome, whereas the pre-weaning strain from formula-fed baby FF3 had 13 GH genes per genome predicted to belong to this GH family. Similarly, strains isolated from babies BF3 and BF4 post-weaning had 11 predicted GH genes, whereas the three strains isolated from infant FF3 were predicted to contain 16, 16, and 18 GH genes per genome, respectively (Table S8).
We next determined if these GH genes differences statistically correlated with breast- and formula-fed groups (Table S8). Significant differences (p < 0.05) were observed between mean numbers of GH genes belonging to the predominant GH families (GH43—higher abundance in FF babies, GH13—higher abundance in BF babies, and GH51—higher abundance in FF babies), and several other GH families, including GH5 (β-glucosidases and β-mannosidases), GH38 (mannosylglycerate hydrolases), and GH36 (α-galactosidases), all more abundant in BF babies. Further analysis of dietary phases suggested significant differences in GH genes between breast- and formula-fed groups during pre-weaning (e.g. families GH43, GH13, GH5, GH38) but not in the post-weaning phase (Table S8).
Because glycosyl hydrolases belonging to distinct GH families may have similar catalytic properties, we next grouped the GH genes for which the predicted enzyme class annotation was available and investigated their abundance (Table S9). The predominant enzyme classes in B. longum strains were non-reducing end α-L-arabinofuranosidases belonging to GH43 and GH51, followed by β-galactosidases (GH2 and GH42), oligo-1,6-glucosidases (GH13), and β-N-acetylhexosaminidases (GH3 and GH20).
The mean numbers of enzyme classes between breast- and formula-fed babies significantly differed (p < 0.05) in the top three above-mentioned predominant enzyme classes as well as several other less abundant ones, including non-reducing end β-L-arabinofuranosidases (GH127 and GH146—higher abundance in BF babies), α-galactosidases (GH36—higher abundance in BF babies), and endo-1,5-α-L-arabinases (GH43—higher abundance in FF babies). Additional analysis of dietary phases indicated significant differences between breast- and formula-fed groups during pre-weaning (e.g. non-reducing end α-L-arabinofuranosidases, β-galactosidases, oligo-1,6-glucosidases as well as α-galactosidases) but not during post-weaning (Table S9).
We next examined the predicted glycosyl hydrolase repertoire of B. infantis strains, with the caveat that the majority of the strains belonging to this subspecies were isolated from a single infant. In contrast to the B. longum group, the most abundant GH family was GH13 (starch), followed by GH42, GH20, and GH38 (Table S8). B. infantis strains also harbored genes predicted to encode members of the GH33 family, which contains exo-sialidases (Milani et al., 2015a). Strains isolated pre-weaning were predicted to contain an average of 34.83 ± 0.4 GH genes per genome, whereas this number was higher for the strains isolated post-weaning (i.e. 43.00 ± 0.00 GH genes). B. infantis strains isolated post-weaning contained families GH1 and GH43 that were absent in the strains isolated pre-weaning. The GH1 family contains enzymes such as β-glucosidases, β-galactosidases, and β-D-fucosidases active on a wide variety of (phosphorylated) disaccharides, oligosaccharides, and sugar–aromatic conjugates (Suzuki et al., 2013). The analysis of enzyme classes in the B. infantis strains suggested that β-galactosidases (GH2 and GH42) were predominant in this group, followed by β-N-acetylhexoaminidases (GH3 and GH20), 4-α-glucanotransferases (GH77), and oligo-1,6-glucosidases (GH13) (Table S9).
Members of the genus Bifidobacterium have previously been shown to contain GH genes involved in metabolism of various HMOs present in breast milk (Garrido et al., 2015, 2016). We identified genes belonging to GH29 and GH95 (α-L-fucosidases found active on fucosylated HMOs (Sela et al., 2012; Garrido et al., 2016)) in all our B. infantis strains, as well as four B. longum strains isolated from formula-fed baby FF3. Furthermore, we found GH20 and GH112 genes (lacto-N-biosidases and galacto-N-biose/lacto-N-biose phosphorylases shown to be involved in degradation of isomeric lacto-N-tetraose (LNT) (Kitaoka, 2012)) in all our B. infantis and B. longum strains (Table S8).
Overall, these findings suggest differences in general carbohydrate utilization at different stages suggesting adaptation of Bifidobacterium to a changing early life nutritional diet, which may be a factor facilitating establishment of these bacteria within individuals during infancy.
Prediction of Gain and Loss of GH Families in B. longum
Given the differences in the carbohydrate utilization profiles between B. longum and B. infantis, we next investigated the acquisition and loss of GH families. For this purpose, we additionally predicted the presence of GH families in type strains B. longum subsp. longum JCM 1217T, B. longum subsp. infantis ATCC 15697T, and B. longum subsp. suis LMG 21814T with dbCAN2 and generated a whole genome SNP tree to reflect gene loss/gain events more accurately (Figure 4 and Table S10). Both B. longum and B. infantis lineages appear to have acquired GH families (when compared to the common ancestor of the phylogenetic group), with the B. longum lineage gaining two GH families (GH121 and GH146) and the B. infantis lineage one GH family (GH33). Within the B. infantis lineage, which also contains the B. suis type strain, the B. infantis taxon has further acquired two and lost five GH families. These findings suggest that the two human-related subspecies have followed different evolutionary paths, which is consistent with our observation of differences between B. longum and B. infantis resulting from phylogenomic analyses. Intriguingly, strain adaptation to the changing host environment (i.e. individual infant gut) seems to be driven by loss of specific GH families (Figure 4). For example, B. infantis strains isolated during pre-weaning and weaning from baby BF2 appear to be missing up to three GH families (GH1, GH43, and GH109) present in strains isolated post-weaning. Lack of family GH43 (containing enzymes involved in metabolism of a variety of complex carbohydrates, including plant-derived polysaccharides) in early life B. infantis strains may explain nutritional preference of this subspecies for an HMO-rich diet. Similarly, we observed differential gene loss events in B. longum strains from individual hosts. For example, all strains isolated from baby BF5 appear to lack GH families GH1, GH29, and GH95. However, strains isolated pre-weaning additionally lacked GH53 family, which includes endogalactanases shown to be involved in liberating galactotriose from type I arabinogalactans in B. longum (Hinz et al., 2005). In contrast, strain B_38 isolated from this infant (BF5) post-weaning appears to have lost families GH136 and GH146. Interestingly, members of family GH136 are lacto-N-biosidases responsible for liberating lacto-N-biose I from LNT, an abundant HMO unique to human milk (Yamada et al., 2017), whereas family GH146 contains β-L-arabinofuranosidases displaying exo-activity on β-linked arabinofuranosyl groups. These events may be linked to dietary changes (withdrawal of breast milk) and/or a shift in the composition of the microbiota post-weaning. Only one B. longum strain was isolated post-weaning from this baby; however, FISH analysis (Figure 1 and Table S1) revealed an increase in the bacteroides group, which might explain the loss of family GH146 by strain B_38 as the founding member of GH146 family, β-L-arabinofuranosidase, was first characterized in Bacteroides thetaiotaomicron (Luis et al., 2018). Overall, the presence of intra-individual and strain-specific GH family repertoires in B. longum suggests their adaptation to host-specific diet. The presence of strains with different GH content at different dietary stages further indicates potential acquisition of new Bifidobacterium strains with nutrient-specific adaptations in response to the changing infant diet.
Prediction of Single Nucleotide Polymorphisms in Glycosyl Hydrolases
Given the intra-strain diversity in the nine babies and the differences in GH repertoires between B. longum and B. infantis, we next sought to examine nucleotide-level differences in glycosyl hydrolase genes between strains in individual infants (Table S11). Unsurprisingly, we did not identify any significant SNPs that may lead to functional changes in GH genes in infants that had the lowest strain diversity (infants BF1, BF3 and FF2) (Table S6). The highest number of GH genes with predicted variants was recorded for B. infantis strains from baby BF2. In total, 52 synonymous variants and 29 missense variants were predicted at 81 different positions in 12 GH genes across strains that showed the highest diversity from the first “reference” isolate, namely one strain isolated during weaning and the three strains isolated post-weaning. Several missense variants, both complex and single, were recorded at several positions in the predominant enzyme classes, i.e. β-galactosidases (EC 3.2.1.23) and β-N-hexosaminidases (EC 3.2.1.52).
Similarly, both synonymous and missense variants were predicted in B. longum strains less closely related to “reference” strains from breast-fed (BF4 and BF5) and formula-fed (FF1, FF3 and FF5) babies. We did not observe any trend in the distribution of SNPs across GH genes in B. longum strains. The number of predicted variants, the number of GH genes with identified mutations, and their enzyme classification differed between individual infants. For example, in baby BF4 9 out of 10 predicted variants (4 synonymous and 5 missense) were identified in an α-xylosidase in a strain isolated post-weaning, whereas in baby FF5 14 synonymous and 10 missense variants were predicted at 24 positions in 7 different GH genes across strains isolated during weaning and post-weaning. Some missense changes do not compromise normal protein function, whereas others can change essential aspects of protein maturation, activity, or stability (Miosge et al., 2015). The presence of missense variants in GH genes of B. longum strains may indicate potential functional differences and provide additional explanation to intra-strain and intra-individual carbohydrate metabolism profiles of these bacteria; however, experimental evidence would be essential to confirm the importance of these predictions.
Phenotypic Characterization of Carbohydrate Utilization
Bifidobacterium longum has previously been shown to metabolize a range of carbohydrates, including dietary and host-derived glycans (Watson et al., 2013; Arboleya et al., 2018). Given the predicted differences in carbohydrate metabolism profiles and to understand strain-specific nutrient preferences, we next determined their glycan fermentation capabilities. We performed growth assays on 49 representative strains from all nine infants, cultured in modified MRS supplemented with selected carbohydrates as the sole carbon source. For these experiments, we chose both plant- and host-derived glycans that we would expect to constitute components of the early life infant diet (Mills et al., 2019). Although all B. longum strains were able to grow on simple carbohydrates (i.e. glucose and lactose), we also observed subspecies-specific complex carbohydrate preferences, consistent with bioinformatic predictions (Figure 5). To represent host-derived carbohydrates, we selected 2′-fucosyllactose (2′-FL) and lacto-N-neotetraose (LNnT) as examples of HMOs found in breast milk. Out of the tested isolates, all B. infantis strains were able to metabolize 2′-FL, as were three B. longum strains isolated from a formula-fed baby FF3 during weaning and post-weaning (Figure 5). These results supported the computational analysis and the identification of genes potentially involved in degradation of fucosylated carbohydrates in the genomes of these isolates (GH29 and GH95). Although bioinformatics identified the presence of genes involved in metabolism of isomeric LNT in all our strains (GH20 and GH112), LNnT metabolism in B. infantis was strain specific, with most strains showing what we considered moderate (above 0.15 difference in OD from time T2) to high growth rates (above 0.25 difference in OD from time T2), with two strains displaying inconsistent growth (Table S12). Out of B. longum strains, B_24 and B_25 (isolated during weaning from breast-fed baby BF3) also showed growth on LNnT, albeit this was inconsistent. In contrast to all other B. longum strains, strain B_25 was not able to metabolize plant-derived arabinose and xylose despite the predicted presence of genes involved in metabolism of monosaccharides (GH43, GH31, GH2). However, it was one of the two strains (out of 49 tested) that showed growth on cellobiose in 2/3 experiments, the other one being the post-weaning B. infantis strain B_19 isolated from baby BF2. Given these interesting results, we performed additional assays using cellobiose as the sole carbon source over 72 h, in which the B. longum strain B_25 showed high growth rate (above 0.25 difference in OD from time T2), whereas the B. infantis B_19 strain did not grow at all (Table S12). In addition, both B. longum and B. infantis strains showed varying degrees of growth performance on mannose, even when analyzing the same strain, whereas none of the tested strains were able to grow on arabinogalactan, pectin, or rhamnose (Figure 5).
Figure 5.
Growth Performance of B. longum Strains on Different Carbon Sources
Heatmap displays the difference in average growth of triplicates between T2 (30 min) and Tend (48 hr). Moderate growth is considered above 0.15 difference in OD from time T2, high growth above 0.25 difference in OD from time T2. Asterisks represent strains for which inconsistent growth was recorded (difference in OD of at least 0.15 between any of the duplicates in the triplicate experiment). See also Table S12.
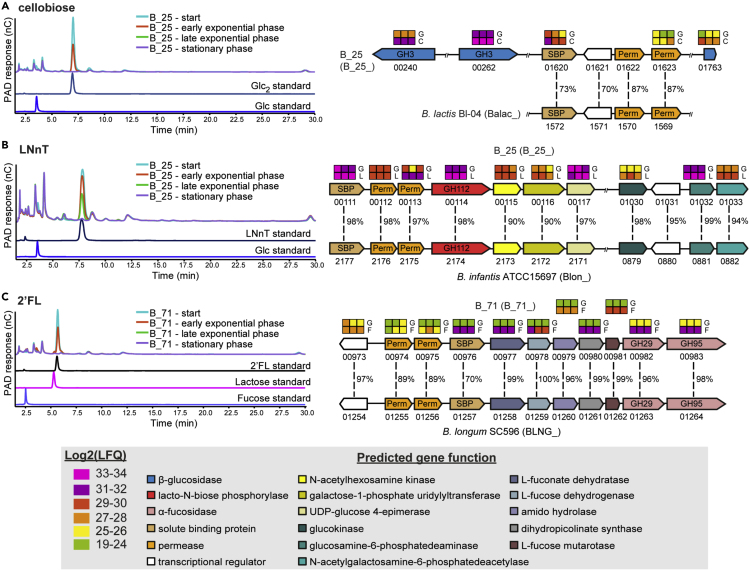
To further characterize strains identified above for putative carbohydrate degradation genes, we performed carbohydrate uptake analysis and proteomics. B. longum strain B_25, from one of the breast-fed identical twins that showed growth on LNnT and cellobiose, and formula-fed strain B_71, which was able to grow on 2′-FL, were chosen. Supernatant from these cultures was initially subjected to high-performance anion-exchange chromatography (HPAEC) to evaluate the carbohydrate-depletion profiles (Figure 6). In all three cases, the chromatograms showed complete utilization of the tested carbohydrates and absence of any respective degradation products in the stationary phase culture. The depletion of cellobiose by B_25 and 2′-FL by B_71 occurred in the early exponential phase, whereas LNnT was still detected in the culture supernatant until the late exponential phase of growth, suggesting that cellobiose and 2′-FL were internalized more efficiently than LNnT. We next determined the proteome of B_25 and B_71 when growing on cellobiose, LNnT and 2′-FL compared with glucose (Figures 6A–6C and Table S13). The top 10 most abundant proteins in the cellobiose proteome of B_25 included three β-glucosidases belonging to GH3 family, as well as a homologue of transport gene cluster previously shown to be upregulated in B. animalis subsp. lactis Bl-04 during growth on cellobiose (Figure 6A and Table S14) (Andersen et al., 2013). Among the three β-glucosidases, B_25_00240 showed 98% sequence identity to the structurally characterized BlBG3 from B. longum, which has been shown to be involved in metabolism of the natural glycosides saponins (Yan et al., 2018). B_25_01763 and B_25_00262 showed 46% identity to the β-glucosidase Bgl3B from Thermotoga neapolitana (Pozzo et al., 2010) and 83% identity to BaBgl3 from B. adolescentis ATCC 15703 (Florindo et al., 2018), respectively, two enzymes previously shown to hydrolyze cello-oligosaccharides. With respect to LNnT metabolism by the same strain, the most abundant proteins were encoded by genes located in two gene clusters (B_25_00111–00117 and B_25_00130-00133) with functions compatible with LNnT import, degradation to monosaccharides, and further metabolism. The gene clusters contain the components of an ABC-transporter (B_25_00111–00113), a predicted intracellular GH112 lacto-N-biose phosphorylase (B_25_00114), an N-acetylhexosamine 1-kinase (B_25_00115) and enzymes involved in the Leloir pathway. All these proteins were close homologues to proteins previously implicated in the degradation of LNT/LNnT by type strain B. infantis ATCC 15697T (Ozcan and Sela, 2018) (Figure 6B and Table S14). Interestingly, all clonal strains isolated from twin babies BF3 and BF4 also contained close homologues of all the above-mentioned genes in their genomes, in some cases identical to those determined in B_25; however, only strain B_25 was able to grow on cellobiose and LNnT. Growth of B_71 on 2′-FL corresponded to increased abundance of proteins encoded by the gene cluster B_71_00973-00983. These proteins showed close homology to proteins described for B. longum SC596 and included genes for import of fucosylated oligosaccharides, fucose metabolism, and two α-fucosidases belonging to the families GH29 and GH95 (Figure 6C and Table S14) (Garrido et al., 2016).
Figure 6.
Carbohydrate Uptake Analysis and Proteomics of B. longum Strains B_25 and B_71
HPAEC-PAD traces showing mono-, di-, and oligo-saccharides detected in the supernatant of either B_25 or B_71 single cultures during growth in mMRS supplemented with (A) cellobiose; (B) LNnT; (C) 2′-FL. The data are representative of biological triplicates. Abbreviations: LNnT, Lacto-N-neotetraose; Glc, glucose; Glc2, cellobiose; 2′-FL, 2′-fucosyllactose. Panel on the right shows (A) cellobiose; (B) LNnT; (C) 2′-FL utilization clusters in B_25 and B_71 and proteomic detection of the corresponding proteins during growth on HMOs. Heatmaps above genes show the LFQ detection levels for the corresponding proteins in triplicates grown on glucose (G); cellobiose (C); LNnT (L); and 2′-FL (F). Numbers between genes indicate percent identity between corresponding genes in homologous gene clusters relative to strains B_25 and B_71. Numbers below each gene show the locus tag in the corresponding genome. Locus tag numbers are abbreviated with the last numbers after the second hyphen (for example B_25_XXXXX). The locus tag prefix for each strain is indicated in parenthesis beside the organism name. See also Tables S13 and S14.
Discussion
High abundance of Bifidobacterium, and B. longum in particular, in early infancy is strongly linked to availability of nutrients (Koenig et al., 2011; Backhed et al., 2015; Yamada et al., 2017). In this study, we aimed to investigate the adaptations of B. longum to the changing infant diet during the early life developmental window. Profiling microbiota composition data (Roger and Mccartney, 2010), genomic diversity of 75 B. longum strains (isolated from infants at different dietary stages), and growth performance on different carbohydrates revealed intra-individual and diet-related differences, which links to strain-level metabolism properties for specific dietary components.
The FISH results corroborate findings of previous studies investigating the infant gut microbiota—inter-individual variability during pre-weaning and weaning, with a shift toward a more adult-like fecal microbiota associated with more complex diet at post-weaning across all samples (Koenig et al., 2011, Mckeen et al., 2019). Bifidobacterium constituted the predominant group in breast-fed infants during pre-weaning and weaning, whereas the composition of microbiota of the formula-fed infants during these stages was more complex.
Our comparative genomic analysis indicates that clonal strains of B. longum can persist in individuals through infancy, for at least 18 months, despite significant changes in diet during weaning, which is consistent with previous reports (Maldonado-Gomez et al., 2016; Odamaki et al., 2018). Concurrently, new strains (that display different genomic content and potential carbohydrate metabolism capabilities) can be acquired, possibly in response to the changing diet. Previously, strain shift in relation to withdrawal of breast milk has been suggested for B. infantis by Vatanen et al. (2019) based on strain-level metagenomic approach. Similarly, Asnicar et al. (2017) suggested that originally acquired maternal strains of B. longum can be replaced at later life stages. Initial vertical acquisition of Bifidobacterium from mother to newborn babies has been well documented (Mikami et al., 2012; Makino et al., 2013; Milani et al., 2015b; Asnicar et al., 2017); however, details of strain transmission events in later life are currently unclear. Work of Odamaki et al. (2018) suggested person-to-person horizontal transmission of a particular B. longum strain between members of the same family, with direct transfer, common dietary sources, or environmental reservoirs, such as family homes (Lax et al., 2014) acting as potential vehicles and routes for strain transmission. Our results showed the presence of clonal strains in identical twins BF3 and BF4, which may have resulted from maternal transfer. However, potential strain transmission between these infants living in the same environment may also occur. Wider studies involving both mothers and twin babies (and other siblings) could provide details on the extent, timing, and location of transmission events between members of the same household.
Another aspect of comparative genomic analysis involved in silico prediction of genes belonging to GH families. This analysis revealed genome flexibility within B. longum, with differences in GH family content between strains belonging to the same subspecies as described previously; B. infantis predominantly enriched in GH families implicated in the degradation of host-derived breast milk-associated dietary components such as HMOs and B. longum containing GH families involved in the metabolism of plant-derived substrates (Milani et al., 2015a; Milani et al., 2016). Previously, Vatanen et al. (2019) suggested that the presence of the HMO gene cluster allowing for intracellular HMO utilization in B. infantis strains, in particular, confers a competitive advantage leading to higher relative abundance of this subspecies in the early life microbiota. Our analysis of B. infantis group identified the presence of glycosyl hydrolases associated with HMO degradation in all isolates and revealed subspecies-specific differences in GH content between pre- and post-weaning strains. Moreover, we observed differences in the number of genes belonging to the most abundant GH families (e.g. GH43) between breast-fed and formula-fed strains at different dietary stages, which can be linked to nutrient availability. Surprisingly, we computationally and phenotypically identified closely related weaning and post-weaning B. longum strains capable of metabolizing HMOs (i.e. 2′-FL) in a formula-fed baby that only received standard non-supplemented (i.e. no prebiotics or synthetic HMOs) formula. The analysis of SNP variants in genes identified as glycosyl hydrolases predicted the presence of missense mutations in both B. longum and B. infantis strains. Given that some missense variants can compromise protein function (Miosge et al., 2015), our results suggest potential functional differences that could further explain intra-strain and intra-individual carbohydrate metabolism profiles of B. longum. However, experimental validation would be essential to confirm the importance of variant predictions.
Recorded phenotypic data support the results of genomic analyses and further highlight differences in carbohydrate utilization profiles between and within B. longum and B. infantis. As highlighted above, the ability of B. infantis to grow on different HMOs may facilitate their early life establishment. Similarly, B. longum preference for plant-based nutrients may influence their ability to persist within individual hosts through significant dietary changes. Differential growth of strains that are genotypically similar on various carbohydrate substrates and the ability of formula-fed strains to metabolize selected HMOs suggest that Bifidobacterium possess an overall very broad repertoire of genes for carbohydrate acquisition and metabolism that may be differentially switched on and off in response to the presence of specific dietary components (Dworkin and Losick, 2001; Slager and Veening, 2016). Another explanation for these results may be a potential influence of the intra-individual environment on epigenetic mechanisms in these bacteria. One potential factor involved in this process may be a cooperative effort supported by cross-feeding activities among Bifidobacterium or between Bifidobacterium and other members of the early life microbiota, e.g. Bacteroides and Eubacterium species (Rios-Covian et al., 2013; Milani et al., 2015a; Schwab et al., 2017; Lawson et al., 2020). Indeed, the FISH analysis revealed the presence of bacteria detected by probes Bac303 (bacteroides) and ER482 (eubacterium) in fecal samples of both breast- and formula-fed infants, with intra-individual variation at different dietary stages. Although B. infantis is principally known as a specialist HMO degrader, we did note growth of one of the B. infantis strains from formula-fed baby FF1 on xylose. However, this growth profile was not consistent between experiments and therefore we did not pursue a fuller characterization. However, future examination of the ability of B. longum subsp. infantis to degrade a wider range of non-HMO carbohydrate sources in early life could provide additional insight into carbohydrate metabolism properties of this subspecies and its role in ecosystem structuring during transition to a more complex diet.
Glycan uptake analysis and proteomic investigation allowed us to determine mechanisms that selected B. longum strains to metabolize different carbohydrates. A common feature, based on the predicted activity of the most abundant proteins detected during grown on the three substrates (cellobiose, LNnT and 2′-FL), was that they were all imported and “selfishly” degraded intracellularly, therefore, limiting release of degradation products that could allow cross-feeding by other gut bacteria. This is in line with the carbohydrate uptake analysis, where no peak for cellobiose, LNnT, and 2′-FL degradation products could be detected. Cellobiose uptake in B_25 occurs via a mechanism similar to B. animalis subsp. lactis Bl-04 (B. lactis) (Andersen et al., 2013); cellobiose hydrolysis appears to be mediated by the activity of three intracellular β-glucosidases, although further confirmatory biochemical characterization of these enzyme is still required. B_25 was observed to utilize LNnT using a pathway similar to that described in B. longum subsp. infantis whereby LNnT is internalized via an ABC-transporter (B_25_00111-00113) followed by intracellular degradation into constituent monosaccharides by a GH112 (B_25_00114) and an N-acetylhexosamine 1-kinase (B_25_00115). LNnT degradation products are further metabolized to fructose-6-phosphate by activities that include B_25_00116-00117 (galactose-1-phosphate uridyltransferase, UDP-glucose 4-epimerase, involved in the Leloir pathway) and B_25_01030-01033 (for metabolism of N-acetylgalactosamine) prior to entering the Bifidobacterium genus-specific fructose-6-phosphate phosphoketolase (F6PPK) pathway (Ozcan and Sela, 2018). B_71 is predicted to deploy an ABC-transporter (B_71_00974-00976) that allows uptake of intact 2′-FL that is subsequently hydrolyzed to L-fucose and lactose by the two predicted intracellular α-fucosidases GH29 (B_71_00982) and GH95 (B_71_00983). L-fucose is further metabolized to L-lactate and pyruvate, via a pathway of non-phosphorylated intermediates that include activities of L-fucose mutarotase (B_71_00981), L-fucose dehydrogenase (B_71_00978), and L-fuconate hydrolase (B_71_00977) as previously described for B. longum subsp. longum SC596 (Garrido et al., 2016). Considering that the proteins encoded by the aforementioned genes are located in the cellobiose, LNnT and 2′-FL gene clusters that share high similarity and similar organization with those found in equivalent systems in other B. longum and B. lactis, it is reasonable to suggest that the gene clusters are related and may be the results of horizontal gene transfer events between B. longum/B. lactis members residing in the infant gut microbiota. Collectively, these data reflect inter- and intra-host phenotypic diversity of B. longum strains in terms of their carbohydrate degradation capabilities and suggest that intra-individual environment may influence epigenetic mechanisms in Bifidobacterium, resulting in differential growth on carbohydrate substrates.
In conclusion, this research provides new insight into distinct genomic and phenotypic abilities of B. longum species and strains isolated from the same individuals during the early life developmental window by demonstrating that subspecies- and strain-specific differences between members of B. longum sp. in infant hosts can be correlated to their adaptation at specific age and diet stages.
Limitations of the Study
Here, we used a combination of bioinformatic approaches and experimental techniques to assess genomic and phenotypic abilities of B. longum species and strains isolated during the early life developmental window. This study, however, is not without its limitations. One important caveat is the small number of B. infantis strains (n = 13) available for analysis and the fact that most of these strains (n = 11) were isolated from a single breast-fed baby (BF2). The examination of these strains provides important insight into the properties of B. infantis during the transition from breastfeeding to more diversified diet; however, it is difficult to assess how representative these results are of wider population. In addition, only one strain isolated from a formula-fed baby was identified as B. infantis, making it impossible to examine properties of members of this subspecies within this dietary group and make comparisons with breast-fed strains. Another important limitation is the fact that our strain collection only contains one bacterial strain per time point. Inclusion of additional strains could contribute further observations on inter-individual diversity of Bifidobacterium in infant hosts and their functional properties. To examine bacterial communities in fecal samples, we revisited and reanalyzed the data generated using FISH, but this technique has a detection limit (~106 bacterial cells (wet weight feces)−1) (Roger and Mccartney, 2010). Thus, FISH allows investigation of important bacterial groups, but fecal samples may contain several organisms at levels below the methodological detection threshold. In addition, this technique does not allow for tracking species-level changes. This limitation could be addressed by the use of comprehensive sequencing methods, such as shotgun metagenomics, combined with advanced computational methods to achieve strain-level resolution. Furthermore, phenotypic investigation of carbohydrate metabolism properties of B. longum revealed inconsistencies in growth of individual strains on certain carbohydrates, including LNnT, cellobiose, and mannose, and we therefore only explored reproducible findings further with proteomics. Previously, variability in growth of B. longum on mannose, even when analyzing the same strain (Bifidobacterium longum NCC2705) has been reported (Parche et al., 2007; Liu et al., 2011). Finally, no metadata on complementary foods during weaning and infant diet post-weaning were available. This information could allow bioinformatic predictions of carbohydrate degradation properties of B. longum to be related to the specific dietary components present in weaning infant foods. Future longitudinal studies could be designed to include these data.
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Lindsay J. Hall (lindsay.hall@quadram.ac.uk).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The draft genomes of 75 B. longum isolates have been deposited to GOLD database at https://img.jgi.doe.gov. The accession number for the draft genomes reported in this paper is GOLD: Gs0145337.
The proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the partner repository with dataset identifier (PRIDE). The accession number for the proteomics data reported in this paper is PRIDE: PXD017277.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We would like to thank Glycom A/S for the kind donation of purified HMOs: 2′-FL and LNnT. The authors would also like to thank Prof Rob Kingsley and Mr Shabhonam Caim for technical support and advice. This work was funded by a Wellcome Trust Investigator Award (no. 100/974/C/13/Z); a BBSRC Norwich Research Park Bioscience Doctoral Training grant no. BB/M011216/1 (supervisor LJH, student MK); an Institute Strategic Programme Gut Microbes and Health grant no. BB/R012490/1 and its constituent projects BBS/E/F/000PR10353 and BBS/E/F/000PR10356; and an Institute Strategic Programme Gut Health and Food Safety grant no. BB/J004529/1 to LJH. LH was in receipt of a Medical Research Council Intermediate Research Fellowship in Data Science (UK MED-BIO, grant no. MR/L01632X/1). PBP and SLLR are grateful for support from The Research Council of Norway (FRIPRO program, PBP: 250479), as well as the European Research Commission Starting Grant Fellowship (awarded to PBP; 336355 - MicroDE). The funding bodies did not contribute to the design of the study, collection, analysis, and interpretation of data or in writing the manuscript.
Author Contributions
LJH, LH, ALM, and MK designed the overall study. ALM provided the unique B. longum strain collection and extracted the DNA. ALM and LCR performed the FISH microbiota profiling studies. MK prepared the DNA for WGS, performed all genomic analysis and visualization, as well as growth studies. MK also reanalyzed the FISH data. SLLR, PBP, LJH, and MK planned metabolomics and proteomics studies. MK prepared samples for metabolomics and proteomics. SLLR and MK performed the metabolomics and proteomics experiments and SLLR analyzed and visualized the resulting data. LJH and MK analyzed the data, with input and discussion from LH and drafted the manuscript. SLLR, PBP, LH, and ALM provided further edits and co-writing of the final version. All authors read and approved the final manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: August 21, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101368.
Supplemental Information
COG categories legend, COG categories identified in all B. longum samples, as well as in specific groups: B. infantis, B. longum, isolates from breast-fed infants, isolates from formula-fed infants (separate tabs)
Strains (1%v/v) were grown in modified (m)MRS (pH 6.8) supplemented with cysteine HCl at 0.05% and 2% (w/v) of selected carbohydrates, except for pectin and mucin (1%). Experiments were performed in triplicates; the data are presented as difference in OD between T2 (30min) and Tend (48h) for growth on 12 selected carbohydrate sources and T2 (30min) and Tend (72h) for an additional experiment on cellobiose (separate tabs)
References
- Ahmed S., Luis A.S., Bras J.L., Ghosh A., Gautam S., Gupta M.N., Fontes C.M., Goyal A. A novel alpha-L-arabinofuranosidase of family 43 glycoside hydrolase (Ct43Araf) from Clostridium thermocellum. PLoS One. 2013;8:e73575. doi: 10.1371/journal.pone.0073575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen J.M., Barrangou R., Abou Hachem M., Lahtinen S.J., Goh Y.J., Svensson B., Klaenhammer T.R. Transcriptional analysis of oligosaccharide utilization by Bifidobacterium lactis Bl-04. BMC Genomics. 2013;14:312. doi: 10.1186/1471-2164-14-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arboleya S., Bottacini F., O’connell-Motherway M., Ryan C.A., Ross R.P., Van Sinderen D., Stanton C. Gene-trait matching across the Bifidobacterium longum pan-genome reveals considerable diversity in carbohydrate catabolism among human infant strains. BMC Genomics. 2018;19:33. doi: 10.1186/s12864-017-4388-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asnicar F., Manara S., Zolfo M., Truong D.T., Scholz M., Armanini F., Ferretti P., Gorfer V., Pedrotti A., Tett A., Segata N. Studying vertical microbiome transmission from mothers to infants by strain-level metagenomic profiling. mSystems. 2017;2 doi: 10.1128/mSystems.00164-16. e00164–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F., Roswall J., Peng Y., Feng Q., Jia H., Kovatcheva-Datchary P., Li Y., Xia Y., Xie H., Zhong H. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17:852. doi: 10.1016/j.chom.2015.05.012. [DOI] [PubMed] [Google Scholar]
- Bokulich N.A., Chung J., Battaglia T., Henderson N., Jay M., Li H.L., Lieber A.D., Wu F., Perez-Perez G.I., Chen Y. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci. Transl. Med. 2016;8:343ra82. doi: 10.1126/scitranslmed.aad7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broekaert W.F., Courtin C.M., Verbeke K., van de Wiele T., Verstraete W., Delcour J.A. Prebiotic and other health-related effects of cereal-derived arabinoxylans, arabinoxylan-oligosaccharides, and xylooligosaccharides. Crit. Rev. Food Sci. Nutr. 2011;51:178–194. doi: 10.1080/10408390903044768. [DOI] [PubMed] [Google Scholar]
- Chaplin A.V., Efimov B.A., Smeianov V.V., Kafarskaia L.I., Pikina A.P., Shkoporov A.N. Intraspecies genomic diversity and long-term persistence of Bifidobacterium longum. PLoS One. 2015;10:e0135658. doi: 10.1371/journal.pone.0135658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das U.N. Breastfeeding prevents type 2 diabetes mellitus: but, how and why? Am. J. Clin. Nutr. 2007;85:1436–1437. doi: 10.1093/ajcn/85.5.1436. [DOI] [PubMed] [Google Scholar]
- de Aguero M.G., Ganal-Vonarburg S.C., Fuhrer T., Rupp S., Uchimura Y., Li H., Steinert A., Heikenwalder M., Hapfelmeier S., Sauer U. The maternal microbiota drives early postnatal innate immune development. Science. 2016;351:1296–1301. doi: 10.1126/science.aad2571. [DOI] [PubMed] [Google Scholar]
- Dogra S., Sakwinska O., Soh S.E., Ngom-Bru C., Bruck W.M., Berger B., Brussow H., Lee Y.S., Yap F., Chong Y.S. Dynamics of infant gut microbiota are influenced by delivery mode and gestational duration and are associated with subsequent adiposity. mBio. 2015;6 doi: 10.1128/mBio.02419-14. e02419–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin J., Losick R. Linking nutritional status to gene activation and development. Genes Dev. 2001;15:1051–1054. doi: 10.1101/gad.892801. [DOI] [PubMed] [Google Scholar]
- Feng Q., Liang S.S., Jia H.J., Stadlmayr A., Tang L.Q., Lan Z., Zhang D.Y., Xia H.H., Xu X.Y., Jie Z.Y. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat. Commun. 2015;6:6528. doi: 10.1038/ncomms7528. [DOI] [PubMed] [Google Scholar]
- Florindo R.N., Souza V.P., Manzine L.R., Camilo C.M., Marana S.R., Polikarpov I., Nascimento A.S. Structural and biochemical characterization of a GH3 beta-glucosidase from the probiotic bacteria Bifidobacterium adolescentis. Biochimie. 2018;148:107–115. doi: 10.1016/j.biochi.2018.03.007. [DOI] [PubMed] [Google Scholar]
- Forbes J.D., Azad M.B., Vehling L., Tun H.M., Konya T.B., Guttman D.S., Field C.J., Lefebvre D., Sears M.R., Becker A.B. Association of exposure to formula in the hospital and subsequent infant feeding practices with gut microbiota and risk of overweight in the first year of life. Jama Pediatr. 2018;172:e181161. doi: 10.1001/jamapediatrics.2018.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido D., Barile D., Mills D.A. A molecular basis for bifidobacterial enrichment in the infant gastrointestinal tract. Adv. Nutr. 2012;3:415S–421S. doi: 10.3945/an.111.001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido D., Ruiz-Moyano S., Kirmiz N., Davis J.C., Totten S.M., Lemay D.G., Ugalde J.A., German J.B., Lebrilla C.B., Mills D.A. A novel gene cluster allows preferential utilization of fucosylated milk oligosaccharides in Bifidobacterium longum subsp longum SC596. Sci. Rep. 2016;6:35045. doi: 10.1038/srep35045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido D., Ruiz-Moyano S., Lemay D.G., Sela D.A., German J.B., Mills D.A. Comparative transcriptomics reveals key differences in the response to milk oligosaccharides of infant gut-associated bifidobacteria. Sci. Rep. 2015;5:13517. doi: 10.1038/srep13517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hald S., Schioldan A.G., Moore M.E., Dige A., Laerke H.N., Agnholt J., Knudsen K.E.B., Hermansen K., Marco M.L., Gregersen S., Dahlerup J.F. Effects of arabinoxylan and resistant starch on intestinal microbiota and short-chain fatty acids in subjects with metabolic syndrome: a randomised crossover study. PLoS One. 2016;11:e0159223. doi: 10.1371/journal.pone.0159223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikkila M.P., Saris P.E. Inhibition of Staphylococcus aureus by the commensal bacteria of human milk. J. Appl. Microbiol. 2003;95:471–478. doi: 10.1046/j.1365-2672.2003.02002.x. [DOI] [PubMed] [Google Scholar]
- Hinz S.W., Pastink M.I., van den Broek L.A., Vincken J.P., Voragen A.G. Bifidobacterium longum endogalactanase liberates galactotriose from type I galactans. Appl. Environ. Microbiol. 2005;71:5501–5510. doi: 10.1128/AEM.71.9.5501-5510.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinose H., Yoshida M., Fujimoto Z., Kaneko S. Characterization of a modular enzyme of exo-1,5-alpha-L-arabinofuranosidase and arabinan binding module from Streptomyces avermitilis NBRC14893. Appl. Microbiol. Biotechnol. 2008;80:399–408. doi: 10.1007/s00253-008-1551-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip S., Chung M., Raman G., Chew P., Magula N., Devine D., Trikalinos T., Lau J. Breastfeeding and maternal and infant health outcomes in developed countries. Evid. Rep. Technol. Assess. (Full Rep.) 2007;153:1–186. [PMC free article] [PubMed] [Google Scholar]
- James K., Motherway M.O., Bottacini F., van Sinderen D. Bifidobacterium breve UCC2003 metabolises the human milk oligosaccharides lacto-N-tetraose and lacto-N-neo-tetraose through overlapping, yet distinct pathways. Sci. Rep. 2016;6:38560. doi: 10.1038/srep38560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama T. Host-derived glycans serve as selected nutrients for the gut microbe: human milk oligosaccharides and bifidobacteria. Biosci. Biotechnol. Biochem. 2016;80:621–632. doi: 10.1080/09168451.2015.1132153. [DOI] [PubMed] [Google Scholar]
- Kitaoka M. Bifidobacterial enzymes involved in the metabolism of human milk oligosaccharides. Adv. Nutr. 2012;3:422S–429S. doi: 10.3945/an.111.001420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig J.E., Spor A., Scalfone N., Fricker A.D., Stombaugh J., Knight R., Angenent L.T., Ley R.E. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. U S A. 2011;108:4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson M.A.E., O'neill I.J., Kujawska M., Gowrinadh Javvadi S., Wijeyesekera A., Flegg Z., Chalklen L., Hall L.J. Breast milk-derived human milk oligosaccharides promote Bifidobacterium interactions within a single ecosystem. ISME J. 2020;14:635–648. doi: 10.1038/s41396-019-0553-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lax S., Smith D.P., Hampton-Marcell J., Owens S.M., Handley K.M., Scott N.M., Gibbons S.M., Larsen P., Shogan B.D., Weiss S. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science. 2014;345:1048–1052. doi: 10.1126/science.1254529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Wang S., Xu B., Guo Y., Zhao J., Liu W., Sun Z., Shao C., Wei X., Jiang Z. Proteomics analysis of Bifidobacterium longum NCC2705 growing on glucose, fructose, mannose, xylose, ribose, and galactose. Proteomics. 2011;11:2628–2638. doi: 10.1002/pmic.201100035. [DOI] [PubMed] [Google Scholar]
- Luis A.S., Briggs J., Zhang X., Farnell B., Ndeh D., Labourel A., Basle A., Cartmell A., Terrapon N., Stott K. Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides. Nat. Microbiol. 2018;3:210–219. doi: 10.1038/s41564-017-0079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magne F., Hachelaf W., Suau A., Boudraa G., Mangin I., Touhami M., Bouziane-Nedjadi K., Pochart P. A longitudinal study of infant faecal microbiota during weaning. FEMS Microbiol. Ecol. 2006;58:563–571. doi: 10.1111/j.1574-6941.2006.00182.x. [DOI] [PubMed] [Google Scholar]
- Makino H., Kushiro A., Ishikawa E., Kubota H., Gawad A., Sakai T., Oishi K., Martin R., Ben-Amor K., Knol J., Tanaka R. Mother-to-infant transmission of intestinal bifidobacterial strains has an impact on the early development of vaginally delivered infant's microbiota. PLoS One. 2013;8:e78331. doi: 10.1371/journal.pone.0078331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino H., Kushiro A., Ishikawa E., Muylaert D., Kubota H., Sakai T., Oishi K., Martin R., Ben Amor K., Oozeer R. Transmission of intestinal Bifidobacterium longum subsp. longum strains from mother to infant, determined by multilocus sequencing typing and amplified fragment length polymorphism. Appl. Environ. Microbiol. 2011;77:6788–6793. doi: 10.1128/AEM.05346-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado-Gomez M.X., Martinez I., Bottacini F., O'callaghan A., Ventura M., van Sinderen D., Hillmann B., Vangay P., Knights D., Hutkins R.W., Walter J. Stable engraftment of Bifidobacterium longum AH1206 in the human gut depends on individualized features of the resident microbiome. Cell Host Microbe. 2016;20:515–526. doi: 10.1016/j.chom.2016.09.001. [DOI] [PubMed] [Google Scholar]
- Marcobal A., Sonnenburg J.L. Human milk oligosaccharide consumption by intestinal microbiota. Clin. Microbiol. Infect. 2012;18:12–15. doi: 10.1111/j.1469-0691.2012.03863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R., Makino H., Cetinyurek Yavuz A., Ben-Amor K., Roelofs M., Ishikawa E., Kubota H., Swinkels S., Sakai T., Oishi K. Early-life events, including mode of delivery and type of feeding, siblings and gender, shape the developing gut microbiota. PLoS One. 2016;11:e0158498. doi: 10.1371/journal.pone.0158498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattarelli P., Bonaparte C., Pot B., Biavati B. Proposal to reclassify the three biotypes of Bifidobacterium longum as three subspecies: Bifidobacterium longum subsp. longum subsp. nov., Bifidobacterium longum subsp. infantis comb. nov. and Bifidobacterium longum subsp. suis comb. nov. Int. J. Syst. Evol. Microbiol. 2008;58:767–772. doi: 10.1099/ijs.0.65319-0. [DOI] [PubMed] [Google Scholar]
- Mckeen S., Young W., Mullaney J., Fraser K., Mcnabb W.C., Roy N.C. Infant complementary feeding of prebiotics for the microbiome and immunity. Nutrients. 2019;11:364. doi: 10.3390/nu11020364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami K., Kimura M., Takahashi H. Influence of maternal bifidobacteria on the development of gut bifidobacteria in infants. Pharmaceuticals (Basel) 2012;5:629–642. doi: 10.3390/ph5060629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani C., Lugli G.A., Duranti S., Turroni F., Mancabelli L., Ferrario C., Mangifesta M., Hevia A., Viappiani A., Scholz M. Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut. Sci. Rep. 2015;5:15782. doi: 10.1038/srep15782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani C., Mancabelli L., Lugli G.A., Duranti S., Turroni F., Ferrario C., Mangifesta M., Viappiani A., Ferretti P., Gorfer V. Exploring vertical transmission of bifidobacteria from mother to child. Appl. Environ. Microbiol. 2015;81:7078–7087. doi: 10.1128/AEM.02037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani C., Turroni F., Duranti S., Lugli G.A., Mancabelli L., Ferrario C., van Sinderen D., Ventura M. Genomics of the genus Bifidobacterium reveals species-specific adaptation to the glycan-rich gut environment. Appl. Environ. Microbiol. 2016;82:980–991. doi: 10.1128/AEM.03500-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills S., Stanton C., Lane J.A., Smith G.J., Ross R.P. Precision nutrition and the microbiome, Part I: current state of the science. Nutrients. 2019;11:923. doi: 10.3390/nu11040923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miosge L.A., Field M.A., Sontani Y., Cho V., Johnson S., Palkova A., Balakishnan B., Liang R., Zhang Y., Lyon S. Comparison of predicted and actual consequences of missense mutations. Proc. Natl. Acad. Sci. U S A. 2015;112:E5189–E5198. doi: 10.1073/pnas.1511585112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'sullivan A., Farver M., Smilowitz J.T. The influence of early infant-feeding practices on the intestinal microbiome and body composition in infants. Nutr. Metab. Insights. 2015;8:1–9. doi: 10.4137/NMI.S29530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odamaki T., Bottacini F., Kato K., Mitsuyama E., Yoshida K., Horigome A., Xiao J.Z., Van Sinderen D. Genomic diversity and distribution of Bifidobacterium longum subsp longum across the human lifespan. Sci. Rep. 2018;8:85. doi: 10.1038/s41598-017-18391-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oki K., Akiyama T., Matsuda K., Gawad A., Makino H., Ishikawa E., Oishi K., Kushiro A., Fujimoto J. Long-term colonization exceeding six years from early infancy of Bifidobacterium longum subsp. longum in human gut. BMC Microbiol. 2018;18:209. doi: 10.1186/s12866-018-1358-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszak T., An D.D., Zeissig S., Vera M.P., Richter J., Franke A., Glickman J.N., Siebert R., Baron R.M., Kasper D.L., Blumberg R.S. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Garcia J.A., Kloosterman N., Alvarez L., Tobarra-Sanchez E., Carceles-Alvarez A., Pastor-Valero R., Lopez-Hernandez F.A., Sanchez-Solis M., Claudio L. Full breastfeeding and obesity in children: a prospective study from birth to 6 years. Child Obes. 2018;14:327–337. doi: 10.1089/chi.2017.0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan E., Sela D.A. Inefficient metabolism of the human milk oligosaccharides lacto-N-tetraose and lacto-N-neotetraose shifts Bifidobacterium longum subsp. infantis physiology. Front. Nutr. 2018;5:46. doi: 10.3389/fnut.2018.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C., Bik E.M., Digiulio D.B., Relman D.A., Brown P.O. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parche S., Amon J., Jankovic I., Rezzonico E., Beleut M., Barutcu H., Schendel I., Eddy M.P., Burkovski A., Arigoni F., Titgemeyer F. Sugar transport systems of Bifidobacterium longum NCC2705. J. Mol. Microbiol. Biotechnol. 2007;12:9–19. doi: 10.1159/000096455. [DOI] [PubMed] [Google Scholar]
- Pokusaeva K., Fitzgerald G.F., van Sinderen D. Carbohydrate metabolism in bifidobacteria. Genes Nutr. 2011;6:285–306. doi: 10.1007/s12263-010-0206-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzo T., Pasten J.L., Karlsson E.N., Logan D.T. Structural and functional analyses of beta-glucosidase 3B from Thermotoga neapolitana: a thermostable three-domain representative of glycoside hydrolase 3. J. Mol. Biol. 2010;397:724–739. doi: 10.1016/j.jmb.2010.01.072. [DOI] [PubMed] [Google Scholar]
- Renz H., Brandtzaeg P., Hornef M. The impact of perinatal immune development on mucosal homeostasis and chronic inflammation. Nat. Rev. Immunol. 2012;12:9–23. doi: 10.1038/nri3112. [DOI] [PubMed] [Google Scholar]
- Rios-Covian D., Arboleya S., Hernandez-Barranco A.M., Alvarez-Buylla J.R., Ruas-Madiedo P., Gueimonde M., de Los Reyes-Gavilan C.G. Interactions between Bifidobacterium and Bacteroides species in cofermentations are affected by carbon sources, including exopolysaccharides produced by bifidobacteria. Appl. Environ. Microbiol. 2013;79:7518–7524. doi: 10.1128/AEM.02545-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riviere A., Selak M., Lantin D., Leroy F., de Vuyst L. Bifidobacteria and butyrate-producing colon bacteria: importance and strategies for their stimulation in the human gut. Front. Microbiol. 2016;7:979. doi: 10.3389/fmicb.2016.00979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberfroid M.B. Inulin-type fructans: functional food ingredients. J. Nutr. 2007;137:2493S–2502S. doi: 10.1093/jn/137.11.2493S. [DOI] [PubMed] [Google Scholar]
- Roger L.C., Costabile A., Holland D.T., Hoyles L., Mccartney A.L. Examination of faecal Bifidobacterium populations in breast- and formula-fed infants during the first 18 months of life. Microbiol. SGM. 2010;156:3329–3341. doi: 10.1099/mic.0.043224-0. [DOI] [PubMed] [Google Scholar]
- Roger L.C., Mccartney A.L. Longitudinal investigation of the faecal microbiota of healthy full-term infants using fluorescence in situ hybridization and denaturing gradient gel electrophoresis. Microbiol. SGM. 2010;156:3317–3328. doi: 10.1099/mic.0.041913-0. [DOI] [PubMed] [Google Scholar]
- Schwab C., Ruscheweyh H.J., Bunesova V., Pham V.T., Beerenwinkel N., Lacroix C. Trophic interactions of infant bifidobacteria and eubacterium hallii during L-fucose and fucosyllactose degradation. Front. Microbiol. 2017;8:95. doi: 10.3389/fmicb.2017.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela D.A., Chapman J., Adeuya A., Kim J.H., Chen F., Whitehead T.R., Lapidus A., Rokhsar D.S., Lebrilla C.B., German J.B. The genome sequence of Bifidobacterium longum subsp infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl. Acad. Sci. U S A. 2008;105:18964–18969. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela D.A., Garrido D., Lerno L., Wu S., Tan K., Eom H.J., Joachimiak A., Lebrilla C.B., Mills D.A. Bifidobacterium longum subsp. infantis ATCC 15697 alpha-fucosidases are active on fucosylated human milk oligosaccharides. Appl. Environ. Microbiol. 2012;78:795–803. doi: 10.1128/AEM.06762-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela D.A., Mills D.A. Nursing our microbiota: molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol. 2010;18:298–307. doi: 10.1016/j.tim.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y., Forster S.C., Tsaliki E., Vervier K., Strang A., Simpson N., Kumar N., Stares M.D., Rodger A., Brocklehurst P. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature. 2019;574:117–121. doi: 10.1038/s41586-019-1560-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivan A., Corrales L., Hubert N., Williams J.B., Aquino-Michaels K., Earley Z.M., Benyamin F.W., Lei Y.M., Jabri B., Alegre M.L. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slager J., Veening J.W. Hard-wired control of bacterial processes by chromosomal gene location. Trends Microbiol. 2016;24:788–800. doi: 10.1016/j.tim.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiemsma L.T., Michels K.B. The role of the microbiome in the developmental origins of health and disease. Pediatrics. 2018;141:e20172437. doi: 10.1542/peds.2017-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Murakami A., Yoshida K. Motif-guided identification of a glycoside hydrolase family 1 alpha-L-arabinofuranosidase in Bifidobacterium adolescentis. Biosci. Biotechnol. Biochem. 2013;77:1709–1714. doi: 10.1271/bbb.130279. [DOI] [PubMed] [Google Scholar]
- Tang W.H.W., Kitai T., Hazen S.L. Gut microbiota in cardiovascular health and disease. Circ. Res. 2017;120:1183–1196. doi: 10.1161/CIRCRESAHA.117.309715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thongaram T., Hoeflinger J.L., Chow J., Miller M.J. Human milk oligosaccharide consumption by probiotic and human-associated bifidobacteria and lactobacilli. J. Dairy Sci. 2017;100:7825–7833. doi: 10.3168/jds.2017-12753. [DOI] [PubMed] [Google Scholar]
- Turnbaugh P.J., Ley R.E., Mahowald M.A., Magrini V., Mardis E.R., Gordon J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Turroni F., Foroni E., Pizzetti P., Giubellini V., Ribbera A., Merusi P., Cagnasso P., Bizzarri B., De'angelis G.L., Shanahan F. Exploring the diversity of the bifidobacterial population in the human intestinal tract. Appl. Environ. Microbiol. 2009;75:1534–1545. doi: 10.1128/AEM.02216-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turroni F., Peano C., Pass D.A., Foroni E., Severgnini M., Claesson M.J., Kerr C., Hourihane J., Murray D., Fuligni F. Diversity of bifidobacteria within the infant gut microbiota. PLoS One. 2012;7:e36957. doi: 10.1371/journal.pone.0036957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatanen T., Plichta D.R., Somani J., Munch P.C., Arthur T.D., Hall A.B., Rudolf S., Oakeley E.J., Ke X., Young R.A. Genomic variation and strain-specific functional adaptation in the human gut microbiome during early life. Nat. Microbiol. 2019;4:470–479. doi: 10.1038/s41564-018-0321-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M., O'flaherty S., Claesson M.J., Turroni F., Klaenhammer T.R., van Sinderen D., O'toole P.W. Genome-scale analyses of health-promoting bacteria: probiogenomics. Nat. Rev. Microbiol. 2009;7:61–71. doi: 10.1038/nrmicro2047. [DOI] [PubMed] [Google Scholar]
- Viborg A.H., Sorensen K.I., Gilad O., Steen-Jensen D.B., Dilokpimol A., Jacobsen S., Svensson B. Biochemical and kinetic characterisation of a novel xylooligosaccharide-upregulated GH43 beta-D-xylosidase/alpha-L-arabinofuranosidase (BXA43) from the probiotic Bifidobacterium animalis subsp lactis BB-12. AMB Express. 2013;3:56. doi: 10.1186/2191-0855-3-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wampach L., Heintz-Buschart A., Hogan A., Muller E.E.L., Narayanasamy S., Laczny C.C., Hugerth L.W., Bindl L., Bottu J., Andersson A.F. Colonization and succession within the human gut microbiome by Archaea, bacteria, and Microeukaryotes during the first year of life. Front. Microbiol. 2017;8:738. doi: 10.3389/fmicb.2017.00738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson D., Motherway M.O., Schoterman M.H.C., van Neerven R.J.J., Nauta A., Van Sinderen D. Selective carbohydrate utilization by lactobacilli and bifidobacteria. J. Appl. Microbiol. 2013;114:1132–1146. doi: 10.1111/jam.12105. [DOI] [PubMed] [Google Scholar]
- Yamada C., Gotoh A., Sakanaka M., Hattie M., Stubbs K.A., Katayama-Ikegami A., Hirose J., Kurihara S., Arakawa T., Kitaoka M. Molecular insight into evolution of symbiosis between breast-fed infants and a member of the human gut microbiome Bifidobacterium longum. Cell Chem. Biol. 2017;24:515–524.e5. doi: 10.1016/j.chembiol.2017.03.012. [DOI] [PubMed] [Google Scholar]
- Yan S., Wei P.C., Chen Q., Chen X., Wang S.C., Li J.R., Gao C. Functional and structural characterization of a beta-glucosidase involved in saponin metabolism from intestinal bacteria. Biochem. Biophys. Res. Commun. 2018;496:1349–1356. doi: 10.1016/j.bbrc.2018.02.018. [DOI] [PubMed] [Google Scholar]
- Yanokura E., Oki K., Makino H., Modesto M., Pot B., Mattarelli P., Biavati B., Watanabe K. Subspeciation of Bifidobacterium longum by multilocus approaches and amplified fragment length polymorphism: description of B. longum subsp. suillum subsp. nov., isolated from the faeces of piglets. Syst. Appl. Microbiol. 2015;38:305–314. doi: 10.1016/j.syapm.2015.05.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
COG categories legend, COG categories identified in all B. longum samples, as well as in specific groups: B. infantis, B. longum, isolates from breast-fed infants, isolates from formula-fed infants (separate tabs)
Strains (1%v/v) were grown in modified (m)MRS (pH 6.8) supplemented with cysteine HCl at 0.05% and 2% (w/v) of selected carbohydrates, except for pectin and mucin (1%). Experiments were performed in triplicates; the data are presented as difference in OD between T2 (30min) and Tend (48h) for growth on 12 selected carbohydrate sources and T2 (30min) and Tend (72h) for an additional experiment on cellobiose (separate tabs)
Data Availability Statement
The draft genomes of 75 B. longum isolates have been deposited to GOLD database at https://img.jgi.doe.gov. The accession number for the draft genomes reported in this paper is GOLD: Gs0145337.
The proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the partner repository with dataset identifier (PRIDE). The accession number for the proteomics data reported in this paper is PRIDE: PXD017277.