Abstract
Perception of sound is initiated by mechanically gated ion channels at the tips of stereocilia. Mature mammalian auditory hair cells require transmembrane channel-like 1 (TMC1) for mechanotransduction, and mutations of the cognate genetic sequences result in dominant or recessive heritable deafness forms in humans and mice. In contrast, zebrafish lateral line hair cells, which detect water motion, require Tmc2a and Tmc2b. Here, we use standard and multiplex genome editing in conjunction with functional and behavioral assays to determine the reliance of zebrafish hearing and vestibular organs on Tmc proteins. Surprisingly, our approach using multiple mutant alleles demonstrates that hearing in zebrafish is not dependent on Tmc1, nor is it fully dependent on Tmc2a and Tmc2b. Hearing however is absent in triple-mutant zebrafish that lack Tmc1, Tmc2a and Tmc2b. These outcomes reveal a striking resemblance of Tmc protein reliance in the vestibular sensory epithelia of mammals to the maculae of zebrafish. Moreover, our findings disclose a logic of Tmc use where hearing depends on a complement of Tmc proteins beyond those employed to sense water motion.
Introduction
Generating an internal representation of the external world requires an assortment of sensory cells (1–3) and sensory transduction proteins (4–6). When stimuli are very different, such as electromagnetic radiation versus chemical molecules, entirely different cell types are required to capture and encode them (7). In contrast, when stimuli are similar but meaningful to distinguish for organismal survival, evolutionary mechanisms have allowed some sensory systems to diversify the proteins between cells to receive and encode information instead of generating an entirely new cell type for each related stimulus (4–6).
Hair cells are sensory receptors that encode diverse mechanical stimuli. In mammals, cochlear hair cells transduce sound stimuli into electrical responses; however, in zebrafish, hair cells of the maculae transduce both sound stimuli and linear accelerations. In the zebrafish ear, hair cells encode pressure waves with a frequency range of 100–4000 Hz (8,9). Larval zebrafish have a fully functioning auditory system (10) with a similar over-all neural architecture to that of mammals (11). The larval zebrafish has two maculae: the anterior macula, or utricle, and the posterior macula, or saccule. During the first week of development, the posterior macula plays a larger role in hearing than does the anterior macula, but both maculae seem to participate (9,12). In contrast, hair cells of the lateral line, which are housed in neuromasts, detect a different régime of mechanical stimuli to serve as water motion detectors, sensing velocity, acceleration or water vibrations (13–15). If and how hair cells vary at the molecular level in different systems is an important question.
Transmembrane channel-like (Tmc) proteins are an evolutionarily conserved family whose members can participate in mechanotransduction in vertebrates and invertebrates (Supplementary Material, Figure S1) (16–19). Insight into TMC1’s involvement in human deafness initially came through investigation of two consanguineous families from India that had members with nonsyndromic hearing impairment. Initial studies identified a recessive deafness form linked to 9q11–q21 to define the locus DFNB7 (20). This segment of the human genome is syntenic to a region of the mouse genome that is known to contain a deafness locus called deafness (dn), which is associated with a recessive form of heritable deafness (21,22). Using positional cloning, the gene mutated in DFNB7/11, dn, and DFNA36 (a dominant, nonsyndromic deafness form found within the same interval as DFNB7/11) was identified as TMC1 (23). Mouse and human genomes each contain a related Tmc1 paralog termed Tmc2. The zebrafish genome contains three Tmc1 and Tmc2 orthologs, tmc1, tmc2a and tmc2b, and each of the cognate mRNAs is present in the larval and adult ear (24).
In mammals, TMC1 and TMC2 proteins localize to the sites of transduction at the tips of stereocilia (25,26). The mechanoelectrical transduction channel properties are changed by mutation in Tmc1 (27,28). Mutational and structural studies of TMC1 support the hypothesis that this protein is a pore-forming component of sensory transduction channels in mammalian auditory hair cells (29,30). Moreover, it was recently demonstrated that TMC1 from green sea turtles and TMC2 from parakeets are pore-forming subunits of mechanosensitive ion channels in vitro (31). In addition, an increasing expression gradient of TMC1 protein from the murine cochlear apex to the base is a major factor in the apical-basal gradient in channel conductance (32).
Recently, we demonstrated that Tmc2a and Tmc2b are necessary for normal mechanotransduction in the lateral line system of zebrafish (18). Here, we investigate the requirement of Tmcs for hearing in zebrafish to answer two salient questions. First, do all vertebrates require Tmc1 for auditory hair cell function? Second, do hair cells that encode mechanical information for different sensory systems use the same set of Tmc proteins? By answering these questions, we gain insight into the Tmc proteins used by auditory hair cells and how hair cells in different sensory systems may be modified to receive and encode diverse stimuli.
Results
Tmc1 is not obligatory for hearing
Since hearing in humans (23) and mice (21,33) is dependent on TMC1 and the orthologous gene’s mRNA is present in the inner ear of zebrafish (24), we used transcription activator-like effector nuclease (TALEN)-mediated genome editing to generate a mutation in the 5th exon of tmc1 (Fig. 1A–E; Tables 1 and 2) to test if this gene is necessary for hearing in zebrafish and to address an essential question: do all vertebrates require Tmc1 to hear? The resulting frameshift mutation, tmc1cwr5, leads to an opal (UGA) stop codon that resides upstream of all but one of the predicted transmembrane domains, deleting 682 out of 990 residues and is expected to produce a nonfunctional protein. To assay for hearing, we used the acoustic startle reflex, also termed the C-start reflex, of larval zebrafish. Starting at 5-days post-fertilization (dpf), zebrafish larvae exhibit a stereotypical behavior in response to sudden and loud acoustic stimulation, the C-start reflex, where the zebrafish bend to form a ‘C’ shape and then dart away from the stimulus (34,35). All larval tmc1cwr5 mutants responded similarly to control animals in the C-start reflex assay, which was conducted using a high-speed camera collecting images at a frame rate of 1000 frames/s (Fig. 1G; Movies 1 and 2). Importantly, the movie demonstrates that the reflex was elicited soon after the stimulus was applied, but well before the waves of water that were also prompted by the stimulus reached the larva, suggesting that the water motion’s action on the lateral line was not what was evoking the response. To obtain additional support for these data, we generated a 2nd mutant allele in tmc1 using clustered regularly interspaced short palindromic repeats (CRISPR), tmc1cwr4. The product is predicted to lack the 605 C-terminal amino acids of the wild-type protein and be devoid of all but two transmembrane domains (Fig. 1A and F; Tables 1 and 2), rendering it nonfunctional. A normal startle reflex was observed for all tmc1cwr4 mutants (Fig. 1G; Movie 3). The schematics shown in Figure 1E and F depict the expected effects of the mutations on the protein products. However, unpredicted changes in splicing, such as exon skipping or use of cryptic splice sites that avoid a frameshift mutation, are possible (36). While either of these scenarios is conceivable, the use of two distinct mutations in different exons of the same gene diminishes the likelihood that bypassing of a frameshift can explain the results. To determine if mutation of tmc1 in either the tmc1cwr4 or the tmc1cwr5 mutants increased the expression of tmc2a or tmc2b, resulting in genetic compensation of mechanotransduction, we performed quantitative real-time PCR on mutant and control animals. Expression of tmc2a mRNA or tmc2b mRNA in the tmc1 mutants did not increase and was similar to controls (Supplementary Material, Figure S2). These results indicate that genetic compensation by tmc2a or tmc2b is not responsible for the C-start reflex observed in the tmc1 mutants.
Figure 1.
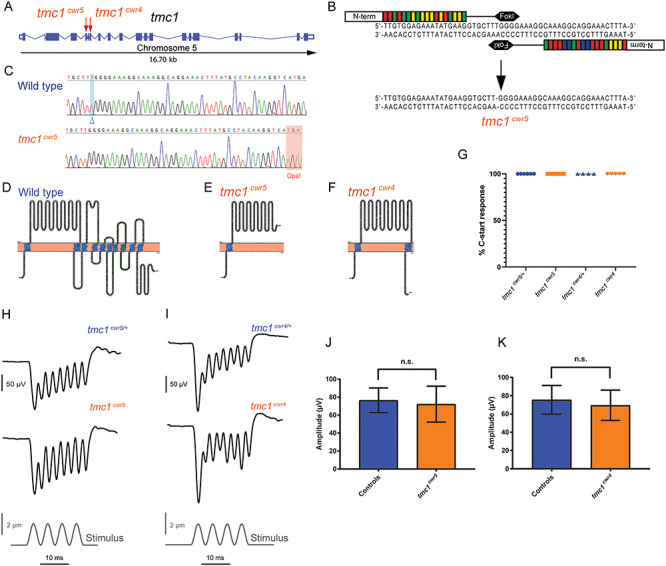
Mutation of tmc1 and behavioral and electrophysiological evaluation of mutants. (A) Schematic diagram illustrating the tmc1 genomic locus in zebrafish with exons and splice sites displayed. Red arrows mark the targeted exons, 5 and 7, and associated mutagenized alleles, tmc1cwr5 and tmc1cwr4, which were generated via TALEN or CRISPR, respectively. (B) Segment of exon 5 subjected to TALEN-mediated genome editing. Two recombinant TALENs associated with corresponding half-sites to enable FokI dimerization and DNA cleavage. Genomic editing deleted one nucleotide yielding a frameshift mutation. This mutant allele is termed tmc1cwr5. (C) Sequencing results of mutagenized and control loci. Blue highlight and blue delta indicate deleted nucleotide absent in the mutant. Red highlight denotes the opal mutation that was generated by TALEN targeting in tmc1cwr5. Topographical representations of wild-type Tmc1 protein (D) and mutated forms associated with alleles tmc1cwr5 (E) and tmc1cwr4 (F). Amino acids of putative transmembrane domains are labeled in blue, and the TMC domain is in green. (G) Graph of the mean percentages of mutant and control zebrafish that exhibited a C-start response when presented with vibrational stimuli ± standard error of the mean (SEM). Each data point represents the percentage of positive responses a larva had in a trial of 20 stimuli (ntmc1cwr5/+ = 6, ntmc1cwr5 = 9, ntmc1cwr4/+ = 4, ntmc1cwr4 = 5). Stimulus-evoked microphonic potentials measured from the otic vesicles of tmc1cwr5 (H), tmc1cwr4 (I) and control animals. Graphs of mean microphonic potentials from otic vesicles of tmc1cwr5 (J), tmc1cwr4 (K) and control animals. Mean potential of tmc1cwr5 ± SEM = 72 ± 7.1 μV (n = 8), mean potential of heterozygous siblings = 76 ± 4.8 μV (n = 8) (P = 0.3823); mean potential of tmc1cwr4 ± SEM = 69 ± 7.4 μV (n = 5), mean potential of wild-type or heterozygous siblings = 75 ± 6.4 μV (n = 6) (P = 0.9307). Statistical significance determined by Mann–Whitney test.
Table 1.
Genotypes
| Gene | Allele | Exon | Type | Target (5′ → 3′) | Genotype | DNA change | aa preserved/ total aa | DNA change description |
|---|---|---|---|---|---|---|---|---|
| tmc1 | cwr4 | 7 | Small deletion | TGGGCTGGTCATGGTTCCAG | tmc1cwr4 | 8-bp deleted | 385/990 | −8 bp at c.1155–1162 (−CATGGTTC) |
| cwr5 | 5 | Small deletion | Left: TTGTGGAGAAATATGAAG; Right: CAAAGGCAGGAAACTTTA | tmc1cwr5 | 1-bp deleted | 308/990 | −1 bp at c.926 (−T) | |
| tmc2a | cwr3 | 9 | Small deletion | AGGTCCCAATGCCCACCATG | tmc2acwr3 | 1-bp deleted/1-bp deleted | 349/916 | −1 bp at c.1050 (−C); −1 bp at c.1052 (−T) |
| cwr6 | Indel | tmc2acwr6 | 2-bp inserted/3-bp deleted | 350/916 | −3/+2 bp at c.1053–1055 (−GGT/+TG) | |||
| tmc2b | cwr1 | 3 | Small deletion | Left: TAGCAACACTTAGGAACA; Right: GGAGACTCAAGTGCTTGA | tmc2bcwr1 | 7-bp deleted | 145/892 | −7 bp at c.438–444 (−GCGCATG) |
| cwr2 | 7 | Small deletion | GGCCGAATCCTGCTCCTCTC | tmc2bcwr2 | 5-bp deleted | 266/892 | −5 bp at c.799–703 (−GAGGA) | |
| cwr8 | 6 | Small deletion | GGACTAAACCTTGTTCTCTT | tmc2bcwr8 | 5-bp deleted | 236/892 | −5 bp at c.710–714 (−TCTTC) |
Table 2.
Mutants
| Mutant fish | Allele | Putative Zebrafish International Resource Center (ZIRC) name |
|---|---|---|
| tmc1 single mutants | cwr4 | tmc1cwr4 |
| cwr5 | tmc1cwr5 | |
| tmc2a single mutant | cwr3 | tmc2acwr3 |
| tmc2b single mutant | cwr1 | tmc2bcwr1 |
| tmc2b tmc2a double mutant | cwr2 cwr3 | tmc2bcwr2 tmc2acwr3 |
| tmc2b tmc1 tmc2a triple mutant | cwr8 cwr4 cwr6 | tmc2bcwr8 tmc1cwr4 tmc2acwr6 |
One possible explanation for these surprising findings is that the zebrafish’s Tmc1 independence is transitory, and adults require Tmc1 to hear. This would be akin to what is observed in the mouse cochlea, where hair cells of the early postnatal mouse use TMC2 for mechanotransduction before gradually transitioning to TMC1 as the animal ages (19,27). To explicitly test this notion, we examined older zebrafish to determine if hearing is impacted in the adult tmc1cwr5 or tmc1cwr4 mutants, which are viable into adulthood. A normal C-start reflex was observed in the adult tmc1 mutants (Supplementary Material, Figure S3; Movies 4 and 5). In all, these observations suggest that Tmc1 is not required for hearing by larval or adult zebrafish, as determined by the acoustic startle reflex assay.
Next, we examined mechanotransduction in the ear of tmc1 mutant larvae by measuring stimulus-evoked microphonic potentials of the maculae (Fig. 1H–K). In the larval ear, the posterior macula contributes more to stimulus-evoked and sound-evoked microphonic potentials than does the anterior macula (9,12). To determine a larval age range of zebrafish where the electrophysiological characteristics of the maculae are relatively stable, we determined the mean microphonic potentials from 7 to 10 dpf and demonstrated that they were similar for wild-type zebrafish (Supplementary Material, Figure S4). The mean microphonic potentials of control fish and each tmc1 mutant (tmc1cwr5 or tmc1cwr4) were comparable (Fig. 1H–K). These findings indicate that Tmc1 is not a major contributor to mechanotransduction in the maculae, as the bulk of ions enter the hair cells without the need of this protein.
To affirm these data visually, we imaged the uptake of fluorescent molecule 4-Di-2-ASP (Fig. 2), a cationic styryl dye that passes through the hair cell’s mechanotransduction channel and can serve as a quantitative indicator of channel function (37,38). After direct injection of 4-Di-2-ASP into otocysts, live hair cells of anterior maculae of each tmc1 mutant (tmc1cwr5 or tmc1cwr4) and control zebrafish permitted passage of similar levels of 4-Di-2-ASP, supporting the conclusion that Tmc1 is not obligatory for mechanotransduction in the maculae. Comparable results were obtained when FM1-43FX, a fixable fluorescent molecule with the capacity to pass through the mechanotransduction channel (39,40), was injected into the ears of larval zebrafish with mutations in tmc1 and controls (Fig. 3). The results from our approach using two alleles are surprising since TMC1 has been suggested to form the mechanotransduction channel pore in vertebrate cochlear hair cells (29). These data demonstrate that Tmc1 is not required for the majority of zebrafish auditory hair cells to function but do not preclude Tmc1 from playing a subtle role in mechanotransduction.
Figure 2.
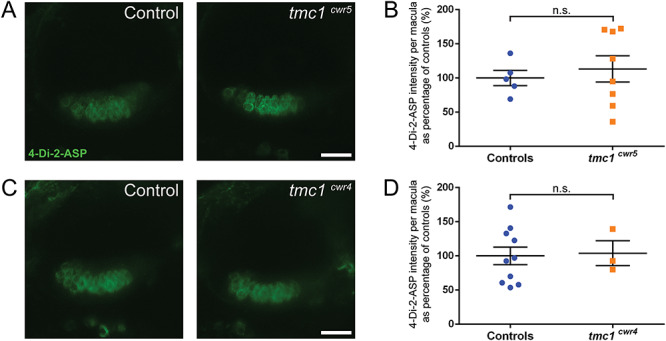
Tmc1 is not required for mechanotransduction in hair cells of the anterior macula as determined by 4-Di-2-ASP dye uptake. (A, C) Confocal micrographs of 4-Di-2-ASP fluorescence in hair cells of the anterior maculae of controls (left) and mutants (right). Controls are tmc1cwr5/+ (A) and wild-type larvae (C). (B, D) Plots of 4-Di-2-ASP fluorescence intensities of anterior maculae in tmc1 mutants. 6–8 dpf larvae were used. The control groups included both wild-type and heterozygous siblings. Each data point is the fluorescence intensity of one macula as a percentage of the average value of the control group. Mean ± SEM is displayed. (B) Intensity of tmc1cwr5 controls = 100% ± 11% (n = 5); intensity of tmc1cwr5 mutants = 113% ± 19% (n = 8). (D) Intensity of tmc1cwr4 controls = 100% ± 13% (n = 10); intensity of tmc1cwr4 mutants = 104% ± 18% (n = 3). Statistical significance determined by two-tailed unpaired t-test. Scale bar = 20 μm.
Figure 3.
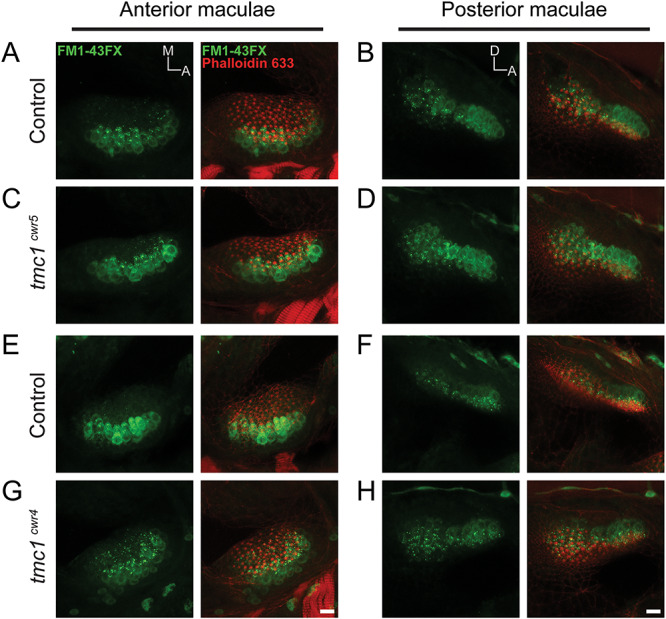
Tmc1 is not required for mechanotransduction in hair cells of the anterior and posterior maculae as determined by FM1-43FX dye entry. Confocal images of hair cells from anterior (A, C, E, G) and posterior (B, D, F, H) maculae of control (tmc1cwr5/+, A, B or tmc1cwr4/+, E, F) and experimental (tmc1cwr5, C, D or tmc1cwr4, G, H) larvae at 7–8 dpf loaded with FM1-43FX (green) dye and labeled with phalloidin (red). A, anterior; M, medial; D, dorsal. Scale bar = 10 μm.
Mutations of tmc2a and tmc2b result in attenuated mechanotransduction in maculae
We next asked whether hair cells that encode mechanical information for different sensory systems within the zebrafish use the same set of Tmc proteins. Since Tmc1 is not an absolute requirement for hearing in zebrafish, we searched for other candidate Tmcs that may be necessary for hearing. Recently, we demonstrated that mechanotransduction by the lateral line of larval zebrafish is dependent on Tmc2a and Tmc2b, with certain subgroups of hair cells absolutely dependent on Tmc2b and others requiring both Tmc2a and Tmc2b (18). This finding established that water motion detection relies on particular complements of Tmc2 proteins depending on the anatomical position of the hair cell on the surface of the animal and the direction the cell faces (18). However, hair cells of the lateral line serve a different function than those of the ear. The mechanical stimuli that impinge on neuromasts of the lateral line and those imposed on maculae are markedly different in duration and quality (8,13–15), and this variation may require different Tmc proteins.
To determine if the hair cells of the maculae absolutely require the same Tmc2 module of proteins (Tmc2a and Tmc2b) as that of the lateral line, whose transcripts are also present in the inner ear, we monitored the acoustic startle reflex of larvae that lacked functional versions of both of these proteins. Interestingly, the acoustic startle reflex of tmc2bcwr2 tmc2acwr3 double mutants (Tables 1 and 2) was inconsistent, with 39% of the provided stimuli being reacted to with a C-start reflex, whereas controls reacted to 100% of stimuli (Fig. 4A and C; Movies 6 and 7). Importantly, the startle reflex could have not been mediated by the lateral line because there is no detectable mechanotransduction in this system in these double mutants (18). This intriguing result suggests that mechanotransduction is functional, albeit attenuated, in the ear of tmc2 double mutants.
Figure 4.
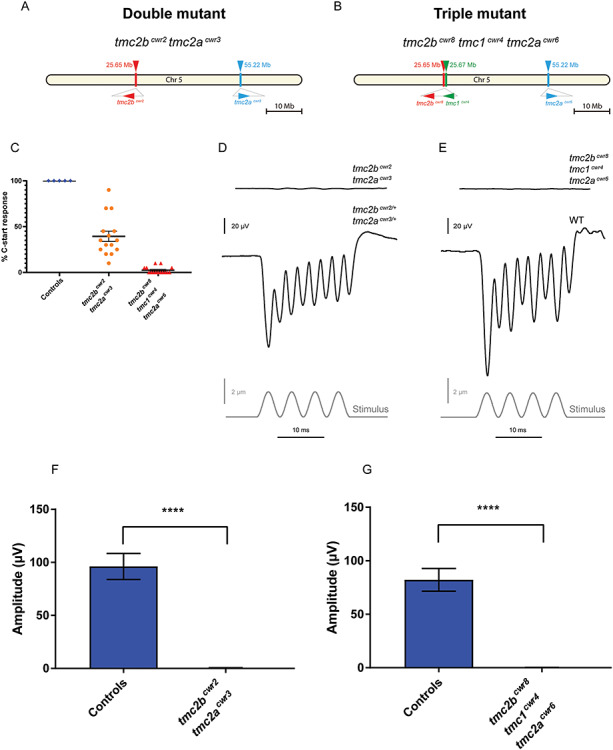
Behavioral and hair cell electrophysiological characteristics of tmc2bcwr2 tmc2acwr3 double and tmc2bcwr8 tmc1cwr4 tmc2acwr6 triple mutants. Schematics of chromosome 5 with relative positions of tmc genes and associated mutant alleles for the tmc2bcwr2 tmc2acwr3 double mutant (A) and the tmc2bcwr8 tmc1cwr4 tmc2acwr6 triple mutant (B). (C) Graph of the mean percentages of mutant and control zebrafish that exhibited a C-start response when presented with vibrational stimuli ± SEM. Each data point represents the percentage of positive responses a larva had in a trial of 20 stimuli (ntmc2bcwr2/+tmc2acwr3/+ = 5 (controls), ntmc2bcwr2tmc2acwr3 = 15, ntmc2bcwr8tmc1cwr4tmc2acwr6 = 15). Tmc2bcwr2 tmc2acwr3 double mutant = 39 ± 5.6%, tmc2bcwr8 tmc1cwr4 tmc2acwr6 triple mutant = 2.0 ± 0.96%. Representative recordings of microphonic potentials measured from the ears of (D) double- and (E) triple-mutant animals. (F, G) Graphs of mean microphonic potentials from otocysts of 7–10 dpf larvae are displayed. (F) Mean potential of tmc2bcwr2 tmc2acwr3 double mutants ± SEM = 0.42 ± 0.07 μV (n = 10), mean potential of heterozygotes (tmc2bcwr2/+ tmc2acwr3/+) = 96 ± 3.5 μV (n = 12). (G) No potentials were observed in tmc2bcwr8 tmc1cwr4 tmc2acwr6 triple mutants (n = 10), mean potential of wild-type controls = 82 ± 3.4 μV (n = 10). Mann–Whitney test, ****P < 0.0001.
To explore this hypothesis, we next probed the quality of mechanotransduction in the ear of the tmc2bcwr2 tmc2acwr3 double mutant (18) by measuring stimulus-evoked microphonic potentials from the otic vesicle. Surprisingly, we did not detect potentials of significant magnitude in the double mutant (Fig. 4D and F). Since the double-mutant zebrafish could respond in the acoustic startle reflex assay, though inconsistently, we thought the microphonic potentials may be too weak to detect or the recording electrode may not be proximal to any hair cells that still function, which could be obscured by the otolith that overlays most of the hair cells (41).
To search the maculae for hair cells that may have residual mechanotransduction capacity in the absence of Tmc2a and Tmc2b, we monitored the ears of double-mutant larvae by visualizing the uptake of cationic styryl dye 4-Di-2-ASP in hair cells after direct injection of the dye into the otic vesicles. We noted a ~2.4-fold reduction in fluorescence in the anterior maculae of tmc2bcwr2 tmc2acwr3 double mutants relative to controls (Fig. 5A and B). To obtain additional support for these findings, we introduced the fixable fluorescent molecule FM1-43FX into the ear. Similar results were observed for FM1-43FX uptake studies in tmc2bcwr2 tmc2acwr3 double mutants, in both anterior and posterior maculae (Fig. 5C and D). Overall, these results indicate that normal mechanotransduction in the maculae requires the Tmc2a–Tmc2b module. Reduced mechanotransduction in the double mutant signifies that hearing uses additional proteins beyond those of the lateral line where the double mutant shows no microphonic potential and no 4-Di-2-ASP or FM1-43FX uptake by neuromast hair cells (18).
Figure 5.
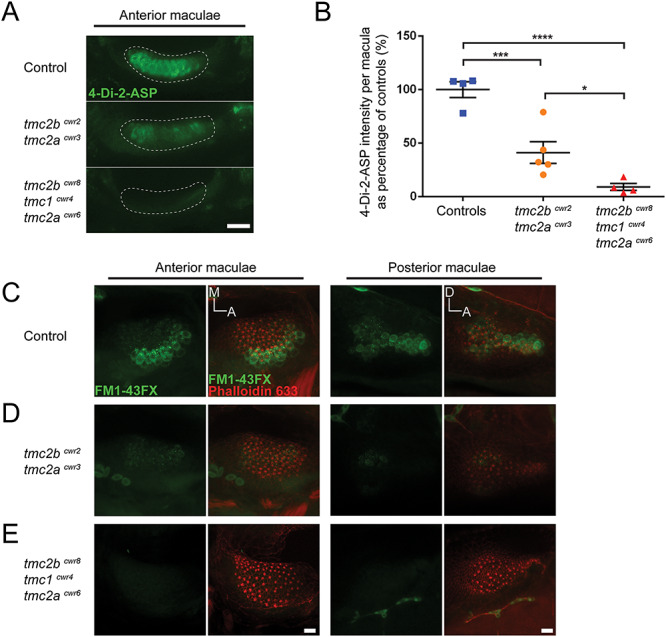
Loss of auditory mechanotransduction in the tmc2bcwr8 tmc1cwr4 tmc2acwr6 triple mutant but attenuation in the tmc2bcwr2 tmc2acwr3 double mutant as determined by fluorescent molecule entry. (A) Confocal micrographs of living hair cells from anterior maculae labeled with 4-Di-2-ASP to assess mechanotransduction function of control (tmc2bcwr8/+ tmc1cwr4/+ tmc2acwr6/+; Top), tmc2bcwr2 tmc2acwr3 double-mutant (Middle) and tmc2bcwr8 tmc1cwr4 tmc2acwr6 triple-mutant (Bottom) larvae at 8 dpf. (B) Graph of 4-Di-2-ASP fluorescence intensities of anterior maculae in double and triple mutants. 6–8 dpf larvae were used. Each data point is the fluorescence intensity of one macula as a percentage of the average value of the control group. Mean ± SEM is displayed. Tmc2bcwr2 tmc2acwr3 double mutants = 41 ± 10% (n = 5), tmc2bcwr8 tmc1cwr4 tmc2acwr6 triple mutants = 9.2 ± 3.3% (n = 4), tmc2bcwr8/+ tmc1cwr4/+ tmc2acwr6/+ control larvae = 100 ± 7.3% (n = 4). Significance determined by one-way ANOVA: ****P < 0.0001, ***P < 0.001 and *P < 0.05. (C–E) Micrographs for qualitative comparisons of FM1-43FX uptake (green) by hair cells of the anterior and posterior maculae: (C) control (tmc2bcwr8/+ tmc1cwr4/+ tmc2acwr6/+), (D) tmc2bcwr2 tmc2acwr3 double mutant and (E) tmc2bcwr8 tmc1cwr4 tmc2acwr6 triple mutant. F-actin of hair cells, including that of stereocilia, in maculae has been counter labeled with phalloidin (red) to reveal the total number of hair cells. A, anterior; M, medial; D, dorsal. Scale bar = 20 μm in A and 10 μm in E.
Mutation of tmc1, tmc2a and tmc2b results in deafness
The residual mechanotransduction observed in the maculae of tmc2bcwr2 tmc2acwr3 double mutants compelled us to identify the proteinaceous source of the diminished contribution. Our first candidate was Tmc1. Although we demonstrated it to have no major effect on hearing in the tmc1 single mutants (Fig. 1), Tmc1 could provide latent mechanotransduction function that had been masked by the presence of Tmc2a and Tmc2b.
To test the hypothesis that Tmc1 allows for residual mechanotransduction in the tmc2bcwr2 tmc2acwr3 double mutant, we edited all three genes simultaneously using multiplex genome editing (42,43) because they are located on the same chromosome. Breeding single and double mutants to generate a triple mutant would be challenging because it would rely on recombination of mutant alleles that are proximal on a chromosome, and, therefore, such a fish would be the product of very rare genetic events. Using multiplex genome editing with CRISPR, we generated a triple mutant, tmc2bcwr8 tmc1cwr4 tmc2acwr6 (Figs 4B and 6; Tables 1 and 2). All three mutations result in stop codons upstream of most of the predicted transmembrane domains of each gene and are therefore anticipated to result in nonfunctional proteins. Acoustic startle reflex tests on the tmc2bcwr8 tmc1cwr4 tmc2acwr6 triple mutants demonstrated that these larvae are deaf (Fig. 4C; Movie 8). Specifically, nearly all (98%) of the triple mutants had no C-start reflex. On rare occasions, a larva did respond, but this was likely due to touch receptors that were still responsive in the animal. Controls reacted to 100% of provided stimuli (Fig. 4C). We noted that 100% (10/10) of the tmc2bcwr8 tmc1cwr4 tmc2acwr6 triple mutants responded to touch stimuli, demonstrating that the cognate proteins are not required for detecting touch. Next, we explicitly evaluated the quality of mechanotransduction in the ear’s maculae by determining microphonic potentials and performing dye uptake studies. In the tmc2bcwr8 tmc1cwr4 tmc2acwr6 triple mutant, no microphonic potentials were observed (Fig. 4E and G), nor did the hair cells pass cationic styryl dyes 4-Di-2-ASP or FM1-43FX (Fig. 5A–C, E). Finally, we examined the anterior and posterior maculae to determine if the triple mutant had altered hair cell viability compared to controls. The anterior and posterior maculae had minor reductions in hair cell numbers (Fig. 7). Overall, these results indicate that in the absence of Tmc2a and Tmc2b, Tmc1 permits attenuated mechanotransduction, allowing reduced levels of cationic styryl dyes to pass and facilitating hearing, albeit inconsistently, indicating a nuanced role for Tmc1 in zebrafish hearing.
Figure 6.
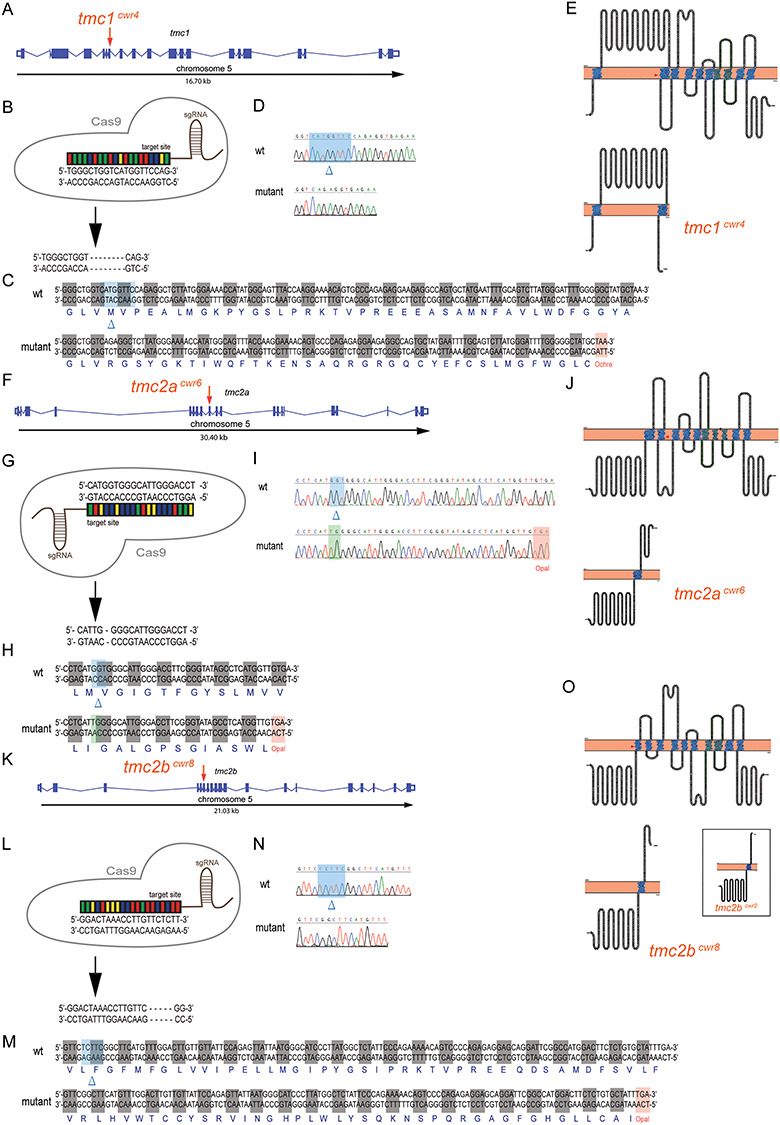
Multiplex genome editing using CRISPR to disrupt tmc1, tmc2a and tmc2b. Graphical representations of tmc1 (A), tmc2a (F) and tmc2b (K) genomic loci of zebrafish. Putative exons and splice sites are displayed. Red arrows mark the targeted exons. Segments of tmc1 exon 7 (B), tmc2a exon 9 (G) and tmc2b exon 6 (L) were subjected to genome editing. Engineered CRISPR single-guide RNAs (sgRNAs) bind target sites to enable DNA cleavage. Mutagenesis deleted eight nucleotides from tmc1 (B), deleted three and inserted two nucleotides from tmc2a (G) and deleted five nucleotides from tmc2b (L) to yield frameshift mutations. (C, H, M) Amino acid sequences of wild-type and mutant proteins. Sequencing results of mutagenized and control loci from tmc1 (D), tmc2a (I) and tmc2b (N). Blue highlights and blue deltas indicate deleted nucleotides. Red highlights denote the stop codons that were generated near the CRISPR-targeting sites. Green highlight marks the site of inserted nucleotides. (E, J, O) (Top) Topographical representations of the Tmc1 (E), Tmc2a (J) and Tmc2b (O) proteins. Arrowheads indicate points of introduced mutations. Amino acids of transmembrane domains are labeled in blue, and the TMC domains are in green. (Bottom) Schematics of predicted truncated polypeptides produced by each mutant allele are displayed. Inset, in O, displays mutant protein associated with tmc2bcwr2 produced in Chou et al 2017, emphasizing its similarity to the protein associated with tmc2bcwr8.
Figure 7.
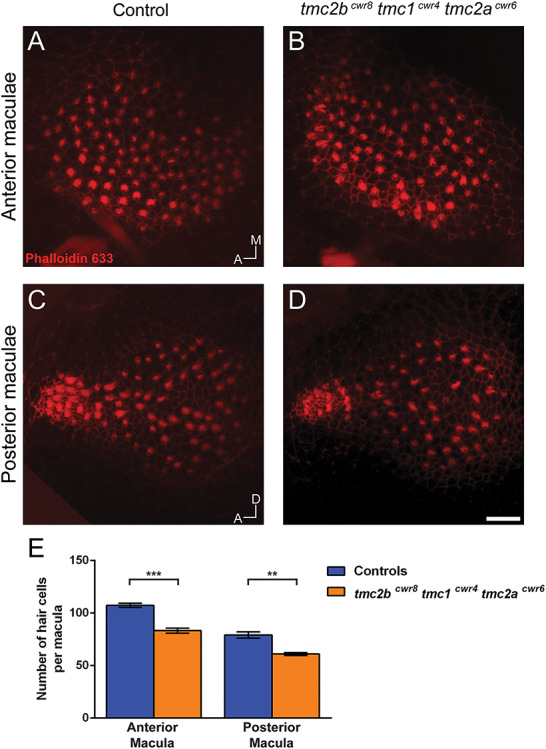
Quantitation of hair cell numbers in tmc2bcwr8 tmc1cwr4 tmc2acwr6 triple-mutant larvae. Confocal micrographs of anterior (A, B) and posterior (C, D) maculae from a tmc2bcwr8 tmc1cwr4 tmc2acwr6 triple mutant and a control (tmc2bcwr8/+ tmc1cwr4/+ tmc2acwr6/+) labeled with phalloidin 633. (E) Graph of enumerated cells of 8-dpf larvae. Mean ± SEM per anterior macula: 107 ± 2.0 (n = 3) for the control and 83 ± 2.4 (n = 4) for the triple mutant. Two-tailed unpaired t-test, ***P = 0.0008. Mean ± SEM per posterior macula: 79 ± 3.0 (n = 3) for the control and 61 ± 1.3 (n = 4) for the triple mutant. **P = 0.0017. A, anterior; M, medial; D, dorsal. Scale bar = 10 μm.
Mutation of tmc2a alone, but not tmc2b alone, reduces mechanotransduction of auditory hair cells
We know that the Tmc2a–Tmc2b module is required for normal auditory hair cell mechanotransduction; however, the contribution of the individual Tmc2a protein to auditory hair cell mechanotransduction is not known. To extricate Tmc2a’s potential contribution from that of Tmc2b in microphonic potentials from auditory hair cells, we characterized the tmc2acwr3 single mutant. The tmc2acwr3 single mutant displayed a reduction in mean microphonic potentials (23.6 ± 3.4 μV; n = 5) relative to controls (80.3 ± 6.9 μV; n = 25) (Fig. 8A and C), suggesting that Tmc2a participates in carrying a significant proportion of ions for mechanotransduction. As noted in our previous paper (18), mutation of tmc2b did not noticeably impact mechanotransduction in the maculae (Fig. 8B and D). These findings agree with expression data where tmc2a mRNA is more abundant in the ear than that of tmc2b (24). However, our data reveal a partial reliance on Tmc2a and Tmc2b for microphonic potentials that could not be predicted by expression data alone. Specifically, in the wild-type configuration, most current is carried by Tmc2a. However, in the absence of Tmc2a, most of the remaining current is carried by Tmc2b. When Tmc2a and Tmc2b are absent, a modicum of Tmc1 activity is present and detectable.
Figure 8.
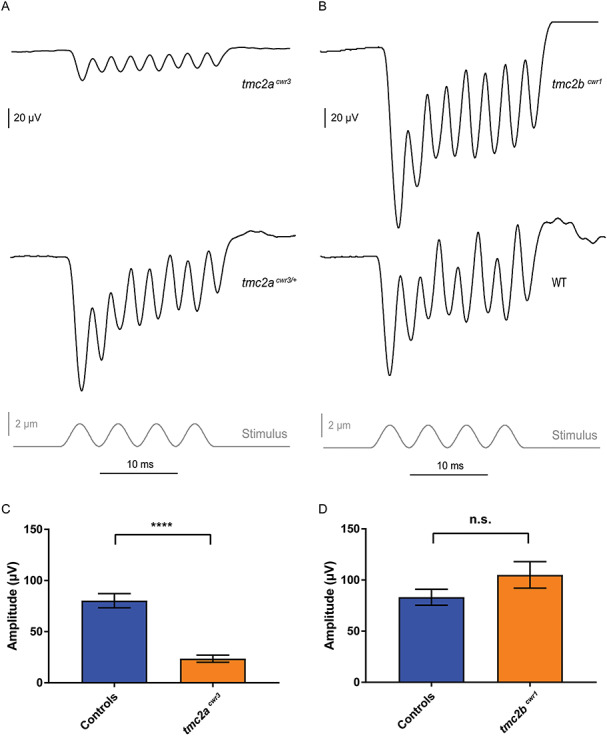
Electrophysiological evaluation of tmc2a and tmc2b single mutants. Stimulus-evoked microphonic potentials measured from the otic vesicles of tmc2acwr3 (A) and tmc2bcwr1 (B) animals with controls. Graphs of mean microphonic potentials from otic vesicles of tmc2acwr3 (C) and tmc2bcwr1 (D) animals with controls. Mean microphonic potential of tmc2acwr3 ± SEM = 23.6 ± 3.4 μV (n = 5), mean microphonic potential of wild-type and heterozygous siblings = 80.3 ± 6.9 μV (n = 25; Mann–Whitney test, ****P < 0.0001); mean microphonic potential of tmc2bcwr1 ± SEM = 105 ± 12.9 μV (n = 6), mean microphonic potential of wild-type and heterozygous siblings = 83.1 ± 7.8 μV (n = 14; P = 0.13).
Discussion
Using a molecular genetics approach that involved seven distinct mutant alleles of three tmc genes, either separately or in strategic combinations, we addressed two central questions pertaining to the mechanotransduction process for hearing in vertebrates. Do all vertebrates require Tmc1 to hear? Hearing in mammals has been shown to depend on TMC1, and this protein is considered a pore-forming subunit in this class of animals (29). Using two zebrafish mutants generated by two different methodologies, we show that Tmc1 is not necessary for hearing (Fig. 1) but instead contributes subtly. We detected a minor role for Tmc1 only in a zebrafish strain where both tmc2a and tmc2b were mutated (Figs 4 and 5). These findings demonstrate that Tmc1 is not required for all vertebrates to hear or for all auditory hair cells to transduce. They also suggest that the bulk of ion entry for auditory mechanotransduction requires Tmc2 protein isoforms where they may serve as pore-forming subunits. In zebrafish, Tmc1 may instead play an ancillary role, perhaps modifying the quality of mechanotransduction in the ear as a pore-forming subunit or as a protein closely associated with the channel. Moreover, Tmc1 may function in a subpopulation of hair cells based on the cell’s position within a sensory epithelium or on its hair bundle’s orientation. Alternatively, Tmc1’s role in other hair cell-containing organs may be more pronounced. Specifically, two hair cell-containing organ types in which Tmc1 may play a prominent role are the lagena and the crista. The lagena is an otolithic organ that appears 11 dpf during development whereas the three cristae are present in the embryonic stage and reside in each ear to detect angular acceleration. Finally, there are two pathways by which sound can stimulate the ear of an adult fish: the direct pathway, where particle motion directly stimulates the ear through the skull, or the indirect pathway, where pressure waves indirectly stimulate the internal ear through the swim bladder (44,45). It is possible that Tmc1 may play a more prominent role in either of these pathways, which awaits determination.
The lack of a significant role for Tmc1 displays what may be a fundamental principle in the evolution of the auditory system. Hearing in fish is macular in origin whereas in mice it is cochlear. Mature cochlear hair cells in the mouse depend on TMC1 for mechanotransduction, but the macular cells of zebrafish depend on Tmc2 proteins. It was unknown if hearing was strictly dependent on Tmc1 because of the unique qualities that this protein bestows upon hair cells or if the macular organ tends to retain the use of Tmc2 proteins for function, be it auditory or vestibular. We show here the latter is the case, and Tmc2a and Tmc2b are the main Tmc components of mechanotransduction in auditory hair cells of zebrafish. Since the high-frequency extreme of zebrafish hearing is much lower than that of mammalian hearing, TMC1 may have evolved to be the main TMC protein in the cochlea because it is better equipped to capture and encode exceptionally high-frequency stimuli that permit mammalian survival.
Interestingly, however, the Tmc protein reliance in the vestibular system of mammals and the maculae of zebrafish is remarkably similar. In mice that lack TMC1, the vestibular ocular reflex, transduction currents and the uptake of FM1-43 by utricular hair cells are all normal (19), akin to what we observe in zebrafish (Figs 1–3). In mice that lack TMC2, transduction currents of utricular hair cells are reduced, again similar to what we see in zebrafish that lack both Tmc2a and Tmc2b where there are reductions in the uptake of FM1-43FX and 4-Di-2-ASP. Overall, these findings show that the maculae of mammals and zebrafish are similar at the molecular level with regard to their Tmc dependencies (Fig. 9). These findings also imply that Tmc proteins in zebrafish and mammals function similarly in mechanotransduction. Therefore, studies analyzing zebrafish Tmcs can lead to a better understanding of their roles in mammals, including how these proteins may function in the human ear. Specifically, future studies using the unique combination of attributes of the zebrafish model system (optical clarity, reverse genetics and transgenesis) may contribute distinctively to our understanding of TMC proteins in human disease. Along those lines, here we have created a zebrafish model for recessive human deafness form DFNB7/11. The tmc2bcwr8 tmc1cwr4 tmc2acwr6 triple mutant is deaf. This animal model should allow for the elucidation of pathophysiological processes that result in DFNB7/11 deafness for which there is limited access in humans.
Figure 9.
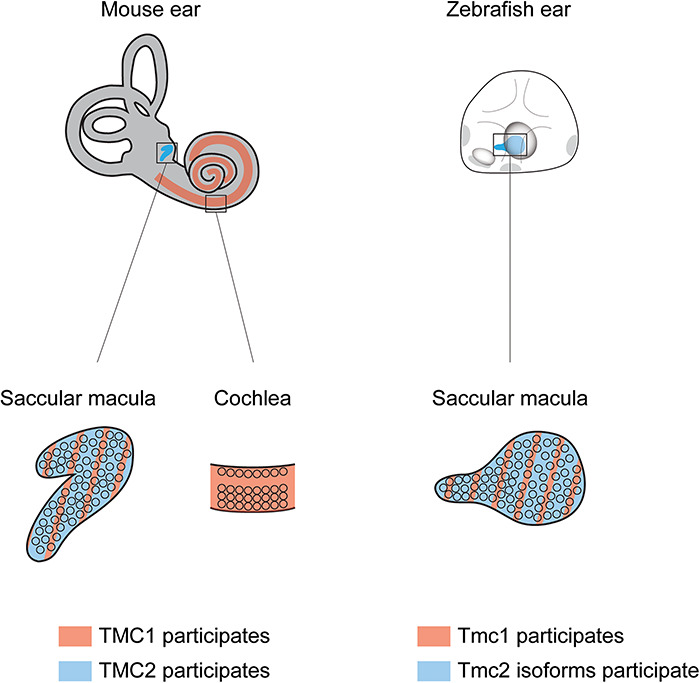
Macular hair cells of mammals and fish use similar Tmcs for mechanotransduction. In the mouse inner ear, cochlear hair cells are dependent on TMC1, but vestibular hair cells use both TMC1 and TMC2 for normal mechanotransduction. The macular hair cells of the zebrafish inner ear also use Tmc1 and Tmc2 isoforms (Tmc2a and Tmc2b) for normal mechanotransduction.
Sensory cells for vision, olfaction and taste—among others—discriminate similar types of stimuli within each respective sensory system using different transduction proteins (4–6). However, it is not clear how hair cells responsible for hearing in the otic vesicle and those that detect water motion in the lateral line may diverge so as to capture and encode the disparate mechanical régimes that are imposed on these two systems. In other words, do hair cells that encode mechanical information for different sensory systems use the same set of transduction proteins? By examining tmc2bcwr2 tmc2acwr3 double and tmc2bcwr8 tmc1cwr4 tmc2acwr6 triple mutants, we demonstrate that the double mutant has attenuated hearing attributable to diminished mechanotransduction, but the triple mutant is deaf owing to extinguished mechanotransduction (Figs 4 and 5). Since it is known that lateral line function is wholly dependent on Tmc2a and Tmc2b, our results demonstrate that normal hearing uses at least one protein beyond that used by the lateral line, Tmc1 (Fig. 10). These results offer the possibility that the mechanotransduction process is modified for distinct sensory systems, such as those that respond to sound waves versus water flow. Our findings support a hypothesis in which hair cells use a molecular strategy similar to that of other sensory systems: variation in transduction proteins, Tmcs, enables sensory cells, hair cells, to capture and encode a vast array of inputs. The specific qualities that differ between Tmc proteins are unknown and revealing them may require new empirical schemes.
Figure 10.
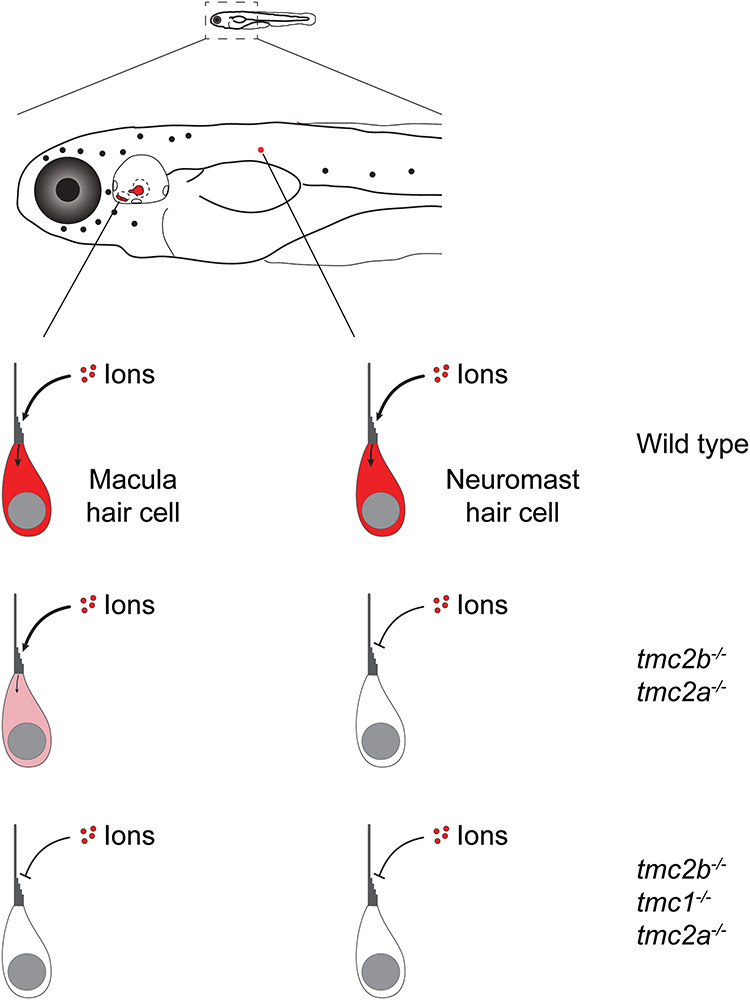
Schematic displaying the Tmc dependencies of maculae and posterior lateral line neuromasts. Mutation of tmc2a and tmc2b results in the loss of mechanotransduction in the lateral line, but residual function is retained in the maculae. Triple mutants show no mechanotransduction in the maculae.
Materials and Methods
Ethical approval
Protocols for housing and handling of zebrafish were approved by Case Western Reserve University’s Institutional Animal Care and Use Committee and conform to the principles and regulations as described in the Editorial by (46). Zebrafish were anesthetized with 3-aminobenzoic acid ethyl ester methanesulfonate (Sigma-Aldrich) at 168 mg l−1. Zebrafish were euthanized with 3-aminobenzoic acid ethyl ester methanesulfonate at 672 mg l−1.
Zebrafish
Wild-type Tübingen (Tü), tmc2bcwr1 single-mutant (18) and tmc2bcwr2 tmc2acwr3 double-mutant (18) zebrafish were used in this study.
Mutation of the tmc1 locus with TALENs
To identify a suitable region of exon 5 of the tmc1 gene (ENSDARG00000056386) for targeting by transcription activator-like (TAL) effector nucleases (TALENs), the ZiFiT Targeter computational program was used (47,48). Specificity of the tmc1 TALEN target site was confirmed by the NCBI BLAST tool using the zebrafish genomic DNA database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). TALEN expression vectors were constructed with the fast ligation-based automatable solid-phase high-throughput (FLASH) assembly protocol (49–51). Biotinylated α units, extension units, termination units and the expression vectors used in this study are listed in order for tmc1-Left-TALEN (TAL376, TAL187-TAL33-TAL52, TAL305, JDS74) and tmc1-Right-TALEN (TAL373, TAL12-TAL246-TAL230, TAL336, JDS74). Designed vectors were confirmed by DNA sequencing using primers oSQT1 (5′ AGTAACAGCGGTAGAGGCAG 3′), oSQT3 (5′ ATTGGGCTACGATGGACTCC 3′) and oJS2980 (5′ TTAATTCAATATATTCATGAGGCAC 3′). Sequence-confirmed expression vectors carrying either tmc1-Left-TALEN or tmc1-Right-TALEN were linearized with PmeI and were used as templates for in vitro transcription reactions (Ambion mMessage mMACHINE Ultra T7 kit; Life Technologies Inc.). RNAs were purified by lithium chloride precipitation, and the quality and quantity of each were verified (Agilent 2100 Bioanalyzer, Agilent Technologies; Nanodrop 2000, Thermo Scientific). RNAs that encode each half site of tmc1-TALEN at 150 ng/μl were mixed with phenol red and 1× Danieau’s solution (58 mm NaCl, 0.7 mm KCl, 0.4 mm MgSO4, 0.6 mm Ca(NO3)2, and 5.0 mm HEPES pH 7.6) and then coinjected into zebrafish embryos at the one-cell stage to create gene-specific mutations. Embryos injected with tmc1-TALEN RNAs were collected individually and subjected to mutation detection at the tmc1 TALEN target site using high resolution melting analysis (HRMA) with primers tmc1 TALEN1 HRM F1 (5′ TCTGTCTCAGGGAGTCGCAG 3′) and tmc1 TALEN1 HRM R1 (5′ ACTCGGATTCATACTCCTCACC 3′). The mutations in tmc1 carried by founder fish and F1 fish were identified using sequencing primers tmc1 TALEN1 F1 (5′ TTGTGTCCCAATCTGATGCG 3′) and tmc1 TALEN R (5′ CTTCATCTCCCACGGGATGC 3′). The tmc1cwr5 single mutant, which has a 1-bp deletion that results in a frameshift, was used in this work.
Mutation of tmc1, tmc2a and tmc2b with CRISPR/Cas9
CRISPR target sites were selected using the CHOPCHOP web tool (http://chopchop.cbu.uib.no). sgRNAs were synthesized using T7 RNA polymerase (MEGAscript, Invitrogen). Tmc1 (ENSDARG00000056386) was targeted with an sgRNA complementary to exon 7 (5′ TGGGCTGGTCATGGTTCCAG 3′, tmc1-CRISPR11). Tmc2a (ENSDARG00000033104) was targeted with an sgRNA complementary to exon 9 (5′ AGGTCCCAATGCCCACCATG 3′ , tmc2a-CRISPR4). Tmc2b (ENSDARG00000030311) was targeted with an sgRNA complementary to exon 6 (5′ GGACTAAACCTTGTTCTCTT 3′, tmc2b-CRISPR2). Successfully targeted loci were identified by HRMA (52) and DNA sequencing. RNAs that encode tmc1-CRISPR11, tmc2a-CRISPR4 and tmc2b-CRISPR2 at 120 ng/μl were mixed with Cas9 mRNA at 200 ng/μl, phenol red and 1× Danieau’s solution and then injected into zebrafish embryos at the one-cell stage to create gene-specific mutations. Embryos injected with tmc1-CRISPR11, tmc2a-CRISPR4 and tmc2b-CRISPR2 were collected individually and subjected to mutation detection at each target site, using HRMA with primers tmc1 CRISPR HRMA Forward (5′ ATTCTTGAGGTGGATGTATGGC 3′), tmc1 CRISPR HRMA Reverse (5′ ATAGAGTCCACATGCAGAACGA 3′), tmc2a CR4 HRMA Forward (5′ CAGGGTTACTGCAAGTACTCAG 3′), tmc2a CR4 HRMA Reverse (5′ GCAATGAATGAAAGAGGGACTC 3′), tmc2b CR2 HRMA Forward (5′ CTTGTAGGCCATTTTGGATCATC 3′) and tmc2b CR2 HRMA Reverse (5′ ACACAACACAACATACATTCCCA 3′). The mutations in each gene carried by founder fish and F1 fish were identified using sequencing primers tmc1 CR10–11 seq Forward Long (5′ TCCACATGCAGAACGAAGACA 3′), tmc1 CR10–11 seq Reverse Long (5′ GAGTCGCAGGAGTTTGTGGA 3′), tmc2a CR4 seq Forward (5′ GAGACCGGAAATCTCGTGCC 3′), tmc2a CR4 seq Reverse (5′ CCTTTTGTCACTGAGGGACAAC 3′), tmc2b CR2 seq Forward (5′ CAAGACGGCTTGCATTCCCTGG 3′) and tmc2b CR2 seq Reverse (5′ GTCCATGGCCGAATCCTGCTCC 3′). The tmc2bcwr8 tmc1cwr4 tmc2acwr6 triple mutant, the tmc1cwr4 single mutant and the tmc2acwr3 single mutant were used for assays in this work. Because tmc1, tmc2a and tmc2b all reside on chromosome 5, generation of triple-mutant zebrafish was achieved by coinjection of one-cell stage zebrafish embryos with Cas9 RNA and sgRNAs that target each of the respective genes. This process is termed multiplex genome editing (42,43).
Acoustic startle reflex assay
Larval zebrafish were placed into individual Petri dishes positioned on a white background whereas adult fish were placed in a small breeding tank. Controls were wild-type or heterozygous siblings. Fish were placed in 1–1.5 cm of water for larvae and in 15–20 cm of water for adults. The startle stimulus was provided by a tap on the Petri dish or tank wall using a metal probe. The stimulus was only applied when the fish were relatively still and not touching the container walls. Larval experiments were performed at 5–10 dpf. To avoid habituation, an interval of 2 minutes was given between stimuli. Each larval fish was presented with 20 vibrational stimuli, and the mean percentages of zebrafish that exhibited a C-start response were graphed. Adult fish were presented with 10 vibrational stimuli per fish. Specimens were recorded as positive if they exhibited the characteristic C-bend turn associated with the acoustic startle response from stimuli (35,53) and as negative if there was no response to stimuli. The apparatus for the startle response filming consisted of a mounted camera and lighting fixture centered around a Petri dish or a tank containing the fish. Videos were captured at 1000 frames/s (Fastec IL5 camera with a Nikon IF aspherical 24–85 mm lens). Recordings were performed in a dark room with infrared LED illumination or with ambient light. Videos were processed using ImageJ.
Real-time polymerase chain reaction (RT-PCR)
RNA from four 6-dpf wild-type or tmc1cwr5 mutant zebrafish were separately extracted (RNeasy Mini Kit; Qiagen). 400 ng of total RNA from each sample was reverse transcribed to produce cDNA (SuperScript III First Strand Synthesis System for RT-PCR Kit; Invitrogen). The relative quantity of each mRNA was obtained with the comparative threshold cycle (Ct) method. PCR amplification was performed for a 20 μl reaction solution [10 μl of 2× RT2 SYBR Green ROX Fast Mastermix (Qiagen), 4 μl of forward primer (1 μM), 4 μl of reverse primer (1 μM), and 2 μl of diluted template cDNA (400 ng/μl)] in a PCR machine (StepOnePlus real-time PCR system; Applied Biosystems). All reactions were performed in quintuplicate. The PCR conditions were as follows: 95°C for 10 min then 40 amplification cycles (95°C for 15 s, 60°C for 1 min). As an internal control, primers for GAPDH were used to amplify in parallel. In all cases, a reverse transcriptase negative control was included. Primer sequences: GAPDH RT-PCR F1, 5′ GTGGAGTCTACTGGTGTCTTC 3′; GAPDH RT-PCR R1, 5′ GTGCAGGAGGCATTGCTTACA 3′; Tmc2a RT-PCR F3, 5′ ATGGCATGAACTTGGTCCTC 3′; TMC2a RT-PCR R3, 5′ CAGTTTTCCGAGGAATGGAC 3′; Tmc2b RT-PCR F5, 5′ CTCTTCGGCTTCATGTTTGG 3′; and Tmc2b RT-PCR R5, 5′ GGGAATAGAGCCATAAGGGATG 3′. The relative quantity of each mRNA was obtained with the comparative threshold cycle (Ct) method, with the mean wild-type control value set at 1.0. Ct values were obtained for each reference gene with Ct defined as the threshold cycle of the PCR at which the amplified product was first detected. ΔCt is the difference in Ct values between target genes and the GAPDH internal control gene. ΔΔCt represents the difference between ΔCts from wild-type zebrafish and the tmc1 mutant. The fold difference in a target gene’s expression between wild-type and tmc1 mutant zebrafish was calculated as 2−ΔΔCt.
Measurement of inner ear microphonic potentials
Zebrafish larvae, age 7–10 dpf, were anesthetized at room temperature (~22°C) with 0.612 mm ethyl 3-aminobenzoate methanesulfonic acid dissolved in a standard bath solution (120 mm NaCl, 2 mm KCl, 10 mm HEPES, 2 mm CaCl2, 0.7 mm NaH2PO4, and adjusted to pH 7.3). Anesthetized larvae were mounted laterally in the recording chamber with low-melting-point agarose (IBI Scientific Inc.). We used an upright microscope (BX51WI; Olympus) equipped with a swing nosepiece and 4 × 0.13 and 40 × 0.6 NA objectives to visualize the larvae. Circulation and heart rate were monitored visually to verify that each larva was viable before, during and after recording. We used a Grasshopper3 CMOS camera (Point Grey Inc.) and manufacturer-provided software to view and capture images. The recording pipette was made from borosilicate glass with a resistance of 3–6 MΩ when filled with the bath solution. We inserted the pipette into the otic vesicle where it was kept near both maculae for recording. A glass stylus, tip diameter of ~7 μm, was placed on the skin above the otic vesicle to vibrate the ear. The 200 Hz sinusoidal command stimulus was generated by jClamp (Scisoft) (18,54–56), power amplified (ENV 800, Piezosystem Jena) and delivered by a piezo actuator (PA 4/12, Piezosystem Jena). Pipette placement was controlled by a set of micromanipulators (MPC-325; Sutter Instrument). The microphonic potentials were recorded with a PC-505B amplifier (Warner Instruments), SIM983 amplifier (set at 20 ×, Stanford Research) and a PCI-6221 digitizer (National Instruments) using jClamp in current-clamp mode. The sampling rate for recordings was 5 kHz. Responses had a frequency of 400 Hz due to the 2f response (57,58) and were low pass filtered at 1000 Hz. All traces shown and used in analysis are the average of at least 1000 trials. Mean amplitude as reported in graphs was calculated as the average amplitude of all 8 peaks for each trace.
Dye injection into otocyst
At 6–8 dpf, zebrafish larvae were anesthetized in 0.612 mm ethyl 3-aminobenzoate methanesulfonic acid in fish water and mounted laterally in a Petri dish (Becton Dickinson) in low-melting-point agarose (Promega). Approximately 1 nl of 200 μM 4-Di-2-ASP (Sigma-Aldrich) or 50 μM FM1-43FX (Invitrogen) mixed with phenol red to 0.125% (Sigma Life Science) was injected into the otic capsule (59,60). The larvae were imaged (4-Di-2-ASP) or fixed (FM1-43FX) immediately after injection. To reduce experimental variation, the same injection needle was used to inject experimental and control larvae.
Imaging and quantification of 4-Di-2-ASP intensity
For imaging, the larvae were anesthetized in 0.612 mm ethyl 3-aminobenzoate methanesulfonic acid in fish water and mounted laterally on glass-bottom dishes (MatTek) in 1.0% (wt/vol) low-melting-point agarose (Promega). Z-stack images of the entire anterior maculae were collected on a confocal microscope (TCS SP8, Leica) with a 40×/1.3 NA oil-immersion objective. A laser wavelength of 488 nm was used for 4-Di-2-ASP excitation.
To quantitate 4-Di-2-ASP uptake in anterior maculae hair cells, the outlines of entire anterior maculae (ROIs) were defined manually with the help of the bright-field images taken simultaneously with the fluorescent images using software (LAS X, Leica). The mean fluorescence intensity within each ROI was measured from the maximum projection of each z-stack series. After background subtraction, the 4-Di-2-ASP intensities of individual maculae were normalized to the mean intensity of their corresponding control group (Figs 2B, D and 5B).
Imaging of FM1-43FX
Fixation was performed as described previously (18). For imaging, fixed zebrafish samples were mounted in antifade mounting medium (VECTASHIELD; Vector Laboratories). Z-stack images of the entire anterior or posterior maculae were collected on a confocal microscope (TCS SP8, Leica) with a 40×/1.3 NA oil-immersion objective. Excitation wavelengths of 488 nm for FM1-43FX and 633 nm for Alexa Fluor 633 phalloidin were used.
Enumeration of mutant hair cells
Confocal images of the anterior and posterior maculae labeled with Alexa Fluor 633 phalloidin were used for hair cell enumeration. This process was carried out manually, and all hair cells of both maculae were scored for one ear of each zebrafish.
Statistics and software
All statistical analyses were performed using GraphPad Prism 7. Data are reported as mean ± SEM. Comparisons between groups were tested by Mann–Whitney test, Student’s t-test or ANOVA with Holm-Sidak post hoc testing depending on data distribution.
To make predictions of transmembrane domains of the zebrafish Tmc proteins, we used computational software PHILLIUS (http://www.yeastrc.org/philius/) (61), OCTOPUS (http://octopus.cbr.su.se/) (62) and HMMTOP (http://www.enzim.hu/hmmtop/) (63). Topographical representations of proteins were generated with PROTTER software (http://wlab.ethz.ch/protter/start/).
Author contributions
Gene editing was carried out by S.Z., S-W.C., and R.D. Imaging was performed by Z.C. and S.Z. Electrophysiology experiments were implemented by K.K. Experimental plans were developed by B.M., R.S., S-W.C., Z.C., S.Z., and K.K. M.D. and S.W. performed C-start reflex tests. Paper was written by B.M. All authors commented on and contributed to the final version of the manuscript.
Supplementary Material
Acknowledgements
We thank H. Nguyen for zebrafish husbandry and the members of our laboratory who provided discussion and comments on this manuscript. This work is dedicated to Neil Peart of Rush.
Funding
National Institutes of Health (DC009437 to B.M., DC015016 to R.S.) and the Center for Clinical Research and Technology at University Hospitals Cleveland Medical Center (to B.M.).
Conflict of Interest. The authors declare no conflict of interest.
References
- 1. Ramon y Cajal S. and Castro F.D. (1933) Elementos de técnica micrográfica del sistema nervioso. Tipografia artistica, Madrid. [Google Scholar]
- 2. Ramón y Cajal S. (1894) Croonian lecture: la fine structure des centres nerveux. Proc. R. Soc., 55, 444–468. [Google Scholar]
- 3. Helmholtz H.v. and Ellis A.J. (1885) On the Sensations of Tone as a Physiological Basis for the Theory of Music. Longmans, Green, London. [Google Scholar]
- 4. Wald G., Brown P.K. and Smith P.H. (1955) IODOPSIN. J. Gen. Physiol., 38, 623–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Buck L. and Axel R. (1991) A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell, 65, 175–187. [DOI] [PubMed] [Google Scholar]
- 6. Hoon M.A., Adler E., Lindemeier J., Battey J.F., Ryba N.J. and Zuker C.S. (1999) Putative mammalian taste receptors: a class of taste-specific GPCRs with distinct topographic selectivity. Cell, 96, 541–551. [DOI] [PubMed] [Google Scholar]
- 7. Kandel E.R. (2013) Principles of Neural Science. McGraw-Hill, New York. [Google Scholar]
- 8. Higgs D.M., Souza M.J., Wilkins H.R., Presson J.C. and Popper A.N. (2002) Age- and size-related changes in the inner ear and hearing ability of the adult zebrafish (Danio rerio). J. Assoc. Res. Otolaryngol., 3, 174–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yao Q., DeSmidt A.A., Tekin M., Liu X. and Lu Z. (2016) Hearing assessment in zebrafish during the first week postfertilization. Zebrafish, 13, 79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bhandiwad A.A., Raible D.W., Rubel E.W. and Sisneros J.A. (2018) Noise-induced hypersensitization of the acoustic startle response in larval zebrafish. J. Assoc. Res. Otolaryngol., 19, 741–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vanwalleghem G., Heap L.A. and Scott E.K. (2017) A profile of auditory-responsive neurons in the larval zebrafish brain. J. Comp. Neurol., 525, 3031–3043. [DOI] [PubMed] [Google Scholar]
- 12. Lu Z. and DeSmidt A.A. (2013) Early development of hearing in zebrafish. J. Assoc. Res. Otolaryngol., 14, 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Engelmann J., Hanke W., Mogdans J. and Bleckmann H. (2000) Hydrodynamic stimuli and the fish lateral line. Nature, 408, 51–52. [DOI] [PubMed] [Google Scholar]
- 14. Voigt R., Carton A.G. and Montgomery J.C. (2000) Responses of anterior lateral line afferent neurones to water flow. J. Exp. Biol., 203, 2495–2502. [DOI] [PubMed] [Google Scholar]
- 15. Montgomery J., Carton G., Voigt R., Baker C. and Diebel C. (2000) Sensory processing of water currents by fishes. Philos. Trans. R. Soc. Lond. B Biol. Sci., 355, 1325–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang Y.V., Aikin T.J., Li Z. and Montell C. (2016) The basis of food texture sensation in drosophila. Neuron, 91, 863–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guo Y., Wang Y., Zhang W., Meltzer S., Zanini D., Yu Y., Li J., Cheng T., Guo Z., Wang Q. et al. (2016) Transmembrane channel-like (tmc) gene regulates Drosophila larval locomotion. Proc. Natl. Acad. Sci. USA., 113, 7243–7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chou S.W., Chen Z., Zhu S., Davis R.W., Hu J., Liu L., Fernando C.A., Kindig K., Brown W.C., Stepanyan R. et al. (2017) A molecular basis for water motion detection by the mechanosensory lateral line of zebrafish. Nat. Commun., 8, 2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kawashima Y., Geleoc G.S., Kurima K., Labay V., Lelli A., Asai Y., Makishima T., Wu D.K., Della Santina C.C., Holt J.R. et al. (2011) Mechanotransduction in mouse inner ear hair cells requires transmembrane channel-like genes. J. Clin. Invest., 121, 4796–4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jain P.K., Fukushima K., Deshmukh D., Ramesh A., Thomas E., Lalwani A.K., Kumar S., Plopis B., Skarka H., Srisailapathy C.R. et al. (1995) A human recessive neurosensory nonsyndromic hearing impairment locus is potential homologue of murine deafness (dn) locus. Hum. Mol. Genet., 4, 2391–2394. [DOI] [PubMed] [Google Scholar]
- 21. Steel K.P. and Bock G.R. (1980) The nature of inherited deafness in deafness mice. Nature, 288, 159–161. [DOI] [PubMed] [Google Scholar]
- 22. Bock G.R. and Steel K.P. (1983) Inner ear pathology in the deafness mutant mouse. Acta Otolaryngol., 96, 39–47. [DOI] [PubMed] [Google Scholar]
- 23. Kurima K., Peters L.M., Yang Y., Riazuddin S., Ahmed Z.M., Naz S., Arnaud D., Drury S., Mo J., Makishima T. et al. (2002) Dominant and recessive deafness caused by mutations of a novel gene, TMC1, required for cochlear hair-cell function. Nat. Genet., 30, 277–284. [DOI] [PubMed] [Google Scholar]
- 24. Maeda R., Kindt K.S., Mo W., Morgan C.P., Erickson T., Zhao H., Clemens-Grisham R., Barr-Gillespie P.G. and Nicolson T. (2014) Tip-link protein protocadherin 15 interacts with transmembrane channel-like proteins TMC1 and TMC2. Proc. Natl. Acad. Sci. USA., 111, 12907–12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kurima K., Ebrahim S., Pan B., Sedlacek M., Sengupta P., Millis B.A., Cui R., Nakanishi H., Fujikawa T., Kawashima Y. et al. (2015) TMC1 and TMC2 localize at the site of mechanotransduction in mammalian inner ear hair cell stereocilia. Cell Rep., 12, 1606–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mahendrasingam S. and Furness D.N. (2019) Ultrastructural localization of the likely mechanoelectrical transduction channel protein, transmembrane-like channel 1 (TMC1) during development of cochlear hair cells. Sci. Rep., 9, 1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pan B., Geleoc G.S., Asai Y., Horwitz G.C., Kurima K., Ishikawa K., Kawashima Y., Griffith A.J. and Holt J.R. (2013) TMC1 and TMC2 are components of the mechanotransduction channel in hair cells of the mammalian inner ear. Neuron, 79, 504–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Beurg M., Goldring A.C. and Fettiplace R. (2015) The effects of Tmc1 Beethoven mutation on mechanotransducer channel function in cochlear hair cells. J. Gen. Physiol., 146, 233–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pan B., Akyuz N., Liu X.P., Asai Y., Nist-Lund C., Kurima K., Derfler B.H., Gyorgy B., Limapichat W., Walujkar S. et al. (2018) TMC1 forms the pore of mechanosensory transduction channels in vertebrate inner ear hair cells. Neuron, 99, 736–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ballesteros A., Fenollar-Ferrer C. and Swartz K.J. (2018) Structural relationship between the putative hair cell mechanotransduction channel TMC1 and TMEM16 proteins. Elife, 7, e338433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jia Y., Zhao Y., Kusakizako T., Wang Y., Pan C., Zhang Y., Nureki O., Hattori M. and Yan Z. (2020) TMC1 and TMC2 proteins are pore-forming subunits of mechanosensitive ion channels. Neuron, 105, 310–321. [DOI] [PubMed] [Google Scholar]
- 32. Beurg M., Cui R., Goldring A.C., Ebrahim S., Fettiplace R. and Kachar B. (2018) Variable number of TMC1-dependent mechanotransducer channels underlie tonotopic conductance gradients in the cochlea. Nat. Commun., 9, 2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vreugde S., Erven A., Kros C.J., Marcotti W., Fuchs H., Kurima K., Wilcox E.R., Friedman T.B., Griffith A.J., Balling R. et al. (2002) Beethoven, a mouse model for dominant, progressive hearing loss DFNA36. Nat. Genet., 30, 257–258. [DOI] [PubMed] [Google Scholar]
- 34. Starr C.J., Kappler J.A., Chan D.K., Kollmar R. and Hudspeth A.J. (2004) Mutation of the zebrafish choroideremia gene encoding Rab escort protein 1 devastates hair cells. Proc. Natl. Acad. Sci. USA., 101, 2572–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kimmel C.B., Patterson J. and Kimmel R.O. (1974) The development and behavioral characteristics of the startle response in the zebra fish. Dev. Psychobiol., 7, 47–60. [DOI] [PubMed] [Google Scholar]
- 36. Anderson J.L., Mulligan T.S., Shen M.C., Wang H., Scahill C.M., Tan F.J., Du S.J., Busch-Nentwich E.M. and Farber S.A. (2017) mRNA processing in mutant zebrafish lines generated by chemical and CRISPR-mediated mutagenesis produces unexpected transcripts that escape nonsense-mediated decay. PLoS Genet., 13, e1007105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Owens K.N., Santos F., Roberts B., Linbo T., Coffin A.B., Knisely A.J., Simon J.A., Rubel E.W. and Raible D.W. (2008) Identification of genetic and chemical modulators of zebrafish mechanosensory hair cell death. PLoS Genet., 4, e1000020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gleason M.R., Nagiel A., Jamet S., Vologodskaia M., Lopez-Schier H. and Hudspeth A.J. (2009) The transmembrane inner ear (Tmie) protein is essential for normal hearing and balance in the zebrafish. Proc. Natl. Acad. Sci. USA., 106, 21347–21352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gale J.E., Marcotti W., Kennedy H.J., Kros C.J. and Richardson G.P. (2001) FM1-43 dye behaves as a permeant blocker of the hair-cell mechanotransducer channel. J. Neurosci., 21, 7013–7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Meyers J.R., MacDonald R.B., Duggan A., Lenzi D., Standaert D.G., Corwin J.T. and Corey D.P. (2003) Lighting up the senses: FM1-43 loading of sensory cells through nonselective ion channels. J. Neurosci., 23, 4054–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Inoue M., Tanimoto M. and Oda Y. (2013) The role of ear stone size in hair cell acoustic sensory transduction. Sci. Rep., 3, 2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Minkenberg B., Wheatley M. and Yang Y. (2017) CRISPR/Cas9-enabled multiplex genome editing and its application. Prog. Mol. Biol. Transl. Sci., 149, 111–132. [DOI] [PubMed] [Google Scholar]
- 43. Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science, 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Popper A.N. and Fay R.R. (1993) Sound detection and processing by fish: critical review and major research questions. Brain Behav. Evol., 41, 14–38. [DOI] [PubMed] [Google Scholar]
- 45. Emde G.v.d., Mogdans J. and Kapoor B.G. (2004) The Senses of Fish: Adaptations for the Reception of Natural Stimuli. Kluwer; Narosa Pub, Boston, New Delhi. [Google Scholar]
- 46. Grundy D. (2015) Principles and standards for reporting animal experiments in The Journal of Physiology and Experimental Physiology. J. Physiol., 593, 2547–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sander J.D., Zaback P., Joung J.K., Voytas D.F. and Dobbs D. (2007) Zinc Finger Targeter (ZiFiT): an engineered zinc finger/target site design tool. Nucleic Acids Res., 35, W599–W605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sander J.D., Maeder M.L., Reyon D., Voytas D.F., Joung J.K. and Dobbs D. (2010) ZiFiT (zinc finger Targeter): an updated zinc finger engineering tool. Nucleic Acids Res., 38, W462–W468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Reyon D., Maeder M.L., Khayter C., Tsai S.Q., Foley J.E., Sander J.D. and Joung J.K. (2013) Engineering customized TALE nucleases (TALENs) and TALE transcription factors by fast ligation-based automatable solid-phase high-throughput (FLASH) assembly. Curr. Protoc. Mol. Biol., 103, 12.16.1-12.16.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Reyon D., Tsai S.Q., Khayter C., Foden J.A., Sander J.D. and Joung J.K. (2012) FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol., 30, 460–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cade L., Reyon D., Hwang W.Y., Tsai S.Q., Patel S., Khayter C., Joung J.K., Sander J.D., Peterson R.T. and Yeh J.R. (2012) Highly efficient generation of heritable zebrafish gene mutations using homo- and heterodimeric TALENs. Nucleic Acids Res., 40, 8001–8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dahlem T.J., Hoshijima K., Jurynec M.J., Gunther D., Starker C.G., Locke A.S., Weis A.M., Voytas D.F. and Grunwald D.J. (2012) Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet., 8, e1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Eaton R.C., Bombardieri R.A. and Meyer D.L. (1977) The Mauthner-initiated startle response in teleost fish. J. Exp. Biol., 66, 65–81. [DOI] [PubMed] [Google Scholar]
- 54. Santos-Sacchi J., Kakehata S. and Takahashi S. (1998) Effects of membrane potential on the voltage dependence of motility-related charge in outer hair cells of the Guinea-pig. J. Physiol., 510, 225–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhao H.B. and Santos-Sacchi J. (1999) Auditory collusion and a coupled couple of outer hair cells. Nature, 399, 359–362. [DOI] [PubMed] [Google Scholar]
- 56. Ricci A.J., Bai J.P., Song L., Lv C., Zenisek D. and Santos-Sacchi J. (2013) Patch-clamp recordings from lateral line neuromast hair cells of the living zebrafish. J. Neurosci., 33, 3131–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Corey D.P. and Hudspeth A.J. (1983) Analysis of the microphonic potential of the bullfrog's sacculus. J. Neurosci., 3, 942–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lin S.Y., Vollrath M.A., Mangosing S., Shen J., Cardenas E. and Corey D.P. (2016) The zebrafish pinball wizard gene encodes WRB, a tail-anchored-protein receptor essential for inner-ear hair cells and retinal photoreceptors. J. Physiol., 594, 895–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Corey D.P., Garcia-Anoveros J., Holt J.R., Kwan K.Y., Lin S.Y., Vollrath M.A., Amalfitano A., Cheung E.L., Derfler B.H., Duggan A. et al. (2004) TRPA1 is a candidate for the mechanosensitive transduction channel of vertebrate hair cells. Nature, 432, 723–730. [DOI] [PubMed] [Google Scholar]
- 60. Erickson T., Morgan C.P., Olt J., Hardy K., Busch-Nentwich E., Maeda R., Clemens R., Krey J.F., Nechiporuk A., Barr-Gillespie P.G. et al. (2017) Integration of Tmc1/2 into the mechanotransduction complex in zebrafish hair cells is regulated by transmembrane O-methyltransferase (Tomt). Elife, 6, e28474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Reynolds S.M., Kall L., Riffle M.E., Bilmes J.A. and Noble W.S. (2008) Transmembrane topology and signal peptide prediction using dynamic bayesian networks. PLoS Comput. Biol., 4, e1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Viklund H. and Elofsson A. (2008) OCTOPUS: improving topology prediction by two-track ANN-based preference scores and an extended topological grammar. Bioinformatics, 24, 1662–1668. [DOI] [PubMed] [Google Scholar]
- 63. Tusnady G.E. and Simon I. (2001) The HMMTOP transmembrane topology prediction server. Bioinformatics, 17, 849–850. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


