Abstract
Summary
Estimating efficacy of gene–target-disease associations is a fundamental step in drug discovery. An important data source for this laborious task is RNA expression, which can provide gene–disease associations on the basis of expression fold change and statistical significance. However, the simply use of the log-fold change can lead to numerous false-positive associations. On the other hand, more sophisticated methods that utilize gene co-expression networks do not consider tissue specificity. Here, we introduce Transcriptome-driven Efficacy estimates for gene-based TArget discovery (ThETA), an R package that enables non-expert users to use novel efficacy scoring methods for drug–target discovery. In particular, ThETA allows users to search for gene perturbation (therapeutics) that reverse disease-gene expression and genes that are closely related to disease-genes in tissue-specific networks. ThETA also provides functions to integrate efficacy evaluations obtained with different approaches and to build an overall efficacy score, which can be used to identify and prioritize gene(target)–disease associations. Finally, ThETA implements visualizations to show tissue-specific interconnections between target and disease-genes, and to indicate biological annotations associated with the top selected genes.
Availability and implementation
ThETA is freely available for academic use at https://github.com/vittoriofortino84/ThETA.
Contact
vittorio.fortino@uef.fi
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
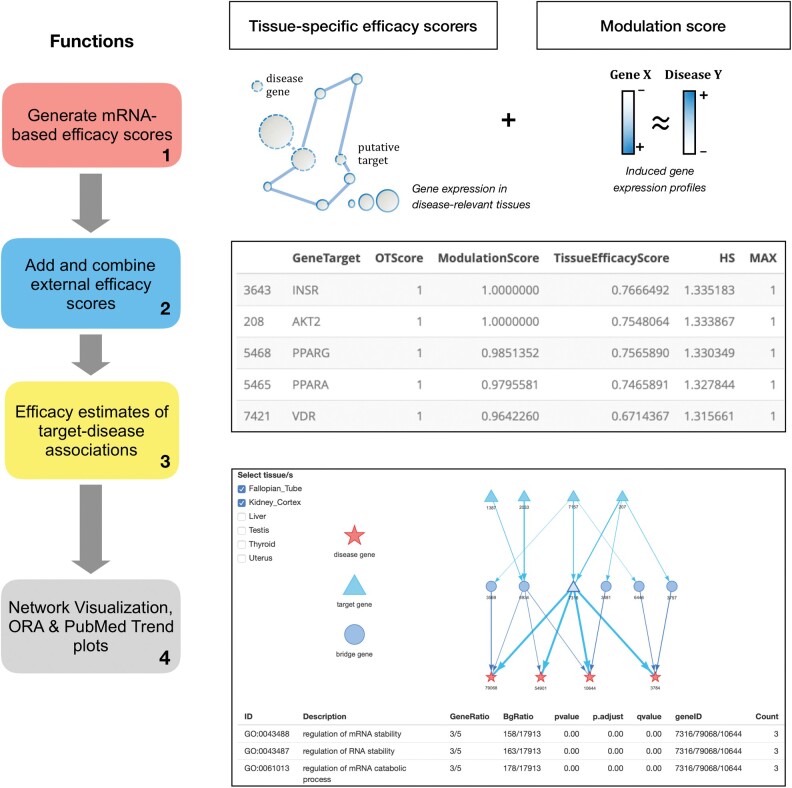
In order to minimize the risk of drug development failures, academic and industrial research has focused on target-based drug discovery approaches. This has led to several computational methods to score gene(target)–disease association upon efficacy estimates calculated from different data sources, ranging from scientific publications to omics databases (Koscielny et al., 2017; Nguyen et al., 2017; Piñero et al., 2017). We have recently proposed two transcriptome-driven approaches to identify and score gene–disease associations (Failli et al., 2019), namely tissue-specific efficacy (TSE) and modulation (MOD) scores. The first method identifies genes that are closely related to disease-genes (genes with genetic variants that associate with disease risk) in tissue-specific gene co-expression networks. The second method estimates the likelihood of a gene perturbation (e.g. knockout or know-down) resulting in specific reversion of disease gene-expression profiles. As we have previously reported, these methods can considerably increase the true positive rate of known target–disease associations (Failli et al., 2019). Here, we introduce ThETA, an R package that easily facilitates performing these efficacy scoring methods. In particular, ThETA provides functions (i) to tailor the workflow of the proposed scoring methods, (ii) to integrate these novel scores with efficacy estimates available on the Open Targets Platform and generate an overall efficacy score that can be used to prioritize target–disease associations. Moreover, ThETA provides visualization tools to depict tissue-specific network paths linking top targets (or genes) and disease-genes, to visualize biological annotations associated to set of selected gene targets. An example of workflow that R-users can implement with the ThETA package is depicted in Figure 1.
Fig. 1.
An overview of the functions provided by ThETA. (1) ThETA generates target(gene)–disease association scores by using two novel mRNA-based scoring methods. (2) ThETA adds and combines efficacy scores retrieved from alternative drug–target discovery platforms (e.g. Open target platform). The table aligned with the steps 2 and 3 indicates the top-ranked targets for Type 2 Diabetes after using the harmonic sum as prioritization score. (3) ThETA compiles efficacy estimates for all annotated disease–gene pairs, and it (4) provides an R-shiny application to display selected drug targets in tissue-specific networks. The tissue-specific gene networks include three different types of node: known disease-genes (red stars), novel targets (light blue triangles) and bridge genes (blue circles), which connect putative targets to known disease-genes. (Color version of this figure is available at Bioinformatics online.)
2 Methods and features
This section describes the main features of the R package ThETA.
2.1 Compiling transcriptome-driven efficacy scores
The R package ThETA provides the implementation of two transcriptome-based efficacy scoring methods, namely TSE and MOD scores, respectively. By traversing existing tissue/disease specific networks, the tissue-specific scoring (TSE) method detects gene targets that are closely related to disease-genes in disease-relevant tissues. While, the MOD score estimates the likelihood of a gene perturbation (e.g. knockout and knockdown) to result in specific reversion of disease gene-expression profiles. More details on the TSE and MOD scores can be found in our previous study (Failli et al., 2019). In order to compile the TSE score, the user has to provide the following data inputs: tissue-specific gene-expression data, gene–disease pairs from genome-wide association studies, and human protein–protein interaction (PPI) network. Additionally, ThETA provides three pre-computed datasets for this purpose, including data retrieved from GTEx (Ardlie, 2015), DisGeNET (Piñero et al., 2017) and StringDB (Franceschini et al., 2013). In addition, ThETA includes two datasets representing pre-computed node centrality scores from the human PPI network and disease–tissue association scores. These five datasets allow users to rapidly compile TSE scores. However, users still have the possibility to specify different input data and cut-off values for the selection, e.g. for the most significant disease–tissue associations. The MOD score requires lists of up- and down-regulated genes induced by disease and gene perturbations (e.g. gene knockout, knockdown, etc.). For this task, ThETA included gene lists retrieved from Enrichr (Kuleshov et al., 2016). Details of the input data format are included in the Supplementary Material document called ‘Walkthrough ThETA’. Moreover, known target–disease associations from the DrugBank database (Wishart et al., 2006, 2018), the Therapeutic Target Database (Chen et al., 2002; Wang et al., 2020) and the Comparative Toxicogenomics Database (Davis et al., 2017, 2019) are provided in order to allow users to assess the accuracy of the compiled efficacy estimates. These databases include pairwise associations on drugs, molecular targets and diseases.
2.2 Uploading and combining external efficacy estimates
Many different drug–target discovery platforms, such as Open Targets (Koscielny et al., 2017) and DisGeNET, provide efficacy scores for drug–target disease associations. These scores, which are freely available for download from their respective web sites, can be integrated with the efficacy estimates provided by ThETA in order to define more robust efficacy estimates for the prioritization of disease–target associations. Currently, ThETA implements two integration methods: the harmonic sum proposed by the authors of Open Targets (https://docs.targetvalidation.org/getting-started/scoring) and the max function. The max simply considers the maximum value across different efficacy estimates. While, the harmonic sum aggregates individual efficacy scores, sorted by descending score i.e. from higher to lower values.
2.3 Compiling tissue-specific networks and biological annotations for selected gene targets
An important novelty presented by ThETA is the use of tissue-specific information for the evaluation of genes as drug targets. Indeed, it is acknowledged that drugs modulating tissue-specific targets are more likely to succeed in phase 3 of clinical trials, and that by targeting tissue specificity there are opportunities to identify drug targets with improved efficacy and safety (Ryaboshapkina and Hammar, 2019). Therefore, given the importance of tissue specificity of drug targets, ThETA includes an interactive visualization tool, based on R-shiny (Chang et al., 2017), to display tissue-specific gene networks highlighting genes and pathways that connect putative targets with the genetic loci that underlie disease susceptibility, or simply disease-genes (see Fig. 1). In these graph structures, the selected genes are distinguished from the disease-genes and the so-called bridge genes, which connects genetic variations associated with diseases and selected targets. Another important feature of the presented R package is the possibility to compile extensive biological annotations. By using enrichplot (Yu, 2018) and clusterProfiler (Yu, 2018) R-packages, ThETA can compile different biological annotations, including KEGG (Kanehisa et al., 2019; Kanehisa and Goto, 2000), GO (Ashburner et al., 2000; The Gene Ontology Consortium, 2019) and REACTOME (Fabregat et al., 2018; Jassal et al., 2020), linked to selected targets. In more detail, given a target, it selects all genes in the shortest pathways connecting that target to known disease-genes, within relevant tissue-specific networks, and compiles corresponding biological annotations. Moreover, ThETA can be used to further explore the genes that are closely related to selected targets by using Random Walk with Restart (Fang and Gough, 2014).
3 Conclusion
ThETA offers a user-friendly toolbox in R for the computation of mRNA-driven efficacy estimates of disease–target associations. It allows the user to customize the selection of disease-relevant tissues and the estimation of tissue-specific and MOD scores. Comprehensive datasets are included to facilitate easy adaption of the methods. Moreover, different visualization and biological annotation tools are provided to conduct biological interpretations on putative drug targets. Finally, the R package ThETA provides tutorial vignettes including extensive examples on how to use its functions.
Financial Support: none declared.
Conflict of Interest: Dr M.F. and Dr J.P. have been working at University of Eastern Finland for Business Finland funded project that explores commercialization of drug–target prioritization technologies. Dr J.P. is an employee of Blueprint Genetics Ltd.
Supplementary Material
Contributor Information
Mario Failli, Institute of Biomedicine, University of Eastern Finland, Kuopio 70210, Finland; Department of Chemical, Materials and Industrial Engineering, University of Naples 'Federico II', Naples 80125, Italy.
Jussi Paananen, Institute of Biomedicine, University of Eastern Finland, Kuopio 70210, Finland; Blueprint Genetics Ltd, Finland.
Vittorio Fortino, Institute of Biomedicine, University of Eastern Finland, Kuopio 70210, Finland; Blueprint Genetics Ltd, Finland.
References
- Ardlie K.G. et al. ; The GTEx Consortium. (2015) Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science, 348, 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M. et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, W. et al. (2017) Shiny: web application framework for R. R package version, 1(5). [Google Scholar]
- Chen X. et al. (2002) TTD: therapeutic target database. Nucleic Acids Res., 30, 412–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A.P. et al. (2017) The comparative toxicogenomics database: update 2017. Nucleic Acids Res., 45, D972–D978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A.P. et al. (2019) The comparative toxicogenomics database: update 2019. Nucleic Acids Res., 47, D948–D954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregat A. et al. (2018) The reactome pathway knowledgebase. Nucleic Acids Res., 46, D649–D655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Failli M. et al. (2019) Prioritizing target-disease associations with novel safety and efficacy scoring methods. Sci. Rep., 9, 9852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H., Gough J. (2014) The “dnet” approach promotes emerging research on cancer patient survival. Genome Med., 6, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini A. et al. (2013) STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res., 41, D808–D815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jassal B. et al. (2020) The reactome pathway knowledgebase. Nucleic Acids Res., 48, D498–D503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res., 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. et al. (2019) New approach for understanding genome variations in KEGG. Nucleic Acids Res., 47, D590–D595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koscielny G. et al. (2017) Open Targets: a platform for therapeutic target identification and validation. Nucleic Acids Res., 45, D985–D994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov M.V. et al. (2016) Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res., 44, W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D.-T. et al. (2017) Pharos: collating protein information to shed light on the druggable genome. Nucleic Acids Res., 45, D995–D1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñero J. et al. (2017) DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res., 45, D833–D839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryaboshapkina M., Hammar M. (2019) Tissue-specific genes as an underutilized resource in drug discovery. Sci. Rep., 9, 7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium (2019) The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res., 47, D330–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. et al. (2020) Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res., 48, D1031–D1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D.S. et al. (2006) DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res., 34, D668–D672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D.S. et al. (2018) DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res., 46, D1074–D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, G. (2018). enrichplot: visualization of functional enrichment result. R package version 112.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.