Figure 7.
Correlation of X-ray and fluorescence data in three dimensions using the beamline B24 platform
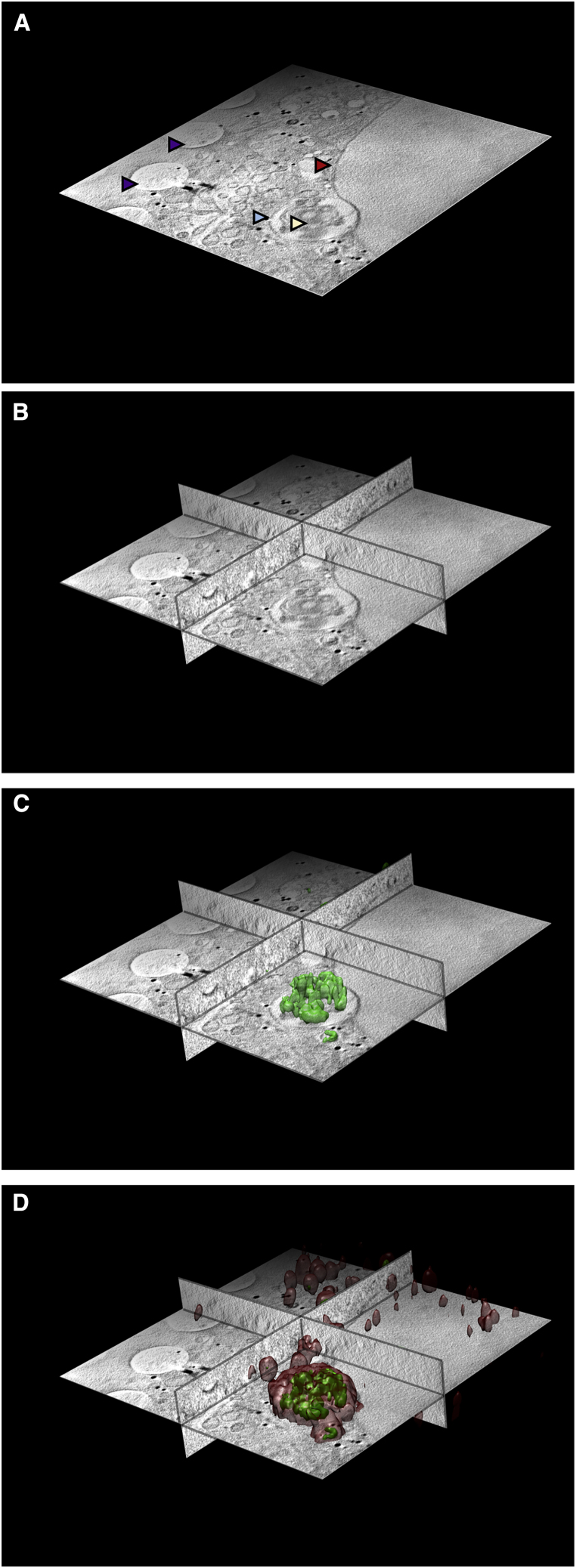
All the data shown here are from a U2OS cell vitrified 4 h AI with high titers of reovirus T3D. Viral components are fluorescently labeled with Alexa488 (green fluorescence), and there is also an endogenously expressed reporter molecule (Galectin-3) that is accumulated in vesicles carrying concentrated reovirus outer capsids and that carries the mCherry fluorophore (red fluorescence).
(A) Z slice from the middle of a soft X-ray absorption tomogram (from beamline B24 using the 40 nm objective) of a U2OS cell with the nucleus on the right side of the figure (red arrow; the two leaves of the nuclear membrane are seen clearly) and the cytoplasmic area with a substantial multi-vesicular body/endolysosome (pale blue arrow), containing folded carbon-dense material given the increased X-ray absorption in that structure (pale yellow arrow). In the thinner areas of the distal cytoplasmic region, several holes can be seen as part of the support film used for cell attachment (deep purple arrows).
(B) Ortho-slices of (A), showing a representative portion of the volumetric data.
(C) 3D rendering of the cryoSIM recorded structures displaying green fluorescence within and around the endolysosome.
(D) 3D semi-transparent rendering of all cytoplasmic vesicles that contain red Galectin-3 fluorescence (data also collected on the cryoSIM) and are seen as an indication of viral presence in those vesicles. All images were generated with Chimera (Pettersen et al., 2004).