Abstract
Negative checkpoint regulators (NCRs) temper the T cell immune response to self-antigens and limit the development of autoimmunity. Unlike all other NCRs that are expressed on activated T lymphocytes, V-type immunoglobulin domain-containing suppressor of T-cell activation (VISTA) is expressed on naïve T cells. We report an unexpected heterogeneity within the naïve T cell compartment in mice, where loss of VISTA disrupted the major quiescent naïve T cell subset and enhanced self-reactivity. Agonistic VISTA engagement increased T cell tolerance by promoting antigen-induced peripheral T cell deletion. Although a critical player in naïve T cell homeostasis, the ability of VISTA to restrain naïve T cell responses was lost under inflammatory conditions. VISTA is therefore a distinctive NCR of naïve T cells that is critical for steady-state maintenance of quiescence and peripheral tolerance.
Checkpoint regulation of T cell function is governed by coinhibitory molecules (e.g., CTLA-4, VISTA, LAG-3, TIM-3, and TIGIT), which act in concert to fine-tune T cell response and fate (1). The importance of these negative checkpoint regulators (NCRs) has been clearly established for cancer and infectious diseases (2), but because NCRs are expressed only after T cell activation, it has not yet been determined if they play a role within the naïve T cell compartment to maintain quiescence or response to self-antigen (1–4). Quiescent T cells make up the over-whelming majority of T lymphocytes in the periphery. Maintaining T cell quiescence and tempering self-reactivity are active processes necessary for survival of an individual. Quiescence regulation is controlled by a diverse set of transcriptional regulators, including forkhead box (FOX) proteins, Kruppel like factors (KLFs), and APRO (Tob1) family members (5–7). Through control of cellular state and cell cycle arrest, these transcription factors (TFs) reduce the resources necessary to maintain the vast repertoire of resting T cells, of which only an extremely limited frequency will be clonally selected by antigen during the lifetime of the host. Impaired function or deletion of these intracellular mediators can lead to T cell activation and a breakdown in self-tolerance (2–4, 8–10). Therefore, quiescence and tolerance are functionally linked. Although insights into the intracellular mediators that control naïve T cell quiescence are being realized, the checkpoint regulators expressed on T cells that regulate quiescence are yet to be described.
V-type immunoglobulin domain-containing suppressor of T-cell activation (VISTA) is a member of the B7 family that is distinct from other negative checkpoint molecules in that it is constitutively expressed on naïve T cells. Mice deficient in VISTA show an enhanced frequency of antigen-experienced memory CD4+ CD44hi T cells, heightened cytokine production, and an increased propensity to develop autoimmunity (11–14). In this regard, genetic deletion of VISTA in the 2D2 myelin oligodendrocyte glycoprotein (MOG)–specific CD4+ T cell receptor (TCR) transgenic (Tg) mouse model of spontaneous autoimmunity results in greatly enhanced inflammatory disease and diminished survival (13). Taken together, these observations support the hypothesis that VISTA deficiency results in a breakdown of self-tolerance and the development of inflammatory T cell self-reactive responses. That VISTA is expressed on naïve T cells and lost upon immunization (12, 13) further suggests that its impact on controlling self-tolerance is within the naïve T cell subset.
Results
VISTA deficiency disrupts the naïve T cell repertoire by reducing quiescence and enhancing T cell activation
VISTA has been shown to act as a coinhibitory receptor on resting CD4+ T cells that negatively regulates T cell activation (12, 13, 15). VISTA-deficient CD4+ T cells exhibit enhanced proliferation and effector responses to anti-CD3 and antigenic stimulation in vitro (15). VISTA−/− mice have heightened antitumor responses to autologous tumors and are more susceptible to death resulting from ConA-induced hepatitis (12, 13, 15). Although the steady-state percentage of CD4+ T cells was not enhanced in VISTA−/− mice, two groups in-dependently reported an increase in “antigen-experienced” CD44hi CD62Llo CD4+ T cells in the spleens and peripheral blood of VISTA−/− mice (12, 13). Under conditions of conditional VISTA deficiency within the CD4+ T cell compartment, we observed a similar increase in the frequency of antigen-experienced CD4+ T cells, suggesting that the intrinsic loss of VISTA was sufficient for the rise of this activated T cell subset (fig. S1A) (12, 13). That VISTA is expressed on >97% of naïve T cells (fig. S1B) and is lost under inflammatory conditions suggests that its impact on controlling T cell responses is intrinsic to the naïve T cell subset. On the basis of these findings, we interrogated the naïve CD4+ T cell compartment to determine if VISTA altered the steady state to influence their differentiation to antigen-experienced CD44hi cells.
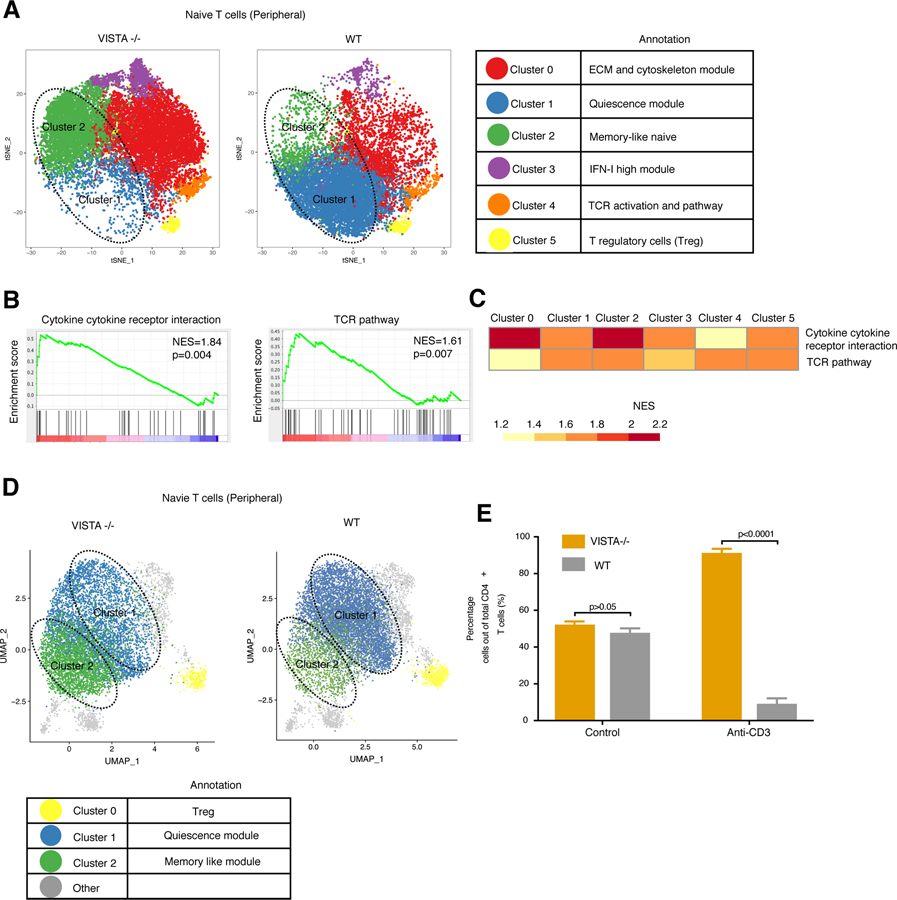
We deleted VISTA in the CD4+ T cell compartment using CD4-Cre mice (hereafter referred to as CD4-Cre-VISTA−/−) and performed single-cell RNA-sequencing (scRNA-seq) to examine the role of VISTA in naïve CD4+ T cell transcriptional heterogeneity. scRNA-seq analysis of sorted, naïve (CD44lo CD62Lhi) CD4+ T cells from CD4-Cre-VISTA−/− mice versus their littermate wild-type (WT) controls revealed a shift in the transcriptional phenotype and heterogeneity within the T cell compartment (Fig. 1A; fig. S1, C and D; and table S1). The most significant phenotypic shift was observed for clusters 1 and 2 (described below). Cluster 1 represents a population of quiescent T cells marked by an up-regulation of Klf2 and its effectors, which include Ccr7 (fig. S1 and table S1). This module was reported to be critical for the active maintenance of T cell quiescence and inhibition of proliferation (7, 9, 16). There is also data supporting the importance of KLF2 in regulating thymocyte trafficking (17, 18). This cluster also included antiproliferative genes such as Klf6, which (similar to Klf2) up-regulates the negative cell cycle regulator p21 (16, 19, 20). Btg1 and Btg2 are members of the Tob gene family, which have critical antiproliferative functions and whose member Tob1 is well described in regulating T cell quiescence and anergy (21–23). Both genes were defining markers for cluster 1, which is predominant in the WT and lost in the VISTA−/− naïve CD4+ T cell population. VISTA deficiency reduced the abundance of naïve T cells in cluster 1 by more than 10-fold compared with WT CD4+ T cells. Cluster 2, a population augmented by fivefold in VISTA−/− CD4+ naive T cells, was marked by an up-regulation of a stem-cell memory-like program defined by the distinct up-regulation of Tcf7, Bcl2, and Il7r (table S1). Of note, one of the defining genes for this cluster was Klf3, which has been reported to antagonize KLF2 function (24). In addition, this cluster had a higher expression of the costimulatory receptors Slamf6 and Ifngr1, which suggests that these cells are transcriptionally poised for better effector cell responses (25–27) and that VISTA may play an intrinsic role in maintaining naïve T cell identity and homeostasis. Cluster 0 (enhanced by 2.5-fold by loss of VISTA) was defined by an up-regulation of extracellular matrix interaction pathways and genes such as actin (Actg1 and Actb) and Cnn2, which mediate cytoskeletal rearrangements (tables S1 and S2). The role of these pathways has been now appreciated in sustaining the immunological synapse and driving T cell effector function and is supported by pathway analysis of this cluster (28, 29). We observed an up-regulation of TCR pathways in this cluster compared with the remaining population, supporting an overall enhanced abundance of greater TCR signaling in VISTA−/− T cells (table S2).
Fig. 1. Intrinsic VISTA deficiency alters heterogeneity in the naïve CD4+ T cell pool.
(A to C) scRNA-seq was performed on naïve CD4+ T cells from WT mice and CD4-Cre-VISTA−/− mice (for which VISTA deficiency is restricted to the CD4+ T cell compartment). (A) t-Distributed stochastic neighbor embedding (t-SNE) plot showing the cluster distribution of FACS-sorted single naïve (>99%) (CD62Lhi CD44lo) CD4+ T cells (sorted on the basis of the 20% lowestCD44− from the negative gate) from CD4-Cre-VISTA−/− and WT littermates. Each dot corresponds to one single cell, colored according to cell cluster. The biological annotation of each cluster is shown in the table on the right. The dashed circles indicate the quiescent T cell cluster (cluster 1) and memory-like naïve T cell cluster (cluster 2). (B) Gene set enrichment analysis (GSEA) pathway enrichment plot indicating the representative gene sets enriched in VISTA−/− versus WT CD4+ T cells. Normalized enrichment score (NES) and P values are shown for each gene set. P values were calculated by Kolmogorov-Smirnov test. (C) Heatmap showing GSEA analysis as performed in (B) for each cluster. The NES is shown for each gene set across clusters. (D) Uniform manifold approximation and projection (UMAP) plot showing the cluster distribution of FACS-sorted single naïve (>99%) (CD62Lhi CD44lo) CD4+ T cells from VISTA−/− and WT mice. For (A) to (D), data are representative of two independent experiments with at least three mice per group. (E) Ratios of recovered WT versus VISTA−/− CD4+ T cells 5 days after coadoptive transfer into Rag1−/− hosts within vivo anti-CD3 stimulation or control immunoglobulin G (IgG). Data are representative of four independent experiments with at least four mice per group. Each bar indicates the mean value, and each error bar refers to one standard deviation (SD). Student’s t tests were performed to compare WT with VISTA deficiency (VISTA−/−) under each condition (i.e., no treatment versus anti-CD3).
Two independent groups previously showed that VISTA deficiency or targeting may affect induced regulatory T cell (Treg) induction and stability (30, 31). There were no significant differences in the abundance of the CD44lo Treg cluster (cluster 5) or a cluster defined by enhanced TCR activation–associated transcriptional differences (cluster 4). As stated above, as with the other clusters, there was enhanced TCR signaling imparted by VISTA deficiency (Fig. 1B and tables S2 and S3). In all of the clusters, we observed a significant up-regulation in multiple TCR signaling and cytokine response pathways in VISTA-deficient cells (Fig. 1, B and C, and table S2). Clusters 3 to 5 accounted for less than 7% of the total naïve T cell population and showed no significant differences in abundance between WT and VISTA−/− groups (not discussed in detail).
Given that loss of VISTA reduced quiescent T cells and altered the naïve CD4 T cell repertoire at the gene expression level, we hypothesized that VISTA maintains the epigenetic program for naïve T cell quiescence. We used the assay for transposase-accessible chromatin using sequencing (single-cell ATAC-seq) (32) on naïve CD4 T cells from CD4-Cre-VISTA−/− mice or littermate controls. The cell population changes observed were in agreement with those predicted by our scRNA-seq studies. For example, a significant reduction in quiescent T cells (cluster 1) and an increase in memory-phenotype cells (cluster 2) in the VISTA−/− naïve CD4+ T cells were seen (Fig. 1D; fig. S1, E to Q; and table S4). As has been reported by several studies of T cells poised to respond to TCR signaling (33, 34), cluster 2 had an enhancement in the accessibility of multiple TCR effectors (Nr4a1, Cd247, Jun, Fos, Lat, Nr4a1, Dgka, and Nfkb1) as well as Cd4, Icos, and Cd40lg (Fig. 1D; fig. S1, H to P; and table S4). There is accumulating evidence suggesting that memory cells have significantly greater chromatin accessibility to TCR effector genes (35, 36). Of note, we also observed enhanced accessibility to genes up-regulated in the VISTA−/− memory-phenotype cluster, such as Tcf7, Ifngr1, Bcl2, and Il7ra, which supports the suggestion that VISTA deficiency epigenetically primes the naïve T cell repertoire toward a more TCR-responsive memory-like phenotype. Although memory regulators such as Lef1, Zbtb20, and Runx3 were not differentially expressed between the quiescent and memory-like clusters at the mRNA level, they had greater chromatin accessibility in the memory-like cluster cells enhanced by VISTA deficiency (table S4). On the other hand, the quiescent cluster representing the majority of WT naïve CD4+ T cells was defined by enhanced chromatin accessibility to Klf2, Klf6, Btg1, and Btg2, which are the defining markers of quiescent cells by scRNA-seq analysis (Fig. 1, A and D; fig. S1, H, K, and L; and table S4). In addition, this cluster also had an enhancement in the accessibility of important quiescence factors like Foxp1, Foxo-1, and Runx1 (37–39) (table S4).
One alternative possibility was that the reduced quiescence state of cluster 2 and its expansion in the absence of VISTA were a consequence of increased autoreactive TCR repertoires in this cluster. To address this hypothesis, we performed single-cell TCR sequencing paired with gene expression to resolve the TCR sequences of each cell per cluster. In addition, we extensively reviewed the literature and manually curated a database of more than 50 TCR Vβ CDR3 sequences from CD4+ T cells in multiple models of autoimmunity matching different autoantigen specificities because certain Vβ genes were associated with autoreactivity (40) (fig. S2A). We found less than 100 cells across all clusters (out of 40,000 total cells) in the naïve CD4 T cell repertoire that matched these TCR sequences (40–42). Because this analysis was not sufficient to capture the landscape of autoreactivity in the naïve CD4+ T cell population, we performed single-cell TCR sequencing on CD44hi CD4+ T cells (10× genomics, paired α and β chain) because these cells have reacted to self-antigen in an unimmunized mouse (fig. S2B) (43). In this analysis, only the Vβ TCR repertoire with full length and productivity were chosen. We performed CDR3-region sequence alignment (44) and chose the Vβ CDR3 sequences that were fully matched between CD44lo and CD44hi cells. We identified a total 4971 “potentially” autoreactive CD44lo cells with 1606 unique TCRs. Then, we quantified the fraction of autoreactive T cells in each CD44lo cluster (clusters having less than 1000 cells were excluded). The fraction of autoreactive T cells was almost evenly distributed among all CD44lo clusters in both VISTA−/− and WT mice (fig. S2B). Because CD44hi CD4+ T cells in an unimmunized mouse may not represent the prototypical autoreactive repertoire seen in autoimmune disease, we performed single-cell TCR sequencing on CD4+ T cells sorted by fluorescence-activated cell sorting (FACS) from B6 Fas lpr mice, an established lupus model for which autoreactive T cells have been reported (45). We also performed the same sequencing procedure on CD4+ T cells from Bim-deficient mice, which fail to delete autoreactive T cells during negative selection (46). This allowed us to generate full CDR3 sequences for autoreactive TCRs from two independent models of autoimmunity. In both TCR sequence datasets, there was an overlap in the TCR sequences (around 4 to 5%) for each of the naïve CD4 T cell clusters (fig. S2, C to F). However, there was no impact of VISTA loss on the percentage or distribution of the autoreactive T cells. Our interpretation is that the changes in the clusters imposed by the loss of VISTA are not due to changes in TCR specificity of the constituting cells but rather mostly due to a change in the cell state.
To determine if the phenotypic changes in the naïve T cell repertoire were imparted by VISTA deficiency at the mature T cell stage in the periphery or were a consequence of a potential role that VISTA plays in thymocyte development, analysis of the impact of VISTA deficiency on thymocyte heterogeneity was studied. VISTA is constitutively expressed on naive CD4+ T cells and also on single-positive thymocytes (fig. S3, A and B). Flow cytometric analysis of thymocyte subset percentages did not show any impact of intrinsic VISTA deficiency on the thymocyte numbers or frequency (fig. S3B). In addition, scRNA-seq of thymocytes from VISTA−/− or WT littermates did not reveal any differences in heterogeneity of the thymic repertoire (fig. S3, C to E, and table S5). Analysis of the CD4+ lineage differentiation trajectory from double-positive thymocytes to the naïve peripheral T cell stage using the well-established Monocle algorithm (47) did not elucidate an impact of VISTA deficiency on the route of CD4+ T cell differentiation (fig. S3, G and H). This suggests that VISTA deficiency did not alter the differentiation route of thymocytes to mature T cells and exclusively exerted an impact on naïve T cell fate in the peripheral compartment.
A series of experiments was performed to gain insights into whether expression of KLF2 was correlated with the expression of other quiescence factors in T cells and, in addition, correlated with VISTA expression. The data presented show that VISTA deficiency results in a global reduction of quiescence regulators such as Klf2, Klf6, Gimap5, and Tob gene family members Btg1 and Btg2 (9, 16, 19–23, 48). Therefore, there was sufficient evidence to suggest that VISTA is necessary for the expression of multiple quiescence regulators. To more directly address if KLF2 expression was coregulated with the other quiescence factors, a KLF2 reporter mouse (18) was used. Using this system, we sought to examine whether higher KLF2 expression on naïve CD4+ T cells recapitulated the naïve T cell quiescence phenotype (cluster 1). KLF2hi and KLF2lo naïve CD4+ T cells were electronically cell sorted (on the basis of the 20% highest and lowest expression) and subjected to deep RNA-seq analysis. KLF2hi CD4+ T cells were highly enriched for genes that define the quiescence cluster of naïve T cells (cluster 1), closely mirroring their profile with regard to differential gene expression (fig. S4A). Therefore, higher expression of KLF2hi is correlated with the heightened expression of other quiescence factors. KLF2hi CD4+ T cells additionally up-regulated several established quiescence regulators such as Tob1, Foxp1, Foxo1, and Tgfbr2 (fig. S4B) (6, 21). Next, we examined the relationship between VISTA and KLF2 expression, a defining cluster 1 TF and marker. As such, flow cytometric analysis revealed a strong direct correlation between VISTA and KLF2 expression, because increased KLF2 expression (KLF2hi) also showed higher VISTA expression (fig. S4, C and D). Of note, KLF2hi CD4+ had significantly higher VISTA mRNA expression. The same RNA-seq analysis was conducted for VISTAhi versus VISTAlo naïve T cells, and VISTAhi CD4+ T cells showed greater KLF2 (as well as Klf6, Btg1, and Btg2). This bidirectional relationship (Pearson correlation coefficient of 0.87) between KLF2 and VISTA is presented as a correlation plot (fig. S4E). Taken together, these data provide compelling evidence using flow cytometry and RNA-seq that VISTA regulates KLF2, an important TF with roles in T cell quiescence. Given the relationship between VISTA and KLF2, we posit that enhanced VISTA expression on the naïve T cell compartment (CD44lo CD62Lhi) correlates with greater quiescence and the naïve phenotype (fig. S4F). We sought to track the impact of graded VISTA expression on naïve T cells. Using deep RNA-seq, we found that VISTAhi naïve T cells display a more quiescent T cell state than the VISTAlo or VISTA−/− T cells at the global gene expression level (fig. S4G). In summary, there was significant up-regulation of Klf2, Klf6, and Slfn2 and a dramatic up-regulation of Foxp1 in VISTAhi T cells, all critical effectors of T cell quiescence (8, 37). However, VISTAlo T cells expressed higher levels of Nr4a1, Myc (inhibited by Klf2) (9), Pdcd1, Ctla4, Cd5, Cd6, Cd2, Nfkb1, Lck, and Nfatc1, all indicative of enhanced TCR signaling, enhanced activity, and reduced quiescence.
Because the percentage of CD44hi memory-phenotype (MP) CD4+ T cells is enhanced in steady-state unimmunized VISTA−/− mice (fig. S1A) and VISTA deficiency skews the naïve CD4+ T cells toward a less quiescent memory-like phenotype at both the transcriptional and epigenetic levels (Fig. 1 and fig. S1), we investigated how VISTA-deficiency influenced the expansion of the naïve CD4+ T cells toward CD44hi MP cells by scRNA-seq profiling of this population (fig. S5). At least for the CD4+ T cell lineage, conversion of naïve (CD62Lhi CD44lo) T cells to CD44hi MP cells requires antigen encounter (self but not commensal antigen) and sufficient TCR stimulation (43, 49). Examination of the VISTA-deficient CD44hi MP T cells revealed that the most dramatic enhancement caused by VISTA deficiency was a more than threefold increase in T helper 1 cell (TH1) effector phenotype cells (cluster 1) (fig. S5, A to C). Cells in cluster 1 up-regulate the TH1 master TF Tbx21 (T-bet) as well as the characteristic TH1 effector molecules Ifng, Ccl5, and Cxcr3 (table S6) (50). Globally, there was an up-regulation of effector molecules Ifng, Ccl5, and Cxcr3, in addition to costimulatory molecules such as Cd7, Cd40lg, Cd69, and Ly6c (fig. S5 and table S6). On the other hand, there was more than a threefold reduction of a coinhibitory module group of cells (cluster 5) in the antigen-experienced repertoire defined by up-regulation of multiple checkpoint regulators such as PD-1, LAG-3, TIGIT, CD73 (Nt5e), FR4 (Izumo1r), BTLA, and Nrp1 and other regulators of T cell dysfunction such as c-Maf, NFATc, Tox, and Tox2 (fig. S5E) (51). Indeed, analysis of the whole T cell population supported this because VISTA−/− had markedly reduced expression of the coinhibitory regulators (fig. S5F). At the cell-state level, we observed that CD4-Cre-VISTA−/− MP CD4+ T cells up-regulate greater downstream TCR activation genes as marked by significant global up-regulation of the AP-1 and JNK TF network (Jun, Junb, Jund, and Fos) as well as Cd69, Nr4a1 (Nur77), and nuclear factor kB (NFkB) pathway effectors (fig. S5D). The majority of these genes had a greater chromatin accessibility in the VISTA−/− naïve CD44lo cells (fig. S1 and table S4). Heightened TCR signaling in VISTA−/− cells and the heightened expression of TCR activation genes and other markers of T cell activation would be predicted by the work by Paul and colleagues who showed that TCR antigen encounter and costimulation are essential for establishment of MP CD4+ T cells in un-immunized mice and that this population expresses high levels of T-bet and interferon-γ (IFN-γ) (49). It is also in agreement with reports that stronger TCR signals favor TH1 polarization (52) and that VISTA-deficiency on naïve T cells enhances production of IFN-γ and other TH1 cytokines upon TCR stimulation in vitro (12). Therefore, intrinsic VISTA expression on the naïve T cell is necessary for restraining T cell activation and maintaining quiescence, and VISTA-deficiency engenders a TH1 proinflammatory phenotype in the absence of the appropriate costimulatory or immunizing signals.
Loss of quiescence has been repeatedly correlated with reduced tolerance susceptibility of T cells (7). Given the enhanced activation phenotype of the VISTA−/− T cells and the reduction in the quiescent cluster, we hypothesized that peripheral, TCR-induced, deletional tolerance of VISTA−/− T cells may be impaired. To this end, we used a coadoptive transfer system whereby naïve VISTA−/− and WT CD4+ T cells are transferred at equal ratios into T cell– deficient Rag−/− hosts and administered anti-CD3 monoclonal antibody (mAb) or control mAb (Fig. 1E). It has been shown that under these conditions, TCR signaling by anti-CD3 induces the deletion of T cells in vivo (53, 54). In the absence of anti-CD3 stimulation, we could not detect a marked difference between WT and VISTA−/− CD4+ T cell numbers, indicating no notable advantage of VISTA deficiency on homeostatic T cell expansion or survival (Fig. 1E and fig. S4G). Only when anti-CD3 stimulation was provided could we detect a marked enhancement in the recovered numbers of VISTA−/− CD4+ T cells compared with the number of WT CD4+ T cells (Fig. 1E and fig. S5G). This supports the scRNA-seq data on the naïve T cell phenotype, establishing that VISTA deficiency leads to a loss in quiescence and a reduced susceptibility to TCR-induced deletion. T cells from CD4-Cre × VISTAfl/fl mice also recapitulated this phenotype, establishing that the resistance to anti-CD3 deletion was T cell intrinsic (fig. S5H). These data suggest that loss of quiescence affects the fate of TCR-triggered T cells in vivo.
Agonistic anti-VISTA mAbs enhance TCR-dependent peripheral T cell deletion
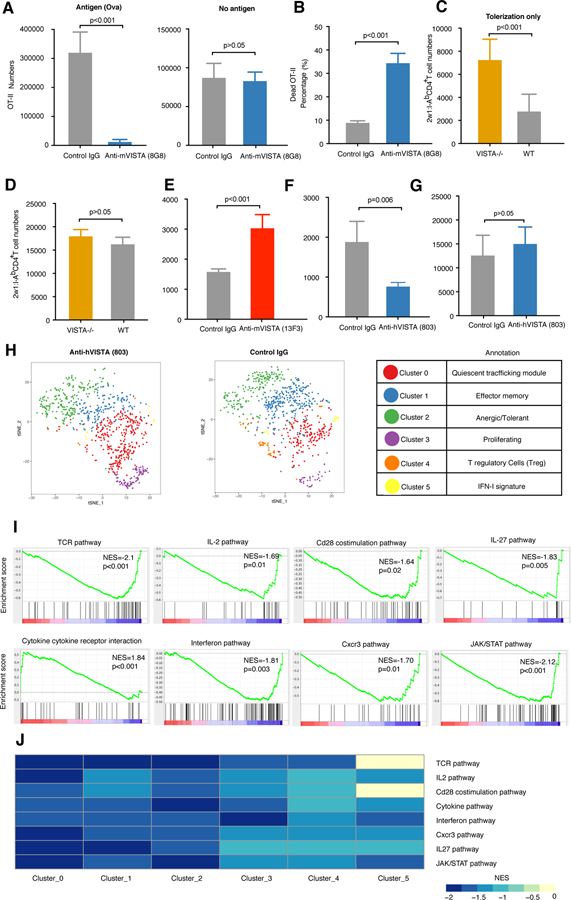
Given that we observed the functional impact of VISTA deficiency to be a reduced susceptibility to anti-CD3–induced deletion, we hypothesized that antibody-based activation of VISTA would enhance TCR-induced T cell deletion. Chen and colleagues introduced a class of anti-VISTA “agonists” and showed in multiple systems, including graft-versus-host disease (GVHD), that the agonist antibodies suppressed T cell immune responses [reviewed in (55)]. We developed both anti-mouse–specific (clone 8G8) and anti-human–specific (clone 803) VISTA agonists to assess the impact of VISTA engagement on T cell fate on TCR engagement. The isotype and functional properties of the anti-VISTA clones used in this study are detailed in table S7 (fig. S6, A to E). These mAbs are specific for mouse or human VISTA, and both suppress GVHD. In addition, mouse anti-VISTA (anti-mVISTA) (8G8) has demonstrated immunosuppressive properties in multiple murine models of inflammation and autoimmunity (fig. S6, B to E). To assess the impact of anti-VISTA agonist on tolerogen-induced T cell deletion, naïve OVA-specific transgenic CD4+ T cells (OT-II) were adoptively transferred to antigen-bearing hosts (Act-Ova) or antigen-deficient B6 controls and treated with anti-mVISTA (8G8) (56, 57) (Fig. 2A and fig. S6F). This system provides a TCR engagement signal (OVA) but no overt inflammatory signal, which are both known to promote tolerance induction (58, 59). Administration of anti-VISTA overwhelmingly reduced the frequency of OT-II T cells in the Act-Ova– expressing hosts but not in B6 hosts. Furthermore, there was an enhanced percentage of dead OT-II T cells, suggesting that anti-VISTA enhanced tolerogenic T cell death (Fig. 2B and fig. S6, G and H). VISTA has been reported to participate in the uptake and clearance of apoptotic cells (14). We therefore investigated whether the change in percentage of dead cells may be due to a role for agonistic anti-VISTA (8G8) on dead cell clearance of OT-II cells. The impact of anti-VISTA on the uptake of apoptotic thymocytes by macrophages was assessed by flow cytometry (fig. S6I). Although VISTA-deficiency had a significant impact on the uptake of apoptotic cells (14), anti-VISTA (8G8) did not demonstrate any significant inhibitory activity. Therefore, the enhanced death of OT-II T cells observed in 8G8-treated Act-OVA mice is likely due to enhanced cell death caused by augmented deletional tolerance with no impact on clearance. It remained possible that anti-VISTA may have enhanced antigen-induced T cell tolerance by direct killing of other VISTA+ immune populations, most prominently antigen-presenting cells. However, examination of the abundance of various immune populations after anti-VISTA treatment revealed no significant impact of the antibody on their numbers or frequency (fig. S6J). There were no significant reductions in the frequency T cell or various myeloid populations upon treatment of mice with either VISTA agonist or antagonist antibodies under steady-state conditions (fig. S6J). These populations include CD11b+ myeloid cells, neutrophils, monocytes, and dendritic cells.
Fig. 2. Agonistic anti-VISTA antibodies augment T cell tolerance, which is abrogated by VISTA deficiency.
(A) Recovered numbers of OT-II CD4+ T cells in the spleen of anti-mVISTA (8G8)–treated or hamster IgG control–treated mice transferred into Act-Ova (left) or B6 hosts (right) 48 hours after adoptive transfer. (B) Percentage of dead OT-II CD4+ T cells out of total recovered OT-II cells from anti-mVISTA (8G8)–treated or hamster IgG control–treated mice. Data are representative of two independent experiments with five mice per group. (C) Two doses of 2w1s peptide (100 µg) were intravenously injected into WT or VISTA−/− mice on days 0 and 3, respectively, followed by tetramer enrichment and 2w1s:I-Ab cell number quantification on day 7. (D) 2w1s peptide (100 µg) with 5 ug of LPS was intravenously injected into WT or VISTA−/− mice on day 0, followed by tetramer enrichment and 2w1s:I-Ab cell number quantification on day 7. (E) Two doses of 2w1s peptide were intravenously injected into antagonistic anti-mVISTA (13F3)–treated or hamster IgG control–treated mice on days 0 and 3, respectively, followed by tetramer enrichment and 2w1s:I-Ab cell number quantification on day 7. (F) Two doses of 2w1s peptide were intravenously injected into agonistic anti-hVISTA (803)–treated or isotype control–treated mice on days 0 and 3, respectively, followed by tetramer enrichment and 2w1s:I-Ab cell number quantification on day 7. Data are representative of four independent experiments with at least eight mice per group [(C) to (F)]. (G) 2w1s peptide (100 ug) with 5 ug of LPS was intravenously injected into control IgG–treated or agonistic anti-hVISTA (803)–treated mice on day 0, followed by tetramer enrichment and 2w1s:I-Ab cell number quantification on day 7. Data are representative of two independent experiments with eight mice per group. (H) t-SNE plot showing the cluster distribution of 2w1:I-Ab peptide–induced CD4+ T cells from agonistic anti-hVISTA (803)–treated and control IgG–treated mice. Each dot corresponds to one single cell, colored according to cell cluster. Biological annotation of each cluster is shown in the table on the right. (I) GSEA pathway enrichment plot indicating the representative gene sets depleted in agonistic anti-hVISTA (803)–treated versus isotype control–treated mice. NESs and P values are shown for each gene set. P values were calculated by Kolmogorov-Smirnov test. (J) Heatmap showing GSEA analysis as performed (I) for each cluster. NESs are shown for each gene set across clusters. Sequencing data are representative of two independent repeats with at least two samples from pooled mice per group. For all bar plots [(A) to (G)], each bar indicates the mean value and each error bar refers to one SD; P values were calculated by Student’s t test.
To prove that VISTA-induced T cell loss was due to a direct effect of anti-VISTA antibody binding to T cells, OT-II T cells expressing human VISTA (hVISTA) were specifically targeted with an anti-hVISTA antibody and transferred into WT host mice. hVISTA-expressing OT-II cells were obtained subsequent to interbreeding with hVISTA knock-in (KI) mice. Extensive validation of hVISTA lineage and expression levels and the specificity and affinity of the anti-hVISTA mAb (803) in hVISTA KI mice were evaluated (fig. S7 and materials and methods). Flow cytometric analysis of hVISTA expression on myeloid and T cell subsets in hVISTA KI mice revealed similar levels of expression to murine VISTA in WT mice (fig. S7, A to C). The specificity of anti-hVISTA (803) was validated in multiple systems (fig. S7, B and C) and also in a Jurkat cell line transduced to express hVISTA versus WT Jurkat cells (fig. S7D). In contrast to VISTA deficiency, anti-hVISTA induced profound reductions in hVISTA CD4+ T cells with anti-CD3 tolerization (Fig. 1E and fig. S7E). Similar to the anti-mVISTA clone 8G8, anti-hVISTA (803) reduced the frequency of adoptively transferred OT-II T cells expressing hVISTA upon administration of soluble OVA peptide but not in the absence of peptide (fig. S7F). These findings show that targeting hVISTA exclusively on the T cell surface, together with TCR engagement, results in a selective reduction of targeted cells. As has been observed with anti-mVISTA (8G8), there was no impact of anti-hVISTA (803) on the abundance of the different immune lineages (fig. S7G). To confirm that the augmented T cell tolerance induced by agonistic anti-VISTA and antigen was mediated by targeting the donor antigen-specific CD4+ T cells, and not due to potential indirect effects of targeting the VISTA+ myeloid cells, we adoptively transferred hVISTA OT-II cells in the presence of OVA and exclusively targeted the host using anti-mVISTA (8G8), sparing the donor hVISTA OT-II cells. In this case, there was no impact on donor antigen-specific T cell numbers. These findings indicate that targeting the T cell compartment is necessary and sufficient for the augmented tolerance by anti-VISTA agonists (fig. S7H).
VISTA regulates the fate of tolerized, endogenous antigen-specific T cells
Our findings suggest that VISTA regulates the fate of TCR-engaged T cells in vivo. To rigorously test this hypothesis, we studied the impact of VISTA targeting on the fate of endogenous, antigen-specific CD4+ T cells under tolerogenic conditions using soluble peptide-loaded major histocompatibility complex (pMHC)– based tetramer enrichment systems (60–62). Analysis of tetramer-positive CD4+ T cells revealed that upon the administration of soluble 2w1s antigen, VISTA deficiency enhanced the number of 2w1s:I-Ab–specific CD4+ T cells by more than twofold under conditions of tolerance induction (Fig. 2C). We tested whether VISTA blockade would recapitulate the outcome observed with VISTA deficiency on 2w1s: I-Ab response to antigen under tolerogenic conditions. To do this, we used the anti-VISTA antagonist antibody (13F3), a well-established VISTA-blocking clone we previously reported (30, 63). Like VISTA deficiency, anti-VISTA blockade increased the number of 2W1s:I-Ab specific T cells upon tolerogenic peptide administration (Fig. 2E). By contrast, agonistic anti-VISTA imparted about a twofold reduction in the number of 2w1s: I-Ab–specific CD4+ T cells under the same tolerogenic conditions (Fig. 2F). The opposing impacts of VISTA deficiency and anti-VISTA agonist on T cell tolerization were also observed using endogenous CD4+ MOG:I-Ab endogenous T cells under conditions of MOG peptide administration (fig. S8, A and B). All studies thus far evaluated the impact of anti-VISTA on T cells under conditions of exclusive TCR engagement and in the absence of inflammation. When mice were immunized with 2w1s peptide and lipopolysaccharide (LPS), there was no impact of VISTA deficiency on the number of endogenous 2W1s-specific T cells (Fig. 2D). Similarly, agonistic anti-VISTA failed to impart a significant impact on antigen-specific T cells under inflammatory conditions (Fig. 2G). This presents evidence that VISTA engagement is important for restraining T cell expansion under tolerogenic conditions and that inflammation can supersede the impact of VISTA on T cell fate.
In addition to deletion, peripheral CD4+ T cell tolerance is regulated by multiple other mechanisms. Therefore, we investigated whether VISTA affected the emergence of antigen-specific anergic and/or regulatory antigen-specific CD4+ T cells. T cell activation under conditions that lack costimulation induced a state of hyporesponsiveness marked by proliferation arrest and markedly diminished effector cytokine production upon restimulation (64). It was previously reported that anergic CD4+ T cells up-regulated the two surface markers CD73 (Nt5e) and FR4 (Izumo1r) (65, 66). Indeed, single-cell analysis of total CD44hi CD4+ T cells from unimmunized mice validates the existence of this naturally anergic CD4+ (CD44hi Foxp3−) T cell population and also identifies multiple additional regulators that participate in T cell anergy such as NFATc1 and Nrp1 (fig. S9A). This population, as do the majority of CD4+ T cells, express significant levels of VISTA, so VISTA was therefore not a defining marker for this cluster. To our knowledge, this presents the first full transcriptional profile of naturally anergic CD4+ T cells. We investigated whether agonistic anti-VISTA would reduce antigen-specific CD4+ T cell numbers under tolerogenic conditions by enhancing the number of anergic cells. Analysis of the percentage of anergic 2w1s: I-Ab–specific CD4+ T cells by CD73hi FR4hi Foxp3− staining did not show a differential impact of anti-VISTA treatment (fig. S9, B to D). As expected from the phenotypic analysis, anti-VISTA also did not enhance the percentage of anergic cells by cytokine-responsiveness [interleukin-2 (IL-2) and IFN-γ] to in vitro restimulation with phorbol 12-myristate 13-acetate (PMA) and ionomycin (fig. S9E). Of note, we did report similar percentages of anergic antigen-specific CD4+ T cells to those previously published for antigen-induced tolerance of the 2w1s-specific repertoire (65).
An additional mechanism of peripheral tolerance is through the emergence of antigen-specific Foxp3+ Tregs and the inhibition of effector T cell expansion and function (67). We asked whether anti-VISTA would change the number of antigen-specific 2w1s: I-Ab– specific Foxp3+ CD4+ Tregs and thereby suppress T cell expansion. Analysis of the percentage of Foxp3+ CD4+ Tregs did not show any impact of anti-VISTA (fig. S9D). These results suggest that VISTA engagement or blockade did not overtly change anergy induction under tolerogenic conditions.
We performed high-resolution scRNA-seq gene expression profiling to assess the impact of anti-VISTA treatment on tolerized 2w1s:I-Ab–specific CD4+ T cells, which revealed insights into repertoire heterogeneity and cell state at the single-cell level (Fig. 2H and fig. S10A). Surprisingly, there was no significant impact of VISTA agonistic targeting or deficiency on the heterogeneity of the antigen-specific repertoire (Fig. 2H; fig. S10, A and B; and table S8). However, this analysis yielded two important observations. First, anti-VISTA reduced T cell clonal expansion of the 2w1s:I-Ab repertoire in all clusters (fig. S10C). Second, analysis of pathway activity revealed that VISTA triggering resulted in a global reduction (>80% of the repertoire) in TCR signaling pathways, such as CD28 costimulation, CXCR3 signaling, and cytokine interactions, suggesting a major impact on the global state of tolerized T cells (Fig. 2, I and J). On the other hand, VISTA deficiency promoted an up-regulation of proliferation pathways and globally enhanced cellular transcription and translation (tables S9 and S10). This indicates that VISTA engagement under tolerogenic conditions imparts an immunosuppressive phenotype to augment T cell tolerance in addition to reducing tolerized T cell numbers, which is in support of data with transgenic systems (Fig. 2, A and B, and figs. S6G and S7F).
Sustained expression of VISTA under tolerogenic, but not inflammatory, conditions
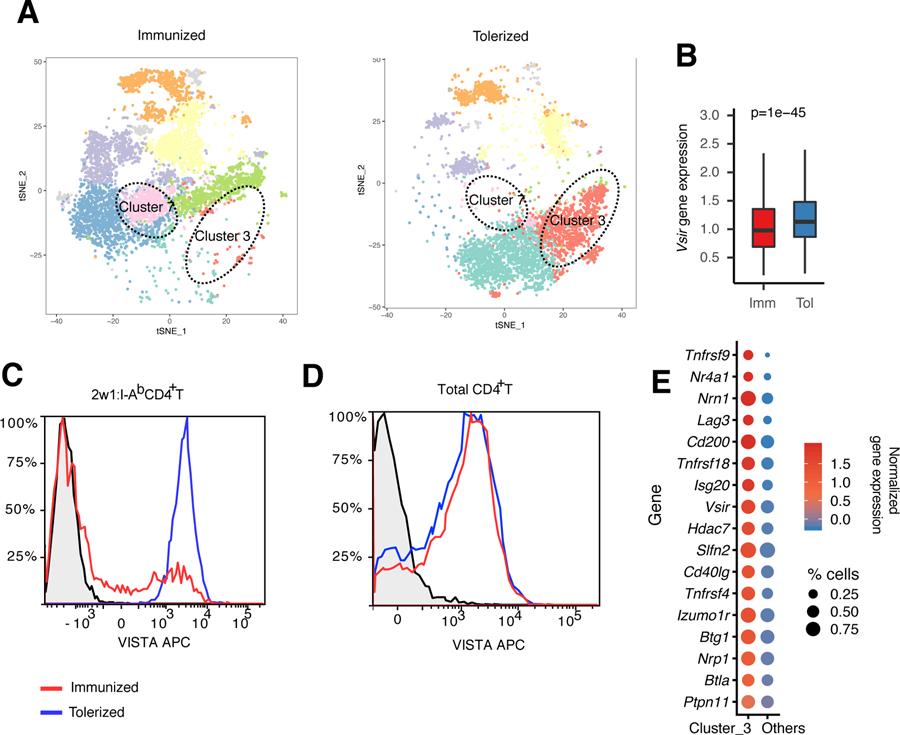
Our data suggest that VISTA engagement renders naïve T cells more susceptible to antigen-induced death and down-regulates pathways of TCR signaling. This may support the argument that sustained VISTA expression would prohibit T cell activation. In addition, we found that VISTA engagement on naïve T cells is abolished under inflammatory conditions (Fig. 2, D and G). We therefore investigated whether TCR engagement under inflammatory (antigen with LPS) versus tolerogenic (antigen only) conditions affected VISTA expression on endogenous antigen-specific CD4+ T cells using scRNA-seq of 2w1s:I-Ab–specific CD4+ T cells. In the inflammation setting, we observed a global transcriptional down-regulation of VISTA in tetramer+ cells (Fig. 3, A and B, and table S11), which was supported by flow cytometric analysis (Fig. 3C). There was no change in VISTA expression on total CD4+ T cells, suggesting that the impact was only on antigen-specific T cells (Fig. 3D). As expected, pathways of proliferation, CD28 costimulation, and antigen response were significantly up-regulated under inflammatory conditions (table S12). Furthermore, a major cluster of cells with a tolerant transcriptional phenotype (cluster 3) was exclusive to tolerization and one of the defining markers for this cluster was VISTA (Fig. 3, A and E, and table S11). This cluster included known regulators of T cell suppression and dysfunction such as FR4 (Izumo1r), LAG-3 (Lag3), BTLA (Btla), SHP-2 (Ptpn11), Neuropilin-1 (Nrp1), Slfn2, and Nr4a1 (1, 65, 68). These molecules were expressed in addition to tumor necrosis factor receptor superfamily molecules, which mirrors the profile of T cell dysfunction previously reported (69). This suggests a potential consequence of VISTA expression under conditions of tolerance but minimal consequence under productive costimulation of T cells.
Fig. 3. VISTA expression is reduced under inflammatory, but not tolerogenic, conditions in vivo.
To provide a model of antigen-specific stimulation for the antigen-specific CD4+ T cell repertoire studies in Fig. 2, C57BL/6 mice were either immunized using 2w1s peptide with LPS or tolerized using 2w1s peptide only. Then, the phenotype of 2w1s:I-Ab–specific CD4+ T cells was analyzed using scRNA-seq and flow cytometry. (A) The t-SNE plot shows the cluster distribution of 2w1:I-Ab peptide–induced CD4+ T cells from immunized (2w1s peptide with LPS) and tolerized (2w1s peptide) mice. Each dot corresponds to one single cell, colored according to cell cluster. The dashed circles indicate the anergic T cell cluster (cluster 3) and T follicular helper T cell cluster (cluster 7). (B) Boxplot depicting the difference in Vsir gene, which encodes VISTA, expression between immunized and tolerized CD4+ T cells across all clusters. The center line refers to the median value for Vsir gene expression. The whisker indicates the 25th to the 75th percentile of Vsir gene, which encodes VISTA, expression. P values were calculated by Wilcoxon rank sum test. (C and D) Flow cytometric analysis of 2w1s:I-Ab–specific CD4+ T cells (C) or total CD4+ T cells (D) under the same tolerization versus immunization conditions presented in (A). The black plot line and shaded region indicate staining of VISTA knockout CD4+ T cells as a biological control. APC, allophycocyanin. (E) Bubble plot showing the average Z-transformed normalized expression of coinhibitory module genes in cluster 3 versus other clusters. The size of each bubble indicates the fraction of cells expressing the represented gene. Data are representative of two independent experiments with at least three mice per group [(A) to (E)].
VISTA targeting induces systemic tolerance and T cell deletion
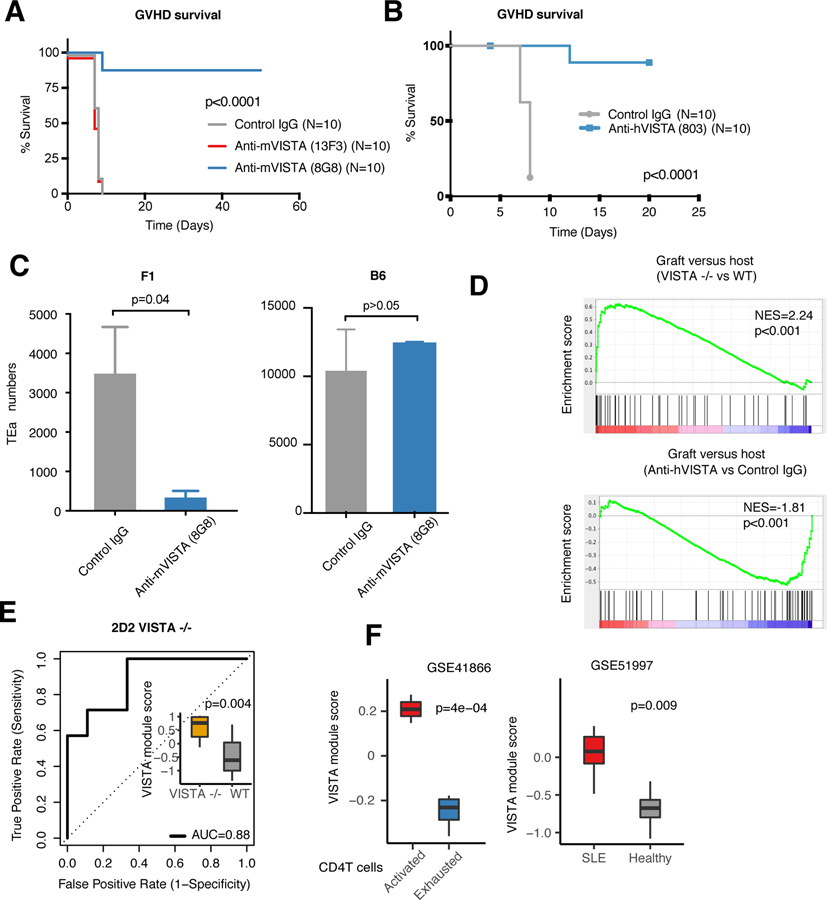
That the targeting of VISTA with anti-VISTA mAbs at the time of donor T cell transfer can ablate the development of GVHD supports the hypothesis that anti-VISTA agonism can induce antigen-specific T cell tolerance (12, 15, 70). We confirmed and expanded this data with both agonistic anti-mouse (8G8) and anti-human (803) VISTA clones (Fig. 4, A and B). Agonistic targeting of VISTA exclusively on the donor T cells arrested the development of GVHD (Fig. 4B). Of note, anti-VISTA blockade (13F3) did not affect alloreactive T cell responses or mouse survival, clearly distinguishing the activities of different anti-VISTA mAbs (Fig. 4A). Under the same GVHD conditions, we investigated the fate of alloreactive CD4+ T cells targeted with agonistic anti-VISTA by using the TEa TCR transgenic CD4+ T cells which recognize I-E⍺ (residues 52 to 68) peptide in the context of I-Ab (71). Similar to experiments presented in Fig. 2 (OT-II), after 24 hours, we noted a significant (>75%) reduction in the number of TEa cells when transferred to anti-VISTA–treated, antigen-bearing F1 hosts. However, no reduction was observed when transferred into B6 mice, indicating that both TCR and VISTA engagement were required for VISTA-mediated deletion (Fig. 4C). The loss of TEa CD4+ T cells mediated by antigen and anti-VISTA was not due to the altered localization of T cells in other tissues (fig. S11, A and B). In support of these in vivo observations, scRNA-seq analysis revealed a marked up-regulation of GVHD pathway mediators in VISTA−/− T cells, which were subsequently down-regulated using agonistic anti-VISTA treatment (Fig. 4D). We developed a VISTA deficiency–associated gene signature to reflect intrinsic VISTA-induced pathway activity change in T cells (see materials and methods and table S13). This signature was validated using bulk RNA sequencing of TCR transgenic VISTA−/− T cells (Fig. 4E) (13). The VISTA module was then applied to a well-documented dataset of CD4+ T cell exhaustion versus activation. In this setting, the activated T cells presented a notably higher VISTA module score compared with exhausted T cells, indicating that VISTA−/− related pathways were up-regulated in the activated T cells (Fig. 4F). In addition, VISTA deficiency was also reported to exacerbate autoimmune murine lupus (11, 14). Indeed, peripheral T cells from systemic lupus erythematosus (SLE) patients from two independent datasets presented a higher VISTA module score compared with that of healthy donors (Fig. 4G and fig. S12A). Similarly, peripheral T cells from rheumatoid arthritis (RA) patients presented an even higher VISTA module score (fig. S12B). This evidence supports a broad regulatory role for VISTA in suppressing T cell self-reactivity and autoimmune manifestations. It may also suggest that VISTA could represent a potential diagnostic biomarker for such inflammatory diseases.
Fig. 4. VISTA targeting induces systemic tolerance and T cell deletion, whereas VISTA deletion imparts an autoimmune-associated gene signature.
(A) C57BL/6 bone marrow (BM) and splenocytes (107 each) were transferred to lethally irradiated BALB/c recipient mice and treated with antagonist (clone 13F3, red) or agonist (clone 8G8, blue) anti-mVISTA antibodies (200 µg per mouse) on day 0 to induce acute GVHD. Survival was monitored for the indicated time periods. (B) C57BL/6 BM and hVISTA-expressing splenocytes (107 each) were transferred to lethally irradiated BALB/c recipient mice, and anti-hVISTA agonist (clone 803, blue) or IgG control (200 µg per mouse) was administered on day 0 to induce GVHD. Survival was monitored. In all survival experiments, P values were calculated by log rank test. Data from the GVHD models are representative of two independent experiments with at least 10 mice per group. (C) TEa CD4+ T cells (2 × 106 cells per mouse) were transferred into 650-centigray-irradiated F1 hosts (left) or age-and-gender matched C5/B6 hosts (right) and intravenously treated with an anti-mVISTA antibody (clone 8G8) or control IgG followed by analysis of cell numbers on day 2 posttransfer. Data show the mean numbers of recovered TEa T cells for each treatment as percentages of CD4+ T cells. Data are representative of four independent experiments with five mice per group. Each bar refers to the mean value, and each error bar refers to one SD; all P values were calculated by Student’s t test. (D) GSEA pathway enrichment plot indicating the GVHD gene set enriched in VISTA−/− versus WT (top) and anti-hVISTA (clone 803)–treated versus control IgG–treated mice (bottom, obtained from Fig. 2 data). NESs and P values are shown for each gene set. P values were calculated by Kolmogorov-Smirnov test. (E) Receiver operating characteristic (ROC) curves for predicting VISTA mutation status in 2D2 transgenic CD4+ T cells using VISTA-deficiency module score as the predictor. VISTA-deficiency module defines the gene signature resulting from loss of VISTA on the naïve T cell. The inset is a boxplot depicting the difference in the VISTA module scores for 2D2 transgenic CD4+ T cells in VISTA−/− and WT mice. P values were calculated by Wilcoxon rank sum test. (F) Boxplots showing the difference in VISTA module scores between exhausted versus activated CD4+ T cells (left) and CD4+ T cells from SLE patients and healthy individuals (right). P values were calculated by Wilcoxon rank sum test.
Concluding remarks
We report a distinct role for VISTA as a negative checkpoint that regulates naïve T cell quiescence and optimal peripheral T cell tolerance. The genetic loss of VISTA in T cells markedly altered the cell state and heterogeneity of mature naïve T cells but had no discernible impact on the steady-state heterogeneity or differentiation trajectory of thymocytes. These findings show that VISTA plays a constitutive function in maintaining naïve T cell identity exclusively outside of the thymus. The disruption of T cell quiescence owing to the loss of VISTA was inextricably linked to undermining peripheral T cell tolerance to antigen in polyclonal, transgenic, and endogenous antigen-specific T cell systems. The function of VISTA in vivo could be amplified using anti-VISTA agonists, which augmented T cell tolerance induction in the same systems in part by enhancing peripheral T cell death under costimulation-deficient conditions. Surprisingly, we did not observe a significant impact of VISTA loss or targeting on other modes of T cell suppression, such as anergy induction. A highly important note is that the function of VISTA is relegated to controlling naïve T cell fate because its impact as well as its expression are all but obliterated under inflammatory conditions (e.g., LPS, CFA, and poly-IC) in which CTLA-4, LAG3, and PD-1 play prominent immunoregulatory roles under inflammatory states. However, under tolerogenic conditions, VISTA expression was sustained. Unlike all other coinhibitory molecules expressed after T cell activation, VISTA presents the first of a class of NCRs critical for maintaining naïve T cell quiescence, directing naïve T cell responses to antigen, and peripheral T cell tolerance. In addition, VISTA represents a specific NCR that can be targeted by both agonists and antagonists to impart opposing outcomes on T cell fate.
These insights explain the impact of VISTA loss on exacerbating T cell–directed immune aggression in multiple mouse models such as SLE, GVHD, and experimental autoimmune encephalomyelitis. The gene signature of VISTA loss was predictive in multiple human autoimmune diseases (e.g., lupus and RA), suggesting the therapeutic potential of VISTA agonistic targeting. Our work also presents a high-resolution profile of the earliest stages of thymocyte and T cell differentiation and the landscape of T cell responses under tolerizing versus immunizing settings with antigen.
One of the important remaining questions is the identity of the regulatory networks that constitutively maintain the expression of VISTA in naïve T cells and distinguish it from other established NCRs. Previous work demonstrated that the TFs p53 and HIF1⍺ bind the VISTA promoter and up-regulate VISTA expression (14, 72). Analysis of ImmGen datasets of mature CD4+ T cells shows a direct correlation between VISTA and p53 (fig. S13A). Analysis of the Encyclopedia of DNA Elements (ENCODE) database (73) for TF binding revealed multiple potential TFs to the VISTA promoter. We screened the TFs expressed in T cells out of these putative regulators and found that Fos, JunD, and NFkB all have binding sites in the VISTA gene (fig. S13B and table S14). Because VISTA expression is reduced on T cells responding to antigen under conditions of inflammation (Fig. 3), we examined the expression of these potential regulators under tolerization versus inflammatory (immunizing) conditions using the same RNA-seq dataset used in Fig. 3. Jund and Fos were significantly up-regulated under inflammatory conditions, thereby showing an inverse relationship with VISTA expression (fig. S13C). Indeed, this finding was supported by analysis of independent datasets in the ImmGen database (fig. S13, D and E). This would imply that they are potential TF repressors of VISTA expression. More extensive studies will determine the regulatory networks that distinguish inhibitory checkpoint expression and activity.
Materials and methods
Mice and cell lines
Eight- to 10-week-old C57BL/6 mice WT were purchased from Charles River (Wilmington, MA). B6N.129S5(B6)-Vsirtm1Lex/Mmucd (VISTA KO) mice were obtained from the Mutant Mouse Regional Resource Centers (www.mmrrc.org; stock no. 031656-UCD) and were fully backcrossed onto the C57BL/6 and BALB/c backgrounds. VISTAfl/fl mice were bred and screened of VISTAfl/fl mice as described previously (14). Conditional deletion of VISTA in the CD4+ T cell compartment was achieved by crossing VISTAfl/fl mice to hemizygous B6.Cg-Tg (Cd4-cre)1Cwi/BfluJ mice (stock no. 022071). Cre-positive mice were compared with Cre-negative littermate controls. Deletion of VISTA on CD4+ T cells and thymocytes was further confirmed by flow cytometry. Rag1−/− (B6. 129S7-Rag1tm1Mom/J) gender-matched 6- to 8-week-old mice were purchased from Charles River. Inducible-deletion of VISTA was achieved by crossing CD4-Cre ERT2 (Jackson Laboratory) to VISTAfl/fl. Human VISTA KI mice were generated by GenOway (Lyon, France) by knocking in a single copy of human VISTA cDNA (GenBank accession no. NM_022153.2) as an in-frame fusion with the 3′ end of the murine signal peptide coding sequence located in the exon 3. This approach resulted in the deletion of part of the exon 3 coding sequence and led to the disruption of the murine gene (fig. S7A). Homologous recombination was done in the C57BL/6-derived ES line. Mouse chimeras were then bred with C57BL/6 Cre deleter mice [Jackson, B6.C-Tg (CMV-cre)1Cgn/J] to excise the neomycin selection cassette (Neo) and to generate heterozygous mice carrying the Neo-excised humanized KI allele. Subsequently, mice were bred to generate homozygous human VISTA KI mice. hVISTA expression and mVISTA deletion were validated by PCR and flow cytometry using fluorophore-conjugated anti-hVISTA [clone 803 (represented in Fig. 2 and fig. S7)] and anti-mVISTA (clone 13F3). Rag2−/− OT-II were bred onto B6-Ly5.1/Cr (B6.SJL-PtprcaPepcb/BoyCrCrl, Charles River) for detection using congenic marker. Gender-matched littermates were then used in the adoptive transfer experiments. For specific experiments, hVISTA KI homozygous mice were interbred with Rag2−/− OT-II B6-Ly5.1. Act-mOva mice were purchased from Jackson Laboratory (C57BL/6-Tg(CAG-OVA)916Jen/J, stock no. 005145) and were gender-matched with the donor mice. KLF2-GFP mice were developed, bred, and screened at the University of Minnesota (Stephen Jameson lab) (17, 18). TEa transgenic mice [B6.Cg-Tg(Tcra,Tcrb)3Ayr/J] expressing GFP were bred and screened in-house (74). CB6F1/J (C57BL/6 × Balb/c) (100007), NZBWF1/J (100008), B6.MRL-Faslpr (000482), Bcl2l11tm1.1Ast Bim-deficient (004525) mice were all purchased from Jackson Laboratory. K/BxN transgenic mice were bred and screened in-house. 2D2 TCR transgenic mice were purchased from Jackson Laboratory and bred onto VISTA−/− B6 background (13). Mice were maintained under specific-pathogen–free conditions in the Dartmouth Center for Comparative Medicine and Research. The Animal Care and Use Committee of Dartmouth College approved all animal experiments. For experiments involving thymocytes, gender- and age-matched littermates from 3- to 4-week-old mice were used. Both male and female mice were used in independent experiments. A Jurkat cell line expressing VISTA was generated using the construct pEF1a-hVISTA-IRES-ZsGreen1. The pEF1-IRES-ZsGreen1 construct was initially purchased from TakaraBio (cat. 631976), and hVISTA sequence was cloned. The Jurkat cell line (ATCC, TIB-152, clone E6–1) was then transfected with the construct using cell line Nucleofector (Lonza, VCA-1003) following the manufacturer’s protocol. A stable pool was then generated, and hVISTA expression on this cell line compared with control Jurkat cells transfected with empty vector was assessed using fluorophore-conjugated anti-VISTA clone 803.
Antibodies
Antibodies for mouse and human VISTA were generated as described previously (63, 75). Female C57BL/6 mice were immunized with human VISTA–Ig fusion protein emulsified in complete Freund’s adjuvant (CFA). They were boosted 4 weeks later with protein in incomplete Freund’s adjuvant (IFA) and then 6 weeks later with A20 cells overexpressing VISTA–red fluorescent protein. Finally, they were boosted with VISTA-Ig fusion protein without the adjuvant. Four days after the last boost, spleens from immunized mice were provided to APS Ltd. Hybridomas and antibodies were generated by APS Ltd. Hybridoma clones that produced VISTA-specific antibodies were selected after limiting dilution and screened by both ELISA and flow cytometry methods. Anti-hVISTA clone 803 was humanized into a full IgG2 human antibody by Aragen Biosciences. To demonstrate specificity of the clone anti-hVISTA 803, 106 peripheral blood mononuclear cells (PBMCs) were stained with 5 µg/ml of anti-hVISTA 803 in the presence of 10 µg of soluble VISTA-Ig. In addition, the Jurkat cell line was stably transfected with human VISTA, and staining was compared with control vector–transfected Jurkat cells. Primary immune cell subsets from human peripheral blood and multiple mouse tissues were stained with both hVISTA and mVISTA antibody clones to demonstrate specificity (fig. S7). Anti-hVISTA 803 is a chimeric human IgG2 antibody. Both antagonist anti-mVISTA clone 13F3 and agonist anti-mVISTA clone 8G8 are hamster IgG clones, and monoclonal hamster IgG (BioXCell, Lebanon, NH) was used as their control.
Adoptive cell transfer
For all experiments involving adoptive transfer, naïve CD4+ T cells from donor age- and sex-matched mice were purified using a naïve CD4+ T cell isolation kit (Miltenyi). For experiments involving Ly5.1 WT and CD4-Cre × VISTAfl/fl T cell transfers (Fig. 1 and fig. S5) of naïve CD4+ T cells, the donor cells were mixed at a 1:1 ratio (validated by flow cytometry) and then a total of 1 × 106 cells were adoptively co-transferred by intravenous tail vein injection into recipient Rag1−/− hosts. Mice were then injected with 5 µg of either hamster anti-CD3e or hamster IgG control (BioXcell). Cells were recovered on day 5 posttransfer, and ratios were quantified by flow cytometry using congenic markers. Inducible deletion of VISTA was achieved by i.p. injection of tamoxifen, as recommended by Jackson Laboratory (https://www.jax.org/research-and-faculty/resources/cre-repository/tamoxifen). Briefly, three injections of tamoxifen were required before full deletion of VISTA on the CD4 T cell compartment was observed by flow cytometry. Cells were then isolated as described and adoptively transferred. For experiments involving OT-II and hVISTA OT-II adoptive transfers, single group transfers (3 × 106 cells per mouse) of congenically discordant CD45.1+ OT-II cells into either Act-Ova or B6 were performed and treated with either anti-VISTA (200 µg) or IgG control followed by cell recovery and quantification 48 hours after transfer. TEa transgenic CD4+ T cells (2 × 106) were transferred into cGy 650 irradiated F1 hosts, and mice were either treated with anti-mVISTA (8G8) or hamster IgG control (200 µg/mouse) followed by cell recovery 48 hours after transfer. For TEa quantification in multiple tissues, cells were transferred under the same conditions and isolated from each of the aforementioned tissues. Isolation from spleen and lymph nodes followed the standard procedure (61, 76). Isolation from liver and lung tissues required Percoll density centrifugation, whereas isolating lymphocytes from small intestine followed the described procedure (77). Isolation from bone marrow was performed as described (78). TEa cell numbers were then quantified by flow cytometry using GFP expression in addition to Thy1.1 (clone OX-7) and CD4 staining.
Acute GVHD model
For the mVISTA treatment experiments, 10-week-old BALB/c recipients and C57BL/6 donor mice were purchased from Charles River. Recipient mice were subjected to total body irradiation (TBI) emanating from a cesium-137 source twice at 450 centigray (cGy) at D0 (9:30 a.m. and 1:30 p.m.) before transfer. Donor mice were euthanized, and bone marrow was harvested by flushing femur and tibia with HBSS. Red blood cells were lysed using ACT solution, and a single-cell suspension of splenocytes and BM cells was prepared and counted. Recipient mice received 10 million bone marrow cells and 10 million spleen cells along with 200 ug of control IgG or anti-mVISTA agonist clone 8G8 or antagonist clone 13F3. Cells and antibodies were administered by tail vein intravenous injection. Mice were weighed regularly to monitor disease progression. Mice were euthanized when they showed signs of morbidity. For anti-hVISTA experiment, the same procedure was applied with the exception that hVISTA splenocytes and WT BM cells were intravenously injected and the mice were either treated with anti-hVISTA 803 or IgG2 control.
Antigen tolerization and immunization
As described previously (58, 62), intravenous injection of soluble 2w1s:I-Ab (EAWGALAN-WAVDSA) antigen was used to induce antigen-specific T cell tolerance, whereas injection of antigen in the presence of LPS adjuvant was used to provide an immunizing inflammatory condition. To induce T cell tolerance, two doses of 100 µg of 2w1s peptide (Genscript Corp) were intravenously injected on days 0 and 3, followed by analysis on day 7. For MOG antigen tolerization, MOG35–55 peptide (200 µg) was intravenously injected on day 0, followed by analysis on day 7. For immunization, mice were intravenously injected on day 0 with 2w1s (100 µg) and LPS (5 µg), followed by analysis on day 7. In the 2w1s peptide tolerization scRNA-seq (Figs. 2 and 3), cells were analyzed 72 hours post intravenous injection. Anti-VISTA or IgG control treatments (200 µg per mouse) were injected on day 0.
Tetramer enrichment
Staining with tetramer and enrichment for antigen-specific endogenous T cell quantification were performed as described previously (61, 62, 76). Spleen and lymph nodes (inguinal, axillary, brachial, cervical, mesenteric, and periaortic) were harvested for each mouse. A single-cell suspension was prepared in 200 µl of Fc-block supplemented sorter buffer (Fc block + 2% BSA, 0.05% sodium azide). PE-conjugated 2w1s or MOG35–55 tetramers (MBL international) was added at a concentration of 20 nM, and the cells were incubated for 1 hour at RT, followed by washing with 15 µl of ice-cold sorter buffer (PBS + 2% BSA, 0.1% sodium azide). The tetramer-stained cells were then resuspended in a volume of 200 µl of sorter buffer, mixed with 50 µl of anti-PE antibody conjugated magnetic microbeads (Miltenyi Biotec), and incubated on ice for 20 min, followed by two washes with 10 ml of sorter buffer. The cells were then resuspended in 3 ml of sorter buffer and passed over a magnetized LS column (Miltenyi Biotech). The column was washed with 3 ml of sorter buffer three times and then removed from the magnetic field. The bound cells were eluted by pushing 5 ml of sorter buffer through the column with a plunger. The resulting enriched fractions were resuspended in 0.1 ml of sorter buffer; a small volume was removed for cell counting, and the rest of the sample was stained with a cocktail of fluorochrome-labeled antibodies specific for B220, CD19 CD11b, CD11c, F4/80, CD3, CD8, NK1.1, CD4, and CD44. Quantification of the number of 2w1s:I-Ab cells per mouse followed the protocol described (76).
Flow cytometry and staining
Gentle manual dissociation of splenocytes and lymph node cells to single-cell suspensions was performed as described previously (61, 76). For thymocyte staining for analysis and sorting (fig. S3), thymi were collected in 5 ml of HBSS supplemented with collagenase/DNase I (Worthington) and homogenized gently then incubated for 15 minutes (37°C, 5% CO2). Cells were then washed and stained for flow cytometric analysis or sorting. For all flow cytometry experiments, T cells were stained with a fixable live-dead stain (Invitrogen) in PBS followed by surface antibody staining in FACS buffer (PBS with 0.5% BSA and 0.1% sodium azide). For intracellular cytokine staining (fig. S9), cells were incubated for 4 hours at 37°C in RPMI-1640 medium plus 10% FBS in the presence of 10 ng/ml PMA (Sigma-Aldrich), 1 µM ionomycin (EMD Chemicals), and 10 µg/ml brefeldin A (Sigma-Aldrich). Cells were then stained for surface markers as described. Intracellular staining was preformed using the eBioscience Cytofix/Cytoperm kit (Thermofisher). Surface-stained cells were then stained with anti–IL-2 (JESS-5H4) and anti–IFN-γ (XMG1.2). The anergic phenotype of 2w1s:I-Ab CD4+ T cells and Foxp3+ Treg quantification were performed as previously described (65). Briefly, tetramer enrichment was performed as described above followed by the same surface-staining procedure for tetramer experiments in addition to staining for CD73 (eBioTY/11.8), FR4 (eBio12A5). Stained cells were then treated with eBioscience Foxp3 fixation and permeabilization buffer sets (Thermofisher) following the manufacturer’s instructions and stained for Foxp3 (FJK-16s). Anergic 2w1s:I-Ab CD4+ T cells were quantified as tet+ CD44hi Foxp3− CD73hi FR4hi. Foxp3+ thymocytes were also stained using the same kit in addition to VISTA and CD4 surface staining. For naïve T cell and thymocyte VISTA surface staining, clone MIH-63 (Biolegend) was used. Samples were collected on MACSQuant Analyzer 10 (Miltenyi Biotec) and analyzed using Flow-Logic Software 7.2 (Miltenyi Biotec). For experiments using OT-II CD4+ T cells (Fig. 2 and figs. S6 and S7), the donor T cells were stained with Vβ5 (MR9–4) and CD45.1 (A20) in addition to CD4 staining. Dead cells were calculated as the percentage of total OT-II recovered using viability dye (Near IR, Invitrogen). For analysis of frequencies of different immune populations after anti-VISTA treatment (figs. S6J and S7G), the following antibody clones were used: CD11b (M1/70), Ly6G (1A8), Ly6C (HK1.4), CD3 (17A2), CD4 (RM4–5), CD8 (53–6.7), TCRb (H57–597), F4/80 (BM8), CD19 (6D5), NK1.1 (PK136), CD11c (N418), and Siglec H (551). Neutrophils were identified as CD11b+ Ly6G+ Ly6C−, whereas monocytes were identified as CD11b+ Ly6C+ Ly6G−. Macrophages were gated as CD11b+ F4/80+ cells. CD4+ and CD8+ T cells were pregated on CD3+ TCRb+ live cells. For dendritic cells (DCs), spleens were digested and processed as described previously (79) and a lineage gating was added (CD19 CD3 Ly6G NK1.1). Conventional DCs were defined as Lineage−ve CD11c+ live cells, whereas plasmacytoid DCs (pDCs) were defined as Lineage−ve CD11cint Siglec H+ live cells.
Flow sorting for single-cell sequencing and total RNA-seq
For scRNA-seq experiments depicted in Fig. 1A and scATAC-seq in Fig. 1D: Cells were stained with CD4 (clone RM4–5), CD62L (MEL-14) and CD44 (IM-7), and lineage/Dump (Lin) gate (CD11b, CD11c, NK1.1, CD19, F4/80, CD8, B220) for 20 min on ice, washed, and then flow-sorted using FACS-ARIA II (BD Biosciences) for CD4+ CD44lo (lowest 20% CD44−) CD62Lhi Lin− cells into 96-well plates. This same procedure was applied for fig. S5 except that cells were sorted based on CD4+ CD44hi Lin−ve. For thymocyte sorting in fig. S3, cells were stained with CD4, CD8 (clone: 53–6.7), and lineage (CD11b, CD11c, NK1.1, CD19, F4/80). For scRNA-seq of 2w1s:I-Ab, cells were first stained and enriched with tetramer, then flow sorted using the staining panel (CD4, CD44, CD8, 2w1s-Tet) for CD4+ CD44hi Tet+ CD8− Lin− and lineage was defined as (CD11c, B220, CD19, CD11b, F4/80, NK1.1). For experiments depicted in fig. S3, A to C, CD4+ CD44lo CD62Lhi from KLF2-GFP mice were sorted based on GFP reporter expression with KLF2hi defined by 20% highest expression and the KLF2lo defined as 20% lowest (positive) expression. For sorting of VISTAhi versus VISTAlo, cells were stained according to the same procedure, in addition to VISTA (clone MIH-63). Purity was validated by using subsequent flow cytometry and scRNA-seq analysis in which nonspecific cells were excluded.
Single-cell RNA sequencing and normalization
Droplet-based 5′-end scRNA-seq was performed by the 10x Genomics platform, and libraries were prepared by the Chromium Single Cell 5′ Reagent kit according to the manufacturer’s protocol (10x Genomics, CA, USA). The Cell Ranger Single-Cell Software Suite (10x Genomics) was used to perform barcode processing and transcript counting after alignment to the mm10 reference genome with default parameters. The Seurat R package (80) was applied to filter out low-quality cells, normalize gene expression profiles, and cluster cells. Cells expressing >10% mitochondrial gene counts or expressing less than 500 genes were discarded using the FilterCells function. Then, the NormalizeData function was applied to normalize and log transform the raw counts for each cell on the basis of its library size.
Single-cell unsupervised clustering
The normalized expression matrices of naïve CD4+ T cells, CD4+ CD44hi MP T cells, CD4+ thymocytes, and 2w1s:I-Ab specific CD4+ T cells were processed by filtering the nonexpressed genes separately. The unsupervised clustering was applied in each dataset as follows: (i) Top variant genes with dispersion higher than 0.5 and average expression higher than 0.15 were selected and used as the input for principal components analysis (PCA) to reflect the major biological variation in the data. (ii) The top 15 PCs were chosen for t-SNE dimension reduction by the RunTSNE function and unsupervised clustering. Specifically, the FindClusters function was used to cluster the cells. (iii) After the cell clusters were determined, marker genes for each cluster were identified by the FindAllMarkers function with the default parameter. The biological annotation of each cluster was further described by the marker gene function reported in the literature and the pathways specifically associated with the cluster (see “Pathway enrichment analysis”) or the representation of the marker gene expression in the ImmGen database, which has a clear description of different CD4+ T subsets (81). We examined the expression pattern of Z-transformed average gene expression of cluster marker genes in ImmGen CD4+ T cells. The ImmGen CD4+ T cell lineage with highest expression level of the cluster marker genes was chosen as the annotation for the CD4+T cell cluster.
Identification of autoreactive T cells
Droplet-based 5′-end single-cell TCR sequencing (scTCR-seq) was performed by the 10x Genomics platform, and libraries were prepared by the Chromium Single Cell Immune Profiling Solution kit according to the manufacturer’s protocol (10x Genomics, CA, USA). The Cell Ranger Single-Cell Software Suite VDJ pipeline (10x Genomics) was used to perform barcode processing and consensus TCR annotation after alignment to the mm10 reference genome with default parameters. The annotated TCR sequences of naïve CD4+ T cells and CD4+ CD44hi MP T cells from VISTA−/− and WT mice and of CD4+ T cells from Fas lpr and Bim-deficient mice were processed by filtering out nonproductive TCRs. To identify the autoreactive CD4+ T cells, the Vβ CDR3 sequences in naïve CD4+ T cells were aligned with the Vβ CDR3 sequences in CD4+ CD44hi MP T cells and CD4+ T cells from Fas lpr and Bim-deficient mice. The pairwiseAlignment function in the Biostring R package (44) with parameter “type = local, gapOpening = 10, gapExtension = 4” was used for sequence matching.
Developmental trajectory inference
To determine the potential lineage differentiation between VISTA−/− and WT, Monocle (version 2) (82) algorithm was used with scRNA thymus double-positive, single-positive, and naïve CD4+ T cells raw counts matrix as the input. The newCellDataset function was used to build a CellDataSet object with the parameter “expressionFamily = negbinomial.” Then, differential gene expression analysis was performed using the differentialGeneTest function with the parameter “fulModelFormulStr = ~Cluster_assign, reducedModelFomulaStr = ~batch.” Specifically, “Cluster_assign” refers to the cluster identification of the scRNA and batch refers to the batch experiment number during which the scRNA was sequenced. Moreover, the dimension reduction was performed by the reduceDimension function with the parameter “max_components = 2, reducedModelFormulaStr = ~ batch, method = DDRTree.” The differentiation trajectory was then inferred with the default setting of Monocle.
Pathway enrichment analysis
The differentially expressed genes between different cell clusters or different VISTA perturbations (e.g. VISTA−/− or anti-VISTA treatments) were ranked on the basis of the average log-fold change. To annotate the pathways that were involved in the differentially expressed genes, pathway gene sets were downloaded from the C2 category of the Molecular Signatures Database (MSigDB v6.2) database (83). Furthermore, gene sets with less than 10 effective genes (i.e., the number of genes presented in a gene expression dataset) were discarded. The preranked gene set enrichment analysis (GSEA) software was used to calculate the enrichment of each pathway in the genes that are most informative in each gene list.
2D2 CD4+ T cell isolation for total RNA-seq
Naïve CD4+ T cells were isolated from 4- to 8-week-old asymptomatic male and female 2D2 transgenic mice on the VISTA−/− or WT background (13) using the naïve CD4 T cell isolation kit (Miltenyi Biotec). Cell purity (~96 to 98%) and viability were assessed by flow cytometry.
Calculation of VISTA module score
Differential gene expression analysis was performed for each gene between grouped CD4-Cre VISTA−/− and WT naïve CD4+ T cells using Wilcoxon rank sum test. A P value of <0.05 was used as the threshold to determine the statistical significance, and the log-fold change was used to determine if the gene was up- or down-regulated in the VISTA−/− naïve CD4+ T cells (table S13). The VISTA gene module was defined as the combination of significantly up- and down-regulated genes in VISTA−/− naïve CD4+ T cells. In a given gene expression dataset, the VISTA module score was first calculated as the average gene expression difference of up- and down-regulated genes in the module and then Z-transformed into normal distribution. A higher VISTA module score indicated a higher chance of VISTA deficiency in a given CD4+ T cell. Validation of the VISTA module score was performed in an independent 2D2 transgenic CD4+ T cells RNA-seq dataset. Area under the ROC (AUC) was used as a metric for evaluating the accuracy of the VISTA module score in capturing VISTA deficiency. For each CD4+ T cell sample, a threshold was set beginning with the lowest score; all samples with a score higher the threshold were predicted to be VISTA−/−, and all samples below the threshold were predicted to be WT. The sensitivity and specificity were then calculated for each threshold by comparing the predicted VISTA−/− with the actual VISTA−/−.
RNA-seq alignment for KLF2hi versus KLF2lo, VISTAhi versus VISTAlo naïve CD4+ T cells, and 2D2 VISTA−/− CD4+ T cells total RNA-seq
Sequencing was performed on a NextSeq 500 (Illumina) instrument to obtain an average of raw 100-bp single end reads per sample. Raw .bcl files were demultiplexed using the Illumina bcl2fastq2 pipeline. The quality of the fastq files was examined with the FastQC software (www.bioinformatics.babraham.ac.uk/projects/fastqc). Raw fastq files were trimmed using the software Trimmomatic by setting the parameter “SLIDINGWINDOW: 4:15 LEADING: 3 TRAILING: 3 MINLEN: 36.” The trimmed fastq files were than aligned to the mouse mm10 reference genome and normalized to obtain transcripts per kilobase million (TPM) for each RNA-seq sample using the software Salmon with the parameter “-l A” (84).
Single-cell ATAC sequencing and normalization
CellRanger-atac v1.1 was used to generate fastq files (mkfast) and to demultiplex, align to the mouse mm10 genome, and call peaks using the “count” pipeline (http://software.10xgenomics.com/single-cell/overview/welcome). Peak count matrices were aggregated using the “aggr” function and normalized to sequencing depth. Cells with peak counts higher than 5000 were kept for further analyses. To further examine the quality of the scATAC-seq, The fragment file, which records the full list of all unique fragments across all cells, was used for quality control. Specifically, the fraction of fragments in total peaks was calculated by the number of fragments that mapped to the peak region divided by the total number fragments in each cell. The blacklist fragments ratio was calculated by the number of fragments that mapped to the blacklist region versus the number of fragments that mapped to peak region (85). As recommended by Stuart et al. (86), cells having total number of fragments in peaks higher than 1000 and fraction of peaks that located in the peak higher than 15% and blacklist ratio lower than 2.5% were considered as good cells. 99.36% of cells passed the quality control.
The generated peak matrix was binarized, and then we performed the term frequency– inverse document frequency (“TF-IDF”) transformation as suggested by Cusanovich et al. (87). We first divided each peak in each cell by the total number of accessibility of peaks in the cell (the “term frequency”) and then multiplied these values by the inverse accessibility of the peaks across cells (the “inverse document frequency”).
Single-cell ATAC unsupervised clustering
Peaks having at least 100 reads across cells were considered variable peaks for unsupervised clustering. The TF-IDF matrix was used as input to conduct SVD to return LSI components. These steps were performed in the RunLSI function in Seurat. We retained 50 dimensions and created a new Seurat object. The clusters were identified using Seurat’s SNN graph clustering using the FindClusters function and visualized using the RunUMAP function (86). To identify cluster markers, the binarized peak matrix was used as input to create a CellDataSet object through the newCellDataset function with the parameter “expressionFamily = binomialff.” Then, the differentialGeneTest function with the parameter “fulModelFormulStr = ~Cluster_ assign” was used to identify the marker peaks for each cluster. “Cluster_assign” refers to the cluster identification of the scATAC seq (87). To further confirm the statistical significance of marker peaks, the FindMarkers function in Seurat was used to perform the likelihood ratio test with the parameter “test.use =”LR”, laten.vars=”peak_region_fragments.” The marker peaks function was annotated on the basis of its nearest gene function. The biological annotation of each cluster was further described by the markers peak associated gene function reported in the literature and the calculated gene activity associated with the cluster (see “Gene activity calculation”). The Signac package was used for peak profile visualization [(86); https://satijalab.org/signac/]. Specifically, the CoveragePlot function grouped the peaks in each cluster and normalized the peaks by sequencing depth and number of cells in each cluster for visualization.
Gene activity calculation
The Cicero package was used to calculate gene activity scores (GA scores) as previously described (88). The binary filtered peak counts matrix was used to build a CellDataSet object with the parameter “expressionFamily = binominalff().” Then a cicero_cds object was made using the function make_cicero_cds with the parameter “reduced_coordinates = UMAP_ coords.” Specifically, the UMAP_coords was generated by previous dimension reduction step. The run_cicero function was used to calculate the coaccessed peak-to-peak links across all cells with default parameters in the mouse mm10 genome. The build_gene_activity_ matrix and normalize_gene_activities were used to calculate and normalize the gene activity scores for each cell. For visualization convenience, the GA score was transformed into log10(GA score*1000 + 1).
Identification of VISTA expression regulators
All ENCODE transcription factor ChIP-seq bigWig files were accessed and downloaded from the ENCODE official website (https://www.encodeproject.org) (89). With a threshold of P value 0.05, the TIP probabilistic method (90) was used to determine the potential transcription factors that bind VISTA in each cell line.
ELISA for anti-mVISTA isotype determination
Anti-VISTA antibodies were diluted in PBS and coated overnight on an ELISA plate (RND). The blocking step was performed using PBS (1% BSA). This was followed by incubation with anti-hamster clones IgG1, IgG2, IgG2/3, and IgG2/3/4 (BD Biosciences). Anti-mouse IgG1-HRP followed by TMB substrate solution was used for detection.
Assessment of anti-VISTA agonist suppressive properties
NZBW/F1 lupus
Twenty-four–week-old NZB/W F1 mice were treated three times a week with either anti-mVISTA 8G8 or control IgG (200 µg). Proteinuria levels (mg/dl) were recorded weekly using Chemstrip test trips (Roche Diagnostics).
ConA acute hepatitis
Con A (Sigma-Aldrich) was dissolved in PBS and administered in a total volume of (15 mg/kg) 300 µl by intravenous tail vein injection. Mice received anti-VISTA 8G8 or control IgG (200 µg) through intraperitoneal injections 3 hours before Con A injection. Mice were monitored for survival.
K/BxN arthritis
Mice were injected with 100 µl of K/BxN serum on days 0 and 2. Anti-VISTA 8G8 or control IgG (200 µg) were given every 3 days starting day 0. Clinical scoring was done as previously described (91).
Imiquimod induced psoriasis
Fifty mg of 3.5% IMQ cream was prepared by diluting the 5% IMQ cream (Taro Pharmaceuticals, New York, NY) using the vehicle cream (Vanicream; Pharmaceutical Specialties, Cleveland, GA). Imiquimod was applied to the ear of mice daily. At day 14, anti-mVISTA 8G8 or control IgG (200 µg) were administered intraperitoneally every other day. Ear thickness was measured by using an Ozaki caliper (model G-A1–0.4N) (Neill-Lavielle Supply, Louisville, KY).
Impact of anti-VISTA on uptake of apoptotic cells
The procedure followed the following references (92, 93). Thymocytes (single-cell suspension of 107 cells/ml) were isolated from C57BL/6 WT mice and exposed to dexamethasone under cell culture conditions (0.1 µM) in complete DMEM for 24 hours to induce apoptosis. This procedure allowed for ~100% apoptosis, which was assessed by annexin V/Propidium idodide (PI) staining followed by flow cytometric analysis. After apoptosis induction, thymocytes were washed twice, then resuspended in pH rodo dye (final concentration of 20 ng/ml) and incubated for 30 min at RT. This is a pH-sensitive dye that emits red dye only in lower pH, such as that located in a phagosome (pH ~5), so it was used to distinguish engulfed thymocytes from unengulfed controls. Thymocytes (1 × 106 cells) were incubated with thioglycolate-elicited peritoneal macrophages (2 × 105 cells) for 90 min in the presence of anti-VISTA (8G8) or control IgG (10 µg/ml). Phagocytosis was determined using flow cytometry by measuring the positive pHrodo-containing (CD11b+ F4/80+) macrophages. VISTA−/− peritoneal macrophages were used as control on the basis of previous work (1).
Supplementary Material
ACKNOWLEDGMENTS
We thank E. J. Wherry and C. M. Burns for the careful review of and insightful comments on the manuscript. Flow cytometry and flow sorting experiments were carried out in DartLab (G. Ward), the Immune Monitoring and Flow Cytometry Shared Resource at the Norris Cotton Cancer Center at Dartmouth, with NCI Cancer Center Support Grant 5P30 CA023108-37. RNA-sequencing experiments were carried out at Dartmouth Medical School in the Genomics Shared Resource (by F. Kolling IV), which was established by equipment grants from the NIH and NSF and is supported in part by a Cancer Center Core Grant (P30CA023108) from the National Cancer Institute.
Funding: Research was supported by NIH grants R01AR070760 (R.J.N.), R01CA214062 (R.J.N.), 1R21CA227996-01A1 (C.C.), RR180061 (C.C.), R01 HL56067 (B.R.B.), R01 HL 11879 (B.R.B.), and R37 AI34495 (B.R.B.) and Cancer Prevention and Research institute of Texas grant RR180061 (C.C.).
Competing interests: R.J.N. is an inventor on patent applications (10035857, 9631018, 9217035, 8501915, 8465740, 8236304, and 8231872) submitted by Dartmouth College, and patent applications (9890215 and 9381244) submitted by Kings College London and Dartmouth College and a co-founder of ImmuNext, a company involved in the development of VISTA-related assets. These applications cover the use of VISTA targeting for modulation of the immune response.
Footnotes
SUPPLEMENTARY MATERIALS
science.sciencemag.org/content/367/6475/page/suppl/DC1 Figs. S1 to S13
Data and materials availability: scRNA-seq data were deposited and are available under BioProject accession numbers PRJNA587711, PRJNA587742, PRJNA587790, PRJNA587769, and PRJNA587564. scATAC-seq data were deposited under BioProject accession number PRJNA587562. All antibodies and mice are available under a Material Transfer Agreement by contacting R.J.N.
REFERENCES AND NOTES
- 1.Chen L, Flies DB, Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat. Rev. Immunol 13, 227–242 (2013). 10.1038/nri3405; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pardoll DM, The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 12, 252–264 (2012). 10.1038/nrc3239; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahn E et al. , Role of PD-1 during effector CD8 T cell differentiation. Proc. Natl. Acad. Sci. U.S.A 115, 4749–4754 (2018). 10.1073/pnas.1718217115; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noel PJ, Boise LH, Thompson CB, Regulation of T cell activation by CD28 and CTLA4. Adv. Exp. Med. Biol 406, 209–217 (1996). 10.1007/978-1-4899-0274-0_22; [DOI] [PubMed] [Google Scholar]
- 5.Kuo CT, Veselits ML, Leiden JM, LKLF: A transcriptional regulator of single-positive T cell quiescence and survival. Science 277, 1986–1990 (1997). 10.1126/science.277.5334.1986; [DOI] [PubMed] [Google Scholar]
- 6.Tzachanis D et al. , Tob is a negative regulator of activation that is expressed in anergic and quiescent T cells. Nat. Immunol 2, 1174–1182 (2001). 10.1038/ni730; [DOI] [PubMed] [Google Scholar]
- 7.Yusuf I, Fruman DA, Regulation of quiescence in lymphocytes. Trends Immunol 24, 380–386 (2003). 10.1016/S1471-4906(03)00141-8; [DOI] [PubMed] [Google Scholar]
- 8.Berger M et al. , An Slfn2 mutation causes lymphoid and myeloid immunodeficiency due to loss of immune cell quiescence. Nat. Immunol 11, 335–343 (2010). 10.1038/ni.1847; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buckley AF, Kuo CT, Leiden JM, Transcription factor LKLF is sufficient to program T cell quiescence via a c-Myc-dependent pathway. Nat. Immunol 2, 698–704 (2001). 10.1038/90633; [DOI] [PubMed] [Google Scholar]
- 10.Gorelik L, Flavell RA, Abrogation of TGFb signaling in T cells leads to spontaneous T cell differentiation and autoimmune disease. Immunity 12, 171–181 (2000). 10.1016/S1074-7613(00)80170-3; [DOI] [PubMed] [Google Scholar]
- 11.Ceeraz S et al. , VISTA deficiency accelerates the development of fatal murine lupus nephritis. Arthritis Rheumatol 69, 814–825 (2017). 10.1002/art.40020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flies DB et al. , Coinhibitory receptor PD-1H preferentially suppresses CD4+ T cell-mediated immunity. J. Clin. Invest 124, 1966–1975 (2014). 10.1172/JCI74589; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L et al. , Disruption of the immune-checkpoint VISTA gene imparts a proinflammatory phenotype with predisposition to the development of autoimmunity. Proc. Natl. Acad. Sci. U.S.A 111, 14846–14851 (2014). 10.1073/pnas.1407447111; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoon KW et al. , Control of signaling-mediated clearance of apoptotic cells by the tumor suppressor p53. Science 349, 1261669 (2015). 10.1126/science.1261669; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flies DB, Higuchi T, Chen L, Mechanistic assessment of PD-1H coinhibitory receptor-induced T cell tolerance to allogeneic antigens. J. Immunol 194, 5294–5304 (2015). 10.4049/jimmunol.1402648; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu J, Lingrel JB, KLF2 inhibits Jurkat T leukemia cell growth via upregulation of cyclin-dependent kinase inhibitor p21WAF1/CIP1. Oncogene 23, 8088–8096 (2004). 10.1038/sj.onc.1207996; [DOI] [PubMed] [Google Scholar]
- 17.Carlson CM et al. , Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature 442, 299–302 (2006). 10.1038/nature04882; [DOI] [PubMed] [Google Scholar]
- 18.Weinreich MA et al. , KLF2 transcription-factor deficiency in T cells results in unrestrained cytokine production and upregulation of bystander chemokine receptors. Immunity 31, 122–130 (2009). 10.1016/j.immuni.2009.05.011; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benzeno S et al. , Cyclin-dependent kinase inhibition by the KLF6 tumor suppressor protein through interaction with cyclin D1. Cancer Res 64, 3885–3891 (2004). 10.1158/0008-5472.CAN-03-2818; [DOI] [PubMed] [Google Scholar]
- 20.Narla G et al. , In vivo regulation of p21 by the Kruppel-like factor 6 tumor-suppressor gene in mouse liver and human hepatocellular carcinoma. Oncogene 26, 4428–4434 (2007). 10.1038/sj.onc.1210223; [DOI] [PubMed] [Google Scholar]
- 21.Baranzini SE, The role of antiproliferative gene Tob1 in the immune system. Clin. Exp. Neuroimmunol 5, 132–136 (2014). 10.1111/cen3.12125; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corjay MH, Kearney MA, Munzer DA, Diamond SM, Stoltenborg JK, Antiproliferative gene BTG1 is highly expressed in apoptotic cells in macrophage-rich areas of advanced lesions in Watanabe heritable hyperlipidemic rabbit and human. Lab. Invest 78, 847–858 (1998). [PubMed] [Google Scholar]
- 23.Rouault JP et al. , BTG1, a member of a new family of antiproliferative genes. EMBO J 11, 1663–1670 (1992). 10.1002/j.1460-2075.1992.tb05213.x; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hart GT, Hogquist KA, Jameson SC, Krüppel-like factors in lymphocyte biology. J. Immunol 188, 521–526 (2012). 10.4049/jimmunol.1101530; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Curtsinger JM, Agarwal P, Lins DC, Mescher MF, Autocrine IFN-γ promotes naive CD8 T cell differentiation and synergizes with IFN-α to stimulate strong function. J. Immunol 189, 659–668 (2012). 10.4049/jimmunol.1102727; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eisenberg G et al. , Soluble SLAMF6 receptor induces strong CD8+ T-cell effector function and improves anti-melanoma activity in vivo. Cancer Immunol. Res 6, 127–138 (2018). 10.1158/2326-6066.CIR-17-0383; [DOI] [PubMed] [Google Scholar]
- 27.Miller BC et al. , Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat. Immunol 20, 326–336 (2019). 10.1038/s41590-019-0312-6; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumari S, Mak M, Poh Y, Tohme M, Watson N, Melo M, Janssen E, Dustin M, Kamm R, Geha R, Irvine DJ, Cytoskeletal tension actively sustains the T cell immunological synapse. bioRxiv 437236 [Preprint] 8 October 2018. 10.1101/437236 [DOI]
- 29.Tsopoulidis N et al. , T cell receptor-triggered nuclear actin network formation drives CD4+ T cell effector functions. Sci. Immunol 4, eaav1987 (2019). 10.1126/sciimmunol.aav1987; [DOI] [PubMed] [Google Scholar]
- 30.Le Mercier I et al. , VISTA regulates the development of protective antitumor immunity. Cancer Res 74, 1933–1944 (2014). 10.1158/0008-5472.CAN-13-1506; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Q, He J, Flies DB, Luo L, Chen L, Programmed death one homolog maintains the pool size of regulatory T cells by promoting their differentiation and stability. Sci. Rep 7, 6086 (2017). 10.1038/s41598-017-06410-w; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buenrostro JD et al. , Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490 (2015). 10.1038/nature14590; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashouri JF, Weiss A, Endogenous Nur77 is a specific indicator of antigen receptor signaling in human T and B cells. J. Immunol 198, 657–668 (2017). 10.4049/jimmunol.1601301; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Conley JM, Gallagher MP, Berg LJ, T cells and gene regulation: The switching on and turning up of genes after T cell receptor stimulation in CD8 T cells. Front. Immunol 7, 76 (2016). 10.3389/fimmu.2016.00076; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akondy RS et al. , Origin and differentiation of human memory CD8 T cells after vaccination. Nature 552, 362–367 (2017). 10.1038/nature24633; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Youngblood B et al. , Effector CD8 T cells dedifferentiate into long-lived memory cells. Nature 552, 404–409 (2017). 10.1038/nature25144; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng X et al. , Transcription factor Foxp1 exerts essential cell-intrinsic regulation of the quiescence of naive T cells. Nat. Immunol 12, 544–550 (2011). 10.1038/ni.2034; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ouyang W, Li MO, Foxo: In command of T lymphocyte homeostasis and tolerance. Trends Immunol 32, 26–33 (2011). 10.1016/j.it.2010.10.005; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong WF et al. , Runx1 deficiency in CD4+ T cells causes fatal autoimmune inflammatory lung disease due to spontaneous hyperactivation of cells. J. Immunol 188, 5408–5420 (2012). 10.4049/jimmunol.1102991; [DOI] [PubMed] [Google Scholar]
- 40.Zhou G et al. , Identification of systemically expanded activated T cell clones in MRL/lpr and NZB/W F1 lupus model mice. Clin. Exp. Immunol 136, 448–455 (2004). 10.1111/j.1365-2249.2004.02473.x; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Madi A et al. , T-cell receptor repertoires share a restricted set of public and abundant CDR3 sequences that are associated with self-related immunity. Genome Res 24, 1603–1612 (2014). 10.1101/gr.170753.113; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tikochinski Y et al. , A shared TCR CDR3 sequence in NOD mouse autoimmune diabetes. Int. Immunol 11, 951–956 (1999). 10.1093/intimm/11.6.951; [DOI] [PubMed] [Google Scholar]
- 43.Kim KS et al. , Dietary antigens limit mucosal immunity by inducing regulatory T cells in the small intestine. Science 351, 858–863 (2016). 10.1126/science.aac5560; [DOI] [PubMed] [Google Scholar]
- 44.Pagès H, Aboyoun P, Gentleman R, DebRoy S, Biostrings: Efficient manipulation of biological strings. R package version 2.54.0 (Bioconductor, 2019). 10.18129/B9.bioc.Biostrings [DOI] [Google Scholar]
- 45.Craft J, Peng S, Fujii T, Okada M, Fatenejad S, Autoreactive T cells in murine lupus: Origins and roles in autoantibody production. Immunol. Res 19, 245–257 (1999). 10.1007/BF02786492; [DOI] [PubMed] [Google Scholar]
- 46.Bouillet P et al. , BH3-only Bcl-2 family member Bim is required for apoptosis of autoreactive thymocytes. Nature 415, 922–926 (2002). 10.1038/415922a; [DOI] [PubMed] [Google Scholar]
- 47.Trapnell C et al. , The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol 32, 381–386 (2014). 10.1038/nbt.2859; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aksoylar HI, Lampe K, Barnes MJ, Plas DR, Hoebe K, Loss of immunological tolerance in Gimap5-deficient mice is associated with loss of Foxo in CD4+ T cells. J. Immunol 188, 146–154 (2012). 10.4049/jimmunol.1101206; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kawabe T et al. , Memory-phenotype CD4+ T cells spontaneously generated under steady-state conditions exert innate TH1-like effector function. Sci. Immunol 2, eaam9304 (2017). 10.1126/sciimmunol.aam9304; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu J, Yamane H, Paul WE, Differentiation of effector CD4 T cell populations. Annu. Rev. Immunol 28, 445–489 (2010). 10.1146/annurev-immunol-030409-101212; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blank CU et al. , Defining ‘T cell exhaustion’. Nat. Rev. Immunol 19, 665–674 (2019). 10.1038/s41577-019-0221-9; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tao X, Constant S, Jorritsma P, Bottomly K, Strength of TCR signal determines the costimulatory requirements for Th1 and Th2 CD4+ T cell differentiation. J. Immunol 159, 5956–5963 (1997). [PubMed] [Google Scholar]
- 53.Esplugues E et al. , Control of TH17 cells occurs in the small intestine. Nature 475, 514–518 (2011). 10.1038/nature10228; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hirsch R, Eckhaus M, Auchincloss H Jr., Sachs DH, Bluestone JA, Effects of in vivo administration of anti-T3 monoclonal antibody on T cell function in mice. I. Immunosuppression of transplantation responses. J. Immunol 140, 3766–3772 (1988). [PubMed] [Google Scholar]
- 55.ElTanbouly MA, Croteau W, Noelle RJ, Lines JL, VISTA: A novel immunotherapy target for normalizing innate and adaptive immunity. Semin. Immunol 42, 101308 (2019). 10.1016/j.smim.2019.101308; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barnden MJ, Allison J, Heath WR, Carbone FR, Defective TCR expression in transgenic mice constructed using cDNA-based a- and b-chain genes under the control of heterologous regulatory elements. Immunol. Cell Biol 76, 34–40 (1998). 10.1046/j.1440-1711.1998.00709.x; [DOI] [PubMed] [Google Scholar]
- 57.Ehst BD, Ingulli E, Jenkins MK, Development of a novel transgenic mouse for the study of interactions between CD4 and CD8 T cells during graft rejection. Am. J. Transplant 3, 1355–1362 (2003). 10.1046/j.1600-6135.2003.00246.x; [DOI] [PubMed] [Google Scholar]
- 58.Kearney ER, Pape KA, Loh DY, Jenkins MK, Visualization of peptide-specific T cell immunity and peripheral tolerance induction in vivo. Immunity 1, 327–339 (1994). 10.1016/1074-7613(94)90084-1; [DOI] [PubMed] [Google Scholar]
- 59.Mueller DL, Mechanisms maintaining peripheral tolerance. Nat. Immunol 11, 21–27 (2010). 10.1038/ni.1817; [DOI] [PubMed] [Google Scholar]
- 60.Altman JD et al. , Phenotypic analysis of antigen-specific T lymphocytes. Science 274, 94–96 (1996). 10.1126/science.274.5284.94; [DOI] [PubMed] [Google Scholar]
- 61.Legoux FP et al. , CD4+ T cell tolerance to tissue-restricted self antigens is mediated by antigen-specific regulatory T cells rather than deletion. Immunity 43, 896–908 (2015). 10.1016/j.immuni.2015.10.011; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moon JJ et al. , Naive CD4+ T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity 27, 203–213 (2007). 10.1016/j.immuni.2007.07.007; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang L et al. , VISTA, a novel mouse Ig superfamily ligand that negatively regulates T cell responses. J. Exp. Med 208, 577–592 (2011). 10.1084/jem.20100619; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fathman CG, Lineberry NB, Molecular mechanisms of CD4+ T-cell anergy. Nat. Rev. Immunol 7, 599–609 (2007). 10.1038/nri2131; [DOI] [PubMed] [Google Scholar]
- 65.Kalekar LA et al. , CD4+ T cell anergy prevents autoimmunity and generates regulatory T cell precursors. Nat. Immunol 17, 304–314 (2016). 10.1038/ni.3331; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martinez RJ et al. , Arthritogenic self-reactive CD4+ T cells acquire an FR4hiCD73hi anergic state in the presence of Foxp3+ regulatory T cells. J. Immunol 188, 170–181 (2012). 10.4049/jimmunol.1101311; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Josefowicz SZ, Lu LF, Rudensky AY, Regulatory T cells: Mechanisms of differentiation and function. Annu. Rev. Immunol 30, 531–564 (2012). 10.1146/annurev.immunol.25.022106.141623; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu X et al. , Genome-wide analysis identifies NR4A1 as a key mediator of T cell dysfunction. Nature 567, 525–529 (2019). 10.1038/s41586-019-0979-8; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chihara N et al. , Induction and transcriptional regulation of the co-inhibitory gene module in T cells. Nature 558, 454–459 (2018). 10.1038/s41586-018-0206-z; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Flies DB, Wang S, Xu H, Chen L, Cutting edge: A monoclonal antibody specific for the programmed death-1 homolog prevents graft-versus-host disease in mouse models. J. Immunol 187, 1537–1541 (2011). 10.4049/jimmunol.1100660; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grubin CE, Kovats S, deRoos P, Rudensky AY, Deficient positive selection of CD4 T cells in mice displaying altered repertoires of MHC class II-bound self-peptides. Immunity 7, 197–208 (1997). 10.1016/S1074-7613(00)80523–3; pmid: 9285405 [DOI] [PubMed] [Google Scholar]
- 72.Deng J et al. , Hypoxia-induced VISTA promotes the suppressive function of myeloid-derived suppressor cells in the tumor microenvironment. Cancer Immunol. Res 7, 1079–1090 (2019). 10.1158/2326-6066.CIR-18-0507; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.ENCODE Project Consortium, An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). 10.1038/nature11247; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gonzalez M et al. , The balance between donor T cell anergy and suppression versus lethal graft-versus-host disease is determined by host conditioning. J. Immunol 169, 5581–5589 (2002). 10.4049/jimmunol.169.10.5581; [DOI] [PubMed] [Google Scholar]
- 75.Lines JL et al. , VISTA is an immune checkpoint molecule for human T cells. Cancer Res 74, 1924–1932 (2014). 10.1158/0008-5472.CAN-13-1504; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Legoux FP, Moon JJ, Peptide:MHC tetramer-based enrichment of epitope-specific T cells. J. Vis. Exp 2012, e4420 (2012). 10.3791/4420; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Couter CJ, Surana NK, Isolation and flow cytometric characterization of murine small intestinal lymphocytes. J. Vis. Exp 2016, e54114 (2016). 10.3791/54114; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Amend SR, Valkenburg KC, Pienta KJ, Murine hind limb long bone dissection and bone marrow isolation. J. Vis. Exp 2016, e53936 (2016). 10.3791/53936; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dong MB, Rahman MJ, Tarbell KV, Flow cytometric gating for spleen monocyte and DC subsets: Differences in autoimmune NOD mice and with acute inflammation. J. Immunol. Methods 432, 4–12 (2016). 10.1016/j.jim.2015.08.015; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R, Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420 (2018). 10.1038/nbt.4096; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Heng TS, Painter MW; Immunological Genome Project Consortium, The Immunological Genome Project: Networks of gene expression in immune cells. Nat. Immunol 9, 1091–1094 (2008). 10.1038/ni1008-1091; [DOI] [PubMed] [Google Scholar]
- 82.Qiu X et al. , Single-cell mRNA quantification and differential analysis with Census. Nat. Methods 14, 309–315 (2017). 10.1038/nmeth.4150; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Subramanian A et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A 102, 15545–15550 (2005). 10.1073/pnas.0506580102; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C, Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). 10.1038/nmeth.4197; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Amemiya HM, Kundaje A, Boyle AP, The ENCODE Blacklist: Identification of problematic regions of the genome. Sci. Rep 9, 9354 (2019). 10.1038/s41598-019-45839-z; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stuart T et al. , Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). 10.1016/j.cell.2019.05.031; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cusanovich DA et al. , A single-cell atlas of in vivo mammalian chromatin accessibility. Cell 174, 1309–1324.e18 (2018). 10.1016/j.cell.2018.06.052; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pliner HA et al. , Cicero predicts cis-regulatory DNA interactions from single-cell chromatin accessibility data. Mol. Cell 71, 858–871. e8 (2018). 10.1016/j.molcel.2018.06.044; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sloan CA et al. , ENCODE data at the ENCODE portal. Nucleic Acids Res 44, D726–D732 (2016). 10.1093/nar/gkv1160; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cheng C, Min R, Gerstein M, TIP: A probabilistic method for identifying transcription factor target genes from ChIP-seq binding profiles. Bioinformatics 27, 3221–3227 (2011). 10.1093/bioinformatics/btr552; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Monach P et al. , The K/BxN mouse model of inflammatory arthritis: Theory and practice. Methods Mol. Med 136, 269–282 (2007). 10.1007/978-1-59745-402-5_20; [DOI] [PubMed] [Google Scholar]
- 92.Miksa M, Komura H, Wu R, Shah KG, Wang P, A novel method to determine the engulfment of apoptotic cells by macrophages using pHrodo succinimidyl ester. J. Immunol. Methods 342, 71–77 (2009). 10.1016/j.jim.2008.11.019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Suzuki J, Denning DP, Imanishi E, Horvitz HR, Nagata S, Xk-related protein 8 and CED-8 promote phosphatidylserine exposure in apoptotic cells. Science 341, 403–406 (2013). 10.1126/science.1236758; [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.