Abstract
Loss of pod shattering is one of the most important domestication-related traits in legume crops. The non-shattering phenotypes have been achieved either by disturbed formation of abscission layer between the valves, or by loss of helical tension in sclerenchyma of endocarp, that split open the pods to disperse the seeds. During domestication, azuki bean (Vigna angularis) and yard-long bean (Vigna unguiculata cv-gr. Sesquipedalis) have reduced or lost the sclerenchyma and thus the shattering behavior of seed pods. Here we performed fine-mapping with backcrossed populations and narrowed the candidate genomic region down to 4 kbp in azuki bean and 13 kbp in yard-long bean. Among the genes located in these regions, we found MYB26 genes encoded truncated proteins in azuki bean, yard-long bean, and even cowpea. As such, our findings indicate that independent domestication on the two legumes has selected the same locus for the same traits. We also argue that MYB26 could be a target gene for improving shattering phenotype in other legumes, such as soybean.
Keywords: legume, azuki bean, yard-long bean, cowpea, secondary wall thickening, MYB26, pod shattering, domestication
Introduction
One of the most important phenotypic changes during crop domestication is loss of seed shattering (Doubley et al., 2006). In legumes, this has been achieved by loss of pod shattering via malformation of dehiscence zone (Dong et al., 2014) or loss of helical shape change in the pod (Suanum et al., 2016; Murgia et al., 2017). However, this trait still needs to be improved as shattering problems sometimes force farmers to lose more than 20% of their annual yield (Philbrook and Oplinger, 1989). Thus, elucidating genetic mechanisms of shattering is important to reduce harvest loss.
Among the angiosperms, legumes have evolved a unique and sophisticated manner of shattering (Fahn and Zohary, 1955). Legume seeds are enclosed in seed pods, which explosively split open with its two valves twisting helically away from each other upon maturity (Armon et al., 2011). This mechanism is much more complicated compared to cereals, where matured seeds freely fall by abscission layer developed in pedicels (stalk of individual flower) (reviewed by Dong and Wang, 2015). Brassica and Arabidopsis are more similar to legumes in that the seeds are embedded in the siliques that also spring open upon maturity (Spence et al., 1996). However, their siliques do not exhibit any helical shape change as legume pods do. As such, knowledge obtained from rice and Arabidopsis cannot be simply applied to understand the shattering of legumes.
The key of helical shape change of legume seed pods is the development of a thick sclerenchyma with a bilayer structure on the endocarp (inner surface of the pod). In this bilayer structure, cellulose microfibrils of the outer and inner layers run at ±45° from the longitudinal axis of seed pods (Erb et al., 2013). As the matured seed pods dry, the microfibrils shrink in perpendicular directions and generate the helical force to blow the seeds off (Erb et al., 2013). In contrast, the endocarp of Arabidopsis, which develops cell layers with thickened secondary walls, is inflexible (Spence et al., 1996). As the matured siliques dry, the pericarp tissues shrink but the endocarp cell layers do not, generating tension to spring open the siliques from the dehiscence zone at the valve margins (Spence et al., 1996).
However, no specific genes have been identified for the sclerenchyma formation, despite several quantitative trait loci (QTL) analyses and genome-wide association studies locating several loci involved in legumes’ pod shattering (Dong et al., 2014; Suanum et al., 2016; Murgia et al., 2017; Lo et al., 2018; Rau et al., 2019). In soybean, the domestication-type allele of SHAT1-5 disturbs only dehiscence zone formation and does not affect helical shape change (Dong et al., 2014). Fiber content in seed pods, which is related to sclerenchyma formation, is reduced in common bean (Murgia et al., 2017; Parker et al., 2019; Rau et al., 2019), but the responsible gene is not cloned yet. An exception is Pdh1 gene in soybean, which reduces the helical force of the sclerenchyma when disrupted (Funatsuki et al., 2014). However, the plants with non-functional PDH1 protein still initiate sclerenchyma formation, so PDH1 seems involved in later steps in sclerenchyma development.
Contrary to soybean, azuki bean [Vigna angularis (Willd.) Ohwi et Ohashi] and yard-long bean [Vigna unguiculata (L.) Walp. cv-gr. Sesquipedalis E. Westphal] seem disrupted in initiating sclerenchyma formation. In azuki bean, an important legume crop in East Asia, the seed pods of domesticated accessions normally form a dehiscence zone, but the pericarps are thinner and tenderer and show little helical shape change (Isemura et al., 2007; Kaga et al., 2008). In yard-long bean, which is often cultivated in Southeast Asia, pods are very long (60–100 cm) and do not show any helical shape change at all (Kongjaimun et al., 2012). In addition, the pods are extremely tender, and thus young pods are favored as vegetables (Kongjaimun et al., 2013; Suanum et al., 2016). Therefore, we consider azuki bean and yard-long bean are the best materials to isolate the gene for initiating sclerenchyma formation.
Thus, in this study, we performed fine-mapping to identify the responsible locus for pod shattering and pod tenderness in azuki bean and yard-long bean, respectively. Interestingly, we have previously revealed that the QTLs controlling pod shattering in azuki bean and pod tenderness in yard-long bean are co-localized around 8–10 cM on the linkage group 7 (LG7) (Kongjaimun et al., 2013), which corresponds to Chr7 in azuki bean (Sakai et al., 2015) and Chr5 in cowpea (Lonardi et al., 2019). To narrow down the candidate region, we developed backcrossed populations and DNA markers based on the genome sequence of azuki bean (Sakai et al., 2015), cowpea (Lonardi et al., 2019), and the wild cowpea (sequenced in this study). Our efforts identified MYB26 transcription factor as the only candidate.
Materials and Methods
Plant Materials and Growth Condition
All the accessions used in this study were provided by NARO Genebank (Tsukuba, Japan) (Figure 1 and Supplementary Figure 1).
FIGURE 1.
Pod phenotypes of parental accessions and BC3F5 plants with wild-type phenotypes in genetic background of domesticated genotypes. (a,d) Wild azuki bean (JP107881). (b,e) BC3F5 plant with shattering phenotype. (c,f) Domesticated azuki bean (JP81481). (g,j) Wild cowpea (JP81610). (h,k) BC3F5 plants with hard pod phenotype. (i,l) Yard-long bean (JP89093). (a–c) Dried mature pods. (d–f,j–l) Cross sections of pods stained with phloroglucinol. (g–i) Young pods.
For azuki bean, we started from the BC1F2 plants derived from a cross between domesticated azuki bean (JP81481, the recurrent parent) and a wild relative, Vigna nepalensis Tateishi & Maxted (JP107881) (Isemura et al., 2007). Of them, we selected one BC1F2 plant where the locus for pod shattering is fixed with the wild-type allele but most of other loci are fixed with domestication-type alleles and crossed again to the recurrent parent. We further backcrossed the obtained BC2F1 plants to obtain BC3F1 plants. We selfed them and obtained BC3F2 seeds from those with pod shattering phenotypes. For BC3F3, BC3F4, and BC3F5 populations, we kept selecting and selfing those with recombination within the candidate region. We also included plants that were heterozygous throughout the candidate region for obtaining more recombination and some that are homozygous in the same region to reconfirm the relationship of genotype and phenotype.
For yard-long bean, we started from BC1F2 plants derived from a cross between yard-long bean (JP89083, the recurrent parent) and a wild cowpea, Vigna unguiculata subsp. dekindtiana (Harms) Verdc. (JP81610) (Kongjaimun et al., 2013). Of them, we selected one BC1F2 plant that are fixed with wild-type allele at the locus for pod tenderness but are fixed with domestication-type at most other loci. The selected BC1F2 plants were backcrossed to the recurrent parent and further backcrossed to obtain BC3F1 plants. We selfed them and obtained BC3F2 seeds from those with hard pods phenotype. We kept selecting and selfing up to BC3F5 populations as done in azuki bean.
All the plants were grown from July through November in a field located in Tsukuba city, Japan, except BC3F5 plants of yard-long bean that were grown in a greenhouse from September through January.
Observation of Sclerenchymal Tissue
We observed sclerenchymal tissues in seed pods by staining lignin with the Wiesner (phloroglucinol–HCl) reaction (Pormar et al., 2002). We harvested three undried mature seed pods per plant, sliced them to 200-μm sections with a Plant Microtome MTH-1 (Nippon Medical & Chemical Instruments Co., Ltd., Osaka, Japan), incubated in phloroglucinol–HCl solution for 1 min and observed with stereoscopic microscope SZX7 (OLYMPUS, Tokyo) for BC3F5 plants of azuki bean and yard-long bean. Phloroglucinol–HCl solution was prepared by dissolving 1 g of phloroglucin in 50 ml ethanol and adding 25 ml of concentrated hydrochloric acid.
Phenotyping
For azuki bean populations, we evaluated pod shattering by counting the number of twists in a pod, as described in Isemura et al. (2007). For BC3F2 and BC3F3, we harvested five pods per plant before shattering and measured the length. We then incubated the pods at 60°C to let them totally dry. Unless the pods dehisced, we manually opened the pods and counted the number of twists and calculated the number of twists/cm. For BC3F4 and BC3F5, we harvested 10 pods per plant, let them dry, and evaluated the rate of shattering pods before counting the number of twists.
For yard-long bean, we manually evaluated the tenderness of five young pods per plant by binarizing hard or soft.
We also measured 100 seed weight on BC3F2 and BC3F5 plants of the azuki bean population and BC3F4 and BC3F5 plants of the yard-long bean population.
Sequencing and Assembly of the Wild Cowpea Genome
We sequenced the whole genome of wild cowpea (JP81610) with RSII sequencer (Pacific Biosciences, Menlo Park, CA, United States), as we have done previously for azuki bean (Sakai et al., 2015). DNA was isolated from 1 g of unexpanded leaves with the cetyl trimethylammonium bromide (CTAB) method and purified with Genomic Tip 20/G (Qiagen K. K. Tokyo). The extracted DNA was sheared into 20-kb fragments using g-TUBE (Covaris, MA, United States) and converted into 20-kb SMRTbell template libraries. The library was size selected for a lower cutoff of 10 kb with BluePippin (Sage Science, MA, United States). Sequencing was performed on the PacBio RS II using P6 polymerase binding and C4 sequencing kits with 360 min acquisition. In total, 21 SMRT cells were used to obtain ∼26.4 Gb of raw reads.
In total, 4 million PacBio reads were used for de novo assembly with Canu v1.6 under the default settings (corOvlErrorRate = 0.2400, obtOvlErrorRate = 0.0450, utgOvlErrorRate = 0.0450, corErrorRate = 0.3000, obtErrorRate = 0.0450, utgErrorRate = 0.0450, cnsErrorRate = 0.0750). About 23.5x error corrected and trimmed reads longer than 1,000 bp were assembled to contigs. Assembled contigs were polished by PacBio raw reads by using the arrow in GenomicConsensus v2.3.2 (Pacific Biosciences of California, Inc.).
Repeat detection was conducted by RepeatMasker ver. 4.0 (Smit et al., 2013)1. A de novo repeats library of wild cowpea genome constructed by RepeatModeler ver. 1.0.11 (see footnote 1) and the Fabaceae repeats library in RepBase24.02 (Smit and Hubley, 2008)2 were used for the prediction.
Ab initio gene prediction was done with AUGUSTUS (version 3.3.2) (Stanke et al., 2008). A set of gene annotation information of recently published cowpea genome was used for training AUGUSTUS (Lonardi et al., 2019). We trained a new model twice using 1,000 high-confidential genes selected based on abundance of annotations of domains, pathway networks, and gene ontology information. BUSCO v3 (Waterhouse et al., 2017) was used to evaluate protein sequences of annotated genes.
Single-nucleotide polymorphisms (SNPs) and short insertions/deletions (indels) were detected by using MUMmer v3.23 (Kurtz et al., 2004). Genome alignment of our wild cowpea assembly and the reference cowpea genome (Lonardi et al., 2019) was conducted by the nucmer command with the following options: –maxgap = 500 –mincluster = 100. Then one-to-one alignment was extracted with delta-filter command with an option: −1. Based on the alignments, SNPs and indels were reported by the show-snps command.
Raw sequence data, the assembled sequences, and gene annotations are all available from DNA Data Bank of Japan (DDBJ)3 under the BioProject ID PRJDB8129.
Genotyping
We extracted DNA from the seeds as described by Kamiya and Kiguchi (2003) and genotyped them by fragment analysis with capillary electrophoresis for simple sequence repeats (SSRs) and INDELs as described by Isemura et al. (2007) or by directly sequencing SNP sites (see section “Direct Sequencing” below). The information of the markers we used is summarized in Supplementary Table 1.
We designed primers by Primer3 (Untergasser et al., 2012) to amplify polymorphic sites between the domesticated azuki bean and V. nepalensis that are available in Vigna Genome Server (VigGS4; Sakai et al., 2016) and those between the domesticated cowpea genome in Legume Information System5 and the wild cowpea genome sequenced in this study. Parameters for designing primers on Primer36 were 20–30 bp in length, 55°C–65°C in annealing temperature, and 100–350 bp and >700 bp in expected length of amplified fragments for fragment analysis and direct sequencing, respectively.
For azuki bean, the markers we used were as follows:
BC3F2: CEDG064 and SPD01-SPD07
BC3F3: CEDG064, SPD03, SPD04, SPD08, SPD09
BC3F4: CEDG064 and SPD08–SPD11
BC3F5: CEDG064, SPD10, SPD11 and the sequencing primers for Vigan.07G034400
For yard-long bean, the markers we used were as follows:
BC3F4: VuPT01–VuPT06
BC3F5: VuPT03, VuPT04, VuPT07–VuPT13 and the sequencing primers for Vigun05g273300, Vigun05g273400, and Vigun05g0723500.
Direct Sequencing
To sequence the (potentially) polymorphic sites, we amplified the template DNA with AmpliTaq Gold 360 Master Mix (Thermo Fisher Scientific K. K., Tokyo), performed sequencing reaction with BigDye Terminator v3.1 (Thermo Fisher Scientific K. K., Tokyo), and sequenced with ABI Genetic Analyzer 3130xl (Thermo Fisher Scientific K. K., Tokyo), according to the provider’s protocol.
Determining Transcribed Sequences of Vigan.07g34400 and Vigun05g273500
To sequence both the domesticated and the wild alleles of Vigan.07g034400.01 and Vigun05g273500 genes, we sequenced the transcribed sequences of both loci. We extracted total RNA from 100 mg of flower buds right before flowering of all the parental accessions using RNeasy Plant Mini Kit (QIAGEN) with RNase-free DNase I (Invitrogen). Total RNA of 1 μg was converted into first-strand cDNA with Super Script II Reverse Transcriptase (Invitrogen) and Oligo(dT)12-18 Primer (Invitrogen) following the manufacturer’s instructions. The cDNA sequences of Vigan.07g034400.01 and Vigun05g273300.01 were then amplified and sequenced with the primers (Supplementary Table 1) and then transferred to the direct-sequencing protocol described above.
Results
Anatomical Analysis of Pod Sclerenchyma
To confirm that azuki bean and yard-long bean have lost or reduced sclerenchyma in seed pods, we observed cross sections of seed pods stained with phloroglucinol–HCl and found a clear-cut difference between the wild and domesticated accessions. In the wild azuki bean and cowpea, which have shattering phenotypes, thick layers of sclerenchyma (∼0.3 mm) were formed on the endocarps of seed pods (Figure 1). However, in the domesticated azuki bean and yard-long bean, which are both non-shattering, the sclerenchyma layer was slightly formed (<0.1 mm) or not formed at all, respectively (Figure 1).
We also evaluated the correlation between the thickness of pod sclerenchyma and helical shape change in seed pods (number of twists/cm) in the BC3F5 plants of azuki bean and yard-long bean. As a result, all the shattering plants in azuki bean population formed thicker sclerenchyma (0.15–0.20 mm) and showed a stronger helical shape change (0.30–0.43) than the non-shattering plants did (0.00–0.08 mm and 0.00–0.05, respectively) (Figure 1 and Supplementary Figure 1). The correlation coefficient between the sclerenchyma thickness and the helical shape change was 0.92. In the yard-long bean population, all the plants with hard pod phenotype formed sclerenchyma (0.16–0.22 mm) and showed little helical shape change (0.09–0.12), whereas those with soft pod phenotype did not form sclerenchyma or show helical shape change at all (Figure 1 and Supplementary Figure 1). The correlation coefficient between the sclerenchyma thickness and the helical shape change was 0.99.
In addition, we observed cross sections of seed pods in a wild soybean and three domesticated soybean cultivars and found thick sclerenchyma layers in all the accessions (Supplementary Figure 2).
Fine-Mapping Pod Shattering in Azuki Bean
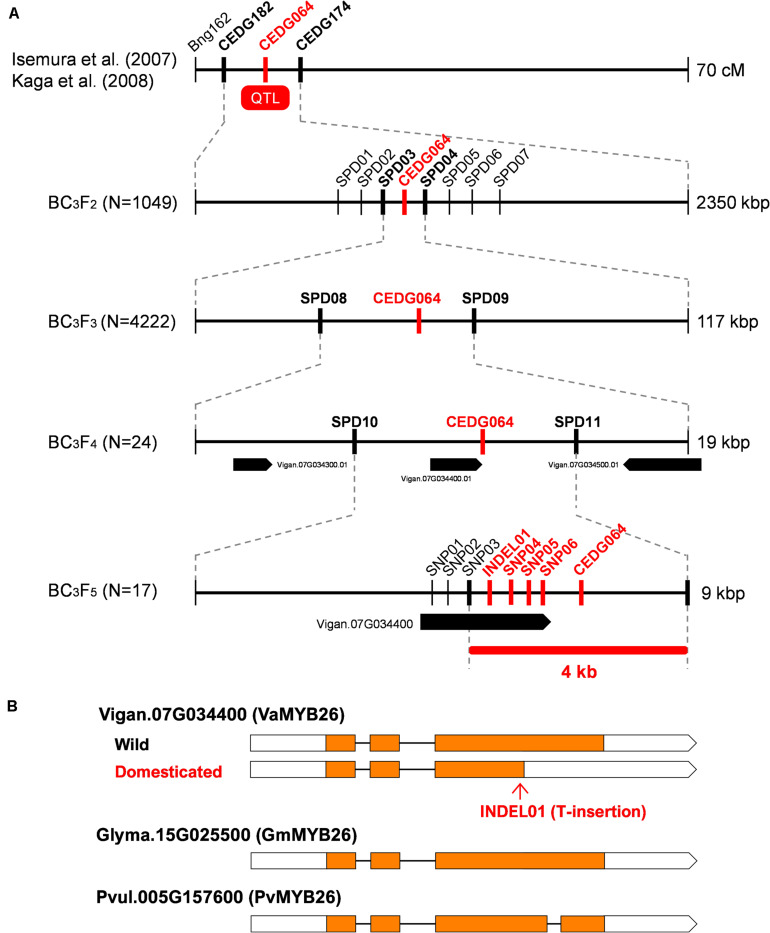
We previously mapped the QTL for pod shattering in between CEDG182 and CEDG174 on LG7, which is around 1.4–4.0 Mbp in Chr7 of the reference azuki bean genome (Sakai et al., 2015) (Figure 2). To more finely locate the genetic factor for pod shattering, we genotyped 1,049 BC3F2 plants and selected 238 for phenotyping. The obtained phenotype and genotype data revealed the pod shattering factor was completely linked with CEDG064 and located in between SPD03 and SPD04 (Figure 2 and Supplementary Table 2). We selected 46 plants out of the 238 (Supplementary Table 2), selfed them, and obtained 4,222 BC3F3 seeds.
FIGURE 2.
Fine-mapping of pod shattering factor in azuki bean. (A) Process of fine mapping. Vertical lines indicate DNA marker sites. Red markers indicate those with complete linkage with the phenotype in the mapping population. Bold black markers indicate those neighboring the candidate region. (B) A schematic of the wild-type and domesticated-type alleles of Vigan.07g034400. The MYB26 orthologues of soybean (Glyma.15G025500) and common bean (Pvul.005G157600) are also shown. Boxes indicate exons, whereas lines indicate introns. Coding DNA sequences (CDS) are filled with yellow.
Of the BC3F3 seeds, we selected 53 that had recombination between SPD03 and SPD04. Phenotyping and genotyping on these 53 revealed that pod shattering factor was still completely linked with CEDG064 and was in between SPD08 and SPD09 (Figure 2 and Supplementary Table 3), which is about 19 kb long and contained three genes (Figure 2).
Since this region seemed to have a relatively higher recombination rate, we expected it would be possible to further locate the pod shattering factor. Thus, of the 53 BC3F3 plants, we selected 24 plants (Supplementary Table 3) and selfed them to obtain BC3F4 seeds. We then cultivated 1–7 plants/line (81 in total; Supplementary Table 4) for further genotyping and phenotyping (Figure 2).
As a result, we again found CEDG064 was completely linked with the phenotype, but we also obtained 29 recombinants between SPD10 and CED064 and five between CEDG064 and SPD11 (Supplementary Table 4). Thus, the pod shattering factor was located in a 9-kb region between SPD10 and SPD11, where there is only one gene Vigan.07g034400 (Figure 2), which encoded VaMYB26, an ortholog of AtMYB26.
To see if there were any loss-of-function mutations in VaMYB26 in azuki bean, we searched the polymorphism data between wild and domesticated azuki bean provided by VigGS. The database showed that there were six SNPs (SNP01-SNP06) and one INDEL (INDEL01) within its open reading frame (ORF). Of these, all the SNPs were in the introns or in the 3′-untranslated region (UTR), but the INDEL was within the coding DNA sequence (CDS) (a T insertion 4 bp before the stop codon in the domesticated azuki bean). Since the wild azuki bean did not have this T insertion, the CDS could be 405 bp (125 aa) longer (Supplementary Figure 3). Interestingly, a BLAST search of MYB26 gene revealed that the longer version is widely conserved across plant taxa (Figure 2). Thus, the VaMYB26 in the domesticated azuki bean seemed to have an immature stop codon.
To confirm the polymorphisms identified in the database, we determined the genomic and the transcribed sequences of VaMYB26 locus of both parents and found all the polymorphisms in the ORF were true (Supplementary Figure 3). We also sequenced the 17 lines of BC3F5, which we obtained from the selected BC3F4 plants (those with fixed genotypes between SPD03 and SPD09; Supplementary Table 5), to further confirm the relationship between the genotypes and the phenotype. Of the 17, one had recombination between SNP03 and INDEL01, but none had recombination between INDEL01 and CEDG064 (Supplementary Table 5). Thus, the genotypes at INDEL01, SNP04–SNP06, and CEDG064 were completely linked with the pod shattering phenotype, locating the pod shattering factor within the 4-kb region between SNP03 and SPD11 (Figure 2 and Supplementary Table 5).
Assembly and Annotation of Wild Cowpea Genome
Though the reference genome of cowpea had been released (Lonardi et al., 2019), it was yet not easy to obtain polymorphic markers because the genome sequence of the wild cowpea was not available. Thus, to facilitate our fine-mapping the factor of pod tenderness, we sequenced and assembled the genome of V. unguiculata subsp. dekindtiana (JP81610), which is a wild cowpea accession used to develop the mapping population of yard-long bean. We used PacBio RSII and obtained 4,413,480 raw reads with an average read length of 6.0 kbp and N50 length of 10.6 kbp, covering 44.9X of the estimated genome size (585.8 Mbp) (Lonardi et al., 2019) (Supplementary Figure 4 and Supplementary Table 6). The assembly produced 4,285 contigs covering 488.5 Mbp (83.4%) of the cowpea genome, with the contig NG50 of 438.6 kbp and the maximum contig length of 2.6 Mbp, respectively (Supplementary Table 7). Of the assembled sequences, 45.7% composed of transposable elements, of which LTR retrotransposons share the largest content (20.5% of the assembly) (Supplementary Table 8). Our ab initio gene prediction detected 36,061 protein-coding genes, and 27,345 of those were non-repeat related (Supplementary Table 9). The annotated genes of the wild cowpea contained 93.2% (85.6% complete and 7.6% partial) of the 1,440 plant genes in BUSCO v3 (Waterhouse et al., 2017).
We aligned the genome sequences of the wild cowpea to those of the reference cowpea (Lonardi et al., 2019) and identified 5,661,319 SNPs and 1,626,169 INDELs. As expected, we detected fewer SNPS and INDELs around centromeric and pericentromeric regions but many in chromosome arms, which were presumably gene-rich regions (Supplementary Figure 5).
Fine-Mapping Pod Tenderness Factor in Yard-Long Bean
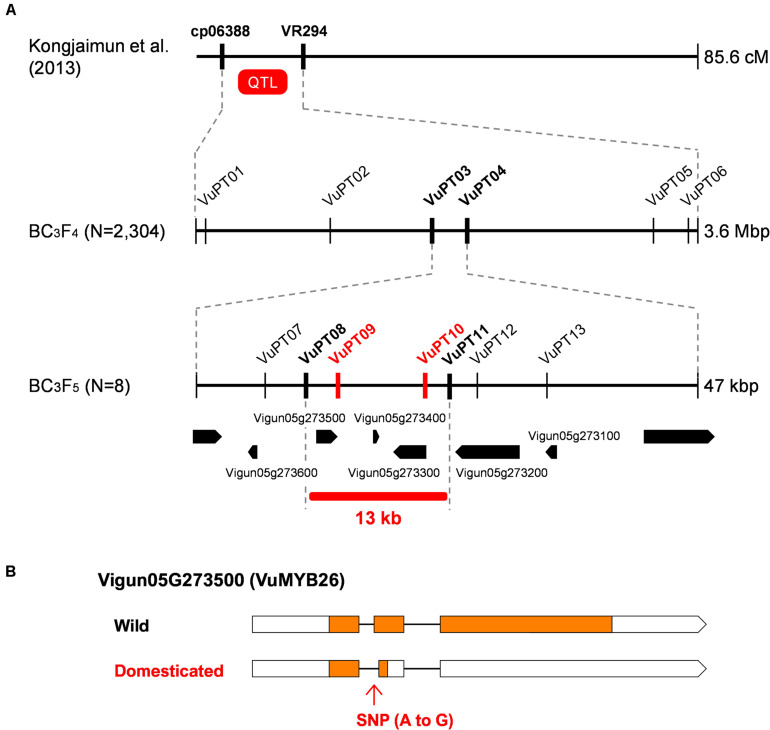
The QTL for pod tenderness was previously located between cp06388 and VR294 on LG7 (Kongjaimun et al., 2013) (Figure 3), which corresponds to about 47.5–45.5 Mbp region in Chr5 of the cowpea genome (Vigna unguiculata v1.0, NSF, UCR, USAID, DOE-JGI)7. To more finely locate the pod tenderness factor, we designed more markers based on the SNPs and INDELs we identified above, genotyped 2,304 BC3F4 seeds, and selected 195 for phenotyping (Supplementary Table 10).
FIGURE 3.
Fine-mapping of pod tenderness factor in yard-long bean. (A) Process of fine-mapping. Vertical lines indicate DNA marker sites. Red markers indicate those with complete linkage with the phenotype in the mapping population. Bold black markers indicate those neighboring the candidate region. (B) A schematic of the wild-type and domesticated-type alleles of Vigun05g273500. Boxes indicate exons, whereas lines indicate introns. Coding DNA sequences (CDS) are filled with yellow.
The obtained data of genotype and phenotype located the pod tenderness factor between VuPT03 and VuPT04, which was about 47 kbp (Figure 3 and Supplementary Table 10). Although no marker was completely linked with the phenotype, eight plants had recombination between this region (Figure 3 and Supplementary Table 10).
Of the 195 BC3F4 plants we tested, we selfed the eight plants and obtained BC3F5 for further mapping. As a result, we located the candidate regions in between VuPT08 and VuPT11, which was 13 kbp long and contained three genes, Vigun05g27300, Vigun05g273400, and Vigun05g273500. Of note, Vigun05g273500 encoded VuMYB26 (Figure 3 and Supplementary Table 11).
According to the SNPs and INDELs data, Vigun05g273500 had an A to G substitution which might disrupt the junction site of the first intron and the second exon (Supplementary Figure 6). On the other hand, Vigun05g27300 seemed to have only synonymous SNPs, and Vigun05g273400 was a non-coding gene. All the SNPs were confirmed by directly sequencing the ORFs of these genes. Interestingly, the ORF sequences of the three genes were exactly the same between the reference cowpea and yard-long bean, which means yard-long bean and the reference cowpea share the domestication-type allele in this locus (Supplementary Figure 6).
To test whether the A to G substitution disrupted splicing, we sequenced cDNAs of this locus prepared from both parents. As a result, we found, in the domestication-type allele, the junction site of the first intron and the second exon had shifted downstream by 8 bp, which could result in a frameshift and an immature stop codon in the middle of the second exon (Supplementary Figure 6). This product would encode a protein of 60 aa, which would be 305 aa shorter than that of the wild-type allele.
Seed Size Increase by Loss of Sclerenchymal Tissue
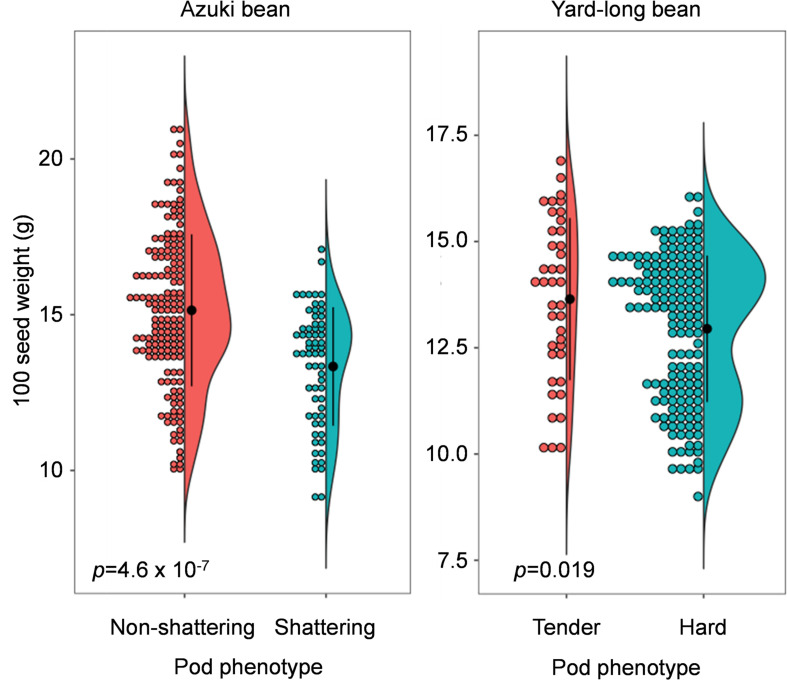
Because Murgia et al. (2017) suggested the pod shattering phenotypes load an energy cost on plants and limit seed size, we measured 100 seed weight of the mapping population (BC3F2 of azuki bean and BC3F4 of yard-long bean).
As expected, those with domestication-type trait produced larger seeds compared to those with the wild-type trait (Figure 4). In the azuki bean population, the seeds of non-shattering plants were 15.4 ± 2.4 g/100 seeds, whereas the seeds of shattering plants were 13.3 ± 1.9 g/100 seeds. In the yard-long bean population, the seeds of tender-pod plants were 13.6 ± 1.9 g/100 seeds, whereas the seeds of hard-pod plants were 12.9 ± 1.7 g/100 seeds. The following t-test revealed that in both cases, the seeds of the plants with the domestication-type phenotypes produced significantly larger seeds than the others (p = 4.6 × 10–7 for azuki bean and p = 0.019 for yard-long bean).
FIGURE 4.
Dot-violin plot of 100 seed weight in azuki bean and yard-long bean mapping population. The bin of dot plots is 0.2 g. The black dots and vertical lines in the violin plots indicate the averages and standard deviations, respectively. The p-values of the t-test are also indicated in the plots.
We also measured 100 seed weight of BC3F5 plants and observed the same trend, though the differences were not significant (Supplementary Figure 7). In azuki bean, the seeds of non-shattering plants were 9.7 ± 1.4 g/100 seeds, whereas the seeds of shattering plants were 8.7 ± 1.2 g/100 seeds. In yard-long bean, the seeds of tender-pod plants were 14.2 ± 1.5 g/100 seeds, whereas the seeds of hard-pod plants were 12.2 ± 1.2 g/100 seeds.
Discussion
In this study, we identified Vigan.07g034400 (Figure 2) and Vigun05g273500 (Figure 3) are the best candidate genes for sclerenchyma formation in the seed pods of azuki bean and yard-long bean, respectively (Figure 1). The domestication-type alleles of both loci encode truncated proteins due to immature stop codons (Supplementary Figures 3, 6), leading to a reduction or loss of sclerenchyma, which is required to generate the helical tension for splitting open legume pods (Figure 1 and Supplementary Figure 1). Although our primary goal of the fine-mapping in yard-long bean was to identify a gene for the extreme tenderness of its seed pod, the SNP we identified was in common between yard-long bean and the reference cowpea (Supplementary Figure 6). Thus, the domestication-type allele of Vigun05g273500 is not the specific allele to yard-long bean but is the one which has originally been selected for resistance to pod shattering during domestication of cowpea (Lush and Evans, 1981).
Vigan.07g034400 and Vigun05g273500 are orthologous to each other and to Arabidopsis transcription factor MYB26, which is strongly expressed in the anther and promotes secondary wall thickening in the endothelium (Yang et al., 2007). Interestingly, this secondary wall thickening is critical for anther dehiscence and thus for releasing the matured pollen (Yang et al., 2007). However, this gene is hardly expressed in siliques and is not involved in pod dehiscence (Yang et al., 2007). On the other hand, MYB26 is expressed in seed pods in many legume species according to the expression database [VigGS, the Phaseolus vulgaris Gene Expression Atlas (PvGEA)8 and Soybean eFP Browser9 ]. In addition, Di Vittori et al. (2020) has recently revealed in common bean that reduced expression of MYB26 is a key for non-shattering genotypes. Thus, in legumes, MYB26 plays a crucial role in the development of pod sclerenchyma and has been a common target of human selection among the three independently domesticated legumes, azuki bean, cowpea, and common bean. This fact suggests the importance of MYB26 in pod shattering, despite there are usually multiple options to achieve a domestication-related trait (Doubley et al., 2006; Takahashi et al., 2019).
Our results also indicate that the accumulation of some more mutations is necessary to achieve the extreme tenderness of seed pod in yard-long bean. Indeed, the QTL analysis on pod tenderness (Kongjaimun et al., 2013) and pod fiber content (Suanum et al., 2016) identified a few other QTLs than the locus we identified here. Thus, such mutations had been selected for the complete loss of pod sclerenchyma during the secondary domestication in Asia (Faris, 1965).
In addition, we have to admit that availability of whole-genome sequences greatly facilitated the study. Although the lower coverage and shorter read length of our assembly resulted in ∼10 times lower contiguity than the reference cowpea genome (Lonardi et al., 2019), its accuracy was good enough as all the SNPs between the two assemblies, which we tried to use as markers, were confirmed to be true (Supplementary Tables 5, 11 and Supplementary Figures 3, 6). We also reemphasize the power of map-base cloning, which is a classic, time-consuming, but reliable approach to isolate responsible genes.
We also consider the knowledge obtained in this study is applicable to improve soybean or other legume crops. As described before, soybean has reduced shattering ability by disturbing dehiscence zone formation between the valves (Dong et al., 2014) and by reducing helical tension of the sclerenchyma (Funatsuki et al., 2014). However, it still forms thick sclerenchyma in the seed pods (Supplementary Figure 2), which causes shattering problems especially under drought conditions (Philbrook and Oplinger, 1989). Thus, loss-of-function mutations or genome-editing in MYB26 genes may reduce sclerenchyma and enforce resistance to shattering. In addition, our findings indicate that loss or reduction of sclerenchyma may result in a slight (∼10%) increase in seed size (Figure 4 and Supplementary Figure 7). Murgia et al. (2017) also reported that common bean plants with non-shattering genotypes produce slightly larger seeds than those with shattering genotypes. This might be because developing sclerenchyma loads an energy cost for plants that could be allocated for seed production (Murgia et al., 2017), or pod hardness limits space for seed enlargement. In any case, MYB26 may serve not only for reducing harvest loss but also for directly increasing seed size.
Conclusion
Our map-based cloning approach with support of whole-genome sequences identified MYB26 as an only candidate for the development of pod sclerenchyma, which generates helical tension of legumes’ shattering pods. Our findings suggest that MYB26 might be a good target to improve resistance to pod shattering in soybean and other legumes.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material.
Author Contributions
YT, PS, AKo, and KN planned the study. YT, AKo, CM, YK, TI, HSai, AKa, NT, and KN performed the experiments and collected the data. MK, HSak, KS, KT, AS, MS, and TH performed the genome sequencing, de novo assembly, and gene annotation. YT, AKa, HSak, MK, and KN analyzed the data. KN wrote the manuscript. All the authors contributed to the article and approved the submitted version.
Conflict of Interest
TH is currently employed by the company Sentan Pharma Inc., his contribution to this study was made while he was working for his former affiliation, OIAS.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by JSPS KAKENHI Grant Numbers 23688002, 13J09808, and 26850006. It was also partially supported by Research Supporting Program of the Advanced Analysis Center, National Agriculture and Food Research Organization (NARO), and the Genebank Project, NARO. The manuscript has been released as a preprint at bioRxiv (Takahashi et al., 2019).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00748/full#supplementary-material
References
- Armon S., Efrati E., Kupferman R., Sharon E. (2011). Geometry and mechanics in the opening of chiral seed pods. Science 333 1726–1730. 10.1126/science.1203874 [DOI] [PubMed] [Google Scholar]
- Di Vittori V., Bitocchi E., Rodriguez M., Alseekh S., Bellucci E., Nanni L., et al. (2020). Pod indehiscence in common bean is associated to the fine regulation of PvMYB26 and a non-functional abscission layer. bioRxiv [Preprint] 10.1101/2020.04.02.021972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Wang Y. Z. (2015). Seed shattering: from models to crops. Front. Plant Sci. 6:476. 10.3389/fpls.2015.00476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Yang X., Liu J., Wang B. H., Liu B. L., Wang Y. Z. (2014). Pod shattering resistance associated with domestication is mediated by a NAC gene in soybean. Nat. Commun. 5:3352. [DOI] [PubMed] [Google Scholar]
- Doubley F. F., Gaut B. S., Smith B. D. (2006). The molecular genetics of crop domestication. Cell 127 1309–1321. 10.1016/j.cell.2006.12.006 [DOI] [PubMed] [Google Scholar]
- Erb R. M., Sander J. S., Grisch R., Studart A. R. (2013). Self-shaping composites with programmable bioinspired microstructures. Nat. Commun. 4:1712. [DOI] [PubMed] [Google Scholar]
- Fahn A., Zohary M. (1955). On the pericardial structure of the legumen, its evolution and relation to dehiscence. Phytomorphology 5 99–111. [Google Scholar]
- Faris D. G. (1965). The origin and evolution of the cultivated forms of Vigna sinensis. Can. J. Genet. Cytol. 7 433–452. 10.1139/g65-058 [DOI] [Google Scholar]
- Funatsuki H., Suzuki M., Hirose A., Inaba H., Yamada T., Hajika M., et al. (2014). Molecular basis of a shattering resistance boosting global dissemination of soybean. Proc. Natl. Acad. Sci. U.S.A. 111 17797–17802. 10.1073/pnas.1417282111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isemura T., Kaga A., Konishi S., Ando T., Tomooka N., Han O. K., et al. (2007). Genome dissection of traits related to domestication in azuki bean (Vigna angularis) and comparison with other warm-season legumes. Ann. Bot. 100 1053–1071. 10.1093/aob/mcm155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaga A., Isemura T., Tomooka N., Vaughan D. A. (2008). The genetics of domestication of the azuki bean (Vigna angularis). Genetics 178 1013–1036. 10.1534/genetics.107.078451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya M., Kiguchi T. (2003). Rapid DNA extraction method from soybean seeds. Breed. Sci. 53 277–279. 10.1270/jsbbs.53.277 26081539 [DOI] [Google Scholar]
- Kongjaimun A., Kaga A., Tomooka N., Somta P., Vaughan D. A., Srinives P. (2012). The genetics of domestication of yardlong bean, Vigna unguiculata (L.) Walp. ssp. unguiculata cv.-gr. Sesquipedalis. Ann. Bot. 109 1185–1200. 10.1093/aob/mcs048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kongjaimun A., Somta P., Tomooka N., Kaga A., Vaughan D. A., Srinives P. (2013). QTL mapping of pod tenderness and total soluble solid in yardlong bean [Vigna unguiculata (L.) Walp. subsp. unguiculata cv.-gr. sesquipedalis]. Euphytica 189:217 10.1007/s10681-012-0781-2 [DOI] [Google Scholar]
- Kurtz S., Pillippy A., Delcher A. L., Smoot M., Shumway M., Sntonescu C., et al. (2004). Versatile and open software for comparing large genomes. Genome Biol. 5:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo S., Muñoz-Amatriaín M., Boukar O., Herniter I., Cisse N., Guo Y. N., et al. (2018). Identification of QTL controlling domestication-related traits in cowpea (Vigna unguiculata L. Walp). Sci. Rep. 8:6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonardi S., Muñoz-Amatriaín M., Liang Q., Shu S., Wanamaker S. I., Lo S., et al. (2019). The genome of cowpea (Vigna unguiculata [L.] Walp.). Plant J. 98 767–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lush W. M., Evans L. T. (1981). The domestication and improvement of cowpeas (Vigna unguiculatea (L.) Walp.). Euphytica 30 579–587. 10.1007/bf00038783 [DOI] [Google Scholar]
- Murgia M. L., Attene G., Rodriguez M., Bitocchi E., Bellucci E., Fois D., et al. (2017). A Comprehensive Phenotypic Investigation of the “Pod-Shattering Syndrome” in Common Bean. Front. Plant Sci. 8:251. 10.3389/fpls.2017.00251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker T. A., Miery Teran J. C. B., Palkovic A., Jernstedt J., Gepts P. (2019). Genetic control of pod dehiscence in domesticated common bean: associations with range expansion and local aridity conditions. biorxiv [Preprint] 10.1101/517516 [DOI] [Google Scholar]
- Philbrook B., Oplinger E. S. (1989). Soybean field losses as influenced by harvest delays. Agron. J. 81 251–258. 10.2134/agronj1989.00021962008100020023x [DOI] [Google Scholar]
- Pormar F., Merino F., Barceló R. (2002). O-4-linked coniferyl and sinapyl alehydes in lignifying cell walls are the main targets of the Wiesner (phloroglucinol-HCl) reaction. Protoplasma 220 17–28. 10.1007/s00709-002-0030-y [DOI] [PubMed] [Google Scholar]
- Rau D., Murgia M. L., Rodriguez M., Bitocchi E., Bellucci E., Fois D., et al. (2019). Genomic dissection of pod shattering in common bean: mutations at non-orthologous loci at the basis of convergent phenotypic evolution under domestication of leguminous species. Plant J. 97 693–714. 10.1111/tpj.14155 [DOI] [PubMed] [Google Scholar]
- Sakai H., Naito K., Ogiso-Tanaka E., Takahashi Y., Iseki K., Muto C., et al. (2015). The power of single molecule real-time sequencing technology in the de novo assembly of a eukaryotic genome. Sci. Rep. 5:16780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H., Naito K., Takahashi Y., Sato T., Yamamoto T., Muto I., et al. (2016). The Vigna Genome Server, ‘VigGS’: a Genomic Knowledge Base of the Genus Vigna Based on High-Quality, Annotated Genome Sequence of the Azuki Bean, Vigna angularis (Willd.) Ohwi & Ohashi. Plant Cell Physiol. 57:e2. 10.1093/pcp/pcv189 [DOI] [PubMed] [Google Scholar]
- Smit A. F. A., Hubley R. (2008). RepeatModeler Open-1.0. Available online at: http://www.repeatmasker.org [Google Scholar]
- Smit A. F. A., Hubley R., Green P. (2013). RepeatMasker Open-4.0. Available online at: http://www.repeatmasker.org [Google Scholar]
- Spence J., Vercher Y., Gates P., Harris N. (1996). ‘Pod shatter’ in Arabidopsis thaliana. Brassica napus and B. juncea. J. Micros. 181 195–203. 10.1046/j.1365-2818.1996.111391.x [DOI] [Google Scholar]
- Stanke M., Diekhans M., Baetsch R., Haussler D. (2008). Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24 637–664. [DOI] [PubMed] [Google Scholar]
- Suanum W., Somta P., Kongjaimun A., Yimram T., Kaga A., Tomooka N., et al. (2016). Co-localization of QTLs for pod fiber content and pod shattering in F 2 and backcross populations between yardlong bean and wild cowpea. Mol. Breed. 36:80. [Google Scholar]
- Takahashi Y., Kongjaimun A., Muto C., Kobayashi Y., Kumagai M., Sakai H., et al. (2019). Genetic factor for twisting legume pods identified by fine-mapping of shattering-related traits in azuki bean and yard-long bean. bioRxiv [Preprint] 10.1101/774844 [DOI] [Google Scholar]
- Untergasser A., Cutcultache I., Koressaar T., Faircloth B. C., Remm M., Rozen S. G. (2012). Primer3—new capabilities and interfaces. Bioinformatics 40:e115. 10.1093/nar/gks596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse R. M., Seppey M., Simão F. A., Manni M., Ioannidis P., Klioutchnikov G., et al. (2017). BUSCO Applications from Quality Assessments to Gene Prediction and Phylogenomics. Mol. Biol. Evol. 35 543–548. 10.1093/molbev/msx319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Xu Z., Song J., Conner K., Vizcay Barrena G., Wilson Z. A. (2007). Arabidopsis MYB26/MALE STERILE35 regulates secondary thickening in the endothecium and is essential for anther dehiscence. Plant Cell 19 534–548. 10.1105/tpc.106.046391 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material.