ABSTRACT
Lactic acid bacteria (LAB) are present in foods, the environment and the animal gut, although fermented foods (FFs) are recognized as the primary niche of LAB activity. Several LAB strains have been studied for their health-promoting properties and are employed as probiotics. FFs are recognized for their potential beneficial effects, which we review in this article. They are also an important source of LAB, which are ingested daily upon FF consumption. In this review, we describe the diversity of LAB and their occurrence in food as well as the gut microbiome. We discuss the opportunities to study LAB diversity and functional properties by considering the availability of both genomic and metagenomic data in public repositories, as well as the different latest computational tools for data analysis. In addition, we discuss the role of LAB as potential probiotics by reporting the prevalence of key genomic features in public genomes and by surveying the outcomes of LAB use in clinical trials involving human subjects. Finally, we highlight the need for further studies aimed at improving our knowledge of the link between LAB-fermented foods and the human gut from the perspective of health promotion.
Keywords: food microbiome, human microbiome, lactic acid bacteria, probiotics
Lactic acid bacteria are widespread in food and many strains have a well-established role as probiotics; however, their occurrence, genome-wide diversity and role in the human gut and gut health is still understudied.
INTRODUCTION
The lactic acid bacteria (LAB) group is phylogenetically located in the Clostridia branch of Gram-positive bacteria and includes non-sporing cocci, coccobacilli or rods, and aero-tolerant anaerobes, with a molar DNA base composition of less than 50% G + C (Pot et al. 1994). LAB are among the most widely studied microorganisms worldwide. Given the important role that LAB play in different biotechnological processes, it is not surprising that they have received much attention from the scientific community for decades. A search for the term ‘lactic acid bacteria’ in the title, keywords and abstract in the scientific database Scopus (www.scopus.com) (Burnham 2006) returned approximately 32,700 documents at the time of this review (May 2020). In addition, using ‘lactic acid bacteria’ AND ‘food’, ‘lactic acid bacteria’ AND ‘gut’ or ‘lactic acid bacteria’ AND ‘environment’ as search terms, 11,800, 1,500 and 1,700 documents can be retrieved, respectively, which clearly indicates that food is the most widely studied environment in association with LAB.
Although LAB exhibit considerable species and strain diversity and can play a significant role in different ecosystems, food remains their major source and preferred activity niche. This is mainly because the fermentation activity of LAB has been associated with foods and studied in fermented foods (FFs) since early 1900s. LAB activity in FFs can be basically considered a transformation of raw materials to edible food products with different characteristics. Food fermentation is actually an ancient process that was used as a strategy for food preservation, dating back to 10,000 years ago when agriculture and farming were introduced (Cordain et al. 2005). Food fermentation can be aerobic, such as alkaline fungal fermentation, and anaerobic, such as alcoholic and lactic acid fermentation by yeast and LAB, respectively (Nout 2014).
Some LAB strains are also considered potential probiotics, and many are commercialized in probiotic preparations and/or functional foods. In addition, they are also members of the gut microbiome of human and animal hosts, although their origin, role and potential activities are still widely discussed.
In this review, we discuss the occurrence of LAB species in both food and the human gut. Moreover, we assess the availability and information retrievable from available genomic and metagenomic data for LAB from food and humans. Finally, we discuss the effect of LAB on the gut microbiome on the basis of the currently available results from clinical trials and highlight future perspectives for exploiting the currently available genome-wide data that can help bridge the gap between food and the gut microbiome and can improve our understanding of the potential of FFs as vehicles for probiotic LAB.
LAB diffusion and phylogenetic diversity
LAB are widely distributed in nutrient-rich habitats associated with food, plants, soil, animals and human hosts (Duar et al. 2017b; Wels et al. 2019). In recent years, the availability of a very large number of genomes of isolates from different sources has allowed comparative and evolutionary studies. Advancements made over the last few years were reviewed by (Duar et al. 2017b). In this work, the lifestyles of Lactobacillus sensu lato (i.e. including lactobacilli and related pediococci) were deduced by combining phylogenomic data with information about metabolism and data from the literature. The > 200 species (Sun et al. 2015) were first grouped in main clades based on the phylogeny according to Zheng et al. (2015a) and then assigned to three main clusters: free living (i.e. associated with plant material or the environment without relying on a eukaryotic host), host adapted (i.e. specialized for living in association with eukaryotic hosts, with adaptive traits that facilitate persistence), and nomadic (i.e. with a dynamic, generalist lifestyle that involves both environmental and host niches, with no signs of specialization) (Martino et al. 2016). Interestingly, lifestyle allocation overlaps with phylogenetic grouping at both the species and subspecies levels, suggesting the occurrence of adaptive genomic evolution in different niches (Duar et al. 2017). To elaborate, the Lb. brevis, Lb. buchneri, Lb. collinoides, Lb. perolens, Lb. sakei, and Lb. vaccinostercus groups were composed of species rarely found in animals and human hosts and therefore considered free living. Among the groups found to be nomadic, those species that, although not strictly autochthonous, exhibited adaptation to niches associated with humans or animals that could contribute to their persistence are of interest. These species could adapt to the gut and persist for at least a limited duration (Duar et al. 2017b). This is the case for Lb. casei/paracasei (Cai et al. 2007, 2009; Broadbent et al. 2012), Lb. plantarum (Siezen et al. 2010; Martino et al. 2016), and Lb. rhamnosus (Ribbera et al. 2013; Ceapa et al. 2015, 2016). Lb. amylovorus, Lb. iners, Lb. johnsonii, Lb. reuteri, Lb. ruminis, and Lb. salivarius were found to be adapted to vertebrate hosts, although some of them are also relevant in food fermentation (Vogel et al. 1999; Zheng et al. 2015b). The Lb. delbrueckii group comprised two main subclusters, one adapted to insects (e.g. Lb. bombicola, Lb. apis) and the other one to vertebrates (e.g. Lb. johnsonii, Lb. gasseri). Finally, the vertebrate gut was proposed as the real habitat of Lb. helveticus, despite its wide use in cheese production. Notably, host-adapted species or strains may have high ecological fitness in their respective hosts and therefore may be highly competitive when administered as probiotics (Duar et al. 2017a,b). On the other hand, species that did not undergone joint evolution with the host may be more appropriate for stimulating the immune system (Duar et al. 2017b). Notably, the lifestyle of pediococci remains unknown (Duar et al. 2017b), a problem that also exists for other LAB species that diverged from lactobacilli. The genus Streptococcus includes several pathogenic species, but the main food-related species, Streptococcus thermophilus, must have followed a divergent evolutionary path from that of its pathogenic relatives, and its genome has adapted to a well-defined and constant ecological niche, milk (Bolotin et al. 2004). This led to the loss of virulence factors and genes involved in the utilization of different carbohydrates, with the organism adapting to an environment in which the main carbohydrate source is lactose (Bolotin et al. 2004). Within the Lactococcus genus, Lc. lactis is of primary importance in the food industry. Lc. lactis taxonomy is currently based on phenotypic differentiation of two subspecies, lactis and cremoris. While the cremoris phenotype was found exclusively in dairy products and related environments, strains within the lactis subspecies have been isolated from different sources, including plants, vegetables and dairy environments (Wels et al. 2019). However, in Lc. lactis subsp. cremoris, a discrepancy between phenotype and genomic clustering was observed, and studies have shown that some strains with a cremoris genotype show a phenotype more similar to that of the lactis subspecies (Wels et al. 2019).
Other important LAB members are part of the family Leuconostocaceae, that includes heterofermentative microbes belonging to the genera Leuconostoc, Weissella, Oenococcus and Fructobacillus. Oenococcus and Fructobacillus were originally assigned to Leuconostoc genus, but were reclassified later, while Weissella includes several species previously classified as Lactobacillus or Leuconostoc spp. The genus Leuconostoc have been isolated from different environments, including plant material, roots, clinical sources and fermented foods, mainly vegetables and dairy, as well as chilled raw meat, where they may act as spoilage agents (Holland and Liu 2011).
Taxonomic classification has been traditionally based on phenotypic traits and sugar metabolism profiling, and subsequently coupled with 16S rRNA gene sequencing. However, the introduction of novel species, together with the widespread of genomic technologies, highlighted that the current LAB taxonomy should be revised. Indeed, several works based on genomic comparison showed that genetic similarity within Lactobacillus genus is as low as the value usually found for different orders or even classes (Sun et al. 2015; Parks et al. 2018; Salvetti et al. 2018). In addition, members of other genera (e.g. Pediococcus, Leuconostoc, Weissella, Oenococcus) were shown to be intermixed among Lactobacillus species (Sun et al. 2015; Salvetti et al. 2018). Therefore, the Lactobacillus genus was proposed to be separated into 10 to 16 different genera (Pot et al. 2019). More recently, Zheng et al. (2020) showed that Lactobacillaceae and Leuconostocaceae families should be merged and suggested a reclassification of the genera included in these two families. Specifically, the emended Lactobacillus genus should incorporate only those species included in the Lb. delbrueckii group, while they proposed 25 novel genera enclosing other Lactobacillus species.
Although the urgent need for a reclassification was frequently highlighted and endorsed by an expert committee organised by the Lactic Acid Bacteria Industrial Platform (LABIP), a final decision was not taken yet. The renaming might have a strong impact on industry, consumers, regulators, as well as on the scientific and medical communities (Pot et al. 2019).
FERMENTED FOODS, PROBIOTIC LAB AND FUNCTIONAL FOODS
By transforming carbohydrates provided by the raw materials to mainly lactic acid, LAB have contributed to food quality and safety for decades, although this has occurred with highly variable degrees of human awareness. In fact, knowledge of the actual contribution and potential of LAB in food fermentation has evolved over time, from popular but uninformed use of fermentation to well-thought-out selection and application of LAB as starter cultures for the food industry.
FFs can be split into at least two major categories: (i) industrial and (ii) artisanal. In the first case, appropriately selected LAB cultures are employed as starter cultures to assure the technological outcome of the fermentation and in some other cases are used as specialized ‘adjuncts’ that are able to perform specific metabolic activities that support aroma production or texture development or add further value to the food product (Burns et al. 2012). The selected LAB cultures are meant to help achieve high reproducibility, quality and safety in highly controlled fermentation. Conversely, artisanal food fermentation is usually carried out with no starter or with naturally selected cultures. In the absence of starter addition, the LAB of environmental origin available in the raw materials can take guide fermentation and assist in product manufacturing and in obtaining the final FF. Although the composition of natural starter cultures is considerably influenced by the specific product and type of fermentation, these cultures are composed mainly of various species and strains of LAB that are specifically and naturally selected by the manufacturing process and whose composition is heavily influenced by raw materials and technological as well as environmental conditions. The spontaneously selected LAB in natural starter cultures are selected by a series of inoculation and refreshment steps in a traditional back-slopping procedure, where part of the fermented matrix of a previous manufacturing process is used as a natural starter in the fermentation process on the following day. The high diversity of FFs available across the globe is mirrored by the equally high microbial diversity of LAB employed daily in food fermentation.
The wide variety of raw material-microbe combinations results in thousands of different FFs and fermented beverages (Marco et al. 2017). Milk, meat, fish, vegetables, cereals and legumes can be fermented to obtain a variety of end products of high quality. Although the industrial use of selected LAB cultures has improved speed and quality standards, the number of FFs available and their associated microbial diversity has reduced. However, many countries across the world are currently promoting the use of FFs, especially traditional foods, for both their hedonic (Xiang et al. 2019) and health-promoting properties (Chilton, Burton and Reid 2015).
Functional foods deliver additional or enhanced benefits over and above their basic nutritional benefits (Bell et al. 2018). LAB can contribute to rendering a FF functional, via both their presence and specific activities. While transforming raw materials through fermentation, LAB activity can indirectly confer several properties to FFs, making them valuable products for human health. In fact, beyond lactic and other acids, some metabolic activities and products can be developed during fermentation and confer interesting potential health-promoting properties to FFs (Şanlier, Gökcen and Sezgin 2019). Several observational studies have been performed to support this hypothesis and have linked the consumption of FFs (mostly yogurt) with beneficial effects on weight management (Mozaffarian et al. 2011), cardiovascular disease and type 2 diabetes (Chen et al. 2014; Tapsell 2015). Moreover, a link between FF consumption and mood and brain activity is also emerging (Tillisch et al. 2013; Aslam et al. 2018).
Several functional foods are recognized as such because they contain and deliver probiotic microorganisms. Many species and strains of LAB are regarded as probiotics, which are ‘live microorganisms that, when administered in adequate amounts, confer a health benefit on the host’, according to the definition proposed in 2001 by an expert panel working on behalf of the Food and Agriculture Organization of the United Nations and the World Health Organization (FAO/WHO) and subsequently endorsed by the International Scientific Association for Probiotics and Prebiotics (ISAPP) in the consensus statement of 2014 (Hill et al. 2014). The ISAPP confirmed that the term ‘probiotic’ for food and food supplements should be used under certain conditions, including the administration of a minimum of 1 × 109 CFU/day, a full genomic characterization of the probiotic strain and a history of safe use (Hill et al. 2014). Although a limited number of claims of health benefits of LAB have been approved, the probiotics market is thriving and is expected to grow further (Global Market Insight 2018). Probiotic strains are defined and potentially selected based on well-established criteria determined by the FAO and WHO (Araya et al. 2002). Strain identification, safety, stress tolerance and epithelial adherence capabilities are among the principal tests for screening probiotic strains (Pereira et al. 2018).
Owing to their food origin, some LAB species (mostly Lactobacillus and Streptococcus spp.) have a generally recognized as safe (GRAS) status according to the U.S. Food and Drug Administration (FDA) (https://www.accessdata.fda.gov/scripts/fdcc/?set=GRASNotices). In Europe, the concept of Qualified Presumption of Safety (QPS) was developed in 2007 by the EFSA to assist the safety assessment of microorganisms deliberately introduced into the food chain. The main difference between the GRAS and QPS concepts is that the former is generally limited to a specific application of a microorganism, while QPS refers to its generic safety in all possible uses. The QPS status evaluation is based on four points: taxonomy, scientific knowledge, the safety assessment (presence of virulence factors, production of toxins, antimicrobial resistance, reported cases of infection) and the expected end usage. When a species is included in QPS list, all the strains of that species will not need a full safety assessment (Sanders et al. 2010; Bourdichon, Laulund and Tenning 2019). Twenty-four Lactobacillus species, besides Lactococcus lactis, Streptococcus thermophilus and some Leuconostoc and Pediococcus species gained QPS status. Notably, no Weissella and Enterococcus spp. are included in this list (https://www.efsa.europa.eu/en/topics/topic/qualified-presumption-safety-qps). However, few cases of septicaemia induced by lactobacilli are also reported, but this typically occurs only in patients with pre-existing health problems, such as immunocompromised (O'Callaghan and O'Toole 2013).
Among LAB, at least ten species of Lactobacillus and Lactococcus lactis have been shown to exhibit probiotic properties, and their importance as health-promoting bacteria together with novel non-LAB species and strains has been recently reviewed (Douillard and de Vos 2019).
Therefore, an additional benefit of FFs is that they are natural sources of LAB, and as such, they can be regarded as ‘naturally potential’ functional foods. Regardless of the origin of the raw material, be it milk, vegetable or even meat, FFs can contain high loads of live LAB at the end of fermentation and in the final product. This does not apply simply to any FF. In fact, many foods obtained through fermentation do not contain live bacteria because they are inactivated by heat, as in the case of bakery products, or are physically removed, as in the case of alcoholic beverages (Rezac et al. 2018). Nevertheless, fermented milks, cheeses, fermented vegetables, meats, etc., do contain a considerable amount of live bacteria at consumption, which increases the number of microbes in the diet by up to 10 000-fold (Lang, Eisen and Zivkovic 2014). Diets rich in FFs offer remarkable microbial exposure in contrast with highly processed foods provided in societies with a high level of westernization and hygienic practices. Rezac et al. (2018) surveyed the amount of live LAB occurring in a variety of FFs at retail and found loads ranging between 105 and 109 CFU/g or ml, with dairy products containing the highest levels. Such high amounts of live LAB are therefore ingested with FFs and reach the human gastrointestinal tract (GIT). What happens after ingestion depends on the specific genetic and functional traits of the LAB strains and on their ability to resist the stress conditions to which they are exposed. High concentrations of pepsin and low pH (<3) are the principal barriers in the stomach, while bile and pancreatin are the typical adversities encountered in the small intestine. However, if they are able to endure to such stress factors, these bacteria can reach the colon and join the complex environment of the gut microbiome (see below).
Fermented foods as source of microbial metabolites
Overall, the numerous enzymatic activities that can be carried out during food fermentation by LAB can change the biochemical composition of foods, releasing bioactive compounds that can provide health-promoting properties that the same matrix would not display without fermentation (Marco et al. 2017). Indeed, some LAB strains may exert health-promoting activity even if inactivated. The term ‘postbiotic’ was recently coined, indicating microbial metabolites or components of bacterial cell walls released in a matrix from which microbes are removed or inactivated and conferring health benefits when administered in sufficient amounts (Aguilar-Toalá et al. 2018). Such compounds include β-galactosidase for improved lactose digestion; conjugated linoleic acid, bioactive peptides and polyamines; and phenolic compound derivatives for oxidative stress improvement (Marco et al. 2017). LAB can also produce exopolysaccharides (EPSs) with potential cholesterol-lowering, antidiabetic, antioxidant, and immunomodulatory properties (Nampoothiri et al. 2017; Şanlier, Gökcen and Sezgin 2019). Several LAB produce B-group vitamins during fermentation and can effectively increase vitamin levels (LeBlanc et al. 2011). For example, Lb. casei KNE-1 was shown to synthetize thiamine (B1) and riboflavin (B2) in fermented milk drinks, while some strains of S. thermophilus, Lb. delbrueckii and Lb. amylovorus can be used to produce yogurts or fermented milks that are naturally rich in folate (B9), which is particularly important during pregnancy (Linares et al. 2017). In addition, some Lc. lactis strains can produce menaquinone (K2) in cheese and kefir (Walther et al. 2013). Other strains can produce neuroactive molecules, among which gamma-aminobutyric acid (GABA) is the most well studied. GABA acts as a neurotransmitter in mammals and performs additional functions, such as lowering blood pressure, relaxing muscles, and reducing psychological stress (Pessione and Cirrincione 2016; Şanlier, Gökcen and Sezgin 2019). The ability to produce GABA is recognized in several bacteria of gut origin (Wall et al. 2014), but fermented products rich in GABA have also been developed using specific strains of Lb. casei, Lb. plantarum, S. thermophilus, Lb. brevis and Lc. lactis as starters in fermented dairy products, legumes, cereals, and chocolate (Pessione and Cirrincione 2016; Linares et al. 2017).
Moreover, LAB can release biologically active peptides via proteolysis (Linares et al. 2017). The most well-studied peptides are antihypertensive peptides that can regulate blood pressure through inhibition of angiotensin-I-converting enzyme (ACE) and have been proposed as natural alternatives to antihypertensive drugs. ACE-inhibiting peptides are found mainly in fermented dairy products and fermented vegetables or legumes and are produced by several LAB used as starter cultures in FFs, including strains of Lb. helveticus, Lb. casei, Lb. delbrueckii, Lb. plantarum, Lc. lactis, and S. thermophilus (Shakerian et al. 2015; Li et al. 2017). In addition, peptides with different activities, such as anti-inflammatory, antioxidant, immunomodulatory, and antimicrobial activities, have also been identified in FFs (Pessione and Cirrincione 2016).
Another class of health-promoting molecules produced in FFs is conjugated fatty acids derived from bioconversion of linoleic acid (conjugated linoleic acid, CLA). CLA is naturally present in ruminant milk due to the activity of rumen bacteria, but the amount is by far sufficient to show some effects (Linares et al. 2017). Indeed, several LAB are known to produce CLA in milk products (Lc. lactis, Lb. acidophilus, Lb. casei, Lb. plantarum, Lb. rhamnosus, Lb. delbrueckii), and their use as starter or adjunct cultures may be a promising strategy for the production of enriched biofunctional foods (Linares et al. 2017).
Finally, LAB may reduce the presence of anti-nutritional compounds in FFs. An example is the phytase activity of some LAB. Phytic acid is present in several foods of vegetable origin, including cereals and legumes, and is considered an anti-nutrient substance since it can form complexes that chelate various minerals, thus reducing their bioavailability (Sharma et al. 2020). Phytase-producing LAB are able to hydrolyse phytates and release minerals. Different strains of Lb. plantarum, Lb. amylovorus, and Lb. acidophilus have been used for fermentation of sourdough from wheat, rye and oat; soy-based products; and beer and were able to reduce phytate concentrations in the fermented matrix (Sharma et al. 2020).
Besides exerting health-promoting activities and producing beneficial metabolites, some LAB strains are recognized as the main producers of biogenic amines (BA) in fermented foods from amino acids decarboxylation (Barbieri et al. 2019). The consumption of products containing high levels of BAs, depending on individual sensitivity or the concomitant assumption of specific drugs or ethanol, can cause headache, heart palpitations, vomiting, diarrhea and hypertensive crises (Barbieri et al. 2019). Moreover, several LAB strains have been shown to carry out genes responsible for antibiotic or antimicrobial resistance, that might be transferred to pathogens or GIT microbes (Campedelli et al. 2019). Therefore, in-depth and rigorous genomic characterization of food-related LAB strains is desirable to identify the presence of potentially dangerous activities.
LAB prevalence and diversity in fermented foods
Fermentation has been traditionally used as an empirical method to improve food stability, and in recent years, it has been used to enhance the flavour, texture, and functional properties of food (Dimidi et al. 2019). LAB from several genera are commonly predominant in FFs, but other bacteria (e.g. propionibacteria and acetic acid bacteria), as well as fungi, also contribute to specific food fermentation processes.
S. thermophilus, Lc. lactis, Leuconostoc species, and several Lactobacillus species are the LAB most commonly found in FFs, either as naturally occurring bacteria or deliberately added as starter cultures. These species are among the most common commercially used bacteria, contributing to the production of yogurt, kefir, cheese and other dairy products; sauerkraut, kimchi and pickles; cured meat and fish; sourdough-based baked products; and many other traditional fermented foodstuffs around the world (Tamang et al. 2020). The main metabolic activity of interest for food production is the ability of these bacteria to carry out lactic acid fermentation, an anaerobic process that converts pyruvate molecules from glycolysis to lactic acid (homolactic fermentation) or lactic acid and other compounds, such as acetic acid, ethanol, and CO2 (heterolactic fermentation). These species can also activate several secondary metabolic processes that lead to the production of flavour compounds or typical textures. Combination of these metabolic processes leads to hundreds of different products, some of which are globally widespread, while many others are locally produced, often according to a traditional manufacturing practice (Chilton, Burton and Reid 2015; Tamang et al. 2020). Different food matrices can be considered specific ecological niches in which well-adapted LAB species finalize the fermentation process. In recent years, hundreds of studies have described microbial dynamics during the fermentation of different foodstuffs by high-throughput sequencing (HTS), extensively reviewed elsewhere (Ercolini 2013; De Filippis, Parente and Ercolini 2017, 2018b). Most of these studies are based on amplicon sequencing of taxonomically relevant genes and merely provide a survey of the microbial diversity occurring during food fermentation. Most of these studies have been collected in the FoodMicrobionet repository (http://www.foodmicrobionet.org; Parente et al. 2016; Parente et al. 2019). FoodMicrobionet contains data on microbial taxonomic composition from 44 HTS studies on food microbial ecology, including 29 datasets on food fermentation. To date, this repository includes a total of 2234 samples from food or food environments covering dairy, meat, fruits, vegetables, cereal-based foods, and ready-to-eat foods, with 806 samples of FF products. The samples are labelled according to the FoodEx classification (http://www.efsa.europa.eu/en/data/data-standardisation). Due to the availability of an app built with the Shiny R package, even inexperienced users can easily explore data, access external resources, filter samples based on multiple predefined criteria, aggregate samples and bacterial taxa, extract the taxonomic composition of specific groups of samples, and use them in comparative studies. We considered 806 samples spanning multiple FF matrices, extracted the prevalence of different LAB genera and species (collated in taxonomic groups, as defined by Salvetti et al. 2018), and grouped them according to the type of product or ripening time (Fig. 1). The niche specificity of Lactobacillus species is highlighted: Lb. delbrueckii group is prevalent mainly in dairy products, while the Lb. plantarum and Lb. sakei groups showed 100% prevalence in fermented vegetables and meat samples, respectively. Lb. buchneri group (including Lb. buchneri, Lb. sanfranciscensis, Lb. brevis) prevailed in sourdough, where Lb. sanfranciscensis is a well-known member of the microbial community (Ripari, Gänzle and Berardi 2016). Among other LAB genera, Weissella is found exclusively in naturally leavened sourdough, while Streptococcus and Lactococcus are found in cheeses and kefir. In addition, while most fresh and short-ripened cheeses contain thermophilic LAB, such as Streptococcus, high variability in LAB composition was found in ripened cheeses, in which mesophilic lactobacilli and Lactococcus are also present (Fig. 1). Some commonly consumed dairy products with a simple and defined microbiota structure (i.e. yogurt) are obviously not considered in Fig. 1 as they have not been studied by HTS approaches. LAB are often deliberately used for inoculation to start fermentation, as either selected commercial cultures or natural starters obtained according to a back-slopping procedure. Nevertheless, several artisanal products are fermented without the addition of starter microbes, but they arise from raw materials or from the facility environment, equipment and tool surfaces. Indeed, the food processing environment harbours a resident and complex microbiota that can be transferred to the product and represent a primary source of beneficial LAB (Montel et al. 2014; Stellato et al. 2015; Bokulich et al. 2016). Unfortunately, taxonomic identification at the species level is often not achievable with common amplicon-based HTS technologies, and many studies have reported genus-level identification (Fig. 1). This is a substantial limitation considering the wide species and subspecies diversity existing within LAB and the specific roles that these microbes can play during food production. This limitation may be overcome using a complex shotgun HTS approach, which remains underexploited in food-related microbiome studies. The use of metagenomics can be of invaluable importance for the identification of microbial genes and pathways leading to the production of metabolites associated with the typical sensorial profile of specific FFs, as well as for detecting potential health-related activities (De Filippis, Parente and Ercolini 2018b). In fact, in addition to producing lactic acid during fermentation, LAB confer important desirable properties to FFs. By degradation of carbohydrates, proteins and lipids, LAB can synthesize molecules positively associated with the flavours of FFs or modify the texture of some products by proteolysis, lipolysis or EPS production (Galle and Arendt 2014; Di Monaco et al. 2015; De Filippis et al. 2016; Gänzle and Ripari 2016; McAuliffe, Kilcawley and Stefanovic 2019). Nevertheless, it should be pointed out that identification by HTS does not imply that the microbes are alive at the moment of consumption, since these studies are usually based on DNA, which may be derived from dead or inactive cells. Viable counts of LAB in several FFs have also been reported (Rezac et al. 2018). It is estimated that between 108 and 1012 CFU of bacteria may be ingested daily with the consumption of FFs (Derrien and van Hylckama Vlieg 2015). The quantities of S. thermophilus and Lb. delbrueckii in commercial yogurts and fermented milk vary from 104 to 109 CFU/ml, while the abundance of lactobacilli in cheeses ranges from 109 to 103 CFU/g, decreasing during ripening (Rezac et al. 2018). The levels of LAB in fermented sausages were reported to vary according to the origin, with fermented sausages from Europe showing higher counts than those from the US (<106 vs 108 CFU/g), which is probably associated with the more artisanal manufacturing process used for European products (Rezac et al. 2018). Therefore, the real amount of LAB ingested through a specific FF may also be extremely variable according to the geographical origin, manufacturing process (e.g. artisanal vs industrial; presence, length, and conditions of ripening; etc.), time and type of storage, and use of inactivation steps before consumption. For example, bread and other baked goods are usually cooked, while several fermented vegetables are pasteurized before commercialization to improve stability. However, the low levels of live microbes in the final product do not preclude a positive functional role. Indeed, several LAB may produce vitamins or other bioactive molecules in situ or inactivate anti-nutritional factors, thus exerting a positive health effect even if not alive at the time of consumption (Linares et al. 2017; see above).
Figure 1.
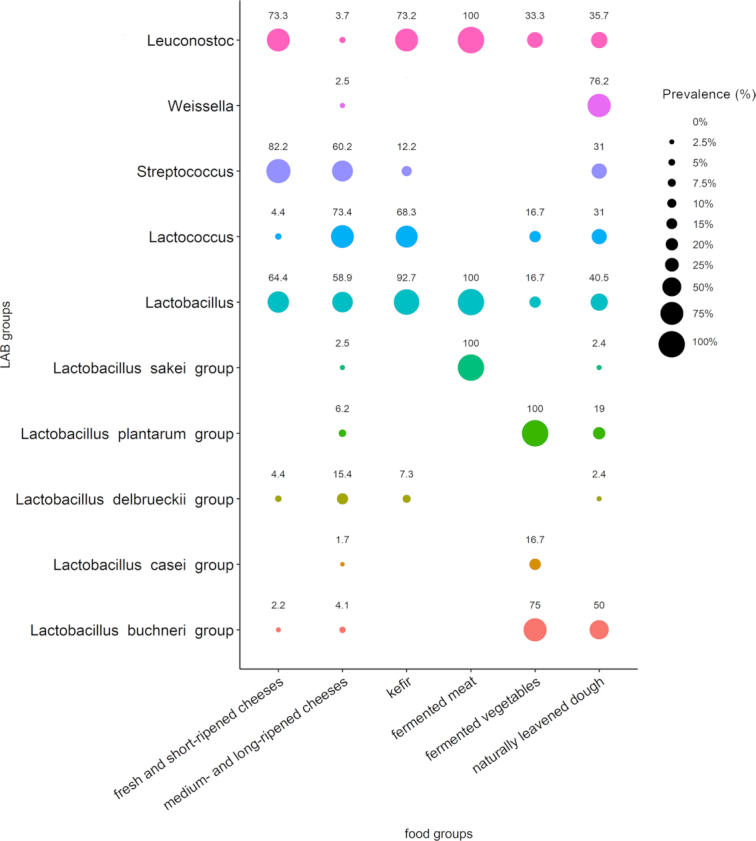
Bubble plot showing prevalence (% of samples) of LAB genera and species in different fermented foods, as obtained from 16S rRNA gene sequencing studies reported in FoodMicrobionet (Parente et al. 2019). A taxon was considered present if its relative abundance was > 0.5%. For lactobacilli, species were grouped into taxonomic groups, as reported by Salvetti et al. (2018).
THE GUT MICROBIOME
The gut microbiome is among the most complex known microbial communities and among the most well studied. It comprises a very large variety of microbial strains belonging to species of bacteria, archaea, fungi and viruses that live in close relationship with the human host and whose combined genome harbours at least 100 times as many genes as the human genome (Bäckhed et al. 2005; Belkaid and Hand 2014). Members of the gut microbiome can influence host health through the production of a wide variety of beneficial or detrimental metabolites, and such molecules can be derived from both metabolic intermediates of the host and dietary precursors (Holmes et al. 2012; De Filippis et al. 2018a; Roager and Dragsted 2019). The most recent research advances have shown a high potential impact of the gut microbiome on the regulation of the equilibrium between health and disease, which is due to both the composition and functions of the microbiome. Complexity in composition is one of the main features of the gut microbiome, and microbial richness in terms of both species and genes has been linked to health (Cotillard et al. 2013; Le Chatelier et al. 2013; Vangay et al. 2018). In addition to differences in microbial composition based on health, lifestyle and geography (Almeida et al. 2019; Nayfach et al. 2019; Pasolli et al. 2019), the gut microbiome is characterized by high inter-individual variability (Truong et al. 2017). However, a large proportion of bacterial species are present in each individual and likely constitute a resilient microbial community (Aguirre de Cárcer 2018). In light of these considerations, the possible role of probiotics appears even more challenging, as after overcoming the barriers presented by the stomach and small intestine, probiotics encounter an army of hundreds of different species and strains to compete with, which affects their chances of exerting their beneficial effects. While the host-specific microbial community can be considered a resident microbiome, the microbes that we ingest and that reach the colon can be regarded as a transient microbiome, the composition of which depends on the type of exposure and on the type of food in the case of food-borne microorganisms (Derrien and van Hylckama Vlieg 2015). Indeed, the gut microbiome is exposed daily to microbes from the external environment, which are mainly of food origin. Probiotic LAB strains can be part of such a transient community and are supposed to perform their activity during passage through the gut and in the presence of the other members of the gut community.
Currently, it remains unknown which fraction of the food microbiome is actively transferred to the intestine and what type of activity those strains can exert in such a complex ecosystem. In fact, there is little literature on the prevalence of LAB in FF consumers and non-consumers. In addition, the possibility that non-probiotic food-borne LAB can also be transferred to the gut to a certain extent is currently underexplored, as are the actual activities that LAB can carry out in the gut as a complementary transient microbial community.
LAB in the human gut
LAB are also widespread in other nutrient-rich environments, among which the human host is of great interest due to the potential functional properties of these bacteria (Soomro, Masud and Anwaar 2002; Duar et al. 2017b). The human microbiome interacts continuously with microbes originating from external environments, including food-origin sources. Probiotic LAB species and strains can constitute a portion of this transient microbiome and perform their activities during their transition in the gut, in addition to non-probiotic LAB that can potentially be transferred into the gut to a certain extent. Despite the availability of abundant nutrients, LAB present in the gut have to deal with a challenging scenario that involves hundreds of different bacterial and non-bacterial species sharing the same habitat (Pessione 2012).
Despite the long-term efforts in characterizing LAB, characterization of the effective contribution of these bacteria to the human microbiome remains a major challenge, and contradictory results have been reported in the literature (Walter 2008; Pessione 2012; George et al. 2018). Going back a hundred years, seminal studies identified lactobacilli, which we focus on here due to their prevalence in the LAB literature, among the most prevalent and abundant microorganisms in the human gut (Tannock 1999). At that time, techniques to cultivate anaerobic organisms were not yet available, which likely led to overestimation of the more easily cultivable microbes such as lactobacilli, while most of the gut (anaerobic) microbes remained undetected, a problem that despite continuous technological advancements has not yet been fully resolved (Almeida et al. 2019; Nayfach et al. 2019; Pasolli et al. 2019). For a long time, lactobacilli were considered to be numerically relevant members of the microbiome (Walter 2008), but most of the research conducted in the last few decades found that these bacteria are subdominant and therefore represent instead a small fraction of the overall microbiome composition. When using total anaerobic culturing techniques, the amount of lactobacilli rarely exceeded 108 CFU/g and accounted for an average of 106 CFU/g of intestinal content (Mitsuoka 1992; Walter et al. 2001; Dal Bello et al. 2003; Walter 2008), which represented a small fraction (< 0.01%) of the total count assuming that the intestinal content can reach up to 1012 CFU/g (O'Hara and Shanahan 2006). However, cultivation-based approaches may also be affected by biases because while lactobacilli can be easily isolated from food, isolation from human stool samples is more difficult since bifidobacteria are much more abundant and share similar nutritional requirements (Quartieri et al. 2016). On the one hand, findings obtained from culture-based approaches were confirmed in multiple studies by culture-independent methods (Walter 2008), which included, for example, fluorescent in situ hybridization (FISH) in combination with fluorescence microscopy (Harmsen et al. 2002), quantitative real-time PCR (Rinttilä et al. 2004), and high-throughput analysis of 16S rRNA gene amplicon sequencing data (Suau et al. 1999; Hayashi, Sakamoto and Benno 2002; Hold et al. 2002; Eckburg et al. 2005). The FISH data showed a relative abundance for the Lactobacillus/Enterococcus group ranging from 0.01–1.8% (Flint 2006; Louis et al. 2007). However, the results of other studies differed from these ones, instead finding that LAB occurrence may not be negligible in the human gut. This was mainly driven by advancements in 16S rRNA and metagenomics methodologies (Quince et al. 2017b), which made it possible to obtain largely unbiased perspectives on the relative importance of LAB in the context of the other members of the gut microbiota (Heeney, Gareau and Marco 2018). Approximately 5% and 13% of the sequences from 16S rRNA libraries were attributed to lactobacilli by (Frank et al. 2007) and (Hayashi et al. 2005), respectively. Using 16S rRNA data, Lactobacillus were estimated to constitute on average 6% of the bacterial cells in the duodenum (Nistal et al. 2016) and 0.3% in the colon (Almonacid et al. 2017). A longitudinal metagenomic study surveyed lactobacilli in a single person at three timepoints and found 52 subdominant species, 80% of which were detected in a two-year timeframe (Rossi et al. 2016). These results suggested that a relevant LAB population may be harboured in the human gut, with consistent inter-individual variations that may be driven by multiple factors, with the most likely one represented by diet (David et al. 2014) and the ingestion of LAB-enriched foods. Notably, most of this literature was derived from studies on faecal material, while very little is known about the small intestine microbiome due to it being accessible via only invasive procedures (Derrien and van Hylckama Vlieg 2015; El Aidy, van den Bogert and Kleerebezem 2015; Stolaki et al. 2019). This may represent an overlooked scenario because consumption of a dose of 1010 bacterial cells may have a strong influence, at least temporarily, on the microbial composition of the small intestine, since the microbial density in this organ, ranging from 104 to 108 bacteria/ml, is much lower than that in the colon (Derrien and van Hylckama Vlieg 2015).
Along with quantification of the LAB community in the gut, of comparable or even greater importance is the discrimination between resident (defined as autochthonous) and transient (allochthonous) components. This task is not trivial since LAB are continuously administered in the human ecosystem through ingested food and therefore represent a rather peculiar microbial group (Walter 2008; Rossi et al. 2016). Notably, populations of allochthonous species may appear stable if introduced regularly into the habitat (Duar et al. 2017b). Seminal studies on this topic were well summarized by (Walter 2008). Pioneering studies (Lerche 1961; Reuter 1965; Mitsuoka 1969) found transient and resident lactobacilli strains in stool samples, with the latter ones identified as Lb. crispatus, Lb. gasseri, Lb. reuteri, Lb. ruminis, and Lb. salivarius (Mitsuoka 1992; Reuter 2001). Further studies showed that a large fraction of the LAB species found in the gut are probably allochthonous and do not form stable populations, along with other species that can be considered autochthonous members of the microbiome (Tannock, Munro and Harmsen 2000; Walter et al. 2001; Walter 2008). For example, Lb. ruminis and Lb. salivarius were found to be persistent in multiple subjects for more than 18 months (Tannock, Munro and Harmsen 2000). A list of 17 lactobacillus species typically found in the gut was reported by (Walter 2008), comprising Lb. acidophilus, Lb. brevis, Lb. casei, Lb. crispatus, Lb. curvatus, Lb. delbrueckii, Lb. fermentum, Lb. gasseri, Lb. johnsonii, Lb. paracasei, Lb. plantarum, Lb. reuteri, Lb. rhamnosus, Lb. ruminis, Lb. sakei, Lb. salivarius, and Lb. vaginalis, most of which were identified as allochthonous members. This list was similarly reported by (Vaughan et al. 2002 and O'Callaghan and O'Toole 2013) and integrated with other LAB species. Some studies have also verified the colonization abilities of specific LAB strains. As an example, two strains of Lb. mucosae and Lb. reuteri reached higher population levels and were recovered more frequently from faecal samples than a strain of Lb. acidophilus (Frese, Hutkins and Walter 2012). Other studies determined the extent to which a host's persistent gut microbiota influences niche permissivity to transient LAB (Zhang et al. 2016) and how invasion by transient LAB can perturb the stability of microbial ecosystems (Amor, Ratzke and Gore 2019).
The quantities of LAB species that are persistent in the gut may be larger than currently documented. In the aforementioned work (Rossi et al. 2016), more than 40 species were detected in a single person in a two-year timeframe, indicating the need to conduct more and much larger analyses in similar settings.
Some untargeted studies have shown variations in the proportions of LAB in the gut and found positive or negative correlations with disease or chronic conditions. Depletion of intestinal lactobacilli was frequently associated with disease. As summarized in (Heeney, Gareau and Marco 2018), Lactobacillus was depleted under conditions of type 1 diabetes (de Goffau et al. 2014; Alkanani et al. 2015), irritable bowel syndrome (Liu et al. 2017; Zhuang et al. 2017), multiple sclerosis (Chen et al. 2016), human immunodeficiency virus infection (Yang et al. 2016), and prenatal stress (Zijlmans et al. 2015). On the other hand, enrichment of lactobacilli was verified in conditions of Crohn's disease (Wang et al. 2014; Lewis et al. 2017) and rheumatoid arthritis (Zhang et al. 2015). Contradictory findings were reported for type 2 diabetes (Karlsson et al. 2013; Forslund et al. 2015) and obesity (F. S. Teixeira et al. 2013; Ignacio et al. 2016).
Notably, most of the previous research has been devoted to the characterization of lactobacilli, while less attention has been given to other LAB members (Van den Bogert et al. 2013; Mignolet et al. 2016). Additionally, there is a lack of research aimed at assessing the distribution of LAB in the global population. This gap may be bridged by taking advantage of the growing availability of HTS data, as we will show below.
DATABASES AND COMPUTATIONAL TOOLS TO RETRIEVE GENOME-WIDE INFORMATION ON THE PREVALENCE AND FUNCTIONAL DIVERSITY OF LAB FROM DIFFERENT SOURCES
Food and microbiome research can take advantage of the continuous improvement in HTS technology, which has revolutionized the microbial ecology field in the last two decades (Goodwin, McPherson and McCombie 2016). The continuous decrease in sequencing cost has been associated with exponential growth in terms of the number, diversity, and complexity of the sequenced data. Large international consortia have been established to mainly characterize the human microbiome (Human Microbiome Project (Human Microbiome Project Consortium 2012) and MetaHIT (Qin et al. 2010)), along with other initiatives such as the Tara Oceans Program (Sunagawa et al. 2015), the MetaSUB Consortium (MetaSUB International Consortium 2016), and the Earth Microbiome Project (Thompson et al. 2017), while such large efforts are still lacking in the food microbiome field.
Currently, two main approaches can be adopted in the microbiome field. The 16S rRNA gene sequencing method profiles selected organisms or single marker genes (Hamady and Knight 2009). It is the most cost-effective method, and the main output is limited to the generation of taxonomic profiles, typically at the genus level. Complete pipelines have been developed and widely used (Schloss et al. 2009; Bolyen et al. 2019), in addition to additional newly proposed methods (Callahan et al. 2016). More advanced analyses include oligotyping to obtain species- or even strain-level resolution (Eren et al. 2013) and (rough) estimation of functional potentials (Langille et al. 2013). Different repositories with annotated reference sequences have been made available and continuously updated (Pruesse et al. 2007; McDonald et al. 2012; Cole et al. 2014; Yoon et al. 2017). In addition, curated databases dedicated to specific environments of interest have been developed, for both 16S rRNA gene sequences and whole microbial genomes. DAIRYdb (Meola et al. 2019) provides a manually curated repository of 10,290 full-length 16S rRNA gene sequences from prokaryotes tailored for dairy product analyses. In addition, Almeida et al. (2014) developed a curated genome catalogue of 137 microbial species isolated from dairy products. Higher resolution can be obtained by acquiring the entire genomic content of a sample through (shotgun) metagenomic sequencing (Quince et al. 2017b). The large compendium of tools developed for this technique can be grouped into two main approaches, i.e. mapping-based profiling and de novo assembly. Species-level taxonomic profiles can be generated by adopting different mapping-based methods (Sunagawa et al. 2013; Wood and Salzberg 2014; Truong et al. 2015). Additional tools have also been developed to reduce errors by taking advantage of environmental and domain-specific information, which is however a quite overlooked research topic. This is the case of the methodology developed in (Seol et al. 2019) and aimed at reducing errors in terms of false positive rate for the specific identification of LAB and probiotic species.
Recently, attention has also been given to methodologies for metagenomic analysis with strain-level resolution. Different techniques have been proposed and are mainly based on the detection of single-nucleotide variants (SNVs) in the core genes (Costea et al. 2017; Truong et al. 2017), the identification of unique combinations of genes in the pangenome of a species (Scholz et al. 2016), or the use of a combination of these methods (Nayfach et al. 2016). Attempts have also been devoted to resolving multiple strains of the same species in a single sample (Quince et al. 2017a), although this remains an unresolved challenge along with the profiling of low-abundance non-dominant strains (Segata 2018). In addition to providing information on taxonomic composition, metagenomics can also be used for functional profiling (Franzosa et al. 2018). Complementary to mapping-based approaches is de novo metagenomic assembly. This method aims to provide (draft) genomes (defined as metagenome-assembled genomes, MAGs) of the microbial members present in samples. It can be used to expand the set of genomes of known and already studied species, but at the same time, due to its reference-free nature, provides the possibility to identify and characterize unknown members of the microbiome. Notably, MAGs can be integrated with genomes reconstructed from isolates and post-processed with a myriad of procedures based on comparative genomics that are quite standard for genomes from isolates. The idea of obtaining genomes directly from metagenomes is not new (Allen and Banfield 2005); however, this method was rarely applied until a few years ago due to computational challenges that have been addressed only recently. First, raw reads are assembled into contigs, with metaSPAdes (Nurk et al. 2017) and MEGAHIT (Li et al. 2015) representing the two most widely used tools. Then, the contigs are grouped into (draft) genomes through binning, with the popular tools represented by CONCOCT (Alneberg et al. 2014), MetaBAT2 (Kang et al. 2019), and DAS Tool (Sieber et al. 2018). Finally, only genomes of sufficient quality (usually evaluated in terms of completeness and contamination through tools such as CheckM (Parks et al. 2015) and BUSCO (Simão et al. 2015)) are retained and constitute the final set of genomes. Different papers devoted to the reconstruction and characterization of MAGs from large-scale scenarios have been recently published. Of great relevance is the characterization of the human microbiome (Almeida et al. 2019; Nayfach et al. 2019; Pasolli et al. 2019) along with the microbiomes, for examples, from the rumen (Stewart et al. 2018, 2019), non-human primates (Manara et al. 2019), and multiple other environments (Parks et al. 2017). However, similar efforts in the food microbiome field are still lacking.
The growing number of publicly available microbiome datasets enables hypothesis testing for environmental niches as well as meta-analyses across multiple studies. However, different factors prevent the research community from taking full advantage of these resources. These barriers include the need for substantial investment of time, computational resources and specialized bioinformatic expertise as well as inconsistencies in annotation and formatting between individual studies. To overcome these issues, in the last few years, several efforts have been devoted to the creation of resources and databases for the release of different types of microbiome data, which represent invaluable resources that allow the community to integrate newly acquired data with existing data. Comprehensive resources for both 16S rRNA and metagenomic data are represented by MGnify (Mitchell et al. 2020) and QIITA (Gonzalez et al. 2018). These resources integrate both the deposition of sequence data and distribution of products derived from multiple post-processing pipelines. Currently, MGnify (formerly EBI Metagenomics) integrates 214,977 samples spanning 3685 studies and is associated with six main biomes, i.e. the aquatic, food production, human, plant, soil, and wastewater biomes. While more than 40% of the samples are associated with the human microbiome, only a tiny fraction (< 1%) is related to food production systems. QIITA includes an even larger number of samples (i.e. 232,651 public and 137,644 private samples), although this resource is much more focused on 16S rRNA data. Additionally, in this case, very few public studies are associated with food.
Other resources of smaller size have also recently been made available and are focused on the collection, curation, and processing of samples derived from specific biomes and data types. A representative example of a resource focused on 16S rRNA data is FoodMicrobionet (Parente et al. 2019), already introduced above, which aims to retrieve and combine information specifically from food bacterial communities. Another interesting platform, although not specifically food focused, is Integrated Microbial Next Generation Sequencing (IMNGS (Lagkouvardos et al. 2016)). All prokaryotic 16S rRNA datasets available in Sequence Read Archive (SRA), which is the major database with permanent storage and public access to DNA sequencing data (Kodama et al. 2012), are systematically and uniformly screened and processed to build sample-specific sequence databases and OTU-based profiles. This integrative sequence resource can be queried by users through a web interface. It also offers a complete workflow for analysis of the user's own datasets for the sake of comparison with existing data. Other databases specifically focused on the human microbiome are represented by MicrobiomeHD (Duvallet et al. 2017) and HMP16SData (Schiffer et al. 2019). MicrobiomeHD includes 28 datasets from previously published case-control studies on the gut microbiome. OTU tables with associated taxonomic information and metadata for each sample can be easily downloaded. HMP16SData is a Bioconductor (Huber et al. 2015) package that provides count data for both 16S rRNA variable regions, integrated with phylogeny, taxonomy, and public participant data of the Human Microbiome Project (HMP). This is a good example in which, by removing the hurdles of data access and management, researchers with only basic R skills can analyse HMP data in a quick and simple way (Human Microbiome Project Consortium 2012).
Similar efforts to build databases for the human microbiome have also been conducted for shotgun metagenomics data. One example is the curatedMetagenomicData package (Pasolli et al. 2017), which currently includes more than 10,000 metagenomes from approximately 50 studies. This tool provides uniformly processed microbiome data, including bacterial, fungal, archaeal, and viral taxonomic abundances, in addition to quantitative metabolic functional profiles and standardized per-participant metadata. As in the case for HMP16SData, the data resources are accessible to users with minimal bioinformatic knowledge, and integration with the R/Bioconductor environment allows flexibility for researchers to perform novel analyses and methodological development and for integration of resources. Other similar resources that have been developed subsequently are Microbiome Learning Repo (ML Repo (Vangay, Hillmann and Knights 2019)) and Data Repository for Gut Microbiota (GMRepo (Wu et al. 2020)). ML Repo is a public, web-based repository of 33 curated classification and regression tasks from 15 already published datasets. GMRepo contains 58,903 human gut samples (17,618 from metagenomics and 41,285 from 16S rRNA data) spanning 253 datasets associated with 92 main phenotypes. In this case, the collected samples are organized according to their associated phenotypes. This tool is equipped with a graphical query builder, enabling users to make customized, complex and biologically relevant queries to obtain relevant information that is easy to access. Although such resources are related to the human microbiome in wide terms, at the same time they can be of interest for researchers interested in characterizing LAB in the human gut. Example of database developed for different biomes is the TerrestrialMetagenomeDB for terrestrial metagenomes (Corrêa et al. 2020), while similar products have not yet been developed for metagenomic studies from food microbiomes.
AVAILABILITY OF LAB GENOMES IN FOOD AND THE GUT FOR COMPARATIVE STUDIES
Along with the availability of databases to improve the accessibility to raw sequences and their integration with metadata information, access to genome assemblies is also of great relevance. The benchmark in this context is the NCBI Assembly database (Kitts et al. 2016), which provides stable accessioning and data tracking for genome assembly data. Data can be found for different structures, such as sets of unordered contigs or scaffold sequences, bacterial genomes, or more complex structures, such as human genomes. A particular version of an assembly is identified unambiguously, and track changes are kept to identify genome updates. Along with the nucleotide sequences, this resource provides metadata such as assembly names, statistical reports of the assembly, and assembly update history. Users can easily download sequences and annotations through the NCBI Genomes FTP site.
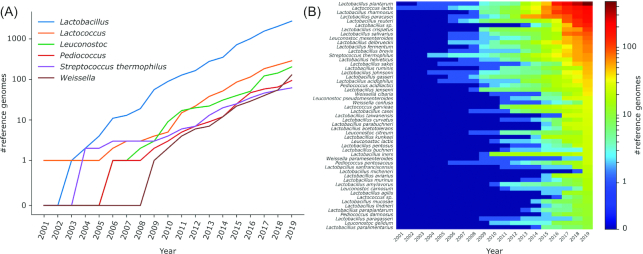
By searching the NCBI Assembly database with the keyword ‘prokaryotes’, we found 223,803 genomes. Filtering by taxonomy, we identified 3525 (1.6%) genomes associated with LAB species, namely, Lactobacillus (N = 2748), Lactococcus (N = 288), Leuconostoc (N = 204), Pediococcus (N = 96), S. thermophilus (N = 62), and Weissella (N = 129) (Fig. 2A and Supplementary Table S1). These genomes were associated with 257 taxa (Fig. 2B and Supplementary Table S2), with Lb. plantarum (N = 473), Lc. lactis (N = 223), Lb. rhamnosus (N = 191), Lb. paracasei (N = 183), and Lb. reuteri (N = 178), representing the most frequently occurring species. The first deposited strain was Lc. lactis subsp. lactis IL1403 in 2001 (Bolotin et al. 2001). The exponential growth of the number of available genomes is represented by the 80% increase in the number of deposited LAB genomes in the last 5 years (Fig. 2). This large number of public genomes represents a fundamental resource for mapping-based computational tools and for comparative genomics. For example, these genomes were used to propose a genome-based reclassification of the genus Lactobacillus (Wittouck, Wuyts and Lebeer 2019; Zheng et al. 2020) due to inconsistencies in the current taxonomy (Wuyts et al. 2017). The same approach was also recently applied to all publicly available bacterial and archaeal genomes (Parks et al. 2019).
Figure 2.
Number of LAB reference genomes in NCBI grouped at (A) genera and (B) species level. In (B), only species with at least 10 genomes deposited in NCBI on December 2019 are shown.
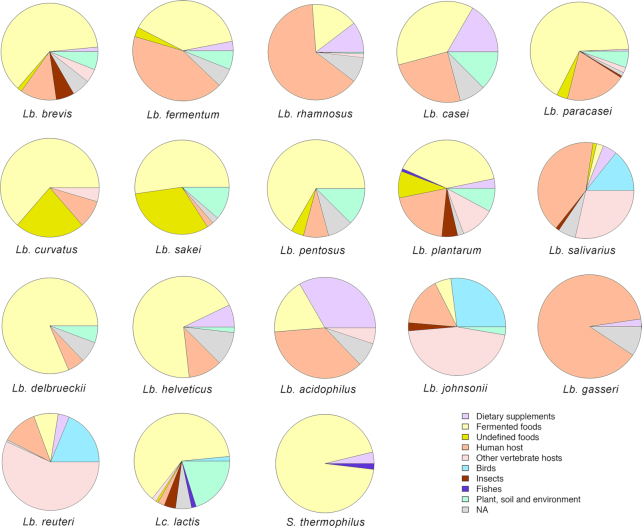
As reported above, LAB are widespread in natural environments. Considering the genomes from NCBI for 18 main LAB species frequently found in FFs or probiotic supplements, we summarized their source of isolation in Fig. 3 (as reported in NCBI or in the linked publications). FFs are the primary source of isolation for several LAB strains. In addition, host-adapted species can be identified. Lb. gasseri, a well-known probiotic species, was mainly isolated from the human infant gut, while Lb. johnsonii, Lb. reuteri, and Lb. salivarius were also retrieved from other animal hosts, both mammals and birds (Fig. 3). Nomadic lactobacilli (i.e. Lb. casei, Lb. paracasei, Lb. plantarum, and Lb. rhamnosus) were isolated from a variety of sources, including human, animal, and insect hosts, as well as soil and plant material (Fig. 3). Indeed, genomic comparisons highlighted that these species usually have a large genome size and a large number of coding sequences, allowing them to adapt and survive in a wide range of environments (Duar et al. 2017).
Figure 3.
Pie charts showing isolation source for public genomes (available on NCBI in December 2019) of 18 selected LAB species, chosen for their importance in fermented foods and/or as probiotic. NA, Not Available. According to the taxonomy update proposed by Zheng et al. (2020), the names of the genera reported would change as follows: Lb. brevis = Levilactobacillus brevis; Lb casei, Lb. paracasei and Lb. rhamnosus = Lacticaseibacillus spp.; Lb. fermentum and Lb. reuteri = Limosilactobacillus spp.; Lb. sakei and Lb. curvatus = Latilactobacillus spp.; Lb. plantarum and Lb. pentosus = Lactiplantibacillus spp.; Lb. salivarius = Ligilactobacillus.
Along with the growing availability of reference genomes from isolated sequences, the number of MAGs retrieved from metagenomic datasets is continuously increasing, and these data can be combined for comparative genomics. To explore the availability of reconstructed genomes from LAB, we considered the large set of MAGs (N = 154,723) retrieved from 9428 human metagenomes that was clustered in 4930 species-level genome bins (SGBs) based on a 5% genetic diversity (Pasolli et al. 2019, 2020). Forty-nine of these SGBs belonged to LAB species, for a total of 830 reconstructed MAGs, grouped as follows: Lactobacillus (37 SGBs, 515 MAGs), Lactococcus (4 SGBs, 49 MAGs), Leuconostoc (3 SGBs, 7 MAGs), Pediococcus (2 SGBs, 5 MAGs), S. thermophilus (243 MAGs), and Weissella (2 SGBs, 11 MAGs) (Supplementary Table S3). These numbers are correlated with the occurrence of such species in the human gut, although the prevalence of low-abundance microbes is underestimated due to the technical impossibility of reconstructing MAGs from metagenomes in such cases. A large majority of the SGBs (44, 89.8%; for a total of 823 MAGs) represent at least partially known SGBs (kSGBs) that include one or more isolate genomes available in public databases. The most extensively reconstructed kSGBs were those of S. thermophilus (243 MAGs), Lb. ruminis (145 MAGs), Lb. mucosae (50 MAGs), Lb. salivarius (42 MAGs), and Lb. rhamnosus (32 MAGs). Only 7 MAGs spanning 5 SGBs were associated with unknown species (kSGBs), defined as SGBs lacking any publicly available genomes from isolate sequencing, which suggests the rarity of as-yet-uncharacterized LAB species in the human microbiome. Notably, reference genomes from human samples were almost entirely absent in the case of species with high prevalence in the gut, such as S. thermophilus and Lc. lactis (Fig. 3). Indeed, more than 90% of the S. thermophilus genomes were derived from FFs (mostly dairy products), while higher heterogeneity was observed for Lc. lactis, which was also found in insects, birds, fish and plant material (Fig. 3). Therefore, integrating isolated genomes with MAGs from large-scale metagenomic datasets can help overcome the lack of genomes from human hosts and represents an actual opportunity to advance the field through comparative genomic analyses of LAB, extensively taking into account different populations and environments of origin.
PROBIOTIC TRAITS OF LAB THAT MAKE THEM RELEVANT FOR THE HUMAN GUT AND THEIR PREVALENCE ACROSS GENOMES
Several LAB species are GRAS, due to their centuries-long history of use and human consumption in FFs, and therefore include most of the probiotic species that are currently available on the market. This has boosted the search for and characterization of novel LAB strains with potential applications as probiotics. Indeed, a Scopus search for ‘probiotic’ and ‘Lactic Acid Bacteria’ returned approximately 4900 documents (December 2019).
The widespread genome sequencing efforts of recent years have led to the availability of hundreds of LAB genomes (Fig. 2), and the new term ‘probiogenomics’ was coined in 2009 (Ventura et al. 2009), describing a discipline aimed at exploring the evolutionary history of commensal and probiotic bacteria and highlighting the genetic bases of their health-promoting activities.
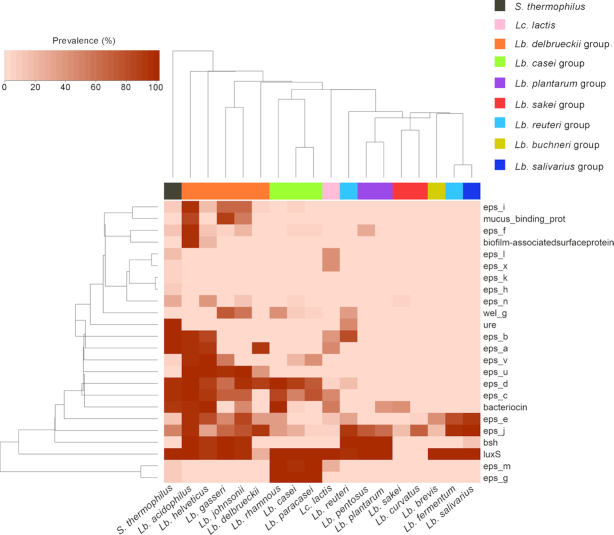
The first desirable feature in a probiotic strain is the ability to survive during passage through the GIT. For the scope of this review, we searched the publicly available genomes of 18 LAB species for 24 genes considered important for the capacity to resist the GIT, adhere to colonic cells, and colonize the intestine and that may be related to the ability of some LAB species commonly found in foods or supplements to reach the GIT and persist in the gut microbiome. A list of the genes and the relevant accession numbers is provided in Supplementary Table S4. Fig. 4 reports the prevalence of these genes in the genomes of 18 LAB species. The genes were predicted using Prokka (Seemann 2014) and were mapped using BlastN against a database containing the genes of interest. A gene was considered present if matched with an identity > 90% over a minimum length > 50%.
Figure 4.
Heat plot showing prevalence of genes involved in resistance to the GIT passage and engraftment in the gut in public genomes (available on NCBI in December 2019) of 18 selected LAB species, chosen for their importance in fermented foods and/or as probiotic. Eps, genes involved in exopolysaccharides production; ure, urease; bsh, bile-salt hydrolase; luxS, S-ribosylhomocysteine lyase. A list of the genes included and their NCBI accession numbers are reported in Supplementary Table S4. Color bar indicates assignment to the different taxonomic groups, as reported by Salvetti et al. (2018). For the taxonomy update proposed by Zheng et al. (2020), see Figure 3 legend.
EPS production is known to protect microbial strains from acid and bile stress. EPSs are high-molecular-weight sugar polymers secreted by microorganisms into the surrounding environment. According to the chemical composition, two types of EPS, homopolysaccharides (HoPSs) and heteropolysaccharides (HePSs), are synthesized by LAB (Zannini et al. 2016). HoPSs are polymers of glucose or fructose, and depending on the type of molecular linkage, these polymers can be α-glucans, β-glucans, or β-fructans, while HePSs comprise two or more different monosaccharide units, mainly D-glucose, D-galactose, and L-rhamnose. Among food-borne LAB, EPS producers have been described in the genera Streptococcus, Leuconostoc, Lactococcus, Pediococcus, Oenococcus, Lactobacillus and Weissella (Zannini et al. 2016). In addition to protecting microorganisms from the GIT environment, some EPSs have been reported to exhibit immunomodulatory and anti-inflammatory properties (Castro-Bravo, Wells and Margolles 2018). The EPS gene cluster in LAB may include several glycosyltransferases, polysaccharide polymerases, a tyrosine kinase (epsC) and its modulator (epsB), a transcriptional regulator (epsA) and a phosphotyrosine phosphatase (epsD), which are differently distributed among different species (Deo, Davray and Kulkarni 2019). These genes are broadly spread in the genomes of the S. thermophilus, Lc. lactis, Lb. casei, and Lb. delbrueckii groups, in which many of them have been identified, but are less frequent in the Lb. reuteri, Lb. brevis, Lb. fermentum, and Lb. curvatus groups (Fig. 4). Several LAB strains with probiotic activities have developed mechanisms to counteract the hostile environment of the GIT, with low pH and the presence of bile acids. Urease activity is one of the mechanisms of defence against acid stress, degrading urea and producing ammonia, which increase the pH in the environment surrounding the microbial cell (Mora and Arioli 2014). Urease was present in the genomes of almost all S. thermophilus strains sequenced but was not detected in public genomes of other LAB species, except for approximately 47% of Lb. reuteri genomes (Fig. 4). In contrast, bile salt hydrolase (bsh) is present in > 90% of the genomes from Lb. reuteri and species within the Lb. plantarum and Lb. delbrueckii groups (Fig. 4). Bile salt hydrolase activity is considered desirable in probiotic strains since it allows the hydrolysis of conjugated bile salts, increasing the possibility of survival of the strain in the GIT (Begley et al. 2006).
Although several probiotic strains can exert a positive health effect without colonization of the GIT, adhesion to intestinal epithelial cells is one of the most commonly screened characteristics during preliminary probiotic characterization, as this property is essential for the competition of the probiotic strain with pathogens for resources and space (Papadimitriou et al. 2015). Colonic epithelial cells are covered by a layer of mucin, a large glycoprotein. Binding to colonic mucin by probiotic bacteria is achieved via a mucus-binding protein (Mub). In addition, the ability of bacterial cells to self-aggregate and form biofilms is considered to influence epithelial adhesion (Papadimitriou et al. 2015; Sanders et al. 2018). The Mub gene was found in 60–90% of the published genomes of Lb. johnsonii, Lb. gasseri, and Lb. acidophilus but was absent in the other LAB species screened, including phylogenetically related species such as Lb. delbrueckii and Lb. helveticus, all belonging to the same taxonomic group. In addition, approximately 94% of the Lb. acidophilus genomes contained a biofilm-associated surface protein. This highlights the high potential of these species to colonize the colonic environment and persist after ingestion. Indeed, these species include most of the commercially available probiotic strains.
Other interesting features that are potentially important for probiotic LAB are the presence of attachment factors, such as fimbriae and pili, and the production of antimicrobial compounds, such as acids, hydrogen peroxide or bacteriocins, which may enhance the ability of the bacteria to compete against other intestinal microbes and could potentially inhibit pathogens (Dobson et al. 2012; Sanders et al. 2019). Bacteriocins are small peptides synthetized ribosomally by a wide range of bacteria and archaea that exert antimicrobial activity against other taxa, either of the same species as the producer or across genera and against which the producer develops specific immunity-related mechanisms. Bacteriocins are a heterogeneous class of peptides with different structures, sizes, types of activity, immunity-related mechanisms, and target cell receptors (Dobson et al. 2012; Chikindas et al. 2018). More than 90% of the genomes of S. thermophilus, Lb. acidophilus, and Lb. helveticus showed genes involved in bacteriocin production, while these genes were present in approximately 40–50% of Lb. sakei, Lb. plantarum, Lb. johnsonii, and Lc. lactis genomes (Fig. 4). However, the importance of bacteriocin production in probiotic bacteria remains controversial (Dobson et al. 2012). Many bacteriocin-producing microorganisms can inhibit pathogens in vitro (Le Blay et al. 2007), but results regarding in vivo efficacy are scarce and often do not match the in vitro activity. For example, the antimicrobial peptide lacticin 3147 produced by an Lc. lactis strain was effective against Listeria monocytogenes in vitro but failed in a mouse model (Dobson et al. 2011). The same result was reported for a strain of Pediococcus acidilactici: a corresponding effect was not observed in vivo despite its activity in the reduction of L. monocytogenes viability by 3 logs in vitro (Dabour et al. 2009). In contrast, some LAB strains are able to synthetize bacteriocins in vivo, showing direct antagonistic activity against pathogens. For example, Corr et al. (2007) demonstrated that oral gavage of Lb. salivarius UCC118 into mice protected them from L. monocytogenes infection. However, we should note that bacteriocins usually show a limited spectrum of action against target bacteria that are phylogenetically close to the producer. Therefore, bacteriocins produced by LAB are usually active against only Gram-positive bacteria.
HEALTH-RELATED ACTIVITIES OF LAB: EVIDENCE FROM IN VIVO TRIALS
Several studies have reported the health-related functional properties of probiotic LAB. Anti-inflammatory and immunomodulatory effects have been proposed for some LAB strains. For example, a strain of Lb. plantarum was suggested to play an anti-inflammatory role due to a specific structure of its teichoic acids (Grangette et al. 2005), while this role was attributed to EPS (Górska et al. 2016), pili (Lebeer et al. 2012) and S-layer proteins (Konstantinov et al. 2008) in strains of Lb. plantarum, Lb. rhamnosus, and Lb. acidophilus, respectively. A strain of Lb. salivarius was able to reduce inflammation and exert a preventive effect on colitis development in mice (Daniel et al. 2006), and the administration of a heat-killed strain of Lb. plantarum ameliorated inflammation and fibrosis in obese rats (Uchinaka et al. 2018). In addition, contact of the growth supernatant of one strain of S. thermophilus with immune cells reduced the release of the inflammatory marker TNF-alpha (Ménard et al. 2004).
The role of LAB in obesity and the related metabolic syndrome remains controversial, and contrasting results have been reported in the literature. The abundance of lactobacilli was found to be higher in obese subjects than in anorexic subjects (Ley et al. 2005) and in type 2 diabetes patients (Larsen et al. 2010; Karlsson et al. 2013). In addition, Drissi et al. (2014) suggested that Lactobacillus species might be differently associated with weight gain or loss; moreover, an enhanced potential for glycolysis, fat digestion, and oxidative stress response in weight gain-associated strains was highlighted when comparing the genomes of the two groups. However, several studies on mouse models have shown that the LAB consumption improved glucose metabolism and hepatic inflammation associated with a high-fat diet (Alard et al. 2016; Park et al. 2017). A similar result was also observed in human clinical trials. Moroti et al. (2012) hypothesized an effect of daily consumption of Lb. acidophilus and B. bifidum strains in reducing glycaemia and cholesterol levels, while Kobyliak et al. (2018b) concluded that supplementation with a multistrain probiotic (containing strains of Lactobacillus, Lactococcus, Bifidobacterium, Acetobacter, Propionibacterium) reduced insulin resistance in type 2 diabetes patients. All these contrasting results highlighted that the role of LAB cannot be generalized and that different species and strains can have a specific effect. Moreover, the overall response is also likely influenced by inter-individual variability.
Most of our knowledge is based on in vitro experiments or animal trials, while the best option to ascertain the possible health benefits of a microbial strain is human randomized controlled trials (Hill et al. 2014; De Filippis et al. 2018a). We surveyed the available literature regarding the effect of dietary intervention with probiotic LAB by searching the Scopus database for documents (only articles written in English) containing the words ‘probiotic’ OR ‘lactic acid bacteria’ AND ‘clinical trial’ OR ‘intervention’ OR ‘treatment’ in the abstract, title or key words. Animal trials were excluded, as well as review articles and studies in which probiotic strains not belonging to the LAB group were exclusively tested. This search identified a total of 95 studies, which are reviewed and reported in Table 1. Probiotic consumption has been extensively tested in the literature in either healthy or diseased populations (Table 1). Most of the studies used multistrain probiotic products, containing 1 to 10 different strains, with high heterogeneity in the amount ingested, ranging mostly from approximately 107 to 1011 cells/day, in single or multiple doses. When multistrain formulations were used, LAB were often administered together with Bifidobacterium spp. strains. In addition, mixed preparations of probiotic strains and prebiotic fibre are often used (symbiotic). Supplementation with probiotic LAB has been proposed for the treatment or improvement of symptoms of several types of diseases, including inflammatory bowel diseases, allergies and intolerance, diabetes and metabolic syndrome, stress and mental disorders, and infant colic. It is important to note that several studies did not report the name of the specific strain(s) used, the viable counts and the number of cells ingested, making comparison across studies impossible. Indeed, many of the probiotic activities were strain specific. For example, two different strains of Lb. acidophilus (LA-5 and NCFM) were tested at a similar dose for a possible role against allergic diseases (asthma and atopic dermatitis, respectively), and only Lb. acidophilus LA-5 was shown to be effective (Table 1).
Table 1.
Human trials involving probiotic LAB administration.
| Probiotic species/strains | Quantity/microbial loads ingested | Participants | Placebo control group | Study design* | Participant characteristics | Length of intervention | Wash out | Gut microbiome analysis method | Variation in gut microbiome detected | Health outcome targeted | Health outcome achieved* | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B. animalis sub. lactis CNCM I-2494, Lb. delbrueckii sub. bulgaricus CNCM I-1632 and CNCM I-1519, S. thermophilus CNCM I-1630, Lc. lactis sub. lactis CNCM I-1631 | Counts and daily amount assumed not reported | 50 | Yes | Double-blind, randomized, placebo-controlled trial | Male (36%), adults (35 ± 11), normal-weight | 2 weeks | No | 16S rRNA gene sequencing | None | IBS symptoms | No | Le Nevé et al. 2019 |
| Bio-25–B. bifidum, Lb. rhamnosus, Lc. lactis, Lb. casei sub. casei, B. breve, S. thermophilus, B. longum sub. longum, Lb. casei sub. paracasei, Lb. plantarum, B. longum sub. infantis; strains not reported | 2 pills/day, 2.5 × 1010 CFU/pill | 8 | No | Not reported | Male (63%), adults (28 ± 2.3), normal-weight | 4 weeks | No | 16S rRNA gene and shotgun sequencing | Increase in Akkermansia, Vagococcus, Enterococcus, Blautia, and Lactococcus | Gut microbiota reconstruction upon antibiotic administration | No | Suez et al. 2018 |
| Dicoflor60®—Lb. rhamnosus GG | 2 capsules/day, 6 × 109 CFU/capsule | 45 | No | Not reported | Male (84%), adults (47 ± 8), normal-weight | 8 weeks | No | 16S rRNA gene sequencing | Decrease in Enterobacteriaceae | Inflammatory status in HIV patients | Yes | Arnbjerg et al. 2018 |
| DUOLAC 7–S. thermophilus KCTC 11870BP, Lb. plantarum KCTC 10782BP, Lb. acidophilus KCTC 11906BP, Lb. rhamnosus KCTC 12202BP, B. lactis KCTC 11904BP, B. longum KCTC 12200BP, B. breve KCTC 12201BP | 2 capsules/day, 5 × 109 CFU/capsule | 17 | Yes | Double-blind, randomized, placebo-controlled trial | Male (0%), obese; age not reported | 8 weeks | No | Real-time qPCR for specific targets | Increase in B. breve, B. lactis and Lb. rhamnosus | Endotoxemia in obese subjects | Yes | Lee et al. 2014 |
| Ecologic® 825–Lb. casei W56, Lb. acidophilus W22, Lb. paracasei W20, B. lactis W51, Lb. salivarius W24, Lc. lactis W19, B. lactis W52, Lb. plantarum W62, B. bifidum W23 | 3 g/day, 7.5 × 106 CFU/g | 15 | Yes | Double-blind, randomized, placebo-controlled trial | Male (50%), adults (26 ± 5), BMI not reported | 4 weeks | No | None | - | Participants behaviour | Yes | Bagga et al. 2019 |
| Ecologic® Barrier—B. bifidum W23, B. lactis W51, B. lactis W52, Lb. acidophilus W37, Lb. brevis W63, Lb. casei W56, Lb. salivarius W24, Lc. lactis W19, Lc. lactis W58 | 4 g/day, 2.5109 CFU/g | 34 | Yes | Placebo-controlled trial | Male (38%), adults (36 ± 12), normal-weight | 8 weeks | No | 16S rRNA gene sequencing | None | Depression | Yes | Chahwan et al. 2019 |
| Ecologic® Barrier—see above | 6 g/day, 2.5 × 109 CFU/g | 12 | Yes | Double-blind, randomized, placebo-controlled trial | Male (92%), adults (61 ± 5), obese | 24 weeks | No | 16S rRNA gene sequencing | Increase in Lb. brevis | Glucose metabolism in type-2 diabetes patients | No | Horvath et al. 2019 |
| Ecologic® Barrier—see above | 2 g/day, 2.5 × 109 CFU/g | 29 | Yes | Double-blind, randomized, placebo-controlled trial | Male (0%), adults (21 ± 1), normal-weight | 4 weeks | No | None | - | Neurocognitive performance | Yes | Papalini et al. 2018 |
| Ecologic® Barrier (see above) | 2 g/day, 2.5 × 109 CFU/g | 20 | Yes | Placebo-controlled trial | Male (25%), adults (20 ± 2), normal-weight | 4 weeks | No | None | - | Depression | Yes | Steenbergen et al. 2015 |
| Ecologic® Barrier (see above) | 1 serving/day, 2.5 × 109 or 1 × 1010 CFU/serving | 30 | Yes | Double-blind, randomized, placebo-controlled trial | Male (0%), adults (56 ± 7), obese | 12 weeks | No | None | - | Iron metabolism in post-menopausa | Yes | Skrypnik et al. 2019 |
| Enterolactis® Plus—Lb. paracasei DG | 2 capsules/day, 2.4 × 1010 CFU/capsule | 30 | Yes | Double-blind, randomized, placebo-controlled, cross-over trial | Male (48%), adults (35 ± 11), normal-weight | 4 weeks | Yes, 4 weeks between treatments | 16S rRNA gene sequencing | Increase in Proteobacteria and Coprococcus; decrease in Blautia | None | - | Ferrario et al. 2014 |
| Enterolactis® Plus—see above | Counts and daily amount assumed not reported | 20 | Yes | Double-blind, randomized, placebo-controlled, cross-over trial | Male (25%), adults (45 ± 13), BMI not reported | 4 weeks | Yes, 4 weeks between treatments | 16S rRNA gene sequencing | Increase in Lactobacillus and Oscillospira, decrease in Ruminococcus | IBS symptoms | Yes | Cremon et al. 2018 |
| GoodBelly StraightShot—Lb. plantarum Lp299v | 1 serving/day, 2 × 1010 CFU/serving | 20 | No | Not reported | Male (100%), adults (63 ± 7), obese | 6 weeks | Yes, 4 weeks | 16S rRNA gene sequencing | Increase in Lactobacillus | Coronary artery disease | Yes | Malik et al. 2018 |
| Heat-killed Lb. gasseri CP2305 | 2 tablets/day, 1 × 1010 CFU/tablet | 31 | Yes | Double-blind, randomized, placebo-controlled trial | Male (68%), adults (25 ± 0.5), normal-weight | 24 weeks | No | 16S rRNA gene sequencing | Decrease in Bifidobacterium, Streptococcus | Stress and anxiety | Yes | Nishida et al. 2019 |
| Heat-killed Lb. paracasei CBA L74 | 1 serving/day, 5.9 × 1011 CFU/serving | 66 | Yes | Double-blind, randomized, placebo-controlled trial | Male (53%), infants (33 ± 9 months) | 12 weeks | No | None | - | Infection disease occurrence in children | Yes | Corsello et al. 2017 |
| Heat-killed Lb. paracasei CBA L74 | 1 serving/day, 5.9 × 1011 CFU/serving | 10 | Yes | Double-blind, randomized, placebo-controlled trial | Male (80%), children (33 ± 9 months) | 12 weeks | No | 16S rRNA gene sequencing | Increase in Lactobacillus, Faecalibacerium, Oscillospira | None | - | Berni Canani et al. 2017 |
| LacClean Gold-S®—B. bifidum KCTC 12199BP, B. lactis KCTC 11904BP, B. longum KCTC 12200BP, Lb. acidophilus KCTC 11906BP, Lb. rhamnosus KCTC 12202BP, S. thermophilus KCTC 11870BP | 2 capsules/day, 5 × 109 CFU/capsule | 25 | Yes | Double-blind, randomized, placebo-controlled trial | Male (44%), adults (46 ± 14), BMI not reported | 4 weeks | No | Real-time qPCR for specific targets | Increase in B. lactis, Lb. rhamnosus, S. thermophilus | IBS symptoms | Yes | Yoon et al. 2014 |
| Lb. acidophilus LA-5, B. animalis subsp. lactis BB-12 | 1 serving/day, 1 × 109 CFU/serving | 23 | Yes | Double-blind, randomized, placebo-controlled trial | Male (53%), adults (52 ± 7), overweight | 6 weeks | No | None | - | Glucose metabolism in type-2 diabetes patients | Yes | Bordalo Tonucci et al.2015 |
| Lb. acidophilus LA-5, Lb. rhamnosus GG, B. animalis subsp. lactis BB-12 | 1 capsule/day, 8.75 × 109 CFU/capsule | 16 | Yes | Double-blind, randomized, placebo-controlled three way cross-over trial | Male (50%), adults (43 ± 20), over-weight | 1 week | Yes, 2 weeks between treatments | Real-time qPCR and 16S rRNA gene sequencing | Increase in Bifidobacterium | Asthma | Yes | McLoughlin et al. 2019 |
| Lb. acidophilus NCFM OR B. lactis Bi-07 | 1 serving/day, 1 × 1010 CFU/serving | 17 | Yes | Double-blind, randomized, placebo-controlled trial | Infants (7–24 months); sex not reported | 8 weeks | No | 16S rRNA gene sequencing | None | Atopic dermatitis | No | Larsen et al. 2011 |
| Lb. acidophilus PBS066, Lb. reuteri PBS072 OR Lb. plantarum PBS067, Lb. rhamnosus LRH020, B. animalis subsp. lactis BL050 | 1 serving/day, 5 × 109 CFU/serving | 50 | Yes | Double-blind, randomized, placebo-controlled trial | Adults (37 ± 13), sex and BMI not reported | 8 weeks | Yes, 4 weeks | None | - | IBS with constipation | Yes | Mezzasalma et al. 2016 |
| Lb. acidophilus, B. bifidum, B. lactis, B. longum; strains not reported | 1 serving/day, 1.5 × 109 CFU/serving | 27 | Yes | Double-blind, randomized, placebo-controlled trial | Male (48%), adults (53 ± 6), BMI not reported | 24 weeks | No | None | - | Metabolic syndrome in pre-diabetic patients | Yes | Kassaian et al. 2019 |
| Lb. acidophilus, B. longum; strains not reported | 2 capsules/day; counts not provided | 37 | Yes | Double-blind, randomized, placebo-controlled trial | Male (30%), adults (44 ± 15), normal-weight | 4 weeks | No | Real-time qPCR for specific targets | None | IBS symptoms | Yes | Cui and Hu 2012 |
| Lb. acidophilus, Lb. casei, Lb. lactis, B. bifidum, B. longum, B. infantis; strains not reported | 2 sachets/day, 3 × 1010 CFU/sachet | 68 | Yes | Double-blind, randomized, placebo-controlled trial | Male (46%), adults (53 ± 9), normal-weight or overweight | 12 weeks | No | Microbial counts | Increase in Bifidobacterium and Lactobacillus | Glucose metabolism in type-2 diabetes patients | Yes | Firouzi et al. 2017 |
| Lb. acidophilus, Lb. plantarum, Lb. fermentum, Lb. gasseri; strains not reported | 0.5 g/day, 5 × 1010 CFU/g | 45 | Yes | Double-blind, randomized, placebo-controlled trial | Male (0%), adults (30 ± 6), normal-weight | 6 weeks | No | None | - | Glucose metabolism in women with gestational diabetes | No | Nabhani et al. 2018 |
| Lb. casei 01 | 1 capsule/day, 1 × 108 CFU/capsule | 22 | Yes | Double-blind, randomized, placebo-controlled trial | Male (0%), adults (41 ± 13), overweight | 8 weeks | No | None | - | Inflammatory status in rheumatoid arthritis | Yes | Vaghef-Mehrabany et al. 2014 |
| Lb. casei Lcr35® | 2 capsules/day, 2 × 108 CFU/capsule | 42 | No | Prospective, randomized, case-controlled study | Male (57%), children (2.3 ± 1.3), normal-weight | 1 week | No | 16S rRNA gene sequencing and plate counts | Increase in Lachnospiraceae, Bacteroides, Ruminococcus and decrease in Enterobacteriaceae, Escherichia, Clostridium | Diarrhea | Yes | Lai et al. 2019 |
| Lb. casei LMG 101/37 P-17 504, Lb. plantarum CECT 4528, B. animalis sub. lactis Bi1 LMG P-17 502, B. breve Bbr8 LMG P-17 501, B. breve Bl10 LMG P-17 500 | 1 sachet/day, 10 × 109 CFU/sachet | 54 | Yes | Double-blind, randomized, placebo-controlled trial | Male (11%), adults (43 ± 10), normal-weight | 6 weeks | No | 16S rRNA gene sequencing and plate counts | Increase in Bifidobacterium, Staphylococcus and lactic acid bacteria | Concomitant celiac disease and IBS | Yes | Francavilla et al. 2019 |
| Lb. casei W56, Lc. lactis W19, Lb. acidophilus W22, B. lactis W52, Lb. paracasei W20, Lb. plantarum W62, B. lactis W51, B. bifidum W23, Lb. salivarius W24 | 1 sachet/day; counts not reported | 20 | No | Not reported | Male (55%), adults (77 ± 10), BMI not reported | 4 weeks | No | Real-time qPCR for specific targets | Increase in Faecalibacterium prausnitzii | Alzheimer's disease | Yes | Leblhuber et al. 2018 |
| Lb. casei Zhang | 1 tablet/day, 1 × 1010 CFU/tablet | 24 | No | Open-label | Male (46%), adults (43 ± 17), normal-weight | 4 weeks | No | 16S rRNA gene sequencing | Positive correlation of Lb. casei with Prevotella, Faecalibacterium, Propionibacterium, Bifidobacterium | None | - | Zhang et al. 2014 |
| Lb. casei, Lb. acidophilus, Lb. rhamnosus, Lb. delbrueckii subsp. bulgaricus, B. breve, B. longum, S. thermophilus; strains not reported | 1 capsule/day, 1.5 × 1010 CFU/capsule | 30 | Yes | Double-blind, randomized, controlled trial | Male (7%), adults (42 ± 2), normal-weight | 8 weeks | No | None | - | Thyroid function | Yes | Talebi et al. 2020 |
| Lb. casei, Lb. rhamnosus, S. thermophilus, B. breve, Lb. acidophilus, B. infantis, Lb. bulgaricus; strains not reported | 1 serving/day, counts not reported | 25 | Yes | Double-blind, randomized, placebo-controlled trial | Male (60%), infants (1.5 ± 0.8 months) | 4 weeks | No | None | - | Crying time in infants with colic | Yes | Kianifar et al. 2014 |
| Lb. casei, Lb. rhamnosus, S. thermophilus, B. breve, Lb. acidophilus, B. longum, Lb delbrueckii sub. bulgaricus; strains not reported | 2 capsules/day, 2 × 108 CFU/capsule | 20 | Yes | Parallel, triple-blind, randomized, controlled trial | Adults (57 ± 1.5), over-weight/obese, sex not reported, | 12 weeks | No | None | - | Metabolic syndrome | Yes | Rabiei et al. 2019 |
| Lb. casei, Lb. rhamnosus, S. thermophilus, B. breve, Lb. acidophilus, B. longum, Lb. bulgaricus; strains not reported | 1 capsule/day, 2 × 108 CFU/capsule | 29 | Yes | Randomized triple-masked controlled trial | Male (45%), children (11 ± 2), obese | 4 weeks | No | Microbial counts | None | Obesity | No | Kelishadi et al. 2014 |
| Lb. gasseri CECT5714, Lb. coryniformis CECT5711 | 1 serving/day, 2 × 109 CFU/serving | 15 | Yes | Double-blind, randomized, placebo-controlled trial | Male (50%), adults (33 ± 10), BMI not reported | 4 weeks | Yes, 2 weeks | None | - | Bowel habits | Yes | Olivares et al. 2006 |
| Lb. gasseri KS-13, B. bifidum G9–1, B. longum MM2 | 2 capsules/day, 1.5 × 109 CFU/capsule | 16 | Yes | Double-blind, randomized, placebo-controlled, cross-over trial | Adults (70 ± 1), sex and BMI not reported | 3 weeks | Yes, 5 weeks between treatments | Real-time qPCR for specific targets and 16S rRNA gene sequencing | Increase in Faecalibacterium prausnitzii | Inflammatory status in older people | Yes | Spaiser et al. 2015 |
| Lb. helveticus Bar13, B. longum Bar33 | 1 serving/day, 1 × 109 CFU/serving | 16 | Yes | Double-blind, randomized, placebo-controlled trial | Male (41%), adults (76 ± 10), BMI not reported | 4 weeks | No | Phylogenetic microarray | Decrease in opportunistic pathogens (Clostridium difficile, Cl. perfringens, Enterococcus faecium) | None | - | Rampelli et al. 2013 |
| Lb. helveticus R0052 | 1 sachet/day, 3 × 109 CFU/sachet | 23 | Yes | Randomized, double-blind, placebo- controlled trial | Infants (3–12 months), sex and BMI not reported | 8 weeks | No | 16S rRNA gene sequencing | Increase in Proteobacteria, decrease in Firmicutes | Inflammatory status | No | De Andrés et al. 2018 |
| Lb. helveticus R0052, B. longum R0175 | 1 sachet/day, 10 × 109 CFU/sachet | 28 | Yes | Double-blind, randomized controlled trial | Male (29%), adults (36 ± 8), normal-weight | 8 weeks | No | None | - | Depression | Yes | Kazemi et al. 2019 |
| Lb. johnsonii LA1 | 2 sachets/day, 2 × 109 CFU/sachet | 48 | Yes | Double-blind, randomized, placebo-controlled trial | Male (54%), adults (32 ± 10), BMI not reported | 24 weeks | No | None | - | Recurrence of Chron's disease after surgery | No | Marteau et al. 2006 |
| Lb. johnsonii LA1 | 1 sachet/day, 1 × 1010 CFU/sachet | 27 | Yes | Double-blind, randomized, placebo-controlled trial | Male (56%), adults (39 ± 15), BMI not reported | 12 weeks | No | None | - | Recurrence of Chron's disease after surgery | No | Van Gossum et al. 2007 |
| Lb. paracasei subsp. paracasei W8 | 1 capsule/day, 1 × 109 OR 1 × 1010 CFU/capsule | 21 | Yes | Three arms, randomized, controlled, crossover study | Male (52%), adults (27 ± 7), normal-weight | 3 days | No | None | - | Energy intake | No | Toksvig Bjerg et al.2014 |
| Lb. plantarum ATTC 202 195 | 1 capsule/day, 1 × 109 CFU/capsule | 2185 | Yes | Double-blind, randomized, placebo-controlled trial | Infants (0–2 months); sex not reported | 8 weeks | No | None | - | Neonatal sepsis | Yes | Panigrahi et al. 2017 |
| Lb. plantarum CCFM8610 | 1 serving/day, 1 × 109 CFU/serving | 29 | Yes | Placebo-controlled trial | Male (40 or 38%), adults (49 ± 15 or 53 ± 14), normal-weight | 8 weeks | No | 16S rRNA and groEL gene sequencing | Increase in microbial diversity, Bacteroidetes and B. pseudocatenulatum, decrease in Firmicutes/Bacteroidetes ratio | Atopic dermatitis | Yes | Fang et al. 2019 |
| Lb. plantarum ECGC 13 110 402 | 2 capsules/day, 2 × 109 CFU/sachet | 23 | Yes | Prospective, randomized, placebo-controlled, parallel-group study | Male (22%), adults (52 ± 11), over-weight | 12 weeks | Yes, 4 weeks | 16S rRNA gene sequencing | None | Blood lipids | Yes | Costabile et al. 2017 |
| Lb. plantarum Lp115 | 80 ml/day, 1.25 107 UFC/ml | 12 | Yes | Not reported | Male (0%), adults (62 ± 4), over-weight | 12 weeks | No | None | - | Metabolic syndrome in postmenopausal | Yes | Barreto et al. 2014 |
| Lb. plantarum MF1298 | 1 capsule/day, 1 × 1010 CFU/capsule | 16 | Yes | Double-blind, randomized, placebo-controlled trial | Male (31%), adults (50 ± 11), normal-weight | 16 weeks | No | None | - | IBS symptoms | No | Farup et al. 2012 |
| Lb. plantarum PS128 (DSM 28 632) | 1 capsule/day, 3 × 1010 CFU/capsule | 36 | Yes | Double-blind, randomized, placebo-controlled trial | Male (100%), children (10 ± 2), normal-weight | 4 weeks | No | None | - | Autism spectrum disorder | Yes | Liu et al. 2019 |
| Lb. reuteri ATTC PTA 6475 | 2 servings/day,5 × 109 CFU/serving | 45 | Yes | Double-blind, randomized, placebo-controlled trial | Male (0%), adults (76 ± 1), normal-weight | 48 weeks | No | None | - | Bone loss in old women | Yes | Nilsson et al. 2018 |
| Lb. reuteri DSM 17 938 | 5 drops/day, 0.2 × 108 CFU/drop | 14 | Yes | Double-blind, randomized, placebo-controlled trial | Male (43%), infants (8 ± 3 months) | 4 weeks | No | None | - | Crying time in infants with colic | No | Nation et al. 2017 |
| Lb. reuteri DSM 17 938 | 5 drops/day, 0.2 × 108 CFU/drop | 25 | Yes | Double-blind, randomized, placebo-controlled trial | Male (60%), infants (1.2 ± 0.8 months) | 3 weeks | No | FISH for specific targets | None | Crying time in infants with colic | Yes | Savino et al. 2010 |
| Lb. reuteri DSM 17 938 | 5 drops/day, 0.2 × 108 CFU/drop | 32 | Yes | Double-blind, randomized, placebo-controlled trial | Male (41%), infants (48 ± 26 months) | 4 weeks | No | Real-time qPCR for specific targets | None | Crying time in infants with colic | Yes | Savino et al. 2018 |
| Lb. reuteri DSM 17 938 | 5 drops/day, 0.2 × 108 CFU/drop | 85 | Yes | Double-blind, randomized, placebo-controlled trial | Male (44%), infants (1.9 ± 0.7 months) | 4 weeks | No | None | - | Crying time in infants with colic | No | Sung et al. 2014 |
| Lb. reuteri DSM 17 938 | 1 serving/day, 1 × 108 CFU/serving | 15 | Yes | Double-blind, randomized, placebo-controlled trial | Male (60%), infants (1 ± 0.4 months) | 3 weeks | No | 16S rRNA gene sequencing | None | Crying time in infants with colic | No | Roos et al. 2013 |
| Lb. reuteri DSM 17 938 | 1 tablet/day, 1 × 108 CFU/tablet | 15 | Yes | Double-blind, randomized, placebo-controlled, cross-over trial | Male (70%), adults, normal-weight | 24 weeks | No | 16S rRNA gene sequencing | Increase in microbial diversity and in Firmicutes | None | - | del Campo et al. 2014 |
| Lb. reuteri DSM17938 | 1 serving/day, 108 OR 1010 CFU/serving | 15 OR 14 | Yes | Double-blind, randomized, placebo-controlled, parallel-group study | Male (80%), adults (65 ± 6), BMI not reported | 12 weeks | No | 16S rRNA gene sequencing | None | Glucose metabolism in type-2 diabetes patients | No | Mobini et al. 2017 |
| Lb. reuteri NCIMB 30 242 | 2 servings/day, from 3 × 109 to 10 × 109 CFU/serving | 10 | No | Randomized, dose-escalation design trial | Adults, normal-weight to obese; sex and age not reported | 4 weeks | No | 16S rRNA gene sequencing | None | Reduction of cholesterol | Yes | Martoni et al. 2015 |
| Lb. rhamnosus 14E4 | 1 serving/day, 1 × 1010 CFU/serving | 14 | Yes | Double-blind, randomized, placebo-controlled, cross-over trial | Male (50%), adults (31 ± 4), normal-weight | 2 weeks | Yes, 1 week | 16S rRNA gene sequencing | Increase in Prevotella and decrease in Bacteroides | None | - | Bautista-Gallego et al. 2019 |
| Lb. rhamnosus GG | 1 serving/day, 4.5 × 109 CFU/serving | 15 | Yes | Double-blind, randomized, placebo-controlled trial | Male (47%), infants (1.3 ± 0.4 months) | 4 weeks | No | None | - | Crying time in infants with colic | No | Pärtty et al. 2015 |
| Lb. rhamnosus GG | 1 serving/day, 6 × 109 CFU/serving | 10 | Yes | Double-blind, randomized, placebo-controlled trial | Male (59%), children (7 ± 2), BMI not reported | 4 weeks | No | DGGE and FISH for specific targets | Increase in Bacteroides | Inflammatory status in cystic fibrosis children | Yes | Bruzzese et al. 2014 |
| Lb. rhamnosus GG | 1 bottle/day, 1.55 × 1011 CFU/bottle | 10 | Yes | Double-blind, randomized, placebo-controlled trial | Male (30%), adults (47 ± 6), normal-weight | 3 weeks | No | Phylogenetic microarray | Increase in Lb. rhamnosus | None | - | Lahti et al. 2013 |
| Lb. rhamnosus GG | 1 serving/day, 1 × 109 CFU/serving | 8 | Yes | Double-blind, randomized, placebo-controlled trial | Infants (6 months); sex not reported | 24 weeks | No | Phylogenetic microarray | None | None | - | Cox et al. 2010 |
| Lb. rhamnosus GG | 4.5 × 107–8.5 × 107/g; day amount not reported | 12 | Yes | Double-blind, randomized, placebo-controlled trial | Male (75%), infants (1–12 months) | 24 weeks | No | 16S rRNA gene sequencing | Increase in Blautia, Roseburia, Coprococcus | Oral tolerance in cow's milk allergic children | Yes | Berni Canani et al. 2016 |
| Lb. rhamnosus GG, B. animalis sub. lactis BB-12 | 1 capsule/day, 1 × 109 CFU/capsule | 207 | Yes | Double-blind, randomized, placebo-controlled trial | Male (0%), adults (32 ± 4.8), from normal-weight to obese | 8 weeks | No | None | - | Gestational diabetes | No | Callaway et al. 2019 |
| Lb. rhamnosus GG, Lb. rhamnosus LC705, B. breve Bb99, Propionibacterium freudenreichii subsp. shermanii JS | 1 capsule/day, 5 × 109 CFU/capsule | 461 | Yes | Double-blind, randomized, placebo-controlled trial | Male (50%), children (1.2 ± 0.3) | 24 weeks | No | None | - | Allergic diseases | No | Kukkonen et al. 2007 |
| Lb. rhamnosus GG, Lb. rhamnosus Lc705, Propionibacterium freudenreichii subsp. shermanii JS, B. animalis subsp. lactis Bb12 | 120 ml/day, 1 × 107 CFU/ml | 43 | Yes | Double-blind, randomized, placebo-controlled trial | Male (9%), adults (50 ± 13), normal-weight | 20 weeks | No | Phylogenetic microarray | None | IBS symptoms | Yes | Kajander et al. 2007 |
| Lb. rhamnosus IMC 501, Lb. paracasei IMC 502 | 1 serving/day, 1 × 109 CFU/serving | 25 | Yes | Double-blind, randomized, placebo-controlled trial | Adults; sex, age and BMI not reported | 12 weeks | Yes, 2 weeks | Microbial counts | Increase in lactobacilli and bifidobacteria | Bowel habits | Yes | Verdenelli et al. 2011 |
| Lb. salivarius CECT5713 | 2 capsules/day, 1 × 108 CFU/capsule | 20 | Yes | Double-blind, randomized, placebo-controlled trial | Male (50%), adults (33 ± 8), BMI not reported | 4 weeks | No | Microbial counts | Increase in lactobacilli | Immune response | Yes | Sierra et al. 2010 |
| Lb. salivarius, strain not reported | 1 serving/day, 2 × 1010 CFU/serving | 27 | Yes | Double-blind, randomized, placebo-controlled trial | Adults (25 ± 5), normal-weight, sex not reported | 16 weeks | No | None | - | Upper respiratory tract infections occurrence in athletes | No | Gleeson et al. 2012 |
| Lb. casei; strain not reported | 1 bottle/day, 6.5 × 109 CFU/bottle | 6 | No | Not reported | Male (33%), children (13 ± 3), normal-weight | 6 weeks | No | Intergenic spacer profiling (IS-pro) | None | None | - | El Manouni el Hassani et al. 2019 |
| Leuconostoc holzapfelii; strain not reported | Not provided | 21 | No | Not reported | Male (43%), adults (28 ± 2.3), normal-weight | 4 weeks | No | 16S rRNA gene sequencing | None | None | - | Yang et al. 2019 |
| S. thermophilus, Lb. acidophilus, B. longum; strains not reported | 3 capsules/day, 3 × 1010 cfu/capsule | 16 | Yes | Double-blind, randomized, placebo-controlled trial | Male (69%), adults (54 ± 11), normal-weight | 12 weeks | No | DGGE | None | Chronic kidney disease | No | Borges et al. 2018 |
| Supherb Bio-25–B. bifidum, Lb. rhamnosus, Lc. lactis, Lb. casei, B. breve, S. thermophilus, B. longum sub. longum, Lb. paracasei, Lb. plantarum and B. longum sub. infantis; strains not reported | 2 pills/day, 2.5 × 1010 CFU/pill | 14 | Yes | Placebo-controlled trial | Male (57%), adults (42 ± 13), normal-weight | 4 weeks | No | Shotgun metagenome, real-time qPCR | Probiotic strains engraftment is dependent on individualized gut microbiome features | None | - | Zmora et al. 2018 |
| Symbiter—Lactobacillus + Lactococcus, Bifidobacterium, Propionibacterium, Acetobacter; species and strains not reported | 10 g/day, 6 × 1010 CFU/g | 31 | Yes | Double-blind, randomized, placebo-controlled trial | Adults (52 ± 2), obese, sex not reported | 8 weeks | No | None | - | Insuline resistance in type-2 diabetes patients | Yes | Kobyliak et al. 2018a |
| Symbiter—see above | 10 g/day, 6 × 1010 CFU/g | 30 | Yes | Double-blind, randomized, placebo-controlled trial | Adults (53 ± 10), obese, sex not reported | 8 weeks | No | None | - | Non-Alcholic Fatty Liver Disease | Yes | Kobyliak et al. 2018b |
| Symprove—Lb. rhamnosus NCIMB 30 174, Lb. plantarum NCIMB 30 173, Lb. acidophilus NCIMB 30 175, Enterococcus faecium NCIMB 30 176 | 1 serving/day, 1 × 1010 CFU/serving | 100 | Yes | Double-blind, randomized, placebo-controlled trial | Male (32%), adults (39 ± 11), BMI not reported | 12 weeks | No | None | - | IBS symptoms | Yes | Sisson et al. 2014 |
| Vivomixx ®—Lb. plantarum DSM 24 730, S. thermophilus DSM 24 731, B. breve DSM 24 732, Lb. paracasei DSM 24 733, Lb. delbrueckii subsp. bulgaricus DSM 24 734, Lb. acidophilus DSM 24 735, B. longum DSM 24 736, B. infantis DSM 24 737 | 2 sachets/day, 4.5 × 1011 CFU/sachet | 9 | Yes | Double-blind, randomized, placebo-controlled trial | Male (100%), adults (45 ± 10), BMI not reported | 24 weeks | No | None | - | Neuroinflammation in HIV patients | Yes | Ceccarelli et al. 2017 |
| VSL#3®—S. thermophilus, B. breve, B. longum, B. infantis, Lb. acidophilus, Lb. plantarum, Lb. paracasei, Lb. delbrueckii; strains not reported | 2 sachets/day, 1.1 × 1011 CFU/sachets | 14 | No | Open-label, single-arm study | Male (0%), adults, age and BMI not reported | 4 weeks | Yes, 4 weeks | 16S rRNA gene sequencing | None | Immune response | Yes | Singh et al. 2018 |
| VSL#3®—see above | 1 capsule/day, 1.1 × 1011 CFU/capsule | 15 | Yes | Randomized, placebo-controlled trial | Adults (50 ± 10), overweight or obese; sex not reported | 6 weeks | No | Microbial counts | None | Cholesterol in obese subjects | Yes | Rajkumar et al. 2014 |
| VSL#3®—see above | 1 capsule/day, 1.1 × 1011 CFU/capsule | 15 | Yes | Double-blind, randomized, placebo-controlled trial | Adults, sex, age and BMI not reported | 8 weeks | No | Phylogenetic microarray | None | Diarrhea-predominant IBS synthomps | Yes | Michail and Kenche 2011 |
| VSL#3®—see above | 2 sachets/day, 9 × 1011 CFU/sachets | 10 | No | Open-label | Male (20%), adults (46 ± 10), BMI not reported | 4 weeks | No | 16S rRNA gene sequencing | Decrease in Bacteroides | IBS symptoms | Yes | Ng et al. 2013 |
| VSL#3®—see above | 1.1 × 1011 CFU/sachets; day amount not reported | 20 | No | Not reported | Male (65%), adults (44 ± 27), BMI not reported | 4 weeks | No | None | - | Liver cirrosis | Yes | Marlicz et al. 2016 |
| VSL#3®—see above | 2 sachets/day, 1.1 × 1011 CFU/sachets | 22 | Yes | Double-blind, randomized, placebo-controlled trial | Male (45%), children (10 ± 2), obese | 16 weeks | No | None | - | Non-Alcholic Fatty Liver Disease in obese children | Yes | Alisi et al. 2014 |
| VSL#3®—see above | 1.1 × 1011 CFU/sachets; day amount not reported | 9 | Yes | Double-blind, randomized, placebo-controlled trial | Male (33%), adolescents (14 ± 2), obese | 16 weeks | No | 16S rRNA gene sequencing | None | Obesity | No | Jones et al. 2018 |
| VSL#3®—see above | 1 sachet/day, 9 × 1011 CFU/sachet | 66 | Yes | Double-blind, randomized, placebo-controlled trial | Male (85%), adults (48 ± 5), BMI not reported | 24 weeks | No | None | - | Recurrence of hepatic encephalopathy in patients with cirrhosis | Yes | Dhiman et al. 2014 |
| Yakult—Lb. casei Shirota | 1 bottle/day, 6.5 × 109 CFU/bottle | 10 | Yes | Double-blind, randomized, placebo-controlled trial | Adults; age, sex and BMI not reported | 20 weeks | No | None | - | Immune response in allergic rhinitis | Yes | Ivory et al. 2008 |
| Yakult—Lb. casei Shirota | 1 bottle/day, 1 × 1011 CFU/bottle | 23 | Yes | Double-blind, randomized, placebo-controlled trial | Male (52%), adults (23 ± 0.4), normal-weight | 8 weeks | No | 16S rRNA gene sequencing | None | Stress-induced abdominal dysfunction | Yes | Kato-Kataoka et al. 2016 |
| Zircombi—B. longum BB536, Lb. rhamnosus HN001 | 1 sachet/day, 4 × 109 CFU/sachet | 23 | Yes | Double-blind, cross-over, randomized, placebo-controlled trial | Male (17%), adults (48 ± 3), normal-weight | 4 weeks | Yes, 2 weeks between treatments | 16S rRNA gene sequencing | Increase in Bifidobacterium, decrease in Enterobacter, Klebsiella, Serratia | Lactose intolerance | Yes | Vitellio et al. 2019 |
| Lb. johnsonii LA1 | 80 ml/day, 1 × 107 CFU/ml | 74 | Yes | Double-blind, cross-over, randomized, placebo-controlled trial | Male (57%), children (12 ± 2), BMI not reported | 3 weeks | No | None | - | Helicobacter pylori infection eradication | Yes | Gotteland et al. 2008 |
*As reported in the original study.
In some cases, independent clinical trials tested the efficacy of the same probiotic formulation targeting an identical health outcome (Table 1). For example, Chahwan et al. (2019), Steenbergen et al. (2015) and colleagues administered a multistrain probiotic preparation (Ecologic® Barrier) containing 4 strains of Lactobacillus spp., 2 strains of Lc. lactis, and 3 strains of Bifidobacterium spp. to adults with depression, and both studies reported an improvement in neurocognitive functions and relief from the symptoms of depression. In contrast, the same formulation was not effective at improving blood glycaemia in type 2 diabetes patients (Horvath et al. 2019). This highlights that a probiotic formulation should be designed to target a well-defined health outcome or population. In two other trials, the consumption of Lb. johnsonii LA1 for 12 or 24 weeks was proposed to reduce the recurrence of Crohn's disease after surgery (van Gossum et al. 2007; Marteau et al. 2006). The investigators considered two different doses (4 × 109 or 1 × 1010 CFU/day), but the treatment was shown to be ineffective in both trials.
In other cases, the results obtained in separate trials were discordant. The same strain of Lb. reuteri (DSM 17 938) was independently examined for its activity against infant colic in five trials, all with similar duration (2–3 weeks) and number of cells ingested per day (108 CFU/day). However, only two of the trials reported a reduction in crying time (Savino et al. 2010, 2018; see Table 1).
Notably, none of the studies presented in this review commented on the rationale for choosing a specific probiotic dosage. The ISAPP provides suggested amounts ranging from 1 × 108 to 1.8 × 1012 CFU once or twice a day, depending on the strain and the type of target population (Guarner et al. 2012). However, this list covers gastrointestinal disorders only and does not take into account the intrinsic variability in the gut microbiota. In general, different dosages should be assessed to understand the effect of the dose-response relationship of probiotic consumption on targeted health outcomes and support the causality of the observed associations. This might at least partly explain the presence of contrasting results observed in clinical trials found in the literature.
Contrasting results might also be related to a subject-specific response associated with individual characteristics of the gut microbiome. In fact, several studies reported that different individuals may respond differently to the same drug, dietary treatment or even probiotic treatment and that this difference may be at least partially related to the individual structure of the gut microbiome (Zmora et al. 2016; De Filippis et al., 2018). According to some recent studies, the baseline microbiome may also influence the possibility of the probiotic strain colonizing the gut and its long-term engraftment and persistence once oral administration is stopped (Maldonado-Gomez et al. 2016; Zmora et al. 2018). Nevertheless, most of the studies found in the literature did not explore the effect of probiotic administration on the gut microbiota composition (Table 1). Moreover, even when this analysis was included, the method used did not allow tracking of the fate of the specific strain except in a few cases (Suez et al. 2018; Zmora et al. 2018). Indeed, probiotic treatment was frequently not able to modify the overall structure of the gut microbiota, and only a few studies reported an increase in the abundance of the microbial genus/genera administered. Finally, interpretation of the effect of probiotic consumption on the composition of the gut microbiota may be particularly complicated due to the lack of consensus around a universally accepted definition of a healthy gut microbiota (Bäckhed et al. 2012; Cani 2018; McBurney et al. 2019).
As reported above, some LAB strains may exhibit health-promoting activity even if inactivated. Some clinical trials demonstrated that the consumption of fermented matrices in which probiotic bacteria were heat-inactivated may still have a positive effect on health (Table 1). For example, consumption of milk fermented by Lb. paracasei CBA L74 and subsequently heat-killed reduced the occurrence of infectious diseases in children and boosted the production of beneficial short-chain fatty acids (Berni Canani et al. 2017; Corsello et al. 2017). In another trial conducted in an adult population, the consumption of inactivated Lb. gasseri CP2305 in tablets reduced anxiety and stress (Nishida et al. 2019).
FERMENTED FOODS AND HUMAN HEALTH
Growing evidence has been provided in the literature regarding the enhanced functional and nutritional properties of FFs. Notably, a large proportion of such foods contain living microorganisms, including LAB, which are genetically similar to the strains used as probiotics. Extensive clinical trials have been conducted to prove the human health-promoting activities of probiotics, as reviewed in the previous section. Epidemiological and clinical studies on FFs have also been similarly conducted, although there are considerably fewer such studies, and a large fraction of them have been well reviewed in (Marco et al. 2017; Fernandez and Marette 2018; Kok and Hutkins 2018).
Consumption of fermented dairy products was inversely correlated with overall mortality (Soedamah-Muthu et al. 2013) and impaired glucose metabolism (Eussen et al. 2016) and linked to overall improvements in health-linked biomarkers (González et al. 2019). Yogurt consumption was inversely associated with type 2 diabetes (Chen et al. 2014; Díaz-López et al. 2016), risks of metabolic syndrome (Babio et al. 2015), risk of colorectal cancer (Pala et al. 2011), and long-term weight gain (Mozaffarian et al. 2011). Multiple randomized, controlled trials showed a higher effectiveness of probiotic yogurts than conventional yogurts in the improving various health outcomes, such as fasting blood glucose levels (Ejtahed et al. 2012) and insulin resistance (Asemi et al. 2013; Nabavi et al. 2015). Different species and strain types present in fermented milk were related to reduced muscle soreness (Iwasa et al. 2013), improved intrinsic brain activity (Tillisch et al. 2013), reduced incidence of fever (Nagata et al. 2016), improved bowel movements (Nagata et al. 2016), reduced risk of cardiovascular disease (Sonestedt et al. 2011), beneficial effects on metabolism (Fernandez and Marette 2018), and positive changes in health and mood (Baars 2019). High consumption of cultured milk lowered the risk of developing bladder cancer (Larsson et al. 2008). Combined consumption of cheese, yogurt, and fermented milk was inversely associated with diabetes (Sluijs et al. 2012). Consumption of fermented kimchi decreased insulin resistance, increased insulin sensitivity (An et al. 2013) and reduced the prevalence of asthma and atopic dermatitis (Park and Bae 2016; Kim, Ju and Park 2017). In a randomized controlled study, kimchi also improved fasting blood glucose levels and other health outcomes (Kim et al. 2011; Choi et al. 2013). Kefir-fermented milk was associated with a large six-month increase in hip bone mineral density among osteoporotic patients (Tu et al. 2015), and Chungkookjang improved multiple parameters associated with obesity (Byun et al. 2016). The risk of high blood pressure was reduced by the consumption of fermented products such as miso and natto (Nozue et al. 2017). Total cholesterol levels were improved by consumption of fermented soy (Cavallini et al. 2016) and Kochujang (Lim et al. 2015). Signs of irritable bowel syndrome were attenuated by consumption of low-FODMAP rye bread (Laatikainen et al. 2016) and lacto-fermented sauerkraut (Nielsen et al. 2018). Clinical trials on FFs have also been conducted in animal models. Examples in mice are represented by the alleviation of atopic dermatitis through cream cheese (Kim, Kim and Kim 2019) and the recovery from antibiotic-induced gut dysbiosis with gut barrier function enhancement through a mixture of Lactobacillus species isolated from traditional FFs (Shi et al. 2018). Focusing specifically on the impact of FFs on gastrointestinal health, there is still limited evidence of the effectiveness of these foods (Mota de Carvalho et al.2018; Dimidi et al. 2019; Wu et al. 2019). Kefir is the FF most commonly investigated in this scenario, with evidence of a beneficial role for lactose malabsorption and Helicobacter pylori eradication (Dimidi et al. 2019). In addition, a recent observational study carried out in the frame of the American Gut project, highlighted that fermented plant consumers showed higher faecal levels of beneficial conjugated linoleic acid (CLA) compared with non-consumers, besides increased abundance of several LAB species and other health-associated taxa (e.g. Faecalibacterium prausnitzii, Prevotella spp., Eubacterium spp.) in their gut microbiome. Interestingly, CLA dietary intake was not different between the two groups, suggesting an effect of the gut microbiome in the biosynthesis (Taylor et al. 2020).
CONCLUSIONS
FFs are widespread worldwide and can be considered as primary reservoirs of live bacteria that can potentially reach the gut microbiome and eventually impact host health. Although many speculations about the food-gut axis exist, there is only limited evidence supporting a direct effect of FF consumption on the gut microbiome and on the possible transfer of LAB from FFs to the host gut microbiome. Well-designed clinical trials and broad population studies are necessary to understand whether ingested LAB are effectively able to engraft in the human gut and become permanent members of the gut microbiome. As discussed, the ability of certain strains to reach and colonize the human gut will strongly depend on their genomic capacity to counteract the barriers that they will face (i.e. low pH and bile salts), as well as the specific composition of the host gut microbiome. In future years, probiotic research will surely benefit from recent advances in genomics and metagenomics and the development of new bioinformatic algorithms and analysis tools. To develop a robust knowledge of the LAB-food-gut axis, comparative genomic studies will be needed to compare the diversity and functions of LAB from food and gut environments. In addition, improved availability of LAB genomes from gut isolates will also be important for better understanding the genomic features (if any) that can be pivotal for the adaptation of LAB to the gut environment. Meanwhile, FFs will continue to serve as inexhaustible sources of potential probiotic LAB strains and as natural dietary sources of live LAB cells with a promising role in human gut health.
Supplementary Material
Contributor Information
Francesca De Filippis, Department of Agricultural Sciences, University of Naples Federico II, via Università, 100, 80055, Portici (NA), Italy; Task Force on Microbiome Studies, Corso Umberto I, 40, 80100, Napoli, Italy.
Edoardo Pasolli, Department of Agricultural Sciences, University of Naples Federico II, via Università, 100, 80055, Portici (NA), Italy; Task Force on Microbiome Studies, Corso Umberto I, 40, 80100, Napoli, Italy.
Danilo Ercolini, Department of Agricultural Sciences, University of Naples Federico II, via Università, 100, 80055, Portici (NA), Italy; Task Force on Microbiome Studies, Corso Umberto I, 40, 80100, Napoli, Italy.
FUNDING
This study was supported by the project MASTER (Microbiome Applications for Sustainable food systems through Technologies and Enterprise). This project has received funding from the European Union's Horizon 2020 research and innovation programme under grant agreement No 818368. The study was also supported by the Italian Ministry of Agricultural, Food, Forestry and Tourism Policies, with the project JPI HDHL-INTIMIC—Knowledge Platform of Food, Diet, Intestinal Microbiomics and Human Health, granted by with ID 790.
This manuscript reflects only the authors’ views and the European Commission is not responsible for any use that may be made of the information it contains.
Conflicts of Interest
None declared.
REFERENCES
- Aguilar-Toalá JE, Garcia-Varela R, Garcia HSet al. Postbiotics: An evolving term within the functional foods field. Trends Food Sci Technol. 2018;75:105–14. [Google Scholar]
- Aguirre de Cárcer D. The human gut pan-microbiome presents a compositional core formed by discrete phylogenetic units. Sci Rep. 2018;8:14069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alard J, Lehrter V, Rhimi Met al. Beneficial metabolic effects of selected probiotics on diet-induced obesity and insulin resistance in mice are associated with improvement of dysbiotic gut microbiota. Environ Microbiol. 2016;18:1484–97. [DOI] [PubMed] [Google Scholar]
- Alisi A, Bedogni G, Baviera Get al. Randomised clinical trial: The beneficial effects of VSL#3 in obese children with non-alcoholic steatohepatitis. Aliment Pharmacol Ther. 2014;39:1276–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkanani AK, Hara N, Gottlieb PAet al. Alterations in intestinal microbiota correlate with susceptibility to type 1 diabetes. Diabetes. 2015;64:3510–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen EE, Banfield JF. Community genomics in microbial ecology and evolution. Nat Rev Microbiol. 2005;3:489–98. [DOI] [PubMed] [Google Scholar]
- Almeida A, Mitchell AL, Boland Met al. A new genomic blueprint of the human gut microbiota. Nature. 2019;568:499–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida M, Hébert A, Abraham ALet al. Construction of a dairy microbial genome catalog opens new perspectives for the metagenomic analysis of dairy fermented products. BMC Genomics. 2014;15:1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almonacid DE, Kraal L, Ossandon FJet al. 16S rRNA gene sequencing and healthy reference ranges for 28 clinically relevant microbial taxa from the human gut microbiome. PLoS One. 2017;12:e0176555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alneberg J, Bjarnason BS, de Bruijn Iet al. Binning metagenomic contigs by coverage and composition. Nat Methods. 2014;11:1144–6. [DOI] [PubMed] [Google Scholar]
- Amor DR, Ratzke C, Gore J. Transient invaders can induce shifts between alternative stable states of microbial communities. bioRxiv. 2019:659052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An S-Y, Lee MS, Jeon JYet al. Beneficial effects of fresh and fermented kimchi in prediabetic individuals. Ann Nutr Metab. 2013;63:111–9. [DOI] [PubMed] [Google Scholar]
- Araya M, Morelli L, Reid Get al. Guidelines for the evaluation of probiotics in food. In: Joint FAO/WHO Working Group Report on Drafting Guidelines for the Evaluation of Probiotics in Food. 2002;1–11. [Google Scholar]
- Arnbjerg CJ, Vestad B, Hov JRet al. Effect of Lactobacillus rhamnosus GG supplementation on intestinal inflammation assessed by PET/MRI scans and gut microbiota composition in HIV-infected individuals. J Acquir Immune Defic Syndr. 2018;78:450–7. [DOI] [PubMed] [Google Scholar]
- Asemi Z, Samimi M, Tabassi Zet al. Effect of daily consumption of probiotic yoghurt on insulin resistance in pregnant women: a randomized controlled trial. Eur J Clin Nutr. 2013;67:71–4. [DOI] [PubMed] [Google Scholar]
- Aslam H, Green J, Jacka FNet al. Fermented foods, the gut and mental health: a mechanistic overview with implications for depression and anxiety. Nutr Neurosci. 2018:1–13. [DOI] [PubMed] [Google Scholar]
- Baars T. The impact of raw fermented milk products on perceived health and mood among Dutch adults. Nutrition & Food Science. 2019;49:1195–206. [Google Scholar]
- Babio N, Becerra-Tomás N, Martínez-González MÁet al. Consumption of yogurt, low-fat milk, and other low-fat dairy products is associated with lower risk of metabolic syndrome incidence in an elderly Mediterranean population. J Nutr. 2015;145:2308–16. [DOI] [PubMed] [Google Scholar]
- Bagga D, Aigner CS, Reichert JLet al. Influence of 4-week multi-strain probiotic administration on resting-state functional connectivity in healthy volunteers. Eur J Nutr. 2019;58:1821–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri F, Montanari C, Gardini Fet al. Biogenic amine production by Lactic Acid Bacteria: a review. Foods. 2019;8:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto FM, Colado Simão AN, Morimoto HKet al. Beneficial effects of Lactobacillus plantarum on glycemia and homocysteine levels in postmenopausal women with metabolic syndrome. Nutrition. 2014;30:939–42. [DOI] [PubMed] [Google Scholar]
- Bautista-Gallego J, Ferrocino I, Botta Cet al. Probiotic potential of a Lactobacillus rhamnosus cheese isolate and its effect on the fecal microbiota of healthy volunteers. Food Res Int. 2019;119:305–14. [DOI] [PubMed] [Google Scholar]
- Begley M, Hill C, Gahan CGM. Bile salt hydrolase activity in probiotics. Appl Environ Microbiol. 2006;72:1729–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell V, Ferrao J, Pimentel Let al. One health, fermented foods, and gut microbiota. Foods. 2018;7:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berni Canani R, De Filippis F, Nocerino Ret al. Specific signatures of the gut microbiota and increased levels of butyrate in children treated with fermented cow's milk containing heat-killed Lactobacillus paracasei CBA L74. Appl Environ Microbiol. 2017;83:e01206–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berni Canani R, Sangwan N, Stefka ATet al. Lactobacillus rhamnosus GG-supplemented formula expands butyrate-producing bacterial strains in food allergic infants. ISME J. 2016;10:742–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich NA, Lewis ZT, Boundy-Mills Ket al. A new perspective on microbial landscapes within food production. Curr Opin Biotechnol. 2016;37:182–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin A, Quinquis B, Renault Pet al. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat Biotechnol. 2004;22:1554–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin A, Wincker P, Mauger Set al. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 2001;11:731–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E, Rideout JR, Dillon MRet al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordalo Tonucci L, Olbrich Dos Santos KM, Licursi de Oliveira Let al. Clinical application of probiotics in type 2 diabetes mellitus: a randomized, double-blind, placebo-controlled study. Clin Nutr. 2017;36:85–92. [DOI] [PubMed] [Google Scholar]
- Borges NA, Carmo FL, Stockler-Pinto MBet al. Probiotic supplementation in chronic kidney disease: a double-blind, randomized, placebo-controlled trial. J Ren Nutr. 2018;28:28–36. [DOI] [PubMed] [Google Scholar]
- Bourdichon F, Laulund S, Tenning P. Inventory of microbial species with a rationale: a comparison of the IDF/EFFCA inventory of microbial food cultures with the EFSA Biohazard Panel qualified presumption of safety. FEMS Microbiol Lett. 2019;366:fnz048. [DOI] [PubMed] [Google Scholar]
- Broadbent JR, Neeno-Eckwall EC, Stahl Bet al. Analysis of the Lactobacillus casei supragenome and its influence in species evolution and lifestyle adaptation. BMC Genomics. 2012;13:533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruzzese E, Callegari ML, Raia Vet al. Disrupted intestinal microbiota and intestinal inflammation in children with cystic fibrosis and its restoration with Lactobacillus GG: a randomised clinical trial. PLoS One. 2014;9:e87796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham JF. Scopus database: a review. Biomed Digit Libr. 2006;3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns P, Cuffia F, Milesi Met al. Technological and probiotic role of adjunct cultures of non-starter lactobacilli in soft cheeses. Food Microbiol. 2012;30:45–50. [DOI] [PubMed] [Google Scholar]
- Byun M-S, Yu O-K, Cha Y-Set al. Korean traditional Chungkookjang improves body composition, lipid profiles and atherogenic indices in overweight/obese subjects: a double-blind, randomized, crossover, placebo-controlled clinical trial. Eur J Clin Nutr. 2016;70:1116–22. [DOI] [PubMed] [Google Scholar]
- Bäckhed F, Fraser CM, Ringel Yet al. Defining a healthy human gut microbiome: current concepts, future directions, and clinical applications. Cell Host Microbe. 2012;12:611–22. [DOI] [PubMed] [Google Scholar]
- Bäckhed F, Ley RE, Sonnenburg JLet al. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–20. [DOI] [PubMed] [Google Scholar]
- Cai H, Rodriguez BT, Zhang Wet al. Genotypic and phenotypic characterization of Lactobacillus casei strains isolated from different ecological niches suggests frequent recombination and niche specificity. Microbiology. 2007;153:2655–65. [DOI] [PubMed] [Google Scholar]
- Cai H, Thompson R, Budinich MFet al. Genome sequence and comparative genome analysis of Lactobacillus casei: insights into their niche-associated evolution. Genome Biol Evol. 2009;1:239–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJet al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway LK, McIntyre HD, Barrett HLet al. Probiotics for the prevention of gestational diabetes mellitus in overweight and obese women: Findings from the SPRING double-blind randomized controlled trial. Diabetes Care. 2019;42:364–71. [DOI] [PubMed] [Google Scholar]
- Campedelli I, Mathur H, Salvetti Eet al. Genus-wide assessment of antibiotic resistance in Lactobacillus spp. Appl Environ Microbiol. 2019;85:e01738–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani PD. Human gut microbiome: hopes, threats and promises. Gut. 2018;67:1716–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Bravo N, Wells JM, Margolles Aet al. Interactions of surface exopolysaccharides from Bifidobacterium and Lactobacillus within the intestinal environment. Front Microbiol. 2018;9:2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallini DCU, Manzoni MSJ, Bedani Ret al. Probiotic soy product supplemented with isoflavones improves the lipid profile of moderately hypercholesterolemic men: a randomized controlled trial. Nutrients. 2016;8:E52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceapa C, Davids M, Ritari Jet al. The variable regions of Lactobacillus rhamnosus genomes reveal the dynamic evolution of metabolic and host-adaptation repertoires. Genome Biol Evol. 2016;8:1889–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceapa C, Lambert J, van Limpt Ket al. Correlation of Lactobacillus rhamnosus genotypes and carbohydrate utilization signatures determined by phenotype profiling. Appl Environ Microbiol. 2015;81:5458–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli G, Brenchley JM, Cavallari ENet al. Impact of high-dose multi-strain probiotic supplementation on neurocognitive performance and central nervous system immune activation of HIV-1 infected individuals. Nutrients. 2017;9:E1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahwan B, Kwan S, Isik Aet al. Gut feelings: A randomised, triple-blind, placebo-controlled trial of probiotics for depressive symptoms. J Affect Disord. 2019;253:317–26. [DOI] [PubMed] [Google Scholar]
- Chen J, Chia N, Kalari KRet al. Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci Rep. 2016;6:28484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Sun Q, Giovannucci Eet al. Dairy consumption and risk of type 2 diabetes: 3 cohorts of US adults and an updated meta-analysis. BMC Med. 2014;12:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikindas ML, Weeks R, Drider Det al. Functions and emerging applications of bacteriocins. Curr Opin Biotechnol. 2018;49:23–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilton SN, Burton JP, Reid G. Inclusion of fermented foods in food guides around the world. Nutrients. 2015;7:390–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi IH, Noh JS, Han J-Set al. Kimchi, a fermented vegetable, improves serum lipid profiles in healthy young adults: randomized clinical trial. J Med Food. 2013;16:223–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Fish JAet al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:D633–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordain L, Eaton SB, Sebastian Aet al. Origins and evolution of the Western diet: health implications for the 21st century. Am J Clin Nutr. 2005;81:341–54. [DOI] [PubMed] [Google Scholar]
- Corr SC, Li Y, Riedel CUet al. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc Natl Acad Sci USA. 2007;104:7617–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrêa FB, Saraiva JP, Stadler PFet al. TerrestrialMetagenomeDB: a public repository of curated and standardized metadata for terrestrial metagenomes. Nucleic Acids Res. 2020;48:D626–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsello G, Carta M, Marinello Ret al. Preventive effect of cow's milk fermented with Lactobacillus paracasei CBA L74 on common infectious diseases in children: a multicenter randomized controlled trial. Nutrients. 2017;9:E669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costabile A, Buttarazzi I, Kolida Set al. An in vivo assessment of the cholesterol-lowering efficacy of Lactobacillus plantarum ECGC 13110402 in normal to mildly hypercholesterolaemic adults. PLoS One. 2017;12:e0187964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costea PI, Munch R, Coelho LPet al. metaSNV: A tool for metagenomic strain level analysis. PLoS One. 2017;12:e0182392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotillard A, Kennedy SP, Kong LCet al. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500:585–8. [DOI] [PubMed] [Google Scholar]
- Cox MJ, Huang YJ, Fujimura KEet al. Lactobacillus casei abundance is associated with profound shifts in the infant gut microbiome. PLoS One. 2010;5:e8745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremon C, Guglielmetti S, Gargari Get al. Effect of Lactobacillus paracasei CNCM I-1572 on symptoms, gut microbiota, short chain fatty acids, and immune activation in patients with irritable bowel syndrome: A pilot randomized clinical trial. United European Gastroenterol J. 2018;6:604–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui S, Hu Y. Multistrain probiotic preparation significantly reduces symptoms of irritable bowel syndrome in a double-blind placebo-controlled study. Int J Clin Exp Med. 2012;5:238–44. [PMC free article] [PubMed] [Google Scholar]
- Dabour N, Zihler A, Kheadr Eet al. In vivo study on the effectiveness of pediocin PA-1 and Pediococcus acidilactici at inhibiting Listeria monocytogenes. Int J Food Microbiol. 2009;133:225–33. [DOI] [PubMed] [Google Scholar]
- Dal Bello F, Walter J, Hammes WPet al. Increased complexity of the species composition of lactic acid bacteria in human feces revealed by alternative incubation condition. Microb Ecol. 2003;45:455–63. [DOI] [PubMed] [Google Scholar]
- Daniel C, Poiret S, Goudercourt Det al. Selecting lactic acid bacteria for their safety and functionality by use of a mouse colitis model. Appl Environ Microbiol. 2006;72:5799–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RNet al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Andrés J, Manzano S, García Cet al. Modulatory effect of three probiotic strains on infants' gut microbial composition and immunological parameters on a placebo-controlled, double-blind, randomised study. Benef Microbes. 2018;9:573–84. [DOI] [PubMed] [Google Scholar]
- De Filippis F, Genovese A, Ferranti Pet al. Metatranscriptomics reveals temperature-driven functional changes in microbiome impacting cheese maturation rate. Sci Rep. 2016;6:21871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippis F, Parente E, Ercolini D. Metagenomics insights into food fermentations. Microb Biotechnol. 2017;10:91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippis F, Parente E, Ercolini D. Recent past, present, and future of the food microbiome. Annu Rev Food Sci Technol. 2018b;9:589–608. [DOI] [PubMed] [Google Scholar]
- De Filippis F, Vitaglione P, Cuomo Ret al. Dietary interventions to modulate the gut microbiome—how far away are we from precision medicine. Inflamm Bowel Dis. 2018a;24:2142–54. [DOI] [PubMed] [Google Scholar]
- de Goffau MC, Fuentes S, van den Bogert Bet al. Aberrant gut microbiota composition at the onset of type 1 diabetes in young children. Diabetologia. 2014;57:1569–77. [DOI] [PubMed] [Google Scholar]
- del Campo R, Garriga M, Pérez-Aragón Aet al. Improvement of digestive health and reduction in proteobacterial populations in the gut microbiota of cystic fibrosis patients using a Lactobacillus reuteri probiotic preparation: a double blind prospective study. J Cyst Fibros. 2014;13:716–22. [DOI] [PubMed] [Google Scholar]
- Deo D, Davray D, Kulkarni R. A diverse repertoire of exopolysaccharide biosynthesis gene clusters in Lactobacillus revealed by comparative analysis in 106 sequenced genomes. Microoorganisms. 2019;7:444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M, van Hylckama Vlieg JET. Fate, activity, and impact of ingested bacteria within the human gut microbiota. Trends Microbiol. 2015;23:354–66. [DOI] [PubMed] [Google Scholar]
- Dhiman RK, Rana B, Agrawal Set al. Probiotic VSL#3 reduces liver disease severity and hospitalization in patients with cirrhosis: a randomized, controlled trial. Gastroenterology. 2014;147:1327–37. [DOI] [PubMed] [Google Scholar]
- Dimidi E, Cox SR, Rossi Met al. Fermented foods: definitions and characteristics, impact on the gut microbiota and effects on gastrointestinal health and disease. Nutrients. 2019;11:E1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Monaco R, Torrieri E, Pepe Oet al. Effect of sourdough with exopolysaccharide (EPS)-producing lactic acid bacteria (LAB) on sensory quality of bread during shelf life. Food Bioprocess Technol. 2015;8:691–701. [Google Scholar]
- Dobson A, Cotter PD, Ross RPet al. Bacteriocin production: a probiotic trait? Appl Environ Microbiol. 2012;78:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson A, Crispie F, Rea MCet al. Fate and efficacy of lacticin 3147-producing Lactococcus lactis in the mammalian gastrointestinal tract. FEMS Microbiol Ecol. 2011;76:602–14. [DOI] [PubMed] [Google Scholar]
- Douillard FP, de Vos WM. Biotechnology of health-promoting bacteria. Biotechnol Adv. 2019;37:107369. [DOI] [PubMed] [Google Scholar]
- Drissi F, Merhej V, Angelakis Eet al. Comparative genomics analysis of Lactobacillus species associated with weight gain or weight protection. Nutr Diabetes. 2014;4:e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duar RM, Frese SA, Lin XBet al. Experimental evaluation of host adaptation of Lactobacillus reuteri to different vertebrate species. Appl Environ Microbiol. 2017a;83:e00132–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duar RM, Lin XB, Zheng Jet al. Lifestyles in transition: evolution and natural history of the genus Lactobacillus. FEMS Microbiol Rev. 2017b;41:S27–48. [DOI] [PubMed] [Google Scholar]
- Duvallet C, Gibbons SM, Gurry Tet al. Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat Commun. 2017;8:1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-López A, Bulló M, Martínez-González MAet al. Dairy product consumption and risk of type 2 diabetes in an elderly Spanish Mediterranean population at high cardiovascular risk. Eur J Nutr. 2016;55:349–60. [DOI] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CNet al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejtahed HS, Mohtadi-Nia J, Homayouni-Rad Aet al. Probiotic yogurt improves antioxidant status in type 2 diabetic patients. Nutrition. 2012;28:539–43. [DOI] [PubMed] [Google Scholar]
- El Aidy S, van den Bogert B, Kleerebezem M. The small intestine microbiota, nutritional modulation and relevance for health. Curr Opin Biotechnol. 2015;32:14–20. [DOI] [PubMed] [Google Scholar]
- El Manouni El Hassani S, de Boer NKH, Jansen FMet al. Effect of daily intake of Lactobacillus casei on microbial diversity and dynamics in a healthy pediatric population. Curr Microbiol. 2019;76:1020–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercolini D. High-throughput sequencing and metagenomics: moving forward in the culture-independent analysis of food microbial ecology. Appl Environ Microbiol. 2013;79:3148–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Maignien L, Sul WJet al. Oligotyping: Differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol Evol. 2013;4, DOI: 10.1111/2041-210X.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eussen SJPM, van Dongen MCJM, Wijckmans Net al. Consumption of dairy foods in relation to impaired glucose metabolism and type 2 diabetes mellitus: the Maastricht Study. Br J Nutr. 2016;115:1453–61. [DOI] [PubMed] [Google Scholar]
- Fang Z, Lu W, Zhao Jet al. Probiotics modulate the gut microbiota composition and immune responses in patients with atopic dermatitis: a pilot study. Eur J Nutr. 2019; [Epub ahead of print] DOI:10.1007/s00394-019-02061-x. [DOI] [PubMed] [Google Scholar]
- Farup PG, Jacobsen M, Ligaarden SCet al. Probiotics, symptoms, and gut microbiota: what are the relations? A randomized controlled trial in subjects with irritable bowel syndrome. Gastroenterol Res Pract. 2012;2012:214102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez MA, Marette A. Novel perspectives on fermented milks and cardiometabolic health with a focus on type 2 diabetes. Nutr Rev. 2018;76:16–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrario C, Taverniti V, Milani Cet al. Modulation of fecal Clostridiales bacteria and butyrate by probiotic intervention with Lactobacillus paracasei DG varies among healthy adults. J Nutr. 2014;144:1787–96. [DOI] [PubMed] [Google Scholar]
- Firouzi S, Majid HA, Ismail Aet al. Effect of multi-strain probiotics (multi-strain microbial cell preparation) on glycemic control and other diabetes-related outcomes in people with type 2 diabetes: a randomized controlled trial. Eur J Nutr. 2017;56:1535–50. [DOI] [PubMed] [Google Scholar]
- Flint HJ. The significance of prokaryote diversity in the human gastrointestinal tract. In: Logan NA, Lappin-Scott HM, Oyston PCF (eds). Prokaryotic diversity: Mechanisms and significance, Cambridge (UK): Cambridge University Press, 2006, 65–90. [Google Scholar]
- Forslund K, Hildebrand F, Nielsen Tet al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528:262–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francavilla R, Piccolo M, Francavilla Aet al. Clinical and microbiological effect of a multispecies probiotic supplementation in celiac patients with persistent IBS-type symptoms: a randomized, double-blind, placebo-controlled, multicenter trial. J Clin Gastroenterol. 2019;53:e117–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DN, St. Amand AL, Feldman RAet al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci. 2007;104:13780–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa EA, McIver LJ, Rahnavard Get al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat Methods. 2018;15:962–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frese SA, Hutkins RW, Walter J. Comparison of the colonization ability of autochthonous and allochthonous strains of lactobacilli in the human gastrointestinal tract. Advances in Microbiology. 2012;2:399. [Google Scholar]
- Galle S, Arendt EK. Exopolysaccharides from sourdough lactic acid bacteria. Crit Rev Food Sci Nutr. 2014;54:891–901. [DOI] [PubMed] [Google Scholar]
- George F, Daniel C, Thomas Met al. Occurrence and dynamism of lactic acid bacteria in distinct ecological niches: A Multifaceted Functional Health Perspective. Front Microbiol. 2018;9:2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson M, Bishop NC, Oliveira Met al. Effects of a Lactobacillus salivarius probiotic intervention on infection, cold symptom duration and severity, and mucosal immunity in endurance athletes. Int J Sport Nutr Exerc Metab. 2012;22:235–42. [DOI] [PubMed] [Google Scholar]
- Global Market Insight . Probiotics market growth - Industry size, share research 2018–2024. 2018. [Google Scholar]
- Gonzalez A, Navas-Molina JA, Kosciolek Tet al. Qiita: rapid, web-enabled microbiome meta-analysis. Nat Methods. 2018;15:796–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González S, Fernández-Navarro T, Arboleya Set al. Fermented dairy foods: impact on intestinal microbiota and health-linked biomarkers. Front Microbiol. 2019;10:1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17:333–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotteland M, Andrews M, Toledo Met al. Modulation of Helicobacter pylori colonization with cranberry juice and Lactobacillus johnsonii La1 in children. Nutrition. 2008;24:421–6. [DOI] [PubMed] [Google Scholar]
- Grangette C, Nutten S, Palumbo Eet al. Enhanced antiinflammatory capacity of a Lactobacillus plantarum mutant synthesizing modified teichoic acids. Proc Natl Acad Sci USA. 2005;102:10321–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarner F, Khan AG, Garish Jet al. World Gastroenterology Organisation Global Guidelines: Probiotics and Prebiotics October 2011. J Clin Gastroenterol. 2012;46:468–81. [DOI] [PubMed] [Google Scholar]
- Gänzle M, Ripari V. Composition and function of sourdough microbiota: From ecological theory to bread quality. Int J Food Microbiol. 2016;239:19–25. [DOI] [PubMed] [Google Scholar]
- Górska S, Sandstrőm C, Wojas-Turek Jet al. Structural and immunomodulatory differences among lactobacilli exopolysaccharides isolated from intestines of mice with experimentally induced inflammatory bowel disease. Sci Rep. 2016;6:37613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Knight R. Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome Res. 2009;19:1141–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmsen HJM, Raangs GC, He Tet al. Extensive set of 16S rRNA-based probes for detection of bacteria in human feces. Appl Environ Microbiol. 2002;68:2982–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H, Sakamoto M, Benno Y. Phylogenetic analysis of the human gut microbiota using 16S rDNA clone libraries and strictly anaerobic culture-based methods. Microbiol Immunol. 2002;46:535–48. [DOI] [PubMed] [Google Scholar]
- Hayashi H, Takahashi R, Nishi Tet al. Molecular analysis of jejunal, ileal, caecal and recto-sigmoidal human colonic microbiota using 16S rRNA gene libraries and terminal restriction fragment length polymorphism. J Med Microbiol. 2005;54:1093–101. [DOI] [PubMed] [Google Scholar]
- Heeney DD, Gareau MG, Marco ML. Intestinal Lactobacillus in health and disease, a driver or just along for the ride? Curr Opin Biotechnol. 2018;49:140–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C, Guarner F, Reid Get al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014;11:506–14. [DOI] [PubMed] [Google Scholar]
- Hold GL, Pryde SE, Russell VJet al. Assessment of microbial diversity in human colonic samples by 16S rDNA sequence analysis. FEMS Microbiol Ecol. 2002;39:33–9. [DOI] [PubMed] [Google Scholar]
- Holland R, Liu S-Q. Lactic Acid Bacteria: Leuconostoc spp. In: Fox PF, McSweeney PLH (eds). Encyclopedia of Dairy Science (Second Edition), United States: Elsevier Academic Press, 2011, 138–42. [Google Scholar]
- Holmes E, Li JV, Marchesi JRet al. Gut microbiota composition and activity in relation to host metabolic phenotype and disease risk. Cell Metab. 2012;16:559–64. [DOI] [PubMed] [Google Scholar]
- Horvath A, Leber B, Feldbacher Net al. Effects of a multispecies synbiotic on glucose metabolism, lipid marker, gut microbiome composition, gut permeability, and quality of life in diabesity: a randomized, double-blind, placebo-controlled pilot study. Eur J Nutr. 2019; [Epub ahead of print] DOI:10.1007/s00394-019-02135-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W, Carey VJ, Gentleman Ret al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 2015;12:115–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium . Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignacio A, Fernandes MR, Rodrigues VAAet al. Correlation between body mass index and faecal microbiota from children. Clin Microbiol Infect. 2016;22:258.e1–8. [DOI] [PubMed] [Google Scholar]
- Ivory K, Chambers SJ, Pin Cet al. Oral delivery of Lactobacillus casei Shirota modifies allergen-induced immune responses in allergic rhinitis. Clin Exp Allergy. 2008;38:1282–9. [DOI] [PubMed] [Google Scholar]
- Iwasa M, Aoi W, Mune Ket al. Fermented milk improves glucose metabolism in exercise-induced muscle damage in young healthy men. Nutr J. 2013;12:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RB, Alderete TL, Martin AAet al. Probiotic supplementation increases obesity with no detectable effects on liver fat or gut microbiota in obese Hispanic adolescents: a 16-week, randomized, placebo-controlled trial. Pediatr Obes. 2018;13:705–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajander K, Krogius-Kurikka L, Rinttilä Tet al. Effects of multispecies probiotic supplementation on intestinal microbiota in irritable bowel syndrome. Aliment Pharmacol Ther. 2007;26:463–73. [DOI] [PubMed] [Google Scholar]
- Kang DD, Li F, Kirton Eet al. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ. 2019;7:e7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson FH, Tremaroli V, Nookaew Iet al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99–103. [DOI] [PubMed] [Google Scholar]
- Kassaian N, Feizi A, Aminorroaya Aet al. Probiotic and synbiotic supplementation could improve metabolic syndrome in prediabetic adults: A randomized controlled trial. Diabetes Metab Syndr. 2019;13:2991–6. [DOI] [PubMed] [Google Scholar]
- Kato-Kataoka A, Nishida K, Takada Met al. Fermented milk containing Lactobacillus casei strain Shirota preserves the diversity of the gut microbiota and relieves abdominal dysfunction in healthy medical students exposed to academic stress. Appl Environ Microbiol. 2016;82:3649–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazemi A, Noorbala AA, Azam Ket al. Effect of probiotic and prebiotic vs placebo on psychological outcomes in patients with major depressive disorder: A randomized clinical trial. Clin Nutr. 2019;38:522–8. [DOI] [PubMed] [Google Scholar]
- Kelishadi R, Farajian S, Safavi Met al. A randomized triple-masked controlled trial on the effects of synbiotics on inflammation markers in overweight children. J Pediatr (Rio J). 2014;90:161–8. [DOI] [PubMed] [Google Scholar]
- Kianifar H, Ahanchian H, Grover Zet al. Synbiotic in the management of infantile colic: a randomised controlled trial. J Paediatr Child Health. 2014;50:801–5. [DOI] [PubMed] [Google Scholar]
- Kim EK, An S-Y, Lee M-Set al. Fermented kimchi reduces body weight and improves metabolic parameters in overweight and obese patients. Nutr Res. 2011;31:436–43. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Ju S-Y, Park YK. Kimchi intake and atopic dermatitis in Korean aged 19–49 years: The Korea National Health and Nutrition Examination Survey 2010–2012. Asia Pac J Clin Nutr. 2017;26:914–22. [DOI] [PubMed] [Google Scholar]
- Kim J-H, Kim K, Kim W. Cream cheese-derived Lactococcus chungangensis CAU 28 modulates the gut microbiota and alleviates atopic dermatitis in BALB/c mice. Sci Rep. 2019;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitts PA, Church DM, Thibaud-Nissen Fet al. Assembly: a resource for assembled genomes at NCBI. Nucleic Acids Res. 2016;44:D73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobyliak N, Abenavoli L, Mykhalchyshyn Get al. A multi-strain probiotic reduces the fatty liver index, cytokines and aminotransferase levels in NAFLD patients: Evidence from a randomized clinical trial. J Gastrointestin Liver Dis. 2018. a;27:41–9. [DOI] [PubMed] [Google Scholar]
- Kobyliak N, Falalyeyeva T, Mykhalchyshyn Get al. Effect of alive probiotic on insulin resistance in type 2 diabetes patients: Randomized clinical trial. Diabetes Metab Syndr. 2018b;12:617–24. [DOI] [PubMed] [Google Scholar]
- Kodama Y, Shumway M, Leinonen Ret al. The Sequence Read Archive: explosive growth of sequencing data. Nucleic Acids Res. 2012;40:D54–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok CR, Hutkins R. Yogurt and other fermented foods as sources of health-promoting bacteria. Nutr Rev. 2018;76:4–15. [DOI] [PubMed] [Google Scholar]
- Konstantinov SR, Smidt H, de Vos WMet al. S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc Natl Acad Sci USA. 2008;105:19474–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukkonen K, Savilahti E, Haahtela Tet al. Probiotics and prebiotic galacto-oligosaccharides in the prevention of allergic diseases: a randomized, double-blind, placebo-controlled trial. J Allergy Clin Immunol. 2007;119:192–8. [DOI] [PubMed] [Google Scholar]
- Laatikainen R, Koskenpato J, Hongisto S-Met al. Randomised clinical trial: low-FODMAP rye bread vs. regular rye bread to relieve the symptoms of irritable bowel syndrome. Aliment Pharmacol Ther. 2016;44:460–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagkouvardos I, Joseph D, Kapfhammer Met al. IMNGS: A comprehensive open resource of processed 16S rRNA microbial profiles for ecology and diversity studies. Sci Rep. 2016;6:33721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahti L, Salonen A, Kekkonen RAet al. Associations between the human intestinal microbiota, Lactobacillus rhamnosus GG and serum lipids indicated by integrated analysis of high-throughput profiling data. PeerJ. 2013;1:e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai HH, Chiu CH, Kong MSet al. Probiotic Lactobacillus casei: effective for managing childhood diarrhea by altering gut microbiota and attenuating fecal inflammatory markers. Nutrients. 2019;11:E1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille MGI, Zaneveld J, Caporaso JGet al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang JM, Eisen JA, Zivkovic AM. The microbes we eat: abundance and taxonomy of microbes consumed in a day's worth of meals for three diet types. PeerJ. 2014;2:e659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen N, Vogensen FK, Gøbel Ret al. Predominant genera of fecal microbiota in children with atopic dermatitis are not altered by intake of probiotic bacteria Lactobacillus acidophilus NCFM and Bifidobacterium animalis subsp. lactis Bi-07. FEMS Microbiol Ecol. 2011;75:482–96. [DOI] [PubMed] [Google Scholar]
- Larsen N, Vogensen FK, van den Berg FWJet al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One. 2010;5:e9085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson SC, Andersson S-O, Johansson J-Eet al. Cultured milk, yogurt, and dairy intake in relation to bladder cancer risk in a prospective study of Swedish women and men. Am J Clin Nutr. 2008;88:1083–7. [DOI] [PubMed] [Google Scholar]
- Lebeer S, Claes I, Tytgat HLPet al. Functional analysis of Lactobacillus rhamnosus GG pili in relation to adhesion and immunomodulatory interactions with intestinal epithelial cells. Appl Environ Microbiol. 2012;78:185–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc JG, Laiño JE, Juarez del Valle Met al. B-Group vitamin production by lactic acid bacteria – current knowledge and potential applications. J Appl Microbiol. 2011;111:1297–309. [DOI] [PubMed] [Google Scholar]
- Le Blay G, Lacroix C, Zihler Aet al. In vitro inhibition activity of nisin A, nisin Z, pediocin PA-1 and antibiotics against common intestinal bacteria. Lett Appl Microbiol. 2007;45:252–7. [DOI] [PubMed] [Google Scholar]
- Leblhuber F, Steiner K, Schuetz Bet al. Probiotic supplementation in patients with Alzheimer's dementia - an explorative intervention study. Curr Alzheimer Res. 2018;15:1106–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Chatelier E, Nielsen T, Qin Jet al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–6. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Bose S, Seo JGet al. The effects of co-administration of probiotics with herbal medicine on obesity, metabolic endotoxemia and dysbiosis: a randomized double-blind controlled clinical trial. Clin Nutr. 2014;33:973–81. [DOI] [PubMed] [Google Scholar]
- Le Nevé B, Derrien M, Tap Jet al. Fasting breath H2 and gut microbiota metabolic potential are associated with the response to a fermented milk product in irritable bowel syndrome. PLoS One. 2019;14:e0214273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerche M. Isolierung und differenzierung anaerober Lactobacillaceae aus dem Darm erwachsener Menschen (Beitrag zum Lactobacillus bifidus-Problem). Zentralblatt fur Bakeriologie, Parasitenkunde, Infektionskrankheiten und Hygiene. 1961;180:324–56. [Google Scholar]
- Lewis JD, Chen EZ, Baldassano RNet al. Inflammation, Antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn's disease. Cell Host Microbe. 2017;22:247. [DOI] [PubMed] [Google Scholar]
- Ley RE, Bäckhed F, Turnbaugh Pet al. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Kwok L-Y, Mi Zet al. Characterization of the angiotensin-converting enzyme inhibitory activity of fermented milks produced with Lactobacillus casei. J. Dairy Sci. 2017;100:9495–507. [DOI] [PubMed] [Google Scholar]
- Li D, Liu C-M, Luo Ret al. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–6. [DOI] [PubMed] [Google Scholar]
- Lim J-H, Jung E-S, Choi E-Ket al. Supplementation with Aspergillus oryzae-fermented kochujang lowers serum cholesterol in subjects with hyperlipidemia. Clin Nutr. 2015;34:383–7. [DOI] [PubMed] [Google Scholar]
- Linares DM, Gómez C, Renes Eet al. Lactic acid bacteria and Bifidobacteria with potential to design natural biofunctional health-promoting dairy foods. Front Microbiol. 2017;8:846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H-N, Wu H, Chen Y-Zet al. Altered molecular signature of intestinal microbiota in irritable bowel syndrome patients compared with healthy controls: A systematic review and meta-analysis. Dig Liver Dis. 2017;49:331–7. [DOI] [PubMed] [Google Scholar]
- Liu YW, Liong MT, Chung YEet al. Effects of Lactobacillus plantarum PS128 on children with autism spectrum disorder in Taiwan: a randomized, double-blind, placebo-controlled trial. Nutrients. 2019;11:E820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis P, Scott KP, Duncan SHet al. Understanding the effects of diet on bacterial metabolism in the large intestine. J Appl Microbiol. 2007;102:1197–208. [DOI] [PubMed] [Google Scholar]
- Maldonado-Gómez MX, Martínez I, Bottacini Fet al. Stable engraftment of Bifidobacterium longum AH1206 in the human gut depends on individualized features of the resident microbiome. Cell Host Microbe. 2016;20:515–26. [DOI] [PubMed] [Google Scholar]
- Malik M, Suboc TM, Tyagi Set al. Lactobacillus plantarum 299v supplementation improves vascular endothelial function and reduces in inflammatory biomarkers in men with stable coronary artery disease. Circ Res. 2018;123:1091–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manara S, Asnicar F, Beghini Fet al. Microbial genomes from non-human primate gut metagenomes expand the primate-associated bacterial tree of life with over 1000 novel species. Genome Biol. 2019;20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco ML, Heeney D, Binda Set al. Health benefits of fermented foods: microbiota and beyond. Curr Opin Biotechnol. 2017;44:94–102. [DOI] [PubMed] [Google Scholar]
- Marlicz W, Wunsch E, Mydlowska Met al. The effect of short term treatment with probiotic VSL#3 on various clinical and biochemical parameters in patients with liver cirrhosis. J Physiol Pharmacol. 2016;67:867–77. [PubMed] [Google Scholar]
- Marteau P, Lémann M, Seksik Pet al. Ineffectiveness of Lactobacillus johnsonii LA1 for prophylaxis of postoperative recurrence in Crohn's disease: a randomised, double blind, placebo controlled GETAID trial. Gut. 2006;55:842–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino ME, Bayjanov JR, Caffrey BEet al. Nomadic lifestyle of Lactobacillus plantarum revealed by comparative genomics of 54 strains isolated from different habitats. Environ Microbiol. 2016;18:4974–89. [DOI] [PubMed] [Google Scholar]
- Martoni CJ, Labbé A, Ganopolsky JGet al. Changes in bile acids, FGF-19 and sterol absorption in response to bile salt hydrolase active L. reuteri NCIMB 30242. Gut Microbes. 2015;6:57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuliffe O, Kilcawley K, Stefanovic E. Symposium review: Genomic investigations of flavor formation by dairy microbiota. J Dairy Sci. 2019;102:909–22. [DOI] [PubMed] [Google Scholar]
- McBurney MI, Davis C, Fraser CMet al. Establishing what constitutes a healthy human gut microbiome: state of the science, regulatory considerations, and future directions. J Nutr. 2019;149:1882–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich Jet al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLoughlin R, Berthon BS, Rogers GBet al. Soluble fibre supplementation with and without a probiotic in adults with asthma: A 7-day randomised, double blind, three way cross-over trial. EBioMedicine. 2019;46:473–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meola M, Rifa E, Shani Net al. DAIRYdb: a manually curated reference database for improved taxonomy annotation of 16S rRNA gene sequences from dairy products. BMC Genomics. 2019;20:560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MetaSUB International Consortium . The Metagenomics and Metadesign of the Subways and Urban Biomes (MetaSUB) International Consortium inaugural meeting report. Microbiome. 2016;4:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezzasalma V, Manfrini E, Ferri Eet al. A randomized, double-blind, placebo-controlled trial: the efficacy of multispecies probiotic supplementation in alleviating symptoms of irritable bowel syndrome associated with constipation. Biomed Res Int. 2016;2016:4740907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michail S, Kenche H. Gut microbiota is not modified by randomized, double-blind, placebo-controlled trial of VSL#3 in diarrhea-predominant irritable bowel syndrome. Probiotics Antimicrob Proteins. 2011;3:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignolet J, Fontaine L, Kleerebezem Met al. Complete genome sequence of Streptococcus salivarius HSISS4, a human commensal bacterium highly prevalent in the digestive tract. Genome Announc. 2016;4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell AL, Almeida A, Beracochea Met al. MGnify: the microbiome analysis resource in 2020. Nucleic Acids Res. 2020;48:570–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuoka T. Comparative studies on lactobacilli from the faeces of man, swine and chickens. Zentralbl Bakteriol Orig. 1969;210:32–51. [PubMed] [Google Scholar]
- Mitsuoka T. The Human Gastrointestinal Tract. In: Wood BJB (ed.). The Lactic Acid Bacteria Volume 1: The Lactic Acid Bacteria in Health and Disease. Boston, MA: Springer US, 1992, 69–114. [Google Scholar]
- Mobini R, Tremaroli V, Ståhlman Met al. Metabolic effects of Lactobacillus reuteri DSM 17938 in people with type 2 diabetes: A randomized controlled trial. Diabetes Obes Metab. 2017;19:579–89. [DOI] [PubMed] [Google Scholar]
- Montel MC, Buchin S, Mallet Aet al. Traditional cheeses: rich and diverse microbiota with associated benefits. Int J Food Microbiol. 2014;177:136–54. [DOI] [PubMed] [Google Scholar]
- Mora D, Arioli S. Microbial urease in health and disease. PLoS Pathog. 2014;10:e1004472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroti C, Souza Magri LF, de Rezende Costa Met al. Effect of the consumption of a new symbiotic shake on glycemia and cholesterol levels in elderly people with type 2 diabetes mellitus. Lipids Health Dis. 2012;11:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mota de Carvalho N, Costa EM, Silva Set al. Fermented foods and beverages in human diet and their influence on gut microbiota and health. Fermentation. 2018;4:90. [Google Scholar]
- Mozaffarian D, Hao T, Rimm EBet al. Changes in diet and lifestyle and long-term weight gain in women and men. N Engl J Med. 2011;364:2392–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménard S, Candalh C, Bambou JCet al. Lactic acid bacteria secrete metabolites retaining anti-inflammatory properties after intestinal transport. Gut. 2004;53:821–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabavi S, Rafraf M, Somi M-Het al. Probiotic yogurt improves body mass index and fasting insulin levels without affecting serum leptin and adiponectin levels in non-alcoholic fatty liver disease (NAFLD). J Funct Foods. 2015;18:684–91. [Google Scholar]
- Nabhani Z, Hezaveh SJG, Razmpoosh Eet al. The effects of synbiotic supplementation on insulin resistance/sensitivity, lipid profile and total antioxidant capacity in women with gestational diabetes mellitus: A randomized double blind placebo controlled clinical trial. Diabetes Res Clin Pract. 2018;138:149–57. [DOI] [PubMed] [Google Scholar]
- Nagata S, Asahara T, Wang Cet al. The effectiveness of Lactobacillus beverages in controlling infections among the residents of an aged care facility: A randomized placebo-controlled double-blind trial. Ann Nutr Metab. 2016;68:51–9. [DOI] [PubMed] [Google Scholar]
- Nampoothiri KM, Beena DJ, Vasanthakumari DSet al. Health benefits of exopolysaccharides in fermented foods. In: Frias J, Martinez-Villaluenga C, Peñas E (eds.). Fermented Foods in Health and Disease Prevention. United States:Elsevier Academic Press, 2017, 49–62. [Google Scholar]
- Nation ML, Dunne EM, Joseph SJet al. Impact of Lactobacillus reuteri colonization on gut microbiota, inflammation, and crying time in infant colic. Sci Rep. 2017;7:15047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayfach S, Rodriguez-Mueller B, Garud Net al. An integrated metagenomics pipeline for strain profiling reveals novel patterns of bacterial transmission and biogeography. Genome Res. 2016;26:1612–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayfach S, Shi ZJ, Seshadri Ret al. New insights from uncultivated genomes of the global human gut microbiome. Nature. 2019;568:505–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SC, Lam EF, Lam TTet al. Effect of probiotic bacteria on the intestinal microbiota in irritable bowel syndrome. J Gastroenterol Hepatol. 2013;28:1624–31. [DOI] [PubMed] [Google Scholar]
- Nielsen ES, Garnås E, Jensen KJet al. Lacto-fermented sauerkraut improves symptoms in IBS patients independent of product pasteurisation-a pilot study. Food Funct. 2018;9:5323–35. [DOI] [PubMed] [Google Scholar]
- Nilsson AG, Sundh D, Bäckhed Fet al. Lactobacillus reuteri reduces bone loss in older women with low bone mineral density: a randomized, placebo-controlled, double-blind, clinical trial. J Intern Med. 2018;284:307–17. [DOI] [PubMed] [Google Scholar]
- Nishida K, Sawada D, Kuwano Yet al. Health benefits of Lactobacillus gasseri CP2305 tablets in young adults exposed to chronic stress: a randomized, double-blind, placebo-controlled study. Nutrients. 2019;11:E1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nistal E, Caminero A, Herran ARet al. Study of duodenal bacterial communities by 16S rRNA gene analysis in adults with active celiac disease vs non-celiac disease controls. J Appl Microbiol. 2016;120:1691–700. [DOI] [PubMed] [Google Scholar]
- Nout MJR. Food technologies: fermentation. Encyclopedia of Food Safety. 2014;3: 168–77. [Google Scholar]
- Nozue M, Shimazu T, Sasazuki Set al. Fermented soy product intake is inversely associated with the development of high blood pressure: The Japan public health center-based prospective study. J Nutr. 2017;147:1749–56. [DOI] [PubMed] [Google Scholar]
- Nurk S, Meleshko D, Korobeynikov Aet al. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 2017;27:824–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Callaghan J, O'Toole PW. Lactobacillus: Host–Microbe Relationships. Curr Top Microbiol Immunol. 2013;358:119–54. [DOI] [PubMed] [Google Scholar]
- O'Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivares M, Díaz-Ropero MA, Gómez Net al. Oral administration of two probiotic strains, Lactobacillus gasseri CECT5714 and Lactobacillus coryniformis CECT5711, enhances the intestinal function of healthy adults. Int J Food Microbiol. 2006;107:104–11. [DOI] [PubMed] [Google Scholar]
- Pala V, Sieri S, Berrino Fet al. Yogurt consumption and risk of colorectal cancer in the Italian European prospective investigation into cancer and nutrition cohort. Int J Cancer. 2011;129:2712–9. [DOI] [PubMed] [Google Scholar]
- Panigrahi P, Parida S, Nanda NCet al. A randomized synbiotic trial to prevent sepsis among infants in rural India. Nature. 2017;548:407–12. [DOI] [PubMed] [Google Scholar]
- Papadimitriou K, Zoumpopoulou G, Foligné Bet al. Discovering probiotic microorganisms: in vitro, in vivo, genetic and omics approaches. Front Microbiol. 2015;6:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papalini S, Michels F, Kohn Net al. Stress matters: Randomized controlled trial on the effect of probiotics on neurocognition. Neurobiol Stress. 2018;10:100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parente E, Cocolin L, De Filippis Fet al. FoodMicrobionet: A database for the visualisation and exploration of food bacterial communities based on network analysis. Int J Food Microbiol. 2016;219:28–37. [DOI] [PubMed] [Google Scholar]
- Parente E, De Filippis F, Ercolini Det al. Advancing integration of data on food microbiome studies: FoodMicrobionet 3.1, a major upgrade of the FoodMicrobionet database. Int J Food Microbiol. 2019;305:108249. [DOI] [PubMed] [Google Scholar]
- Park S, Bae J-H. Fermented food intake is associated with a reduced likelihood of atopic dermatitis in an adult population (Korean National Health and Nutrition Examination Survey 2012–2013). Nutr Res. 2016;36:125–33. [DOI] [PubMed] [Google Scholar]
- Park S, Ji Y, Jung HYet al. Lactobacillus plantarum HAC01 regulates gut microbiota and adipose tissue accumulation in a diet-induced obesity murine model. Appl Microbiol Biotechnol. 2017;101:1605–14. [DOI] [PubMed] [Google Scholar]
- Parks DH, Chuvochina M, Chaumeil PAet al. Selection of representative genomes for 24,706 bacterial and archaeal species clusters provide a complete genome-based taxonomy. BioRxiv. 2019. [Google Scholar]
- Parks DH, Chuvochina M, Waite DWet al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 2018;36:996–1004. [DOI] [PubMed] [Google Scholar]
- Parks DH, Imelfort M, Skennerton CTet al. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Rinke C, Chuvochina Met al. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol. 2017;2:1533–42. [DOI] [PubMed] [Google Scholar]
- Pasolli E, Asnicar F, Manara Set al. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell. 2019;176:649–62..e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasolli E, De Filippis F, Mauriello IEet al. Large-scale genome-wide analysis links lactic acid bacteria from food with the gut microbiome. Nature Commun. 2020;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasolli E, Schiffer L, Manghi Pet al. Accessible, curated metagenomic data through ExperimentHub. Nat Methods. 2017;14:1023–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira GV de M, de Melo Pereira GV, de Oliveira Coelho Bet al. How to select a probiotic? A review and update of methods and criteria. Biotechnol Adv. 2018;36:2060–76. [DOI] [PubMed] [Google Scholar]
- Pessione E, Cirrincione S. Bioactive molecules released in food by lactic acid bacteria: encrypted peptides and biogenic amines. Front Microbiol. 2016;7:876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessione E. Lactic acid bacteria contribution to gut microbiota complexity: lights and shadows. Front Cell Infect Microbiol. 2012;2:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pot B, Ludwig W, Kersters Ket al. Taxonomy of Lactic Acid Bacteria. In: De Vuyst L, Vandamme EJ (eds). Bacteriocins of Lactic Acid Bacteria. Boston: Springer, 1994, 13–90. [Google Scholar]
- Pot B, Salvetti E, Mattarelli Pet al. The potential impact of the Lactobacillus name change: The results of an expert meeting organised by the Lactic Acid Bacteria Industrial Platform (LABIP). Trends Food Sci Technol. 2019;94:105–13. [Google Scholar]
- Pruesse E, Quast C, Knittel Ket al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pärtty A, Lehtonen L, Kalliomäki Met al. Probiotic Lactobacillus rhamnosus GG therapy and microbiological programming in infantile colic: a randomized, controlled trial. Pediatr Res. 2015;78:470–5. [DOI] [PubMed] [Google Scholar]
- Qin J, Li R, Raes Jet al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quartieri A, Simone M, Gozzoli Cet al. Comparison of culture-dependent and independent approaches to characterize fecal bifidobacteria and lactobacilli. Anaerobe. 2016;38:130–7. [DOI] [PubMed] [Google Scholar]
- Quince C, Delmont TO, Raguideau Set al. DESMAN: a new tool for de novo extraction of strains from metagenomes. Genome Biol. 2017a;18:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince C, Walker AW, Simpson JTet al. Shotgun metagenomics, from sampling to analysis. Nat Biotechnol. 2017b;35:833–44. [DOI] [PubMed] [Google Scholar]
- Rabiei S, Hedayati M, Rashidkhani Bet al. The effects of synbiotic supplementation on Body Mass Index, metabolic and inflammatory biomarkers, and appetite in patients with metabolic syndrome: a triple-blind randomized controlled trial. J Diet Suppl. 2019;16:294–306. [DOI] [PubMed] [Google Scholar]
- Rajkumar H, Mahmood N, Kumar Met al. Effect of probiotic (VSL#3) and omega-3 on lipid profile, insulin sensitivity, inflammatory markers, and gut colonization in overweight adults: a randomized, controlled trial. Mediators Inflamm. 2014;2014:348959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampelli S, Candela M, Severgnini Met al. A probiotics-containing biscuit modulates the intestinal microbiota in the elderly. J Nutr Health Aging. 2013;17:166–72. [DOI] [PubMed] [Google Scholar]
- Reuter G. The Lactobacillus and Bifidobacterium microflora of the human intestine: composition and succession. Curr Issues Intest Microbiol. 2001;2:43–53. [PubMed] [Google Scholar]
- Reuter G. Untersuchungen uber die Zusammensetzung und die Beeinflussbarkeit der menschlichen Magen-und Darmflora unter besonderer Berucksichtigung der Laktobazillen. Ernahrungsforsch Ber Mitt. 1965;10:429–35. [Google Scholar]
- Rezac S, Kok CR, Heermann Met al. Fermented foods as a dietary source of live organisms. Front Microbiol. 2018;9:1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbera A, Jï HM, Kant Ret al. Comparative genomic and functional analysis of Lactobacillus casei and Lactobacillus rhamnosus strains marketed as probiotics. Appl Environ. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinttilä T, Kassinen A, Malinen Eet al. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J Appl Microbiol. 2004;97:1166–77. [DOI] [PubMed] [Google Scholar]
- Ripari V, Gänzle MG, Berardi E. Evolution of sourdough microbiota in spontaneous sourdoughs started with different plant materials. Int J Food Microbiol. 2016;232:35–42. [DOI] [PubMed] [Google Scholar]
- Roager HM, Dragsted LO. Diet-derived microbial metabolites in health and disease. Nutr Bull. 2019;44:216–27. [Google Scholar]
- Roos S, Dicksved J, Tarasco Vet al. 454 pyrosequencing analysis on faecal samples from a randomized DBPC trial of colicky infants treated with Lactobacillus reuteri DSM 17938. PLoS One. 2013;8:e56710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi M, Martínez-Martínez D, Amaretti Aet al. Mining metagenomic whole genome sequences revealed subdominant but constant Lactobacillus population in the human gut microbiota. Environ Microbiol Rep. 2016;8:399–406. [DOI] [PubMed] [Google Scholar]
- Salvetti E, Harris HMB, Felis GEet al. Comparative genomics of the genus Lactobacillus reveals robust phylogroups that provide the basis for reclassification. Appl Environ Microbiol. 2018;84:e00993–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders ME, Akkermans LMA, Haller Det al. Safety assessment of probiotics for human use. Gut Microbes. 2010;1:164–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders ME, Benson A, Leeber Set al. Shared mechanisms among probiotic taxa: implications for general probiotic claims. Curr Opin Biotechnol. 2018;49:207–16. [DOI] [PubMed] [Google Scholar]
- Sanders ME, Merenstein DJ, Reid Get al. Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat Rev Gastroenterol Hepatol. 2019;16:605–16. [DOI] [PubMed] [Google Scholar]
- Savino F, Cordisco L, Tarasco Vet al. Lactobacillus reuteri DSM 17938 in infantile colic: a randomized, double-blind, placebo-controlled trial. Pediatrics. 2010;126:e526–33. [DOI] [PubMed] [Google Scholar]
- Savino F, Garro M, Montanari Pet al. Crying time and RORγ/FOXP3 expression in Lactobacillus reuteri DSM17938-treated infants with colic: a randomized trial. J Pediatr. 2018;192:171–7. [DOI] [PubMed] [Google Scholar]
- Schiffer L, Azhar R, Shepherd Let al. HMP16SData: Efficient access to the Human Microbiome Project through Bioconductor. Am J Epidemiol. 2019;188:1023–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin Tet al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz M, Ward DV, Pasolli Eet al. Strain-level microbial epidemiology and population genomics from shotgun metagenomics. Nat Methods. 2016;13:435–8. [DOI] [PubMed] [Google Scholar]
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. [DOI] [PubMed] [Google Scholar]
- Segata N. On the road to strain-resolved comparative metagenomics. mSystems. 2018;3:e00190–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol D, Jhang SY, Kim Het al. Accurate and strict identification of probiotic species based on coverage of whole-metagenome shotgun sequencing data. Front Microbiol. 2019;10:1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakerian M, Razavi SH, Ziai SAet al. Proteolytic and ACE-inhibitory activities of probiotic yogurt containing non-viable bacteria as affected by different levels of fat, inulin and starter culture. J Food Sci Technol. 2015;52:2428–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N, Angural S, Rana Met al. Phytase producing lactic acid bacteria: Cell factories for enhancing micronutrient bioavailability of phytate rich foods. Trends in Food Sci Technol. 2020;96:1–12. [Google Scholar]
- Shi Y, Zhao X, Zhao Jet al. A mixture of Lactobacillus species isolated from traditional fermented foods promote recovery from antibiotic-induced intestinal disruption in mice. J Appl Microbiol. 2018;124:842–54. [DOI] [PubMed] [Google Scholar]
- Sieber CMK, Probst AJ, Sharrar Aet al. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat Microbiol. 2018;3:836–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra S, Lara-Villoslada F, Sempere Let al. Intestinal and immunological effects of daily oral administration of Lactobacillus salivarius CECT5713 to healthy adults. Anaerobe. 2010;16:195–200. [DOI] [PubMed] [Google Scholar]
- Siezen RJ, Tzeneva VA, Castioni Aet al. Phenotypic and genomic diversity of Lactobacillus plantarum strains isolated from various environmental niches. Environ Microbiol. 2010;12:758–73. [DOI] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis Pet al. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–2. [DOI] [PubMed] [Google Scholar]
- Singh A, Sarangi AN, Goel Aet al. Effect of administration of a probiotic preparation on gut microbiota and immune response in healthy women in India: an open-label, single-arm pilot study. BMC Gastroenterol. 2018;18:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisson G, Ayis S, Sherwood RAet al. Randomised clinical trial: A liquid multi-strain probiotic vs. placebo in the irritable bowel syndrome - a 12 week double-blind study. Aliment Pharmacol Ther. 2014;40:51–62. [DOI] [PubMed] [Google Scholar]
- Skrypnik K, Bogdański P, Sobieska Met al. The effect of multistrain probiotic supplementation in two doses on iron metabolism in obese postmenopausal women: a randomized trial. Food Funct. 2019;10:5228–38. [DOI] [PubMed] [Google Scholar]
- Sluijs I, Forouhi NG, Beulens JWJet al. The amount and type of dairy product intake and incident type 2 diabetes: results from the EPIC-InterAct Study. Am J Clin Nutr. 2012;96:382–90. [DOI] [PubMed] [Google Scholar]
- Soedamah-Muthu SS, Masset G, Verberne Let al. Consumption of dairy products and associations with incident diabetes, CHD and mortality in the Whitehall II study. Br J Nutr. 2013;109:718–26. [DOI] [PubMed] [Google Scholar]
- Sonestedt E, Wirfält E, Wallström Pet al. Dairy products and its association with incidence of cardiovascular disease: the Malmö diet and cancer cohort. Eur J Epidemiol. 2011;26:609–18. [DOI] [PubMed] [Google Scholar]
- Soomro AH, Masud T, Anwaar K. Role of lactic acid bacteria (LAB) in food preservation and human health-a review. Pakistan Journal of Nutrition. 2002;1:20–4. [Google Scholar]
- Spaiser SJ, Culpepper T, Nieves C Jret al. Lactobacillus gasseri KS-13, Bifidobacterium bifidum G9-1, and Bifidobacterium longum MM-2 ingestion induces a less inflammatory cytokine profile and a potentially beneficial shift in gut microbiota in older adults: a randomized, double-blind, placebo-controlled, crossover study. J Am Coll Nutr. 2015;34:459–69. [DOI] [PubMed] [Google Scholar]
- Steenbergen L, Sellaro R, van Hemert Set al. A randomized controlled trial to test the effect of multispecies probiotics on cognitive reactivity to sad mood. Brain Behav Immun. 2015;48:258–64. [DOI] [PubMed] [Google Scholar]
- Stellato G, De Filippis F, La Storia Aet al. Coexistence of lactic acid bacteria and potential spoilage microbiota in a dairy processing environment. Appl Environ Microbiol. 2015;81:7893–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart RD, Auffret MD, Warr Aet al. Assembly of 913 microbial genomes from metagenomic sequencing of the cow rumen. Nat Commun. 2018;9:870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart RD, Auffret MD, Warr Aet al. Compendium of 4,941 rumen metagenome-assembled genomes for rumen microbiome biology and enzyme discovery. Nat Biotechnol. 2019;37:953–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolaki M, Minekus M, Venema Ket al. Microbial communities in a dynamic in vitro model for the human ileum resemble the human ileal microbiota. FEMS Microbiol Ecol. 2019;95:8. [DOI] [PubMed] [Google Scholar]
- Suau A, Bonnet R, Sutren Met al. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol. 1999;65:4799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suez J, Zmora N, Zilberman-Schapira Get al. Post-antibiotic gut mucosal microbiome reconstitution is impaired by probiotics and improved by autologous FMT. Cell. 2018;174:1406–23. [DOI] [PubMed] [Google Scholar]
- Sunagawa S, Coelho LP, Chaffron Set al. Structure and function of the global ocean microbiome. 2015. [DOI] [PubMed]
- Sunagawa S, Mende DR, Zeller Get al. Metagenomic species profiling using universal phylogenetic marker genes. Nat Methods. 2013;10:1196–9. [DOI] [PubMed] [Google Scholar]
- Sung V, Hiscock H, Tang MLet al. Treating infant colic with the probiotic Lactobacillus reuteri: double blind, placebo controlled randomised trial. BMJ. 2014;348:g2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Harris HMB, McCann Aet al. Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat Commun. 2015;6:8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Şanlier N, Gökcen BB, Sezgin AC. Health benefits of fermented foods. Crit Rev Food Sci Nutr. 2019;59:506–27. [DOI] [PubMed] [Google Scholar]
- Talebi S, Karimifar M, Heidari Zet al. The effects of synbiotic supplementation on thyroid function and inflammation in hypothyroid patients: A randomized, double-blind, placebo-controlled trial. Complement Ther Med. 2020;48:102234. [DOI] [PubMed] [Google Scholar]
- Tamang JP, Cotter P, Endo Aet al. Fermented foods in a global age: east meets west. Compr Rev Food Sci F. 2020;19:184–217. [DOI] [PubMed] [Google Scholar]
- Tannock GW, Munro K, Harmsen HJMet al. Analysis of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillus rhamnosus DR20. Appl Environ Microbiol. 2000;66:2578–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannock GW. Analysis of the intestinal microflora: a renaissance. Antonie Van Leeuwenhoek. 1999;76:265–78. [PubMed] [Google Scholar]
- Tapsell LC. Fermented dairy food and CVD risk. Br J Nutr. 2015;113:S131–5. [DOI] [PubMed] [Google Scholar]
- Taylor BC, Lejzerowicz F, Poirel Met al. Consumption of fermented foods is associated with systematic differences in the gut microbiome and metabolome. mSystems. 2020;5:e00901–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira TFS, ŁM Grześkowiak, Salminen Set al. Faecal levels of Bifidobacterium and Clostridium coccoides but not plasma lipopolysaccharide are inversely related to insulin and HOMA index in women. Clin Nutr. 2013;32:1017–22. [DOI] [PubMed] [Google Scholar]
- Thompson LR, et al. The Earth Microbiome Project Consortium. A communal catalogue reveals Earth's multiscale microbial diversity. Nature. 2017;551:457–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillisch K, Labus J, Kilpatrick Let al. Consumption of fermented milk product with probiotic modulates brain activity. Gastroenterology. 2013;144:1394–401., 1401.e1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toksvig Bjerg A, Kristensen M, Ritz Cet al. Lactobacillus paracasei subsp paracasei L. casei W8 suppresses energy intake acutely. Appetite. 2014;82:111–8. [DOI] [PubMed] [Google Scholar]
- Truong DT, Franzosa EA, Tickle TLet al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods. 2015;12:902–3. [DOI] [PubMed] [Google Scholar]
- Truong DT, Tett A, Pasolli Eet al. Microbial strain-level population structure and genetic diversity from metagenomes. Genome Res. 2017;27:626–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu M-Y, Chen H-L, Tung Y-Tet al. Short-term effects of kefir-fermented milk consumption on bone mineral density and bone metabolism in a randomized clinical trial of osteoporotic patients. PLoS One. 2015;10:e0144231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchinaka A, Azuma N, Mizumoto Het al. Anti-inflammatory effects of heat-killed Lactobacillus plantarum L-137 on cardiac and adipose tissue in rats with metabolic syndrome. Sci Rep. 2018;8:8156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaghef-Mehrabany E, Alipour B, Homayouni-Rad Aet al. Probiotic supplementation improves inflammatory status in patients with rheumatoid arthritis. Nutrition. 2014;30:430–5. [DOI] [PubMed] [Google Scholar]
- Van den Bogert B, Boekhorst J, Herrmann Ret al. Comparative genomics analysis of Streptococcus isolates from the human small intestine reveals their adaptation to a highly dynamic ecosystem. PLoS One. 2013;8:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vangay P, Hillmann BM, Knights D. Microbiome Learning Repo (ML Repo): A public repository of microbiome regression and classification tasks. Gigascience. 2019;8, giz042:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vangay P, Johnson AJ, Ward TLet al. US immigration westernizes the human gut microbiome. Cell. 2018;175:962–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gossum A, Dewit O, Louis Eet al. Multicenter randomized-controlled clinical trial of probiotics (Lactobacillus johnsonii, LA1) on early endoscopic recurrence of Crohn's disease after ileo-caecal resection. Inflamm Bowel Dis. 2007;13:135–42. [DOI] [PubMed] [Google Scholar]
- Vaughan EE, de Vries MC, Zoetendal EGet al. The intestinal LABs. Antonie Van Leeuwenhoek. 2002;82:341–52. [PubMed] [Google Scholar]
- Ventura M, O'Flaherty S, Claesson MJet al. Genome-scale analysis of health-promoting bacteria: probiogenomics. Nat Rev Microbiol. 2009;7:61–71. [DOI] [PubMed] [Google Scholar]
- Verdenelli MC, Silvi S, Cecchini Cet al. Influence of a combination of two potential probiotic strains, Lactobacillus rhamnosus IMC 501® and Lactobacillus paracasei IMC 502® on bowel habits of healthy adults. Lett Appl Microbiol. 2011;52:596–602. [DOI] [PubMed] [Google Scholar]
- Vitellio P, Celano G, Bonfrate Let al. Effects of Bifidobacterium longum and Lactobacillus rhamnosus on gut microbiota in patients with lactose intolerance and persisting functional gastrointestinal symptoms: a randomised, double-blind, cross-over study. Nutrients. 2019;11:E886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel RF, Knorr R, Müller MRAet al. Non-dairy lactic fermentations: the cereal world. Antonie Van Leeuwenhoek. 1999;76:403–11. [PubMed] [Google Scholar]
- Wall R, Cryan JF, Ross RPet al. Bacterial neuroactive compounds produced by psychobiotics. Adv Exp Med Biol. 2014;817:221–39. [DOI] [PubMed] [Google Scholar]
- Walter J, Hertel C, Tannock GWet al. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2001;67:2578–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J. Ecological role of lactobacilli in the gastrointestinal tract: implications for fundamental and biomedical research. Appl Environ Microbiol. 2008;74:4985–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther B, Karl JP, Booth SLet al. Menaquinones, bacteria, and the food supply: the relevance of dairy and fermented food products to vitamin K requirements. Adv Nutr. 2013;4:463–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Chen L, Zhou Ret al. Increased proportions of Bifidobacterium and the Lactobacillus group and loss of butyrate-producing bacteria in inflammatory bowel disease. J Clin Microbiol. 2014;52:398–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wels M, Siezen R, van Hijum Set al. Comparative genome analysis of Lactococcus lactis indicates niche adaptation and resolves genotype/phenotype disparity. Front Microbiol. 2019;10:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittouck S, Wuyts S, Lebeer S. Towards a genome-based reclassification of the genus Lactobacillus. Appl Environ Microbiol. 2019;85:e02155–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Sun C, Li Yet al. GMrepo: a database of curated and consistently annotated human gut metagenomes. Nucleic Acids Res. 2020;48:D545–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Wan J, Choe Uet al. Interactions between food and gut microbiota: Impact on human health. Annu Rev Food Sci Technol. 2019;10:389–408. [DOI] [PubMed] [Google Scholar]
- Wuyts S, Wittouck S, De Boeck Iet al. Large-scale phylogenomics of the Lactobacillus casei group highlights taxonomic inconsistencies and reveals novel clade-associated features. mSystems. 2017;2:e00061–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang H, Sun-Waterhouse D, Waterhouse GINet al. Fermentation-enabled wellness foods: A fresh perspective. Food Science and Human Wellness. 2019;8:203–43. [Google Scholar]
- Yang J, McDowell A, Kim EKet al. Consumption of a Leuconostoc holzapfelii-enriched synbiotic beverage alters the composition of the microbiota and microbial extracellular vesicles. Exp Mol Med. 2019;51:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Poles MA, Fisch GSet al. HIV-induced immunosuppression is associated with colonization of the proximal gut by environmental bacteria. AIDS. 2016;30:19–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JS, Sohn W, Lee OYet al. Effect of multispecies probiotics on irritable bowel syndrome: a randomized, double-blind, placebo-controlled trial. J Gastroenterol Hepatol. 2014;29:52–9. [DOI] [PubMed] [Google Scholar]
- Yoon SH, Ha SM, Kwon Set al. Introducing EzBioCloud: A taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol. 2017;67:1613–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannini E, Waters DM, Coffey Aet al. Production, properties, and industrial food application of lactic acid bacteria-derived exopolysaccharides. Appl Microbiol Biotechnol. 2016;100:1121–35. [DOI] [PubMed] [Google Scholar]
- Zhang C, Derrien M, Levenez Fet al. Ecological robustness of the gut microbiota in response to ingestion of transient food-borne microbes. ISME J. 2016;10:2235–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wang L, Guo Zet al. 454 pyrosequencing reveals changes in the faecal microbiota of adults consuming Lactobacillus casei Zhang. FEMS Microbiol Ecol. 2014;88:612–22. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhang D, Jia Het al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med. 2015;21:895–905. [DOI] [PubMed] [Google Scholar]
- Zheng J, Ruan L, Sun Met al. A genomic view of Lactobacilli and Pediococci demonstrates that phylogeny matches ecology and physiology. Appl Environ Microbiol. 2015a;81:7233–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Wittouck S, Salvetti Eet al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int J Syst Evol Microbiol. 2020; DOI:10.1099/ijsem.0.004107. [DOI] [PubMed] [Google Scholar]
- Zheng J, Zhao X, Lin XBet al. Comparative genomics Lactobacillus reuteri from sourdough reveals adaptation of an intestinal symbiont to food fermentations. Sci Rep. 2015b;5:18234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X, Xiong L, Li Let al. Alterations of gut microbiota in patients with irritable bowel syndrome: A systematic review and meta-analysis. J Gastroenterol Hepatol. 2017;32:28–38. [DOI] [PubMed] [Google Scholar]
- Zijlmans MAC, Korpela K, Riksen-Walraven JMet al. Maternal prenatal stress is associated with the infant intestinal microbiota. Psychoneuroendocrinology. 2015;53:233–45. [DOI] [PubMed] [Google Scholar]
- Zmora N, Zeevi D, Korem Tet al. Taking it personally: Personalized utilization of the human microbiomein health and disease. Cell Host Microbe. 2016;19:12–20. [DOI] [PubMed] [Google Scholar]
- Zmora N, Zilberman-Schapira G, Suez Jet al. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell. 2018;174:1388–405. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.