As a positive-sense single-strand RNA virus, coronavirus (CoV) possesses some of the largest genomes among RNA viruses, ∼30 Kb in size and encodes more than two-dozen proteins that ensure a long-lasting parasitic cellular life leveraging on both informational inheritability and operational integrity by constantly changing the underlying molecular constituents toward harmony with those of the hosts whose genomes harbor over 20,000 genes and 2–3 Gb in sizes. We, taking the advantage of the unprecedented accumulation of genomic sequences, interrogate mutation spectra of SARS-CoV-2 as a whole or of the major clades in details using comprehensive genomic tools and structure chemistry principles. Two key mechanisms are associated with variable mutation patterns (permutations); one takes the advantage of protein-coding rules to maintain cellular homeostasis including composition dynamics of the host RNA and protein reservoirs and the other concerns strand-biased replication to fine-tuning these mutation spectra that are attributable to the strands and the round of replication. The former is supported by both global sweeping of amino acids for distinct chemical and structural characteristics and local fitness mutation-selection for catalytic specificity and structural subtleties, and the latter is validated when altered mutation spectra among phylogenetic hierarchies becomes comprehensible. In this context, SARS-CoV-2 is extraordinarily different from both SARS-CoV and MERS-CoV, whose both G + C and A + G contents have been drifting toward the low ends, a signature of diminishing selective pressure, approaching those of the deteriorated, parasitic, and less pathogenic human CoVs, such as hsaCov-229E, hsaCov-OC43, hsaCov-HKU1, and hsaCov-NL63. With such trends and principles, genotypic variations can be analyzed in details to associate with phenotypic variables including both molecular anomalies and clinical symptoms. These mechanisms provide novel guidance for genome analysis of RNA viruses and shed lights on rational designing of targeted drugs, vaccines and diagnostics.
Dedication
This essay is what I owe my former graduate student and colleague Dr. Xiaowei Zhang, who had a pair of magic hands for challenging experiments and a short yet productive scientific career; his thesis stories had not been fully published and a part of them is narrated here contributing valuable insights for the fight against global pandemics of COVID-19.
A primer to RNA genomics: DNA is the chosen one by the RNA World
At the very end of the RNA World, the Queen of the Macromolecules – RNA designated one of its two roles, operational (some scientists prefer the word catalytic) and informational, as another Crown to the King of the Macromolecules – DNA. DNA, double-stranded deoxyribonucleic acids, have been playing this informational role by choosing its corresponding four building blocks – nucleotides A, T, G, and C – to those of the RNA, A, U, G, C, and transferred the genetic code to the scrambles finally to produce proteins for stable inheritance. By transferring the informational role into DNA, dwellers of the RNA World compensated two key changes for their genomes; one is to pair the two strands by using the DNA building blocks, deoxyribonucleotides, and the other is to inherit A, G, and C but to replace U with T in informational context. From 150 or so structure-diverse candidate nucleosides [1], collectively present in all extant life forms since the Dawn of Life, to find four backbone nucleotides was not hard, but the choice of T is intelligent: it has a larger molecular weight, due to a methyl group in its thymine ring as compared to uracil, than that of U (Table 1). As a result, the molecular weight difference between G + C and A + T contents in DNA is reduced to 1 Dalton, whereas this difference in RNA is 15 Daltons. The transfer of informational role to DNA also means giving up the magic power of the G-U pairing, the so-called Wobble base pairing (Table 2) [2]. U in RNA provides an extraordinary base-pairing power from its versatile role in physiochemical operations and the differences between G and A or U and C are 16 Daltons and 1 Dalton, respectively. For highly effective synthesis of genetic materials, a single Dalton disparity of the synthetic machinery may lead to diversification as vast as all life forms on earth if given enough time. This subtle difference has at least two indications. One is that the physical dimension within the two categories of nucleobases, purines and pyrimidines, is largely distinguishable, and frequent exchanges within the group lead to tremendous variability within predictable permutations. The other is that there is seemingly slight but significant disparity between the two hydrogen-bonding paired bases, G + C and A + U, in weight, and such a difference certainly becomes significant when amino acid constituents of the catalytic pocket of RNA-dependent RNA polymerases (RdRPs), as well as the larger operational entity, RTC itself, vary due to mutation-altered structure and conformation [3].
Table 1.
Molecular weights of RNA and DNA nucleosides
| DNA | RNA | |
|---|---|---|
| G + C | G + C | 494.4 | 526.4 (32D) |
| G + C > A + T | 494.4 > 493.4 (1D) | NA |
| G + C > A + U | NA | 526.4 > 511.4 (15D) |
| T > C | U > C | 242.2 > 227.2 (15D) | 244.2 > 243.2 (1D) |
| G > A | G > A | 267.2 > 251.2 (16D) | 283.2 > 267.2 (16D) |
Note: Only MWs of nucleosides are calculated and U is 1 Daltons heavier than C. The MW differences between nucleosides compared are shown in parentheses. D, Dalton.
Table 2.
Free energy calculations on Watson-Crick and wobble basepairs
| Basepairing | ΔG°1 (kcal/mol) | d1 (A˚) | ΔG°2 (kcal/mol) | d2 (A˚) |
|---|---|---|---|---|
| C:G | −5.53 | 2.94 | −0.58 | 5.50 |
| U:A | −4.42 | 2.96 | −0.72 | 5.73 |
| U:G | −4.45 | 3.75 | −0.87 | 5.78 |
| U:U | −5.82 | 3.80 | −1.17 | 5.62 |
| U:C | −0.37 | 3.64 | 0.01 | 5.51 |
Note: The table is a simplified version from [2].
The association between genome parameters and molecular mechanisms are essential for understanding viral RNA biology
The fundamental mechanistic differences between DNA and RNA genomes, other than their building blocks, mostly lie in the way they are replicated and how their damages are repaired; both replication and damage repair may alter their primary sequences. In the DNA genome, the two strands of the DNA double helix are equivalent in that mutations in one strand are inherited in the paired opposite strand. In other words, C-to-T mutation in the Watson strand is the same as G-to-A mutation in the Crick, and together they are checked and error-corrected through repairing mechanisms and passed on to the next generation faithfully (e.g., mutation rates of DNA viruses: 10−6 to 10−8 mutations per base per generation; those of RNA viruses: 10−3 to 10−5 mutations per base per generation [4]). In the RNA World, such as in the case of CoV, its positive-sense RNA genome is replicated and transcribed subsequently without pairing to the opposite strand so that errors made in replication are passed on to the next generation without serious sequence-checking surveillance [5]. Therefore, the DNA rules, such as different damage repair systems, may not be applicable to the RNA World, at least in the case of CoV.
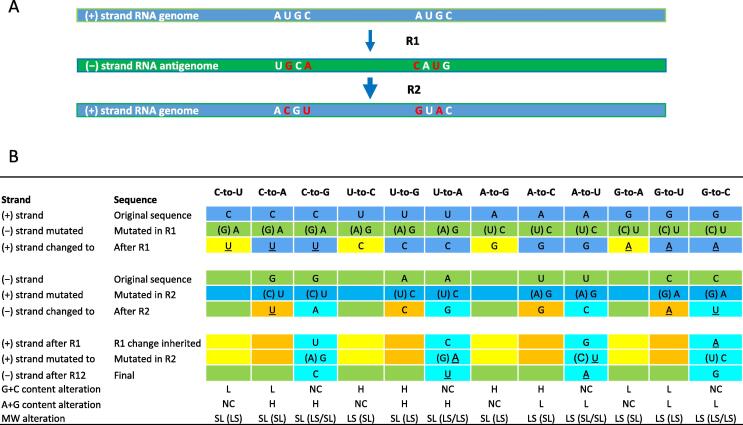
Let us walk through how CoV generates its mutations (Figure 1). We start with two basic rules. First, mutations among all kingdoms of life follow a single universal rule since creation: among the two mutation types, transition (Ts, changes between the two purines of pyrimidines) and transversion (Tv, between the purines and pyrimidines), the transitional type occurs more often than the other due to replication-associated errors and the transversional type is always less than half of the transitional type in number, assumed to be a result of repair errors. The ratios of Ts-to-Tv mutations are 2.0:1.0 in humans (representing DNA genomes) and ∼2.5:1.0 in CoVs (Table 3). This number is expected to be in the same order of magnitude to all RNA viruses and it indicates a stronger mutation pressure (or tolerance toward mutation trajectory) over selection as compared to DNA genomes. A recent measurement of influenza A viruses (a segmented positive-sense single-strand RNA virus), based on a cell culture assay, has narrowed down the mutation rates to an overall mutation rate of 1.8 × 10–4 substitutions per nucleotide per round of copying or s/n/r for PR8 (H1N1) and 2.5 × 10–4 s/n/r for Hong Kong 2014 (H3N2) and a transitional bias of 2.7–3.6 [6]. In virome studies or the history of virology, there has not been a single case like the current COVID-19 pandemics that allow researchers and physicians to chase after such large viral and infected human populations in such a continuous way and an enormous scale. There have been a limited number of studies on avian flu viruses but not intensively for CoVs since the previous two serious CoV outbreaks are relatively short-lived. Second, as a single-strand RNA virus, CoV genome does not have a stable intermediate double-strand structure, and instead, has a positive-sense genome as template to make negative-sense antigenomes of full or shorter length, via its replication or transcription machinery (Figure 1A). Given this basic knowledge, we can now scheme out mutation spectra for CoVs that include phylogenetic clades and clade clusters; a clade cluster usually contains multiple clades that share similar mutation spectra so that they can be analyzed together. A key point to be aware of is the fact that a single variation is capable of separate clades from each other but shared mutation spectrum may still keep its momentum.
Figure 1.
Mutation mechanism and spectrum of CoVs
A. The horizontal bars display how the RNA genome mutates in the process of replication. The RNA synthesis begins from a positive-sense strand (labeled as plus-strand genome, blue), and two sets of four nucleotides, A, U, G, and C are used to represent two sequence contexts. The first round of synthesis happens by using the plus-strand as template and the mutated replicate is a negative-sense strand (labeled as minus-strand genome, green). Four mutations occur in the first synthesis (R1), U-to-G, C-to-A, A-to-C, and G-to-U, due to mismatching between A and G or U and C; all are transitional. These mutations are carried over to the next synthesis (R2) without further sequence change so that they are dominant permutations in a CoV-specific mutation spectrum: U-to-C, C-to-U, A-to-G, and G-to-A. B. a summary of all possible permutations in a CoV mutation spectrum; it is true for all RNA genomes and some may start with a negative sense genome, such as in the case of influenza viruses. The strands where mutations occur are labeled with plus or minus signs in parentheses and the three rounds of mutations are highlighted with yellow (R1), orange (R2), and blue (R12). We assume that abasic nucleotides are one of the repair mechanisms, leading to transversional permutations, and when mutations happen in different strands during synthesis, the intermediate nucleotides are labeled in the parentheses. Permutations lead to lower G + C content are underlined and labeled as L. Permutations lead to higher G + C content are labeled as H. Four permutations that do not vary G + C content are labeled NC (no change). Purine sensitive permutations are also scored in a similar way to the G + C content row. The molecular weight altering consequence are scored as small-to-large or SL and vice versa. The intermediates are also indicated in the parentheses. For instance, a G-to-U mutation as a permutation of the mutation spectrum happens when the positive-sense strand (+) is synthesized (the second-round replication or R2) where G supposed to pair with C but altered as U so that the mechanism has to be a C-by-U replacement that is a size increase. Another more complicated instance is G-to-C mutation, where it has to go through a first R1 mutation G-to-A (a negative sense strand mutation) and then a U-to-C mutation (a positive-sense strand mutation and thus labeled as R12 for double mutations of the positive-sense strand) follows to complete the permutation. CoV, coronavirus; R12, R1+R2; L, low; H, high; NC, no change; SL, small-to-large; LS, large-to-small; MW, molecular weight.
Table 3.
A snapshot statistics of transition (Ts) and transversion (Tv) variations of SARS-CoV-2
| Cutoff |
Number of mutations per genome |
All | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11+ | |||
| 1 | Ts | 42 | 156 | 235 | 247 | 414 | 731 | 1098 | 1288 | 1357 | 1111 | 1704 | 8383 |
| Tv | 19 | 50 | 72 | 88 | 147 | 291 | 412 | 518 | 552 | 463 | 952 | 3564 | |
| Ts/Tv | 2.21 | 3.12 | 3.26 | 2.81 | 2.82 | 2.51 | 2.67 | 2.49 | 2.46 | 2.40 | 1.79 | 2.35 | |
| 2 | Ts | 53 | 154 | 184 | 226 | 328 | 583 | 894 | 993 | 942 | 770 | 938 | 6065 |
| Tv | 19 | 38 | 56 | 52 | 130 | 233 | 299 | 371 | 356 | 294 | 446 | 2294 | |
| Ts/Tv | 2.79 | 4.05 | 3.29 | 4.35 | 2.52 | 2.50 | 2.99 | 2.68 | 2.65 | 2.62 | 2.10 | 2.64 | |
| 3 | Ts | 50 | 128 | 156 | 207 | 288 | 510 | 743 | 806 | 733 | 559 | 677 | 4857 |
| Tv | 20 | 31 | 42 | 57 | 104 | 190 | 260 | 295 | 284 | 220 | 321 | 1824 | |
| Ts/Tv | 2.50 | 4.13 | 3.71 | 3.63 | 2.77 | 2.68 | 2.86 | 2.73 | 2.58 | 2.54 | 2.11 | 2.66 | |
Note: The median numbers of mutations per genome are 6–7, which are slightly different among clades. Data are downloaded from the National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences/China National Center for Bioinformation in May, 2020. The data are not an up-to-date collection so that it provides only a snapshot of the reality in passing. Ts, transition; Tv, transversion. We set cutoffs (cutoffs 1–3) to extract mutations identified in at least 1, 2, or 3 CoV genome sequences.
RNA genomes have 12 permutations (Figure 1B), which are more to be aware of than those of DNA genomes, whose C-to-T permutation in one strand is equivalent to A-to-G in the opposite strand. As mutations happen, a C-to-U mutation occurs in the process of negative-sense strand synthesis (namely the first replication cycle or R1) where the template sequence C, which is supposed to pair up with G, mismatches with the non-canonical purine A, and it is this particular untidy action by the CoV RTCs (reasons for this type of action will be discussed in detail later in this session) that leads to a U in the same position on the newly synthesized positive-sense strand. This appears rather irrational for DNA synthesis where the newly synthesized double helix is subjected to a much tidier (with roughly 1000-times more stringency) repair system – mismatch DNA repair – to fix such an obvious erroneous process [7]. Following the same principle, the R1 permutations that are all Ts mutations, including C-to-U, A-to-G, U-to-C, and G-to-A, should happen in the same way. The second set of permutations includes all Tv mutations but can be divided into two groups. The first group (R2) includes permutations that change both G + C and A + G contents: G-to-U, C-to-A, U-to-G, A-to-C, and the second group (R12) are those only altering A + G content: A-to-U, G-to-C, U-to-A, and C-to-G. The second set of permutations may be related to DNA repair mechanisms such as base excision repair (BER), which removes abasic (apurinic/apyrimidinic, AP) sites [8], [9]. Note that there are not only timing and sequence alteration issues here but also concerns on strand-specificity in addition to structural principles where C-to-U mutation is realized by a G-by-A replacement and its counterpart, such as G-to-A is different by such structural measures. Since full-length negative-sense strand is always a minor population in the viral cellular life cycle and only full-length positive-sense strands are to be assembled into viral particles or virons, the third issue concerns copy-number sensitivity, where the number of positive-sense strands are expected to be 50–100 fold more than that of negative-sense strands within a host cell [10].
There are more to be discussed on the definition of a mutation spectrum. First, among the 12 permutations, the theoretical Ts/Tv ratio is actually 1 (4 R1 permutations):2 (4 R2 and 4 R12 permutations) and there would be, in theory, more Tv permutations than Ts permutations if every mutation occurs by equal chances. In reality, this ratio is determined by order of synthesis and specificity and governed by structural or conformational variables of the viral RTCs. Second, there is a hidden mechanism where the predominant mutations should have mostly been gone through the Ts-mutation intermediates, C-by-U or G-by-A replacement and the reverse (Figure 1B). For instance, a R1-derived C-to-U mutation is a G-by-A replacement carried by the negative-sense strand and its offspring, the positive-sense viral genome, harbors the expected U. Another example is the R2-derived G-to-U, the same G-by-A replacement occurs once a C-to-A mutation occurred on the negative strand due to a repair error. We should expect the fact that when C-to-U becomes the dominant permutation in a viral genome, the permutation G-to-U must lead the permutation U-to-G if selection (often referring to changes classified into synonymous and non-synonymous; the latter by and large indicates amino acid alteration and thus functional alteration) is not strong enough to override this effect. However, in the case of R12-derived permutations, the first change often is not the same transitional changes as the second. For instance, the R12-derived U-to-A and A-to-U permutations do not follow the C-to-U and G-to-U routes but go through a U-by-C or A-by-G and a G-by-A or C-by-U double replacements, respectively. Therefore, the mechanistic Ts/Tv ratio is both strand-specific and order-sensitive. Apparently, other qualitative and even quantitative (more likely statistical) parameters have to be introduced in order to solve this puzzle completely. Obviously, mathematical models and related algorithms, which theorize such permutation dynamics, are of essence for computer-based simulation studies. Third, in order to predict mechanistic principles, where the variability of permutations in a given mutation spectrum fits certain empirical rules, the three sets of permutations and their fractions must be mapped and associated to structure-centric and conformation-centric changes of CoV-specific RTCs and other related dynamic constituents. Nevertheless, the rationales are two-fold, one is related to mutation specificity and the other to strand specificity that includes the order of mutation occurrence.
The mutation spectrum with 12 permutations and their patterns appears characteristics of SARS-CoV-2 and their closely-related relatives
Are the frequencies of permutations in viral mutation spectra predictable? The answer is yes and no. Let us go through the positive side of the story first. The trend of these mutation spectra is highly predictable once mutations are classified in a logical way, simply by combining mechanistic and statistical means. Among RdRPs, the substrate-specificity is known to be governed by its catalytic center, whose key amino acid residues are highly conserved and not easily to be altered [11]. RdRPs (CoV-RdRP, nonstructural protein 12 or nsp12) contain a 500–600-residue catalytic module with distinct palm, finger, and thumb domains forming a right-handed “pocket”. Since there are seven polymerase catalytic motifs (A to G) are in the palm-finger domains of RdRPs, the substrate-specificity is of vast yet subtle structural and conformational variations. In addition, other nsps, such as nsp7 and nsp8 are known to be part of the RTCs [3], [12]. If all relevant mutations keep accumulating, such as the case of SARS-CoV-2, we will be able to associate precisely most varied amino acid sequences to enzymatic functions and even virus-centric symptoms of infected patients. The negative side of the story has to do with how mutations are mapped to structure and conformation related to enzymatic function, and certainly, wet-bench efforts are required to validate proposals, conjectures, and assumptions, which are long-term and yet limited by in depth biomedical characterization of the virus and its genes as well as their products.
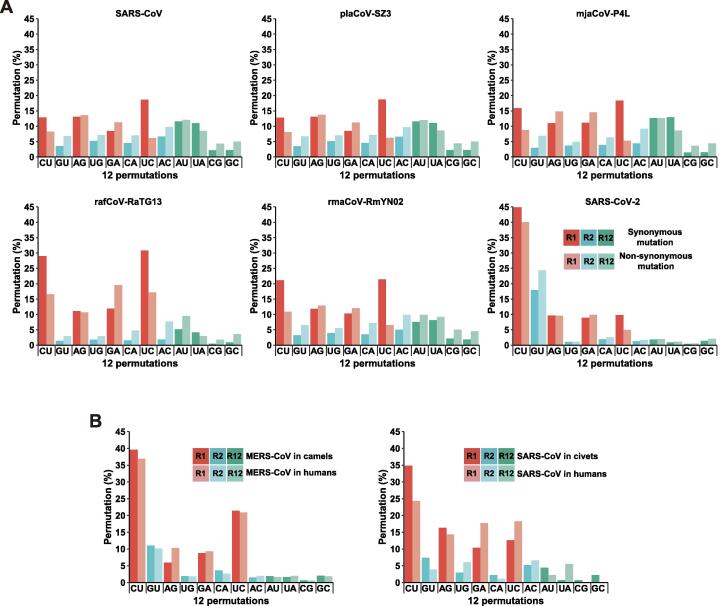
We proceed our discussion by examining discrete examples that cover a series of mutation spectra of human-infecting CoVs and their closely-related known and implicated natural and/or intermediate hosts (Figure 2A). Before getting into the details, two population genetics principles have to be clarified, i.e., within-population (for lack of better term, it is defined as variations based on a collection of CoV genomes from both humans and the true intermediate hosts in a single outbreak) and between-population variations (those of CoV genomes from multiple outbreaks of a minimal or the same lineage, such as within the lineage of betacoronaviruses), and we calculate within-population permutations based on sequence alignment of all SARS-CoV-2 genomes and what referenced to the SARS-CoV-2 reference genome but not isolated from COVID-19 patients (such as other mammals, bats and pangolins) are classified as between-population variations. Mutation spectra of SARS-CoV-2, containing a snap-shot total and the non-synonymous mutation fraction of it, shows typical patterns of their permutations. Clearly, all R1 permutations are dominant with a trend where stronger C-to-U and weaker A-to-G exceed the reverse pair, U-to-C and G-to-A, respectively. Among R2 permutations, G-to-U and C-to-A are both dominant over the opposing pairs due to the similar mechanism to R1 permutations C-to-U and A-to-G but happen during the positive-sense strand synthesis. This C-to-U dominance appears rather universal to all mammalian CoVs in terms of within-population permutations but not among between-population permutations; we believe this is determined by the highly conserved mammal-infecting CoV RdRPs. Similarly, of R12 permutations, the A-to-U and G-to-C pair occurs more frequently than the other pair, U-to-A and C-to-G. These trends of permutation variability as compositional signatures are very much preserved for the non-synonymous mutations since the newly generated within-population mutations have not yet been subjected to strong or long-term selections. Data from the two previous CoV evasions are also very informative (Figure 2B), where within-population (dominated by R1 and R2 permutations) and between-population (R12 permutations increase over time due to selection) variations are more obvious as compared to what between SARS-CoV-2 and its close relatives that are neither true natural nor intermediate hosts.
Figure 2.
Selected mutation spectra of human-infecting CoVs and their true and closely related intermediate hosts
A. Mutation spectra of SARS-CoVs (starting from the left) and a civet intermediate host (pla-betaCoV-SZ3), three CoVs closely related to SARS-CoV-2, one from a pangolin (mja-betaCoV-P4L) and two from bats (raf-betaCoV-RaTG13 and rma-betaCoV-RmNY02), and SARS-CoV-2. The data from SARS-CoV-2 are a collection from public databases with 12,642 full-length high-quality genome sequences. The corresponding lower panels are permutations based on nonsynonymous mutations. Note that all SARS-CoV-2 data show clear C-to-U dominance in R1 permutations and G-to-U dominance in the R2 permutations; both share the same mechanism of a G-by-A replacement on the synthesis of negative-sense and positive-sense strands, respectively. B. The within-population mutation spectra of SARS-CoV, MERS-CoV and their mammalian co-hosts. The permutations are calculated based on public collections with a limited number of individual sequences. SARS-CoV data contain 105 and 18 genomes from human and within-population civets, respectively; MERS-CoV data have 248 and 182 genomes from human and camels, respectively.
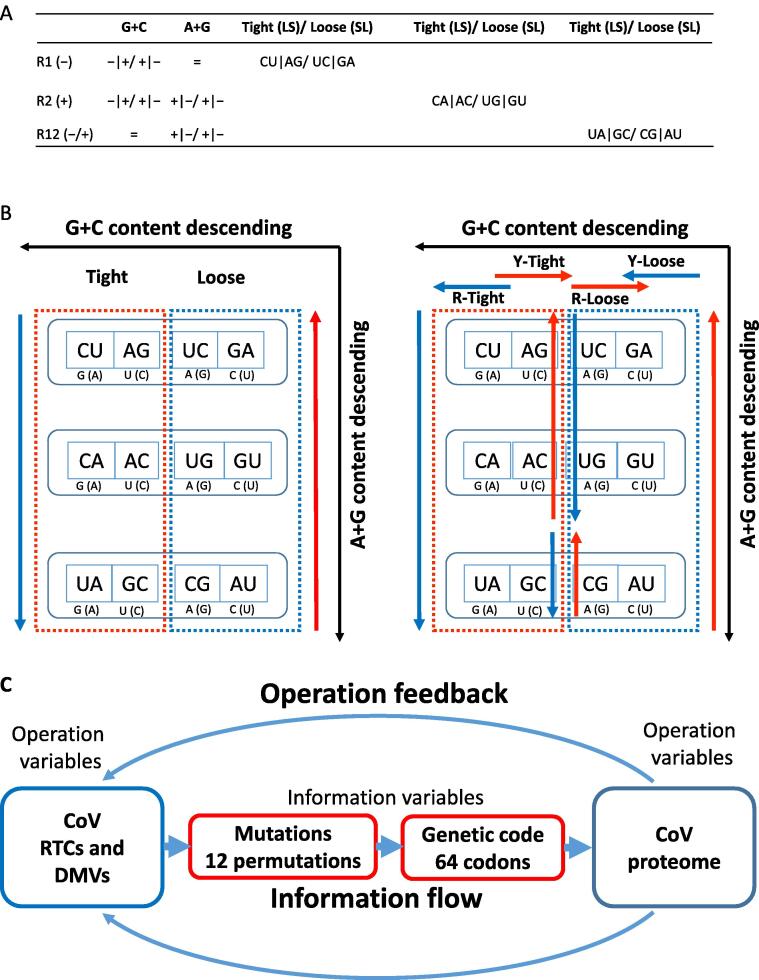
We summarize this within-population SARS-CoV-2 spectrum into a table (Figure 3A) and further assume that there is a mechanistic explanation for it based on physiochemical features of the virus-specific replication machinery. Assigning all 12 permutations to the different features, such as nucleobase-specific size and hydrogen-bounding as well as nucleotide composition dynamics, we divide the permutations into two categories, composition-centric and structure-centric. In the composition-centric category, for instance, neither C-to-U and A-to-G nor U-to-C and G-to-A alter A + G or purine content but G + C content. In other words, if a RNA virus, such as CoV, needs to have a better (an ideal one is balanced at 50%) purine content, the permutations for change are these four (see discussion in the next session). Similarly, the best permutations for constant G + C content are A-to-U and G-to-C as well as the reverse pairs.
Figure 3.
Mutation spectra of CoVs and proposed molecular mechanisms
A. This table listed all permutations and their impact on G + C and purine contents and possible influences on the RTCs. R1, first replication that synthesizes the negative-sense strand; R2, the second replication that synthesizes the positive-sense strand after a mutation happens in the negative strand; R12, mutation proceeds a previous one, such as R2 after R1 that synthesizes the positive-sense strand. The positive-sense and negative-sense strands are indicated in parentheses with plus and minus signs, respectively. The “=” and “+/-” signs depict no change and change toward high or low nucleotide composition parameters, respectively. Vertical bars (|) separate two parallel permutations and slash signs (/) mean one or the other. Note that the R1 and R12 permutations are not sensitive to purine and GC contents, respectively, but R2 changes are sensitive to both. The common words “loose” and “tight” or their ordered combinations suggest possible mechanisms for structural and/or conformational characteristics of RTCs (replicase-transcriptase or replication-transcription complexes) related to binding statuses to the nucleotide substrates. B. A further illustration as to how conformational and/or structural characteristics relate to permutations, referred as mechanistic models. We have two sets for two different models. The first model (the left panel) divides 12 permutations into 3 rows (R1, R2, and R12) and 2 columns (left and right). Base on mutation mechanisms, the left 6 permutations are G-by-A and C-by-U replacements, i.e., they are mechanistically large-to-small transitional switches, and the right 6 permutations are the opposite small-to-large. This two-division model suggests that the tight (G-by-A and U-by-C replacements) status leads to excess U and the loose status (A-by-G and C-by-U) leads to A surplus. The right panel describes a four-division model where purine-centric (R-) and pyrimidine-centric (Y-) tight and loose model dictates a more intricate permutation variability. C. A schematic chart to emphasize that information flow and operation feedback are both directional. CoV takes the advantage of ample variations generated by its replication–transcription complexes (RTCs; in a formation known as double-membrane vesicles or DMVs) in 12 permutations and functional variations is implemented though the genetic code in its proteome that includes mutation-generating machinery itself, which are promptly tested by survival in the host.
In the structure-centric category, all 12 permutations are evaluated based on spatial parameters, i.e., RdRP-related structure-conformation indications (Figure 3B). Here, we propose two models for structure-conformation constraints. One is a two-parameter model where a binary choice for substrate specificity is made as “tight” (G-to-A and U-to-C are both large-to-small or L-to-S replacement; or simply LS) or “loose” (small-to-large replacement is permitted; SL), i.e., purines or pyrimidines are not distinguished. Another is a four-parameter model where two binary choices have to be made and purines and pyrimidines are treated differently. Obviously, the latter is more realistic but the first is easier to understand and a useful approximation. In the SARS-CoV-2 dataset, the absolute dominant C-to-U permutation, as a major benchmark, is rather obvious. The underlying principle is the proposed specificity principle, the favorite G-by-A replacement, which is a 16-Dalton difference in molecular weight change dictated by the CoV RTC. Similar principle is applicable to A-to-G permutation, where a U-by-C replacement represents a 1-Dalton difference. We have also realized that A-to-G and G-to-A have much stronger size-related discrimination power than C-to-U and U-to-C pairs, and the indication here is that the molecular weight differences is certainly a predominant measure, characteristics of the catalytic pocket and its milieus in both conformational and structural terms. The other pair of permutations, U-to-C and G-to-A are denoted as loose as a larger pocket is demanded for A-by-G and C-by-U replacements (SL). Such assumptions are difficult to prove due to the complex nature of catalytic enzymes and uncertainty of conformational effects of distant amino acid residues, but nevertheless very useful for association of genome compositional dynamics to protein structural dynamics via sequence mutations in the context of permutations. The two-division and four-division models are in complete agreement with our previous genetic code and its codon arrangement models [13], [14], [15], [16].
The two categories represent the compositional (or informational) and structural (or operational) variables as well as their interplays that are interconnected though the genetic code (Figure 3C) [15], [16]. The essence of such relationship is best manifested by CoVs, especially through their transmission and host jumping scenarios. From the SARS-Cov-2 dataset, we observe very little selection but strong mutation tendency toward G + C content decrease, as seen in a much lower A-to-G, higher G-to-U, and even less selection on U-to-C. As mentioned before, within-population and between-population mutations are rather distinct, even though sometimes genetic distances are hard to measure for genomes as small as a few tens of kilo-nucleotides (Knt) and fast-mutating. The two closely-related bat (RaT13G and NY02) and the pangolin CoVs, in contrast, show a rather balanced trend where all four dominant G + C content altering permutations have similar values, which is a typical between-population comparison (mutations are mapped to the SARS-CoV-2 reference genome). In addition, R12 permutations appear also contributing to the G + C content balance in a significant way. For a more distant comparison, SARS-CoV and its civet counterpart (sequence referend to SARS-Cov-2) show almost identical mutation spectra between the two but different from the SARS-Cov-2 cluster that are truly based on within-population variations (Figure 2B). A note to add is that the R1 permutation A-to-G and G-to-A of these two datasets are not as deviated as C-to-U and U-to-C, and the fact is also seen in the within-population permutations in the SARS-CoV-2 dataset. From a structure point of view, molecular weight differences (Table 1) between the two sets of permutations may not be as significant in terms of structural features as what between the two purines, since the C-to-U pair is based on A–G exchange and the A-to-G pair is based on U–C exchange.
Two rules can be drawn clearly from the above observations. First, R1 permutations are always dominant to alter G + C content, flowed by R12 permutations to alter purine content. Second, these permutations have interchangeable pairing scheme, such as C-to-U can pair up with G-to-A or A-to-G, along the path of approximation toward the best fit to what the host genome and ribogenome composition are able to tolerate and compromise. It is tempting to propose that such rules may reflect structural principles as to how RdRPs and their associated proteins evolve to take the advantage of protein-coding complexity in responding to the composition dynamics. What we have not mentioned here is how strand-biased replication and copy number variables affect the mutation landscape of the SARS-CoV-2 data but anticipate clear interpretations based on our understanding of the mechanistic process of replication-transcription within SARS-CoV-2 and among other CoVs.
A history of compositional dynamics among human-infecting coronaviruses
Our decades-long studies have deciphered how mutation spectra relate to DNA polymerase complexity and proposed mechanistic explanation for genome compositional dynamics of the Kingdom of Bacteria [17], [18], [19], [20]. In particular, we conclude that G + C content variation for bacterial DNA genomes is dictated by a DnaE grouping scheme, which confines genomic G + C content into rather fixed boundaries among bacterial phyla [18], [19]. The same principles are mostly applicable to the RNA viruses where the genome compositional dynamics is characteristics of RNA RTCs. Another major point from our studies is a model – the pendulum model, where we propose that compositional variables are interconnected to the codon table or its organizational principles [15], [16].
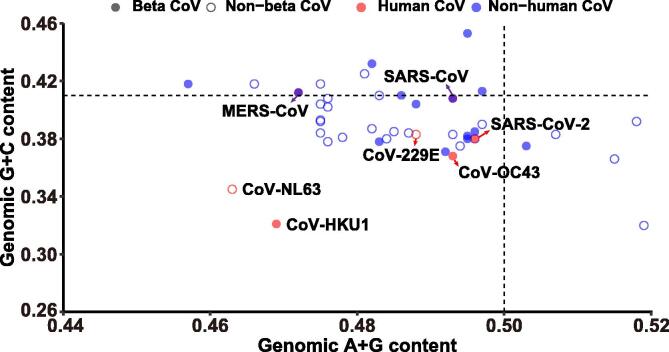
The two essential, apparently simple but extremely informative genomic sequence parameters: G + C and purine contents exhibit an overall compositional homeostasis of genome sequences. The full spectra of the G + C content and purine contents range from 20% to 80% and 40% to 60% [17], [18], [19], respectively. In the case of CoVs, their G + C and purine contents haven both been drifting between even narrower ranges, from 32.0% to 45.3% and from 34.5% to 51.9%, respectively (Figure 4). To cope with host operational systems, mostly those of cellular ribogenomes and proteomes as well as their complicated networking, viral genomes have to shape theirs in whatever possible ways to fit what the hosts have. In general, the genome of bats has a slightly higher G + C content (42.3%) [21] than that of humans (40.9%) [22]. The ribogenome of mammals usually has a higher G + C content than that of the genomic average, which is about ∼49%. Therefore, the genomic G + C content of CoVs has to drift below or above these particular boundaries. In reality, they appear to have G + C contents deviating around a mammalian genomic average and their purine content is also in a narrower range, perhaps due to a limitation of the negatively-correlated G + C content. Several striking observations can be made clearly. First, the most closely related CoVs to SARS-CoV-2 (38.0%, 49.6%) are four in the plot: two from bats (RaTG13, 38.0%, 49.5%; RmYN02, 38.2%, 49.5%) [23], [24], the other from a pangolin (pangolin_Guangxi_P4L_2017, 38.5%, 49.6%5) [25], and the fourth one overlapping completely in the two contents, which comes from a beta-CoV isolated from a vole (Myodes rufocanus, commonly found in the northern provinces of China, including Heilongjiang, Inner Mongolia, Hebei, Shanxi, Xinjiang, and Jilin) in 2014 (RtMruf-CoV-2/JL2014, 38.0%, 49.6%) [26]. The most closely related CoVs from intermediate hosts are those of camels to MERS and civets to SARS, and their compositional contents completely overlap with those of the human viruses since their mutations are truly within-population based on the establishment of intermediate host status. Second, together with the “new comer” SARS-CoV-2, all the “old timer” human CoVs, hasCoV-OC43, hsaCoV-229E, hsaCoV-HKU1, and hsaCoV-NL63 [27], have much lower G + C contents and lower purine contents than the other two “new comers”, MERS-CoV and SARS-CoV, which have G + C contents above the human average. In addition, the G + C content of most human CoVs appear drifting toward a lower value. Our explanations for the two phenomena are two-fold; for higher G + C content of SARS-CoV and MERS-CoV, we assume that SARS-CoV-2 is more advanced than the new comers, in that it not only has an almost optimal purine content but also a lower perhaps close to an optimum of G + C contents best fitting to a virus-host strangle and compromise; for much lower G + C content of the older timer human CoVs, we assume that they have passed many selection hurdles for maintaining their optimal G + C contents so that their compositions, G + C and/or purine contents are free to drift toward lower ends and even absurdity, such as the oldest hsaCoV-NL63. Third, since the dominant permutations (C-to-U|A-to-G and U-to-C|G-to-A) are most insensitive to purine content variation, the closer the position is it to the 50% line, the lesser related mutations would occur in the CoVs nearby. As a result, other permutation types, such as the purine-content sensitive A-to-U and G-to-C as well as the reverse, might be encouraged and discouraged await further exploitation of compositional diversity toward new fitness landscapes. Fourth, none of the currently-identified close relatives of SARS-CoV-2, bats and pangolins, appear to be true intermediate hosts that are capable of passing SARS-CoV-2 onto humans. However, according to this analysis, we are able to propose two scenarios for the outcome of searching for the intermediate mammalian host of SARS-CoV-2. Based on the fact that there are so many closely-related CoVs to the human viruses and these CoVs are so similar among themselves in genome composition parameters, such as their between-population mutation spectra, indirect transmission via wild animals, bats or other mammals, to humans appear unnecessary. The alternative scenario is that even if the assumed intermediate hosts may exist but they are so unpredictable in number of possible species, presumably involvement of large populations and geographic eras, that direct transmission through casual and short-term contacts cannot be easily verified, let alone the fact that the viral genomes keep changing in both humans and animals at the same time and in a very fast pace (https://www.gisaid.org/about-us/mission/; https://bigd.big.ac.cn/ncov/).
Figure 4.
Compositional variations of human and closely-related CoVs
Human CoVs (red solid circles) vs. non-human CoVs (blue solid circles) and betaCoVs (solid circles) vs. non-betaCoVs (open circles) are distinguished. There are three completely overlapping circles: two around the human average G + C content (the horizontal dashed line) are SARS-CoV and MERS-CoV, together with their representative within-population zoonotic counterparts, a civet and a camel, respectively (blue + red = purple); one near the vertical 50% purine content line (dashed) is the SARS-CoV-2 so happens overlapping with a vole CoV (a red solid circle surrounded by a purple open circle). The complete list of the samples and compositional parameters: (1) Alphacoronavirus Bat-CoV/P.kuhlii/Italy/3398–19/2015; 0.404, 0.475; (2)Bat coronavirus 1A; 0.381, 0.478; (3) Bat coronavirus BM48-31/BGR/2008; 0.404,0.488; (4) Bat coronavirus CDPHE15/USA/2006; 0.408, 0.476; (5) Bat coronavirus RaTG13; 0.38, 0.495; (6) Beluga whale coronavirus SW1; 0.392,0.518; (7) BetaCoV_pangolin_Guangxi_P4L_2017; 0.385, 0.496;(8) Betacoronavirus Erinaceus/VMC/DEU/2012; 0.375, 0.503; (9)Bovine coronavirus; 0.371, 0.492; (10) BtMr-AlphaCoV/SAX2011; 0.41, 0.483; (11) BtNv-AlphaCoV/SC2013; 0.418, 0.475; (11) BtRf-AlphaCoV/HuB2013; 0.383, 0.493; (12) BtRf-AlphaCoV/YN2012; 0.378, 0.476; (13) Bulbul coronavirus HKU11-934; 0.387, 0.482; (14) Camel alphacoronavirus; 0.384, 0.487; (15) Canada goose coronavirus strain Cambridge_Bay_2017; 0.384, 0.475; (16) Ferret coronavirus; 0.39, 0.497; (17) hsa229E; 0.383, 0.488; (18) hsaHKU1; 0.321, 0.469; (19) hsaNL63; 0.345, 0.463; (20) hsaOC43; 0.368, 0.493; (21) Lucheng Rn rat coronavirus; 0.402,0.476; (22) MERS-CoV; 0.412, 0.472; (23) Miniopterus bat coronavirus HKU8; 0.418, 0.466; (24) Mink coronavirus strain WD1127; 0.375, 0.494; (25) Munia coronavirus HKU13-3514; 0.425,0.481; (26) NL63-related bat coronavirus; 0.392, 0.475; (27) Pipistrellus bat coronavirus HKU5; 0.432, 0.482; (28) Rat coronavirus Parker; 0.413, 0.497; (29) Rhinolophus bat coronavirus HKU2; 0.393, 0.475; (30) Rodent coronavirus isolate RtMruf-CoV-2/JL2014; 0.38, 0.496; (31) Rousettus bat coronavirus; 0.453, 0.495; (32) Rousettus bat coronavirus HKU10; 0.385,0.485; (33) Rousettus bat coronavirus HKU9; 0.41, 0.486; (34) Severe acute respiratory syndrome coronavirus 2; 0.38, 0.496; (35) SARS-CoV-2; 0.408, 0.493; (36) Shrew coronavirus isolate Shrew-CoV/Tibet2014; 0.366, 0.515; (37) Thrush coronavirus HKU12-600; 0.38, 0.484; (38) Turkey coronavirus; 0.383,0.507; (39) Tylonycteris bat coronavirus HKU4; 0.378, 0.483; (40) Wencheng Sm shrew coronavirus; 0.32, 0.519; (41) bat RmYN02; 0.382, 0.495; (42) mouse MHV A59; 0.418, 0.457.
In summary, once we place a viral genome on a three-dimensional space, several pillars drive its compositional and structural parameters to fit the cellular niche of its best host. Compositional parameters are permutations propelled by the RTCs and tailored to different strands. Strand specificity is also associated with order of synthesis and number of synthesized copies, which also relates to sensitivity of G + C and purine content alterations. The four R1 permutations vary dramatically, such as in the case of SARS-CoV-2, brutally forcing G + C content to decrease while maintaining a balanced purine content and the four R12 permutations as minor variables are seen as fine-tuned purine content. The four R2 permutations serve as the most content- and structure-sensitive set for best compositional and structural buffering, whose underlying structural parameters and their underlying mechanisms are more variable and complex to be deciphered. The signature low G + C content discussed in the literature represents as relaxed selection in cellular environment for parasitic lifestyles, especially for unicellular organisms, such as the best-known malaria parasite, Plasmodium falciparum, and some of its relatives [28], [29], [30]; however, the opposite is also true for its other relative, P. vivax, that has been increasing genomics G + C content toward higher values [31], [32]. Composition variability is also observed among virus-host comparative studies and falls into a similar category and a recent study has pointed that virus codon usage bias tends to be more similar to that of symptomatic hosts than that of natural hosts [33]. Nevertheless, the interaction between viral and host genomes, as well as other cellular omics are believed to be more complicated than the current thinking.
What do we expect when CoVs become frequent visitors of our shared world?
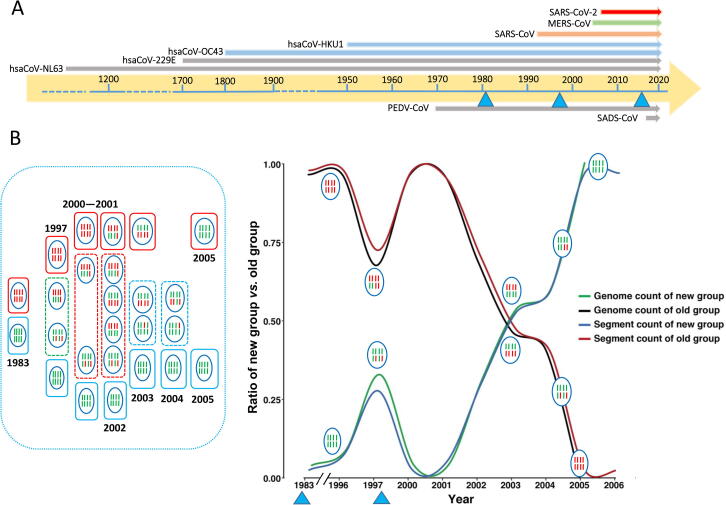
The three recent CoV outbreaks in human and domestic animals within the past three decades have demonstrated that a new wave of CoV infections has come to human communities and neighborhoods (Figure 4 and Figure 5A) [34], [35], [36], [37], [38], [39], [40]. This observation is benchmarked by the much lower G + C content and close-to-optimum purine content exhibited by SARS-CoV-2 and its closely related natural hosts, bats, and other possible mammal hosts, such as pangolins. Specially, the lower G + C content of these CoVs indicates less selective pressure in adaptation to cellular, physiological, and pathological environments of its adaptive hosts. This trend was not observed in SARS-CoV and MERS-CoV as well as their corresponding intermediate hosts. Although we have yet to pin down the true natural host of SARS-CoV-2 as a single species or single population, its trails together with those of its close relatives are rather clear. It most likely has a recently-recombined genome in coping with relatively-frequent host-jumping events. It may come from a single species of CoV harbored by mixed bat populations that live, may seasonally migrate locally, not far from human habitats. Most importantly, the current virus species and its clades may all be inoculated from the same habitats sporadically within a period of several months in late 2019 since they have not subjected to strong selective pressure from human immune systems and populations.
Figure 5.
The very strong El Niño events and viral outbreaks of CoVs and AIVs A. The recent and historic CoV outbreaks are displayed over time based on assumptions that these CoV outbreaks are not isolated events. The three recently-recorded El Niño events (blue triangles) are 1982–1983, 1997–1998, 2015–2016. Two porcine CoV outbreaks are also indicated (below the horizontal arrowed bar for time), PEDV (porcine epidemic diarrhea virus, an alphacoronavirus) and SADS-CoV (swine acute diarrhea syndrome, an alphacoronavirus) [56]. B. A historic study based on HP H5N1 AIV genome sequences in segments (blue oval circles with 8 solid bars; red, segments of the Old group; green, segments of the New group). Two genome types once had dwelled in a near-by islands (red and blue frames), the New (red) and the Old (green) groups. They were carried to China through the flyway by migratory birds seen as sampling the local populations (shared territories, green dashed frames) until the mixing population (1/4 over each other) were destroyed by the strong El Niño event in 1997–1998 (red dashed frames). The process started again after 2001 and reached its half way in 2003. The complete takeover of Old group’s territory happened after 2005. This takeover had struggled in a time period of ∼ 10 years, with matching point around 2003 (blue dashed frames). An obvious slow-down point is around 1997, a showdown by nature, the yet-strongest El Niño event in the recent history, and it was within the recovery phase, 2003 when HP H5N1 and SARS-CoV outbreaks both happened in China. This plot is reproduced based on data from reference [35], where genome segments of the old and the new groups are counted as percentages of the total.
The question now to be addressed is where are the habitats and why. In a past intensive study of SARS-CoV and highly-pathogenic avian influenza virus H5N1 together from 2003 to 2006, we have learnt two relevant lessons about the biology of RNA viruses [37], [38], [39], [40], [41], [42], [43]. The first is about SARS-CoV and its habitat story. It is unquestionable that the epidemics started in the Guangdong province where both civets and humans harbored the same population of CoVs, which were identified over a period of a few months [38], [39], [40], [41], [42], [43], [44]. The striking fact is that most complete and numerous ORF8-defective CoVs were found in Guangdong in the early phase of the 2003 outbreak and an only form found outside the province was a minimum-defective CoV with a 29-nt deletion of ORF8 in the mid-phase of the outbreak (several other slightly larger deletions in odd numbers, such as 53-nt and 87-nt, symmetric to the same site were also identified from CoV isolates in Guangdong; Jun Yu unpublished data). This phenomenon suggests that SARS-CoV exhibited defectiveness when infecting humans and a deleted form allowed the virus to escape a host-defense element and to gain ability for a short-term transmission in the middle of the epidemic struggles among infected humans. A note to add is that a similar deletion in principle has also been identified in ORF8 of SARS-CoV-2 in Singapore [44]. These are useful clues for understanding the infection processes and immune responses at cellular and molecular levels of SARS CoV-2 and COVID-19.
The second is an avian flu story about sequence studies from a historic collection, in particular the highly-pathogenic (HP) H5N1 found in China [37], [38]. In this study, we sequenced (139 isolates), analyzed (189 isolates) HP H5N1 genomes, and discovered several important facts. The first observation suggests that there had been two groups of H5N1 AIVs, one termed the Old Group and the other the New. It took a 23-year period (1983–2006) for the New Group to slowly replaced the Old and became prevalent in China (Figure 5B). Mechanisms of this slow takeover are multifold. The first is re-assortment of the segmented viral genomes, where the New had replaced the Old one or a few at a time over these years until absolute dominance. This process appeared so vivid that the strongest 1997–1998 El Niño had shown its mark in this as seen a delayed timing of the increasing AIVs of the New group [30], [31]. El Niño and La Niña are two opposing global climate patterns with distinction among events based on oceanic surface temperature changes, which are natural parts of the climate system and have strong impact on wildlife and ecosystems worldwide, especially the unusual warming and cooling of surface waters in the eastern Pacific Ocean (https://www.ncdc.noaa.gov/cag/). There have been three very strong El Niño events in the past, 1982–1983, 1997–1998, and 2015–2016 and every one of them appears relevant to our observations and discussion here [45], [46], [47], [48]. For instance, the New group of HP H5N1 AIVs started to emerge after the first event and the rise of them delayed by the second event, and the third may be linked to other AIVs, such as the recently-reported prevalent H6 types [49]. Second, the reasons why the New Group had replaced the Old are its potency of infection rather than specificity to any particular hosts [50], [51], [52], [53], [54], [55], [56] and multiple environmental factors that encourage the change, such as distinct yet understood migration networks and flyways [53], [54]. Third, all these elements point to a multidisciplinary, mammoth and concerted effort to understand all major zoonotic and human viruses as well as their hosts in a broader scope and larger landscape, which must include biodiversity [55], ecology, geography, genetics, cell biology and physiopathology of both viruses and their possible hosts.
What behind these observations is an assumption that there was a distant source pool for the viral genomes and it was its slow takeovers, the Old by the New, that had been spreading out by the seasonal migrating birds over time. In other words, what we had sampled in China was a mirrored process of HP AIV Old-by-New takeover over time in the source viral genome pool afar not the real propagation of AIV HP H5N1 in China. We did at the time started vaccine development [56], [57], together with other biological and cellular studies but called it quits as uncertainty about other deterministic factors that may delay the next outbreak. That thought came out more than 10 years before the 2015–2016 El Niño peak, but now we are right in its recover phase 4 to 5 years after. Nonetheless, the lesson learnt here is what we scrutinize on the sequence dataset of SARS-CoV-2 may not provide any clue about how the CoVs are mutating and changing to gain access to human hosts in the bat populations, and some longitudinal studies on bat and suspected mammal (such as pangolins and rodents) populations are most urgent. We certainly need to compare notes on AIV and CoV studies since they may be deeply related in terms of shared habitats, seasonal outbreaks, similarity in RNA biology and cell biology.
Conclusion
CoVs once prevalent among wild bat species have completed their course in preparing their genomes to be able to freely jump over any compositional and structural hurdles, as particularly focused in this discussion, and they may now be ready to evade many mammalian species constantly in addition to bats and humans. A full-spectrum CoV defense plan is of importance to all nations, including scientific and medical communities, which are undoubtedly pushed to the forefront. Our actions in series are desperately needed in the fields of genomics, proteomics and bioinformatics. First, we need to propose and practice a knowledgebase-centric protocol (including thorough annotation, authentic dataset, error assessment, interactive display, visualization, etc.) so that data not only can be shared freely by all experts and laymen but also digested in correct and professional ways. Second, we need to understand and associate mutations (in terms of synonymous/nonsynonymous mutations, permutations, mutation spectra, etc.) to genes and protein structures, as well as clinical parameters and data (such as pathology and symptoms) by developing mathematical models and bioinformatic algorithms. Of course, large-scale genomics data (such as studies on genomes of related wild animals) and datasets (high-quality for in-depth analysis) should be collected and housed by other databases/knowledgebases for multi-disciplinary research activities. Third, we should make a full list of projects on viral biology, especially remove host-associated species barriers, including both wild and domestic animals as research subjects. Finally, cellular and animal studies should all be welcome to provide vital information for vaccine and drug designs.
In a broader scope, our ultimate search for the origin of SARS-CoV-2 may not easily succeed as the virus is still propagating and evading new territories – they are everywhere already. From the current collection of genomes and mutations, we have yet to paint a portrait of the single genome and what it gives rise to, the offspring clades; they may not from a single virus, as it seems at this point of time, but a population that we have sampled in a long period of time that could be months. It is up to the viral genome source pools as what they are now and in the years to come. What we need now is to be prepared in two fronts; one is to be ready for the next wave by the end of this year and the other is to gain as much information as possible from the current pandemics. Special attentions are needed to start wild life surveys for CoVs even though activities of similar kinds have been carried on after the SARS-CoV outbreak [58]. Another version of SARS-CoV-2 will reemerge, and we may not have to wait another 17 years for sure. Both bats and migrating birds are to be targeted for the surveys and a special focus should be the broader territories of Southeast Asia. A new international organizational supporting model may be needed across nations as a major task force to fight the AIVs and CoVs together.
Competing interests
The author declares no competing interests
Acknowledgments
Acknowledgments
The author likes to acknowledge Xufei Teng, Qianpeng Li, and Dr. Yanan Chu for technical support, and Drs. Zhang Zhang, Shuhui Song, Jingfa Xiao, Lina Ma, Lili Hao, and Meng Zhang for helpful discussion and critical reading of this manuscript. This work is supported by the National Natural Science Foundation of China (NSFC, Grant No. 31671350), Key programs of the Chinese Academy of Sciences (Grant No. QYZDY-SSW-SMC017).
ORCID
0000-0002-2702-055X (Jun Yu)
Handled by Fangqing Zhao
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China.
References
- 1.Shi H., Wei J., He C. Where, when, and how: context-dependent functions of rna methylation writers, readers, and erasers. Mol Cell. 2019;74:640–650. doi: 10.1016/j.molcel.2019.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vendeix F.A., Munoz A.M., Agris P.F. Free energy calculation of modified base-pair formation in explicit solvent: a predictive model. RNA. 2009;15:2278–2287. doi: 10.1261/rna.1734309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Posthuma C.C., te Velthuis A.J., Snijder E.J. Nidovirus RNA polymerases: complex enzymes handling exceptional RNA genomes. Virus Res. 2017;234:58–73. doi: 10.1016/j.virusres.2017.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drake J.W., Charlesworth B., Charlesworth D., Crow J.F. Rates of spontaneous mutation. Genetics. 1998;148:1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ogando N.S., Ferron F., Decroly E., Canard B., Posthuma C.C., Snijder E.J. The curious case of the nidovirus exoribonuclease: its role in rna synthesis and replication fidelity. Front Microbiol. 2019;10:1813. doi: 10.3389/fmicb.2019.01813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pauly M.D., Procario M.C., Lauring A.S. A novel twelve class fluctuation test reveals higher than expected mutation rates for influenza A viruses. Elife. 2017;6:e26437. doi: 10.7554/eLife.26437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iyer R.R., Pluciennik A., Burdett V., Modrich P.L. DNA mismatch repair: functions and mechanisms. Chem Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 8.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 9.Antoniali G., Malfatti M.C., Tell G. Unveiling the non-repair face of the Base Excision Repair pathway in RNA processing: A missing link between DNA repair and gene expression? DNA Repair (Amst) 2017;56:65–74. doi: 10.1016/j.dnarep.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 10.Sawicki S.G., Sawicki D.L., Siddell S.G. A contemporary view of coronavirus transcription. J Virol. 2007;81:20–29. doi: 10.1128/JVI.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jia H., Gong P. A structure-function diversity survey of the RNA-dependent RNA polymerases from the positive-strand rna viruses. Front Microbiol. 2019;10:1945. doi: 10.3389/fmicb.2019.01945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu J. A content-centric organization of the genetic code. Genomics Proteomics Bioinformatics. 2007;5:1–6. doi: 10.1016/S1672-0229(07)60008-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao J.-F., Yu J. A scenario on the stepwise evolution of the genetic code. Genomics Proteomics Bioinformatics. 2007;5:143–151. doi: 10.1016/S1672-0229(08)60001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Z., Yu J. On the organizational dynamics of the genetic code. Genomics Proteomics Bioinformatics. 2011;9:21–29. doi: 10.1016/S1672-0229(11)60004-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Z., Yu J. The pendulum model for genome compositional dynamics: from the four nucleotides to the twenty amino acids. Genomics Proteomics Bioinformatics. 2012;10:175–180. doi: 10.1016/j.gpb.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao X.Q., Zhang Z., Yan J.W., Yu J. GC content variability of eubacteria is governed by the pol III alpha subunit. Biochem Biophys Res Commun. 2007;356:20–25. doi: 10.1016/j.bbrc.2007.02.109. [DOI] [PubMed] [Google Scholar]
- 18.Hu J., Zhao X., Zhang Z., Yu J. Compositional dynamics of guanine and cytosine content in prokaryotic genomes. Res Microbiol. 2007;158:363–370. doi: 10.1016/j.resmic.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 19.Wu H., Zhang Z., Hu S., Yu J. On the molecular mechanism of GC content variation among eubacterial genomes. Biol Direct. 2012;7:2. doi: 10.1186/1745-6150-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu H., Fang Y., Yu J., Zhang Z. The quest for a unified view of bacterial land colonization. ISME J. 2014;8:1358–1369. doi: 10.1038/ismej.2013.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasai F., O’Brien P.C., Ferguson-Smith M.A. The bat genome: GC-biased small chromosomes associated with reduction in genome size. Chromosoma. 2013;122:535–540. doi: 10.1007/s00412-013-0426-9. [DOI] [PubMed] [Google Scholar]
- 22.Piovesan A., Pelleri M.C., Antonaros F., Strippoli P., Caracausi M., Vitale L. On the length, weight and GC content of the human genome. BMC Res Notes. 2019;12:106. doi: 10.1186/s13104-019-4137-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou H., Chen X., Hu T., Li J., Song H., Liu Y. A novel bat coronavirus closely related to SARS-CoV-2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Curr Biol. 2020;30:2196–2203.e3. doi: 10.1016/j.cub.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao K., Zhai J., Feng Y., Zhou N., Zhang X., Zou J.J. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature. 2020;583:286–289. doi: 10.1038/s41586-020-2313-x. [DOI] [PubMed] [Google Scholar]
- 26.Wu Z., Lu L., Du J., Yang L., Ren X., Liu B. Comparative analysis of rodent and small mammal viromes to better understand the wildlife origin of emerging infectious diseases. Microbiome. 2018;6:178. doi: 10.1186/s40168-018-0554-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forni D., Cagliani R., Clerici M., Sironi M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;25:35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Videvall Plasmodium parasites of birds have the most AT-rich genes of eukaryotes. Microb Genomics. 2018;4:e000150. doi: 10.1099/mgen.0.000150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ca I., Nad L., Eric L. Tail wags the Dog? Functional gene classes driving genome-wide GC content in Plasmodium spp. Genome Biol Evol. 2019;11:497–507. doi: 10.1093/gbe/evz015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamilton W.L., Antoine C., Otto T.D., Mihir K., Fairhurst R.M., Rayner J.C. Extreme mutation bias and high AT content in Plasmodium falciparum. Nucleic Acids Res. 2017;45:1889–1901. doi: 10.1093/nar/gkw1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goel P., Singh G.P. Divergent pattern of genomic variation in Plasmodium falciparum and P. vivax. F1000 Research. 2016;5:2763. [Google Scholar]
- 32.Nikbakht H., Xia X., Hickey D.A., Golding B. The evolution of genomic GC content undergoes a rapid reversal within the genus Plasmodium. Genome. 2014;57:507–511. doi: 10.1139/gen-2014-0158. [DOI] [PubMed] [Google Scholar]
- 33.Chen F., Wu P., Deng S., Zhang H., Hou Y., Hu Z. Dissimilation of synonymous codon usage bias in virus–host coevolution due to translational selection. Nat Ecol Evol. 2020;4:589–600. doi: 10.1038/s41559-020-1124-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou P., Fan H., Lan T., Yang X.L., Shi W.F., Zhang W. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556:255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olival K.J., Hosseini P.R., Zambrana-Torrelio C., Ross N., Bogich T.L., Daszak P. Host and viral traits predict zoonotic spillover from mammals. Nature. 2017;546:646–650. doi: 10.1038/nature22975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X.W. Beijing Institute of Genomics, Chinese Academy of Sciences; 2006. Large-scale sequencing and analysis of avian influenza viruses. A Ph.D. thesis. [Google Scholar]
- 38.Zhu Q.Y., Qin E.D., Wang W., Yu J., Liu B.H., Hu Y. Fatal infection with influenza A (H5N1) virus in China. N Engl J Med. 2006;354:2731–2732. doi: 10.1056/NEJMc066058. [DOI] [PubMed] [Google Scholar]
- 39.Hu J., Wang J., Xu J., Li W., Han Y., Li Y. Evolution and variation of the SARS-CoV genome. Genomics Proteomics Bioinformatics. 2003;1:216–225. doi: 10.1016/S1672-0229(03)01027-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bi S., Qin E., Xu Z., Li W., Wang J., Hu Y. Complete genome sequences of the SARS-CoV: the BJ Group (Isolates BJ01-BJ04) Genomics Proteomics Bioinformatics. 2003;1:180–192. doi: 10.1016/S1672-0229(03)01023-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiu R.W., Chim S.S., Tong Y.K., Fung K.S., Chan P.K., Zhao G.P. Tracing SARS-coronavirus variant with large genomic deletion. Emerg Infect Dis. 2005;11:168–170. doi: 10.3201/eid1101.040544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chinese S.M.E.C. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303:1666–1669. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- 43.Song H.D., Tu C.C., Zhang G.W., Wang S.Y., Zheng K., Lei L.C. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc Natl Acad Sci U S A. 2005;102:2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su YCF, Anderson DE, Young BE, Linster M, Zhu F, Jayakumar J, et al. Discovery and genomic characterization of a 382-nucleotide deletion in ORF7b and ORF8 during the early evolution of SARS-CoV-2. mBio 2020;11:e01610-20. [DOI] [PMC free article] [PubMed]
- 45.Changnon S.A. Oxford University Press; New York: 2000. El Niño, 1997–1998: the climate event of the century. [Google Scholar]
- 46.Newbery D.M., Clutton-Brock T.H., Prance G.T. Imperial College Press; London: 2000. Changes and disturbance in tropical rainforest in South-East Asia. [Google Scholar]
- 47.United Nations Development Programme (UNDP), United Nations Economic and Social Commission for Asia and the Pacific (ESCAP), United Nations Office for the Coordination of Humanitarian Affairs (OCHA), Regional Integrated Multi-Hazard Early Warning System for Africa and Asia (RIMES), (APCC) tACC (2017), 'Enhancing Resilience to Extreme Climate Events: Lessons from the 2015-2016 El Niño Event in Asia and the Pacific'.
- 48.Anyamba A., Chretien J.P., Britch S.C., Soebiyanto R.P., Small J.L., Jepsen R. Global disease outbreaks associated with the 2015–2016 El Nino Event. Sci Rep. 2019;9:1930. doi: 10.1038/s41598-018-38034-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu C., Li X., Zhu C., Zhou F., Tang W., Wu D. Co-circulation of multiple reassortant H6 subtype avian influenza viruses in wild birds in eastern China, 2016–2017. Virol J. 2020;17:62. doi: 10.1186/s12985-020-01331-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Imai H., Dinis J.M., Zhong G., Moncla L.H., Lopes T.J.S., McBride R. Diversity of influenza A(H5N1) viruses in infected humans, Northern Vietnam, 2004–2010. Emerg Infect Dis. 2018;24:1128–1238. doi: 10.3201/eid2407.171441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Borremans B., Faust C., Manlove K.R., Sokolow S.H., Lloyd-Smith J.O. Cross-species pathogen spillover across ecosystem boundaries: mechanisms and theory. Philos Trans R Soc Lond B Biol Sci. 2019;374:20180344. doi: 10.1098/rstb.2018.0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Geoghegan J.L., Holmes E.C. Predicting virus emergence amid evolutionary noise. Open Biol. 2017;7 doi: 10.1098/rsob.170189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tian H., Zhou S., Dong L., Van Boeckel T.P., Cui Y., Newman S.H. Avian influenza H5N1 viral and bird migration networks in Asia. Proc Natl Acad Sci U S A. 2015;112:172–177. doi: 10.1073/pnas.1405216112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Olsen B., Munster V.J., Wallensten A., Waldenstrom J., Osterhaus A.D., Fouchier R.A. Global patterns of influenza a virus in wild birds. Science. 2006;312:384–388. doi: 10.1126/science.1122438. [DOI] [PubMed] [Google Scholar]
- 55.Hosseini P.R., Mills J.N., Prieur-Richard A.H., Ezenwa V.O., Bailly X., Rizzoli A. Does the impact of biodiversity differ between emerging and endemic pathogens? The need to separate the concepts of hazard and risk. Philos Trans R Soc Lond B Biol Sci. 2017;372 doi: 10.1098/rstb.2016.0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang L., Zhu Q., Qin E., Yu M., Ding Z., Shi H. Inactivated SARS-CoV vaccine prepared from whole virus induces a high level of neutralizing antibodies in BALB/c mice. DNA Cell Biol. 2004;23:391–394. doi: 10.1089/104454904323145272. [DOI] [PubMed] [Google Scholar]
- 57.Qin E., Shi H., Tang L., Wang C., Chang G., Ding Z. Immunogenicity and protective efficacy in monkeys of purified inactivated Vero-cell SARS vaccine. Vaccine. 2006;24:1028–1034. doi: 10.1016/j.vaccine.2005.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]