Abstract
Sinonasal undifferentiated carcinoma (SNUC) is an aggressive malignancy harboring IDH2 R172 mutations in >80% cases. We explored the potential of genome-wide DNA methylation profiling to elucidate tumor biology and improve the diagnosis of sinonasal undifferentiated carcinoma and its histologic mimics. Forty-two cases, including sinonasal undifferentiated, large cell neuroendocrine, small cell neuroendocrine, and SMARCB1-deficient carcinomas and olfactory neuroblastoma, were profiled by Illumina Infinium Methylation EPIC array interrogating >850,000 CpG sites. The data were analyzed using a custom bioinformatics pipeline. IDH2 mutation status was determined by the targeted exome sequencing (MSK- IMPACTTM) in most cases. H3K27 methylation level was assessed by the immunohistochemistry-based H-score. DNA methylation-based semi-supervised hierarchical clustering analysis segregated IDH2 mutants, mostly sinonasal undifferentiated (n = 10) and large cell neuroendocrine carcinomas (n = 4), from other sinonasal tumors, and formed a single cluster irrespective of the histologic type. t-distributed stochastic neighbor embedding dimensionality reduction analysis showed no overlap between IDH2 mutants, SMARCB1-deficient carcinoma and olfactory neuroblastoma. IDH2 mutants demonstrated a global methylation phenotype and an increase in repressive trimethylation of H3K27 in comparison to IDH2 wild-type tumors (p < 0.001). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis showed no difference in pathway activation between IDH2-mutated sinonasal undifferentiated and large cell neuroendocrine carcinomas. In comparison to SMARCB1-deficient, IDH2-mutated carcinomas were associated with better disease-free survival (p = 0.034) and lower propensity for lung metastasis (p = 0.002). ARID1A mutations were common in small cell neuroendocrine carcinoma but not among IDH2 mutants (3/3 versus 0/18 and p < 0.001). IDH2 mutations in sinonasal carcinomas induce a hypermethylator phenotype and define a molecular subgroup of tumors arising in this location. IDH2- mutated sinonasal undifferentiated carcinoma and large cell neuroendocrine carcinoma likely represent a phenotypic spectrum of the same entity, which is distinct from small cell neuroendocrine and SMARCB1-deficient sinonasal carcinomas. DNA methylation-based analysis of the sinonasal tumors has potential to improve the diagnostic accuracy and classification of tumors arising in this location.
Keywords: sinonasal undifferentiated carcinoma, IDH2, DNA methylation
INTRODUCTION
Sinonasal undifferentiated carcinoma is a rare, aggressive malignancy of the sinonasal tract first described in 1986 as a high-grade epithelial malignancy with or without neuroendocrine differentiation [1]. Novel sequencing technologies helped elucidate the genetic underpinnings of this entity [2,3]. We detected oncogenic hotspot IDH2 R172 mutations in 82% sinonasal undifferentiated carcinoma but also in large cell neuroendocrine carcinoma and non-intestinal type adenocarcinoma [2]. IDH1/2 hotspot variants are known to occur in various malignancies including glioma [4], chondrosarcoma [5], intrahepatic cholangiocarcinoma [6], acute myeloid leukemia [7], angioimmunoblastic T-cell lymphoma [8] and solid papillary breast carcinoma with reverse polarity [9]. The putative oncogenic effect of IDH1/2 mutations is mediated via abnormally increased levels of the “oncometabolite” (R)-2-hydroxyglutarate, which increase repressive trimethylation of H3K9 and H3K27 [9–11] and induce acquisition of a hypermethylator phenotype [12]. These epigenetic changes, in consequence, significantly impair gene expression leading to block in cellular differentiation [10,11].
Sinonasal undifferentiated carcinoma is a diagnosis of exclusion and must be distinguished from a variety of morphologically similar tumors, such as SMARCB1-deficient carcinoma, nuclear protein in testis (NUT) carcinoma, large cell neuroendocrine carcinoma, small cell neuroendocrine carcinoma, olfactory neuroblastoma, Ewing sarcoma, melanoma or lymphoma [13–15]. The relative rarity of sinonasal undifferentiated carcinoma and the fact that the Schneiderian epithelium has enormous potential to give rise to clinically and histologically similar high-grade/poorly-differentiated carcinomas have historically resulted in pathologists often using this diagnosis for a spectrum of sinonasal poorly-differentiated carcinomas lacking substantial evidence of glandular, squamous or definitive neuroendocrine differentiation. This is best reflected in previous studies on NUT carcinoma and SMARCB1-deficient sinonasal carcinomas, the two molecularly defined malignancies which had been commonly misdiagnosed as sinonasal undifferentiated carcinoma due to their respective clinical and morphologic overlap with this entity [16–18].
There is a growing body of literature on using genome-wide DNA methylation profiling as a tool to refine tumor diagnosis and classification based on the epigenetic tumor signature [19]. This approach has been particularly successfully exploited in central nervous system malignancies leading to a re-definition of several entities such as, for example, atypical teratoid/rhabdoid tumor [20] and primitive neuroectodermal tumor of the central nervous system [21]. More recently, large sets of diagnostically optimized DNA methylome data on brain tumors were employed to design a machine-learning classifier algorithm as a robust diagnostic tool for any primary tumor arising in this location [22].
In this study, we performed genome-wide DNA methylation on a histologically diverse sinonasal tumor cohort including sinonasal undifferentiated carcinoma and its histologic mimics with the two specific aims: (1) to examine the effects of IDH2 R172 mutations on the tumor methylome in sinonasal carcinomas, and (2) to explore the potential of genome-wide DNA methylation profiling to elucidate tumor biology and improve pathologic diagnosis of sinonasal tumors, focusing in particular on high-grade sinonasal carcinomas.
MATERIALS AND METHODS
Patients and tissue samples
The study was approved by the Institutional Research Board of Memorial Sloan Kettering Cancer Center. A total of 62 formalin-fixed paraffin embedded sinonasal tumor samples (including the primary and metastasis in one case) from 61 patients diagnosed from 1995–2018 included 16 sinonasal undifferentiated carcinomas, 15 SMARCB1-deficient sinonasal carcinomas, 6 large cell neuroendocrine carcinomas, 6 poorly-differentiated carcinomas with focal glandular/acinar features, 4 poorly-differentiated carcinomas with neuroendocrine and glandular differentiation, 4 olfactory neuroblastomas 3 intestinal type adenocarcinomas, 3 small cell neuroendocrine carcinomas, 2 combined small cell neuroendocrine carcinomas and squamous cell carcinomas, 2 non-intestinal type adenocarcinoma, and a single HPV-related multiphenotypic sinonasal carcinoma. The experimental summary is provided in Supplementary Table 1.
Pathologic diagnostic criteria
Pathology slides were reviewed by 3 head and neck pathologists (SD, BX, RG) and diagnoses were rendered according to the World Health Organization classification [23]. In brief, a diagnosis of sinonasal undifferentiated carcinoma was made only in entirely undifferentiated epithelial tumors without any morphologic evidence of glandular or squamous differentiation, absent or focal and weak immunopositivity for neuroendocrine markers (synaptophysin, chromogranin and/or INSM1), negative NUT, retained Baf-47 (INI-1), and negative p16 immunostain and/or negative HPV RNA in situ hybridization (Supplementary Methods), [2]. Large cell neuroendocrine carcinomas were characterized by medium to large-sized tumor cells with coarse or specked chromatin, prominent nucleoli, and positive neuroendocrine and cytokeratin markers expression. Olfactory neuroblastoma diagnosis required neuroendocrine markers expression, positive S-100 in sustentacular pattern, and absent or focal (<25%) cytokeratin expression. SMARCB1-deficient carcinoma diagnosis was based on the nuclear loss of Baf-47 (INI-1) protein [18]. Tumors were given descriptive diagnoses if they did not belong to any definitive category. In two cases, SN_14 and SN_31, diagnoses were modified as described in Supplementary Methods.
DNA extraction and targeted massive parallel sequencing
Genomic DNA was extracted from formalin-fixed paraffin embedded tumor sections and a targeted hybridization capture-based massive parallel sequencing was performed by Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT™), a clinically validated molecular assay interrogating somatic variants in 279–468 cancer-related genes as previously described [2,24,25]. Genetic analysis of 34 cases rendered by MSK-IMPACT™ (n=30) or IDH1/2 Sanger sequencing (n=4) were previously reported [2]. Additional 23 cases including 13 clinical and 10 research samples were analyzed by MSK-IMPACT™ using 279 (n=2), 410 (n=5) or 468 (n=16) gene panel. Matched normal tissue DNA (n=14) or unmatched pool normal DNA (n=9) were used for the analysis. Oncogenic potential of somatic genetic alterations was determined by OncoKB [26]. In 5 cases no sequencing was performed and the IDH2/SMARCB1 status was determined by immunohistochemistry as described below.
DNA methylation profiling
DNA methylation profiling was performed in 42 cases with available material for testing (Supplementary Table S1). The Illumina EPIC Array 850 Bead-Chip (850K) array was used for a genome-wide methylation profiling to determine the DNA methylation status of >850,000 CpG sites (Illumina, San Diego, CA) as described previously [27]. The raw idats generated from iScan was processed and analyzed using Bioconductor R package Minfi [28]. All the Illumina EPIC array probes were normalized using quantile normalization and corrected for background signal. Samples were checked for their quality using mean detection p values (p value <0.05). Thirty-nine samples passed the quality criteria and were used for the subsequent analysis. To identify the differentially methylated CpG probes, the samples were grouped in two ways; (1) grouped based on the IDH2 mutation status to observe the global methylation differences between wild-type and IDH2-mutated tumors, and (2) grouped based on the histologic categories of the tumors. Beta values were generated and probes with FDR cutoff (q<0.05) were considered as the most significantly variably methylated probes. Beta values <0.2 designated hypomethylation and Beta value >0.8 designated hypermethylation. Heatmaps were produced using the semi-supervised hierarchical clustering of the top 10,000 significantly differentially methylated probes across the samples. Red represented hypermethylation and blue represented hypomethylation. For each differential methylation of CpG islands based on IDH2 status and histologic category, semi-supervised hierarchical clustering of samples was analyzed using t-distributed stochastic neighbor embedding method [29] to show the natural lower dimensional clustering of the samples and was applied on the 10,000 most significantly differentially methylated probes obtained using Minfi package [28]. Previously published DNA methylation data by Capper et al. were used for comparison [22].
Pathway Analysis
Significantly differentially methylated probes between sinonasal undifferentiated carcinoma vs. large cell neuroendocrine carcinoma groups were used to identify the enriched signaling pathways in KEGG database using R package ClusterProfiler [30]. Dot plot of pathways represents the ratio of genes (x-axis) against the KEGG signaling pathways (y-axis) identifying the ratio of genes enriched in each signaling pathway, with color indicating the level of significance and size represents the number of genes in each pathway.
Immunohistochemistry
Immunohistochemistry for mutated IDH2 (clone 11C8B1, 1:2000 dilution, catalogue # 26408; NewEast Biosciences, Malvern, PA) was performed on a Leica-Bond-3 automated stainer platform (Leica, Buffalo Grove, IL), employing a secondary polymeric detection kit (Refine, Leica) and a heat-based antigen retrieval method using a high pH retrieval buffer (ER2, Leica, 30 minutes). Positive cases rendered a moderate to strong granular cytoplasmic staining [31]. immunohistochemistry for H3K27me3 (clone C36B11, 1:200 dilution, catalogue # 9733, Cell Signaling Technology, Danvers, MA), and INSM1 (clone A-8 clone, 1:250 dilution, catalogue # SC-271408, Santa Cruz Biotechnology, Dallas, TX) were performed on the Ventana Benchmark Ultra platform (Ventana Medical Systems Inc., Tucson, AZ, United States) using a streptavidin-biotin-peroxidase secondary (iView, Ventana). Ancillary studies performed as part of the diagnostic work-up at the time of clinical diagnosis are included in Supplementary Table 2 and details on antibodies and probes are included in Supplementary Table 3. H-score was determined based on the intensity of nuclear staining and the proportion of labeled tumor cells as previously described [32]. In brief, the nuclear staining intensity was graded as 0 (no staining), 1 (weak), 2 (moderate), 3 (strong) and used in the formula: (% of positive cells, intensity 3 × 3) + (% of positive cells, intensity 2 × 2) + (% of positive cells, intensity 1 × 1) = H-score.
Statistics for clinical data analyses
Statistical analyses were performed using the SPSS software 24.0 (IBM Corporation, New York, NY, U.S.). Clinical-pathological-molecular comparisons were made using appropriate statistical tests, i.e. Fisher’s exact test for qualitative variables and two-tailed Student’s t test for continuous variables. Prognostic value relative to the molecular status was determined using Log rank test to obtain disease-free survival, disease-specific survival, or overall survival. P value <0.05 was considered significant.
RESULTS
1. DNA methylation-based clustering analysis of sinonasal tumors
DNA methylation profiling rendered successful results in 39 samples including 10 sinonasal undifferentiated carcinoma, 4 large cell neuroendocrine carcinoma, 5 SMARCB1-deficient carcinomas, 4 olfactory neuroblastomas, 5 poorly-differentiated carcinomas with focal glandular/acinar differentiation, 3 poorly-differentiated carcinomas with neuroendocrine and glandular differentiation, 2 small cell neuroendocrine carcinoma, 2 combined small cell neuroendocrine-squamous cell carcinomas, 2 intestinal type adenocarcinomas, and 2 non-intestinal type adenocarcinomas. We identified 4 distinct clusters using the hierarchical clustering. The analysis showed that IDH2-mutated carcinomas formed a distinct cluster irrespective of the histologic type and included sinonasal undifferentiated carcinoma, large cell neuroendocrine carcinoma, one poorly-differentiated carcinoma with focal glandular/acinar differentiation, and a single poorly-differentiated non-intestinal type adenocarcinoma. Except for the latter case (SN_11), which exhibited a somewhat distinct methylation signature possibly associated with its morphology i.e. glandular differentiation, the remaining IDH2 mutants, including all sinonasal undifferentiated carcinomas and large cell neuroendocrine carcinomas, formed a compact cluster suggesting largely similar epigenetic signature, which separated them from other tumors (Figure 1A). Other well-defined methylation classes included SMARCB1-deficient carcinoma (n=5) and olfactory neuroblastoma (n=4), while moderately-differentiated intestinal type adenocarcinoma formed a distinct subgroup within their branch. Other histological entities clustered in the last group. Interestingly, the case initially diagnosed as high-grade non-intestinal type adenocarcinoma (SN_18) clustered more closely with poorly-differentiated carcinoma with neuroendocrine and glandular differentiation (Figure 1A). Immunohistochemistry work-up with neuroendocrine markers chromogranin and INSM1 supported neuroendocrine features in an otherwise entirely gland-forming carcinoma (Figure 1B) confirming the accuracy of the epigenetic signature. t-distributed stochastic neighbor embedding analysis was consistent with the DNA methylation-based clustering analysis and no overlap was observed between IDH2 mutants, olfactory neuroblastoma, and SMARCB1-deficient carcinoma, while high-grade non-intestinal type adenocarcinoma overlapped with other poorly-differentiated carcinomas with neuroendocrine and glandular differentiation (Figure 1C). We also compared our cohort to other malignancies typically considered in the differential diagnosis of sinonasal undifferentiated carcinoma and showed that IDH2 mutated sinonasal carcinomas clearly separated from all other IDH2 wild-type tumors of epithelial or mesenchymal origin, including lymphoma, melanoma and Ewing sarcoma. Interestingly, SMARCB1-deficient carcinomas were in a close proximity of atypical/teratoid rhabdoid tumor-MYC subgroup (Supplementary Figure 1).
Figure 1.
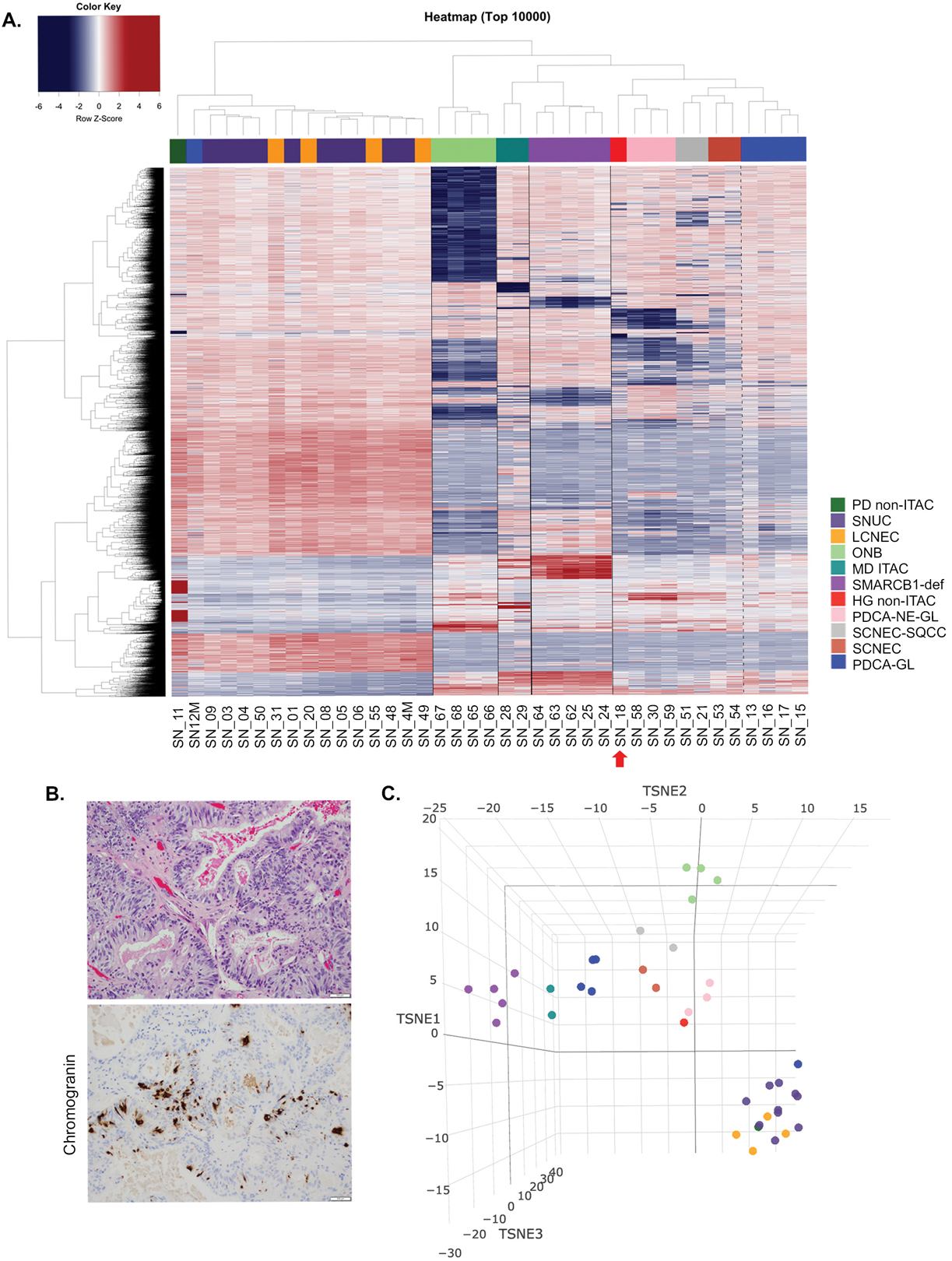
DNA methylation-based classification of the sinonasal tumors. Semi-supervised hierarchical clustering analysis of a histologically diverse cohort of the sinonasal tumors (n=39) based on the top 10,000most variably methylated probes. IDH2 R172 mutant tumors formed a single cluster of IDH2-mutated carcinomas including sinonasal undifferentiated carcinoma and large cell neuroendocrine carcinoma. The other two distinct clusters are seen in olfactory neuroblastoma (n=4) and SMARCB1-deficient sinonasal carcinomas (n=5). Despite the small sample size, moderately-differentiated intestinal type adenocarcinoma (n=2) also showed a distinct methylation fingerprint and separation from other tumor categories. Dotted line separates carcinomas with neuroendocrine differentiation from poorly-differentiated carcinoma with focal glandular/acinar differentiation. A single case of a high-grade non-intestinal type adenocarcinoma (SN_18, red arrow) clustered with poorly-differentiated carcinoma with neuroendocrine and glandular differentiation (A). High-grade non-intestinal type adenocarcinoma (SN_18) displayed an entirely glandular growth pattern (H&E, top) and neuroendocrine differentiation was supported by positive chromogranin immunohistochemistry (200x magnification, B). t-SNE analysis of the sinonasal tumors was consistent with the DNA methylation-based clustering analysis as depicted in the heatmap in Figure 1A. IDH2 mutants are represented as the largest cluster in the lower right. Each tumor group is color-coded according to legend in Figure 1A (C).
Abbreviations: PD non-ITAC= poorly-differentiated carcinoma non-intestinal type adenocarcinoma, SNUC=sinonasal undifferentiated carcinoma, LCNEC= large cell neuroendocrine carcinoma, ONB=olfactory neuroblastoma, MD ITAC=moderately-differentiated intestinal type adenocarcinoma, SMARCB1-def=SMARCB1-deficient sinonasal carcinoma, HG non-ITAC=high-grade non-intestinal type adenocarcinoma, PDCA-NE-GL=poorly-differentiated carcinoma with neuroendocrine and glandular differentiation, SCNEC-SQCC=combined small cell neuroendocrine carcinoma and squamous cell carcinoma, SCNEC=small cell neuroendocrine carcinoma, PDCA-GL=poorly-differentiated carcinoma with focal glandular/acinar differentiation.
2. DNA methylation and H3K27 methylation status of IDH2-mutated sinonasal carcinomas
All IDH2 mutated carcinomas displayed a global hypermethylation pattern and segregated from IDH2 wild-type sinonasal tumors (Figure 2A, Supplementary Figure 2). IDH2 mutants (n=15) showed higher H-score of H3K27me3 than IDH2 wild-type carcinomas (n=8), (239 ±12 vs. 136±14, two-tailed Student’s t test, p<0.001) consistent with the increase in repressive trimethylation of H3K27 (Figure 2). The pathway analysis of IDH2 mutant sinonasal undifferentiated carcinoma and large cell neuroendocrine carcinomas identified enrichment for common cancer signaling pathways including MAPK signaling pathway, Hippo signaling pathway and mTOR signaling pathway that were shared among these tumors (Supplementary Figure 3).
Figure 2.
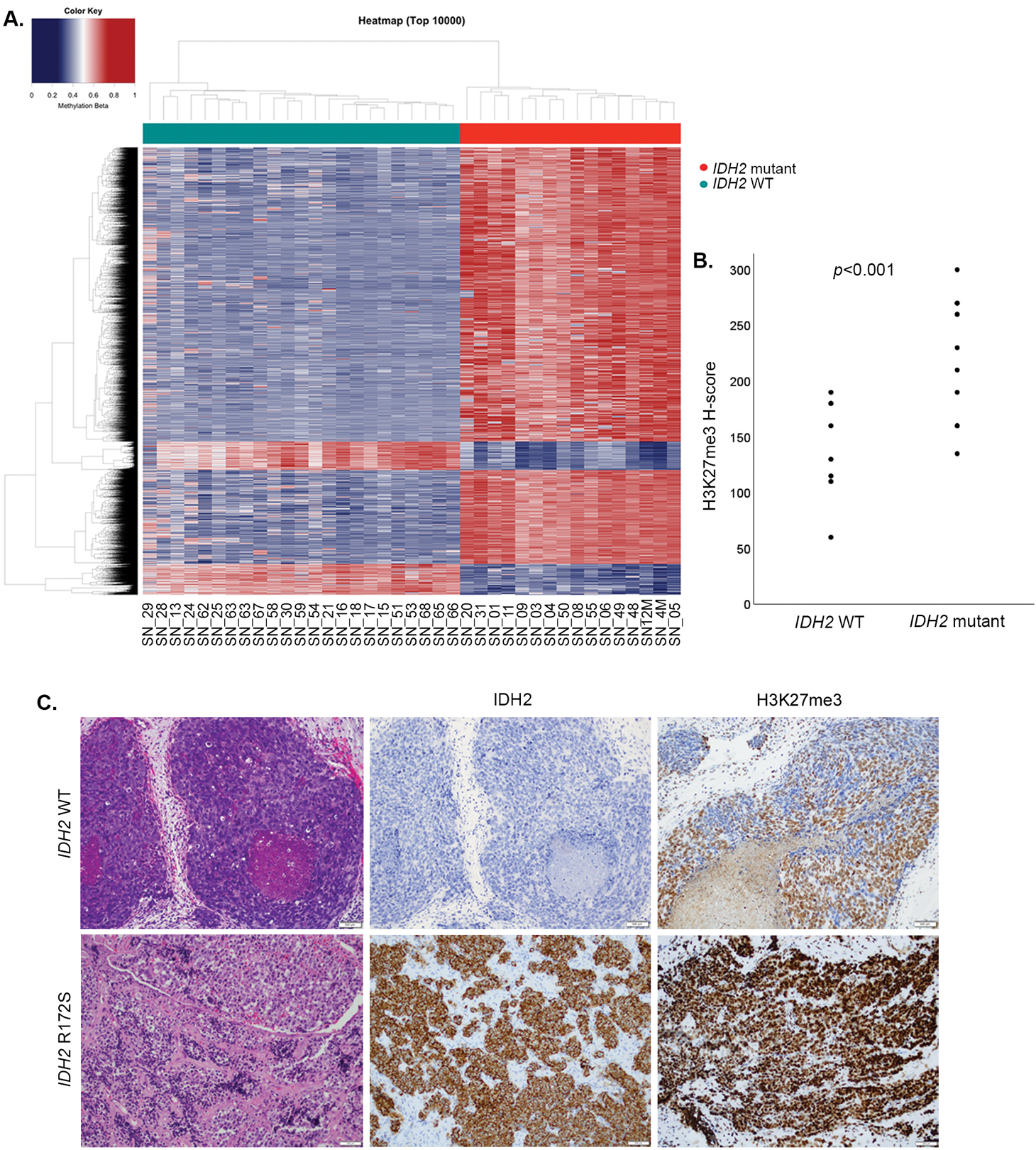
IDH2 R172 mutations in sinonasal carcinomas induce a hypermethylator phenotype and increase histone H3K27 trimethylation. Heatmap of semi-supervised hierarchical clustering analysis of the top 10,000 most variably methylated probes in IDH2 wild-type and IDH2 R172 mutated sinonasal tumors depicts a clear segregation of the two cancer methylomes and supports a global methylation in IDH2 R172 mutants (A). Scattered plots depict H-scores of H3K27me3 immunohistochemistry, which was significantly higher in IDH2 R172 mutants consistent with the induced histone H3K27 trimethylation (B). IDH2 wild-type SNUC (SN_07, H&E, left) was negative for mutant IDH2 (11C8B1) immunostain and showed patchy positive labeling for H3K27me3 (H-score 150). In contrast, IDH2 R172S mutant SNUC (SN_50, H&E, left) was mutant IDH2 immunopositive and showed an increased labeling for H3K27me3 (H-score 260, 200x magnification, C).
Abbreviations: SNUC=sinonasal undifferentiated carcinoma, WT=wild-type.
3. Molecular characteristics of sinonasal large cell and small cell neuroendocrine carcinomas
IDH2 R172 hotspot mutations were detected in 88% (14/16) sinonasal undifferentiated carcinoma and 83% (5/6) large cell neuroendocrine carcinoma (Supplementary Table 1). Similar to IDH2-mutated sinonasal undifferentiated carcinoma, in large cell neuroendocrine carcinoma IDH2 variants co-existed with a median 4 (range 2–6) oncogenic mutations, most commonly oncogenic TP53 variants (60%, 3/5, Figure 3, Supplementary Table 4) [2]. One IDH2-mutated large cell neuroendocrine carcinoma (SN_83) harbored CECR2-FANCA t(16;22)(q24.3;q11.21) rearrangement resulting in a fusion of CECR2 exons 1–7 to FANCA exons 42–43, likely representing an inactivating alteration of one copy of FANCA. We compared IDH2 mutants with IDH2 wild-type carcinomas with neuroendocrine differentiation, including poorly-differentiated carcinoma with neuroendocrine and glandular differentiation (n=4), combined small cell neuroendocrine carcinoma and squamous cell carcinoma (n=2), and small cell neuroendocrine carcinoma (n=3). This IDH2 wild-type subset harbored frequent ARID1A (56%, 5/9) mutations, including all 3 small cell neuroendocrine carcinoma, while no ARID1A alterations was detected in any of the IDH2 mutants (3/3 vs. 0/18, p=0.0008, Fisher’s exact test). IDH2 wild-type carcinomas with neuroendocrine differentiation also harbored recurrent TP53 mutations (33%, 3/9), and most (56%, 5/9) had alterations in Wnt pathway genes including CTNNB1 (33%, 3/9), AMER1 (22%, 2/9) and APC (11%, 1/9, Figure 3A). INSM1 immunohistochemistry was performed in 28 cases (Supplementary Table 2) to assess neuroendocrine features/differentiation and significant differences in INSM1 H-scores were observed across the histologic types, sinonasal undifferentiated carcinoma, large cell neuroendocrine carcinoma and small cell neuroendocrine carcinoma (Figure 3C).
Figure 3.
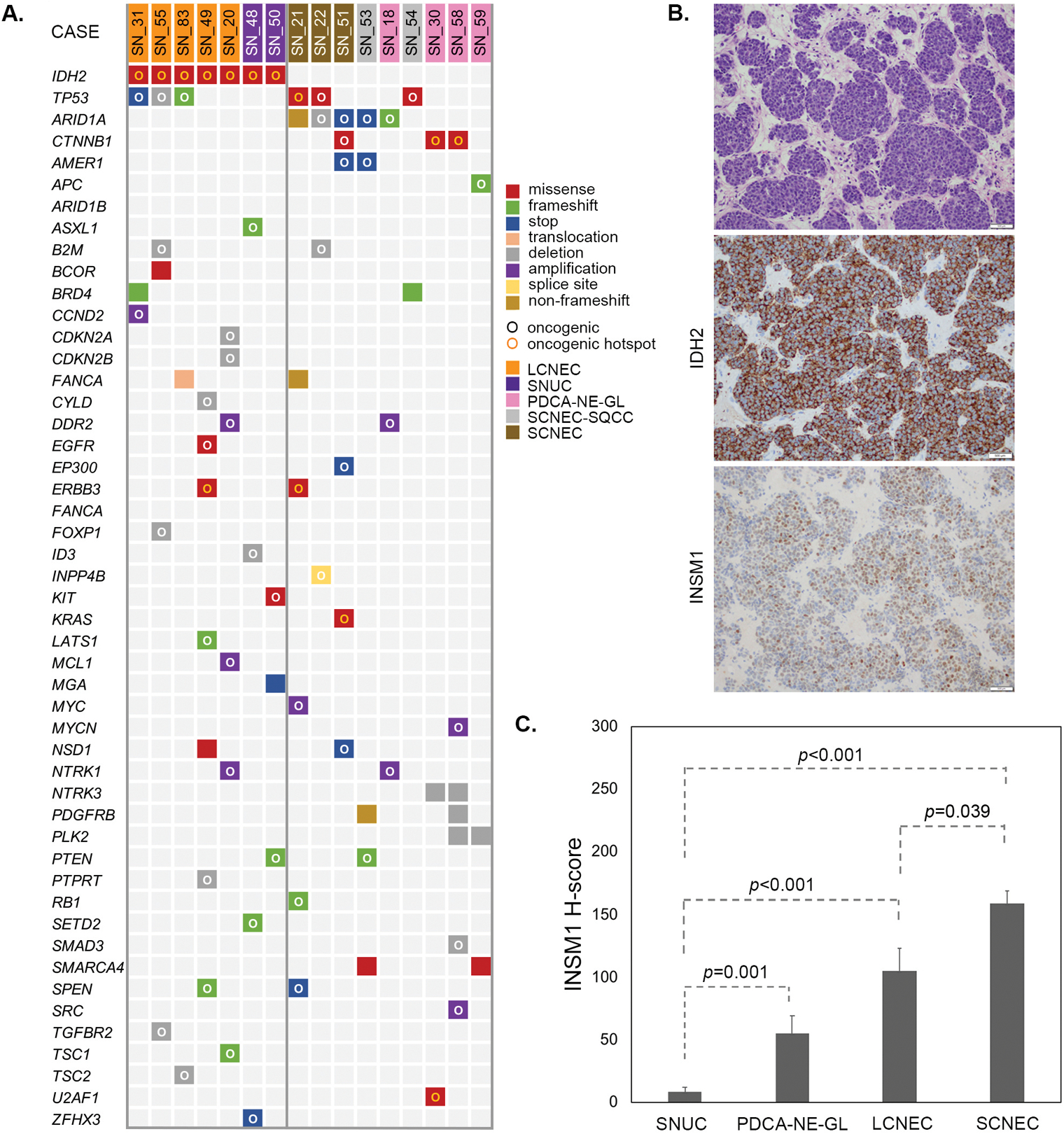
IDH2 mutant large cell neuroendocrine carcinoma is genetically similar to the IDH2 mutant SNUC and is distinct from small cell neuroendocrine carcinoma, which harbor recurrent ARID1A mutations. Oncoprint summarizes all oncogenic and select recurrent somatic alterations in large cell neuroendocrine carcinoma and other carcinomas with neuroendocrine features detected by MSK-IMPACT™. Oncogenic potential of the genetic alterations is defined by the OncoKB ([26], www.cBioPortal.org). SNUC cases previously published are not included (A). IDH2 R172S mutated large cell neuroendocrine carcinoma (SN_49, H&E, top) was immunopositive for IDH2 (11C8B1) and INSM1 (H-score 100, 200x magnification), (B). INSM1 immunoexpression defined by H-score was distinct among sinonasal carcinomas with neuroendocrine features showing the lowest expression in SNUC and the highest expression in small cell neuroendocrine carcinoma. Error bars represent standard deviation (C).
Abbreviations: SNUC=sinonasal undifferentiated carcinoma, PDCA-NE-GL=poorly-differentiated carcinoma with neuroendocrine and glandular differentiation, LCNEC= large cell neuroendocrine carcinoma, SCNEC=small cell neuroendocrine carcinoma.
4. Clinical features of IDH2 mutant and SMARCB1-deficient sinonasal carcinomas
Outcome analysis was performed on 53 of 57 sinonasal carcinomas with available follow-up data. Olfactory neuroblastomas were excluded from the survival analysis due to their remarkably distinct clinical behavior and outcome [33]. Men (n=34, 64%) presented at a younger age (median age 47, range 24–95 years) than women (median age 63, range 30 to 83, two-tailed Student’s t test, p<0.001, Table 1). Most patients presented at stage T4 (79%), a minority had positive neck lymph nodes (19%), and distant metastasis were rare at presentation (6%). The majority (53%) was treated with multimodal therapy including surgery, chemotherapy and radiation. Comparisons were made between the molecular subsets, IDH2 mutant, SMARCB1-deficient and IDH2/SMARCB1 wild-type carcinomas. In comparison to IDH2 mutants, SMARCB1-deficient cases were associated with poorer disease-free survival (Log rank test, p=0.034), and showed a non-significant trend towards shorter disease-specific survival (Log rank test, p=0.334, Figure 4). The 3-year disease-specific survival and 3-year disease-free survival were 60% and 32% in the entire cohort, 74% and 53% in IDH2 mutants, 54% and 10% in SMARCB1-deficient tumors, and 50% and 27% in the IDH2/SMARCB1 wild-type cases. Notably, SMARCB1-deficient tumors had a significantly higher propensity for lung metastasis than IDH2 mutants and IDH2/SMARCB1 wild-type (Fisher’s exact test, p=0.002 and p=0.011, respectively, Table 1).
Table 1.
Clinical characteristics of the sinonasal carcinomas.
| All patients (n=53) | IDH2 mutant (n=18) | SMARCB1-def (n=15) | IDH2/SMARCB1 WT (n=20) | p value | ||
| Sex | ||||||
| Men | 34 (64%) | 12 (67%) | 12 (80%) | 10 (50%) | ||
| Age, median (range, years) | 47 (24–95) | 48 (37–69) | 47 (24–95) | 47 (28–80) | 0.001 | |
| Women | 19 (36%) | 6 (33%) | 3 (20%) | 10 (50%) | ||
| Age, median (range, years) | 63 (30–83) | 51 (43–83) | 66 (53–79) | 64 (30–81) | ||
| T stage | ||||||
| Tis-T3 | 11 (21%) | 16 (33%) | 2 (13%) | 3 (15%) | ||
| T4 | 42 (79%) | 12 (67%) | 13 (87%) | 17 (85%) | ||
| N stage | ||||||
| N0 | 43 (81) | 13 (72%) | 10 (67%) | 20 (100%) | ||
| N1-N2 | 10 (19%) | 5 (28%) | 5 (33%) | 0 | ||
| M stage | ||||||
| M0 | 50 (94%) | 18 (100%) | 14 (93%) | 18 (90%) | ||
| M1 | 3 (6%) | 0 | 1 (7%) | 2 (10%) | ||
| Clinical stage | ||||||
| 0 | 1 (2%) | 1 (6%) | 0 | 0 | ||
| I | 0 | 0 | 0 | 0 | ||
| II | 6 (11%) | 2 (11%) | 1 (7%) | 3 (15%) | ||
| III | 1 (2%) | 1 (6%) | 0 | 0 | ||
| IV | 45 (85%) | 14 (78%) | 14 (93%) | 17 (85%) | ||
| IVA | 16 (30%) | 6 (33%) | 2 (13%) | 8 (40%) | ||
| IVB | 26 (49%) | 8 (44%) | 11 (73%) | 7 (35%) | ||
| IVC | 3 (6%) | 0 | 1 (7%) | 2 (10%) | ||
| Treatment | ||||||
| SxCRT | 28 (53%) | 9 (50%) | 8 (53%) | 11 (55%) | ||
| CRT | 11 (21%) | 5 (28%) | 2 (13%) | 4 (20%) | ||
| SxRT | 8 (15%) | 2 (11%) | 2 (13%) | 4 (20%) | ||
| SxC | 1 (2%) | 0 | 1 (7%) | 0 | ||
| Sx | 3 (6%) | 1 (6%) | 1 (7%) | 1 (5%) | ||
| C | 2 (4%) | 1 (6%) | 1 (7%) | 0 | ||
| Recurrence | ||||||
| All | 35 (66%) | 10 (56%) | 13 (87%) | 12 (60%) | ||
| Local | 22 (46%) | 4 (22%) | 8 (53%) | 10 (50%) | ||
| Regional | 13 (25%) | 5 (28%) | 4 (27%) | 4 (20%) | ||
| Distant | 23 (43%) | 6 (33%) | 9 (60%) | 8 (40%) | ||
| Metastatic sites | ||||||
| Bone | 10 (19%) | 4 (22%) | 3 (20%) | 3 (15%) | ||
| Lung | 8 (15%) | 0 | 7 (47%) | 1 (5%) | 0.002 | 0.011 |
| Neck LN | 7 (13%) | 4 (22%) | 2 (13%) | 1 (5%) | ||
| Liver | 5 (9%) | 3 (17%) | 1 (7%) | 1 (5%) | ||
| Brain | 4 (8%) | 1 (6%) | 3 (20%) | 0 | ||
| Abdominal LN | 3 (6%) | 1 (6%) | 1 (7%) | 1 (5%) | ||
| Mediastinal LN | 3 (6%) | 0 | 3 (20%) | 0 | ||
| Adrenal | 2 (4%) | 1 (6%) | 0 | 1 (5%) | ||
| Parotid | 1 (2%) | 1 (6%) | 0 | 0 | ||
| Soft palate | 1 (2%) | 1 (6%) | 0 | 0 | ||
| Skin | 1 (2%) | 1 (6%) | 0 | 0 | ||
| Mandible | 1 (2%) | 0 | 1 (7%) | 0 | ||
| Pleura | 1 (2%) | 0 | 0 | 1 (5%) | ||
p values compare age of all men and women in the cohort and frequency of lung metastasis between IDH2 mutant and SMARCB1-deficient cases (left) and between SMARCB1-deficient and IDH2/SMARCB1 WT carcinomas (right).
Abbreviations: SMARCB1-def=SMARCB1-deficient sinonasal carcinoma, WT=wild-type, Sx=surgery, C=chemotherapy, RT=radiation therapy, LN=lymph nodes.
Figure 4.
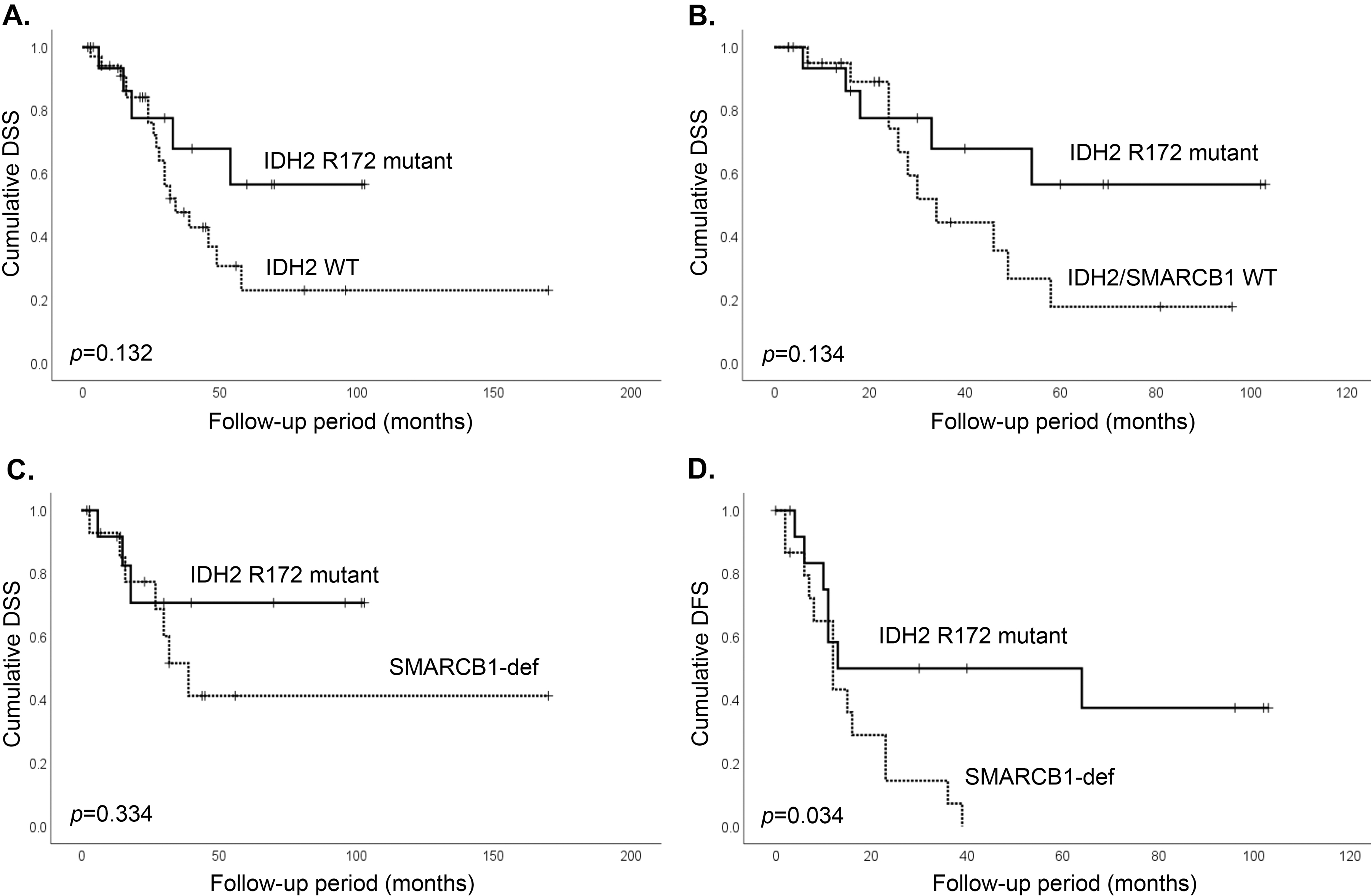
Outcome of the sinonasal carcinomas based on IDH2/SMARCB1 mutation status. IDH2 mutated sinonasal carcinomas show non-significant trend in better disease-specific survival in comparison to all IDH2 wild-type carcinomas (A), IDH2/SMARCB1 wild-type carcinomas (B), and SMARCB1-deficient sinonasal carcinomas (C), but a significantly better disease-free survival than SMARCB1-deficient sinonasal carcinomas (D), (Log Rank test).
Abbreviations: DSS=disease-specific survival, DFS=disease-free survival, WT=wild-type, SMARCB1-def= SMARCB1-deficient sinonasal carcinoma.
DISCUSSION
In the present study, we demonstrated that DNA methylation-based analysis (1) separated sinonasal tumors into the respective histologic/molecular groups, and (2) defined IDH2 mutants as a unique molecular subset of sinonasal carcinomas. IDH2 mutant sinonasal carcinomas displayed a distinctive hypermethylation pattern and an increase in repressive trimethylation of H3K27 suggesting that the putative oncogenic effects of IDH2 mutations in the sinonasal tract might be similar to the established role of IDH1/2 oncogenic variants in other tumor types [11,12]. Our genetic, epigenetic and immunoexpression analyses suggested that IDH2-mutated sinonasal undifferentiated carcinoma and IDH2-mutated large cell neuroendocrine carcinoma represent phenotypic variants of the same entity. IDH2-mutated sinonasal undifferentiated carcinoma/large cell neuroendocrine carcinoma can be distinguished from small cell neuroendocrine carcinoma, which frequently harbors ARID1A mutations, and from molecularly and clinically distinct SMARCB1-deficient sinonasal carcinomas.
Our DNA methylation-based clustering analysis revealed a remarkable epigenetic similarity between IDH2-mutated sinonasal undifferentiated carcinoma and IDH2-mutated large cell neuroendocrine carcinoma, which are currently defined as separate entities by the World Health Organization classification [23]. Sinonasal large cell neuroendocrine carcinoma is one of the two subtypes of neuroendocrine carcinoma of the sinonasal tract; the other one is small cell neuroendocrine carcinoma [23]. Due to their rarity, there are no specific management guidelines for these sinonasal cancers and treatment options would be typically extrapolated from the regimens for their pulmonary histologic counterparts [34–36]. Therefore, an accurate pathologic diagnosis and recognition of neuroendocrine differentiation in a high-grade sinonasal carcinoma is considered crucial and would impact treatment decision-making. Distinguishing large cell neuroendocrine carcinoma and sinonasal undifferentiated carcinoma microscopically can be difficult because of commonly shared microscopic features such as necrosis and brisk mitotic activity, large tumor cells with prominent nucleoli and lobulated/nested growth pattern [23,37]. While large cell neuroendocrine carcinoma labels diffusely and/or strongly positive for neuroendocrine marker(s), only focal and weak positive neuroendocrine marker expression is accepted for the diagnosis of sinonasal undifferentiated carcinoma [38]. In our cohort, large cell neuroendocrine carcinoma and sinonasal undifferentiated carcinoma were clinically similar, affected mostly men and presented as a large destructive mass [37]. Most notably, we demonstrated that their similarities extend beyond the microscopy level. Both of these tumors harbored highly recurrent IDH2 R172 mutations, appeared epigenetically indistinguishable, while the pathway analysis supported their functional similarity. A significantly higher expression of INSM1 [39] supported the neuroendocrine differentiation in large cell neuroendocrine carcinoma and reflected its phenotypic distinction at the protein level. These findings together suggest that IDH2-mutated sinonasal undifferentiated carcinoma and IDH2-mutated large cell neuroendocrine carcinoma likely represent a phenotypic spectrum of the same entity. IDH2-mutated sinonasal undifferentiated carcinoma/large cell neuroendocrine carcinomas were also distinct from small cell neuroendocrine carcinoma, which harbored recurrent ARID1A mutations. The absence of ARID1A somatic variants in IDH2 sinonasal mutants raised a possibility that IDH2 and ARID1A somatic mutations could be mutually exclusive and activate biologically redundant oncogenic pathways in high-grade carcinomas in this location.
SMARCB1-deficient carcinoma was another molecularly defined entity, which displayed a unique methylome signature. Given their aggressive biology and undifferentiated morphology observed in the first described cases [40, 41] the current World Health Organization classification refers to SMARCB1-deficient carcinomas in the context of sinonasal undifferentiated carcinoma as a type of undifferentiated carcinoma with rhabdoid features [23]. Subsequent studies showed that these tumors can be morphologically and immunophenotypically heterogeneous [18,42,43] and as such can be distinguished from sinonasal undifferentiated carcinoma. We have demonstrated that SMARCB1-deficient carcinoma stands out not only epigenetically but also clinically by having a significantly higher propensity for lung metastasis among all sinonasal carcinomas and a shorter disease-free survival than IDH2 mutants.
We observed a separation of the olfactory neuroblastoma methylation cluster from the IDH2 mutants as well as from all other tumor categories. We have not identified IDH2 R172 mutations in 9 olfactory neuroblastomas included in this and the prior study although none of these cases were Hyams grade 4 [2]. Our experience is in line with de la Vega et al. and Topcagic et al. [44,45], but in contrast to others reporting IDH2 R172 variants in the minority of olfactory neuroblastoma cases [46,47]. The current controversies in the literature regarding the detection of IDH2 mutations in this tumor type could be due to the high degree of interobserver variability in diagnosing olfactory neuroblastoma, especially in the presence of high-grade tumor features [23,38,48]. Indeed, Capper et al. performed a DNA methylation-based profiling of 66 sinonasal tumors diagnosed as olfactory neuroblastoma. About 10% of their cases designated as “sinonasal tumors with IDH2 mutation” separated from the core olfactory neuroblastoma, most were high-grade, cytokeratin positive and negative or sparsely positive for chromogranin and S-100 [49]. Similarly, Classe et al. identified IDH2 mutations in a subset they termed “basal” olfactory neuroblastoma, which were also high-grade, had relatively lower chromogranin and higher cytokeratin expression, and were mostly S-100 negative. “Basal” type had predilection for male sex and were clinically relatively more aggressive than “neural” olfactory neuroblastoma [47]. Based on these features, it is possible that some of the reported IDH2-mutated olfactory neuroblastomas could have been designated as sinonasal undifferentiated carcinoma or large cell neuroendocrine carcinoma by other head and neck pathologists. Interestingly, however, none of the 8 sinonasal undifferentiated carcinomas in the article by Capper et al. clustered with the “sinonasal tumors with IDH2 mutation” nor the tumors defined as this entity formed a distinct cluster [49]; this might suggest that most “sinonasal undifferentiated carcinoma” diagnoses were assigned to histologically/molecularly diverse IDH2 wild-type tumors, which further emphasizes the complexity of differential diagnosis in this location.
t-distributed stochastic neighbor embedding analysis of an extended data set showed that all of our tested entities can be distinguished from other small round cell tumors including lymphoma, melanoma or Ewing sarcoma. In addition, the high-grade non-intestinal type adenocarcinoma, which clustered with poorly-differentiated carcinoma with neuroendocrine and glandular differentiation illustrated the potential of methylation-based analysis to refine sinonasal tumor diagnosis and raised a question of the significance of neuroendocrine differentiation in a subset of high-grade non-intestinal type adenocarcinomas [50]. These results provide a strong rationale for assembling larger data sets and designing the DNA methylation-based classifier for the sinonasal tumors, similarly to what has been accomplished in brain tumors [22] and bone sarcomas [19]. Nevertheless, although emerging novel next-generation sequencing technologies have proven an enormous potential for better classification and risk stratification of various malignancies [19–22,49], the pathologic diagnosis, as accurate as possible, rendered by subspecialized surgical pathologists is an important pre-requisite for optimal experimental design and valid interpretation of the results.
A potential limitation of our study is a lack of methylation analysis data on IDH2 wild-type examples of sinonasal undifferentiated carcinoma and large cell neuroendocrine carcinoma due to their extreme rarity. In the absence of the hypermethylator phenotype, it is unlikely that these tumors would cluster with their IDH2 mutant counterparts and might rather form a subgroup within IDH2 wild-type tumors. However, several important conclusions can be drawn from our genome-wide methylation profiling study of sinonasal undifferentiated carcinoma and its histologic mimics. In sinonasal carcinomas, IDH2 R172 somatic mutations induce a hypermethylator phenotype and define a molecular subgroup of carcinomas arising in this location. Future revisions of the current World Health Organization categories (1) sinonasal neuroendocrine carcinoma including large cell neuroendocrine carcinoma and small cell neuroendocrine carcinoma, and (2) sinonasal undifferentiated carcinoma, including SMARCB1-deficient carcinoma will have to consider their molecular characteristics for improved re-classification. DNA methylation-based classifier for the sinonasal tumors could help refine the classification of a variety of rare malignancies and provide a solution to the historical problem of differential diagnosis in this location.
Supplementary Material
ACKNOWLEDGEMENTS
We wish to thank Tessara Baldi, Brian Tedino (Department of Pathology, MSKCC), Caitlin Bourque (Sloan Kettering Institute) for the technical support, and Zoran Perak for the assistance with the figures preparation. Research reported in this publication was supported by the Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute under award number P30CA008748 (to SD) and in part by the Friedberg Charitable Foundation (to MS).
Footnotes
Disclosure Statement:
No competing financial interests exist for all contributory authors. Research reported in this publication was supported by the Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute under award number P30CA008748 (to SD), and in part by the Friedberg Charitable Foundation (to MS).
REFERENCES
- 1.Frierson HF Jr., Mills SE, Fechner RE, Taxy JB, Levine PA. Sinonasal undifferentiated carcinoma. An aggressive neoplasm derived from schneiderian epithelium and distinct from olfactory neuroblastoma. Am J Surg Pathol 1986; 10: 771–779. [PubMed] [Google Scholar]
- 2.Dogan S, Chute DJ, Xu B, Ptashkin RN, Chandramohan R, Casanova-Murphy J, et al. Frequent IDH2 R172 mutations in undifferentiated and poorly-differentiated sinonasal carcinomas. J Pathol 2017; 242: 400–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jo VY, Chau NG, Hornick JL, Krane JF, Sholl LM. Recurrent IDH2 R172X mutations in sinonasal undifferentiated carcinoma. Mod Pathol 2017; 30: 650–659. [DOI] [PubMed] [Google Scholar]
- 4.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med 2009; 360: 765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol 2011; 224: 334–343. [DOI] [PubMed] [Google Scholar]
- 6.Borger DR, Tanabe KK, Fan KC, Lopez HU, Fantin VR, Straley KS, et al. Frequent mutation of isocitrate dehydrogenase (IDH) 1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist 2012; 17: 72–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med 2009; 361: 1058–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cairns RA, Iqbal J, Lemonnier F, Kucuk C, de Leval L, Jais JP, et al. IDH2 mutations are frequent in angioimmunoblastic T-cell lymphoma. Blood 2012; 119: 1901–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiang S, Weigelt B, Wen HC, Pareja F, Raghavendra A, Martelotto LG, et al. IDH2 Mutations Define a Unique Subtype of Breast Cancer with Altered Nuclear Polarity. Cancer Res 2016; 76: 7118–7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A,, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 2010; 18: 553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 2012; 483: 474–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 2012; 483: 479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ejaz A, Wenig BM. Sinonasal undifferentiated carcinoma: clinical and pathologic features and a discussion on classification, cellular differentiation, and differential diagnosis. Adv Anat Pathol 2005; 12: 134–143. [DOI] [PubMed] [Google Scholar]
- 14.Iezzoni JC, Mills SE. “Undifferentiated” small round cell tumors of the sinonasal tract: differential diagnosis update. Am J Clin Pathol 2005; 124 Suppl: S110–121. [DOI] [PubMed] [Google Scholar]
- 15.Bell D, Hanna EY, Weber RS, DeMonte F, Triantafyllou A, Lewis JS Jr., et al. Neuroendocrine neoplasms of the sinonasal region. Head Neck 2016; 38 Suppl 1: E2259–2266. [DOI] [PubMed] [Google Scholar]
- 16.Bishop JA, Westra WH. NUT midline carcinomas of the sinonasal tract. Am J Surg Pathol 2012; 36: 1216–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Solomon LW, Magliocca KR, Cohen C, Muller S. Retrospective analysis of nuclear protein in testis (NUT) midline carcinoma in the upper aerodigestive tract and mediastinum. Oral Surg Oral Med Oral Pathol Oral Radiol 2015; 119: 213–220. [DOI] [PubMed] [Google Scholar]
- 18.Agaimy A, Hartmann A, Antonescu CR, Chiosea SI, El-Mofty SK, Geddert H, et al. SMARCB1 (INI-1)-deficient Sinonasal Carcinoma: A Series of 39 Cases Expanding the Morphologic and Clinicopathologic Spectrum of a Recently Described Entity. Am J Surg Pathol 2017; 41: 458–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu SP, Cooper BT, Bu F, Bowman CJ, Killian JK, Serrano J, et al. DNA Methylation-Based Classifier for Accurate Molecular Diagnosis of Bone Sarcomas. JCO Precis Oncol 2017; 2017: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johann PD, Erkek S, Zapatka M, Kerl K, Buchhalter I, Hovestadt V, et al. Atypical Teratoid/Rhabdoid Tumors Are Comprised of Three Epigenetic Subgroups with Distinct Enhancer Landscapes. Cancer Cell 2016; 29: 379–393. [DOI] [PubMed] [Google Scholar]
- 21.Sturm D, Orr BA, Toprak UH, Hovestadt V, Jones DTW, Capper D, et al. New Brain Tumor Entities Emerge from Molecular Classification of CNS-PNETs. Cell 2016; 164: 1060–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D, et al. DNA methylation-based classification of central nervous system tumours. Nature 2018; 555: 469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis JS, Bishop JA, Gillison M et al. In: Tumours of the nasal cavity, paranasal sinuses and skull base In: El-Naggar AK, Chan JKC, Grandis JR, Takata T, Slootweg PJ, editors. World Health Organization Classification of Head and Neck Tumours (4th edn). International Agency for Reseach on Cancer (IARC): Lyon, 2017. p. 18–26. [Google Scholar]
- 24.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn 2015; 17: 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017; 23: 703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol 2017; 2017: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Serrano J, Snuderl M. Whole genome DNA methylation analysis of human glioblastoma using Illumina BeadArrays. Methods Mol Biol. 2018; 1741: 31–51. [DOI] [PubMed] [Google Scholar]
- 28.Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014; 30: 1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Maaten L, Hinton G. Visualizing data using t-SNE. J Mach. Learn Res 2008; 9: 2579–2605. [Google Scholar]
- 30.Yu G, Wang L-G, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012; 16: 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dogan S, Frosina D, Fayad M, de Oliveira TB, Alemar B, Rosenblum M, et al. The role of a monoclonal antibody 11C8B1 as a diagnostic marker of IDH2-mutated sinonasal undifferentiated carcinoma. Mod Pathol 2019; 32: 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Detre S, Jotti GS, Dowsett M. A” quickscore” method for immunohistochemical semiquantitation: validation for oestrogen receptor in breast carcinomas. J Clin Pathol 1995; 48: 876–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su SY, Bell D, Hanna EY. Esthesioneuroblastoma, neuroendocrine carcinoma, and sinonasal undifferentiated carcinoma: differentiation in diagnosis and treatment. Int Arch Otorhinolaryngol 2014; 18: S149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perez-Ordonez B, Caruana SM, Huvos AG, Shah JP. Small cell neuroendocrine carcinoma of the nasal cavity and paranasal sinuses. Hum Pathol 1998; 29: 826–832. [DOI] [PubMed] [Google Scholar]
- 35.Rossi G, Cavazza A, Marchioni A, Longo L, Migaldi M, Sartori G, et al. Role of chemotherapy and the receptor tyrosine kinases KIT, PDGFRa, PDGFRb, and Met in large-cell neuroendocrine carcinoma of the lung. J Clin Oncol 2005; 23: 8774–8785. [DOI] [PubMed] [Google Scholar]
- 36.Babin E, Rouleau V, Vedrine PO, Toussaint B, de Raucourt D, Malard O, et al. Small cell neuroendocrine carcinoma of the nasal cavity and paranasal sinuses. J Laryngol Otol 2006; 120: 289–297. [DOI] [PubMed] [Google Scholar]
- 37.Thompson ED, Stelow EB, Mills SE, Westra WH, Bishop JA. et al. Large Cell Neuroendocrine Carcinoma of the Head and Neck: A Clinicopathologic Series of 10 Cases With an Emphasis on HPV Status. Am J Surg Pathol 2016; 40: 471–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barnes E Nasal Cavity, Paranasal Sinuses, and Nasopharynx In: Barnes E, editor. Surgical Pathology of the Head and Neck, (3rd edn). Informa Healthcare Inc.: New York, USA, 2009; 343–422. [Google Scholar]
- 39.Rooper LM, Bishop JA, Westra WH. INSM1 is a Sensitive and Specific Marker of Neuroendocrine Differentiation in Head and Neck Tumors. Am J Surg Pathol 2018; 42: 665–671. [DOI] [PubMed] [Google Scholar]
- 40.Agaimy A, Koch M, Lell M, Semrau S, Dudek W, Wachter DL, et al. SMARCB1(INI1)-deficient sinonasal basaloid carcinoma: a novel member of the expanding family of SMARCB1-deficient neoplasms. Am J Surg Pathol 2014; 38: 1274–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bishop JA, Antonescu CR, Westra WH. SMARCB1 (INI-1)-deficient carcinomas of the sinonasal tract. Am J Surg Pathol 2014; 38: 1282–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cotzia P, Ptashkin RN, Gounder MM, Pfister DG, Berger MF, Ladanyi M, et al. Genetic and Histologic Spectrum of SMARCB1-deficient Carcinomas of the Head and Neck Including Sinonasal Tract, Thyroid and Skin. Lab Investigation 2018; 98: 474–475. [Google Scholar]
- 43.Kakkar A, Antony VM, Pramanik R, Sakthivel P, Singh CA, Jain D. SMARCB1 (INI1)–deficient sinonasal carcinoma: a series of 13 cases with assessment of histologic patterns. Hum Pathol 2019; 83: 59–67. [DOI] [PubMed] [Google Scholar]
- 44.de la Vega LL, McHugh JB, Cani AK, Kunder K, Walocko FM, Liu C-J, et al. Comprehensive Molecular Profiling of Olfactory Neuroblastoma Identifies Potentially Targetable FGFR3 Amplifications. Mol Cancer Res 2017; 15: 1551–1557. [DOI] [PubMed] [Google Scholar]
- 45.Topcagic J, Feldman R, Ghazalpour A, Swensen J, Gatalica Z, Vranic S. Comprehensive molecular profiling of advanced/metastatic olfactory neuroblastomas. PLoS One 2018; 13: e0191244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gay LM, Kim S, Fedorchak K, Kundranda M, Odia Y, Nangia C, et al. Comprehensive Genomic Profiling of Esthesioneuroblastoma Reveals Additional Treatment Options. Oncologist 2017; 22: 834–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Classe M, Yao H, Mouawad R, Creighton CJ, Burgess A, Allanic F, et al. Integrated Multi-omic Analysis of Esthesioneuroblastomas Identifies Two Subgroups Linked to Cell Ontogeny. Cell Rep 2018; 25: 811–821 e815. [DOI] [PubMed] [Google Scholar]
- 48.Cohen ZR, Marmor E, Fuller GN, DeMonte F. Misdiagnosis of olfactory neuroblastoma. Neurosurg Focus 2002; 12: e3. [DOI] [PubMed] [Google Scholar]
- 49.Capper D, Engel NW, Stichel D, et al. DNA methylation-based reclassification of olfactory neuroblastoma. Acta Neuropathol 2018; 136: 255–271. [DOI] [PubMed] [Google Scholar]
- 50.Stelow EB, Jo VY, Mills SE, Carlson DL. A histologic and immunohistochemical study describing the diversity of tumors classified as sinonasal high-grade nonintestinal adenocarcinomas. Am J Surg Pathol 2011; 35: 971–980. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


