Abstract
Abnormal brain development manifests itself at different spatial scales. However, whether abnormalities at the cellular level can be diagnosed from network activity measured with functional magnetic resonance imaging (fMRI) is largely unknown, yet of high clinical relevance. Here a putative mechanism reported in neurodevelopmental disorders, that is, excitation-to-inhibition ratio (E:I), was chemogenetically increased within cortical microcircuits of the mouse brain and measured via fMRI. Increased E:I caused a significant “reduction” of long-range connectivity, irrespective of whether excitatory neurons were facilitated or inhibitory Parvalbumin (PV) interneurons were suppressed. Training a classifier on fMRI signals, we were able to accurately classify cortical areas exhibiting increased E:I. This classifier was validated in an independent cohort of Fmr1y/− knockout mice, a model for autism with well-documented loss of parvalbumin neurons and chronic alterations of E:I. Our findings demonstrate a promising novel approach towards inferring microcircuit abnormalities from macroscopic fMRI measurements.
Keywords: DREADD, excitation: inhibition balance, fMRI, functional connectivity, hypoconnectivity
Introduction
Complex behavior results from the interplay of distributed yet anatomically connected neuronal populations which form brain-wide networks. Abnormal brain development as in autism spectrum disorders (ASD) often affects the structural and functional connectivity (FC) within these networks at different spatial scales: at the cellular and (micro)circuit levels, neurodevelopmental disorders are associated with dysregulated synaptogenesis, cell excitability, synaptic transmission, and plasticity (Penagarikano et al. 2011; Bulow et al. 2019), as well as aberrant axonal migration and abnormal functioning of cortical (micro)circuits (Contractor et al. 2015); at the network level, long-range functional connectivity and macroscopic neuronal dynamics are altered (Haberl et al. 2015; Zerbi et al. 2018). Mechanistically linking disease mechanisms across scales is an important goal for neuropsychiatry, particularly, to identify whether certain cellular disease pathways map onto specific alterations of large-scale network activity, which are accessible in human patients and might thus allow to infer the underlying pathobiology of an individual patient.
An abnormal increase of the excitation-to-inhibition (E:I) ratio within specific cortical circuits (Rubenstein and Merzenich 2003) has been hypothesized as a putative disease mechanism underlying ASD pathology and its core behavioral symptoms (Silverman et al. 2010; Kazdoba et al. 2014). In transgenic mouse models of ASD, elevated E:I has been observed either because excitatory transmission is increased (Gibson et al. 2008) or because inhibitory transmission is reduced, for example, due to the loss of fast-spiking parvalbumin (PV) GABAergic interneurons (Penagarikano et al. 2011; Wohr et al. 2015; Vogt et al. 2017). However, it is unknown whether such an imbalance at the microcircuit level propagates across scales and whether macroscopic markers of brain activity signal these abnormalities with anatomical specificity.
Here we acutely increase microcircuit E:I within one brain region and test whether this well-controlled manipulation causes an anatomically specific change of neuronal dynamics and macroscopic connectivity of the mouse brain. We used chemogenetics (Rogan and Roth 2011; Michaelides and Hurd 2016) to manipulate cell activity of primary somatosensory cortex (SSp) while simultaneously imaging brain activity using resting-state functional magnetic resonance imaging (rsfMRI). rsfMRI is a noninvasive method used to measure spontaneous slow-frequency fluctuations of the blood-oxygen level-dependent (BOLD) signal (an indirect estimate of neuronal activity) in the absence of a task and across the entire brain (Fox and Raichle 2007). A growing body of evidence suggests that rsfMRI has great potential for translation, as it detects abnormalities in long-range network correlations in human patients as well as homologous networks in mouse models expressing similar transgenic mutations (Strand et al. 2007; Smucny et al. 2014; Shofty et al. 2019). Here, we show that experimentally increasing E:I in primary somatosensory cortex (SSp) by either 1) facilitating excitatory neurons or 2) suppressing inhibitory PV interneurons both caused a reduction of long-range functional connectivity between SSp and anatomically connected areas. Increasing E:I changed the dynamics of BOLD fluctuations, an effect that was most pronounced in the targeted SSp. We used the BOLD time series measured during experimentally controlled chemogenetic suppression of PV interneurons to train a classifier to detect acutely overexcited brain areas. We applied this classifier to an independent cohort of Fmr1y/− knockout mice, a well-characterized model for ASD (Haberl et al. 2015; Zerbi et al. 2018) which suffers from PV interneuron depletion and exhibits increased cortical E:I due to the loss of inhibitory inputs (Antoine et al. 2019). Importantly, the classifier identified specific brain regions in somatosensory and prefrontal cortices which exhibited an overexcitation phenotype based on their BOLD dynamics. This new finding indicates that chemogenetically perturbing microcircuit abnormalities in wild-type mice allows us to build “computational sensors” for detecting E:I imbalances from noninvasive macroscopic rsfMRI measurements in a disease model. Our results demonstrate a promising novel approach toward inferring microcircuit abnormalities from macroscopic measurements which potentially opens new opportunities for investigating disease mechanisms across species.
Materials and Methods
All experiments and procedures were conducted following the Swiss federal ordinance for animal experimentation and approved by Zurich Cantonal Veterinary Office (ZH238/15). C57BL/6J mice were obtained from Charles River Laboratories (Germany), while Pvalbtm1(cre)Arbr (PVCre) mice were derived from in-house breeding (first generation obtained from the Jackson Laboratory). All mice were kept in standard housing under 12-h light/dark cycle with food and water provided ad libitum throughout the whole experiment. A total of 29 male C57BL/6J mice were used in the experiment, aged 9 ± 1 weeks and weighing 25.1 ± 1.8 g (mean ± standard deviation [SD]) at the day of the surgery. A total of 19 PVCre mice were utilized in this experiment, aged 11 ± 1.6 weeks and weighing 26.1 ± 3.8 g at the day of the surgery.
Procedures for Wild-Type (hM3Dq) Mice Study
Transfection Procedures for “wt-hSyn-hM3Dq” and “wt-CAMK-hM3Dq” Mice
Each mouse was initially anesthetized using a mixture of midazolam (5 mg/mL; Sintetica, Switzerland), fentanyl (50 mcg/mL; Actavis AG, Switzerland), and medetomidine (1 mg/mL; Orion Pharma, Finland). Upon anesthesia induction, mice were placed on a heating pad, and the temperature was kept at 35 °C (Harvard Apparatus, United States). Following shaving and cleaning, an incision along the midline of scalp was made. The right primary somatosensory cortex was targeted at the coordinates of +0.5 mm AP (anterior–posterior), −3.0 mm ML (medio–lateral), and −2.25 mm DV (dorso–ventral) relative to bregma using a drill and microinjection robot (Neurostar, Germany) with a 10-ul NanoFil syringe and 34-G beveled needle (World Precision Instruments, Germany). Nine hundred fifty nanoliters of ssAAV8-hSyn-hM3Dq-mCherry (n = 13) of a physical titer ≥5.4 × 1012 vg/mL (*vector genomes; fluorometric quantification) was injected at the rate of 0.06 ul/min. The virus was provided by Viral Vector Core Facility of the Neuroscience Centre Zurich (http://www.vvf.uzh.ch/en.html). Upon the injection, the needle was left in place for 10 min and then slowly withdrawn. C57BL/6J control mice (n = 9) underwent the same surgical procedures, where the needle was kept in place for 5 min but without any viral injections. Subsequently, mice were given an anesthesia antidote consisting of Temgesic (0.3 mg/mL; Reckitt Benckiser AG, Switzerland), annexate (0.1 mg/mL; Swissmedic, Switzerland), and antisedan (0.1 mg/mL; Orion Pharma, Finland) and left to fully recover. Following the surgery, ketoprofen (10 mg/kg; Richter Pharma AG, Austria) was subcutaneously injected daily for at least 3 days to reduce any postoperative pain. Animals were given 3–4 weeks to fully recover from the surgery and to allow for expression of the transgene prior to the scanning session.
Another cohort of 13 C57BL/6J mice was injected with 950 nL of ssAAV8-mCAMKIIα-hM3D-mCherry of a physical titer ≥1.8 × 1012 vg/mL following identical procedures as outlined above. Supplementary Figure 7 illustrates the extension of viral expression at the targeted region.
Electrophysiological Recordings
The electrophysiological data used for verification of neuronal activity were acquired in four wt-hSyn-hM3Dq animals and two wild-type animals that have not received a viral designer receptor exclusively activated by designer drug (DREADD) injection, that is, controls. Briefly, animals were anesthetized with isoflurane to match conditions used in imaging studies (2–4% for induction, 0.5% for data collection), and their body temperature was maintained using a heating pad. A small craniotomy was performed over the right and left somatosensory cortices, and the brain was covered with silicone oil. A small trepanation was performed over the cerebellum, and a silver wire was placed in contact with the CSF to serve as reference electrode. For hM3Dq validation, one silicone probe (Atlas Neurotechnologies; E16-100-S1-L6NT, 16 sites, 100-μm spacing, iridium oxide contacts) was implanted in each hemisphere. After implantation, we waited 20–30 min in order to allow the recording to stabilize. After stabilization, the broadband voltage was amplified and digitally sampled at a rate of 30 kHz or 48 kHz using one of the two commercial extracellular recording systems (Intan or Axona). The raw voltage traces were filtered off-line in order to separate the multiunit activity (MUA) (band-pass filter 0.46–6 kHz) using a second-order Butterworth filter. Subsequently, for each electrode, a threshold was applied to the high-pass filtered data to isolate multiunits and reject background noise (4.5 times the standard deviation across the recording session, specified per electrode). The number of spikes was counted in 10-s windows. As an additional control, we lowered the detection threshold which did not change the results, unless it was decreased below the background noise. Recording sessions lasted for 45 min. At 15 min following the start of the recordings (“baseline”) 30-μg/kg clozapine was injected (intravenously). In order to combine data across mice, the activity at sites with clear multiunit activity was expressed in percent of the baseline value, that is, each 10-s bin of MUA activity was divided by the average spike rate during the 15-min preinjection baseline (100%). All multiunits were then combined from the injected or control region.
MRI Setup and Animal Preparation
Two MRI sessions were performed on a 7-T Bruker BioSpec scanner equipped with a PharmaScan magnet, with two coil setups optimized for the two different acquisition sequences, that is, arterial spin labeling (ASL) and resting-state fMRI (rsfMRI). First, the ASL was obtained utilizing a receiver-only surface coil, coupled with a volume resonator for radio-frequency (rf) transmission. For rsfMRI measurements, a high signal-to-noise ratio (SNR) receive-only cryogenic coil (Bruker BioSpin AG, Fällanden, Switzerland) was used in combination with a linearly polarized room temperature volume resonator for rf transmission.
Standardized anesthesia protocols and animal monitoring procedures were utilized for both MRI sessions (Zerbi et al. 2018). Briefly, mice were initially anesthetized with 4% isoflurane in 1:4 O2 to air mixture for 4 min to allow for endotracheal intubation and tail vein cannulation. Mice were positioned on an MRI-compatible support, equipped with hot water-flowing bed to keep the temperature of the animal constant throughout the entire measurement (36.6 ± 0.5 °C). The animals were fixed with ear bars and mechanically ventilated via a small animal ventilator (CWE, Ardmore, United States) at the rate of 80 breaths per min, with 1.8-mL/min flow with isoflurane at 2%. Subsequently, a bolus containing a mixture of medetomidine (0.05 mg/kg) and pancuronium (0.25 mg/kg) was injected via the cannulated vein and isoflurane lowered at 1%. Five minutes following the bolus injection, a continuous infusion of medetomidine (0.1 mg/kg/h) and pancuronium (0.25 mg/kg/h) was started while isoflurane was further reduced to 0.5%. Animal preparation took on average 15.5 ± 2.7 min and all animals fully recovered within 10 min after the measurement.
Arterial Spin Labeling (ASL)
Brain perfusion was measured using an established arterial spin labeling (ASL) method using a flow-sensitive alternating inversion recovery (FAIR) sequence (Leithner et al. 2008; Zerbi et al. 2014). Briefly, a two-segment spin echo was used with following parameters: echo time (TE) = 12.47 ms, recovery time (TR) = 13 000 ms, image matrix = 128 × 96, field of view = 20 × 20 mm, slice thickness = 1 mm, and spatial resolution = 0.156 × 0.208 × 1 mm/pixel. Inversion parameters include inversion time = 40 ms, inversion slab thickness = 4 mm, and slice margin = 1.5 mm. Sixteen images at different inversion times (50 ms to 3 s) were obtained for T1 calculations, resulting in a scan time of 15 min (referred to as “baseline”). After the first baseline acquisition, clozapine was intravenously injected at 10–30 μg/kg, and three more sessions were acquired (i.e., Post 1, Post 2, and Post 3, respectively), resulting in a total scan time of 1 h. A total of 26 animals (5 controls, 8 wt-hM3Dq, and 13 CAMK-hM3Dq mice) were scanned. Due to a subjective placement of a 1-mm-thick slice over the somatosensory cortex prior to the scan start, there may be slight differences across subjects in slice position relative to the bregma. However, we made sure that every brain slice of every subject covered virally transfected SSp.
Local changes in cerebral blood flow (CBF) could be considered as an indirect marker of neural activity (Juepter and Weiller 1995). Taking this into account, CBF is expected to increase at the site of DREADD injection, where we observed strongest increases in firing rates, but not elsewhere. We chose our regions of interest (ROIs) based on this analogy, and they included (i) right primary somatosensory cortex (SSp), as a DREADD injection site (main region of interest); (ii) left primary somatosensory cortex, as an anatomically connected region; and (iii) left striatum (dorsomedial part) (CP), a control region without any anatomical projections to the right primary somatosensory area, according to Allen Mouse Connectivity Atlas (Oh et al. 2014).
Resting-State fMRI (rsfMRI)
Acquisition parameters were the following: repetition time (TR) = 1 s, echo time (TE) = 15 ms, flip angle = 60°, matrix size = 90 × 50, in-plane resolution = 0.2 × 0.2 mm2, number of slices = 20, slice thickness = 0.4 mm, and 3600 volumes for a total scan of 60 min. Clozapine was intravenously injected 15 min after the scan start at the doses of 10 μg/kg or 30 μg/kg. A total of 40 C57BL/6J animals (13 controls and 14 wt-hM3Dq and 13 CAMK-hM3Dq mice) were scanned. These include eight C57BL/6J animals that were scanned twice (four controls and four wt-hM3Dq mice), once with 10 μg/kg dose of clozapine and another with a 30 μg/kg dose of clozapine (Supplementary Table 1 and Supplementary figure 8).
Data Preprocessing
Data was preprocessed using an already established pipeline for removal of artifacts from the time series (Zerbi et al. 2015; Sethi et al. 2017). Briely, each 4D dataset was normalized in a study-specific EPI template (Advanced Normalization Tools [ANTs] v2.1, picsl.upenn.edu/ANTS) and fed into MELODIC (Multivariate Exploratory Linear Optimized Decomposition into Independent Components) to perform within-subject spatial independent component analysis (ICA) with a fixed dimensionality estimation (number of components set to 60). The procedure included motion correction and in-plane smoothing with a 0.3-mm kernel. FSL-FIX study-specific classifier, obtained from an independent dataset of 15 mice, was used to perform a “conservative” removal of the variance of the artifactual components (Griffanti et al. 2014). Subsequently, the dataset was despiked, band-pass filtered (0.01–0.25 Hz), and finally normalized into Australian Mouse Brain Mapping Consortium (AMBMC) template (www.imaging.org.au/AMBMC) using ANTs. The choice of our band-pass filter is based on the recent literature where it is stated that the frequency distribution of the fMRI signal yields the maximum amplitude between 0.1 Hz and 0.2 Hz under medetomidine anesthesia (Pan et al. 2013; Grandjean et al. 2014). Each dataset was split into 4 parts of 900 data points (equivalent of 15 min of scanning). The difference between the baseline (first 15 min of scan) and the rest of the bins are further referred to as ∆1, ∆2, and ∆3.
Seed-to-Seed Analysis
The “somatosensory network” in the mouse was defined anatomically using the tracer-based axonal projection pattern of SSp from the Allen Mouse Brain Connectivity Atlas (experiment No. 114290938) and contained contralateral primary SSp, right supplementary somatosensory area (SSs), bilateral somatomotor area (MO), bilateral temporal association cortices (TEa), as well as ipsilateral caudoputamen (CP) and thalamus (TH [parafascicular nucleus]) (Grandjean et al. 2017). BOLD time series from these regions, apart from thalamus, were extracted using a voxel cube of 3 mm. The thalamus was excluded because a previous study from our lab has shown a rather limited FC of corticothalamic pathways (Grandjean et al. 2017) detected by rsfMRI. Correlation Z-scored matrices were calculated using FSLNets (FMRIB Analysis Group).
Regional Homogeneity (ReHo)
Voxel-wise regional homogeneity (ReHo) maps were computed using AFNI. ReHo was calculated for a given voxel and its 19 nearest neighbors and smoothened by Gaussian kernel with full width at half maximum (FWHM) equal to 10.
Voxel-Mirrored Homotopic Connectivity (VMHC)
VMHC measures the similarity between any pair of symmetric interhemispheric voxels by computing Pearson’s correlation coefficient between the time series of each voxel and that of its exact symmetrical interhemispheric counterpart. VMHC was computed for all the four time-series bins and normalized to the baseline for each mouse.
Functional Connectome Analysis
Whole-brain correlation matrices were obtained from a parcellation scheme of 130 regions using the Allen Mouse Brain ontology (Oh et al. 2014). In total, 65 regions of interest (ROIs) per hemisphere were considered, including regions from the isocortex, hippocampal formation, cortical subplate, striatopallidum, thalamus, midbrain, and hindbrain. Cohen’s d effect size was calculated, using the time series obtained from these whole-brain correlation matrices.
Statistical Analysis
Prior to performing statistical tests, within-subject normalization to the baseline was implemented. FSL general linear model (GLM) was used to perform statistical comparison between controls and wt-hM3Dq mice for the rsfMRI data (including seed-to-seed analysis, ReHo, VMHC, connectome analysis). We performed nonparametric permutation testing with 5000 permutations, using family-wise error correction with threshold-free cluster enhancement (TFCE). Statistical significance was defined as P < 0.05. CBF differences were statistically tested using linear mixed models implemented in SPSS24 (IBM, United States). In order to account for possible within-subject variance during scan preparation, all the statistical analysis included six covariates: 1) mouse age at the time of surgery; 2) bodyweight at the time of the scan; 3) total scan preparation time, that is, the time between the first anesthesia induction to the start of the resting-state scan; 4) bolus time, that is, the time from the bolus injection to the start of the continuous anesthetic infusion (isoflurane at 0.5%); 5) the number of ICA components rejected using FSL-FIX; and 6) clozapine dose. These covariates were implemented in all the statistical tests for all types of analysis performed.
Procedures for PVCre (hM4Di) Mice Study
Transfection Procedures for “PVCre-hSyn-hM4Di” Mice
The right primary somatosensory cortex of PVCre mice (mice expressing Cre recombinase in parvalbumin-expressing neurons) was unilaterally targeted with 950 nL of ssAAV8-hSyn1-dlox-hM4Di-mCherry(rev)-dlox-WPRE-hGHp(A) (n = 19) of a physical titer ≥5.4 × 1012 vg/mL (*vector genomes; fluorometric quantification) at the rate of 0.06 μL/min. The same procedures for animal preparation, anesthesia, and coordinates were used as already described for wild-typemice.
In Vivo Electrophysiology
Four PVCre-hSyn-hM4Di mice underwent in vivo electrophysiological recordings following the same procedures as described for wild-type mice (see Electrophysiological Recordings). The only difference was in the location of the control probe (Atlas Neurotechnologies, E32-100-S1-L6NT, 32 sites, 100-μm spacing, iridium oxide contacts), which was implanted into the right (ipsilateral to the DREADD injection site) striatum.
RsfMRI Acquisition
Preparation and anesthesia used for the scanning of the PVCre-hSyn-hM4Di mice were identical to the procedures outlined for wild-type mice (see MRI Setup and Animal Preparation). The two acquisitions only differed in the length of the scan. The PVCre-hSyn-hM4Di mice were scanned for 45 min where clozapine was intravenously injected at 15 min at the dose of 30 mg/mL.
Classification of Univariate BOLD Time Series
Note that all code for analysis of univariate BOLD dynamics can be found at https://github.com/benfulcher/hctsa_DREADD.
Data Processing and Feature Computation
We analyzed univariate BOLD time series measured from three brain regions: 1) right SSp (injected region); 2) left SSp (contralateral region); and 3) visual cortex (control region). Time-series measurements were split into three 15-min durations and labeled according to four classes of experiments: 1) wt-hSyn-hM3Dq (14 mice); 2) wt-CAMK-hM3Dq (13 mice); 3) PVCre-hSyn-hM4Di (19 mice); and 4) “sham” controls (13 mice). To understand which time-series properties distinguish different experimental conditions, we converted each univariate time series to a vector of 7873 interpretable properties (or features) using v0.9.6 of the hctsa toolbox (Fulcher et al. 2013; Fulcher and Jones 2017). This toolbox contains implementations of time-series features developed across different scientific disciplines, including summaries of the distribution of values in the data; its autocorrelation structure; stationarity, entropy, and predictability; parameters and goodness of time-series model fits; and methods from the physical nonlinear time-series analysis literature (see Fulcher and Jones, 2017, for a full list). In each brain region, measured time-series data were then represented as a 108 (experiments) × 7873 (time-series features) data matrix. Features with well-behaved, real-numbered outputs across the whole dataset and were nonconstant within all the three groups were retained for further analysis (7279 features). Each time series was labeled by its experimental condition (wt-hSyn-hM3Dq, wt-CAMK-hM3Dq, PVCre-hSyn-hM4Di, or sham) and time-point (baseline, “t2,” and “t3”).
Classification
We used the feature-based representations of BOLD time series in each brain area as the basis for classifying the different experimental conditions. We focused our analysis on the ∆1 time period, first subtracting time-series features computed at baseline and then normalizing these feature differences using an outlier-robust sigmoidal transformation (Fulcher et al. 2013). A linear support vector machine classification model was learned on the normalized feature matrix for a given brain area, using inverse probability class reweighting to accommodate class imbalance. A measure of discriminability of a pair of classes was quantified as the balanced classification accuracy using 10-fold stratified cross-validation. Balanced accuracy was computed as the arithmetic mean of sensitivity and specificity to account for the small class imbalance: 13 sham controls and 14 wt-hSyn-hM3Dq or 13 wt-CAMK-hM3Dq or 19 PVCre-hSyn-hM4Di mice. To reduce variance in the random partition of data into 10 folds, we repeated this procedure 50 times (with each iteration yielding a balanced accuracy value) and summarized the resulting distribution of balanced accuracies as its mean and standard deviation.
In smaller samples, there is a greater probability that optimistic classification results can be obtained by chance. To quantify this, we evaluated the statistical significance of our classification results relative to random assignments of class labels to data. This was achieved by computing a null distribution of the same performance metric used above (mean across 50 repeats of 10-fold cross-validated balanced accuracy) for 5000 random class label assignments to time series. The statistical significance of a given classification result was then estimated as a permutation test (as the proportion of 5000 null samples with a mean balanced classification rate exceeding that of the true assignment of class labels).
Low-Dimensional Projections
As explained above, each BOLD signal was represented as a feature vector or, equivalently, as a point in a high-dimensional feature space. To aid the visualization of the class structure of data in this space, we projected normalized time series × feature data matrices into a lower-dimensional space that captures the maximal variance in the full feature space using principal component analysis. In these plots, individual time series are points in the space, providing an intuitive visualization of the structure of the time-series dataset.
Interpreting Differences in Univariate Dynamics
We aimed to determine which properties of the univariate BOLD dynamics in the injected brain area best discriminated 1) wt-hSyn-hM3Dq from controls; 2) wt-CAMK-hM3Dq from controls; 3) PVCre-hSyn-hM4Di from controls; and 4) wt-hSyn-hM3Dq from wt-CAMK-hM3Dq and also from PVCre-hSyn-hM4Di mice. Focusing on the first time period after baseline, we first computed the difference in each feature value between this first time period and the baseline period (∆1). We then tested whether these relative feature values differed between groups using a Wilcoxon rank-sum test. Statistical correction for the large number of hypothesis tests was performed as the false discovery rate (Benjamini and Hochberg 1995), setting a significance threshold of q = 0.05.
Predicting Fmr1y/− Mice Using a Classifier Trained on PVCre-hSyn-hM4Di
Time series from 46 cortical ROIs (23 in each hemisphere) obtained from Fmr1y/− (n = 44) and wild-type littermates (n = 39) was obtained from 2 of our previous studies (Zerbi et al. 2018; Zerbi et al. 2019b). The classification proceeded in two stages. In the training phase, a classification model was trained on normalized features of rsfMRI time series in SSp (right hemisphere) during the ∆1 time period, as described above. That is, the trained classifier learns features of the BOLD dynamics to accurately predict whether a new time series is PVCre-hSyn-hM4Di or control. In the testing phase, the trained model was used to predict labels of rsfMRI time series in Fmr1y/− dataset. We identified PVCre-hSyn-hM4Di with the Fmr1y/− knockout condition and quantified prediction performance as balanced accuracy (%) as above, evaluated in each brain region independently. P values were estimated for each brain area using permutation testing with 5 × 106 null samples generated by independent shuffling of labels and corrected across brain regions (focusing just on the right hemisphere) using the false discovery rate method of Benjamini and Hochberg (Benjamini and Hochberg 1995).
Histological Evaluation of Transfection
DREADD’s viral expression (for wt-hSyn-hM3Dq, PVCre-hSyn-hM4Di, and wt-CAMK-hM3Dq) was confirmed by mCherry staining using standard immunohistochemistry protocols. Briefly, after the last MRI session, mice were deeply anesthetized using a mixture of ketamine (100 mg/kg; Graeub), xylazine (10 mg/kg; Rompun, Bayer), and acepromazine (2 mg/kg; Fatro S.p.A) and transcardially perfused with 4% paraformaldehyde (PFA, pH = 7.4). The brains were postfixed in 4% PFA for 1.5 h at 4 °C and then placed overnight in 30% sucrose solution. Brains were frozen in a tissue mounting fluid (Tissue-Tek O.C.T Compound, Sakura Finetek Europe B.V.) and sectioned coronally in 40-μm-thick slices using a cryostat (MICROM HM 560, Histocom AG, Switzerland). Free-floating slices were first permeabilized in 0.2% Triton X-100 for 30 min and then incubated overnight in 0.2% Triton X-100, 2% normal goat serum, and rabbit anti-mCherry (1:1000, Ab167453, Abcam) at 4 °C under continuous agitation (100 rpm/min). The next-day sections were incubated for 1 h in 0.2% Triton X-100, 2% normal goat serum, goat antirabbit Alexa Fluor 546 (1:300, A11035, Life Technologies), and Nissl (1:300, NeuroTrace 660, Thermo Molecular Probes) or DAPI (1:300, Sigma-Aldrich) at room temperature under continuous agitation. Afterward, slices were mounted on the SuperFrost slides where they were left to air-dry and later coverslipped with Dako Fluorescence mounting medium (Agilent Technologies). Confocal laser-scanning microscope (Leica, SP8, Center for Microscopy and Image Analysis, UZH) and Zeiss Slide Scanner (Zeiss Axio scan, Z1, Center for Microscopy and Image Analysis, UZH) were used to detect the viral expression. Microscopy protocol included a tile scan with a 10× or a 20× objective, a pixel size of 1.2 μm, and an image size of 1024 × 1024 pixels. Images were preprocessed and analyzed using ImageJ-Fiji and Zeiss Zen Blue, respectively.
Results
The E:I balance was experimentally increased in the right SSp (Supplementary Figure 1). Adult mice were transfected with excitatory designer receptors exclusively activated by designer drugs (hM3Dq DREADDs), which were tuned to primarily target pyramidal neurons (wt-CAMK-hM3Dq and wt-hSyn-hM3Dq, n = 13; n = 10). Control mice were sham operated (weight- and age-matched littermate controls, n = 9; Supplementary Table 1). Three to four weeks after surgery, mice were lightly anesthetized, and electrophysiology or macroscopic brain activity was recorded before and after activating the DREADDs with a low dose of clozapine.
In vivo electrophysiology recordings in wt-hSyn-hM3Dq mice, used primarily to ensure successful DREADD activation, confirmed that activating the DREADDs increases net excitation within the targeted right SSp but not in a control area, that is, its homotopic counterpart (left SSp). Multiunit activity was recorded under isoflurane anesthesia (0.5%) from multicontact electrode arrays (Fig. 1A–D). During the baseline period (15 min), the average basal firing rate was stable in both brain areas. However, upon clozapine injection, the average firing rate in the right SSp increased steadily over time while activity in the left SSp remained virtually unchanged. Thirty minutes after injection, the difference in averaged multiunit activity between left and right SSp reached 11% (Fig. 1A–D). We further investigated the effects of clozapine on neuronal activity in mice that did not express DREADDs. With clozapine injection, the average firing rate in the right SSp as well as in a control region remained unchanged (Supplementary Figure 2). This suggests that clozapine administered at our concentration has no effect on neuronal activity in the absence of DREADD.
Figure 1.
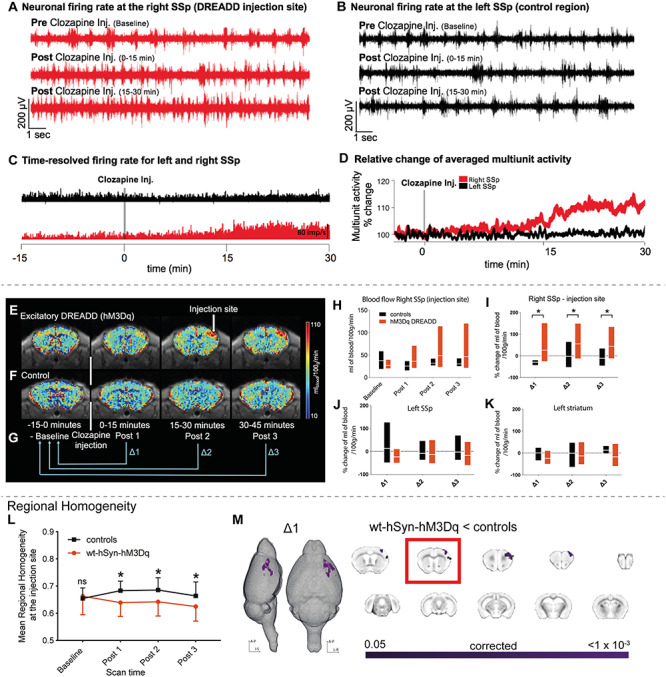
Local changes in neural activity induced by activating hSyn-hM3Dq. (A) Neuronal firing rate before and after clozapine injection in the right SSp, indicating a steady increase in firing rate upon DREADD activation. (B) As A but for the left SSp (control area) indicating no change in the neuronal firing rate after clozapine injection. (C) Time-resolved firing rate in millisecond bins for the right (red) and left (black) SSp. A steady increase in firing rate occurs in the right SSp once clozapine is injected, while no change occurs in the left SSp. (D) Relative change of averaged multiunit activity recorded in the right and left SSp. (E), (F) Cerebral blood flow maps were collected every 15 min for wt-hSyn-hM3Dq and control mice (displayed in their native space), respectively. (G) Clozapine is injected 15 min after the start of the experiment. These 15 min are referred to as the baseline period (−15 to 0 min). The rest of the experiment is 45 min long and divided into three periods, that is, Post 1 (0–15 min), Post 2 (15–30 min), and Post 3 (30–45 min). For all analyses performed in this experiment (unless otherwise stated), Post data was expressed relative to baseline (Post 1/Post 2/Post 3—baseline) and is referred to as ∆1, ∆2, and ∆3, respectively. (H) Comparison of blood flow (mL of blood/100 g/min) between controls and wt-hSyn-hM3Dq mice over time (measured at the injection site). (I) Percentage change in blood flow over time between the controls (n = 7) and wt-hSyn-hM3Dq (n = 7) mice (measured at the injection site). Linear mixed models indicate a significant main groups × scan time effect (F2,21 = 5.657; P = 0.04; asterisk indicates significant between-group independent samples post hoc t-test at Post 1 (t12 = 3.23; P = 0.07), at Post 2 (t12 = 2.66; P = 0.02), and at Post 3 (t12 = 2.35; P = 0.04). (J) Percentage change in blood flow over time measured in left SSp. (K) Percentage change in blood flow over time measured in the left lateral striatum. (L) Average regional homogeneity (ReHo) change over time at the injection site. Repeated measures ANOVA (control [n = 13]; wt-hSyn-hM3Dq [n = 14]) showed a significant groups × scan time effect (F1.93,75 = 11.8; P < 0.001; asterisk indicates significant between-group independent samples post hoc t-test at Post 1 (t25 = −2.57; P = 0.01), at Post 2 (t25 = −2.28; P = 0.03), and at Post 3 (t25 = −2.1; P = 0.05). (M) Significant decrease in ReHo in wt-hSyn-hM3Dq mice versus controls for ∆1 depicted as a 3D image and in coronal slices (TFCE-corrected). The slice marked with a red rectangle shows the injectionsite.
Next, we used noninvasive fMRI to test whether activation of hM3Dq-DREADD in wt-hSyn-hM3Dq would elicit anatomically specific changes in cerebral blood flow (CBF) which reflects glucose metabolism and has been shown to be an indirect marker of neural activity at the cell population level (Juepter and Weiller 1995; Chen et al. 2008). After administering clozapine, wt-hSyn-hM3Dq mice showed an increase in CBF around the injection site (Fig. 1E,H). The increase was significantly higher than in sham-operated mice (controls) scanned with identical procedures for all time points up to 45 min (linear mixed model: F2,21 = 5.657, P = 0.04; Fig. 1F,I). No changes in CBF were observed in other brain regions, that is, the contralateral somatosensory cortex and ipsilateral striatum (Fig. 1J,K). Together our electrophysiological and CBF results show that activating the DREADDs caused microcircuit excitation which was anatomically specific to the injectedarea.
Activating hM3Dq-DREADD in wt-hSyn-hM3Dq Mice Decreases Short-Range Connectivity Selectively Within Somatosensory and Somatomotor Cortices
We next tested whether hM3Dq-DREADD also caused a local disruption of functional connectivity (FC) measured with rsfMRI. To this end, we calculated the regional homogeneity (ReHo) index, a commonly used metric to assess the “synchronization” of BOLD fluctuations across neighboring voxels. We chose ReHo because it has been shown to reflect alterations occurring in various psychiatric diseases including depression, schizophrenia, and autism (Paakki et al. 2010; Iwabuchi et al. 2015). Activation of hM3Dq-DREADD significantly reduced regional homogeneity only at the injection site when compared with controls (blue cluster in Fig. 1L,M; P < 0.05, TFCE-corrected). These changes occurred within the first 15 min after clozapine injection and were maintained for the whole scan period (Supplementary Figure 3A). No other area exhibited similar changes. Note that the controls exhibited a slight increase of ReHo after the clozapine injection. Since the animals were only slightly anesthetized, it is likely that they responded to the injection with an increase of general arousal which has been shown to affect rsfMRI connectivity (Zerbi, Floriou-Servou, et al. 2019a).
Activated DREADD in wt-hSyn-hM3Dq Mice Reduces Functional Connectivity Between Somatosensory Cortex and Monosynaptically Connected Cortical Areas
Does an increase in local microcircuit excitation also affect long-range FC as quantified by correlating spontaneous BOLD fluctuations between areas? To address this question, we measured FC between 130 brain regions as identified by the Allen Brain Reference Atlas Common Coordinate Framework (CCFv3) at every 1-min interval (see Methods). We then extracted the effect size using the standardized difference between the group means (Cohen’s d, wt-hSyn-hM3Dq vs. control mice) between each pair of regions. During the first 15 min (baseline), Cohen’s d varied on average between −0.2 and +0.2 (null-to-small effect) and did not exhibit an appreciable spatial or temporal pattern. However, immediately after clozapine injection, we noticed a rapid reduction in FC between a number of connections in the somatosensory and somatomotor cortices, which remained hypoconnected for the whole scan session (Fig. 2A,B).
Figure 2.
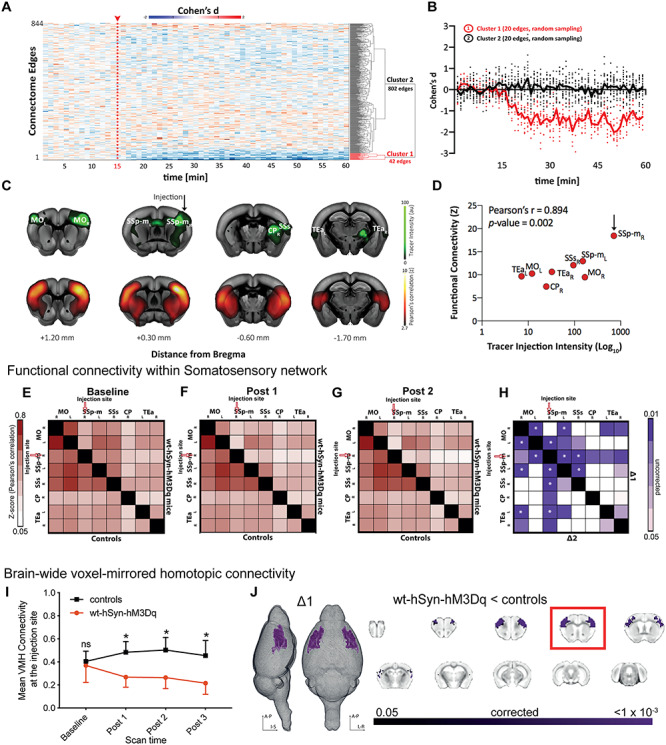
Long-range connectivity changes induced by activating hSyn-hM3Dq. (A) Effect-size analysis (Cohen’s d) of functional connectivity is calculated for single edges (n = 844). Only cluster 1 shows a decrease in the effect size after clozapine injection. (B) Averaged effect size (Cohen’s d) over time for 20 edges of cluster 1 and 20 randomly selected edges of cluster 2. (C) Coronal brain slices depicting tracer data after unilateral injection in SSp (green, obtained from Allen Mouse Brain Connectivity Atlas, Injection ID: 114290938) and averaged voxel-wise Pearson’s correlation (Fisher’s z-transform) during the baseline period from a seed in the right SSp. (D) Pearson’s correlation between functional connectivity of the target region (right SSp) and tracer injection intensity obtained from images depicted in C. (E–G) Matrices represent averaged and z-transformed Pearson’s correlation of all control animals (lower triangular matrix) and all wt-hSyn-hM3Dq animals (upper triangular matrix) for the baseline period, Post 1 and Post 2, respectively. (H) The upper triangle of the matrix shows a reduction in FC in wt-hSyn-hM3Dq (n = 14) mice compared with controls (n = 13) during ∆1 (i.e., Post 1-baseline), while the lower triangle shows the same set of comparisons in ∆2 (i.e., Post 2-baseline). Color coding reflects randomized permutation testing, P < 0.05, uncorrected. Asterisks indicate significant differences that survived FDR correction for multiple comparisons. MO, somatomotor cortex; SSp-m, primary somatosensory cortex (mouth); SSs, supplementary somatosensory cortex; CP, caudoputamen; TEa, temporal association cortex; R, right; L, left. (I) Change over time of the mean brain-wide VMHC (symmetric connectivity) at the injection site. Repeated measures ANOVA (controls [n = 13]; wt-hSyn-hM3Dq [n = 14]) indicates significant groups × scan time effect (F1.67,41.9 = 21.18; P < 0.001; asterisk indicates significant between-group independent samples post hoc t-test at Post 1 (t24 = −6.01; P = 10−5), at Post 2 (t24 = −5.93; P = 10−5), and at Post 3 (t24 = −5.22; P = 10−5). (J) Significant decrease (TFCE-corrected) in symmetric connectivity depicted on 3D images and coronal slices for the ∆1 time point. The slice marked with a red rectangle is the injection slice.
We next quantified specific changes in FC within the somatosensory network, that is, areas which are monosynaptically connected to the chemogenetic target, right SSp according to the Allen Mouse Brain Connectivity Atlas (Oh et al. 2014). This anatomically defined network includes contralateral primary SSp, bilateral somatomotor cortex (MO), ipsilateral supplementary somatosensory cortex (SSs), bilateral association areas (TEa), and ipsilateral caudoputamen (CP) (Fig. 2C). We found high FC between all of these structures at baseline (Fig. 2C) and a strong structure–function correspondence (Fig. 2D). To increase the power of our analysis, the BOLD time series of the somatosensory network regions were binned into four intervals of 15 min each. Pearson’s correlations were calculated between regions and are summarized in FC matrices (Fig. 2E–G and Supplementary Figure 4A). At baseline, FC did not show differences between groups (Fig. 2E). By contrast, after clozapine injection (Fig. 2F–H), we observed a reduction in FC between right SSp and nearly all other areas of the network in wt-hSyn-hM3Dq mice, including corticostriatal circuitry. Significant group differences (permutation testing, P < 0.05, FDR corrected) were found between the right SSp and left SSp/left MO, as well as between left and right MOp, and were maintained for the duration of the scan session (Fig. 2H, Supplementary Figure 4B). We also performed similar analyses in a small cohort of animals using 1 mg/kg CNO which resulted in FC decrease within MO regions in wt-hSyn-hM3Dq mice (Supplementary Figure 5).
We further complemented our analysis by assessing the similarity between the BOLD time series of symmetric voxels in the left and right hemispheres. This index has been shown to identify pathological connectivity patterns with high sensitivity as compared with other FC metrics (Hahamy et al. 2015). In line with our previous findings, injecting clozapine caused a significant reduction of symmetric connectivity in wt-hSyn-hM3Dq mice but not in controls which occurred only between the left and right primary SSp and left and right MO (P < 0.05; TFCE-corrected) (Fig. 2I,J). In summary, our analyses demonstrate that increasing local cortical excitation causes a reduction in local connectivity that propagates into long-range FC reduction exclusively within the targeted network.
Activated DREADD in wt-CAMK-hM3Dq Mice Reduces Functional Connectivity
The above results were obtained using human synapsin-1 as the promoter for the viral injection. Human synapsin-1 promoter is not solely specific toward pyramidal neurons but can also transfect inhibitory interneurons. To ensure specificity for pyramidal neurons, we repeated the above experiments in a separate cohort of wild-type mice (n = 13) using the CAMKII promoter for AAV hM3Dq-DREADD delivery (Supplementary Figure 7). Firstly, we measured CBF to confirm reliable DREADD activation at the injection site. After administering clozapine, wt-CAMK-hM3Dq mice showed a significant increase in CBF around the injection site (Fig. 3A) as compared with controls (repeated measures ANOVA: F(1.0, 20.0) = 7.3; P = 0.004). This increase was limited to the injection site and not observed in other brain regions (Fig. 3B,C). Regional homogeneity as well as symmetric connectivity metrics significantly decreased at the injection site after DREADD activation in wt-CAMK-hM3Dq mice versus controls (repeated measures ANOVA: F(1.3, 25.2) = 7.74, P = 0.006; F(1.16, 21.98) = 8.21, P = 0.007, respectively; Fig. 3D,E), and there was pronounced underconnectivity within the somatosensory network (Fig. 3F). Importantly, we replicated our results obtained with human synapsin-1 promoter and confirmed that increasing E:I by enhancing net excitatory synaptic transmission causes a robust reduction of FC within an anatomically connected network.
Figure 3.
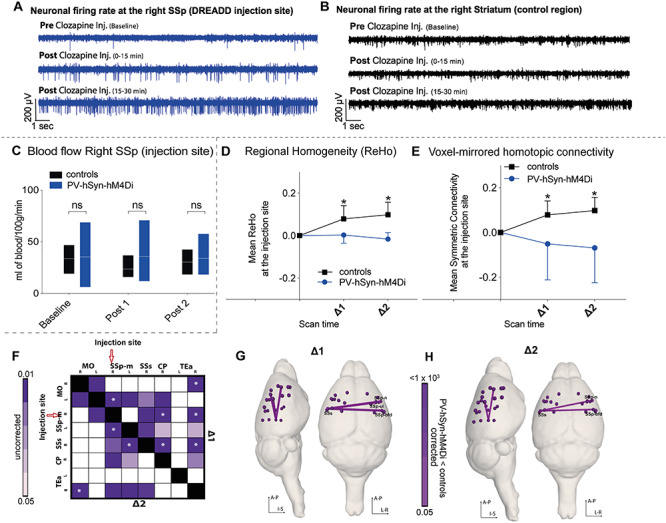
Changes induced by activating CAMK-hM3Dq in wild-type mice. (A) Comparison of blood flow (mL of blood/100 g/min) over time between controls and wild-type mice injected with excitatory (hM3Di) DREADD on CAMKII promoter (measured at the injection site). We observe a significant repeated measures ANOVA groups × scan time effect (F(1.0, 20.0) = 7.3; P = 0.004; asterisk indicates significant between-group independent samples post hoc t-test at Post 1 (t9.8 = 2.7; P = 0.02) and at Post 2 (t9.4 = 2.1; P = 0.05) in blood flow in wt-CAMK-hM3Dq mice (n = 13) after clozapine injection (similar to wt-hSyn-hM3Dq from Fig. 2H). (B,C) Blood flow in the left SSp and left CP does not significantly differ between controls and wt-CAMK-hM3Dq mice. (D) Change over time of mean regional homogeneity at the injection site (right SSp). There is a significant decrease (repeated measures ANOVA significant groups × scan time effect: F(1.3, 25.2) = 7.74, P = 0.006; asterisk indicates significant between-group independent samples post hoc t-test at Post 1 [t23 = −3.46; P = 0.002] and at Post 2 [t23 = −3.49; P = 0.002]) in mean connectivity in wt-CAMK-hM3Dq (n = 13) mice as compared with controls (n = 13). (E) Change over time of mean symmetric connectivity at the injection site. There is a significant decrease (repeated measures ANOVA groups × scan time effect: F(1.16, 21.98) = 8.21, P = 0.007; asterisk indicates significant between-group independent samples post hoc t-test at Post 1 (t24 = 3.85; P = 0.001) and at Post 2 [t24 = 4.26; P = 10−5]) in mean connectivity in wt-CAMK-hM3Dq (n = 13) mice as compared with controls (n = 13). (F) Matrix represents statistically significant differences during ∆1 and ∆2 time period among three brain regions (MO, somatomotor cortex; SSp, somatosensory cortex; SSs, supplementary somatosensory cortex; R, right; L, left) between the controls and wt-CAMK-hM3Dq mice. The asterisk within ROIs indicates a statistically significant difference that survived correction of multiple comparisons.
Changing E:I Balance by Inhibiting GABAergic Parvalbumin Neurons
Changes in E:I have been proposed as a principle mechanism that underlies symptoms of neurodevelopmental disorders, irrespective of whether primarily excitatory or inhibitory synapses are affected. Next, we tested whether suppressing the activity of GABAergic PV interneurons—as reported in several mouse models for ASD (Penagarikano et al. 2011; Wohr et al. 2015; Ajram et al. 2017)—would cause similar macroscopic hypoconnectivity as observed above. Nineteen Pvalbtm1(cre)Arbr (PVCre) were transfected with an inhibitory hM4Di-DREADD. Clozapine activation reduced PV cell activity which resulted in overexcitation of right SSp but not a control region. Accordingly, the average firing rate in right SSp went up by 20% following clozapine injection (Fig. 4A,B and Supplementary Figure 6A,B), and local cerebral blood flow increased moderately (repeated measures ANOVA: F1.4,17.9 = 2.45; P = 0.1; Fig. 4C). Our rsfMRI analysis confirmed the expected reduction in local synchronization around the injection site (Fig. 3D) and also marked underconnectivity which was most pronounced between right and left SSp (Fig. 3E–H) in PVCre-hSyn-hM4Di mice as compared with controls. In summary, our results reveal that shifting E:I toward excitation by inhibiting PV interneurons leads to a reduction of local and long-range FC, which is qualitatively similar to that observed during direct system excitation.
Figure 4.
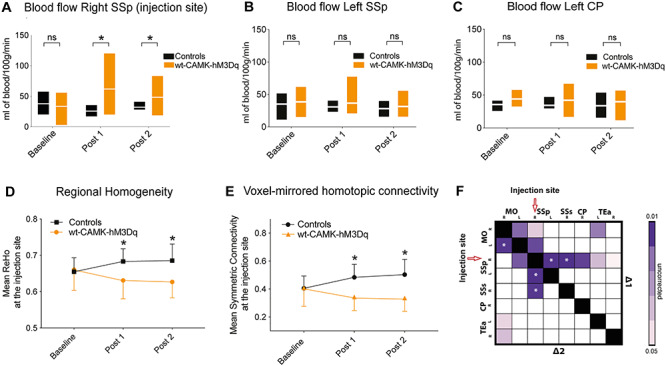
Changes induced by activating hSyn-hM4Di in PVCre mice. (A) Neuronal firing rate at the right SSp DREADD injection site of the PVCre-hSyn-hM4Di mice before and after clozapine injection, indicating a steady increase in firing rate on DREADD activation. (B) Similar to figure A but for right caudoputamen, indicating no change in the neuronal firing rate after clozapine injection. (C) Comparison of blood flow (mL of blood/100 g/min) between controls (n = 7) and PVCre-hSyn-hM4Di (n = 13) mice over time (measured at the injection site). No significant differences observed between groups (repeated measures ANOVA: F1.4,17.9 = 2.45; P = 0.1). (D) Normalized average regional homogeneity (ReHo) change over time at the injection site. Repeated measures ANOVA indicates significant groups × scan time effect: F1,25 = 14.01; P = 0.001; asterisk indicates significant between-group independent samples post hoc t-test at ∆1 (t30 = −4.17; P = 10−5) and at ∆2 (t30 = −7.15; P = 10−6). (E) Normalized averaged symmetric connectivity over time at the injection site. Repeated measures ANOVA indicates significant groups × scan time effect: F1.2, 21.9 = 8.21; P = 0.007; asterisk indicates significant between-group independent samples post hoc t-test at ∆1 (t29 = −3.52; P = 0.001) and at ∆2 (t29 = −5.78; P = 10−5). (F) Seed-to-seed analysis indicates reduced FC (randomized permutation testing, P < 0.05, uncorrected) between controls (n = 13) and PVCre-hSyn-hM4Di (n = 19) mice. (G,H) Whole-brain connectome analysis shows a significant interhemispheric reduction between somatosensory cortices for ∆1 and ∆2 between PVCre-hSyn-hM4Di (n = 14) mice and controls (n = 13). Regions affected are as follows: SSp-m, primary somatosensory area, mouth; SSp-ul, primary somatosensory area, upper limb; SSs, supplementary somatosensory area; SSp-bdf, primary somatosensory area, barel field; SSp-n, primary somatosensory area, nose.
Altered E:I Balance Changes Local BOLD Dynamics
Our previous analyses demonstrated that increasing E:I in right SSp affects its FC with other brain regions. We next aimed to understand how microcircuit E:I shapes the fluctuations of spontaneous BOLD regional activity. As depicted in Figure 4A, we focused our analysis on three regions of interest: 1) right SSp (the injection site); 2) left SSp (its homotopic counterpart); and 3) left visual cortex (a control region). The dynamics of BOLD time series were characterized by an interdisciplinary library of over 7000 features (using the software package hctsa (Fulcher and Jones 2017)), which capture distinct properties of the BOLD signal, including spectral power in different frequency bands, temporal entropy, and many more (Fulcher et al. 2013; Fulcher and Jones 2017; Sethi et al. 2017) (Fig. 5B; see Methods for details).
Figure 5.
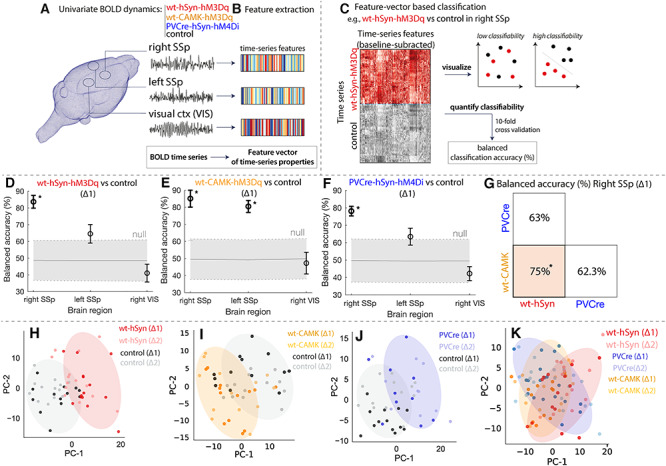
DREADD manipulations lead to characteristic changes in univariate BOLD dynamics. Classification of BOLD signal dynamics in three brain regions (the injected right SSp, left SSp, and VIS) across four conditions (wt-hSyn-hM3Dq, wt-CAMK-hM3Dq PVCre-hSyn-hM4Di, and control). A schematic of the approach is depicted in A–C: (A) BOLD dynamics are measured from each brain region as a univariate time series (a 15-min time series per time window and experiment), which was (B) converted to a set of properties (a “feature vector”) using hctsa. (C) For a given region and pair of classes, we used the features of each time series (relative to baseline) as the basis for classification, which was visualized using a low-dimensional principal component projection and quantified as the 10-fold cross-validated balanced classification accuracy (%). Classification results in each brain region at ∆1 are shown for (D) wt-hSyn-hM3Dq (n = 14) versus control (n = 13), (E) wt-CAMK-hM3Dq (n = 13) versus control (n = 13), and (F) PVCre-hSyn-hM4Di (n = 10) versus control (n = 13), revealing significant discriminability in the right SSp for wt-hSyn-hM3Dq and PVCre-hSyn-hM4Di versus control (permutation test, P < 0.05, annotated as “*”). There is a consistent trend of high discriminability in the injected region (right SSp), followed by the contralateral region (left SSp), and lowest discriminability in the control region (VIS). (G) Classification results in the right SSp at ∆1 are shown for wt-hSyn-hM3Dq versus wt-CAMK-hM4Di, wt-hSyn-hM3Dq versus PVCre-hSyn-hM4Di, and wt-CAMK-hM4Di versus PVCre-hSyn-hM4Di. High discriminability is only present for wt-hSyn-hM3Dq versus wt-CAMK-hM4Di (permutation test, P < 0.05, annotated as “*”). BOLD time series in right SSp are visualized in two-dimensional principal component spaces in (H–K) for the same three pairs of classes as in (D–G). Time series with similar properties are close in the space, revealing a visual depiction of the discriminability of wt-hSyn-hM3Dq, wt-CAMK-hM3Dq, and PVCre-hSyn-hM4Di relative to control (H–J), but a relative lack of discriminability in K. Shaded ellipses (∆1) have been added to guide the eye, and time series from each class are labeled for ∆1 and ∆2.
Based on the change of these features from baseline to activated DREADDs, we trained linear support-vector machine (SVM) classifiers to distinguish: 1) wt-hSyn-hM3Dq versus controls, 2) wt-CAMK-hM3Dq versus control, 3) PVCre-hM4Di versus controls, 4) wt-hSyn-hM3Dq versus wt-CAMK-hM3Dq, 5) wt-CAMK-hM3Dq versus PVCre-hSyn-hM4Di, and 6) wt-hSyn -hM3Dq versus PVCre-hSyn-hM4Di as shown in Figure 5C.
Our classification results reveal that BOLD dynamics of the injected right SSp at ∆1 are differentiated from control mice in wt-hSyn-hM3Dq (Fig. 5D), wt-CAMK-hM3Dq (Fig. 5E), and PVCre-hSyn-hM4Di (Fig. 5F) with high accuracy (≥84% balanced accuracy after 10-fold cross-validation), significantly exceeding chance levels (P ≤ 1 × 10−3, permutation test). Qualitatively similar results were obtained at ∆2 (see Supplementary Results). DREADD activation had a similarly large effect on the accuracy of classifying BOLD dynamics in the left SSp but only for wt-CAMK-hM3Dq (81%, P ≤ 1 × 10−3), while for the other two groups, the effect was much smaller—wt-hSyn-hM3Dq, 64% (P = 0.1), and PVCre-hSyn-hM4Di, 69% (P = 0.03)—and finally the weakest for the control region, VIS, in all groups wt-hSyn-hM3Dq, 43% (P = 0.7); wt-CAMK-hM3Dq, 45% (P = 0.7); and PVCre-hSyn-hM4Di, 54% (P = 0.3).
To aid visualization, we constructed principal component projections of each dataset (including data at ∆2), shown for wt-hSyn-hM3Dq versus control, wt-CAMK-hM3Dq versus control, and PVCre-hSyn-hM4Di versus control in Figure 5H–J, respectively. These plots reveal a clear distinction between DREADD-activated and control mice on the basis of their BOLD dynamics in rightSSp.
We next sought to investigate the properties of BOLD time series between the three DREADD manipulated groups at the right SSp during ∆1. As shown in Figure 5G, BOLD dynamics at the right SSp between wt-hSyn-hM3Dq and wt-CAMK-hM3Dq are clearly distinct (75% balanced accuracy, P = 0.01). BOLD dynamics between wt-hSyn-hM3Dq and PVCre-hSyn-hM4Di and wt-CAMK-hM3Dq and PVCre-hSyn-hM4Di, in the right SSp, are not significantly distinguishable (63% balanced accuracy, P = 0.09, and 62% balanced accuracy, P = 0.1, respectively). Moreover, the clear discrimination of BOLD dynamics in principal components space is mainly absent (visible as strongly overlapping distributions in Fig. 5K). Properties of BOLD time series were also unable to significantly distinguish DREADD manipulations in left SSp and visual control region during ∆1 (Supplementary Figure 9A,B).
Since wt-hSyn-hM3Dq and PVCre-hSyn-hM4Di increase E:I through different mechanisms, we next sought to further compare the effect of their manipulations on BOLD dynamics; we computed a similarity measure between changes in BOLD time-series properties caused by both manipulations relative to controls (as a Spearman’s ρ in right SSp and left SSp relative to the control region across all time-series features; see Supplementary Figure 10). We found overall agreement between wt-hSyn-hM3Dq and PVCre-hSyn-hM4Di in right SSp (ρ = 0.59) and left SSp (ρ = 0.47), suggesting that the temporal properties of BOLD dynamics are more sensitive to the net overexcitation common to both DREADD cohorts than to the underlying synaptic transmission pathway within the cortical microcircuit (see Supplementary Material for an analysis of BOLD time-series features that are individually informative ofE:I).
Fmr1y/− Exhibit Similar Local BOLD Dynamics to DREADD-Manipulated Mice
Our previous analysis yielded a classifier for predicting an acute microcircuit E:I increase from macroscopic BOLD dynamics. Next, we asked whether such a classifier can also detect abnormal BOLD dynamics in a transgenic disease model of ASD. We used rsfMRI data from Fmr1y/− knockout mice that mimic fragile X syndrome, a neurodevelopmental disorder characterized by cognitive impairment, epileptic seizures, physical abnormalities, and hypersensitivity. Previous studies in Fmr1y/− mice report a reduction in cortical PV cell density, specifically within somatosensory region, followed by cortical hyperexcitability (Selby et al. 2007; Gibson et al. 2008; Wohr et al. 2015; Nomura et al. 2017). These findings indicate that the disease mechanism of Fmr1y/− mice affects the net excitability of cortical microcircuits in a similar manner as chemogenetically suppressing PVCre activity, but it is unknown whether these circuit-level alterations cause analogous alteration of macroscopic rsfMRI dynamics. To test this hypothesis, we trained a classifier for PVCre-hSyn-hM4Di rsfMRI time series in the right SSp during ∆1 as above (Figs 4E and 5A). We then applied this classifier to rsfMRI data from 46 cortical ROIs (23 from each hemisphere) in Fmr1y/− mice (n = 44) and wild-type littermates (Fmr1y/+, n = 39) and assigned predicted labels to each brain region of Fmr1y/− and Fmr1y/+ independently (Fig. 5B). As shown in Figure 5C, regional BOLD dynamics of Fmr1y/− versus wild-type controls could be significantly classified in 10 different areas of the right hemisphere (balanced classification accuracy, P < 0.05, FDR-corrected) using a classifier trained on PVCre-hSyn-hM4Di data. Corresponding to the brain area in which the DREADDs classifier was trained, regions of the SSp displayed the highest classifiability. Similar results were obtained in the left hemisphere, resulting in a Spearman’s ρ = 0.69 (P = 3 × 10−4) between the left and right hemispheres (Fig. 5E,F). We next tested whether regional variation in classification accuracy was related to differences in PV cell density using data from qBrain (Kim CK et al. 2017a). As shown in Figure 6D, classification accuracy is only weakly correlated with PV cell density, ρ = 0.33 (P = 0.1), but is highest in SSp regions with the highest PV cell density (dark green in Fig. 6D). Interestingly, the classifier identified several prefrontal areas (bright green in Fig. 6D) which exhibited abnormal rsfMRI dynamics according to the classifier despite low PV cell density. This finding is in line with former reports of abnormal prefrontal cortex function in Fmr1y/− mice (Paul et al. 2013; Routh et al. 2017). Our results demonstrate that a classifier trained on a controlled chemogenetic manipulation causing an acute shift of the E:I balance is generalizable to Fmr1y/−, indicating a strong degree of similarity between cortical BOLD dynamics resulting from PVCre-hSyn-hM4Di and Fmr1y/− knockout.
Figure 6.
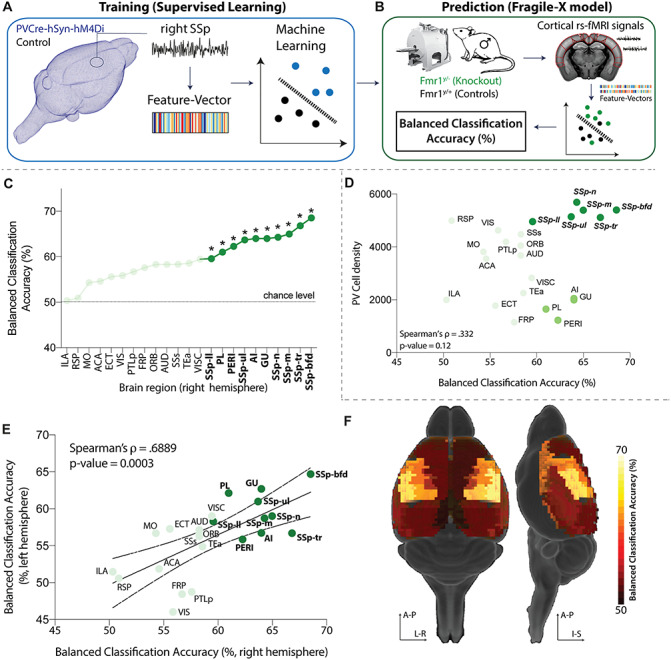
Fmr1y/− exhibit similar local BOLD dynamics as DREADD-manipulated mice. (A) The classifier was trained on features of right SSp BOLD time series to distinguish PVCre-hSyn-hM4Di from control mice. (B) The trained classifier was used to predict the identity of cortical BOLD time series measured from Fmr1y/− (knockout) and Fmr1y/+ (control) mice, identifying predictions of PVCre-hSyn-hM4Di as Fmr1y/−. Predictability in each brain region was assessed as balanced classification accuracy (%); P values were estimated using a permutation test. (C) Classification results are shown for the right hemisphere, highlighting regions with significant classification accuracy (P < 0.05, FDR-corrected) as bold, dark green, and marked with asterisks. (D) Scatter plot of balanced classification accuracy and PV cell density. (E) Balanced classification accuracy is plotted for each region in the right and left hemispheres, exhibiting a strong positive correlation, Spearman’s ρ = 0.69 (P = 3 × 10−4). (F) Balanced classification accuracy visualized on a 3D brain.
Discussion
It has been a long-standing question how disease mechanisms at the cellular level map onto abnormal dynamics at the level of macroscopic brain networks. Here we show in three independent cohorts that acutely increasing the microcircuit E:I balance in somatosensory cortex via chemogenetics causes distinct changes of macroscopic brain dynamics and severe functional underconnectivity between the manipulated region of interest (SSp) and monosynaptically connected brain areas. These macroscopic changes were observed irrespective of whether we enhanced excitatory transmission or selectively suppressed inhibitory PV neurons of the same cortical circuits. Our results constitute the first direct evidence that macroscopic BOLD dynamics reflect the state of underlying cortical microcircuits. Furthermore, we show that different cellular mechanisms can cause common pathophysiological abnormalities and converge on highly similar alterations of neural dynamics at the network level. In particular, upon activating the DREADDs, there was a marked reduction of FC which was temporally locked to our stimulation paradigm and spatially specific to the injected area and its monosynaptically connected neighbors. Reduced FC has been frequently reported in human fMRI studies of neurodevelopmental disorders, such as autism and schizophrenia (Uhlhaas and Singer 2006; Minshew and Keller 2010; Gao and Penzes 2016; Kraguljac et al. 2016; Wang et al. 2018), as well as in relevant mouse models of these disorders (CNTNAP2, Fmr1y/−, En2) (Bertero et al. 2018; Zerbi et al. 2018). Many of these mouse models have been shown to exhibit an increased E:I (Rubenstein and Merzenich 2003; Gibson et al. 2008; Contractor et al. 2015). Additionally, they suffer from reduced PV interneuron expression (Penagarikano et al. 2011; Vogt et al. 2017). Depletion of this specific cell type has been hypothesized to cause aberrant patterns of neural activity and connectivity (Penagarikano et al. 2011; Wohr et al. 2015), as fast-spiking PV interneurons target the proximal regions of pyramidal cells (Hu et al. 2014; Safari et al. 2017) and synchronize firing within pyramidal cell populations (Engel and Singer 2001; Bartos et al. 2007; Cardin et al. 2009; Sohal et al. 2009; Roux and Buzsaki 2015; Cardin 2018). This hypothesis has been recently confirmed by Marissal and colleagues, who found that DREADD-induced suppression of PV activity causes the disinhibition of pyramidal neurons and leads to mass desynchronization of spiking activity (Marissal et al. 2018). Congruent with these previous empirical findings and recent theoretical models (Lee et al. 2018), our data suggest that inhibiting PV neurons reduces the synchronization of neuronal ensembles as indicated by the reduction of FC, despite higher net cortical activity.
Interactions among different types of neurons within a microcircuit should be balanced for proper functioning. Lee and colleagues (Lee et al. 2018) have recently built a computational model of the sensory cortex microcircuit and demonstrated the importance of connectivity homeostasis between neuronal types (i.e., inhibitory and excitatory cells) for producing synchronized oscillatory behavior with maximum frequency tuning capacity. Any deviation from this optimal configuration, such as overexcitation of pyramidal neurons, breaks this synchrony and impairs the communication among neuronal ensembles, which at the macroscale may be perceived as a reduction in FC. In line with this theoretical framework, we observed a reduction in FC when DREADDs were used to activate pyramidal neurons. We replicated this principle finding across three independent cohorts, using either clozapine or CNO to activate the DREADDs (see Supplementary Discussion) and using either human synapsin (hSyn) or CAMKII as a promoter in the AAV constructs. While hSyn promoter is active in both excitatory and inhibitory neurons, CAMKII generates a clear bias toward gene expression in excitatory cortical neurons (up to 85%) even though it might also transfect a small number of inhibitory neurons. Importantly, our electrophysiological results confirmed that activating excitatory DREADDs in the wt-hSyn-hM3Dq mice caused a significant net increase of neuronal firing rate, and our CBF measurements confirmed that such an increase was anatomically specific and most pronounced within the targeted SSp. Activating the excitatory DREADDs caused overall a stronger reduction of FC in wt-hSyn-hM3Dq mice than in PVCre-hM4Di mice, which might result from a higher number of transfected neurons: mouse cortex contains 12% GABAergic neurons, of which approximately one-quarter are PV (Feldmeyer et al. 2018) resulting in a rather low absolute number of transfected neurons and smaller DREADD-induced effects (Grayson et al. 2016).
Our results also characterize a potential role of local E:I balance in shaping the BOLD dynamics of individual brain regions. Specifically, we showed that the chemogenetic manipulation of the cellular excitability causes specific changes of local BOLD features that we could distinguish from neurotypical BOLD dynamics with high accuracy (balanced accuracy >80%) and anatomical specificity, irrespective of whether we increased excitatory transmission (wt-hSyn-hM3Dq and wt-CAMK-hM3Dq cohort) or suppressed inhibition (PVCre-hSyn-hM4Di cohort). In a cohort of wt-CAMK-hM3Dq mice, we detected BOLD dynamics with a high accuracy in the contralateral left SSp; this could be due to DREADD-transfected transcallosal excitatory neurons. Moreover, high classification accuracy was also observed in the dynamics of the right SSp between the two excitatory DREADD cohorts (wt-hSyn-hM3Dq and wt-CAMK-hM3Dq). This difference can be attributed to the two promoters and their distinct transfection affinities toward excitatory and inhibitory neurons.
The E:I BOLD signature learned from acutely modulating cell activity could be detected in two independent cohorts of Fmr1y/− mice, a well-characterized mouse model of ASD. We chose Fmr1y/− mice for external validation because they are known to have deficits in local feedback inhibition due to reduced PV interneuron density (Selby et al. 2007; Cea-Del Rio and Huntsman 2014). This leads to increased excitability and decreased synchrony of pyramidal cell firing through gamma oscillations (Gibson et al. 2008; Wohr et al. 2015) as well as substantially reduced FC in corticocortical and corticostriatal circuits (Zerbi et al. 2018; Zerbi et al. 2019b). Interestingly, above chance-level detection accuracies of BOLD dynamics characterizing overexcitation in Fmr1y/− mice were found in regions which normally exhibit high PV cell densities (Selby et al. 2007) providing further indirect evidence that the specific features of abnormal BOLD dynamics are related to the depletion of PV neurons in this mouse model. Fmr1y/− mice exhibit depletion of PV interneurons resulting in chronic overexcitation of the cortex, while PV interneurons in our PVCre-hSyn-hM4Di mice were only acutely manipulated. Despite these differences between the two models, we observed high classification accuracies in distinguishing Fmr1y/− mice from a statistical model trained on the PVCre DREADD manipulation. This correspondence strongly suggests that both acute and chronic overexcitation caused by PV depletion or inhibition share a common BOLD dynamical signature.
Our findings demonstrate that a chemogenetically inspired and experimentally validated classifier can be used as a computational sensor which uses BOLD dynamics to infer whether a specific brain area exhibits increased microcircuit E:I, for example, due to abnormal PV neuron activity.
It is unclear whether this discovery is generalizable to the entire cortex since changes in microstructural properties such as cytoarchitecture, E:I, size, density, and laminar distribution of distinct cell types vary continuously through mouse cortex (Kim Y et al. 2017b; Fulcher et al. 2019). These variations in spatial gradients shape functional specialization along the cortex and influence the intrinsic time scale that can be measured in BOLD dynamics in different cortical areas (Sethi et al. 2017). Nevertheless, we show above chance-level detection accuracy in the prefrontal cortex of Fmr1y/− mice that are also attributed to an increased E:I balance from previous studies (Paul et al. 2013). These results provide a new framework for mechanistically linking microscale circuit dynamics to macroscopic fMRI-based brain activity at rest. Thus far, this approach is unique because it uses causal microcircuit alterations to train a classifier that can detect similar alterations from distinct datasets obtained from large-scale imaging techniques. Our framework goes beyond describing long-range connectional aberrations caused by neurodevelopmental disorders and aims at providing deeper insights into how distinct microcircuit dynamics relate to large-scale, noninvasive imaging measurements which can be obtained across different species, including in human patients. Better linking macroscopic alteration to the underlying neurobiology is a prerequisite for advancing neuroimaging as a tool in the context of clinical decision-making.
Funding
ETH Research (grant ETH-38 16-2 to N.W.). ETH Career Seed (grant SEED-42 54 16-1 to V.Z.); SNSF AMBIZIONE (PZ00P3_173984/1 to V.Z.); European Research Council (ERC Advanced) (grant BRAINCOMPATH, project 670 757 to F.H.).
Notes
We thank the team of the EPIC animal facility for providing animal care. We thank Jean-Charles Paterna from the Viral Vector Facility (VVF) of the Neuroscience Center Zurich, a joint competence center of ETH Zurich and University of Zurich for producing viral vectors and viral vector plasmids. We thank Amalia Floriou Servou and Mattia Privitera for their useful feedback regarding immunohistochemistry.
Conflict of Interest: None declared.
Author Contributions
M.M. performed viral injections, conducted the fMRI experiments, analyzed the data, performed immunohistochemistry, and wrote the paper. B.D.F. performed machine-learning analysis and wrote the paper. C.L. conducted electrophysiology experiments, analyzed the data, and wrote the paper (results, method). F.H. supervised electrophysiology and provided feedback on the study. M.R. supervised fMRI experiments and provided feedback on the study. V.Z and N.W. designed the study, supervised fMRI experiments, analyzed data, and wrote the paper.
Supplementary Material
References
- Ajram LA, Horder J, Mendez MA, Galanopoulos A, Brennan LP, Wichers RH, Robertson DM, Murphy CM, Zinkstok J, Ivin G et al. 2017. Shifting brain inhibitory balance and connectivity of the prefrontal cortex of adults with autism spectrum disorder. Transl Psychiatry. 7:e1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoine MW, Langberg T, Schnepel P, Feldman DE. 2019. Increased excitation-inhibition ratio stabilizes synapse and circuit excitability in four autism mouse models. Neuron. 101(648–661):e644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartos M, Vida I, Jonas P. 2007. Synaptic mechanisms of synchronized gamma oscillations in inhibitory interneuron networks. Nat Rev Neurosci. 8:45–56. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: A practical and powerful Approach to multiple testing. J R Stat Soc B Methodol. 57(1):289–300. [Google Scholar]
- Bertero A, Liska A, Pagani M, Parolisi R, Masferrer ME, Gritti M, Pedrazzoli M, Galbusera A, Sarica A, Cerasa A et al. 2018. Autism-associated 16p11.2 microdeletion impairs prefrontal functional connectivity in mouse and human. Brain. 141:2055–2065. [DOI] [PubMed] [Google Scholar]
- Bulow P, Murphy TJ, Bassell GJ, Wenner P. 2019. Homeostatic intrinsic plasticity is functionally altered in Fmr1 KO cortical neurons. Cell Rep. 26(1378–1388):e1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardin JA. 2018. Inhibitory interneurons regulate temporal precision and correlations in cortical circuits. Trends Neurosci. 41:689–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardin JA, Carlen M, Meletis K, Knoblich U, Zhang F, Deisseroth K, Tsai LH, Moore CI. 2009. Driving fast-spiking cells induces gamma rhythm and controls sensory responses. Nature. 459:663–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cea-Del Rio CA, Huntsman MM. 2014. The contribution of inhibitory interneurons to circuit dysfunction in fragile X syndrome. Front Cell Neurosci. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JJ, Wieckowska M, Meyer E, Pike GB. 2008. Cerebral blood flow measurement using fMRI and PET: a cross-validation study. Int J Biomed Imaging. 2008:516359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contractor A, Klyachko VA, Portera-Cailliau C. 2015. Altered neuronal and circuit excitability in fragile X syndrome. Neuron. 87:699–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel AK, Singer W. 2001. Temporal binding and the neural correlates of sensory awareness. Trends Cogn Sci. 5. [DOI] [PubMed] [Google Scholar]
- Feldmeyer D, Qi G, Emmenegger V, Staiger JF. 2018. Inhibitory interneurons and their circuit motifs in the many layers of the barrel cortex. Neuroscience. 368:132–151. [DOI] [PubMed] [Google Scholar]
- Fox MD, Raichle ME. 2007. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat Rev Neurosci. 8:700–711. [DOI] [PubMed] [Google Scholar]
- Fulcher BD, Jones NS. 2017. Hctsa: A computational framework for automated time-series Phenotyping using massive feature extraction. Cell Syst. 5(527–531):e523. [DOI] [PubMed] [Google Scholar]
- Fulcher BD, Little MA, Jones NS. 2013. Highly comparative time-series analysis: the empirical structure of time series and their methods. J R Soc Interface. 10(83):20130048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcher BD, Murray JD, Zerbi V, Wang XJ. 2019. Multimodal gradients across mouse cortex. Proc Natl Acad Sci USA. 116(10):4689–4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Penzes P. 2016. Common mechanisms of excitatory and inhibitory imbalance in schizophrenia and autism Spectrum disorders. Curr Mol Med. 15(2):146–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson JR, Bartley AF, Hays SA, Huber KM. 2008. Imbalance of neocortical excitation and inhibition and altered UP states reflect network hyperexcitability in the mouse model of fragile X syndrome. J Neurophysiol. 100:2615–2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandjean J, Schroeter A, Batata I, Rudin M. 2014. Optimization of anesthesia protocol for resting-state fMRI in mice based on differential effects of anesthetics on functional connectivity patterns. Neuroimage. 102(Pt 2):838–847. [DOI] [PubMed] [Google Scholar]
- Grandjean J, Zerbi V, Balsters JH, Wenderoth N, Rudin M. 2017. Structural basis of large-scale functional connectivity in the mouse. J Neurosci. 37(34):8092–8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson DS, Bliss-Moreau E, Machado CJ, Bennett J, Shen K, Grant KA, Fair DA, Amaral DG. 2016. The rhesus monkey connectome predicts disrupted functional networks resulting from Pharmacogenetic inactivation of the amygdala. Neuron. 91:453–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffanti L, Salimi-Khorshidi G, Beckmann CF, Auerbach EJ, Douaud G, Sexton CE, Zsoldos E, Ebmeier KP, Filippini N, Mackay CE et al. 2014. ICA-based artefact removal and accelerated fMRI acquisition for improved resting state network imaging. Neuroimage. 95:232–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberl MG, Zerbi V, Veltien A, Ginger M, Heerschap A, Frick A. 2015. Structural-functional connectivity deficits of neocortical circuits in the Fmr1−/y mouse model of autism. Sci Adv. 1(10):e1500775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahamy A, Behrmann M, Malach R. 2015. The idiosyncratic brain: distortion of spontaneous connectivity patterns in autism spectrum disorder. Nat Neurosci. 18:302–309. [DOI] [PubMed] [Google Scholar]
- Hu H, Gan J, Jonas P. 2014. Interneurons. Fast-spiking, parvalbumin(+) GABAergic interneurons: from cellular design to microcircuit function. Science. 345:1255263. [DOI] [PubMed] [Google Scholar]
- Iwabuchi SJ, Krishnadas R, Li C, Auer DP, Radua J, Palaniyappan L. 2015. Localized connectivity in depression: a meta-analysis of resting state functional imaging studies. Neurosci Biobehav Rev. 51:77–86. [DOI] [PubMed] [Google Scholar]
- Juepter M, Weiller C. 1995. Review: does measurement of regional cerebral blood flow reflect synaptic activity?—implications for PET and fMRI. Neuroimage. 2:148–156. [DOI] [PubMed] [Google Scholar]
- Kazdoba TM, Leach PT, Silverman JL, Crawley JN. 2014. Modeling fragile X syndrome in the Fmr1 knockout mouse. Intractable Rare Dis Res. 3:118–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CK, Ye L, Jennings JH, Pichamoorthy N, Tang DD, Yoo AW, Ramakrishnan C, Deisseroth K. 2017a. Molecular and circuit-dynamical identification of top-down neural mechanisms for restraint of reward seeking. Cell. 170(1013–1027):e1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Yang GR, Pradhan K, Venkataraju KU, Bota M, Garcia Del Molino LC, Fitzgerald G, Ram K, He M, Levine JM et al. 2017b. Brain-wide maps reveal stereotyped cell-type-based cortical architecture and subcortical sexual dimorphism. Cell. 171(456–469):e422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraguljac NV, White DM, Hadley N, Hadley JA, Ver Hoef L, Davis E, Lahti AC. 2016. Aberrant hippocampal connectivity in unmedicated patients with schizophrenia and effects of antipsychotic medication: a longitudinal resting state functional MRI study. Schizophr Bull. 42:1046–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Shin D, Gross SP, Cho KH. 2018. Combined positive and negative feedback allows modulation of neuronal oscillation frequency during sensory processing. Cell Rep. 25(1548–1560):e1543. [DOI] [PubMed] [Google Scholar]
- Leithner C, Gertz K, Schrock H, Priller J, Prass K, Steinbrink J, Villringer A, Endres M, Lindauer U, Dirnagl U et al. 2008. A flow sensitive alternating inversion recovery (FAIR)-MRI protocol to measure hemispheric cerebral blood flow in a mouse stroke model. Exp Neurol. 210:118–127. [DOI] [PubMed] [Google Scholar]
- Marissal T, Salazar RF, Bertollini C, Mutel S, De Roo M, Rodriguez I, Muller D, Carleton A. 2018. Restoring wild-type-like CA1 network dynamics and behavior during adulthood in a mouse model of schizophrenia. Nat Neurosci. 21:1412–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelides M, Hurd YL. 2016. Chemogenetics: DREADDs, In: Pfaff D, Volkow N, editors. Neuroscience in the 21st Century. New York, NY: Springer, pp. 2847–2856. [Google Scholar]
- Minshew NJ, Keller TA. 2010. The nature of brain dysfunction in autism: functional brain imaging studies. Curr Opin Neurol. 23:124–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Musial TF, Marshall JJ, Zhu Y, Remmers CL, Xu J, Nicholson DA, Contractor A. 2017. Delayed maturation of fast-spiking interneurons is rectified by activation of the TrkB receptor in the mouse model of fragile X syndrome. J Neurosci. 37:11298–11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S, Wang Q, Lau C, Kuan L, Henry AM et al. 2014. A mesoscale connectome of the mouse brain. Nature. 508:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paakki JJ, Rahko J, Long X, Moilanen I, Tervonen O, Nikkinen J, Starck T, Remes J, Hurtig T, Haapsamo H et al. 2010. Alterations in regional homogeneity of resting-state brain activity in autism spectrum disorders. Brain Res. 1321:169–179. [DOI] [PubMed] [Google Scholar]
- Pan WJ, Thompson GJ, Magnuson ME, Jaeger D, Keilholz S. 2013. Infraslow LFP correlates to resting-state fMRI BOLD signals. Neuroimage. 74:288–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul K, Venkitaramani DV, Cox CL. 2013. Dampened dopamine-mediated neuromodulation in prefrontal cortex of fragile X mice. J Physiol. 591:1133–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penagarikano O, Abrahams BS, Herman EI, Winden KD, Gdalyahu A, Dong H, Sonnenblick LI, Gruver R, Almajano J, Bragin A et al. 2011. Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell. 147:235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogan SC, Roth BL. 2011. Remote control of neuronal signaling. Pharmacol Rev. 63:291–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routh BN, Rathour RK, Baumgardner ME, Kalmbach BE, Johnston D, Brager DH. 2017. Increased transient Na(+) conductance and action potential output in layer 2/3 prefrontal cortex neurons of the fmr1(−/y) mouse. J Physiol. 595:4431–4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux L, Buzsaki G. 2015. Tasks for inhibitory interneurons in intact brain circuits. Neuropharmacology. 88:10–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein JLR, Merzenich MM. 2003. Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav. 255–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safari MS, Mirnajafi-Zadeh J, Hioki H, Tsumoto T. 2017. Parvalbumin-expressing interneurons can act solo while somatostatin-expressing interneurons act in chorus in most cases on cortical pyramidal cells. Sci Rep. 7:12764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby L, Zhang C, Sun QQ. 2007. Major defects in neocortical GABAergic inhibitory circuits in mice lacking the fragile X mental retardation protein. Neurosci Lett. 412:227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi SS, Zerbi V, Wenderoth N, Fornito A, Fulcher BD. 2017. Structural connectome topology relates to regional BOLD signal dynamics in the mouse brain. Chaos. 27:047405. [DOI] [PubMed] [Google Scholar]
- Shofty B, Bergmann E, Zur G, Asleh J, Bosak N, Kavushansky A, Castellanos FX, Ben-Sira L, Packer RJ, Vezina GL et al. 2019. Autism-associated Nf1 deficiency disrupts corticocortical and corticostriatal functional connectivity in human and mouse. Neurobiol Dis. 130:104479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JL, Yang M, Lord C, Crawley JN. 2010. Behavioural phenotyping assays for mouse models of autism. Nat Rev Neurosci. 11:490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smucny J, Wylie KP, Tregellas JR. 2014. Functional magnetic resonance imaging of intrinsic brain networks for translational drug discovery. Trends Pharmacol Sci. 35:397–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal VS, Zhang F, Yizhar O, Deisseroth K. 2009. Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature. 459:698–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand AD, Aragaki AK, Baquet ZC, Hodges A, Cunningham P, Holmans P, Jones KR, Jones L, Kooperberg C, Olson JM. 2007. Conservation of regional gene expression in mouse and human brain. PLoS Genet. 3:e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlhaas PJ, Singer W. 2006. Neural synchrony in brain disorders: relevance for cognitive dysfunctions and pathophysiology. Neuron. 52:155–168. [DOI] [PubMed] [Google Scholar]
- Vogt D, Cho KKA, Shelton SM, Paul A, Huang ZJ, Sohal VS, Rubenstein JLR. 2017. Mouse Cntnap2 and human CNTNAP2 ASD alleles cell autonomously regulate PV+ cortical interneurons. Cereb Cortex. 28(11):3868–3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zhan Y, Zhang Y, Lyu L, Lyu H, Wang G, Wu R, Zhao J, Guo W. 2018. Abnormal long- and short-range functional connectivity in adolescent-onset schizophrenia patients: a resting-state fMRI study. Prog Neuropsychopharmacol Biol Psychiatry. 81:445–451. [DOI] [PubMed] [Google Scholar]
- Wohr M, Orduz D, Gregory P, Moreno H, Khan U, Vorckel KJ, Wolfer DP, Welzl H, Gall D, Schiffmann SN et al. 2015. Lack of parvalbumin in mice leads to behavioral deficits relevant to all human autism core symptoms and related neural morphofunctional abnormalities. Transl Psychiatry. 5:e525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbi V, Floriou-Servou A, Markicevic M, Vermeiren Y, Sturman O, Privitera M, von Ziegler L, Ferrari KD, Weber B, De Deyn PP et al. 2019a. Rapid reconfiguration of the functional connectome after chemogenetic locus coeruleus activation. Neuron. 103(702–718):e705. [DOI] [PubMed] [Google Scholar]
- Zerbi V, Grandjean J, Rudin M, Wenderoth N. 2015. Mapping the mouse brain with rs-fMRI: an optimized pipeline for functional network identification. Neuroimage. 123:11–21. [DOI] [PubMed] [Google Scholar]
- Zerbi V, Ielacqua GD, Markicevic M, Haberl MG, Ellisman MHA, Frick A, Rudin M, Wenderoth N. 2018. Dysfunctional autism risk genes cause circuit-specific connectivity deficits with distinct developmental trajectories. Cereb Cortex. 28:2495–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbi V, Jansen D, Wiesmann M, Fang X, Broersen LM, Veltien A, Heerschap A, Kiliaan AJ. 2014. Multinutrient diets improve cerebral perfusion and neuroprotection in a murine model of Alzheimer's disease. Neurobiol Aging. 35:600–613. [DOI] [PubMed] [Google Scholar]
- Zerbi V, Markicevic M, Gasparini F, Schroeter A, Rudin M, Wenderoth N. 2019b. Inhibiting mGluR5 activity by AFQ056/Mavoglurant rescues circuit-specific functional connectivity in Fmr1 knockout mice. Neuroimage. 191:392–402. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


