ABSTRACT
As organisms are constantly exposed to the damaging effects of oxidative stress through both environmental exposure and internal metabolic processes, they have evolved a variety of mechanisms to cope with this stress. One such mechanism is the highly conserved p38 MAPK (p38K) pathway, which is known to be post-translationally activated in response to oxidative stress, resulting in the activation of downstream antioxidant targets. However, little is known about the role of p38K transcriptional regulation in response to oxidative stress. Therefore, we analyzed the p38K gene family across the genus Drosophila to identify conserved regulatory elements. We found that oxidative stress exposure results in increased p38K protein levels in multiple Drosophila species and is associated with increased oxidative stress resistance. We also found that the p38Kb genomic locus includes conserved AP-1 and lola-PT transcription factor consensus binding sites. Accordingly, over-expression of these transcription factors in D. melanogaster is sufficient to induce transcription of p38Kb and enhances resistance to oxidative stress. We further found that the presence of a putative lola-PT binding site in the p38Kb locus of a given species is predictive of the species' survival in response to oxidative stress. Through our comparative genomics approach, we have identified biologically relevant putative transcription factor binding sites that regulate the expression of p38Kb and are associated with resistance to oxidative stress. These findings reveal a novel mode of regulation for p38K genes and suggest that transcription may play as important a role in p38K-mediated stress responses as post-translational modifications.
KEY WORDS: Gene regulation, Comparative genomics, p38 MAPK
Summary: A comparative genomics approach reveals conserved transcription factor consensus sites in the p38 MAPK gene family that regulate stress responses in Drosophila.
INTRODUCTION
Organisms encounter oxidative stress, which damages a variety of cellular structures, both through external environmental exposure and from the internal release of oxygen radicals during ATP synthesis. Thus, organisms have evolved a variety of mechanisms to counteract it. One such mechanism is the highly conserved p38 mitogen activated protein kinase (p38K) pathway, which consists of a phosphorylation cascade of upstream kinases that leads to the dual phosphorylation of the p38K protein on a TXY motif. This activated p38K can phosphorylate target proteins in the cytoplasm and can also translocate to the nucleus, where it can phosphorylate downstream transcription factors to induce the expression of antioxidant genes (Krishna and Narang, 2008; McCubrey et al., 2006; Peti and Page, 2013), allowing the cell to appropriately respond to the oxidative stress.
Though much is known about the role of post-translational regulation in the p38K-mediated oxidative stress response, the transcriptional regulation of p38K during stress conditions is not well understood. In order to address this question, we utilized a comparative genomics approach using the genus Drosophila to identify conserved putative transcription factor binding sites that play a role in mediating p38K gene expression during oxidative stress.
In the fruit fly Drosophila melanogaster, there are three p38K genes: p38Ka, p38Kb and p38Kc. Flies are viable following the loss of any one of these p38K genes; however, p38Ka–p38Kb double knockouts are lethal (Vrailas-Mortimer et al., 2011, 2012; Chen et al., 2010; Shinzawa et al., 2009; Cully et al., 2010; Park et al., 2009; Craig et al., 2004), suggesting that these genes are at least partially redundant. Both p38Ka and p38Kb play a role in resistance to heat shock, starvation and oxidative stress (Vrailas-Mortimer et al., 2011; Craig et al., 2004; Inoue et al., 2001). Furthermore, both of these genes play a role in the immune response, with p38Ka regulating the gut DUOX system (Ha et al., 2009), while p38Kb is involved in bacterial phagocytosis (Chen et al., 2010) and antiviral immunity (West and Silverman, 2018). In addition, these genes have unique roles in the body. p38Ka plays a role in cardiac function (Na et al., 2013), while p38Kb is a regulator of lifespan, locomotor functions and circadian rhythms (Vrailas-Mortimer et al., 2011; Vrailas-Mortimer et al., 2014). Interestingly, p38Kc has distinct functions in the cell in regulating innate immunity through the Ddc pathway (Davis et al., 2008) and intestinal oxidative stress and lipid homeostasis (Chakrabarti et al., 2014). The three p38K genes have been hypothesized to have arisen through gene duplication, and the differences in function suggest that evolutionary pressures may be playing a role in the divergence of these genes. As these genes play differing roles in the cell, they may have distinct genetic regulatory elements that control the expression of p38K during different developmental or homeostatic cellular responses.
In order to identify these potential regulatory elements, we utilized the sequenced genomes of 22 species with known evolutionary relationships within the Drosophila genus to identify conserved transcription factor binding consensus motifs upstream of the p38K family genes. We found that diverse species of Drosophila express high levels of p38K protein during oxidative stress, and that there are conserved Activator Protein-1 (AP-1) and longitudinals lacking (lola)-PT putative transcription factor binding sites that regulate p38Kb expression under normal and oxidative stress conditions. The levels of p38Kb expression and the presence of these consensus motifs are also predictors of resistance to oxidative stress. These data suggest that transcriptional regulation of p38Kb may play as important a role in oxidative stress resistance in addition to the better-described post-translational modification, and may boost the effectiveness of post-translational regulation of p38Kb in regulating stress responses.
MATERIALS AND METHODS
Sequences
Sequences corresponding to mitogen activated protein kinase (MAPK) family genes were downloaded from FlyBase (flybase.org) and NCBI. Accession numbers are given for the DNA sequences and protein sequences used in Table S1.
Phylogenetic trees
Phylogenetic trees for the MAPK family were constructed using amino acid sequences. Sequences were aligned using the ClustalW algorithm with the Gonnet Protein Weight Matrix implemented in MEGA (v.5.2.2; Tamura et al., 2011), and all positions containing gaps and missing data were eliminated. Initial trees were obtained using the neighbor-joining method. The phylogenetic relationships among the sequences were inferred using the maximum likelihood method based on the Jones–Taylor–Thornton model (Jones et al., 1992), and the trees with the highest log-likelihood are shown. Each tree was tested with 1000 bootstrap replications (Felsenstein, 1985), and the given values are the percentage of trees in which the associated sequences clustered together.
Phylogenetic trees for the Drosophila p38K gene family and ITS2 (the second internal transcribed spacer) were constructed using nucleic acid sequences. Sequences were aligned using the ClustalW algorithm with the IUB DNA Weight Matrix implemented in MEGA, and all positions containing gaps and missing data were eliminated. Pairwise distances were estimated using the maximum composite likelihood approach and initial trees were obtained using the neighbor-joining method. The phylogenetic relationships among the sequences were inferred using the maximum likelihood method based on the Tamura–Nei model (Tamura and Nei, 1993), and the trees with the highest likelihood are shown. Each tree was tested with 1000 bootstrap replications (Felsenstein, 1985), and the given values are the percentage of trees in which the associated sequences clustered together.
The output from these analyses was used to draw trees using FigTree v.1.4.
Chromosome synteny
The regions of interest from the Muller B and E elements across Drosophila species were accessed through GBrowse on FlyBase (https://flybase.org/). Synteny was determined manually.
Genetic conservation analysis
Pairwise dN and dS values [the rate of substitutions at non-synonomous sites (dN) and synonomous sites (dS)] for p38K family genes across the Drosophila species were computed in MEGA (v.5.2.2) using the Nei–Gojobori model. Values were plotted in R.
Drosophila stocks
Drosophila melanogaster OregonR, y[1] w[*]; P{w[+mC]=UAS-lola.4.7}3 (#28828) and P{w[+mC]=UAS-lola.4.7}1, w[*] (#28831) were obtained from the Bloomington Drosophila Stock Center, Indiana University. w−;;MHC-GAL4 was used as reported in Vrailas-Mortimer et al. (2011) and w−;UAS-Jun; UAS-Fos is described in Franciscovich et al. (2008). Drosophila pseudoobscura (strain 14011-0121.94), Drosophila virilis (strain 15010-1051.87), Drosophila simulans (strain 14021-0251.195), Drosophila yakuba (strain 14021-0261.00), Drosophila mauritiana (strain 14021-0241.151) and Drosophila ananassae (strain 14024-0371.13) were obtained from the University of California San Diego Drosophila Species Stock Center, which is now the National Drosophila Species Stock Center, Cornell University.
Predicting transcription factor binding sites
Putative transcription factor binding sites were predicted using iMotifs (Piipari et al., 2010). The TomTom Motif Comparison Tool v.4.9.1 (MEME web server; Gupta et al., 2007) compared the resulting predicted sites with all known experimentally validated Drosophila transcription factor binding sites. The size of the letter corresponds to the program's confidence in that nucleotide for that specific position.
qRT-PCR
Drosophila melanogaster lines were reared at 25°C on a 12 h:12 h light:dark cycle on standard fly food media. Virgin female flies were collected at eclosion and fed standard fly food media and aged for 1 week. Flies were then transferred to food mixed with 20 mmol l−1 paraquat (Sigma). Food was changed every 3–4 days. Surviving flies were collected when 30% of controls had died. Thoraxes from 6 flies were dissected and pooled. RNA was extracted using Trizol (Thermo Fisher Scientific) and reverse transcribed using SuperScript IV (Thermo Fisher Scientific). qRT-PCR was performed using SYBR green (Thermo Fisher Scientific). Primers designed to amplify p38Kb or the housekeeping gene Arp88 were utilized. To compare p38Kb expression across genotypes, the ΔCt value for each replicate was calculated by subtracting the Ct value of Arp88 from the Ct value of p38Kb. We tested the hypothesis that AP-1 and lola-PT transcription factor over-expression would increase the amount of p38Kb transcript using one-tailed Student's t and Wilcoxon signed-rank tests (implemented in R).
Lifespan
AP-1 and lola-PT flies and outcrossed controls were reared at 25°C on a 12 h:12 h light:dark cycle on standard fly food media. Flies were collected at eclosion and lifespan was assayed daily. Food was changed every 3–4 days. Differences in survival were assayed using the log-rank test. Pairwise comparisons between samples were made using the pairwise log-rank test and P-values were adjusted using the false discovery rate. These tests were performed using the survival package in R.
Paraquat survival
We found that the efficacy of the paraquat was batch dependent (Table S2). Therefore, all experiments were performed with biological replicates and controls using the same batch of paraquat. Virgin females from each species were collected at eclosion and transferred to fly food media mixed with 20 mmol l−1 paraquat (Sigma). Drosophila simulans and D. pseudoobscura were reared at 21°C on a 12 h:12 h light:dark cycle. Drosophila melanogaster, D. virilis, D. yakuba, D. ananassae and D. mauritiana were reared at 25°C on a 12 h:12 h light:dark cycle on standard fly food media. Food was changed every 3–4 days. Survival was scored daily.
AP-1 and lola-PT flies and outcrossed controls were reared at 25°C in a 12 h:12 h light:dark cycle on standard fly food media. Virgin female flies were collected at eclosion and aged 1 week and then transferred to standard fly food media mixed with either 10 mmol l−1 or 20 mmol l−1 paraquat. Food was changed every 3–4 days. Survival was scored daily. Differences in survival were assayed using the Log-Rank test. Pairwise comparisons between samples were made using the Pairwise Log-Rank test and P-values were adjusted using the false discovery rate. These tests were performed using the survival package in R.
Immunoblots
Three thoraxes from adult female flies were homogenized in 1× Laemmli buffer. Immunoblots were performed as described in Vrailas-Mortimer et al. (2011). Membranes were probed with mouse anti-total p38 MAPK (1:1000, Cell Signaling Technologies, 9212), rabbit phosphor-p38 MAPK (1:1000, Cell Signaling Technologies, 9211) and rabbit anti-alpha tubulin (1:5000, Cell Signaling Technologies, 2125S) and either goat anti-mouse HRP (1:20,000, Jackson Labs, 115-035-003) or goat anti-rabbit HRP (1:40,000, Life Technologies, G-21234). The p38K antibodies have been previously validated in D. melanogaster (Vrailas-Mortimer et al., 2011). Membranes were then developed using SuperSignal West Femto kit (Thermo Fisher Scientific) or Pierce ECL (Thermo Fisher Scientific) and exposed on autoradiography film. All immunoblots were performed at least in triplicate. Densitometry was performed using Adobe Photoshop. For each replicate, the densitometry value of each p38K band was normalized to its corresponding α-tubulin control. These normalized values were then fitted to linear regression models with each blot treated as a separate blocking factor and compared using ANOVA (implemented in R).
Interaction analysis
To test for a correlation between body size and survival on paraquat, we calculated the Pearson's product-moment correlation between body size of female flies (approximated by thorax length; Markow and O'Grady, 2007) and the median survival following paraquat exposure. In an attempt to uncouple the phenotypic correlation from phylogenetic relationships, we used the phylogenetically independent contrasts approach (as implemented in the caper R package) to calculate the Spearman rho correlation between body size and median survival contrasts. To test for a correlation between p38K protein levels and survival on paraquat, we calculated the Spearman rho correlation between phylogenetically independent contrasts for the change in p38K protein levels in flies fed paraquat and the median survival following paraquat exposure. Analyses were performed in R and correlation data were plotted using ggplot2 in R.
To test the relationship between p38K protein levels, paraquat exposure and the presence of predicted transcription factor binding sites, we used a model in which paraquat exposure and site presence were independent variables and p38K level was the dependent variable. These data were compared using the analysis of variance of aligned rank transformed data method (implemented using the ARTool package in R). Interaction plots were generated using ggplot2 in R.
To test the relationship between the presence of predicted transcription factor binding sites and survival on paraquat, the data were compared using the analysis of variance of aligned rank transformed data method (ARTool package in R). Data were plotted using ggplot2 in R.
RESULTS
Analysis of MAPK family genes across taxa
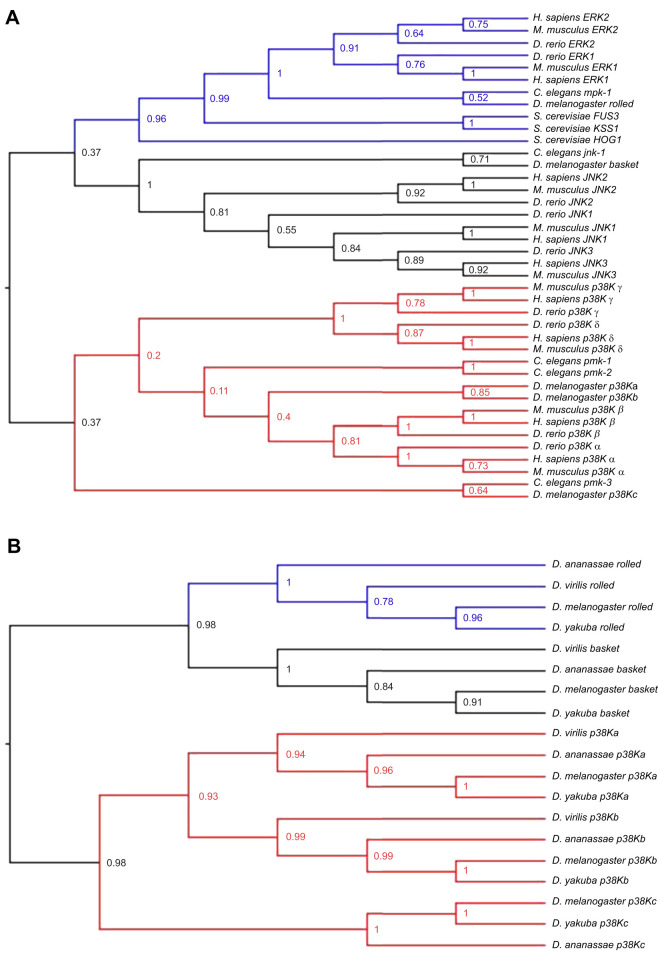
As p38K is closely related to the other MAPK family members extracellular regulated kinase (ERK) and c-Jun N-terminal kinase (JNK), we first analyzed the relationship between the three MAPK families ERK, JNK and p38K across taxa (Table S3) as well as within a subset of the sequenced Drosophila species that are representative of the genus (Table S3). While humans have multiple ERK and JNK family members, D. melanogaster only has one ortholog for each of these families (rolled and basket, respectively). In addition, vertebrates have four p38K genes while D. melanogaster have three p38K orthologs (p38Ka, p38Kb and p38Kc). We found that all three MAPK families emerged very early in animal evolution before the split between vertebrates and invertebrates (Fig. 1A). In addition, the ERK and JNK families evolved from a more recent ancestral gene, whereas the p38K family diverged earlier (Fig. 1A). Within the p38K family, the three D. melanogaster p38K genes are not orthologous to the distinct vertebrate p38K genes, suggesting that the family members have evolved since the split between vertebrates and invertebrates (Fig. 1A). Analysis of a subset of Drosophila species revealed that these relationships are also conserved across the genus, with the species having single orthologs of ERK and JNK and multiple members of the p38K family (Fig. 1B). Furthermore, in D. melanogaster, the p38Kb gene resides on the second chromosome, while both p38Ka and p38Kc reside in close proximity on the third chromosome, suggesting that p38Ka and p38Kc may be the result of a tandem gene duplication.
Fig. 1.
Phylogenetic analysis of the MAPK family. (A) The evolutionary relationship between the ERK1/2 (blue), JNK (black) and p38 (red) MAPK genes from Homo sapiens, Mus musculus, Danio rerio, Drosophila melanogaster, Caenorhabditis elegans and Saccharomyces cerevisiae. Note that HOG1 is the Saccharomyces cerevisiae homolog of both JNK and p38. (B) The evolutionary relationship between the ERK1/2 (blue), JNK (black) and p38 (red) MAPK genes from representative species in the genus Drosophila. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches.
Comparison of p38K genes in Drosophila
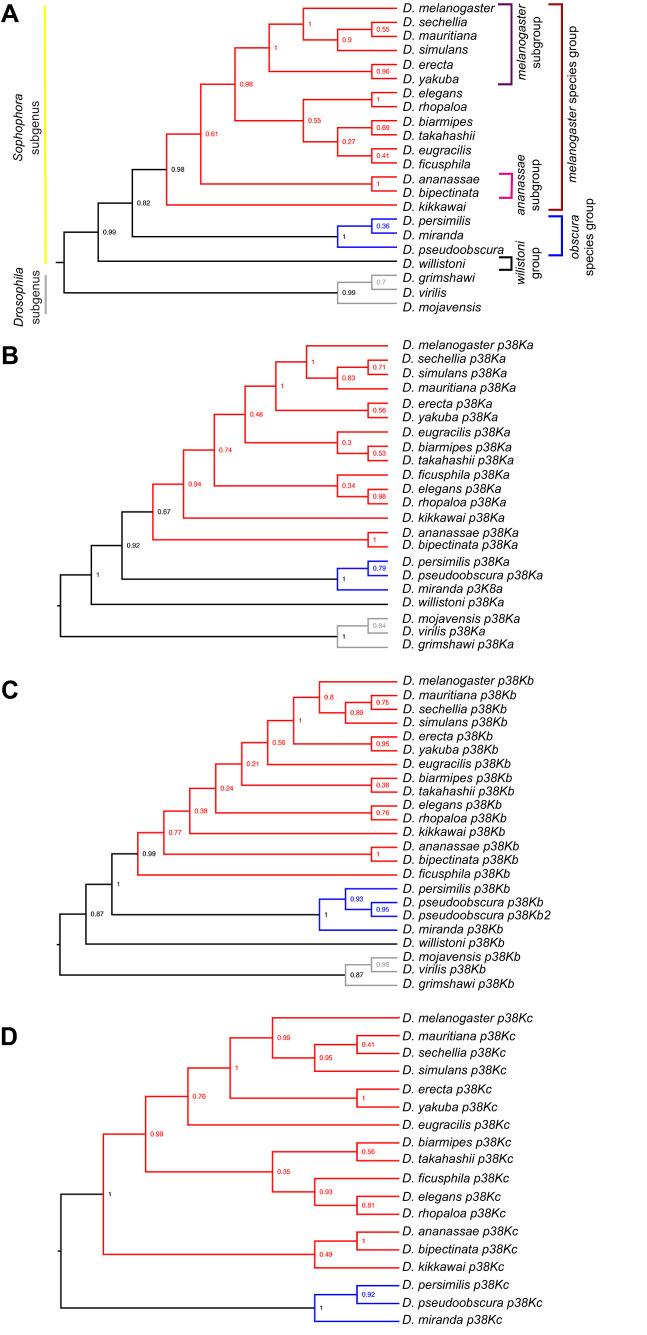
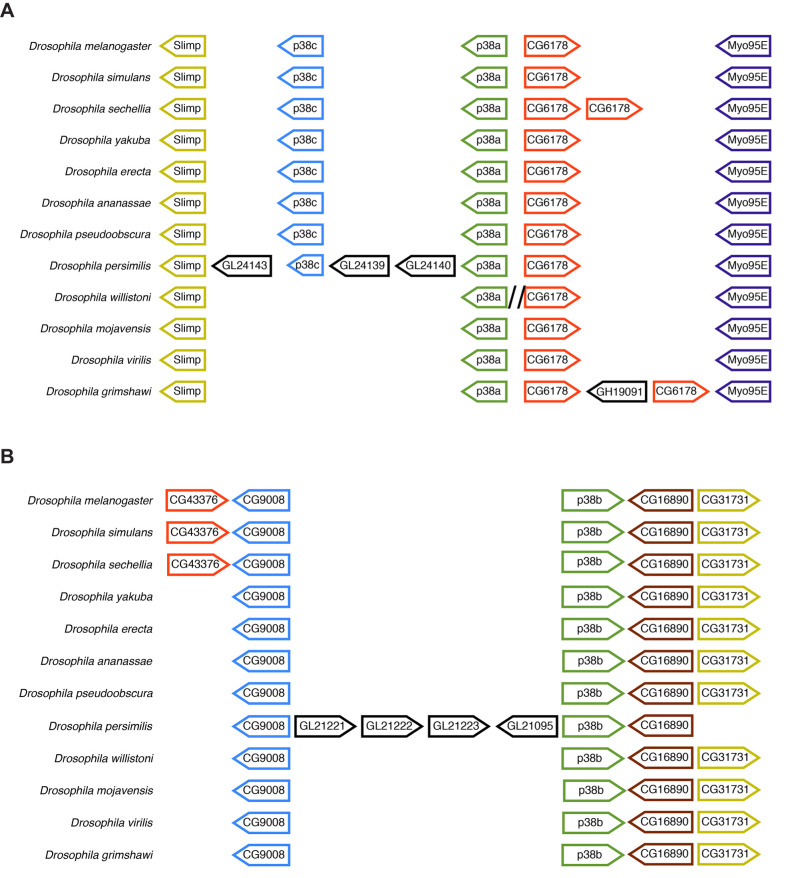
In order to identify conserved genetic regulatory elements, we next compared the genomic locus of the p38K genes across 22 sequenced Drosophila species. We found that across species, each p38K gene forms a distinct cluster (Fig. S1). All three of the p38K genes mostly cluster along taxonomic lines as determined by molecular phylogeny based on ITS2 (Fig. 2A). Interestingly, unlike the other Drosophila species, D. pseudoobscura has two p38Kb genes (Fig. 2C), suggesting a recent duplication of p38Kb in this species. As not all of the 22 sequenced Drosophila species have fully annotated genomes, we focused on 12 species that are fully sequenced and annotated to analyze the genomic architecture of the p38K loci. We found that the synteny of the p38Ka and p38Kc genes is highly conserved (Fig. 3A), with their proximity suggesting that p38Kc most likely arose from a tandem duplication event of p38Ka, after the divergence of the willistoni group (Fig. S1; Fig. 3A). Furthermore, in D. persimilis, p38Kc is truncated as compared with the p38Kc gene in other Drosophila species leading to a shortened p38Kc protein. Interestingly, D. persimilis also has three additional genes (GL24143, GL24139, GL24140) that have appeared near the p38Kc locus, disrupting its close proximity to p38Ka (Fig. 3A). Both D. sechellia and D. grimshawi have independent duplications of a neighboring gene (CG6178), and D. grimshawi also has a new gene (GH19091) inserted into this region.
Fig. 2.
Phylogenetic analysis of each p38K gene in Drosophila. The evolutionary relationships of the (A) second internal transcribed spacer (ITS2), (B) p38Ka, (C) p38Kb and (D) p38Kc genes in 22 sequenced species of Drosophila. Red denotes the melanogaster species group, blue denotes the obscura group, black denotes the willistoni group, and gray represents the Drosophila subgenus. Note that D. pseudoobscura is the only species with two copies of the p38Kb gene. p38Kc is only found in species that arose after the split of the obscura group from D. willistoni. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches.
Fig. 3.
Synteny of the p38K genes in Drosophila. (A) p38Ka and p38Kc lie in close proximity to each other. Drosophila persimilis has additional genes present at this locus as well as a truncated p38Kc gene. Drosophila sechellia has a duplication of CG6178 as does D. grimshawi, which also has an additional gene inserted into the locus. Backslashes indicate additional inserted DNA with no predicted genes. (B) p38Kb synteny is also highly conserved. Note the appearance of CG43376 within the melanogaster subgroup. Drosophila persimilis has an insertion of four genes within this locus and is lacking the CG31731 gene. Drosophila pseudoobscura has two p38Kb genes, though the second p38Kb gene is found on a different chromosome.
The synteny of the p38Kb locus is also highly conserved (Fig. 3B). The melanogaster subgroup has an additional gene (CG43376) inserted into this region. Furthermore, much like the p38Ka and p38Kc locus, D. persimilis has an insertion of four genes (GL21221, GL21222, GL21223 and GL21095) as well as loss of the gene CG31731 in this region (Fig. 3B). Though D. pseudoobscura has two p38Kb genes (Fig. 2C), these genes reside on different chromosomes, with p38Kb on the second chromosome (Muller element E) and p38Kb 2 on the fourth chromosome (Muller element B).
The p38K genes are under purifying selection
As the p38K genes are conserved across Drosophila species, we investigated whether these genes show evidence of selection. We analyzed the dN/dS ratio for each of the p38K genes in 22 different species of Drosophila (Fig. 4). Both p38Ka and p38Kb are under strong purifying selection with average dN/dS ratios of 0.0741 and 0.0479, respectively (Fig. 4). This suggests that amino acid substitutions in the predicted protein sequences are not well tolerated in these genes and that they have evolved important functions that when disrupted lead to decreased fitness. Interestingly, the p38Ka and p38Kb proteins are also very similar to each other. For example, in D. melanogaster, the two proteins are 78.4% identical and 94.5% similar at the amino acid level. Therefore, the limited amino acid differences between the p38Ka and p38Kb proteins may be important for their independent functions.
Fig. 4.

The p38K genes are under purifying selection. Plot of the dN (substitutions at non-synonomous sites) and dS (substitutions at synonomous sites) values for the p38K family genes p38Ka, p38Kb and p38Kc. Each spot represents a pairwise comparison between two species.
We also found that p38Kc is under purifying selection but has a higher dN/dS ratio (dN/dS ratio of 0.3346; Fig. 4). These data suggest that p38Kc is less constrained than p38Ka and p38Kb, which allowed not only the diversity in amino acid sequences across species but also potentially the evolution of new functions. Furthermore, as p38Kc is under selective pressure, this suggests that these novel functions are also critical for the well-being of the organism.
Identification of conserved transcription factor consensus motifs for p38Ka and p38Kb
Though the D. melanogaster p38Ka and p38Kb genes are strikingly similar to each other and have some redundant functions (Vrailas-Mortimer et al., 2011; Chen et al., 2010; Shinzawa et al., 2009), these genes have also been shown to have some independent functions (Vrailas-Mortimer et al., 2011; Chen et al., 2010; Shinzawa et al., 2009; Cully et al., 2010; Park et al., 2009; Craig et al., 2004; Vrailas-Mortimer et al., 2014). We have also previously reported that the p38Kb protein is highly expressed in muscle and brain, whereas the p38Ka protein is expressed at lower levels in these tissues (Vrailas-Mortimer et al., 2011), which may contribute to the specific functions of each of these genes. Therefore, we analyzed 1 kb upstream of each gene for putative transcription factor binding sites in the 12 Drosophila species. We found that p38Ka has four consensus transcription factor binding sites (Fig. 5A; Table S4). Three of these motifs are mostly conserved across Drosophila species within the melanogaster group. The first site is a consensus homeobox binding site, which can be bound by members of the Hox family of transcription factors. The other two conserved binding sites are consensus sequences for two isoforms of the transcription factor lola: lola-PO and lola-PK. The final consensus binding site (motif 3) is for an unknown transcription factor and is found only in D. erecta.
Fig. 5.

Putative transcription factor binding sites upstream of p38Ka and p38Kb. Analysis of the 1 kb upstream region for (A) p38Ka and (B) p38Kb reveals conserved transcription factor motifs, including longitudinals lacking (lola) and activator protein-1 (AP-1), which are linked to MAPK signaling pathways and/or oxidative stress.
In comparison, p38Kb has five consensus transcription factor binding sites (Fig. 5B; Fig. S2A–F and Table S4) that are conserved across the melanogaster and obscura species groups. Similar to p38Ka, p38Kb has two consensus sites for isoforms of the lola transcription factor: lola-PT and lola-PO. p38Kb also has a widely conserved site which corresponds to the AP-1 transcription factor binding site. The final two predictive motifs represent binding sites for unknown transcription factors. Motif 4 is conserved within the obscura group and also in the more distantly related species D. takahashii. Motif 5 may be a binding site that has been repeatedly gained or lost over evolutionary history within the melanogaster species group. Interestingly, Motif 5 has been lost in D. melanogaster but retained in its closely related sister species D. simulans, D. mauritiana and D. sechellia. The putative lola-PO binding site, in contrast, is restricted to D. melanogaster and its sister species. Furthermore, we identified both AP-1 and lola-PT consensus binding sites in most of the species analyzed.
Both the p38Ka and p38Kb loci include putative binding sites for transcription factors that play a role in development (the Hox family and AP-1; Perkins et al., 1990; Riesgo-Escovar and Hafen, 1997; Bataillé et al., 2015; Enriquez et al., 2010). Though animals are viable following loss of either p38Ka or p38Kb, double-knockout animals are inviable (Vrailas-Mortimer et al., 2011; Chen et al., 2010; Shinzawa et al., 2009; Cully et al., 2010; Craig et al., 2004), suggesting that p38K signaling may play an important role in developmental processes much like in mammalian systems (Keren et al., 2006). In addition to its role in development, AP-1, a heterodimer of the transcription factors jun and fos, also plays a role in immune function (Kim et al., 2005; Tokusumi et al., 2009) and oxidative stress response (Milton et al., 2011). Interestingly, p38Kb has been shown to phosphorylate jun (Sano et al., 2005), suggesting that p38Kb expression may be regulated through a feedback loop.
lola is a key regulator of axon guidance (Giniger et al., 1994; Madden et al., 1999; Crowner et al., 2002). However, lola has also been shown to act in the ovary to regulate programmed cell death in both developmental and starvation conditions (Bass et al., 2007). Furthermore, the ModENCODE project demonstrated that lola is highly expressed in response to adult exposure to the oxidizing agent paraquat (Brown et al., 2014; Celniker et al., 2009), though the role of specific isoforms of lola in oxidative stress are not well characterized.
AP-1 and lola-PT regulate the expression of p38Kb in D. melanogaster
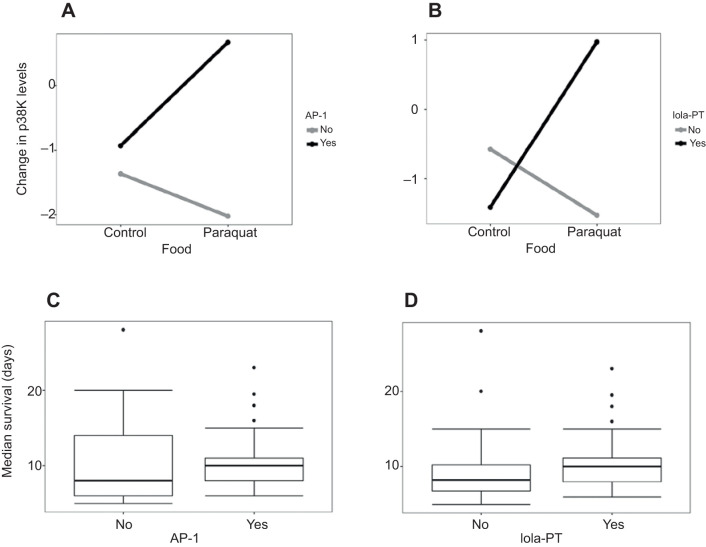
Oxidative stress is known to induce the phosphorylation and thus activation of p38K proteins in a variety of systems including D. melanogaster. However, the presence of both the AP-1 and lola isoform sites suggests that p38K transcription may also be regulated by oxidative stress. Therefore, we tested whether these conserved transcription factor consensus binding sites are important for the regulation of the p38Kb gene. We focused on the putative AP-1 and lola-PT binding sites as they are exclusive to p38Kb and have links to MAPK signaling pathways and/or oxidative stress. We over-expressed either AP-1 or lola-PT in the muscle using the MHC-GAL4 driver as p38Kb has been shown to act in this tissue to regulate viability and the oxidative stress response (Vrailas-Mortimer et al., 2011). We found that over-expression of AP-1 is able to induce expression of p38Kb under normal conditions, but not when the flies are exposed to the oxidizing agent paraquat (Fig. 6A,B). Over-expression of lola-PT induced p38Kb expression in both normal and oxidative stress conditions (Fig. 6C,D). These data suggest that both AP-1 and lola-PT are capable of mediating p38Kb transcription and that lola-PT may be playing a role in the oxidative stress response by promoting the transcription of p38Kb during stress conditions. As transgenic over-expression of p38Kb in the muscle leads to increased lifespan (Vrailas-Mortimer et al., 2011), we next tested the effect of AP-1 and lola-PT on lifespan. We found that over-expression of neither AP-1 nor lola-PT alters lifespan (Fig. S2G,H, Table S2; χ2=1.8, P=0.4 and χ2=5.5, P=0.06, respectively), suggesting that the levels of increased p38Kb transcription induced by AP-1 or lola-PT are not sufficient to extend lifespan. Over-expression of p38Kb is also protective against oxidative stress exposure (Vrailas-Mortimer et al., 2011). Therefore, we assessed the effects of AP-1 and lola-PT over-expression on resistance to paraquat. We found that over-expression of AP-1 leads to increased survival in response to both high (20 mmol l−1) and low (10 mmol l−1) paraquat exposure (Fig. 6E,G; Table S2). Interestingly, lola-PT over-expression also leads to increased survival in response to oxidative stress but only in response to low-dose paraquat exposure (Fig. 6F,H; Table S2). These data suggest that AP-1 and lola-PT are able to induce sufficient levels of p38Kb transcript to provide a readily available pool of p38K protein that can be activated and induce the oxidative stress response. However, lola-PT-mediated protection is limited in terms of the degree of oxidative stress exposure.
Fig. 6.
AP-1 and lola-PT mediate p38Kb transcription and oxidative stress resistance. (A,B) Levels of p38Kb transcription in response to over-expression of AP-1 under control conditions (A) and with exposure to 20 mmol l−1 paraquat (B). (C,D) Levels of p38Kb transcription in response to over-expression of lola-PT under control conditions (C) and with exposure to 20 mmol l−1 paraquat (D). Findings (means±s.e.m.) for A–D are based on 3 biological replicates for each genotype; each replicate consists of 6 individual flies. Significance was determined by one-tailed Student's t and Wilcoxon signed-rank tests. (E–H) Resistance to 20 mmol l−1 paraquat (E,F) or 10 mmol l−1 paraquat (G,H) of AP-1 (E,G) or lola-PT (F,H) over-expressing flies (red). Controls are outcrossed GAL4 (black) and transgene (gray) flies. In E and F the number of animals and replicates per genotype was as follows: MHC:AP-1 n=120, 8 replicates; w1118:AP-1 n=145, 10 replicates; MHC:lola-PT n=123, 8 replicates; w1118:lola-PT n=106, 7 replicates; and MHC:w1118 n=75, 5 replicates. In G and H, the number of animals and replicates per genotype was as follows: MHC:AP-1 n=90, 9 replicates; w1118:AP-1 n=66, 7 replicates; MHC:lola-PT n=98, 10 replicates; w1118:lola-PT n=68, 7 replicates; and MHC:w1118 n=75, 5 replicates. Differences in survival were assayed using the log-rank test.
Assessing the relationship between p38K and survival across species
To explore the evolution of the oxidative stress response across Drosophila, we tested the resistance of seven species to exposure to the oxidizing agent paraquat. We found significant differences in survival between the different species (Fig. 7A; Table S2). Interestingly, D. virilis, the largest species in our study (Markow and O'Grady, 2007), did not have the longest survival time on paraquat (Fig. 7A; Table S2). Accordingly, we found that body size and paraquat resistance are only weakly correlated between species (Fig. S2I; R=0.44, P=0.4529) or when considering phylogenetically independent contrasts (Fig. S2J; R=−0.4, P=0.6), suggesting that body size is not a major determinant of stress resistance.
Fig. 7.
Oxidative stress is correlated with p38K protein levels. (A) Kaplan–Meier plot of species survival during exposure to 20 mmol l−1 paraquat. The number of animals and replicates per genotype was as follows: D. melanogaster n=199, 11 replicates; D. mauritiana n=207, 10 replicates; D. ananassae n=212, 7 replicates; D. simulans n=211, 9 replicates; D. pseudoobscura n=200, 12 replicates; D. yakuba n=209, 9 replicates; and D. virilis n=245, 11 replicates. Differences in survival were assayed using the log-rank test. (B) log2 fold-change in levels of p38K protein during paraquat exposure based on 3 biological replicates of each of the 7 species. (C) Scatterplot of the correlation between phylogenetically independent contrasts of p38K protein levels and median survival time. Phylogenetically independent contrasts were calculated from survival data of 3 biological replicates of each of 7 species as in A. The scatterplot is based on the 6 calculated contrasts. Correlation values were determined by Spearman rho correlation. (D) Median survival of Drosophila species with decreased, unchanged or increased levels of p38K during paraquat exposure. Data are shown as quartiles in box plots (center line indicates median and error bars are the interquartile range). Different lowercase letters indicate significance groups with P<0.05 as determined by analysis of variance of aligned rank transformed data testing.
We found that, of the studied species, D. pseudoobscura showed the longest survival in response to paraquat (Fig. 7A; Table S2). This finding is especially interesting as D. pseudoobscura has two copies of the p38Kb gene, suggesting that p38Kb may be important for stress resistance and that both copies may contribute to this response. To examine the potential role of p38K, flies were exposed to either control food or food mixed with 20 mmol l−1 paraquat and collected at early and late time points based on the average survival time for that species. We performed immunoblots to analyze changes in the levels of p38K protein. We observed a decrease in p38K protein levels with time in flies fed on control food across the species (Fig. S2K; P=0.026), but no significant differences between species (P=0.40), suggesting that, in general, p38K protein levels decline with age in Drosophila species.
When flies were subjected to prolonged paraquat exposure, we found an increase in the levels of p38K in D. melanogaster, D. mauritiana and D. pseudoobscura (log2 fold-change greater than 0.5), a lack of change in D. simulans and D. virilis (log2-fold change between −0.5 and 0.5), and decreased levels in D. yakuba and D. ananassae (log2 fold-change less than −0.5; Fig. 7B).
We next tested whether the changes in p38K protein levels are associated with species survival when exposed to paraquat. To account for the potential role of phylogenetic relationships in survival, we compared phylogenetically independent contrasts for changes in p38K protein levels and species survival. We found that there is a moderate positive correlation between the change in p38K levels and paraquat resistance (Fig. 7C; R=0.77, P=0.1). When we categorized the species by changes in total p38K as increased, unchanged or decreased, we found that decreased levels of p38K are associated with poor survival on paraquat when compared with those species with neutral or increased p38K (Fig. 7D; P=0.00429).
It has been well documented that oxidative stress induces the activation of p38K through its phosphorylation (McCubrey et al., 2006). We performed western blots on our samples using an anti-phosphorylated p38K antibody and found that, across species and conditions, there is a linear relationship between the change in levels of total and phosphorylated forms of p38K (Fig. S2L–N).
Paraquat resistance and p38K protein levels across Drosophila taxa are associated with AP-1 and lola-PT consensus sites
As over-expression of either AP-1 or lola-PT is able to induce p38Kb transcription in D. melanogaster, we next tested whether having a conserved AP-1 and/or lola-PT site is important for paraquat resistance. Therefore, we tested seven species of Drosophila with different combinations of these sites (Fig. 5B) for oxidative stress resistance. Two of these species, D. simulans and D. mauritiana, are sister species of D. melanogaster that have both the putative AP-1 and lola-PT binding sites as well as a conserved motif (motif 4) not found in D. melanogaster. Drosophila yakuba is also a member of the melanogaster subgroup and has the AP-1 consensus site but not the lola-PT site. Drosophila pseudoobscura is a more distantly related species and has two p38Kb genes, both of which have predicted AP-1 and lola-PT binding sites. Finally, we tested two species that do not have the conserved AP-1 or lola-PT sites, D. ananassae and D. virilis, the latter of which is the most distantly related of the species. In order to assess the roles of the five predicted transcription factor binding sites, we used ANOVA on aligned rank transformed data to test for interactions between the presence of a predictive transcription factor binding site and p38K protein levels. We first tested the relationship between p38K levels and the presence of a consensus lola-PO, motif 4 or motif 5 site, which are either also present in the p38Ka locus or have no known association with oxidative stress. We found, across species, no significant interaction with any of these three consensus binding sites and p38K protein levels under any conditions (lola-PO, P=0.14149; motif 4, P=0.10614; and motif 5, P=0.52630).
Though p38K levels tend to decrease across time (Fig. S2K), we found that the presence of a consensus AP-1 site is associated with a smaller decrease in p38K levels under standard conditions (Fig. 8A; P=0.037). In addition, the AP-1 site is also associated with increased levels of p38K in flies fed on paraquat-treated food (Fig. 8A; P=0.037); however, there is no interaction between AP-1 site presence and food treatment (Fig. 8A; P=0.175), suggesting that AP-1 regulation of p38K levels is not responsive to paraquat treatment, in line with our qPCR results (Fig. 6A,B). The presence of a putative lola-PT binding site leads to increased levels of p38K on paraquat but not control food (Fig. 8B), and significantly interacts with treatment (Fig. 8B; P=0.013), suggesting that lola-PT regulates p38K levels in response to oxidative stress. We found that the presence of a putative AP-1 or lola-PT binding site shows a positive trend toward increasing median survival on paraquat food, although these effects fall just short of significance (Fig. 8C,D; P=0.098 for AP-1, P=0.069 for lola-PT). These data suggest that AP-1 plays a role in the normal regulation of p38Kb transcription, whereas lola-PT acts in response to oxidative stress to increase p38Kb transcription, and that both modes of regulation can contribute to p38K-mediated oxidative stress resistance.
Fig. 8.
The presence of putative AP-1 and lola-PT binding sites is associated with p38K protein levels. (A) The presence of an AP-1 site is associated with increased levels of p38K protein but does not interact with food treatment. (B) The presence of a lola-PT site is associated with increased p38K levels during paraquat treatment and interacts with paraquat treatment. (C,D) Median survival of Drosophila species with (C) AP-1 and (D) lola-PT sites. In box plots, the center line indicates the median and error bars are the interquartile range. Experiments were conducted on seven species.
DISCUSSION
A common theme in biology is the repurposing of signaling pathways to perform new cellular functions. Evolutionary pressures can act on these signaling pathways in various ways, leading to sequence divergence, gene duplications or the creation/loss of regulatory elements. In order to understand how these pressures may have influenced an organism's ability to respond to oxidative stress, we utilized the power of the Drosophila genome sequencing projects, which have resulted in the sequencing of 22 Drosophila species, 12 of which have been fully annotated. We undertook a comparative genomics approach to analyze the genomic architecture of the conserved p38K genes across the genus Drosophila, which represents 30–40 million years of evolutionary history (Obbard et al., 2012). In addition, analysis of the 1 kb upstream region revealed loss and gain of putative transcription factor binding sites for both the p38Ka and p38Kb loci. We found that p38Ka and p38Kb share lola-PO consensus binding sites, which likely plays a role in regulating the expression of both p38Ka and p38Kb in shared functions. Additionally, each gene has a specific set of consensus transcription factor binding sites that may play a role in regulating the expression of these genes for specific functions.
We had previously generated a D. melanogaster p38Kb mutant (p38KbΔ25) in which 299 bp of the upstream regulatory region were deleted (Vrailas-Mortimer et al., 2011), including both the AP-1 and lola-PT putative transcription factor binding sites. We found that this was a partial loss of function mutation, reducing the levels of p38Kb protein in the muscle. In addition, null mutations of both p38Ka (Δp38Ka) and p38Kb (p38KbΔ45) result in lethality, whereas combining the null p38Ka mutation with our p38Kb partial loss of function mutation results in viable adults (p38K DKO). Though p38Ka null mutants were not sensitive to oxidative stress, when combined with the p38Kb partial loss of function mutation, these p38K DKO animals had increased sensitivity to oxidizing agents (Vrailas-Mortimer et al., 2011). These data suggest that this upstream regulatory region is critical for expression of p38Kb and that loss of this expression leads to oxidative stress sensitivity. As our bioinformatics approach identified consensus AP-1 and lola-PT binding sites in this upstream region, we tested whether these sites were biologically relevant regulatory elements in vivo using the genetically amenable species D. melanogaster.
We found that the p38Kb putative transcription factor binding sites for both AP-1 and lola-PT are functional, as over-expression of AP-1 and lola-PT induce expression of p38Kb under control conditions. In addition, we also found that these consensus sites play different roles in the regulation of p38Kb as only over-expression of lola-PT was able to further induce expression of p38Kb in the presence of the oxidizing agent paraquat. Intriguingly, we found that flies over-expressing AP-1 that were allowed to age for a week after eclosion before being exposed to paraquat had increased resistance compared with those over-expressing lola-PT. These data suggest that AP-1 may be able to transcribe sufficient amounts of p38Kb pre-exposure to result in a protective effect once the animal encounters an oxidizing environment. Thus, AP-1 and lola-PT are playing unique roles in the regulation of p38Kb expression and this may, in turn, help the organism as it navigates environmental changes. To determine whether these sites play similar roles in other Drosophila species, we tested how species with differing combinations of the AP-1 and lola-PT consensus binding sites respond to oxidative stress. Similar to our results with D. melanogaster, we found a significant interaction between the presence of an AP-1 consensus site and increased levels of p38K protein regardless of the oxidative stress state, whereas the presence of a lola-PT consensus binding site shows a significant interaction with p38K protein levels only in the presence of oxidative stress.
Furthermore, we found a positive trend between the presence of these consensus sites and increased survival. However, these results were not significant, likely due to the limited number of species identified with different combinations of predicted AP-1 and lola-PT binding sites that we could test. Overall, our data suggest that AP-1 and lola-PT act in a similar manner across species to regulate p38Kb expression. Much of what is known about how p38K proteins regulate stress responses has focused on the role of post-translational modifications – in particular, the dual phosphorylation of p38K (Krishna and Narang, 2008; Peti and Page, 2013; Chang and Karin, 2001) – and little is known about the role of transcriptional regulation in this process. We found that over-expression of either AP-1 or lola-PT is sufficient to increase p38Kb transcription under non-stress conditions and confer resistance to subsequent oxidative stress. These findings suggest that transcriptional regulation of p38Kb can also play an important role in regulating the oxidative stress response, and the transcription of p38Kb in response to oxidative stress may complement the effects of p38Kb phosphorylation in promoting resistance. However, more research will be necessary to understand the relationship between induced p38Kb transcription and subsequent phosphorylation and how this supports the oxidative stress response.
Our results suggest that we can successfully utilize the genomes of sequenced species to identify biologically relevant regulatory elements that may give insights into how different species have evolved mechanisms for responding to selective pressures. Furthermore, as little is known about the transcriptional regulation of other MAPK, our comparative genomics approach may be utilized to identify conserved transcription factor binding sites that may play important roles in regulating these MAPK and potentially other key signaling genes in response to a variety of cellular or environmental stimuli.
Supplementary Material
Acknowledgements
We would like to acknowledge the Bloomington Drosophila Stock Center (NIH P40OD018537), and the Drosophila Species Stock Center for providing fly stocks used in this study. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: N.T.M., A.D.V.-M.; Methodology: N.T.M., A.D.V.-M.; Formal analysis: S.M.R., K.W., B.O.-P., N.T.M., A.D.V.-M.; Investigation: S.M.R., K.W., B.O.-P.; Resources: A.D.V.-M.; Writing - original draft: S.M.R., A.D.V.-M.; Writing - review & editing: N.T.M., A.D.V.-M.; Visualization: N.T.M., A.D.V.-M.; Supervision: S.B., N.T.M., A.D.V.-M.; Project administration: A.D.V.-M.; Funding acquisition: S.B., N.T.M., A.D.V.-M.
Funding
A.D.V.-M. was funded by start-up funds from the University of Denver, a Knoebel Center for the Study of Aging pilot grant, start-up funds and a New Faculty Initiative Grant from Illinois State University and the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Number R15AR070505. S.M.R. was supported by a Knoebel Center for the Study of Aging pilot grant to A.D.V.-M. and S.B. N.T.M. was funded by start-up funds and a Pre-tenure Faculty Initiative Grant from Illinois State University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at https://jeb.biologists.org/lookup/doi/10.1242/jeb.221622.supplemental
References
- Bass B. P., Cullen K. and McCall K. (2007). The axon guidance gene lola is required for programmed cell death in the Drosophila ovary. Dev. Biol. 304, 771-785. 10.1016/j.ydbio.2007.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bataillé L., Frendo J.-L. and Vincent A. (2015). Hox control of Drosophila larval anatomy; the alary and thoracic alary-related muscles. Mech. Dev. 138, 170-176. 10.1016/j.mod.2015.07.005 [DOI] [PubMed] [Google Scholar]
- Belozerov V. E., Ratkovic S., McNeill H., Hilliker A. J. and McDermott J. C. (2014). In vivo interaction proteomics reveal a novel p38 mitogen-activated protein kinase/Rack1 pathway regulating proteostasis in Drosophila muscle. Mol. Cell. Biol. 34, 474-484. 10.1128/MCB.00824-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. B., Boley N., Eisman R., May G. E., Stoiber M. H., Duff M. O., Booth B. W., Wen J., Park S., Suzuki A. M. et al. (2014). Diversity and dynamics of the Drosophila transcriptome. Nature 512, 393-399. 10.1038/nature12962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker S. E., Dillon L. A. L., Gerstein M. B., Gunsalus K. C., Henikoff S., Karpen G. H., Kellis M., Lai E. C., Lieb J. D., MacAlpine D. M. et al. (2009). Unlocking the secrets of the genome. Nature 459, 927-930. 10.1038/459927a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S., Poidevin M. Lemaitre B. (2014). The Drosophila MAPK p38c regulates oxidative stress and lipid homeostasis in the intestine. PLoS Genet. 10, e1004659 10.1371/journal.pgen.1004659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L. and Karin M. (2001). Mammalian MAP kinase signalling cascades. Nature 410, 37-40. 10.1038/35065000 [DOI] [PubMed] [Google Scholar]
- Chen J., Xie C., Tian L., Hong L., Wu X. and Han J. (2010). Participation of the p38 pathway in Drosophila host defense against pathogenic bacteria and fungi. Proc. Natl. Acad. Sci. USA 107, 20774-20779. 10.1073/pnas.1009223107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig C. R., Fink J. L., Yagi Y., Ip Y. T. and Cagan R. L. (2004). A Drosophila p38 orthologue is required for environmental stress responses. EMBO Rep. 5, 1058-1063. 10.1038/sj.embor.7400282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowner D., Madden K., Goeke S. and Giniger E. (2002). Lola regulates midline crossing of CNS axons in Drosophila. Dev. Camb. Engl. 129, 1317-1325. [DOI] [PubMed] [Google Scholar]
- Cully M., Genevet A., Warne P., Treins C., Liu T., Bastien J., Baum B., Tapon N., Leevers S. J. and Downward J. (2010). A role for p38 stress-activated protein kinase in regulation of cell growth via TORC1. Mol. Cell. Biol. 30, 481-495. 10.1128/MCB.00688-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. M., Primrose D. A. and Hodgetts R. B. (2008). A member of the p38 mitogen-activated protein kinase family is responsible for transcriptional induction of Dopa decarboxylase in the epidermis of Drosophila melanogaster during the innate immune response. Mol. Cell. Biol. 28, 4883-4895. 10.1128/MCB.02074-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enriquez J., Boukhatmi H., Dubois L., Philippakis A. A., Bulyk M. L., Michelson A. M., Crozatier M. and Vincent A. (2010). Multi-step control of muscle diversity by Hox proteins in the Drosophila embryo. Dev. Camb. Engl. 137, 457-466. 10.1242/dev.045286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783 10.1111/j.1558-5646.1985.tb00420.x [DOI] [PubMed] [Google Scholar]
- Franciscovich A. L., Mortimer A. D. V., Freeman A. A., Gu J. and Sanyal S. (2008). Overexpression screen in Drosophila identifies neuronal roles of GSK-3β/shaggy as a regulator of AP-1-dependent developmental plasticity. Genetics 180, 2057-2071. 10.1534/genetics.107.085555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giniger E., Tietje K., Jan L. Y. and N Y. (1994). Jan, lola encodes a putative transcription factor required for axon growth and guidance in Drosophila. Dev. Camb. Engl. 120, 1385-1398. [DOI] [PubMed] [Google Scholar]
- Gupta S., Stamatoyannopoulos J. A., Bailey T. L. and Noble W. S. (2007). Quantifying similarity between motifs. Genome Biol. 8, R24 10.1186/gb-2007-8-2-r24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha E.-M., Lee K.-A., Seo Y. Y., Kim S.-H., Lim J.-H., Oh B.-H., Kim J. and Lee W.-J. (2009). Coordination of multiple dual oxidase-regulatory pathways in responses to commensal and infectious microbes in drosophila gut. Nat. Immunol. 10, 949-957. 10.1038/ni.1765 [DOI] [PubMed] [Google Scholar]
- Inoue H., Tateno M., Fujimura-Kamada K., Takaesu G., Adachi-Yamada T., Ninomiya-Tsuji J., Irie K., Nishida Y., Matsumoto K. and Drosophila MAPKKK A. (2001). D-MEKK1, mediates stress responses through activation of p38 MAPK. EMBO J. 20, 5421-5430. 10.1093/emboj/20.19.5421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. T., Taylor W. R. and Thornton J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Bioinformatics 8, 275-282. 10.1093/bioinformatics/8.3.275 [DOI] [PubMed] [Google Scholar]
- Keren A., Tamir Y. and Bengal E. (2006). The p38 MAPK signaling pathway: a major regulator of skeletal muscle development. Mol. Cell. Endocrinol. 252, 224-230. 10.1016/j.mce.2006.03.017 [DOI] [PubMed] [Google Scholar]
- Kim T., Yoon J., Cho H., Lee W.-B., Kim J., Song Y.-H., Kim S. N., Yoon J. H., Kim-Ha J. and Kim Y.-J. (2005). Downregulation of lipopolysaccharide response in Drosophila by negative crosstalk between the AP1 and NF-κB signaling modules. Nat. Immunol. 6, 211-218. 10.1038/ni1159 [DOI] [PubMed] [Google Scholar]
- Krishna M. and Narang H. (2008). The complexity of mitogen-activated protein kinases (MAPKs) made simple. Cell. Mol. Life Sci. CMLS 65, 3525-3544. 10.1007/s00018-008-8170-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden K., Crowner D. and Giniger E. (1999). LOLA has the properties of a master regulator of axon-target interaction for SNb motor axons of Drosophila. Dev. Biol. 213, 301-313. 10.1006/dbio.1999.9399 [DOI] [PubMed] [Google Scholar]
- Markow T. A. and O'Grady P. M. (2007). Drosophila biology in the genomic age. Genetics 177, 1269-1276. 10.1534/genetics.107.074112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCubrey J. A., Lahair M. M. and Franklin R. A. (2006). Reactive oxygen species-induced activation of the MAP kinase signaling pathways. Antioxid. Redox Signal 8, 1775-1789. 10.1089/ars.2006.8.1775 [DOI] [PubMed] [Google Scholar]
- Milton V. J., Jarrett H. E., Gowers K., Chalak S., Briggs L., Robinson I. M. and Sweeney S. T. (2011). Oxidative stress induces overgrowth of the Drosophila neuromuscular junction. Proc. Natl. Acad. Sci. USA 108, 17521-17526. 10.1073/pnas.1014511108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na J., Musselman L. P., Pendse J., Baranski T. J., Bodmer R., Ocorr K. and Cagan R. (2013). A Drosophila model of high sugar diet-induced cardiomyopathy. PLoS Genet. 9, e1003175 10.1371/journal.pgen.1003175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obbard D. J., Maclennan J., Kim K.-W., Rambaut A., O'Grady P. M. and Jiggins F. M. (2012). Estimating divergence dates and substitution rates in the Drosophila phylogeny. Mol. Biol. Evol. 29, 3459-3473. 10.1093/molbev/mss150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.-S., Kim Y.-S. and Yoo M.-A. (2009). The role of p38b MAPK in age-related modulation of intestinal stem cell proliferation and differentiation in Drosophila. Aging 1, 637-651. 10.18632/aging.100054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins K. K., Admon A., Patel N. and Tjian R. (1990). The Drosophila Fos-related AP-1 protein is a developmentally regulated transcription factor. Genes Dev. 4, 822-834. 10.1101/gad.4.5.822 [DOI] [PubMed] [Google Scholar]
- Peti W. and Page R. (2013). Molecular basis of MAP kinase regulation. Protein Sci. Publ. Protein Soc. 22, 1698-1710. 10.1002/pro.2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piipari M., Down T. A., Saini H., Enright A. and Hubbard T. J. P. (2010). iMotifs: an integrated sequence motif visualization and analysis environment. Bioinformatics 26, 843-844. 10.1093/bioinformatics/btq026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesgo-Escovar J. R. and Hafen E. (1997). Common and distinct roles of DFos and DJun during Drosophila development. Science 278, 669-672. 10.1126/science.278.5338.669 [DOI] [PubMed] [Google Scholar]
- Sano Y., Akimaru H., Okamura T., Nagao T., Okada M. and Ishii S. (2005). Drosophila activating transcription factor-2 is involved in stress response via activation by p38, but not c-Jun NH(2)-terminal kinase. Mol. Biol. Cell 16, 2934-2946. 10.1091/mbc.e04-11-1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzawa N., Nelson B., Aonuma H., Okado K., Fukumoto S., Miura M. and Kanuka H. (2009). p38 MAPK-dependent phagocytic encapsulation confers infection tolerance in Drosophila. Cell Host Microbe. 6, 244-252. 10.1016/j.chom.2009.07.010 [DOI] [PubMed] [Google Scholar]
- Tamura K. and Nei M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10, 512-526. [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M. and Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731-2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokusumi T., Sorrentino R. P., Russell M., Ferrarese R., Govind S. and Schulz R. A. (2009). Characterization of a lamellocyte transcriptional enhancer located within the misshapen gene of Drosophila melanogaster. PLoS ONE 4 e6429 10.1371/journal.pone.0006429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrailas-Mortimer A., del Rivero T., Mukherjee S., Nag S., Gaitanidis A., Kadas D., Consoulas C., Duttaroy A. and Sanyal S. (2011). A muscle-specific p38 MAPK/Mef2/MnSOD pathway regulates stress, motor function, and life span in Drosophila. Dev. Cell 21, 783-795. 10.1016/j.devcel.2011.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrailas-Mortimer A., Gomez R., Dowse H. and Sanyal S. (2012). A survey of the protective effects of some commercially available antioxidant supplements in genetically and chemically induced models of oxidative stress in Drosophila melanogaster. Exp. Gerontol. 47, 712-722. 10.1016/j.exger.2012.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrailas-Mortimer A. D., Ryan S. M., Avey M. J., Mortimer N. T., Dowse H. and Sanyal S. (2014). p38 MAP kinase regulates circadian rhythms in Drosophila. J. Biol. Rhythms 29, 411-426. 10.1177/0748730414555183 [DOI] [PubMed] [Google Scholar]
- West C. and Silverman N. (2018). p38b and JAK-STAT signaling protect against Invertebrate iridescent virus 6 infection in Drosophila. PLoS Pathog. 14, e1007020 10.1371/journal.ppat.1007020 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.