Abstract
Background
Despite the public health importance of toxocariasis/toxascariasis, only a few species of these ascaridoid parasites from wild canine and feline carnivores have been studied at the molecular level so far. Poor understanding of diversity, host distribution and the potential (zoonotic) transmission of the ascaridoid species among wild animals negatively affects their surveillance and control in natural settings. In this study, we updated previous knowledge by profiling the genetic diversity and phylogenetic relationships of ascaridoid species among eleven wild canine and feline animals on the basis of a combined analysis of the ribosomal internal transcribed spacer region (ITS) gene and the partial mitochondrial cytochrome c oxidase subunit 2 (cox2) and NADH dehydrogenase subunit 1 (nad1) genes.
Results
In total, three genetically distinct ascaridoid lineages were determined to be present among these wild carnivores sampled, including Toxocara canis in Alopex lagopus and Vulpes vulpes, Toxocara cati in Felis chaus, Prionailurus bengalensis and Catopuma temmincki and Toxascaris leonina in Canis lupus, Panthera tigris altaica, Panthera tigris amoyensis, Panthera tigris tigris, Panthera leo and Lynx lynx. Furthermore, it was evident that T. leonina lineage split into three well-supported subclades depending on their host species, i.e. wild felids, dogs and wolves and foxes, based on integrated genetic and phylogenetic evidence, supporting that a complex of T. leonina other than one species infecting these hosts.
Conclusions
These results provide new molecular insights into classification, phylogenetic relationships and epidemiological importance of ascaridoids from wild canids and felids and also highlight the complex of the taxonomy and genetics of Toxascaris in their wild and domestic carnivorous hosts.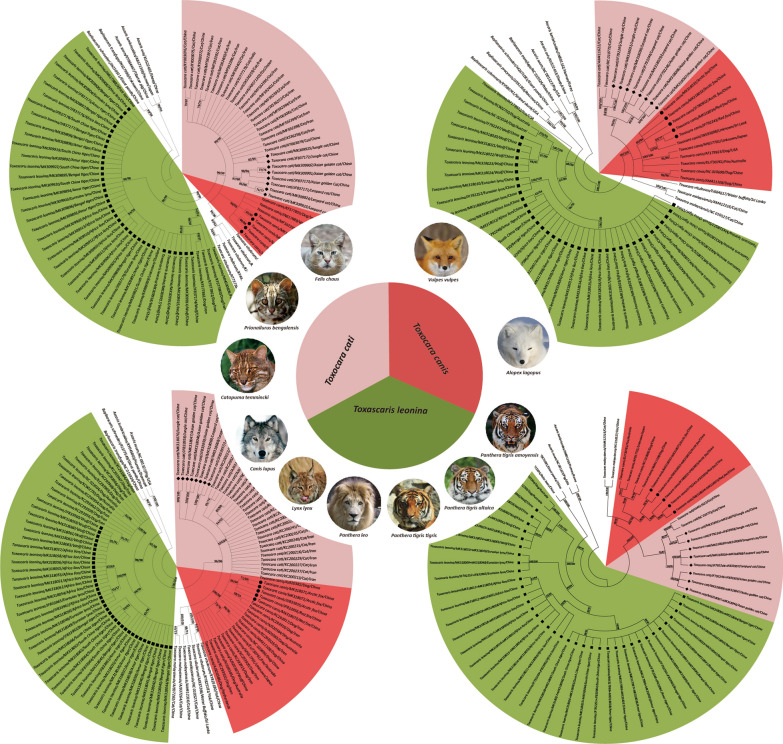
Keywords: Toxocara, Toxascaris, Wild canids and felids, Nuclear ITS, Mitochondrial DNA, Phylogeny
Introduction
Ascaridoid nematodes of the genera Toxocara (Toxocaridae) and Toxascaris (Ascarididae) are the most common intestinal parasites among carnivores of the families Canidae and Felidae [1, 2]. Several species among them, such as Toxocara canis Werner, 1782, T. cati Schrank, 1788, T. malaysiensis Gibbons et al., 2001 and Toxascaris leonina Linstow, 1902 are regarded as zoonotic or potentially zoonotic parasites and pose threats to public health [3–8]. These parasites (at the adult stage) usually live in the small intestines of domestic and/or wild definitive hosts and cause significant health problems. The clinical symptoms include diarrhea, emesis, stunted growth, abdominal discomfort, and even life-threatening blockage of intestinal obstruction [9]. The definitive hosts of T. canis are dogs (Canis familiaris), dingoes (Canis dingo), wolfs (Canis lupus), coyotes (Canis latrans), jackals (Canis aureus), red foxes (Vulpes vulpes), arctic foxes (Vulpes lagopus), fennecs (Megalotis zerda) and a few feline species, while the definitive hosts for T. cati include cats (Felis catus), wild cats (Felis silvestris), servals (Felis serwal), cheetahs (Actinomyx jubatus), lynxes (Lynx lynx), pumas (Puma concolor), lions (Panthera leo), American leopards (Panthera onca), tigers (Panthera tigris), ocelots (Leopardus pardalis) and other felines [1]. Like T. cati, the congenetic T. malaysiensis infects felids [10]. However, T. leonina is defined to infect both feline and canine species [11, 12]. Humans are accidental hosts for Toxocara spp. and become infected through ingestion of infective eggs from the environment or articles contaminated with infected animal feces, with clinical forms including visceral larva migrans (VLM), ocular larva migrans (OLM), eosinophilic meningoencephalitis (EME), covert toxocariasis (CT) and neurological toxocariasis (NT) [2, 3].
Traditional approaches for the specific identification of ascaridoids within the genera Toxocara and Toxascaris rely on morphological characteristics, host preference and geographical distributions. However, these criteria are not always sufficient to distinguish the closely related and/or morphologically similar ascaridoid species, particularly when the larval and egg stages are involved [1, 12, 13]. Moreover, the diversity patterns of life-cycles (e.g. Toxocara) may complicate this procedure [4]. In such context, obtaining a more efficient and reliable approach to species identification has become crucial for clinical diagnosis and epidemiological investigation, and achieving this goal is foreseeable only through utilization of molecular methodologies [13]. Increased studies demonstrated that the nuclear ribosomal DNA (rDNA) and/or mitochondrial DNA (mtDNA) are valuable markers and have been often used to achieve the identification of parasites to species in Toxocara and Toxascaris [5, 6, 11, 12, 14–17]. For example, Jacobs et al. [18] used the second internal transcribed spacer sequence (ITS2) of rDNA to distinguish between T. canis, T. cati and T. leonina from foxes, cats and dogs in Australia. Likewise, based on the sequences of the internal transcribed spacers (ITS, ITS1 plus ITS2), Li et al. [15] developed a species-specific PCR tool for diagnosis of four ascaridoid species of dogs and cats including T. canis, T. cati, T. malaysiensis and T. leonina in China, Malaysia, Australia, England and the Netherlands. More recently, an ITS1-based PCR-RFLP was also employed for molecular determination of T. cati from wild felids in Argentina [19]. Compared to rDNA, however, mtDNA appears to be more variable both between and within species and is therefore more sensitive for genetic analysis of related ascaridoid species of Toxocara and Toxascaris [11, 16, 20]. In fact, the mitochondrial cytochrome c oxidase subunits 1 and 2 (cox1 and cox2) as well as NADH dehydrogenase subunits 1 and 4 (nad1 and nad4) genes have proven to be useful in resolving relationships among and between Toxocara spp. and Toxascaris spp. from dogs, cats and cattle [11, 12, 14, 16, 21, 22]; importantly, such mtDNA-derived evidence supports the rDNA-based conclusion that T. malaysiensis is another valid ascaridoid species found in cats [23]. Nevertheless, considering that single rDNA or mtDNA loci only allow limited inference of molecular analyses [24] and that current sampling and studies mostly focus on domestic animal-originated Toxocara and Toxascaris [4, 12], it would be essential and urgent to develop a combined analysis of nuclear and mitochondrial data for more accurate and robust branches in ascaridoids from a wider host-range that includes wild canids and felids. Unfortunately, until now there has been still very limited molecular information available for these ascaridoids [11, 12, 14, 25].
In the present study, ongoing epidemiological surveys on potential helminthic zoonoses, centred in the southwestern zoos of China, expanded previous investigations by including specimens of Toxocara and Toxascaris in 11 species of carnivores from Canidae and Felidae. Our aim was to characterise ascaridoids from an assemblage of diverse hosts relative to currently defined lineages of Toxocara and Toxascaris [1, 26], facilitating (i) identification of these isolates at species level by genetical analysis of the complete nuclear ITS and partial mitochondrial cox2 and nad1 markers; and (ii) determination of levels of genetic variation among these ascaridoids by comparisons among those documented in dogs and cats as well as other canids and felids which are available in public databases. This study should provide new molecular data for genetic diversity and phylogenetic relationships of ascaridoids from wild carnivorous animals and these results presented here also serve as molecular technical support for accurate diagnosis and better control of these parasites.
Methods
Sample collection and DNA extraction
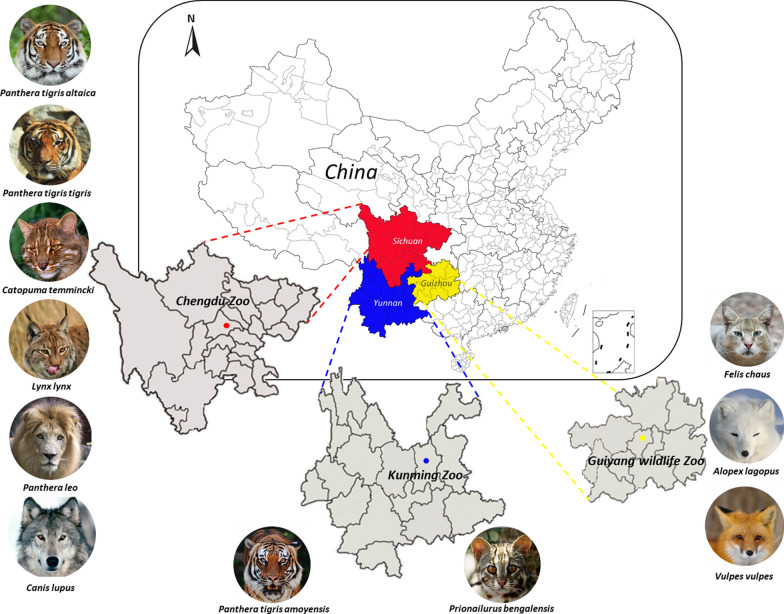
During April 2011 to November 2018, a total of 50 adult ascaridoids were collected from 11 captive wild canine and feline animals after routine treatment with pyrantel pamoate, including the arctic fox (Alopex lagopus), red fox (Vulpes vulpes) and jungle cat (Felis chaus) in Guiyang Wildlife Zoo (Guizhou, China), South China tiger (Panthera tigris amoyensis) and leopard cat (Prionailurus bengalensis) in Kunming Zoo (Yunnan, China), and wolf (Canis lupus), Asian golden cat (Catopuma temmincki), Eurasian lynx (Lynx lynx), Bengal tiger (Panthera tigris tigris), Amur tiger (Panthera tigris altaica) and African lion (Panthera leo) in the Chengdu Zoo (Sichuan, China) (Table 1, Fig. 1). Each individual ascaridoid specimen was washed thoroughly in physiological saline solution and subjected to morphological identification according to taxonomic keys [27, 28]. Subsequently, all worm samples were stored at − 70 °C until required for DNA extraction. The genomic DNA (gDNA) of each isolate was extracted according to the method described by Jacobs et al. [18]. The DNA concentration and purity were measured by micro-spectrophotometry (NanoDrop ND-2000; Thermo Fisher Scientific, Wilmington, DE, USA).
Table 1.
Summary information of canid and felid hosts and their ascaridoid nematodes sampled in this study
| Host species | No. of sampled | |
|---|---|---|
| Hosts | Parasitea | |
| Wolf (Canis lupus) | 3 | 6 (1/2/3) |
| Arctic fox (Alopex lagopus) | 2 | 3 (1/2) |
| Red fox (Vulpes vulpes) | 1 | 2 |
| Asian golden cat (Catopuma temmincki) | 2 | 3 (1/2) |
| South China tiger (Panthera tigris amoyensis) | 3 | 5 (1/2/2) |
| Eurasian lynx (Lynx lynx) | 2 | 3 (1/2) |
| African lion (Panthera leo) | 3 | 10 (2/2/6) |
| Jungle cat (Felis chaus) | 2 | 2 (1/1) |
| Amur tiger (Panthera tigris altaica) | 1 | 6 |
| Bengal tiger (Panthera tigris tigris) | 2 | 7 (3/4) |
| Leopard cat (Prionailurus bengalensis) | 3 | 3 (1/1/1) |
aParasite numbers sampled from each individual are shown in parentheses
Fig. 1.
Sampling map in China showing the locations (coloured circles) of Toxocara/Toxascaris spp. collected from 11 wild canine and feline animal species housed in Chengdu Zoo of Sichuan Province (red), Kunming Zoo of Yunnan Province (blue) and Guiyang Wildlife Zoo of Guizhou Province (yellow)
PCR amplification
The complete nuclear ITS sequence and partial sequences of the mitochondrial cox2 and nad1 were amplified by polymerase chain reaction (PCR) using the following primers: ITS (forward: 5′-GTA GGT GAA CCT GCG GAA GGA TCA TT-3′ and reverse: 5′-TTA GTT TCT TTT CCT CCG CT-3′); cox2 (forward: 5′-CAC CAA CTC TTA AAA TTA TC-3′ and reverse: 5′-TTT TCT AGT TAT ATA GAT TGR TTT YAT-3′) [22]; nad1 (forward: 5′-TTC TTA TGA GAT TGC TTT T-3′ and reverse: 5′-TAT CAT AAC GAA AAC GAG G-3′) [16]. All reactions were carried out in 50 μl reaction volume with 25 μl 2× Taq PCR MasterMix (Tiangen Biotech Co., Beijing, China), 3 μl gDNA, 16 μl sterile water and 3 μl of each primer (10 pmol/μl; TaKaRa Biotech, Dalian, China). The following cycle conditions were used in a Mastercycler Gradient 5331 Thermocycler (Eppendorf, Hamburg, Germany): 94 °C for 5 min; 35 cycles at 94 °C for 30 s, followed either by 60 °C for 30 s for ITS, 44 °C for 30 s for cox2 or 46 °C for 30 s for nad1, then 72 °C for 60 s; with a final extension step of 72 °C for 10 min. For each amplification, samples without parasite and host DNA were also included as negative controls to detect possible contamination. The PCR fragments were separated in 1% agarose gels, visualized using ethidium bromide and photographed by a Bio-Rad ChemiDoc XRS (Bio-Rad Laboratories, Hercules, CA, USA). The corrected gel-isolated amplicons were column-purified with the TIANgel Midi Purification Kit (Tiangen) and sub-cloned into Escherichia coli (DH5a) using the vector pMD-19T (TaKaRa). Positive clones were screened and were sequenced in-house on an ABI PRISMTM 377XL DAN sequencer (Invitrogen Biotechnology Co. Ltd., Shanghai, China). To ensure maximum accuracy, an individual clone from one specimen was sequenced four times independently using the M13F and M13R primers. The consensus sequences were deposited in the GenBank database under the accession numbers JF837169-JF837179 and MK309890-MK309928 (ITS), JF780952-JF792252 and MK317996-MK318034 (cox2) and JF833955-JF833965 and MK318035-MK318073 (nad1).
Sequence analysis and phylogeny
The ITS, cox2 and nad1 sequences of ascaridoid species in the present study were initially edited with BioEdit (Ibis Biosciences, Carlsbad, USA) and subjected to separate sequence similarity search using BLAST (NCBI; http://blast.ncbi.nlm.nih.gov/Blast.cgi) to identify the nearest phylogenetic neighbours, and then aligned with ClustalX 1.83 [29]. All reference sequences from GenBank with parasite species, host species and geographical origins are listed in Table 2. During alignment, a codon-guided protein alignment was used for manual adjustment of the nucleotide alignments of cox2 and nad1. Pairwise comparisons of sequence identities (I) were calculated among and within species using the formula I = M/L, where M is the number of alignment positions at which the two sequences have a same base, and L is the total number of alignment positions included in the two sequences. Given almost identical nucleotide sequences of the ITS region and cox2 and nad1 genes in worms from the same host species in this study, one representative ascaridoid identified here was also selected and compared with T. canis, T. cati, T. malaysiensis, T. vitulorum and T. leonina as well as Baylisascaris spp. and Ascaris spp. for further detection of synonymous and non-synonymous mutations in the mitochondrial cox2 and nad1 genes according to their corresponding amino acid alignments, followed by calculation of genetic distances between them using a distance matrix based on the Kimura 2-parameter (K2P) model in MEGA 6.1 [30]. For phylogenetic analyses, two different methods, namely Bayesian inference (BI) (MrBayes 3.1.2 [31]) and maximum likelihood (ML) (PHYML 3.0 [32]) were carried out based on the refined sequence alignment datasets by the online Gblocks server (http://molevol.cmima.csic.es/castresana/Gblocks_server.html). For the BI analysis, the general time reversible (GTR) including gamma-distributed rate variation (+G) and a proportion of invariable sites (+I) (GTR + G + I; ITS) and gamma-distributed rate variation (GTR + G; cox2, nad1 and cox2 and nad1) was determined as the best-fit nucleotide substitution model using the Bayesian information criteria (BIC) test in jModeltest 2.1.6 [33], and the trees were constructed using four independent Markov chain runs for 10,000,000 (ITS), 100,000 (cox1), 50,000 (nad1) or 10,000,000 (cox1 and nad1) metropolis-coupled MCMC generations with every 10,000th (ITS), 100th (cox1), 50th (nad1) or 10,000th (cox1 and nad1) tree being sampled; when the average standard deviation of the split frequencies dropped below 0.01, 25% initial trees were discarded as “burn-in” and the remaining trees were used to calculate posterior probabilities and visualised graphically with TreeviewX (http://darwin.zoology.gla.ac.uk/~rpage/treeviewx/). The ML computations were performed using PHYML 3.0 under the GTR model for ITS and GTR + I for cox2, nad1 and cox2 and nad1. Both Baylisascaris spp. and Ascaris spp. were used as reference outgroups in phylogenetic analyses. In addition, given recent studies showing a genetic separation among T. leonina depending on their host origins, these newly generated and previously published sequences of the nuclear and mitochondrial genes of T. leonina were also concatenated and used to test the assumption using the same phylogenetic methods as described above.
Table 2.
Summary information of Toxocara/Toxascaris and other related ascaridoid species used for molecular identification in the present study
| Parasites | Stages | Host species | Geographical origin | Living conditions | GenBank accession number | References | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| ITS | cox2 | nad1 | ITS | cox2 | nad1 | |||||
| Toxascaris leonina | Adult | South China tiger | China | China | China | Zoo | JF837178 | JF792251 | JF833964 | This study |
| Adult | African lion | China | China | China | Zoo | JF837176 | JF792249 | JF833962 | This study | |
| Adult | Wolf | China | China | China | Zoo | JF837174 | JF792247 | JF833960 | This study | |
| Adult | Eurasian lynx | China | China | China | Zoo | JF837179 | JF792252 | JF833965 | This study | |
| Adult | Bengal tiger | China | China | China | Zoo | JF837177 | JF792250 | JF833963 | This study | |
| Adult | Amur tiger | China | China | China | Zoo | JF837175 | JF792248 | JF833961 | This study | |
| Adult | Wolf | China | China | China | Zoo | MK309918 | MK318024 | MK318063 | This study | |
| Adult | Wolf | China | China | China | Zoo | MK309917 | MK318023 | MK318062 | This study | |
| Adult | Wolf | China | China | China | Zoo | MK309916 | MK318022 | MK318061 | This study | |
| Adult | Wolf | China | China | China | Zoo | MK309915 | MK318021 | MK318060 | This study | |
| Adult | Wolf | China | China | China | Zoo | MK309914 | MK318020 | MK318059 | This study | |
| Adult | Amur tiger | China | China | China | Zoo | MK309890 | MK317996 | MK318035 | This study | |
| Adult | Amur tiger | China | China | China | Zoo | MK309891 | MK317997 | MK318036 | This study | |
| Adult | Amur tiger | China | China | China | Zoo | MK309892 | MK317998 | MK318037 | This study | |
| Adult | Amur tiger | China | China | China | Zoo | MK309893 | MK317999 | MK318038 | This study | |
| Adult | Amur tiger | China | China | China | Zoo | MK309894 | MK318000 | MK318039 | This study | |
| Adult | African lion | China | China | China | Zoo | MK309905 | MK318011 | MK318050 | This study | |
| Adult | African lion | China | China | China | Zoo | MK309913 | MK318019 | MK318058 | This study | |
| Adult | African lion | China | China | China | Zoo | MK309912 | MK318018 | MK318057 | This study | |
| Adult | African lion | China | China | China | Zoo | MK309911 | MK318017 | MK318056 | This study | |
| Adult | African lion | China | China | China | Zoo | MK309910 | MK318016 | MK318055 | This study | |
| Adult | African lion | China | China | China | Zoo | MK309907 | MK318013 | MK318052 | This study | |
| Adult | African lion | China | China | China | Zoo | MK309906 | MK318012 | MK318051 | This study | |
| Adult | Eurasian lynx | China | China | China | Zoo | MK309904 | MK318010 | MK318049 | This study | |
| Adult | African lion | China | China | China | Zoo | MK309908 | MK318014 | MK318053 | This study | |
| Adult | African lion | China | China | China | Zoo | MK309909 | MK318015 | MK318054 | This study | |
| Adult | Bengal tiger | China | China | China | Zoo | MK309895 | MK318001 | MK318040 | This study | |
| Adult | Bengal tiger | China | China | China | Zoo | MK309896 | MK318002 | MK318041 | This study | |
| Adult | Bengal tiger | China | China | China | Zoo | MK309897 | MK318003 | MK318042 | This study | |
| Adult | Bengal tiger | China | China | China | Zoo | MK309898 | MK318004 | MK318043 | This study | |
| Adult | Bengal tiger | China | China | China | Zoo | MK309899 | MK318005 | MK318044 | This study | |
| Adult | Bengal tiger | China | China | China | Zoo | MK309900 | MK318006 | MK318045 | This study | |
| Adult | South China tiger | China | China | China | Zoo | MK309919 | MK318025 | MK318064 | This study | |
| Adult | South China tiger | China | China | China | Zoo | MK309920 | MK318026 | MK318065 | This study | |
| Adult | South China tiger | China | China | China | Zoo | MK309921 | MK318027 | MK318066 | This study | |
| Adult | Eurasian lynx | China | China | China | Zoo | MK309903 | MK318009 | MK318048 | This study | |
| Adult | South China tiger | China | China | China | Zoo | MK309922 | MK318028 | MK318067 | This study | |
| Adult | African lion | China | China | China | Zoo | JN617988 | – | – | Unpublished | |
| Adult | Red fox | USA | USA | – | – | – | AF179922 | – | Nadler and Hudspeth [22] | |
| Adult | Dog | – | Australia | Australia | Domestication | – | NC_023504 | NC_023504 | Liu et al. [35] | |
| Adult | Dog | Iran | – | – | Domestication | KF577860-62 | – | – | Unpublished | |
| Adult | Fox | USA | – | – | Zoo | MH030606 | – | – | Hoberg et al. [67] | |
| Adult | Unknown | Thailand | – | – | – | KR999999 | – | – | Unpublished | |
| Adult | Dog | – | – | Iran | In the wild | – | – | KC293969, KC293947-69 | Fogt-Wyrwas et al. [12] | |
| Adult | Fox | Poland | – | – | In the wild | HM800922 | – | – | Fogt-Wyrwas et al. [12] | |
| Adult | Dog | Iran | – | – | Domestication | KF577860-62 | – | – | Fogt-Wyrwas et al. [12] | |
| Toxocara cati | Adult | Jungle cat | China | China | China | Zoo | JF837172 | JF792245 | JF833958 | This study |
| Adult | Asian golden cat | China | China | China | Zoo | JF837173 | JF792246 | JF833959 | This study | |
| Adult | Leopard cat | China | China | China | Zoo | JF837171 | JF792244 | JF833957 | This study | |
| Adult | Jungle cat | China | China | China | Zoo | MK309925 | MK318031 | MK318070 | This study | |
| Adult | Asian golden cat | China | China | China | Zoo | MK309902 | MK318008 | MK318047 | This study | |
| Adult | Asian golden cat | China | China | China | Zoo | MK309901 | MK318007 | MK318046 | This study | |
| Adult | Leopard cat | China | China | China | Zoo | MK309924 | MK318030 | MK318069 | This study | |
| Adult | Leopard cat | China | China | China | Zoo | MK309923 | MK318029 | MK318068 | This study | |
| Adult | Cat | Japan | Domestication | AB571303 | – | – | Arizono et al. [68] | |||
| Adult | Cat | – | China | China | In the wild | – | NC_010773 | NC_010773 | Li et al. [69] | |
| Adult | Cat | China | – | – | Domestication |
KY003075-76, KY003079-81 |
– | – | He et al. [70] | |
| Egg | Cat | Iran | – | – | In the wild | MF592392-99, MF592400-02 | – | – | Unpublished | |
| Adult | Cat | Iran | – | – | In the wild | JX536257-59 | – | – | Unpublished | |
| Adult | Cat | India | – | – | Domestication | KJ777179 | – | – | Unpublished | |
| Adult | Cat | – | – | Iran | Domestication | – | – | KC200213-17, KC200222-23, KC200225-27, KC200229-32, KC200235, KC200237, KC200239-41, KC200247 | Mikaeili et al. [21] | |
| Egg | Cat | – | – | Iran | Domestication | – | – | KC200246 | Mikaeili et al. [21] | |
| Adult | Cat | – | – | UK | Domestication | – | – | AJ937261 | Li et al. [16] | |
| Adult | Cat | – | China | China | Domestication | – | AM411622 | AM411622 | Li et al. [69] | |
| Adult | Cat | – | – | Australia | – | – | – | AJ937262 | Li et al. [16] | |
| Toxocara canis | Adult | Red fox | China | China | China | Zoo | JF837170 | JF792243 | JF833956 | This study |
| Adult | Arctic fox | China | China | China | Zoo | JF837169 | JF780952 | JF833955 | This study | |
| Adult | Red fox | China | – | – | Zoo | MK309928 | MK318034 | MK318073 | This study | |
| Adult | Arctic fox | China | – | – | Zoo | MK309926 | MK318032 | MK318071 | This study | |
| Adult | Arctic fox | China | – | – | Zoo | MK309927 | MK318033 | MK318072 | This study | |
| Adult | Fox | – | Australia | Australia | In the wild | – | EU730761 | EU730761 | Jex et al. [71] | |
| Unknown | Unknown | – | Japan | Japan | Unknown | – | AP017701 | AP017701 | Unpublished | |
| Unknown | Unknown | – | Sri Lanka | – | Unknown | – | JN593098 | – | Wickramasinghe et al. [72] | |
| Adult | Wolf | China | – | – | Zoo | JN617989 | – | – | Unpublished | |
| Adult | Dog | – | USA | – | – | – | AF179923 | – | Nadler and Hudspeth [22] | |
| Adult | Dog | – | China | China | Domestication | – | NC_010690 | NC_010690 | Li et al. [69] | |
| Egg | Dog | Iran | – | – | In the wild | MF592391 | – | – | Choobineh et al. [73] | |
| Adult | Dog | Iran | – | – | In the wild | KF577855 | – | – | Unpublished | |
| Adult | Dog | – | – | Iran | In the wild | – | – | KC293915-17, KC293920-23 | Mikaeili et al. [21] | |
| Adult | Dog | – | – | Australia | In the wild | – | – | AJ920383-85 | Li et al. [16] | |
| Adult | Dog | – | – | China | In the wild | – | – | AJ920382 | Li et al. [16] | |
| Larvae | Dog | – | – | Netherlands | – | – | – | AJ920386 | Li et al. [16] | |
| Adult | Cat | – | – | China | Domestication | – | – | AJ920387 | Li et al. [16] | |
| Toxocara malaysiensis | Adult | Cat | – | China | China | Domestication | – | AM412316 | AM412316 | Li et al. [16] |
| Adult | Cat | China | – | – | Domestication | AM231609 | – | – | Li et al. [16] | |
| Adult | Cat | – | China | Domestication | – | – | AJ937263-65 | Li et al. [16] | ||
| Toxocara vitulorum | Adult | Yak | India | – | – | Domestication | KJ777180 | – | – | Unpublished |
| Adult | Cattle | India | – | – | – | KJ777181 | – | – | Unpublished | |
| Adult | Mithun calf | India | – | – | – | KJ777182 | – | – | Unpublished | |
| Adult | Cattle | USA | – | – | Domestication | KT737382 | – | – | Unpublished | |
| Adult | European bison | Germany | – | – | – | KY442062 | – | – | Unpublished | |
| Egg | Water buffalo | – | – | Sri Lanka | – | – | – | AJ937266 | Li et al. [16] | |
| Adult | Yak | – | – | China | Domestication | – | – | KY825180-81 | Li et al. [74] | |
| Adult | Water buffalo | – | Sri Lanka | – | Domestication | – | FJ664617 | – | Wickramasinghe et al. [75] | |
| Baylisascaris transfuga | Adult | Unknown | China | – | – | Zoo | JN617990 | – | – | Unpublished |
| Adult | Polar bear | – | China | China | Zoo | – | NC_015924 | NC_015924 | Xie et al. [37] | |
| Baylisascaris columnaris | Adult | Striped skunk | – | USA | In the wild | – | KY580741 | – | Choi et al. [76] | |
| Baylisascaris procyonis | Adult | Raccoon | – | China | China | Zoo | – | JF951366 | JF951366 | Xie et al. [33] |
| Baylisascaris schroederi | Adult | Giant panda | – | China | China | Zoo | – | HQ671081 | HQ671081 | Xie et al. [37] |
| Adult | Giant panda | – | – | China | Zoo | – | – | FJ377549 | Unpublished | |
| Adult | Giant panda | China | – | Zoo | JN210911 | – | – | Lin et al. [77] | ||
| Ascaris ovis | Adult | Sheep | – | China | China | – | – | KU522453 | KU522453 | Unpublished |
| Adult | Sheep | China | – | – | – | KU522455 | – | – | Unpublished | |
| Ascaris suum | Adult | Tibetan pig | China | – | – | Domestication | KY964447 | – | – | Li et al. [78] |
| Adult | Pig | – | USA | USA | Domestication | – | NC_001327 | NC_001327 | Wolstenholme et al. [79] | |
| Ascaris lumbricoides | Adult | Human | – | Korea | Korea | Domestication | – | JN801161 | JN801161 | Park et al. [80] |
| Adult | Human | – | Denmark | Denmark | Domestication | – | KY045803 | KY045803 | Unpublished | |
| Adult | Human | Japan | – | – | Domestication | AB571300 | – | – | Arizono et al. [68] | |
Results
Sequence characterization
The DNA sequences representing the ITS (856–975 bp) region and cox2 (582 bp) and nad1 (366 bp) genes were generated for all 50 ascaridoid isolates. Sequence alignments showed common insertions/deletions (indels) in the ITS region with 314 variable sites including 313 parsimony-informative and one singleton sites; while compared to the ITS, both cox2 and nad1 genes appeared more conserved in either length or base composition with 135 parsimony-informative sites for cox2 and 93 variable sites for nad1 (no singleton sites for both genes; Additional file 1: Table S1). The mean A + T contents of cox2 and nad1 were 66.5% and 68.6%, respectively, a typical mitochondrial nucleotide feature in nematodes (AT bias). BLAST searches using either ITS, cox2 or nad1 sequence all assigned the 50 ascaridoid isolates into two groups, and one group had highest nucleotide identities to representative reference sequences for specimens of Toxocara (97.6–99.6% for ITS, 93.0–99.8% for cox2 and 91.3–98.6% for nad1) and another had highest nucleotide identities to representative reference sequences for specimens of Toxascaris (100% for ITS, 98.5% for cox2 and 98.9% for nad1). Within the Toxocara group, 5 of 13 ascaridoids shared the highest nucleotide identity with T. canis (99.6–99.9%; GenBank accession numbers JN617989 for ITS, JN593098 for cox2 and KC293917 for nad1) and the remaining 8 ascaridoids exhibited high nucleotide identities with T. cati (89.3–94.5%; GenBank accession numbers KY003079 for ITS, AM411622 for cox2 and KC200223 for nad1). For the Toxascaris group, all 37 ascaridoids showed high nucleotide identities with T. leonina (90.7–96.8%; GenBank accession numbers KR999999 for ITS, KC902750 for cox2 and KC293956 for nad1). Following the identity comparisons, the conserved and genus-specific nucleotide sites of ITS, cox2 and nad1were also identified by adding the congeneric species T. canis, T. cati, T. malaysiensis and T. vitulorum as well as T. leonina and other related species, Baylisascaris spp. and Ascaris spp. It was evident that both cox2 and nad1 exhibited more-stringent nucleotide sequence conservation than that of ITS. Thus, we mainly focused on the conserved sites in these two mitochondrial genes and detected their variable sites in the same regions as well, in order to determine if the base conservations were Toxocara and Toxascaris-specific and if there were non-synonymous substitutions apparent in these two genes by respective comparisons of their protein sequences in representative specimens. As shown in Additional file 2: Figure S1 and Additional file 3: Figure S2, we found that among the conserved base sites of cox2 9 were Toxocara-specific (50:G, 215:A, 376:A, 377:C, 439:G, 507:T, 563:T, 577:T and 579:G) and 3 were Toxascaris-specific (271:G, 273:A and 498:A); likewise, among the conserved base sites of nad1 3 were Toxocara-specific (71:C, 273:T and 276:G) and 3 were Toxascaris-specific (62:A, 262:G and 263:T). Among the variable base sites of cox2 and nad1, however, 13 (cox2) and 8 (nad1) were found to be unique for Toxocara spp. (in red), and 3 (cox2) and 5 (nad1) were unique for Toxascaris spp. (in blue). Importantly, among these variable sites 6 were confirmed to be non-synonymous substitutions based on respective protein alignments of cox2 and nad1, which lead to a total of 6 amino-acid changes, including 25:K/R/S(Val/Arg/Ser) → C(Ilu), 135:G(Gly) → S(Ser) and 167:V/I(Val/Ilu) → L(Leu) in cox2 and 25:V/F(Val/Phe) → I(Ilu), 89:L(Leu) → M(Met) and 104:L/F/I/C(Leu/Phe/Ilu/Cys) → V(Val) in nad1 (see Additional file 2: Figure S1, Additional file 3: Figure S2).
Evolutionary distance analysis
The evolutionary distances among the 11 representative specimens of Toxocara/Toxascaris and with other closely related ascaridoids were estimated and are shown in Additional file 4: Table S2 and Additional file 5: Table S3. Our analysis showed that among these Toxocara/Toxascaris isolates their evolutionary distances varied depending on the different genetic markers used. For example, the ascaridoid species in the Asian golden cat showed a minimum intraspecific evolutionary distance (0.036) with that in the jungle cat in ITS-based analysis while the value changed into 0.108 in the cox2 and 0.124 in the nad1 data. Nevertheless, the three gene datasets consistently placed ascaridoid species in either the arctic fox or red fox close to T. canis in the domestic dog, ascaridoid species in either the jungle cat, leopard cat or Asian golden cat close to T. cati in the domestic cat, and ascaridoid species in either the wolf, South China tiger, Eurasian lynx, African lion, Amur tiger or Bengal tiger close to T. leonina in the domestic dog or wild fox with the minimum intraspecific evolutionary distances of 0.002–0.004 for ITS, 0.003 for cox2 and 0.007–0.010 for nad1, 0.006–0.010 for ITS, 0.008–0.012 for cox2 and 0.003–0.017 for nad1, and 0.000–0.003 for ITS, 0.007–0.010 for cox2 and 0.003–0.010 for nad1, respectively, in accordance with conclusions of our identity analysis. More significant divergence was found in comparisons to T. vitulorum with 0.096–0.264 (ITS), 0.093–0.131 (cox2) and 0.127–0.193 (nad1), Ascaris spp. with 0.192–0.303 (ITS), 0.091–0.149 (cox1) and 0.153–0.181 (nad1) and Baylisascaris spp. with 0.190–0.307 (ITS), 0.097–0.173 (cox1) and 0.123–0.185 (nad1) (see Additional file 4: Table S4, Additional file 5: Table S3).
Phylogenetic analysis
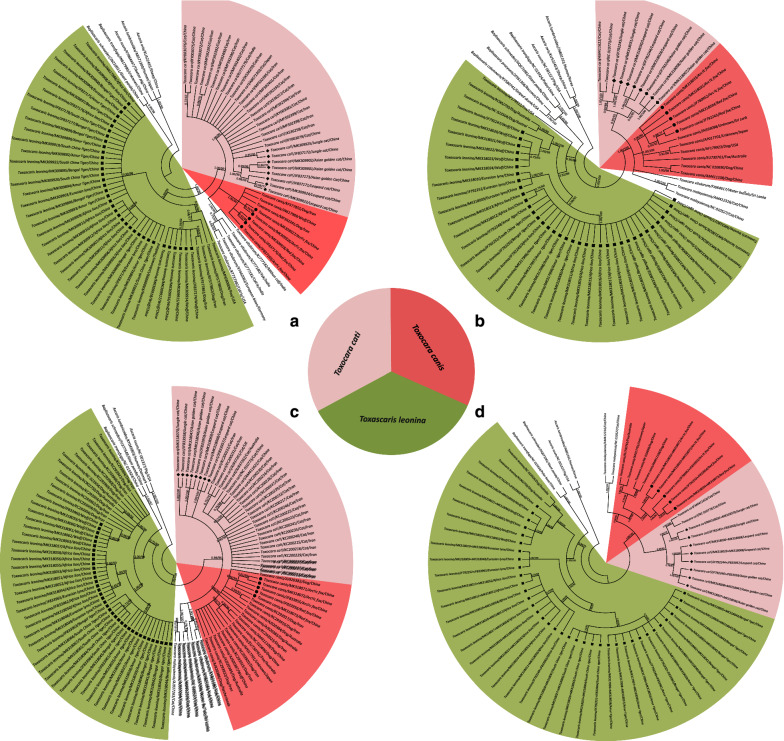
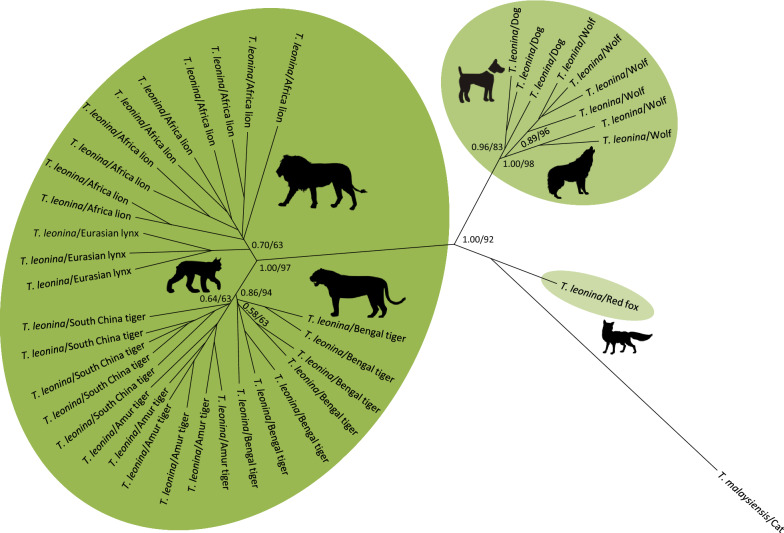
The phylogeny of the 50 specimens of Toxocara/Toxascaris and their relationships with other related ascaridoid species were inferred on the basis of the respective sequences of ITS, cox2 and nad1 as well as a combination of cox2 and nad1 using both BI and ML methods, and their corresponding tree topologies are showed in Fig. 2. Although four identical trees (BI/ML) topologically differed from each other because of genes and reference species included here, all analyses yielded a consistent, robust phylogenetic resolution for these 50 ascaridoid isolates and their congeneric species in the genera Toxocara in the family Toxocaridae and Toxascaris in the family Ascarididae. In total, three unequivocal clades including all ascaridoid isolates identified here were demonstrated, suggesting varying patterns of broad to relatively narrow host range. Among them, one clade placed the ascaridoid isolates of the arctic fox and red fox together with the dog/wolf/fox T. canis and showed a sister relationship with another clade that contained the ascaridoid isolates of the jungle cat, leopard cat and Asian golden cat as well as the cat T. cati, with almost maximum support values for three tree topologies (bootstrap values: 1.00/99/, 1.00/100, 0.98/96 or 1.00/100); we named these two clades as T. canis (red sectors) and T. cati (pink sectors) lineages, respectively (Fig. 2). In other words, the ascaridoids from the arctic fox and red fox were identified as T. canis and the ascaridoids from the jungle cat, leopard cat and Asian golden cat were identified as T. cati in this study. Combined with the congeneric T. malaysiensis and T. vitulorum lineages, these species showed a sister-group relationship in the genus Toxocara within Toxocaridae. Within the third clade, although the ascaridoid isolates of the wolf, South China tiger, Eurasian lynx, African lion, Amur tiger and Bengal tiger clustered with the dog/fox T. leonina with high statistical supports (bootstrap values: 1.00/99, 0.90/88, 1.00/98 or 1.00/100) and were referred as T. leonina lineage (green sectors), this lineage was more closely related to species of the Toxocaridae than other species of the Ascarididae in both ITS- and cox2-based analyses (Fig. 2a, b), in contrast with a close relationship between T. leonina and Baylisascaris spp. based on nad1 and cox2 plus nad1 analyses (Fig. 2c, d). Moreover, within the T. leonina lineage, it was notable that these T. leonina representatives appeared to belong to three distinct subclades depending to their host species, i.e. T. leonina from wild felids (such as lions, tigers and lynxes) in one subclade and T. leonina from canid hosts in another two subclades, including the dog/wolf-T. leonina subclade and fox-T. leonina subclade. Similar phylogenetic relationships were also supported when using a combination of the nuclear and mitochondrial data of T. leonina (Fig. 3), suggesting a cryptic speciation of T. leonina. For the inter-relationships of Baylisascaris spp. and Ascaris spp. and both with Toxocara spp. including T. canis, T. cati, T. malaysiensis and T. vitulorum, the phylogenetic topologies were in agreement with previously proposed molecular phylogenies of the ascaridoids based on nuclear and mitochondrial DNA data [33–37], confirming the phylogenetic stability of these paraphyletic groups characterised in this study.
Fig. 2.
Phylogenetic relationships of Toxocara/Toxascaris spp. from wild and domestic canine and feline carnivores and other related ascaridoid species. The phylogenetic topologies were inferred on the basis of nuclear ITS (a) and mitochondrial cox2 (b) and nad1 (c) as well as a combination of cox2 and nad1 (d) sequences using Bayesian inference (BI) and maximum likelihood (ML) methods. The reference sequences for species of Ascaris were used the outgroups. All species information about ascaridoid nematodes included in this study is summarized in Table 1. Species of Toxocara/Toxascaris recovered from wild and domestic canids and felids were genetically divided into three lineages, i.e. Toxocara canis, T. cati and Toxascaris leonina and indicated by three differently coloured sectors with T. canis in red, T. cati in pink and T. leonina in green. The numbers along the branches show bootstrap values resulting from different analyses in the order BI/ML; values < 50% are not shown
Fig. 3.
Inferred phylogenetic relationships among Toxascaris leonina isolates from different hosts by Bayesian inference (BI) and maximum likelihood (ML) methods based on a combination of the nuclear (ITS) and mitochondrial (cox2 + nad1) nucleotide sequence data, using Toxocara malaysiensis as the outgroup. The numbers along the branches show bootstrap values resulting from different analyses in the order BI/ML; values < 50% are not shown
Discussion
Toxocara and Toxascaris are the most common intestinal parasites of domestic and wild canids and felids and can cause human toxocariasis (mainly involving the species of Toxocara) with an important public health impact [1–4, 7, 8]. Although morphological characteristics and host preferences can be used for species identification, these existing keys and descriptions are often insufficient particularly when differentiation is required within and between Toxocara spp. and Toxascaris spp. and especially their larval and egg stages [1, 12, 13]. Similar problems may be exacerbated in attempts to perform species-specific identification among certain wild animal-derived species. Species of Toxocara and Toxascaris in wild canids and felids are considered not only because these wildlife hosts may have recently adapted to the human-environment due to rapid urbanization leading to increased interactions with people in conservation centers and zoological gardens, but also because little attention has been paid to these ascaridoids due to the limited access to samples [2, 38–41]. Such situations would negatively affect their diagnosis and surveillance. Recent advances in molecular diagnosis provide substantial opportunities for overcoming this problem. Polymerase chain reaction (PCR)-based approaches using genetic markers from rDNA and/or mtDNA have proven to be effective and have been used for large-scale studies on the identification and differentiation of ascaridoids to the species level [5, 6, 12, 15–17]. In this study, the ascaridoids from 11 captive wild canine and feline hosts were identified and characterised by PCR amplifying and genetically analyzing the nuclear ITS and the partial mitochondrial cox2 and nad1 genes.
Fifty ascaridoid isolates in this study were morphologically identified as Toxocara (n = 13) or Toxascaris (n = 37) according to the presence of a post-oesophageal bulbus and the length and shape of the cervical alae [26–28]. Further pairwise comparisons of ITS showed that among the 13 Toxocara isolates the ascaridoids in the arctic fox and red fox were found to share high nucleotide identities with representative references from specimens of T. canis, while the ascaridoids in the jungle cat, leopard cat and Asian golden cat had high nucleotide identities to representative references from specimens of T. cati in the cat. For the 37 Toxascaris isolates that were in the wolf, South China tiger, Eurasian lynx, African lion, Amur tiger and Bengal tiger, ITS identity analysis showed their high nucleotide identities with representative references from specimens of T. leonina. Thus, it can be assumed that there may be three ascaridoid species representing T. canis, T. cati and T. leonina among the ascaridoid isolates studied here. To confirm this assumption, ITS-based phylogenetic analyses (BI/ML) were performed and the results once again showed that the ascaridoids in the arctic fox and red fox grouped with T. canis, the ascaridoids in the jungle cat, leopard cat and Asian golden cat grouped with T. cati, and the ascaridoids in the wolf, South China tiger, Eurasian lynx, African lion, Amur tiger and Bengal tiger grouped with T. leonina, with high bootstrap values (see Fig. 2a), supporting that T. canis and T. leonina can co-occur in wild canids and T. cati and T. leonina can co-occur in wild felids [42–46]. Encouragingly, this conclusion was strengthened by analyses of mitochondrial cox2 and nad1. Both cox2 and nad1 were included because of their utility in identifying and differentiating nematode species among very closely related taxa due to their fast mutation rates, maternal inheritance and lack of recombination [16, 47, 48]. Similar to the nuclear ITS, high nucleotide identities of cox2 and nad1 were also observed between T. canis and the ascaridoids in either the arctic fox and red fox, between T. cati and the ascaridoids in either the jungle cat, leopard cat and Asian golden cat, and between T. leonina and the ascaridoids in either the wolf, South China tiger, Eurasian lynx, African lion, Amur tiger and Bengal tiger (see Additional file 2: Figure S1, Additional file 3: Figure S2). Importantly, consistent strong bootstrap support was evident, on the basis of phylogenetic analyses (MP/ML) of cox2 and nad1 that verified the same species of Toxocara in either the arctic fox and red fox (i.e. T. canis) or the jungle cat, leopard cat and Asian golden cat (i.e. T. cati), and the same species of Toxascaris in the wolf, South China tiger, Eurasian lynx, African lion, Amur tiger and Bengal tiger (i.e. T. leonina) (see Fig. 2b, d). Of course, possible cross-infection could be a confounder for this outcome because of the close or sympatric housing conditions for these captive wild canids and felids. This possibility was refuted because sampling records, particularly for the three species of the Felinae, showed that the jungle cat was housed in the Guiyang Wildlife Zoo (Guizhou, China) and the leopard cat in the Kunming Zoo (Yunnan, China), while the Asian golden cat was kept in the Chengdu Zoo (Sichuan, China). Besides, the captive history also showed that these host species were bred in zoos and no translocations and/or introductions occurred before, suggesting that these Toxocara/Toxascaris spp. were present with the original captive populations of these wild canids and felids and their own infection cycles may have been maintained over time.
Building on the results from integrated molecular evidence, we propose that Toxocara spp. from either wild canids or felids represent the same species but belong to different lineages in the genus Toxocara, i.e. Toxocara sp. of the arctic fox and red fox was within the T. canis lineage and Toxocara sp. of the jungle cat, leopard cat and Asian golden cat was within the T. cati lineage. In fact, since Jacobs et al. [18] first demonstrated the ascaridoid of the fox as T. canis, increased epidemiological studies have produced a consistent conclusion that T. canis is the common ascaridoid of foxes, regardless of fox species and originations [1, 49–52]. For instance, T. canis infections in red foxes have been reported in Great Britain (estimated prevalence: 62%) [45], Denmark (49%) [51], Switzerland (44%) [53], Norway (3.5–41.2%) [50], Ireland (20%) [54], Poland (11%) [55] and Italy (9%) [49]. Moreover, Meijer et al. [56] showed a prevalence of 7% and 30% of T. canis in arctic foxes during two summers (2008 and 2010, respectively) in Sweden. For T. cati, this parasite is commonly found in cats including wild species. Like T. cati in golden cats, jungle cats and leopard cats included here, T. cati infections have been reported in feral cats in Spain (35%) [57], stray cats in Iran (43%) [58] and wild cats in Egypt (59%) [59] and Italy (44%) [60]. Thus, taking into account the fact that canids and felids are natural hosts for T. canis and T. cati, respectively, and combining this with the aforementioned genetic evidence, our proposal that the same Toxocara species parasitizes either the arctic fox and red fox (T. canis) or the jungle cat, leopard cat and Asian golden cat (T. cati) is reasonable. Of course, additional information regarding ultrastructure and complete genomic information of these Toxocara species and other related ascaridoids as well as broader taxonomic comparisons is still required to provide an increasingly precise morphological and molecular basis for species recognition among ascaridoids.
In addition, our phylogenetic analyses placed Toxascaris sp. of the wolf, South China tiger, Eurasian lynx, African lion, Amur tiger and Bengal tiger into the T. leonina lineage (Fig. 2). It is clear that T. leonina is a cosmopolitan and polyxenical parasite in wild and domestic canids and felids [1]. However, recent studies by Fogt-Wyrwas et al. [12], Jin et al. [11] and Xie et al. [61] consistently pointed out that T. leonina may be a species complex. Because combined molecular evidence from ribosomal nuclear DNA strongly showed the separation of T. leonina from different hosts into three distinct clades, i.e. T. leonina isolates from wild felid hosts were in the same clade; T. leonina isolates from canid hosts including dogs and wolves formed another clade; while T. leonina isolates from foxes grouped together in a third clade. Our phylogenies (BI/ML) based on either ITS, cox2, nad1, cox2 plus nad1 or a combination of the nuclear (ITS) and mitochondrial (cox2 + nad1) data supported this split in which T. leonina specimens clustered into three subclades depending on their host species, i.e. foxes, wolfs and dogs and wild felids (including lions, tigers and lynxes; Figs. 2, 3). Such species complex phenomenon was also described in whipworms of primates. Thus, Liu et al. [62] and Hawash et al. [63] proposed that there may be several Trichuris species parasitizing primates, with some species only infecting non-human primates and others infecting both non-human primates and humans, on the basis of comparative mitogenomics. Variation in host range mirrors the interaction of trends in generalisation and specialization, the opportunity for host colonisation and the capacity to establish infection as reflected in outcomes for ecological fitting in sloppy fitness space [64, 65]. Moreover, faunal mixing and the potential for exchange can be also facilitated in zoopark environments that may bring phylogenetically disparate assemblages of hosts and their parasites into proximity [66]. For parasites with a direct life-cycle such as Toxascaris, this establishes the potential for considerable opportunities for host colonisation that may not be apparent in natural settings, and such extensive colonisation processes may further result in the possibility of new disease through the exchange of parasites over time. In addition, there were four species-specific non-synonymous base substitutions detected in cox2 and nad1 genes of T. leonina in the South China tiger, Eurasian lynx, African lion, Amur tiger and Bengal tiger (see Additional file 2: Figure S1, Additional file 3: Figure S2) that were further confirmed to be fixed after homologous comparisons with other congeneric species including that in the wolf.
As a part of epidemiological surveys of captive wild canids and felids in the southwestern zoos of China, this study focused on species diversity among Toxocara/Toxascaris spp. and raised some questions: (i) what routes are being possibly used for parasite introduction and establishment (natural or cross infections); (ii) what approaches are the best choices for surveillance of parasite infections, especially when various cryptic species may be involved; (iii) what strategies are the most reasonable interventions for prevention and control of parasite transport in the space-limited and artificial zoos; and (iv) what issues are the limits and facilitators for zoonotic risk. Given the fact that the recognised host range (defined by opportunity) of one parasite is almost always a subset of the actual host range (defined by capacity to use host-based resources), to some extent, this study also sets a quasi-experimental example to illustrate the roles of isolation and ecological fitting in expansions of host range. Such processes are most often related emerging infectious diseases in the context of ecological disruption, suggesting direct lessons about the potential versus realised host range for Toxocara/Toxascaris spp. and the risk of zoonotic infections. Therefore, it is urgent and necessary to clarify the species of Toxocara/Toxascaris which infect wild and domestic canids and felids in order to uncover transmission routes and develop suitable prevention and control measures.
Conclusions
In the present study, based on the combined analysis of the nuclear and mitochondrial datasets, we suggest that at least three genetically distinct ascaridoid lineages are parasitizing wild canine and feline animals, i.e. T. canis in the arctic fox and red fox, T. cati in the jungle cat, leopard cat and Asian golden cat and T. leonina in the wolf, South China tiger, Eurasian lynx, African lion, Amur tiger and Bengal tiger. Further evidence derived from genetic distance analysis and phylogenies showed that there was a separation of T. leonina from different hosts, followed by the pattern that isolates from wild felids were in one subclade; isolates from dogs and wolves in another subclade and isolates from foxes in a third subclade, supporting a complex of T. leonina infecting these hosts. Of course, additional confirmation by further broad sampling and extensive morphological and genetic comparisons are required. Taken together, the results presented here yield new molecular insights into the classification, phylogenetic relationships and epidemiological importance of ascaridoids from wild canids and felids and also highlight the complex of the taxonomy and genetics of Toxascaris in their wild and domestic carnivorous hosts.
Supplementary information
Additional file 1: Table S1. Nucleotide variability of the nuclear ITS and mitochondrial cox2 and nad1 genes of Toxocara/Toxascaris spp. identified in this study. Abbreviations: C, conserved sites; V, variable sites; Pi, parsimony-informative sites; S, singleton sites.
Additional file 2: Figure S1. A simultaneous alignment of nucleotide and amino acid sequences of partial mitochondrial cox1 genes of 11 representative isolates of Toxocara/Toxascaris identified in this study and other related ascaridoid species.
Additional file 3: Figure S2. A simultaneous alignment of nucleotide and amino acid sequences of partial mitochondrial nad1 genes of 11 representative isolates of Toxocara/Toxascaris identified in this study and other related ascaridoid species.
Additional file 4: Table S2. Estimates of evolutionary distance between ascaridoid species recovered from different host species using the nuclear ITS. Evolutionary distances between 11 wild animals included in this study are highlighted in bold for ITS-based estimates. Given almost identical nucleotide sequences of ITS regions in worms from the same host species, 11 representative specimens were used to calculate evolutionary distances using a maximum composite likelihood model.
Additional file 5: Table S3. Estimates of evolutionary distance between ascaridoid species recovered from different host species using the mitochondrial cox2 (below diagonal) and nad1 (above diagonal). Evolutionary distances between 11 wild animals included in this study are highlighted in bold for cox2- and nad1-based estimates, respectively. Given almost identical nucleotide sequences of cox2 or nad1 gene in worms from the same host species, 11 representative specimens were used to calculate evolutionary distances using a maximum composite likelihood model.
Acknowledgements
We thank the staff of the Veterinary Departments of Guiyang Wildlife Zoo (Guizhou, China), Kunming Zoo (Yunnan, China) and Chengdu Zoo (Sichuan, China) for their assistance in collecting parasite materials; and Dr. Joseph F. Urban from USDA-ARS, Beltsville Human Nutrition Research Center, Diet, Genomics and Immunology Laboratory for reviewing the manuscript and advice.
Abbreviations
- ITS
internal transcribed spacer region
- ITS2
second internal transcribed spacer region
- cox2
cytochrome c oxidase subunit 2
- cox1
cytochrome c oxidase subunit 1
- nad1
NADH dehydrogenase subunit 1
- nad4
NADH dehydrogenase subunit 4
- rDNA
nuclear ribosomal DNA
- mtDNA
mitochondrial DNA
- gDNA
genomic DNA
- VLM
visceral larva migrans
- OLM
ocular larva migrans
- EME
eosinophilic meningoencephalitis
- CT
covert toxocariasis
- NT
neurological toxocariasis
- PCR
polymerase chain reaction
- K2P
Kimura 2-parameter
- BI
Bayesian inference
- ML
maximum likelihood
- GTR
general time reversible
- indels
insertions/deletions
Authors’ contributions
YX and GYY conceived and designed the experiments. YX, YXL and XBG performed the experiments, including PCR and sequencing. YX, YXL, XBG, YJL and LW achieved the data analysis. XZ, RH and XRP contributed reagents/materials/analysis tools. YX and GYY wrote the initial manuscript. All authors critically revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Sichuan International Science and Technology Innovation Cooperation/Hong Kong, Macao and Taiwan Science and Technology Innovation Cooperation Project, Sichuan, China (grant no. 2019YFH0065), Key Open Laboratory of Conservation Biology of Rare Animals in Giant Panda National Park, State Forestry and Grassland Administration (no. KLSFGAGP2020.014) and the High-level Scientific Research Foundation for the Introduction of Talents of Sichuan Agricultural University (no. 03120322). The funders had no role in design, decision to publish, or preparation of the manuscript.
Availability of data and materials
Data supporting the conclusions of this article are included within the article and its additional files. Nucleotide sequences reported in this article are available in the GenBank database under the accession numbers JF837169-JF837179 and MK309890-MK309928 for ITS, JF780952-JF792252 and MK317996-MK318034 for cox2 and JF833955-JF833965 and MK318035-MK318073 for nad1.
Ethics approval and consent to participate
This study was approved by the Animal Ethics Committee of Sichuan Agricultural University (Sichuan, China; approval no. SYXK 2014-187) and the Wildlife Management and Animal Welfare Committee of China, and all procedures involving animals in the present study were in strict accordance with the Guide for the Care and Use of Laboratory Animals (National Research Council, Bethesda, MD, USA) and the recommendations in the ARRIVE guidelines (https://www.nc3rs.org.uk/arrive-guidelines).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yue Xie, Yingxin Li and Xiaobin Gu contributed equally to this work
Contributor Information
Yue Xie, Email: xyue1985@gmail.com.
Yingxin Li, Email: liyx1031@outlook.com.
Xiaobin Gu, Email: guxiaobin198225@126.com.
Yunjian Liu, Email: liuyunjianjishengchong@hotmail.com.
Xuan Zhou, Email: zhouxuan198866@163.com.
Lu Wang, Email: wanglu1770870@hotmail.com.
Ran He, Email: ranhe1991@hotmail.com.
Xuerong Peng, Email: pxuerong@aliyun.com.
Guangyou Yang, Email: guangyou1963@aliyun.com.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13071-020-04254-4.
References
- 1.Okulewicz A, Perec-Matysiak A, Buńkowska K, Hildebrand J. Toxocara canis, Toxocara cati and Toxascaris leonina in wild and domestic carnivores. Helminthologia. 2012;49:3–10. [Google Scholar]
- 2.Ma G, Holland CV, Wang T, Hofmann A, Fan CK, Maizels RM, Hotez PJ, Gasser RB. Human toxocariasis. Lancet Infect Dis. 2018;18:e14–e24. doi: 10.1016/S1473-3099(17)30331-6. [DOI] [PubMed] [Google Scholar]
- 3.Moreira GM, Telmo Pde L, Mendonça M, Moreira AN, McBride AJ, Scaini CJ, Conceição FR. Human toxocariasis: current advances in diagnostics, treatment, and interventions. Trends Parasitol. 2014;30:456–464. doi: 10.1016/j.pt.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Holland C. Knowledge gaps in the epidemiology of Toxocara: the enigma remains. Parasitology. 2017;144:81–94. doi: 10.1017/S0031182015001407. [DOI] [PubMed] [Google Scholar]
- 5.Gasser RB. A perfect time to harness advanced molecular technologies to explore the fundamental biology of Toxocara species. Vet Parasitol. 2013;193:353–364. doi: 10.1016/j.vetpar.2012.12.031. [DOI] [PubMed] [Google Scholar]
- 6.Chen J, Zhou DH, Nisbet AJ, Xu MJ, Huang SY, Li MW, et al. Advances in molecular identification, taxonomy, genetic variation and diagnosis of Toxocara spp. Infect Genet Evol. 2012;12:1344–1348. doi: 10.1016/j.meegid.2012.04.019. [DOI] [PubMed] [Google Scholar]
- 7.Hajipour N. A survey on the prevalence of Toxocara cati, Toxocara canis and Toxascaris leonina eggs in stray dogs and cats’ faeces in Northwest of Iran: a potential risk for human health. Trop Biomed. 2019;36:143–151. [PubMed] [Google Scholar]
- 8.Macpherson CN. The epidemiology and public health importance of toxocariasis: a zoonosis of global importance. Int J Parasitol. 2013;43:999–1008. doi: 10.1016/j.ijpara.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 9.Lee AC, Schantz PM, Kazacos KR, Montgomery SP, Bowman DD. Epidemiologic and zoonotic aspects of ascarid infections in dogs and cats. Trends Parasitol. 2010;26:155–161. doi: 10.1016/j.pt.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 10.Le TH, Anh NT, Nguyen KT, Nguyen NT, Thuy do TT, Gasser RB. Toxocara malaysiensis infection in domestic cats in Vietnam—an emerging zoonotic issue? Infect Genet Evol. 2016;37:94–98. doi: 10.1016/j.meegid.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 11.Jin YC, Li XY, Liu JH, Zhu XQ, Liu GH. Comparative analysis of mitochondrial DNA datasets indicates that Toxascaris leonina represents a species complex. Parasit Vectors. 2019;12:194. doi: 10.1186/s13071-019-3447-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fogt-Wyrwas R, Dabert M, Jarosz W, Rząd I, Pilarczyk B, Mizgajska-Wiktor H. Molecular data reveal cryptic speciation and host specificity in Toxascaris leonina (Nematoda: Ascarididae) Vet Parasitol. 2019;266:80–83. doi: 10.1016/j.vetpar.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Gasser R, Zhu XQ, Hu M, Jacobs DE, Chilton NB. Molecular genetic characterization of members of the genus Toxocara—taxonomic, population genetic and epidemiological considerations. In: Holland CV, Smith HV, editors. Toxocara: the enigmatic parasite. Wallingford: CABI Publishing; 2006. pp. 18–31. [Google Scholar]
- 14.Fogt-Wyrwas R, Jarosz W, Rzad I, Pilarczyk B, Mizgajska-Wiktor H. Variability in sequences of mitochondrial cox1 and nadh1 genes in Toxocara canis, Toxocara cati, and Toxascaris leonina (Nematoda: Toxocaridae) from different hosts. Ann Parasitol. 2016;62:48. [Google Scholar]
- 15.Li M, Lin R, Chen H, Sani R, Song H, Zhu X. PCR tools for the verification of the specific identity of ascaridoid nematodes from dogs and cats. Mol Cell Probes. 2007;21:349–354. doi: 10.1016/j.mcp.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 16.Li MW, Lin RQ, Song HQ, Sani RA, Wu XY, Zhu XQ. Electrophoretic analysis of sequence variability in three mitochondrial DNA regions for ascaridoid parasites of human and animal health significance. Electrophoresis. 2008;29:2912–2917. doi: 10.1002/elps.200700752. [DOI] [PubMed] [Google Scholar]
- 17.Zhu X, Jacobs D, Chilton N, Sani R, Cheng N, Gasser R. Molecular characterization of a Toxocara variant from cats in Kuala Lumpur, Malaysia. Parasitology. 1998;117:155–164. doi: 10.1017/s0031182098002856. [DOI] [PubMed] [Google Scholar]
- 18.Jacobs DE, Zhu X, Gasser RB, Chilton NB. PCR-based methods for identification of potentially zoonotic ascaridoid parasites of the dog, fox and cat. Acta Trop. 1997;68:191–200. doi: 10.1016/s0001-706x(97)00093-4. [DOI] [PubMed] [Google Scholar]
- 19.Vega R, Prous CG, Krivokapich S, Gatti G, Brugni N, Semenas L. Toxocariasis in Carnivora from Argentinean Patagonia: species molecular identification, hosts, and geographical distribution. Int J Parasitol Parasites Wildl. 2018;7:106–110. doi: 10.1016/j.ijppaw.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu M, Gasser RB. Mitochondrial genomes of parasitic nematodes—progress and perspectives. Trends Parasitol. 2006;22:78–84. doi: 10.1016/j.pt.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 21.Mikaeili F, Mirhendi H, Mohebali M, Hosseini M, Sharbatkhori M, Zarei Z, et al. Sequence variation in mitochondrial cox1 and nad1 genes of ascaridoid nematodes in cats and dogs from Iran. J Helminthol. 2015;89:496–501. doi: 10.1017/S0022149X14000133. [DOI] [PubMed] [Google Scholar]
- 22.Nadler SA, Hudspeth DS. Phylogeny of the Ascaridoidea (Nematoda: Ascaridida) based on three genes and morphology: hypotheses of structural and sequence evolution. J Parasitol. 2000;86:380–393. doi: 10.1645/0022-3395(2000)086[0380:POTANA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 23.Li MW, Zhu XQ, Gasser RB, Lin RQ, Sani RA, Lun ZR, et al. The occurrence of Toxocara malaysiensis in cats in China, confirmed by sequence-based analyses of ribosomal DNA. Parasitol Res. 2006;99:554–557. doi: 10.1007/s00436-006-0194-z. [DOI] [PubMed] [Google Scholar]
- 24.Li Y, Niu L, Wang Q, Zhang Z, Chen Z, Gu X, et al. Molecular characterization and phylogenetic analysis of ascarid nematodes from twenty-one species of captive wild mammals based on mitochondrial and nuclear sequences. Parasitology. 2012;139:1329–1338. doi: 10.1017/S003118201200056X. [DOI] [PubMed] [Google Scholar]
- 25.Sheng ZH, Chang QC, Tian SQ, Lou Y, Zheng X, Zhao Q, et al. Characterization of Toxascaris leonina and Tococara canis from cougar (Panthera leo) and common wolf (Canis lupus) by nuclear ribosomal DNA sequences of internal transcribed spacers. Afr J Microbiol Res. 2012;6:3545–3549. [Google Scholar]
- 26.Jenkins EJ. Toxocara spp. in dogs and cats in Canada. Adv Parasitol. 2020;109:641–653. doi: 10.1016/bs.apar.2020.01.026. [DOI] [PubMed] [Google Scholar]
- 27.Taylor MA, Coop RL, Wall RL. Veterinary parasitology. Oxford: Blackwell Publishing; 2007. [Google Scholar]
- 28.Warren G. Studies on the morphology and taxonomy of the genera Toxocara Stiles, 1905 and Neoascaris Travassos, 1927. Zool Anz. 1970;185:393–442. [Google Scholar]
- 29.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 33.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li K, Yang F, Abdullahi A, Song M, Shi X, Wang M, et al. Sequence analysis of mitochondrial genome of Toxascaris leonina from a South China Tiger. Korean J Parasitol. 2016;54:803–807. doi: 10.3347/kjp.2016.54.6.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu GH, Zhou DH, Zhao L, Xiong RC, Liang JY, Zhu XQ. The complete mitochondrial genome of Toxascaris leonina: comparison with other closely related species and phylogenetic implications. Infect Genet Evol. 2014;21:329–333. doi: 10.1016/j.meegid.2013.11.022. [DOI] [PubMed] [Google Scholar]
- 36.Xie Y, Niu L, Zhao B, Wang Q, Nong X, Chen L, et al. Complete mitochondrial genomes of chimpanzee-and gibbon-derived Ascaris isolated from a zoological garden in southwest China. PLoS ONE. 2013;8:e82795. doi: 10.1371/journal.pone.0082795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie Y, Zhang Z, Wang C, Lan J, Li Y, Chen Z, et al. Complete mitochondrial genomes of Baylisascaris schroederi, Baylisascaris ailuri and Baylisascaris transfuga from giant panda, red panda and polar bear. Gene. 2011;482:59–67. doi: 10.1016/j.gene.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 38.Bradley CA, Altizer S. Urbanization and the ecology of wildlife diseases. Trends Ecol Evol. 2007;22:95–102. doi: 10.1016/j.tree.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fryxell JM, Sinclair AR, Caughley G. Wildlife ecology, conservation, and management. New York: Wiley; 2014. [Google Scholar]
- 40.Mackenstedt U, Jenkins D, Romig T. The role of wildlife in the transmission of parasitic zoonoses in peri-urban and urban areas. Int J Parasitol Parasites Wildl. 2015;4:71–79. doi: 10.1016/j.ijppaw.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie Y, Hoberg EP, Yang Z, Urban JF, Yang G. Ancylostoma ailuropodae n. sp. (Nematoda: Ancylostomatidae), a new hookworm parasite isolated from wild giant pandas in Southwest China. Parasites Vectors. 2017;10:277. doi: 10.1186/s13071-017-2209-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Antolová D, Reiterová K, Miterpáková M, Stanko M, Dubinský P. Circulation of Toxocara spp. in suburban and rural ecosystems in the Slovak Republic. Vet Parasitol. 2004;126:317–324. doi: 10.1016/j.vetpar.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 43.Criado-Fornelio A, Gutierrez-Garcia L, Rodriguez-Caabeiro F, Reus-Garcia E, Roldan-Soriano M, Diaz-Sanchez M. A parasitological survey of wild red foxes (Vulpes vulpes) from the province of Guadalajara, Spain. Vet Parasitol. 2000;92:245–251. doi: 10.1016/s0304-4017(00)00329-0. [DOI] [PubMed] [Google Scholar]
- 44.Popiołek M, Szczęsna J, Nowak S, Mysłajek RW. Helminth infections in faecal samples of wolves Canis lupus L. from the western Beskidy Mountains in southern Poland. J Helminthol. 2007;81:339–344. doi: 10.1017/s0022149x07821286. [DOI] [PubMed] [Google Scholar]
- 45.Smith G, Gangadharan B, Taylor Z, Laurenson M, Bradshaw H, Hide G, et al. Prevalence of zoonotic important parasites in the red fox (Vulpes vulpes) in Great Britain. Vet Parasitol. 2003;118:133–142. doi: 10.1016/j.vetpar.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 46.Torres J, Garcia-Perea R, Gisbert J, Feliu C. Helminth fauna of the Iberian lynx, Lynx pardinus. J Helminthol. 1998;72:221–226. doi: 10.1017/s0022149x00016473. [DOI] [PubMed] [Google Scholar]
- 47.Blouin MS, Yowell CA, Courtney CH, Dame JB. Substitution bias, rapid saturation, and the use of mtDNA for nematode systematics. Mol Biol Evol. 1998;15:1719–1727. doi: 10.1093/oxfordjournals.molbev.a025898. [DOI] [PubMed] [Google Scholar]
- 48.Hu M, Chilton NB, Gasser RB. The mitochondrial genomics of parasitic nematodes of socio-economic importance: recent progress, and implications for population genetics and systematics. Adv Parasitol. 2004;56:134–213. doi: 10.1016/s0065-308x(03)56003-1. [DOI] [PubMed] [Google Scholar]
- 49.Magi M, Macchioni F, Dell’Omodarme M, Prati M, Calderini P, Gabrielli S, et al. Endoparasites of red fox (Vulpes vulpes) in central Italy. J Wildl Dis. 2009;45:881–885. doi: 10.7589/0090-3558-45.3.881. [DOI] [PubMed] [Google Scholar]
- 50.Mørk T, Ims RA, Killengreen ST. Rodent population cycle as a determinant of gastrointestinal nematode abundance in a low-arctic population of the red fox. Int J Parasitol Parasites Wildl. 2019;9:36–41. doi: 10.1016/j.ijppaw.2019.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saeed I, Maddox-Hyttel C, Monrad J, Kapel CM. Helminths of red foxes (Vulpes vulpes) in Denmark. Vet Parasitol. 2006;139:168–179. doi: 10.1016/j.vetpar.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 52.Stien A, Voutilainen L, Haukisalmi V, Fuglei E, Mørk T, Yoccoz N, et al. Intestinal parasites of the Arctic fox in relation to the abundance and distribution of intermediate hosts. Parasitology. 2010;137:149–157. doi: 10.1017/S0031182009990953. [DOI] [PubMed] [Google Scholar]
- 53.Reperant LA, Hegglin D, Fischer C, Kohler L, Weber J-M, Deplazes P. Influence of urbanization on the epidemiology of intestinal helminths of the red fox (Vulpes vulpes) in Geneva, Switzerland. Parasitol Res. 2007;101:605–611. doi: 10.1007/s00436-007-0520-0. [DOI] [PubMed] [Google Scholar]
- 54.Strube C, Heuer L, Janecek E. Toxocara spp. infections in paratenic hosts. Vet Parasitol. 2013;193:375–389. doi: 10.1016/j.vetpar.2012.12.033. [DOI] [PubMed] [Google Scholar]
- 55.Borecka A, Gawor J, Zieba F. A survey of intestinal helminths in wild carnivores from the Tatra National Park, southern Poland. Ann Parasitol. 2013;59:169–172. [PubMed] [Google Scholar]
- 56.Meijer T, Mattsson R, Angerbjörn A, Osterman-Lind E, Fernández-Aguilar X, Gavier-Widén D. Endoparasites in the endangered Fennoscandian population of arctic foxes (Vulpes lagopus) Eur J Wildl Res. 2011;57:923–927. [Google Scholar]
- 57.Millán J, Casanova JC. High prevalence of helminth parasites in feral cats in Majorca Island (Spain) Parasitol Res. 2009;106:183–188. doi: 10.1007/s00436-009-1647-y. [DOI] [PubMed] [Google Scholar]
- 58.Zibaei M, Sadjjadi SM, Sarkari B. Prevalence of Toxocara cati and other intestinal helminths in stray cats in Shiraz, Iran. Trop Biomed. 2007;24:39–43. [PubMed] [Google Scholar]
- 59.Radwan NA, Khalil AI, El Mahi RA. Morphology and occurrence of species of Toxocara in wild mammal populations from Egypt. Comp Parasitol. 2009;76:273–283. [Google Scholar]
- 60.Napoli E, Anile S, Arrabito C, Scornavacca D, Mazzamuto MV, Gaglio G, et al. Survey on parasitic infections in wildcat (Felis silvestris silvestris Schreber, 1777) by scat collection. Parasitol Res. 2016;115:255–261. doi: 10.1007/s00436-015-4742-2. [DOI] [PubMed] [Google Scholar]
- 61.Xie Y, Li H, Wang C, Li Y, Liu Y, Meng X, et al. Characterization of the complete mitochondrial genome sequence of the dog roundworm Toxascaris leonina (Nematoda, Ascarididae) from China. Mitochondrial DNA B Resour. 2019;4:3517–3519. doi: 10.1080/23802359.2019.1675545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu GH, Gasser RB, Nejsum P, Wang Y, Chen Q, Song HQ, et al. Mitochondrial and nuclear ribosomal DNA evidence supports the existence of a new Trichuris species in the endangered françois’ leaf-monkey. PLoS ONE. 2013;8:e66249. doi: 10.1371/journal.pone.0066249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hawash MB, Andersen LO, Gasser RB, Stensvold CR, Nejsum P. Mitochondrial genome analyses suggest multiple Trichuris species in humans, baboons, and pigs from different geographical regions. PLoS Negl Trop Dis. 2015;9:e0004059. doi: 10.1371/journal.pntd.0004059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Agosta SJ, Janz N, Brooks DR. How specialists can be generalists: resolving the “parasite paradox” and implications for emerging infectious disease. Zoologia. 2010;27:151–162. [Google Scholar]
- 65.Araujo SB, Braga MP, Brooks DR, Agosta SJ, Hoberg EP, von Hartenthal FW, et al. Understanding host-switching by ecological fitting. PLoS ONE. 2015;10:e0139225. doi: 10.1371/journal.pone.0139225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xie Y, Zhao B, Hoberg EP, Li M, Zhou X, Gu X, et al. Genetic characterisation and phylogenetic status of whipworms (Trichuris spp.) from captive non-human primates in China, determined by nuclear and mitochondrial sequencing. Parasites Vectors. 2018;11:516. doi: 10.1186/s13071-018-3100-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hoberg EP, Burek-Huntington K, Beckmen K, Camp LE, Nadler SA. Transuterine infection by Baylisascaris transfuga: neurological migration and fatal debilitation in sibling moose calves (Alces alces gigas) from Alaska. Int J Parasitol Parasites Wildl. 2018;7:280–288. doi: 10.1016/j.ijppaw.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arizono N, Yoshimura Y, Tohzaka N, Yamada M, Tegoshi T, Onishi K, et al. Ascariasis in Japan: is pig-derived Ascaris infecting humans. Jpn J Infect Dis. 2010;63:447–448. [PubMed] [Google Scholar]
- 69.Li MW, Lin RQ, Song HQ, Wu XY, Zhu XQ. The complete mitochondrial genomes for three Toxocara species of human and animal health significance. BMC Genomics. 2008;9:224. doi: 10.1186/1471-2164-9-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.He X, Lv MN, Liu GH, Lin RQ. Genetic analysis of Toxocara cati (Nematoda: Ascarididae) from Guangdong province, subtropical China. Mitochondrial DNA A DNA Mapp Seq Anal. 2018;29:132–135. doi: 10.1080/24701394.2016.1258404. [DOI] [PubMed] [Google Scholar]
- 71.Jex AR, Waeschenbach A, Littlewood DTJ, Hu M, Gasser RB. The mitochondrial genome of Toxocara canis. PLoS Negl Trop Dis. 2008;2:e273. doi: 10.1371/journal.pntd.0000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wickramasinghe S, Yatawara L, Agatsuma T. Toxocara canis (Ascaridida: Nematoda): mitochondrial gene content, arrangement and composition compared with other Toxocara species. Sri Lankan J Infect Dis. 2014;4:90–98. doi: 10.1016/j.molbiopara.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 73.Choobineh M, Mikaeili F, Sadjjadi S, Ebrahimi S, Iranmanesh S. Molecular characterization of Toxocara spp. eggs isolated from public parks and playgrounds in Shiraz, Iran. J Helminthol. 2019;93:306–312. doi: 10.1017/S0022149X18000354. [DOI] [PubMed] [Google Scholar]
- 74.Li K, Lan Y, Luo H, Zhang H, Liu D, Zhang L, et al. Prevalence, associated risk factors, and phylogenetic analysis of Toxocara vitulorum infection in yaks on the Qinghai Tibetan plateau, China. Korean J Parasitol. 2016;54:645–652. doi: 10.3347/kjp.2016.54.5.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wickramasinghe S, Yatawara L, Rajapakse R, Agatsuma T. Toxocara vitulorum (Ascaridida: Nematoda): mitochondrial gene content, arrangement and composition compared with other Toxocara species. Mol Biochem Parasitol. 2009;166:89–92. doi: 10.1016/j.molbiopara.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 76.Choi Y, Mason S, Ahlborn M, Zscheile B, Wilson E. Partial molecular characterization of the mitochondrial genome of Baylisascaris columnaris and prevalence of infection in a wild population of striped skunks. Int J Parasitol Parasites Wildl. 2017;6:70–75. doi: 10.1016/j.ijppaw.2017.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lin Q, Li H, Gao M, Wang X, Ren W, Cong M, et al. Characterization of Baylisascaris schroederi from Qinling subspecies of giant panda in China by the first internal transcribed spacer (ITS-1) of nuclear ribosomal DNA. Parasitol Res. 2012;110:1297–1303. doi: 10.1007/s00436-011-2618-7. [DOI] [PubMed] [Google Scholar]
- 78.Li K, Luo H, Zhang H, Mehmood K, Shahzad M, Zhang L, et al. Analysis of the internal transcribed spacer region of Ascaris suum and Ascaris lumbricoides derived from free range Tibetan pigs. Mitochondrial DNA A DNA Mapp Seq Anal. 2018;29:624–628. doi: 10.1080/24701394.2017.1331348. [DOI] [PubMed] [Google Scholar]
- 79.Wolstenholme DR, Okimoto R, Macfarlane JL. Nucleotide correlations that suggest tertiary interactions in the TV-replacement loop-containing mitochondrial tRNAs of the nematodes, Caenorhabditis elegans and Ascaris suum. Nucleic Acids Res. 1994;22:4300–4306. doi: 10.1093/nar/22.20.4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Park YC, Kim W, Park JK. The complete mitochondrial genome of human parasitic roundworm, Ascaris lumbricoides. Mitochondrial DNA. 2011;22:91–93. doi: 10.3109/19401736.2011.624608. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Nucleotide variability of the nuclear ITS and mitochondrial cox2 and nad1 genes of Toxocara/Toxascaris spp. identified in this study. Abbreviations: C, conserved sites; V, variable sites; Pi, parsimony-informative sites; S, singleton sites.
Additional file 2: Figure S1. A simultaneous alignment of nucleotide and amino acid sequences of partial mitochondrial cox1 genes of 11 representative isolates of Toxocara/Toxascaris identified in this study and other related ascaridoid species.
Additional file 3: Figure S2. A simultaneous alignment of nucleotide and amino acid sequences of partial mitochondrial nad1 genes of 11 representative isolates of Toxocara/Toxascaris identified in this study and other related ascaridoid species.
Additional file 4: Table S2. Estimates of evolutionary distance between ascaridoid species recovered from different host species using the nuclear ITS. Evolutionary distances between 11 wild animals included in this study are highlighted in bold for ITS-based estimates. Given almost identical nucleotide sequences of ITS regions in worms from the same host species, 11 representative specimens were used to calculate evolutionary distances using a maximum composite likelihood model.
Additional file 5: Table S3. Estimates of evolutionary distance between ascaridoid species recovered from different host species using the mitochondrial cox2 (below diagonal) and nad1 (above diagonal). Evolutionary distances between 11 wild animals included in this study are highlighted in bold for cox2- and nad1-based estimates, respectively. Given almost identical nucleotide sequences of cox2 or nad1 gene in worms from the same host species, 11 representative specimens were used to calculate evolutionary distances using a maximum composite likelihood model.
Data Availability Statement
Data supporting the conclusions of this article are included within the article and its additional files. Nucleotide sequences reported in this article are available in the GenBank database under the accession numbers JF837169-JF837179 and MK309890-MK309928 for ITS, JF780952-JF792252 and MK317996-MK318034 for cox2 and JF833955-JF833965 and MK318035-MK318073 for nad1.