Abstract
Despite great advancement in genetic typing, phenotyping is still an indispensable tool for categorization of bacteria. Certain amino acids may be essential for bacterial survival, growth, pathogenicity or toxin production, which prompts the idea that the intrinsic ability to utilize single amino acid under live-or-die situation could be a basis for differentiation of bacteria species. In this study, we determined the single amino acid consumption profiles of 7 bacterial species, and demonstrated that most bacteria have species-specific pattern of amino acid consumption. We also discovered that bacterial strains from different hosts, toxigenicity, and antibiotic-resistance presented distinct preference for certain amino acids. Taken altogether, the amino acid consumption profiles showed potential to be a novel tool complementary to study not only bacterial categorization but also biochemical characteristics of the bacteria such that its phenotyping can be used to uncover strategies for nutritional, pharmaceutical, taxonomic, and evolutionary aspects of bacterial researches.
Subject terms: Bacteria, Bacteriology
Introduction
Bacterial identification is crucial in a wide variety of applications including disease diagnosis, food safety, and environmental monitoring. In general, two distinct methods are used for bacterial identification; genotyping and the phenotyping. Genotypic methods such as DNA sequencing, refer to discrimination of bacterial strains based on their genetic content1. On the other hand, phenotypic traits are based on the characteristics expressed by bacterial strains, which comprise morphological, physiological, and biochemical features. For example, Analytical Profile Index (API) and Biolog system are commonly used phenotypic methods which rely on the utilization and metabolism of simple chemical compounds2,3. Although genotyping has recently become mainstream for bacterial strain typing, phenotypic methods still play an important role in many cases and provide crucial insights into bacterial strains. For instance, about two-thirds of genes from bacterial to mammalian cells have been shown to possess biochemical function, but only a small fraction of these genes are known to be associated with phenotypes4,5. Therefore, more phenotypic information is desirable in understanding the roles of those inexplicable genes6,7 and to discover their resultant behaviors in relations to pathology, epidemiology, and ecology.
Amino acids (AAs) are critical substances for nitrogen and carbon metabolism in bacteria. Different bacteria may have different abilities to utilize AAs depending on their genetic and adopted differences as well as the nutritional availability from the environment, rendering significant variations in their phenotypes even when they belong to the same species. So far, bacteria utilize AAs in a species-dependent manner have been reported elsewhere. For example, Escherichia coli can use serine (Ser), aspartate (Asp), cysteine (Cys), glycine (Gly), glutamate (Glu), and alanine (Ala) as the sole carbon and nitrogen sources during aerobic growth, while they consume Ser, Asp, Cys, and asparagine (Asn) under anaerobic conditions8. All these utilizable AAs are also chemoattractants for E. coli. In contrast, E. coli try to avoid high concentrations of other AAs, such as valine (Val) and leucine (Leu), which inhibit cell growth and therefore are considered as chemorepellents8. Different Pseudomonas species utilize AAs differently; for example, most P. aeruginosa strains can use lysine (Lys), while P. fluorescens, P. stutzeri, and P. alcaligenes do not. However, all Pseudomonas spp. have some characteristics in common, they usually consume Ala, Glu, and Asp but not methionine (Met), Cys, and threonine (Thr)9. Staphylococcus aureus often required between 3 and 12 AAs for growth, with proline (Pro), arginine (Arg), Val, and Cys being most frequently required10,11. In addition, Arg, Cys, and phenylalanine (Phe) are necessary for its enterotoxin production12. Asp, glutamine (Gln), Gly, Pro, Ser, Ala, Arg, and Asn can be used as sole carbon and nitrogen sources by Salmonella enterica in germinating alfalfa exudates13, the utilization of these AAs may be contributed to bacterial fitness during plant colonization. Toxigenic and nontoxigenic strains of Pasteurella multocida can grow in a specially designed minimal medium14, the study concluded that Met, Cys, and Glu were strictly required for growth while Leu was not necessary but stimulated growth. The previous study of amino acid auxotrophies revealed that Arg, histidine (His), Met, tryptophan (Trp), isoleucine (Ile), Leu, and Val were essential for Enterococcus faecalis15. In addition, Glu and Gln were found to be an important hub in metabolism. Although these kinds of information above are not rare, they are mostly sporadic with different experimental conditions and many cultured with multiple AAs or concurrent nutritional supplements.
Different bacterial species require different types of AAs for growth, development of resistance to a hostile environment, or toxin production, which means certain AAs are essential for some bacteria but may not be necessary for others. These facts pointed to a possibility to employ AA utilization as a phenotypic mechanism to distinguish bacterial species, especially further down to strains, types, toxigenicity, and other characteristic properties. However, a comprehensive study of simple or essential AA utilization among various bacterial strains is still lacking. For measuring AA utilization of bacteria, a method that can accurately quantitate AAs is needed. Flow-injection analysis (FIA) with electrochemical detection using copper nanoparticle-plated screen-printed carbon electrode (Cun SPE) has been successfully demonstrated for sensitive determination of all native AAs16,17. Compared with tag-based methods18, this electrochemical detection method can directly detect AAs without any chromophore or fluorophore groups. Moreover, this method is more economical and easier than spectroscopic methods such as nuclear magnetic resonance (NMR) and Raman spectroscopy19,20. Thus, this CunSPE detection method can be a practical tool in the measurement of AA utilization for bacterial differentiation.
The aims of this study were to exam the hypothesis that the ability of different bacteria species to utilize single AA under minimal nutritional condition (live or die) is distinct enough to comprise a new method for novel classification/typing of bacterial species, and for study of animal tropisms in the same bacterium, differentiation of toxigenicity in the same bacterium, and differentiation of the same bacterial species with or without antimicrobial resistance. Our findings may provide pivotal phenotypic information in terms of AA dependency and may uncover strategies for nutritional, pharmaceutical, taxonomic, and evolutionary aspects of bacterial researches.
Results
Differential utilization preference of 20 AAs among bacterial species
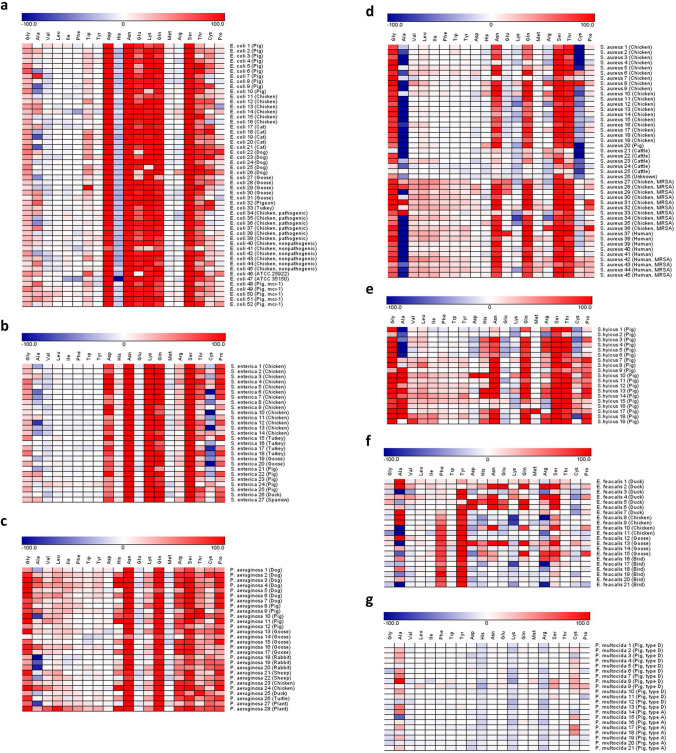
The 7 bacterial species, namely E. coli, S. enterica, P. aeruginosa, S. aureus, S. hyicus, E. faecalis, and P. multocida, showed distinctive consumption profiles (Fig. 1). The consumption levels of AA can be conveniently categorized into high (> 80%), intermediate (20%-80%), low (< 20%), and negative (biotransformation) (< 0%). Both E. coli and S. enterica (Fig. 1a,b) have consistently high consumption of Asp, Asn, Lys, Gln, and Ser. In addition, a high consumption of Glu and a slight negative consumption of His were observed in E. coli. In most P. aeruginosa strains (Fig. 1c), Gly, Asn, Gln, Arg, Ser, and Pro were highly consumed. S. aureus and S. hyicus (Fig. 1d,e) exhibited a similar pattern in highly consuming Gly, Asn, Gln, Ser, and Thr. The E. faecalis (Fig. 1f) had an inconsistent consumption pattern in comparison with other bacterial species; only Phe and tyrosine (Tyr) were found to have a relatively steady utilization. The consumption of most AAs was low in P. multocida (Fig. 1g); only a slight negative consumption of His, Lys, and Arg was consistently observed. Overall, nearly all strains of the 7 species have low consumption of Val, Leu, Ile, and Met.
Figure 1.
Heat map showing AA consumption profiles of 7 bacterial species. The heat map shows the consumption of 20 AAs by 213 strains from 7 bacterial species including (a) E. coli, (b) S. enterica, (c) P. aeruginosa, (d) S. aureus, (e) S. hyicus, (f) E. faecalis, and (g) P. multocida . The color gradients, which goes from white to red and white to blue, represent the level of positive consumption (0–100%) and negative consumption (0 to -100%) of AAs, respectively.
Differentiation of bacterial species by AA consumption profiles
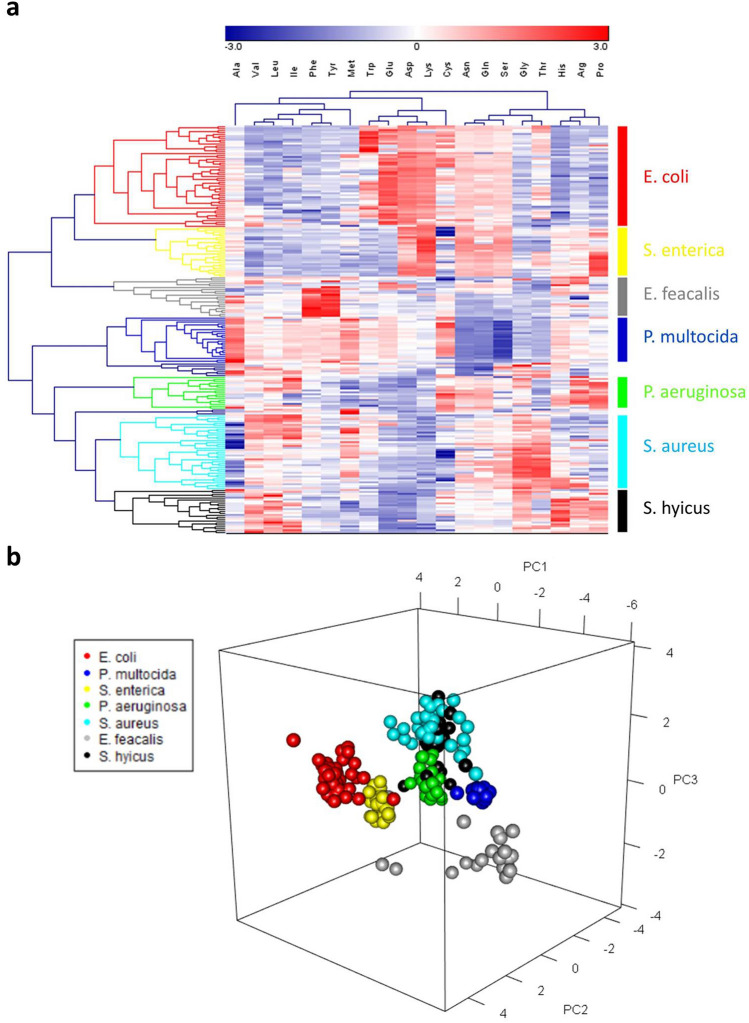
Hierarchical clustering analysis (HCA) on all 213 strains (Fig. 2a) revealed 7 clusters corresponded to the 7 bacterial species. All 52 strains of E. coli and all 27 strains of S. enterica were independently clustered with Glu being a significant factor to distinguish these two clusters. All P. multocida strains formed an individual cluster mainly due to the relatively low consumption of Asn, Gln, and Ser. Almost all strains of E. faecalis were grouped according to their specific consumption of Phe and Tyr. Most strains of P. aeruginosa were clustered together based on their ability to specifically consume Arg and Pro, whereas some of the strains were distributed to S. hyicus cluster. S. aureus cluster contained most of the S. aureus strains, but also contained some strains of S. hyicus and vice versa. Overall, HCA indicated that AA consumption pattern successfully categorized 100% of E. coli (52/52), S. enterica (27/27), and P. multocida strains (21/21), 90% of E. faecalis strains (19/21), 80% S. aureus strains (36/45), 62% of P. aeruginosa strains (17/28), and 52% of S. hyicus strains (10/19).
Figure 2.
Clustering of bacterial strains by AA consumption profiles. (a) Hierarchical clustering and heat map of differentially consumed AAs in 213 strains. The clusters represent seven bacterial species are evident. Red represents high relative consumption and green represents low relative consumption as shown in colored scale bar above the heat map. (b) 3-D view of principal component analysis of 213 bacterial strains. Each spot colored according to different species indicates individual strain.
According to the results of HCA, 11 AAs (Gly, His, Tyr, Asp, Asn, Glu, Lys, Gln, Ser, Thr, and Pro) showing higher variance among the 7 bacterial species were selected to perform principal component analysis (PCA) for further validation to differentiate/classify bacterial species (Fig. 2b). In order to confirm whether the selected principal components (PCs) were representative enough of AA consumption data, eigenvalues and the cumulative proportion of the PCs were checked (Supplementary Fig. 1). The eigenvalues of PC1, PC2, and PC3 were all more than 1 and in combination can explain 80% of the variation, suggesting that the first 3 PCs in our PCA had an adequate reliability. The results (Fig. 2b) demonstrated that 6 species (except for S. hyicus which could not be readily distinguished from the S. aureus group) were nicely separated into 6 main groups.
Distinction by hosts
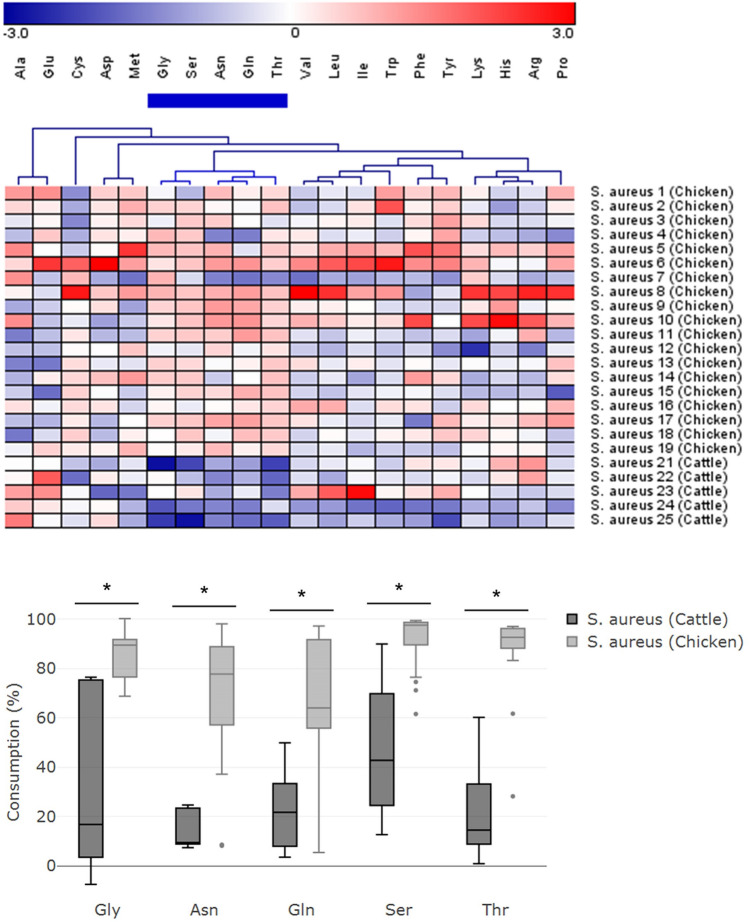
Consumption patterns were also analyzed based on the host origin for each bacterial species. In S. aureus, disparate consumption patterns were observed between the strains from chicken and cattle (both groups were not methicillin-resistant strains). The heat map and hierarchical clustering indicated that isolates from the cattle distinctively utilize Gly, Ser, Asn, Gln, and Thr lower than those from chicken with statistically significant difference (P < 0.05) (Fig. 3). The median [IQR] of consumption of Gly, Ser, Asn, Gln, and Thr were 17% [3 to 75], 43% [24 to 70], 9% [9 to 23], 22% [8 to 33], and 14% [9 to 33], respectively in the strains from cattle, while they were 89% [77 to 92], 98% [90 to 99], 78% [57 to 89], 64% [56 to 92], and 93% [88 to 96], respectively in the strains from chicken.
Figure 3.
Comparative consumption of AAs among S. aureus strains with different host origins. The heat map shows AAs consumption by S. aureus strains from chicken (S. aureus strain 1–19) and from cattle (S. aureus strain 21–25). Hierarchical clustering of AAs indicated that the green cluster of cattle origin strains have a relatively lower AA consumption compared with strains from chicken, and the consumption of all five AAs in the green cluster were significantly different (P < 0.05) between two groups.
Distinction by antibiotic-resistance
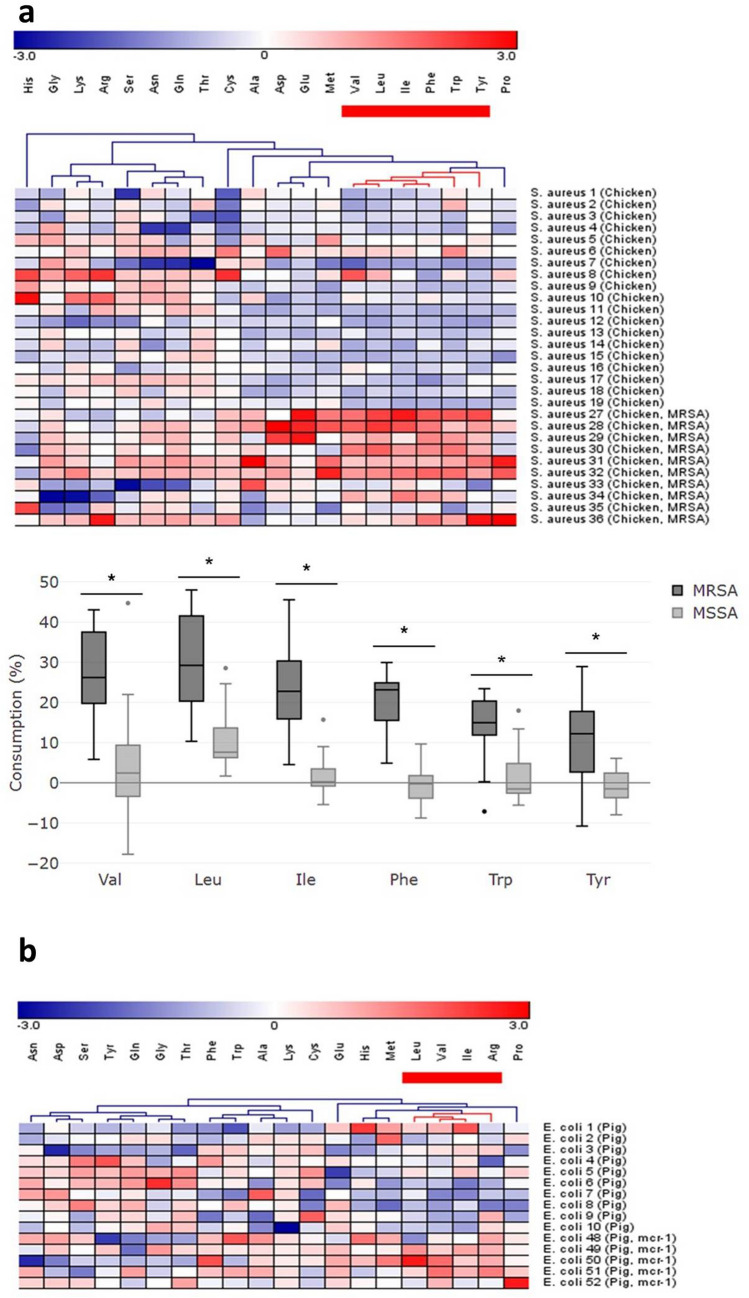
The difference in AA consumption was observed between methicillin-resistant and non-resistant S. aureus strains. Methicillin-resistant S. aureus (MRSA) utilized comparatively higher amounts of Val, Leu, Ile, Phe, Trp, and Tyr than methicillin-sensitive S. aureus (MSSA) (P < 0.05) (Fig. 4a). Median [IQR] of consumption of Val, Leu, Ile, Phe, Trp, and Tyr were 26% [20 to 38], 29% [20 to 42], 23% [16 to 30], 23% [16 to 25], 15% [12 to 20] and 12% [3 to 18], respectively in MRSA, while they were 2% [-3 to 9], 8% [6 to 14], 0% [-1 to 4], 0% [-4 to 2], -1% [-3 to 5] and -1% [-4 to 2], respectively in MSSA. Differential consumption was also observed in E. coli colistin-resistant (mcr-1 positive) strains where they utilized higher Leu, Val, Ile and Arg upon compared with non-resistant E. coli strains (Fig. 4b); however, only Val and Arg were significantly higher (P < 0.05) (Supplementary Fig. 2).
Figure 4.
Comparative consumption of AAs among strains with and without antibiotic resistance. (a) The heat map shows AAs consumption by MSSA (S. aureus strain 1–19) and MRSA (S. aureus strain 27–36). Hierarchical clustering of AAs indicated that the red cluster in MRSA has a relatively higher AA consumption compared with MSSA, and the consumption of six AAs in the red cluster were significantly different (P < 0.05) between two groups. (b) The heat map of E. coli strains with and without mcr-1 gene(E. coli strain 1–10 and strain 48–52) and hierarchical clustering of AAs indicated that four AAs in the red cluster were consumed in different levels.
Distinction by toxigenicity
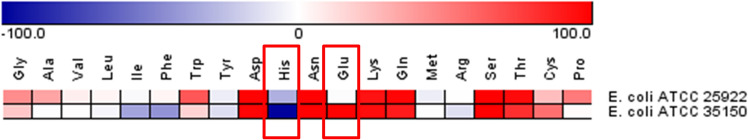
The non-toxigenic strains of wild-type E. coli (ATCC 25922) were compared with Shiga toxin-producing E. coli (ATCC 35150) (Fig. 5). The His and Glu were the most distinct differences. Wild-type E. coli had a slight negative consumption of His and almost no consumption of Glu, whereas Shiga toxin-producing E. coli had almost 100% negative and positive consumption of His and Glu, respectively.
Figure 5.
Comparative consumption of AAs between E. coli strains with and without toxigenicity. Consumption patterns of wild-type E. coli (ATCC 25922) and Shiga toxin-producing E. coli (ATCC 35150) are shown by heat map. These two strains have a big difference in consuming His and Glu.
Decision tree strategy based on specific AA utilization for bacterial differentiation
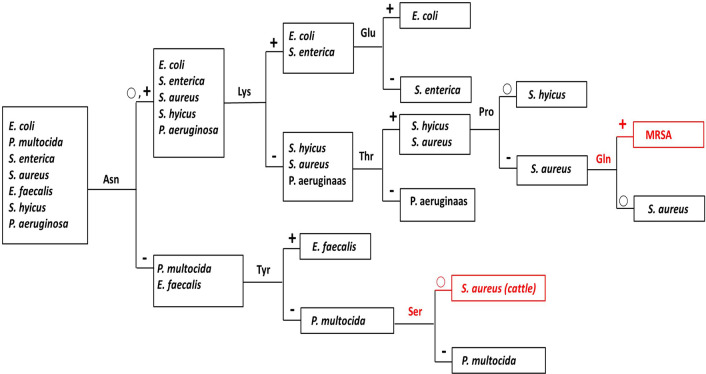
Based on the overall analysis of AA utilization, we discovered that 8 AAs, namely Asn, Lys, Tyr, Glu, Thr, Ser, Pro, and Gln can be used to make a decision tree for quick differentiation of these 7 bacteria species in a blind test situation (Fig. 6). These 7 bacterial species could be successfully differentiated in 4 serial decision making processes by using 50% or 75% as the cut-off consumption level (Table 1 and Fig. 6). For instance, an unknown bacterial strain is first tested by Asn, if the consumption is over 50%, it is further tested by Lys; and if the consumption is over 75%, then it can further be tested for Glu, if the Glu consumption is again over 75%, then this test bacterium is determined to be E. coli, otherwise (if the Glu consumption is less than 75%) the bacterium is deemed to be S. enterica. The decision tree can also differentiate MRSA versus chicken MSSA by the utilization level of Gln.
Figure 6.
Decision tree for differentiation of bacterial species. A flow chart of the identification process based on the consumption level of 8 AAs can efficiently identify 7 bacterial species, cattle S. aureus and MRSA. − indicates the consumption is less than 50%; ○ indicates the consumption is from 50 to 75%; + indicates the consumption is more than 75%.
Table 1.
Average consumption of AAs. The average AA consumption profiles of E. coli (n = 52), P. multocida (n = 21), S. enterica (n = 27), P. aeruginosa (n = 28), S. aureus (n = 20), cattle S. aureus (n = 5), MRSA (n = 10), E. faecalis (n = 21), and S. hyicus (n = 19). − indicates the average consumption is less than 50%; ○ indicates the average consumption is from 50 to 75%; + indicates the average consumption is more than 75%.
| Gly | Ala | Val | Leu | Ile | Phe | Trp | Tyr | Asp | His | |
|---|---|---|---|---|---|---|---|---|---|---|
| E. coli 1–52 | − | − | − | − | − | − | − | − | + | − |
| P. multocida 1–21 | − | − | − | − | − | − | − | − | − | − |
| S. enterica 1–27 | − | − | − | − | − | − | − | − | ○ | − |
| P. aeruginosa 1–28 | ○ | − | − | − | − | − | − | − | − | − |
| S. aureus 1–20 | + | − | − | − | − | − | − | − | − | − |
| S. aureus 21–25 | − | − | − | − | − | − | − | − | − | − |
| S. aureus 27–36 | + | − | − | − | − | − | − | − | − | − |
| E. feacalis 1–21 | − | − | − | − | − | ○ | − | + | − | − |
| S.hyicus 1–19 | + | − | − | − | − | − | − | − | − | − |
| Asn | Glu | Lys | Gln | Met | Arg | Ser | Thr | Cys | Pro | |
|---|---|---|---|---|---|---|---|---|---|---|
| E. coli 1–52 | + | + | + | + | − | − | + | ○ | − | − |
| P. multocida 1–21 | − | − | − | − | − | − | − | − | − | − |
| S. enterica 1–27 | + | − | + | + | − | − | + | − | − | ○ |
| P. aeruginosa 1–28 | + | − | − | + | − | ○ | + | − | − | ○ |
| S. aureus 1–20 | ○ | − | − | ○ | − | − | + | + | − | − |
| S. aureus 21–25 | − | − | − | − | − | − | ○ | − | − | − |
| S. aureus 27–36 | + | − | − | + | − | − | + | + | − | − |
| E. feacalis 1–21 | − | − | − | − | − | − | − | − | − | − |
| S.hyicus 1–19 | ○ | − | − | ○ | − | − | + | + | − | ○ |
Discussion
In this study, an AAs consumption profile was established for phenotypic categorization of bacteria (Fig. 1). Different from other common bacterial phenotyping methods such as API or the Biolog system, which are based on detection of enzymatic activity related to biological transformation of carbohydrates, proteins, or organic acids, our method directly and quantitatively analyzes the consumption levels of specific AAs. The method described here can afford a more focused fingerprinting of bacterial metabolism of AAs and complement other phenotyping methods. The ease and simplicity of this method allow rapid and continuous detection of AAs without any derivatization or complex sample processing. For reliable detection, we injected 1.5 mM Gly as an internal standard before and after the detection of each AA (Supplementary Fig. 3), and set the error range of the internal standard within 15%. With this precision of quantitative analysis, the exact difference in consumption levels and thus their phenotypic properties, can be explicitly distinguished among bacterial species. However, the major drawback of our method is that the bacteria must survive and grow in liquid media (broth cultures); for those that can only grow on agar plates, gas phase analysis21,22 might be an alternative.
By determining each consumption level for the 20 AAs, the utilization phenotype and nutritional requirements of bacteria can be understood. Among 7 bacterial species, 5 species, namely E. coli, S. enterica, P. aeruginosa, S. aureus, and S. hyicus, showed consistently high consumption of 3 neutral polar AAs (Ser, Asn, and Gln). All 3 AAs play a crucial role in bacterial metabolism; for instance, Ser can be catabolized by L-serine dehydratase to produce pyruvate and ammonia, which is important for bacteria to regulate nitrogen balance and maintain extracellular pH23,24. Likewise, Gln and Asn are required as nitrogen donors for many important biosynthetic reactions in bacterial cells, including nucleic acid synthesis and transamidation reactions25,26. Previous works also demonstrated that Ser, Asn, and Gln could be rapidly utilized and metabolized by E. coli and other small intestinal bacteria27,28. All 7 species in our study showed low consumption of branched-chain amino acids (BCAAs) (Val, Leu, and Ile). Although BCAAs have been reported to play an important role in bacterial protein synthesis and maintenance of central metabolism29–31, studies in pig small intestine revealed that BCAAs are largely degraded by intestinal mucosal cells instead of the microbiota, which may suggest that either the bacteria could not utilize a great amount of BCAAs or a small amount of BCAAs would be sufficient to support the growth of these bacteria32,33. Thus, significant consumption of extracellular BCAAs could not be observed in our current study. Both Glu and Asp were largely consumed by E. coli in the current study; this may be related to the presence of glutamate transporters importing extracellular Glu and Asp into the cells34. Several L-glutamate transport systems have been identified in E. coli, such as a sodium-dependent transport system for Glu, a binding protein-dependent Glu/Asp transport system, and a proton symport system for Glu and Asp35–37. High levels of Lys were consumed only by E. coli and S. enterica; the possible reason might be that lysine decarboxylase’s activity is enhanced for tolerating the acidic environment caused by the metabolic byproducts produced from cultivating E. coli and S. enterica38,39. The observation that Gly was largely consumed by S. aureus and S. hyicus might be related to the formation of pentaglycine cross bridge, which is required for linking glycan chains into a two- or three-dimensional network in the biosynthesis of the peptidoglycan cell wall, and the absence of a pentaglycine cross bridge would inhibit the survival of S. aureus cells40–42. Collectively, the preferences for AAs are consistent enough such that categorization of bacteria by their intrinsic ability to utilize a simple form of nutrient becomes a feasible new way to complement genetic typing. Further validation by HCA and PCA pointed out the critical AAs with prominent effect on bacterial differentiation (Fig. 2). Most clusters of bacterial species identified by both analyses were well-separated except for the S. hyicus and S. aureus. Both bacteria belong to the genus Staphylococcus with high genetic similarity and biochemical properties43. In fact, detection of S. hyicus by API 20 has been reported to show low specificity44, thus, it is not totally unique that they shared similar AA consumption patterns. Nevertheless, a greater consumption of Arg and Pro by most S. hyicus strains in comparison to that by S. aureus strains was observed (Fig. 1d,e), which may carry biochemical significance associated with their coagulase productivity. S. aureus and S. hyicus are classified as coagulase-positive and coagulase-variable staphylococci, respectively, based on their ability to produce extracellular coagulase that can convert fibrinogen to fibrin45. Studies have suggested that Arg is an important alternative substrate to coagulase-negative staphylococci46, hence, consumption of Arg by most S. hyicus strains may possibly be one factor contributing to the variance in coagulase production. While this speculation requires further elucidation, our AA consumption profiles showed great potential for studying the biochemical characteristics of bacteria.
Some of the AA utilization determined in our study were not in agreement with those in previous reports. For instance, Met and Glu were regarded as necessary elements for P. multocida growth14, however, we did not observe the consumption of these AAs in our study. Similarly, a large amount of Arg was not consumed by S. aureus10. The discrepancy might be due to several reasons. First, the nutritional environment of the culture media may influence the metabolic state of the bacterial cells, resulting in different bacterial phenotypes47. In this study, the minimal nutritional medium (MNM) used for determining AA consumption only contained very minimal amounts of glucose and salt for energy source and for maintaining osmotic pressure, respectively. The nutritional sources in MNM are relatively poor in relation to media used in other studies, which possibly forced the change in bacterial metabolism. Second, the AA requirements are different during each growth phase: bacteria in the exponential phase need more AAs for cell growth and division than those in the stationary phase. Third, specific substances are required when bacteria utilize AAs,for example, S. aureus S-6 needs biotin as a growth factor when Glu is used as carbon source48. One interesting observation here was that some AAs presented negative consumption, which technically was due to a larger electrochemical current value of the samples in comparison to the positive control (Supplementary Fig. 3). Taking E. coli for example, a slight negative consumption of His was consistently observed in each strain. This phenomenon may suggest that certain AA metabolite(s) which could also produce electrochemical signals were formed upon utilization of His; further identification of metabolites is needed to confirm this deduction. It is noteworthy that the negative value was not merely a measurement of consumption but likely the sum of processes involving both the consumption and production of electroactive metabolite(s), which taken together might be better represented by the term “biotransformation.” These observations imply that the nutritional requirement of a bacterium is the result of complex evolutionary processes that cannot be deciphered or is beyond the understanding of current knowledge. Therefore, this study provided a scenario that indicates how different bacterial species react to its survival in a nutrient-scarce situation. In fact, a partial parallel study using only a single AA without MNM also showed very similar results (Supplementary Fig. 4).
Other potential applications of the current method were noticed during the study. First, S. aureus showed distinctive consumption profiles between different hosts, i.e., cattle and chicken (Fig. 3). Almost no AAs were consumed to a significant degree by S. aureus isolated from mastitis cow milk. Although a previous study suggested that strains from milk required several AAs for growth, milk is normally deficient in free AAs49 so alternative sources of nitrogen might be available for sustained growth of these isolates. Next, a relatively higher level of BCAA consumption was observed in MRSA and colistin-resistant E. coli compared to that in non-resistant strains (Fig. 4). Antibiotic-resistant strains were suggested to have reduced metabolism due to defective central metabolic pathways of glucose or AA (e.g. Ala)50,51. It is feasible that BCAAs, which also have a significant role in maintaining central metabolism, showed higher utilization by antibiotic-resistant strains. Interestingly, we found that human S. aureus and chicken MRSA strains had similar specific AAs consumption (Fig. 1d; whether the similarity of AA consumption among hosts represents a higher possibility of disease spreading (cross-host movement remains to be elucidated, and is certainly worth further study. Finally, most of the E.coli isolates in this study consumed a high level of Glu and a prominent difference in the consumption of His and Glu was found between the Shiga toxin-producing and wild-type E. coli (Fig. 5). E. coli mutant strains have been reported to take up Glu at a much faster rate than wild-type strains, which can be attributed to changes in cell permeability52. The glutamate decarboxylase (GAD) system was also reported to play an important role in the resistance to multiple environmental stresses53. Collectively, through these preliminary observations, we believe that the AA consumption profile holds reasonably good potential as a tool, complementary to other bacterial typing methods, for studying specific bacterial characteristics/traits. A larger-scale investigation is warranted to strengthen the power of the above claims.
Based on the consumption profiles, we were able to create a simple and straightforward strategy for differentiation of bacterial species and/or strains with just a few AAs (Fig. 6). The prerequisite is to decide on the adequate consumption level for differentiation: for instance, 50% or 70% of consumption as the cut-off points. Although the efficacy could be improved by larger sample numbers, this prospective strategy should benefit bacterial identification and research on bacterial metabolism.
In conclusion, we applied FIA-ED to quantitatively determine single AA consumption profiles of bacteria and revealed their characteristic preferences in consuming AAs. AA phenotyping might be a potential way to differentiate bacterial species and with larger-scale investigations, might also reveal a feasible way to study/categorize the development of host tropisms, toxin production, and drug resistance. Additionally, this AA phenotyping determined not only the nutritional requirements but also in part the metabolism of AAs by bacteria (such as in the negative consumption). Therefore, bacterial strains may be categorized as “specific AAs utilizing strains” or “non-utilizable strains,” which places emphasis on both the nutritional and/or metabolic phenotyping regardless of their original species. Once the relationships between AA utilization and its characteristic roles in pathogenesis, toxogenesis, or resistance can be established, this categorization will be a more powerful and informative way to describe a bacterium. Expanding on the current study will offer more phenotypic information, which can afford researchers a new perspective in analyzing microbiological data derived from biochemical and molecular biological techniques. Specific relationships acquired from this kind of study could shed more light on bacterial metabolism, bacterial adaptation to host, and development of toxigenicity and antibiotic-resistance. They may also help develop novel strategies for treating infections. For example, offering specific AA(s) that are highly or differentially utilized by the resistant strains may alter their bacterial metabolism and thereby their antibiotic susceptibility51. Furthermore, it might be possible to develop media formulation to selectively inhibit or enhance the growth of specific bacterial strains for targeted studies.
Methods
Bacterial strains
A total of 213 strains in 7 bacterial species and different hosts were collected for this study. All strains were identified to gene level by 16S rRNA gene sequencing. Bacterial species include Escherichia coli (52 strains), Pasteurella multocida (21 strains), Salmonella enterica (27 strains), Pseudomonas aeruginosa (28 strains), Staphylococcus aureus (45 strains), Enterococcus faecalis (21 strains), and Staphylococcus hyicus (19 strains). All clinical isolates were isolated from diseased animals in outbreak farms or veterinary hospitals, and the standard strains (ATCC 25922 and ATCC 35150) were purchased from American Type Culture Collection (Manassas). The information of all strains used in this work was listed in Supplementary Table 1.
Preparation of amino acids (AAs)
All AAs except for Gly were purchased from Applichem GmbH (Darmstadt, Germany). Gly was purchased from GERBU Biotechnik GmbH (Heidelberg, Germany). All AAs were dissolved in MNM to a final concentration of 3 mM (Tyr was first dissolved at 55 °C). MNM was prepared by adding 0.243 g K2HPO4, 0.234 g C6H12O6 and 0.491 g NaCl to 1,000 mL of deionized water (1.4 mM K2HPO4, 1.4 mM C6H12O6 and 8.4 mM NaCl), and then filtered by 0.22 μm syringe filters with nylon membrane (Pall Corp., USA).
Culture medium and growth condition
All bacterial strains were first recovered from its freeze-dried or frozen condition, and inoculated in Tryptic soy agar (TSA) for S. enterica, S. aureus, S. hyicus, E. faecalis and P. aeruginosa, in Luria agar (LA) for E. coli, and in Todd Hewitt agar for P. multocida, and incubated at 37 °C overnight (18 h). On the next day, a single colony was picked and inoculated in 5 mL of Tryptic soy broth (TSB) for S. enterica, S. aureus, S. hyicus, E. faecalis and P. aeruginosa, in Luria broth (LB) for E. coli, and in Brain heart infusion (BHI) broth for P. multocida, and incubated at 37 °C under shaking (180 rpm) for 18 ± 1 h. After that, 400 μL of the cultured broth was subcultured in 10 mL of respective broth and incubated at 37 °C under shaking (180 rpm). Bacterial cells were harvested by centrifugation (3,500 rpm, 10 min) when OD600 reaches 0.8. The pellet was washed by 5 mL of the MNM twice for removing the original broth that would interfere with the electrochemical signal (see below). The washed pellet was subsequently grown in tube each containing 3 mM of single AA solution. The bacterial suspension was adjusted to 1 × 10@@@9 CFU/mL and then incubated at 37 °C, 180 rpm for 18 h. After growth, the bacterial cultures were centrifuged (3,500 rpm, 10 min) and the supernatant was used for AA analysis.
Electrochemical analysis of amino acids
Flow-injection analysis (FIA) with electrochemical detection (FIA-ED) was used to conduct chronoamperometric experiments (i-t) and quantification of AA as previously described17. In brief, the FIA system consists of a Hitachi L-6200 intelligent pump drive, a Shimadzu SIL-10A Auto Injector with an interconnecting Teflon tube and a specially designed electrochemical flow cell (Zensor R&D, Taichung, Taiwan) incorporating a three-electrode system consisting of a working electrode (Cun-SPE, copper nanoparticle-plated screen-printed electrode, geometric area of 0.2 cm@@@2), an Ag/AgCl reference electrode, and a platinum auxiliary electrode (geometric area of 0.07 cm@@@2). The detection condition using FIA system were: mobile phase: 0.01 M, pH 7 phosphate buffered saline (PBS), analytical potential: 0.15 V vs. Ag/AgCl, scan rate: 0.05 V/s, flow rate 0.5 mL/min, injection volume 20 μL. The internal standard is 1.5 mM Gly. Chronoamperometric signals for quantification were analyzed by CHI1021C electrochemical analyzer with CHI system software (CH Instruments, Austin, TX, USA). For determination of AA utilization, the percentage of AA consumption (A%) were determined by the equation
where N is the electrochemical current value of the negative control, in which bacterial strain was grown in the medium containing only MNM; P is the current value of the positive control, which contained 3 mM of each AA solution without bacterial inoculation; T is the average current value of replicate samples containing both bacteria and AA solution.
Statistical analysis
Genesis software version 1.8.1 was used to create heat maps and HCA54. In HCA, each AA consumption data was first standardized by z-score normalization and then used to perform average linkage distance hierarchical clustering. PCA was performed using the prcomp function in R version 3.5.3 to investigate similarities between individual samples based on correlation55. The rgl package was used to create interactive three-dimensional visualization of data. To assess the difference between groups, SPSS software (v.25) was used to perform the Mann–Whitney U test for non-normally distributed data. The data is represented as the median with interquartile range (IQR), and box plots were created for visualization of the data using R.
Supplementary information
Acknowledgements
We thank Dr. Jeremy J.W. Chen and Dr. Mingchang Chih for helpful discussions. This work is supported by the Ministry of Science and Technology, Taiwan, R.O.C. (Grant No. MOST 106-2313-B-005-040).
Author contributions
C.C.C. conceived and designed the study. H.C.K and C.H.L provided and identified clinical isolates from animals and humans. Y.K.L. carried out the experiments and analyzed the experimental data. C.C.C., C.H.L., and Y.K.L. contributed to the final version of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-69686-5.
References
- 1.Li W, Raoult D, Fournier PE. Bacterial strain typing in the genomic era. FEMS Microbiol. Rev. 2009;33:892–916. doi: 10.1111/j.1574-6976.2009.00182.x. [DOI] [PubMed] [Google Scholar]
- 2.Shayegani M, Maupin PS, McGlynn DM. Evaluation of the API 20E system for identification of nonfermentative Gram-negative bacteria. J. Clin. Microbiol. 1978;7:539–545. doi: 10.1128/jcm.7.6.539-545.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller JM, Rhoden DL. Preliminary evaluation of Biolog, a carbon source utilization method for bacterial identification. J. Clin. Microbiol. 1991;29:1143–1147. doi: 10.1128/jcm.29.6.1143-1147.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bassett DE, et al. Genome cross-referencing and XREFdb: implications for the identification and analysis of genes mutated in human disease. Nat. Genet. 1997;15:339–344. doi: 10.1038/ng0497-339. [DOI] [PubMed] [Google Scholar]
- 5.Rubin GM, et al. Comparative genomics of the eukaryotes. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson CS, et al. Cross-phenotype association tests uncover genes mediating nutrient response in Drosophila. BMC Genom. 2016;17:867. doi: 10.1186/s12864-016-3137-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung VG, Spielman RS. The genetics of variation in gene expression. Nat. Genet. 2002;32:522–525. doi: 10.1038/ng1036. [DOI] [PubMed] [Google Scholar]
- 8.Yang Y, et al. Relation between chemotaxis and consumption of amino acids in bacteria. Mol. Microbiol. 2015;96:1272–1282. doi: 10.1111/mmi.13006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sivolodskii EP. Application of the profiles of amino acid utilization as the sole carbon and nitrogen sources for pseudomonad taxonomy. Microbiology. 2009;78:711–716. [Google Scholar]
- 10.Zhu Y, et al. Staphylococcus aureus biofilm metabolism and the influence of arginine on polysaccharide intercellular adhesin synthesis, biofilm formation, and pathogenesis. Infect. Immun. 2007;75:4219–4226. doi: 10.1128/IAI.00509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Somerville GA, Proctor RA. At the crossroads of bacterial metabolism and virulence factor synthesis in Staphylococci. Microbiol. Mol. Biol. Rev. 2009;73:233–248. doi: 10.1128/MMBR.00005-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Onoue Y, Mori M. Amino acid requirements for the growth and enterotoxin production by Staphylococcus aureus in chemically defined media. Int. J. Food Microbiol. 1997;36:77–82. doi: 10.1016/s0168-1605(97)01250-6. [DOI] [PubMed] [Google Scholar]
- 13.Kwan G, et al. Few differences in metabolic network use found between Salmonella enterica colonization of plants and typhoidal mice. Front. Microbiol. 2018;9:695. doi: 10.3389/fmicb.2018.00695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jablonski PE, Jaworski M, Hovde CJ. A minimal medium for growth of Pasteurella multocida. FEMS Microbiol. Lett. 1996;140:165–169. doi: 10.1111/j.1574-6968.1996.tb08331.x. [DOI] [PubMed] [Google Scholar]
- 15.Veith N, et al. Using a genome-scale metabolic model of Enterococcus faecalis V583 to assess amino acid uptake and its impact on central metabolism. Appl. Environ. Microbiol. 2015;81:1622–1633. doi: 10.1128/AEM.03279-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu CT, et al. An electrochemical cell coupled with disposable screen-printed electrodes for use in flow injection analysis. Anal. Sci. 2006;22:35–38. doi: 10.2116/analsci.22.35. [DOI] [PubMed] [Google Scholar]
- 17.Zen JM, Hsu CT, Kumar AS, Lyuu HJ, Lin KY. Amino acid analysis using disposable copper nanoparticle plated electrodes. Analyst. 2004;129:841–845. doi: 10.1039/b401573h. [DOI] [PubMed] [Google Scholar]
- 18.Bidlingmeyer BA, Cohen SA, Tarvin TL. Rapid analysis of amino-acids using pre-column derivatization. J. Chromatogr. B. 1984;336:93–104. doi: 10.1016/s0378-4347(00)85133-6. [DOI] [PubMed] [Google Scholar]
- 19.Nord LI, Vaag P, Duus JO. Quantification of organic and amino acids in beer by H-1 NMR spectroscopy. Anal. Chem. 2004;76:4790–4798. doi: 10.1021/ac0496852. [DOI] [PubMed] [Google Scholar]
- 20.Schumerich MV, et al. Amino acid quantification in bulk soybeans by transmission raman spectroscopy. Anal. Chem. 2013;85:11376–11381. doi: 10.1021/ac402284b. [DOI] [PubMed] [Google Scholar]
- 21.Stewart EJ. Growing unculturable bacteria. J. Bacteriol. 2012;194:4151–4160. doi: 10.1128/JB.00345-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calleja M, Elahi A, Caruana DJ. Gas phase electrochemical analysis of amino acids and their fragments. Commun. Chem. 2018;1:48. [Google Scholar]
- 23.Chaussee MS, Somerville GA, Reitzer L, Musser JM. Rgg coordinates virulence factor synthesis and metabolism in Streptococcus pyogenes. J. Bacteriol. 2003;185:6016–6024. doi: 10.1128/JB.185.20.6016-6024.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vining L, Magasanik B. Serine utilization by Klebsiella aerogenes. J. Bacteriol. 1981;146:647–655. doi: 10.1128/jb.146.2.647-655.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilcox M. γ-Glutamyl phosphate attached to glutamine-specific tRNA: a precursor of glutaminyl-tRNA in bacillus subtilis. Eur. J. Biochem. 1969;11:405–412. doi: 10.1111/j.1432-1033.1969.tb00788.x. [DOI] [PubMed] [Google Scholar]
- 26.Forchhammer K. Glutamine signalling in bacteria. Front. Biosci. 2007;12:70. doi: 10.2741/2069. [DOI] [PubMed] [Google Scholar]
- 27.Dai ZL, Zhang J, Wu G, Zhu WY. Utilization of amino acids by bacteria from the pig small intestine. Amino Acids. 2010;39:1201–1215. doi: 10.1007/s00726-010-0556-9. [DOI] [PubMed] [Google Scholar]
- 28.Dai ZL, et al. L-Glutamine regulates amino acid utilization by intestinal bacteria. Amino Acids. 2013;45:501–512. doi: 10.1007/s00726-012-1264-4. [DOI] [PubMed] [Google Scholar]
- 29.Kaiser JC, Heinrichs DE. Branching out: alterations in bacterial physiology and virulence due to branched-chain amino acid deprivation. mBio. 2018;9:e 01188. doi: 10.1128/mBio.01188-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gesbert G, et al. Importance of branched-chain amino acid utilization in Francisella intracellular adaptation. Infect. Immun. 2015;83:173–183. doi: 10.1128/IAI.02579-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Franco TMA, Blanchard JS. Bacterial branched-chain amino acid biosynthesis: structures, mechanisms, and drugability. Biochemistry. 2017;56:5849–5865. doi: 10.1021/acs.biochem.7b00849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen L, et al. Catabolism of nutritionally essential amino acids in developing porcine enterocytes. Amino Acids. 2009;37:143–152. doi: 10.1007/s00726-009-0268-1. [DOI] [PubMed] [Google Scholar]
- 33.Yang YX, Dai ZL, Zhu WY. Important impacts of intestinal bacteria on utilization of dietary amino acids in pigs. Amino Acids. 2014;46:2489–2501. doi: 10.1007/s00726-014-1807-y. [DOI] [PubMed] [Google Scholar]
- 34.Raunser S, et al. Structure and function of prokaryotic glutamate transporters from Escherichia coli and Pyrococcus horikoshii. Biochemistry. 2006;45:12796–12805. doi: 10.1021/bi061008+. [DOI] [PubMed] [Google Scholar]
- 35.Schellenberg GD, Furlong CE. Resolution of the multiplicity of the glutamate and aspartate transport systems of Escherichia coli. J. Biol. Chem. 1977;252:9055–9064. [PubMed] [Google Scholar]
- 36.Halpern YS, Barash H, Druck K. Glutamate transport in Escherichia coli K-12: nonidentity of carriers mediating entry and exit. J. Bacteriol. 1973;113:51–57. doi: 10.1128/jb.113.1.51-57.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miner KM, Frank L. Sodium-stimulated glutamate transport in osmotically shocked cells and membrane vesicles of Escherichia coli. J. Bacteriol. 1974;117:1093–1098. doi: 10.1128/jb.117.3.1093-1098.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park YK, Bearson B, Bang SH, Bang IS, Foster JW. Internal pH crisis, lysine decarboxylase and the acid tolerance response of Salmonella typhimurium. Mol. Microbiol. 1996;20:605–611. doi: 10.1046/j.1365-2958.1996.5441070.x. [DOI] [PubMed] [Google Scholar]
- 39.Lin J, Lee IS, Frey J, Slonczewski JL, Foster JW. Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J. Bacteriol. 1995;177:4097–4104. doi: 10.1128/jb.177.14.4097-4104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsuhashi M, Dietrich CP, Strominger JL. Incorporation of glycine into the cell wall glycopeptide in Staphylococcus aureus: Role of sRNA and lipid intermediates. Proc. Natl. Acad. Sci. USA. 1965;54:587. doi: 10.1073/pnas.54.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou X, Cegelski L. Nutrient-dependent structural changes in S. aureus peptidoglycan revealed by solid-state NMR spectroscopy. Biochemistry. 2012;51:8143–8153. doi: 10.1021/bi3012115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monteiro JM, et al. The pentaglycine bridges of Staphylococcus aureus peptidoglycan are essential for cell integrity. Sci. Rep. 2019;9:5010. doi: 10.1038/s41598-019-41461-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langlois BE, Parlindungan AK, Harmon RJ, Akers K. Biochemical characteristics of Staphylococcus species of human and bovine origin (1) J. Food Prot. 1990;53:119–126. doi: 10.4315/0362-028X-53.2.119. [DOI] [PubMed] [Google Scholar]
- 44.Zadoks RN, Watts JL. Species identification of coagulase-negative staphylococci: genotyping is superior to phenotyping. Vet. Microbiol. 2009;134:20–28. doi: 10.1016/j.vetmic.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 45.Kloos, W. E. & Schleifer, K.H. Genus IV. Staphylococcus. in Bergey’s Manual of Systematic Bacteriology, Volume 2 (ed. Sneath, P.H.A., Mair, N.S., Sharpe, M.E. and Holt, J.G.) (Williams & Wilkins, Baltimore, 1986).
- 46.Janssens M, et al. The use of nucleosides and arginine as alternative energy sources by coagulase-negative staphylococci in view of meat fermentation. Food Microbiol. 2014;39:53–60. doi: 10.1016/j.fm.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 47.Smith A, et al. The culture environment influences both gene regulation and phenotypic heterogeneity in Escherichia coli. Front. Microbiol. 2018;9:1739. doi: 10.3389/fmicb.2018.01739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mah RA, Fung DYC, Morse SA. Nutritional requirements of Staphylococcus aureus S-6. Appl. Microbiol. 1967;15:866–870. doi: 10.1128/am.15.4.866-870.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lincoln RA, Leigh JA, Jones NC. The amino acid requirements of Staphylococcus aureus isolated from cases of bovine mastitis. Vet. Microbiol. 1995;45:275–279. doi: 10.1016/0378-1135(95)00041-8. [DOI] [PubMed] [Google Scholar]
- 50.Allison KR, Brynildsen MP, Collins JJ. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature. 2011;473:216–220. doi: 10.1038/nature10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peng B, et al. Exogenous alanine and/or glucose plus kanamycin kills antibiotic-resistant bacteria. Cell Metab. 2015;21:249–262. doi: 10.1016/j.cmet.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 52.Halpern YS, Lupo M. Glutamate transport in wild-type and mutant strains of Escherichia coli. J. Bacteriol. 1965;90:1288–1295. doi: 10.1128/jb.90.5.1288-1295.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feehily C, Karatzas KA. Role of glutamate metabolism in bacterial responses towards acid and other stresses. J. Appl. Microbiol. 2013;114:11–24. doi: 10.1111/j.1365-2672.2012.05434.x. [DOI] [PubMed] [Google Scholar]
- 54.Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- 55.R Core Team (2018). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.