Abstract
Blood metabolites of the tryptophan pathway were found to be associated with kidney function and disease in observational studies. In order to evaluate causal relationship and direction, we designed a study using a bidirectional Mendelian randomization approach. The analyses were based on published summary statistics with study sizes ranging from 1,960 to 133,413. After correction for multiple testing, results provided no evidence of an effect of metabolites of the tryptophan pathway on estimated glomerular filtration rate (eGFR). Conversely, lower eGFR was related to higher levels of four metabolites: C-glycosyltryptophan (effect estimate = − 0.16, 95% confidence interval [CI] (− 0.22; − 0.1); p = 9.2e−08), kynurenine (effect estimate = − 0.18, 95% CI (− 0.25; − 0.11); p = 1.1e−06), 3-indoxyl sulfate (effect estimate = − 0.25, 95% CI (− 0.4; − 0.11); p = 6.3e−04) and indole-3-lactate (effect estimate = − 0.26, 95% CI (− 0.38; − 0.13); p = 5.4e−05). Our study supports that lower eGFR causes higher blood metabolite levels of the tryptophan pathway including kynurenine, C-glycosyltryptophan, 3-indoxyl sulfate, and indole-3-lactate. These findings aid the notion that metabolites of the tryptophan pathway are a consequence rather than a cause of reduced eGFR. Further research is needed to specifically examine relationships with respect to chronic kidney disease (CKD) progression among patients with existing CKD.
Subject terms: Chronic kidney disease, Epidemiology, Risk factors
Introduction
The kidneys play a major role in maintaining homeostasis in the human body by regulating the excretion of both endogenous and exogenous molecules below a certain mass, such as many metabolites. The estimated glomerular filtration rate (eGFR) is the most widely used measure of kidney function1. Chronic kidney disease (CKD) is defined as a persistent reduction of eGFR or kidney damage and is associated with a higher risk of comorbidities and mortality2. With an increasing prevalence of currently 10–15% worldwide, it represents a global health burden2,3. Therapeutic options are limited and aim to stop CKD progression, thereby attempting to reduce risk of end-stage kidney disease (ESKD), early death and comorbidities2.
Blood and urine metabolite levels are altered in the presence of CKD, making evaluations of the metabolome of interest in nephrology4–6: while some metabolites may represent filtration markers such as creatinine and thus may allow for improving the estimation of GFR, other metabolites may provide complementary insights into renal physiology by acting as a readout of active tubular secretion or reabsorption. Furthermore, metabolites might be causally related to development and progression of CKD, such as glucose in diabetic nephropathy. A central question in nephrology therefore is whether alterations in metabolite levels represent a cause or a consequence of CKD.
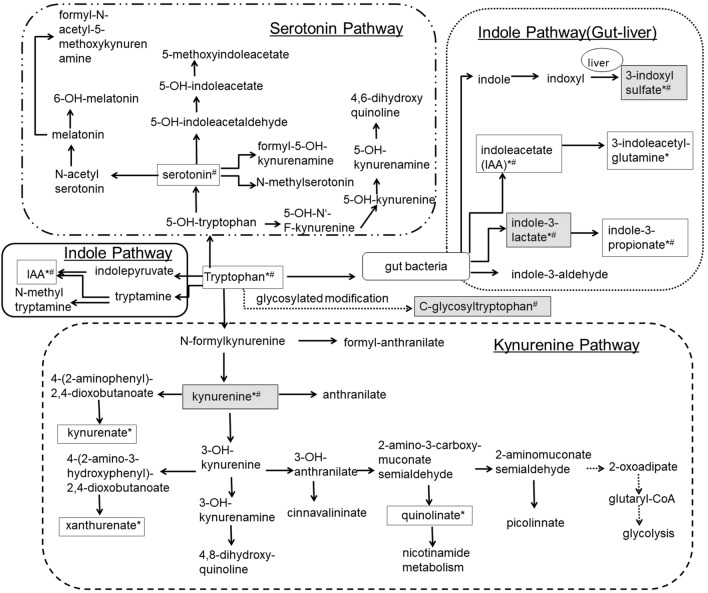
Recent advances in mass methodology7–9 allow for comprehensive studies of the metabolome and its relation to kidney function4,5. A common theme from the reported literature is the association between blood levels of metabolites of the tryptophan pathway (i.e. tryptophan and its down-stream products) and kidney function and disease (Supplementary Table 1). Tryptophan is an essential amino acid in humans10,11. While less than 1% of tryptophan is actually used for protein biosynthesis, the major proportion serves as a biosynthetic precursor of microbial and host metabolites along different pathways, including the serotonin, the kynurenine, and the indole pathways (Fig. 1)10–13.
Figure 1.
Metabolites and sub-pathways of the tryptophan pathway. For Metabolites with a frame necessary data were available and evaluated in this MR study to evaluate the impact of metabolites on eGFR (*) and/or the impact of eGFR on metabolites (#). Furthermore, frames with grey background mark metabolites that show significant results in our analysis.
We previously reported that several ratios of serum metabolite levels containing tryptophan that had been quantified by a targeted measurement technique in a population-based study were significantly associated with eGFR14. In a follow-up project, the serum kynurenine-to-tryptophan ratio was significantly associated with annual change in eGFR and incident CKD15. This specific ratio presumably reflects the enzymatic activity of indoleamine 2,3-dioxygenase and tryptophan 2,3-dioxygenase, responsible for the conversion of tryptophan to kynurenine via N-formylkynurenine. Furthermore, we evaluated nine metabolites of the tryptophan pathway based on non-targeted measurements in a similar study, and identified C-glycosyltryptophan (also known as C-mannosyltryptophan) to be associated with eGFR and CKD, as well as with prospective endpoints of eGFR decline, incident CKD and ESKD16.
Overall, studies evaluating associations between metabolites of the tryptophan pathway and kidney function or disease provide a mixture of significant and non-significant associations. Potential reasons relate to variability in measurement techniques (e.g. liquid chromatography, mass spectrometry) and coverage of measurements (e.g. targeted vs non-targeted, number of metabolites measured) as well as to variability in analysis methods (e.g. comparison of levels between groups, analysis of association with eGFR or incident CKD). Moreover, these findings originate only from observational studies of different designs, sizes and characteristics. Observational associations do not allow for establishing causal relationships between a metabolite and kidney function or disease, and can be limited in their ability to distinguish between cause and consequence6,17.
Experimental studies also support a relationship between metabolites of the tryptophan pathway and kidney function: For example, an excess in the conversion of tryptophan to N-formylkynurenine was shown to promote renal ischemia–reperfusion injury in mice18. Korstanje et al. showed that the knockdown of kynurenine 3-mono-oxygenase gene expression in zebrafish and systemic deletion of the gene in mice led to a proteinuric phenotype19. C-mannosylation as a specific type of glycosylation was shown to be increased under hyperglycemic conditions, prompting a hypothesis of its pathological role in the development of diabetic complications20. Another example, serum 2-(α-mannopyranosyl)-l-tryptophan was found to be an accurate measure of renal function in rats21. Kobayashi et al. finally demonstrated in an untargeted metabolite screen of rats with and without CKD that metabolites of the tryptophan pathway might be used to detect early stages of CKD22. Thus, complementary lines of evidence connect metabolites of the tryptophan pathway to kidney function and disease.
Here, we designed a study to address the following question: Are tryptophan, kynurenine or other metabolites of the tryptophan pathway in blood causally related to kidney function, or does reduced kidney function influence blood metabolite levels? We used a bidirectional Mendelian randomization (MR) approach allowing inference of causality17,23. An extension of this approach conveniently allows for the usage of publicly available summary statistics from published genetic association studies24.
Results
More than 40 metabolites qualified as down-stream products of tryptophan in humans (Fig. 1, Supplementary Information 1). The search for relevant published studies focused on genome-wide association studies (GWAS) of these metabolites. In addition, we searched for GWAS of kidney function. Five published studies including four GWAS of metabolites and one large GWAS of eGFR were identified and used as the basis of this MR analysis (“Methods”, Supplementary Table 2).
Based on published results of selected GWAS, genetic variants were identified as instruments for the respective trait (see “Methods” for details). The number of selected instruments per trait ranged from one instrument to 22 instruments for metabolites and resulted in 73 instruments for eGFR (Supplementary Table 3). Selected instruments for a given trait are not only independent of each other but with few exceptions also across traits (R2 < 0.2, Supplementary Table 3). Whenever possible, the strength of genetic variants was estimated in terms of F statistic and ranged between 95 and 631 for eGFR and between 18 and 135 for metabolites (Supplementary Table 3)25,26.
Mendelian randomization analysis reveals four significant effects
Overall, 18 MR analyses could be conducted (Supplementary Table 3): For the direction: metabolite → kidney function, ten different metabolites of the tryptophan pathway and the kidney function measure eGFR were evaluated. Data from more than one GWAS were available for kynurenine (N = 2) and indole-3-propionate (N = 3). For the other direction: kidney function → metabolite, MR analysis was carried out for eGFR and eight metabolites from one GWAS.
Reported pairwise correlations of metabolites ranged from − 0.24 for tryptophan and C-glycosyltryptophan to 0.72 for kynurenate and xanthurenate, with many of them close to zero (Supplementary Fig. 1).
In the MR analysis with metabolites as exposure and eGFR as the outcome, none of the reported effect estimates reached significance after correction for multiple testing (p > 2.78e−03; Table 1). The effect of xanthurenate on eGFR was nominally significant (effect estimate = − 0.0086; p = 2.9e−02). Conversely, eGFR affected the levels of four metabolites: C-glycosyltryptophan (effect estimate = − 0.16, 95% confidence interval [CI] (− 0.22; − 0.1); p = 9.2e−08), kynurenine (effect estimate = − 0.18, 95% CI (− 0.25; − 0.11); p = 1.1e−06), 3-indoxyl sulfate (effect estimate = − 0.25, 95% CI (− 0.4; − 0.11); p = 6.3e−04) and indole-3-lactate (effect estimate = − 0.26, 95% CI (− 0.38; − 0.13); p = 5.4e−05; Table 1). The negative estimates for all four metabolites indicate that higher levels of metabolites are a consequence of lower eGFR. Further interpretation of effect estimates with regards to their magnitude was not possible due to the nature of metabolite measurements (semi-quantitative, unitless) and different handling of measurements (transformation) in the underlying studies (Supplementary Table 2).
Table 1.
Main results of Mendelian randomization analyses.
| Exposure: metabolite, unitless | Outcome: eGFR, mL/min/1.73 m2 (Pattaro et al. 201690) | |||||||
|---|---|---|---|---|---|---|---|---|
| N SNPs | Main analysis resultsa | Main sensitivity analysis results | ||||||
| Effect Estimate (SE) | 95% CI | p | Egger regression | MR-PRESSO global test p | ||||
| Effect Estimate (SE) | Intercept (SE) | |||||||
| Direction: metabolite → kidney function | ||||||||
| 3-indoxyl sulfate | Rhee et al. 201386 | 1 | − 0.0012 (0.0041) | (− 0.0092; 0.0068) | 7.68E−01 | NA | NA | NA |
| indoleacetate | Shin et al. 201487 | 2 | 0.00001 (0.0375) | (− 0.0735; 0.0735) | 1.00E + 00 | NA | NA | NA |
| indoleacetylglutamine | Long et al. 201789 | 1 | 0.0004 (0.0058) | (− 0.0110; 0.0117) | 9.51E−01 | NA | NA | NA |
| indole-3-lactate | Shin et al. 201487 | 1 | − 0.08 (0.05) | (− 0.18; 0.01) | 8.11E−02 | NA | NA | NA |
| indole-3-propionate | Rhee et al. 201386 | 1 | − 0.0029 (0.0044) | (− 0.0116; 0.0058) | 5.13E−01 | NA | NA | NA |
| indole-3-propionate | Shin et al. 201487 | 2 | 0.01 (0.02) | (− 0.03; 0.06) | 5.14E−01 | NA | NA | NA |
| indole-3-propionate | Long et al. 201789 | 1 | − 0.01 (0.01) | (− 0.03; 0) | 5.73E−02 | NA | NA | NA |
| kynurenate | Long et al. 201789 | 1 | 0.01 (0.01) | (0; 0.02) | 7.74E−02 | NA | NA | NA |
| kynurenine | Rhee et al. 201386 | 1 | <|0.00001| (0.0034)b | (− 0.0067; 0.0067) | 1.00E + 00 | NA | NA | NA |
| kynurenine | Shin et al. 201487 | 5 | − 0.04 (0.04) | (− 0.12; 0.05) | 3.69E−01 | 0.03 (0.16) | − 0.0012 (0.0027) | 2.94E−02 |
| tryptophan | Shin et al. 201487 | 22 | 0.04 (0.04) | (− 0.03; 0.11) | 3.04E−01 | − 0.23 (0.22) | 0.0015 (0.0012) | 8.61E−01 |
| quinolinate | Rhee et al. 201386 | 2 | 0.0026 (0.0032) | (− 0.0038; 0.0089) | 4.29E−01 | NA | NA | NA |
| xanthurenate | Rhee et al. 201386 | 1 | − 0.0086 (0.0039) | (− 0.0162; − 0.0009) | 2.92E−02 | NA | NA | NA |
| Outcome: metabolite, unitless | Exposure: eGFR, mL/min/1.73 m2 (Pattaro et al. 201690) | |||||||
|---|---|---|---|---|---|---|---|---|
| N SNPs | Main analysis resultsa | Main sensitivity analysis results | ||||||
| Effect Estimate (SE) | 95% CI | p | Egger regression | MR-PRESSO global test p | ||||
| Effect Estimate (SE) | Intercept (SE) | |||||||
| Direction: kidney function → metabolite | ||||||||
| 3-indoxyl sulfate | Shin et al. 201487 | 73 | − 0.25 (0.07) | (− 0.4; − 0.11) | 6.26E−04 | − 0.05 (0.27) | − 0.0016 (0.0021) | 2.46E−01 |
| C-glycosyltryptophan | Shin et al. 201487 | 73 | − 0.16 (0.03) | (− 0.22; − 0.1) | 9.22E−08 | − 0.14 (0.12) | − 0.0002 (0.0009) | 1.49E−01 |
| indoleacetate | Shin et al. 201487 | 73 | − 0.05 (0.07) | (− 0.19; 0.09) | 5.03E−01 | − 0.19 (0.26) | 0.0011 (0.0020) | 1.20E−02 |
| indole-3-lactate | Shin et al. 201487 | 73 | − 0.26 (0.06) | (− 0.38; − 0.13) | 5.43E−05 | − 0.35 (0.24) | 0.0007 (0.0018) | 3.00E−04 |
| indole-3-propionate | Shin et al. 201487 | 73 | − 0.05 (0.08) | (− 0.2; 0.1) | 5.26E−01 | − 0.18 (0.29) | 0.0010 (0.0022) | 1.02E−01 |
| kynurenine | Shin et al. 201487 | 73 | − 0.18 (0.04) | (− 0.25; − 0.11) | 1.07E−06 | − 0.18 (0.13) | 0.0001 (0.0010) | 3.17E−01 |
| serotonin | Shin et al. 201487 | 73 | − 0.13 (0.08) | (− 0.28; 0.02) | 7.85E−02 | 0.03 (0.28) | − 0.0013 (0.0021) | 2.27E−02 |
| tryptophan | Shin et al. 201487 | 73 | 0.04 (0.02) | (− 0.01; 0.09) | 1.08E−01 | 0.02 (0.09) | 0.0002 (0.0006) | 3.20E−02 |
Note of caution: Because of unitless metabolite levels and differences in applied transformations of traits in underlying genome-wide association studies, effect estimates and confidence intervals are limited in their interpretation and comparison.
SE standard error, CI confidence interval. Association results marked in bold passed significance threshold corrected for multiple testing: 0.05/(10 + 8) = 2.78E−03 (A: 10 Metabolites B: 8 Metabolites).
aThe Wald ratio estimator was used for effect estimation if one single instrument was available, and the inverse variance weighted estimation method if more than one instrument was available.
bDue to numerical constraints, no more exact estimate can be provided; estimated effect is within the range of − 0.00001 and + 0.0001. NA: Respective analysis was not feasible due to small number of instruments (< 3).
Sensitivity analyses support a role of low eGFR in elevated blood metabolites of the tryptophan pathway
As outlined in the “Methods” section, additional analyses were performed to evaluate the robustness of the results and to ensure the absence of any major violations of the assumptions underlying MR (Supplementary Table 4, Supplementary Fig. 2). Overall, the results were consistent. Focussing on significant observations:
For C-glycosyltryptophan and kynurenine, all estimates of the effects using additional estimation methods were direction-consistent showing little variation (range of effect estimates: − 0.14; − 0.21, Supplementary Table 4, Supplementary Fig. 3). Neither the estimate of Egger intercept nor other statistics indicated any violations of the MR assumptions. Similarly, there was also no indications for any violations for 3-indoxyl sulfate, although the range of effect estimates was wider (− 0.05; − 0.29).
In contrast, the heterogeneity test (p = 4.3E−04) as well as the MR-PRESSO test (p = 3.0E−04) indicated some violations of underlying assumptions for indole-3-lactate (Supplementary Table 4). Since MR-PRESSO implicated two SNPs (marked in Supplementary Fig. 3-D) as outliers, an additional analysis was conducted after exclusion of these two SNPs. The outlier-corrected effect estimate of eGFR on indole-3-lactate, however, differed only marginally from the estimate of the main analysis (effect estimates: − 0.24 vs − 0.26, respectively) as well as results of other methods (range of effect estimates: − 0.23; − 0.48) and therefore supported the relationship despite the indications for a violation of the MR assumption that instruments are only associated with the outcome through the exposure.
In addition to sensitivity analyses using statistical methods, we assessed the validity of the results from a biological perspective. Besides the potential issues caused by the inclusion of pleiotropic SNPs, we also addressed the potential issue of the inclusion of weak instruments or instruments with spurious associations by utilizing differently defined subsets of SNPs (“Methods”, Supplementary Table 5). The results of these additional sensitivity analyses were quite similar in comparison to the main analysis (Supplementary Table 4), supporting the validity of the main results on the relationship of eGFR with these four blood metabolites.
Discussion
Here, we carried out a bidirectional two-sample MR analysis to study whether blood metabolite levels of the tryptophan pathway causally affect the kidney function measure eGFR, or whether kidney function affects levels of these metabolites in blood. After correction for multiple testing, the principal finding of this study indicates that lower eGFR causes higher levels of the four metabolites kynurenine, C-glycosyltryptophan, 3-indoxyl sulfate and indole-3-lactate in blood, whereas the opposite analysis did not support a causal role of these metabolites on eGFR.
Kynurenine is synthesized from tryptophan via N-formylkynurenine10,13,27. Levels of kynurenine are elevated in ESKD patients and a toxic effect has been discussed28,29. Several cross-sectional and case–control studies reported increased levels of kynurenine in patients with reduced kidney function thereby supporting our finding16,30–35.
In contrast to kynurenine, levels of tryptophan are depleted in patients with reduced kidney function33,36–38. This might indicate an increased activity of enzymes converting tryptophan to kynurenine via N-formylkynurenine18,27,37–44. Indeed, observational studies in humans reported an association between the tryptophan/kynurenine ratio and lower eGFR15,31,38. Our results do not indicate a causal relationship between tryptophan and kidney function of any direction. As there are no genetic association results for metabolite ratios such as the tryptophan/kynurenine ratio available, no MR analysis could be conducted with respect to this reported observation.
C-glycosyltryptophan (or C-mannosyltryptophan) results from a post-translational modification of tryptophan by linking a sugar via a carbon–carbon bond45,46. Certain post-translational modifications such as carbamylation have been linked to different chronic conditions including renal failure47. Studies in humans showed consistently increased levels of C-glycosyltryptophan in people with decreased eGFR16,48–52. Moreover, studies of humans and animal models suggested C-glycosyltryptophan to be a good indicator of renal function with more favourable properties compared to serum creatinine16,21,51,53. Thus, our finding that eGFR has an impact on blood levels of C-glycosyltryptophan is supported by published results. Although there is no literature to support the opposite direction in general, some prospective studies in humans (population-based or diabetic cohorts) showed that higher levels of C-glycosyltryptophan are associated with an increased risk for eGFR decline, even after adjustment for baseline eGFR16,50,54. In addition, Sekula et al. reported that the significant risk for ESKD in CKD patients was attenuated after adjustment for baseline eGFR and lost significance after adjustment for baseline measured GFR16. While our study did not indicate a causal relationship of C-glycosyltryptophan on eGFR in general, higher levels of C-glycosyltryptophan may still be of relevance in patients with advanced CKD.
The other two metabolites, 3-indoxyl sulfate and indole-3-lactate, belong to the indole pathway and are synthesized by gut bacteria11,55,56. Disturbances of the microbiome have been associated with many chronic diseases such as CKD11,57–66, and a bidirectional relationship between host and microbiome has been reported56,60,67,68.
3-Indoxyl sulfate, also known as indican, is a known uremic toxin with highly elevated levels in ESKD patients28,29,44,69–71. Its toxic effect on kidney function as well as other organs and pathways is still being discussed6,72. Several observational studies on humans with and/or without CKD reported associations between higher levels of 3-indoxyl sulfate and lower eGFR16,30,32,33,35,73–76. Increased levels of 3-indoxyl sulfate were also observed in rats with autosomal dominant polycystic kidney disease (ADPKD) versus control rats77. All considered, the consistent results of these studies support our finding on the impact of eGFR on 3-indoxyl sulfate but not on its discussed toxic effect on kidney function.
In contrast, there is not much data available for indole-3-lactate. In a previous association study, higher levels of indole-3-lactate were found to be cross-sectionally associated with lower eGFR16. In an elderly population, indole-3-lactate was found to be associated not only with muscle composition but also with the renal function markers serum creatinine and blood urea nitrogen78. Quite recently, a study group reported that increased levels of indole-3-lactate were found to be associated with lower eGFR in a cohort of patients with ADPKD79. Altogether, study results support a causal relationship of low eGFR on blood indole-3-lactate levels.
Strengths of this MR study are the availability of multiple metabolites within a specific pathway, the use of various publicly available datasets based on rigorously conducted GWAS meta-analyses, the conduct of statistically as well as biologically motivated sensitivity analyses, and consistent results pointing into one direction. The analysis of summary statistics also has some disadvantages and depends upon the quality and power of included studies. All studies incorporated in this MR study are observational and vary with respect to several characteristics:
Besides differences in general study characteristics and in analysis strategies (e.g. transformation of traits), groups used different laboratory platforms (genetics, metabolomics) with measurements of varying quality (e.g. semi-quantitative measurements), limiting their comparability. It also prevented pooling of studies to gain additional power. Furthermore, interpretation of effect estimates is thus hampered as well as their comparison to reported associations. Here, interpretation of results is restricted to the direction of effects.
Furthermore, the coverage of evaluated metabolites varied over time because of advances in metabolite quantification. In combination with only partial reporting and sharing of analysis results, the search for relevant GWAS for studies such as ours is limited in itself. In consequence, the MR analyses were restricted to a subset of metabolites of the tryptophan pathway. Still, analyses could be conducted for several metabolites and data from more than one source was partially available to evaluate relationship for some metabolite-kidney function combinations.
The population sizes of selected studies were quite different, with the sample size for eGFR being much larger than the currently largest reported studies for the metabolites (Supplementary Table 2), which may raise concern about differential statistical power for bidirectional MR. However, the tight link between the genome and the metabolome does not require a similar study size as needed for the evaluation of a complex trait such as eGFR, as evidenced in large F statistics of the selected metabolite instruments. Since levels of metabolites are often influenced by a single gene, the low number of instruments for metabolites is therefore not unexpected. In contrast, we used several SNPs as instruments for eGFR reflecting its complex nature.
A limitation inherent to all MR-studies is that effect estimates might be biased in the presence of weak instruments that do not reflect lifelong exposures to altered levels of the factor of interest such as eGFR or a metabolite strongly enough. To ensure inclusion of strong instruments, we addressed this challenge by selecting only genome-wide significant genetic variants80. In addition, we evaluated the strength of instruments by estimating the F statistics whenever possible. Instruments of eGFR were stronger than those of metabolites because of the large sample size of the eGFR GWAS, but metabolite instruments still had large F statistics. Sensitivity analyses restricting instruments to SNPs with the very strongest or – in case of eGFR – strong and replicated associations supported the main findings based on all identified instruments.
Another concern in MR-studies is the presence of pleiotropy, such that genetic instruments are associated with the studied outcome directly or via other paths. Here, we also observed that there were few instances in which the same or a closely linked genetic variant was selected as instrument for more than one metabolic trait. For metabolites, this is possible even in the absence of pleiotropy, because, for instance, a genetic variant in an enzyme can be associated both with its metabolic substrate and its product. We addressed this potential limitation by using a variety of additional, more robust estimation methods81 as well as by using subsets of SNPs thereby reducing risk of pleiotropy. However, sensitivity analyses could be conducted only if a sufficient number of instruments for the respective exposure was available. Although we cannot completely rule out any pleiotropy, the results of our sensitivity analyses supported the main finding that eGFR is related to blood levels of tryptophan metabolites.
Lastly, our findings are based on summary statistics from population-based cohorts with relatively few individuals with advanced CKD. While our results do not indicate that metabolite levels of the tryptophan pathway are causally related to eGFR, our data are not sufficient to address the question whether these levels are related to CKD progression in patients with pre-existing CKD. Moreover, limited data did not allow to address other traits of kidney function (e.g. urinary albumin-to-creatinine ratio, UACR).
In summary, our study supports that lower eGFR causes higher blood levels of four metabolites of the tryptophan pathway kynurenine, C-glycosyltryptophan, 3-indoxyl sulfate, and indole-3-lactate, whereas higher levels of these metabolites did not cause lower eGFR. Based on our MR study, these findings thus aid the notion that elevations of metabolites of the tryptophan pathway are a consequence rather than a cause of reduced eGFR. Further research is needed to specifically examine these relationships with respect to CKD progression among patients with existing CKD.
Methods
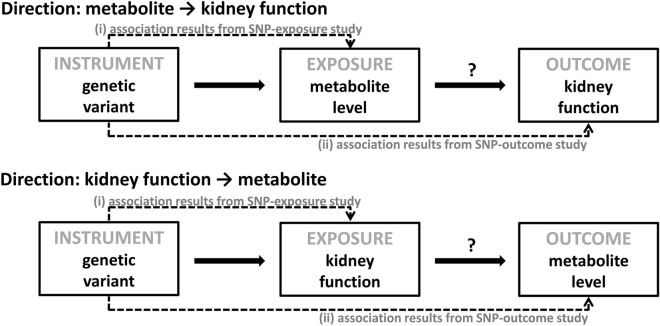
A bidirectional MR approach was used to infer both directions, whether metabolite levels in blood causally affect kidney function, or whether kidney function levels causally affect metabolite levels in blood using publicly available genetic association summary data (two-sample MR)17,23,24. The underlying idea of the MR method is to use genetic variants as instruments to assign participants of a study to exposure groups in order to evaluate the causal relationship of this exposure with an outcome82. In this MR study, exposures and outcomes of interest for the direction: metabolite → kidney function, are blood metabolite levels of the tryptophan pathway and the kidney function marker eGFR, respectively (Fig. 2). For the other direction: kidney function → metabolite, the assignment is reversed. Since germline genetic variants are randomly assigned during gamete formation and represent a non-modifiable, lifelong exposure, the assignment to exposure groups using such variants as instruments conceptually corresponds to the randomization of participants in a clinical trial. For example, persons can be exposed to genetically determined higher or lower blood tryptophan levels over the course of their life. In order to represent a valid instrument, a genetic variant needs to fulfil three assumptions: (1) it must be associated with the exposure, (2) it must not be associated with confounders of the exposure-outcome relationship, and (3) it must only be associated with the outcome through the exposure17.
Figure 2.
Analytical approach of Mendelian Randomization applied in this study.
Tryptophan pathway
Because of the research question addressing not just one metabolite but a whole pathway, a biology-centered literature search to identify metabolites of the tryptophan pathway in humans was conducted. Briefly, the basic tryptophan metabolism structure was obtained from the KEGG database83, and then each reaction was cross-checked in the Reactome database (tryptophan catabolism, URL with https://doi.org/10.3180/REACT_916.2; serotonin and melatonin biosynthesis, URL with StableID: R-HSA-209931; Nicotinate metabolism, URL with StableID: R-HSA-196807)10,84,85. While some typical metabolites related with tryptophan lack of direct reactions, we assembled all obtained information into Fig. 1. As a result, over 40 metabolites that qualified as downstream products of tryptophan were identified. A summarizing description of the pathway can be found in Supplementary Information S1.
Selected studies
MR analyses were based on publicly available research results that reported on the association between genome-wide genetic variants with either metabolites of interest or eGFR, a measure of kidney function.
Since we were interested in several metabolites (Fig. 1), we evaluated genetic screens of many metabolites. Based on the results of a systematic literature search in December of 20175, we identified studies that met the following pre-defined criteria: (1) evaluation of serum or plasma samples, (2) total population size of > 1,500 participants, (3) evaluation of at least one metabolite of interest, and (4) publicly available genome-wide association summary statistics. After exclusion of studies that were part of a more comprehensive study, four studies were selected (Supplementary Table 6)86–89. All four studies reported association results for a varying number of metabolites of interest (range: 1–13). Some metabolites were available in more than one study.
Regarding eGFR as a measure of kidney function, we used the most comprehensive results from a GWAS meta-analysis of eGFR published by the Chronic Kidney Disease Genetics (CKDGen) consortium in 2016 that were available at the time our MR study started90.
Basic characteristics of all five included studies are summarized in Supplementary Table 2. The number of single study groups contributing to the selected summary statistics varied greatly (range: 1–48). Some of the single study groups contributed to both, GWAS of metabolites and of eGFR. For example, data from the KORA F4 study were included into the GWAS of metabolites of Shin et al. (~ 23% of total study population) as well as into the GWAS of eGFR where its proportion to the whole study population is quite negligible (< 3%)87,90.
Selection of instruments
Several steps were required to prepare data for the different analyses (Supplementary Information 2). Briefly, genetic instruments were SNPs with an association p-value < 5 × 10–8 (i.e. genome-wide significant) in the GWAS of the respective exposure that were also present in the GWAS of the respective outcome. In case several SNPs were available for an exposure, only independent SNPs were considered (linkage disequilibrium R2 < 0.2). After alignment and harmonization of data from different sources, data for ten metabolites (exposures) from three studies were available for the analysis addressing the direction: metabolite → kidney function86,87,89. For the other direction: kidney function → metabolite, only the study by Shin et al. contributed data of eight metabolites (outcomes)87, because the studies by Rhee et al. and Long et al. did not publish full genome-wide association summary statistics (Supplementary Table 2)86,89. Regarding eGFR, data for the analysis of both directions were available90.
Statistical analysis
All analyses were conducted using R: A Language and Environment for Statistical Computing (https://www.R-project.org) in combination with the R package TwoSampleMR (MR-Base)81.
MR analysis was conducted for all exposures with at least one available instrument (Supplementary Table 3). In the main analysis, the effect for each instrument was estimated with the Wald ratio estimator and combined with the inverse variance weighted (IVW) meta-analysis method when several instruments were available80. To correct for multiple testing, statistical significance was defined using a Bonferroni correction that acknowledges the conduct of 18 MR analyses (metabolite → kidney function: 10; kidney function → metabolite: 8): 0.05/(10 + 8) = 2.78E−03. While in the main analysis, all selected instruments were used, the analyses were repeated in a sensitivity analysis with instruments that were selected based on a more stringent pruning (LD R2 < 0.001).
For all MR analyses utilizing more than three instruments, sensitivity analyses were conducted in order to evaluate the robustness of results and to assess different potential violations of the assumptions underlying MR, using additional estimation methods as provided by the TwoSampleMR-package such as median-based estimation and Egger regression81. In addition, the MR-PRESSO method was applied to detect outliers and to provide an outlier-corrected effect estimate91. An overview of all estimation methods is provided in Supplementary Information 3.
Furthermore, we annotated all selected genetic instruments for their associations with other complex traits and diseases using the comprehensive SNiPA database as well as GeneATLAS92,93. This annotation allowed for the evaluation of potential pleiotropy from a non-statistical perspective. Based on the obtained annotations, we defined two subsets of SNPs to minimize the risk of bias due to pleiotropy (Supplementary Table 5, Selection 1 [less stringent] and 2 [more stringent]). In addition, we used subsets of SNPs with very strong associations with the respective trait (p < 10–20 and p < 10–10) to evaluate potential bias due to the inclusion of weaker instruments (Supplementary Table 5, Selection 3). For eGFR as exposure, we specifically used another subset of eGFR instruments that was restricted to replicated SNPs to minimize the chance of including SNPs with false-positive associations (Supplementary Table 5, Selection 4). Respective MR analyses were conducted using the Wald ratio estimator (1 instrument) or the IVW estimator (> 1 instrument).
Finally, we excluded single variants selected for several traits or in LD (R2 > 0.2) with other variants selected for other traits in a separate sensitivity analysis whenever possible.
Supplementary information
Acknowledgements
We thank Gabi Kastenmüller, Corresponding Author of Shin et al. (PMID: 24816252), for the additional input on their work and Ulla T. Schultheiss for improving the language. The presented study is based on publicly available data. There was no dedicated funding for this study. YC was supported by a scholarship of the Chinese Research Council. The work of AK was supported by DFG KO 3598/5-1 (Heisenberg Professorship). The work of YL was supported by the German Research Foundation (KO 3598/4-2). Open access funding provided by Projekt DEAL.
Author contributions
P.S. and A.K. designed the study; Y.C., Y.L., P.B. and P.S. did literature searches and data analyses; Y.C., C.L., A.K. and P.S. interpreted results, all authors drafted/revised the manuscript and approved its final version.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-69559-x.
References
- 1.Stevens LA, Coresh J, Greene T, Levey AS. Assessing kidney function–measured and estimated glomerular filtration rate. N. Engl. J. Med. 2006;354:2473–2483. doi: 10.1056/NEJMra054415. [DOI] [PubMed] [Google Scholar]
- 2.Eckardt KU, et al. Evolving importance of kidney disease: from subspecialty to global health burden. Lancet. 2013;382:158–169. doi: 10.1016/S0140-6736(13)60439-0. [DOI] [PubMed] [Google Scholar]
- 3.Levin A, et al. Global kidney health 2017 and beyond: a roadmap for closing gaps in care, research, and policy. Lancet. 2017;390:1888–1917. doi: 10.1016/S0140-6736(17)30788-2. [DOI] [PubMed] [Google Scholar]
- 4.Hocher B, Adamski J. Metabolomics for clinical use and research in chronic kidney disease. Nat. Rev. Nephrol. 2017;13:269–284. doi: 10.1038/nrneph.2017.30. [DOI] [PubMed] [Google Scholar]
- 5.Kottgen A, Raffler J, Sekula P, Kastenmuller G. Genome-wide association studies of metabolite concentrations (mGWAS): relevance for nephrology. Semin. Nephrol. 2018;38:151–174. doi: 10.1016/j.semnephrol.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Rhee EP. A systems-level view of renal metabolomics. Semin. Nephrol. 2018;38:142–150. doi: 10.1016/j.semnephrol.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beckonert O, et al. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat. Protoc. 2007;2:2692–2703. doi: 10.1038/nprot.2007.376. [DOI] [PubMed] [Google Scholar]
- 8.Evans A, et al. High resolution mass spectrometry improves data quantity and quality as compared to unit mass resolution mass spectrometry in high-throughput profiling metabolomics. Metabolomics. 2014;4:1. [Google Scholar]
- 9.Johnson CH, Ivanisevic J, Siuzdak G. Metabolomics: beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell. Biol. 2016;17:451–459. doi: 10.1038/nrm.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peters JC. Tryptophan nutrition and metabolism: an overview. Adv. Exp. Med. Biol. 1991;294:345–358. doi: 10.1007/978-1-4684-5952-4_32. [DOI] [PubMed] [Google Scholar]
- 11.Keszthelyi D, Troost FJ, Masclee AA. Understanding the role of tryptophan and serotonin metabolism in gastrointestinal function. Neurogastroenterol. Motil. 2009;21:1239–1249. doi: 10.1111/j.1365-2982.2009.01370.x. [DOI] [PubMed] [Google Scholar]
- 12.Zhu W, et al. Quantitative profiling of tryptophan metabolites in serum, urine, and cell culture supernatants by liquid chromatography-tandem mass spectrometry. Anal. Bioanal. Chem. 2011;401:3249–3261. doi: 10.1007/s00216-011-5436-y. [DOI] [PubMed] [Google Scholar]
- 13.Strasser B, Gostner JM, Fuchs D. Mood, food, and cognition: role of tryptophan and serotonin. Curr. Opin. Clin. Nutr. Metab. Care. 2016;19:55–61. doi: 10.1097/MCO.0000000000000237. [DOI] [PubMed] [Google Scholar]
- 14.Goek ON, et al. Serum metabolite concentrations and decreased GFR in the general population. Am. J. Kidney Dis. 2012;60:197–206. doi: 10.1053/j.ajkd.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 15.Goek ON, et al. Metabolites associate with kidney function decline and incident chronic kidney disease in the general population. Nephrol. Dial Transplant. 2013;28:2131–2138. doi: 10.1093/ndt/gft217. [DOI] [PubMed] [Google Scholar]
- 16.Sekula P, et al. A metabolome-wide association study of kidney function and disease in the general population. J Am Soc Nephrol. 2016;27:1175–1188. doi: 10.1681/ASN.2014111099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sekula P, Del Greco MF, Pattaro C, Kottgen A. Mendelian randomization as an approach to assess causality using observational data. J. Am Soc. Nephrol. 2016;27:3253–3265. doi: 10.1681/asn.2016010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohib K, et al. Indoleamine 2,3-dioxygenase expression promotes renal ischemia-reperfusion injury. Am. J. Physiol. Renal. Physiol. 2008;295:F226–234. doi: 10.1152/ajprenal.00567.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korstanje R, et al. Loss of Kynurenine 3-mono-oxygenase causes proteinuria. J. Am. Soc. Nephrol. 2016;27:3271–3277. doi: 10.1681/ASN.2015070835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ihara Y, et al. Increased expression of protein C-mannosylation in the aortic vessels of diabetic Zucker rats. Glycobiology. 2005;15:383–392. doi: 10.1093/glycob/cwi012. [DOI] [PubMed] [Google Scholar]
- 21.Takahira R, et al. Tryptophan glycoconjugate as a novel marker of renal function. Am. J. Med. 2001;110:192–197. doi: 10.1016/S0002-9343(00)00693-8. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi T, et al. Exploration of novel predictive markers in rat plasma of the early stages of chronic renal failure. Anal. Bioanal. Chem. 2014;406:1365–1376. doi: 10.1007/s00216-013-7472-2. [DOI] [PubMed] [Google Scholar]
- 23.Lawlor DA, Harbord RM, Sterne JAC, Timpson N, Smith GD. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat. Med. 2008;27:1133–1163. doi: 10.1002/sim.3034. [DOI] [PubMed] [Google Scholar]
- 24.Pierce BL, Burgess S. Efficient design for Mendelian randomization studies: subsample and 2-sample instrumental variable estimators. Am J Epidemiol. 2013;178:1177–1184. doi: 10.1093/aje/kwt084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wuttke M, et al. A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat. Genet. 2019;51:957–972. doi: 10.1038/s41588-019-0407-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burgess S, Dudbridge F, Thompson SG. Combining information on multiple instrumental variables in Mendelian randomization: comparison of allele score and summarized data methods. Stat. Med. 2016;35:1880–1906. doi: 10.1002/sim.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saito K, et al. Mechanism of increases in L-kynurenine and quinolinic acid in renal insufficiency. Am. J. Physiol. Renal. Physiol. 2000;279:F565–572. doi: 10.1152/ajprenal.2000.279.3.F565. [DOI] [PubMed] [Google Scholar]
- 28.Vanholder R, et al. Review on uremic toxins: classification, concentration, and interindividual variability. Kidney Int. 2003;63:1934–1943. doi: 10.1046/j.1523-1755.2003.00924.x. [DOI] [PubMed] [Google Scholar]
- 29.Mair RD, Sirich TL, Meyer TW. Uremic toxin clearance and cardiovascular toxicities. Toxins. 2018;10:1. doi: 10.3390/toxins10060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kobayashi T, et al. A metabolomics-based approach for predicting stages of chronic kidney disease. Biochem. Biophys. Res. Commun. 2014;445:412–416. doi: 10.1016/j.bbrc.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 31.Schefold JC, et al. Increased indoleamine 2,3-dioxygenase (IDO) activity and elevated serum levels of tryptophan catabolites in patients with chronic kidney disease: a possible link between chronic inflammation and uraemic symptoms. Nephrol. Dial Transplant. 2009;24:1901–1908. doi: 10.1093/ndt/gfn739. [DOI] [PubMed] [Google Scholar]
- 32.Rhee EP, et al. Metabolite profiling identifies markers of uremia. J. Am. Soc. Nephrol. 2010;21:1041–1051. doi: 10.1681/ASN.2009111132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toyohara T, et al. Metabolomic profiling of uremic solutes in CKD patients. Hypertens. Res. 2010;33:944–952. doi: 10.1038/hr.2010.113. [DOI] [PubMed] [Google Scholar]
- 34.Rhee EP, et al. A combined epidemiologic and metabolomic approach improves CKD prediction. J. Am. Soc. Nephrol. 2013;24:1330–1338. doi: 10.1681/ASN.2012101006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen DQ, et al. Gene and protein expressions and metabolomics exhibit activated redox signaling and wnt/beta-catenin pathway are associated with metabolite dysfunction in patients with chronic kidney disease. Redox. Biol. 2017;12:505–521. doi: 10.1016/j.redox.2017.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rhee EP, Thadhani R. New insights into uremia-induced alterations in metabolic pathways. Curr. Opin. Nephrol. Hy. 2011;20:593–598. doi: 10.1097/MNH.0b013e32834b8a1d. [DOI] [PubMed] [Google Scholar]
- 37.Breit M, Weinberger KM. Metabolic biomarkers for chronic kidney disease. Arch. Biochem. Biophys. 2016;589:62–80. doi: 10.1016/j.abb.2015.07.018. [DOI] [PubMed] [Google Scholar]
- 38.Debnath S, et al. Tryptophan metabolism in patients with chronic kidney disease secondary to type 2 diabetes: relationship to inflammatory markers. Int. J. Tryptophan. Res. 2017;10:1178646917694600. doi: 10.1177/1178646917694600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pawlak K, Domaniewski T, Mysliwiec M, Pawlak D. The kynurenines are associated with oxidative stress, inflammation and the prevalence of cardiovascular disease in patients with end-stage renal disease. Atherosclerosis. 2009;204:309–314. doi: 10.1016/j.atherosclerosis.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 40.Koenig P, et al. Enhanced degradation of tryptophan in patients on hemodialysis. Clin. Nephrol. 2010;74:465–470. doi: 10.5414/cnp74465. [DOI] [PubMed] [Google Scholar]
- 41.Kaden J, Abendroth D, Volp A, Marzinzig M. Dynamics and diagnostic relevance of Kynurenine serum level after kidney transplantation. Ann. Transplant. 2015;20:327–337. doi: 10.12659/AOT.893721. [DOI] [PubMed] [Google Scholar]
- 42.42de Vries, L. V. et al. The tryptophan/kynurenine pathway, systemic inflammation, and long-term outcome after kidney transplantation. Am J Physiol Renal Physiol313, F475-F486, doi:10.1152/ajprenal.00690.2016 (2017). [DOI] [PubMed]
- 43.Nelp MT, et al. Immune-modulating enzyme indoleamine 2,3-dioxygenase is effectively inhibited by targeting its apo-form. Proc. Natl. Acad. Sci. U S A. 2018;115:3249–3254. doi: 10.1073/pnas.1719190115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saucedo AL, et al. Chronic kidney disease and the search for new biomarkers for early diagnosis. Curr. Med. Chem. 2018;25:3719–3747. doi: 10.2174/0929867325666180307110908. [DOI] [PubMed] [Google Scholar]
- 45.Furmanek A, Hofsteenge J. Protein C-mannosylation: facts and questions. Acta Biochim. Pol. 2000;47:781–789. doi: 10.18388/abp.2000_3996. [DOI] [PubMed] [Google Scholar]
- 46.Manabe S, Marui Y, Ito Y. Total synthesis of mannosyl tryptophan and its derivatives. Chemistry. 2003;9:1435–1447. doi: 10.1002/chem.200390163. [DOI] [PubMed] [Google Scholar]
- 47.Gillery P, Jaisson S. Post-translational modification derived products (PTMDPs): toxins in chronic diseases? Clin. Chem. Lab. Med. 2014;52:33–38. doi: 10.1515/cclm-2012-0880. [DOI] [PubMed] [Google Scholar]
- 48.Shah VO, et al. Plasma metabolomic profiles in different stages of CKD. Clin. J. Am. Soc. Nephrol. 2013;8:363–370. doi: 10.2215/CJN.05540512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Niewczas MA, et al. Uremic solutes and risk of end-stage renal disease in type 2 diabetes: metabolomic study. Kidney Int. 2014;85:1214–1224. doi: 10.1038/ki.2013.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Solini A, et al. Prediction of declining renal function and albuminuria in patients with type 2 diabetes by metabolomics. J. Clin. Endocrinol. Metab. 2016;101:696–704. doi: 10.1210/jc.2015-3345. [DOI] [PubMed] [Google Scholar]
- 51.Sekula P, et al. From discovery to translation: characterization of C-mannosyltryptophan and pseudouridine as markers of kidney function. Sci. Rep. 2017;7:17400. doi: 10.1038/s41598-017-17107-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haukka JK, et al. Metabolomic profile predicts development of microalbuminuria in individuals with type 1 diabetes. Sci. Rep. 2018;8:13853. doi: 10.1038/s41598-018-32085-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yonemura K, Takahira R, Yonekawa O, Wada N, Hishida A. The diagnostic value of serum concentrations of 2-(alpha-mannopyranosyl)-L-tryptophan for normal renal function. Kidney Int. 2004;65:1395–1399. doi: 10.1111/j.1523-1755.2004.00521.x. [DOI] [PubMed] [Google Scholar]
- 54.Niewczas MA, et al. Circulating modified metabolites and a risk of ESRD in patients with type 1 diabetes and chronic kidney disease. Diabetes Care. 2017;40:383–390. doi: 10.2337/dc16-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Devlin AS, et al. Modulation of a circulating uremic solute via rational genetic manipulation of the gut microbiota. Cell Host. Microbe. 2016;20:709–715. doi: 10.1016/j.chom.2016.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gao J, et al. Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front. Cell Infect. Microbiol. 2018;8:13. doi: 10.3389/fcimb.2018.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ramezani A, Raj DS. The gut microbiome, kidney disease, and targeted interventions. J. Am. Soc. Nephrol. 2014;25:657–670. doi: 10.1681/ASN.2013080905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Crespo-Salgado J, et al. Intestinal microbiota in pediatric patients with end stage renal disease: a Midwest Pediatric Nephrology Consortium study. Microbiome. 2016;4:50. doi: 10.1186/s40168-016-0195-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wing MR, Patel SS, Ramezani A, Raj DS. Gut microbiome in chronic kidney disease. Exp. Physiol. 2016;101:471–477. doi: 10.1113/EP085283. [DOI] [PubMed] [Google Scholar]
- 60.Al Khodor S, Shatat IF. Gut microbiome and kidney disease: a bidirectional relationship. Pediatr Nephrol. 2017;32:921–931. doi: 10.1007/s00467-016-3392-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Armani RG, et al. Gut microbiome in chronic kidney disease. Curr Hypertens Rep. 2017;19:29. doi: 10.1007/s11906-017-0727-0. [DOI] [PubMed] [Google Scholar]
- 62.Koppe L, Fouque D, Soulage CO. Metabolic abnormalities in diabetes and kidney disease: role of uremic toxins. Curr. Diab. Rep. 2018;18:97. doi: 10.1007/s11892-018-1064-7. [DOI] [PubMed] [Google Scholar]
- 63.Yang CY, Tarng DC. Diet, gut microbiome and indoxyl sulphate in chronic kidney disease patients. Nephrology (Carlton) 2018;23(Suppl 4):16–20. doi: 10.1111/nep.13452. [DOI] [PubMed] [Google Scholar]
- 64.Snelson M, Kellow NJ, Coughlan MT. Modulation of the gut microbiota by resistant starch as a treatment of chronic kidney diseases: evidence of efficacy and mechanistic insights. Adv. Nutr. 2019;10:303–320. doi: 10.1093/advances/nmy068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roager HM, Licht TR. Microbial tryptophan catabolites in health and disease. Nat Commun. 2018;9:3294. doi: 10.1038/s41467-018-05470-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu IW, et al. Gut microbiota as diagnostic tools for mirroring disease progression and circulating nephrotoxin levels in chronic kidney disease: discovery and validation study. Int. J. Biol. Sci. 2020;16:420–434. doi: 10.7150/ijbs.37421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Evenepoel P, Poesen R, Meijers B. The gut-kidney axis. Pediatr. Nephrol. 2017;32:2005–2014. doi: 10.1007/s00467-016-3527-x. [DOI] [PubMed] [Google Scholar]
- 68.Mahmoodpoor F, Rahbar Saadat Y, Barzegari A, Ardalan M, Zununi Vahed S. The impact of gut microbiota on kidney function and pathogenesis. Biomed, Pharmacother. 2017;93:412–419. doi: 10.1016/j.biopha.2017.06.066. [DOI] [PubMed] [Google Scholar]
- 69.Kikuchi K, et al. Metabolomic analysis of uremic toxins by liquid chromatography/electrospray ionization-tandem mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed Life Sci. 2010;878:1662–1668. doi: 10.1016/j.jchromb.2009.11.040. [DOI] [PubMed] [Google Scholar]
- 70.Niwa T. Uremic toxicity of indoxyl sulfate. Nagoya J. Med. Sci. 2010;72:1–11. [PMC free article] [PubMed] [Google Scholar]
- 71.Duranton F, et al. Normal and pathologic concentrations of uremic toxins. J. Am. Soc. Nephrol. 2012;23:1258–1270. doi: 10.1681/ASN.2011121175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leong SC, Sirich TL. Indoxyl Sulfate-Review of Toxicity and Therapeutic Strategies. Toxins. 2016;8:1. doi: 10.3390/toxins8120358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barreto FC, et al. Serum indoxyl sulfate is associated with vascular disease and mortality in chronic kidney disease patients. Clin. J. Am. Soc. Nephrol. 2009;4:1551–1558. doi: 10.2215/CJN.03980609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lin CJ, et al. p-Cresylsulfate and indoxyl sulfate level at different stages of chronic kidney disease. J. Clin. Lab. Anal. 2011;25:191–197. doi: 10.1002/jcla.20456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu B, et al. Serum metabolomic profiling and incident CKD among African Americans. Clin. J. Am. Soc. Nephro. 2014;9:1410–1417. doi: 10.2215/Cjn.11971113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu JZ, et al. Clinical and metabolomic risk factors associated with rapid renal function decline in sickle cell disease. Am J Hematol. 2018;93:1451–1460. doi: 10.1002/ajh.25263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Toyohara T, et al. Metabolomic profiling of the autosomal dominant polycystic kidney disease rat model. Clin. Exp. Nephrol. 2011;15:676–687. doi: 10.1007/s10157-011-0467-4. [DOI] [PubMed] [Google Scholar]
- 78.Lustgarten MS, Fielding RA. Metabolites related to renal function, immune activation, and carbamylation are associated with muscle composition in older adults. Exp. Gerontol. 2017;100:1–10. doi: 10.1016/j.exger.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim K, Trott JF, Gao G, Chapman A, Weiss RH. Plasma metabolites and lipids associate with kidney function and kidney volume in hypertensive ADPKD patients early in the disease course. BMC Nephrol. 2019;20:66. doi: 10.1186/s12882-019-1249-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 2013;37:658–665. doi: 10.1002/gepi.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hemani G, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7:1. doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Smith GD, Ebrahim S. 'Mendelian randomization': can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 83.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ruddick JP, et al. Tryptophan metabolism in the central nervous system: medical implications. Expert Rev. Mol. Med. 2006;8:1–27. doi: 10.1017/S1462399406000068. [DOI] [PubMed] [Google Scholar]
- 85.Magni G, et al. Enzymology of NAD`+ homeostasis in man. Cell Mol Life Sci. 2004;61:19–34. doi: 10.1007/s00018-003-3161-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rhee EP, et al. A genome-wide association study of the human metabolome in a community-based cohort. Cell Metab. 2013;18:130–143. doi: 10.1016/j.cmet.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shin SY, et al. An atlas of genetic influences on human blood metabolites. Nat. Genet. 2014;46:543–550. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Draisma HHM, et al. Genome-wide association study identifies novel genetic variants contributing to variation in blood metabolite levels. Nat. Commun. 2015;6:7208. doi: 10.1038/ncomms8208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Long T, et al. Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nat. Genet. 2017;49:568–578. doi: 10.1038/ng.3809. [DOI] [PubMed] [Google Scholar]
- 90.Pattaro C, et al. Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nat. Commun. 2016;7:1. doi: 10.1038/ncomms10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018;50:693–698. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Arnold M, Raffler J, Pfeufer A, Suhre K, Kastenmuller G. SNiPA: an interactive, genetic variant-centered annotation browser. Bioinformatics. 2015;31:1334–1336. doi: 10.1093/bioinformatics/btu779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Canela-Xandri O, Rawlik K, Tenesa A. An atlas of genetic associations in UK Biobank. Nat. Genet. 2018;50:1593–1599. doi: 10.1038/s41588-018-0248-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.