Abstract
Detection of nitric oxide (NO) in biological systems is challenging due to both physicochemical properties of NO and limitations of current imaging modalities and probes. Magnetic resonance imaging (MRI) could be applied for studying NO in living tissue with high spatiotemporal resolution, but there is still a need for chemical agents that effectively sensitize MRI to biological NO production. To develop a suitable probe, we studied the interactions between NO and a library of manganese complexes with various oxidation states and molecular structures. Among this set, the manganese(III) complex with N,N′-(1,2-phenylene)bis(5-fluoro-2-hydroxybenzamide) showed favorable changes in longitudinal relaxivity upon addition of NO-releasing chemicals in vitro while also maintaining selectivity against other biologically relevant reactive nitrogen and oxygen species, making it a suitable NO-responsive contrast agent for T1-weighted MRI. When loaded with this compound, cells ectopically expressing nitric oxide synthase (NOS) isoforms showed MRI signal decreases of over 20% compared to control cells and were also responsive to NOS inhibition or calcium-dependent activation. The sensor could also detect endogenous NOS activity in antigen-stimulated macrophages and in a rat model of neuroinflammation in vivo. Given the key role of NO and associated reactive nitrogen species in numerous physiological and pathological processes, MRI approaches based on the new probe could be broadly beneficial for studies of NO-related signaling in living subjects.
Keywords: nitric oxide, magnetic resonance imaging, contrast agent, inflammation, manganese, brain
Graphical Abstract
Nitric oxide (NO) is a gaseous reactive nitrogen species that acts as an important signaling molecule in diverse biological contexts.1,2 In cells, NO is generated by oxidation of L-arginine to L-citrulline, catalyzed by a class of enzymes known as nitric oxide synthases (NOS). An inducible, calcium-independent NOS isoform (iNOS) is associated with the large transient production of NO at sites of inflammation, while two constitutively expressed calcium-dependent isoforms, endothelial NOS (eNOS) and neuronal NOS (nNOS), regulate NO dynamics involved in vascular reactivity and aspects of neuronal function.3
Chemical probes for imaging NO dynamics in vivo can provide important capability for studying physiological and pathological processes in both laboratory and clinical settings,4 but measuring NO in living tissue is challenging due to the short lifetime5 and diffusibility of NO in biological media.6 In vivo imaging of NO production has been demonstrated using probes detectable by optical imaging, photoacoustic tomography, and electron paramagnetic resonance,7–9 but these techniques all provide limited capability for noninvasive deep tissue imaging, in comparison with medical imaging modalities. Magnetic resonance imaging (MRI) offers a powerful combination of unlimited depth penetration and high spatiotemporal resolution, and NO-responsive MRI contrast agents could thus be especially powerful tools for measuring NO over wide fields of view in contexts like neuroinflammation.10,11 Previous efforts to measure NO in MRI have not demonstrated the detection of biologically produced NO, however.12–16
To identify an improved basis for the detection of biogenic NO by molecular MRI, we turned to manganese-based paramagnetic complexes, which provide effective contrast enhancement in longitudinal relaxation time (T1)-weighted MRI scans.17 Manganese ions are known to interact with reactive nitrogen species,18 suggesting that manganese-based contrast agents could function as MRI-detectable probes for NO. Water-soluble manganese(III) porphyrins are among the best studied ligands for reactive nitrogen species, in some cases displaying differential affinity for NO and HNO.19–21 We therefore hypothesized that NO-selective sensors could be identified from among manganese chelates that incorporate porphyrins or porphyrin-like structures. We further reasoned that interactions between NO and the manganese centers of such structures could produce longitudinal relaxivity (r1) changes by altering the average water occupancy (q) or residence time (τM) at axial coordination sites, providing a possible mechanism for NO-dependent modulation of T1 relaxation effects. In this study, we describe the selection of an aromatic manganese complex that functions as a NO sensor in vitro and in vivo, and we discuss its characterization, validation, and initial application in animals.
RESULTS
Evaluation of Candidate NO Sensors.
We assembled a library of 10 manganese complexes that differ in molecular structure and oxidation state of the metal ion (Figure 1A). Relaxation rate enhancement by each agent was investigated in the presence or absence of 2.5 equiv of the NO releasing agent 2-(N,N-diethylamino)-diazenolate-2-oxide diethylammonium salt (DEA/NO)22 in 25 mM 3-(N-morpholino)-propanesulfonic acid (MOPS) buffer at pH 7.4 and ambient temperature. Measurements were also performed in the presence of other biologically relevant reactive nitrogen and oxygen species, including H2O2, , and (Figure S1). For most of the candidate compounds, the NO donor induced a reduction in T1 relaxation rate (R1), corresponding to a decrease in r1 (Figure 1B). The greatest relative changes (Δr1/r1) were observed for the Schiff base complex MnL5 (−29 ± 5%) and the phenylenediamido complex MnL1F (−22 ± 3%) (Figure 1C). Other phenylenediamido complexes, MnL1Me and MnL1, showed weaker NO donor-dependent r1 decreases. Interestingly, these complexes differed substantially in their sensitivity to H2O2. MnL5 reacted instantly with hydrogen peroxide, resulting in color changes and r1 increases on the order of 30%, while MnL1F exhibited a much smaller increase (Δr1/r1 = +7%) (Figure S1). MnL1F also responded minimally to (Δr1/r1 = −5%) and (Δr1/r1 = −3%) and displayed no apparent sensitivity to dioxygen (t-test p = 0.73, n = 3) (Figure S2). Transverse relaxivity (r2) of MnL1F was likewise insensitive in response to DEA/NO, , , H2O2, and O2 (Figure S3) (all effects not significant with t-test p ≥ 0.36, n = 3).
Figure 1.
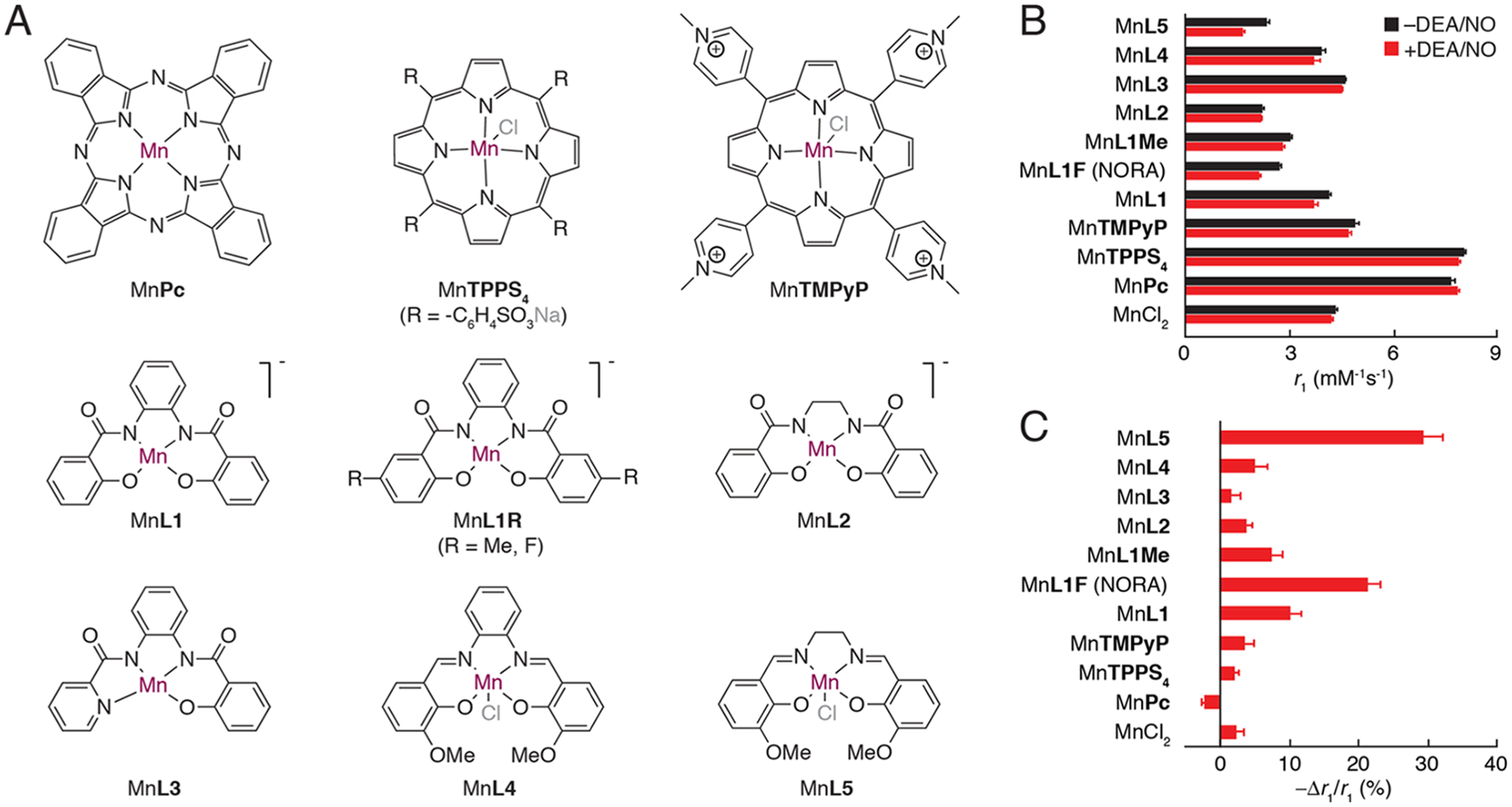
Candidate manganese-based NO sensors. (A) Molecular structures of manganese complexes with tetradentate ligands (counter ions denoted in gray). (B) T1 relaxivity values (r1) for each complex, as well as MnCl2, were measured at 7 T and 22 °C after 60 min treatment with (red) or without (black) the NO donor DEA/NO in 2.5:1 molar excess. (C) Relative response to NO treatment, quantified by −Δr1/r1 for each compound. MnL5 shows the largest response but is also responsive to H2O2 (see Figure S1); MnL1F was thus selected as the most effective NO-responsive agent (NORA). All error bars denote s.e.m. of three independent measurements.
Mechanistic Characterization of NO Dependence.
The combination of relatively strong Δr1/r1 responses to NO and much smaller responses to H2O2 and other species led us to select MnL1F as a candidate nitric oxide-responsive agent (henceforth NORA) for MRI. The NO-dependent r1 change exhibited by this compound, from 2.7 ± 0.0 to 2.1 ± 0.1 mM−1 s−1, is consistent with the possibility that NO axially coordinates the manganese ion and thus reduces solvent accessibility of the metal center.23 Such an interaction has been observed crystallographically in a similar manganese complex.24 To evaluate whether this mechanism explains the behavior of NORA, we studied the interactions between the compound and NO in water using variable temperature 17O-NMR studies.25 Upon addition of DEA/NO to NORA in 17OH2 (1 vol %), the bandwidth of the 17OH2 peak decreased by a factor of 3, as expected in the axial NO-binding model (Figure 2). Further evidence in support of a direct interaction between NO and NORA was obtained using cyclic voltammetry, which revealed an increase in the peak cathodic to anodic current density ratio (jp,c/jp,a) from unity to 1.4 upon addition of 1 equiv of DEA/NO. These results indicate the influence of NO on redox transitions of NORA’s manganese ion (Figure S4).
Figure 2.
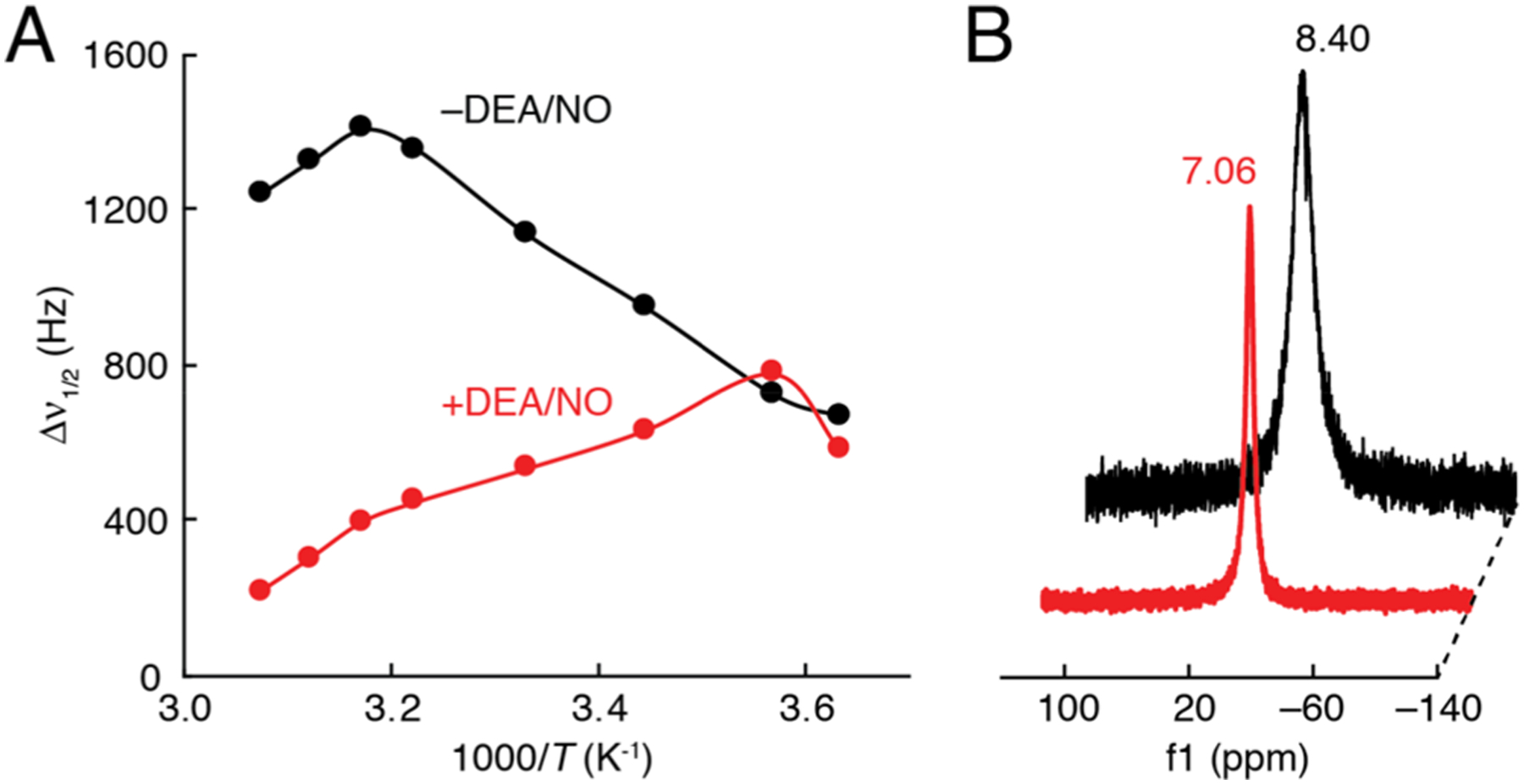
17O-NMR of NORA in the absence and presence of DEA/NO. (A) Temperature-dependent 17O-NMR linewidth (Δν1/2) at 11.7 T of 200 μM NORA in the absence (black) and presence (red) of 300 μM DEA/NO in 1 vol % 17O-enriched water. (B) 17O-NMR of NORA in the absence and presence of DEA/NO, illustrating the NO donor-dependent difference in linewidth at 297 K.
Ultraviolet–visible spectroscopy showed negligible effects of the NO donor on the spectrum of NORA (Figure S5), demonstrating that the covalent structure of the complex remains stable and unmodified in the presence of NO. Repeated measurements of the relaxivity of the NO–NORA complex reveal no significant changes on the time scale of hours. Effective r1 values were 2.1 ± 0.1 mM−1 s−1 1 h after exposure to DEA/NO and 2.0 ± 0.4 mM−1 s−1 4 h later. Given that DEA/NO has a reported half-life of about 4 min,26 the consistency of r1 is unlikely to reflect a competition between NO dissociation and further NO adduct formation. NORA also displays good solution stability, with a solubility of approximately 5 mM in MOPS buffer.
Detection of NOS-Catalyzed NO Production in Cells.
We next investigated the possibility of applying NORA for detecting biological NO production in cells using T1-weighted MRI. FreeStyle 293-F cells were transiently transfected with genes encoding either iNOS or green fluorescent protein (GFP control), incubated for 30 min with 20 μM NORA, rinsed repeatedly, and then imaged as cell pellets in a 7 T MRI scanner (Figure 3A). As expected from previous evidence for the cell permeability of phenylenediamido complexes,27 cells incubated with NORA experienced considerable labeling, resulting in up to ~33% higher R1 than untreated cells. While GFP-expressing cells exhibited a mean R1 difference of 0.162 ± 0.006 s−1, cells expressing iNOS displayed a ΔR1 of only 0.039 ± 0.007 s−1 (Table S2), a very significant difference with t-test p = 10−5 (n = 4). Approximately proportional results were obtained in the presence of varying concentrations of NORA (Figure S6), and the integrity of NORA under the incubation conditions was verified by mass spectrometric analysis of labeled cell pellets (Figure S7).
Figure 3.
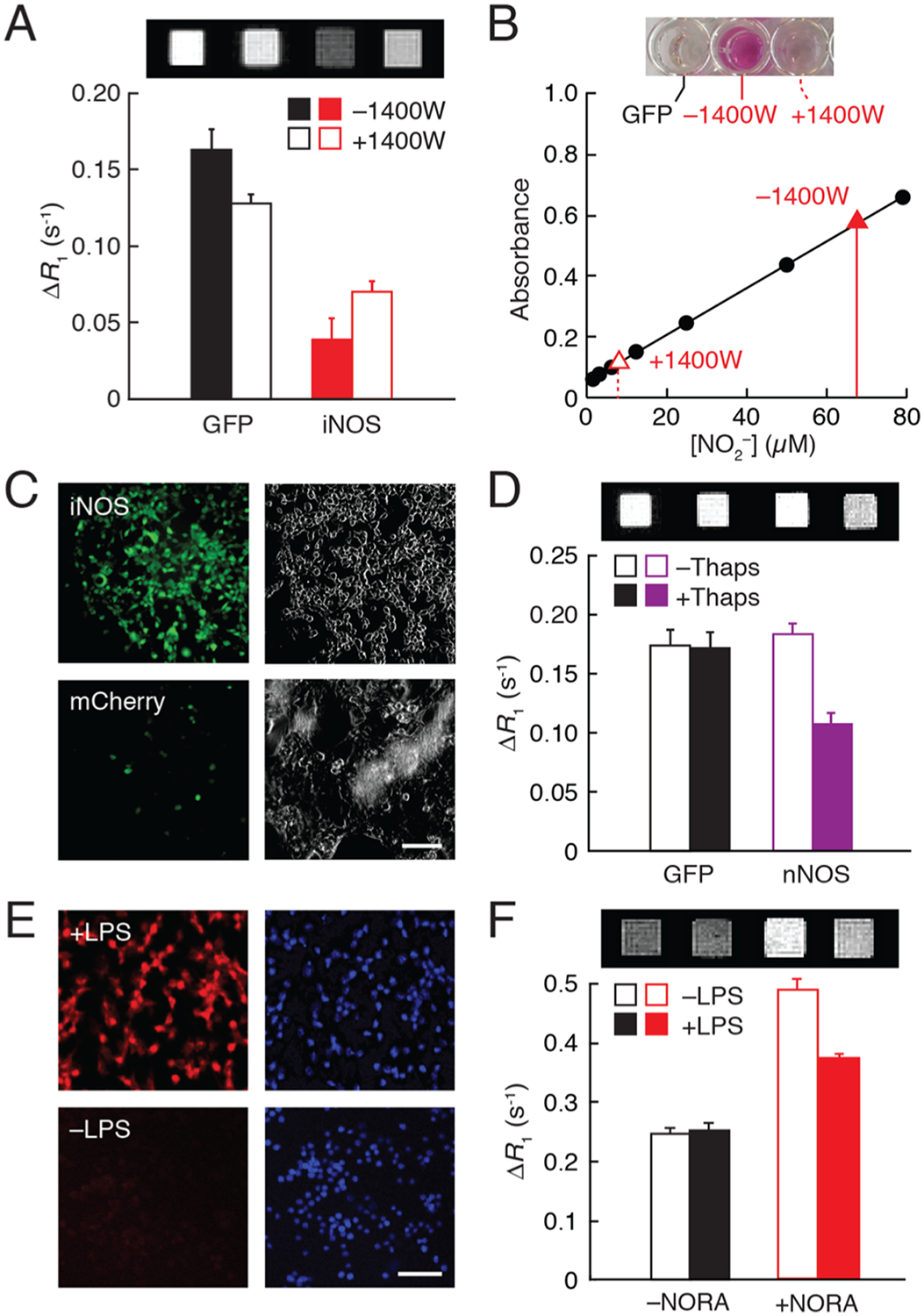
Measurement of NO production in cells. (A) Relaxation rate differences at 7 T and 22 °C, with respect to unlabeled cells, exhibited by 20 μM NORA-labeled cells transfected with GFP or iNOS and untreated or treated with 100 μM of iNOS inhibitor 1400 W. Corresponding T1-weighted images of cell pellets at the top. (B) Griess assay of nitrite production by iNOS-expressing cells in the presence or absence of 1400 W (red) with respect to calibration points (black). Images of assay wells from GFP- and iNOS-expressing cells ±1400 W at the top. (C) Micrographs of iNOS-expressing cells (top) or control mCherry-expressing cells (bottom) treated with the NO indicator DAF-FM (left). Scattering signal (right) reveals both stained and unstained cells. (D) Relaxation rate differences with respect to vehicle-treated cells exhibited by GFP- or nNOS-expressing cells after treatment with 20 μM NORA in the presence or absence of 1 μM thapsigargin (Thaps). T1-weighted MRI scans shown at the top. (E) Micrographs of macrophage cells treated with 5 μM NO indicator DAR-4M AM (left) in the presence (top) and absence (bottom) of LPS stimulation. Nuclear (DAPI) staining shown at the right. (F) R1 differences from buffer for macrophages incubated without or with NORA in the presence and absence of LPS; corresponding images at the top. All error bars represent s.e.m. (n ≥ 2). Scale bars = 50 μm.
In each case, the iNOS dependence of R1 was consistent with the expected effect of NO production on the relaxivity of the contrast agent, but elemental analysis of NORA-incubated cells revealed that the effective r1 of intracellular NORA, as well as its apparent modulation by NO production (Table S2), was substantially larger than values observed in vitro. To examine the basis for this mismatch, we measured the r1 of NORA in 20% cell lysate. Relaxivities of 4.5 ± 0.2 and 2.8 ± 0.2 mM−1 s−1 were observed in the absence and presence of 150 μM DEA/NO, respectively, indicating a 36% NO-dependent response. Similar results were obtained when NORA was mixed with 40 mg/mL bovine serum albumin in place of cell lysate. These results suggest that interaction between NORA and cytosolic macromolecules enhances the efficacy of the probe as a contrast agent and NO sensor inside cells, consistent with earlier reports that the interaction of Mn(III) porphyrins with proteins also boosts r1 at high fields.28
To confirm that the R1 changes observed with NORA in cells indeed required a NO-responsive contrast agent, we repeated the experiment of Figure 3A on cells incubated with MnTMPyP in place of NORA (Figure S8). The R1 difference between iNOS- and GFP-expressing cells incubated with MnTMPyP was only 11 ± 1%, compared with 21 ± 2% for cells labeled with NORA, consistent with the substantially lower NO sensitivity of MnTMPyP in the in vitro data of Figure 1C. To ascertain moreover that iNOS activity was required for these results, cells expressing the enzyme were treated with the selective inhibitor N-([3-(aminomethyl)-phenyl]methyl) ethanimidamide dihydrochloride (1400 W) before being incubated with NORA (Figure 2A). NORA produced approximately 2-fold greater relaxation enhancement in 1400 W-treated cells than in untreated cells (t-test, p = 0.007, n = 4), consistent with the expected effects of iNOS-dependent NO production on NORA relaxivity.29
To obtain independent measurements of NO production in these experiments, we applied the Griess test, which quantifies the buildup of the spontaneous NO oxidation product via an optical readout. Results of this assay confirmed iNOS-dependent increases in NO and 1400 W-dependent suppression of iNOS consistent with the MRI results (Figure 3B).30 Failure of 1400 W treatment to completely suppress NO production according to the Griess test might explain why iNOS-expressing cells display lower R1 than GFP-expressing cells, even in the presence of 1400 W (Figure 3A). The intracellular origin of NO in these experiments was further verified using the cytosolic fluorescent probe 4-amino-5-methylamino-2′,7′-difluorescein (DAF-FM), which exhibits a ~160-fold increase in quantum yield upon interaction with NO.31 DAF-FM staining results showed considerably more fluorescence in iNOS-transfected cells than in control cells expressing the fluorescent protein mCherry (Figure 3C), again consistent with the NORA-based sensing of NO within cells. Finally, to rule out the possibility that NO-dependent differences in the cellular retention of NORA contributed to the results of Figure 3A, we performed elemental analysis of NORA-labeled cells under different conditions and found no significant differences in the manganese content between iNOS- and GFP-expressing cells or due to inhibition with 1400 W (t-test, p values ≥ 0.24, n = 3) (Table S2).
We next evaluated the ability of NORA to enable MRI-based detection of calcium-dependent NO generation in cells transfected with nNOS. To stimulate nNOS activity, cells were treated for 30 min with 1 μM thapsigargin, which promotes calcium release from intracellular stores.32 Prior to stimulation, we observed comparable R1 values in nNOS- and GFP-expressing cells incubated with NORA (Figure 3D). Stimulation of the cells with thapsigargin caused a significant decrease in relaxation rate, by 38 ± 1%, with respect to unstimulated GFP-expressing cells (t-test, p = 0.0002, n = 3). Again, these results could not be explained by differences in NORA retention (t-test, p = 0.07, n = 3) (Table S2). The results are thus consistent with the expected effect of calcium-dependent nNOS-mediated NO production on NORA relaxivity. Griess test results further confirmed that the lesser change in R1 measured from stimulated nNOS-expressing cells compared with iNOS-expressing cells results from lesser NO production.
We also examined the potential toxicity of NORA to cells using a viability assay. Tests for acute toxicity and long-term viability of HEK293 cells incubated with NORA show no evidence of ill health with respect to controls (Figure S9). These results confirm that NORA is compatible with normal cellular functions, but they do not rule out that NORA could perturb NO-mediated signaling functions due to its NO binding mechanism.
MRI Measurement of NO Production by Activated Macrophages.
To examine whether NORA could be used to quantify NO production associated with the physiological functions of unengineered cells, we tested NORA’s ability to detect endogenous iNOS activity in RAW 264.7 macrophage cells. Expression of iNOS was induced by treating the macrophages with the bacterial antigen lipopolysaccharide (LPS).33 We confirmed NO production in this paradigm by incubating the macrophages with 5 μM fluorescent NO indicator diaminorhodamine-4M acetomethoxyester (DAR-4M AM) (Figure 3E).34 Macrophages treated with 10 ng/μL LPS exhibited substantially higher DAR-4M-mediated fluo rescence compared to control cells, indicative of NO production. Identically treated macrophages were then incubated with 20 μM NORA for 1 h, rinsed twice, and then scanned at 7 T and 22 °C. The R1 of NORA-labeled cells was significantly lower (t-test, p = 0.009, n = 3) after LPS treatment, compared with control treatment, indicative again of NO production and the correspondingly expected reduction of NORA relaxivity (Figure 3F). Based on the mean R1 values recorded from RAW cells alone (0.23 s−1), cells plus NORA (0.51 s−1), and cells plus NORA after LPS stimulation (0.38 s−1), the r1 values of NORA in cell lysate suggest that an average of ~60 μM contrast agent was present in labeled macrophages and that LPS addition resulted in complete saturation of this agent with ligands produced during a 60 min stimulation period.
In Vivo NO Detection in a Rodent Neuroinflammation Model.
We next applied NORA in live rat brains to evaluate its sensitivity in a model of neuroinflammation.35 To induce acute inflammation, LPS was infused locally into the rat hippocampus approximately 22 h before the administration of NORA (Figure 4A). NO production in this model was assessed using immunohistochemical visualization of 3-nitrotyrosine, a reaction product of NO derivatives such as peroxynitrite with nearby proteins36 (Figure 4B). NORA-dependent T1 contrast and relaxation rates were then evaluated by MRI at 9.4 T in vivo. T1 contrast was sharply brightened in LPS-treated brain areas infused with the contrast agent, but brightening was considerably greater in the presence of the iNOS inhibitor 1400 W (Figure 4C) and also in sham-treated animals that did not receive LPS, compared with animals that received LPS and NORA but not the iNOS inhibitor. Differences could be quantified using R1 values observed in the NORA-labeled regions in each condition (Figure 4D and Figure S10). Mean R1 values observed after LPS treatment were 0.75 ± 0.05 s−1 in the absence and 0.93 ± 0.05 s−1 in the presence of 1400 W and 1.07 ± 0.10 s−1 in sham-treated animals (Figure 4E). We performed a one-way ANOVA followed by posthoc Tukey’s tests for differences among the three treatment groups. R1 values in the two control conditions were significantly higher (Tukey’s test, p ≤ 0.03, n = 3), consistent with the expected suppressive effect of NO binding on NORA relaxivity and R1 enhancement.
Figure 4.
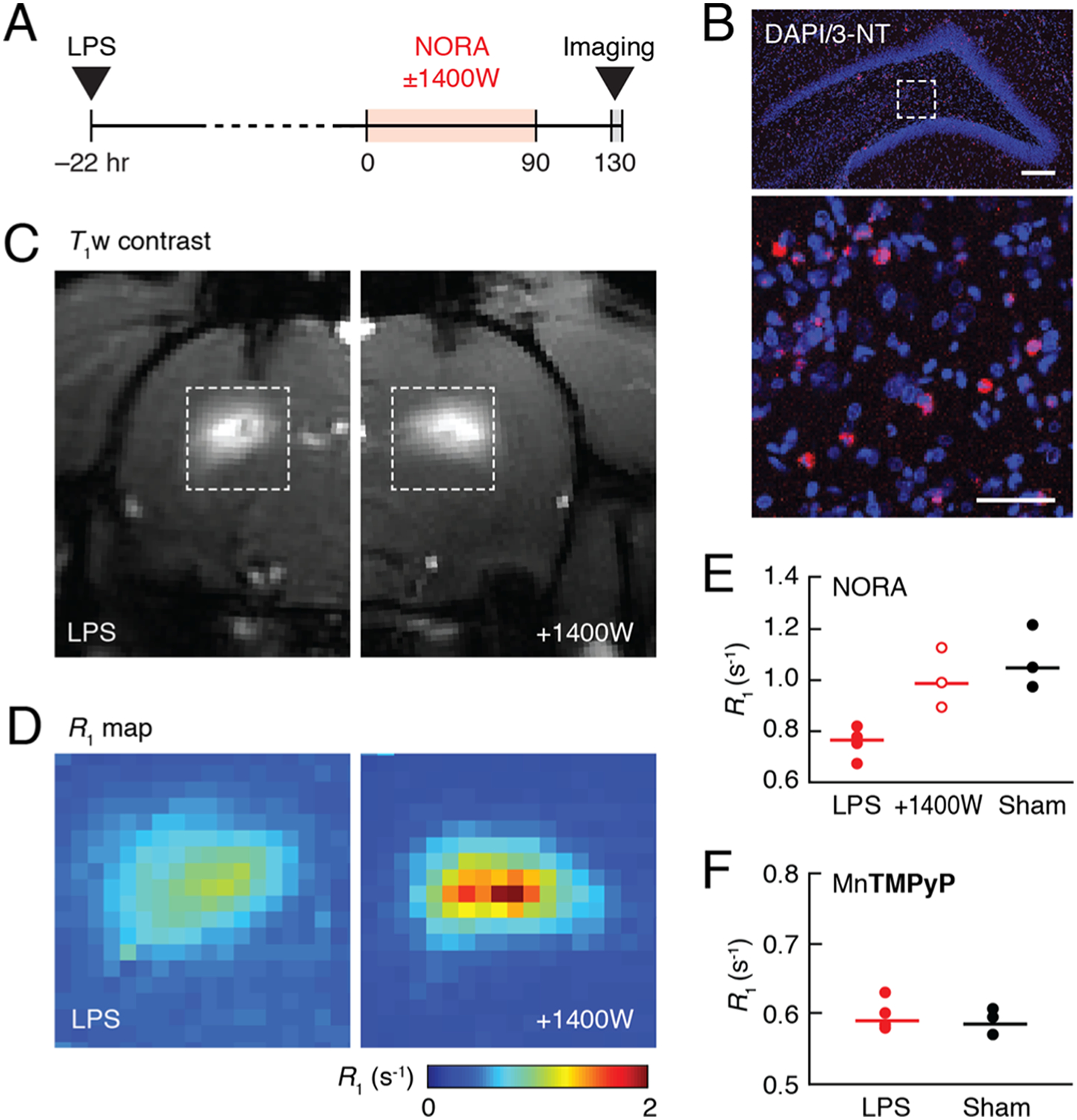
Detection of NO production in a neuroinflammation model in rats. (A) Experimental timeline. (B) Coronal section of rat brain treated with LPS was stained with 3-nitrotyrosine (3-NT) and DAPI. The image at the top shows the hippocampal brain region where LPS was injected. The magnified view on the bottom corresponds to the boxed region at the top and reveals cells costained with anti-3-NT antibody (red) and nuclei (blue) stained with DAPI. Scale bars = 200 μm (top), 50 μm (bottom). (C) Representative T1-weighted images of rats treated with LPS and infused with NORA in the absence (left) or presence (right) of the iNOS inhibitor 1400 W. (D) R1 maps of LPS and NORA-treated regions corresponding to the boxes in (C), averaged over animals in groups without (left) or with (right) 1400 W treatment (n = 4 and n = 3, respectively). (E) Average R1 values observed in NORA-enhanced regions of rats treated with LPS alone, LPS and 1400 W, or sham surgery. (F) Average R1 values observed in rat brain regions enhanced with the control contrast agent MnTMPyP after either LPS or sham treatment. Dot plots indicate mean values (horizontal lines) and measurements from each subject (dots).
When interpreted in light of the NORA r1 values observed in cell culture, the R1 differences between conditions enable an approximate quantification of the NO production rates observed in vivo. The NORA-dependent R1 difference of 0.32 s−1 between LPS and sham-treated animals is consistent with the binding of 190 μM NO to NORA over an exposure time of ~130 min. This implies a NO generation rate of roughly 1.5 μM/min, assuming that NORA captures NO stoichiometrically and irreversibly as it is produced. As a further test of the specificity of these results, we performed a comparison between LPS and sham-treated animals injected with the comparatively NO-insensitive compound MnTMPyP in place of NORA and found no appreciable difference between R1 values observed under the two conditions (Figure 4F). This control suggests that the stimulus-associated differences reported in Figure 4E do not arise from LPS-dependent effects on contrast agent labeling or washout rate.
DISCUSSION
Our data indicate that the manganese complex NORA functions as a NO-responsive T1 contrast agent for MRI, suitable for measurement of NO production in vivo. NORA-dependent responses in MRI are comparable with established chromometric and fluorometric methods for NO quantification and represent an integration of NO flux according to a mechanism analogous to NO adduct formation by metal-based fluorescent probes.37 Although the NO-dependent r1 change exhibited by NORA in buffer is modest, considerable responses to biogenic NO production were observed in cells, reflective of a relaxivity enhancement experienced by the probe in biological contexts. Application of 20 μM NORA thus enables biologically relevant monitoring of endogenous NO generation in a variety of cellular systems, including macrophages previously reported to produce ~0.5 μM peak steady-state NO levels at the cell surface.38 In addition, NORA can be injected into living animals, where it enables signatures of inflammation to be recognized and quantified in deep tissue, based on contrast differences with respect to controls.
A special challenge of detecting NO in biological systems arises from the very low steady-state concentrations of this analyte that are present in vivo. NO concentrations are often reported to be in the nanomolar range,39,40 whereas paramagnetic MRI contrast agents like NORA typically need to be applied at concentrations above 10 μM. Estimates of physiological NO production are nevertheless possible under these circumstances because of NORA’s ability to respond irreversibly to NO generation, as assessed on a time scale of hours. Given a typical biological lifetime of about 0.1 s for NO,5 even a low steady-state concentration of 10 nM would require generation of about 100 nM NO per second. If NORA competed effectively with biological quenching pathways for NO binding under such circumstances, 60 μM NORA could become saturated in about 10 min, a time frame shorter than the experiments we report here. Such kinetic considerations help explain the successful operation of NORA in vitro and in vivo.
This analysis assumes that relaxivity changes exhibited by NORA arise from NO binding, as opposed to extraneous environmental changes such as interactions with other reactive nitrogen and oxygen species. In practice, the moderate sensitivity NORA displays toward and (3–5% Δr1 in buffer) suggests the possibility that high levels of these reagents could be mistaken for NO production in some contexts. Conversely, high levels of hydrogen peroxide production, which produces r1 changes that are opposite in polarity to NO (Figure S1), could lead to underestimates of the level of NO or reactive nitrogen complexation with NORA. Another species that could influence the response properties of NORA is peroxynitrite, which is formed in vivo by the reaction of biogenic NO with superoxide anions.41 Given the dismutase activity of manganese complexes similar to NORA,42 we consider it unlikely that superoxide, and hence peroxynitrite, would accumulate in the presence of the micromolar probe concentrations required for effective molecular imaging. If NORA–peroxynitrite complexes do form and exhibit different r1 from NORA–NO, however, they could also complicate efforts to quantify NO production rates. These caveats should be considered in interpreting results we present here.
It is notable that NORA responds to NO binding with a reduction in r1, rather than an increase. Although a turn-on probe might have advantages in some contexts, the turn-off mechanism displayed by NORA provides two distinct benefits. First, assuming that the ambient NO level is low in the absence of stimuli, the MRI signal level in the presence of NORA is highest under basal conditions, meaning that the signal-to-noise ratio for subsequent detection of stimulus-driven NO release is greater than for corresponding turn-on probes. Second, again assuming that the basal NO level is low, the presence of a comparatively more visible turn-off probe makes it easier to quantify probe concentrations in vivo. We used this property to estimate NO production levels here, in particular by comparing R1 values observed in the presence versus absence of stimulation, and have similarly exploited turn-off behavior with other T1 probes to quantify other analytes in molecular fMRI experiments in vivo.43,44 With a turn-on probe, quantification would be more difficult because of ambiguity as to whether the absence of an R1 change from background results from low probe concentrations or low analyte concentrations. The problem of in vivo quantification could be further addressed by using an alternative means for estimating probe concentrations, such as complementary imaging or postmortem analytical approaches; such efforts would also benefit from further characterization of NORA’s pharmacokinetic properties.
Future work could lead to improved versions of NORA and applications in vivo. Most obviously, it could be desirable to further enhance the probe’s NO sensitivity. The fact that interactions between NORA and cell lysate components substantially increase the compound’s r1 and dynamic range could suggest strategies for such an effort. For instance, molecular characterization of NORA’s behavior in the cytosolic milieu could be performed, and pendant modifications to the probe could be designed to emulate whatever interactions are implicated. Similar modifications might also improve probe specificity for NO compared with other reactive oxygen and nitrogen species. In addition, improved methods for wide-field delivery could be employed. For delivery to the brain, both intracerebrospinal fluid delivery45 and intravascular injection combined with blood–brain barrier disruption46 might be especially effective. Intravenous infusions could also enable the probe to reach regions outside the brain, where NO imaging might be valuable as well. Finally, the potential for dynamic NO detection using NORA or its derivatives in vivo could be explored. Even though the probe appears to bind NO irreversibly, the rates of contrast change observed during image series acquisition could provide quantitative estimates of NO production as a function of time as a means for studying the time course of neuroinflammation or related processes. Results we present here suggest that such experiments could already be plausible in rodent models. Following more complete characterization of sensitivity, distribution, and toxicity in animals, similar applications in human subjects may also eventually be possible.
METHODS
Synthesis.
Complexes MnPc, MnTMPyP, MnTPPS4 were purchased from Merck KGaA (Darmstadt, Germany). All information regarding the synthesis and characterization of manganese complexes and related compounds are reported in the Supporting Information.
Evaluation of Responses to Reactive Oxygen and Nitrogen Species.
All experiments were carried out in 25 mM MOPS buffer (pH 7.4) in the presence of 100 mM KCl as the supporting electrolyte at 22 °C. Relaxivities of all manganese complexes were measured at 7 T and 22 °C in the absence and presence of DEA/NO (150 μM), NaNO2 (150 μM), NaNO3 (150 μM), or H2O2 (150 μM). All samples were treated with the relevant analytes for 1 h at 22 °C. To study interactions between NORA and dioxygen, buffer was degassed by purging with an argon stream for 20 h under an argon atmosphere. The +O2 sample was then generated by bubbling the degassed sample with O2 for 1 h.
17O-NMR.
Samples were prepared in 25 mM MOPS (pH 7.4) buffer in the presence of 100 mM KCl as the supporting electrolyte and 1 vol % 17OH2. The variable temperature spectra (277–323 K) were recorded on a VARIAN Inova-500 NMR spectrometer with an Oxford Instruments (Abingdon, United Kingdom) superconducting magnet.
Cyclic Voltammetry.
Cyclic voltammetry experiments were conducted using a Bio-Logic (Seyssinet-Pariset, France) VMP-3 multichannel potentiostat. Solutions were prepared in 25 mM MOPS (pH 7.4) and 100 mM KCl and analyzed in a 10 mL cell with Teflon cap using a glassy carbon working electrode (Pine, 5 mm outer diameter), a saturated calomel reference, and a platinum counter electrode. Solution resistance was compensated. Prior to analysis, the solution was purged for 10 min under 1 atm argon.
In Vitro Magnetic Resonance Imaging.
MRI data were acquired on a 7 T Bruker (Ettlingen, Germany) Biospec system using a T1-weighted 2D gradient echo sequence (echo time, TE = 5 ms, repetition time, TR = 100 ms, flip angle, FA = 65°). Longitudinal relaxivity (r1) measurements were acquired using a 2D spin echo sequence (TE = 11 ms, TR = 125, 200, 400, 800, 1500, 3000, and 5000 ms), with an in-plane resolution of 200 × 200 μm2 and 2 mm slice thickness. R1 maps and values were generated using Karl Schmidt’s MRI Analysis Calculator Plugin for ImageJ 1.52a (National Institutes of Health, Bethesda, MD) or MATLAB (Mathworks, Natick, MA) scripts written in house. Stimulus-dependent R1 changes in cells were calculated as ΔR1/R1 = [R1 (incubated cells) − R1 (naive cells)]/R1 (naive cells). Statistical comparisons between paired conditions were performed using Student’s t-test, and all error bars indicate uncertainty in estimates of mean values by reporting the standard error of the mean (s.e.m.) from multiple measurements.
DNA Assembly.
The plasmids pCWori-nNOS and pCWori-iNOS containing rat nNOS and human iNOS constructs, respectively, were a generous gift from the lab of Dr. Thomas Poulos at the University of California, Irvine. The nNOS and iNOS genes were individually cloned into the pIRESpuro3 mammalian vector (TakaraBio, Kusatsu, Japan). An inverse polymerase chain reaction was performed to create the backbone. Inserts were amplified from the pCWori backbone using primers with overhangs ranging from 24–30 nucleotides on each end. A single-step, isothermal, ligation-free procedure was used to integrate the inserts. The entire length of the coding strand was verified using Sanger sequencing.
Cell Culture Experiments.
293-F FreeStyle cells (Thermo Fisher, Waltham, MA) were maintained and passaged according to the manufacturer’s protocol. To obtain nNOS- or iNOS-expressing cells, high-purity DNA of pIRESpuro3-nNOS and pIRESpuro3-iNOS was obtained using QIAGEN’s Maxiprep kit (Venlo, Netherlands). DNA was introduced into the cells using 293Fectin (Thermo Fisher) following the manufacturer’s protocols. N-[[3-(Aminomethyl)-phenyl]methyl]-ethanimidamide dihydrochloride (100 μM, 1400 W, Tocris, Bristol, UK) was added as necessary. To estimate the catalytic activity of iNOS-expressing cells, the supernatant was collected 48 h post-transfection. The amount of nitrite was measured using the Griess reagent (Promega, Madison, WI).
For relaxation rate measurements in cell pellets, cells expressing GFP, nNOS, or iNOS and treated or not treated with 1400 W were incubated with 20 μM contrast agent for 30 min. In most experiments, after incubation, cells were pelleted, washed twice with Hank’s balanced salt solution (HBSS) and aliquoted into a 384-well plate. In the case of nNOS- or GFP-expressing cells, after incubation with NORA, cells were pelleted and suspended in the FreeStyle 293 expression medium (Thermo Fisher) containing 1 μM thapsigargin or 0.1% dimethyl sulfoxide (Merck). After a 30 min incubation, these cells were washed twice with HBSS, pelleted, and then transferred into a 384-well plate. Plates were imaged using a 7 T MRI scanner. A spin echo pulse sequence was used with parameters described above in the “In Vitro Magnetic Resonance Imaging“ section and R1 values were fitted using MATLAB. Effective r1 values were obtained by normalizing the R1 values by metal quantification results obtained using inductively coupled plasma-mass spectrometry (ICP-MS).
Cellular Toxicity and Viability Assays.
HEK293 cells were incubated with NORA (0–20 μM) solution in media for 60 min and then washed. Control cells were either untreated (naive) or treated with saponin (positive control). To assess acute toxicity and membrane disruption, a subset of cells was incubated with 4 μM ethidium homodimer III (Biotium, Fremont, CA) and assayed for fluorescence at 530 and 620 nm. A higher fluorescence ratio (530/620) indicates increased cell penetration and intercalation into DNA as a result of cell membrane disruption. To assess long-term viability, a 3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide (MTT) assay (Life Technologies, Carlsbad, CA) was performed. Cells were incubated in the MTT reagent for 4 h at 37 °C to generate formazan crystals, which were then solubilized in sodium dodecyl sulfate solution for 2 h at 37 °C and assayed for optical density at 570 nm. Higher absorption corresponds to higher metabolic activity, indicative of cell health.
Mass Spectrometry of Cell Samples.
iNOS-expressing cells incubated with 20 μM NORA for 30 min were centrifuged and washed. The pellet was freeze-dried and the resulting residue was suspended in acetonitrile (100 μL). The supernatant was concentrated to dryness using a speed vacuum concentrator (300 g, 10 mbar) and redissolved in 10 μL of acetonitrile. Two microliters of the sample and 2 μL of α-cyano-4-hydroxycinnamic acid (MALDI matrix) were codried and the mass spectrum was recorded on a Bruker Omniflex MALDI-TOF instrument. ICP-MS was performed by first digesting cells (5 × 106) in 200 μL of concentrated nitric acid and heated at 60 °C for 14 h. The resulting clear solution was heated further until all volatiles evaporated. The residue was dissolved in 2% nitric acid solution in ICP-grade water and the manganese contents were determined by ICP-MS (Agilent 7900).
Macrophage Stimulation Experiments.
RAW 264.7 murine macrophage cells (ATCC, Manassas, VA) were maintained and passaged according to the manufacturer’s protocol. To induce iNOS expression, cells were treated with 10 ng/μL LPS (Escherichia coli O111:B4, MilliporeSigma, Burlington, MA). Prior to immunocyto-chemistry, RAW 264.7 cells were grown on specialized glass coverslips (Thermo Fisher) and treated with LPS overnight. Media were then aspirated and cells were incubated with fresh media containing 5 μM DAR-4 M AM (Enzo Life Sciences, Farmingdale, NY) for 30 min at 37 °C. Cells were washed three times with HBSS, fixed with a solution of 4% paraformaldehyde in standard phosphate-buffered saline (PBS), permeabilized using PBS with 0.1% Triton X-100 (MilliporeSigma), stained with 1 μg/mL diamidino-2-phenylindole (DAPI, Thermo Fisher), and preserved on glass slides with ProLong Diamond Antifade Mountant (Thermo Fisher). Slides were imaged using a confocal microscope (Zeiss, Oberkochen, Germany).
To visualize the biological production of nitric oxide using NORA, RAW 264.7 cells were grown on 15 cm tissue culture-treated dishes (MilliporeSigma). When cells were about 80% confluent, they were stimulated with LPS. These cells were then incubated with 20 μM NORA for 1 h at 37 °C. Cells were subsequently washed in HBSS, centrifuged, plated into 384-well plates, and imaged as pellets on a 7 T MRI scanner.
Intracranial LPS Infusion.
Adult male Sprague–Dawley rats (300–350 g) were purchased from Charles River Laboratories (Wilmington, MA). After arrival, animals were housed and maintained on a 12 h light/dark cycle and permitted ad libitum access to food and water. All procedures were carried out in strict compliance with National Institutes of Health guidelines, under oversight of the Committee on Animal Care at the Massachusetts Institute of Technology.
Animals were anesthetized with isoflurane (3% for induction, 2% for maintenance) with a water heating pad (Braintree Scientific, Braintree, MA) to keep the body temperature at 37 °C. Animals were then fixed in a stereotaxic frame, and topical lidocaine was applied before a 3 cm lateral incision extending from bregma to lambda was made, exposing the skull. Craniotomies (0.5 mm) were drilled bilaterally over the hippocampus, 4 mm anterior and 3.7 mm lateral to bregma. 22G Guide cannulae (Plastics One, Roanoke, VA) were implanted and sealed into place with cold cure dental cement (A-M Systems, Sequim, WA). 28G Infusion cannulae were lowered to 3 mm below the surface of the skull through each craniotomy and 10 μL of LPS or artificial cerebrospinal fluid (for sham treatment) was infused over 10 min. Ten minutes after the injection, infusion cannulae were slowly removed from the brain. Animals were given buprenorphine (0.1 mg/kg IP), disconnected from isoflurane, and monitored until full recovery from anesthesia.
In Vivo Imaging.
Approximately 22 h after LPS infusion, NORA was delivered and MRI was performed. Animals were anesthetized with isoflurane again and positioned into the stereotaxic frame. 26G Infusion cannulae were lowered through the guide cannulae to the same coordinates as LPS infusion in the prior day and animals were infused with 10 μL of 2 mM NORA, 2 mM control agent MnTMPyP, or 2 mM NORA mixed with 100 μM 1400 W over a period of 90 min followed by a 10 min rest before cannula withdrawal. Although relatively high contrast agent concentrations were used for these experiments, we expect that considerably lower doses could be used, as dictated by the minimum levels of such agents that can generally be detected in vivo (~10 μM). Sham-treated animals had their guide cannulae implanted at this stage and 1400 W-treated animals also received 25 mg/kg 1400 W intraperitoneally 1 h prior to NORA infusion. Animals were then transferred and fixed into a custom rat imaging cradle, maintained under 2% isoflurane anesthesia, and kept warm using a recirculating water heating pad for the duration of imaging.
All MRI images were acquired on a 9.4 T BioSpec scanner (Bruker, Billerica, MA) using a transmit-only 70 cm inner diameter linear volume coil (Bruker) and a 2 cm diameter receive-only surface coil (Doty Scientific, Columbia, SC). Scanner operation was controlled using the ParaVision 6.01 software (Bruker). Multislice T1-weighted fast low-angle-shot MRI images were acquired to evaluate the spread of the contrast agent in the hippocampus with TE = 4 ms, TR = 200 ms, field of view (FOV) = 30 × 18 mm, in-plane resolution 200 × 200 μm, and five coronal slices with slice thickness = 1 mm. The slice with the maximal contrast signal was chosen for high-resolution mapping. Forty minutes after the agent infusion, R1 and R2 maps were measured using a rapid acquisition with refocused echoes (RARE) pulse sequence with multiple echo times. Parameters were as follows: FOV = 30 × 18 mm, slice thickness = 1 mm, in-plane resolution = 200 × 200 μm, and RARE factor = 2, with eight TR values (175, 300, 500, 750, 1000, 1500, 3000, and 5000 ms) and eight TE values (10, 30, 50, 70, 90, 110, 130, and 150 ms). The total scan time was 366 s.
R1 maps were calculated by voxel-level fitting of the MRI signal at TE = 10 ms to M(TR) = M0(1 − exp{−TR × R1}) using nonlinear regression. Contrast agent-infused regions were defined as those with R1 ≥ 0.5 s−1. Mean R1 values in LPS ± 1400 W or sham-treated brain regions were calculated by averaging R1 values over the agent-infused region at each injection site and then combining data across animals. NORA-bound NO concentrations N were estimated by comparing measurements obtained in the presence versus absence of LPA. Explicit knowledge of the concentration of the contrast agent is not required for this analysis, under the assumptions that NO produced in the absence of stimulation is low. In each case, , where R10 is the T1 relaxation rate in the absence of contrast agent, c+ and c− are the concentrations of contrast agent bound versus unbound to NO, and and are the corresponding relaxivities of these two forms. In our experiments, both stimulated and unstimulated brain areas contained the same unknown postinjection concentration of contrast agent ctot = c+ + c−. Since c+ = N by definition, . The R1 difference between conditions with and without stimulation is therefore , where Δr1 is the relaxivity difference between bound and unbound versions of the contrast agent, so N = ΔR1/Δr1. We performed these calculations using a Δr1 value of 1.7 mM−1 s−1 measured for NORA ± DEA/NO in cell lysate; this value may differ somewhat from the true Δr1 exhibited in vivo due to differences in temperature and chemical environment. All quantitative data analyses were performed using MATLAB. Statistical comparisons were performed using Student’s t-test, unless otherwise specified.
Immunohistochemistry.
Brains treated with LPS or control injections were harvested and fixed in paraformaldehyde (PFA, 4% v/v in PBS) for 48 h at 4 °C. These brains were then sectioned on a vibratome (VT1200, Leica Biosystems, Wetzlar, Germany) into 50 μm-thick free-floating slices and stored in PBS at 4 °C. Selected slices were permeabilized in PBS + 0.3% Triton X (MilliporeSigma, Burlington, MA) for 15 min at room temperature and then blocked with 10% v/v donkey serum (MilliporeSigma) for 1 h at room temperature. The slices were then probed with rabbit anti-nitrotyrosine antibody (1:100) (MilliporeSigma) solution in PBS + 0.1% Triton X-100 (PBST) overnight at 4 °C. Slices were then washed with PBST thrice and probed with 1:500 donkey anti-rabbit IgG (H + L) tagged with Alexa Fluor 594 (Thermo Fisher) for 2 h at room temperature. Slices were finally washed thrice in PBS and mounted on glass slides with ProLong Gold Antifade Mountant with DAPI (Thermo Fisher). Slides were imaged on a confocal microscope (LSM 710, Carl Zeiss, Oberkochen, Germany).
We did not perform postmortem analysis to determine the subcellular localization profile of our imaging agents. Our previous studies of chemically related agents in cells and in vivo, however, indicate that they tend to be stably retained in cells and tissue over time periods well over an hour, and that their subcellular distribution includes a mix of cytosolic, membranous, and nuclear content.27,47
Supplementary Material
ACKNOWLEDGMENTS
The authors are grateful for access to equipment made available through the MIT Department of Chemistry Instrumentation Facility and the laboratory of professor Yogi Surendranath. This work was supported by NIH grants U01-NS090451, U01-NS103470, and UF1-NS107712 to A.J. S.G. was supported by an HHMI International Graduate Fellowship and a Shelly Razin Fund Fellowship from the McGovern Institute for Brain Research. M.L.P. was supported by NIH grant F32-GM130071.
KEY ABBREVIATIONS
- eNOS
endothelial nitric oxide synthase
- GFP
green fluorescent protein
- iNOS
inducible nitric oxide synthase
- T1
longitudinal relaxation time
- R1
longitudinal relaxation rate
- r1
longitudinal relaxivity
- MRI
magnetic resonance imaging
- nNOS
neuronal nitric oxide synthase
- NO
nitric oxide
- NORA
nitric oxide-responsive agent
- NOS
nitric oxide synthase
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acssensors.0c00322.
Supplementary tables, figures, and methods (PDF)
The authors declare no competing financial interest.
Contributor Information
Ali Barandov, Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, United States.
Souparno Ghosh, Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, United States.
Nan Li, Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, United States.
Benjamin B. Bartelle, Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, United States
Jade I. Daher, Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, United States
Michael L. Pegis, Department of Chemistry, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, United States;.
Hannah Collins, Department of Chemistry, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, United States.
Alan Jasanoff, Department of Biological Engineering, Department of Brain & Cognitive Sciences, and Department of Nuclear Science & Engineering, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, United States;.
REFERENCES
- (1).Heinrich TA; da Silva RS; Miranda KM; Switzer CH; Wink DA; Fukuto JM Biological nitric oxide signalling: chemistry and terminology. Br. J. Pharmacol 2013, 169, 1417–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Liaudet L; Soriano FG; Szabó C Biology of nitric oxide signaling. Crit. Care Med 2000, 28, N37–N52. [DOI] [PubMed] [Google Scholar]
- (3).Alderton WK; Cooper CE; Knowles RG Nitric oxide synthases: structure, function and inhibition. Biochem. J 2001, 357, 593–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Nagano T; Yoshimura T Bioimaging of nitric oxide. Chem. Rev 2002, 102, 1235–1270. [DOI] [PubMed] [Google Scholar]
- (5).Thomas DD; Liu X; Kantrow SP; Lancaster JR Jr. The biological lifetime of nitric oxide: implications for the perivascular dynamics of NO and O2. Proc. Natl. Acad. Sci. U. S. A 2001, 98, 355–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Hunter RA; Storm WL; Coneski PN; Schoenfisch MH Inaccuracies of nitric oxide measurement methods in biological media. Anal. Chem 2013, 85, 1957–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Yoshimura T; Yokoyama H; Fujii S; Takayama F; Oikawa K; Kamada H In vivo EPR detection and imaging of endogenous nitric oxide in lipopolysaccharide-treated mice. Nat. Biotechnol 1996, 14, 992–994. [DOI] [PubMed] [Google Scholar]
- (8).Reinhardt CJ; Zhou EY; Jorgensen MD; Partipilo G; Chan J A Ratiometric Acoustogenic Probe for in Vivo Imaging of Endogenous Nitric Oxide. J. Am. Chem. Soc 2018, 140, 1011–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Pluth MD; Tomat E; Lippard SJ Biochemistry of mobile zinc and nitric oxide revealed by fluorescent sensors. Annu. Rev. Biochem 2011, 80, 333–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Jacobs AH; Tavitian B; the INMiND Consortium. Noninvasive molecular imaging of neuroinflammation. J. Cereb. Blood Flow Metab 2012, 32, 1393–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).di Pulli B; Chen JW Imaging Neuroinflammation - from Bench to Bedside. J. Clin. Cell Immunol 2014, 05, 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Di Salle F; Barone P; Hacker H; Smaltino F; d’Ischia M Nitric oxide-haemoglobin interaction: a new biochemical hypothesis for signal changes in fMRI. Neuroreport 1997, 8, 461–464. [DOI] [PubMed] [Google Scholar]
- (13).Fichtlscherer B; Mülsch A MR imaging of nitrosyl-iron complexes: experimental study in rats. Radiology 2000, 216, 225–231. [DOI] [PubMed] [Google Scholar]
- (14).Liu G; Li Y; Pagel MD Design and characterization of a new irreversible responsive PARACEST MRI contrast agent that detects nitric oxide. Magn Reson Med 2007, 58, 1249–1256. [DOI] [PubMed] [Google Scholar]
- (15).Suchý M; Li AX; Liu Y; Feng Q; Bartha R; Hudson RHE Preliminary evaluation of PARACEST MRI agents for the detection of nitric oxide synthase. Can. J. Chem 2016, 94, 715–722. [Google Scholar]
- (16).Daryaei I; Randtke EA; Pagel MD A biomarker-responsive T2ex MRI contrast agent. Magn. Reson. Med 2017, 77, 1665–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Botta M; Carniato F; Esteban-Gómez D; Platas-Iglesias C; Tei L Mn(II) compounds as an alternative to Gd-based MRI probes. Future Med. Chem 2019, 11, 1461–1483. [DOI] [PubMed] [Google Scholar]
- (18).Wayland BB; Olson LW; Siddiqui ZU Nitric oxide complexes of manganese and chromium tetraphenylporphyrin. J. Am. Chem. Soc 1976, 98, 94–98. [Google Scholar]
- (19).Martí MA; Bari SE; Estrin DA; Doctorovich F Discrimination of nitroxyl and nitric oxide by water-soluble Mn(III) porphyrins. J. Am. Chem. Soc 2005, 127, 4680–4684. [DOI] [PubMed] [Google Scholar]
- (20).Spasojević I; Batinić-Haberle I; Fridovich I Nitrosylation of manganese(II) tetrakis(N-ethylpyridinium-2-yl)porphyrin: a simple and sensitive spectrophotometric assay for nitric oxide. Nitric Oxide 2000, 4, 526–533. [DOI] [PubMed] [Google Scholar]
- (21).Yu C-H; Su YO Electrocatalytic reduction of nitric oxide by water-soluble manganese porphyrins. J. Electroanal. Chem 1994, 368, 323–327. [Google Scholar]
- (22).Liang H; Nacharaju P; Friedman A; Friedman JM Nitric oxide generating/releasing materials. Future Sci. OA 2015, 1, FSO54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Caravan P; Ellison JJ; McMurry TJ; Lauffer RB Gadolinium(III) chelates as MRI contrast agents: Structure, dynamics, and applications. Chem. RevChem Rev 1999, 99, 2293–2352. [DOI] [PubMed] [Google Scholar]
- (24).Olmstead MM; Eroy-Reveles AA; Mascharak PK [Mn(bpb)(DMAP)(NO)], an {Mn-NO} nitrosyl with Z’ = 8. Acta Crystallogr. Sect. E Struct. Rep. Online 2011, 67, m1451–m1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Lieb D; Zahl A; Shubina TE; Ivanović-Burmazović I Water exchange on manganese(III) porphyrins. Mechanistic insights relevant for oxygen evolving complex and superoxide dismutation catalysis. J. Am. Chem. Soc 2010, 132, 7282–7284. [DOI] [PubMed] [Google Scholar]
- (26).Ferrero R; Rodríguez-Pascual F; Miras-Portugal MT; Torres M Comparative effects of several nitric oxide donors on intracellular cyclic GMP levels in bovine chromaffin cells: correlation with nitric oxide production. Br. J. Pharmacol 1999, 127, 779–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Barandov A; Bartelle BB; Gonzalez BA; White WL; Lippard SJ; Jasanoff A Membrane-Permeable Mn(III) Complexes for Molecular Magnetic Resonance Imaging of Intracellular Targets. J. Am. Chem. Soc 2016, 138, 5483–5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Yushmanov VE; Tominaga TT; Borissevitch IE; Imasato H; Tabak M Binding of manganese and iron tetraphenylporphine sulfonates to albumin is relevant to their contrast properties. Magn. Reson. Imaging 1996, 14, 255–261. [DOI] [PubMed] [Google Scholar]
- (29).Garvey EP; Oplinger JA; Furfine ES; Kiff RJ; Laszlo F; Whittle BJR; Knowles RG 1400W is a slow, tight binding, and highly selective inhibitor of inducible nitric-oxide synthase in vitro and in vivo. J. Biol. Chem 1997, 272, 4959–4963. [DOI] [PubMed] [Google Scholar]
- (30).Tsikas D Analysis of nitrite and nitrate in biological fluids by assays based on the Griess reaction: appraisal of the Griess reaction in the L-arginine/nitric oxide area of research. J. Chromatogr. B Analyt. Technol. Biomed Life Sci 2007, 851, 51–70. [DOI] [PubMed] [Google Scholar]
- (31).Kojima H; Urano Y; Kikuchi K; Higuchi T; Hirata Y; Nagano T Fluorescent Indicators for Imaging Nitric Oxide Production. Angew. Chem. Int. Ed. Engl 1999, 38, 3209–3212. [DOI] [PubMed] [Google Scholar]
- (32).Rogers TB; Inesi G; Wade R; Lederer WJ Use of thapsigargin to study Ca2+ homeostasis in cardiac cells. Biosci. Rep 1995, 15, 341–349. [DOI] [PubMed] [Google Scholar]
- (33).MacMicking J; Xie QW; Nathan C Nitric oxide and macrophage function. Annu. Rev. Immunol 1997, 15, 323–350. [DOI] [PubMed] [Google Scholar]
- (34).Kojima H; Hirotani M; Nakatsubo N; Kikuchi K; Urano Y; Higuchi T; Hirata Y; Nagano T Bioimaging of nitric oxide with fluorescent indicators based on the rhodamine chromophore. Anal. Chem 2001, 73, 1967–1973. [DOI] [PubMed] [Google Scholar]
- (35).Espinosa-Oliva AM; de Pablos RM; Herrera AJ Intracranial injection of LPS in rat as animal model of neuroinflammation. Methods Mol. Biol 2013, 1041, 295–305. [DOI] [PubMed] [Google Scholar]
- (36).Ahsan H 3-Nitrotyrosine: A biomarker of nitrogen free radical species modified proteins in systemic autoimmunogenic conditions. Hum. Immunol 2013, 74, 1392–1399. [DOI] [PubMed] [Google Scholar]
- (37).Lim MH; Lippard SJ Metal-based turn-on fluorescent probes for sensing nitric oxide. Acc. Chem. Res 2007, 40, 41–51. [DOI] [PubMed] [Google Scholar]
- (38).Jiang S; Cheng R; Wang X; Xue T; Liu Y; Nel A; Huang Y; Duan X Real-time electrical detection of nitric oxide in biological systems with sub-nanomolar sensitivity. Nat. Commun 2013, 4, 2225. [DOI] [PubMed] [Google Scholar]
- (39).Hall CN; Garthwaite J What is the real physiological NO concentration in vivo? Nitric Oxide 2009, 21, 92–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Bohlen HG Is the real in vivo nitric oxide concentration pico or nano molar? Influence of electrode size on unstirred layers and NO consumption. Microcirculation 2013, 20, 30–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Pacher P; Beckman JS; Liaudet L Nitric oxide and peroxynitrite in health and disease. Physiol. Rev 2007, 87, 315–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Faulkner KM; Liochev SI; Fridovich I Stable Mn(III) porphyrins mimic superoxide dismutase in vitro and substitute for it in vivo. J. Biol. Chem 1994, 269, 23471–23476. [PubMed] [Google Scholar]
- (43).Lee T; Cai LX; Lelyveld VS; Hai A; Jasanoff A Molecular-level functional magnetic resonance imaging of dopaminergic signaling. Science 2014, 344, 533–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Hai A; Cai LX; Lee T; Lelyveld VS; Jasanoff A Molecular fMRI of Serotonin Transport. Neuron 2016, 92, 754–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Iliff JJ; Lee H; Yu M; Feng T; Logan J; Nedergaard M; Benveniste H Brain-wide pathway for waste clearance captured by contrast-enhanced MRI. J. Clin. Invest 2013, 123, 1299–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Poon C; McMahon D; Hynynen K Noninvasive and targeted delivery of therapeutics to the brain using focused ultrasound. Neuropharmacology 2017, 120, 20–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Barandov A; Bartelle BB; Williamson CG; Loucks ES; Lippard SJ; Jasanoff A Sensing intracellular calcium ions using a manganese-based MRI contrast agent. Nat. Commun 2019, 10, 897. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


