Abstract
BACKGROUND AND PURPOSE:
In this study, we used power analysis to calculate required sample sizes to detect group-level changes in quantitative neuroanatomical estimates derived from MRI scans obtained from multiple imaging centers. Sample size estimates were derived from (i) standardized 3T image acquisition protocols and (ii) nonstandardized clinically acquired images obtained at both 1.5 and 3T as part of the multicenter Human Epilepsy Project. Sample size estimates were compared to assess the benefit of standardizing acquisition protocols.
METHODS:
Cortical thickness, hippocampal volume, and whole brain volume were estimated from whole brain T1-weighted MRI scans processed using Freesurfer v6.0. Sample sizes required to detect a range of effect sizes were calculated using (i) standard t-test based power analysis methods and (ii) a nonparametric bootstrap approach.
RESULTS:
A total of 32 participants were included in our analyses, aged 29.9 ± 12.62 years. Standard deviation estimates were lower for all quantitative neuroanatomical metrics when assessed using standardized protocols. Required sample sizes per group to detect a given effect size were markedly reduced when using standardized protocols, particularly for cortical thickness changes <.2 mm and hippocampal volume changes <10%.
CONCLUSIONS:
The use of standardized protocols yielded up to a five-fold reduction in required sample sizes to detect disease-related neuroanatomical changes, and is particularly beneficial for detecting subtle effects. Standardizing image acquisition protocols across scanners prior to commencing a study is a valuable approach to increase the statistical power of multicenter MRI studies.
Keywords: Brain morphometry, multisite studies, power analysis, quantitative neuroanatomy
Introduction
Multicenter studies are widely used in neuroimaging research, primarily due to the potential for increased recruitment. The benefit of increased sample size in multicenter studies may be offset by the increased variability in quantitative estimates derived from MRI-based neuroimaging due to differences in MRI scanner hardware, image acquisition protocols, and other site-specific factors such as variability in local site QA policies regarding image quality.1–3 For multisite studies, it is important to characterize and adjust for site-related differences in order to improve our ability to reliably detect neuroanatomical changes.
Although a number of postprocessing methods have been developed and applied to multicenter imaging data to correct for site-related differences in morphometric estimates,4–9 standardizing image acquisition protocols prior to imaging is likely to be useful for ameliorating unwanted site-related effects. The value of standardized image acquisition protocols is largely recognized by the research neuroimaging community,10,11 yet few studies have explicitly quantified the benefit of standardized imaging for multicenter studies.1,12,13 In this study, we used statistical power analysis techniques applied to quantitative morphometric estimates obtained from individuals who have been imaged using both standardized and nonstandardized image acquisition protocols as part of the multicenter Human Epilepsy Project. This allowed us to estimate and directly compare sample sizes required to detect morphometric changes when using standardized and nonstandardized image acquisition protocols. We specifically applied these sample size estimation techniques to estimates of hippocampal volume, cortical thickness, and brain volume. Changes in these brain metrics are associated with the neurobiology of a number of diseases or adverse health conditions, as well as healthy aging (Table 1). Following from this, evaluation of these morphometric properties may be relevant for treatment planning. For example, reduced hippocampal volume is a marker of hippocampal sclerosis in epilepsy patients,14 and individuals with this tissue pathology are often amenable to surgical intervention. A further potential use of quantitative imaging metrics derived from MRI data are as enrollment criteria for clinical trials, with a goal to “enriching” the trial population to increase the likelihood of enrolling participants who will benefit from the intervention; this strategy has gained some traction in dementia intervention trials.15
Table 1.
A selection of reported changes in hippocampal volume, cortical thickness, and brain volume in a variety of diseases or conditions relative to healthy controls
| Neuroanatomical Measure | Disease State/Condition | Reported Effect Size |
|---|---|---|
| Hippocampal volume | Temporal lobe epilepsy | 10–33%14 |
| Alzheimer’s disease | 22%30 | |
| Mild cognitive impairment | 14%30 | |
| Stroke | 19–32%31 | |
| Depression | 19%32 | |
| Healthy aging (age 40 to 75) | 16%33 | |
| Cortical thickness | Alzheimer’s disease | 11.7% (.5 mm)34 |
| Mild cognitive impairment | 3.7% (.16 mm)34 | |
| Attention-deficit hyperactivity disorder | 1.9% (.09 mm)29 | |
| Schizophrenia | 3.5% (.09 mm)35 | |
| Brain volume | Chronic alcoholism | 5.9%36 |
| Schizophrenia | 1.5%37 | |
| Childhood lead exposure | 1.2%38 | |
| Alzheimer’s disease | .98% per year15 |
All estimates in patient groups are reduced relative to healthy controls. Similarly, healthy aging is associated with reduced hippocampal volume between ages 45 and 60 and >75 years old. Percentage changes are either provided as reported or estimated from reported absolute values as .
We hypothesized that the across-subject standard deviation of (i) cortical thickness, (ii) hippocampal volume, and (iii) total brain volume, when calculated using standardized research imaging protocols, would be smaller than when estimated using nonstandardized clinical imaging protocols in data obtained from the same set of individuals. We expected that sample size estimates obtained using standard power analysis methods would demonstrate a substantive improvement when using a standardized image acquisition protocols compared with nonstandardized clinically acquired imaging.
Methods
Subject Recruitment and MRI Acquisition
The Human Epilepsy Project (HEP) study is a prospective multicenter study of newly diagnosed epilepsy, with enrollment running from 2012 to 2017. A subset of participants had imaging available, acquired as part of their clinical epilepsy evaluation, in addition to HEP-specific research imaging. Participants with both a clinically acquired (nonstandardized) 3D T1-weighted whole brain scan and a standardized 3D T1-weighted whole brain acquisition were used in this study. The average time between the standardized HEP scan and unstandardized clinical MRI scan was 8.5 months. For the standardized image acquisitions, specific parameters vary by scanner make and model across sites, but all scans were obtained on 3T MRI scanners with a 1 mm3 voxel size. Image acquisition parameters for the standardized acquisition were obtained from MRI scanner protocols provided by the Alzheimer’s Disease Neuroimaging Initiative (ADNI, http://adni.loni.usc.edu/methods/documents/mri-protocols/);10 for HEP, we modified the voxel size for T1-weighted MRI scans to 1 mm isotropic (Table 2). Nonstandardized clinical imaging was obtained as part of an individual’s work up for epilepsy assessment, typically prior to enrollment in HEP. Clinical imaging was obtained using 1.5 T scanners (N = 15) and 3T scanners (N = 17). A range of acquisition parameters were used in the clinical acquisitions, for example, in-plane voxel size ranged from .35 to 1.2 mm2 and slice thickness ranged from .7 to 1.9 mm. See Table 3 for image acquisition parameters for nonstandardized clinical image acquisitions.
Table 2.
Image acquisition parameters for participants imaged using standardized research image acquisition protocols
| Scanner Manufacturer | Model | Field Strength | Number of Subjects | Acquisition | Repetition Time (ms) | Echo Time (ms) | Inversion Time (ms) | Flip Angle (°) |
|---|---|---|---|---|---|---|---|---|
| Siemens | Allegra | 3T | 19 | MPRAGE | 2500 | 3.93 | 900 | 8 |
| Philips | Achieva | 3T | 3 | TFE | 2500 | 3.03 | 900 | 8 |
| Siemens | TrioTrim | 3T | 1 | MPRAGE | 2500 | 3.03 | 900 | 8 |
| GE | Discovery MR750 |
3T | 1 | FSPGR | 8.152 | 3.17 | n/a | 11 |
| Philips | Achieva | 3T | 4 | TFE | 8.073 | 3.68 | n/a | 6 |
| Philips | Ingenia | 3T | 1 | TFE | 2550 | 74.71 | n/a | 90 |
| Philips | Ingenia | 3T | 1 | TFE | 4800 | 298.69 | n/a | 90 |
| Siemens | Verio | 3T | 1 | MPRAGE | 2500 | 2.99 | 900 | 8 |
n/a = not applicable; MPRAGE = magnetization prepared rapid gradient echo; TFE = turbo field echo; FSPGR = fast spoiled gradient echo; GE = General Electric.
Table 3.
Image acquisition parameters for participants imaged using nonstandardized clinical imaging
| Site | Manufacturer | Model | Field Strength (T) | Acquisition (mm3) | Voxel Size (ms) | Repetition Time | Echo Time (ms) | Inversion Time (ms) | Flip Angle (°) |
|---|---|---|---|---|---|---|---|---|---|
| Site 1 | Philips | Intera | 1.5 | FFE | 1 × 1 × 1 | 3.8 | 1.7 | n/a | 8 |
| Site 1 | GE | Signa HDxt | 1.5 | GRE | .35 × 1 × .35 | 5.412 | 1.664 | n/a | 30 |
| Site 1 | Siemens | Avanto | 1.5 | MPRAGE | 1.5 × .86 × .86 | 1900 | 3.99 | 950 | 12 |
| Site 1 | Siemens | Avanto | 1.5 | MPRAGE | .86 × .86 × 1 | 2100 | 3.79 | 1100 | 12 |
| Site 1 | Siemens | TrioTrim | 3 | MPRAGE | 1 × 1 × 1 | 1360 | 2.15 | 800 | 15 |
| Site 1 | Siemens | Biograph mMR | 3 | MPRAGE | 1 × 1 × 1 | 2100 | 2.79 | 900 | 8 |
| Site 1 | Siemens | Biograph mMR | 3 | MPRAGE | 1 × 1 × 1 | 2100 | 2.79 | 900 | 8 |
| Site 1 | Philips | Ingenia | 1.5 | FFE | .98 × 1.3 × .98 | 7.1 | 3.328 | n/a | 8 |
| Site 1 | Philips | Intera | 1.5 | FFE | .86 × 1.1 × .86 | 20 | 4.599 | n/a | 15 |
| Site 1 | Philips | Ingenia | 1.5 | FFE | .89 × 1.3 × .89 | 7 | 3.289 | n/a | 8 |
| Site 1 | Siemens | TrioTrim | 3 | MPRAGE | 1 × 1 × 1 | 1360 | 2.15 | 800 | 15 |
| Site 1 | Siemens | TrioTrim | 3 | MPRAGE | 1 × 1 × 1 | 1360 | 2.15 | 800 | 15 |
| Site 1 | Siemens | Skyra | 3 | MPRAGE | .9 × 1 × 1 | 2100 | 3.17 | 900 | 8 |
| Site 1 | Siemens | Biograph mMR | 3 | MPRAGE | 1 × 1 × 1 | 2300 | 2.98 | 900 | 9 |
| Site 1 | GE | Signa HDxt | 1.5 | GRE | 1.2 × 1.9 × 1.9 | 12.364 | 5.072 | 450 | 12 |
| Site 1 | Philips | Achieva | 3 | FFE | 1 × .93 × .93 | 8 | 3.686 | n/a | 8 |
| Site 1 | Siemens | Biograph mMR | 3 | MPRAGE | 1 × 1 × 1 | 1360 | 2.19 | 800 | 15 |
| Site 1 | Siemens | Prisma | 3 | MPRAGE | 1.1 × 1.1 × 1.1 | 2220 | 3.22 | 1100 | 9 |
| Site 1 | Siemens | Aera | 1.5 | MPRAGE | .97 × .97 × .97 | 2300 | 2.27 | 900 | 8 |
| Site 1 | Siemens | Avanto | 1.5 | MPRAGE | 1.1 × 1.1 × 1.1 | 2730 | 3.4 | 1000 | 7 |
| Site 2 | Philips | Ingenia | 3 | FFE | .89 × .83 × .83 | 9 | 4.163 | n/a | 8 |
| Site 2 | Philips | Ingenia | 3 | FFE | .89 × .83 × .83 | 8.5 | 3.873 | n/a | 8 |
| Site 3 | Siemens | Verio | 3 | MPRAGE | .48 × 1.5 × .45 | 1900 | 2.93 | 900 | 9 |
| Site 4 | GE | Signa HDxt | 1.5 | GRE | .51 × .51 × 1 | 14.72 | 6.368 | 450 | 13 |
| Site 5 | GE | Signa HDxt | 1.5 | GRE | .43 × .70 × .43 | 10 | 4.072 | 600 | 13 |
| Site 6 | Philips | Achieva | 1.5 | FFE | 1 × 1 × 1 | 12 | 2.398 | n/a | 20 |
| Site 6 | GE | Signa HDxt | 1.5 | GRE | .47 × .47 × 1.4 | 11.2 | 5.008 | n/a | 12 |
| Site 6 | Philips | Achieva | 1.5 | FFE | .92 × 1.2 × .92 | 12 | 2.399 | n/a | 20 |
| Site 6 | Philips | Achieva | 3 | FFE | .94 × .94 × 1 | 5.5 | 1.503 | n/a | 8 |
| Site 7 | Siemens | TrioTrim | 3 | MPRAGE | .48 × 1 × .48 | 1860 | 2.94 | 1000 | 8 |
| Site 7 | Siemens | TrioTrim | 3 | MPRAGE | .53 × 1.14 × .53 | 1800 | 2.94 | 1000 | 8 |
| Site 7 | Siemens | TrioTrim | 3 | MPRAGE | .48 × 1 × .48 | 1800 | 2.94 | 1000 | 8 |
n/a = not applicable; MPRAGE = magnetization prepared rapid gradient echo; FFE, GRE = gradient echo sequences; GE = General Electric.
Image Processing
All scans were visually inspected for image quality prior to analysis and poor quality scans were excluded from the analysis, following a qualitative image quality evaluation system we developed for a prior study.16 We investigated cortical thickness, hippocampal volume, and brain volume (supratentorial volume) as the three key morphometric measures of interest using default image processing routines provided as part of Freesurfer v6.0.17,18 Across-subject standard deviation was calculated for each of the three measures and for the standardized and unstandardized protocols. The across-subject morphometric estimates were tested for normality using the Shapiro-Wilk normality test implemented in R.19
Power Analyses
One of the primary goals of power analyses is to estimate the number of subjects required in a study to minimize the likelihood of a false negative finding; in the context of neuroanatomical imaging studies, the goal is to estimate the number of subjects per group to scan in order to detect an existing difference in brain structure between subject groups. Sample sizes were estimated using (i) standard power analysis methods derived from Student’s t-test and (ii) a novel bootstrap-based nonparametric approach, described below. The bootstrap-based approach was utilized to accommodate for potential non-normal distributions of morphometric parameters. Standard power analysis methods were used as implemented in the “power.t.test” function distributed as part of the R software package.20 The formula for sample size n per group required for a well-powered study is:
where σ is the sample standard deviation, δ is the target effect size, α is the false positive rate, β is the false negative rate, and z is the quantile function for the normal distribution.21
Assumptions for these analyses include (i) the morphometric estimates are normally distributed and (ii) patient and control groups have similar variability, characterized by their standard deviation. For cortical thickness analyses, target effect sizes were varied from .05 to .5 mm. Two-sided analyses were used, with power (= 1 − β, where β is the false negative rate) set at .8 and the false positive rate α = .05. The required sample size to detect the cortical thickness effect sizes was calculated using standard deviation of cortical thickness values estimated from (i) standardized image acquisitions and (ii) nonstandardized image acquisitions. Similar analyses were undertaken with hippocampal volume and brain volume estimates; for the volumetric analyses, sample sizes required to detect changes between 5 and 20% in mean volume were calculated.
Sample Size Estimation Using Nonparametric Bootstrapping
An estimate of required sample sizes to obtain an adequately powered study can be provided using bootstrapping. This alternative to parametric methods may be preferable when the population distribution of the morphometric parameter of interest is unknown and potentially non-normal. Code for the following approach is provided at https://github.com/hpardoe/bootstrap-power. Sample size was estimated as follows:
A range of target sample sizes per group was specified that encompassed the likely final target sample size; in the current study this was determined based on the results of the parametric power analyses.
For each sample size n in the range of values, two samples were simulated. These may be thought of as a hypothetical control sample and diagnostic group sample. The first sample was created by sampling n values with replacement from the morphometric dataset of interest. The second sample was simulated as per the first, with a prespecified effect size added.
Differences between the two samples were tested using a Mann-Whitney test.
If the P-value of the Mann-Whitney test was <.05, the comparison is recorded as a true positive; if P > .05 the comparison was recorded as a false negative finding.
Steps 2 to 4 were iterated (number of iterations = 5,000) in order to estimate the false negative rate β (Type II=error rate) and power (1 − β) for each sample size.
The output of Step 5 is a numeric table with the estimated power for each hypothetical sample size. The sample size required to obtain power = .8 is estimated by linear interpolation between the sequential points in the table that span power = .8.
We reported the relationship between sample size and morphometric effect sizes using both traditional power analyses and the bootstrapping approach is described above.
Image Acquisition Parameters and Sample Size Estimates for Nonstandardized Clinical Imaging
We analyzed the clinical imaging dataset to investigate how variability in clinical image acquisition parameters affected sample size estimates. For this analysis, we selected a single effect size per morphometric parameter and compared the required sample size to detect this effect while creating subgroups based on the image acquisition parameter of interest. Sample sizes were calculated using the nonparametric bootstrap approach. For cortical thickness, the effect size was set to .1 mm; for hippocampal volume, effect size = 200 mm3 (5%); and for brain volume the effect size = 5.1 × 105 mm3 (5%). The image acquisition parameters investigated were:
Magnetic field strength, 1.5 versus 3T.
Voxel anisotropy, defined as the ratio between the maximum and minimum voxel size. For our dataset, this was equivalent to the ratio between the slice thickness (maximum voxel dimension) and the in-plane voxel length (minimum voxel dimension). Anisotropic voxels have an anisotropy >1; isotropic voxels have an anisotropy value = 1. For this analysis, the clinical imaging dataset was subdivided into two groups by ranking the voxel anisotropies and separating by the median anisotropy value.
Slice thickness. As per the voxel anisotropy analysis, the clinical imaging dataset was subdivided into two groups by ranking the slice thickness and separating by the median value.
Results
We identified 32 HEP participants who had both a research whole brain T1-weighted MRI with standardized image acquisition parameters and a clinical whole brain T1-weighted acquisition with unstandardized image acquisition parameters (9 males, 23 females, age 29.9 ± 12.6 years). These participants were a subset of 88 HEP participants who had both clinical and research imaging as part of the HEP study. Four participants were excluded because their research protocol MRI scan deviated from the HEP imaging protocol, and a further two participants did not have a research scan. Further reasons for excluding participants based on their clinical imaging included high slice thickness T1-weighted imaging that precluded morphometric analysis (n = 44), postcontrast T1-weighted imaging (n = 4) or limited brain coverage (n = 2). Morphometric estimates are summarized in Table 4. Five of the eight morphometric estimates showed evidence for nonnormal distributions as indicated with a Shapiro-Wilk test P < .05, comprising research and clinical cortical thickness estimates, left and right hippocampal volumes estimated using research imaging and right hippocampal volume estimated using clinical imaging (Table 4). These findings justify the additional use of the nonparametric boostrap power analyses.
Table 4.
Quantitative neuroanatomical properties estimated using standardized and nonstandardized imaging
| Image Acquisition Protocol | Cortical Thickness (mm) | Hippocampal Volume (mm3) | Brain Volume (mm3) | |||
|---|---|---|---|---|---|---|
| Mean ± Standard Deviation | Coefficient of Variation | Mean ± Standard Deviation | Coefficient of Variation | Mean ± Standard Deviation | Coefficient of Variation | |
| Standardized 1 mm isotropic | 2.5 ± .1 mm* | .04 | 3890 ± 443 mm3 (left)* | .11 | 9.96 ± .93 × 105 | .09 |
| 4077 ± 599 mm3 (right)* | .14 | |||||
| Clinical nonstandardized | 2.5 ± .3 mm* | .12 | 3928 ± 665 mm3 (left) | .17 | 1.02 ± .12 × 106 | .12 |
| 4093 ± 829 mm3 (right)* | .2 | |||||
Note that across-subject standard deviation estimates for cortical thickness, hippocampal volume, and brain volume were all reduced when using a multicenter standardized acquisition relative to nonstandardized clinically acquired data.
Morphometric estimates with Shapiro-Wilk test P < .05, indicating nonnormal distribution of values.
Figure 1 demonstrates the number of subjects required per group to detect a range of cortical thickness changes ranging from .05 to .5 mm. Our data show that the use of a standardized image acquisition protocol results in a five-fold reduction in the number of participants required to detect a cortical thickness difference of .1 mm between subject groups.
Fig 1.
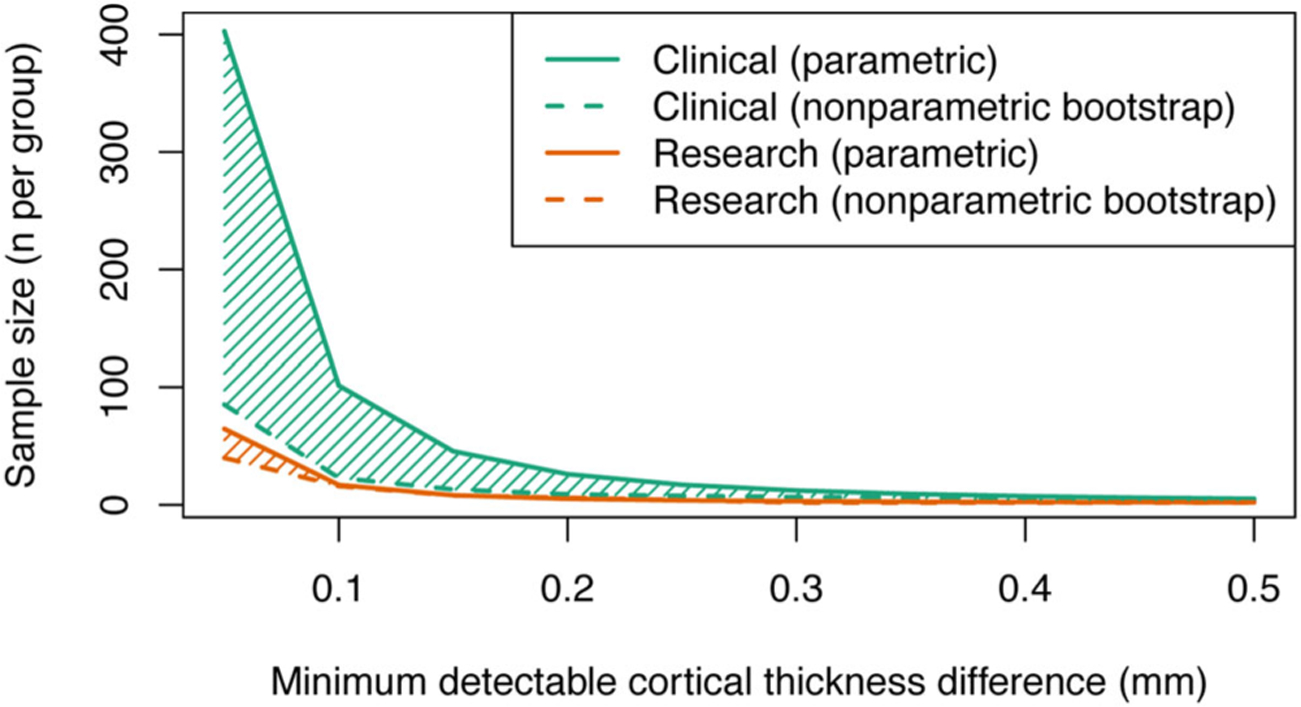
Standardization of image acquisition protocols improves statistical power for multicenter cortical thickness studies. The figure demonstrates a substantive reduction in required sample size when using a standardized image acquisition protocol (orange lines) compared with a nonstandardized protocol (green lines). The solid lines show sample size estimates obtained from conventional power analysis techniques that assume values are sampled from a normal distribution, dashed lines indicate sample size estimates obtained using a nonparametric bootstrap approach.
Power analyses of hippocampal and brain volumes were carried out to determine the minimum number of subjects required to detect a hypothetical 5 to 20% volume change. Analyses were conducted separately for left and right hippocampi and sample size estimates for each side were subsequently averaged (Figure 2). A similar plot showing the relationship between sample size and brain volume is shown in Figure 3.
Fig 2.

The use of a standardized image acquisition protocol improves statistical power for detection of hippocampal volume changes in multicenter imaging studies. The plot shows that the number of subjects required per group is less for a given effect size (hippocampal volume difference) when using a standardized protocol (orange lines) compared to a nonstandardized protocol (green lines). As an example, to detect a 200 mm3 volume change (5%) requires approximately 110 subjects for a standardized protocol and approximately 220 subjects for a nonstandardized protocol.
Fig 3.
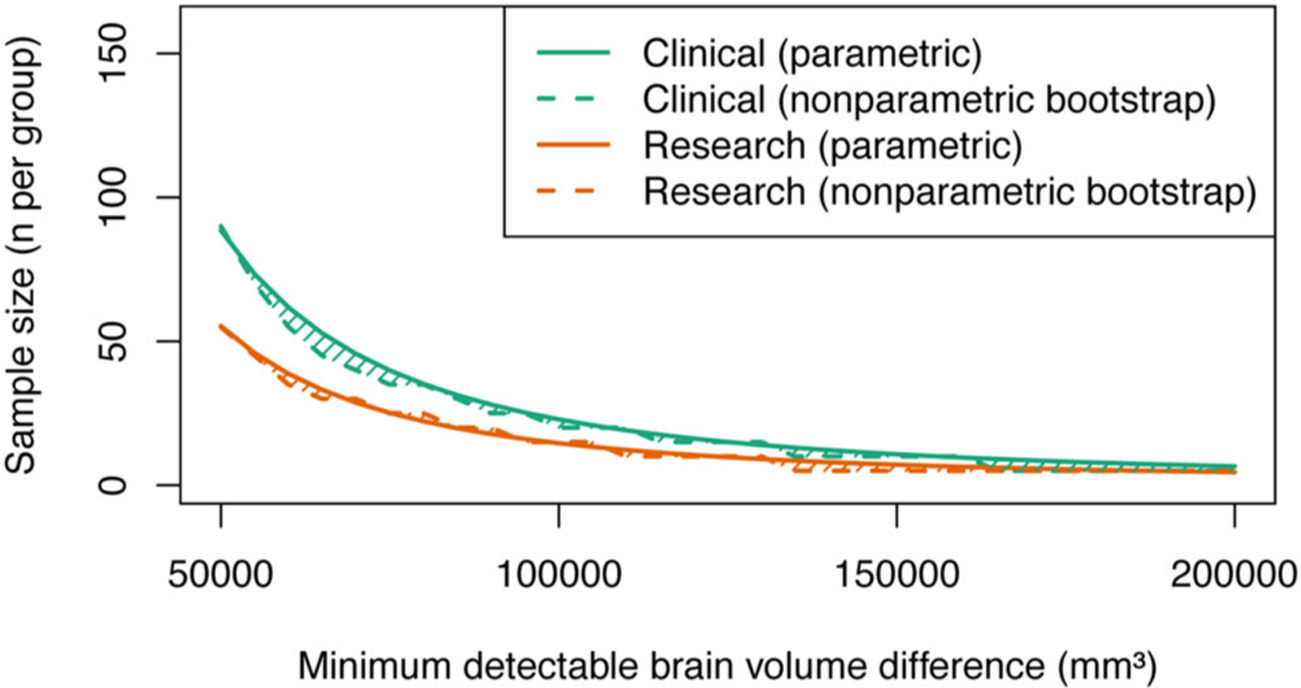
The use of a standardized image acquisition protocol improves statistical power for detection of brain volume changes in multicenter imaging studies. The plot shows that the number of subjects required per group is less for a given effect size when using a standardized protocol (orange lines) compared to a nonstandardized protocol (green lines). As an example, to detect a 50,000 mm3 volume change (~5%) requires approximately 60 subjects for a standardized protocol and approximately 90 subjects for a nonstandardized protocol.
Subdividing the clinical imaging dataset based on field strength yielded N = 15 participants imaged at 1.5T and N = 17 participants imaged at 3T. When the groups were split based on voxel anisotropy, the lower group had an average anisotropy of 1.01 ± .03 (mean ± SD) and the upper group had an average anisotropy of 1.87 ± .67. The low slice thickness participants had an average slice thickness of .96 ± .08 mm and the high slice thickness group had an average thickness of 1.2 ± .25 mm. Both cortical thickness and hippocampal volume estimates showed a substantive decrease in required sample sizes when using 3T isotropic imaging with low slice thickness (Figure 4).
Fig 4.
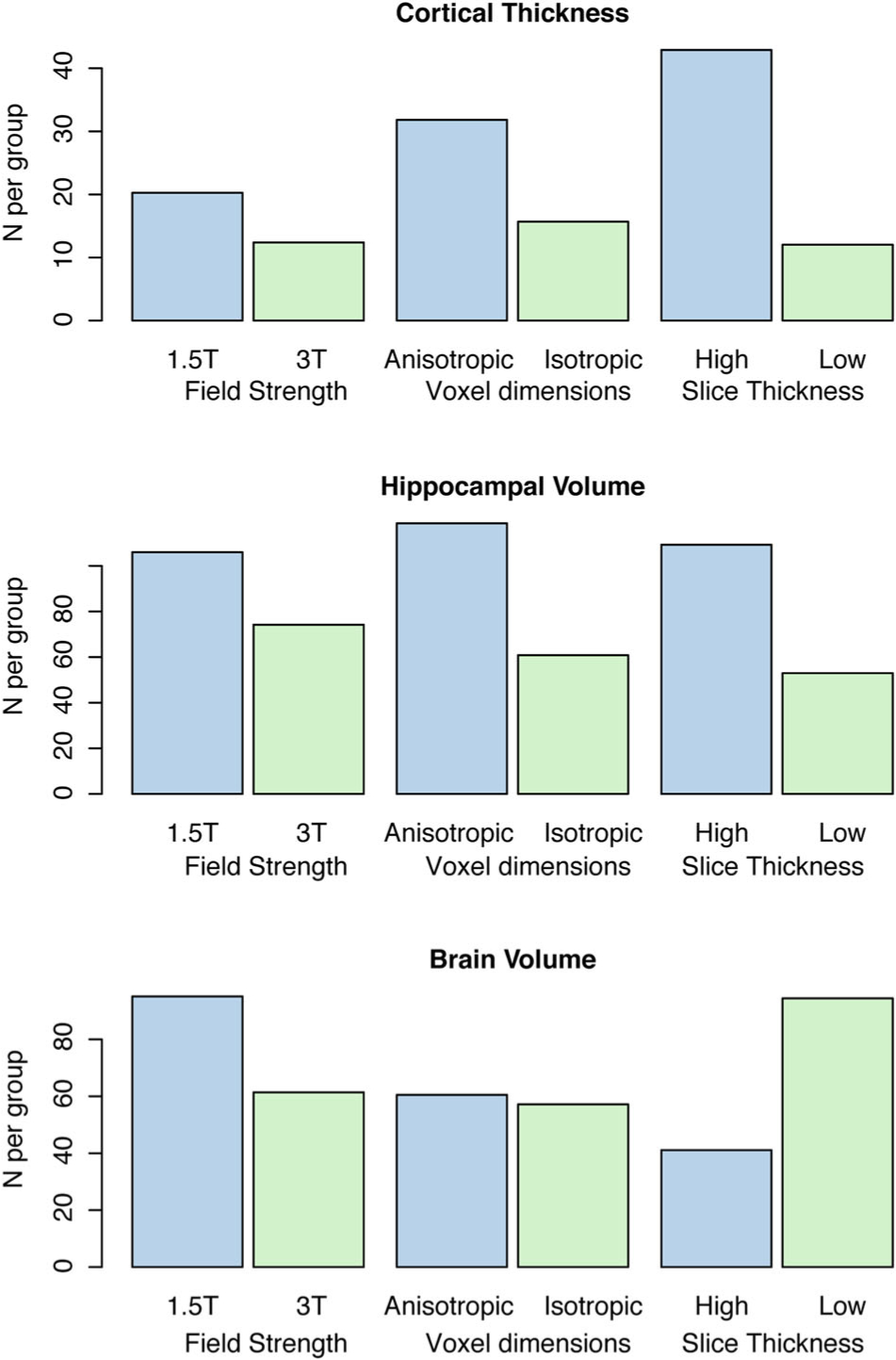
The relationship between image acquisition parameters and sample size estimates obtained using nonstandardized clinical imaging. The figure shows that 3T imaging with isotropic voxel size and low slice thickness allows lower sample sizes and, therefore, higher power for detection of changes in cortical thickness and hippocampal volume.
Discussion
Standardizing image acquisition protocols in a multicenter setting is expected to decrease scanner-related variance in quantitative morphometric estimates and, therefore, increase statistical power. Here, we use power analyses to quantify the benefit of standardizing protocols by estimating required sample sizes for a range of biologically plausible effect sizes in analyses of cortical thickness, hippocampal thickness, and supratentorial volume. Our findings will be useful for optimizing the design of future multicenter studies in terms of cost effectiveness, particularly in scenarios where recruitment may be difficult or morphometric brain changes are likely to be subtle.
We found that standardized protocols yield a strikingly smaller (over two-fold decrease) of standard deviation in cortical thickness, when compared against nonstandardized clinical scans. A more modest decrease in variability is observed in volumetric measures. The greatest benefit for standardizing sequences occurs when investigating subtle changes, for example, cortical thickness differences of less than .3 mm or hippocampal volume changes of less than 400 mm3 (10% change in volume). Our analysis of the clinical imaging dataset indicated that both cortical thickness and hippocampal volume estimates have a substantive reduction in variability and associated improvement in power when 3T imaging is used relative to 1.5T; isotropic voxel sizes are used relative to anisotropic voxels; and lower slice thickness is used compared with high slice thickness acquisitions. We wish to note that the investigation of these image acquisition parameters was largely driven by the available data in our study and, therefore, should not be considered a comprehensive analysis. Notably, we did not consider variability in parameters that are varied to manipulate image contrast properties, namely echo time (TE), repetition time (TR), inversion time (TI), and flip angle, since these were inconsistent across subjects and are difficult to compare between scanner manufacturers. Variations in the parameters that we did investigate were not made in isolation and were not made prospectively; therefore, there may be significant collinearities between the image acquisition parameters under consideration.
Finally, for the analyses of clinical imaging parameters, subjects were not matched based on participant demographics (eg, age and sex) or epilepsy-related factors such as etiology. These potential sources of error may explain the counterintuitive finding that lower slice thickness acquisitions require a larger number of participants for analyses of brain volume relative to higher slice thickness acquisitions (Figure 4). We also wish to note that power analyses are designed to minimize the likelihood of making a false negative finding (Type II error). They are uninformative regarding the most accurate method for measuring morphometric properties. A morphometric technique can have both poor accuracy and low variability. If only the variability of the measure is taken into account via analyses similar to those presented in this study, a future researcher may draw the incorrect conclusion that a method that requires fewer participants is superior to a method that requires more.
A vast number of published studies use the morphometric estimates that we investigated in this study, precluding a systematic analysis of effect sizes associated with various diseases. However, we believe the range of effect sizes analyzed in our work is broadly representative of those observed in a variety of neurological disorders. A summary of reported effect sizes is provided in Table 1.
Previous studies have shown that variable acquisition protocols, scanner make and model, coil configurations, and even variability in site QA policies regarding acceptable image quality may introduce variability in quantitative neuroanatomical estimates in multicenter imaging studies.1–3,6,13,16 To our knowledge, this is the first study employing power analysis techniques to explicitly quantify the benefit of standardized image acquisition protocols versus nonstandardized protocols to determine whether the variability from these confounds can be mitigated. For the HEP study, standardization was implemented by centrally distributing scanner-specific image acquisition protocols from the imaging core. This process ensured that image acquisition parameters were largely consistent across sites, although it is possible that minor deviations from the specified parameters existed due to site-specific factors, for example, variations in scanner software versions. Nonstandardized clinical imaging parameters were not dictated by the HEP study team and were decided by protocols developed by the individual imaging or epilepsy centers. For many individual sites, the goal for epilepsy imaging is to obtain scans suitable for radiological assessment leading to individual diagnosis, not group-level morphometric analyses; therefore, a significant proportion of imaging data was not suitable for morphometric analysis, primarily due to high slice thickness T1-weighted imaging.
Our work contributes to a growing body of literature that characterizes the effect of site-related differences on morphometric estimates. Previous work has shown that site-related differences contribute to systematic differences in all three of the quantitative morphometric estimates investigated in our study including cortical thickness,12,13 hippocampal volume,1,22 and brain volume.1,23 Although our work is derived from an observational study, there are some notable prospective studies that were designed for accurate characterization of between-site effects. An interesting approach that appears to be useful for characterizing between-site differences utilizes a “living phantom,” in which individuals are imaged at a number of participating sites in a multisite study.6,24 There are also a number of proposed postprocessing methods that can be used to ameliorate site-based effects; examples of these include statistical methods to model the effect of the scanner or site in the analysis of morphometric data7,9,25 and intensity normalization of acquired images.26,27 An interesting recent approach applies multitask learning, a machine learning technique, to the problem of identifying disease-specific brain changes in the presence of sources of variability introduced by multiple scanners.28 Existing techniques for postacquisition harmonization of imaging data typically rely on the availability of enough scans per site to estimate site-specific effects. In this context, existing postprocessing methods to harmonize multisite data fail when applied to our clinical imaging dataset because a number of the sites only had a single scan available per MRI scanner. We are not aware of any existing methods that are able to harmonize imaging data acquired under these conditions.
A limitation of this study is that our participants were epilepsy patients rather than healthy controls. Despite this limitation, there is prior evidence that variance in morphometric estimates tends to be similar across diagnostic categories, see for example Table 1, Shaw et al.29 Healthy controls are unlikely to be scanned in a clinical setting and, therefore, we believe this dataset provides important guidance for future studies. An additional limitation is that our power analyses were only done for detecting main effects. Sample size requirements for detecting interactions between explanatory variables will be considerably larger; an example of a relatively common interaction of interest is characterizing the relationship between age and disease status. Both across-subject mean cortical thickness and variance are variable across the cortex and there may be some brain regions that require considerably more participants than the estimates provided in these analyses to detect a given effect size. This may also explain why our reported cortical thickness sample size estimates are lower than the volumetric estimates. Finally, it is noteworthy that the acquisitions in HEP were standardized across platforms; no prospective acquisition harmonization was carried out. However, the HEP research protocol was based on existing MRI protocols from the ADNI study, which was harmonized.10 Although the term “harmonization” is used in different contexts in the neuroimaging literature, in this context we interpret harmonization of acquisition protocols as an iterative process in which image acquisition parameters are optimized to provide imaging metrics, such as contrast to noise ratio, that are within predefined limits across sites. Prospective study-specific harmonization of image acquisition protocols may provide an additional improvement in statistical power over that demonstrated in our analyses.
In summary, we have provided quantitative estimate of the benefit of the use of standardized image acquisition protocols. Up to a five-fold reduction in sample sizes is expected to detect disease-related neuroanatomical changes. Standardizing image acquisition protocols prior to scanning is a valuable approach to increase the statistical power in multicenter MRI studies.
Acknowledgments:
The Human Epilepsy Project (HEP) is supported by The Epilepsy Study Consortium (ESCI), a nonprofit organization dedicated to accelerate the development of new therapies in epilepsy to improve patient care. The funding provided to ESCI to support HEP comes from industry, philanthropy, and foundations (UCB Pharma, Finding A Cure for Epilepsy and Seizures, Pfizer, Lundbeck, The Andrews Foundation, Friends of Faces and others).
Footnotes
Conflict of Interest: The authors have no conflicts of interest to disclose.
References
- 1.Cannon TD, Sun F, McEwen SJ, et al. Reliability of neuroanatomical measurements in a multisite longitudinal study of youth at risk for psychosis. Hum Brain Mapp 2014;35:2424–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jovicich J, Marizzoni M, Sala-Llonch R, et al. Brain morphometry reproducibility in multi-center 3t MRI studies: a comparison of cross-sectional and longitudinal segmentations. Neuroimage 2013;83:472–84. [DOI] [PubMed] [Google Scholar]
- 3.Keshavan A, Paul F, Beyer MK, et al. Power estimation for nonstandardized multisite studies. Neuroimage 2016;134:281–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedman L, Stern H, Brown GG, et al. Test-retest and between-site reliability in a multicenter fMRI study. Hum Brain Mapp 2008;29:958–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gouttard S, Styner M, Prastawa M, Piven J, Gerig G. Assessment of reliability of multi-site neuroimaging via traveling phantom study. Med Image Comput Comput Assist Interv 2008;11:263–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jovicich J, Czanner S, Greve D, et al. Reliability in multi-site structural MRI studies: effects of gradient non-linearity correction on phantom and human data. Neuroimage 2006;30:436–43. [DOI] [PubMed] [Google Scholar]
- 7.Pardoe H, Pell GS, Abbott DF, Berg AT, Jackson GD. Multi-site voxel-based morphometry: methods and a feasibility demonstration with childhood absence epilepsy. Neuroimage 2008;42:611–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schnack HG, van Haren NE, Hulshoff Pol HE, et al. Reliability of brain volumes from multicenter MRI acquisition: a calibration study. Hum Brain Mapp 2004;22:312–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fortin JP, Cullen N, Sheline YI, et al. Harmonization of cortical thickness measurements across scanners and sites. Neuroimage 2018;167:104–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jack CR Jr., Bernstein MA, Fox NC, et al. The alzheimer’s disease neuroimaging initiative (ADNI): MRI methods. J Magn Reson Imaging 2008;27:685–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pearlson G Multisite collaborations and large databases in psychiatric neuroimaging: advantages, problems, and challenges. Schizophr Bull 2009;35:1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schnack HG, van Haren NE, Brouwer RM, et al. Mapping reliability in multicenter MRI: Voxel-based morphometry and cortical thickness. Hum Brain Mapp 2010;31:1967–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han X, Jovicich J, Salat D, et al. Reliability of mri-derived measurements of human cerebral cortical thickness: the effects of field strength, scanner upgrade and manufacturer. Neuroimage 2006;32:180–94. [DOI] [PubMed] [Google Scholar]
- 14.Pardoe HR, Pell GS, Abbott DF, Jackson GD. Hippocampal volume assessment in temporal lobe epilepsy: how good is automated segmentation? Epilepsia 2009;50:2586–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fotenos AF, Snyder AZ, Girton LE, Morris JC, Buckner RL. Normative estimates of cross-sectional and longitudinal brain volume decline in aging and ad. Neurology 2005;64:1032–9. [DOI] [PubMed] [Google Scholar]
- 16.Pardoe HR, Kucharsky Hiess R, Kuzniecky R. Motion and morphometry in clinical and nonclinical populations. Neuroimage 2016;135:177–85. [DOI] [PubMed] [Google Scholar]
- 17.Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci USA 2000;97:11050–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischl B, Salat DH, Busa E, et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 2002;33:341–55. [DOI] [PubMed] [Google Scholar]
- 19.Royston P Remark as r94: a remark on algorithm as 181: The w-test for normality. JRoyal Stat Soc Series C (Applied Statistics) 1995;44:547–51. [Google Scholar]
- 20.R: A Language and Environment for Statistical Computing [computer program]. Vienna, Austria: R Foundation for Statistical Computing; 2018. [Google Scholar]
- 21.Van Belle G Statistical Rules of Thumb. Hoboken, New Jersey: John Wiley & Sons Inc; 2008. [Google Scholar]
- 22.Fennema-Notestine C, Gamst AC, Quinn BT, et al. Feasibility of multi-site clinical structural neuroimaging studies of aging using legacy data. Neuroinformatics 2007;5:235–45. [DOI] [PubMed] [Google Scholar]
- 23.Chu R, Tauhid S, Glanz BI, et al. Whole brain volume measured from 1.5T versus 3T MRI in healthy subjects and patients with multiple sclerosis. J Neuroimaging 2016;26:62–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shinohara RT, Oh J, Nair G, et al. Volumetric analysis from a harmonized multisite brain MRI study of a single subject with multiple sclerosis. AJNR Am J Neuroradiol 2017;38:1501–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chua AS, Egorova S, Anderson MC, et al. Handling changes in mri acquisition parameters in modeling whole brain lesion volume and atrophy data in multiple sclerosis subjects: comparison of linear mixed-effect models. Neuroimage Clin 2015;8:606–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shinohara RT, Sweeney EM, Goldsmith J, et al. Statistical normalization techniques for magnetic resonance imaging. Neuroimage Clin 2014;6:9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fortin JP, Sweeney EM, Muschelli J, Crainiceanu CM, Shinohara RT, Alzheimer’s Disease Neuroimaging Initiative. Removing inter-subject technical variability in magnetic resonance imaging studies. Neuroimage 2016;132:198–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma Q, Zhang T, Zanetti MV, et al. Classification of multi-site MR images in the presence of heterogeneity using multi-task learning. Neuroimage Clin 2018;19:476–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaw P, Lerch J, Greenstein D, et al. Longitudinal mapping of cortical thickness and clinical outcome in children and adolescents with attention-deficit/hyperactivity disorder. Arch Gen Psychiatry 2006;63:540–9. [DOI] [PubMed] [Google Scholar]
- 30.Convit A, De Leon MJ, Tarshish C, et al. Specific hippocampal volume reductions in individuals at risk for alzheimer’s disease. Neurobiol Aging 1997;18:131–8. [DOI] [PubMed] [Google Scholar]
- 31.Werden E, Cumming T, Li Q, et al. Structural mri markers of brain aging early after ischemic stroke. Neurology 2017;89:116–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bremner JD, Narayan M, Anderson ER, Staib LH, Miller HL, Charney DS. Hippocampal volume reduction in major depression. Am J Psychiatry 2000;157:115–8. [DOI] [PubMed] [Google Scholar]
- 33.Scahill RI, Frost C, Jenkins R, Whitwell JL, Rossor MN, Fox NC. A longitudinal study of brain volume changes in normal aging using serial registered magnetic resonance imaging. Arch Neurol 2003;60:989–94. [DOI] [PubMed] [Google Scholar]
- 34.Singh V, Chertkow H, Lerch JP, Evans AC, Dorr AE, Kabani NJ. Spatial patterns of cortical thinning in mild cognitive impairment and alzheimer’s disease. Brain 2006;129:2885–93. [DOI] [PubMed] [Google Scholar]
- 35.Kuperberg GR, Broome MR, McGuire PK, et al. Regionally localized thinning of the cerebral cortex in schizophrenia. Arch Gen Psychiatry 2003;60:878–88. [DOI] [PubMed] [Google Scholar]
- 36.Pfefferbaum A, Lim KO, Zipursky RB, et al. Brain gray and white matter volume loss accelerates with aging in chronic alcoholics: a quantitative MRI study. Alcohol Clin Exp Res 1992;16:1078–89. [DOI] [PubMed] [Google Scholar]
- 37.van Haren NE, Hulshoff Pol HE, Schnack HG, et al. Progressive brain volume loss in schizophrenia over the course of the illness: evidence of maturational abnormalities in early adulthood. Biol Psychiatry 2008;63:106–13. [DOI] [PubMed] [Google Scholar]
- 38.Cecil KM, Brubaker CJ, Adler CM, et al. Decreased brain volume in adults with childhood lead exposure. PLoS Med 2008;5:e112. [DOI] [PMC free article] [PubMed] [Google Scholar]


