Highlights
-
•
We observed the characteristics of specific antibody production of COVID-19 patients.
-
•
For the first time, we studied the specific antibody levels of different populations.
-
•
Detecting specific antibodies is a complementary approach to diagnose COVID-19.
Keywords: Coronavirus, Serology, Nucleic acid, Antibody
Abstract
Corona Virus Disease 2019 (COVID-19) has spread rapidly to more than 215 countries, with over 11.91 million reported cases and more than 540,000 deaths. Rapid diagnosis remains a bottleneck for containing the epidemic. We used an automated chemiluminescent immunoassay to detect serum IgM and IgG antibodies to the 2019-nCoV in 742 subjects, so as to observe the dynamic process of antibody production in COVID-19 disease and seroepidemiology in different populations. Patients with COVID-19 were reactive (positive) for specific antibodies within 3–15 days after onset of symptoms. Specific IgM and IgG levels increased with the progression of the disease. The areas under the receiver operating characteristic curves for IgM and IgG were 0.984 and 1.000, respectively. This antibody detection assay had good sensitivity and specificity. The understanding of the dynamic serological changes of COVID-19 patients and the seroepidemiological situation of the population will be helpful to further control the epidemic of COVID-19.
1. Introduction
Coronaviruses (CoVs) are enveloped single-stranded positive-sense RNA viruses. They often cause respiratory, digestive, and nervous system diseases in humans and other mammals [1]. In the past 20 years, coronaviruses have caused two global epidemics of severe respiratory infections: the severe acute respiratory syndrome (SARS) [2], [3] in 2002–2003 and the Middle East Respiratory Syndrome (MERS) in 2012 [4].
Pneumonia caused by the 2019 novel coronavirus (2019-nCoV) has spread rapidly worldwide. More than 215 countries and regions have reported cases. As of 24:00 on July 7th, 2020, 11.91 million confirmed the Corona Virus Disease 2019 (COVID-19) cases and 540,000 deaths have been reported. Since the outbreak, the China National Health Commission has published the “Diagnosis and Treatment plan of Corona Virus Disease 2019,” with several revisions according to the actual status of the epidemic [5], [6], [7], [8], [9], [10]. In addition to the epidemiological history, clinical signs, and imaging characteristics of viral pneumonia, an important diagnostic criterion for COVID-19 is a positive 2019-nCoV nucleic acid test using nasal and pharyngeal swabs [5], [6], [7], [8], [9]. However, the sensitivity of nucleic acid detection is not ideal. Due to sampling and other determinants, only 30%–50% of the confirmed COVID-19 cases had positive results on the first nucleic acid test after morbidity. In clinical practice, the highly suspicious cases with a first negative test usually were subjected to multiple nucleic acid analyses until a definitive diagnosis was found or evidence of exclusion was found. Detection of IgM and IgG against COVID-19, a fast and convenient method, has been confirmed as the basis for diagnosis of suspected patients with COVID-19, that is the serum specific IgM antibody and IgG antibody positive or the serum specific IgG antibody changes from negative to positive or the recovery period is 4 times or more higher than the acute period.
After viral infection, the host immune system is activated to defend against the virus, with specific antibody production. For the laboratory diagnosis of infectious diseases, the detection of virus-specific antibodies is a sensitive method. However, the antibody production against the 2019-COVID and changes during the COVID-19 progression have not been characterized.
In this study, we determined specific antibody dynamics in COVID-19 patients and seroepidemiology in other populations by using an automated chemiluminescent immunoassay to evaluate antibody production during disease progression and the value of antibody detection for the laboratory diagnosis of COVID-19.
2. Materials and methods
2.1. Study design and participants
A total of 742 subjects were included in the study. According to the “Diagnosis and Treatment plan of Corona Virus Disease 2019” [8], 9 confirmed COVID-19 cases from two government-designated COVID-19 treatment hospitals in Liaoning province were assigned to the COVID-19 group. 225 patients with suspected COVID-19 admitted to the fever clinic were observed in quarantine and excluded of COVID-19 after two negative results by 2019-nCoV nucleic acid testing were assigned to the non-COVID-19 group. Another 222 outpatients with other diseases during the same period, 63 medical staff at the fever clinic, and 223 healthy physical examinees in 2018 were assigned to the other disease group, medical staff group, and health control group, respectively.
2.2. Clinical data collection
According to a unified form, two residents collected clinical data from medical records separately.
2.3. Blood sampling
Venous blood (5 mL) under fasting conditions was collected from all subjects and placed in a yellow-top vacuum tube containing separation gel. After centrifugation, the serum samples were stored at-20 °C.
2.4. 2019-nCoV nucleic acid detection
Nasopharynx/oropharynx swab samples were collected by trained medical staff and tested by a qualified laboratory. Fluorescence reverse transcriptase polymerase chain reaction (RT-PCR) was used to detect the expression of open reading frame 1ab (ORF1ab) and the nucleocapsid protein (NCP) in the 2019-nCoV genome. The CT values for the 2019-nCoV nucleic acid test were interpreted according to the manufacturer's instructions, and suspicious results were recommended for clinical re-sampling and re-examination. For a positive laboratory test result, it is necessary for the 2019-nCoV ORF1ab and the N genes of the same sample to show at least one positive target-specific RT-PCR test result.
2.5. Diagnostic criteria for confirmed, severe, and critical cases of COVID-19
Diagnostic criteria have been described in the “Diagnosis and Treatment plan of Corona Virus Disease 2019 (Tentative fifth revised edition)” [8] published by the China National Health Commission.
2.6. 2019-nCoV IgM and IgG antibody detection
The Chemiluminescence Detection Kit (Shenzhen Yahuilong Biotechnology Co. Ltd., Shenzhen, China) was used to detect the 2019-nCoV IgM and IgG antibodies. Magnetic particle-coated antigens were used, including the mixed recombinant 2019-nCoV full length Spike protein S1 and the full length nucleocapsid protein N. All operations were performed after strict calibration and quality control in accordance with the manufacturer’s instructions. The results have been reported in 30 min after sample loading as relative luminescence intensity (RLU) values. Because the detection antibody was labeled with a luminescent substance, there was a positive correlation between the amount of detected 2019-nCoVIgM or IgG antibody in a sample and RLU, and the concentration of the 2019-nCoV IgM or IgG antibody (AU/mL) was automatically calculated according to RLU and built-in calibration curve. A concentration of 10.0 AU/mL was regarded as reactive (positive).
2.7. Statistical analysis
SPSS version 20.0 (SPSS Inc., Chicago, IL, USA) and GraphPad Prism version 5.01 (GraphPad Software Inc., San Diego, CA, USA) were used for statistical analyses. Quantitative variables are expressed as medians (P99). The normality of variables was tested using the Kolmogorov–Smirnov test. LDS t-tests were used for comparisons among groups. A receiver operating characteristic curve (ROC) was plotted by the SPSS Software to evaluate diagnostic performance. All tests were two-sided and P < 0.05 was considered statistically significant.
3. Results
3.1 Of the nine confirmed cases, four were male and five were female. The age of subjects ranged from 19 years to 57 years. Four patients had diabetes, hypertension, chronic kidney disease and chronic liver disease. Three were mild cases, four were severe cases and two were asymptomatic infection cases. One case had a history of contact with individuals in Wuhan, two cases had no clear epidemiological history and the other six cases were imported from abroad (Table 1 ).
Table 1.
Baseline characteristics of 9 patients with COVID-19.
| Case1 | Case 2 | Case 3 | Case 4 | Case 5 | Case 6 | Case 7 | Case 8 | Case 9 | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Age (years) | 56 | 39 | 57 | 50 | 51 | 47 | 22 | 19 | 46 | |
| Gender | female | male | male | female | male | female | female | male | female | |
| History of epidemiology | N | Y | N | Y | Y | Y | Y | Y | Y | |
| Comorbidities | ||||||||||
| hypertention | Y | N | N | N | N | N | N | N | N | |
| cardiovascular disease | N | N | N | N | N | N | N | N | N | |
| diabetes | N | N | Y | N | N | N | N | N | N | |
| chronice kidney disease | N | Y | N | N | N | N | N | N | N | |
| chronic liver disease | N | Y | N | Y | N | N | N | N | N | |
| Signs and symptoms | ||||||||||
| fever | Y | Y | Y | Y | Y | Y | N | N | Y | |
| dry cough | Y | N | N | Y | Y | Y | N | N | N | |
| dyspnea | Y | N | Y | Y | N | N | N | N | N | |
| pharyngalgia | N | N | N | Y | N | Y | N | N | Y | |
| Heart rate (/min) | 96 | 106 | 106 | 90 | 75 | 89 | 96 | 88 | 89 | |
| Respiratory rate (/min) | 16 | 22 | 30 | 20 | 20 | 18 | 18 | 20 | 18 | |
| Blood pressure mmHg | 157/99 | 134/84 | 134/82 | 132/70 | 115/75 | 135/91 | 90/73 | 120/70 | 142/98 | |
| Clinical type | severe | common | severe | severe | severe | common | asymptomatic | asymptomatic | common | |
3.2 The main laboratory findings for patients with COVID-19 were normal or exhibited slightly lower levels of the white blood cells and lymphocytes, elevated levels of the inflammatory indicators (e.g., interleukin-6, procalcitonin, C-reactive protein, serum amyloid A, and erythrocyte sedimentation rate), and normal myocardial marker levels (Table 2 ).
Table 2.
Laboratory findings of 9 patients with COVID-19.
| Normal range | Case 1 | Case 2 | Case 3 | Case 4 | Case 5 | Case 6 | Case 7 | Case 8 | Case 9 | |
|---|---|---|---|---|---|---|---|---|---|---|
| White blood cell count, ×109/L | 3.5–9.5 | 5.0 | 6.3 | 4.0 | 2.8* | 4.5 | 3.9 | 2.9* | 4.2 | 4.2 |
| Neutrophil count, ×109/L | 1.9–7.2 | 3.7 | 4.1 | 3.4 | 1.3* | 2.8 | 2.8 | 2.3 | 2.7 | 2.8 |
| Lymphocyte count, ×109/L | 1.1–2.7 | 0.9* | 1.7 | 0.4* | 1.0* | 0.4* | 0.7* | 0.6* | 1.1 | 0.9* |
| PaO2/FiO2, mmHg | greater than300 | 257* | 344 | 110* | 216* | 238* | greater than300 | — | — | greater than300 |
| Aspartate aminotransferase, U/L | 5–34 | 30 | 45* | 53 | 20 | 18 | — | 25 | 19 | 21 |
| Alanine aminotransferase, U/L | 0–40 | 22 | 28 | 47* | 38 | 21 | — | 39 | 19 | 22 |
| Creatine kinase, U/L | 29–200 | 60.5 | 354.7* | 183.4 | 36 | 89 | — | 61 | 68 | 47 |
| Creatine kinase MB isoenzyme, U/L | 0–24 | 16.2 | 16.8 | 36.5* | 9 | 9 | — | 1 | 7 | 10 |
| Myoglobin, μg/L | 0–105.7 | 25.5 | 46.9 | 47.8 | — | — | — | — | — | — |
| Troponin I, μg/L | 0–0.04 | 0.00 | 0.01 | 0.01 | — | — | — | — | — | — |
| Interleukin 6, pg/mL | 0–7 | 111.20* | 55.32* | 14.77* | 4.58 | 3.88 | — | 2.12 | 30.1* | 10.3* |
| Procalcitonin, ng/mL | <0.05 | 0.20* | 0.12* | 0.17* | — | 0.33* | — | 0.99* | — | — |
| Serum amyloid A, mg/L | <6.8 | 149* | 67* | 217* | — | — | — | — | — | — |
| C-reactive protein, mg/L | 0.0–8.0 | 29.3* | 27.6* | 129.7* | 31* | 122* | 9.56* | 29* | 0.52 | 32* |
| D-dimer, μg/L (DDU) | 0–252 | 149 | 213 | 5561* | — | — | — | — | — | — |
| Erythrocyte sedimentation rate, mm/h | 0–15 | 39* | 58* | 73* | — | — | — | — | — | — |
| Creatinine, μmol/L | 59–104 | 79.1 | 106.2* | 63.0 | 50* | 83 | — | 39* | 62 | 40* |
* out of the upper or lower limits.
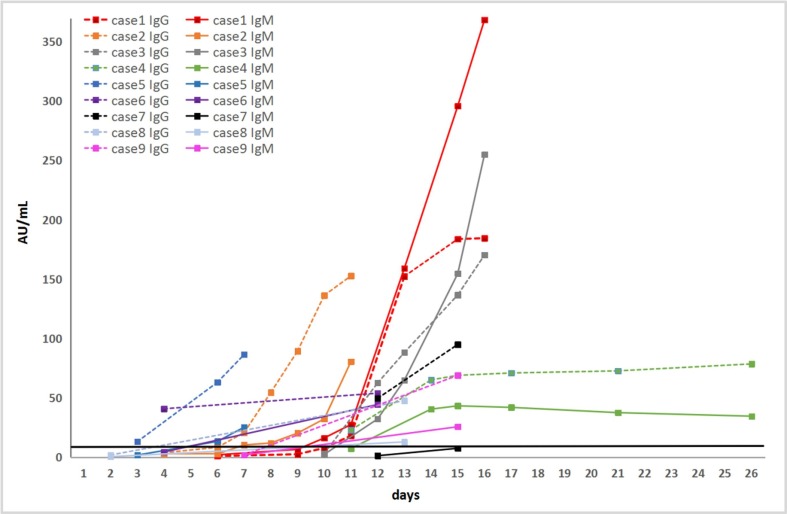
3.3 Anti-2019-nCoV antibody production after the onset of symptoms has been summarized in Fig. 1 and Table 3 . Nine patients with COVID-19 showed reactivity (positive) for specific anti-2019-nCoV antibodies at 3–15 days after onset, and the anti-2019-nCoV IgM and IgG antibody levels increased with the progression of the disease. In one of nine cases, anti-2019-nCoV IgG was detected 1 day after IgM detection, in three of nine cases, anti-2019-nCoV IgM was detected 3–8 days after IgG detection, and in the other four cases, anti-2019-nCoV IgM and IgG production were detected almost on the same day, in the remaining case (case7), the IgG was first detected positive on the 12th day, and then the IgM level showed a tendency to increase, but still did not exceed the positive cutoff value on the 15th day. However, the changes in anti-2019-nCoV IgM and IgG values differed among cases. In seven of nine cases, the values of anti-2019-nCoV IgG was consistently higher than that of IgM. In the other two cases, the values of anti-2019-nCoV IgM became higher than that of IgG only after 2 weeks of onset.
Fig. 1.
Dynamics of antibody production in 9 patients. The black line parallel to the x-axis represents the positive threshold value, 10.0 AU/mL; The x-axis represents the days since the disease morbidity.
Table 3.
Anti-2019-nCoV antibody production.
| Case 1 | Case 2 | Case 3 | Case 4 | Case 5 | Case 6 | Case 7 | Case 8 | Case 9 | |
|---|---|---|---|---|---|---|---|---|---|
| Durations from illness onset to first admission (days) | 7 | 4 | 10 | 1 | 1 | 2 | asymptomatic | asymptomatic | 1 |
| Durations from illness onset to nucleic acid testing (days) | 8 | 5 | 11 | Negative | 1 | 2 | asymptomatic | asymptomatic | 2 |
| Durations from illness onset to anti-2019-nCoV IgM reactive (days) | 10 | 7 | 12 | 14 | 6 | 12 | Negative during observation period | 13 (from positive nucleic acid testing) | 15 |
| Durations from illness onset to anti-2019-nCoV IgG reactive (days) | 11 | 7 | 12 | 11 | 3 | 4 | 12 (from positive nucleic acid testing) | 13 (from positive nucleic acid testing) | 15 |
3.4 In non-COVID-19, other disease, medical staff, and health control groups, a few cases were reactive for 2019-nCoV IgM and IgG, all of which were single reactive for IgM or IgG. For this particular cohort, the sensitivities of IgM and IgG were 88.89% and 100%, and the specificities were both over 97% (Table 4 ).
Table 4.
Anti-2019-nCoV antibody detection in different groups.
| Non-COVID-19 | Other disease | Medical staff | Health control | |
|---|---|---|---|---|
| Number | 225 | 222 | 63 | 223 |
| Age years, median (range) | 35(1–86) | 50(27–85) | 40(25–61) | 59(21–95) |
| Male/female | 124/101 | 62/160 | 7/56 | 77/146 |
| 2019-nCoV IgM reactive | 6 | 2 | 0 | 3 |
| 2019-nCoV IgG reactive | 1 | 2 | 0 | 4 |
| 2019-nCoV IgM median/P99 (AU/mL) | 1.82/19.66* | 0.85/10.99 | 1.37/4.56* | 0.86/11.35 |
| 2019-nCoV IgG median/P99 (AU/mL) | 1.82/8.52 | 1.21/10.52* | 1.27/6.26 | 1.49/11.18 |
| 2019-nCoV RNA | 0 | N/A | N/A | N/A |
| influenza A RNA | 2 | N/A | N/A | N/A |
| influenza B RNA | 2 | N/A | N/A | N/A |
| adenovirus DNA | 4 | N/A | N/A | N/A |
| mycoplasma pneumoniae DNA | 17 | N/A | N/A | N/A |
| Sensitivity (IgM) | 88.89% | 88.89% | 88.89% | 88.89% |
| Sensitivity (IgG) | 100% | 100% | 100% | 100% |
| Specifictity (IgM) | 97.33% | 99.10% | 100.00% | 98.65% |
| Specifictity (IgG) | 99.56% | 99.10% | 100.00% | 98.21% |
| negative predictive values (IgM) | 99.55% | 99.55% | 98.44% | 99.55% |
| positive predictive values (IgM) | 57.14% | 80.00% | 100.00% | 72.73% |
| negative predictive values (IgG) | 100.00% | 100.00% | 100.00% | 100.00% |
| positive predictive values (IgG) | 90.00% | 81.82% | 100.00% | 69.23% |
*compared with health control, P < 0.05.
3.5 Of 225 non-COVID-19 cases, 2 tested positive for the influenza A RNA, 2 tested positive for the influenza B RNA, 4 were positive for the adenovirus DNA, and 17 were positive for the Mycoplasma pneumoniae DNA (Table 4).
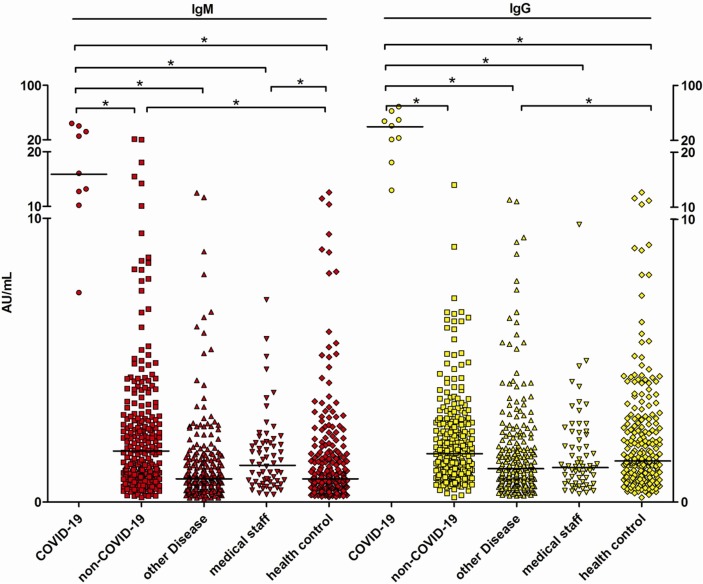
3.6 We also compared the distributions of anti-2019-nCoV antibody levels among groups. The anti-2019-nCoV IgM levels were significantly higher (P < 0.05) in the non-COVID-19 group than those in the healthy control group (Table 4, Fig. 2 ).
Fig. 2.
The anti-2019-nCoV IgM and IgG antibodies distribution in different groups. Each data point represents the antibody level of the participants, the short horizontal line represents the median antibody level of the group, and * represents the difference between the two groups is statistically significant, P < 0.05.
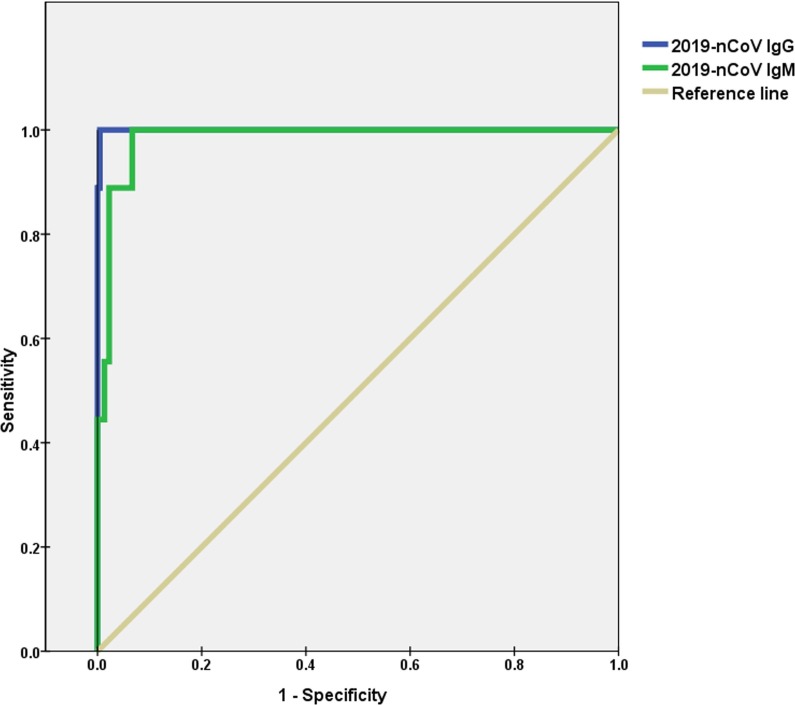
3.7 To further clarify the diagnostic efficacy of specific IgM and IgG antibodies in patients with fever with a suspected COVID-19 (all 234 patients had been proved by testing 2019-nCoV nucleic acid), we generated an ROC curve of anti-2019-nCoV IgM and IgG levels. The areas under the curve were 0.984 and 1.000, and the optimal cut-off values for IgM and IgG were 7.09 AU/mL (when sensitivity is 1.000 and specificity is 0.933) and 10.97 AU/mL (when sensitivity is 1.000 and specificity is 0.996), respectively (Fig. 3 ).
Fig. 3.
ROC curves of anti-2019-nCoV IgM and IgG in the diagnosis of COVID-19.
4. Discussion
With the growing surveillance network and laboratory capacity, the viral genome sequence was announced quickly [11], enabling the development of in vitro diagnostic tests. At present, the main diagnostic method is 2019-nCoV nucleic acid detection by a real-time quantitative fluorescent PCR.
However, as the number of COVID-19 cases increased, physicians have found that the confirmed cases had relatively lower positive rate for 2019-nCoV nucleic acid detection, especially in pharyngeal swab. Missed detection can be explained by the timing of the oropharyngeal or nasopharyngeal specimen collection, improper collection sites, and the infeasibility of standardized clinical nucleic acid testing in some laboratories. Furthermore, the viral load changes during various COVID-19 stages. Therefore, a fast and convenient detection method to distinguish and trace suspicious cases or contacts as early as possible is critical for the prevention of super-transmission events.
Antibodies are the products of the humoral immune response after infection with viruses. While the detection of nucleic acid cannot be used widely and a relatively high false negative rate, specific antibodies to 2019-nCoV can be used to determine whether a suspected patient has been recently infected with 2019-nCoV or not. In case 4, the clinical signs and symptoms was obvious after morbidity and the patient had a definite epidemiologic history, but the nucleic acid tests were negative for 7 times after morbidity. But the positive anti-2019-nCoV IgG and IgM antibody appeared successively and kept increasing from the 11th day of morbidity helped to confirm the diagnosis. The nucleic acid test was keep negative until the patient was discharged from hospital after recovery. The immune response of pathogenic microorganisms is usually stimulated by the increase in IgM after an infection. IgG usually appears 1 week-2 weeks after IgM and increases to high levels, which are maintained in the body for a long time. Because COVID-19 is a new infectious disease and immunological test reagents have just recently been developed, little is known about IgM and IgG antibody production after 2019-nCoV infection.
We detected the dynamics of specific antibodies to 2019-nCoV after the onset of symptoms in all nine confirmed patients. Different from the general rule, in 8 of the nine COVID-19 cases, anti-2019-nCoV IgG antibodies appeared concomitantly with or even earlier than the 2019-nCoV IgM, this phenomenon may be related to the decrease in the number of lymphocytes caused by 2019-nCoV infection and low affinity of the pentameric IgM, but the overall trend of the humoral immune response to 2019-nCoV infection has not yet been fully determined and still need further study on it. The rates of increase in anti-2019-nCoV IgG and IgM antibodies varied among individuals.
2019-nCoV is highly infectious in the general population. Severe cases are prone to rapid progression to an acute respiratory distress syndrome, septic shock, high risk of admission to intensive care units, and even death [12]. Therefore, the development of methods for the close monitoring of patients and the early identification of severe cases is the key to reduce mortality. According to our findings, the time and speed of specific anti-2019-nCoV IgM antibody production were correlated with the disease severity. However, owing to the small number of cases, more research will be needed for confirmation.
In addition to patients with COVID-19, those with fever and non-COVID-19, other diseases, medical staff, and healthy controls were also evaluated. The non-COVID-19 group included patients with several other respiratory viruses, such as influenza A, influenza B, and adenovirus infection. These cases were negative for anti-2019-nCoV-specific antibodies, indicating the high specificity and effectiveness for the differential diagnosis of viral respiratory infections. Among each group, only patients with COVID-19 were positive for both anti-2019-nCoV IgM and IgG antibodies; in other populations, either IgG or IgM antibodies (but not both) were positive in a few cases. However, combined with the 2019-nCoV nucleic acid detection results and clinical data, these were identified as false positive results. COVID-19 has spread to many countries around the world, the main problem at present is the need for a highly sensitive tests to screen suspected cases and prevent false negatives by nucleic acid tests; the low false-positive rates for antibody testing are acceptable. In the meantime, for patients with symptoms for a week or more, simultaneous positive anti-2019-nCoV IgM and IgG results can improve the assay specificity.
Compared with RNA test, the operation requirement of serum antibody detection in clinical laboratory is lower, fast (30 min) and high throughput. The disadvantage of nucleic acid detection is the existence of relative high false negative rate, and serological antibody detection has the advantage of high sensitivity, so the combination of the two will be a good diagnostic means. It can be inferred that after the future epidemic situation has been controlled to a certain extent, as a convenient method, antibody detection is still necessary to make differential diagnosis of other respiratory pathogens infection.
It must be emphasized that independent results of specific antibodies testing should not be used as a diagnostic criteria, especially when the epidemiological history is unclear, and must be combined with the patient's morbidity time and clinical signs.
Little is known about specific antibody production during the course of COVID-19 infection or about antibody production in patients with fever and non-COVID-19, other diseases, special contact (e.g., medical staff), and the healthy population. This study provides a detailed analysis of antibody production in the course of COVID-19 infection as well as basic data for specific antibodies in different populations. Our results provide a basis for the rapid screening of suspected cases by serological testing to curb the rapid progression of the epidemic globally. On the day this manuscript was submitted, the China National Health Commission published the new edition of the “Diagnosis and Treatment plan of Corona Virus Disease 2019” [10], which suggested that positive anti-2019-nCoV IgM and IgG results could be used as one of standard for diagnosis, further supporting our findings.
This study had some limitations. First, only nine confirmed COVID-19 cases were included. Although the dynamic process of anti-2019-nCoV antibody production and its relationship with disease progression have been carefully observed, a large sample size will be needed for verification. Second, changes in anti-2019-nCoV antibody levels were only tracked for 2 days to 26 days after morbidity. However, increasing trends in anti-2019-nCoV IgG and IgM antibody production were generally observed. Third, 2019-nCoV nucleic acid testing was not been performed in every group, and asymptomatic infections may be missed in other disease groups, impacting the evaluation of the diagnostic efficacy of antibodies. Considering that the Liaoning Province, where this study was conducted, is a low-level epidemic area, the potential for asymptomatic infection is expected to be low.
5. Conclusion
COVID-19 now is an urgent global medical issue that caused widely concerned. We analyzed Chinese population to study the trend of specific antibody and seroepidemiology in different population. In this paper, we first characterized specific antibody production in patients with COVID-19 and evaluated the diagnostic value of antibodies in various populations, providing key information for doctors in the process of diagnosis and treatment. As a useful complement to nucleic acid detection, the detection of specific anti-2019-nCoV antibodies will enable a more comprehensive, rapid, and accurate diagnostic approach to effectively distinguish between COVID and non-COVID-19 and curb the rapid spread of 2019-nCoV globally.
6. Ethics approval and consent to participate
The study was conducted in accordance with the International Coordinating Council for Clinical Trials and the Helsinki Declaration and was approved by the Hospital Ethics Review Committee (Ethics No 2020PS038K). Informed consent from patients was exempted.
7. Funding statement
This work was supported by Scientific Research Projects Related to Prevention and Control of Coronavirus Disease 2019 (COVID-19) of China Medical University; the 345 talent project of Shengjing Hospital of China Medical University; the National Science and Technology Major Project of China [2018ZX10302205]; the Guangdong Province Major key projects of industrial technology [201902010003].
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
We thank Prof. Qijun Wu (Department of Epidemiology, Shengjing Hospital of China Medical University) for his guidance in statistical analysis.
References
- 1.Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ksiazek T.G., Erdman D., Goldsmith C.S. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 3.Rota P.A., Oberste M.S., Monroe S.S. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 4.Zaki A.M., van Boheemen S., Bestebroer T.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 5.China National Health Commission. Novel coronavirus pneumonia Diagnosis and Treatment Program (Trial Third Edition). http://www.nhc.gov.cn/yzygj/s7653p/202001/f492c9153ea9437bb587ce2ffcbee1fa.shtml.
- 6.China National Health Commission. Novel coronavirus pneumonia Diagnosis and Treatment Program (Trial Forth Edition). http://www.nhc.gov.cn/yzygj/s7653p/202001/4294563ed35b43209b31739bd0785e67.shtml.
- 7.China National Health Commission. Novel coronavirus pneumonia Diagnosis and Treatment Program (Trial Fifth Edition). http://www.nhc.gov.cn/yzygj/s7653p/202002/3b09b894ac9b4204a79db5b8912d4440.shtml.
- 8.China National Health Commission. Novel coronavirus pneumonia Diagnosis and Treatment Program (Revised version of the trial fifth edition). http://www.nhc.gov.cn/yzygj/s7653p/202002/d4b895337e19445f8d728fcaf1e3e13a.shtml.
- 9.China National Health Commission. Novel coronavirus pneumonia Diagnosis and Treatment Program (Trial Sixth Edition). http://www.nhc.gov.cn/yzygj/s7653p/202002/8334a8326dd94d329df351d7da8aefc2.shtml.
- 10.China National Health Commission. Novel coronavirus pneumonia Diagnosis and Treatment Program (Trial Seventh Edition). http://www.nhc.gov.cn/yzygj/s7653p/202003/46c9294a7dfe4cef80dc7f5912eb1989.shtml.
- 11.Ning D, Yang X, Ye L, et al. Genomic and protein structure modelling analysis depicts the origin and infectivity of 2019-nCoV, a new coronavirus which caused a pneumonia outbreak in Wuhan, China, bioRxiv https://doi.org/10.1101/2020.01.20.913368.
- 12.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China, Lancet. 395 (2020) 497–506. [DOI] [PMC free article] [PubMed]