Abstract
The remobilization and resorption of plant nutrients is considered as a crucial aspect of the seasonal senescence of plant organs. In leaves, the mechanisms responsible for the relocation of valuable compounds are well understood while the related processes in roots are still being debated. Some research indicates that remobilization in roots occurs, while other studies have not found evidence of this process. Considering that the total biomass of fine roots is equal to or greater than that of leaves, clarifying the conflicting reports and ambiguities may provide critical information on the circulation of chemical elements in forest ecosystems. This study provides new information concerning the basis for remobilization processes in roots by combining physiological data with gene expression and protein levels. We suggest that, as in leaves, molecular mechanisms involved in nitrogen (N) resorption are also activated in senescent roots. An analysis of N concentration indicated that N levels decreased during the senescence of both organs. The decrease was associated with an increase in the expression of a glutamine synthetase (GS) gene and a concomitant elevation in the amount of GS—one of the most important enzymes in N metabolism. In addition, significant accumulation of carbohydrates was observed in fine roots, which may represent an adaptation to unfavorable weather conditions that would allow remobilization to occur rather than a rapid death in response to ground frost or cold. Our results provide new insights into the senescence of plant organs and clarify contentious topics related to the remobilization process in fine roots
Keywords: carbohydrates, fine roots, glutamine synthetase, nitrogen, Populus trichocarpa, remobilization, seasonal variation
Introduction
Senescence is the last stage of plant ontogenetic development and can have a major influence on a wide spectrum of ecological processes, ranging from litter formation to nutrient cycling. The senescence process is largely mediated by programmed cell death (PCD), regardless of whether the organs in question are leaves, roots or petals (van Doorn and Woltering 2004, Sobieszczuk-Nowicka et al. 2018, Wojciechowska et al. 2018a). The PCD, however, is thought to represent an adaptation to the intermediate state between living and dead cells (Thomas et al. 2003). The irreversible senescence that occurs in leaves is associated with dramatic changes in gene expression, the degradation of macromolecules and a decrease in protein synthesis (Liu et al. 2008, Avila-Ospina et al. 2014). The global changes in gene expression vary and are represented by both increases and decreases in transcript levels. Mechanisms that regulate the network responsible for proper management of the degradation and remobilization of cellular material during leaf senescence have been identified. Thus, plant constituents are preserved rather than lost. In order to maximize resource efficiency across many growing seasons, elements (especially nitrogen [N]) are translocated from the senescing tissues to developing seeds, newly formed leaves or storage organs (Buchanan-Wollaston and Ainsworth 1997, Avila-Ospina et al. 2014, 2015).
In plants, the senescence process is coordinated at both a structural and physiological level that can explicitly link plant traits to changes in vitality. For example, morphologically, yellowing is commonly associated with leaf senescence. It is directly related to chlorophyll levels and the conversion of chloroplasts into gerontoplasts (Avice and Etienne 2014, Sobieszczuk-Nowicka et al. 2018). Considering that more than 70% of all leaf proteins are present in chloroplasts, degradation of those structures is then associated with the release of a large pool of N (Liu et al. 2008). Hence, changes in chloroplast structure are highly correlated with the initial stages of senescence, when mechanisms associated with remobilization are activated. Increasing evidence suggests that this activation includes the release of free amino acids during the process of protein degradation and conversion into ammonia. Due to the toxicity of that compound, it is rapidly incorporated into glutamate by the amination of 2-oxoglutarane. This reaction is catalyzed by glutamate dehydrogenase (GDH) (Liu et al. 2008). Subsequently, glutamate is transformed into glutamine, which has the potential to be mobilized and relocated through the phloem sap. Glutamine synthetase (GS), one of the most important enzymes in N metabolism, is involved in this reaction (Liu et al. 2008, Avila-Ospina et al. 2015). Based on cellular localization, there are two main isoforms of GS: cytosolic (GS1) and chloroplastic (GS2) (Zhang et al. 2017). GS1 plays a fundamental role in glutamine synthesis, which enables the relocalization of N from senescent tissues to locations where it can be preserved or utilized (Liu et al. 2008, Castro-Rodríguez et al. 2011, Zhang et al. 2017). In contrast, GS2 is required for the reassimilation of ammonia generated during photorespiration (Miflin and Habash 2002, Liu et al. 2008, Castro-Rodríguez et al. 2011, Zhang et al. 2017). Although a well-defined set of steps is involved in N remobilization from leaves, the pattern, magnitude and factors involved in N remobilization from roots during senescence are weakly defined and supported by research.
Among all compounds that are relocated and recycled during senescence, N and carbon (C) (due to the high concentration in plant tissues) are critically important. There are reports indicating that starch is degraded and transformed into sucrose, the main form in which C is transported in plants (Cerasoli et al. 2004). Despite the importance of carbohydrate redistribution for nutrient conservation, the molecular mechanism of C translocation from absorptive roots undergoing senescence has received little attention.
In contrast to the considerable progress that has been made in elucidating the senescence process in leaves and flower petals (Agüera et al. 2010, Shibuya et al. 2011, 2013, 2014, Shibuya 2012, Avila-Ospina et al. 2015, Springer et al. 2015, Sobieszczuk-Nowicka et al. 2018), much less attention has been given to the belowground component of plant biomass. As the annual biomass production of fine roots is equal to or even greater than the biomass of leaves, the senescence and death of fine roots is important from the standpoint of the cycling of chemical elements (Gill and Jackson 2008, Brassard et al. 2009). Though traditionally defined as roots <2 mm in diameter, it is increasingly recognized that fine roots are not a homogeneous entity as they include both absorptive roots and transport roots (Bagniewska-Zadworna et al. 2012, 2014, McCormack et al. 2015, Zadworny et al. 2015, Wojciechowska et al. 2018b). The absorptive roots belong to the first two orders of roots, which are characterized by the highest absorptive capacity, high N concentration and respiration rate, and often mycorrhizal colonization (Eissenstat et al. 2000, Pregitzer et al. 2002, McCormack et al. 2015). Thus, the senescence process of the short-lived absorptive roots may provide important information for nutrient recycling, as well as understanding of plant adaptation to autumn and winter seasons. There is a premise that the death of absorptive roots may be a passive process where the provision of sugars and defense compounds is stopped, making the roots an easy target for pathogens (Yanai and Eissenstat 1997, Eissenstat and Volder 2005). Our recent studies, however, have indicated that the senescence and death process in these roots is active and genetically regulated, and represents another example of PCD in plants (Bagniewska-Zadworna et al. 2014, Wojciechowska et al. 2018b). The fact that absorptive roots undergo a genetically regulated death process emphasizes the premise that their nutrients, which are either limited in the environment or difficult to absorb, would be targeted for relocation so that they can be recycled. No details on the mechanisms associated with relocation and remobilization of valuable compounds from senescing absorptive roots are available, and the information that does exist is variable or conflicting (Kunkle et al. 2009, Zadworny et al. 2015). Identification of the mechanisms responsible for the senescence of absorptive roots represents the first step to understanding how roots die and how nutrient resorption from the entire root system can be incorporated into the measurement of nutrient turnover at the whole-plant level.
Therefore, an experiment was designed to analyze the senescence process in leaves and absorptive fine roots, with particular emphasis placed on the regulation of nutrient remobilization. We specifically expected that N and C concentrations would decline with senescence and the same dominant genetic cues (i.e. GS) would be closely associated with changes of N in both leaf and root.
Materials and methods
Plant material and growth condition
All experiments were performed on Populus trichocarpa (Torr. & A. Gray ex Hook.). Seeds, provided by the FLORPAK Młynki Seed Store, Poland, were placed on 1% agar. After germination, the seedlings (~1–2 cm in length) were planted in soil in a seed-starting system and grown for 2 months in a plant growth chamber (Conviron GR96) at 18 °C day/14 °C night temperature and a 16 h day/8 h night photoperiod. The 2-month-old plants were then removed from the seed-starting system, along with a clod of dirt in order to prevent injuring the root system, and re-planted in rhizotrons consisting of underground boxes (50 × 30 cm) made from two transparent polycarbonate plates held 3 cm apart by thick-walled plastic tubing to provide sufficient room for root growth. The rhizotrons were filled with soil obtained from a forest where Populus species naturally grow. The plants were watered with an automated system. The bottom of each rhizotron contained a drainage hole to avoid hypoxic, flooding conditions and to ensure that the soil was aerated. Rhizotrons were placed in containers, in a semi-open, greenhouse located at the Institute of Dendrology, Polish Academy of Sciences, in Kórnik, Poland (52°14′40″N and 17°06′27″E).
Leaf and root samples for each biological replicate were harvested from at least three individual plants. During sampling, the rhizotron windows were pulled out, opened and then the harvested roots were divided into individual orders using a steel scalpel, taking into account that tip-ended roots are first order (Pregitzer et al. 2002). Leaf and root samples were collected three times during the growing season based on morphological and anatomical indications of senescence as described by Wojciechowska et al. (2018b). The senescence stages of leaves and roots selected for analysis are presented in Table 1.
Table 1.
Senescence stages of roots and leaves.
| Variant | Abbreviation | Characteristic features |
|---|---|---|
| Control leaves | LC | Green leaves without senescence symptoms |
| First stage of leaf senescence | LS1 | Yellowing leaves in which chlorophyll level had decreased by ~40% |
| Second stage of leaf senescence | LS2 | Yellow leaves in which chlorophyll level had decreased by ~60% |
| Control roots | RC | White roots without senescence symptoms |
| First stage of root senescence | RS1 | Roots which had changed in color from white to brown |
| Second stage of root senescence | RS2 | Roots which had changed in color from brown to dark brown or almost black. Shrinkage was also visible in most fine roots |
Quantitative determination of N, C and carbohydrates
The analysis of N, C and carbohydrates was conducted during the course of the growing season on leaves and the first three root orders. Fine roots were divided into three groups based on their order (first, second—absorptive roots, without secondary growth, third—transport roots, longer and mostly with secondary structure) immediately after harvesting. Root order was assigned according to the morphometric approach where distal roots represent first-order roots (Pregitzer et al. 2002). This grouping was used to assess the relocation of the studied elements from lower to higher order roots. The material (Table 1) was collected at the same time of day (morning) to avoid any daily fluctuations in the level of the studied elements/compounds. The samples were dried at 65 °C for 3 days and ground to a powder in a Retsch MM 200 mill (Retsch, Haan, Germany). Quantitative determination of N and C concentrations was performed using an Elemental Combustion System CHNS-O 4010 (Costech Instruments, Pioltello/Valencia, Italy/USA).
Carbohydrate levels were measured as described by Oleksyn et al. (2000), and the level of soluble carbohydrates and starch was analyzed. Sugars were extracted from the ground material with a solution of methanol–chloroform–water. Starch concentration was measured by converting the starch to glucose with amyloglucosidase followed by oxidation with a peroxidase–glucose oxidase complex. A UV-1700 Pharma Spec (Shimadzu, Kyoto, Japan) spectrophotometer was used to determine the concentration of soluble carbohydrates (wavelength of λ = 625 nm) and starch (wavelength of λ = 450 nm). The concentration of the studied carbohydrates was determined using glucose standards and is presented as a percentage of dry mass.
Protein extraction, gel electrophoresis and immunoblots
Protein extraction was performed according to the method described by Szuba et al. (2013). Proteins were dissolved in a buffer containing 7 M urea, 2 M thiourea, 40 mM dithiothreitol, 0.5% carrier ampholytes and 4% CHAPS, and protein concentration was measured with a 2-D Quant kit (GE Healthcare, Piscataway, NJ, USA). Proteins were separated by SDS-PAGE on 4–20% Mini-PROTEAN TGX precast gels (Bio-Rad Laboratories, Inc., Grand Junction, CO, USA), with an equal amount of protein (20 μg) loaded in each lane. Protein transfer from the gel to a polyvinylidene fluoride (PVDF) membrane was conducted using a Trans-Blot® Turbo™ (Bio-Rad). Rabbit, anti-GS antibodies (AS08 295, Agrisera, Vännäs, Sweden) were used. The GS antibodies were able to bind to both GS1 and GS2 isoenzymes. Antibodies were diluted 1:10,000 in 2% skimmed milk powder. Incubation with primary antibodies was carried out overnight at 4 °C. Antibodies were washed from the PVDF membrane with phosphate-buffered saline (PBS) (Sigma-Aldrich St. Louis, MO, USA), followed by PBS with Tween-20 (PBST) (Sigma-Aldrich). Incubation with secondary antibodies was conducted using antibodies conjugated to horseradish peroxidase, goat, anti-rabbit (Agrisera) diluted 1:10,000 in 2% skimmed milk powder. After 1 h incubation with secondary antibodies, the PVDF membranes were washed in PBS and PBST and then incubated in Clarity western ECL substrate chemiluminescent detection reagent (Bio-Rad) for 5 min prior to image acquisition in a G-BOX CHEMI XR5 (Syngene, Cambridge, UK).
Reverse transcription-quantitative PCR analysis of gene expression
RNA isolation was performed using leaves and first-order roots. RNA isolation and cDNA synthesis were performed as described by Wojciechowska et al. (2018b). All of the reverse transcription-quantitative PCR (RT-qPCR) analyses of gene expression were conducted utilizing three technical replicates from each of independent three biological replicates of each variant. RT-qPCR analyses were conducted in 96-well plates in a CFX96 Touch Real-Time PCR Detection System (Bio-Rad) using the following amplification program: denaturation by a hot start at 95 °C for 10 min, followed by 40 cycles of a two-step program (denaturation at 95 °C for 15 s and annealing/extension at 60 °C for 1 min). The DNA sequences for GS1.1, GS1.2, GS1.3, GS2 and primer design were selected based on the sequences reported by Castro-Rodríguez et al. (2011). Primers for the amplification of references genes were designed in Primer3 software (The Whitehead Institute for Biomedical Research, Cambridge, MA, USA). The sequences of the primer pairs used in the RT-qPCR analyses are listed in Table 2. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH), β-tubulin and ubiquitin were selected as reference genes as they exhibited the most stable expression in all sample types and time points. The method utilized to determine the relative level of expression was described by Bagniewska-Zadworna and Stelmasik (2015).
Table 2.
Sequence of gene-specific primers used in the RT-qPCR analysis.
| Gene | Primer |
|---|---|
| GS1.1 | F-5′ATGGTTGTCTGTCAATTTGTTTGCC-3′ |
| R-5′CCAGCAAGAGTTTTATTTAGATTAG-3′ | |
| GS1.2 | F-5′GGAATTGAGTATTGGAAGATGATGG-3′ |
| R-5′TATGTTCATAAATGATCAACAGCC-3′ | |
| GS1.3 | F-5′TGGAAACCATAAGAGATCACCACC-3′ |
| R-5′GAAGAGGCAATTCTTGTACCAAG-3′ | |
| GS2 | F-5′GGAGCATCACTTGGATCTAGATGG -3′ |
| R-5′CAAAACCCAAGAGTAAAAAGGTCC-3′ | |
| β-Tubulin | F-5′TTCTCCTGAACATGGCAGTG-3′ |
| R-5′CCACACAACGTGAAATCCAG-3′ | |
| GAPDH | F-5′CAATGAATGGGGCTACAGGT-3′ |
| R-5′CATGAATCAGCTGCACATCC-3′ | |
| Ubiquitin | F-5′AGGAACGCGTTGAGGAGAAG-3′ |
| R-5′TATAABCAAAAACCGCCCCTG-3′ |
Microarray analysis
Total RNA was isolated in triplicate from each sample of first-order roots and from leaves using an RNeasy Plant Mini kit (Qiagen, Germantown, MD, USA). RNA quantity and quality were measured using a NanoDrop1000 (ThermoFisher Scientific, Carlsbad, CA, USA). cRNA synthesis and microarray hybridization to an Affymetrix GeneChip Poplar Genome Array (A-AFFY-131) were performed at the Laboratory of Microarray Analysis (Institute of Biochemistry and Biophysics Polish Academy of Science, Warsaw, Poland) according to the provided Affymetrix protocol. The normalized data were statistically analyzed using GeneSpringGX7 13.1 software (Agilent Technologies, Inc., Santa Cara, CA, USA). Statistical analysis was performed using a one-way ANOVA with a corrected P-value cut off ≤0.05 and a Benjamini Hochberg correction.
Statistical analysis
Statistical relationships were considered significant at P ≤ 0.05. Root and leaf biochemical traits were log10 transformed to meet the assumption of normality. However, figures show the non-transformed data. In the quantitative analyses of N, C and C:N, as well as in the analyses of GS gene expression, one-way ANOVA and Duncan’s post-hoc test were performed using Statistica 12.0 software (StatSoft Poland Inc., Tulsa, OH, USA).
Results
Quantitative assessment of N, C and the C:N ratio
Considering the ambiguous data on N relocation during root senescence, we determined the N and C concentration [%] to dispel doubts regarding the relocation of two main elements. Moreover, to check that remobilization is another universal feature of senescence, we also performed those and the rest of the analyses for leaves. Over the course of the growing season, there was a general decrease in N concentration in both leaves and the first three root orders (Figure 1A and B). In contrast to N, no significant seasonal or senescence effect on the level of C was observed in either leaves or roots (Figure 1C and D). However, due to the changes in N, there was a distinct increase in the C:N ratio in senescing leaves and the first three orders of fine roots (P ≤ 0.05; Figure 1E and F). Our results demonstrate that N was relocated during senescence in both studied organs.
Figure 1.

Quantitative analysis of N and C concentrations and the C:N ratio in absorptive roots (A, C, E) and leaves (B, D, F) of P. trichocarpa during the course of the growing season. (A, B) Quantitative analysis of N concentration [%]; (C, D) quantitative analysis of C concentration [%]; (E, F) quantitative analysis of the C:N ratio (RC: root control, RS1: first stage of root senescence, RS2: second stage of root senescence, LC: leaf control, LS1: first stage of leaf senescence and LS2: second stage of leaf senescence). Bars sharing the same letter are not significantly different (P ≤ 0.05). Values represent the mean ± standard error (SE).
Expression of genes involved in N remobilization
The effect of the senescence processes on the expression of genes involved in N metabolism was studied by analyzing the expression of three genes (GS1.1, GS1.2 and GS1.3) encoding a cytosolic form of GS. In addition, a gene encoding a chloroplastic isoform of the GS2 protein was also examined in leaves (Figure 2A and C). Results of the RT-qPCR analyses indicated that leaf senescence induced the expression of GS1.1 and GS1.3. A statistically significant increase in GS1.1 expression, relative to non-senescent leaves, was observed only during the second stage of senescence (LS2: yellow leaves), whereas GS1.3 was up-regulated in both the first (LS1: yellowing leaves) and second stage of senescence (Figure 2A). GS1.2 exhibited a different pattern of expression than GS1.1 and GS1.3 (Figure 2A). GS2 expression in leaves decreased over the course of the growing season and senescence (Figure 2A). A significant increase in GS1.1 expression was observed at the first stage of root senescence (RS1: brown roots), and the elevated expression was maintained at the second stage of this process (RS2: dark brown roots with shrinkage) (Figure 2B). In contrast, the expression of GS1.2 and GS1.3 exhibited the opposite trend to GS1.1, with the highest level of expression was observed in non-senescent, control roots (RC: white roots, without any visible symptoms of senescence) (Figure 2B).
Figure 2.

Analysis of the expression of GS. (A, B) Relative expression of GS in leaves (A) and roots (B) of Populus trichocarpa; (C, D) distribution of GS in leaves (C) and fine roots (D) (LC: leaf control, LS1: first stage of leaf senescence, LS2: second stage of leaf senescence, RC: root control, RS1: first stage of root senescence and RS2: second stage of root senescence). Bars sharing the same letter are not significantly different (P ≤ 0.05). Values represent the mean ± SE.
Immunoblot analysis of glutamine synthetase
We have checked also whether the level of GS is increased during senescence. Using immunoblots, we detected the changed content of this protein over the course of the growing season in both leaves and roots (Figure 2C and D). Two forms of GS were detected in leaves, cytosolic (GS1, 40 kDa) and chloroplastic (GS2, 45 kDa). Both isoforms were observed in green leaves, with the level of GS2 being slightly greater than that of GS1 (Figure 2C). In the LS1 stage of senescence, the level of GS2 significantly decreased while the level of GS1 was only slightly lower than in the LC stage. In fully yellow leaves (LS2), the GS2 form was almost undetectable, whereas the level of GS1 increased significantly, relative to the LC stage (Figure 2C). In contrast, a trend in the level of GS1 increasing was observed at the first stage of root senescence (RS1), relative to non-senescent, control roots (RC), but its level was significantly lower at the second stage of senescence (RS2) (Figure 2D). This experiment together with the GS expression analysis provides evidence of the presence a similar process involved in the relocation of N in both examined organs.
Quantitative assessment of carbohydrate levels during the growing season
We tested whether similar to how N is relocated during senescence, plants also remobilize C from starch and soluble carbohydrates in order not to lose those compounds, which may be used in the future as an energy source. The concentration of starch decreased significantly in senescing leaves, relative to non-senescent leaves (Figure 3A). In contrast, the concentration of starch increased in all examined orders of senescent roots, relative to non-senescent roots (Figure 3B). In addition, differences in carbohydrate levels were also observed between leaves and roots. Statistically significant increases in soluble carbohydrate concentration were observed in the first three orders of fine roots (Figure 3D), whereas soluble carbohydrate levels were stable in leaves and no statistically significant changes were observed during the senescence process (Figure 3C).
Figure 3.

Quantitative analysis of carbohydrate levels in leaves (A, C) and in fine roots (B, D) of P. trichocarpa during the course of the growing season. (A, B) Quantitative analysis of starch concentration [%]; (C, D) quantitative analysis of the concentration of soluble carbohydrate [%] (LC: leaf control, LS1: first stage of leaf senescence, LS2: second stage of leaf senescence, RC: root control, RS1: first stage of root senescence and RS2: second stage of root senescence). Bars sharing the same letter are not significantly different (P ≤ 0.05). Values represent the mean ± SE.
Analysis of the expression of genes related to N remobilization and carbohydrate metabolism
Microarray analyses were performed to assess the expression of genes related to the process of the remobilization of chemical elements and nutrients during senescence of leaves and the first-order fine roots. This was done to identify genes other than GS1 that are involved in N remobilization. Additionally, due to the unexpected results obtained in the quantitative assessment of carbohydrate levels, it was deemed important to characterize the expression of genes involved in sugar metabolism in senescing leaves and first order of fine roots. Within the 56,055 transcripts present in the microarray, a total of 1348 differentially expressed genes (DEGs) were identified during the senescence process in leaves (one-way ANOVA, P ≤ 0.001, fold change ≥2). A total of 1898 DEGs were identified in first-order roots during the course of senescence (one-way ANOVA, P ≤ 0.001, fold change ≥2).
Among this large pool of DEGs, only these involved in N remobilization were subjected to further, in-depth analysis (Figure 4). In addition to GS1, the expression of which was confirmed by RT-qPCR analysis to be significantly increased during leaf senescence, an additional 20 genes were examined (Figure 4). Among those 20 selected genes, the majority of them were associated with amino acid transport (16 genes) and were up-regulated. A gene encoding GDH was also found to increase in expression during senescence. In contrast, two genes encoding glutamate synthetase were down-regulated, an NADH-dependent glutamate synthetase and a ferredoxin-dependent glutamate synthetase (Figure 4). In the case of roots, the number of genes associated with N remobilization was less pronounced (Figure 4). In addition to GS1, two genes encoding GDH were identified and both were down-regulated. The expression of six genes that encode N compound transporters (amino acid transporters—four genes, ammonium transporter—one gene and nitrate transporter [NRT]—one gene) were also examined. Results indicated that a gene encoding a high-affinity NRT and another gene encoding an amino acid permease were both up-regulated, while the others were down-regulated (Figure 4).
Figure 4.
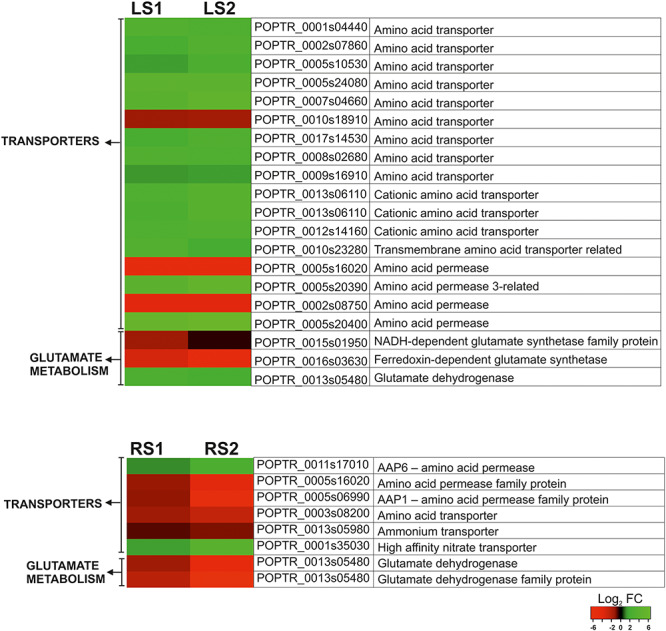
Heat map illustrating the fold changes (log2 basis) in the expression of the selected genes associated with N remobilization during leaf and root senescence.
To gain insight into which genes related to carbohydrate metabolism are different, regulated functional classification of the DEGs was performed using the database for annotation, visualization and integrated discovery. The most abundant gene ontology (GO) categories identified in senescing roots were those related to carbohydrate metabolic process (GO: 0005975). Thus, genes involved in sugar metabolism during the senescence of roots and leaves were examined further.
A total of 25 genes related to carbohydrate metabolism were identified, whose expression changed during leaf senescence. Among these DEGs, 13 were up-regulated and 12 were down-regulated (Figure 5). Similarly, the senescence of first-order absorptive roots was associated with the differential expression of 20 genes involved in carbohydrate metabolism. Among these DEGs, 11 were up-regulated and 9 were down-regulated (Figure 5). The DEGs could be divided into four functional groups in both leaves and roots—starch metabolism, sucrose metabolism, sugar transporters and hexose metabolism (Figure 5).
Figure 5.
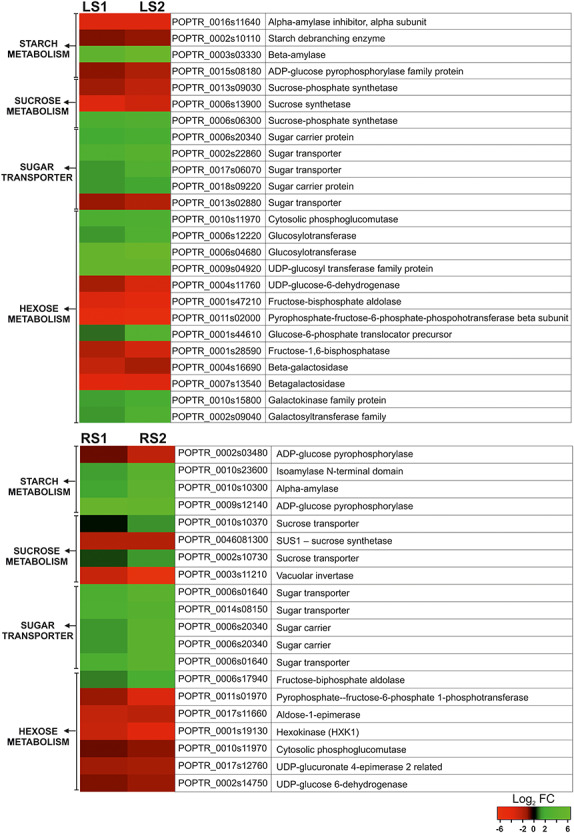
Heat map illustrating the fold changes (log2 basis) in the expression of the selected genes associated with carbohydrate metabolism during leaf and root senescence.
Overall microarray analysis results indicate a large change in expression of genes associated with the remobilization of N and carbohydrates. These changes are in agreement with the quantitative analyses of N and sugars and confirm the N relocations and also indicate a different carbohydrates economy.
Discussion
The seasonal death of ephemeral plant organs is generally studied as a sequence of senescence-associated processes. The majority of research thus far has concentrated on elucidating this process in leaves or flower petals (Pérez-Rodríguez and Valpuesta 1996, Otegui et al. 2005, Liu et al. 2008, Agüera et al. 2010, Guiboileau et al. 2012, Shibuya 2012, Avila-Ospina et al. 2015). As a result, the ultrastructural, physiological and molecular changes associated with leaf and petal senescence have been identified and described as being a typical PCD (Yen and Yang 1998, Quirino et al. 2000, van Doorn and Woltering 2008, Shibuya et al. 2011, Sobieszczuk-Nowicka et al. 2018, Wojciechowska et al. 2018a). Despite the numerous studies on senescence that have been reported in the literature, only a small number of them have focused on the senescence process in absorptive roots (Bagniewska-Zadworna et al. 2014, Wojciechowska et al. 2018b).
Given that senescence is characterized by the intensification of catabolic and a decrease in anabolic processes, questions regarding the degradation and remobilization of valuable macromolecules and nutrients need to be addressed (Guo et al. 2004). Macromolecules are hydrolyzed and reduced to smaller, more mobile components that can be transported through the conductive tissue to other parts of the plant, such as developing seeds (Lemaıˆtre et al. 2008, Guiboileau et al. 2012). Remobilization mechanisms are activated at the beginning of the senescence to avoid losing these valuable macromolecules and their constituent elements. Mechanisms related to recycling N are well understood in senescent leaves (Liu et al. 2008). Studies in herbaceous plants (Diaz et al. 2008, Agüera et al. 2010, 2012, Avila-Ospina et al. 2015) have described a sequence of events associated with remobilization; including a decrease in chlorophyll, reduction in protein and N levels, and an increase in GS1 or asparagine synthetase activity. The occurrence of these events suggests that N can be translocated from senescent tissues through phloem sap to other plant organs (Lemaıˆtre et al. 2008, Guiboileau et al. 2012); however, the regulation of N remobilization and transfer in tree roots is poorly understood and it is possible that the mechanism may be different.
In the present study, we examined and compared factors regulating the translocation process in both leaves and absorptive roots of P. trichocarpa. Quantitative analyses showed that in both organs, concentration of N decreased during senescence. In leaves, this result is in line with literature, which showed such a relationship for Arabidopsis thaliana (Diaz et al. 2008), Hordeum vulgare (Avila-Ospina et al. 2015) and Helianthus annuus (Agüera et al. 2010, 2012). Remobilization of N in the absorptive roots of trees is more ambiguous. The quantitative analysis of N concentration in fine roots of different species that has been conducted previously provided contradictory information. Results of studies performed on the fine roots of Quercus robur are in agreement with the results obtained in the present study, suggesting that remobilization of N from fine roots may occur at the end of the growing season (Zadworny et al. 2015). This premise is supported by the observed decrease in N concentration in the first three orders of fine roots with a simultaneous significant increase of N in higher orders (from the fourth to sixth) of roots. Higher order roots exhibit a less ephemeral nature and have a longer lifespan than lower order roots (Xia et al. 2010, Jia et al. 2011, Zadworny et al. 2015). Nambiar (1987), however, suggested that N translocation is not a crucial process in trees, while Kunkle et al. (2009) reported an increase in N levels in dead roots of Populus tremuloides, Betula alleghaniensis, Acer rubrum and Acer saccharum, relative to the levels in living roots of these species. Results obtained by Kunkle et al. (2009) suggest that the relocation of N during senescence does not occur. However, their conclusion may arise from the colonization of dead roots by microorganisms containing high levels of chitin or due to the existence of significant variability in the classification of fine roots (e.g. as all roots with diameter lower than 2 mm), without dividing them based on their function as transport or absorptive roots (McCormack et al. 2015). Data may also have been collected without identifying senescence stages based on morphological or anatomical factors, as was described by Wojciechowska et al. (2018b). Considering the total biomass of fine, absorptive roots, the lack of active mechanisms related to N remobilization would result in the loss of a huge amount of N (Jackson et al. 1997).
To obtain additional evidence confirming remobilization in studied organ, additional molecular analysis has been conducted. A genetic analysis performed by Castro-Rodríguez et al. (2011) identified in Populus three groups of duplicated genes that encode GS1—GS1.1, GS1.2 and GS1.3—and one gene encoding GS2. These genes were shown to exhibit organ-specific and seasonal-dependent patterns of expression (Castro-Rodríguez et al. 2011). In our study, GS1.1 was up-regulated in both senescent organs—leaves and absorptive roots. Moreover, in leaves, expression of GS1.3 was also increased. Those duplicated genes may be involved in N metabolism, suggested function of GS1.1 is glutamine biosynthesis, whereas the role of protein encoding by GS1.3 is N remobilization (Castro-Rodríguez et al. 2011). The other isoform, GS1.2 exhibited their highest expression in roots at the beginning of the growth season (RC) that could be associated with the primary assimilation of N from soil and/or lignin biosynthesis (Castro-Rodríguez et al. 2011). In addition to GS1, the chloroplastic isoform of GS (GS2) was principally expressed in green leaves (LC). This result is consistent with our previous results, where observed chloroplasts began to transform into gerontoplasts in the LS1 stage of senescence, and most of these organelles were in advanced stages of degradation at the LS2 stage of senescence (Wojciechowska et al. 2018b). Analyses of gene expression are compatible with protein analyses where increased GS1 was observed during senescence both organs. Such results during leaf senescence were observed in several species; however, for fine roots, this is the first such documentation, which supports hypothesis about resorption of N from the most distal senescent absorptive roots in Populus.
In addition to genes that encode GS1, the microarray analyses identified the differential expression of other genes related to N metabolism. Among these genes, N compound transporters constituted a large group, which was identified in both organs. In leaves, the majority of those genes were up-regulated, whereas in roots, only two genes encoding N transporters were characterized by increased expression. This included a gene that encodes a protein belonging to a large family of high-affinity NRT that may play multifunctional roles in nitrate uptake and transport throughout the plant (Bai et al. 2013). Studies performed on Arabidopsis indicated that NRT1.7 and NRT2.5 NRTs have contributed to the remobilization of nitrate from the source leaves to the sink organs (Fan et al. 2009, Lezhneva et al. 2014, Wu et al. 2014). Moreover, there is a premise that other NRTasNRT1.6 and NRT1.5 — that are up-regulated during leaf senescence may play a role in senescence and they could also be involved in nitrate and ammonium remobilization (Havé et al. 2017). Given the broad range of roles of NRT, which may act as either a nitrate sensor, a signal transducer or a transporter, it is hard to interpret which role it may play in senescent roots, and this issue requires further analysis. Moreover in leaves, up-regulation of the gene encoding GDH, which functions in the transfer of remobilized N, was noticed. A similar finding was reported in Arabidopsis and Nicotiana (Bernhard and Matile 1994, Masclaux et al. 2000, Guo et al. 2004, Li et al. 2017).
The mechanisms underlying N translocation in trees are not well understood. Guiboileau et al. (2012) reported that autophagy is an essential aspect of the relocation of N from senescing leaves to developing seeds. Studies with Arabidopsis mutants double mutants atg + salicylic acid defective lines have demonstrated that plants with impaired autophagy machinery accumulate N in leaves, especially under low nitrate availability. In the previous article (Wojciechowska et al. 2018b), we documented an increase in processes associated with autophagy in senescing roots and leaves of P. trichocarpa. Up-regulation of autophagy genes, as well as presence of micro- and macroautophagy, was observed in the same stage of fine roots senescence as increasing expression of GS1 genes and decreasing concentration of N. Based on this research, it seems likely that autophagy may also be involved in N remobilization in both leaves and roots of P. trichocarpa.
In contrast to N, C concentrations in senescing leaves and roots remained stable in our study. A slight decrease in C concentration was observed, however, during leaf senescence in H. vulgare (Avila-Ospina et al. 2015). A more important parameter related to the regulation of cellular metabolism during the senescence of plant organs is the C:N ratio (Chen et al. 2015). Neither C nor N alone, but rather the C:N ratio, plays a crucial role in several processes in Arabidopsis such as the regulation of seedling growth, remobilization of storage lipids, the expression of photosynthetic genes and natural senescence (Martin et al. 2002). A similar change in the C:N ratio during the vegetative season was observed in both leaves and roots, while the C:N ratio increased during senescence. An initiation of senescence has been reported to be induced by high C and low N availability in plant tissues (Wingler et al. 1998, Aoyama et al. 2014).
Significant differences between the examined organs were observed in carbohydrate metabolism during senescence. In the present study, starch concentration decreased rapidly in leaves during senescence. A previously reported ultrastructure analysis also revealed a decreasing number of starch granules in yellowing leaves of P. trichocarpa (Wojciechowska et al. 2018b). Similarly, senescing leaves of H. annuus (Agüera et al. 2012), Oryza sativa (Muthukumaran and Rao 2013) and A. thaliana (Diaz et al. 2008) were also characterized by a decrease in starch levels. Studies of starch degradation suggest that autophagy may play an important role in the breakdown of this carbohydrate. Wang et al. (2013) demonstrated that silencing of ATG genes reduces leaf starch degradation, resulting in an excessive accumulation of starch in atg mutants. These observations support the premise that the increase of autophagy activity observed in senescing leaves and roots of P. trichocarpa may play a multifunctional role in degradation, N remobilization and starch degradation (Wojciechowska et al. 2018a, 2018b, Guiboileau et al. 2012, Wang et al. 2013). Microarray analysis in senescent leaves revealed several genes associated with starch metabolism, which is in line with quantitative analyses of starch concentration, e.g. significant up-regulation of the gene encoding a β-amylase, which is associated with starch degradation (Lin et al. 1988, Kaplan and Guy 2004). In addition to β-amylase, the down-regulation of genes encoding starch debranching enzymes and ADP-glucose pyrophosphorylase was observed, indicating that starch biosynthesis is inhibited or completely stopped during senescence (Kubo et al. 1999, Ballicora et al. 2004). An inverse relationship is observed in absorptive roots, where the concentration of starch increased. This was surprising, however, it may be elucidated by the fact that N relocation is dependent on an adequate supply of carbohydrates to provide energy for active senescence processes and for the conversion of N to glutamine. In contrast to leaves, expression of the ADP-glucose pyrophosphorylase gene was strongly up-regulated during absorptive root senescence. Similarly, expressions of genes encoding α-amylase and the isoamylase N-terminal domain, which regulate starch degradation, were also increased. In addition to initiating starch degradation, α-amylases are also involved in abiotic stress response, such as cold acclimation—a process that includes an increase in soluble sugars and hexose-phosphates in the cytosol (Ristic and Ashworth 1993, Hurry et al. 1995). A large body of research has demonstrated that sugars accumulate in cells and tissues to increase osmotic pressure and cold tolerance (Sasaki et al. 1996, Klemens et al. 2013, Tarkowski and Van den Ende 2015). An increase in sugar levels could help to prevent damage to roots by temporarily occurring freezing temperature conditions (even in fall), and thus extend their life span, thereby prolonging the senescence process until nutrients can be transferred to higher root orders. Microarray analyses also revealed several genes associated with sucrose metabolism in both organs. The primary mobile sugar in plants—sucrose—is transported from its source (synthesizing) organs to sink organs through the phloem, even at times over long distances (Lemoine 2000). Sucrose is known to inhibit the onset of senescence, which has been documented in Brassica oleracea branches (Irving and Joyce 1995), Lilium and Dianthus caryophyllus petals, and Asparagus officinalis spears (Hoeberichts et al. 2007, Arrom and Munné-Bosch 2012, Park 2016). During leaf senescence, several genes involved in the sucrose metabolism were identified. Sucrose synthetase is involved in the reversible conversion of sucrose and UDP into fructose and UDP-glucose. Similar to our results, changes in the expression levels of genes that encode a sucrose synthetase were also documented during the senescence of leaves of Nicotiana tabacum (Li et al. 2017). In senescent roots, the sucrose synthetase (SUS1) gene was down-regulated. A gene encoding a vacuolar invertase, which is responsible for converting sucrose into glucose and fructose inside a vacuole, was also down-regulated. However, the up-regulation of genes encoding sucrose transporters was observed, as well as the elevated expression of five genes encoding sugar carriers, collectively implying an increase in sugar relocation. In senescent leaves, the microarray analysis identified five genes encoding sugar transporters. Expression of four of them was up-regulated, which suggests that sugars are actively translocated out of senescing cells and tissues.
Conclusion
The senescence of plant organs is a precisely controlled process that allows plants to relocate valuable nutrients from senescent organs to other locations rather than lose them to the environment. The results of the present study provide evidence that in both leaves and roots, the process of N resorption is activated during senescence. This premise is supported by the analysis of N concentration and the molecular analysis of GS levels, a key enzyme in N remobilization. To our knowledge, this is the first confirmation that relocation of N during the senescence of absorptive fine roots is regulated at the molecular level. Significant changes in carbohydrate metabolism–gene expression were observed in both leaves and roots during senescence. Only in fine roots, however, was the accumulation of sugars observed. This may be related to the need to cold acclimate and increase the tolerance of roots to freezing temperatures, so that the remobilization of nutrients to higher order roots can be completed—something that could not occur if the roots died quickly due to freezing injury. The specific mechanisms responsible for the remobilization of nutrients during senescence and the functional role of carbohydrates during this process are not well understood and remain a critical priority for future research.
Acknowledgments
The authors thank Michael Luke McCormack and Marcin Zadworny for critical reading of the manuscript.
Conflict of interest
None declared.
Funding
This work was supported by grant no. 2012/07/E/NZ9/00194 to A.B.-Z. from the National Science Centre, Poland and by grant no. 2016/23/N/NZ3/00073 to N.W. from the National Science Centre, Poland.
References
- Agüera E, Cabello P, de la Haba P (2010) Induction of leaf senescence by low nitrogen nutrition in sunflower (Helianthus annuus) plants. Physiol Plant 138:256–267. [DOI] [PubMed] [Google Scholar]
- Agüera E, Cabello P, de la Mata L, Molina E, de la Haba P (2012) Metabolic regulation of leaf senescence in sunflower (Helianthus annuus L.) plants In: Nagata T. (ed) Senescence. InTech, pp 51–68. [Google Scholar]
- Aoyama S, Huarancca Reyes T, Guglielminetti L, Lu Y, Morita Y, Sato T, Yamaguchi J (2014) Ubiquitin ligase ATL31 functions in leaf senescence in response to the balance between atmospheric CO2 and nitrogen availability in Arabidopsis. Plant Cell Physiol 55:293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrom L, Munné-Bosch S (2012) Sucrose accelerates flower opening and delays senescence through a hormonal effect in cut lily flowers. Plant Sci 188–189:41–47. [DOI] [PubMed] [Google Scholar]
- Avice J-C, Etienne P (2014) Leaf senescence and nitrogen remobilization efficiency in oilseed rape (Brassica napus L.). J Exp Bot 65:3813–3824. [DOI] [PubMed] [Google Scholar]
- Avila-Ospina L, Marmagne A, Talbotec J, Krupinska K, Masclaux-Daubresse C (2015) The identification of new cytosolic glutamine synthetase and asparagine synthetase genes in barley (Hordeum vulgare L.), and their expression during leaf senescence. J Exp Bot 66:2013–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila-Ospina L, Moison M, Yoshimoto K, Masclaux-Daubresse C (2014) Autophagy, plant senescence, and nutrient recycling. J Exp Bot 65:3799–3811. [DOI] [PubMed] [Google Scholar]
- Bagniewska-Zadworna A, Stelmasik A (2015) Root heterogeneity and developmental stage determine the pattern of cellulose synthase and cinnamyl alcohol dehydrogenase gene expression profiles during xylogenesis in Populus trichocarpa (Torr. Et Gray). Int J Plant Sci 176:458–467. [Google Scholar]
- Bagniewska-Zadworna A, Byczyk J, Eissenstat DM, Oleksyn J, Zadworny M (2012) Avoiding transport bottlenecks in an expanding root system: xylem vessel development in fibrous and pioneer roots under field conditions. Am J Bot 99:1417–1426. [DOI] [PubMed] [Google Scholar]
- Bagniewska-Zadworna A, Stelmasik A, Minicka J (2014) From birth to death — Populus trichocarpa fibrous roots functional anatomy. Biol Plant 58:551–560. [Google Scholar]
- Bai H, Euring D, Volmer K, Janz D, Polle A (2013) The nitrate transporter (NRT) gene family in poplar. PLoS One 8:e72126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballicora MA, Iglesias AA, Preiss J (2004) ADP-glucose pyrophosphorylase: a regulatory enzyme for plant starch synthesis. Photosynth Res 79:1–24. [DOI] [PubMed] [Google Scholar]
- Bernhard WR, Matile P (1994) Differential expression of glutamine synthetase genes during the senescence of Arabidopsis thaliana rosette leaves. Plant Sci 98:7–14. [Google Scholar]
- Brassard BW, Chen HYH, Bergeron Y (2009) Influence of environmental variability on root dynamics in northern forests. Crit Rev Plant Sci 28:179–197. [Google Scholar]
- Buchanan-Wollaston V, Ainsworth C (1997) Leaf senescence in Brassica napus: cloning of senescence related genes by subtractive hybridisation. Plant Mol Biol 33:821–834. [DOI] [PubMed] [Google Scholar]
- Castro-Rodríguez V, García-Gutiérrez A, Canales J, Avila C, Kirby EG, Cánovas FM (2011) The glutamine synthetase gene family in Populus. BMC Plant Biol 11:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerasoli S, Scartazza A, Brugnoli E, Chaves MM, Pereira JS (2004) Effects of partial defoliation on carbon and nitrogen partitioning and photosynthetic carbon uptake by two-year-old cork oak (Quercus suber) saplings. Tree Physiol 24:83–90. [DOI] [PubMed] [Google Scholar]
- Chen D, Wang S, Xiong B, Cao B, Deng X (2015) Carbon/nitrogen imbalance associated with drought-induced leaf senescence in Sorghum bicolor. PLoS One 10e:0137026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz C, Lemaître T, Christ A, Azzopardi M, Kato Y, Sato F, Morot-Gaudry J-F, Le Dily F, Masclaux-Daubresse C (2008) Nitrogen recycling and remobilization are differentially controlled by leaf senescence and development stage in Arabidopsis under low nitrogen nutrition. Plant Physiol 147:1437–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenstat DM, Volder A (2005) The efficiency of nutrient acquisition over the life of a root In: BassiriRad H (ed) Nutrient acquisition by plants: an ecological perspective. Springer-Verlag, Berlin-New York, pp 185–220. [Google Scholar]
- Eissenstat DM, Wells CE, Yanai RD, Whitbeck JL (2000) Building roots in a changing environment: implications for root longevity. New Phytol 147:33–42. [Google Scholar]
- Fan S-C, Lin C-S, Hsu P-K, Lin S-H, Tsay Y-F (2009) The Arabidopsis nitrate transporter NRT1.7, expressed in phloem, is responsible for sourceto-sink remobilization of nitrate. Plant Cell 21:2750–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill RA, Jackson RA (2008) Global patterns of root turnover for terrestrial ecosystems. New Phytol 147:13–31. [Google Scholar]
- Guiboileau A, Yoshimoto K, Soulay F, Bataillé M-P, Avice J-C, Masclaux-Daubresse C (2012) Autophagy machinery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis. New Phytol 194:732–740. [DOI] [PubMed] [Google Scholar]
- Guo Y, Cai Z, Gan S (2004) Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ 27:521–549. [Google Scholar]
- Havé M, Marmagne A, Chardon F, Masclaux-Daubresse C (2017) Nitrogen remobilization during leaf senescence: lessons from Arabidopsis to crops. J Exp Bot 68:2513–2529. [DOI] [PubMed] [Google Scholar]
- Hoeberichts FA, Doorn WG, Vorst O, Hall RD, Wordragen MF (2007) Sucrose prevents up-regulation of senescence-associated genes in carnation petals. J Exp Bot 58:2873–2885. [DOI] [PubMed] [Google Scholar]
- Hurry VM, Strand A, Tobiaeson M, Gardestrom P, Oquist G (1995) Cold hardening of spring and winter wheat and rape results in differential effects on growth, carbon metabolism, and carbohydrate content. Plant Physiol 109:697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving DE, Joyce DC (1995) Sucrose supply can increase longevity of broccoli (Brassica oleracea) branchlets kept at 22°C. Plant Growth Regul 17:251–256. [Google Scholar]
- Jackson RB, Mooney HA, Schulze ED (1997) A global budget for fine root biomass, surface area, and nutrient contents. PNAS 94:7362–7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia S, Wang Z, Li X, Zhang X, McLaughlin NB (2011) Effect of nitrogen fertilizer, root branch order and temperature on respiration and tissue N concentration of fine roots in Larix gmelinii and Fraxinus mandshurica. Tree Physiol 31:718–726. [DOI] [PubMed] [Google Scholar]
- Kaplan F, Guy CL (2004) β-Amylase induction and the protective role of maltose during temperature shock. Plant Physiol 135:1674–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemens PAW, Patzke K, Deitmer J, et al. (2013) Overexpression of the vacuolar sugar carrier AtSWEET16 modifies germination, growth, and stress tolerance in Arabidopsis. Plant Physiol 163:1338–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A, Fujita N, Harada K, Matsuda T, Satoh H, Nakamura Y (1999) The starch-debranching enzymes isoamylase and pullulanase are both involved in amylopectin biosynthesis in rice endosperm. Plant Physiol 121:399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkle JM, Walters MB, Kobe RK (2009) Senescence-related changes in nitrogen in fine roots: mass loss affects estimation. Tree Physiol 29:715–723. [DOI] [PubMed] [Google Scholar]
- Lemaître T, Gaufichon L, Boutet-Mercey S, Christ A, Masclaux-Daubresse C (2008) Enzymatic and metabolic diagnostic of nitrogen deficiency in Arabidopsis thaliana Wassileskija accession. Plant Cell Physiol 49:1056–1065. [DOI] [PubMed] [Google Scholar]
- Lemoine R. (2000) Sucrose transporters in plants: update on function and structure. Biochim Biophys Acta Biomembr 1465:246–262. [DOI] [PubMed] [Google Scholar]
- Lezhneva L, Kiba T, Feria-Bourrellier A-B, Lafouge F, BoutetMercey S, Zoufan P, Sakakibara H, Daniel-Vedele F, Krapp A (2014) The Arabidopsis nitrate transporter NRT2.5 plays a role in nitrate acquisition and remobilization in nitrogen-starved plants. Plant J 80:230–241. [DOI] [PubMed] [Google Scholar]
- Li W, Zhang H, Li X et al. (2017) Intergrative metabolomic and transcriptomic analyses unveil nutrient remobilization events in leaf senescence of tobacco. Sci Rep 7:12126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TP, Spilatro SR, Preiss J (1988) Subcellular localization and characterization of amylases in Arabidopsis leaf. Plant Physiol 86:251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wu YH, Yang JJ, Liu YD, Shen FF (2008) Protein degradation and nitrogen remobilization during leaf senescence. J Plant Biol 51:11–19. [Google Scholar]
- Martin T, Oswald O, Graham IA (2002) Arabidopsis seedling growth, storage lipid mobilization, and photosynthetic gene expression are regulated by carbon:nitrogen availability. Plant Physiol 128:472–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux C, Valadier MH, Brugière N, Morot-Gaudry JF, Hirel B (2000) Characterization of the sink/source transition in tobacco (Nicotiana tabacum L.) shoots in relation to nitrogen management and leaf senescence. Planta 211:510–518. [DOI] [PubMed] [Google Scholar]
- McCormack ML, Dickie IA, Eissenstat DM et al. (2015) Redefining fine roots improves understanding of below-ground contributions to terrestrial biosphere processes. New Phytol 207:505–518. [DOI] [PubMed] [Google Scholar]
- Miflin BJ, Habash DZ (2002) The role of glutamine synthetase and glutamate dehydrogenase in nitrogen assimilation and possibilities for improvement in the nitrogen utilization of crops. J Exp Bot 53:979–987. [DOI] [PubMed] [Google Scholar]
- Muthukumaran M, Rao AVB (2013) Starch metabolism during leaf senescence in two rice varieties on exposure to aluminium. Nat Environ Pollut Technol 12:703–708. [Google Scholar]
- Nambiar S. (1987) Do nutrients translocate from fine roots? Can J For Res 17:913–918. [Google Scholar]
- Oleksyn J, Zytkowiak R, Karolewski P, Reich PB, Tjoelker MG (2000) Genetic and environmental control of seasonal carbohydrate dynamics in trees of diverse Pinus sylvestris populations. Tree Physiol 20:837–847. [DOI] [PubMed] [Google Scholar]
- Otegui MS, Noh Y-S, Martínez DE, Vila Petroff MG, Staehelin LA, Amasino RM, Guiamet JJ (2005) Senescence-associated vacuoles with intense proteolytic activity develop in leaves of Arabidopsis and soybean. Plant J 41:831–844. [DOI] [PubMed] [Google Scholar]
- Park M-H. (2016) Sucrose delays senescence and preserves functional compounds in Asparagus officinalis L. Biochem Biophys Res Commun 480:241–247. [DOI] [PubMed] [Google Scholar]
- Pérez-Rodríguez J, Valpuesta V (1996) Expression of glutamine synthetase genes during natural senescence of tomato leaves. Physiol Plant 97:576–582. [Google Scholar]
- Pregitzer KS, Deforest J, Burton A, Allen M, Ruess R (2002) Fine roots architecture of nine north American trees. Ecol Monogr 72:293–309. [Google Scholar]
- Quirino BF, Noh YS, Himelblau E, Amasino RM (2000) Molecular aspects of leaf senescence. Trends Plant Sci 5:278–282. [DOI] [PubMed] [Google Scholar]
- Ristic Z, Ashworth EN (1993) Changes in leaf ultrastructure and carbohydrates in Arabidopsis thaliana L. (Heyn) cv. Columbia during rapid cold acclimation. Protoplasma 172:111–123. [Google Scholar]
- Sasaki H, Ichimura K, Oda M (1996) Changes in sugar content during cold acclimation and deacclimation of cabbage seedlings. Ann Bot 78:365–369. [Google Scholar]
- Shibuya K. (2012) Molecular mechanisms of petal senescence in ornamental plants. J Japan Soc Hort Sci 81:140–149. [Google Scholar]
- Shibuya K, Shimizu K, Yamada T, Ichimura K (2011) Expression of autophagy-associated ATG8 genes during petal senescence in Japanese morning glory. J Japan Soc Hort Sci 80:89–95. [Google Scholar]
- Shibuya K, Niki T, Ichimura K (2013) Pollination induces autophagy in petunia petals via ethylene. J Exp Bot 64:1111–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya K, Shimizu K, Niki T, Ichimura K (2014) Identification of a NAC transcription factor, EPHEMERAL1, that controls petal senescence in Japanese morning glory. Plant J 79:1044–1051. [DOI] [PubMed] [Google Scholar]
- Sobieszczuk-Nowicka E, Wrzesiński T, Bagniewsla-Zadworna A, Kubala S, Rucińska-Sobkowiak R, Polcyn W, Misztal L, Mattoo AK (2018) Physio-genetic dissection of dark-induced leaf senescence and timing its reversal in barley. Plant Physiol 178:654–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer A, Acker G, Bartsch S, Bauerschmitt H, Reinbothe S, Reinbothe C (2015) Differences in gene expression between natural and artificially induced leaf senescence in barley. J Plant Physiol 176:180–191. [DOI] [PubMed] [Google Scholar]
- Szuba A, Wojakowska A, Lorenc-Plucińska G (2013) An optimized method to extract poplar leaf proteins for two-dimensional gel electrophoresis guided by analysis of polysaccharides and phenolic compounds. Electrophoresis 34:3234–3243. [DOI] [PubMed] [Google Scholar]
- Tarkowski ŁP, Van den Ende W (2015) Cold tolerance triggered by soluble sugars: a multifaceted countermeasure. Front Plant Sci 6:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas H, Ougham HJ, Wagstaff C, Stead AD (2003) Defining senescence and death. J Exp Bot 54:1127–1132. [DOI] [PubMed] [Google Scholar]
- Doorn WG, Woltering EJ (2004) Senescence and programmed cell death: substance or semantics? J Exp Bot 55:2147–2153. [DOI] [PubMed] [Google Scholar]
- Doorn WG, Woltering EJ (2008) Physiology and molecular biology of petal senescence. J Exp Bot 59:453–480. [DOI] [PubMed] [Google Scholar]
- Wang Y, Yu B, Zhao J et al. (2013) Autophagy contributes to leaf starch degradation. Plant Cell 25:1383–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler A, Schaewen A, Leegood RC, Lea PJ, Quick PW (1998) Regulation of leaf senescence by cytokinin, sugars, and light. Plant Physiol 116:329–335. [Google Scholar]
- Wojciechowska N, Sobieszczuk-Nowicka E, Bagniewska-Zadworna A (2018a) Plant organ senescence - regulation by manifold pathways. Plant Biol (Stuttg) 20:167–181. [DOI] [PubMed] [Google Scholar]
- Wojciechowska N, Marzec-Schmidt K, Kalemba EM, Zarzyńska-Nowak A, Jagodziński AM, Bagniewska-Zadworna A (2018b) Autophagy counteracts instantaneous cell death during seasonal senescence of the fine roots and leaves in Populus trichocarpa. BMC Plant Biol 18:260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Qin Z, Fan L, Xue C, Zhou X, Xin M, Du Y (2014) Involvement of CsNRT1.7 in nitrate recycling during senescence in cucumber. J Plant Nutr Soil Sc 177:714–721. [Google Scholar]
- Xia M, Guo D, Pregitzer KS (2010) Ephemeral root modules in Fraxinus mandshurica. New Phytol 188:1065–1074. [DOI] [PubMed] [Google Scholar]
- Yanai R, Eissenstat DM (1997) Root life span, efficiency, and turnover In: Waisel Yet al (eds) Plant roots: The hidden half, 3rd ed. Marcel Dekker, New York, NY, pp 221–238. [Google Scholar]
- Yen C-H, Yang C-H (1998) Evidence for programmed cell death during leaf senescence in plants. Plant Cell Physiol 39:922–927. [Google Scholar]
- Zadworny M, McCormack ML, Rawlik K, Jagodziński AM (2015) Seasonal variation in chemistry, but not morphology, in roots of Quercus robur growing in different soil types. Tree Physiol 35:644–652. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Xiong S, Wei Y, Meng X, Wang X, Ma X (2017) The role of glutamine synthetase isozymes in enhancing nitrogen use efficiency of N-efficient winter wheat. Sci Rep 7:1000. [DOI] [PMC free article] [PubMed] [Google Scholar]


