Abstract
Recent studies have resulted in deeper understanding of a variety of telomere maintenance mechanisms as well as plausible models of telomere evolution. Often overlooked in the discussion of telomere regulation and evolution is the synthesis of the DNA strand that bears the 5’-end (i.e. the C-strand). Herein I describe a scenario for telomere evolution that more explicitly accounts for the evolution of the C-strand synthesis machinery. In this model, CST (CTC1-STN1-TEN1), the G-strand-binding complex that regulates primase-Pol α-mediated C-strand synthesis, emerges as a key player and evolutionary link. Itself arising from RPA, CST not only coordinates telomere synthesis, but also gives rise to the POT1-TPP1 complex that became part of shelterin and that regulates telomerase in G-strand elongation.
Keywords: Telomere, CST, POT1-TPP1, telomerase, primase-Pol α
Evolution of linear chromosomes: time to consider the making of the telomere C-strand?
A distinguishing feature of eukaryotes is the partitioning of genomes into linear chromosomes. This in turn necessitates the elaboration of special terminal structures (i.e., telomeres) that can be distinguished from abnormal chromosome breaks [1]. The standard telomere system in most organisms consists of numerous copies of a short DNA repeat and a protein assembly that binds specifically to the repeats and stabilizes the chromosome ends against degradation, fusion, and recombination. Owing to the end replication problem, telomere DNA suffers progressive shortening with each cell division [2]. To compensate for the loss, most organisms utilize a cellular reverse transcriptase called telomerase to add de novo repeats onto the 3’ ends of shortened telomeres [3, 4]. Thus, the two essential attributes of the telomere are its ability to confer stability and to replenish itself. The origins and adaptive advantages of linear chromosomes and telomeres (vis-à-vis circular chromosomes in eubacteria and archaea) have been the subject of much speculation and discussion. An idea that has strong evolutionary implication posits the invasion of group II intron into the eukaryotic genome as serving a pivotal role in shaping the genome [5, 6]. Group II introns are mobile genetic elements that spread throughout the genome by reverse splicing. These elements are believed to give rise to present-day introns and to necessitate the elaboration of the mRNA splicing machinery. Another proposed consequence of group II intron invasion is none other than the linearization of circular chromosomes through breaks within an intron followed by strand invasion of the ends into two other introns [5]. This linearization, followed by a series of evolutionary inventions that include the emergence of telomerase and telomere proteins capable of synthesizing and stabilizing short sequence repeats at chromosome ends, eventually culminates in the current canonical telomere system. This proposal for the origin of linear chromosomes and telomeres does not invoke any inherent advantage for linear genomes, but rather envisions such genomes as a consequence of “host-parasite” interactions, both with respect to the ingestion of an alpha-proteobacterial cell by an archaeal cell, and the resulting invasion of archaeal genome by the proteobacterial group II intron. Notably, such host-parasite interactions are gaining increasing recognition as the driver of evolutionary complexity, of which the origin of linear genomes is but one of many examples [7].
While the proposed model of telomere evolution is interesting and compelling, it (and most other evolutionary models) does not consider one significant aspect of telomere maintenance, namely the elongation of the DNA strand that bears the 5’ end. In the present-day telomere system, this strand is referred to as the C-strand by virtue of its sequence composition, and is synthesized by the primase-Pol α complex under the regulation of the CST (CTC1-STN1-TEN1) complex. How this aspect of telomere maintenance emerged during evolution, and how components of this system co-evolved with the G-strand elongation machinery is largely overlooked. Yet it should be pointed out that both primase-Pol α and CST are unique to eukaryotes (i.e., absent from archaea and eubacteria), and thus specific to organisms with linear genomes. It is therefore possible that they are as integral to the evolution of telomeres as telomerase. In this article, I propose a model of telomere evolution that gives prominent considerations to the roles of CST, primase-Pol α, and telomere C-strand synthesis. First, I outline the key pathways of telomere protection and maintenance as well as associated factors in present-day organisms. This is followed by a brief review of a plausible path of telomere evolution based on group II intron-induced linearization of chromosomes and the progression from recombination-based to telomerase-based telomere maintenance mechanism [5]. Finally, in the context of this framework, I propose specific scenarios for the evolution of primase-Pol α, CST, and C-strand synthesis. I discuss how these factors could have originated and could have given rise to key players in the G-strand synthesis and protection systems. I will emphasize the advantage of this more elaborate view to (i) provide a step-wise account of the emergence of modern telomere system, (ii) rationalize unexpected similarities between the G-strand and C-strand synthesis pathways; and (iii) re-conceptualize the “variant” telomere machinery found in Drosophila and budding yeasts, two taxa with unusual telomere maintenance mechanisms.
Telomere structures and maintenance: The modern state
To date, the structure and regulation of telomeres have been characterized primarily in mammals and a few model organisms belonging to distinct clades. While each system exhibits interesting variations, the basic structure of telomeres and the key telomere maintenance factors are, with few exceptions, conserved among these systems. In this section, I describe the most conserved elements of telomere structure and maintenance, which include telomerase and primase-Pol α and are shared by organisms in almost all major eukaryotic branches (e.g., all five eukaryotic supergroups [8]). In addition, I describe an alternative, recombination-based telomere maintenance pathway that is relevant to models of telomere evolution.
In most organisms, telomere DNA consists of a few to thousands of copies of a short repeat. The repeat sequence is typically rich in G residues on the 3’-termini bearing strand, and this “G-strand” often protrudes beyond the 5’-end of the complementary “C-strand” to form a G-overhang or G-tail. The main protein complex that coats both the duplex telomeres and G-tails is named shelterin (Fig. 1), and it can be divided structurally into three modules: the duplex telomere-binding factors and associated polypeptide (named TRF1, TRF2, and RAP1 in humans); the G-tail binding factors (POT1 and TPP1 in humans), and the bridge factors (TIN2 in humans) [1, 9]. Of particular importance in the discussion herein is the G-tail binding module, which plays a pivotal role in regulating the activity of the G-strand elongating enzyme, namely telomerase (see below). Another important and widely conserved complex at telomeres is CST (CTC1-STN1-TEN1) [10, 11]. This complex, like POT1-TPP1 (PT), has high affinity for G-tails, but instead of promoting G-strand elongation, CST plays a key role in repressing telomerase activity and stimulating C-strand synthesis by primase-Pol α (see below). Thus, the two G-strand binding complexes that emerged in evolution mediate complementary functions in maintaining telomere repeats at chromosome ends. Both CST and PT contain multiple OB fold domains and utilize some of these domains for DNA binding, but the general view is that they are not evolutionarily related (Fig. 2A). For example, CST bears substantial structural similarity to RPA (RPA1-RPA2-RPA3), the ubiquitous trimeric ssDNA-binding complex in eukaryotes, whereas PT does not seem to be related to RPA (Fig. 2A and [10, 12]) (see below for a more detailed comparison of the structural organizations of these complexes).
Figure 1. The prevalence of the standard telomere protection and maintenance system and the main regulatory complexes at telomeres.
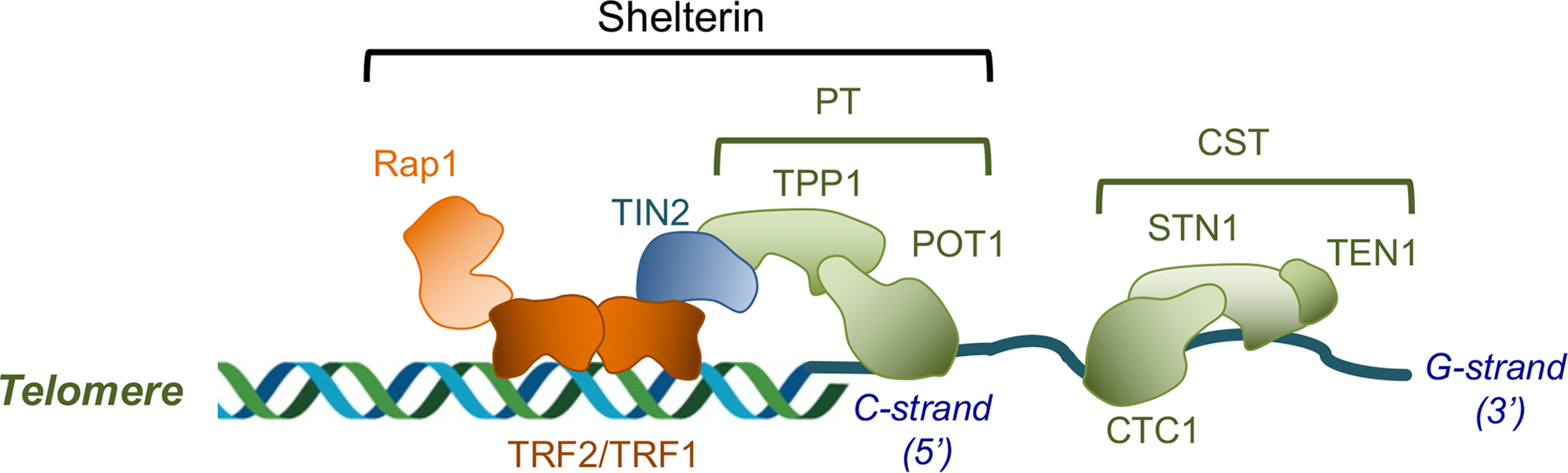
The compositions and subunit arrangements of the shelterin and CST complexes are illustrated.
Figure 2. Telomere G- and C-strand elongation mechanisms and the domain structures of RPA, CST, and PT.
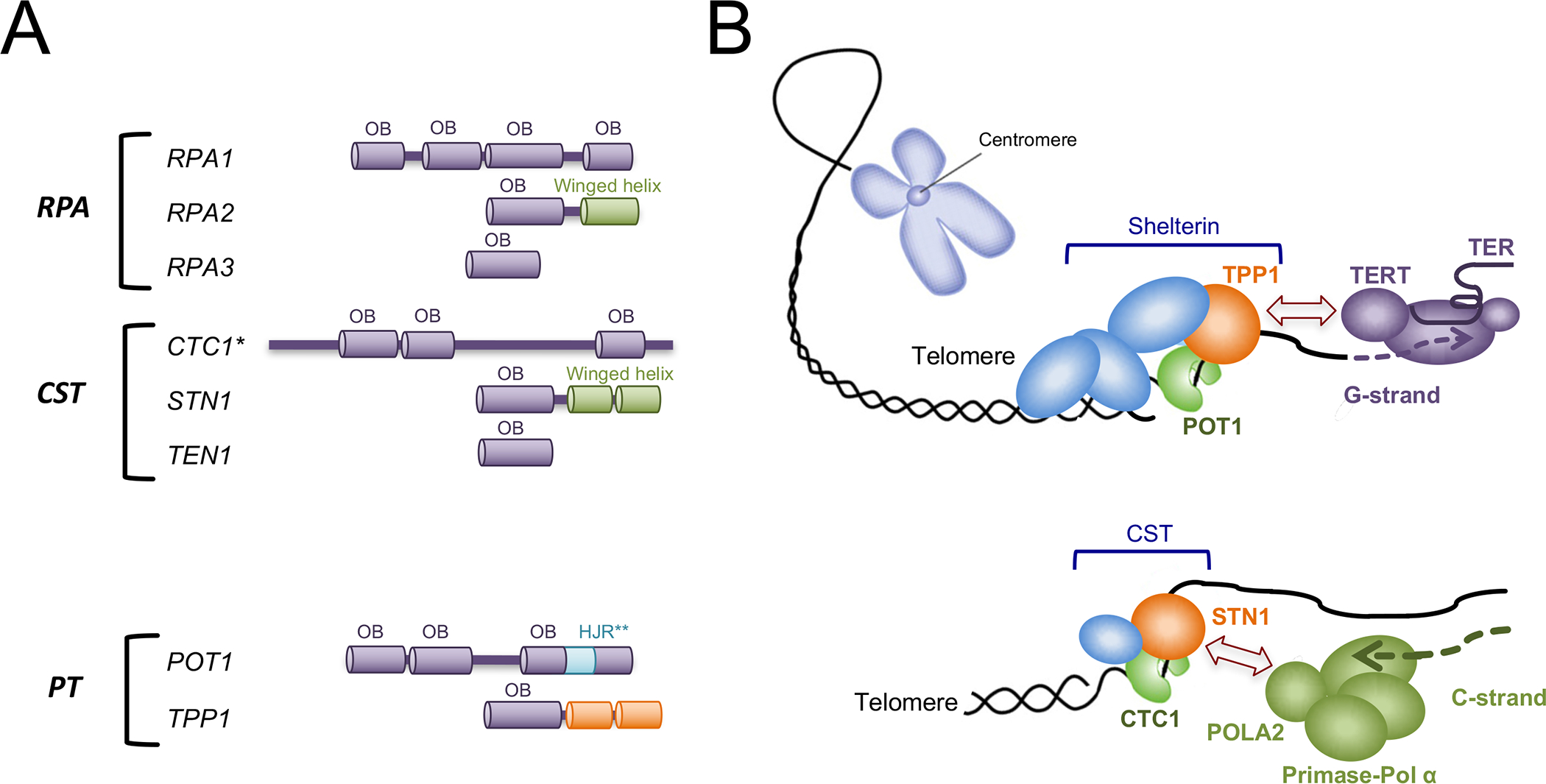
(A) The domain organizations of the RPA, CST, and POT1-TPP1 subunits are illustrated. (*) The number and distribution of CTC1 OB folds have not been determined. Most bioinformatics analysis indicates the existence of two N-terminal OB folds and one C-terminal OB fold (For example, [53, 73]). (**) The insertion of a HJR (Holiday Junction Resolvase) like domain in POT1 is not a conserved feature of this family of proteins; it is true for the human orthologue, but not for TEBPα [74, 75].
(B) The G-strand of telomeres is extended by telomerase under the control of the shelterin complex. More specifically, the TPP1 subunit of shelterin utilizes its N-terminal OB fold to recruit and to enhance the processivity of telomerase. TERT and TER are respectively the catalytic reverse transcriptase and the RNA template component of the telomerase complex. The C-strand of telomeres is synthesized by the primase-Pol α complex under the control of the CST complex. This regulation appears to be primarily mediated through an interaction between the N-terminal OB fold of STN1 and the POLA2 subunit of primase-Pol α.
Mechanistically, the telomere G- and C-strand synthesis machines are quite distinct, and how they are regulated also appear distinct (Fig. 2B). Telomerase is a ribonucleoprotein bearing an integral RNA component (TER) that directs the catalytic reverse transcriptase subunit (TERT) to synthesize the cognate telomere G-strand. Although many means of regulating telomerase function have been described, arguably the most important regulatory factor is TPP1: it utilizes an N-terminal OB fold domain to bind the N-terminal TEN domain of TERT, and this interaction serves to recruit telomerase to telomere ends and to stimulate telomerase processivity (i.e., its ability to add long tracts of telomere repeats) [13, 14] (Fig. 2B, top). The C-strand synthesis machine, primase-Pol α, is a bi-functional enzyme that uses sequentially two separate active sites to synthesize an RNA-DNA chimera [15–17], which is subsequently processed such that the RNA segment is removed and the DNA segment ligated to the original telomere 5’ end. Notably, C-strand synthesis has received considerably less attention than that given to telomerase, and steps governing RNA removal and ligation remain poorly understood [18]. However, recent studies provide strong support for a critical role of CST in this pathway: it is crucial for C-strand synthesis in vivo, most likely by utilizing an N-terminal OB fold of STN1 to bind and stimulate primase-Pol α at telomeres [17, 19, 20] (Fig. 2B, bottom). Primase-Pol α is also critical for chromosomal replication, by generating the primers for both leading and lagging strand synthesis. However, this function of primase-Pol α at replication origins and replication forks does not depend on CST [21]. Rather, CST has a specialized role in genome-wide replication by helping the cells to overcome replication stress at G-rich regions, and this function is evidently connected to the DNA repair protein RAD51 [22, 23].
The outline sketched above is likely to apply to organisms belonging to many taxa. For example, in the ciliates Oxytricha, the PT orthologue TEBPα/β is likely to modulate telomere end structure and may control the activity of telomerase [24, 25]. In fission yeast, although other interactions between shelterin and telomerase have been described, an important regulatory step has been attributed to Tpz1 (equivalent to TPP1)-Trt1 (equivalent to TERT) interaction [26]. With respect to C-strand synthesis, notwithstanding many gaps in our knowledge, the critical function of both CST and primase-Pol α subunits have been established not only in mammals, but also in plants and budding yeast [27, 28]. Moreover, even though experimental evidence that highlights the STN1 OB fold in C-strand synthesis exists only for mammals and budding yeast, the singular role of this domain is buttressed by its greater degree of evolutionary conservation vis-à-vis other CST domains [29]. STN1 is also, unlike CTC1 and TEN1, almost universally present in all eukaryotic genomes that have received careful scrutiny.
An alternative to the canonical telomere maintenance system described above is recombination-based telomere maintenance. The recombination pathways are not utilized by normal organisms, but instead found in telomerase-null mutants or telomerase-negative cancer cells. Even though these mechanisms differ in detail, they share the characteristic of relying on a subset of recombination and repair proteins. In budding and fission yeast telomerase null mutants, recombination mechanisms that enable the amplification of subtelomeric repeats or terminal repeats are often activated, and this allows the mutants to bypass replicative senescence [30, 31]. Likewise, in about 10–15 % of cancer cells, telomerase is absent, and a recombination pathway (ALT) that allows telomere repeats to spread from one chromosome end to others provides the basis for achieving replicative immortality [32]. Recent studies point to substantial mechanistic similarities between ALT and break-induced replication (BIR), including e.g., the conservative nature of DNA synthesis and the functional requirement for DNA polymerase δ [33, 34]. However, other features of ALT appear to be different from BIR, such as the differential effects of reducing or eliminating the Sgs1/BLM family of helicases [35, 36]. Notably for both BIR and ALT, the bulk of evidence indicates that the 3’ end which successfully invades another duplex to initiate DNA synthesis is most likely extended by Pol δ [33, 37]. However, the mechanisms and polymerase responsible for the conversion of the extended ssDNA into dsDNA remain quite obscure. Interestingly, while PT has not been implicated in ALT, a recent study suggests a positive regulatory role for CST in the ALT pathway. In particular, knocking down CST reduced the levels of C-circles, which is one of the most accurate markers of ALT activity [38, 39].
The origin of telomeres and the transition from recombination-based telomere elongation to telomerase: The G-strand-centric view
de Lange recently proposed a model for the origin and maintenance of ancestral telomeres (Fig. 3) based on (Fig. 3, step i) the invasion of group II introns into the genomes of the ancestral eukaryotes, (step ii) stabilization of DNA breaks in group II introns through the formation of D-loops with interstitial copies of the introns—leading to the linearization of the genome, and (step iii) the use of D-loop mediated recombinational telomere extension to replenish telomere loss [5]. This ancient system eventually gave rise to the present-day system (Fig. 3, step iv) through two major developments: the emergence of the telomerase that synthesizes short telomere repeats, and the elaboration of proteins capable of recognizing and protecting these short repeats. This model is consistent with other compelling propositions on eukaryotic genome evolution. For example, the invasion of group II intron into the genome of the early eukaryotes can explain the prevalence of introns and the similarities between the group II splicing and the mRNA splicing machinery [6, 40]. In addition, it is entirely plausible to imagine an evolutionary path from group II intron to telomerase; phylogenetic analysis indicates that the catalytic subunit of telomerase (TERT) most likely shares a common ancestor with the Penelope-like elements (PLE) of retrotransposons [41, 42], which in turn exhibit some similarities to group II introns. Moreover, the D-loop mediated recombinational extension mechanism proposed for the ancient telomeres shares mechanistic similarities with ALT, the backup pathway for present-day telomeres. Both pathways are initiated by the invasion of DNA 3’ ends into homologous sites followed by 3’ end extension and strand displacement. Thus, it could be argued that ALT recapitulates features of ancient telomere maintenance. The group II intron hypothesis is far from the only possible explanation for the origin of linear genomes. Indeed bacteria are known to harbor linear plasmids and chromosomes and it has been suggested that insertion of a circular chromosome into a linear retroplasmid or a linear bacterial chromosome could have given rise to the primordial eukaryotic chromosomes [43].
Figure 3. A model for the origin and evolution of telomeres.
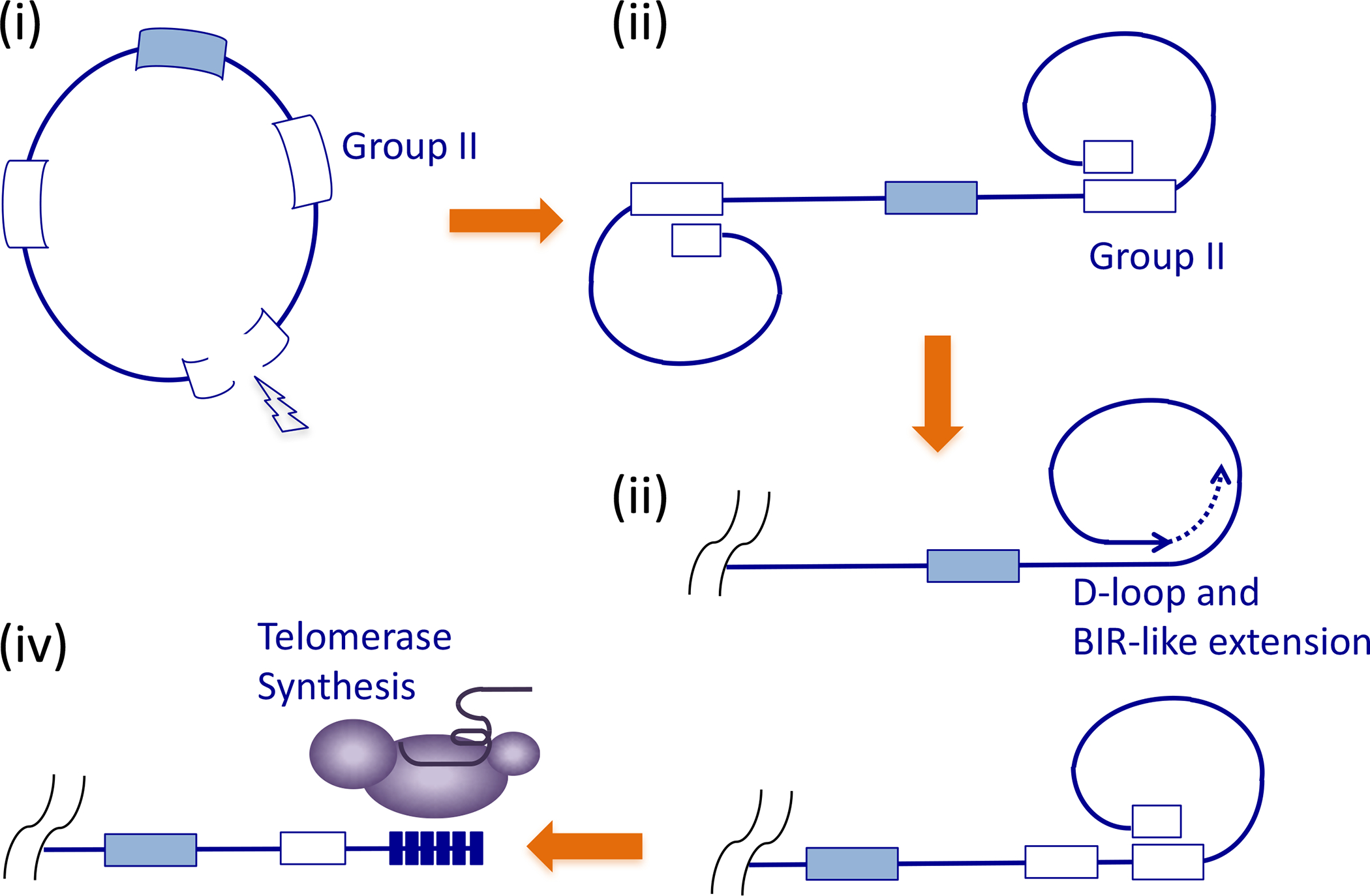
The model postulates the invasion of group II introns into the genome of the ancestral eukaryotes (i). Double strand breaks in one the introns may lead the formation of D-loops between the two broken ends and two interstitial introns, resulting in the linearization of the genome (ii). In this ancestral state, terminal DNA loss may be compensated by the periodic activation of D-loop mediated, BIR-like, telomere extension (iii). At some point, the ancient group II intron reverse transcriptase or a relative may have acquired the ability to utilize an unlinked RNA template and to synthesize the short telomere repeat sequence (iv).
Adding C-strand synthesis to the mix
It is notable that in the discussion of the ancestral telomere system, as well as that of the transition from the ancestral to modern system, the mechanism that mediates the synthesis of the C-strand, or the strand that contains the 5’ end, is largely ignored. This may be partly attributable to our incomplete understanding of BIR. While there is substantial evidence that following strand invasion, DNA polymerase δ is responsible for elongating the 3’ terminus, the priming and polymerization activities responsible for the synthesis of the complementary strand remain obscure. However, the balance of evidence suggests that all three replicative polymerases are involved [44], and the lack of an obvious alternative priming mechanism suggest that primase-Pol α may again be enlisted for this purpose. In this regard, it is instructive to consider the origin of primase-Pol α in the context of archaea/eukaryote evolution. Notably, while the primase subunits in these two domains of life are related, Pol α is exclusive to eukaryotes and organisms with telomeres [45]. In contrast, Pol δ, another replicative DNA polymerase, exhibits substantial similarities to an archaeal Pol B and is most likely derived from an ancestral protein in LAECA (last archaeal eukaryal common ancestor) [46, 47]. Forterre has proposed that Pol α and other eukaryote-specific proteins to which it binds (including CST subunits) may be introduced in a proto-eukaryotic lineage at the same time as telomeres [45]. Since unlike telomerase, primase-Pol α is promiscuous in regard to the copying of any DNA sequence, it is well suited to carrying out C-strand synthesis in both the ancient, D-loop-based system of telomere elongation and the modern telomerase-based system. Moreover, because Pol α probably originated as a viral protein [46], it is tempting to suggest that the ancestral eukaryotes may have co-opted this polymerase to solve the problem of telomere C-strand synthesis. In this view, then, primase-Pol α predates telomerase at telomeres and must have successfully negotiated the transition from the ancient to the modern telomere system.
The Pol α-first proposition in turn has significant implications for the origin of the CST and POT-TPP1 complex. First, since CST is closely connected to Pol α, this ssDNA-binding complex may have arrived at telomeres concurrent with or shortly after Pol α. CST is likely to share a common origin with RPA, the major ssDNA-binding complex in eukaryotes—crystallographic analysis revealed a high degree of structural similarity between the two smaller subunits of each complex (i.e., STN1~RPA2 and TEN1~RPA3) [48]. CST is thus paralogous to RPA, but unlike RPA, which has archaeal homologs [49], is exclusively found in eukaryotes. CST subunits are also arguably the most widely conserved telomere proteins, being present even in Drosophila (see below), which has lost the canonical modern telomere sequence [50]. It is thus reasonable to postulate an ancient origin for this ssDNA-binding complex, through the duplication of the RPA genes and neofunctionalization. In accordance with the need of the ancient CST to interact with group II intron or related sequences, the present-day CST complexes do not exhibit strong recognition specificity for telomere repeats per se – but they do show a preference for G-rich sequences [23, 51]. Several CST complexes have also been shown to exhibit low affinity or unstable binding to non-telomeric and non-G-rich sequences [52, 53]. This relatively broad DNA-binding specificity is compatible with the finding that in mammals, CST has a genome-wide function in helping cells overcome replication stress [22, 23]. Whether this genome-wide function emerged prior to or subsequent to CST’s telomere function is difficult to resolve.
What about PT and the rest of the shelterin complex? These factors have clearly evolved to protect the modern telomeres and regulate telomerase. Both TRF1/2 and POT1-TPP1 exhibit rather strict sequence preferences for the telomere repeats; even single base substitutions can dramatically reduce binding affinity of these ds and ss telomere binding proteins [54, 55]. It is thus more plausible to envision the arrival of PT and shelterin after the transition to the modern telomere sequence. In other words, the timeline of telomere arrival is likely to be the following: primase-Pol α/CST → telomerase → shelterin. This in turn raises interesting questions concerning the evolution of telomerase regulatory mechanisms. For example, in the pre-shelterin era, could telomerase recruitment or activity be regulated by any telomere components? In addition, why did shelterin use PT rather than other subunits as the main control switch for telomerase activity? Below I argue based on structural and function considerations that CST may have served the telomerase regulatory function in the pre-shelterin era, and CST may have given rise to the PT module of shelterin through gene duplication and functional specialization.
CST and PT as ancient paralogs: evidence and evolutionary implications
The evolutionary relationship between CST and PT has been controversial. CST was initially believed to be confined to budding yeast, which lacks shelterin. In budding yeast, the largest subunit of CST is replaced by Cdc13, which has a different structural organization from that of CTC1 [12]. Like shelterin, the budding yeast CST is a key mediator of telomere protection. It was thus suggested that Cdc13 and POT1 are perhaps orthologues, given their shared function in telomere protection and utilization of OB folds for ssDNA-binding. However, the subsequent discovery of CST subunits in most organisms that harbor shelterin made this idea untenable [11]. Instead, the two complexes are now generally thought to have independent origins.
Some recent experimental and conceptual developments, however, make it worthwhile to revisit the idea of an evolutionary connection between CST and PT. First, in silico analysis of CTC1 uncovered significant structural similarities between it and both the POT1 and RPA1 family members (Box 1). Second, recent functional studies of STN1 suggests unexpected similarities between it and TPP1 [20]. Both STN1 and TPP1 consist of an N-terminal OB fold followed by a non-OB domain (winged-helix for STN1 and undetermined for TPP1). Remarkably, STN1 was shown to utilize its N-terminal OB fold to bind and stimulate primase-Pol α activity, much as TPP1 uses its N-terminal OB fold to bind and stimulate telomerase activity. Thus, there are structural as well as functional similarities between the two large subunits of CST and the PT complex.
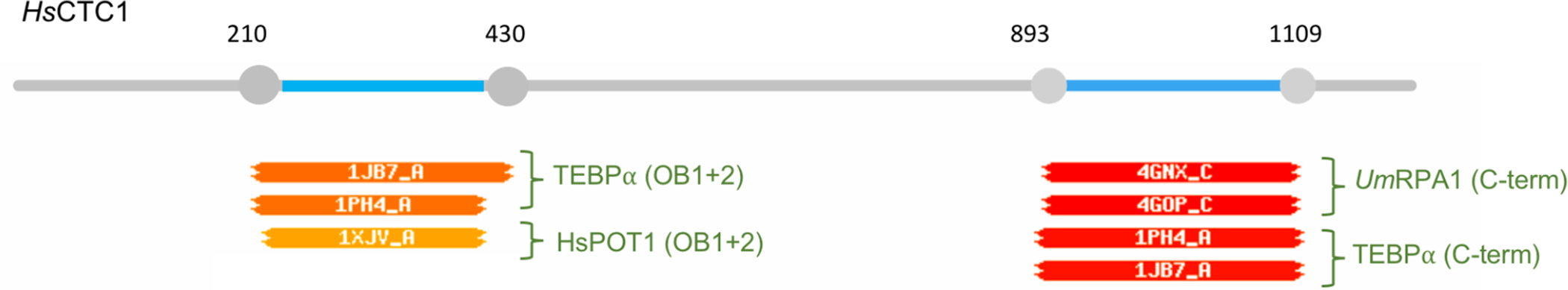
Text Box 1: Relationships between CTC1, POT1 and RPA1.
HHPred analysis of human CTC1 suggests homologies to both RPA1 and POT1. When CTC1 was used as the query against the PDB_mmCIF70 database, two of its regions were predicted to resemble structures in the database (Fig. I). The two highest scoring hits for amino acids 210–430 are the first two OB folds of TEBPα and human POT1, whereas the two highest scoring hits for amino acids 890–1100 are the C-terminus of U. maydis RPA1 and that of TEBPα, respectively. All matches have probability values of greater than 90%. These findings not only reinforce the long-postulated kinship between RPA1 and CTC1, but also suggest a common ancestry for CTC1 and POT1.
Fig. I.
Given the similarities between CST and PT, as well as their co-existence in individual organisms, it is tempting to hypothesize a paralogous relationship for these complexes. If one accepts this hypothesis, as well as the hierarchical sequence of protein arrival at telomeres sketched above, then the following scenario for telomere evolution can be envisaged (Fig. 4, Key Figure). At the primordial, group II intron-capped chromosome ends, telomeres are replenished by a BIR-like, intra-chromosomal D-loop mediated elongation mechanism in which the 3’ end strand is lengthened by Pol δ. Shortly thereafter, Pol α was enlisted from a virus to facilitate the synthesis of the 5’ telomere strand, and the lack of pre-existing primers for this strand necessitated a strong and stable interaction between Pol α and primase. In addition, duplication of the ancient RPA and functional specialization of the new paralog resulted in a telomere-specific RPA (i.e., CST) that regulates primase-Pol α. When telomerase first emerged to add short repeats to telomere 3’ end (before the evolution of the shelterin complex), CST is well positioned to regulate this new polymerase at telomeres (and to fulfill its original function of controlling primase-Pol α). In fact, If CST were to use the same protein surface to bind both telomerase and primase-Pol α, this could provide a means for switching from G- and C-strand synthesis, thereby ensuring that the de novo synthesized DNA is mostly duplex. However, because telomerase and primase-Pol α are structurally quite distinct, it may be difficult to achieve optimal regulations of both polymerases using a single complex. As the cells enlist new proteins to coat and protect the short telomere repeat sequence, the ancient CST may have given birth to PT (while losing the TEN1-equivalent subunit in the process). This then allows for subfunctionalization of the two ssDNA binding complexes for dedicated promotion of C- and G-strand synthesis. One significant advantage of this model is that it allows for step-wise, gradual evolution of the G- and C-strand maintenance machinery. Another advantage is that it accounts for the apparently more general role of CST in promoting both telomerase- and recombination-based telomere maintenance. As noted above, CST (but not PT) has been implicated in ALT, which exhibits similarities to the proposed BIR-based telomere maintenance mechanism at ancient telomeres; the role of CST in ALT may thus be due to the retention of an ancient function.
Figure 4 (Key Figure). An expanded, C-strand-centric model for the evolution of the telomere protection and maintenance machinery based on potential kinship between CST and PT.
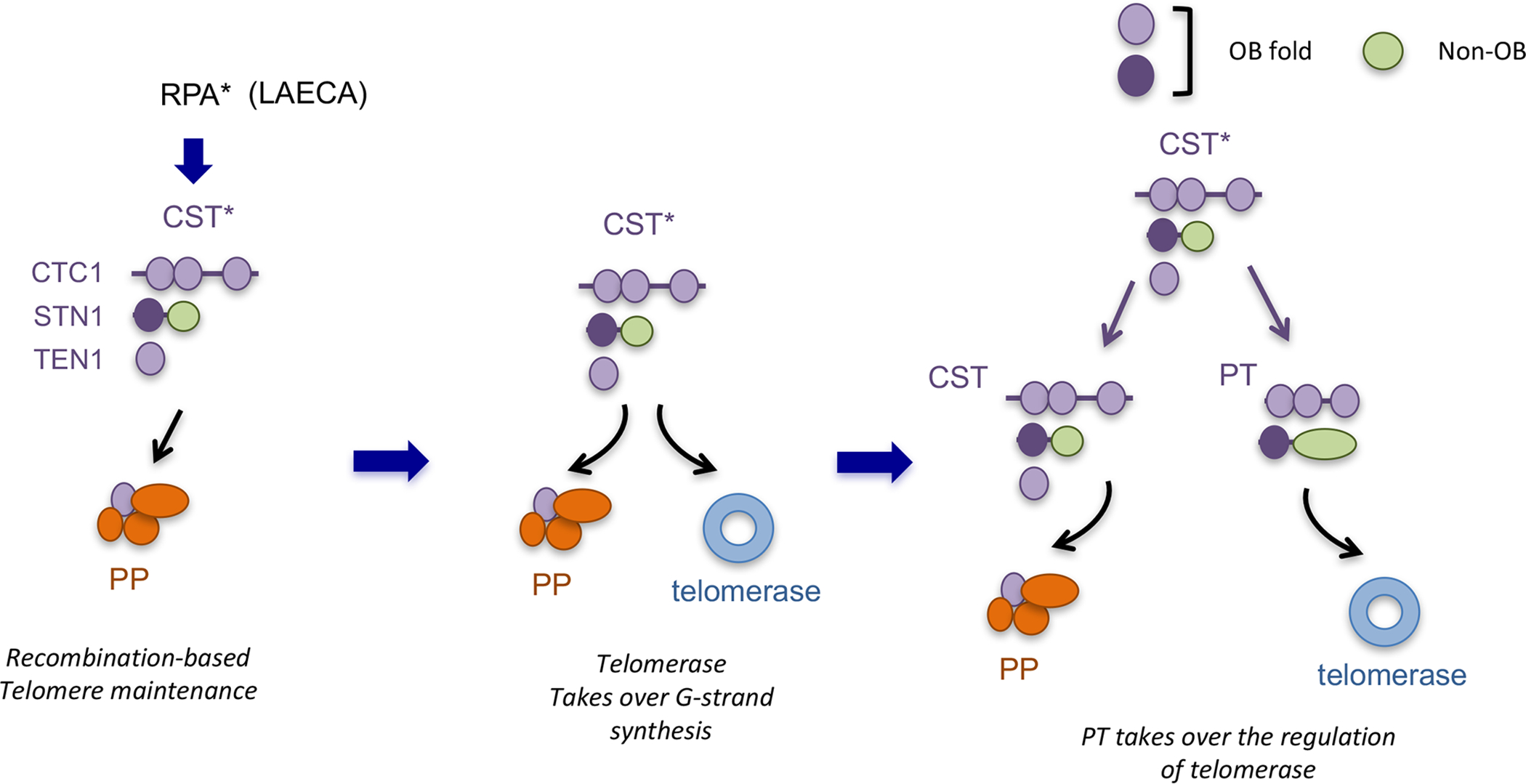
The following steps of telomere evolution are proposed. (i) In the proto-eukaryotic organism, the linearization of the genome by group II intron is initially accompanied by the D-loop mediated, BIR-like extension of the strand bearing the 3’ end. In addition, Pol α was enlisted from a virus to mediate C-strand synthesis, Concurrently, the archaeal-eukaryal RPA may have duplicated and given rise to the proto-CST complex (CST*) capable of regulating primase-Pol α. (ii) Subsequently, telomerase evolved from group II intron or a related reverse transcriptase to mediate the synthesis of short telomere repeats in cooperation with telomerase RNA. The proto-CST (CST*) has the necessary localization and DNA-binding property to also regulate telomerase at this stage of telomere evolution. (iii) Finally, the proto-CST underwent another round of duplication to give rise to PT–the proto-TEN1 was either not duplicated or lost during evolution. CST and PT underwent sub- and neofunctionalization such that PT acquired the ability to interact with other shelterin proteins and to specialize in the recruitment and activation of telomerase.
It is worth noting that once the main players of the telomere machinery have emerged, their physical proximity and functional connection would likely encourage the evolution of additional physical interactions. In the ciliate Tetrahymena, the telomerase holoenzyme evidently contains the CST complex, which suggests a potential mechanism for coordinating G- and C-strand synthesis [56]. Another interesting case is found in rodents, where POT1 has experienced gene duplication and functional specialization. While POT1a mediates telomere protection, POT1b helps to recruit CST, providing another potential linkage between the G- and C-strand maintenance machinery [57]. Similar interaction between human shelterin and CST has also been reported [58, 59].
Reversion back to the ancient ways?
The postulated history of telomeres and the evolutionary kinship between CST and PT allows one to re-conceptualize two exceptions to the general theme of telomere regulation sketched earlier. The first exception is found in the Saccharomycotina subphylum of budding yeast (Text Box 2) [12]. In this taxon, most of the shelterin components including POT1 and TPP1 have been lost, and the activities of both telomerase and primase-Pol α are regulated by the fungal CST complex. The budding yeast telomeres thus resemble an ancient state, before the arrival of the shelterin complex. The second exception is found in Drosophila and represents an even more radical departure from the standard system (Text Box 3). In Drosophila, telomeres are capped by retrotransposons, and few of the telomere proteins share any similarity with shelterin subunits [60, 61]. Nevertheless, recent studies suggest that the telomere retrotransposons are regulated by a Drosophila CST orthologue named MTV [50]. The Drosophila telomere system is thus akin to a reversion to an even more ancient state, before the invention of telomerase.
Text Box 2: The atypical telomere machinery in budding yeast.
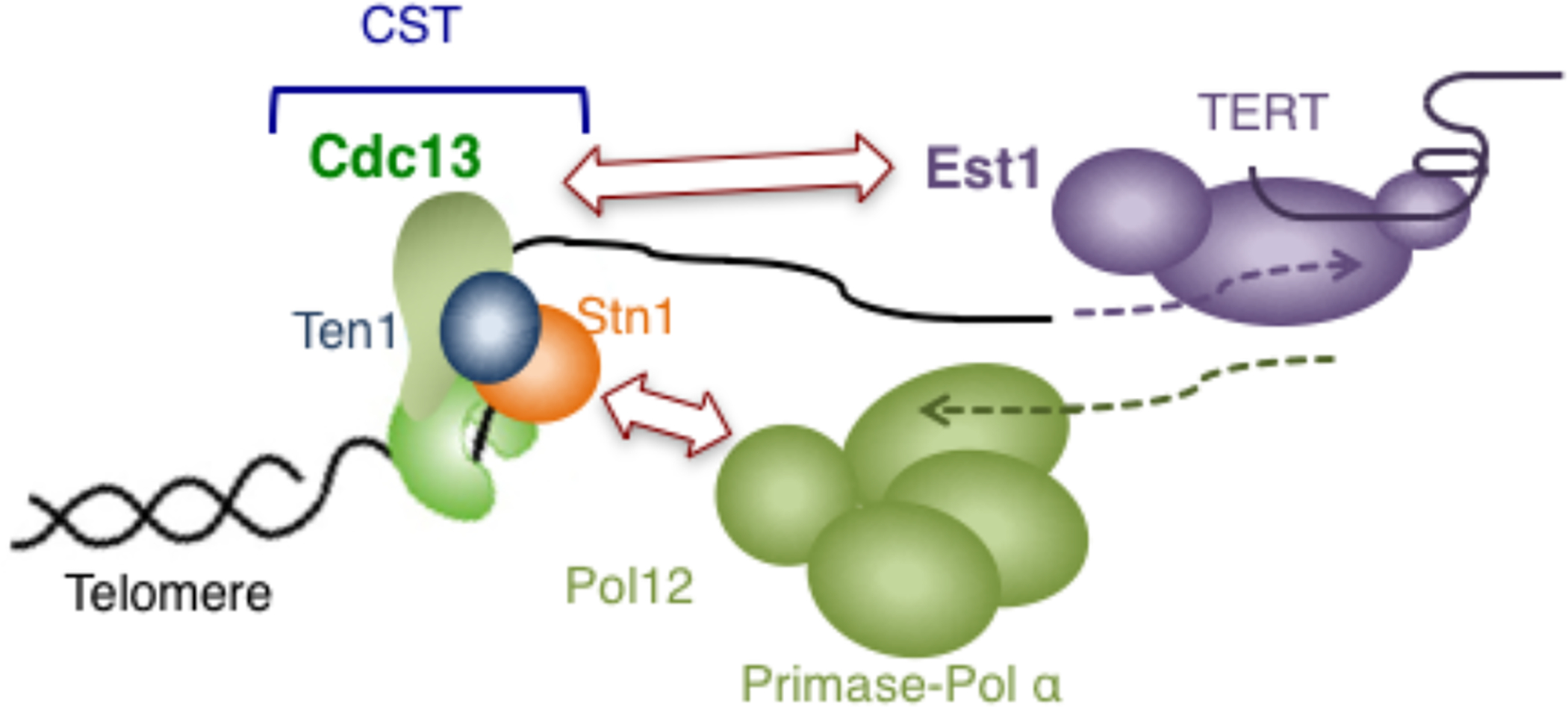
In the yeasts that belong to the Saccharomycotina subphylum, even though the C-strand synthesis mechanism evidently conforms to the general theme, the G-strand synthesis machine, or telomerase, is regulated differently, owing to the absence of PT as well as most other components of shelterin [12]. Instead of relying on TPP1-TERT interaction, S. cerevisiae utilizes two alternative telomere and telomerase components to mediate telomerase recruitment, namely Cdc13, the CTC1 equivalent in the fungal CST complex, and Est1, a fungi-specific telomerase component [65] (Fig. II). Curiously, budding yeast contains a miniaturized TPP1 named Est3 that consists solely of an OB fold domain [66, 67]. Est3, instead of being part of the telomere nucleoprotein complex, is a telomerase component that is essential for telomere maintenance in vivo and optimal telomerase activity in vitro. The “migration” of Est3 from telomeres to telomerase as well as the loss of shelterin in budding yeast may be related to the other distinguishing feature of telomeres in this subphylum: the extraordinarily variable telomere repeat sequences. Whereas telomere repeats in most lineages are short (i.e., 8 bp or shorter), regular, and relatively stable, those in Saccharomycotina can be long (i.e., as long as 25 bp), irregular, and different between closely related species. An evolutionary hypothesis that invokes alterations in telomere /telomerase RNA template sequence, as well as the failure of shelterin to evolve the requisite, alternative sequence specificity for binding telomeres, has been proposed to account for the deviations of Saccharomycotina telomeres [12]. It is worth noting that upon the loss of PT and shelterin in this fungal lineage, the task of controlling telomerase was taken up by CST, the C-strand synthesis regulator. Given that the CST complex was already present at telomeres and has a more flexible sequence recognition property, it was perhaps more facile for budding yeast to enlist this ancient paralog to serve the purpose of telomere protection and telomerase regulation in lieu of PT. The budding yeast telomere system, in which the CST complex regulates both telomerase and primase-Pol α, is thus akin to a reversion back to an ancient state, before the arrival of the shelterin complex. It is worth noting that an alternative interpretation of the close link between CST and telomerase in budding yeast invokes regulation of telomerase by the telomere replication complex, of which CST is a component [68].
Fig. II.
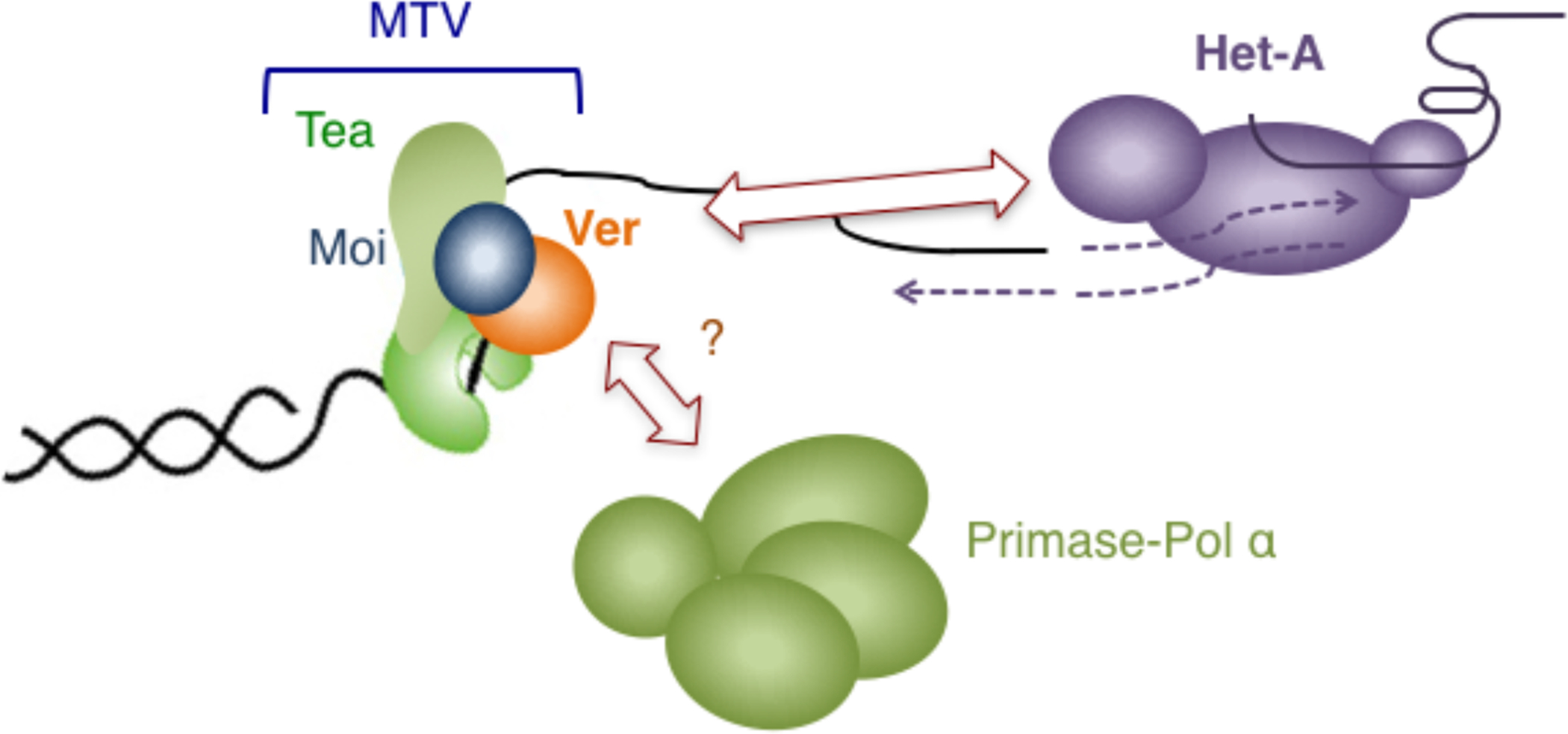
Text Box 3: The atypical telomere machinery in Drosophila.
In Drosophila, key elements of the standard telomere system are lost, including the short telomere repeat unit, the shelterin complex, and telomerase (Fig. III). Instead, the Drosophila chromosome ends are capped by end-specific non-LTR retrotransposons such as Het-A and TART, and the maintenance of these sequences by the corresponding retrotransposition machinery [60]. Given the lack of simple sequence repeats at chromosome ends, the Drosophila telomere capping complex proteins (collectively named terminin) must recognize and bind telomere DNA in a non-sequence-specific manner, and indeed no component of terminin (e.g., HOAP, HipHop) appears to be related to any shelterin protein [61]. Notably, however, a CST-like complex named MTV (Moi-Tea-Ver) was recently characterized and shown to be essential for telomere stability and to bind ssDNA (single-stranded DNA) in a non-sequence-specific manner [50]. Moreover, MTV appears to promote the formation or localization of retrotransposon RNP at telomeres, thereby playing a critical role in telomere maintenance [69]. Even though the mechanisms of Het-A and TART retrotransposition have not been subjected to detailed investigation, in the case of similar non-LTR retrotransposons, the encoded reverse transcriptase is evidently responsible for synthesizing both strands of DNA that is inserted into the target site. Hence one may view the retrotransposon RNP as carrying out the task of both telomerase and primase-Pol α in telomere maintenance. By this argument, Drosophila, like budding yeast, evidently use CST/MTV to control the synthesis of both strands of telomeres. MTV also mediates functions in telomere protection; depleting or disrupting MTV subunits caused telomere fusions or telomere DNA damage response [50, 70, 71]. Whether primase-Pol α plays a role in telomere maintenance in Drosophila and whether MTV also regulates this polymerase is unclear. It may be speculated that the CST complex in the ancestor of Drosophila, because of its affinity for telomeres and its flexible DNA binding properties, was adept at being modified (into MTV) to serve critical functions at the radically altered, retrotransposon-based Drosophila telomeres. The Drosophila telomere system is thus akin to a reversion to an even more ancient state, before the invention of telomerase-mediated telomere maintenance.
Fig. III.
It is important to not take the notion of reversion literally—evolution moves only forward in time. The larger lesson from these two exceptional cases is instead that the loss of one paralog (i.e., PT) may be more easily accommodated through the re-acquisition of a lost property by the retained paralog (i.e., CST). Clearly, more studies will be necessary to assess the validity of this hypothesis. It will also be interesting in the future to explore similar cases of compensating for the loss of one paralog by modifying the other.
Conclusions and speculations
An overarching theme that emerges from the current discussion of telomere machinery is the power of mobile genetic elements and gene duplication in shaping the genome. Indeed, there are other interesting illustrations of these themes (e.g., plant telomerase RNA [62] and fungal Cdc13 paralogs [63, 64]) that could not be covered in this article. While the evolutionary events proposed herein occurred quite far in the past, the model is not entirely refractory to experimental interrogation. For example, the postulated evolutionary relationship between CTC1 and POT1 may be investigated by structural and biochemical analysis of CTC1. Additional studies of TPP1 C-terminus structure could also reveal unexpected similarity to STN1. Moreover, the hypothesized scenario may be bolstered by resemblances between the TPP1-TERT and STN1-primase-Pol α interfaces, which have not been determined. Beyond these telomere-related issues, the notion that the Pol α was originally enlisted to solve the C-strand synthesis problem could also have implications for the evolution of the chromosomal replication machinery. For example, the low fidelity of Pol α and the lack of an associated proof-reading nuclease could be less problematic if the initial function of this polymerase was confined to telomeres where the precise sequence is less critical. When Pol α was enlisted to synthesize primers at replication origins and to initiate Okazaki fragments, it may have been necessary then to devise an Okazaki fragment maturation system that largely eliminates the DNA segments produced by Pol α [45]. Putting Pol α and CST at the inception of linear chromosomes and telomeres may thus help to explain current features of genome maintenance.
Highlights.
When linear chromosomes first emerged in eukaryotic genome evolution, they may have been capped by group II introns and replenished through a recombination pathway. Through a series of transitions and evolutionary inventions, this ancient telomere system was replaced by the modern system comprising short repetitive sequences, protective proteins, and dedicated telomere-synthesis machinery.
The two key complexes responsible for telomere C-strand synthesis (i.e., CST and primase-Pol α) are both unique to eukaryotes and may have evolved or been enlisted early to promote telomere maintenance. The biochemical properties of CST and primase-Pol α make them suitable for acting in both the ancient and modern telomere system.
Structural and functional comparisons suggest that CST may have evolved from an archaeal-eukaryal RPA complex, and may have in turn given birth to the POT1-TPP1 complex that regulates telomerase-mediated G-strand synthesis.
By placing CST and primase-Pol α near the origin of telomeres, prior to the emergence of telomerase, one can envision a step-wise, hierarchical model of telomere evolution involving gradual replacement of elements of the ancient system. This model can also better rationalize atypical telomere systems found in selected budding yeast and insects.
Outstanding questions.
Does CTC1 share sufficient structural similarities to POT1 to support a common evolutionary origin for these two families of proteins?
Can further exploration of CST and POT1-TPP1 in various organisms (e.g., in deep branches of eukaryotes) reveal greater structural and functional similarities that reinforce a paralogous relationship between these two ssDNA-binding complexes?
Can a better understanding of Drosophila telomere maintenance mechanisms, in particular the regulation of telomere-specific retrotransposition by the MTV complex, uncover hidden similarities between the regulation of telomere G-and C-strand synthesis?
Do CST orthologues in different taxa contribute differently to their telomere-specific and genome-wide replication functions? Could these differences provide insights on the origin and evolution of this complex?
Glossary
- Shelterin
A widely conserved complex that protects chromosome ends and regulates telomere DNA synthesis. It contains proteins that bind duplex telomere repeats, single strand telomeres, as well as bridging proteins.
- Primase-Pol α
A bi-functional polymerase that plays a critical role in chromosome replication as well as telomere C-strand synthesis. Pol α is specific to eukaryotes, and the only replicative DNA polymerase capable of initiating DNA synthesis owing to its association with primase.
- Telomerase
A special reverse transcriptase responsible for lengthening the G-strand of telomeres to compensate for incomplete end replication. It does so through reverse transcription of an integral RNA component that specifies the synthesis of telomere G-strand, and it likely shares common ancestry with the Penelope-like element (PLE) of retrotransposon.
- CST (CTC1-STN1-TEN)
An RPA (RPA1-RPA2-RPA3)-like ssDNA-binding complex with a preference for G-rich sequences, including but not limited to telomere repeats. CST mediates multiple and variable functions in telomere maintenance and genome-wide replication. One of its most conserved function is to promote telomere C-strand synthesis by simulating the activity of primase-Pol α.
- POT1-TPP1
Two components of the shelterin complex; together this heterodimer recognizes the telomere G-strand with high affinity and sequence-specificity. In addition to protecting telomeres, POT1-TPP1 plays a key role in regulating telomerase-mediated G-strand elongation.
- ALT (alternative lengthening of telomere)
A recombination-based pathway for telomere maintenance that has been described in various telomerase-negative cells. It shares mechanistic similarities with break-induced replication (e.g., the requirement for Pol δ).
- Group II Intron
A mobile genetic element that propagates by reverse splicing. The invasion of group II intron into the ancestral eukaryotic genome is believed to account for the origin of introns. The group II intron reverse transcriptase also exhibits similarities to telomerase reverse transcriptase.
References
- 1.de Lange T (2009) How telomeres solve the end-protection problem. Science 326 (5955), 948–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olovnikov A (1973) A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J Theor Biol 41 (1), p181–90. [DOI] [PubMed] [Google Scholar]
- 3.Autexier C and Lue NF (2006) The Structure And Function Of Telomerase Reverse Transcriptase. Annu Rev Biochem 75, 493–517. [DOI] [PubMed] [Google Scholar]
- 4.Wu RA et al. (2017) Telomerase Mechanism of Telomere Synthesis. Annu Rev Biochem 86, 439–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Lange T (2015) A loopy view of telomere evolution. Front Genet 6, 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koonin EV (2006) The origin of introns and their role in eukaryogenesis: a compromise solution to the introns-early versus introns-late debate? Biol Direct 1, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koonin EV (2016) Viruses and mobile elements as drivers of evolutionary transitions. Philos Trans R Soc Lond B Biol Sci 371 (1701). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burki F (2014) The eukaryotic tree of life from a global phylogenomic perspective. Cold Spring Harb Perspect Biol 6 (5), a016147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jain D and Cooper JP (2011) Telomeric strategies: means to an end. Annu Rev Genet 44, 243–69. [DOI] [PubMed] [Google Scholar]
- 10.Giraud-Panis MJ et al. (2010) CST meets shelterin to keep telomeres in check. Mol Cell 39 (5), 665–76. [DOI] [PubMed] [Google Scholar]
- 11.Price CM et al. (2010) Evolution of CST function in telomere maintenance. Cell Cycle 9 (16), 3157–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lue NF (2010) Plasticity of telomere maintenance mechanisms in yeast. Trends Biochem Sci 35 (1), 8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nandakumar J and Cech TR (2013) Finding the end: recruitment of telomerase to telomeres. Nat Rev Mol Cell Biol 14 (2), 69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hockemeyer D and Collins K (2015) Control of telomerase action at human telomeres. Nat Struct Mol Biol 22 (11), 848–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baranovskiy AG and Tahirov TH (2017) Elaborated Action of the Human Primosome. Genes (Basel) 8 (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pellegrini L (2012) The Pol alpha-primase complex. Subcell Biochem 62, 157–69. [DOI] [PubMed] [Google Scholar]
- 17.Lue NF et al. (2014) The CDC13-STN1-TEN1 complex stimulates Pol alpha activity by promoting RNA priming and primase-to-polymerase switch. Nat Commun 5, 5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chow TT et al. (2012) Early and late steps in telomere overhang processing in normal human cells: the position of the final RNA primer drives telomere shortening. Genes Dev 26 (11), 1167–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng X et al. (2017) CTC1-mediated C-strand fill-in is an essential step in telomere length maintenance. Nucleic Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ganduri S and Lue NF (2017) STN1-POLA2 interaction provides a basis for primase-pol alpha stimulation by human STN1. Nucleic Acids Res 45 (16), 9455–9466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bell SP and Dutta A (2002) DNA replication in eukaryotic cells. Annu Rev Biochem 71, 333–74. [DOI] [PubMed] [Google Scholar]
- 22.Stewart JA et al. (2012) Human CST promotes telomere duplex replication and general replication restart after fork stalling. EMBO J 31 (17), 3537–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chastain M et al. (2016) Human CST Facilitates Genome-wide RAD51 Recruitment to GC-Rich Repetitive Sequences in Response to Replication Stress. Cell Rep 16 (5), 1300–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Froelich-Ammon SJ et al. (1998) Modulation of telomerase activity by telomere DNA-binding proteins in Oxytricha. Genes Dev 12 (10), 1504–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paeschke K et al. (2005) Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat Struct Mol Biol 12 (10), 847–54. [DOI] [PubMed] [Google Scholar]
- 26.Armstrong CA et al. (2014) Telomerase activation after recruitment in fission yeast. Curr Biol 24 (17), 2006–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Derboven E et al. (2014) Role of STN1 and DNA polymerase alpha in telomere stability and genome-wide replication in Arabidopsis. PLoS Genet 10 (10), e1004682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grossi S et al. (2004) Pol12, the B subunit of DNA polymerase alpha, functions in both telomere capping and length regulation. Genes Dev 18 (9), 992–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao H et al. (2007) RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol 14 (3), 208–14. [DOI] [PubMed] [Google Scholar]
- 30.Lue NF and Yu EY (2017) Telomere recombination pathways: tales of several unhappy marriages. Curr Genet 63 (3), 401–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lundblad V (2002) Telomere maintenance without telomerase. Oncogene 21 (4), 522–31. [DOI] [PubMed] [Google Scholar]
- 32.Pickett HA and Reddel RR (2015) Molecular mechanisms of activity and derepression of alternative lengthening of telomeres. Nat Struct Mol Biol 22 (11), 875–80. [DOI] [PubMed] [Google Scholar]
- 33.Dilley RL et al. (2016) Break-induced telomere synthesis underlies alternative telomere maintenance. Nature 539 (7627), 54–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roumelioti FM et al. (2016) Alternative lengthening of human telomeres is a conservative DNA replication process with features of break-induced replication. EMBO Rep 17 (12), 1731–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lydeard JR et al. (2010) Sgs1 and exo1 redundantly inhibit break-induced replication and de novo telomere addition at broken chromosome ends. PLoS Genet 6 (5), e1000973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sobinoff AP et al. (2017) BLM and SLX4 play opposing roles in recombination-dependent replication at human telomeres. EMBO J 36 (19), 2907–2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson MA et al. (2013) Pif1 helicase and Poldelta promote recombination-coupled DNA synthesis via bubble migration. Nature 502 (7471), 393–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cesare AJ and Reddel RR (2010) Alternative lengthening of telomeres: models, mechanisms and implications. Nat Rev Genet 11 (5), 319–30. [DOI] [PubMed] [Google Scholar]
- 39.Huang C et al. (2017) The human CTC1/STN1/TEN1 complex regulates telomere maintenance in ALT cancer cells. Exp Cell Res 355 (2), 95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lambowitz AM and Belfort M (2015) Mobile Bacterial Group II Introns at the Crux of Eukaryotic Evolution. Microbiol Spectr 3 (1), MDNA3-0050-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gladyshev EA and Arkhipova IR (2011) A widespread class of reverse transcriptase-related cellular genes. Proc Natl Acad Sci U S A 108 (51), 20311–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arkhipova IR et al. (2003) Retroelements containing introns in diverse invertebrate taxa. Nat Genet 33 (2), 123–4. [DOI] [PubMed] [Google Scholar]
- 43.Arkhipova IR (2012) Telomerase, retrotransposons, and evolution In Telomerases-chemistry, biology, and clinical applications (Lue NF and Autexier C eds), pp. 265–299, John Wiley and Sons. [Google Scholar]
- 44.Lydeard JR et al. (2010) Break-induced replication requires all essential DNA replication factors except those specific for pre-RC assembly. Genes Dev 24 (11), 1133–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Forterre P (2013) Why are there so many diverse replication machineries? J Mol Biol 425 (23), 4714–26. [DOI] [PubMed] [Google Scholar]
- 46.Filee J et al. (2002) Evolution of DNA polymerase families: evidences for multiple gene exchange between cellular and viral proteins. J Mol Evol 54 (6), 763–73. [DOI] [PubMed] [Google Scholar]
- 47.Villarreal LP and DeFilippis VR (2000) A hypothesis for DNA viruses as the origin of eukaryotic replication proteins. J Virol 74 (15), 7079–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun J et al. (2009) Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres. Genes Dev 23 (24), 2900–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chedin F et al. (1998) Novel homologs of replication protein A in archaea: implications for the evolution of ssDNA-binding proteins. Trends Biochem Sci 23 (8), 273–7. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Y et al. (2016) MTV, an ssDNA Protecting Complex Essential for Transposon-Based Telomere Maintenance in Drosophila. PLoS Genet 12 (11), e1006435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hom RA and Wuttke DS (2017) Human CST Prefers G-Rich but Not Necessarily Telomeric Sequences. Biochemistry 56 (32), 4210–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lue NF et al. (2013) The telomere capping complex CST has an unusual stoichiometry, makes multipartite interaction with G-Tails, and unfolds higher-order G-tail structures. PLoS Genet 9 (1), e1003145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miyake Y et al. (2009) RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol Cell 36 (2), 193–206. [DOI] [PubMed] [Google Scholar]
- 54.Croy JE and Wuttke DS (2006) Themes in ssDNA recognition by telomere-end protection proteins. Trends Biochem Sci 31 (9), 516–25. [DOI] [PubMed] [Google Scholar]
- 55.Court R et al. (2005) How the human telomeric proteins TRF1 and TRF2 recognize telomeric DNA: a view from high-resolution crystal structures. EMBO Rep 6 (1), 39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang J et al. (2015) Structure of Tetrahymena telomerase reveals previously unknown subunits, functions, and interactions. Science 350 (6260), aab4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu P et al. (2012) Telomeric 3’ overhangs derive from resection by Exo1 and Apollo and fill-in by POT1b-associated CST. Cell 150 (1), 39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wan M et al. (2009) OB fold-containing protein 1 (OBFC1), a human homolog of yeast Stn1, associates with TPP1 and is implicated in telomere length regulation. J Biol Chem 284 (39), 26725–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen LY et al. (2012) The human CST complex is a terminator of telomerase activity. Nature 488 (7412), 540–4. [DOI] [PubMed] [Google Scholar]
- 60.Pardue ML and DeBaryshe PG (2011) Retrotransposons that maintain chromosome ends. Proc Natl Acad Sci U S A 108 (51), 20317–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raffa GD et al. (2011) Terminin: a protein complex that mediates epigenetic maintenance of Drosophila telomeres. Nucleus 2 (5), 383–91. [DOI] [PubMed] [Google Scholar]
- 62.Xu H et al. (2015) A transposable element within the Non-canonical telomerase RNA of Arabidopsis thaliana modulates telomerase in response to DNA damage [corrected]. PLoS Genet 11 (6), e1005281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lue NF and Chan J (2013) Duplication and functional specialization of the telomere-capping protein Cdc13 in Candida species. J Biol Chem 288 (40), 29115–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Steinberg-Neifach O et al. (2015) Combinatorial recognition of a complex telomere repeat sequence by the Candida parapsilosis Cdc13AB heterodimer. Nucleic Acids Res 43 (4), 2164–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pennock E et al. (2001) Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104 (3), 387–96. [DOI] [PubMed] [Google Scholar]
- 66.Yu EY et al. (2008) A proposed OB-fold with a protein-interaction surface in Candida albicans telomerase protein Est3. Nat Struct Mol Biol 15 (9), 985–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee J et al. (2008) The Est3 protein associates with yeast telomerase through an OB-fold domain. Nat Struct Mol Biol 15 (9), 990–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Greider CW (2016) Regulating telomere length from the inside out: the replication fork model. Genes Dev 30 (13), 1483–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang L et al. (2014) Coordination of transposon expression with DNA replication in the targeting of telomeric retrotransposons in Drosophila. EMBO J 33 (10), 1148–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Raffa GD et al. (2010) Verrocchio, a Drosophila OB fold-containing protein, is a component of the terminin telomere-capping complex. Genes Dev 24 (15), 1596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cicconi A et al. (2017) The Drosophila telomere-capping protein Verrocchio binds single-stranded DNA and protects telomeres from DNA damage response. Nucleic Acids Res 45 (6), 3068–3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Baldauf SL (2003) The deep roots of eukaryotes. Science 300 (5626), 1703–6. [DOI] [PubMed] [Google Scholar]
- 73.Gu P and Chang S (2013) Functional characterization of human CTC1 mutations reveals novel mechanisms responsible for the pathogenesis of the telomere disease Coats plus. Aging Cell 12 (6), 1100–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen C et al. (2017) Structural insights into POT1-TPP1 interaction and POT1 C-terminal mutations in human cancer. Nat Commun 8, 14929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rice C et al. (2017) Structural and functional analysis of the human POT1-TPP1 telomeric complex. Nat Commun 8, 14928. [DOI] [PMC free article] [PubMed] [Google Scholar]


