Abstract
Nanocarrier-mediated protein delivery is a promising strategy for fundamental research and therapeutic applications. However, the efficacy of current platforms for delivery into cells is limited by endosomal entrapment of delivered protein cargo, with concomitantly inefficient access to the cytosol and other organelles, including the nucleus. We report here a robust, versatile polymeric-protein nanocomposite (PPNC) platform capable of efficient (≥90%) delivery of proteins to the cytosol. We synthesized a library of guanidinium-functionalized poly(oxanorborneneimide) (PONI) homopolymers with varying molecular weights to stabilize and deliver engineered proteins featuring terminal oligoglutamate “E-tags”. The polymers were screened for cytosolic delivery efficiency using imaging flow cytometry, with cytosolic delivery validated using confocal microscopy, and activity of the delivered proteins demonstrated through functional assays. These studies indicate that the PPNC platform provides highly effective and tunable cytosolic delivery over a wide range of formulations, making them robust agents for therapeutic protein delivery.
Keywords: protein delivery, polymer nanocarrier, direct cytosolic delivery, poly(oxanorbornene imide), protein engineering
Graphical Abstract
INTRODUCTION
Proteins play a critical role in many cellular processes, including cell signaling,1,2 metabolism,3,4 and development.5,6 As a result, many disease states are linked to protein dysfunction or low levels of functional protein in the cell.7,8,9 Several protein therapeutics have been approved by the FDA and utilized in clinical trials to combat diseases including cystic fibrosis,10 cancers,11 and hemophilia.12 Recombinant proteins have also gained traction in recent years for treating human disease at a genetic level through CRISPR-Cas9,13,14 zinc finger nucleases,15 and other nuclease-guided techniques.16 Recently, these approaches have shown efficacy for systemic administration in vivo, opening the door for a new range of therapies against human disease.17,18,19
The vast majority of potential applications for protein therapeutics require access to the cytosol, either for their activity or as a gateway to other organelles such as the nucleus.20,21 The first barrier is the cell membrane that is virtually impermeable to large, highly charged species like exogenous proteins.21 This barrier can be bypassed through use of carriers or tags that induce endosomal uptake. These strategies, however, lead to entrapment of payload in the endosomal/lysosomal pathway, with concomitantly low levels of active protein present in the cytosol.22,23,24,25 The development of robust carrier systems for direct cytosolic protein delivery that are effective under biologically-relevant conditions is critical for the advancement of protein-based therapeutics to in vitro, ex vivo, in vivo and clinical settings.26,27,28
Recent studies have shown that protein-nanoparticle co-engineering provides vectors that can deliver proteins directly into the cytosol.29 These systems use supramolecular assembly of oligo(glutamate)-tagged (E-tagged) proteins with arginine-functionalized gold nanoparticles to deliver a range of proteins in vitro.17,29,30 More recently, these vectors have been used to deliver CRISPR-Cas9 machinery to macrophages in vivo and edit them with a high degree of specificity.17 These self-assembled nanoscale platforms deliver protein cargo directly to the cytosol through a membrane fusion-type mechanism, avoiding issues associated with endosomal entrapment. The design, however, features two key limitations. First, clearance of the gold core from biological systems is quite slow, providing concerns over long-term effects.11 Second, the isotropic gold particles employed feature inherent design space limitations relative to soft materials that impose concomitant restrictions on self-assembly and hence function..31
Preorganization of charge on the nanoparticle surface was considered to be a key factor in enabling direct cytosolic delivery.30,32 We hypothesized that the ‘semi-arthritic’ backbone of poly(oxanorbornene)imide (PONI) polymers33 could replicate key functional aspects of the nanoparticle delivery systems and provide similar self-assembly and delivery capabilities while allowing for greater flexibility of design.34,35 We report here the direct cytosolic delivery of three E-tagged proteins across a wide range of working formulations using rationally designed cationic guanidinium-functionalized PONI homopolymers (Figure 1). These polymers self-assemble with E-tagged proteins to form discrete polymer-protein nanocomposites (PPNCs) that are stable under serum-containing conditions. The role of polymer length on overall delivery efficiency was established using flow cytometry of assemblies formed with E-tagged green fluorescent protein (GFP-E20), with the best results observed using 27K and 60K Mw polymers. The ability of the PPNCs to deliver GFP-E20 to the cytosol was quantified using imaging flow cytometry, where significantly higher (over 90%) efficiency was found using the 60K Mw PONI polymer. The generalizability of this approach was demonstrated by the delivery of multiple E-tagged proteins, ranging from a dimeric fluorescent protein (tdTomato-E10) and a catalytically active endonuclease enzyme (Cre recombinase. Cre-E10) which retained its nuclease capability through the delivery process.
Figure 1.
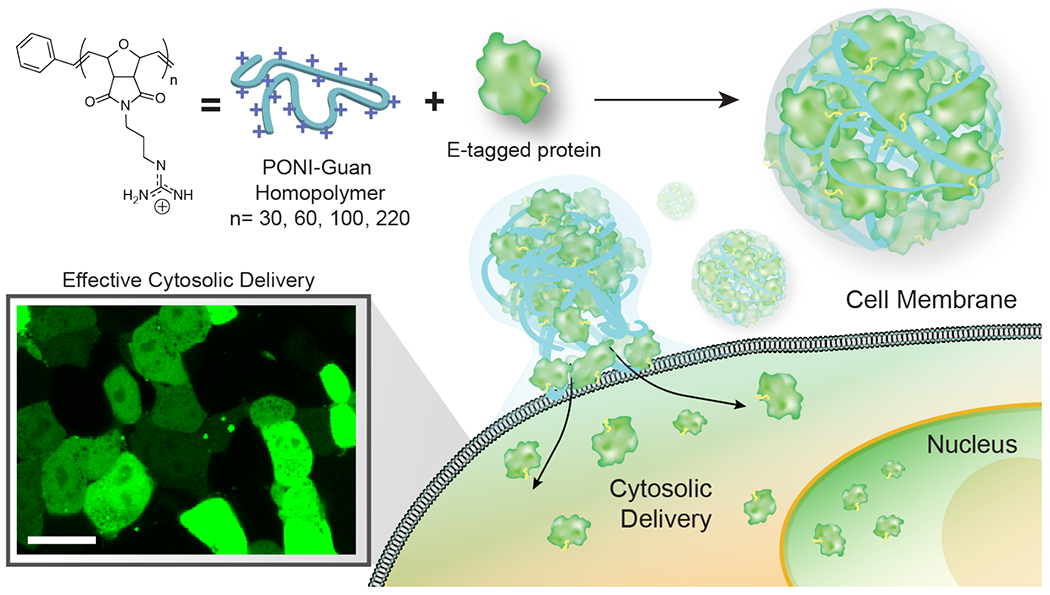
Schematic depiction of PPNCs formation between PONI polymers and E-tagged protein. PPNCs were generated through self-assembly between the two components, with competent PPNCs delivering protein cargo directly to the cytosol. Scale bars = 100 nm.
RESULTS AND DISCUSSION
Design and synthesis of polymers.
A library of guanidinium-functionalized homopolymers with varying molecular weight, ranging from ~8K to ~60K was prepared (Figure 2). Polymers were generated using ring-opening metathesis polymerization (ROMP).36 Norbornene/oxanorbornene-based polymers feature conformational restrictions reminiscent of the arranged surface of our previously reported arginine-functionalized gold nanoparticles.37,38,39 Guanidinium moieties were selected as monomer functional groups because they have been previously shown to interact with proteins without causing denaturation, and to promote direct cytosolic entry.30 PONI polymers were generated through polymerization of the protected monomer (Figure 2a) using 3rd generation Grubbs catalyst at room temperature in dichloromethane (CH2Cl2) (details of the synthetic procedure are provided in Supporting Information). The molecular weight and PDI of Boc-protected PONI were measured using gel permeation chromatography (GPC, polystyrene-calibrated) in tetrahydrofuran. Deprotection of Boc-protected PONI was performed in acidic condition to provide the PONI homopolymer, which is highly soluble in aqueous media. The purity of PONI was confirmed by proton nuclear magnetic resonance (1H-NMR). (Figure 2a and Supporting Figure S1–S8).
Figure 2.
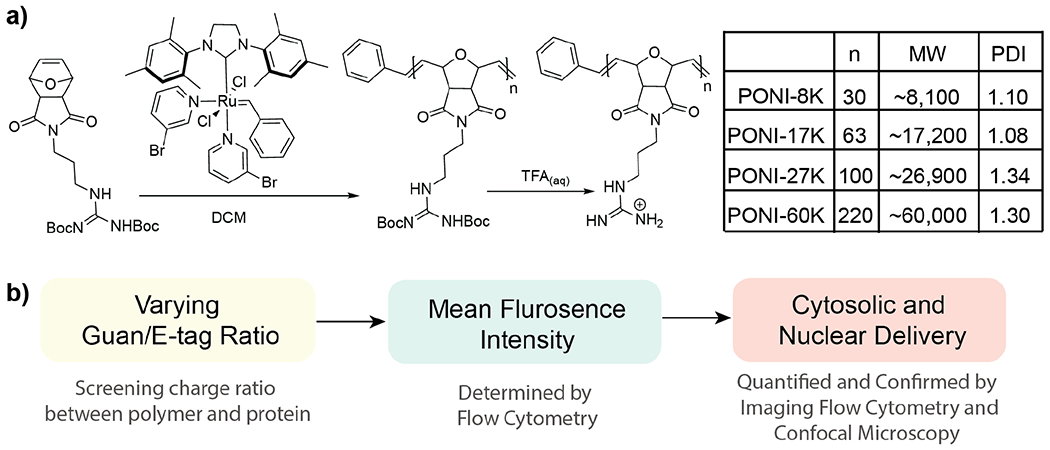
(a) PONI-Guan homopolymer library prepared with a series of molecular weights ranging from ~8K to ~60K, with accompanying PDI. Values were determined by GPC. (b) Experimental overview for determining delivery effectiveness. Total MFI values narrow the screening window for subsequent imaging flow cytometry. GFP delivery was evaluated through cytosolic distribution and nuclear localization.
Polymer-protein PPNC formulation and characterization.
A series of PONI-GFP self-assembled structures were prepared and characterized for size and stability. GFPs featuring a 20-mer glutamate tag (E20-tag) at the C-terminus were expressed as previously reported.29,30 Polymeric-protein nanocomposites (PPNCs) were then generated by mixing parametric ratios of GFP-E20 and PONI polymer, followed by incubation for 10 min at ambient temperature. Formulations are classified based on charge (Guan/E-tag) ratio of total polymeric guanidinium (Guan) to E-tag. The formation of discrete nanocomposites was observed using transmission electron microscopy (TEM, Figure 3a), with distribution quantified using dynamic light scattering (DLS) and characterized by zeta potential. (Figure 3b and Supporting Figures S11 and S16) Notably, when prepared in media containing 10% fetal bovine serum (FBS), PPNCs maintained a constant size without significant aggregation over a 24-hour period (Figure S12).
Figure 3.
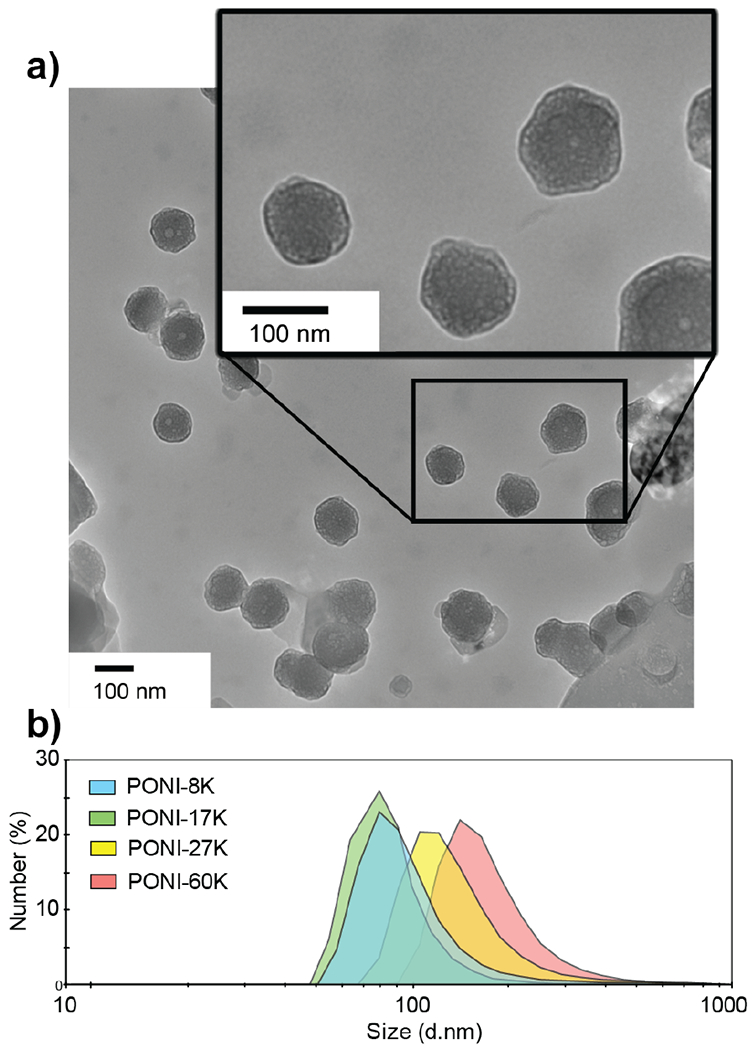
(a) Representative TEM images of PPNC structures reveal an average size of ~100-200 nm. PPNCs were formulated using PONI-60K with GFP-E20 at a Guan/E-tag ratio of 10. (b) Average size is confirmed by representative DLS spectrum (number) in PBS. Additional DLS data is available in Figure S11
Quantification of cellular uptake/association of PPNCs using flow cytometry.
Cellular uptake of PPNCs was measured in HEK-293T cells using standard flow cytometry. The mean fluorescence intensity (MFI) values were used to assess uptake/association based on Guan/E-tag charge ratio. In these studies, GFP+ populations included cells that had internalized GFP-E20, as well as cells to which GFP-E20-PPNCs had simply associated with the cell surface. In brief, formulations were prepared as described in Supporting Information (Table S1–S4) and incubated with HEK-293T cells for 24 hours. All deliveries were performed in media containing 10% FBS. Negligible levels of toxicity were observed for all polymers upon incubation with mammalian cells at relevant working conditions (Figure S9–S10). After delivery, cells were trypsinized, fixed with 2% paraformaldehyde (PFA) solution, and resuspended in modified flow buffer prior to flow cytometry experiments (see SI). A summary of HEK-293T/PPNC association efficiencies for PONI homopolymers is shown in Figure 4, where MFI represents the GFP+ percentage of the cell population. Interestingly, PPNCs prepared from higher molecular weight PONI polymers (17K and above) showed increased MFI at Guan/E-tag charge ratios up to ~50, above which the association diminished significantly. Higher molecular weight PONI polymers also displayed a broader range of potentially effective charge ratios. The highest MFI was observed with PONI-60K with an MFI ~1.3 times larger than the next best PPNC formulation (PONI-27K).
Figure 4.
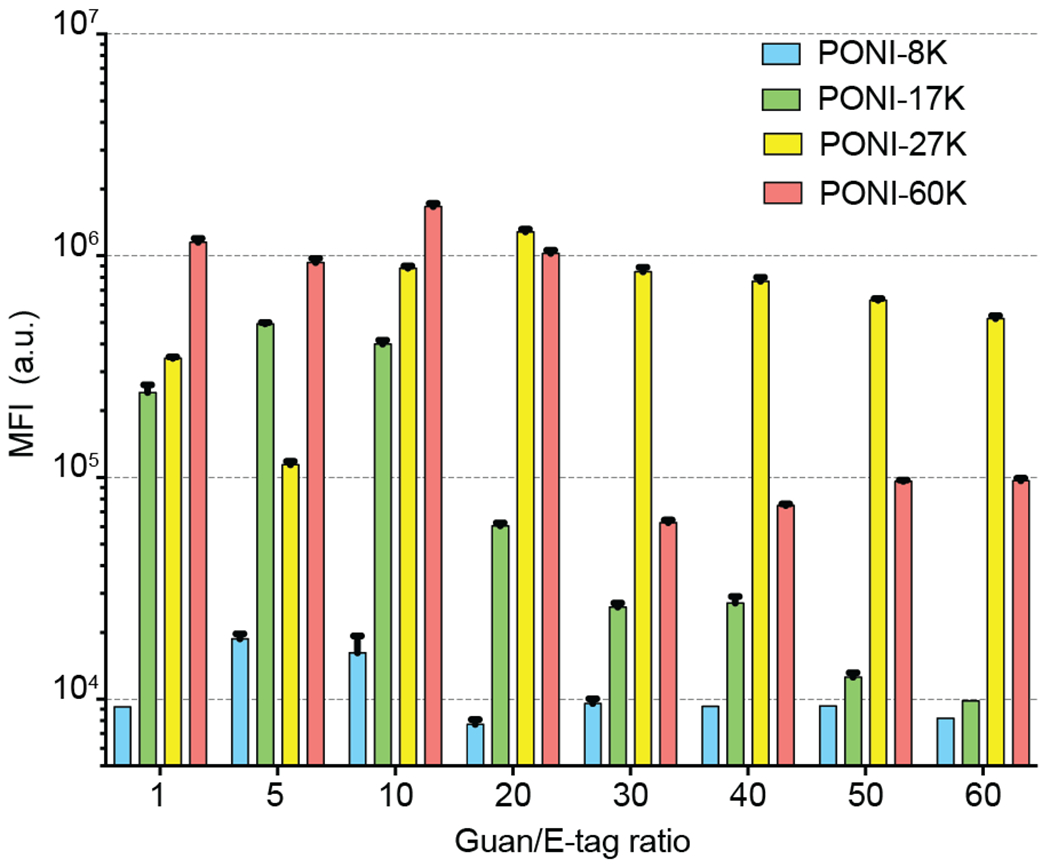
Overall MFI values for cell populations after incubation with PPNCs at varying Guan/E-tag ratios, as determined by flow cytometry (n=5). Higher molecular weight polymers exhibit significantly increased MFI values for all formulations.
Quantification of nuclear localization by flow imaging cytometry.
The efficiency of cytosolic delivery was assessed using imaging flow cytometry,40 allowing quantification of cytosolic GFP-E20 relative to surface-bound and endosomally-entrapped species. This technique provides large data sets that yield enhanced statistical power for cell analyses.41 Results of this analysis are displayed in Figure 5, along with representative images of non-delivered, endosomally-entrapped, and nuclear-localized cells.
Figure 5.
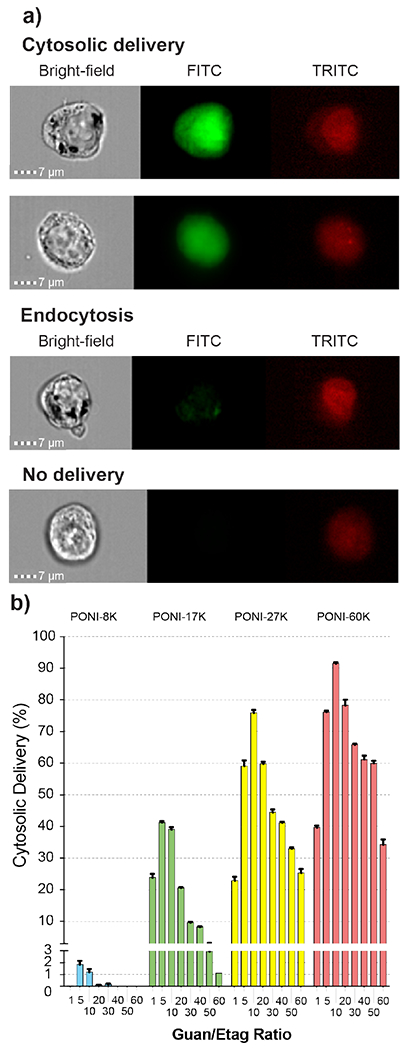
(a) Representative imaging flow cytometry images of cytosolic delivery, endocytosis, and undelivered cells (n=5). Channels displayed are bright-field, 488 nm (FITC, for GFP), and 633 nm (TRITC, for DRAQ5 nuclear stain). All images at 60x magnification. Scale bar = 7μm. (b) Percentages of total cell populations with cytosolic delivery of GFP, as determined by flow imaging cytometry. Note the larger range of effective conditions for polymers with higher molecular weight.
A robust flow imaging cytometry method was developed to reliably determine delivery efficiency at the single cell level. GFP will freely diffuse from the cytosol into the nucleus due to its small (~27 kDa) size.42 Nuclear localization was thus used as a metric to distinguish cytosolic delivery of GFP, as described in the SI. These criteria were applied to the data as a series of gates, as visualized and discussed in Figure S13 and isolated the cell population with GFP-E20 delivered to the cytosol, as quantitatively displayed in Figure 5. Delivery efficiency was also compared with a commercially-available transfection kit under serum-free conditions (Figure S14). Cytosolic delivery was confirmed by confocal microscopy and revealed delivery consistent with the results obtained by imaging flow cytometry. Demonstrated in Figure 6a, both PONI-27K and PONI-60K demonstrated effective delivery with GFP fluorescence signal evenly distributed throughout the cytosol and the nucleus.
Figure 6.
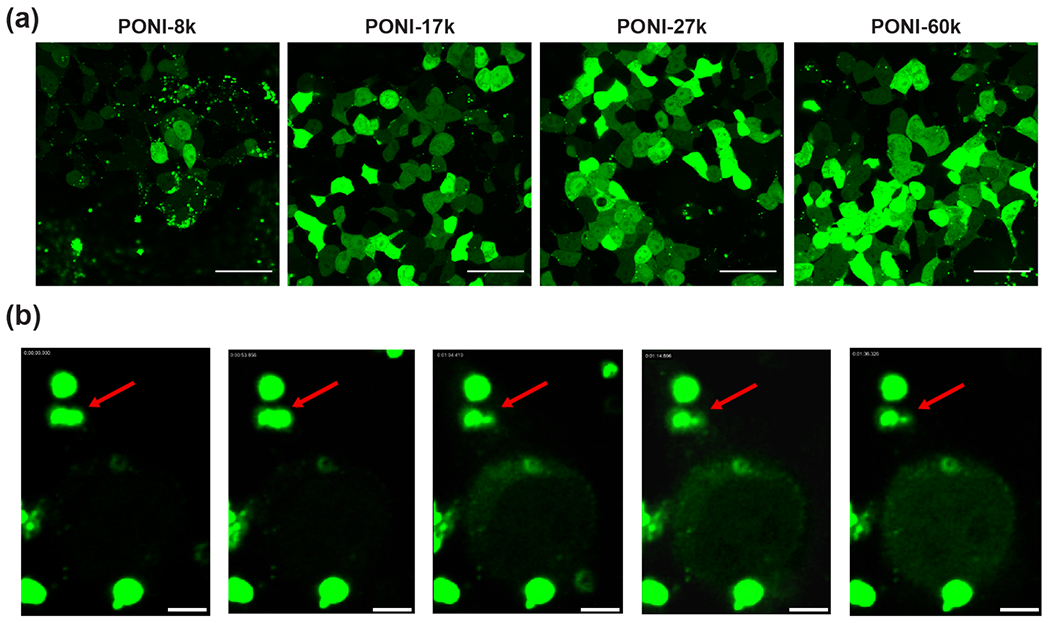
(a) Representative confocal images of PPNC-mediated delivery of GFP-E20. PPNCs were formed at a Guan/E-tag ratio of 10 and incubated with HEK-293T cells for 24h in the presence of 10% FBS. Scale bars = 50 μm. (b) Still frames of delivery event from time-lapse imaging of GFP-E20 delivery to HeLa. T=0 corresponds to ~5 hours following initial incubation of PPNCs with cultured cells. Delivery event occurs within ~40 sec. Red arrow marks PPNC responsible for delivery event; other fluorescent assemblies are extracellular. Scale bars = 5 μm.
We further performed time-lapse imaging of delivery 5 h after initial incubation of PPNCs with HeLa cells. Notably, we observed a similar delivery phenomenon as in our previous reports; a single PPNC electrostatically associated with the cell membrane swiftly delivers its cargo inside, followed by green fluorescence spreading throughout the cytosol and nucleus, all within ~40 seconds (Figure 6b and Supporting Video S1). This rate of fluorescence spread is too fast for endosomal uptake and escape for a protein with the size and charge of GFP, supporting the concept that delivery with the PPNC platform undergoes a membrane-fusion type mechanism, as opposed to an endocytosis-dependent pathway.43
We further investigated the delivery mechanism using three common small molecule inhibitors of cellular uptake. Cells were pretreated with either nystatin (an inhibitor of caveolae-mediated endocytosis), chlorpromazine (an inhibitor of clathrin-mediated endocytosis), or methyl-β-cyclodextrin (a cholesterol depletion agent).
We found that while nystatin and chlorpromazine pretreatment affected delivery only negligibly, pretreatment with methyl-β-cyclodextrin shut down delivery almost entirely (Figure S15). This result further supports the contention that delivery is dependent on membrane flexibility and the presence of cholesterol. In combination with our other results, these data support the ability of PPNC vehicles to deliver their protein cargo directly to the cytosol.
Delivery of tdTomato and functional Cre recombinase.
We further demonstrated the generalizability of the PPNC delivery platform through delivery of an E-tagged dimeric protein. We observed that delivery efficiency was reliant solely on Guan/Etag ratio and was independent of E-tag length for lengths of E0 (wild-type), E10 and E20 (Figure S16). tdTomato-E10, a ~55 kDa (pI: ~6.3) was chosen as our model protein to provide consistent E-tag content.44 As shown in Figure 7a, efficient PPNC-mediated delivery of tdTomato-E10 was evident at the same charge ratio as GFP-E20, reformulated for the shortened E-tag length.
Figure 7.
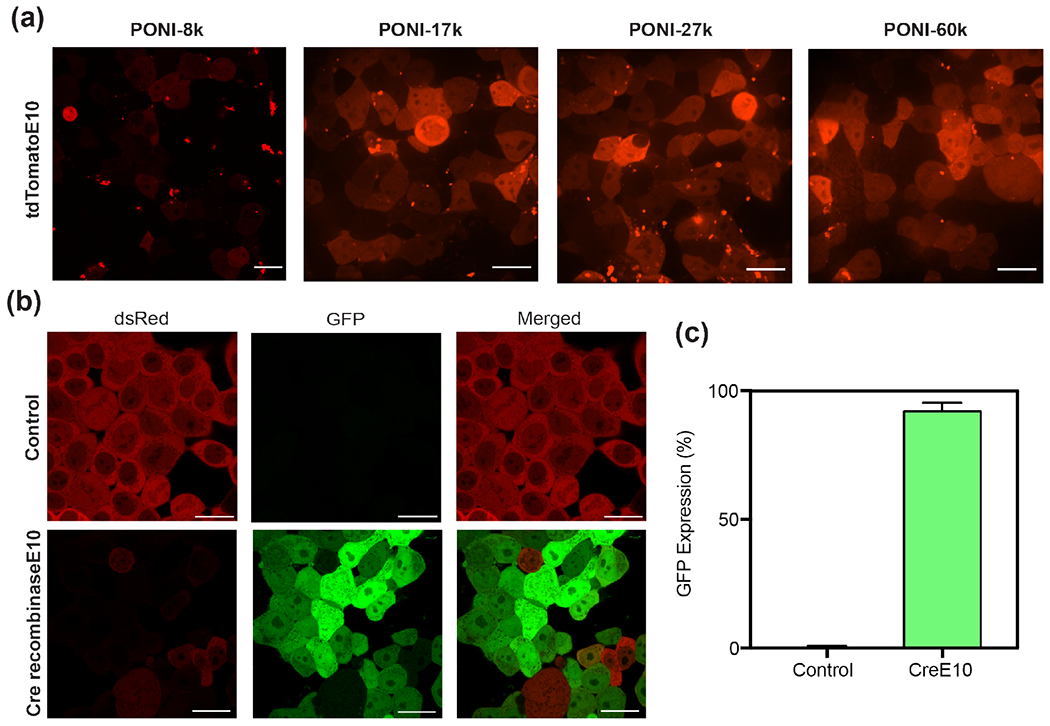
Representative confocal images (40x) of (a) tdTomato-E10 delivery to HEK-293T using PONI-27k after 24h PPNC incubation, and (b) Cre-reporter HEK-293T cells after 24h PPNC incubation and an additional 24h growth period. Scale bars=20 μm. (c) Percentage of GFP+ cells, determined by analysis through ImageJ. Additional confocal images used for analysis are available in Figure S17.
We investigated if a functional enzyme would retain its enzymatic activity through the delivery process. The tyrosine recombinase enzyme Cre recombinase (~38.5 kDa, pI: ~9.6) has been extensively utilized as both a model and therapeutic protein in genetics research. The small size of Cre recombinase allows it to freely diffuse into the nucleus once present in the cytosol.42 There, Cre recombinase catalyzes a site-specific recombination event between two DNA LoxP recognition sites. We employed a Cre-reporter HEK-293T cell line to provide a fluorescent readout of enzymatic activity.30 These cells express dsRed fluorescent protein prior to recombination. Cre-mediated recombination excises the dsRed DNA element and proceeding STOP codon and triggers the expression of GFP. We performed PPNC-mediated delivery of functional Cre-E10 to engineered HEK-293T using PONI-27K at a Guan/E-tag ratio of 10. PONI-27K was used in these studies as a representative delivery vehicle for its broad effective window. Media was changed after 24 hours, and cells were incubated an additional 24 hours to allow for total genetic recombination. Cells were analyzed by confocal microscopy and revealed a shift in fluorescence from dsRed to GFP corresponding to >90% of cells (Figure 7b and 7c). This result confirms the capability of PPNC delivery platforms to transport functional protein cargo directly to the cytosol and, indirectly, the nucleus.
CONCLUSION
In summary, we present here a robust method for direct cytosolic protein delivery using rationally designed cationic homopolymers and co-engineered E-tagged proteins. PPNCs demonstrate high versatility in their ability to deliver protein to the cytosol and nucleus under conditions varied by charge. This platform immediately provides a useful tool for protein delivery in ex vivo cell culture, for therapeutic studies, or in situ cell biology applications. PPNCs demonstrated efficient cytosolic delivery under a wide range of working formulations, as well as complete stability under serum-containing conditions, making them promising vehicles for protein delivery in vivo. We anticipate that these versatile PPNC systems will provide a functionalizable platform for therapeutic translation, with significant medical and diagnostic applications.
Supplementary Material
ACKNOWLEDGMENT
The authors would like to specially thank the Flow Cytometry Core Facility at UMass Amherst and Dr. Amy S. Burnside for her help establishing flow cytometric assays. The authors would also like to acknowledge Dr. James Chambers and the Light Microscopy Core Facility at UMass Amherst. Dr. Rubul Mout and Dr. Federica Scaletti were instrumental in preliminary development of this delivery system, and the authors would like to gratefully acknowledge their contributions. pCSCMV:tdTomato was a gift from Gerhart Ryffel (Addgene plasmid # 30530), and pJJH1320 was a gift from Jurgen Heinisch (Addgene plasmid # 36915).
Funding Sources
This research was supported by the NIH (EB022641 to V.M.R and GM008515 to D.C.L), and the NSF (CHE-1808199).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
REFERENCES
- 1.Casey PJ Protein lipidation in cell signaling. Science 1995, 268 (5208), 221. [DOI] [PubMed] [Google Scholar]
- 2.Chang VT; Fernandes RA; Ganzinger KA; Lee SF, Siebold C; McColl J; Jönsson P; Palayret M, Harlos K, Coles CH; Jones EY; Lui Y; Huang E; Gilbert RJC; Klenerman D; Aricescu AR; Davis SJ Initiation of T cell signaling by CD45 segregation at ‘close contacts’. Nat. Immunol 2016, 17 (5), 574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cairns RA; Harris IS; Mak TW Regulation of cancer cell metabolism. Nat. Rev. Cancer 2011, 11 (2), 85. [DOI] [PubMed] [Google Scholar]
- 4.Verdin E; Ott M 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat. Rev. Mol. Cell Biol, 2015, 16 (4), 258. [DOI] [PubMed] [Google Scholar]
- 5.Pearce EL; Pearce EJ Metabolic pathways in immune cell activation and quiescence. Immunity 2013, 38 (4), 633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker A; Thakur BK; Weiss JM; Kim HS; Peinado H; Lyden D Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell, 2016, 30 (6), 836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aguzzi A; O’Connor T Protein aggregation diseases: pathogenicity and therapeutic perspectives. Nat. Rev. Drug Disc, 2010, 9, 237–248. [DOI] [PubMed] [Google Scholar]
- 8.Kohl S; Zobor D; Chiang WC; Weisschuh N; Staller J; Menendez IG; Chang S; Beck SC; Garrido MG; Sothilingam V; Seeliger MW; Stanzial F; Benedicenti F; Inzana F; Héon E; Vincent A; Beis J; Strom TM; Rudolph G; Roosing S; Hollander, Anneke ID; Cremers FPM; Lopez I; Ren H; Moore AT; Webster AR; Michaelides M; Koenekoop RK; Zrenner E; Kaufman RJ; Tsang SH; Wissinger B; Lin JH Mutations in the Unfolded Protein Response Regulator ATF6 Cause the Cone Dysfunction Disorder Achromatopsia. Nat. Genet 2015, 47 (7), 757–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cuanalo-Contreras K; Mukherjee A; Soto C Role of protein misfolding and proteostasis deficiency in protein misfolding diseases and aging. Int. J. Cell Biol 2013. 2013, 638083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mullard A 2012 FDA drug approvals. Nat. Rev. Drug Discov 2013, 12, 87. [DOI] [PubMed] [Google Scholar]
- 11.Bobo D; Robinson KJ; Islam J; Thurecht KJ; Corrie SR Nanoparticle-based medicines: a review of FDA-approved materials and clinical trials to date. Pharm. Res 2016, 33 (10), 2373. [DOI] [PubMed] [Google Scholar]
- 12.Hartmann J; Croteau SE 2017 Clinical trials update: Innovations in hemophilia therapy. Am. J. Hematol 2016, 91 (12), 1252. [DOI] [PubMed] [Google Scholar]
- 13.Hsu PD; Lander ES; Zhang F Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014, 157 (6), 1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu C; Wan T; Wang H; Zhang S; Ping Y; Cheng Y A Boronic Acid-Rich Dendrimer with Robust and Unprecedented Efficiency for Cytosolic Protein Delivery and CRISPR-Cas9 Gene Editing. Sci. Adv 2019, 5 (6), eaaw8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaj T; Gersbach CA; Barbas CF III ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013, 31 (7), 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soutschek J; Akinc A; Bramlage B; Charisse K; Constien R; Donoghue M; Elbashir S; Geick A; Hadwiger P; Harborth J; John M; Kesavan V; Lavine G; Pandey RK; Racie T; Rajeev KG; Rohl I; Toudjarska I; Wang G; Wuschko S; Bumcrot D; Koteliansky V; Limmer S; Manoharan M; Vornlocher H-P Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature 2004, 432 (7014) 173. [DOI] [PubMed] [Google Scholar]
- 17.Lee Y-W; Mout R; Luther DC; Liu Y; Castellanos-Garcia L; Burnside AS; Ray M; Tonga GY; Hardie J; Nagaraj H; Das R; Phillips EL; Tay T; Vachet RW; Rotello VM In vivo editing of macrophages through systemic delivery of CRISPR-Cas9-ribonucleoprotein-nanoparticle nanoassemblies. Adv. Therap 2019, 1900041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee K; Conboy M; Park HM; Jiang F; Kim HJ; Dewitt MA; Mackley VA; Chang K; Rao A; Skinner C; Shobha T; Mehdipour M; Liu H; Huang W-C; Lan F; Bray NL; Li S; Corn JE; Kataoka K; Doudna JA; Conboy I; Murthy N Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nat. Biomed. Eng 2017, 1 (11), 889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu H; Zhang L; Tong S; Lee CM; Deshmukh H; Bao G Spatial Control of in vivo CRISPR–Cas9 Genome Editing via Nanomagnets. Nat. Biomed. Eng 2019, 3 (2), 126–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill R; Cautain B; de Pedro N; Link W Targeting nucleocytoplasmic transport in cancer therapy. Oncotarget. 2014. 5 (1) 11–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scaletti F; Hardie J; Lee Y-W; Luther D; Ray M; Rotello VM Protein delivery into cells using inorganic nanoparticle-protein supramolecular assemblies. Chem Soc Rev. 2018, 47 (10) 3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghosh P; Yang X; Arvizo R; Zhu ZJ; Agasti SS; Mo Z, Rotello VM Intracellular delivery of a membrane-impermeable enzyme in active form using functionalized gold nanoparticles. J. Am. Chem. Soc 2010, 132 (8), 2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu C; Haque F; Jasinski DL; Binzel DW; Shu D; Guo P Favorable biodistribution, specific targeting and conditional endosomal escape of RNA nanoparticles in cancer therapy. Cancer Lett. 2018, 414, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang K-L; Higuchi Y; Kawakami S; Yamashita F; Hashida M Efficient Gene Transfection by Histidine-Modified Chitosan through Enhancement of Endosomal Escape. Bioconjug. Chem 2010, 21 (6), 1087–1095. [DOI] [PubMed] [Google Scholar]
- 25.Smith SA; Selby LI; Johnston APR; Such GK The endosomal escape of nanoparticles: toward more efficient cellular delivery. Bioconjug. Chem 2019, 30 (2), 263–272. [DOI] [PubMed] [Google Scholar]
- 26.Lee Y-W; Luther DC; Kretzmann JA; Burden A; Jeon T; Zhai S; Rotello VM Protein delivery into the cell cytosol using non-viral nanocarriers. Theranostics 2019, 9 (11) 3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lv J; Fan Q; Wang H; Cheng Y Polymers for Cytosolic Protein Delivery. Biomaterials. 2019, 218, 119358. [DOI] [PubMed] [Google Scholar]
- 28.Liu C; Shen W; Li B; Li T; Chang H; Cheng Y Natural Polyphenols Augment Cytosolic Protein Delivery by a Functional Polymer. Chem Mater. 2019, 31 (6), 1956. [Google Scholar]
- 29.Mout R; Ray M; Yesilbag Tonga G; Lee YW; Tay T; Sasaki K; Rotello VM Direct cytosolic delivery of CRISPR/Cas9-ribonucleoprotein for efficient gene editing. ACS Nano 2017, 11 (3), 2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mout R; Ray M; Tay T; Sasaki K; Yesilbag Tonga G; & Rotello VM General strategy for direct cytosolic protein delivery via protein-nanoparticle co-engineering. ACS Nano 2017, 11 (6), 6416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicolas J; Mura S; Brambilla D; Mackiewicz N; Couvreur P Design, functionalization strategies and biomedical applications of targeted biodegradable/biocompatible polymer-based nanocarriers for drug delivery. Chem. Soc. Rev 2013, 42 (3), 1147. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Z; Shen W; Ling J; Yan Y; Hu J; Cheng Y The Fluorination Effect of Fluoroamphiphiles in Cytosolic Protein Delivery. Nat. Commun 2018, 9 (1), 1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Y; Chen J; Zhu W; Zhang K Unique Post-Functionalization Method for ROMP Polymers Based on Triazolinedione Alder-Ene Chemistry. Polymer. 2015, 74, 16–20. [Google Scholar]
- 34.Li Z; Zhang K; Ma J; Cheng C; Wooley KL Facile Syntheses of Cylindrical Molecular Brushes by a Sequential RAFT and ROMP “Grafting-through” Methodology. J. Polym. Sci. Part A Polym. Chem 2009, 47 (20), 5557–5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Som A; Reuter A; Tew GN Protein Transduction Domain Mimics: The Role of Aromatic Functionality. Angew. Chemie Int. Ed 2012, 51 (4), 980–983. [DOI] [PubMed] [Google Scholar]
- 36.Bielawski CW; Grubbs RH Living ring-opening metathesis polymerization. Prog. Polym. Sci 2007, 32 (1), 1. [Google Scholar]
- 37.Mout R; Moyano DF; Rana S; Rotello VM Surface Functionalization of Nanoparticles for Nanomedicine. Chem. Soc. Rev 2012, 41 (7), 2539–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pesek SL; Li X; Hammouda B; Hong K; Verduzco R Small-Angle Neutron Scattering Analysis of Bottlebrush Polymers Prepared via Grafting-Through Polymerization. Macromolecules 2013, 46 (17), 6998–7005. [Google Scholar]
- 39.Sgolastra F; Backlund CM; Ilker Ozay E; deRonde BM; Minter LM; Tew GN Sequence Segregation Improves Non-Covalent Protein Delivery. J. Control. Release 2017, 254, 131–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Basiji DA; Ortyn WE; Liang L; Venkatachalam V; Morrissey P Cellular image analysis and imaging by flow cytometry. Clin. Lab. Med 2007, 27 (3), 653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henriksen M; Miller B; Newmark J; Al-Kofahi Y; Holden E Laser scanning cytometry and its applications: a pioneering technology in the field of quantitative imaging cytometry. Methods Cell biol. 2011, 102, 159. [DOI] [PubMed] [Google Scholar]
- 42.Timney BL; Raveh B; Mironska R; Trivedi JM; Kim SJ; Russel D; Wente SR; Sali A; Rout MP Simple rules for passive diffusion through the nuclear pore complex. J. Cell Biol 2016, 215 (1), 57–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lippincott-Schwartz J; Snapp E; Kenworthy A Studying Protein Dynamics in Living Cells. Nat. Rev. Mol. Cell Bio 2001, 2, 444–456. [DOI] [PubMed] [Google Scholar]
- 44.Lee Y-J; Erazo-Oliveras A; Pellois J-P Delivery of Macromolecules into Live Cells by Simple Co-Incubation with a Peptide. Chembiochem. 2010, 11 (3), 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


