Abstract
Purpose
To evaluate the diagnostic utility of two convolutional neural networks (CNNs) for severity staging of anterior cruciate ligament (ACL) injuries.
Materials and Methods
In this retrospective study, 1243 knee MR images (1008 intact, 18 partially torn, 77 fully torn, and 140 reconstructed ACLs) from 224 patients (mean age, 47 years ± 14 [standard deviation]; 54% women) were analyzed. The MRI examinations were performed between 2011 and 2014. A modified scoring metric was used. Classification of ACL injuries using deep learning involved use of two types of CNN, one with three-dimensional (3D) and the other with two-dimensional (2D) convolutional kernels. Performance metrics included sensitivity, specificity, weighted Cohen κ, and overall accuracy, and the McNemar test was used to compare the performance of the CNNs.
Results
The overall accuracies for ACL injury classification using the 3D CNN and 2D CNN were 89% (225 of 254) and 92% (233 of 254), respectively (P = .27), and both CNNs had a weighted Cohen κ of 0.83. The 2D CNN and 3D CNN performed similarly in classifying intact ACLs (2D CNN, sensitivity of 93% [188 of 203] and specificity of 90% [46 of 51] vs 3D CNN, sensitivity of 89% [180 of 203] and specificity of 88% [45 of 51]). Classification of full tears by both networks was also comparable (2D CNN, sensitivity of 82% [14 of 17] and specificity of 94% [222 of 237] vs 3D CNN, sensitivity of 76% [13 of 17] and specificity of 100% [236 of 237]). The 2D CNN classified all reconstructed ACLs correctly.
Conclusion
Two-dimensional and 3D CNNs applied to ACL lesion classification had high sensitivity and specificity, suggesting that these networks could be used to help nonexperts grade ACL injuries.
Supplemental material is available for this article.
© RSNA, 2020
Summary
The deep learning pipeline in this study may lend toward diagnostic worklist prioritization, standardization, and generalizability in assessing anterior cruciate ligament lesions, in addition to improved point-of-care communication with patients, by those who are nonexperts in grading anterior cruciate ligament injuries.
Key Points
■ Two convolutional neural networks (CNNs), each with respective two-dimensional (2D) and three-dimensional (3D) convolutional kernels, achieved a high overall accuracy of 92% and 89%, respectively, and each achieved a weighted Cohen κ value of 0.83 for anterior cruciate ligament (ACL) severity staging.
■ All reconstructed ACLs were correctly classified by the 2D model (sensitivity of 100%, specificity of 100%), while the 3D CNN displayed similar performance (sensitivity of 97%, specificity of 100%).
■ Sensitivity and specificity of the 2D and 3D models for intact classification were 93% and 90%, and 89% and 88%, respectively.
Introduction
The anterior cruciate ligament (ACL) is the most commonly injured ligament in the knee (1,2). ACL injuries increase the risk of developing posttraumatic knee osteoarthritis and the need for total knee replacement (3–6). Currently, MRI is the most effective imaging modality for distinguishing structural properties of the ACL in relation to adjacent musculoskeletal structures (7–10). Several multigrading scoring systems have been developed to standardize reporting of knee joint abnormalities using MRI (11,12). The Whole-Organ Magnetic Resonance Imaging Score (WORMS) is among the most widely used semiquantitative MRI scoring systems for assessing knee osteoarthritis (13–15) and takes into account several chondral, bony, and ligamentous compartments in the knee (15,16). The Anterior Cruciate Ligament OsteoArthritis Score (ACLOAS) has been shown to offer increased longitudinal and cross-sectional reliability of ACL staging (17). The aforementioned grading metrics are susceptible to interrater variability, especially pertaining to torn fibers and mucoid degeneration (10,18).
Deep learning methods have recently shown potential to serve as an aid for clinicians with limited time or experience in osteoarthritis grading of the knee menisci and cartilage (9). Four other applications of deep learning have also resulted in binary models for distinguishing an intact ACL from a fully torn ACL (19–22). These neural networks possess large domains for learning and are versatile for new tasks using pretrained weights, producing inferences in seconds. The specific mode of learning depends on the architecture, but the most successful algorithms are known to learn shallow features and high-level features with many convolutions (23,24). However, deep learning has yet to be applied to predict multigrade, semiquantitative lesion severity for the ACL.
In this study, we propose a deep learning–based pipeline to isolate the ACL region of interest in the knee, detect ACL abnormalities, and stage lesion severity using three-dimensional (3D) and previously reported two-dimensional (2D) convolutions with MR images. This is a proof-of-concept study to show that hierarchical image analysis methods work with 3D convolutional neural networks (CNNs) with the goal of hierarchical staging and comparison with the 2D network. Additionally, to the best of our knowledge, this is the first instance of multiclass ACL severity staging using deep learning, in which reconstructed, fully torn, partially torn, and intact ACLs are graded in accordance with semiquantitative scoring. This deep learning pipeline would lend toward standardization and generalizability in assessing ACL lesions for clinicians with limited time as well as those with limited experience reading knee MR images.
Materials and Methods
This work was sponsored by GE Healthcare and National Institutes of Health/National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIH/NIAMS).
Patient Datasets
This retrospective study was conducted according to regulations from the Committee for Human Research at all institutions, and all patients provided informed consent. Authors had full control of the data. A total of 1243 knee MRI studies (1008 intact, 18 partially torn, 77 fully torn, and 140 reconstructed ACLs) in 224 unique patients (mean age, 47 years ± 14 [standard deviation]; mean body mass index, 24.58 kg/m2 ± 3.60; 121 women) were obtained from three prior research studies that evaluated joint degeneration in osteoarthritis and after ACL injury between 2011 and 2014. Patients were excluded based on concurrent use of an investigational drug, history of fracture, total knee replacement in the study knee, and any contraindications to MRI. Patients were recruited in the osteoarthritis group if they reported knee pain, aching, or stiffness on most days per month during the past year, reported use of medication for knee pain on most days per month during the past year, or exhibited any possible radiologic sign of knee osteoarthritis (Kellgren-Lawrence grade > 0), and age of 36 years or older. Patients were included in the control group if they had no knee pain or stiffness in either knee and if no use of medications for knee pain in the last year was reported, and if no radiologic evidence of osteoarthritis on either knee was noted (Kellgren-Lawrence grade 0). Portions of this dataset were used in prior studies connected with grants NIH/NIAMS P50AR060752 and NIH/NIAMS R01AR046905. These studies had different aims, such as identifying differences in relaxation time between loading and unloading in patients with osteoarthritis, identifying cartilage change after ACL injury, and classifying knee meniscus and cartilage abnormalities (9).
Patients in the ACL study were enrolled at three sites: University of California, San Francisco (San Francisco, Calif), Mayo Clinic (Rochester, Minn), and Hospital for Special Surgery (New York City, NY). Patients (n = 77) underwent anatomic single-bundle ACL reconstruction by board-certified, fellowship-trained orthopedic surgeons. Only soft-tissue grafts were used, including the hamstring (either allograft or autograft) or the posterior tibialis (allograft). No special sequences were used for metal artifact suppression. Hamstring, patellar tendon, and allograft ACL reconstructions typically do not cause metal artifacts along the intra-articular course of the graft, which allows evaluation of ACL graft degeneration or retears. Moreover, metal artifacts related to ACL reconstruction are owing to the use of metallic interference screws or endobuttons, although many reconstructions typically make use of nonmetallic interference screws and have no associated artifact. All patients underwent a standard postoperative rehabilitation protocol.
MRI Acquisition
In all imaging studies from all three sites, 3D fast spin-echo-Cube proton density–weighted sagittal oblique sequences with the following parameters were used: repetition time msec/echo time msec, 1500/26.69; field of view, 14 cm; matrix, 384 × 384; slice thickness, 0.5 mm; echo train length, 32; bandwidth, 50.0 kHz; number of excitations, 0.5; and acquisition time, 10.5 minutes. All images were acquired with five 3-T MRI scanners (GE Healthcare, Waukesha, Wis) and eight surface coils.
Ground Truth Image Grading
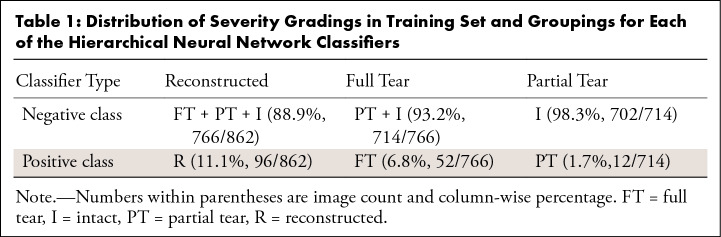
Between 2011 and 2014, five board-certified radiologists, each with more than 5 years of training, graded a nonoverlapping section of the dataset. The radiologists were blinded with respect to both number and type of lesion. Intraobserver agreement assessment was conducted by three additional board-certified radiologists currently involved with the study. All readers were trained by one senior musculoskeletal radiologist (T.M.L.), who read at least 20 imaging studies with each of the other two radiologists (radiology residents, each with more than 2 years of training) in two imaging sessions. During these readings, the WORMS and ACLOAS grading systems were explained, and readers were asked to grade lesions of cartilage, menisci, bone marrow, ligaments, and synovium under supervision with direct feedback. This training was followed by independent assessment of 60 randomly chosen studies from the dataset. The ACL was graded using the original WORMS grading scale. The WORMS grade was then placed into one of four classes, including intact (grade 0), partial tear (grade 1), full tear (grade 2), and reconstructed (grade 3) to generalize newer ACL scoring systems (ie, ACLOAS) for use in the deep learning pipeline. The distribution of partial tears was small compared with that of full tears because the inclusion criteria in one of the studies was the presence of a fully torn ACL.
Deep Learning Pipeline
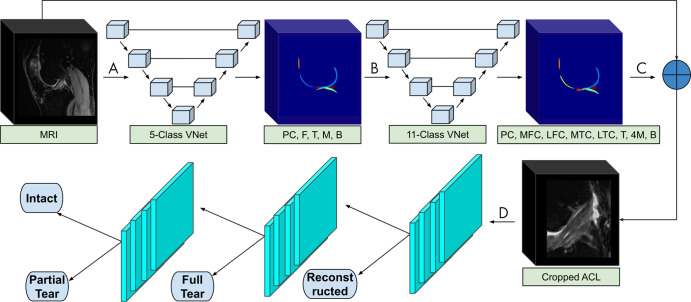
Our framework consists of a deep learning segmentation that categorizes the knee into 11 distinct anatomic components, followed by an image cropping to isolate the ACL, and a 3D CNN to classify lesion severity (9) (Fig 1). The segmentation and cropping for ACL localization are described in Appendix E1 (supplement). The CNN was developed through a hierarchical approach; specifically, three cascaded models were built to classify reconstructed ligaments, full tears, partial tears, and intact ACLs. We further compared the hierarchical approach to a single four-class model with the same parameters and saw superior performance using the hierarchical approach. The same hierarchical classification network was implemented with a 2D CNN for comparison with the 3D CNN (19).
Figure 1:
The segmentation and classification pipeline begins with A, the input of a full MRI volume into a three-dimensional V-Net, which segments the knee into patellar cartilage (PC), femur (F), tibia (T), meniscus (M), and background (B). B, The five-class segmentation is then input to a second V-Net that further categorizes the knee into 11 compartments, including PC, medial femoral condyle (MFC), lateral femoral condyle (LFC), medial tibial condyle (MTC), lateral tibial condyle (LTC), four meniscal horns (4M), T, and B. C, The 11-class segmentation is used to determine the anterior cruciate ligament (ACL) boundaries of the original input MRI. D, The cropped ACL volume is input to three hierarchical convolutional neural networks (either two- or three-dimensional), which each detect reconstructed, fully torn, partially torn, and intact ACLs.
Classification of the 3D and 2D CNNs
The 3D CNN was developed in TensorFlow (Google, Mountain View, Calif) and the 2D CNN in PyTorch (Facebook, Menlo Park, Calif). All computations were performed on NVIDIA GeForce GTX Titan X graphics processing units (Santa Clara, Calif).
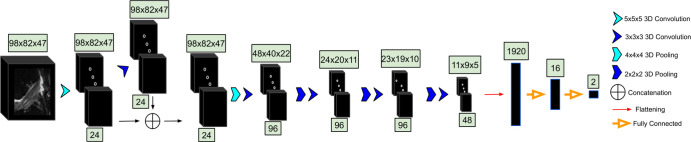
3D CNN.—The cropped ACL volumes were input into a CNN consisting of 3D convolutional kernels (9). The network was built with six layers, including one skip connection after the first convolution, to preserve initial features and mitigate overfitting (Fig 2). Training was performed over 100 epochs with the following parameters: an Adam optimizer, a learning rate of 10−5, empirically weighted cross-entropy loss to account for class imbalances, and a batch size of eight images. Three-dimensional translations and zooming were applied to all classes for augmentation of the training set. Rotations were not applied to preserve ligament fiber angles for model learning.
Figure 2:
The MRI is input to the three-dimensional (3D) convolutional neural network. In the first layer, convolutional kernels are applied to the entire volume. The second layer contains a concatenation with the first, followed by four additional 3D convolutions. The convolutional output is then flattened and input to two dense layers. The number beneath each set of blocks denotes the number of convolutions applied to the input; the number above represents the dimensions of the output after convolving the input.
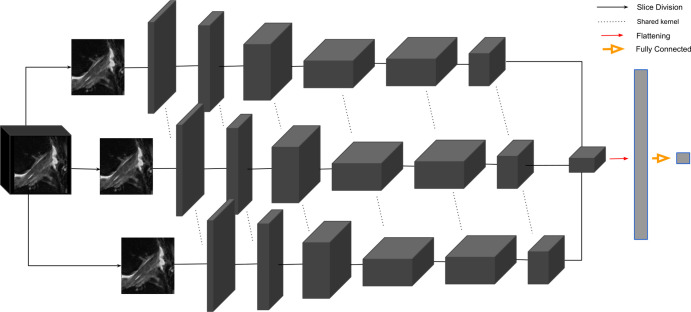
2D CNN.—Performance of the 3D CNN was compared with the MRNet, a 2D CNN (19). In the MRNet, each slice of the input 3D volume is passed through an AlexNet to extract features (25) (Fig 3). The MRNet was pretrained on the ImageNet dataset and was additionally trained with the same training sets as the 3D CNN using transfer learning. The MRNet pools features within each slice and among all slices in the volume to produce a classification probability. The same image augmentations and loss functions as the 3D CNN were used to ensure correct comparison.
Figure 3:
The two-dimensional convolutional neural network, MRNet, feeds each slice of an MRI into an AlexNet with shared parameters across each kernel. The weights are pooled as they pass into a final convolution and a subsequent fully dense flattening layer. The output of the dense layer is a single probability for the input MRI. The full parameters of the architecture are reported by Bien et al (19).
Learning Strategy
The deep learning classifier first differentiated the reconstructed ligaments (grade 3) from grades 0–2. The remaining studies were then analyzed for detecting full-thickness tears (grade 2). Partial tear lesions (grade 1) were further classified apart from intact ligaments (grade 0). The total of 1243 images (1008 control, 235 injured) from 224 patients were split into training (70%), validation (10%), and test sets (20%) for each grade, preserving the distributions of age, sex, and body mass index. The studies in each split were from distinct, nonoverlapping patients.
Statistical Analysis
The training set was used to train each of the CNNs with back propagation. The validation set served to evaluate model performance at each training epoch, and the testing set was blind to the model until after training to serve as the final metric of performance. For each CNN, we report overall accuracy and linear-weighted Cohen κ (26), as well as sensitivity and specificity for each severity score. The McNemar test was used to determine statistical significance between the two classifiers for sensitivity, specificity, and overall accuracy; however, the Fisher exact test was used instead if the number of patients with differing test results was less than 20 (27). Two-sample t tests were used to compare training, validation, and test set demographics. Python (version 3.6.5; Python Software Foundation, Beaverton, Ore) was used for all statistical analysis. P values less than .05 were considered significant.
Results
The intrareader agreement assessment for ACL grading resulted in linear-weighted κ values of 0.66–0.78 among each pair of radiologists. A random sample of 17 ACL volumes resulted in a mean ± standard deviation intersection over union of 0.89 ± 0.06. There were no statistically significant differences between patients in the training, validation, and test sets regarding age (P = .24), body mass index (P = .17), or sex (P = .85).
Performance Metrics of the 2D and 3D CNNs
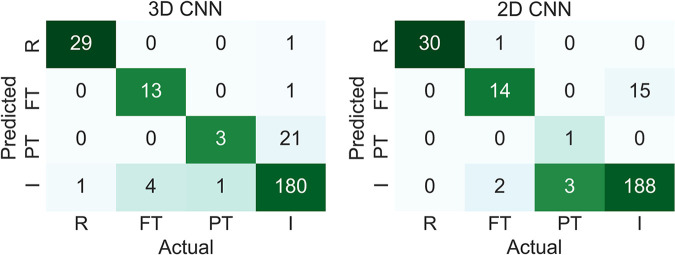
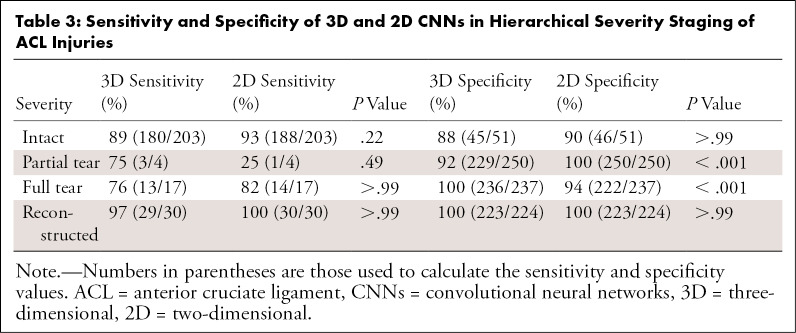
The 3D CNN and 2D CNN had overall accuracy of 89% (225 of 254) and 92% (233 of 254), respectively (P = .27), and both CNNs had a linear-weighted κ of 0.83 for ACL staging. Table 1 displays the distribution of grades for model training, while Table 2 contains the total grades and demographics for the training, validation, and test sets. As seen in Figure 4, both models performed highest on reconstructed ACL classification. The sensitivity (2D CNN, 100% [30 of 30]; 3D CNN, 97% [29 of 30]) and specificity (2D CNN, 100% [223 of 224]; 3D CNN, 100% [223 of 224]) in reconstructed classification was not significantly different between 2D CNN and 3D CNN (sensitivity, P > .99; specificity, P > .99). The 2D CNN demonstrated higher sensitivity and specificity than the 3D CNN in detecting intact ACLs, with sensitivity of 93% (188 of 203) with 2D CNN versus sensitivity of 89% (180 of 203) with 3D CNN and specificity of 90% (46 of 51) with 2D CNN versus specificity of 88% (45 of 51) with 3D CNN (Table 3). In fully torn ACLs, the sensitivity of the 2D CNN was similar to that of the 3D CNN (82% [14 of 17] and 76% [13 of 17], respectively; P > .99), although the specificity of the 3D CNN was higher than that of the 2D CNN (100% [236 of 237] and 94% [222 of 237], respectively; P < .001).
Table 1:
Distribution of Severity Gradings in Training Set and Groupings for Each of the Hierarchical Neural Network Classifiers
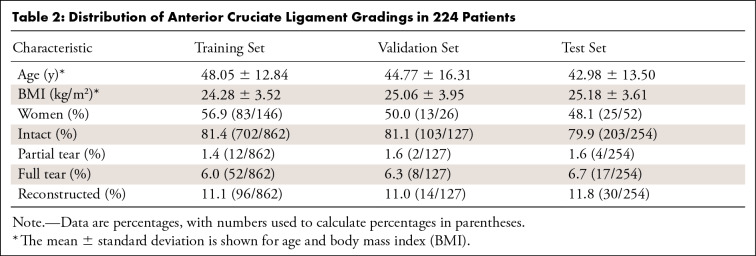
Table 2:
Distribution of Anterior Cruciate Ligament Gradings in 224 Patients
Figure 4:
Confusion matrices for three-dimensional (3D) and two-dimensional (2D) convolutional neural networks (CNNs) of reconstructed (R), fully torn (FT), partially torn (PT), and intact (I) anterior cruciate ligaments.
Table 3:
Sensitivity and Specificity of 3D and 2D CNNs in Hierarchical Severity Staging of ACL Injuries
Examples of Correct and Incorrect Classifications
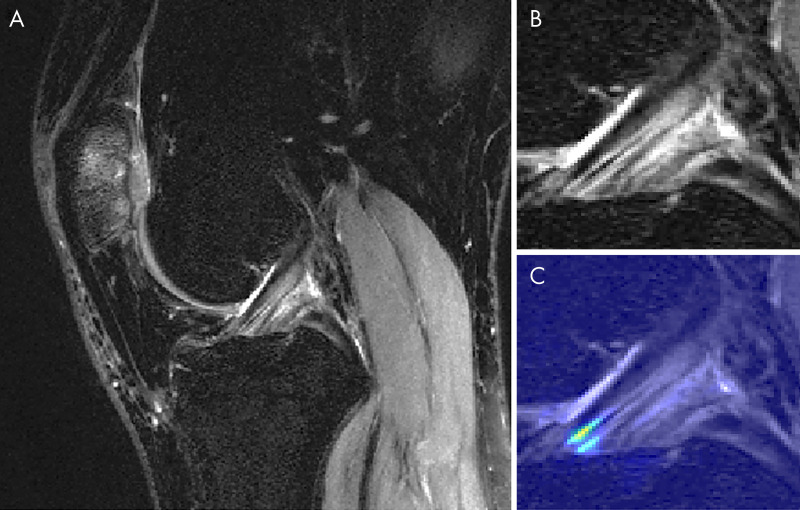
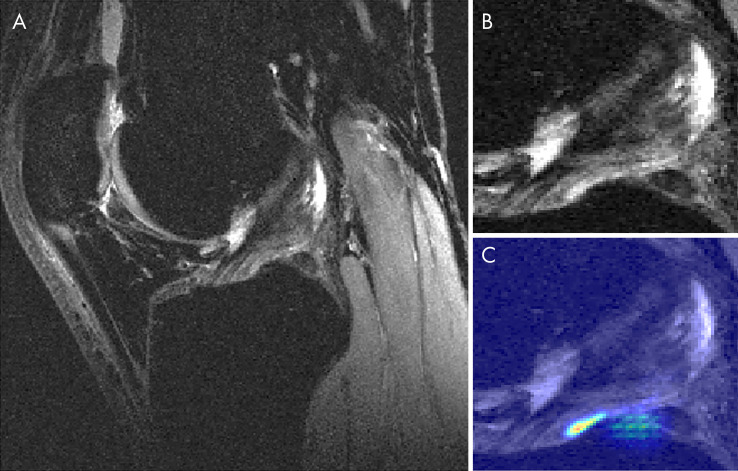
Figure 5 displays a knee with intact ACL that was input into the pipeline, followed by localization of the ACL and the corresponding saliency map generated by the model’s classification weighting. Saliency maps were generated from the rectified linear unit output of the 3D CNN in its last dense layer. This ACL was correctly classified and possesses a high intensity on the inferior portion of the ligament’s saliency. An incorrectly classified intact ACL, predicted to be partially torn, is seen in Figure 6. The model placed a high intensity on a sagittal view with overlapping ACL and femur signal. The resulting saliency possessed large weight on a portion of the joint posterior to the ligament and also has speckles of noise further posterior. The 3D CNN took less than 1 second to classify a single ACL that went through all three hierarchical classifiers.
Figure 5:
Sagittal MRI views of, A, a correctly classified knee with an intact anterior cruciate ligament (ACL) and, B, its ACL localization with, C, overlaid saliency map. The saliency demonstrates the anterior-inferior portion of the ACL as being of high importance for model classification.
Figure 6:
Sagittal MRI views of, A, an incorrectly classified knee with an intact anterior cruciate ligament (ACL) and, B, its ACL localization with, C, overlaid saliency map. The model predicted a partial tear in this knee. The model misplaces a relatively high probability mapping on a slice with ACL that is obstructed by signal from the femur. Additionally, the saliency intensity is posterior to the actual ligament; speckles of noise are also present in the inferior-posterior portion of the saliency. This intact ACL exhibited focal fluid collection posterior to the ligament on a separate sagittal view, which may have led to the misclassification.
Discussion
In this work we present a fully automated ACL segmentation and classification framework that provides hierarchical severity staging of the ACL using deep learning architectures. We compare the performance of a 3D CNN with a 2D CNN in ACL lesion classification. A higher overall accuracy was observed with the 2D model.
Four groups have previously used deep learning frameworks for ACL lesion classification tasks, the first being the original MRNet by Bien et al (19). Their dataset consisted of 319 ACL tears from a total of 1370 examinations. Their MRNet displayed 97% sensitivity and 76% specificity for fully torn ACLs. The MRNet we built demonstrated a higher specificity than sensitivity. The discrepancy may be owing to difficulty in generalizing the MRNet to images from other institutions. Liu et al (20) approached the binary ACL tear classification task using three cascaded deep learning architectures (28–30). The cascaded model of Liu et al (20) achieved 96% sensitivity and specificity but was not statistically significant when compared with radiologist grading. In addition, this group used a relatively small number of images (350) for training, validation, and testing, which may have led to overfitting. Chang et al (21) applied CNNs with residual blocks on 260 volumes in the coronal plane, which is a more difficult cross-section to use to grade the ACL. Most recently, Germann et al (22) used CNNs to classify ACL tears in MRI studies from 59 institutions, resulting in overall and in-house performance metrics lower than those of fellowship-trained faculty musculoskeletal radiologists. Compared with these four prior studies, our pipeline classifies ACLs using 3D convolutional kernels in a hierarchical sequence. We implemented a hierarchical classifier because the ACLs possess an ordered sequence of injury severity, increasing from intact to reconstructed. A stepwise approach beginning sequentially with the most severe classification can enhance accuracy because decisions are less complicated if they are binary.
MRI studies are volumetric and 3D; therefore, 3D convolutional kernels can learn 3D features that 2D convolutions cannot. The ground truth we are using is on the patient level on all the 3D volumes, which is a scalable design because it does not require pixel- or slice-level annotation. Thus, supervised feature learning can occur exclusively in 3D models, as opposed to 2D models. However, 3D models are more complex, with higher parameter spaces that are more likely to overfit with small sample sizes, unlike 2D models, which have lower parameter space convolutional filters and typically perform well for general image classification problems. Using a 3D CNN did not outperform a 2D model, which may be because the 2D CNN utilized transfer learning from ImageNet, which is a dataset of 14 million 2D images that is not compatible for training a 3D CNN. The pretrained 2D CNN without transfer learning would perform worse had we not trained it with our MR images. However, our goal was to compare transfer learning in a 2D CNN with a 3D CNN possessing no transfer learning, to compare the benefits of 3D spatial relations with those of transfer learning.
Although we evaluate a hierarchical severity staging classifier, the partial tear class has little clinical relevance without subcategorization because partial tears denote a wide spectrum of injury, some of which require surgery and others that do not. Our limited sample size of partial tears did not allow for substratification, but one of the primary end goals of this study is to classify subcategories of partial tears ranging from intact to fully torn. The limited number of partial tears in validation and testing sets should be considered when externalizing the partial tear results, as many more cases are needed to draw significant conclusions.
Our method contains other limitations, beginning with the use of MRI as the reference standard. The grades used for model training are dependent on subjective assessment by a radiologist. Using arthroscopy as an additional standard of comparison may improve the ground truth labels for the ACLs, thereby increasing the model’s capability to learn accurately. However, the model learning is limited because early model building notably could not subclassify mucoid degeneration within intact ligaments. Furthermore, our sample of patients was not balanced among all gradings, which we addressed by using a weighted cross-entropy loss function during training for both 3D and 2D CNN models. Another limitation of our study was that we split our dataset into training, validation, and testing sets according to patient. This may lead to correlations among multiple images from the same patient, which are nonindependent observations. Accounting for such correlations in training neural networks is not common; most models are built using image augmentations to increase training capacity. Thus, even if each patient had solely one image, data augmentation would yet lend toward correlation discrepancies. For this reason, dividing by images without preserving patient splits would offer little correlation benefit for the model, which would instead display falsely elevated accuracy by inferring on the same patients used for training.
Deep learning can provide relatively fast classification and visualization of ACL lesions, which may facilitate clinical translation. Both the 2D and 3D architectures displayed a relatively high degree of sensitivity and specificity for intact, fully torn, and reconstructed ACLs, which may warrant clinical value of deep learning as a tool for standardizing and generalizing ACL severity staging for clinicians with limited experience with knee MRI.
APPENDIX
Acknowledgments
Acknowledgment
The authors would like to acknowledge Jinhee J. Lee for contributions in statistical analysis.
Contract grant sponsor: GE Healthcare; National Institutes of Health (NIH)/National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), contract grant number: K99AR070902 (V.P.), R61AR073552 (S.M., V.P.), P50AR060752 (S.M.).
Disclosures of Conflicts of Interest: N.K.N. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: disclosed no relevant relationships. Other relationships: This work was sponsored by GE Healthcare and National Institutes of Health/National Institute of Arthritis and Musculoskeletal and Skin Diseases. The funders had no editorial input into the project, and the authors had full control of the data. Author was not paid/supported directly by any of these groups. I.F. disclosed no relevant relationships. B.A. Activities related to the present article: institution received grant from GE Healthcare. Activities not related to the present article: disclosed no relevant relationships. Other relationships: disclosed no relevant relationships. R.S. Activities related to the present article: institution supported by GE Healthcare grant. Activities not related to the present article: disclosed no relevant relationships. Other relationships: disclosed no relevant relationships. R.T. disclosed no relevant relationships. F.C. disclosed no relevant relationships. T.M.L. Activities related to the present article: institution received grants from GE Healthcare and NIH. Activities not related to the present article: disclosed no relevant relationships. Other relationships: disclosed no relevant relationships. V.P. Activities related to the present article: institution received grant from GE Healthcare. Activities not related to the present article: disclosed no relevant relationships. Other relationships: disclosed no relevant relationships. S.M. Activities related to the present article: institution received grants from GE Healthcare and NIH. Activities not related to the present article: author paid consultant for Smith Research; institution received grants from Samumed and GE Healthcare. Other relationships: institution (University of California) receives money from patents (issued and licensed).
Abbreviations:
- ACL
- anterior cruciate ligament
- ACLOAS
- Anterior Cruciate Ligament OsteoArthritis Score
- CNN
- convolutional neural network
- 3D
- three-dimensional
- 2D
- two-dimensional
- WORMS
- Whole-Organ Magnetic Resonance Imaging Score
References
- 1.Spindler KP, Wright RW. Anterior cruciate ligament tear. N Engl J Med 2008;359(20):2135–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnston JT, Mandelbaum BR, Schub D, et al. Video analysis of anterior cruciate ligament tears in professional American football athletes. Am J Sports Med 2018;46(4):862–868. [DOI] [PubMed] [Google Scholar]
- 3.Hunter DJ, Lohmander LS, Makovey J, et al. The effect of anterior cruciate ligament injury on bone curvature: exploratory analysis in the KANON trial. Osteoarthritis Cartilage 2014;22(7):959–968. [DOI] [PubMed] [Google Scholar]
- 4.Prodromos CC, Han Y, Rogowski J, Joyce B, Shi K. A meta-analysis of the incidence of anterior cruciate ligament tears as a function of gender, sport, and a knee injury–reduction regimen. Arthroscopy 2007;23(12):1320–1325.e6. [DOI] [PubMed] [Google Scholar]
- 5.Brophy RH, Gill CS, Lyman S, Barnes RP, Rodeo SA, Warren RF. Effect of anterior cruciate ligament reconstruction and meniscectomy on length of career in National Football League athletes: a case control study. Am J Sports Med 2009;37(11):2102–2107. [DOI] [PubMed] [Google Scholar]
- 6.Suter LG, Smith SR, Katz JN, et al. Projecting lifetime risk of symptomatic knee osteoarthritis and total knee replacement in individuals sustaining a complete anterior cruciate ligament tear in early adulthood. Arthritis Care Res (Hoboken) 2017;69(2):201–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shakoor D, Guermazi A, Kijowski R, et al. Cruciate ligament injuries of the knee: a meta-analysis of the diagnostic performance of 3D MRI. J Magn Reson Imaging 2019;50(5):1545–1560. [DOI] [PubMed] [Google Scholar]
- 8.Ai T, Zhang W, Priddy NK, Li X. Diagnostic performance of CUBE MRI sequences of the knee compared with conventional MRI. Clin Radiol 2012;67(12):e58–e63. [DOI] [PubMed] [Google Scholar]
- 9.Pedoia V, Norman B, Mehany SN, Bucknor MD, Link TM, Majumdar S. 3D convolutional neural networks for detection and severity staging of meniscus and PFJ cartilage morphological degenerative changes in osteoarthritis and anterior cruciate ligament subjects. J Magn Reson Imaging 2019;49(2):400–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li K, Du J, Huang LX, Ni L, Liu T, Yang HL. The diagnostic accuracy of magnetic resonance imaging for anterior cruciate ligament injury in comparison to arthroscopy: a meta-analysis. Sci Rep 2017;7(1):7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hunter DJ, Guermazi A, Lo GH, et al. Evolution of semi-quantitative whole joint assessment of knee OA: MOAKS (MRI Osteoarthritis Knee Score). Osteoarthritis Cartilage 2011;19(8):990–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brandt KD, Fife RS, Braunstein EM, Katz B. Radiographic grading of the severity of knee osteoarthritis: relation of the Kellgren and Lawrence grade to a grade based on joint space narrowing, and correlation with arthroscopic evidence of articular cartilage degeneration. Arthritis Rheum 1991;34(11):1381–1386. [DOI] [PubMed] [Google Scholar]
- 13.Yang X, Li Z, Cao Y, et al. Efficacy of magnetic resonance imaging with an SPGR sequence for the early evaluation of knee cartilage degeneration and the relationship between cartilage and other tissues. J Orthop Surg Res 2019;14(1):152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong Z, Chen J, Zhang S, et al. Intra-articular injection of autologous adipose-derived stromal vascular fractions for knee osteoarthritis: a double-blind randomized self-controlled trial. Int Orthop 2019;43(5):1123–1134. [DOI] [PubMed] [Google Scholar]
- 15.Peterfy CG, Guermazi A, Zaim S, et al. Whole-organ magnetic resonance imaging score (WORMS) of the knee in osteoarthritis. Osteoarthritis Cartilage 2004;12(3):177–190. [DOI] [PubMed] [Google Scholar]
- 16.Kretzschmar M, Lin W, Nardo L, et al. Association of physical activity measured by accelerometer, knee joint abnormalities, and cartilage T2 measurements obtained from 3T magnetic resonance imaging: data from the Osteoarthritis Initiative. Arthritis Care Res (Hoboken) 2015;67(9):1272–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roemer FW, Frobell R, Lohmander LS, Niu J, Guermazi A. Anterior Cruciate Ligament OsteoArthritis Score (ACLOAS): longitudinal MRI-based whole joint assessment of anterior cruciate ligament injury. Osteoarthritis Cartilage 2014;22(5):668–682. [DOI] [PubMed] [Google Scholar]
- 18.Crawford R, Walley G, Bridgman S, Maffulli N. Magnetic resonance imaging versus arthroscopy in the diagnosis of knee pathology, concentrating on meniscal lesions and ACL tears: a systematic review. Br Med Bull 2007;84(1):5–23. [DOI] [PubMed] [Google Scholar]
- 19.Bien N, Rajpurkar P, Ball RL, et al. Deep-learning-assisted diagnosis for knee magnetic resonance imaging: development and retrospective validation of MRNet. PLoS Med 2018;15(11):e1002699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu F, Guan B, Zhou Z, et al. Fully automated diagnosis of anterior cruciate ligament tears on knee MR images by using deep learning. Radiol Artif Intell 2019;1(3):e180091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang PD, Wong TT, Rasiej MJ. Deep learning for detection of complete anterior cruciate ligament tear. J Digit Imaging 2019;32(6):980–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Germann C, Marbach G, Civardi F, et al. Deep convolutional neural network-based diagnosis of anterior cruciate ligament tears: performance comparison of homogenous versus heterogeneous knee MRI cohorts with different pulse sequence protocols and 1.5-t and 3-t magnetic field strengths. Invest Radiol 2020 Mar 10 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cui Y, Song Y, Sun C, Howard A, Belongie S. Large scale fine-grained categorization and domain-specific transfer learning. 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition 2018; 4109–4118. [Google Scholar]
- 24.Anumanchipalli GK, Chartier J, Chang EF. Speech synthesis from neural decoding of spoken sentences. Nature 2019;568(7753):493–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. In: Proceedings of the 25th International Conference on Neural Information Processing Systems. Red Hook, NY: Curran Associates, 2012:1097–1105. [Google Scholar]
- 26.Warrens MJ. Cohen’s linearly weighted kappa is a weighted average. Adv Data Anal Classif 2012;6(1):67–79. [Google Scholar]
- 27.Zhou XH, Obuchowski NA, McClish DK. Statistical methods in diagnostic medicine. Wiley, 2011. [Google Scholar]
- 28.Han S, Pool J, Tran J, Dally W. Learning both weights and connections for efficient neural network. In: Proceedings of the 25th International Conference on Neural Information Processing Systems. Red Hook, NY: Curran Associates, 2015:1135–1143. [Google Scholar]
- 29.Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely connected convolutional networks. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR)2017; 2261–2269.
- 30.Redmon J, Divvala S, Girshick R, Farhadi A. You only look once: unified, real-time object detection. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR)2016; 779–788.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.